PP2-A5 is an Arabidopsis TIR-lectin two-domain protein that confers defense properties against Tetranychus urticae by affecting gene expression and hormonal signaling.
Abstract
Plant immunity depends on fast and specific transcriptional reprogramming triggered by the perception of biotic stresses. Numerous studies have been conducted to better understand the response of plants to the generalist herbivore two-spotted spider mite (Tetranychus urticae). However, how plants perceive mites and how this perception is translated into changes in gene expression are largely unknown. In this work, we identified a gene induced in Arabidopsis (Arabidopsis thaliana) upon spider mite attack that encodes a two-domain protein containing predicted lectin and Toll/Interleukin-1 receptor domains. The gene, previously named PP2-A5, belongs to the Phloem Protein2 family. Biotic assays showed that PP2-A5 confers tolerance to T. urticae. Overexpression or knockout of PP2-A5 leads to transcriptional reprogramming that alters the balance of hormone accumulation and corresponding signaling pathways. The nucleocytoplasmic location of this protein supports a direct interaction with regulators of gene transcription, suggesting that the combination of two putative signaling domains in a single protein may provide a novel mechanism for regulating gene expression. Together, our results suggest that PP2-A5 improves the ability to defend against T. urticae by participating in the tight regulation of hormonal cross talk upon mite feeding. Further research is needed to determine the mechanism by which this two-domain protein functions and to clarify its molecular role in signaling following a spider mite attack.
Plants and phytophagous pests coexist as foes sharing more than 100 million years of evolution. Among plant feeders, spider mites (Acari, Tetranychidae) represent the most important family of phytophagous mites, with over 1,000 host species (Vacante, 2016). In particular, the two-spotted spider mite (Tetranychus urticae) is an extremely polyphagous species found worldwide that feeds on nearly 1,100 documented host plants, including about 150 agronomically important crops (Migeon and Dorkeld, 2018). It is considered one of the most significant agricultural threats, since it has a short life cycle, high offspring production, and an extraordinary ability to develop pesticide resistance (Van Leeuwen and Dermauw, 2016; Rioja et al., 2017; Agut et al., 2018). Under increased temperatures and drought stress associated with climate change, T. urticae shortens its life cycle, produces more generations per year, and appears earlier in the season and on a wider range of hosts (Ximénez-Embún et al., 2017). Spider mites pierce individual mesophyll parenchymatic cells using stylets to suck their nutrients without damaging epidermal cells and produce severe chlorosis and the consequent reduction in crop yield (Park and Lee, 2002; Bensoussan et al., 2016). Mite feeding causes cellular degradation and cytoplasmic leakage, releasing compounds to the apoplast. The documented salivary secretions and the deposited enzymatically active feces on the leaf surface provide a battery of putative elicitors (damage/herbivore-associated molecular patterns) to induce specific plant defenses (Santamaría et al., 2015; Jonckheere et al., 2016, 2018; Villarroel et al., 2016). In addition, microbe-associated molecular patters released with mite gut-associated endosymbionts exert additional effects on host-plant defenses, and eggs and silk balls may act as an additional source of elicitors/effectors (Rioja et al., 2017; Staudacher et al., 2017; Santamaria et al., 2018a). Plants recognize these signals and trigger a cascade of short-term responses that eventually result in the production of molecules with defense properties (Santamaria et al., 2018a). Previous findings indicate that T. urticae infestation leads to changes in cytosolic Ca2+ influx, protein phosphorylation, and the generation of reactive oxygen species (ROS), followed by the induction of a conserved set of genes associated with jasmonic acid (JA), salicylic acid (SA), and ethylene (ET) biosynthesis (Rioja et al., 2017; Agut et al., 2018). These early events regulate the expression of direct defenses and the emission of volatiles to attract natural enemies, mainly phytoseiids (Arimura et al., 2002; Ament et al., 2004; Schweighofer et al., 2007; Agut et al., 2015, 2018; Santamaría et al., 2017, 2018a).
In recent years, T. urticae has become a model within chelicerate herbivores, with its genome sequenced and a broad range of tools and protocols developed (Grbić et al., 2011; Santamaría et al., 2012b; Cazaux et al., 2014; Suzuki et al., 2017a, 2017b). Functional studies performed with T. urticae-Arabidopsis (Arabidopsis thaliana) as a model plant-herbivore system have provided valuable information on their interaction and the specific responses from both the plant and the arthropod side. Zhurov et al. (2014) analyzed the natural genetic variation in resistance to T. urticae among 26 Arabidopsis accessions, identifying Bla-2 and Kondara (Kon) as accessions at the opposing ends of the susceptibility spectrum. Transcriptome analysis highlighted the importance of the induction of the JA biosynthesis pathway and JA-regulated accumulation of indole glucosinolates (IGs) in the reciprocal plant-mite interaction. Concomitant with these plant responses was the overexpression of mite genes involved in the detoxification of xenobiotics. Schweighofer et al. (2007) demonstrated that the Arabidopsis phosphatase AP2C1, a negative regulator of some mitogen-activated protein kinases, modulated JA and ET levels and participated in the innate immunity to pests and pathogens, since ap2c1 mutants displayed enhanced resistance to spider mites. Recently, several genes/proteins have been described as key players of Arabidopsis defense against T. urticae. Santamaría et al. (2017) characterized a new protein termed MATI (Mite Attack Triggered Immunity) involved in the Arabidopsis response to mite feeding that modulates JA and SA levels and participates in redox homeostasis to avoid oxidative damage and cell death. Likewise, the ABSCISIC ACID INSENSITIVE4 transcription factor has been defined as a crucial component of chloroplast retrograde signaling that regulates Arabidopsis response to spider mite infestation (Barczak-Brzyżek et al., 2017). In addition, the relationship between ROS-metabolizing systems and mite-triggered Arabidopsis-induced responses has been studied. Four ROS-related genes encoding proteins putatively involved in the hydrogen peroxide balance and in the degradation of ascorbate showed effects on ROS metabolism and JA and SA signaling pathways and, consequently, on plant defenses against spider mites (Santamaría et al., 2018b). The participation of two Kunitz trypsin inhibitors (KTI), AtKTI4 and AtKTI5, in Arabidopsis defense has also been demonstrated. T. urticae inflicted more leaf damage in kti4 and kti5 mutant lines than in wild-type plants, and in parallel, the mite performance was improved after feeding on these mutants (Arnaiz et al., 2018). Probably, the inhibition of the proteolysis mediated by AtKTIs decreased mite access to essential amino acids and, consequently, disrupted important mite physiological events.
Plant responses to spider mite infestation have also been studied in crops, since mite populations do not perform equally on all potential hosts (Agrawal et al., 2002). The battery of plant defenses, either constitutive or inducible, depends on the plant species. Using different omics approaches, comparative analyses performed in mite-infested versus noninfested leaves of tomato (Solanum lycopersicum), lima bean (Phaseolus lunatus), and Citrus and Vitis spp. have increased our understanding of signaling networks leading to physiological alterations affected by T. urticae (Arimura et al., 2000; Maserti et al., 2011; Agut et al., 2015; Martel et al., 2015; Díaz-Riquelme et al., 2016). Comparison of defense responses between tomato and Arabidopsis revealed that JA and secondary metabolites such as phenylpropanoids, flavonoids, and terpenoids represented a conserved core of mite-induced defenses. Species-specific differences in some JA-regulated downstream responses were also detected, highlighting the relevant role of IGs in Arabidopsis and protease inhibitors in tomato (Martel et al., 2015). Moreover, comparisons of the transcriptome or proteome of maize (Zea mays) and barley (Hordeum vulgare) leaves upon the combination of T. urticae infestation and water limitation illustrated the complexity of biotic-abiotic cross talk under variable environmental conditions and emphasized the difficulties of predicting consequences on crop production (Dworak et al., 2016; Santamaria et al., 2018c). Other mechanisms of plant defense are related to physical/chemical barriers. High leaf trichome density in raspberry (Rubus iadaeus) impaired the movement of spider mites and had deterrent effects on egg deposition (Karley et al., 2016). In addition, the presence of phenolic compounds in Chrysanthemum × morifolium, flavonoids in Citrus spp., and terpenoids in cucumber (Cucumis sativus) and Citrus spp. correlated with cultivar resistance to T. urticae (Kielkiewicz and van de Vrie, 1990; Balkema-Boomstra et al., 2003; Agut et al., 2014, 2015; Kos et al., 2014).
Spider mites have the ability to manipulate plant defenses (Glas et al., 2014; Alba et al., 2015; Wybouw et al., 2015). T. urticae salivary peptides identified by Villarroel et al. (2016) suppressed Nicotiana benthamiana defenses downstream of SA and promoted the mite’s performance. The suppression of defenses not only benefits herbivore communities, but it can also generate ecological costs when mite performance is promoted (Sarmento et al., 2011; Ataide et al., 2016). In spite of the wide knowledge of the plant-mite interaction, gaps still exist concerning spider mite signal perception. The identification of plant receptors, together with other new elements and regulators involved in the plant cascade of defenses, is needed for a more complete view of the plant-spider mite interaction.
