Centric diatoms contain several fucoxanthin-chlorophyll protein light-harvesting complexes that differ in oligomeric state and the presence of the photoprotective subunits Lhcx1 and Lhcx6_1.
Abstract
The ecological success of diatoms, key contributors to photosynthesis, is partly based on their ability to perfectly balance efficient light harvesting and photoprotection. Diatoms contain higher numbers of antenna proteins than vascular plants for light harvesting and for photoprotection. These proteins are arranged in fucoxanthin-chlorophyll protein (FCP) complexes. The number of FCP complexes, their subunit composition, and their interactions in the thylakoid membranes remain elusive in different diatoms. We used the recently available genome sequence of the centric diatom Cyclotella cryptica to analyze gene sequences for putative light-harvesting proteins in C. meneghiniana, and to elucidate the FCP complex composition. We analyzed two pools of FCP complexes that were trimeric (FCPa) and nonameric (FCPb). FCPa was composed of four different trimeric subtypes. Two different nonameric FCPb complexes were present. All were distinguished by their polypeptide composition and partly by pigmentation. With use of a milder purification method, two fractions composed of different FCP complexes were isolated. One was enriched in FCPs incorporating the photoprotective subunit Lhcx1, such as the newly identified nonameric FCPb2 and the major trimeric FCPa4 complex, which are predetermined to be involved in energy-dependent nonphotochemical quenching. The other fraction contained mainly FCPs that were devoid of Lhcx1, FCPa3, and FCPb1. Both fractions also included small amounts of trimeric FCPa complexes with the centric diatom-specific Lhcx protein, Lhcx6_1, as subunit. Thus, the antenna organization of centric diatoms, as well as the distribution of different photoprotective Lhcx proteins, differs from that of other diatoms, as well as from plants.
Diatoms are unicellular, eukaryotic algae that contribute about 25% of worldwide primary production (Falkowski et al., 1998). In addition, they have attracted increasing interest in biotechnology because of their high content in unsaturated fatty acids (Barka et al., 2016; Traller et al., 2016). Despite this importance and their ecological success, our knowledge about fundamental biosynthesis pathways is far behind that of vascular plants. Although their basic reactions are often comparable, their light-harvesting systems used in photosynthesis differ significantly. Pigmentation, and also the macromolecular organization of light-harvesting complexes that serve the photosystems, is fundamentally different, as is the thylakoid membrane arrangement.
The distant relationship to vascular plants is because diatoms evolved via a so-called secondary endosymbiosis, whereby an eukaryotic photosynthetic microorganism related to present day red algae was engulfed by a heterotrophic unicellular eukaryotic host and became the later plastid (Cavalier-Smith, 2013; Keeling, 2013), the only organelle that remained from the former red alga. Thus, four membranes envelope the plastids of diatoms. Thylakoids are arranged in a specific three-banded structure, running along the whole plastid. Early electron microscopy and more recent work have elucidated that this peculiar thylakoid stacking is accompanied by an enrichment of PSII complexes in the inner four thylakoid membranes, whereas PSI is enriched in the outer two peripheral membranes (Pyszniak and Gibbs, 1992; Flori et al., 2017). Diatoms contain a large set of antenna proteins, assembling into fucoxanthin (Fx)-chlorophyll (Chl) protein (FCP) complexes. These antenna proteins are nuclear encoded like the light-harvesting proteins of vascular plants (Armbrust et al., 2004; Bowler et al., 2008). Based on sequence homologies, most of the antenna proteins fall into three groups, named Lhcf, Lhcr, and Lhcx. These three families all contain many members; some are even encoded by several genes and/or gene copies (Eppard and Rhiel, 1998; Armbrust et al., 2004; Bowler et al., 2008). All different proteins were proved to be expressed (Lepetit et al., 2007, 2010; Grouneva et al., 2011), but information about their precise function is limited. So far, functional assignment was only done for some members of each group, but is principally assumed to hold for all members: Whereas the main light-harvesting proteins (Lhcf) seem to serve both photosystems, Lhcr are connected to PSI, and the Lhcx proteins, which are related to the LhcSR (Lhc stress related) proteins of green algae, are involved in photoprotection [Fig. 1; for review see Büchel (2015)]. In addition, information about FCP complex formation is also limited, and neither their distribution between the core and peripheral thylakoid membranes nor their specific association with photosystems is known, except for the Lhcrs that serve PSI (Veith and Büchel, 2007; Veith et al., 2009; Lepetit et al., 2010; Grouneva et al., 2011). Even less is known in the case of additional, putative antenna proteins that are not classified as Lhcf, Lhcr, or Lhcx, like, for example, the products of the so-called FCP genes in Thalassiosira pseudonana (Mock et al., 2017).
Figure 1.
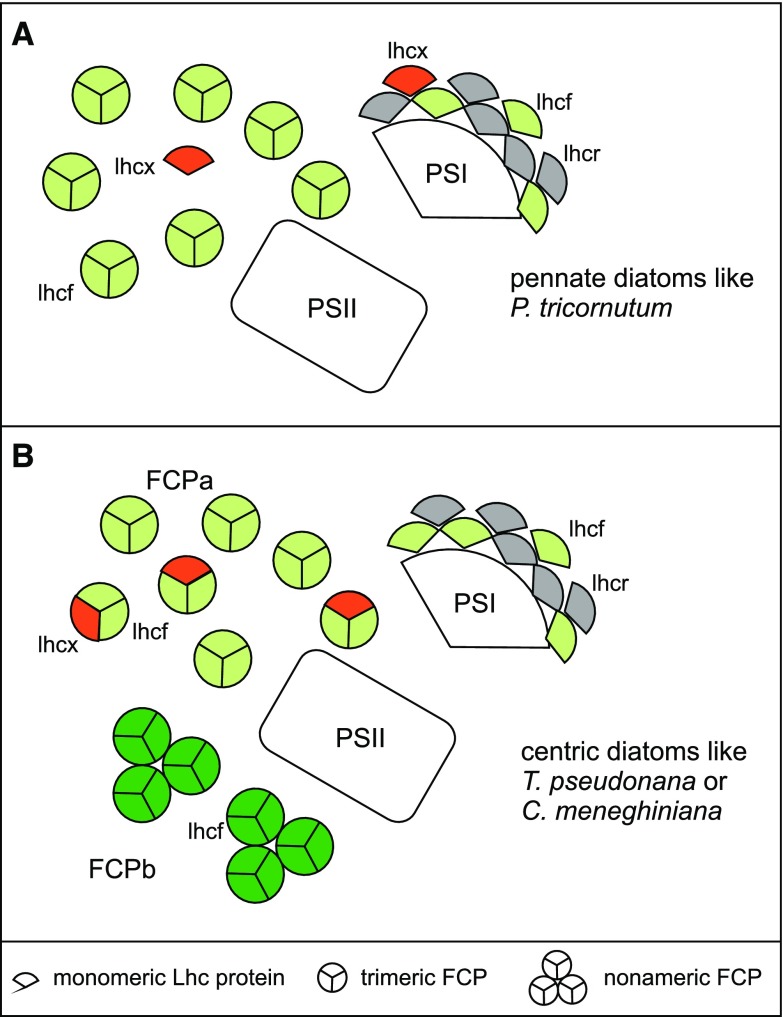
Scheme of the antenna organization of diatoms. Lhcf (green), Lhcr (gray), and Lhcx (red) arrangement in pennate diatoms is shown in (A) and the organization in centric diatoms is depicted in (B). Color coding only relates to the three protein families, not to individual members, and the amount of proteins shown does not reflect the actual numbers. Antenna proteins that do not belong to these major three groups are omitted. A, For P. tricornutum, different trimeric FCP complexes were analyzed that consist of different Lhcf proteins (green; Grouneva et al., 2011; Gundermann et al., 2013). Lhcx (red) have not been detected in specific FCP complexes so far, but coelute in the major FCP pool (Lepetit et al., 2007, 2010). The PSI antenna was shown to consist of Lhcr (gray) proteins, and contains several Lhcf proteins and Lhcx1 in addition (Lepetit et al., 2007; Veith and Büchel, 2007; Grouneva et al., 2011). B, In case of centric diatoms, a trimeric (FCPa) and a nonameric (FCPb) population of FCP complexes consisting of different Lhcf proteins (light green and dark green, respectively) was demonstrated (Beer et al., 2006; Grouneva et al., 2011; Nagao et al., 2013a; Röding et al., 2018), whereby the FCPa trimers were shown to contain Lhcx1 (red) as well. Like in P. tricornutum, the PSI antenna consists of several Lhcr and Lhcf proteins, but does not contain Lhcx1 (Veith et al., 2009; Grouneva et al., 2011).
Diatoms are subdivided into two major groups, the so-called pennate and centric diatoms. This division is not only reflected in their morphology like the symmetry of their cells, but also in physiological and molecular features. Phaeodactylum tricornutum has become the model organism for pennates since its relatively small genome was sequenced in 2008 (Bowler et al., 2008) and tools for transformation became available (Apt et al., 1996). As a model for centric diatoms, Thalassiosira pseudonana was established even earlier, again because of small genome size (Armbrust et al., 2004). However, for in-depth studies on the biochemistry and biophysics of the photosynthetic apparatus, other centric diatoms emerged, namely Chaetoceros gracilis (Nagao et al., 2007, 2010, 2013a, 2013b, 2014; Yokono et al., 2015) and Cycloctella cryptica 1020-1a that was later renamed as C. meneghiniana 1020-1a (Büchel, 2003; Papagiannakis et al., 2005; Beer et al., 2006; Gildenhoff et al., 2010; Gundermann and Büchel, 2012; Ghazaryan et al., 2016). Those species are much more widely used than T. pseudonana (Zhu and Green, 2010; Grouneva et al., 2011, 2016; Dong et al., 2015, 2016), mainly because of better biochemical accessibility and larger cell size that leads to higher yields in protein purifications.
Organization of FCP complexes differs between pennate and centric diatoms (Fig. 1): whereas in pennates trimeric complexes of different Lhcf composition are present, but no higher oligomers of specific polypeptide composition have been isolated so far (Lepetit et al., 2007; Grouneva et al., 2011; Gundermann et al., 2013), in centrics nonameric complexes of nine subunits (FCPb) besides trimeric FCPa’s could be identified (Beer et al., 2006; Grouneva et al., 2011; Nagao et al., 2013a; Röding et al., 2018). Both FCPa and FCPb complexes contain specific Lhcf proteins. Importantly, Lhcx proteins are subunits of the trimeric complexes of centric diatoms as well (Beer et al., 2006; Grouneva et al., 2011). This is not the case for pennates (Grouneva et al., 2011; Gundermann et al., 2013), where Lhcx proteins can be found in the pool of FCPs (Lepetit et al., 2007; Taddei et al., 2018), but no specific association of Lhcx proteins to any trimeric FCP complex has been demonstrated so far. This difference is accompanied by differences in the set of Lhcx proteins: whereas P. tricornutum expresses four different Lhcx proteins (Lhcx1-4), T. pseudonana was shown to have genes coding for five different Lhcx (Lhcx1 and Lhcx2 that are identical, Lhcx4-6 and 6_1). For P. tricornutum as well as for C. meneghiniana Lhcx1 was shown to be essential for the energy-dependent (qE) part of nonphotochemical quenching (NPQ), that is, for photoprotection by converting excess incident energy to heat (Bailleul et al., 2010; Ghazaryan et al., 2016). Lhcx1 is the only Lhcx protein that is highly similar in centrics and pennates. However, some differences were still observed, especially in the lumenal parts of the protein and in the second membrane helix (Büchel, 2014; Supplemental Fig. S1A). Recently it was shown that Lhcx1 in pennates is more involved in short-term protection and thus constitutively present, albeit in higher amounts in high light (HL) grown cells (Taddei et al., 2018). With use of mass spectrometry (MS), Lhcx1 was detected in PSI of pennates (Grouneva et al., 2011), but immunodetection revealed its presence additionally in the pool of FCP and even in PSII fractions (Taddei et al., 2018). In the centric diatom T. pseudonana, Lhcx1 was found in the trimeric FCPa complexes (Grouneva et al., 2011), but only in protein purifications derived from cells grown under HL, whereas immunodetection revealed a more constitutive expression (Zhu and Green, 2010). Concerning the other Lhcx proteins, differences between the two major diatom groups are stronger: For Lhcx3 of pennate diatoms it was recently shown that it promotes the part of qE under prolonged HL conditions that takes place in the antenna complexes, whereby Lhcx3 is accumulated to a much higher extent than Lhcx1, and immunodetection revealed its presence in PSI and the pool of FCPs of HL-grown cells (Taddei et al., 2018). In addition, the same hypothesis was proposed for Lhcx2, because it also accumulates under prolonged HL, albeit less so than Lhcx3 (Lepetit et al., 2013, 2017; Taddei et al., 2016). Lhcx3 was also discussed to be involved in NPQ induced by nitrogen starvation, whereas Lhcx2 enhanced NPQ under iron deprivation. The centric T. pseudonana was shown to contain Lhcx4 in PSI and FCPa’s under HL conditions instead, and to constitutively incorporate Lhcx6_1 in these complexes (Grouneva et al., 2011). In contrast, Lhcx6 of centrics was shown to be induced only under short HL illumination by immunodetection in thylakoids (Zhu and Green, 2010). Nothing is known so far about the precise role of these other Lhcx proteins in centrics.
