Abstract
Integration of unusual domains in plant immune receptors is a widespread mechanism of NLR diversification enabling specific pathogen detection.
Unlike animals, plants lack an adaptive and circulating immune system. Thus, to detect pathogens and generate effective defense responses, plants rely on an elaborate innate immunity that involves different types of immune receptors (Cook et al., 2015). Conserved pathogen-associated molecular patterns are recognized in the extracellular compartment of the host by cell surface-localized receptors. This event triggers the activation of basal immune responses called pathogen-associated molecular pattern-triggered immunity (PTI). During evolution, pathogens have evolved sophisticated virulence strategies to overcome host defense responses. Host-adapted pathogens use an arsenal of virulence factors called effectors that are delivered into the plant cell in order to subvert diverse cellular functions (effector-triggered susceptibility) through interference with PTI signaling (Jones and Dangl, 2006). However, effector activities can turn against the pathogen, as they often betray its presence within the cell. To recognize pathogen effectors, plants use a repertoire of intracellular immune receptors that belong to a superfamily of nucleotide-binding domain and leucine-rich repeat (LRR)-containing proteins (NLRs). NLRs can mediate the specific recognition of pathogen effectors and initiate effector-triggered immunity (ETI). ETI involves transcriptional reprogramming overlapping with transcriptional regulations during PTI and often provokes localized host cell death at infection sites to limit pathogen spread (Jones and Dangl, 2006; Maekawa et al., 2011). Adapted pathogens can evade host recognition by reconfiguring their effector repertoires through various mechanisms including gain and loss of effector genes, modulation of expression, and rapid evolution of effectors by mutation (Arnold and Jackson, 2011; Lo Presti et al., 2015). Therefore, cycles of pathogen-induced effector-triggered susceptibility and plant-mediated ETI are considered as major forces driving plant host-pathogen coevolution (Jones and Dangl, 2006).
NLR proteins belong to STAND (Signal Transduction ATPases with Numerous Domains) P-loop NTPases. Canonical plant NLRs possess a multidomain architecture composed of a central nucleotide-binding site (NBS) and a C-terminal LRR domain. The NBS is believed to function, through nucleotide-dependent conformational changes, as a molecular switch for NLR activation (Takken et al., 2006). Depending on the nature of their N-terminal domain, NLRs can be divided into two main classes: those having a Toll and IL-1 receptor (TIR) domain and those with a coiled-coil (CC) domain (Takken and Goverse, 2012). A third class, based on the presence of the N-terminal RPW8 domain, also can be defined. Over the last two decades, with the cloning of plant NLRs and their associated effectors, molecular characterization of the mechanisms employed by NLRs for specific effector recognition and signaling have been the subject of intensive research.
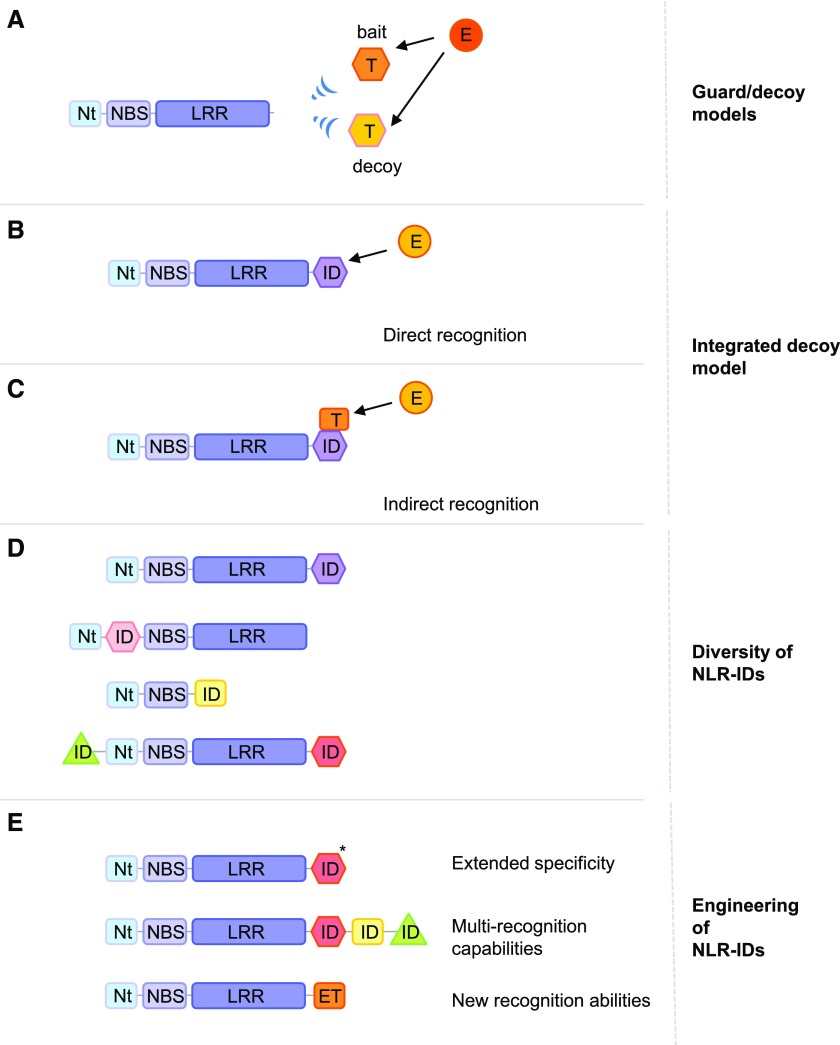
NLR-mediated effector recognition often involves host components that bind to and/or are modified by effectors (Fig. 1). In the guard model, effector interference with a host target (cofactor or bait) is detected by the NLR, acting as a guard of modified self (Dodds and Rathjen, 2010; Maekawa et al., 2011). Many identified guarded host proteins (also referred to as guardees) represent hubs with key immune-related functions, including signaling or the regulation of gene expression, and therefore are commonly targeted by various effectors (Mukhtar et al., 2011; Weßling et al., 2014). An effector decoy model has been proposed for a number of effector-sensing NLRs. In this model, the guarded host protein has no defense role but mimics an operational effector target and, thus, acts as a decoy that lures the pathogen effector and diverts it from its real targets (van der Hoorn and Kamoun, 2008; Lewis et al., 2013; Ntoukakis et al., 2014). Studies have shown that several NLRs can both detect pathogens and initiate downstream signaling, whereas other NLR proteins form heterogenous protein complexes (Césari et al., 2014b; Williams et al., 2014). In the core of these complexes are NLR pairs in which the two members are encoded by genes arranged in a head-to-head orientation with a common promoter region, which strongly suggests their coregulation (Birker et al., 2009; Saucet et al., 2015). In several cases, the two proteins of NLR pairs form a heterocomplex receptor with each partner featuring specific attributes: one detects pathogen effectors (the sensor) while the other functions as an inducer of disease resistance (the transducer), and the signaling activity of the latter is repressed by the sensor (Césari et al., 2014b; Williams et al., 2014). To explain how effectors are recognized by NLR pairs, an extension of the decoy model has been proposed with the integrated decoy hypothesis (Cesari et al., 2014a). Indeed, sensor NLR partners were shown to contain, in addition to their conserved multidomain NLR architecture, unconventional domains that are able to interact physically with their corresponding effectors (Kanzaki et al., 2012; Cesari et al., 2013). Recent studies demonstrated that several of these integrated domains (IDs) act as decoys of effector targets, enabling the sensor NLR to specifically detect pathogens (Fig. 1; Le Roux et al., 2015; Maqbool et al., 2015; Ortiz et al., 2017). Perturbations of the sensor NLR are perceived by the signaling partner, which activates immune signaling (Le Roux et al., 2015; Sarris et al., 2015; Ortiz et al., 2017). Comparative analyses of plant immune receptor architectures suggest that the integration of unusual domains, which potentially serve as baits for pathogen effectors, is not restricted to paired NLRs and represent a widespread mechanism (Kroj et al., 2016; Sarris et al., 2016; Bailey et al., 2018). The identification of NLR-IDs signifies a breakthrough in plant NLR biology pathology, since it has profoundly changed our view of how plant NLRs can function and evolve.