In this work, we characterized the Arabidopsis AT1G65390 (PP2-A5) gene, which consistently showed higher expression levels in the resistant Bla-2 accession relative to the susceptible Kon accession after mite feeding. We demonstrate that this gene encodes a protein contributing to plant protection against spider mites through reprogramming of gene expression related to defense responses.
RESULTS
PP2-A5, a Gene Putatively Involved in Arabidopsis Defense against Spider Mites
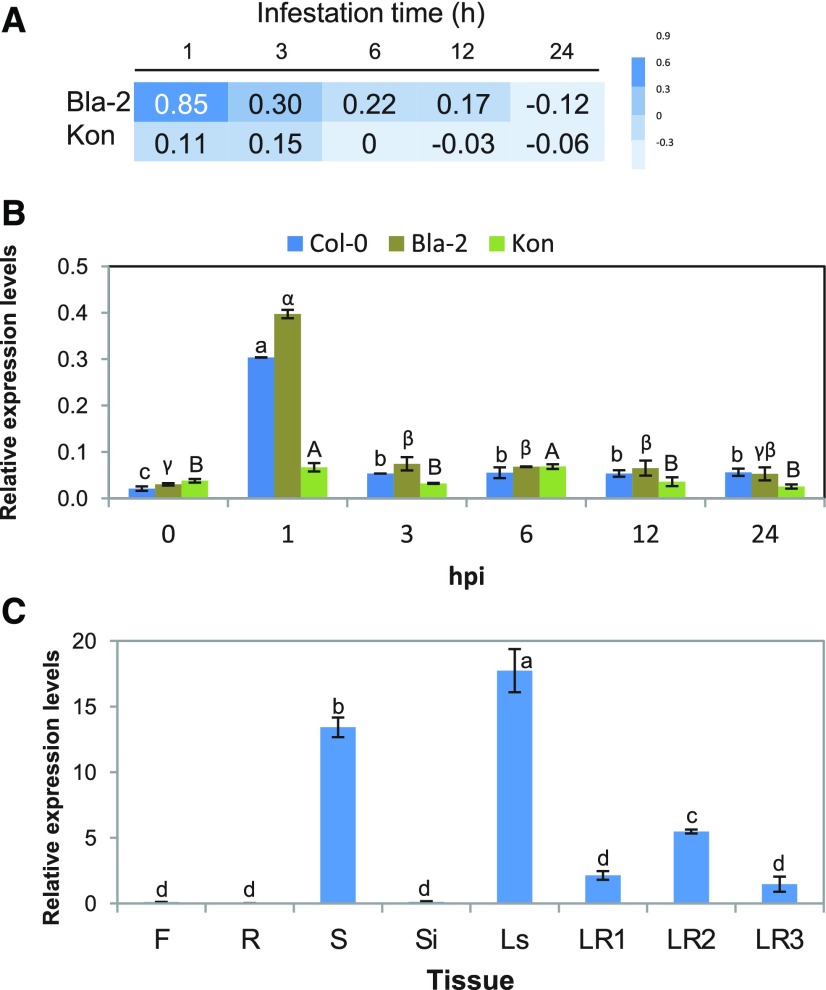
Zhurov et al. (2014) analyzed the transcriptional response to spider mite infestation in the resistant Arabidopsis accession Bla-2 and the susceptible accession Kon, found in Blanes, in northeast Spain, and in Tadjikistan, respectively. Although mites caused 20-fold more damage to Kon than to Bla-2, the mite-induced transcriptional responses of both accessions were very similar, leading to the hypothesis that constitutive differences between them shaped their differential response to mite herbivory. Among a handful of differentially expressed genes (DEGs) between both accessions, AT1G65390, termed PP2-A5, was one of the genes showing the highest level of induction in the resistant Bla-2 accession (7.43-fold change after 1 h of mite feeding) relative to Kon (Fig. 1A). This gene was selected to investigate its potential role in the Arabidopsis biotic stress response. Two isoforms for PP2-A5 had been identified (AT1G65390.1 and AT1G65390.2), and gene expression studies did not detect expression of AT1G65390.2 before or after T. urticae infestation (Supplemental Fig. S1). Thus, the AT1G65390.1 isoform was selected for further studies. Reverse transcription quantitative PCR (RT-qPCR) assays confirmed the previous differential induction in the expression of PP2-A5 between Bla-2 and Kon observed in microarray data (Fig. 1A) and highlighted the high induction of this gene in Col-0 upon spider mite attack (Fig. 1B). The expression of PP2-A5 in different tissues of Col-0 Arabidopsis plants was also determined by RT-qPCR. PP2-A5 mRNA levels were mainly detected in seeds, and in leaves from stems or rosettes at different developmental stages, while it was scarcely detectable in roots, flowers, or siliques (Fig. 1C).
Figure 1.
PP2-A5 (AT1G65390) gene expression in Arabidopsis Bla-2, Kon, and Columbia-0 (Col-0) accessions in response to T. urticae infestation and in different Col-0 tissues. A, Microarray data from the PP2-A5 expression in Arabidopsis Bla-2 and Kon accessions at 1, 3, 6, 12, and 24 h postinfestation represented as log2. B, Microarray validation by RT-qPCR assays of PP2-A5 gene expression at 1, 3, 6, 12, and 24 h postinfestation. Gene expression is referred to as relative expression levels (2^-dCt). C, Relative gene expression levels (2^-dCt) of PP2-A5 in different Arabidopsis Col-0 tissues: flowers (F), roots (R), seeds (S), siliques (Si), leaves from stem (Ls), and leaves from rosettes at 1, 2, and 3 weeks old (LR1, LR2, and LR3). For B and C, data are means ± se of three biological replicates. Different letters indicate significant differences between times postinfestation within each accession or among different tissues (P < 0.05, one-way ANOVA followed by Tukey’s multiple comparisons test).
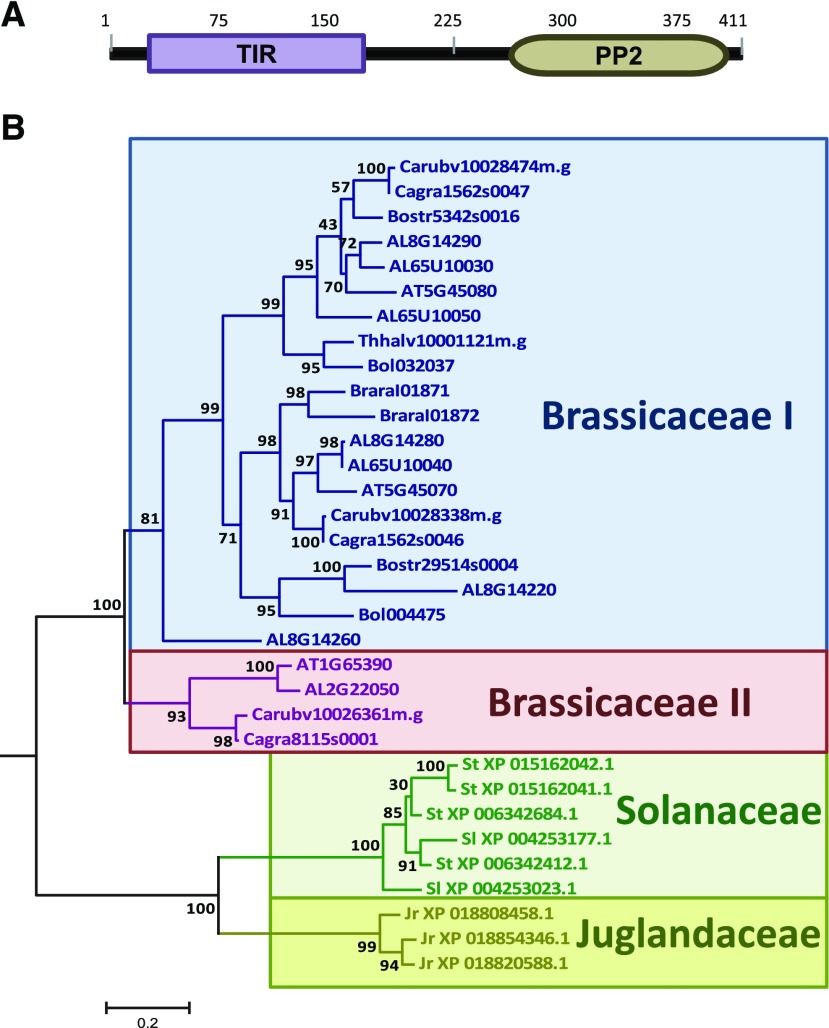
PP2-A5 encodes a predicted protein of 411 amino acids with no signal peptide. This protein belongs to the 30-member Phloem Protein2 (PP2) family in Arabidopsis, which share a conserved PP2-like domain (Dinant et al., 2003). Two domains were predicted in PP2-A5, a Toll/Interleukin-1 receptor (TIR) domain located in the N-terminal region and a PP2 domain in the C-terminal part of the protein (Fig. 2A). Searches in the Phytozome database only identified homologs sharing the two-domain structure of the PP2-A5 protein in Brassicaceae and Solanaceae families. In a wider search using the National Center for Biotechnology Information nonredundant database, proteins with a predicted TIR-PP2 structure were also found in the species Juglans regia, which belongs to the Juglandaceae family. A phylogenetic tree was constructed using protein sequences of PP2-A5 homologs from different species (Fig. 2B). Whereas Solanaceae and Juglandaceae members shared a common ancestor that evolved specifically in each family, Brassicaceae members were located on a different branch of the tree. Among Brassicaceae, two major groups were detected, with PP2-A5 as the only member of Arabidopsis in the smaller Brassicaceae II group. Although searches in the Zhurov et al. (2014) data sets showed induction by mites for some genes encoding proteins from the PP2 family with an additional F-box domain (PP2-A11, PP2-A14, PP2-B6, and PP2-B11), no induction was found for the two members of the Brassicaceae I group that share the PP2-A5 two-domain TIR-PP2 structure, At5g45070 and At5g45080 (PP2-A6 and PP2-A8).
