Altering the nuclear periphery leads to an age-dependent induction of pathogen defense responses in plants, mediated by the signaling molecule, salicylic acid.
Abstract
Defects in the nuclear lamina of animal cell nuclei have dramatic effects on nuclear structure and gene expression as well as diverse physiological manifestations. We report that deficiencies in CROWDED NUCLEI (CRWN), which are candidate nuclear lamina proteins in Arabidopsis (Arabidopsis thaliana), trigger widespread changes in transcript levels and whole-plant phenotypes, including dwarfing and spontaneous cell death lesions. These phenotypes are caused in part by ectopic induction of plant defense responses via the salicylic acid pathway. Loss of CRWN proteins induces the expression of the salicylic acid biosynthetic gene ISOCHORISMATE SYNTHASE1, which leads to spontaneous defense responses in crwn1 crwn2 and crwn1 crwn4 mutants, which are deficient in two of the four CRWN paralogs. The symptoms of ectopic defense response, including pathogenesis marker gene expression and cell death, increase in older crwn double mutants. These age-dependent effects are postulated to reflect an increase in nuclear dysfunction or damage over time, a phenomenon reminiscent of aging effects seen in animal nuclei and in some human laminopathy patients.
The morphology of cellular organelles is closely related to their function and physiological state. For example, mitochondrial cristae result from a complex corrugation of the inner membrane required to accommodate enzymes for cellular respiration. Mitochondria in skeletal or heart muscle, which require high amounts of energy, have stacked cristae, while tissues with lower energy demands, such as liver or kidney, contain mitochondria with less stacked cristae (Kühlbrandt, 2015). This relationship implies that structural components, which provide physical support and define organellar structure, are intimately involved in organelle function. In nuclei, the relationship between morphology and function is particularly important, as nuclei possess genetic information whose expression can be influenced by changes in nuclear structure and organization (Reddy et al., 2008; Schreiber and Kennedy, 2013; Davidson and Lammerding, 2014).
Eukaryotic nuclei are wrapped in a nuclear envelope, a double leaflet consisting of outer and inner nuclear membranes. There are various proteins associated with the envelope essential for nuclear function and morphology. Examples include nucleoporins, which form nuclear pores, Linker of Nucleoskeleton and Cytoskeleton complex proteins, which span the nuclear envelope (Lombardi and Lammerding, 2011; Tapley and Starr, 2013; Tatout et al., 2014; Meier, 2016), and a nuclear lamina (NL) structure underlying the inner nuclear envelope (Aebi et al., 1986). Among these, the NL is a key architectural feature that affects both the morphology and the function of nuclei. In animals, the NL is a reticular structure under the inner nuclear membrane. The animal NL is mainly composed of intermediate filament-like proteins called lamins, which polymerize to form fibrillar networks (Aebi et al., 1986). This structure provides docking sites for chromatin and serves as a physical support for the organelle (Gruenbaum and Foisner, 2015). Most heterochromatin resides near or at the nuclear periphery, which is typically a repressive environment for gene expression (Egecioglu and Brickner, 2011). Studies have demonstrated that tethering genes to the NL results in transcriptional repression in mammalian cells (Reddy et al., 2008). Genomic profiling approaches have defined lamina-associated domains (LADs), characterized by repressive chromatin and genes with low expression levels (Pickersgill et al., 2006; Guelen et al., 2008; Ikegami et al., 2010; Peric-Hupkes et al., 2010; van Steensel and Belmont, 2017). This interaction between the genome and NL can link alterations in NL structure to genomic instability as well as modifications in nuclear morphology. In a well-documented example, certain dominant mutations in the human lamin A gene lead to abnormal nuclear shape, chromatin organization defects, and clinical syndromes, such as premature aging (e.g. Hutchinson-Gilford progeria syndrome; Hutchinson, 1886; Gilford, 1904; Goldman et al., 2004). Also, fibroblasts cultured from Hutchinson-Gilford progeria syndrome patients exhibit repositioned chromosomes within the nucleus and transcriptional misregulation (Csoka et al., 2004; Meaburn et al., 2007). These findings indicate that the NL is essential for proper gene expression (Zheng et al., 2018).
Although plants lack intermediate filaments and lamin orthologs, different classes of nuclear coiled-coil proteins have been identified as putative NL components, based on pioneering studies by Masuda and colleagues on carrot (Daucus carota) Nuclear Matrix Constituent Proteins (NMCPs; Masuda et al., 1993, 1997). In Arabidopsis (Arabidopsis thaliana), NMCP orthologs, called CROWDED NUCLEI (CRWN) proteins, are candidates for the major structural component of a plant NL. The Arabidopsis genome contains four CRWN genes, which are expressed broadly at the transcript level without obvious tissue specificity. The proteins partition into two distinct clades: one containing the CRWN1, CRWN2, and CRWN3 paralogs (NMCP1 clade), and a distinct one including CRWN4 (NMCP2 clade). Among these paralogs, CRWN1 and CRWN4 proteins localize to the nuclear periphery and are therefore the best candidates for NL components, while CRWN2 and CRWN3 are distributed throughout the nucleoplasm (Dittmer et al., 2007; Dittmer and Richards, 2008; Sakamoto and Takagi, 2013). CRWN genes are essential, as a loss of all four genes leads to inviability. CRWN proteins are important for maintenance of nuclear structure and morphology, as many crwn mutants have small and round nuclei (Dittmer et al., 2007; Sakamoto and Takagi, 2013; Wang et al., 2013) compared with wild-type Arabidopsis nuclei, which are elongated in many differentiated cell types (Chytilova et al., 2000; Meier et al., 2016, 2017). Our group previously reported that CRWN genes have roles in specifying the structure of heterochromatin aggregates in interphase (chromocenters), implying that CRWN proteins not only control nuclear morphology but are also able to regulate higher order genome organization (Dittmer et al., 2007; Wang et al., 2013). Whole-plant phenotypes of crwn mutants show that many double and triple crwn mutants are smaller in size and have wrinkled leaves, demonstrating that altered plant nuclear structure can ultimately lead to abnormal growth.
Here, we investigate the effects of crwn mutations on gene expression and elucidate a mechanism through which loss of CRWN proteins causes dwarfism. We show that mutants lacking the full complement of CRWN genes show altered gene expression patterns, characterized by overexpression of transcripts associated with biotic pathogen response. Plants lacking both CRWN1 and CRWN2, or CRWN1 and CRWN4, exhibit ectopic defense responses, including partial resistance against the virulent bacterial pathogen, Pseudomonas syringae pv tomato (strain DC3000). In addition, these crwn genotypes exhibit spontaneous cell death, which is associated with the up-regulation of the salicylic acid (SA)-biosynthesis gene, ISOCHORISMATE SYNTHASE1 (ICS1/SID2), and subsequent SA-dependent defense signaling. Our results demonstrate that nuclear periphery defects in plants, like similar changes in animal cells, can lead to premature cell death.
RESULTS
Three crwn Single Mutants Show Altered Transcriptional Profiles in the Absence of Whole-Plant Phenotypes
We reasoned that the alterations in nuclear size and shape in the crwn mutants could have an effect on transcriptional regulation and/or posttranscriptional processing. To test this prediction, we assessed the effect of crwn mutations on steady-state transcript levels using RNA sequencing (RNA-seq). Four-week-old rosette leaves from five mutants (crwn1, crwn2, crwn4, crwn1 crwn2, and crwn1 crwn4) were used as tissue for the RNA purification and transcriptomic analysis. These five mutants encompass the range of phenotypes displayed by crwn family mutants and involve both the NMCP1 and NMCP2 clades of CRWN paralogs. Mutants lacking either CRWN1 or CRWN4 show abnormal nuclei that fail to elongate in differentiated cells and are reduced in size. The crwn2 mutation by itself does not alter nuclear morphology, but it was included in our analysis because crwn1 crwn2 double mutants have much smaller nuclei and display dwarf phenotypes (Dittmer et al., 2007; Wang et al., 2013), indicating that CRWN1 and CRWN2 have at least partially overlapping function. Similarly, crwn1 crwn4 double mutants are smaller than the wild type or either crwn1 or crwn4 single mutant.
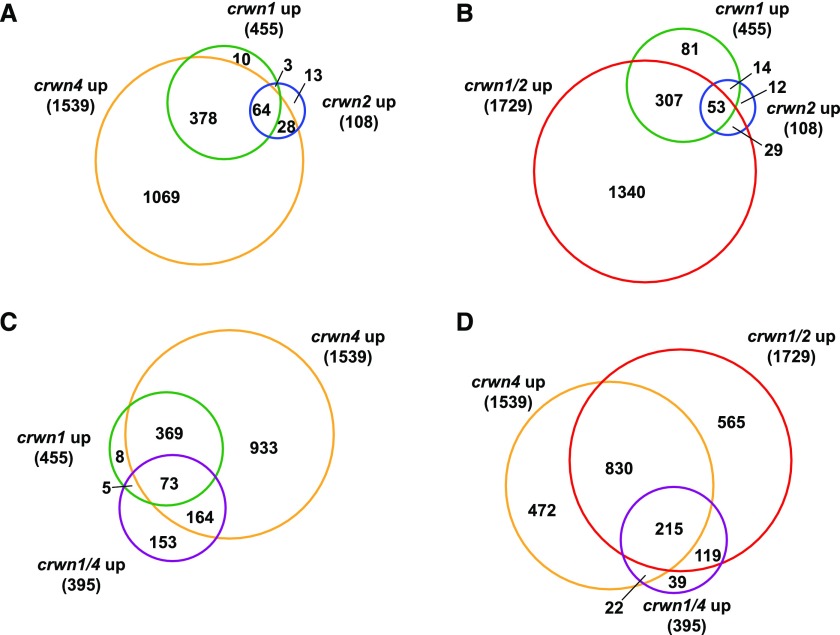
We first compared misregulated genes among the three single mutants using a threshold for misregulation of 2-fold or greater increase or decrease compared with the wild type, with q < 0.01. We discovered that crwn4 mutants displayed the largest number of up-regulated loci, with 1,539 genes, compared to 455 for crwn1 mutants and 108 for crwn2 mutants (Fig. 1A). The number of genes with altered expression scaled with the severity of the abnormal nuclei phenotypes in leaf cells. That is, crwn2 plants have normally shaped nuclei and the least number of up-regulated genes, while crwn1 mutants exhibit smaller, round nuclei and a moderate alteration in transcription (Fig. 1A). The most severe misregulation was seen in the crwn4 mutant (Fig. 1A), which has a more severe effect on nuclear organization, evidenced by a disruption in chromocenter organization and an abnormal nuclear boundary. These results indicate that loss of a single CRWN protein is enough to elicit transcriptomic alterations. The absence of morphological defects on the whole-plant level in crwn single mutants indicates that the changes in steady-state transcript levels are tied directly to alterations in nuclear structure, rather than a result of secondary effects from abnormal growth and development.