As mentioned above, the FCP antenna system of C. meneghiniana 1020-1a is relatively well studied biochemically, in much more detail than the antenna system of T. pseudonana. However, the lack of a sequenced genome hampered a detailed analysis. The pioneering work of Eppard and Rhiel (1998) and Eppard et al. (2000) identified in C. meneghiniana genes for two Lhcf proteins (encoded by Fcp1-3 and by Fcp5, respectively), one Lhcr protein (encoded by Fcp4), and three Lhcx proteins (encoded by Fcp6, 8, 9; by Fcp7 that is almost identical; and by Fcp12), whereby Fcp6, 8, and 9 are almost identical to the Thalassiosira Lhcx1 proteins (Supplemental Fig. S1) and will thus be referred to as Lhcx1 from now on. Specific antibodies against Fcp1-3 (Lhcf-proteins) and Fcp6-9 (Lhcx1 and Fcp7-proteins) allowed the identification of these proteins in the trimeric FCPa complexes, whereas the nonameric FCPb complexes were hypothesized to be built of Fcp5 (another Lhcf protein; Fig. 1; Beer et al., 2006). In C. meneghiniana, FCPa was shown to reduce its fluorescence yield upon aggregation in a pH and diatoxanthin (DT)-dependent way, and thus to contribute to NPQ (Gundermann and Büchel, 2008, 2012; Chukhutsina et al., 2014). In addition, a higher content of Lhcx1 was demonstrated to decrease fluorescence yield (Gundermann and Büchel, 2008). However, CD spectra of FCPa complexes from wild-type and mutants with a reduced Lhcx1 content did not differ, although changes were obvious in thylakoid levels (Ghazaryan et al., 2016). Thus, it was concluded that Lhcx1 is acting in the aggregation of FCPa complexes needed for NPQ, but not in quenching itself.
Despite the analyses described above, important questions remain about the light-harvesting organization in centric diatoms. The pool of FCPa complexes especially is known to be heterogeneous because at least its Lhcx1 content had been shown to depend on the light intensity during growth (Beer et al., 2006). Thus, the pool of FCPa seemed to vary with growth conditions, and the question about the number of different FCPa trimers and their precise subunit composition remains. In addition, nothing is known about the interaction of different FCPa and FCPb complexes in the form of supramolecular assemblies or supercomplexes. Recently the genome of C. cryptica CCMP332 was sequenced (Traller et al., 2016), a species very closely related to C. meneghiniana 1020-1a. Indeed, the latter had only previously been renamed from C. cryptica. This allowed us to analyze the gene sequences for putative light-harvesting proteins in C. meneghiniana, and to elucidate the FCP complex composition in much more detail. Here we present data showing that the pool of FCPa complexes is indeed composed of four types of specific subunits, and also that two different FCPb complexes exist. By sucrose density fractionation, two bands could be isolated that are characterized by a different combination of these FCPa and FCPb complexes. One is enriched in FCPs with Lhcx1 as subunits mainly under high-light conditions, whereas the other contains predominantly FCPs that are devoid of Lhcx1. In addition, trimeric FCPa complexes that constitutively incorporate Lhcx6_1, one of the Lhcx proteins specific for centrics, are found in both. We characterized the specific subunit composition of the different FCP complexes in diatoms in-depth, and the implications for the organization of the PSII antenna system and for NPQ in centric diatoms are discussed.
RESULTS
Genes Encoding Light-Harvesting Proteins in C. meneghiniana
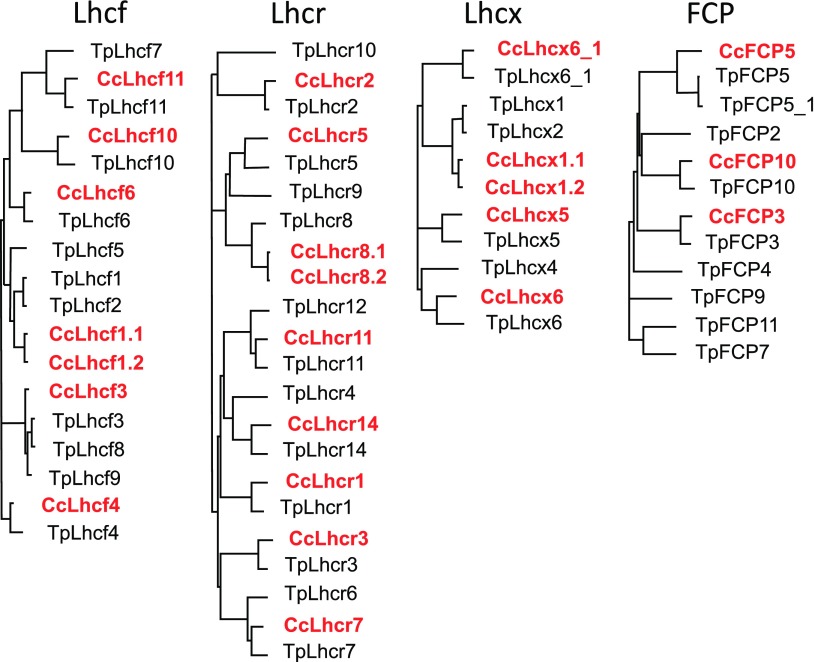
The FCP complexes of the centric diatom C. meneghiniana 1020-1a have been studied before, but an in-depth analysis of the subunit composition was hampered by the lack of a sequenced genome. The recently available genome of C. cryptica allows us to elucidate how many, and which, Lhc genes are present in Cyclotella. This knowledge can be used for further in-depth biochemical analysis of the FCP complex composition of C. meneghiniana 1020-1a. To identify the Lhc genes of Cyclotella we used all known Lhc sequences from T. pseudonana and the few already known from C. meneghiniana 1020-1a (Eppard and Rhiel, 1998). Besides the major groups of Lhcf, Lhcr, and Lhcx present in all diatoms, further so-called FCP genes are annotated for T. pseudonana as well. Most of the Lhc/FCP genes of T. pseudonana and those already known from C. meneghiniana 1020-1a (Eppard and Rhiel, 1998) could be identified in the sequenced Cyclotella genome, but no new, additional Lhc sequences emerged (Table 1). A draw back of the current genome is, however, that the automatically generated gene models have not yet been functionally assigned and further verified by EST (expressed sequence tags) or RNA sequencing data. Thus, in some cases several models exist for one locus because of different intron predictions. When these different models are compared at the protein level, differences occur mostly in either the C-terminus or in the presequences, in the latter case maintaining the mature protein sequence. For the group of Lhcf genes that consists of 11 genes in the case of T. pseudonana (Tp), substantial differences compared with Cyclotella cryptica (Cc) were obvious (Fig. 2): Homologs to TpLhcf5 as well as TpLhcf7 are missing. Two very similar genes termed CcLhcf1.1 and CcLhcf1.2 were found instead of TpLhcf1 and TpLhcf2 that shared more sequence similarity to each other than to either TpLhcf1 or TpLhcf2. CcLhcf1.1 exists in three gene copies (Table 1; Supplemental Fig. S1). Although the copies are located differently, sequences are identical up to 100 bases up- and downstream of the predicted coding region. The same holds for two copies of CcLhcf1.2, whereas the third differs in the 3′-untranslated region. Because C. cryptica and C. meneghiniana are closely related, we expected to find all genes that were previously detected by Eppard and Rhiel (1998). Indeed CcLhcf1.1 is identical to the gene described as Fcp1 in C. meneghiniana (Eppard and Rhiel, 1998), except that it codes for a longer presequence, and CcLhcf1.2 is identical to Fcp2 (Supplemental Fig. S1). However, a gene identical to Fcp3, which codes for a protein that differs only in one amino acid from Fcp2, was missing in the genome of C. cryptica CCMP332. However, the predicted Lhcf1 and Fcp1-3 proteins are identical concerning their mature protein sequence, that is, after cleavage of the presequences (Supplemental Fig. S1), and will further be referred to as Lhcf1.
Table 1. Lhcf, Lhcr, Lhcx, and FCP genes identified in C. cryptica CCMP332 using T. pseudonana and C. meneghiniana 1020-1a gene sequences.
In cases where identical gene models exist at several loci, only one gene name is given. In cases where different gene predictions for one locus are present in the database, the models with the least resemblance to their T. pseudonana counterparts are shown in italics and named ‘a’ and ‘b’. Fcp genes of C. meneghiniana are according to Eppard and Rhiel (1998) and Eppard et al. (2000). Dashes indicate no gene identifier available in database.
| Name | Gene Identifier | Location | Mature Protein |
|---|---|---|---|
| Lhcf | |||
| CcLhcf1.1a | g17931.t1 | g009304_00092:845-1770 | Lhcf1 = Fcp1-3 |
| g25998.t1 | g002362_00144:845-1770 | ||
| g26023.t1 | g002346_00143:1116-2041 | ||
| CcLhcf1.2a | g11743.t1 | g017983_00088:3722-4321 | Lhcf1 = Fcp1-3 |
| g9813.t1 | g021620_00082:470-1069 | ||
| g21457.t1 | g005884_00107:470-1069 | ||
| CcLhcf3b | g27206.t1 | g001521_00091:7-495 | Lhcf3 |
| Fcp5b | Not found | Fcp5 | |
| CcLhcf4 | g4695.t1 | g037374_00040:1988-2593 | Lhcf4 |
| CcLhcf6 | g20180.t1 | g007028_00058:2475-3077 | Lhcf6 |
| CcLhcf6a | — | g007028_00058:2475-3219 | |
| CcLhcf10 | g1413.t1 | g068182_00128:55071-55679 | Lhcf10 |
| CcLhcf11 | g9270.t1 | g022704_00101:7497-8159 | Lhcf11 |
| Lhcr | |||
| CcLhcr1 | g4191.t1 | g039603_00095:12747-13619 | Lhcr1 |
| CcLhcr2 | g17037.t1 | g010304_00068:3475-3987 | Lhcr2 |
| CcLhcr2a | g13001.t1 | g010304_00068:3478-4257 | |
| CcLhcr2b | — | g010304_00068:3238-4373 | |
| CcLhcr3c | g22357.t1 | g005177_00041:3338-4286 | Lhcr3, similar to |
| CcLhcr3a | — | g005177_00041:3112-4460 | Fcp4 |
| CcLhcr3b | — | g005177_00041:3338-4286 | |
| CcLhcr5 | g12115.t1 | g017270_00043:10106-10888 | Lhcr5 |
| CcLhcr5a | g9253.t1 | g017270_00043:10109-11167 | |
| CcLhcr5b | — | g017270_00043:10106-11252 | |
| CcLhcr7 | g15657.t1 | g012035_00070:5433-6183 | Lhcr7 |
| CcLhcr8.1 | g25826.t1 | g00250_00057:1026-1894 | Lhcr8.1 |
| CcLhcr8.2 | g22748.t1 | g004863_00062:1-677 | Lhcr8.2 |
| CcLhcr11 | g10947.t1 | g019362_00072:17991-18731 | Lhcr11 |
| CcLhcr14 | g18288.t1 | g008875_00105:6649-7349 | Lhcr14 |
| Lhcx | |||
| CcLhcx1.1d | g2066.t1 | g059009_00077:10108-10839 | Lhcx1.1 |
| CcLhcx1.2d | g14788.t1 | g013146_00085:3526-4158 | Lhcx1.2 = Fcp6, 8, 9 |
| CcLhcx5e | g21285.t1 | g006023_00084:4427-5140 | Lhcx5, similar to |
| CcLhcx5a | g16306.t1 | g006023_00084:4430-5454 | Fcp12 |
| CcLhcx6 | g14787.t1 | g013146_00085:2016-2768 | Lhcx6 |
| CcLhcx6a | g11298.t1 | g013146_00085:1957-2768 | |
| CcLhcx6_1 | g10346.t1 | g020598_00076:14804-15523 | Lhcx6_1 |
| CcLhcx6_1a | g7882.t1 | g020598_00076:14657-15523 | |
| FCP | |||
| CcFCP3 | g6093.t1 | g025849_00094:2456-3409 | FCP3 |
| CcFCP5 | g12200.t1 | g017153_00078:12827-14314 | FCP5 |
| CcFCP10 | g4274.t1 | g039269_00091:21945-22616 | FCP10 |
Lhcf1 genes are identical to Fcp1-3 except for the N-terminal presequences, where only Lhc1.2 is identical to Fcp2 (see Supplemental Fig. S1).