Figure 2.
Evolutionary and sequence features of PP2-A5 protein. A, Predicted positions of protein domains. B, Phylogram of PP2-A5 homologs. The phylogenetic tree was constructed by the PhyML method using the protein sequences of PP2-A5 homologs. Numbers are approximate likelihood-ratio test values for statistical support. AL, Arabidopsis lyrata; AT, Arabidopsis thaliana; Bol, Brassica oleracea; Bostr, Boechera stricta; Brara, Brassica rapa; Cagra, Capsella grandiflora; Carubv, Capsella rubella; Jr, Juglans regia; Sl, Solanum lycopersicum; St, Solanum tuberosum; Thhalv, Eutrema salsugineum.
Subcellular Localization of PP2-A5
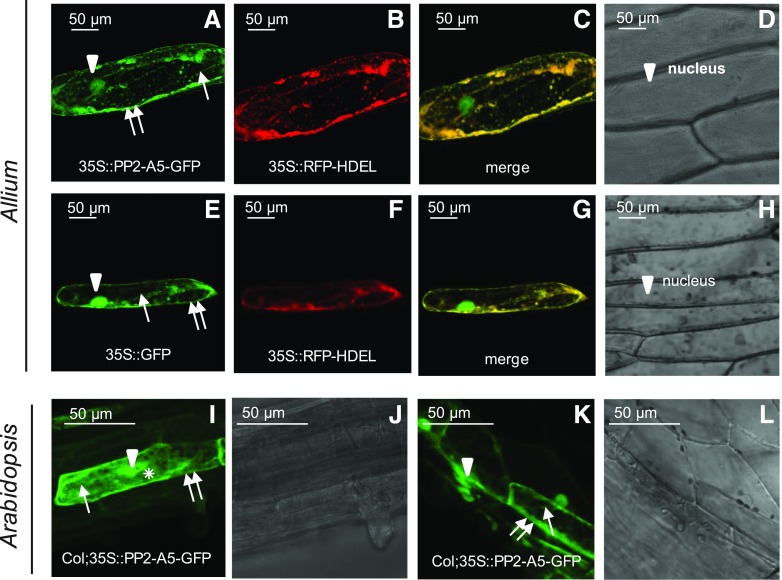
Transient expression assays were conducted to determine the subcellular localization of PP2-A5 using particle bombardment in onion (Allium cepa) epidermal cells and agroinfiltration in N. benthamiana leaves. The protein product corresponding to the open reading frame of PP2-A5 translationally fused to green fluorescent protein (GFP) was detected throughout the entire endoplasmic reticulum (ER) network, which was continuous with the nuclear membrane. This subcellular location was revealed by its colocation with the fluorescence emitted by using the red fluorescent protein (RFP) tagged with the C-terminal ER retrieval motif His-Asp-Glu-Leu (HDEL) construct (35S-RFP-HDEL) as a control for localization in the endomembrane system. The PP2-A5 protein was also detected in the plasma membrane and within the nucleus and in the cytoplasm. Plasmolysis, induced by 1 m mannitol treatment, confirmed the presence of PP2-A5 in the cell-detached plasma membrane, with no fluorescence observed either in the cell wall or in the apoplast (Fig. 3, A–D). As expected, fluorescence was found through the whole cell when the 35S-GFP construct was bombarded into onion cells (Fig. 3, E–H). N. benthamiana agroinfiltration experiments corroborated a similar localization pattern of the fused PP2-A5-GFP protein (nucleus, endomembrane system, and cytoplasm) and of GFP alone used as a control (Supplemental Fig. S2, A–D and I–L). Additionally, the chloroplastic localization suggested by the TAIR database was not observed, as is shown in Supplemental Figure S2, E to H. The chloroplast autofluorescence did not merge with the fluorescence emitted by the GFP detected in the endomembrane system. Furthermore, PP2-A5-GFP-overexpressing lines were created in the Col-0 background to validate the PP2-A5 localization in the whole plant. The stable expression of the 35S-PP2-A5-GFP plasmid in transgenic Col-0 plants confirmed the protein localization in the endomembrane system, nucleus, and cytoplasm of leaf cells. In addition, PP2-A5 was located in the cytosol, as shown in roots and hypocotyls of Arabidopsis transgenic plants (Fig. 3, I–L).
Figure 3.
PP2-A5 subcellular localization. Confocal stacks spanning epidermal onion cells cotransformed with 35S-PP2-A5-GFP and 35S-RFP-HDEL control or 35S-GFP control with 35S-RFP-HDEL control are shown. A to H, Confocal images and projections of the PP2-A5 localization after plasmolysis with 1 m mannitol. Projections are from GFP (A and E), RFP (B and F), merged (C and G), and the corresponding Nomarski snapshots (D and H). I and K, Subcellular localization of PP2-A5 protein in roots (I) and hypocotyl (K) of transgenic Col-PP2-A5-GFP lines. J and L, Observations with Nomarski optics. Arrowheads indicate nucleus location, one arrow indicates ER location, two arrows indicate cytoplasmic membrane location, and the asterisk indicates cytoplasm location. Scale bars are as indicated.
Effects of PP2-A5 on Plant Resistance and Pest Performance
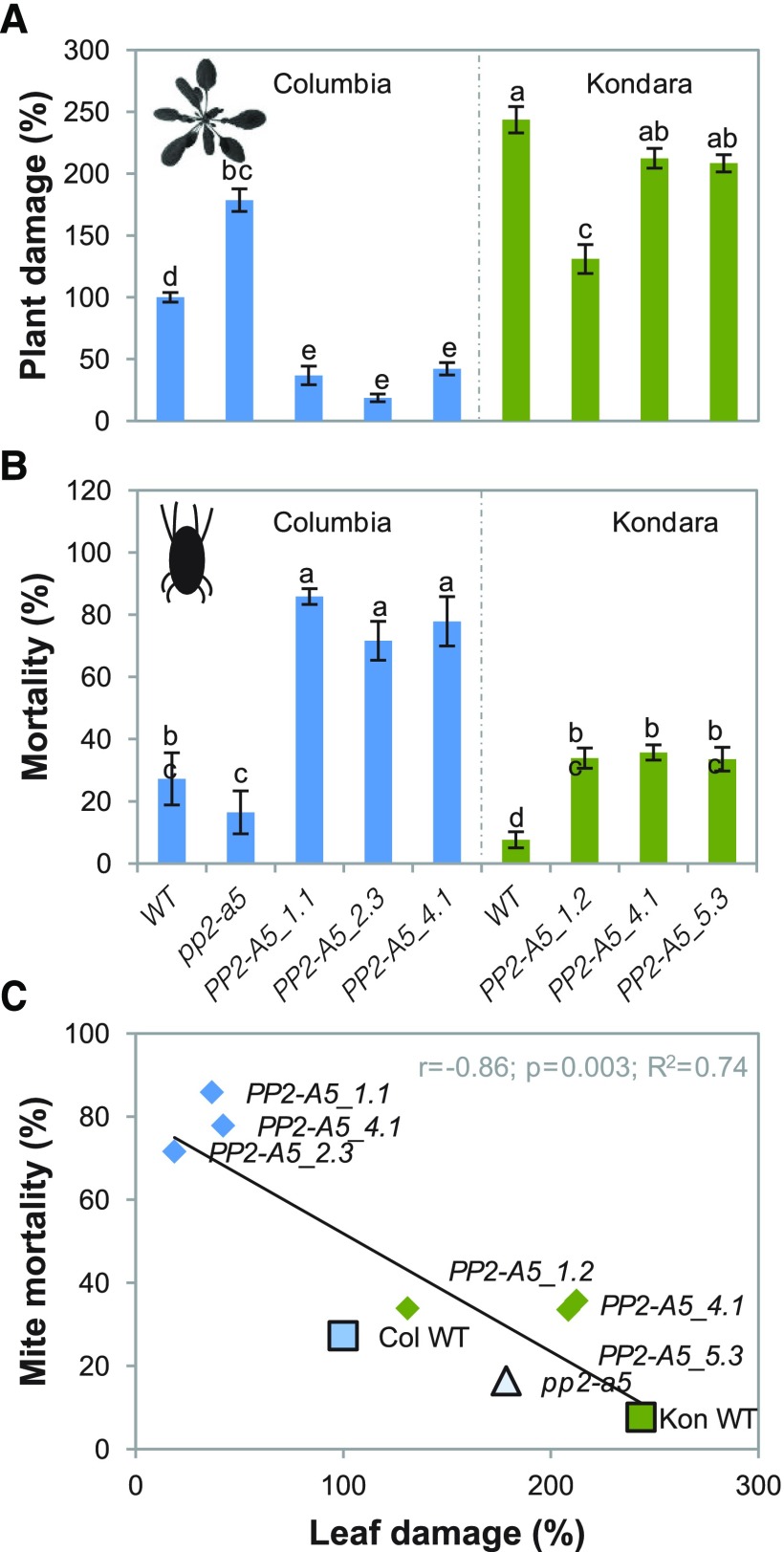
To investigate the role of PP2-A5 in plant defense against mites, PP2-A5-overexpressing Arabidopsis Col-0 and Kon lines (Col-PP2-A5 and Kon-PP2-A5, respectively) were generated, and a Salk line (pp2-a5) was ordered. The characterization of the homozygous pp2-a5 Salk line revealed a loss-of-function allele generated by the insertion of a transfer DNA (T-DNA) in the first exon (Supplemental Fig. S3). The gene expression analysis of transgenic plants constitutively expressing PP2-A5 in Col-0 and Kon backgrounds allowed the selection of overexpressing lines for further studies (Supplemental Fig. S3). No differences in phenotype along the lifespan among the three genotypes were observed. To test the involvement of PP2-A5 in plant responses to mite herbivory, homozygous T-DNA insertion mutants (Col-pp2-a5), overexpressing lines (Col-PP2-A5_1.1, _2.3, and _4.1 and Kon-PP2-A5_1.2, _4.2, and _5.3), as well as the corresponding wild-type plants were infested with spider mites, and plant