Figure 1.
A, NLRs can directly or indirectly detect the presence of pathogen effectors by monitoring the manipulation of their host targets (baits or decoys). B and C, According to the integrated decoy model, integrated domains (IDs) in NLRs behave as decoys of effector targets, enabling the recognition of effector activities. This recognition can be direct (B) or indirect (C). D, Different studies reported the existence of diverse IDs (in sequence and predicted molecular functions), which can be present at various positions within the modular structure of NLRs. E, NLR-IDs can be engineered using different strategies aimed at providing (1) extended specificity (i.e. specific point mutations in IDs enabling the recognition of various allelic forms of a pathogen effector), (2) multirecognition capabilities (by integrating IDs from different NLRs within a single NLR), or (3) new recognition specificities (by integrating previously characterized effector targets that then act as sensors). E, Effector; ET, previously characterized effector target; Nt, N-terminal domain; T, effector target.
In this review, we summarize current knowledge of NLR-IDs with detailed examples, discuss their genetic and functional diversity, and illustrate how the study of NLR function and mode of action has led to advances in plant disease control.
NLRS WITH INTEGRATED DECOYS: AN INGENIOUS PATHOGEN DETECTION MECHANISM
Recent independent studies have provided convincing evidence that IDs enable the specific detection of pathogens by acting as molecular decoys that structurally mimic pathogen true virulence targets to monitor host immunosuppression attempts. How these IDs confer effector recognition and trigger the activation of immune signaling are very intriguing questions. Well-characterized examples of NLR-ID fusions in paired NLRs include the Arabidopsis (Arabidopsis thaliana) RRS1, which carries a WRKY domain (Le Roux et al., 2015), and the RGA5 and Pik-1 proteins from rice (Oryza sativa), both of which integrate a heavy metal-associated (HMA; RATX1) domain (Ortiz et al., 2017). These examples are described in detail below.
The WRKY Domain of RRS1-R
Experimental validation of the integrated decoy model was first provided for the Arabidopsis/Ralstonia solanacearum model. In 2001, Deslandes and colleagues cloned a resistance gene encoding RRS1-R (RESISTANCE TO RALSTONIA SOLANACEARUM1) conferring broad-spectrum resistance to the soil-borne bacterium R. solanacearum, the causal agent of bacterial wilt (Deslandes et al., 1998, 2002). RRS1-R contains at its C terminus a WRKY DNA-binding domain. This domain is conserved in defensive plant WRKY transcription factors that orchestrate biotic stress responses through the recognition of W-box cis-regulatory elements in gene promoters (Rushton et al., 2010). As the first cloned NLR with an extra domain, RRS1-R was initially considered an anomaly in the field. Later, RRS1-R was shown to cooperate genetically and molecularly with a second TIR-NB-LRR, namely RPS4 (RESISTANCE TO PSEUDOMONAS SYRINGAE4; Birker et al., 2009; Narusaka et al., 2009), to recognize effectors from different pathogens. These effectors included R. solanacearum PopP2, a member of the YopJ superfamily of acetyltransferase (Deslandes et al., 2003; Tasset et al., 2010), and AvrRps4, an unrelated effector from leaf-infecting Pseudomonas syringae pv pisi (Hinsch and Staskawicz, 1996; Sohn et al., 2012). Encoded by two coregulated genes present in a head-to-head orientation, RRS1-R and RPS4 TIR-NB-LRRs form a functional receptor recognition/signaling complex through homodimerization and heterodimerization involving their respective TIR domains (Williams et al., 2014). Two recent studies revealed that the RRS1-R/RPS4 NLR complex is activated through the targeting of the RRS1-R WRKY domain by PopP2 and AvrRps4 effectors (Le Roux et al., 2015; Sarris et al., 2015). Catalytically active PopP2 acetylates a key Lys residue (K1221) in the invariant heptad of the WRKY domain of RRS1-R, blocking its binding to W-box DNA sequences. Homology modeling predicts that K1221 acetylation disrupts WRKY domain electrostatic potential at the interface with DNA. In the absence of RRS1-R/RPS4 recognition, PopP2 uses this acetylation strategy to inhibit WRKY DNA-binding activities and transactivation functions needed for defense gene expression and basal resistance. Therefore, the RRS1-R WRKY domain represents a decoy that betrays the defense-suppressing abilities of PopP2 and AvrRps4 on their host virulence targets: the defensive WRKY transcription factors. The direct fusion of a WRKY decoy domain within the RRS1-R/RPS4 NLR complex creates an efficient monitoring system for the indispensable virulence activities of different pathogens. Recently, Ma et al. (2018) demonstrated that, prior to effector detection, the WRKY domain negatively regulates the RPS4-RRS1 complex through specific interactions with an adjacent domain in RRS1. Binding of AvrRps4 to the WRKY domain disrupts these intramolecular interactions, leading to the derepression of RRS1. Therefore, besides its effector-sensing function, the integrated WRKY domain of RRS1 also has an important regulatory role in preventing inappropriate receptor activation in the absence of pathogens.