Lhcf3 encodes for a protein that is identical to Fcp5 except for two amino acid exchanges and a shorter N-terminal presequence in case of Lhcf3 (see Supplemental Fig. S1). Fcp5 is listed as well because both Lhcf3 and Fcp5 were identified by MS in the FCP complexes of C. meneghiniana (Tables 2 and 3), but only CcLhcf3 is present in the C. cryptica CCMP332 genome.
Different predictions for Lhcr3 have different insertions, but are most comparable with Fcp4 (see Supplemental Fig. S1).
Lhcx1.1 codes for a protein that has a one amino acid deletion compared with Lhcx1.2. Lhcx1.2 is identical to Fcp6, 8, 9 and almost identical to Fcp7 (in the protein five amino acids are exchanged, four of them in the presequence; see Supplemental Fig. S1).
Lhcx5 is most similar to Fcp12, but the N terminus of the protein is six amino acids longer, and in total five amino acids are exchanged (four of them in presequence; see Supplemental Fig. S1).
Figure 2.
Phylogenetic trees of Lhcf, Lhcr, Lhcx, and FCP of C. cryptica CCMP332 (in bold red) and T. pseudonana (black). The trees are based on predicted protein sequences including presequences from the respective gene models that show the highest resemblance between the two species listed in Table 1.
Only one CcLhcf3 gene could be detected in the Cyclotella genome, whereas in T. pseudonana the similar TpLhcf3, 8, and 9 genes code for an identical protein (Fig. 2). CcLhcf3 is highly homologous to Fcp5 of C. meneghiniana. Again, the lengths of the presequences deviated, but in addition two amino acids in the mature protein are different as well (Supplemental Fig. S1).
In the case of Lhcr genes, TpLhcr4, 6, 9, 10, and 12 had no putative orthologs in C. cryptica (Fig. 2), whereas two versions of CcLhcr8 could be identified. CcLhcr3 is the only homolog found for the known Fcp4 of C. meneghiniana (Eppard et al., 2000), but in all three different gene models insertions are predicted (Table 1; Supplemental Fig. S1). However, at this locus some uncertainties seem to exist because of bases that were not sequenced. Also, for the FCP genes the number was smaller in C. cryptica than in T. pseudonana, with only FCP3, 5, and 10 present in the former.
Concerning Lhcx, Lhcx4 of T. pseudonana that is closely related to Lhcx6 was missing in C. cryptica (Fig. 2). TpLhcx1 and 2 are replaced by CcLhcx1.1 and CcLhcx1.2 that share more sequence similarity with each other than with the respective Thalassiosira genes. They are identical to the genes described as Fcp6, 8, and 9 of C. meneghiniana, whereby in CcLhcx1.1 one codon is missing. No gene predicted to encode a protein similar to Fcp7 that deviates again mainly in the presequence from Fcp6, 8, and 9 (Supplemental Fig. S1) was found. CcLhcx5 is very similar to Fcp12 and only present in one copy. For CcLhcx6_1 two model predictions exist, whereby one shows a closer resemblance to TpLhcx6_1 (Table 1).
Heterogeneity of the Pools of FCPa and FCPb Complexes
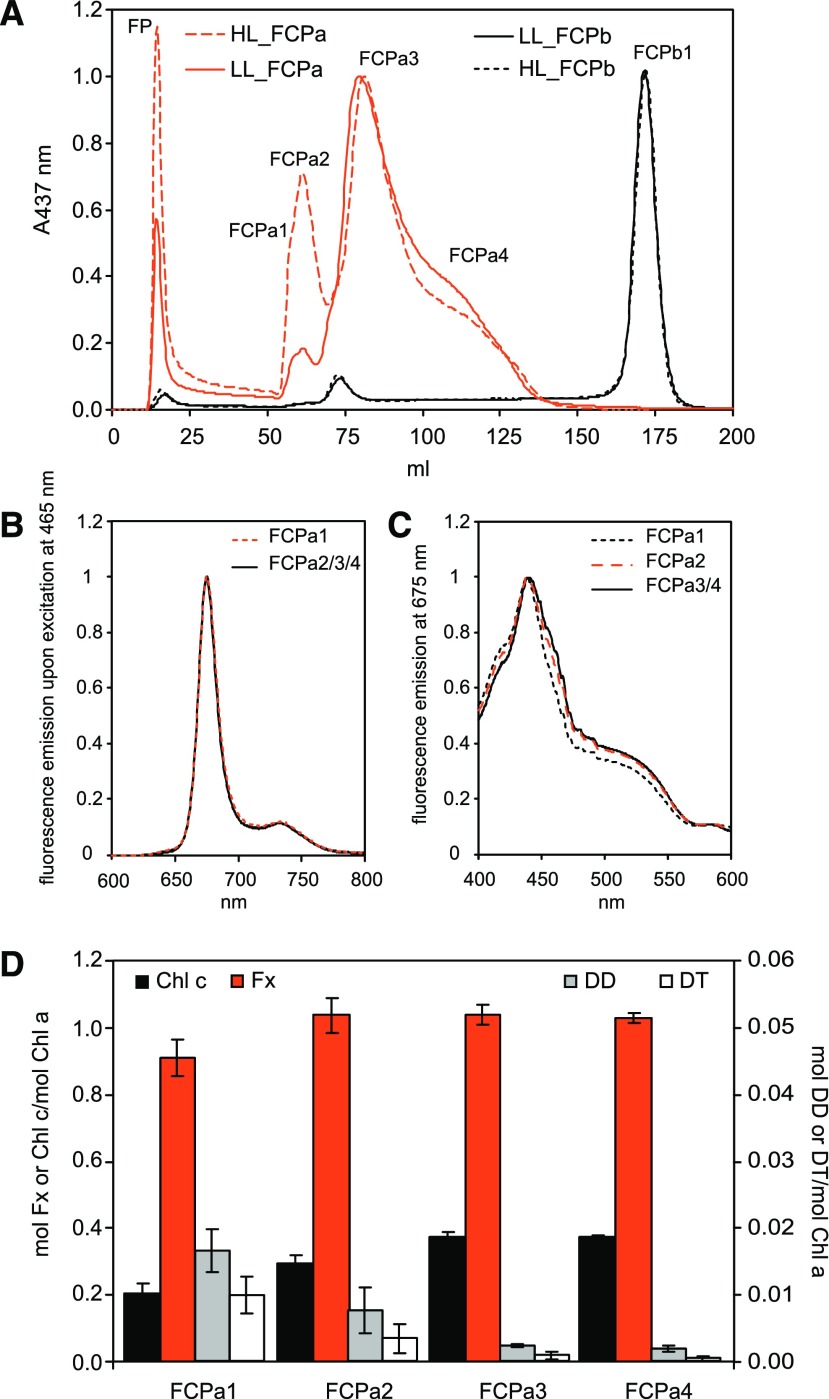
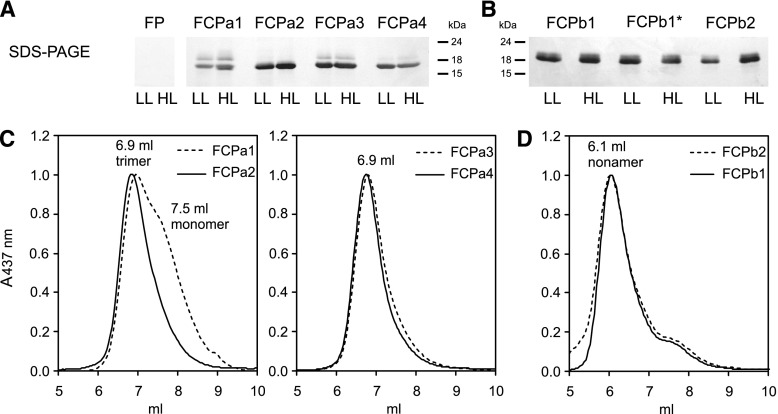
The putative sequences of Lhc genes in Cyclotella enabled the identification of polypeptides by liquid chromatography-electrospray ionization-tandem mass spectrometry (LC-ESI-MS/MS) analysis, and could be used to evaluate the heterogeneity of the pools of FCPa and FCPb complexes in this representative centric diatom. To this end, thylakoids were isolated from cells grown under low light (LL) or under HL conditions. After solubilization, FCP complexes were purified by ion-exchange chromatography (IEX) followed by sucrose density centrifugation to remove coeluted photosystems (Fig. 3, left; also described in Gundermann and Büchel, 2008). The FCPa and FCPb complexes obtained were then subjected to a second IEX for separation of possible subtypes (Fig. 4A). In the case of the nonameric FCPb, only one major band was identified, accompanied by one very small band and a free pigment band, whereas in the case of the trimeric FCPa four bands and shoulders (besides a free pigment band) could be distinguished. Free pigment was identified by its lack of polypeptides on SDS-PAGE (Fig. 5A). It contained all pigments usually found in FCPs, but in different stoichiometries with less Chl c and more Diadinoxanthin (DD) and Diatoxanthin (DT) per Chl a (Supplemental Fig. S2). The four fractions of FCPa (FCPa1-4) were all functional as demonstrated by fluorescence emission spectra recorded upon excitation at 465 nm, exciting preferentially Chl c (Fig. 4B; for HL samples see Supplemental Fig. S3A). No emission of detached Chl c at 636 nm became visible, and in the fluorescence excitation spectra both Chl c at 465 nm and Fx between 475 and 585 nm were demonstrated to transfer energy to Chl a (Fig. 4C; Supplemental Fig. S3B). Fluorescence spectra of the different FCPa complexes were almost identical for FCPa2-4, whereas FCPa1 showed some deviation, because transfer from Chl c around 465 nm was less pronounced in the excitation spectrum. This was because of a lower content of Chl c as shown in Figure 4D and Supplemental Figure S4 and not because of a loss in energy transfer function, because no signal of detached Chl c was present in the emission spectra. Whereas the Chl c/Chl a as well as the Fx/Chl a ratio was identical for FCPa3-4, FCPa1 had significantly less Chl c and Fx per Chl a bound (Supplemental Fig. S4). FCPa2, albeit carrying the same amount of Fx as FCPa3-4, still had slightly less Chl c/Chl a. This might actually be due to some contamination of FCPa2 with FCPa1 because of their imperfect separation on the IEX column. DD and DT were also found in all complexes (Fig. 4D; Supplemental Fig. S4). A slightly higher amount of DT was present in case of FCPa1 and FCPa2 from HL cells. However, the free pigment (FP) fractions were strongly enriched in DD and DT (Supplemental Fig. S2), and in the case of FCPa1-4 amounts decreased with increasing elution volume. Thus, DD- and DT-binding to the complexes was most probably sensitive to the salt concentration on the IEX, and our results are not representative of the in vivo situation. All FCPa subpopulations showed a very similar protein pattern in SDS-PAGE (Fig. 5A). Two typical main polypeptide bands (Beer et al., 2006) were present, whereby the band of lower Mr was more abundant. All complexes were trimeric as shown by their elution at 6.9 ml on gel filtration (Fig. 5C; Supplemental Fig. S5A). Only FCPa1 contained more monomers, eluting at 7.5 ml. Overall the pigment stoichiometries and thus the spectroscopic features, as well as the oligomeric states and protein bands on SDS-PAGE, were almost identical for complexes from HL- and LL-grown cells.