damage was quantified 4 d after mite feeding. Col-PP2-A5-overexpressing lines showed less damage than nontransformed Col-0 wild-type infested plants. Similarly, overexpression of PP2-A5 in Kon (Kon-PP2-A5_1.2, _4.1, and _5.3 lines) resulted in a decrease in the damage in comparison with infested Kon wild-type plants. In contrast, the damaged area in mutant Col-pp2a5 lines was about 2-fold greater than the damage quantified in Col-0 wild-type plants after mite feeding (Fig. 4A). The damage intensity measured as chlorotic area on infested leaves correlated with PP2-A5 gene expression levels in the Col-0 background (Supplemental Fig. S3B). To ensure that the chlorotic area associates with enhanced mite feeding and is not a consequence of a greater cell death, mite performance was determined after feeding on PP2-A5-overexpressing, mutant, and wild-type plants. Mite mortality assays were carried out after feeding on leaves from different Col-0 and Kon genotypes. Results showed that mites that fed on pp2-a5 and wild-type plants had lower mortality rates than the ones that fed on PP2-A5-overexpressing lines (Fig. 4B). Thus, greater leaf damage reflected higher mite survival, indicating that PP2-A5 overexpression conferred resistance to mite herbivory. Figure 4C shows the negative correlation between the mite mortality and the leaf damage quantified in the leaves after spider mite attack. In parallel, developmental rates of mites feeding on the different Col-0 and Kon genotypes were analyzed. Mites feeding on PP2-A5-overexpressing plants tended to show slower developmental rates than mites feeding on control plants, independent of the genetic background, although the differences were not statistically significant. The opposite trend was observed when mites fed on pp2-a5 plants (Supplemental Table S1).
Figure 4.
Plant damage of Col-0 and Kon genotypes infested with T. urticae and mite performance. A, Leaf damage on the Arabidopsis mutant Col-0 Salk line (pp2a5) as well as Col-PP2-A5 (PP2-A5_1.1, PP2-A5-2.3, and PP2-A5_4.1) and Kon-PP2-A5 (PP2-A5_1.2, PP2-A5-4.1, and PP2-A5_5.3) overexpressing lines and Col-0 and Kon wild type (WT), relative to nontransformed Col-0 wild type, 4 d after infestation. Data are means ± se of six independent plants. B, Effects of silencing and overexpression of PP2-A5 in Arabidopsis Col-0 or Kon plants on mite mortality. Data are means ± se of eight independent plants. For A and B, different letters indicate significant differences (P < 0.05, one-way ANOVA followed by Tukey’s test). C, Correlation among mite mortality and leaf damage (P < 0.05, Pearson product moment).
DEGs in Col-PP2A5 or Col-pp2a5 Compared with Col-0 Wild-Type Plants
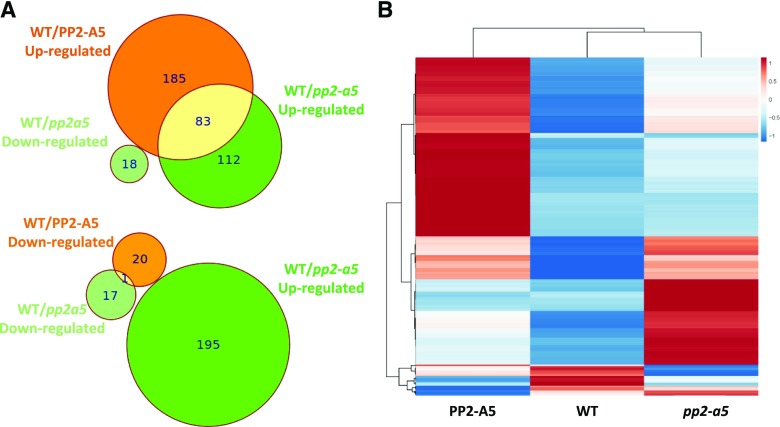
The defense effect of PP2-A5 could result from a direct toxicity of the PP2 domain (lectin activity) or from an indirect effect caused by both the PP2 and TIR domains. To identify changes in gene expression associated with variations in PP2-A5 expression levels and to investigate its effects on the plant’s defense response, an RNA sequencing analysis was performed in 3-week-old plantlets of the three PP2-A5 Col-0 genotypes, PP2-A5_2.3, pp2-a5, and the wild type. DEGs were detected in the wild type/PP2-A5 (289) and in the wild type/pp2-a5 (213) pairwise comparisons (Supplemental Data Set S1). Many more genes were up-regulated than down-regulated in both modified genotypes when compared with the Col-0 wild type. Figure 5A shows the overlap in the DEGs performed by wild type/PP2-A5 and wild type/pp2-a5 pairwise comparisons. The Euler diagrams showed that in addition to the 185 and 112 genes uniquely up-regulated in Col-PP2-A5 and pp2-a5 plants, respectively, 83 up-regulated genes were shared between wild type/PP2-A5 and wild type/pp2-a5 pairwise comparisons. On the contrary, only one gene showed reduced expression in both genotypes. Strikingly, any DEG that was significantly up-regulated in one modified genotype was down-regulated in the other when compared with the wild type. This pattern is reflected in the heat map that shows the relative expression of all DEGs found in the wild type/PP2-A5 and wild type/pp2-a5 comparisons (Fig. 5B), suggesting transcriptional reprogramming of different gene modules when PP2-A5 is overexpressed or knocked out.
Figure 5.
Transcriptional analysis of Arabidopsis plants with different PP2-A5 genotypes. A, Euler diagrams showing the number of induced or repressed DEGs between Col-PP2-A5 or pp2-a5 and the Col-0 wild type (WT). B, Heat map showing normalized color intensity from the expression values of the 334 DEGs found in the Col-PP2-A5 or pp2-a5 and Col-0 wild-type comparisons.