Figure 1.
The patterns of up-regulated genes in crwn mutants demonstrate both synergistic and antagonistic relationships among crwn mutations. The sizes of each circle in the Venn diagrams are proportional to the number of genes up-regulated at least 2-fold in our RNA-seq data (q < 0.01). A, Up-regulated genes in three crwn single mutants, crwn1, crwn2, and crwn4, showing shared transcriptomic alterations. B, Up-regulated genes in the double mutant crwn1 crwn2 and corresponding single mutants, crwn1 and crwn2, showing the synergistic effects of the crwn1 and crwn2 mutations. C, Up-regulated genes in the double mutant crwn1 crwn4 and corresponding single mutants, crwn1 and crwn4, showing the antagonistic effects of these mutations. D, Up-regulated genes in two double mutants, crwn1 crwn2 and crwn1 crwn4, and one single mutant, crwn4, illustrating overlapping profiles. Note that PR1 and PR2 fell in the overlap between crwn1 crwn2 and crwn1 crwn4 mutants (this overlap contains 119 genes), and PR5 was found only in the sector corresponding to the crwn1 crwn2 mutant (region with 565 genes).
Complex Interactions among crwn Mutations Are Revealed by Transcriptomic Profiling
The genes misregulated in each crwn mutant formed sets with a high degree of overlap, suggesting shared functions among CRWN proteins. For example, many of the genes that were up-regulated genes in the crwn1 and crwn2 single mutants were also up-regulated in the crwn4 mutants (Fig. 1A). A similar pattern was observed in the Venn diagrams of down-regulated genes as well as for all misregulated genes (Supplemental Fig. S1).
We explored the overlap of function among CRWN proteins further by analyzing the transcriptomic profiles of the two crwn double mutants, crwn1 crwn2 and crwn1 crwn4, which display dwarf phenotypes. Of the two, crwn1 crwn2 mutants showed more extensive transcriptomic misregulation, exceeding the sum of the transcriptomic profiles of crwn1 and crwn2 single mutants (Fig. 1B). Thus, loss of CRWN1 and CRWN2 leads to a synergistic effect on transcription (Supplemental Fig. S2). This result is consistent with the synergistic phenotypes of crwn1 crwn2 double mutants, which have smaller nuclei and suffer from stunted growth, compared with the wild type and crwn1 or crwn2 single mutants. The transcriptomic patterns reinforce our previous conclusion that CRWN1 and CRWN2 have overlapping functions, but CRWN1, which is more highly expressed than CRWN2 (∼3-fold in wild-type plants in our RNA-seq data set), is sufficient to cover for the loss of its close paralog in crwn2 mutants.
Given the overlap of misregulated genes in crwn1 and crwn4 single mutants, it was surprising to observe that the combination of crwn1 and crwn4 mutations resulted in a smaller perturbation of gene expression compared with crwn4 mutants (Fig. 1C; Supplemental Fig. S2). This result was also unexpected because crwn1 crwn4 mutants have intermediate whole-plant phenotypes and a nuclear size reduction between that of the single mutants and the crwn1 crwn2 double mutant. However, the transcriptomic data demonstrating that crwn1 suppresses the crwn4 mutation are consistent with our previous observations that CRWN1 and CRWN4 have opposing effects on chromocenter aggregation (Wang et al., 2013).
To summarize, our transcriptomic profiling uncovered a complex set of interactions among CRWN genes. On one hand, the gene expression results confirm that CRWN1 and CRWN2, close paralogs in the same clade, have overlapping functions. In contrast, CRWN1 and CRWN4, which lie in distinct clades, appear to have at least partially antagonistic functions.
Misregulated Transcripts in crwn Mutants Are Evenly Distributed on Chromosome Arms But Depleted from Pericentromeric Regions
Next, we determined if genes misregulated in crwn mutants have a nonrandom distribution across the genome. Plant genomes make nonrandom contacts with the nuclear periphery (Bi et al., 2017), as is the case in animals, where a subset of the genome is partitioned into LADs (van Steensel and Belmont, 2017). Therefore, we investigated whether or not particular regions of the folded genome were preferentially affected in crwn mutants. An uneven distribution of misregulated genes could also reflect differential effects on distinct epigenetic compartments in the genome. To investigate these possibilities, we drew chromosomal maps marking the positions of misexpressed transcripts. As shown in Supplemental Figure S3, misregulated loci were evenly distributed across each chromosome arm but significantly depleted from pericentromeric regions. The lack of differentially expressed genes in these regions could indicate that transposable elements (TEs), which are concentrated in heterochromatic regions flanking the centromere, are not derepressed in crwn mutants. However, the paucity of TEs detected might result from a bias in our RNA-seq analysis based on uniquely mapped reads and a gene-based annotation. Accordingly, we used TEtranscripts, an independent program designed to analyze differential expression of TEs, allowing transcripts to be mapped to families of elements (Jin et al., 2015). This supplemental analysis determined that only a small number of TE families showed modest expression changes (Supplemental Tables S1 and S2), and, in most cases, the detected differences reflected changes in transcript levels from genes that contain an embedded TE. For example, transcripts from AT2TE56040, an element in the VANDAL5A subfamily, were ∼16-fold higher in crwn1 crwn2 mutants compared with the wild type (Supplemental Table S1), with no significant overexpression in crwn1 or crwn2. However, inspection of the RNA-seq reads indicated that this apparent overexpression was due to longer transcripts corresponding to AT2G30020, which contains the element within an exon. Taken as a whole, our results indicate that epigenetic silencing of TEs is not significantly reduced in any of the crwn mutants, despite the significant dispersion of chromocenters observed in crwn4 nuclei (Wang et al., 2013).
We then characterized the chromatin states of misregulated genes in crwn mutants to determine if certain epigenetic signatures were overrepresented or underrepresented. For this analysis, we used the nine chromatin categories established by Sequeira-Mendes et al. (2014) to define the landscape of the Arabidopsis epigenome. We found that up-regulated genes in crwn mutants showed a higher proportion of genes in chromatin state 2 compared with either the whole Arabidopsis gene list or the genes that were tested for differential expression (Supplemental Fig. S4). Chromatin state 2 is characterized by active marks (e.g. histone H3K4me2 and H3K4me3) associated with transcription as well as a high level of the repressive nucleosome modification, histone H3K27me3. The overrepresentation of chromatin state 2 coincided with an underrepresentation of states 1 and 3, which are associated with actively expressed genes and transcriptional elongation, respectively. The shift in representation of chromatin states among up-regulated targets suggests that CRWNs might contribute to the modulation of H3K27me-associated chromatin or differentially affect inducible genes that are packaged in chromatin state 2.
Analysis of Gene Ontology of Misregulated Genes Shows Ectopic Defense Responses in crwn Mutants
Next, we asked what kind of genes are misregulated in the RNA-seq profiles of crwn mutants. We found two patterns in this analysis. First, up-regulated genes were characterized by stress-associated Gene Ontology (GO) terms (Table 1). Although we found that both biotic and abiotic stress-related GO terms were identified, we focused on biotic stress because the GO terms with the highest fold enrichment were those related to plant defense (e.g. response to chitin and response to molecule of bacterial origins). The second pattern was an enrichment of genes associated with metabolism and other stress pathways among the down-regulated loci (Supplemental Table S3). Examples of these down-regulated pathways include glucosinolate biosynthesis, wax biosynthesis, and cell wall biogenesis. Together, these two observations suggest that a loss of CRWN1 or CRWN4, or a combination of CRWN paralogs, leads to an induction of defense-related genes and a down-regulation of a subset of metabolic processes.
Table 1. Statistical overrepresentation test for up-regulated genes in crwn mutants.