The HMA Domain of RGA5 and Pik-1
The study of the RGA4/RGA5 receptor NLR pair in rice has enabled significant progress in deciphering the mode of action of paired NLRs. This NLR pair cooperates genetically and physically in the recognition of AVR-PiA and AVR1-CO39, two unrelated effectors of the rice blast fungus Magnaporthe oryzae (Okuyama et al., 2011; Cesari et al., 2013). In the absence of the pathogen, constitutive disease resistance and cell death mediated by RGA4 is repressed by RGA5 through the formation of heteroprotein complexes. The C-terminal part of RGA5 contains an HMA domain, initially found in a cytoplasmic copper chaperone in Saccharomyces cerevisiae, which can interact directly with AVR-PiA and AVR1-CO39, enabling pathogen recognition. Physical association of the AVR-PiA effector with the HMA domain of RGA5 triggers cell death through RGA4 derepression (Césari et al., 2014b). Interestingly, recognition of the AVR-Pik effector of M. oryzae by the unrelated CC-NLR pair Pik-1/Pik-2 in rice also is triggered by direct binding to an HMA domain in Pik-1 that, contrary to RGA5, is integrated between its CC and nucleotide-binding domain regions (Ashikawa et al., 2008; Maqbool et al., 2015). The different locations of the HMA domain in RGA5 and Pik-1 suggests that these domains have been fused to those two unrelated NLRs through independent events (Cesari et al., 2013). Although HMA domain-containing proteins have not been described previously as effector targets, the presence of an HMA domain in the rice Pi21 factor, which is required for full susceptibility to the rice blast fungus (Fukuoka et al., 2009; Zhang et al., 2016), supports the idea that the HMA domains of RGA5 and Pik-1 are decoys for AVR-PiA, AVR1-CO39, AVR-Pik, and functionally related effectors. Determination of the crystal structure of AVR-PikD complexed with a dimer of the Pikp-1 HMA domain revealed that key residues at the interaction interface are required for effector binding and recognition (Maqbool et al., 2015). In addition, variations at binding interfaces between AVR-Pik effector variants and HMA domains of Pik alleles were found to determine recognition specificity. Such recent findings highlight how new receptor specificities arise from natural selection (De la Concepcion et al., 2018). How the binding of effectors to HMA domains can trigger the activation of immune signaling remains unknown. It is hypothesized that effector binding promotes NLR domain rearrangements leading to immune complex activation (Césari et al., 2014b). The binding of AVR-PiA to the RGA5 HMA domain also is necessary for pathogen recognition, but protein-protein interaction analyses revealed that moderate affinity to mutated AVR-PiA proteins still confers recognition (Ortiz et al., 2017). Furthermore, additional sites in RGA5, outside the ID, are suspected to mediate interaction with the effector. Thus, the juxtaposition of integrated decoy domains with NLR sites having additional interacting properties creates a highly resilient surveillance system.
The NOI/RIN4 Domain of Pii-2
Pathogen detection by paired NLR-IDs is not restricted to the direct recognition model. Indeed, IDs also might function in indirect recognition by perceiving modifications of a host protein targeted by an effector. This concept is supported by a study of the unconventional NOI/RIN4 domain of the rice NLR-ID Pii-2 that is hypothesized to monitor, through direct binding, the integrity of the OsExo70-F3 host protein, a target of the M. oryzae effector AVR-Pii (Fujisaki et al., 2017).
NLR-IDS: A MECHANISM OF NLR DIVERSIFICATION
The search for protein domains associated with typical NLR domains in public databases made it possible to identify entire NLR-ID directories and to analyze their structure in many plants. Already present in mosses, NLR-IDs occasionally represent a large proportion of NLRs in terrestrial plants (Kroj et al., 2016; Sarris et al., 2016; Bailey et al., 2018; Stein et al., 2018; Table 1). Kroj et al. (2016) detected 162 hypothetical IDs across 33 genomes by looking for interpro domains using the GreenPhyl database. Although their analysis was not exhaustive due to potential misannotations of the applied databases, they identified a high diversity of IDs (90 different domains). Sarris et al. (2016) reported 265 unique IDs fused to NLRs in 37 genomes of land plants. More recently, Bailey et al. (2018) identified NLR-IDs in nine grass species. Thirty-one types of different domains were represented mainly in these species. By focusing on 13 Oryza species, Stein et al. (2018) described a highly variable structure of genes coding for several hundreds of different NLR-ID proteins. They were able to detect a significant enrichment for these NLR-IDs within pairs of genes arranged in a head-to-head configuration in the genomes. The widespread distribution of NLR-IDs, despite their low abundance in some plant genomes (Table 1), suggests a successful evolutionary mechanism of NLR diversification commonly used by plants to expand their pathogen recognition capabilities, allowing them to cope with highly and rapidly adaptable pathogen-derived molecules. Accordingly, the IDs identified in these studies are derived from proteins that are extremely diverse in sequence and predicted molecular functions. The most frequent domains found in NLR-IDs include the WKRY and BED (BEAF and DREF proteins from Drosophila) zinc finger (Znf-BED) DNA-binding domains and the protein kinase domains. The decoy function of the WRKY domain in the RRS1-R NLR already has been validated (see above; Le Roux et al., 2015; Sarris et al., 2015). The Znf-BED domain was identified originally in transposases and transcription factors (Hayward et al., 2013), but, contrary to RRS1-R, the targeting by pathogen effectors remains to be demonstrated.
Table 1. Overview of NLR-ID repertoires in plant species.
| Reference | No. of Species Investigated | No. of Species with NLR-IDs | No. of NLR-IDs | No. of NLR-IDs per Species | Average Percentage of NLR-IDs (of All NLRs) |
|---|---|---|---|---|---|
| Kroj et al. (2016) | 33 | 23 | 34 | 1 to 16 | 3.5 |
| Sarris et al. (2016) | 36 | 35 | 717 | 1 to 93 | 6.8 |
| Bailey et al. (2018) | 9 | 9 | 331 | 7 to 133 | 7.9 |
| Funk et al. (2018) | 1 | 1 | 24 | 24 | 14 |
| Stein et al. (2018) | 13 | 13 | 446 | – | 8.2 |
However, Kroj et al. (2016) showed that the ZBED NLR protein from rice, containing three BED domains, is required for resistance to M. oryzae. In response to the pathogen, ZBED-overexpressing lines were more resistant, whereas a zbed null mutant showed increased susceptibility. These data strongly suggest that the BED domains in ZBED NLR proteins represents decoys that mimic host BED proteins targeted by M. oryzae effectors. The Xa1 NLR from rice, which confers resistance against isolates of the bacterial blight pathogen Xanthomonas oryzae by recognizing multiple transcription activator-like effectors (TALEs; Yoshimura et al., 1998; Ji et al., 2016), contains a Znf-BED domain in its N-terminal part. The mechanism that allows Xa1 to recognize TALEs remains to be elucidated. It is tempting to speculate that the Xa1 Znf-BED domain also might act as a decoy to lure TALEs that target host Znf-BED proteins for the subversion of host gene expression (Zuluaga et al., 2017).