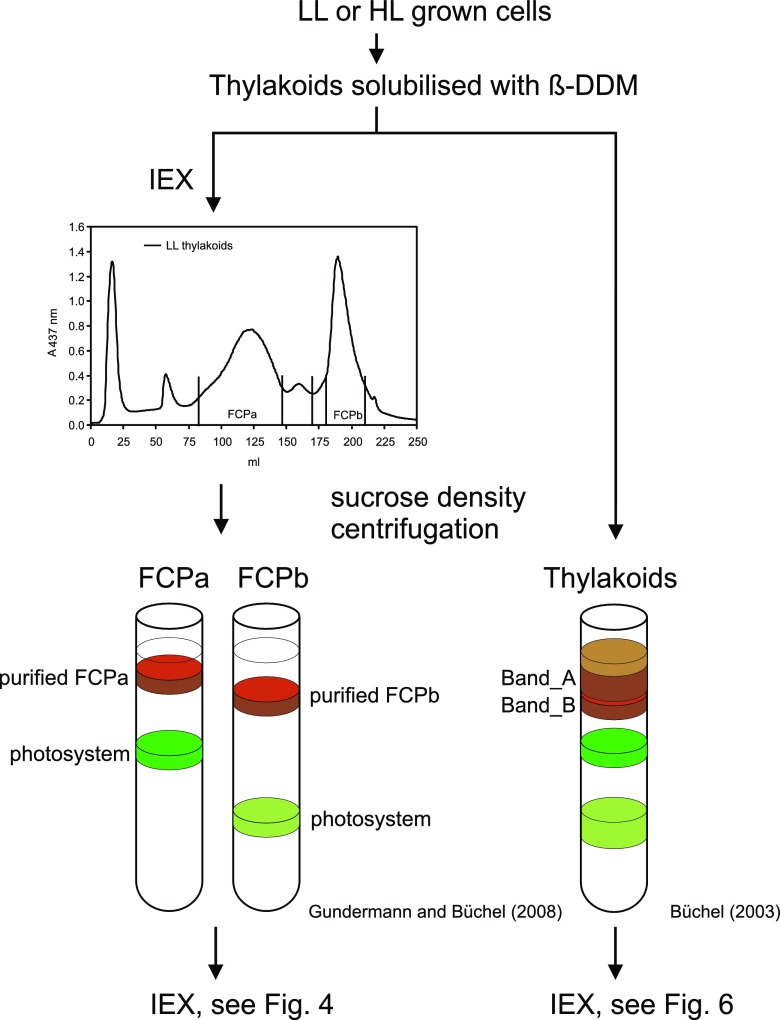
Figure 3.
Schematic representation of the purification methods used. Thylakoid membranes were isolated either from LL- or HL-grown cells and proteins solubilized using a mild detergent (β-DDM [β-dodecyl maltoside]). In one case the solubilized proteins were separated by IEX (left). The pools of FCPa and FCPb were harvested as indicated and subjected to sucrose density centrifugation to separate them from coeluting photosystems. The purified pools of FCPs were then subjected to a second IEX (for results see Fig. 4). In another approach (right), solubilized thylakoid proteins were directly loaded on sucrose density gradients. The two brown bands obtained (Band A and Band B) were then further separated by IEX (for results see Fig. 6)
Figure 4.
Proteins from solubilized thylakoids from C. meneghiniana cells grown under LL or HL were separated by IEX followed by sucrose density centrifugation to yield pure FCPa and FCPb (Fig. 3; Gundermann and Büchel, 2008). A, further separation by an additional IEX run of LL-FCPa (red solid line), HL-FCPa (red dotted line), as well as LL-FCPb (black solid line line) and HL-FCPb (black dotted line). In (B), the fluorescence emission spectra recorded upon excitation at 465 nm are depicted for the subpopulations of LL-FCPa, FCPa1 (red dotted line), and FCPa2-4 (black lines). No differences became obvious in the spectra of the respective HL complexes (Supplemental Fig. S3A). C, fluorescence excitation spectra for LL-FCPa1 (black dotted line), LL-FCPa2 (red dotted line), and LL-FCPa3-4 (black solid lines) recorded at 675 nm, which were again indistinguishable from their HL counterparts (Supplemental Fig. S3B). The four FCPa complexes obtained were also analyzed by HPLC. D, ratios of mol pigment/mol Chl a for Chl c (black bars, left axis), Fx (red bars, left axis), DD (gray bars, right axis), and DT (white bars, right axis) for the subpopulations of LL-FCPa. Data represent means and standard deviations of five to nine biological replicates; for statistical analysis see Supplemental Fig. S4. HL-derived complexes showed similar Chl c, Fx, and DD per Chl a values compared with LL-complexes, whereas DT/Chl a was increased (Supplemental Fig. S4).
Figure 5.
FCPa (A) and FCPb (B) subpopulations prepared by IEX like shown in Fig. 4 (FCPa1-4, FCPb1) and Fig. 6 (FCPb1*, FCPb2) from HL- or LL-grown cells were subjected to SDS-PAGE, as well as the FP band. Samples equivalent to 1 μg Chl were loaded in each lane; molecular weights of the marker bands are given in kDa. In (C), gel filtration runs of the different FCPa subpopulations from LL cells are shown. At left, FCPa1 (dotted line) and FCPa2 (solid line) are shown, whereas at right FCPa3 (dotted line) and FCPa4 (solid line) are depicted. D, comparison of the gel filtration for FCPb1 (solid line) and FCPb2 (dotted line). In (C and D), elution volumes of the main peaks are indicated in the figures. According to Röding et al. (2018) monomers elute at 7.5 ml, trimers at 6.9 ml, and nonamers at 6.1 ml on the same gel filtration system. HL derived complexes showed the same elution patterns compared with LL-derived complexes, and FCPb1* exhibited the same pattern as FCPb1 (Supplemental Fig. S5).
Protein Composition of the Light-Harvesting System of C. meneghiniana
The knowledge of putative Lhc gene sequences enabled mass spectrometric analysis of the purified complexes. Proteins in the range of about 15-25 kDa, that is, the expected size of Lhc proteins, were eluted from the SDS-PAGE and subjected to MS analysis. To identify the proteins, all sequences known from C. meneghiniana as well as those newly identified in C. cryptica were used. Results are presented in Tables 2 and 3 and Supplemental Table S1. Taking the results of all FCPa and FCPb complexes together, Lhcf1 was identified with up to six unique polypeptides especially in FCPa complexes. There it occurred with a high number of total polypeptides, supporting older data that Lhcf1 is one of the major Lhc proteins expressed in C. meneghiniana (Beer et al., 2006). Both Lhcf4 and Lhcf6 were identified at the protein level as well, whereas the gene model denoted as CcLhcf6a could not be verified by MS. Although overall sequence identity is low, Lhcf4 and Lhcf6 share an identical C-terminus (Supplemental Fig. S1). Thus, one peptide common to both was detected in the different FCPa preparations, and in addition up to four and six unique peptides identified Lhcf4 and Lhcf6, respectively (Supplemental Table S1). Both Lhcf3 and Fcp5 were demonstrated by MS analysis (Tables 2 and 3; Supplemental Table S1). This is astonishing, because no gene model for Fcp5 was found in C. cryptica. Because of their high similarity with only two amino acids exchanged, up to three peptides were identified that are specific for both these proteins. In addition, one peptide each was found that was unique for either Fcp5 or Lhcf3, whereby the total number of the peptides unique for Fcp5 (IAQLAFLGQVVTR) was usually higher (Supplemental Table S1). FCPb complexes especially incorporate this subunit, as previously suggested (Beer et al., 2006). Lhcf11 was also proven to be expressed. Thus, almost all predicted Lhcf proteins were identified by MS. The missing Lhcf10 was found as well, but with only one unique peptide (Supplemental Table S1).
Table 2. Overview of the Lhc proteins detected by LC-ESI-MS/MS analysis in the different FCPa complexes isolated from LL- or HL-grown cells.
Only proteins that were identified by at least two unique peptides (UP) are shown. For the sequences of the peptides and their Xcorr- and charge values, see Supplemental Table S1.
| FCPa | FCPa1 | FCPa2 | FCPa3 | FCPa4 | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| UP | Total Peptides | UP | Total Peptides | UP | Total Peptides | UP | Total Peptides | |||||
| LL | HL | LL | HL | LL | HL | LL | HL | |||||
| Lhcf1 | 5 | 259 | 341 | 3 | 16 | 3 | 5 | 411 | 413 | 6 | 368 | 328 |
| Lhcf4/Lhcf6 | 9 | 90 | 129 | 11 | 406 | 426 | 7 | 33 | 61 | 7 | 15 | 15 |
| Fcp5/Lhcf3 | 4 | 127 | 181 | 4 | 123 | 115 | 5 | 40 | 50 | 4 | 20 | 20 |
| Lhcf11 | 0 | 0 | 0 | 3 | 9 | 2 | 0 | 0 | 0 | 0 | 0 | 0 |
| Lhcx1 | 2 | 0 | 6 | 0 | 0 | 0 | 0 | 0 | 0 | 3 | 2 | 9 |
| Lhcx6_1 | 2 | 2 | 3 | 2 | 5 | 5 | 0 | 0 | 0 | 0 | 0 | 0 |
| Lhcr1 | 0 | 0 | 0 | 2 | 6 | 8 | 2 | 2 | 1 | 0 | 0 | 0 |
| FCP10 | 0 | 0 | 0 | 3 | 6 | 10 | 0 | 0 | 0 | 0 | 0 | 0 |
Table 3. Overview of the Lhc proteins detected by LC-ESI-MS/MS analysis in the different FCPb complexes isolated from LL- or HL-grown cells.
Only proteins that were identified by at least two unique peptides (UP) are shown. For the sequences of the peptides and their Xcorr- and charge values, see Supplemental Table S1.
| FCPb | FCPb1 | FCPb1* | FCPb2 | ||||||
|---|---|---|---|---|---|---|---|---|---|
| UP | Total Peptides | UP | Total Peptides | UP | Total Peptides | ||||
| LL | HL | LL | HL | LL | HL | ||||
| Lhcf1 | 3 | 16 | 1 | 3 | 14 | 8 | 3 | 9 | 14 |
| Lhcf4/Lhcf6 | 0 | 0 | 0 | 0 | 0 | 0 | 4 | 5 | 40 |
| Fcp5/Lhcf3 | 5 | 323 | 284 | 5 | 260 | 285 | 4 | 80 | 279 |
| Lhcf11 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 2 |
| Lhcx1 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 3 |
| Lhcx6_1 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 3 |
Concerning Lhcx proteins, Lhcx1 was identified in the FCP complexes (Tables 2 and 3; Supplemental Table S1), whereby Lhcx1.1 and Lhcx1.2 could not be distinguished at the protein level, because the decisive peptide was missing. For the same reason no distinction could be made between Lhcx6_1 and Lhcx6_1a. We will thus refer to Lhcx6_1 for Lhcx6_1a as well, and use Lhcx1 for Lhcx1.1, Lhcx1.2 (that is identical to Fcp6, 8, and 9) from now on, keeping in mind that Fcp7 might be included as well.
Because Lhcr proteins are predicted to serve as direct antennae for PSI, we did not expect to find many in the FCP complexes. Lhcr1, however, was found by MS analysis, albeit with a low number of peptides (Tables 2 and 3; Supplemental Table S1). FCP10 had never been reported before as a constituent of the major FCP complexes but was safely identified in one FCPa fraction.
The complexes used for MS analysis were obtained after long preparation methods; therefore, we cannot rule out that some proteins got lost. However, losses can only be small because no protein signal was found for the FP fraction of IEX on SDS-PAGE (Fig. 5A), and substantial amounts of monomers as a hint for protein complex degeneration was only found on size exclusion chromatography (SEC) in the case of FCPa1 (Fig. 5C).
Protein Composition of FCPa Complexes
All subcomplexes of FCPa were characterized by a specific subunit composition. FCPa3, the most abundant complex under LL conditions, was composed mainly of Lhcf1 (Table 2). Lhcf1 was also the main subunit of FCPa4 and FCPa1, whereas FCPa2 contained Lhcf4 and Lhcf6 instead. Interestingly, Lhcf3 and Fcp5, proteins that are the main constituents of FCPb, were also found in high amounts especially in FCPa1 and FCPa2. FCPa2 had the most heterogeneous protein composition of all subcomplexes, containing in addition small amounts of Lhcf11, Lhcr1, and FCP10. Lhcx1 was present in FCPa1, but only under HL conditions, and was mainly found in FCPa4, that is, the latest fraction from IEX, where it was present in LL-derived samples but showed an increased amount in samples from HL. Therefore, its appearance did not depend on the elution volume like the DT content, confirming that it is a true constituent of the complexes. Lhcx6_1 was identified only in FCPa1 and FCPa2, but in samples from both growth conditions.
In summary, the pool of FCPa complexes consisted mainly of FCPa3 and FCPa4, whereas FCPa1 and FCPa2 were minor constituents. FCPa3 and FCPa4 were very similar in pigmentation and thus also dominated the spectral features published for the pool of FCPa characterized before (Beer et al., 2006). For both, Lhcf1 was the main subunit. The decisive difference was that FCPa4 bound Lhcx1 as well, a subunit that was lacking in FCPa3. The two other FCPa complexes, FCPa1 and FCPa2, showed the strongest deviations: both contained Lhcx6_1 in contrast with the other FCPa complexes. FCPa2 had Lhcf4 and Lhcf6 as major subunits and was the only complex where the total number of the peptide specific for Lhcf3 was higher than the total number of the Fcp5-specific peptide (Supplemental Table S1). FCPa1 was distinguished by a different pigmentation with less Chl c and Fx per Chl a.