| Input | GO Terms (Biological Process Complete) | Total No. of Arabidopsis Genes in the Term | Input Genes No. | Expected No. of Genes | Fold Enrichment | P |
|---|---|---|---|---|---|---|
| Up-regulated genes in crwn1 mutants (444 genes mapped) | Response (rsp.) to chitin | 108 | 19 | 1.76 | 10.76 | 1.19E-10 |
| Rsp. to jasmonic acid | 174 | 18 | 2.84 | 6.33 | 2.68E-06 | |
| Rsp. to wounding | 179 | 17 | 2.93 | 5.81 | 2.66E-05 | |
| Ethylene-activated signaling pathway | 135 | 11 | 2.21 | 4.99 | 4.08E-02 | |
| Rsp. to SA | 164 | 13 | 2.68 | 4.85 | 9.72E-03 | |
| Rsp. to abscisic acid | 413 | 25 | 6.75 | 3.70 | 8.24E-05 | |
| Rsp. to salt stress | 417 | 24 | 6.81 | 3.52 | 3.82E-04 | |
| Protein phosphorylation | 828 | 47 | 13.53 | 3.47 | 6.30E-10 | |
| Rsp. to bacteria (bac.) | 353 | 20 | 5.77 | 3.47 | 5.12E-03 | |
| Cellular rsp. to acid chemical | 346 | 19 | 5.65 | 3.36 | 1.41E-02 | |
| Up-regulated genes in crwn2 mutants (107 genes mapped) | Rsp. to organonitrogen compound | 133 | 6 | 0.52 | 11.53 | 3.42E-02 |
| Rsp. to cold | 281 | 9 | 1.1 | 8.19 | 4.02E-03 | |
| Rsp. to salt stress | 417 | 10 | 1.63 | 6.13 | 1.35E-02 | |
| Rsp. to acid chemical | 896 | 21 | 3.51 | 5.99 | 9.39E-08 | |
| Rsp. to oxygen-containing compound | 1,156 | 22 | 4.52 | 4.86 | 1.51E-06 | |
| Rsp. to inorganic substance | 690 | 12 | 2.7 | 4.45 | 3.88E-02 | |
| Rsp. to hormone | 1,218 | 18 | 4.76 | 3.78 | 2.65E-03 | |
| Up-regulated genes in crwn4 mutants (1,510 genes mapped) | Rsp. to chitin | 108 | 57 | 5.98 | 9.53 | 1.22E-32 |
| Rsp. to molecule of bac. origin | 20 | 8 | 1.11 | 7.22 | 4.54E-02 | |
| Defense rsp. to fungus, incompatible interaction | 37 | 14 | 2.05 | 6.83 | 8.20E-05 | |
| Defense rsp. to bac., incompatible interaction | 40 | 11 | 2.22 | 4.96 | 4.48E-02 | |
| Rsp. to wounding | 179 | 46 | 9.91 | 4.64 | 1.09E-13 | |
| Plant-type hypersensitive rsp. | 67 | 17 | 3.71 | 4.58 | 8.54E-04 | |
| Rsp. to SA | 164 | 39 | 9.08 | 4.29 | 2.75E-10 | |
| Protein autophosphorylation | 115 | 27 | 6.37 | 4.24 | 1.98E-06 | |
| Cellular rsp. to jasmonic acid stimulus | 61 | 14 | 3.38 | 4.14 | 2.67E-02 | |
| Ethylene-activated signaling pathway | 135 | 29 | 7.48 | 3.88 | 3.47E-06 | |
| Up-regulated genes in crwn1 crwn2 mutants (1,686 genes mapped) | Rsp. to chitin | 108 | 62 | 6.68 | 9.27 | 5.61E-35 |
| Rsp. to molecule of bac. origin | 20 | 9 | 1.24 | 7.27 | 1.33E-02 | |
| SA-mediated signaling pathway | 35 | 14 | 2.17 | 6.46 | 1.61E-04 | |
| Defense rsp. to bac., incompatible interaction | 40 | 15 | 2.48 | 6.06 | 1.27E-04 | |
| Plant-type hypersensitive rsp. | 67 | 24 | 4.15 | 5.79 | 3.97E-08 | |
| Systemic acquired resistance | 44 | 15 | 2.72 | 5.51 | 4.24E-04 | |
| Regulation of cell death | 58 | 16 | 3.59 | 4.46 | 2.64E-03 | |
| Regulation of rsp. to biotic stimulus | 49 | 13 | 3.03 | 4.29 | 3.82E-02 | |
| Regulation of rsp. to external stimulus | 53 | 14 | 3.28 | 4.27 | 1.94E-02 | |
| Positive regulation of defense rsp. | 64 | 16 | 3.96 | 4.04 | 9.07E-03 | |
| Up-regulated genes in crwn1 crwn4 mutants (382 genes mapped) | Toxin metabolic process | 54 | 8 | 0.76 | 10.53 | 2.89E-03 |
| Cellular rsp. to SA stimulus | 50 | 7 | 0.7 | 9.95 | 1.91E-02 | |
| Defense rsp. to bac. | 281 | 29 | 3.96 | 7.33 | 5.01E-13 | |
| Rsp. to organonitrogen compound | 133 | 13 | 1.87 | 6.94 | 1.85E-04 | |
| Innate immune rsp. | 239 | 16 | 3.36 | 4.76 | 9.87E-04 | |
| Secondary metabolite biosynthetic process | 256 | 14 | 3.6 | 3.89 | 4.85E-02 | |
| Rsp. to abscisic acid | 413 | 22 | 5.81 | 3.78 | 3.62E-04 | |
| Rsp. to fungus | 490 | 26 | 6.9 | 3.77 | 2.90E-05 | |
| Rsp. to oxidative stress | 364 | 17 | 5.12 | 3.32 | 4.89E-02 | |
| Protein phosphorylation | 828 | 29 | 11.65 | 2.49 | 1.97E-02 |
Two crwn Double Mutants Exhibit DC3000 Resistance and Elevated PR Gene Expression
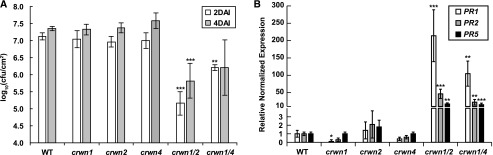
Our analysis of transcriptomic data uncovered an ectopic induction of defense responses in crwn mutants, prompting us to test if there are any defense phenotypes in these mutants. We infected an array of crwn mutants with two commonly studied Arabidopsis pathogens representing both fungi and bacteria: the necrotrophic fungal pathogen Botrytis cinerea or the hemibiotrophic bacterial pathogen P. syringae (DC3000). We did not detect differences among the mutants challenged with B. cinerea; however, we found that the crwn1 crwn2 mutant significantly suppressed DC3000 growth in leaf tissue, compared with the wild type and single crwn mutants (Fig. 2A), while the crwn1 crwn4 mutant exhibited an intermediate phenotype.
Figure 2.
crwn1 crwn2 and crwn1 crwn4 mutants exhibit spontaneous defense responses. A, DC3000 growth in rosette leaves of 22-d-old plants. The two double mutants have elevated resistance against DC3000. Log10 values of colony-forming units (cfu) from each mutant were calculated 2 and 4 d after infection (DAI). Error bars indicate sd (n = 4). Each mutant was compared with the wild type (WT) using Student’s t test (**, P < 0.01 and ***, P < 0.001). B, RT-qPCR data showing expression of three PR genes in 27-d-old plants. Transcript levels of the marker genes are significantly higher in the two double mutants, consistent with their DC3000 resistance phenotype. Error bars indicate se (n = 3). Student’s t tests were performed based on delta-Ct values (*, P < 0.05; **, P < 0.01; and ***, P < 0.001).
These graded disease phenotypes paralleled the expression of Pathogenesis Related (PR) marker genes in uninfected crwn mutants. Reverse transcription quantitative PCR (RT-qPCR) assays showed that PR1, PR2, and PR5 were highly expressed in both crwn1 crwn2 and crwn1 crwn4 double mutants (Fig. 2B), but to a more extreme degree in crwn1 crwn2 mutants. Furthermore, despite showing widespread transcriptional changes, single crwn1 and crwn4 mutants did not show significant up-regulation of PR genes, nor did these genotypes suppress the growth of DC3000. These RT-qPCR results were consistent with our RNA-seq data, which indicated that PR genes are up-regulated in crwn1 crwn2 and crwn1 crwn4 plants but not in crwn single mutants (Fig. 1D). Thus, the PR gene expression pattern indicates that crwn single mutants fail to induce defense responses fully, while the crwn1 crwn2 and crwn1 crwn4 double mutants induce defense responses above a threshold necessary to interfere with pathogen growth.
crwn Double Mutants Exhibit Cell Death Lesions
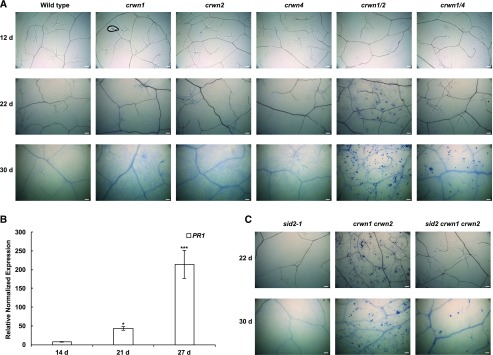
Spontaneous defense responses, such as those observed in crwn double mutants, are also characteristic of lesion-mimic mutants (LMMs), which develop ectopic cell death foci similar to hypersensitive response lesions (Moeder and Yoshioka, 2008). As LMMs trigger premature and prolonged defense responses, they also show many growth phenotypes, such as slower growth and dwarfism (Bowling et al., 1997; Bruggeman et al., 2015). We assayed cell death in the crwn mutants because crwn double mutants display both autonomous defense responses and dwarfism. We found that both crwn1 crwn2 and crwn1 crwn4 double mutants exhibited dead cells in rosette leaves stained with Trypan Blue (Fig. 3A). Consistent with the expression level of the PR genes and the resistance against DC3000, crwn1 crwn2 mutants showed a higher incidence of cell death compared with crwn1 crwn4 plants. Almost no cell death was found in matched tissues from either single crwn mutants or wild-type control samples.
Figure 3.
crwn double mutants exhibit cell death and PR gene expression, which are exacerbated with aging. A, Trypan Blue staining showed that crwn1 crwn2 and crwn1 crwn4 mutants form spontaneous lesions. As plants matured, cell death was more apparent and more frequent. B, PR1 expression in leaves from crwn1 crwn2 mutants of different ages. Expression of PR1 is higher in chronologically older leaves of crwn1 crwn2 mutants, consistent with our observation that older plants develop more lesions. PR1 expression at each stage was normalized to transcript levels in wild-type samples for the housekeeping gene ROC1. Error bars indicate se (n = 3). Student’s t tests were performed based on delta-Ct values (*, P < 0.05 and ***, P < 0.001). C, The sid2-1 mutation suppressed cell death in 22-d-old crwn1 crwn2 mutants; however, 30-d-old plants still exhibited some lesion formation. Bars = 100 mm.
We checked if spontaneous defense phenotypes intensify over time, because young seedlings of crwn1 crwn2 mutants have less wrinkled leaves than older plants. We found no cell death in 12-d-old crwn1 crwn2 mutants but lesions were frequent in 22-d-old crwn1 crwn2 leaves (Fig. 3A). Moreover, cell death was more widespread in 30-d-old crwn1 crwn2 plants (Fig. 3A). A similar age-dependent increase in cell death was observed in crwn1 crwn4 mutants.