Also, the functionality of NLR IDs with predicted catalytically active protein kinase domains needs to be experimentally validated. However, their sensing abilities can be deduced from well-described examples of kinases acting as decoys that interact physically with classical NLRs (e.g. the kinases Pto and PBS5 interacting with the NLRs Prf and RPS5, respectively; Khan et al., 2016). It is noteworthy that, in primitive land plants, the fusion of kinase domains or DUF676 to NBS-LRRs that lack CC or TIR domains has been described (Gao et al., 2018), suggesting that these kind of IDs could ensure the signaling function of these missing domains. Therefore, such IDs in NLR proteins could fulfill either sensor or signaling functions, or both.
Interestingly, there are significant overlaps between IDs and protein domains identified previously as interacting partners of effectors in interactome screens (Mukhtar et al., 2011; Weßling et al., 2014; Sarris et al., 2016), including well-characterized guardees or decoys. Examples are the exocyst complex factor Exo70, required for the recognition of AvrPii by NLR Pii in rice (Fujisaki et al., 2015), and RIN4, a target of multiple effectors that is guarded by RPS2 and RPM1 NLRs in Arabidopsis (Mackey et al., 2002, 2003; Kim et al., 2005). Such overlaps strongly suggest that IDs could act as sensor/decoy by mimicking effector targets. Since many IDs correspond to protein domains with unknown biological activity, they represent promising candidates to uncover host components targeted by effectors and whose participation in various layers of plant immunity has not been assigned yet.
Whether all the putative IDs identified in the whole-genome analyses also serve as sensor/decoy in immune responses remains to be demonstrated. Moreover, detailed investigations on gene structure and function should help to reduce false-positive results among computationally predicted NLR-IDs (Giannakopoulou et al., 2016) and shed light on new resistance mechanisms.
MECHANISMS INVOLVED IN NLR-ID FUSION EVENTS
Within NLR-IDs, the majority of IDs appear as singular N- or C-terminal domains. However, in some cases, the fusion of several domains in the same protein is observed. For example, the AtWRKY19 NLR in Arabidopsis integrates both an N-terminal WRKY domain and a C-terminal kinase domain. In a minority of cases, including rice Pik-1, integration has occurred between the N-terminal signaling domain and the central nucleotide-binding domain of the NLR. These observations indicate that some NLRs can tolerate the integration of sensor domains at various positions in their modular architecture while maintaining their signaling functions. Bailey et al. (2018) recently investigated the evolutionary dynamics of NLR-IDs in the genomes of nine grass species. They concluded that NLR-IDs in grasses were not distributed evenly across their phylogeny, but a specific clade with up to 58% of NLRs containing IDs was observed. In this clade, they highlighted an amino acid sequence motif located immediately upstream of the fusion site, which could play an important role in the integration process. They proposed that DNA transposition and/or ectopic recombination is a major driving force behind domain integration in grasses; repeated independent integration events were observed, suggesting that integration occurred frequently and independently during evolution, giving rise to a high diversity of IDs. Similarly, Brabham et al. (2018) recently showed that orthologs of the RGH2 NLR from species across the grasses were subject to large variation in domain structure, including the presence/absence of an integrated Exo70 domain. These transspecies polymorphisms provided an opportunity to follow the molecular evolution of the Exo70 gene family and to investigate the role of Exo70 as an ID in the RGH2 NLR. This study showed that, upon pathogen pressure, nonintegrated Exo70 genes are under strong purifying selection, whereas they are under relaxed purifying selection when integrated into RGH2. Across the Oryza genomes, the presence of IDs in 17 different NLR subfamilies (from a total of 36) point to multiple and independent acquisition of IDs (Stein et al., 2018).
NLR-IDS: TOWARD THE ELUCIDATION OF ADDITIONAL NLR FUNCTIONS?
Besides the well-documented role of NLRs for innate immunity in both animals and plants, additional functions controlled by NLRs are currently discussed. In animals, NLRs play a role in developmental processes, such as spermatogenesis and fertility, suggesting a control of the reproductive system by NLR proteins (Meunier and Broz, 2017). In plants, inappropriate activation of NLRs caused by mutations or incompatible combinations also impacts development (Chae et al., 2014; Chen et al., 2016; Atanasov et al., 2018; Chakraborty et al., 2018). The integrated decoy model predicts an important role of IDs in pathogen detection. Beyond their function of effector sensor, some IDs might have retained the biological activities of the proteins from which they are derived. Thus, additional functions controlled by NLRs in plants could be revealed by looking at IDs.
In this regard, the integration of a BED domain, one of the most frequently found IDs in plant species, gained attention. This domain appears to be shared by transposases and by proteins that perform critical cellular functions (Aravind, 2000; Hayward et al., 2013). The Znf-BED domain of the Tam3 (Transposase of Activator from maize) transposase has been shown to suppress the DNA TAM3 transposon activity in Anthirrhinum majus by relocalizing the Tam3 transposase out of the nucleus (Zhou et al., 2017). More globally, BED-related IDs could be sensors of cellular homeostasis perturbation by environmental stress that rely in part on DNA transposition control (Negi et al., 2016).
In the case of RRS1, inhibition of its DNA-binding activity provoked by particular mutations in its WRKY domain leads to autoimmunity in an RPS4-dependent manner (Noutoshi et al., 2005; Sohn et al., 2014). Therefore, RRS1-R was initially considered a negative regulator of immune-related genes. The autoimmune phenotype of the RRS1-R slh1 variant is conditioned by low humidity, suggesting that RRS1-R, besides its function in pathogen recognition, also could sense particular environmental disturbances such as drought stress. In response to specific biotic and abiotic stresses, RRS1-R likely acts directly at genomic DNA and, together with RPS4, behaves as a reactive switch for transcriptional and signaling reprogramming. Whether NLR-IDs possess broader sensing functions remains to be determined, but almost certainly the functional characterization of their IDs will help to elucidate potential additional NLR functions.