Differences between LL- and HL-derived FCPa complexes concerned only Lhcx1 subunits: the number of Lhcx1 polypeptides in FCPa4 was increased in complexes from HL, and FCPa1 contained Lhcx1 in addition when isolated from HL cells. HL-FCPa complexes had previously been shown to be enriched in Lhcx1 (Beer et al., 2006). Although single FCPa complexes showed an increase in the relative amount of Lhcx1 when isolated from cells grown under HL, surprisingly enough the Lhcx1-free complex FCPa2 was relatively more abundant in the elution profiles of complexes from cells grown under HL, as was FCPa1 (Fig. 4A).
Characterization of FCPb1
In the case of FCPb, only two bands besides FPs were found during IEX separation (Fig. 4A). The first band was present in amounts too low for thorough analysis. FCPb1 as the major subpopulation showed only one band on SDS-PAGE (Fig. 5B) that consisted almost exclusively of Fcp5/Lhcf3, no matter whether isolated from HL or LL cells (Table 3), in accordance with data published by Grouneva et al. (2011) for T. pseudonana. As previously reported (Beer et al., 2006; Röding et al., 2018) for the pool of FCPb, this complex is a nonamer (Fig. 5D; Supplemental Fig. S5B). It was intact concerning excitation energy transfer (Fig. 6, C and D; Supplemental Fig. S6) and showed all the features already known from previous characterizations: (1) slightly more red shifted Fx molecules as compared with FCPa (Supplemental Fig. S2); (2) slightly blue shifted fluorescence emission maximum (673 nm instead of 675 nm in FCPa; Figs. 4B and 6C); and (3) about the same concentration of Chl c with 0.345 ± 0.011 mol Chl c/mol Chl a (in LL, HL: 0.346 ± 0.007 mol Chl c/mol Chl a) and less Fx (LL: 0.896 ± 0.021 mol Fx/mol Chl a, HL: 0.899 ± 0.013 mol Fx/mol Chl a) than the main FCPa complexes (Fig. 4D). Mass spectrometry revealed no Lhcx proteins in FCPb1 (Table 3).
Figure 6.
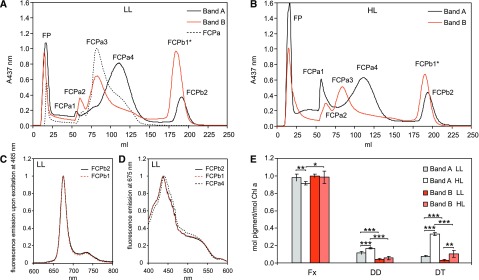
Solubilized thylakoids were separated on sucrose density gradients. The two top brown bands (Band A and Band B) consisting of nonamers and trimers each (Supplemental Fig. S7) were harvested and further separated on an IEX column. The same amount of Chl was loaded for each run. A, elution profiles for LL-derived samples. B, elution profiles for HL-derived samples. The separation of proteins from Bands A is shown with black solid lines and compared with the elution profiles of Band B (red solid lines) as well as to the IEX run to separate different FCPa complexes taken from Fig. 4A including the labeling for FCPa1-4 (black dotted line). Because the elution time of the FCPb fraction of Band A deviated slightly from FCPb1 as shown in Fig. 4, the complex is denoted as FCPb1* here. C and D, fluorescence emission (C) and excitation spectra (D) of LL-FCPb1 (red dotted line, isolated like shown in Fig. 4A), LL-FCPb2 (black solid line), and LL-FCPa4 (black dotted line, taken from Fig. 4C) for comparison, whereby HL-derived complexes exhibited identical spectra (Supplemental Fig. S6). The pigment composition of Band A (LL: gray bars; HL: white bars) and Band B (LL: red bars; HL: light red bars) is depicted in (E). Data represent means and standard deviations of four biological replicates (LL data according to Gundermann and Büchel [2008]), whereby ***P < 0.0001, **P < 0.01, and *P < 0.05; p-values were calculated by unpaired Student’s t test.
Separation of FCP Complexes by Sucrose Density Centrifugation
When solubilized thylakoid membranes were separated directly by sucrose density gradient centrifugation, two brown bands can be distinguished (Fig. 3, right; Büchel, 2003). The top band, Band A, was shown to have strong similarities to FCPa, whereas the bottom Band B obviously contained FCPb as well as FCPa. The mixed composition of Band B is especially obvious if one compares the CD-spectra of Band B (Büchel, 2003) with the CD-spectra of purified FCPa and FCPb (Ghazaryan et al., 2016) and the polypeptide pattern on SDS-PAGE of Band B (Büchel, 2003) with isolated FCPb (Beer et al., 2006). This is intriguing, because it implies that FCPb and certain FCPa complexes either coelute on sucrose density gradients, or are more closely connected in vivo, and the interactions are more resistant to mild solubilization. With the knowledge of the different subpopulations of FCPa, we thus re-examined the composition of Band A and Band B from sucrose density centrifugation.
When the components of Band A were run on an IEX (Fig. 6, A and B), several fractions appeared that could be attributed to FCPa1-4. Surprisingly, Band A additionally contained an FCP complex eluting almost at the same time as the FCPb1 complex described above (Fig. 4), termed FCPb2. Band B was less pronounced on Suc gradients (usually roughly one third of total Chl compared with Band A) and, as expected, it was composed of FCPa and FCPb complexes. The FCPb fraction of Band B (termed FCPb1*) was analyzed as pointed out above and showed no spectroscopic difference to FCPb1 (Supplemental Fig. S6, C and D) and on SDS-PAGE (Fig. 5B), in MS (Table 3) or on gel filtration (Supplemental Fig. S5C), that is, this band is identical to FCPb1. As expected, when FCPb2 of Band A was analyzed, this complex also resembled an FCPb complex. The spectral features of FCPb2 were absolutely similar to FCPb1 (Fig. 6, C and D), as was the nonameric state of the complex (Fig. 5D). However, although FCPb2 consisted mainly of Fcp5 polypeptides (Table 3), differences to FCPb1 were obvious: In addition to the trace amounts of Lhcf1 also found in FCPb1, Lhcf4 and Lhcf6 were present in case of FCPb2. Most importantly, the presence of Lhcx1 and Lhcx6_1 in FCPb2 complexes from HL cells was revealed. Thus, although extremely similar to the FCPb1 complexes concerning pigmentation and oligomeric state, FCPb2 is different concerning polypeptide composition. A complex similar to FCPb2 had not been detected when analyzing the subfractions of FCPa or FCPb, although it was present in fair amounts in Band A (Fig. 6, A and B). However, the first IEX for the separation of the pools of FCPa and FCPb had been sampled very conservatively to avoid major contaminations by photosystems. Therefore, FCPb2 was probably lost during this step. On the other hand, we cannot fully exclude that FCPb2 might have consisted of trimers that artificially aggregated to nonamers only on the IEX column. However, this possibility is extremely unlikely because this would require complete, that is, 100%, aggregation. Otherwise, hexamers, for example, should have been found on the SEC column; in addition, the constituting trimers, including pure Lhcf3/Fcp5 trimers, should have been found during IEX, which have not been detected at all. In addition, nonameric complexes (see arrow in Supplemental Fig. S7) were also visible when analyzing Band A directly by SEC (Supplemental Fig. S7). Thus, both bands contained trimeric as well as nonameric complexes (Supplemental Fig. S7), and simple coelution on density gradients seems unlikely.
The elution profiles of the postpurification by IEX demonstrated the relative amount of complexes (Fig. 6, A and B): whereas Band A consisted mainly of FCPa4, FCPa3 especially was prominent in Band B besides FCPb1. When the IEX runs from LL- and HL-derived samples are analyzed, it also becomes obvious that FCPa1 is found mainly in Band A, whereas FCPa2 is mostly found in Band B (Fig. 6, A and B). Both are slightly enriched in HL-derived samples, but otherwise no major difference between HL- and LL-grown cells could be distinguished. This is consistent with the higher abundance of FCPa1 and FCPa2 in the IEX profiles from prepurified FCPa complexes from HL-grown cultures (Fig. 4A). As expected, the Fx/Chl ratio of the bands was almost similar and did not change with growth light intensity. Concerning the xanthophyll cycle pigments DD and DT, enrichment in Band A compared with Band B was present, whereby growth under HL led to a further increase, especially in the DT/Chl a ratio (Fig. 6E).
As already pointed out, Band A showed a higher content of FCPa4, that is, the complexes that carried the highest amount of Lhcx1, especially under HL. These FCPa were accompanied by FCPa1 and FCPb2, both containing Lhcx1 in HL-derived complexes. In contrast, Band B was mostly composed of complexes devoid of Lhcx1: FCPa2 and the more abundant complexes FCPa3 and FCPb1. Both bands included a small number of trimeric complexes that were more abundant under HL than under LL, and constitutively contained Lhcx6_1 (FCPa1 or FCPa2, respectively). In Band A, the amount of this polypeptide was additionally increased because it occurred in FCPb2 as well.
DISCUSSION
Genes Encoding Light-harvesting Proteins in C. meneghiniana, C. cryptica, and T. pseudonana
To analyze the FCP subunit composition of C. meneghiniana in detail, the putative gene sequences had to be elucidated. So far, only the genomes of C. cryptica, that is, a species closely related to C. meneghiniana, and another centric species, T. pseudonana, are sequenced. The genome of C. cryptica CCMP332, although publicly available, has not been functionally annotated yet regarding protein functions, and intron-exon borders have not been verified by transcript analyses so far. Thus, we searched for Lhc genes using the provided gene models by comparison with the functionally annotated T. pseudonana genes and the few known sequences from C. meneghiniana. Although Cyclotella and Thalassiosira are rather closely related centric diatoms, both belonging to the order Thalassiosirales (e.g., see World register of marine species www.marinespecies.org; Integrated taxonomic information system www.itis.gov; NCBI taxonomy browser www.ncbi.nlm.nih.gov/Taxonomy), differences became obvious (Fig. 2). Mainly the numbers of the different Lhc genes, such as Lhcf, Lhcr, Lhcx, or FCP, were reduced in C. cryptica. For example, Lhcf5 and Lhcr4 could not be detected, and also Lhcx4 was missing in C. cryptica. In most cases the missing genes had close homologs that were present in both species. Thus, they might in fact code for functionally identical proteins and be dispensable. In many cases several copies of one gene exist, which might be because of gene duplications that have taken place recently. This certainly holds for, for example, CcLhcf1, where the gene copies present are identical even up- and downstream of the coding region.
For C. meneghiniana 1020-1a some Lhc genes had been identified earlier (Eppard and Rhiel, 1998; Eppard et al., 2000). All those genes were found in the genome of C. cryptica CCMP332 with some minor deviations that concerned mainly the parts of the genes coding for presequences, but not those encoding the mature proteins. Gene copy numbers especially were slightly different, again arguing for recent gene duplications/losses. In addition, no gene predicted to encode a protein identical to Fcp7 that belongs to the Lhcx1 group could be found. The most surprising deviation was revealed only when testing the expression of the predicted proteins by MS. Eppard and Rhiel (1998) reported 5 to 6 copies of Fcp5, a gene that is closely related to CcLhcf3. Although only one CcLhcf3 gene and no gene for Fcp5 was found in C. cryptica, both proteins were revealed in C. meneghiniana by MS. Thus, Fcp5 in C. meneghiniana 1020-1a most probably derived from a gene duplication of CcLhcf3 that has not taken place in C. cryptica CCMP332.
In summary, Thalassiosira and Cyclotella are closely related species, but still differ in their Lhc sequences. Even more surprising, some differences were found between C. cryptica CCMP332 and C. meneghiniana 1020-1a. This highlights that Lhc genes seem to be highly variable in diatoms and gene duplications and/or losses seem to be common events.
Subunit Composition of Different FCP Complexes of C. meneghiniana
Using the newly derived, more complete set of Lhc sequences, we were able to analyze the subunit composition of the different purified FCP complexes. In total four different FCPa complexes and two FCPb complexes could be distinguished. The major FCPa3 and FCPa4 and about the same amounts of FCPb1 and FCPb2 were accompanied by FCPa1 and FCPa2, which are present only in minor amounts. When looking at the overall occurrence of subunits, Lhcf1 and Lhcf3/Fcp5 were the most abundant proteins, followed by Lhcf4/Lhcf6. Lhcf1 was the major subunit of the very abundant FCPa3 and FCPa4 complexes. Lhcf3 and Fcp5 occurred in every complex at least in small amounts and were the prominent subunits of both FCPb complexes. Lhcf4 and Lhcf6 were also present in almost all complexes (except for FCPb1) and were the main subunits of FCPa2. When the overall protein composition was compared with that of the pools of FCP complexes from T. pseudonana reported by Grouneva et al. (2011), results were very similar, even concerning the rough quantities of the subunits. In the pools of T. pseudonana FCPa and FCPb, however, Lhcf5 and Lhcf7 are also present, proteins for which no genes were found in Cyclotella. Lhcf5 is most closely related to Lhcf1 and might thus be redundant. Lhcf7 is only a minor subunit of FCPa in T. pseudonana, and in addition closely related to Lhcf11. Thus, although centric diatoms express many Lhcf proteins, the major FCP complexes are built predominantly of a small set of subunits. The remaining Lhcf proteins were found in only FCPb2 and FCPa2. In addition, the content of FCPa2 was highly dependent on the light intensity used for growth. Thus, the additional Lhcf proteins like Lhcf11 as well as FCP10 seem to be of regulatory function, whereas Lhcf1, Lhcf3/Fcp5, and Lhcf4/Lhcf6 are the more constitutive light-harvesting proteins.