Paralleling the increased incidence of cell death, PR1 expression was relatively low in leaves from younger 14- and 21-d-old crwn1 crwn2 plants but increased dramatically in 27-d-old plants (Fig. 3B). These results show that perturbation of the nuclear periphery in plant cells leads to age-dependent effects on the whole-plant level, manifested here in the form of cell death and gene expression changes.
crwn1 crwn2 and crwn1 crwn4 Double Mutants Show High Levels of Total SA
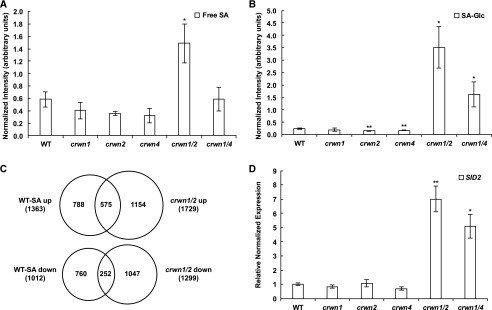
Next, we sought to understand the connection among the diverse responses displayed by the crwn double mutants. SA is a prominent regulator of plant defense responses, including the hypersensitive response, systemic acquired resistance, and reactive oxygen species production (Vlot et al., 2009). Consequently, we investigated the causal connection between the phenotypes of crwn mutants and SA. First, we measured SA abundance in crwn mutants using mass spectrometry coupled with HPLC (HPLC/MS). SA can exist as several derivatives, including SA glucoside and salicylate glucose ester. The glucosylation of SA is known to reduce its toxicity and allows storage of large quantities of the hormone (Dempsey et al., 2011). The level of free SA was more than 2-fold higher in crwn1 crwn2 mutants compared with the wild type or crwn1, crwn2, and crwn4 single mutants (Fig. 4A). For glucosylated SA, the fold change was much higher than that for free SA: we recorded a 14-fold increase of glucosylated SA in crwn1 crwn2 mutants and a more than 6-fold increase in crwn1 crwn4 mutants (Fig. 4B). Thus, we found that both free SA and glucosylated SA are elevated in crwn1 crwn2 mutants, while only glucosylated SA is highly abundant in crwn1 crwn4 mutants. These results are consistent with the intermediate defense phenotypes in crwn1 crwn4 plants compared with crwn1 crwn2 mutants and the lack of significant defense phenotypes in crwn single mutants. These findings suggest that SA is an important mediator of the abnormal gene expression and defense responses exhibited by crwn mutants.
Figure 4.
SA levels in crwn1 crwn2 and crwn1 crwn4 mutants are significantly higher than in wild-type (WT) plants or crwn single mutants. A, HPLC/MS detection of free SA levels from 27-d-old plants. Peak intensities of free SA per fresh sample weight were normalized to the spiked internal control d4-SA. Each mutant was compared with the wild type using Student’s t test, and error bars indicate sd (*, P < 0.05; n = 3). B, HPLC/MS detection of glucosylated SA (SA-Glc, indicating either SA glucoside or salicylate Glc ester) levels from each mutant. The detection method, units, and statistical analysis were identical to those described for A (*, P < 0.05 and **, P < 0.01). C, Overlap of misregulated genes in our RNA-seq data from crwn1 crwn2 mutants and those in microarray data from 1 mm SA-treated wild-type plants (Zhou et al., 2015). For SA-treated wild-type microarray data, genes at least 2-fold increased or decreased with adjusted P < 0.05 were used. For the RNA-seq of crwn1 crwn2 mutants, the criteria described in Figure 1 were used. The significance of the overlap of both up-regulated and down-regulated gene comparisons was assessed by the hypergeometric test; the resulting P values in both cases were far less than 0.001. D, RT-qPCR data showing expression of the SID2 gene in 27-d-old plants. Error bars indicate se (n = 3). Student’s t tests were performed based on delta-Ct values (*, P < 0.05 and **, P < 0.01).
SA Is Responsible for a Significant Portion of Misregulated Genes in crwn1 crwn2 Mutants
To test the prediction that SA overaccumulation is responsible for the altered transcriptome in crwn mutants, we compared our RNA-seq data of crwn1 crwn2 mutants with a microarray data set from SA-treated wild-type plants (Zhou et al., 2015). About 40% of the up-regulated genes in the SA-treated wild-type sample overlapped with the up-regulation profile of crwn1 crwn2 mutants (Fig. 4C). GO term analysis revealed that genes in the overlap of these sets are closely related to bacterial resistance and SA response (Supplemental Table S4). Moreover, 25% of the down-regulated genes in the SA-treated wild-type sample overlapped with those down-regulated in crwn1 crwn2 samples (Fig. 4C). Among the genes down-regulated in both data sets are those featuring the response to jasmonic acid GO term (Supplemental Table S4), reflecting the well-known antagonism between SA and jasmonic acid signaling (Robert-Seilaniantz et al., 2011).
The number of loci misregulated in both SA-treated plants and crwn single mutants was reduced relative to the overlap between SA-treated and crwn1 crwn2 double mutant samples (Supplemental Fig. S5). This pattern is consistent with the fact that the crwn single mutants do not accumulate high levels of SA. However, this explanation is incomplete, as the overlap is relatively modest between the SA-treatment data set and the misregulated loci in crwn1 crwn4 mutants with elevated SA levels. Taken together, our findings indicate that SA signaling accounts for a significant portion of the transcriptomic changes in the crwn1 crwn2 mutants but that crwn mutations cause gene misregulation through additional mechanisms.
SID2 Transcripts Are Abundant in Two crwn Double Mutants
To understand the mechanism responsible for SA accumulation in crwn1 crwn2 and crwn1 crwn4 mutants, we checked for changes in the transcript levels of genes involved in SA biosynthesis. There are two different SA biosynthetic pathways in plants (Rivas-San Vicente and Plasencia, 2011). First, the phenylpropanoid pathway utilizes Phe in the cytoplasm to produce SA. Phenylalanine ammonia lyases (PALs) are important enzymes in this pathway. The second pathway begins with isochorismate in the chloroplast. In the latter pathway, ICS, where ICS1 is a synonym of SID2, are key enzymes responsible for SA biosynthesis (Seyfferth and Tsuda, 2014). SID2 is highly expressed when plants are infected with P. syringae pv maculicola (Wildermuth et al., 2001). We found that crwn1 crwn2 mutants express very high levels of SID2 transcripts, while transcription of PAL1 and PAL4 is reduced by more than half in our RNA-seq data (Supplemental Fig. S6). Using RT-qPCR, we validated that the SID2 transcript level was significantly increased in crwn1 crwn2 mutants, while crwn1 crwn4 mutants exhibited an intermediate level of expression (Fig. 4D). These results are consistent with the total SA levels and the magnitude of defense responses in the crwn mutants. In addition, we found that the genes encoding several positive regulators of SID2 (Seyfferth and Tsuda, 2014), including SARD1 and CBP60g, are highly up-regulated in the transcriptomic profiles of crwn1 crwn2 mutants (Supplemental Fig. S6). These results indicate that loss of CRWN genes leads to ectopic pathogen signaling by ramping up the production of SA via overexpression of SID2.
Blocking Induced SA Biosynthesis Diminishes But Does Not Abolish Lesion Formation
After we observed that abnormally high SA levels were synthesized in crwn double mutants, we introduced a sid2-1 mutation to block SA biosynthesis to determine which crwn phenotypes are dependent on SA. As shown in Supplemental Figure S7, the sid2-1 mutation effectively suppressed SA accumulation in the crwn1 crwn2 background. The lack of SA is consistent with the fact that DC3000 growth in sid2 crwn1 crwn2 mutants was comparable to that in sid2 mutants (Supplemental Fig. S8). Therefore, the sid2-1 allele was epistatic to the crwn mutations for both SA accumulation and bacterial growth phenotypes, indicating that the primary reason for ectopic defense responses in crwn mutants is a high level of SA.
We observed that the sid2-1 mutation also suppressed lesion formation in crwn1 crwn2 mutants, but, in this case, the suppression was incomplete. While no cell death was observed in 22-d-old sid2 crwn1 crwn2 leaves (Fig. 3C), scattered lesions were still evident in 30-d-old leaf tissue. This result demonstrates that some cell death occurs in crwn1 crwn2 mutants in an SA-independent manner.
crwn1 crwn2 Mutants Exhibit SA-Independent Developmental Defects
A number of additional phenotypes exhibited by crwn1 crwn2 mutants are independent of SA accumulation to varying degrees. For example, the wrinkled leaves and dwarf stature of crwn double mutants were only partially suppressed by disruption of SID2 (Supplemental Fig. S9). In particular, sid2 crwn1 crwn2 plants still exhibited some dwarfism, most noticeably in bolt stature (Supplemental Fig. S9C). These results indicate that a large component of the dwarfing syndrome displayed by crwn1 crwn2 mutants is caused by SA overaccumulation, although there is still an SA-independent pathway affecting bolt stature.
Seed shape was also altered in certain crwn mutants, especially crwn1 crwn2. As shown in Supplemental Figure S10, crwn1 crwn2 seeds often had deeply creviced surfaces, and we observed a range of seed morphologies, from normal to smaller and darker seeds. Addition of the sid2-1 allele did not have an appreciable effect on these phenotypes, as seeds harvested from sid2 crwn1 crwn2 mutants were also misshapened.
We also examined nuclei in the sid2 crwn1 crwn2 mutant to determine if the reduced organellar size and abnormal spherical shape characteristic of this genotype might be partially attributable to ectopic SA signaling. As shown in Supplemental Figure S11, elongated anther filament cells of sid2 crwn1 crwn2 mutants retained the small, round nuclear morphology observed in crwn1 crwn2 mutants. Therefore, the sid2-1 mutation had no effect on nuclear size or shape. This finding demonstrates that the nuclear phenotypes caused by loss of CRWN1 and CRWN2 are primary effects, not downstream consequences of elevated SA production in these mutants.