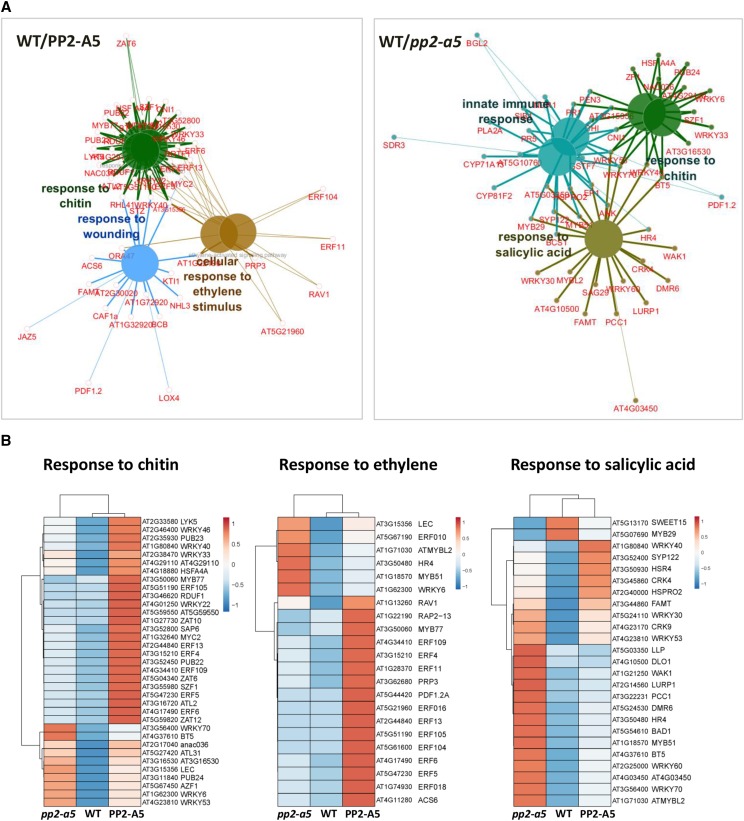
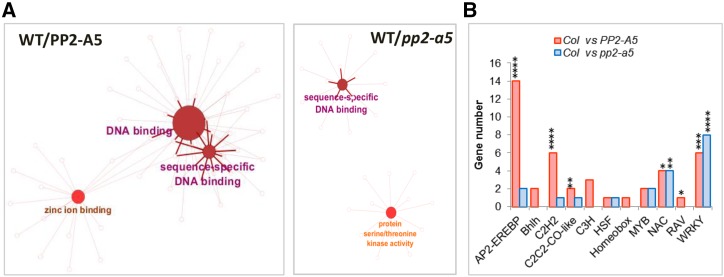
Enrichment Gene Ontology (GO) analyses were performed to identify the biological processes characterized by the DEGs (Fig. 6A; Supplemental Table S2). The number of genes related to the response to chitin was enriched when PP2-A5 was overexpressed or knocked out. However, whereas Col-PP2-A5 plants showed a basal induction of genes related to wounding and ET signaling, pp2-a5 plants had a basal induction of genes related to immune response and SA signaling. A heat map was generated to visualize the relative expression of DEGs included in the response to the chitin GO category. Most of them showed higher expression in the transgenic lines than in the Col-0 wild type (Fig. 6B). Heat maps of the DEGs associated with ET and SA pathways confirmed their differential induction in the Col-PP2-A5 and pp2-a5 genotypes (Fig. 6B). In addition, enrichment GO analyses were performed to reveal the molecular functions overrepresented in DEGs (Fig. 7A; Supplemental Table S3). As expected from the high number of differentially expressed transcription factors, the sequence-specific DNA binding GO term was enriched when PP2-A5 was overexpressed or knocked out. Interestingly, Col-PP2-A5 plants showed a basal induction of genes that encode zinc-binding proteins and pp2-a5 plants had a basal enrichment in the expression of genes encoding protein kinases. These results also point to a modulation of regulatory pathways caused by modified PP2-A5 expression.
Figure 6.
Expression analysis of DEGs from enriched biological processes. A, Functionally grouped network of biological process enriched categories after ClueGO analysis of DEGs between wild type (WT)/PP2-A5 or wild type/pp2-a5. Significant GO terms with more than 10 genes in the comparison and more than 7% of the genes in the category are represented as big nodes. Functionally related groups partially overlap. Only the most significant term in the group was labeled. Edges represent connections between GO term nodes and the gene term nodes associated with each group. B, Heat maps showing normalized color intensity from the expression values of the DEGs found in the wild type/PP2-A5 and wild type/pp2-a5 comparisons related to the enriched biological processes.
Figure 7.
Expression analysis of DEGs from enriched molecular functions. A, Functionally grouped network of molecular function enriched categories after ClueGO analysis of DEGs between wild type (WT)/PP2-A5 or wild type/pp2-a5. GO terms are represented as nodes, and the node size represents the term enrichment significance. Edges represent connections between GO term nodes and gene nodes associated with each group. B, Number of identified DEGs in the wild type/PP2-A5 and wild type/pp2-a5 comparisons for each transcription factor family (AP2-EREBP, Bhlh, C2H2, CC2C2-CO-like, C3H, HSF, Homeobox, MYB, NAC, RAV, and WRKY). Asterisks mark transcription factor enrichments identified by Fisher’s exact test (*, P < 0.05; **, P < 0.01; ***, P < 0.001; and ****, P < 0.0001).
Focusing our attention on transcription factor categories (Fig. 7B), a strong overrepresentation of AP2-EREBP and C2H2 categories was found in the DEGs from the wild type versus Col-PP2-A5 comparison. In addition, the C2C2-CO-like, NAC, RAV, and WRKY categories were also enriched. In contrast, the most overrepresented category in the DEGs from the wild type versus pp2-a5 comparison was the WRKY family of transcription factors. In addition, an enrichment of the NAC transcription factor family was found.
PP2-A5 Affects Plant Defense against Mites through Modulation of Hormonal Signaling
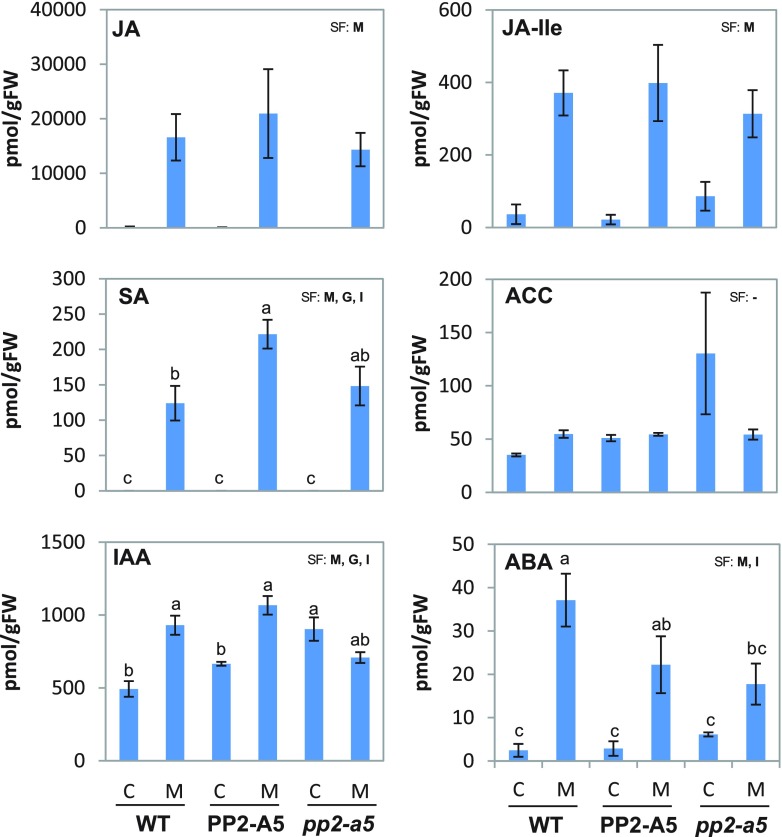
Transcriptional reprogramming caused by PP2-A5 could be related to changes in hormonal contents and their corresponding signaling pathways. A comprehensive study of the hormonal profile in the three Col-0 Arabidopsis genotypes was carried out to understand the role of PP2-A5 (Fig. 8). The results demonstrated that JA, its active form JA-Ile, and SA accumulated in leaf tissues during the first 24 h of spider mite feeding in the three Col-0 backgrounds. No significant differences in the levels of these hormones were observed among the genotypes, except for the SA content, which was significantly higher in infested PP2-A5-overexpressing lines than in wild-type plants after infestation. The levels of 1-aminocyclopropane-1-carboxylic acid (ACC), a precursor of ET, were similar in all genotypes, and they were not altered in response to mite attack. Abscisic acid (ABA) levels were significantly increased in wild-type and Col-PP2-A5 plants after mite feeding but not in pp2-a5 plants. Basal indole-3-acetic acid (IAA) levels were not significantly different between the wild type and PP2-A5 overexpression lines and were higher in the pp2-a5 line. However, whereas IAA content significantly increased in the wild-type and Col-PP2-A5 genotypes upon mite attack, the IAA amount decreased to significantly lower levels in the pp2-a5 genotype than in the other two genotypes. Likewise, the amounts of three IAA precursors increased in infested wild-type and Col-PP2-A5 lines (Supplemental Fig. S4). With respect to the cytokinin content, most of the compounds did not show differences among genotypes or in response to mite treatment. Only N6-(Δ2-isopentenyl)adenine (iP) was accumulated after infestation in all Col-0 backgrounds, while trans-zeatin (tZ) and trans-zeatin-7-glucoside (tZ7G) almost disappeared when mites fed on them (Supplemental Fig. S4).
Figure 8.
Quantification of hormones in the three PP2-A5 Col-0 genotypes infested with T. urticae. Quantifications were determined in 3-week-old-rosettes from PP2-A5_2.3, pp2-a5, and the wild type (WT). Values of JA, JA-Ile, SA, ACC, IAA, and ABA are expressed as pmol of each hormone (JA, JA-Ile, SA, IAA, and ABA) or hormone precursor (ACC) per g fresh weight (FW) and are means ± se of four biological replicates. Significant factors (SF) indicate whether the two independent factors, G (genotype) and M (mite infestation), and/or their interaction, I (G × M), were statistically significant (two-way ANOVA, P < 0.05). When the interaction was significant, Tukey’s posthoc test was performed. Different lowercase letters indicate significant differences between treatments/genotypes. P values of the two-way ANOVA are shown in Supplemental Table S5. C, Control noninfested plants; M, mite-infested plants.