TOWARD THE ENGINEERING OF SYNTHETIC NLR-IDS WITH EXTENDED RECOGNITION CAPABILITIES
The high diversity of NLRs provides plants with versatile options for effective plant immunity. However, on the one hand, the ability of effectors to evolve rapidly and, on the other hand, the necessary fine-tuning of NLR functions to avoid autoimmunity (Box 1) restrict the possibilities to transfer immune receptors into other plant genomes for improved crop protection. Hence, strategies for NLR engineering are of particular interest in plant immunity research. If successful, it could be possible to modify NLR recognition specificity or, alternatively, widen the range of effector recognition while making it less feasible for pathogens to bypass NLRs. One possibility is to alter the structure of NLR proteins itself. For example, a study conducted by Segretin et al. (2014) showed that a few amino acid changes in the LRR domain of R3a in potato (Solanum tuberosum) enabled this NLR to recognize another isoform of AVR3a from Phytophthora infestans. However, direct NLR effector recognition (modified self) is likely to be less tolerant to variations compared with the indirect and integrated decoy models. Therefore, decoy engineering represents a more suitable approach. A proof-of-concept study was performed recently by Kim et al. (2016), demonstrating successfully the modification of the Arabidopsis protein kinase PBS1 that represents a decoy for pathogen-derived proteases. PBS1 is involved in basal immune responses and acts as target/decoy for the effector AvrPphB from P. syringae, which can cleave PBS1 due to its protease activity (Shao et al., 2003; Ade et al., 2007). PBS1 cleavage products are recognized by the NLR protein RPS5 (DeYoung et al., 2012) that, in turn, initiates resistance signaling. For a modified PBS1 decoy variation, the proteolytic target site, which is normally cleaved by AvrPphB, is exchanged with another proteolytic site. This modification enables the cleavage of PBS1 by other effectors, such as AvrRpt2 of P. syringae, NIa protease of Tobacco etch virus, and Nla protease of Turnip mosaic virus. AvrRpt2 also triggers the activation of RPS2 NLR by cleaving a nitrate-induced (NOI) domain present in RIN4, a negative regulator of plant defense that is guarded by multiple NLR proteins (Mackey et al., 2002, 2003; Kim et al., 2005). Such protease cleavage sites are found in IDs present in a subset of NLRs, paving the way for the design of single NLR-IDs combining multiple protease recognition sequences.
The modifications of NLR IDs, as well as the replacement of IDs with other identified effector targets from the host genome, display other promising tools to create novel effector traps. Considering that modifications of NLRs as well as of their IDs can compromise the homocomplex/heterocomplex formation required for their function and trigger either inactivation or autoactivation of the receptor, it is important for this approach to identify which IDs and which NLRs are best suited for fusion manipulation (e.g. ID swapping/shuffling). For this, we need to gain better knowledge of interaction sites within NLR and its IDs but also of the interaction of sensor and signaling NLRs. Furthermore, it is important to determine the layout of effector-ID complexes. Recently, the structures of several complexes have been resolved (Maqbool et al., 2015; Ortiz et al., 2017; Zhang et al., 2017). Determination of the molecular and structural bases of these interactions is crucial for the successful design of synthetic multiple-sensor NLR-IDs made from the juxtaposition of different IDs, giving them extended recognition capabilities.
CONCLUSION
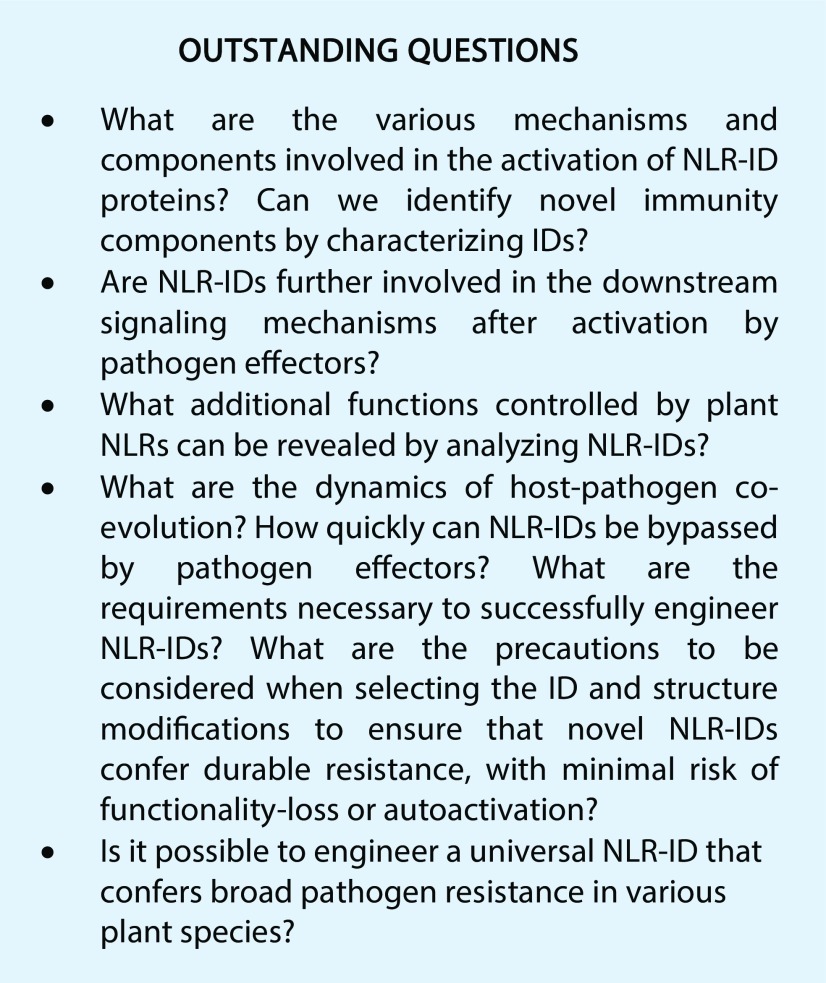
The past 20 years have brought major advances in plant NLR biology. The discovery of NLR-IDs represents a significant step toward a better understanding of the mechanisms involved in the evolution and function of NLRs. During plant evolution, NLR-IDs appeared independently several times and in different configurations. Besides the sensing and regulating functions of IDs, important questions remain unanswered (see Outstanding Questions). There is still a critical need for further in-depth studies to establish the biological functions of IDs fused to NLRs and to elucidate the molecular events that link NLR-ID activation with immunity pathways. Overall, NLR-IDs provide promising tools for the design of new strategies to protect plants against pathogens. Nevertheless, it remains to be determined whether engineered synthetic NLR-IDs can provide sustainable disease resistance, especially in different crop species.
Acknowledgments
We apologize to researchers whose contributions could not be cited more extensively because of space limitations.