DISCUSSION
To investigate the importance of three-dimensional genome organization within the nucleus, we asked if perturbation of the plant nuclear periphery leads to aberrant gene expression patterns. We found that deficiencies of individual NMCP proteins in Arabidopsis, particularly CRWN1 or CRWN4, led to changes in the steady-state levels of a large number of transcripts, and even more transcripts were altered in crwn1 crwn2 double mutants. A large portion of the overexpressed genes were involved in pathogen defense. The proximate cause of much of this misregulation is the overproduction of the defense signaling molecule SA as a consequence of the up-regulation of the SA biosynthetic gene, SID2. A constellation of phenotypes, including elevated bacterial resistance, dwarfism, cell death lesions, and wrinkled leaves, could be either fully or partially suppressed by the null sid2-1 mutation. Using this approach, we also identified consequences of nuclear defects that are independent of SA signaling, such as reduced plant stature, abnormal seed shape, and a baseline level of cell death.
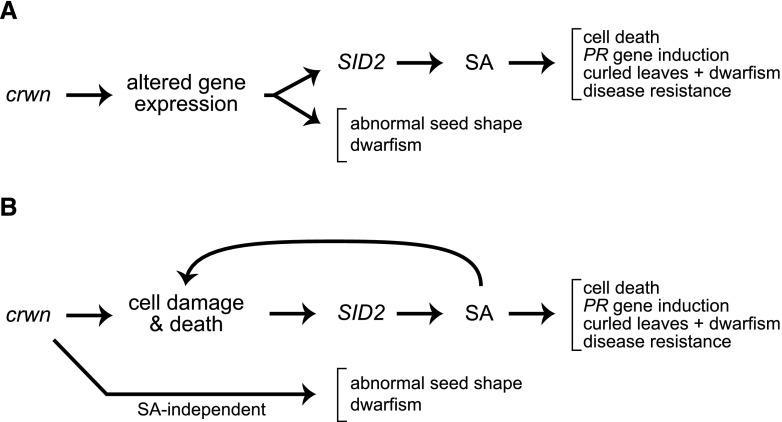
Synthesizing our findings, we propose two models (Fig. 5) to explain this complex response to altering nuclear structure in plants. The initiating event in both scenarios is the nuclear defect caused by genetic ablation of a subset of CRWN nuclear periphery proteins. In the first model (Fig. 5A), these defects lead to expression changes in vanguard genes, including SID2, which leads to SA production and secondary changes to the transcriptome as well as downstream effects on pathogen defense and plant morphology. Several mechanisms might be operating to disrupt gene expression, such as the direct involvement of CRWN proteins in transcriptional regulation (either as part of an NL or as free proteins; see below) or changes in three-dimensional positioning of genes. Support for the latter mechanism was recently reported for mouse embryonic stem cells, where changes in three-dimensional genome structure due to lamin deficiency led to altered gene expression patterns within LADs (Zheng et al., 2018). Furthermore, an increase in trans- versus cis-chromosomal contacts was detected via Hi-C analysis of crwn1 and crwn4 mutants, suggesting that NL changes could reduce local looping interactions or alter other three-dimensional configurations important for transcriptional regulation in interphase plant nuclei (Grob et al., 2014).
Figure 5.
Two models to explain how crwn mutations affect disease signaling and other phenotypes. A, The first model posits that crwn mutations lead to transcriptional changes via the disruption in the direct action of CRWN proteins or epigenetic mechanisms (e.g. through changes in chromatin modification or three-dimensional genomic configuration). In this first model, overexpression of SID2 and subsequent SA accumulation lead to pathogen defense phenotypes and an LMM syndrome. B, In the alternative model, cell damage and/or death due to nuclear dysfunction is the primary effect of crwn mutations. Downstream effects of age-dependent cell damage/death are amplified via an SA-mediated pathway, but an SA-independent pathway also mediates developmental defects.
More localized changes in chromatin could also play a role, mirroring the direct effects that the nuclear periphery exerts on histone modification or chromatin-mediated silencing in animals and fungi (Harr et al., 2015, 2016). Recent work from Schubert and colleagues in Arabidopsis demonstrated a physical and functional connection between CRWN1 and PWWP INTERACTORS OF POLYCOMBS1, a protein associated with the Polycomb Repressive Complex 2 responsible for H3K27 methylation (Mikulski et al., 2017). This mechanism is consistent with our observation that loci packaged in chromatin state 2, featuring both active and repressive (e.g. histone H3K27me3) epigenetic marks, are overrepresented among up-regulated genes in crwn mutants (Supplemental Fig. S4). This result, however, could also reflect the fact that SA-modulated genes tend to be packaged in chromatin state 2; approximately 40% of genes overexpressed in response to SA contained at least one domain of state 2 chromatin (relative to the expected occurrence of approximately 25%; Supplemental Fig. S12). We investigated the possible role of H3K27me3 regulation via chromatin immunoprecipitation (ChIP)/qPCR experiments in different crwn genotypes, using the SA-regulated locus PR1, as well as AGAMOUS (AG) as a control locus. AG is a well-known target of H3K27me3 repression that is not known to be regulated by SA (Zhang et al., 2007). As shown in Supplemental Figure S13, we observed a trend toward reduced H3K27me3 modification at the PR1 locus in crwn1 crwn2 mutants (below the cutoff for significance) but no change in crwn1 mutants. These results are consistent with the increase in PR1 expression in the double but not the single mutant. These findings could be interpreted as evidence that histone H3K27me3 changes, mediated by the loss of CRWN proteins, play a role in PR1 gene induction, but it is possible that chromatin modification changes lie downstream of SA-modulated transcription. It is important to note that the ultimate targets of misregulation in crwn mutants are genes in the chromosome arms, rather than the heterochromatic regions surrounding the centromere or TEs (Fransz et al., 2002). Our results are supported by previous findings from Poulet et al. (2017) showing that crwn1 crwn2 mutants do not induce transcription of transposons, such as TSI, or centromeric repeats.
The second model (Fig. 5B) does not invoke a role for CRWN proteins or nuclear organization in transcriptional control; rather, it posits that the cascade of transcriptional changes lies downstream of cell death or cell damage perception caused by nuclear defects (e.g. leakage [Denais et al., 2016] or abnormal connection to the cytoskeleton). This model explains the age-dependent effects seen in crwn mutants, in which gene expression changes and the incidence of cell death lesions progress in their intensity over time, as a consequence of accumulated cell damage and death. In the second model, SA production lies downstream of this damage and cell death and leads to an amplification and proliferation of cell death in a feed-forward manner. Key support for this model is the low incidence of cell death in the absence of induced SA production in the sid2 crwn1 crwn2 mutant (Fig. 3C). This result suggests that age-dependent cell death/damage precipitated by nuclear changes initiates the subsequent SA-dependent lesion-mimic syndrome, with its characteristic dwarfing, wrinkled leaves, and PR gene induction. The age-dependent effects seen in crwn mutants are consistent with the types of progressive changes in nuclear structure that are associated with both natural aging as well as progeria syndromes caused by human laminopathies (Scaffidi and Misteli, 2006).
Several recent publications shed light on these models and highlight the importance of the plant nucleus in defense signaling. Most relevant is a recently published study claiming that CRWN1 acts as a corepressor, with NAC WITH TRANSMEMBRANE MOTIF1-LIKE9 (NTL9), of PR genes (Guo et al., 2017). These data support aspects of the transcriptional control model (Fig. 5A) in which CRWN proteins can play a direct role in transcriptional regulation. However, this specific corepressor activity for CRWN1 with NTL9 fails to explain the up-regulation of SID2, as NTL9 is an activator of this SA biosynthetic locus (Zheng et al., 2015). An alternative possibility is that CRWN1 interacts with NTL9 to block its activation of SID2 by sequestering the transcription factor at the nuclear periphery. We note that Guo et al. (2017) did not observe a spike in SA production in crwn1 crwn2 mutants, which we demonstrate here (Fig. 4), an observation that can account for PR gene induction as well as the broader changes in gene expression and the elevated bacterial pathogen resistance exhibited by these mutants. The increase in SA levels in crwn double mutants can also explain the observed induction of reactive oxygen species recently noted by Wang et al. (2019), which they associated with their independent observation of the elevated level of cell death.
The role of the nuclear structure in defense signaling has also been brought into focus by studies showing that plant nucleoporins play important roles. For example, Gu et al. (2016) demonstrated that CONSTITUTIVE EXPRESSION OF PR GENES5 (CPR5; Bowling et al., 1997), long known for its roles in systemic acquired resistance, is a nucleoporin that regulates the release of cyclin-dependent kinase inhibitors and the transport of signaling complexes. Furthermore, they showed that cpr5 mutants, which are LMMs exhibiting spontaneous defense response, have round nuclei similar to those of crwn mutants. These findings suggest that the nuclear structural changes that occur in cpr5 and crwn mutants might be mechanistically related. For instance, defects at the nuclear periphery could alter the organization or activity of nuclear pore complexes (Al-Haboubi et al., 2011; Guo et al., 2014). Other recent work points to nuclear pores as a site of defense signaling in plants. Tamura et al. (2017) reported that the nucleoporin mutant nup136 shows compromised resistance against DC3000, while Gu et al. (2016) suggested that the same mutants have enhanced effector-triggered immunity resistance against another bacterial pathogen (Psm/AvrRpt2). Although these two studies suggest different roles of this particular nucleoporin, it is now indisputable that components at the nuclear periphery participate in regulation of plant defense responses.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Arabidopsis (Arabidopsis thaliana) plants were grown on MetroMix 360 soil (Sun Gro Horticulture) under long-day conditions (16 h of light/8 h of dark) at approximately 22°C in environmental growth chambers after 2 d of stratification in a 4°C cold room. All genotypes are in the Columbia background. The crwn mutant material used was described by Wang et al. (2013); the original allele sources are crwn1 (SALK_025347), crwn2 (SALK_076653), and crwn4 (SALK_079296RP), which were acquired from the Arabidopsis Biological Resource Center at Ohio State University. The sid2-1 mutant material originated from the Metraux group (Nawrath and Métraux, 1999). The sid2-1 crwn1 crwn2 triple mutants were generated by crossing sid2-1 (♀) mutants to crwn1 crwn2 (♂) mutants and subsequent segregation. The genotypes of all the mutants used in this study were identified by PCR amplification with primers listed below and reference to marker polymorphisms (Supplemental Table S5).