The FCPa pool was differentiated into four subcomplexes of specific protein composition but almost equal pigmentation, except that the minor complex FCPa1 especially had an enhanced Chl a/(Chl c + Fx) ratio. Different populations of trimeric FCP complexes have also been reported before from the pennate diatom P. tricornutum (Gundermann et al., 2013). In contrast with the complexes of C. meneghiniana, these FCPs are devoid of Lhcx and only contain Lhcf subunits. In P. tricornutum, trimers of Lhcf5 are the most abundant FCP complexes, especially in LL-grown cells, characterized by a low Fx/Chl a ratio. Under HL, more complexes built mainly of Lhcf4 are present, which have a higher Fx/Chl a ratio. Therefore, in pennates not only the abundance of the major complexes but also the pigment stoichiometries changed, in contrast with what was seen here. Note that TpLhcf5/PtLhcf5 and TpLhcf4/PtLhcf4 are rather different despite their names, falling into different subgroups of Lhcf proteins present only in pennate or centric diatoms, respectively (Gundermann et al., 2013). In C. meneghiniana, only the minor FCPa1 and FCPa2 were relatively more abundant in the pool of HL-derived FCPa complexes. According to Chukhutsina et al. (2013), the overall amount of antenna Chl per photosystems is decreased under HL conditions as compared with LL. Thus, most probably in vivo, the amount of the minor complexes FCPa1 and FCPa2 per photosystem will not be increased under HL, but remain constant or become less strongly reduced than that of the major complexes FCPa3 and FCPa4.
As already reported by Grouneva et al. (2011) for T. pseudonana, Lhcx1 and Lhcx6_1 are present in FCPa complexes in C. meneghiniana. In both species the amounts are low, but significant. In C. meneghiniana Lhcx1 was found in the major FCPa4 with an increased amount when complexes were purified from cells grown under HL, and only under the latter condition in the minor FCPa1 and in FCPb2 as well. The other Lhcx protein, Lhcx6_1, was present in FCPa1, FCPa2, and FCPb2. The level of Lhcx6_1 proteins in the FCPa complexes did not change whether they came from HL- or LL-grown cells, but in FCPb2 the subunits were more prominent under HL. For Lhcx4 that is present in T. pseudonana, no gene could be identified in C. cryptica/meneghiniana. TpLhcx4 is most closely related to TpLhcx6 (Fig. 2), but also Lhcx6 is absent from FCPa complexes from C. meneghiniana. However, the same was reported for the pool of FCPa of T. pseudonana (Grouneva et al., 2011). For Lhcx6 an increased expression after short HL stress was demonstrated previously using whole T. pseudonana cells (Zhu and Green, 2010). After prolonged illumination levels dropped again, Lhcx6 might be present only after short-term exposure in the range of hours, but not in HL-grown cells as used here.
It must be emphasized that the purified FCPb2, although spectroscopically indistinguishable from FCPb1 and mainly composed of Lhcf3 and Fcp5 subunits, showed an important difference in binding Lhcx1 and Lhcx6_1 when isolated from cells cultured under HL, in contrast with FCPb1. Thus Lhcx1 is not only a subunit of trimeric FCPa complexes as known before, but also a subunit of a nonameric FCPb complex in centric diatoms. So far only FCPb1 (at that time called FCPb) has been tested for its in vitro ability to quench fluorescence, that is, for its intrinsic capability to contribute to NPQ. Although fluorescence yield is decreased upon aggregation in a manner comparable with FCPa (Elnour et al., 2018), no pH-dependent aggregation and quenching was observed (Gundermann and Büchel, 2008). Because FCPb1 does not contain Lhcx1 that was demonstrated to be indispensable for pH-dependent aggregation, this is not surprising. FCPb2, however, should probably be able to contribute to NPQ in vivo as well, because of its Lhcx1 content.
Purification of Fractions Composed of Different FCP Complexes
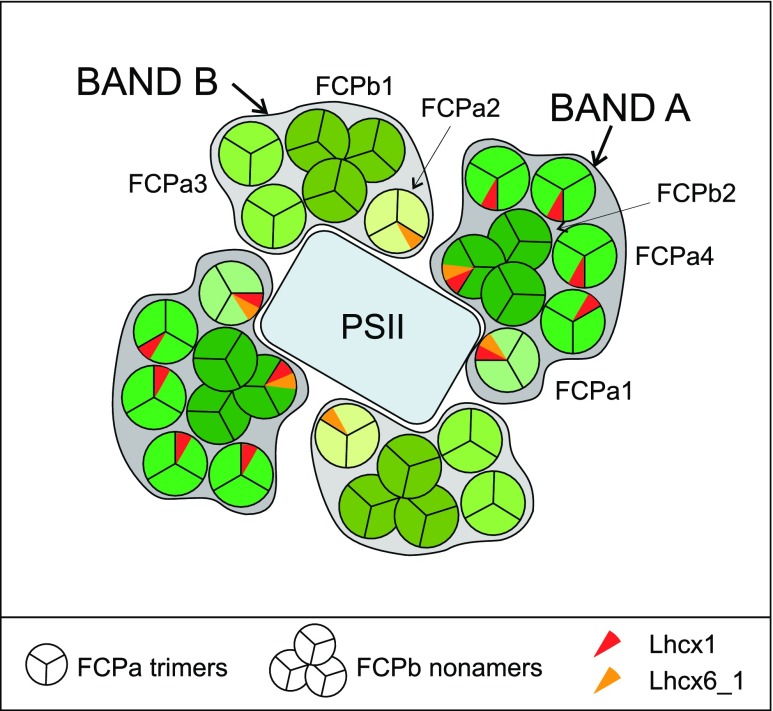
When the mild procedure of sucrose density centrifugation was used for separation of FCPs, two bands were obtained that differed in FCP composition, although comprising FCPa and FCPb complexes in both cases. As known from literature, such preparations also retain good amounts of lipids (Lepetit et al., 2010). Because of the separation principle of density gradients, simple coelution of different nonameric and trimeric complexes of similar protein composition in both bands seems unlikely, and the two bands are thus hypothesized to represent different supramolecular assemblies (Fig. 7). One band is enriched in FCP complexes that incorporate Lhcx1 especially under HL conditions (FCPa4, FCPa1, and FCPb2), and the other contains only little Lhcx1 because of its high content of FCPb1 and FCPa3. Both have a small amount of FCPa complexes that incorporate Lhcx6_1 (FCPa1 or FCPa2, respectively), and FCPb2 also contains Lhcx6_1 under HL conditions. This overall organization is extremely different to pennate diatoms like P. tricornutum, where (1) nonameric FCPs of specific composition are lacking, (2) Lhcx1 has not been found in trimeric FCP complexes so far, and (3) no Lhcx homologous to Lhcx6_1 is present.
Figure 7.
Model of the hypothetical arrangement of FCP complexes around PSII. Note that the actual stoichiometries inside the bands are different, especially with a higher content of FCPa3 and FCPa4 (see Fig. 6). Because Lhcx1 (red) and Lhcx6_1 (yellow) are present in different amounts in complexes from HL- or LL-grown cells (Tables 2 and 3), they are only indicated by small wedges. Note that PSI supercomplexes are built as outlined in Fig. 1B. Only Lhcx1 containing FCPa complexes, that is, mainly FCPa4 (and FCPa1) of Band A, have so far been demonstrated to be involved in the aggregation of complexes needed for NPQ (Gundermann and Büchel, 2012; Chukhutsina et al., 2014).
It has to be emphasized that both methods used here, that is, IEX and sucrose density centrifugation, subdivided the ‘free’ pool of FCP, those FCPs that are easily removed from the photosystems even by mild solubilization. When separating complexes by sucrose density centrifugation, PSI retains a considerable amount of antenna proteins, whereas only little Lhc are found in the PSII band (Veith and Büchel, 2007; Veith et al., 2009). Thus, most of the antenna complexes considered here are probably serving mainly PSII. This is supported because the red shifted Fx molecules as found in FCPb1 and FCPb2, that is, in Band B and Band A, respectively, are predominantly transferring excitation energy to PSII (Chukhutsina et al., 2013). A preferential localization of PSII in the inner core membranes of the typical stack of three thylakoids each has been shown recently (Flori et al., 2017). It is tempting to speculate that the more abundant FCPs of Band A serve PSII that is located in the core membranes. This is in line with the hypothesis of Lepetit et al. (2012) that the core membranes are enriched in monogalactosyl diglycerol. The lipid was shown to be essential for proper function of the enzyme converting DD to DT (Goss et al., 2009). Band A contained significantly more DD and its conversion product DT than Band B, supporting the idea of its preferential localization in the core membranes. Band B might then be more closely arranged to the minor amount of PSII found in the peripheral membranes, but the absence of Lhcx1 would leave these PSII complexes unprotected by the part of NPQ that is related to Lhcx1. Thus, all PSII complexes are probably surrounded by both supramolecular assemblies (Fig. 7), albeit maybe to a different extent in the core and peripheral thylakoid membranes. However, as long as the precise distribution of proteins and lipids between the core and peripheral membranes and the function of the other Lhcx proteins in centric diatoms is unknown, this question has to remain open.
Relevance of the Different FCP Complexes for NPQ
It was reported that in C. meneghiniana only FCPa complexes, but not FCPb, are able to quench fluorescence in a pH- and DT-dependent manner, that is, to contribute to the energy dependent part (qE) of NPQ in vivo (Gundermann and Büchel, 2012; Chukhutsina et al., 2014). This quenching ability was shown to depend on the Lhcx1 content that is increased under HL conditions (Gundermann and Büchel, 2008), whereby Lhcx1 is not the direct quencher but thought to enable the aggregation of FCPa complexes needed for the quenching (Ghazaryan et al., 2016). Thus, the Lhcx1 containing FCPa and FCPb2 complexes in Band A should be the ones involved in NPQ, an idea that is supported by the higher DT content of Band A in comparison with Band B, especially in HL thylakoids. FCPs of Band B then constitute the basic antenna of PSII, involved mainly in light harvesting but not in energy-dependent quenching relying on Lhcx1 and DT. Centric diatoms like Cyclotella show a biphasic development of the energy-dependent part of NPQ over time, termed qE1 and qE2, whereby the latter is dependent on DT accumulation (Grouneva et al., 2008). The first phase was recently shown to be subdivided into qE1a and qE1b, depending on FCPs close to PSII and on aggregated FCPs, respectively (Chukhutsina et al., 2014). FCP complexes containing Lhcx1, for example, FCPa4 and maybe FCPb2 present in Band A, will thus be responsible for qE1b because of aggregation mediated by Lhcx1. Quenching of fluorescence dependent on DT (qE2) is again more likely for Band A complexes, because they are enriched in DT. In this case, no attribution to certain complexes can be made, but at least for FCPa complexes DT-dependent quenching was proven (Gundermann and Büchel, 2012). Whereas the Lhcx1 content is strongly dependent on the illumination condition, Lhcx6_1 seems to be more constitutively present. If Lhcx6_1 is also involved in qE, qE1a might then be because of Lhcx6_1-enriched FCPa1/2 complexes that are present in both bands, which would argue for a localization closer to the core (Fig. 7). The same holds for FCPb2. If one compares the distribution and action of Lhcx proteins of C. meneghiniana with those of P. tricornutum, some similarities but also again differences become obvious: In P. tricornutum Lhcx3, but not Lhcx1, is much more up-regulated under HL conditions, making Lhcx1 the prevalent form under LL conditions (Taddei et al., 2018). Because Lhcx1 was detected in FCPs as well as in both PS fractions, whereas Lhcx3 was only found in FCP and PSI, Taddei et al. concluded that the PSII core quenching (qE2) is more related to Lhcx1 and constitutes a form of ‘basal’ quenching, whereas Lhcx3 (and maybe Lhcx2) is responsible for antenna quenching (qE1) introduced by prolonged HL conditions. Thus, Lhcx1 would play a different role in the two major groups of diatoms: it would be responsible for a basal, core-related quenching in pennate diatoms, but for antenna quenching inducible by prolonged HL in centrics, whereby the latter role is taken by Lhcx3 in pennates. In addition, Lhcx6_1 seems to constitute a quenching possibility that is more closely connected to the core, like Lhcx1 in pennates. However, the function of Lhcx6_1 is completely unknown so far, and this hypothesis will have to be proved in the future.