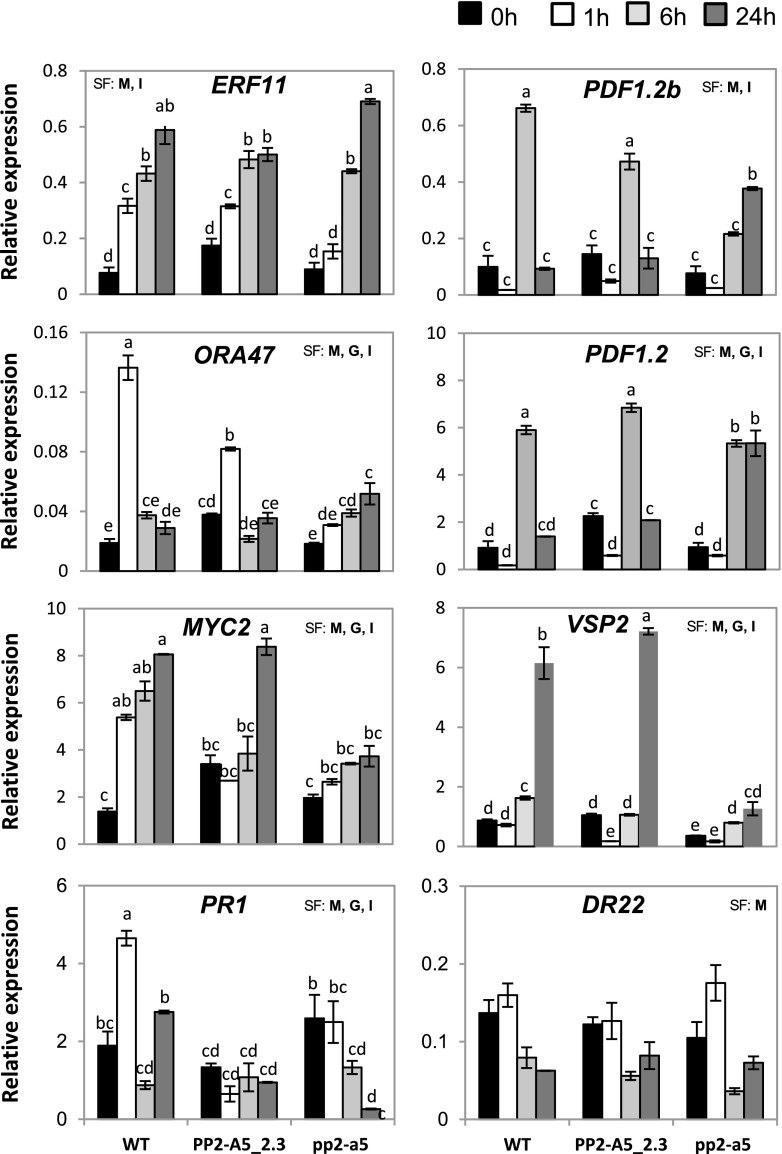
To further analyze the molecular basis of PP2-A5 function, the expression patterns of some genes related to ET/JA/SA and ABA pathways were analyzed 1, 6, and 24 h after mite feeding (Fig. 9). Ethylene-Responsive Transcription Factor11 (ERF11), Octadecanoid-Responsive AP2/ERF-Domain Transcription Factor47 (ORA47), MYC2 Transcription Factor (MYC2), Vegetative Storage2 (VSP2), and Plant Defensins (PDF1.2 and PDF1.2b) were the selected genes to dissect JA and ET/JA pathways. Some changes were observed at the transcriptional level among different PP2-A5 genotypes. Whereas expression of ERF11 peaked at 24 h of infestation and reached higher levels in pp2-a5 than in Col-PP2-A5 plants, messengers of ORA47 were highly accumulated in wild-type and Col-PP2-A5 plants and peaked at 1 h of infestation. MYC2 presented higher basal mRNA levels in Col-PP2-A5 plants than in wild-type or pp2-a5 plants. Interestingly, only Col-PP2-A5 and wild-type plants had increased MYC2 expression, reaching the highest values in both genotypes after 24 h of infestation. Regarding defense genes, PDF1.2 and PDF1.2b had increased expression upon mite feeding. Wild-type and Col-PP2-A5 plants showed the maximum induction of both genes at 6 h of infestation, returning to basal levels at 24 h. On the contrary, the up-regulation of PDF1.2 and PDF1.2b peaked at 24 h of infestation in the pp2-a5 plants. VSP2 showed higher basal expression in wild-type and Col-PP2-A5 plants than in pp2-a5 plants. Whereas a strong induction of VSP2 expression after infestation was found in Col-PP2-A5 and wild-type plants, this was more pronounced in Col-PP2-A5, and a subtle induction was shown in pp2-a5 plants.
Figure 9.
Expression of hormone-related genes upon spider mite feeding on the different plant PP2-A5 genotypes. Arabidopsis leaf tissue was collected 1, 6, and 24 h postinfestation, and expression was quantified by RT-qPCR. Gene expression levels were normalized to ubiquitin gene expression. Error bars represent se of the replicate for each treatment and time point. Data are means ± se of three biological replicates. Significant factors (SF) indicate whether the two independent factors, G (genotype) and M (mite infestation), and/or their interaction, I (G × M), were statistically significant (two-way ANOVA, P < 0.05). When the interaction was significant, Tukey’s posthoc test was performed. Different lowercase letters indicate significant differences between treatments/genotypes. P values of the two-way ANOVA are shown in Supplemental Table S5. WT, Wild type.
The expression of marker genes for SA and ABA pathways, Pathogenesis Related Protein1 (PR1) and Responsive to Dessication22, was also checked. Only minor variations were found for PR1 expression in the PP2-A5 overexpression and knockout lines upon mite feeding, but a significant increase was observed in wild-type plants after 1 h of spider mite infestation. Regarding Responsive to Dessication22, differences in gene expression were not found among the genotypes either at the basal level or in response to mites.
IGs Do Not Differentially Accumulate in Plant Lines with Altered PP2-A5 Levels
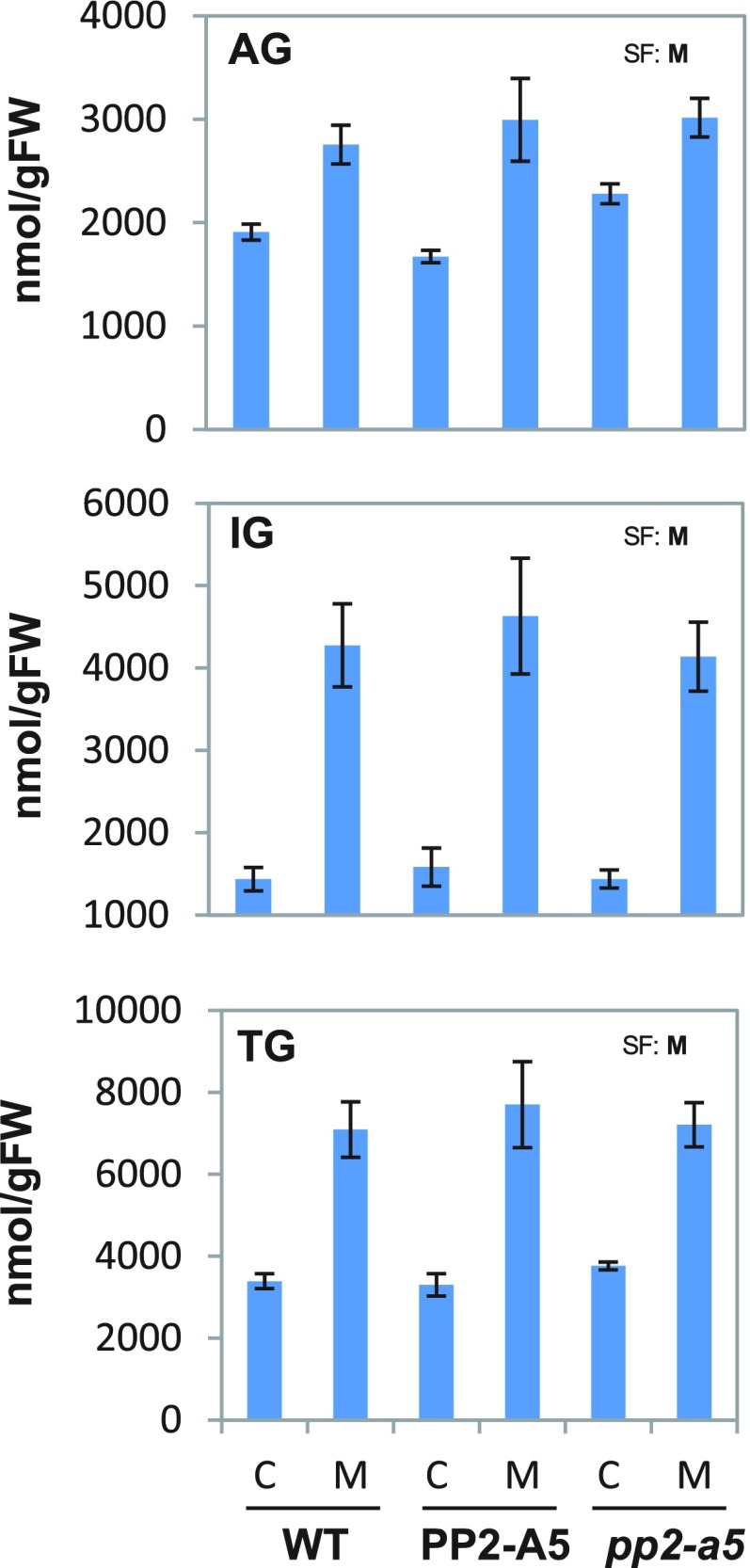
Basal and inducible glucosinolate levels were analyzed in wild-type, pp2-a5, and Col-PP2-A5 plants to determine whether resistance to spider mites conferred by PP2-A5 is due to differential accumulation of these defense compounds. The results showed no significant differences in total glucosinolates, IGs, or aliphatic glucosinolates among genotypes in either basal or mite-induced levels (Fig. 10). However, some differences were shown by individual aliphatic glucosinolates, such as 3msp, which showed equal basal levels in all three genotypes but a lack of induction in the sensitive genotypes wild type and pp2-a5 (Supplemental Figs. S5 and S6). This finding suggests that the resistance phenotype conferred by the overexpression of PP2-A5 might be partially due to higher accumulation of specific defensive metabolites in response to mites. However, IGs (previously described as major defense compounds in Arabidopsis against spider mites; Zhurov et al., 2014) did not show increased accumulation among genotypes either before or after mite herbivory.