RNA-Seq and Bioinformatic Analysis
Three biological replicates from each genotype were used for RNA-seq (National Center for Biotechnology Information [NCBI] SRA BioProject ID PRJNA485018). Rosette leaves from 4-week-old adult plants were used to extract total RNA using TRIzol reagent (Thermo Fisher Scientific) and further purified with the Qiagen RNeasy kit. RNA samples were processed by the Genomic Core Facility at Cornell’s Weill Medical School using a standard Illumina protocol to construct single-stranded mRNA libraries. The Illumina Hi-Seq 2000 platform was used for the sequencing. The resulting 51 nucleotide reads were analyzed using the Tuxedo Suite software (Trapnell et al., 2012). Briefly, Bowtie2 (v. 2.2.2) and Tophat2 (v. 2.1.1) were used to build an index for the TAIR10 Arabidopsis genome to map RNA-seq reads and to identify splice junctions between exons (Arabidopsis Genome Initiative, 2000; Langmead et al., 2009; Kim et al., 2013). Finally, Cufflinks (v. 2.2.1) was used to calculate differential gene expression (Trapnell et al., 2010). GO term analysis was conducted using PANTHER (Mi et al., 2017). The overrepresentation test (released July 15, 2016) was performed with misregulated gene sets against the annotation data set GO biological process complete (GO ontology database released November 30, 2016). Note that the numbers of genes listed for the different GO terms in Table 1 do not match the values in the Venn diagrams in Figure 1 and Supplemental Figure S1 because not all genes in our data set are assigned GO terms in the PANTHER database.
Venn diagrams for all five genotypes in Supplemental Figure S1 were generated using InteractiVenn (Heberle et al., 2015).
For TE analysis, STAR (v. 2.5.3a_modified) was used for index building and read mapping (Dobin et al., 2013). Mapping employed -winAnchorMultimapNmax 100 and -outFilterMultimapNmax 100 options. Next, TEtranscripts (v. 1.5.1) was used to analyze TE expression in alignment files from the STAR output (Jin et al., 2015). The annotation GFF3 file for genes was obtained from TAIR10 and converted to GTF by gffread (Trapnell et al., 2010) with -FTo option. The TE annotation file was provided by the Hammell lab (http://labshare.cshl.edu/shares/mhammelllab/www-data/TEToolkit/TE_GTF/). The GTF file from the Hammell lab grouped TEs by family name. We also modified this file so that reads could be mapped to individual TEs. Read count tables produced by TEtranscripts were analyzed by DESeq2 (Love et al., 2014).
For the comparison between SA-treated wild type and crwn mutants, microarray data from 23-d-old wild-type plants sampled 3 h after 1 mm SA treatment (GSM1496067, 1496075, and 1496083) and water-treated controls (GSM1496065, 1496073, and 1496081) were obtained from the NCBI Gene Expression Omnibus database (series no. GSE61059; Zhou et al., 2015). Genes misregulated more than 2-fold with adjusted P < 0.05 were screened with GEO2R using default options (Edgar et al., 2002; Barrett et al., 2013). The resulting list of misregulated microarray elements were converted to locus identifiers by TAIR Microarray Elements Search and Download tool. These genes were compared with our RNA-seq data from the crwn mutants.
Our chromatin state analysis was based on the study by Sequeira-Mendes et al. (2014) of the epigenetic landscape of the wild-type Arabidopsis genome. We overlapped chromatin states of Arabidopsis genomes onto misregulated genes in crwn mutants by using Intersect the intervals of two data sets under the Operate on Genomic Intervals tool in Galaxy (Afgan et al., 2016). Minimum overlapping was set as 200 bp, which is the length of a nucleosome. Because of this overlapping definition, one gene can exist as more than two states.
To examine the distribution of misregulated genes across the genome, chromosomal maps were drawn based on the Columbia reference genome sequence (TAIR10) with the genoPlotR package (Guy et al., 2010). Pericentromeric regions were marked based on Stroud et al. (2013). Genes depicted on the map are the same as the ones used to draw the Venn diagrams in Figure 1 and Supplemental Figure S1.
Pseudomonas syringae DC3000 Bacterial Growth Assays
DC3000 was grown on King’s B medium (20 g L−1 Proteose Peptone no. 3, 10 mL L−1 glycerol, 0.4 g L−1 MgSO4, 1.5 g L−1 K2HPO4, and 18 g L−1 agar) containing 50 μg mL−1 rifampicin and 50 μg mL−1 kanamycin. Bacterial cells were collected and suspended in 10 mm MgCl2 solution to a final concentration of 105 cells mL−1. The abaxial sides of three fully expanded rosette leaves of 22-d-old plants were infiltrated with the bacterial suspension using a syringe without a needle. Four plants per genotype were used for biological replicates. Infected leaves were harvested 2 and 4 d after inoculation. For the set of infections investigating the effect of sid2 mutations, only 0 and 2 d after inoculation were checked. Bacterial growth experiments were repeated multiple times, with each replicate showing a significant reduction of DC3000 growth in the crwn double mutants.
RNA Extraction and RT-qPCR
Aboveground shoot material, excluding bolts and flowers, was harvested and frozen in liquid nitrogen. Frozen samples were ground using three metal beads or mortars and pestles. The E.Z.N.A. Plant RNA Kit (Omega Bio-tek) was used to extract total RNA, followed by DNase I (New England Biolabs) treatment. cDNA synthesis was performed with either the SuperScript III or SuperScript IV First-Strand Synthesis System (Thermo Fisher Scientific) using oligo(dT) primers. For the RT-qPCR, iTaq Universal SYBR Green Supermix (Bio-Rad) was used. Three biological replicates were tested (plant cohorts grown independently at separate times; one plant per sample), and each replicate consisted of at least three reactions per target gene. The size of amplicons was designed to be approximately 100 bp. A CFX 96 Real-Time System (Bio-Rad) mounted on a C1000 Thermal Cycler (Bio-Rad) was used to detect products. The results were analyzed using the Bio-Rad CFX Manager 3.1. software package.
Cell Death Lesion Detection and Imaging
Fully expanded rosette leaves were used for cell death lesion analysis. Whenever possible, the fifth leaf that emerged from the meristem was used. However, similar patterns were found in all fully expanded leaves of similar size. Leaves were harvested and stained as described (van Wees, 2008). Briefly, leaves were immersed in 2.5 mg mL−1 Trypan Blue-lactophenol solution with 2 volumes of ethanol added. Samples were then boiled for 1 min and left on a shaker at room temperature for 2 to 3 h. Destaining was performed twice with 2.5 g mL−1 chloral hydrate solution. Final samples were stored with 70% (v/v) glycerol and observed with a Leica DM5500B microscope with 10× magnification to check stained foci.
SA Extraction and HPLC/MS Analysis
Extraction of SA was performed as indicated in the protocol from Stingl et al. (2013) with slight modifications. Briefly, leaves were frozen in liquid nitrogen and ground using metal beads. Samples were suspended with 950 μL of ethylacetate:formic acid solution (99:1, v/v). To make the d4-SA internal control solution, d6-SA (#616796; Sigma-Aldrich) was dissolved in methanol. Fifty microliters of 7.5 ng μL−1 d4-SA solution was added to the sample as an internal control. Nine hundred microliters of the supernatants was isolated, and solvents were evaporated with a SpeedVac concentrator (Savant ISS110; Thermo Scientific). The final residues were dissolved in 100 μL of acetonitrile:water (1:1, v/v) solution and filtered with a 0.45-μm MultiScreen HTS HV Filter Plate (MSHVN4510; Millipore).
The analytical reverse-phase separation was performed with a Dionex Ultimate 3000 Series LC system with a Titan C18 UHPLC Column (7.5 cm × 2.1 mm, 1.9 µm; Sigma-Aldrich) at 40°C. The flow rate was set at 0.5 mL min−1. The starting condition was 95% solvent A (0.1% [v/v] formic acid in water) and 5% solvent B (acetonitrile) for 0.5 min, rising to 95% B at 19 min, held for 1 min, followed by 2 min of reequilibration at the starting condition. The separation was sent to an Orbitrap Q-Exactive mass spectrometer (Thermo Fisher Scientific) equipped with a HESI-II heated electrospray ionization source. Samples were analyzed in triplicate under a negative spray mode: voltage, 3,500 V; capillary and auxiliary gas heater temperatures, 300°C; sheath, auxiliary, and sweep gas flow rates of 70, 10, and 5, respectively (arbitrary units). The data were acquired under full-scan mass spectra in mass-to-charge ratio range of 100 to 600, 140,000 full width at half maximum resolution (at mass-to-charge ratio = 200), AGC target 1e6, and maximum injection time of 200 ms.
Nuclear Morphology Phenotyping
We observed nuclear morphology in anther filament cells, which are nonpigmented and contain elongated, endopolyploid nuclei in wild-type plants (Dittmer et al., 2007). Anther filaments were fixed with ethanol:acetic acid solution (3:1, v/v) for 10 min, followed by exposure to 10 μg mL−1 4′,6-diamidino-2-phenylindole staining solution for 15 to 30 s. Samples were immersed in distilled water for 1 min and then observed with an epifluorescence microscope (DM5500; Leica).
ChIP
ChIP was performed based on the protocol from Yamaguchi et al. (2014) with the following modifications. Anti-H3K27me3 antibodies were purchased from Abcam (catalog no. ab6002). Vacuum infiltration of formaldehyde solution was adjusted to 6 min followed by 4 min of incubation on ice. This procedure was repeated three times. Vacuum infiltration of Gly solution was modified to 5 min. Sonication was applied with a Bioruptor UCD-200 (Diagenode) for 30 s with the high-intensity option followed by 30 s of pause; this 1-min cycle was repeated 15 times. For preclearing and immunoprecipitation capture, 45 μL of Dynabeads Protein A (ThermoFisher) was used. To purify the resulting ChIP DNA, the ChIP DNA Clean & Concentrator Kit (Zymo) was used. qPCR was performed as described for our RT-qPCR analysis. Primers are listed in Supplemental Table S5.