CONCLUSION
The FCP complexes of the thylakoid membranes of centric diatoms can be separated into two bands using sucrose density centrifugation, both consisting of trimeric as well as nonameric complexes of different subunit composition. Whereas Band A consists mainly of FCPa4 accompanied by FCPb2 and small amounts of FCPa1, Band B includes mainly FCPb1, FCPa3, and little FCPa2. Whereas Band A thus contains mainly the complexes that accumulate Lhcx1 under HL, this is not the case for Band B. Both bands contain Lhcx6_1 as subunits. The presence of nonameric and trimeric complexes of similar protein and pigment composition in both bands renders simple coelution unlikely, and the bands might thus represent supramolecular assemblies of different solubilization properties. Because of their spectral properties, they most likely serve PSII energetically. Thus, the antenna organization of centric diatoms is not only extremely different from what is known from vascular plants, but also differs significantly from the organization in pennate diatoms: differences concern not only oligomerization but, most importantly, the distribution of different Lhcx proteins known to work in photoprotection.
MATERIALS AND METHODS
FCP Preparation
The centric diatom Cyclotella meneghiniana (Culture Collection Göttingen, strain 1020-1a) was cultured for 7 d in modified artifical sea water medium according to Provasoli et al. (1957) under either LL (40 μmol quanta m−2 s−1) or HL (140 μmol quanta m−2 s−1) conditions with 16-h light and 8-h dark and constant shaking at 120 rpm.
Cells were harvested in the first hour of light by a centrifugation at 4304 g for 3–5 min at 4°C. Thylakoid preparation was performed according to the method described in Beer et al. (2006) by differential centrifugation after breaking the cells using a bead mill. Thylakoids of 0.25 mg Chl a/mL were solubilized with 20 mm β-dodecyl maltoside for 20 min on ice by gentle shaking. FCP complexes were then either isolated by IEX on DEAE-650S Toyopearl (Tosho Bioscience) followed by sucrose density centrifugation in buffer B1a (2 mm KCl, 25 mm Tris, 0.03% [w/v] β-dodecyl maltoside at pH 7.4) according to Gundermann and Büchel (2008) or by sucrose density centrifugation only (Büchel, 2003). In both cases subpopulations of the pool of FCP complexes were then separated by a further IEX run, using the same buffer system as before. For an overview of the preparation procedure see Fig. 3. Concentration of samples was performed using filtration devices with a Mr cutoff of 30 kDa (Centripreps, Merck Millipore) at 1000 g and 4°C. The samples were frozen in liquid nitrogen and stored at −80°C until further use.
Spectroscopy
Chlorophyll content was measured in 90% (v/v) acetone and calculated according to Jeffrey and Humphrey (1975). Absorption spectra were recorded in a Jasco Spectrophotometer (V-550) between 370 and 750 nm with 1-nm bandwidth and 1-cm optical path length. For recording fluorescence spectra, samples were adjusted in the QY maximum of Chl a to around 0.03, which corresponds to roughly 0.3 μg Chl a/ml. Fluorescence spectra were measured using a Jasco Fluorometer (FP-6500) with 3-nm bandwidth for the emission as well as for the excitation side. The photomultiplier was corrected via a calibrated lamp spectrum and the excitation side by a Rhodamin B spectrum. For emission spectra samples were excited at 465 nm to preferentially excite Chl c, and emission was recorded from 600 to 800 nm. For the excitation spectra the sample was excited from 400 to 600 nm and emission was recorded at 675 nm.
Pigment Analysis
Pigments were extracted with 90% (v/v) acetone (final concentration). Precipitated proteins were pelleted at 13,000 g in a precooled table top centrifuge for 10 min at 4°C, and the supernatant was injected into an HPLC system, run as described in Papagiannakis et al. (2005), using a reversed phase column (Lichrosorb RP18, 5 μm, 250 mm × 4 mm).
Protein Analyses
The oligomeric state of the FCP complexes was analyzed as described in Büchel (2003) and Beer et al. (2006) by analytical SEC using Superdex200 (GE Healthcare) as column material and a flow rate of 0.3 mL/min B1a buffer on an ÄKTA purifier P-900 (Amersham Biosciences). Elution was recorded at 437 nm, and the same size calibration was used as described before (Röding et al., 2018).
Proteins were separated by 10% (w/v) SDS-PAGE that contained 0.3% (w/v) piperazine diacrylamide as a cross-linker (Wagner et al., 2004) for LC-ESI-MS/MS analysis. Samples equivalent to 1 μg Chl a were denatured using Rotiload (4:1, v:v). Subsequent to the gel electrophoresis, the gels were silver stained (Heukeshoven and Dernick, 1985). For mass spectrometric analysis, the part of the lanes between about 15 and 25 kDa from HL- or LL-FCPs was excised, gel slices destained for 8 min at room temperature with freshly prepared destaining solution (15 mm K3[Fe(CN)6] and 50 mm NaS2O3), and washed four times with MS-grade water. Proteins in the gel slices were in-gel tryptically digested. The peptide solution was desalted by reversed phase chromatography using Zip-tip pipette tips (Veith et al., 2009) and then subjected to nano LC-ESI-MS/MS using an UltiMate 3000 nano HPLC apparatus (Dionex, now Thermo Fisher Scientific) coupled online with a linear ion trap mass spectrometer (Finnigan LTQ; Thermo Electron) as described previously (Veith et al., 2009). Data were analyzed using the Thermo Electron Corp. Proteome Discoverer software (version 1.4) that includes the SEQUEST algorithm (Link et al., 1999). A new database was created, consisting of the predicted Lhc-related C. cryptica CCMP332 protein sequences including all possible gene models and the translated Fcp sequences from C. meneghiniana (former C. cryptica) strain 1020-1a according to Eppard and Rhiel (1998). A false discovery rate of equal to or less than 1% was applied. The minimum Xcorr for the three charge states was set to 2.0 for +1, 2.5 for +2, and 3.0 for +3. Only proteins that were identified with at least two different unique peptides have been considered further and are listed in the tables.
Bioinformatic Methods
Lhcf, Lhcr, Lhcx, and FCP genes annotated for T. pseudonana in the JGI database (https://genome.jgi.doe.gov/Thaps3/Thaps3.home.html) were used for BLAT (basic local alignment search tool like) search in the C. cryptica CCMP332 genome data (http://genomes.mcdb.ucla.edu/cgi-bin/hgGateway?hgsid=75138&clade=plant&org=C.+cryptica&db=0), searching the translated nucleotide sequences against protein sequences. The published gene sequences for C. meneghiniana 1020-1a (Eppard and Rhiel, 1998) were also used for sequence retrieval. In the C. cryptica genome gene models are based on only automatic Augustus prediction so far. Every hit was checked using Clustal Omega (Sievers et al., 2011). In case of several predictions for the same locus, all possible models were taken into account for the identification of proteins in MS analyses. For calculation of simple phylogenetic trees, Clustal Omega using the default settings was used as well, whereby either of those gene models verified by MS were taken, or the gene models showing the closest resemblance to T. pseudonana genes.
Accession Numbers
Sequence data from this article can be found in the JGI database (https://genome.jgi.doe.gov/Thaps3/Thaps3.home.html) in the case of T. pseudonana and under http://genomes.mcdb.ucla.edu/cgi-bin/hgGateway?hgsid=75138&clade=plant&org=C.+cryptica&db=0 in the case of C. cryptica CCMP332. C. meneghiniana 1020-1a (former C. cryptica 1020-1a) sequences can be found in the GenBank/EMBL data libraries under accession numbers AJ000670 (Fcp1), AJ000545 (Fcp2), AJ000965 (Fcp3), AJ005838 (Fcp4), AJ000967 (Fcp5), AJ000968 (Fcp6), AJ000969 (Fcp7), AJ000970 (Fcp8), AJ000971 (Fcp9), and AJ005839 (Fcp12).
Supplemntal Data
The following supplemental materials are available:
Supplemental Figure S1. Comparison between gene sequences encoding subunits of FCP complexes.
Supplemental Figure S2. Absorption spectra of FCPa4, FCPb1 and free pigment (FP), and HPLC analysis of FP.
Supplemental Figure S3. Fluorescence emission spectra and fluorescence excitation spectra of subpopulations of HL-FCPa.
Supplemental Figure S4. Pigment stoichiometries in mol pigment/mol Chl a in FCPa1-4 from HL and LL grown cells.
Supplemental Figure S5. SEC elution profiles of HL-FCPa1-4, HL-FCPb2, HL- and LL-FCPb1 as well as LL-FCPb1*.
Supplemental Figure S6. Fluorescence spectra of HL-FCPb1, HL-FCPb2, HL-FCPa4, LL-FCPb1 and LL-FCPb1*.
Supplemental Figure S7. SEC elution profile of LL-Band A and LL-Band B taken directly from Suc gradients.
Supplemental Table S1. Results from mass spectrometric analysis of purified FCP complexes.
Acknowledgments
The authors wish to thank Dr. Matthias Schmidt and Kerstin Pieper for the preparation of samples for MS.
Footnotes
This work was supported by the Deutsche Forschungsgemeinschaft (DFG) (grant no. Bu812/10-1).