Figure 10.
Levels of glucosinolates grouped in aliphatic (AG), indole (IG), and total (TG). Values, expressed as nmol g−1 fresh weight (FW), are means ± se of eight biological replicates. Significant factors (SF) indicate whether the two independent factors, G (genotype) and M (mite infestation), and/or their interaction, I (G × M), were statistically significant (two-way ANOVA, P < 0.05). P values of the two-way ANOVA are shown in Supplemental Table S5. C, Control noninfested plants; M, mite-infested plants; WT, wild type.
DISCUSSION
Plant immunity is based on a fast transcriptional reprogramming triggered by the perception of a pathogen or a pest. Differences in the activation of these regulatory networks have consequences on the resistance/susceptibility to the attacker. When we compared the effect of the generalist herbivore T. urticae on several Arabidopsis accessions, major differences in plant damage were found. These results prompted us to analyze variations in the transcriptional reprogramming between a resistant accession, Bla-2, and a susceptible one, Kon (Zhurov et al., 2014). From this analysis, several DEGs were shown to play a role in plant defense against T. urticae, such as the novel gene MATI and several ROS-related genes (Santamaría et al., 2017, 2018b). When searching for additional genes related to signaling, a gene that encodes a protein showing a lectin and a TIR domain was found. This gene was previously named PP2-A5, as it belongs to the PP2 family (Dinant et al., 2003). An in-depth search identified homologous proteins only in the Brassicaceae, Solanaceae, and Juglandaceae plant families. This scattered representation and the family-specific behavior of these proteins suggest a convergent evolutionary feature in which TIR and PP2 domains have been independently recruited. Thus, future studies directed at elucidating the specific function of each PP2-A5 domain are warranted.
As expected for a gene in which expression is induced by a biotic stress, overexpression or silencing of PP2-A5 conferred greater resistance or susceptibility levels, respectively, to the plant. Plants with an increased basal expression of PP2-A5 had less damage and caused a higher mite mortality when infested with T. urticae. Conversely, mutant pp2-a5 plants were more susceptible to mite attack than wild-type plants. The question now is to address how this gene performs its protective role. The defense role of PP2-A5 could be exerted by a toxic effect of the protein itself or by an indirect effect by modulation of signaling pathways. To elucidate this request, the specific function of each PP2-A5 domain should be considered.
The lectin domain has been widely associated with plant defense. Lectins recognize and bind carbohydrates without altering their structure. As plant defense proteins, lectins play a double role as signaling molecules that recognize carbohydrates and as toxic compounds against phytopathogenic insects, fungi, and bacteria (Lannoo and Van Damme, 2014; Bellande et al., 2017). The lectin domain of PP2-A5 belongs to the Nictaba family. Nictaba domains have been implicated in the response to several plant stresses (Vandenborre et al., 2009; Van Holle et al., 2016; Eggermont et al., 2018). The toxic activity of agglutinin, a tobacco (Nicotiana tabacum) Nictaba protein, against lepidopteran insect pests was demonstrated by feeding experiments using transgenic tobacco plants overexpressing or silencing the lectin gene (Vandenborre et al., 2010). Likewise, Arabidopsis PP2-A1, a phloem protein homologous to PP2-A5 that contains only the lectin domain, showed high binding affinity for chitin oligomers and displayed a negative effect on the green peach aphid (Myzus persicae), the Cucurbit aphid borne yellow virus, and various fungal strains (Bencharki et al., 2010; Zhang et al., 2011; Lee et al., 2014). Interestingly, the nucleocytoplasmic localization of the tobacco Nictaba protein has been related to a signaling role. In the nucleus, it interacts with core histone proteins through their O-GlcNAc modifications (Schouppe et al., 2011; Delporte et al., 2014), suggesting an enhancement in the transcription of defense-related genes by chromatin remodeling (Lannoo and Van Damme, 2010). Similarly, the subcellular localization of PP2-A5 in the nucleus and cytoplasm supports that the Nictaba domain of PP2-A5 could function as a signaling molecule that triggers defense responses by interactions with histone proteins or as a toxic compound against T. urticae feeding. Lectins have been shown to bind to the epithelium of the insect gut, triggering their insecticidal activity by an unknown mechanism (Michiels et al., 2010). As T. urticae ingests the contents of mesophyll cells, PP2-A5 would be ingested and could exert its potential toxic activity in the mite gut.
On the other hand, the TIR domain has been found in proteins involved in innate immunity pathways in animals and plants. TIR is considered the signaling domain in plant receptors with a TIR nucleotide binding-leucine rich repeat (NB-LRR) structure, as they can cause cell death autonomously when expressed ectopically in planta (Bernoux et al., 2011; Williams et al., 2014). Likewise, several TIR-only and TIR-NB proteins induced cell death when expressed in tobacco and provided enhanced resistance when overexpressed in Arabidopsis (Nandety et al., 2013). Recently, it was shown that the TIR-only Arabidopsis protein RBA1 triggers cell death in response to the bacterial type III effector HopBA1 (Nishimura et al., 2017). Thus, the TIR domain of PP2-A5 could act in a similar way, as an activator of immune responses. Interestingly, the functionality of TIR domains requires TIR-TIR interactions, leading to high-order assemblies through self-association and homotypic association (Nimma et al., 2017; Zhang et al., 2017). One of these heterodimers (or oligomers) formed by the TIR-NB-LRR proteins RPS4 (Resistance to Pseudomonas syringae 4) and RRS1-R (Resistance to Ralstonia solanacearum 1) acts as an inhibited receptor complex that is activated by direct binding of bacterial effectors to RRS1-R (Williams et al., 2014). The acetyltransferase effector PopP2 acetylates an additional C-terminal WRKY transcription factor domain present in RRS1-R that binds DNA, disrupting the nuclear RRS1-R DNA association and activating RPS4-dependent immunity (Le Roux et al., 2015; Sarris et al., 2015). In addition, the TIR-NB protein TN13 is released from the ER membrane in response to bacterial stimulus and is translocated to the nucleus, where it is important for activating plant defense (Roth et al., 2017). The subcellular localization of PP2-A5, which is compatible with a nucleocytoplasmic protein that may be associated with membrane systems by protein-protein interactions, supports a similar mechanism for PP2-A5 transmitting cytoplasm-membrane signals to the nucleus. In the nucleus, some Arabidopsis TIR-NB-LRR proteins have been demonstrated to interact with transcription factors. One of them, SNC1, is able to interact with the transcriptional corepressor TPR1 to negatively regulate the expression of known defense suppressors and with the transcriptional factor basic helix-loop-helix 84, which also binds to RPS4 to activate defense responses (Zhu et al., 2010; Xu et al., 2014).
Interestingly, overexpression and silencing of PP2-A5 result in differential modular transcriptional reprogramming. Whereas overexpression of PP2-A5 causes an increase in the expression of genes related to ET signaling, mutant pp2-a5 plants were characterized by an increase in the expression of genes involved in the SA response. Although the chitin response category was enriched in both genotypes, different groups of genes were induced by changes in PP2-A5 levels. Transcriptional reprogramming in the PP2-A5 overexpression and mutant lines may result from the nuclear localization of PP2-A5 protein, where it modifies gene transcription modules through binding of the lectin domain to histones or through an activated stage obtained by oligomerization that could interact with transcription factors. In fact, the yeast two-hybrid system has demonstrated the in vitro interaction of PP2-A5 with the transcription factors RVE1, ABSCISIC ACID INSENSITIVE 5, and a predicted basic helix-loop-helix -containing protein (Lumba et al., 2014).