Accession Numbers
Locus identifiers of genes investigated in this study include AT1G67230 (CRWN1), AT1G13220 (CRWN2), AT1G68790 (CRWN3), AT5G65770 (CRWN4), AT1G74710 (ICS1/SID2), AT2G14610 (PR1), AT3G57260 (PR2), AT1G75040 (PR5), AT4G38740 (ROC1), AT5G09810 (ACT7), and AT4G18960 (AG). The RNA-seq data described here are available through NCBI (SRA BioProject ID PRJNA485018).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Mutations in Arabidopsis CRWN genes result in widespread changes in transcript levels.
Supplemental Figure S2. A heat map of transcript level differences of a subset of misregulated genes in different crwn genotypes underscores the synergy among the effects of the crwn1 and crnw2 mutations.
Supplemental Figure S3. Misregulated genes are distributed evenly across the chromosome arms.
Supplemental Figure S4. Chromatin states of genes misregulated in each crwn mutant are shown.
Supplemental Figure S5. SA-regulated genes overlap with misregulated genes in crwn mutants.
Supplemental Figure S6. RNA-seq data showing expression levels of SA biosynthesis genes and genes regulating SA biosynthesis genes.
Supplemental Figure S7. HPLC/MS data demonstrating that the sid2-1 mutation diminishes both SA and glucosylated SA levels.
Supplemental Figure S8. The sid2-1 mutation suppresses DC3000 resistance in crwn1 crwn2 mutants.
Supplemental Figure S9. The sid2-1 mutation partially suppresses the rosette dwarfism of the crwn1 crwn2 mutant, while the lengths of the bolts are not affected.
Supplemental Figure S10. crwn1 crwn2 mutants exhibit abnormal seed shape, which is not suppressed by sid2 mutations.
Supplemental Figure S11. SA overproduction does not explain the nuclear morphology phenotypes of crwn1 crwn2 mutants.
Supplemental Figure S12. Overrepresentation of chromatin state 2 within genes regulated in response to SA treatment.
Supplemental Figure S13. Histone H3 Lys-27 trimethylation levels at PR1 are decreased in crwn1 crwn2 mutants.
Supplemental Table S1. Differential expression of TEs sorted as family in crwn mutants.
Supplemental Table S2. Differential expression of TEs sorted as individual in crwn mutants.
Supplemental Table S3. Statistical overrepresentation test for down-regulated genes in crwn mutants.
Supplemental Table S4. Statistical overrepresentation test for genes misregulated in both crwn1 crwn2 mutants and SA-treated wild-type plants.
Supplemental Table S5. Primers used in this study.
Acknowledgments
Haiyi Wang conducted the original RNA-seq experiment, and we are grateful for her efforts to launch this project. We thank Hyong Woo Choi for help with the bacterial resistance tests and for providing relevant materials for these assays. Molly Hammell generously provided advice on the use of the TE analysis approach that her group designed. We also thank Navid Movahed, Tae Hyung Won, and Shaoqun Zhou for MS analysis and advice, as well as the Boyce Thompson Institute Computational Biology Center for guidance and bioinformatics resources. We gratefully acknowledge the helpful comments of the anonymous referees.
Footnotes
This work was supported by the facilities and financial support of the Boyce Thompson Institute. Partial funding for this project was derived from U.S. National Science Foundation grants to E.J.R. (MCB-0956820 and MCB-1158297). J.C. was funded in part by the Korean Government Scholarship Program for Study Overseas and Cornell University.
Articles can be viewed without a subscription.
References
- Aebi U, Cohn J, Buhle L, Gerace L (1986) The nuclear lamina is a meshwork of intermediate-type filaments. Nature 323: 560–564 [DOI] [PubMed] [Google Scholar]
- Afgan E, Baker D, van den Beek M, Blankenberg D, Bouvier D, Čech M, Chilton J, Clements D, Coraor N, Eberhard C, et al. (2016) The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2016 update. Nucleic Acids Res 44: W3–W10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Haboubi T, Shumaker DK, Köser J, Wehnert M, Fahrenkrog B (2011) Distinct association of the nuclear pore protein Nup153 with A- and B-type lamins. Nucleus 2: 500–509 [DOI] [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408: 796–815 [DOI] [PubMed] [Google Scholar]
- Barrett T, Wilhite SE, Ledoux P, Evangelista C, Kim IF, Tomashevsky M, Marshall KA, Phillippy KH, Sherman PM, Holko M, et al. (2013) NCBI GEO: Archive for functional genomics data sets—update. Nucleic Acids Res 41: D991–D995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi X, Cheng YJ, Hu B, Ma X, Wu R, Wang JW, Liu C (2017) Nonrandom domain organization of the Arabidopsis genome at the nuclear periphery. Genome Res 27: 1162–1173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowling SA, Clarke JD, Liu Y, Klessig DF, Dong X (1997) The cpr5 mutant of Arabidopsis expresses both NPR1-dependent and NPR1-independent resistance. Plant Cell 9: 1573–1584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruggeman Q, Raynaud C, Benhamed M, Delarue M (2015) To die or not to die? Lessons from lesion mimic mutants. Front Plant Sci 6: 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chytilova E, Macas J, Sliwinska E, Rafelski SM, Lambert GM, Galbraith DW (2000) Nuclear dynamics in Arabidopsis thaliana. Mol Biol Cell 11: 2733–2741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csoka AB, English SB, Simkevich CP, Ginzinger DG, Butte AJ, Schatten GP, Rothman FG, Sedivy JM (2004) Genome-scale expression profiling of Hutchinson-Gilford progeria syndrome reveals widespread transcriptional misregulation leading to mesodermal/mesenchymal defects and accelerated atherosclerosis. Aging Cell 3: 235–243 [DOI] [PubMed] [Google Scholar]
- Davidson PM, Lammerding J (2014) Broken nuclei: Lamins, nuclear mechanics, and disease. Trends Cell Biol 24: 247–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dempsey DA, Vlot AC, Wildermuth MC, Klessig DF (2011) Salicylic acid biosynthesis and metabolism. The Arabidopsis Book 9: e0156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denais CM, Gilbert RM, Isermann P, McGregor AL, te Lindert M, Weigelin B, Davidson PM, Friedl P, Wolf K, Lammerding J (2016) Nuclear envelope rupture and repair during cancer cell migration. Science 352: 353–358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dittmer TA, Richards EJ (2008) Role of LINC proteins in plant nuclear morphology. Plant Signal Behav 3: 485–487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dittmer TA, Stacey NJ, Sugimoto-Shirasu K, Richards EJ (2007) LITTLE NUCLEI genes affecting nuclear morphology in Arabidopsis thaliana. Plant Cell 19: 2793–2803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR (2013) STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 29: 15–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R, Domrachev M, Lash AE (2002) Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res 30: 207–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egecioglu D, Brickner JH (2011) Gene positioning and expression. Curr Opin Cell Biol 23: 338–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fransz P, De Jong JH, Lysak M, Castiglione MR, Schubert I (2002) Interphase chromosomes in Arabidopsis are organized as well defined chromocenters from which euchromatin loops emanate. Proc Natl Acad Sci USA 99: 14584–14589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilford H. (1904) Progeria: A form of senilism. Practitioner 73: 188–203 [Google Scholar]
- Goldman RD, Shumaker DK, Erdos MR, Eriksson M, Goldman AE, Gordon LB, Gruenbaum Y, Khuon S, Mendez M, Varga R, et al. (2004) Accumulation of mutant lamin A causes progressive changes in nuclear architecture in Hutchinson-Gilford progeria syndrome. Proc Natl Acad Sci USA 101: 8963–8968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grob S, Schmid MW, Grossniklaus U (2014) Hi-C analysis in Arabidopsis identifies the KNOT, a structure with similarities to the flamenco locus of Drosophila. Mol Cell 55: 678–693 [DOI] [PubMed] [Google Scholar]
- Gruenbaum Y, Foisner R (2015) Lamins: Nuclear intermediate filament proteins with fundamental functions in nuclear mechanics and genome regulation. Annu Rev Biochem 84: 131–164 [DOI] [PubMed] [Google Scholar]
- Gu Y, Zebell SG, Liang Z, Wang S, Kang BH, Dong X (2016) Nuclear pore permeabilization is a convergent signaling event in effector-triggered immunity. Cell 166: 1526–1538.e11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guelen L, Pagie L, Brasset E, Meuleman W, Faza MB, Talhout W, Eussen BH, de Klein A, Wessels L, de Laat W, et al. (2008) Domain organization of human chromosomes revealed by mapping of nuclear lamina interactions. Nature 453: 948–951 [DOI] [PubMed] [Google Scholar]
- Guo T, Mao X, Zhang H, Zhang Y, Fu M, Sun Z, Kuai P, Lou Y, Fang Y (2017) Lamin-like proteins negatively regulate plant immunity through NAC WITH TRANSMEMBRANE MOTIF1-LIKE9 and NONEXPRESSOR OF PR GENES1 in Arabidopsis thaliana. Mol Plant 10: 1334–1348 [DOI] [PubMed] [Google Scholar]