References
- Apt KE, Kroth-Pancic PG, Grossman AR (1996) Stable nuclear transformation of the diatom Phaeodactylum tricornutum. Mol Gen Genet 252: 572–579 [DOI] [PubMed] [Google Scholar]
- Armbrust EV, Berges JA, Bowler C, Green BR, Martinez D, Putnam NH, Zhou S, Allen AE, Apt KE, Bechner M, et al. (2004) The genome of the diatom Thalassiosira pseudonana: Ecology, evolution, and metabolism. Science 306: 79–86 [DOI] [PubMed] [Google Scholar]
- Bailleul B, Rogato A, de Martino A, Coesel S, Cardol P, Bowler C, Falciatore A, Finazzi G (2010) An atypical member of the light-harvesting complex stress-related protein family modulates diatom responses to light. Proc Natl Acad Sci USA 107: 18214–18219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barka F, Angstenberger M, Ahrendt T, Lorenzen W, Bode HB, Büchel C (2016) Identification of a triacylglycerol lipase in the diatom Phaeodactylum tricornutum. Biochim Biophys Acta 1861: 239–248 [DOI] [PubMed] [Google Scholar]
- Beer A, Gundermann K, Beckmann J, Büchel C (2006) Subunit composition and pigmentation of fucoxanthin-chlorophyll proteins in diatoms: Evidence for a subunit involved in diadinoxanthin and diatoxanthin binding. Biochemistry 45: 13046–13053 [DOI] [PubMed] [Google Scholar]
- Bowler C, Allen AE, Badger JH, Grimwood J, Jabbari K, Kuo A, Maheswari U, Martens C, Maumus F, Otillar RP, et al. (2008) The Phaeodactylum genome reveals the evolutionary history of diatom genomes. Nature 456: 239–244 [DOI] [PubMed] [Google Scholar]
- Büchel C. (2003) Fucoxanthin-chlorophyll proteins in diatoms: 18 and 19 kDa subunits assemble into different oligomeric states. Biochemistry 42: 13027–13034 [DOI] [PubMed] [Google Scholar]
- Büchel C. (2014) Fucoxanthin-chlorophyll-proteins and non-photochemical fluorescence quenching of diatoms. In Demmig-Adams B, Garab G, Adams W, Govindjee III, eds, Non-photochemical quenching and energy dissipation in plants, algae and cyanobacteria, Vol 40 Springer Verlag, Dordrecht, pp 259–275 [Google Scholar]
- Büchel C. (2015) Evolution and function of light harvesting proteins. J Plant Physiol 172: 62–75 [DOI] [PubMed] [Google Scholar]
- Cavalier-Smith T. (2013) Symbiogenesis: Mechanisms, evolutionary consequences, and systematic implications. Annu Rev Ecol Evol Syst 44: 145–172 [Google Scholar]
- Chukhutsina VU, Büchel C, van Amerongen H (2013) Variations in the first steps of photosynthesis for the diatom Cyclotella meneghiniana grown under different light conditions. Biochim Biophys Acta 1827: 10–18 [DOI] [PubMed] [Google Scholar]
- Chukhutsina VU, Büchel C, van Amerongen H (2014) Disentangling two non-photochemical quenching processes in Cyclotella meneghiniana by spectrally-resolved picosecond fluorescence at 77K. Biochim Biophys Acta 1837: 899–907 [DOI] [PubMed] [Google Scholar]
- Dong H-P, Dong Y-L, Cui L, Balamurugan S, Gao J, Lu S-H, Jiang T (2016) High light stress triggers distinct proteomic responses in the marine diatom Thalassiosira pseudonana. BMC Genomics 17: 994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong Y-L, Jiang T, Xia W, Dong H-P, Lu S-H, Cui L (2015) Light harvesting proteins regulate non-photochemical fluorescence quenching in the marine diatom Thalassiosira pseudonana. Algal Res 12: 300–307 [Google Scholar]
- Elnour HMAM, Dietzel L, Ramanan C, Büchel C, van Grondelle R, Krüger TPJ (2018) Energy dissipation mechanisms in the FCPb light-harvesting complex of the diatom Cyclotella meneghiniana. Biochim Biophys Acta Bioenerg 1859: 1151–1160 [DOI] [PubMed] [Google Scholar]
- Eppard M, Rhiel E (1998) The genes encoding light-harvesting subunits of Cyclotella cryptica (Bacillariophyceae) constitute a complex and heterogeneous family. Mol Gen Genet 260: 335–345 [DOI] [PubMed] [Google Scholar]
- Eppard M, Krumbein WE, von Haesler A, Rhiel E (2000) Characterization of fcp4 and fcp12, two additional genes encoding light harvesting proteins of Cyclotella cryptica (Bacillariophyceae) and phylogenetic analysis of this complex gene family. Plant Biol 2: 283–289 [Google Scholar]
- Falkowski PG, Barber RT, Smetacek V (1998) Biogeochemical controls and feedbacks on ocean primary production. Science 281: 200–207 [DOI] [PubMed] [Google Scholar]
- Flori S, Jouneau P-H, Bailleul B, Gallet B, Estrozi LF, Moriscot C, Bastien O, Eicke S, Schober A, Bártulos CR, et al. (2017) Plastid thylakoid architecture optimizes photosynthesis in diatoms. Nat Commun 8: 15885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghazaryan A, Akhtar P, Garab G, Lambrev PH, Büchel C (2016) Involvement of the Lhcx protein Fcp6 of the diatom Cyclotella meneghiniana in the macro-organisation and structural flexibility of thylakoid membranes. Biochim Biophys Acta 1857: 1373–1379 [DOI] [PubMed] [Google Scholar]
- Gildenhoff N, Amarie S, Gundermann K, Beer A, Büchel C, Wachtveitl J (2010) Oligomerization and pigmentation dependent excitation energy transfer in fucoxanthin-chlorophyll proteins. Biochim Biophys Acta 1797: 543–549 [DOI] [PubMed] [Google Scholar]
- Goss R, Nerlich J, Lepetit B, Schaller S, Vieler A, Wilhelm C (2009) The lipid dependence of diadinoxanthin de-epoxidation presents new evidence for a macrodomain organization of the diatom thylakoid membrane. J Plant Physiol 166: 1839–1854 [DOI] [PubMed] [Google Scholar]
- Grouneva I, Jakob T, Wilhelm C, Goss R (2008) A new multicomponent NPQ mechanism in the diatom Cyclotella meneghiniana. Plant Cell Physiol 49: 1217–1225 [DOI] [PubMed] [Google Scholar]
- Grouneva I, Rokka A, Aro E-M (2011) The thylakoid membrane proteome of two marine diatoms outlines both diatom-specific and species-specific features of the photosynthetic machinery. J Proteome Res 10: 5338–5353 [DOI] [PubMed] [Google Scholar]
- Grouneva I, Muth-Pawlak D, Battchikova N, Aro E-M (2016) Changes in relative Thylakoid protein abundance induced by fluctuating light in the diatom Thalassiosira pseudonana. J Proteome Res 15: 1649–1658 [DOI] [PubMed] [Google Scholar]
- Gundermann K, Büchel C (2008) The fluorescence yield of the trimeric fucoxanthin-chlorophyll-protein FCPa in the diatom Cyclotella meneghiniana is dependent on the amount of bound diatoxanthin. Photosynth Res 95: 229–235 [DOI] [PubMed] [Google Scholar]
- Gundermann K, Büchel C (2012) Factors determining the fluorescence yield of fucoxanthin-chlorophyll complexes (FCP) involved in non-photochemical quenching in diatoms. Biochim Biophys Acta 1817: 1044–1052 [DOI] [PubMed] [Google Scholar]
- Gundermann K, Schmidt M, Weisheit W, Mittag M, Büchel C (2013) Identification of several sub-populations in the pool of light harvesting proteins in the pennate diatom Phaeodactylum tricornutum. Biochim Biophys Acta 1827: 303–310 [DOI] [PubMed] [Google Scholar]
- Heukeshoven J, Dernick R (1985) Simplified method for silver staining of proteins in polyacrylamide gels and the mechanism of silver staining. Electrophoresis 6: 103–112 [Google Scholar]
- Jeffrey SW, Humphrey GF (1975) New spectrometric equations for determining chlorophyll a, b, c1 and c2 in higher plants, algae and natural phytoplankton. Biochem Physiol Pflanz 167: 191–194 [Google Scholar]
- Keeling PJ. (2013) The number, speed, and impact of plastid endosymbioses in eukaryotic evolution. Annu Rev Plant Biol 64: 583–607 [DOI] [PubMed] [Google Scholar]
- Lepetit B, Volke D, Szabó M, Hoffmann R, Garab G, Wilhelm C, Goss R (2007) Spectroscopic and molecular characterization of the oligomeric antenna of the diatom Phaeodactylum tricornutum. Biochemistry 46: 9813–9822 [DOI] [PubMed] [Google Scholar]
- Lepetit B, Volke D, Gilbert M, Wilhelm C, Goss R (2010) Evidence for the existence of one antenna-associated, lipid-dissolved and two protein-bound pools of diadinoxanthin cycle pigments in diatoms. Plant Physiol 154: 1905–1920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lepetit B, Goss R, Jakob T, Wilhelm C (2012) Molecular dynamics of the diatom thylakoid membrane under different light conditions. Photosynth Res 111: 245–257 [DOI] [PubMed] [Google Scholar]
- Lepetit B, Sturm S, Rogato A, Gruber A, Sachse M, Falciatore A, Kroth PG, Lavaud J (2013) High light acclimation in the secondary plastids containing diatom Phaeodactylum tricornutum is triggered by the redox state of the plastoquinone pool. Plant Physiol 161: 853–865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lepetit B, Gélin G, Lepetit M, Sturm S, Vugrinec S, Rogato A, Kroth PG, Falciatore A, Lavaud J (2017) The diatom Phaeodactylum tricornutum adjusts nonphotochemical fluorescence quenching capacity in response to dynamic light via fine-tuned Lhcx and xanthophyll cycle pigment synthesis. New Phytol 214: 205–218 [DOI] [PubMed] [Google Scholar]
- Link AJ, Eng J, Schieltz DM, Carmack E, Mize GJ, Morris DR, Garvik BM, Yates JR III (1999) Direct analysis of protein complexes using mass spectrometry. Nat Biotechnol 17: 676–682 [DOI] [PubMed] [Google Scholar]
- Mock T, Otillar RP, Strauss J, McMullan M, Paajanen P, Schmutz J, Salamov A, Sanges R, Toseland A, Ward BJ, et al. (2017) Evolutionary genomics of the cold-adapted diatom Fragilariopsis cylindrus. Nature 541: 536–540 [DOI] [PubMed] [Google Scholar]
- Nagao R, Ishii A, Tada O, Suzuki T, Dohmae N, Okumura A, Iwai M, Takahashi T, Kashino Y, Enami I (2007) Isolation and characterization of oxygen-evolving thylakoid membranes and photosystem II particles from a marine diatom Chaetoceros gracilis. Biochim Biophys Acta 1767: 1353–1362 [DOI] [PubMed] [Google Scholar]
- Nagao R, Tomo T, Noguchi E, Nakajima S, Suzuki T, Okumura A, Kashino Y, Mimuro M, Ikeuchi M, Enami I (2010) Purification and characterization of a stable oxygen-evolving Photosystem II complex from a marine centric diatom, Chaetoceros gracilis. Biochim Biophys Acta 1797: 160–166 [DOI] [PubMed] [Google Scholar]
- Nagao R, Takahashi S, Suzuki T, Dohmae N, Nakazato K, Tomo T (2013a) Comparison of oligomeric states and polypeptide compositions of fucoxanthin chlorophyll a/c-binding protein complexes among various diatom species. Photosynth Res 117: 281–288 [DOI] [PubMed] [Google Scholar]
- Nagao R, Yokono M, Akimoto S, Tomo T (2013b) High excitation energy quenching in fucoxanthin chlorophyll a/c-binding protein complexes from the diatom Chaetoceros gracilis. J Phys Chem B 117: 6888–6895 [DOI] [PubMed] [Google Scholar]
- Nagao R, Yokono M, Tomo T, Akimoto S (2014) Control mechanism of excitation energy transfer in a complex consisting of Photosystem II and fucoxanthin chlorophyll a/c-binding protein. J Phys Chem Lett 5: 2983–2987 [DOI] [PubMed] [Google Scholar]
- Papagiannakis E, H M van Stokkum I, Fey H, Büchel C, van Grondelle R (2005) Spectroscopic characterization of the excitation energy transfer in the fucoxanthin-chlorophyll protein of diatoms. Photosynth Res 86: 241–250 [DOI] [PubMed] [Google Scholar]
- Provasoli L, McLaughlin JJA, Droop MR (1957) The development of artificial media for marine algae. Arch Mikrobiol 25: 392–428 [DOI] [PubMed] [Google Scholar]
- Pyszniak AM, Gibbs SP (1992) Immunocytochemical localization of photosystem I and the fucoxanthin-chlorophyll a/c light-harvesting complex in the diatom Phaeodactylum tricornutum. Protoplasma 166: 208–217 [Google Scholar]
- Röding A, Boekema E, Büchel C (2018) The structure of FCPb, a light-harvesting complex in the diatom Cyclotella meneghiniana. Photosynth Res 135: 203–211 [DOI] [PubMed] [Google Scholar]
- Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWillam H, Remmert M, Söding J, et al. (2011) Fast, scalable generation of high‐quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol 7: 539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taddei L, Stella GR, Rogato A, Bailleul B, Fortunato AE, Annunziata R, Sanges R, Thaler M, Lepetit B, Lavaud J, et al. (2016) Multisignal control of expression of the LHCX protein family in the marine diatom Phaeodactylum tricornutum. J Exp Bot 67: 3939–3951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taddei L, Chukhutsina VU, Lepetit B, Stella GR, Bassi R, van Amerongen H, Bouly J-P, Jaubert M, Finazzi G, Falciatore A (2018) Dynamic changes between two LHCX-related energy quenching sites control diatom photoacclimation. Plant Physiol 177: 953–965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traller JC, Cokus SJ, Lopez DA, Gaidarenko O, Smith SR, McCrow JP, Gallaher SD, Podell S, Thompson M, Cook O, et al. (2016) Genome and methylome of the oleaginous diatom Cyclotella cryptica reveal genetic flexibility toward a high lipid phenotype. Biotechnol Biofuels 9: 258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veith T, Büchel C (2007) The monomeric photosystem I-complex of the diatom Phaeodactylum tricornutum binds specific fucoxanthin chlorophyll proteins (FCPs) as light-harvesting complexes. Biochim Biophys Acta 1767: 1428–1435 [DOI] [PubMed] [Google Scholar]
- Veith T, Brauns J, Weisheit W, Mittag M, Büchel C (2009) Identification of a specific fucoxanthin-chlorophyll protein in the light harvesting complex of photosystem I in the diatom Cyclotella meneghiniana. Biochim Biophys Acta 1787: 905–912 [DOI] [PubMed] [Google Scholar]
- Wagner V, Fiedler M, Markert C, Hippler M, Mittag M (2004) Functional proteomics of circadian expressed proteins from Chlamydomonas reinhardtii. FEBS Lett 559: 129–135 [DOI] [PubMed] [Google Scholar]
- Yokono M, Nagao R, Tomo T, Akimoto S (2015) Regulation of excitation energy transfer in diatom PSII dimer: How does it change the destination of excitation energy? Biochim Biophys Acta 1847: 1274–1282 [DOI] [PubMed] [Google Scholar]
- Zhu S-H, Green BR (2010) Photoprotection in the diatom Thalassiosira pseudonana: role of LI818-like proteins in response to high light stress. Biochim Biophys Acta 1797: 1449–1457 [DOI] [PubMed] [Google Scholar]