Footnotes
This work was supported by the Agence Nationale de la Recherche project RADAR (ANR15-CE20-0016-01).
Articles can be viewed without a subscription.
References
- Ade J, DeYoung BJ, Golstein C, Innes RW (2007) Indirect activation of a plant nucleotide binding site-leucine-rich repeat protein by a bacterial protease. Proc Natl Acad Sci USA 104: 2531–2536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravind L. (2000) The BED finger, a novel DNA-binding domain in chromatin-boundary-element-binding proteins and transposases. Trends Biochem Sci 25: 421–423 [DOI] [PubMed] [Google Scholar]
- Arnold DL, Jackson RW (2011) Bacterial genomes: Evolution of pathogenicity. Curr Opin Plant Biol 14: 385–391 [DOI] [PubMed] [Google Scholar]
- Ashikawa I, Hayashi N, Yamane H, Kanamori H, Wu J, Matsumoto T, Ono K, Yano M (2008) Two adjacent nucleotide-binding site-leucine-rich repeat class genes are required to confer Pikm-specific rice blast resistance. Genetics 180: 2267–2276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atanasov KE, Liu C, Erban A, Kopka J, Parker JE, Alcázar R (2018) NLR mutations suppressing immune hybrid incompatibility and their effects on disease resistance. Plant Physiol 177: 1152–1169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey PC, Schudoma C, Jackson W, Baggs E, Dagdas G, Haerty W, Moscou M, Krasileva KV (2018) Dominant integration locus drives continuous diversification of plant immune receptors with exogenous domain fusions. Genome Biol 19: 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birker D, Heidrich K, Takahara H, Narusaka M, Deslandes L, Narusaka Y, Reymond M, Parker JE, O’Connell R (2009) A locus conferring resistance to Colletotrichum higginsianum is shared by four geographically distinct Arabidopsis accessions. Plant J 60: 602–613 [DOI] [PubMed] [Google Scholar]
- Brabham HJ, Hernández-Pinzón I, Holden S, Lorang J, Moscou MJ (2018) An ancient integration in a plant NLR is maintained as a trans-species polymorphism. BioRxiv [Google Scholar]
- Cesari S, Thilliez G, Ribot C, Chalvon V, Michel C, Jauneau A, Rivas S, Alaux L, Kanzaki H, Okuyama Y, et al. (2013) The rice resistance protein pair RGA4/RGA5 recognizes the Magnaporthe oryzae effectors AVR-Pia and AVR1-CO39 by direct binding. Plant Cell 25: 1463–1481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cesari S, Bernoux M, Moncuquet P, Kroj T, Dodds PN (2014a) A novel conserved mechanism for plant NLR protein pairs: The “integrated decoy” hypothesis. Front Plant Sci 5: 606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Césari S, Kanzaki H, Fujiwara T, Bernoux M, Chalvon V, Kawano Y, Shimamoto K, Dodds P, Terauchi R, Kroj T (2014b) The NB-LRR proteins RGA4 and RGA5 interact functionally and physically to confer disease resistance. EMBO J 33: 1941–1959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chae E, Bomblies K, Kim ST, Karelina D, Zaidem M, Ossowski S, Martín-Pizarro C, Laitinen RA, Rowan BA, Tenenboim H, et al. (2014) Species-wide genetic incompatibility analysis identifies immune genes as hot spots of deleterious epistasis. Cell 159: 1341–1351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty J, Ghosh P, Das S (2018) Autoimmunity in plants. Planta 248: 751–767 [DOI] [PubMed] [Google Scholar]
- Chen C, E Z, Lin HX (2016) Evolution and molecular control of hybrid incompatibility in plants. Front Plant Sci 7: 1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook DE, Mesarich CH, Thomma BP (2015) Understanding plant immunity as a surveillance system to detect invasion. Annu Rev Phytopathol 53: 541–563 [DOI] [PubMed] [Google Scholar]
- De la Concepcion JC, Franceschetti M, Maqbool A, Saitoh H, Terauchi R, Kamoun S, Banfield MJ (2018) Polymorphic residues in rice NLRs expand binding and response to effectors of the blast pathogen. Nat Plants 4: 576–585 [DOI] [PubMed] [Google Scholar]
- Deslandes L, Pileur F, Liaubet L, Camut S, Can C, Williams K, Holub E, Beynon J, Arlat M, Marco Y (1998) Genetic characterization of RRS1, a recessive locus in Arabidopsis thaliana that confers resistance to the bacterial soilborne pathogen Ralstonia solanacearum. Mol Plant Microbe Interact 11: 659–667 [DOI] [PubMed] [Google Scholar]
- Deslandes L, Olivier J, Theulieres F, Hirsch J, Feng DX, Bittner-Eddy P, Beynon J, Marco Y (2002) Resistance to Ralstonia solanacearum in Arabidopsis thaliana is conferred by the recessive RRS1-R gene, a member of a novel family of resistance genes. Proc Natl Acad Sci USA 99: 2404–2409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deslandes L, Olivier J, Peeters N, Feng DX, Khounlotham M, Boucher C, Somssich I, Genin S, Marco Y (2003) Physical interaction between RRS1-R, a protein conferring resistance to bacterial wilt, and PopP2, a type III effector targeted to the plant nucleus. Proc Natl Acad Sci USA 100: 8024–8029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeYoung BJ, Qi D, Kim SH, Burke TP, Innes RW (2012) Activation of a plant nucleotide binding-leucine rich repeat disease resistance protein by a modified self protein. Cell Microbiol 14: 1071–1084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodds PN, Rathjen JP (2010) Plant immunity: Towards an integrated view of plant-pathogen interactions. Nat Rev Genet 11: 539–548 [DOI] [PubMed] [Google Scholar]
- Fujisaki K, Abe Y, Ito A, Saitoh H, Yoshida K, Kanzaki H, Kanzaki E, Utsushi H, Yamashita T, Kamoun S, et al. (2015) Rice Exo70 interacts with a fungal effector, AVR-Pii, and is required for AVR-Pii-triggered immunity. Plant J 83: 875–887 [DOI] [PubMed] [Google Scholar]
- Fujisaki K, Abe Y, Kanzaki E, Ito K, Utsushi H, Saitoh H, Białas A, Banfield M, Kamoun S, Terauchi R (2017) An unconventional NOI/RIN4 domain of a rice NLR protein binds host EXO70 protein to confer fungal immunity. BioRxiv [Google Scholar]