- Guo Y, Kim Y, Shimi T, Goldman RD, Zheng Y (2014) Concentration-dependent lamin assembly and its roles in the localization of other nuclear proteins. Mol Biol Cell 25: 1287–1297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guy L, Kultima JR, Andersson SG (2010) genoPlotR: Comparative gene and genome visualization in R. Bioinformatics 26: 2334–2335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harr JC, Luperchio TR, Wong X, Cohen E, Wheelan SJ, Reddy KL (2015) Directed targeting of chromatin to the nuclear lamina is mediated by chromatin state and A-type lamins. J Cell Biol 208: 33–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harr JC, Gonzalez-Sandoval A, Gasser SM (2016) Histones and histone modifications in perinuclear chromatin anchoring: From yeast to man. EMBO Rep 17: 139–155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heberle H, Meirelles GV, da Silva FR, Telles GP, Minghim R (2015) InteractiVenn: A web-based tool for the analysis of sets through Venn diagrams. BMC Bioinformatics 16: 169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchinson J. (1886) Congenital absence of hair and mammary glands with atrophic condition of the skin and its appendages, in a boy whose mother had been almost wholly bald from alopecia areata from the age of six. Med Chir Trans 69: 473–477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikegami K, Egelhofer TA, Strome S, Lieb JD (2010) Caenorhabditis elegans chromosome arms are anchored to the nuclear membrane via discontinuous association with LEM-2. Genome Biol 11: R120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Y, Tam OH, Paniagua E, Hammell M (2015) TEtranscripts: A package for including transposable elements in differential expression analysis of RNA-seq datasets. Bioinformatics 31: 3593–3599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL (2013) TopHat2: Accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol 14: R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kühlbrandt W. (2015) Structure and function of mitochondrial membrane protein complexes. BMC Biol 13: 89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL (2009) Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombardi ML, Lammerding J (2011) Keeping the LINC: The importance of nucleocytoskeletal coupling in intracellular force transmission and cellular function. Biochem Soc Trans 39: 1729–1734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S (2014) Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15: 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuda K, Takahashi S, Nomura K, Arimoto M, Inoue M (1993) Residual structure and constituent proteins of the peripheral framework of the cell-nucleus in somatic embryos from Daucus carota L. Planta 191: 532–540 [Google Scholar]
- Masuda K, Xu ZJ, Takahashi S, Ito A, Ono M, Nomura K, Inoue M (1997) Peripheral framework of carrot cell nucleus contains a novel protein predicted to exhibit a long alpha-helical domain. Exp Cell Res 232: 173–181 [DOI] [PubMed] [Google Scholar]
- Meaburn KJ, Cabuy E, Bonne G, Levy N, Morris GE, Novelli G, Kill IR, Bridger JM (2007) Primary laminopathy fibroblasts display altered genome organization and apoptosis. Aging Cell 6: 139–153 [DOI] [PubMed] [Google Scholar]
- Meier I. (2016) LINCing the eukaryotic tree of life: Towards a broad evolutionary comparison of nucleocytoplasmic bridging complexes. J Cell Sci 129: 3523–3531 [DOI] [PubMed] [Google Scholar]
- Meier I, Griffis AH, Groves NR, Wagner A (2016) Regulation of nuclear shape and size in plants. Curr Opin Cell Biol 40: 114–123 [DOI] [PubMed] [Google Scholar]
- Meier I, Richards EJ, Evans DE (2017) Cell biology of the plant nucleus. Annu Rev Plant Biol 68: 139–172 [DOI] [PubMed] [Google Scholar]
- Mi H, Huang X, Muruganujan A, Tang H, Mills C, Kang D, Thomas PD (2017) PANTHER version 11: Expanded annotation data from Gene Ontology and Reactome pathways, and data analysis tool enhancements. Nucleic Acids Res 45: D183–D189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikulski P, Hohenstatt ML, Farrona S, Smaczniak C, Kaufmann K, Angenent G, Schubert D (2017) PWWP INTERACTOR OF POLYCOMBS (PWO1) links PcG-mediated gene repression to the nuclear lamina in Arabidopsis. bioRxiv, doi:10.1101/220541 [Google Scholar]
- Moeder W, Yoshioka K (2008) Lesion mimic mutants: A classical, yet still fundamental approach to study programmed cell death. Plant Signal Behav 3: 764–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawrath C, Métraux JP (1999) Salicylic acid induction-deficient mutants of Arabidopsis express PR-2 and PR-5 and accumulate high levels of camalexin after pathogen inoculation. Plant Cell 11: 1393–1404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peric-Hupkes D, Meuleman W, Pagie L, Bruggeman SW, Solovei I, Brugman W, Gräf S, Flicek P, Kerkhoven RM, van Lohuizen M, et al. (2010) Molecular maps of the reorganization of genome-nuclear lamina interactions during differentiation. Mol Cell 38: 603–613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickersgill H, Kalverda B, de Wit E, Talhout W, Fornerod M, van Steensel B (2006) Characterization of the Drosophila melanogaster genome at the nuclear lamina. Nat Genet 38: 1005–1014 [DOI] [PubMed] [Google Scholar]
- Poulet A, Duc C, Voisin M, Desset S, Tutois S, Vanrobays E, Benoit M, Evans DE, Probst AV, Tatout C (2017) The LINC complex contributes to heterochromatin organisation and transcriptional gene silencing in plants. J Cell Sci 130: 590–601 [DOI] [PubMed] [Google Scholar]
- Reddy KL, Zullo JM, Bertolino E, Singh H (2008) Transcriptional repression mediated by repositioning of genes to the nuclear lamina. Nature 452: 243–247 [DOI] [PubMed] [Google Scholar]
- Rivas-San Vicente M, Plasencia J (2011) Salicylic acid beyond defence: Its role in plant growth and development. J Exp Bot 62: 3321–3338 [DOI] [PubMed] [Google Scholar]
- Robert-Seilaniantz A, Grant M, Jones JDG (2011) Hormone crosstalk in plant disease and defense: More than just jasmonate-salicylate antagonism. Annu Rev Phytopathol 49: 317–343 [DOI] [PubMed] [Google Scholar]
- Sakamoto Y, Takagi S (2013) LITTLE NUCLEI 1 and 4 regulate nuclear morphology in Arabidopsis thaliana. Plant Cell Physiol 54: 622–633 [DOI] [PubMed] [Google Scholar]
- Scaffidi P, Misteli T (2006) Lamin A-dependent nuclear defects in human aging. Science 312: 1059–1063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber KH, Kennedy BK (2013) When lamins go bad: Nuclear structure and disease. Cell 152: 1365–1375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sequeira-Mendes J, Aragüez I, Peiró R, Mendez-Giraldez R, Zhang X, Jacobsen SE, Bastolla U, Gutierrez C (2014) The functional topography of the Arabidopsis genome is organized in a reduced number of linear motifs of chromatin states. Plant Cell 26: 2351–2366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seyfferth C, Tsuda K (2014) Salicylic acid signal transduction: The initiation of biosynthesis, perception and transcriptional reprogramming. Front Plant Sci 5: 697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stingl N, Krischke M, Fekete A, Mueller MJ (2013) Analysis of defense signals in Arabidopsis thaliana leaves by ultra-performance liquid chromatography/tandem mass spectrometry: Jasmonates, salicylic acid, abscisic acid. Methods Mol Biol 1009: 103–113 [DOI] [PubMed] [Google Scholar]
- Stroud H, Greenberg MV, Feng S, Bernatavichute YV, Jacobsen SE (2013) Comprehensive analysis of silencing mutants reveals complex regulation of the Arabidopsis methylome. Cell 152: 352–364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Fukao Y, Hatsugai N, Katagiri F, Hara-Nishimura I (2017) Nup82 functions redundantly with Nup136 in a salicylic acid-dependent defense response of Arabidopsis thaliana. Nucleus 8: 301–311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tapley EC, Starr DA (2013) Connecting the nucleus to the cytoskeleton by SUN-KASH bridges across the nuclear envelope. Curr Opin Cell Biol 25: 57–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatout C, Evans DE, Vanrobays E, Probst AV, Graumann K (2014) The plant LINC complex at the nuclear envelope. Chromosome Res 22: 241–252 [DOI] [PubMed] [Google Scholar]
- Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L (2010) Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol 28: 511–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L (2012) Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc 7: 562–578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Steensel B, Belmont AS (2017) Lamina-associated domains: Links with chromosome architecture, heterochromatin, and gene repression. Cell 169: 780–791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Wees S. (2008) Phenotypic analysis of Arabidopsis mutants: Trypan blue stain for fungi, oomycetes, and dead plant cells. CSH Protoc 2008: pdb prot4982. [DOI] [PubMed] [Google Scholar]
- Vlot AC, Dempsey DA, Klessig DF (2009) Salicylic acid, a multifaceted hormone to combat disease. Annu Rev Phytopathol 47: 177–206 [DOI] [PubMed] [Google Scholar]
- Wang H, Dittmer TA, Richards EJ (2013) Arabidopsis CROWDED NUCLEI (CRWN) proteins are required for nuclear size control and heterochromatin organization. BMC Plant Biol 13: 200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Liu S, Lu C, La Y, Dai J, Ma H, Zhou S, Tan F, Wang X, Wu Y, et al. (2019) Roles of CRWN-family proteins in protecting genomic DNA against oxidative damage. J Plant Physiol 233: 20–30 [DOI] [PubMed] [Google Scholar]
- Wildermuth MC, Dewdney J, Wu G, Ausubel FM (2001) Isochorismate synthase is required to synthesize salicylic acid for plant defence. Nature 414: 562–565 [DOI] [PubMed] [Google Scholar]
- Yamaguchi N, Winter CM, Wu MF, Kwon CS, William DA, Wagner D (2014) PROTOCOLS: Chromatin immunoprecipitation from Arabidopsis tissues. The Arabidopsis Book 12: e0170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Clarenz O, Cokus S, Bernatavichute YV, Pellegrini M, Goodrich J, Jacobsen SE (2007) Whole-genome analysis of histone H3 lysine 27 trimethylation in Arabidopsis. PLoS Biol 5: e129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng XY, Zhou M, Yoo H, Pruneda-Paz JL, Spivey NW, Kay SA, Dong X (2015) Spatial and temporal regulation of biosynthesis of the plant immune signal salicylic acid. Proc Natl Acad Sci USA 112: 9166–9173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X, Hu J, Yue S, Kristiani L, Kim M, Sauria M, Taylor J, Kim Y, Zheng Y (2018) Lamins organize the global three-dimensional genome from the nuclear periphery. Mol Cell 71: 802–815.e7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou M, Wang W, Karapetyan S, Mwimba M, Marqués J, Buchler NE, Dong X (2015) Redox rhythm reinforces the circadian clock to gate immune response. Nature 523: 472–476 [DOI] [PMC free article] [PubMed] [Google Scholar]