- Fukuoka S, Saka N, Koga H, Ono K, Shimizu T, Ebana K, Hayashi N, Takahashi A, Hirochika H, Okuno K, et al. (2009) Loss of function of a proline-containing protein confers durable disease resistance in rice. Science 325: 998–1001 [DOI] [PubMed] [Google Scholar]
- Funk A, Galewski P, McGrath JM (2018) Nucleotide-binding resistance gene signatures in sugar beet, insights from a new reference genome. Plant J 95: 659–671 [DOI] [PubMed] [Google Scholar]
- Gao Y, Wang W, Zhang T, Gong Z, Zhao H, Han GZ (2018) Out of water: The origin and early diversification of plant R-genes. Plant Physiol 177: 82–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giannakopoulou A, Bialas A, Kamoun S, Vleeshouwers VG (2016) Plant immunity switched from bacteria to virus. Nat Biotechnol 34: 391–392 [DOI] [PubMed] [Google Scholar]
- Hayward A, Ghazal A, Andersson G, Andersson L, Jern P (2013) ZBED evolution: Repeated utilization of DNA transposons as regulators of diverse host functions. PLoS ONE 8: e59940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinsch M, Staskawicz B (1996) Identification of a new Arabidopsis disease resistance locus, RPs4, and cloning of the corresponding avirulence gene, avrRps4, from Pseudomonas syringae pv. pisi. Mol Plant Microbe Interact 9: 55–61 [DOI] [PubMed] [Google Scholar]
- Ji Z, Ji C, Liu B, Zou L, Chen G, Yang B (2016) Interfering TAL effectors of Xanthomonas oryzae neutralize R-gene-mediated plant disease resistance. Nat Commun 7: 13435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones JD, Dangl JL (2006) The plant immune system. Nature 444: 323–329 [DOI] [PubMed] [Google Scholar]
- Kanzaki H, Yoshida K, Saitoh H, Fujisaki K, Hirabuchi A, Alaux L, Fournier E, Tharreau D, Terauchi R (2012) Arms race co-evolution of Magnaporthe oryzae AVR-Pik and rice Pik genes driven by their physical interactions. Plant J 72: 894–907 [DOI] [PubMed] [Google Scholar]
- Khan M, Subramaniam R, Desveaux D (2016) Of guards, decoys, baits and traps: Pathogen perception in plants by type III effector sensors. Curr Opin Microbiol 29: 49–55 [DOI] [PubMed] [Google Scholar]
- Kim MG, da Cunha L, McFall AJ, Belkhadir Y, DebRoy S, Dangl JL, Mackey D (2005) Two Pseudomonas syringae type III effectors inhibit RIN4-regulated basal defense in Arabidopsis. Cell 121: 749–759 [DOI] [PubMed] [Google Scholar]
- Kim SH, Qi D, Ashfield T, Helm M, Innes RW (2016) Using decoys to expand the recognition specificity of a plant disease resistance protein. Science 351: 684–687 [DOI] [PubMed] [Google Scholar]
- Kroj T, Chanclud E, Michel-Romiti C, Grand X, Morel JB (2016) Integration of decoy domains derived from protein targets of pathogen effectors into plant immune receptors is widespread. New Phytol 210: 618–626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Roux C, Huet G, Jauneau A, Camborde L, Trémousaygue D, Kraut A, Zhou B, Levaillant M, Adachi H, Yoshioka H, et al. (2015) A receptor pair with an integrated decoy converts pathogen disabling of transcription factors to immunity. Cell 161: 1074–1088 [DOI] [PubMed] [Google Scholar]
- Lewis JD, Lee AH, Hassan JA, Wan J, Hurley B, Jhingree JR, Wang PW, Lo T, Youn JY, Guttman DS, et al. (2013) The Arabidopsis ZED1 pseudokinase is required for ZAR1-mediated immunity induced by the Pseudomonas syringae type III effector HopZ1a. Proc Natl Acad Sci USA 110: 18722–18727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo Presti L, Lanver D, Schweizer G, Tanaka S, Liang L, Tollot M, Zuccaro A, Reissmann S, Kahmann R (2015) Fungal effectors and plant susceptibility. Annu Rev Plant Biol 66: 513–545 [DOI] [PubMed] [Google Scholar]
- Ma Y, Guo H, Hu L, Martinez PP, Moschou PN, Cevik V, Ding P, Duxbury Z, Sarris PF, Jones JDG (2018) Distinct modes of derepression of an Arabidopsis immune receptor complex by two different bacterial effectors. Proc Natl Acad Sci USA 115: 10218–10227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackey D, Holt BF III, Wiig A, Dangl JL (2002) RIN4 interacts with Pseudomonas syringae type III effector molecules and is required for RPM1-mediated resistance in Arabidopsis. Cell 108: 743–754 [DOI] [PubMed] [Google Scholar]
- Mackey D, Belkhadir Y, Alonso JM, Ecker JR, Dangl JL (2003) Arabidopsis RIN4 is a target of the type III virulence effector AvrRpt2 and modulates RPS2-mediated resistance. Cell 112: 379–389 [DOI] [PubMed] [Google Scholar]
- Maekawa T, Kufer TA, Schulze-Lefert P (2011) NLR functions in plant and animal immune systems: So far and yet so close. Nat Immunol 12: 817–826 [DOI] [PubMed] [Google Scholar]
- Maqbool A, Saitoh H, Franceschetti M, Stevenson CE, Uemura A, Kanzaki H, Kamoun S, Terauchi R, Banfield MJ (2015) Structural basis of pathogen recognition by an integrated HMA domain in a plant NLR immune receptor. eLife 4: e08709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meunier E, Broz P (2017) Evolutionary convergence and divergence in NLR function and structure. Trends Immunol 38: 744–757 [DOI] [PubMed] [Google Scholar]
- Mukhtar MS, Carvunis AR, Dreze M, Epple P, Steinbrenner J, Moore J, Tasan M, Galli M, Hao T, Nishimura MT, et al. (2011) Independently evolved virulence effectors converge onto hubs in a plant immune system network. Science 333: 596–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narusaka M, Shirasu K, Noutoshi Y, Kubo Y, Shiraishi T, Iwabuchi M, Narusaka Y (2009) RRS1 and RPS4 provide a dual Resistance-gene system against fungal and bacterial pathogens. Plant J 60: 218–226 [DOI] [PubMed] [Google Scholar]
- Negi P, Rai AN, Suprasanna P (2016) Moving through the stressed genome: Emerging regulatory roles for transposons in plant stress response. Front Plant Sci 7: 1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noutoshi Y, Ito T, Seki M, Nakashita H, Yoshida S, Marco Y, Shirasu K, Shinozaki K (2005) A single amino acid insertion in the WRKY domain of the Arabidopsis TIR-NBS-LRR-WRKY-type disease resistance protein SLH1 (sensitive to low humidity 1) causes activation of defense responses and hypersensitive cell death. Plant J 43: 873–888 [DOI] [PubMed] [Google Scholar]
- Ntoukakis V, Saur IM, Conlan B, Rathjen JP (2014) The changing of the guard: The Pto/Prf receptor complex of tomato and pathogen recognition. Curr Opin Plant Biol 20: 69–74 [DOI] [PubMed] [Google Scholar]
- Okuyama Y, Kanzaki H, Abe A, Yoshida K, Tamiru M, Saitoh H, Fujibe T, Matsumura H, Shenton M, Galam DC, et al. (2011) A multifaceted genomics approach allows the isolation of the rice Pia-blast resistance gene consisting of two adjacent NBS-LRR protein genes. Plant J 66: 467–479 [DOI] [PubMed] [Google Scholar]
- Ortiz D, de Guillen K, Cesari S, Chalvon V, Gracy J, Padilla A, Kroj T (2017) Recognition of the Magnaporthe oryzae effector AVR-Pia by the decoy domain of the rice NLR immune receptor RGA5. Plant Cell 29: 156–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rushton PJ, Somssich IE, Ringler P, Shen QJ (2010) WRKY transcription factors. Trends Plant Sci 15: 247–258 [DOI] [PubMed] [Google Scholar]
- Sarris PF, Duxbury Z, Huh SU, Ma Y, Segonzac C, Sklenar J, Derbyshire P, Cevik V, Rallapalli G, Saucet SB, et al. (2015) A plant immune receptor detects pathogen effectors that target WRKY transcription factors. Cell 161: 1089–1100 [DOI] [PubMed] [Google Scholar]
- Sarris PF, Cevik V, Dagdas G, Jones JD, Krasileva KV (2016) Comparative analysis of plant immune receptor architectures uncovers host proteins likely targeted by pathogens. BMC Biol 14: 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saucet SB, Ma Y, Sarris PF, Furzer OJ, Sohn KH, Jones JD (2015) Two linked pairs of Arabidopsis TNL resistance genes independently confer recognition of bacterial effector AvrRps4. Nat Commun 6: 6338. [DOI] [PubMed] [Google Scholar]
- Segretin ME, Pais M, Franceschetti M, Chaparro-Garcia A, Bos JI, Banfield MJ, Kamoun S (2014) Single amino acid mutations in the potato immune receptor R3a expand response to Phytophthora effectors. Mol Plant Microbe Interact 27: 624–637 [DOI] [PubMed] [Google Scholar]
- Shao F, Golstein C, Ade J, Stoutemyer M, Dixon JE, Innes RW (2003) Cleavage of Arabidopsis PBS1 by a bacterial type III effector. Science 301: 1230–1233 [DOI] [PubMed] [Google Scholar]
- Sohn KH, Hughes RK, Piquerez SJ, Jones JD, Banfield MJ (2012) Distinct regions of the Pseudomonas syringae coiled-coil effector AvrRps4 are required for activation of immunity. Proc Natl Acad Sci USA 109: 16371–16376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohn KH, Segonzac C, Rallapalli G, Sarris PF, Woo JY, Williams SJ, Newman TE, Paek KH, Kobe B, Jones JD (2014) The nuclear immune receptor RPS4 is required for RRS1SLH1-dependent constitutive defense activation in Arabidopsis thaliana. PLoS Genet 10: e1004655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein JC, Yu Y, Copetti D, Zwickl DJ, Zhang L, Zhang C, Chougule K, Gao D, Iwata A, Goicoechea JL, et al. (2018) Genomes of 13 domesticated and wild rice relatives highlight genetic conservation, turnover and innovation across the genus Oryza. Nat Genet 50: 285–296 [DOI] [PubMed] [Google Scholar]
- Takken FL, Goverse A (2012) How to build a pathogen detector: Structural basis of NB-LRR function. Curr Opin Plant Biol 15: 375–384 [DOI] [PubMed] [Google Scholar]
- Takken FL, Albrecht M, Tameling WI (2006) Resistance proteins: Molecular switches of plant defence. Curr Opin Plant Biol 9: 383–390 [DOI] [PubMed] [Google Scholar]
- Tasset C, Bernoux M, Jauneau A, Pouzet C, Briere C, Kieffer-Jacquinod S, Rivas S, Marco Y, Deslandes L (2010) Autoacetylation of the Ralstonia solanacearum effector PopP2 targets a lysine residue essential for RRS1-R-mediated immunity in Arabidopsis. PLoS Pathog 6: e1001202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Hoorn RA, Kamoun S (2008) From guard to decoy: A new model for perception of plant pathogen effectors. Plant Cell 20: 2009–2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weßling R, Epple P, Altmann S, He Y, Yang L, Henz SR, McDonald N, Wiley K, Bader KC, Gläßer C, et al. (2014) Convergent targeting of a common host protein-network by pathogen effectors from three kingdoms of life. Cell Host Microbe 16: 364–375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams SJ, Sohn KH, Wan L, Bernoux M, Sarris PF, Segonzac C, Ve T, Ma Y, Saucet SB, Ericsson DJ, et al. (2014) Structural basis for assembly and function of a heterodimeric plant immune receptor. Science 344: 299–303 [DOI] [PubMed] [Google Scholar]
- Yoshimura S, Yamanouchi U, Katayose Y, Toki S, Wang ZX, Kono I, Kurata N, Yano M, Iwata N, Sasaki T (1998) Expression of Xa1, a bacterial blight-resistance gene in rice, is induced by bacterial inoculation. Proc Natl Acad Sci USA 95: 1663–1668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Zhao J, Li Y, Yuan Z, He H, Yang H, Qu H, Ma C, Qu S (2016) Transcriptome analysis highlights defense and signaling pathways mediated by rice pi21 gene with partial resistance to Magnaporthe oryzae. Front Plant Sci 7: 1834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang ZM, Ma KW, Gao L, Hu Z, Schwizer S, Ma W, Song J (2017) Mechanism of host substrate acetylation by a YopJ family effector. Nat Plants 3: 17115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H, Hirata M, Osawa R, Fujino K, Kishima Y (2017) Detainment of Tam3 transposase at plasma membrane by its BED-zinc finger domain. Plant Physiol 173: 1492–1501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuluaga P, Szurek B, Koebnik R, Kroj T, Morel JB (2017) Effector mimics and integrated decoys, the never-ending arms race between rice and Xanthomonas oryzae. Front Plant Sci 8: 431. [DOI] [PMC free article] [PubMed] [Google Scholar]