A relevant question to answer is the relationship between PP2-A5 signaling and hormonal pathways. The JA signaling cascade is activated following herbivore attack and is considered crucial to trigger resistance mechanisms (Zhurov et al., 2014; Santamaría et al., 2017). In addition, an SA-parallel pathway and the modulation of JA response via cross talk with other hormones are involved in fine-tuning the defense machinery to achieve appropriate herbivore-specific responses (Erb et al., 2012). In response to spider mite infestation, JA and SA accumulate in Arabidopsis plants (Zhurov et al., 2014). However, whereas plants defective in JA biosynthesis or JA-regulated transcription displayed increased susceptibility to T. urticae attack, plants deficient in SA biosynthesis or downstream signaling did not encounter more plant damage upon spider mite infestation (Zhurov et al., 2014). SA accumulation induced by spider mites in Arabidopsis has been proposed as a strategy to minimize accurate plant defense by hampering JA-induced responses (Santamaría et al., 2017). As expected, JA, JA-Ile, and SA levels were increased upon mite infestation in wild-type plants. Strikingly, the overexpression or silencing of PP2-A5 did not cause alterations in the basal content of these hormones or in their induction, with the exception of a higher increase in SA content in Col-PP2-A5 plants. These results suggest that the modulation of resistance/susceptibility of Arabidopsis to T. urticae is independent of classical spider mite-related hormonal signaling pathways. Interestingly, differences were found in both basal and spider mite-regulated auxin levels. The main functional auxin compound, IAA, had higher basal levels in pp2-a5 plants than in wild-type and PP2-A5 plants. Whereas spider mites induced the accumulation of IAA in wild-type and Col-PP2-A5 plants, pp2-a5 plants had reduced amounts of IAA. Auxin interacts synergistically with JA to promote resistance against necrotrophic pathogens (Shigenaga and Argueso, 2016). In Arabidopsis, the levels of both JA and IAA increase upon infection with Alternaria brassicicola (Qi et al., 2012). Additionally, increased susceptibility to necrotrophic pathogens was displayed by mutants in auxin biosynthesis or transport (Llorente et al., 2008). Conversely, although cytokinins were rapidly induced upon herbivore elicitation (Schäfer et al., 2015), synergistic and antagonistic interactions between cytokinin and SA and between cytokinin and auxin have been demonstrated (Naseem et al., 2012). Although minor differences were found between genotypes for most cytokinins, the significantly high basal level of iP in pp2-a5 plants could be related to its higher susceptibility by negatively affecting auxin signaling. Finally, ABA induction by spider mites was detected in wild-type and Col-PP2-A5 plants but not in pp2-a5 plants. ABA has been shown to be a crucial regulator of herbivore-induced resistance by activating JA-dependent defense responses in Arabidopsis plants challenged with Pieris rapae (Vos et al., 2013). Thus, transcriptomic reprogramming mediated by PP2-A5 alters the hormonal signaling pathways, probably to contribute to a tight regulation of the hormonal cross talk upon mite feeding.
In accordance with the predicted role of these minor differences in hormonal levels, an enrichment of DEGs involved in hormonal responses was found in Col-PP2-A5 and pp2-a5 plants. Even though no changes were found for the ET precursor ACC at the investigated time point, several transcription factors involved in the ET response were up-regulated by PP2-A5 overexpression. Likewise, SA-responsive genes were up-regulated by PP2-A5 silencing in spite of having similar SA levels in pp2-a5 and in wild-type or Col-PP2-A5 plants. The expression of some of the differentially expressed transcription factors displayed an apparent correlation with the induction of JA/ET-responsive genes. Whereas a rapid induction of ERF11 and ORA47, transcription factors of the AP2/ERF family, in wild-type and Col-PP2-A5 plants was accompanied by a maximal induction of PDF1.2 and PDF1.2b levels at 6 h of infestation, the slower induction of these transcription factors in pp2-a5 plants correlates with a delayed induction of both defense genes. Likewise, a higher induction of the expression of MYC2 in response to the mite attack was accompanied by a higher induction of the VSP2-responsive gene. The relationships of ERF11 and ORA47 with JA and ET biosynthesis have been previously reported. Whereas ERF11 is a repressor of ET biosynthesis, ORA47 is an activator of JA biosynthetic genes (Li et al., 2011; Chen et al., 2016). ABA has been proven to activate the JA/MYC2 branch and to suppress the JA/ET/ERFs branch upon herbivory (Kazan and Manners, 2013). Thus, ABA induction upon spider mite feeding in wild-type and Col-PP2-A5 plants would help to elicit JA responses via the JA/MYC2 pathway. Interestingly, different responses were previously reported to two Arabidopsis herbivores. Whereas Spodoptera exigua elicits both JA/MYC2 and JA/ET/ERFs pathways, P. rapae elicits the JA/MYC2 branch, bypassing the JA/ET/ERFs signaling cascade (Rehrig et al., 2014). Regarding T. urticae, plant resistance is associated with an unbalanced expression of JA- and JA/ET-responsive genes, favoring the JA-induced genes (VSP2) against the JA/ET-regulated genes (PDF1.2) and a trend toward lower expression of SA-induced genes (PR1).
The final question was to elucidate how PP2-A5-overexpressing plants exert effects on the spider mite. IGs are central to the Arabidopsis defense to mite herbivory and dramatically increase mite mortality (Zhurov et al., 2014). Basal levels of IGs were similar in the Col-PP2-A5 and pp2-a5 plants and were increased to similar levels in both genotypes upon herbivory. Although minor differences in the levels of certain individual glucosinolates were found, a key role in the differential resistance/susceptibility abilities cannot be attributed to the accumulation of these compounds across the whole leaf. However, as different glucosinolates are produced in different cells and spider mites feed on individual cells, it is still possible that local changes are effective. As glucosinolates have to be degraded to actual bioactive compounds (Wittstock and Burow, 2010), possible alterations in this process caused by the overexpression/silencing of PP2-A5 cannot be discarded. Furthermore, other physical/chemical processes could be related to mite resistance, including the regulation by hormones of a high number of biosynthetic pathways for secondary metabolites. For example, JA is involved in the accumulation of phenylpropanoids, polyketides, terpenoids, and nitrogen-containing compounds (Guo et al., 2018). Additionally, different plant species use alternative defenses to combat spider mite attack (i.e. protease inhibitors contribute to the defense response in tomato plants; Martel et al., 2015). These features support the activation of synergistic responses triggered by T. urticae in Arabidopsis to produce additional defenses to glucosinolates.
In summary, the defensive role of an evolutionarily adapted protein, PP2A5, against herbivores has been demonstrated. The combination of two putative signaling domains in a single protein may contribute to improving the resistance mechanisms against spider mites. Further research is needed to understand the mechanistic process used by this two-domain protein to confer defense against the spider mite. To know the exact role of each single domain, direct-mutagenesis assays combined with the generation of derived-transgenic lines and feeding bioassays will provide a deeper knowledge about their defense properties.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Arabidopsis (Arabidopsis thaliana) Col-0, Kon, and Bla-2 accessions (Nottingham Arabidopsis Seed Collection) were used as wild-type controls. The Arabidopsis T-DNA mutants (SALK_ 132166C, N664027, referred to as pp2-a5 in this article) were obtained from the Arabidopsis Biological Resource Center (https://abrc.osu.edu/) through the European Arabidopsis Stock Centre (http://arabidopsis.info/BasicForm/). T-DNA insertion, T-DNA homozygous status, and gene expression levels of the Salk line was analyzed by conventional PCR and RT-PCR (Supplemental Fig. S3). For soil growth, autoclaved peat moss and vermiculite (2:1, v/v) were used. Seeds were planted and incubated 5 d at 4°C. For in vitro growth, seeds were surface sterilized with 75% (v/v) ethanol, dried, and plated onto petri dishes containing 0.53 g of Murashige and Skoog salts (Sigma-Aldrich), 1% (w/v) Suc, 0.5 g L−1 MES, and 0.4% (w/v) Phytagel (Sigma-Aldrich), adjusted to pH 5.7 with KOH. Plants and plates were then grown in growth chambers (Sanyo MLR-350-H) under control conditions (23°C ± 1°C, greater than 70% relative humidity, and a 16-h/8-h day/night photoperiod).
To generate overexpression lines, PP2-A5 cDNA from Col-0 plants was cloned into pGWB2 (Cauliflower mosaic virus 35S, no tag) and pGWB5 (Cauliflower mosaic virus 35S, C-sGFP) Gateway binary vectors (Nakagawa et al., 2007) using the specific primers included in Supplemental Table S4. The recombinant plasmids were introduced into Arabidopsis Col-0 plants using Agrobacterium tumefaciens floral dip transformation (Clough and Bent, 1998), termed Col-PP2-A5 and Col-PP2-A5-GFP plants in this article. pGWB2 plasmid was also inserted into Arabidopsis Kon plants, termed Kon-PP2-A5 plants in this article. Shoots were regenerated on selective medium containing hygromycin (100 mg L−1), and primary transformants (T0) were allowed to self-fertilize. Plants were then selected and self-fertilized twice more to generate the third generation lines (T3). Homozygous plants with a single-copy insertion and the highest transgene expression levels from different transformation events were selected for our experiments (Supplemental Fig. S3).
Gene Expression Analyses by RT-qPCR
Since two isoforms (AT1G65390.1 and AT1G65390.2) for our gene of interest were found, an initial RT-qPCR analysis was performed to select the more suitable isoform for further studies (Supplemental Fig. S1). In addition, RT-qPCR assays were also used to validate data from transcriptomic analysis, to determine the homozygous status of the Salk mutant lines, and to study PP2-A5 gene expression in major Arabidopsis tissues. Arabidopsis rosettes from Col-0, Bla-2, and Kon accessions were sampled after different times of mite infestation (1, 3, 6, 12, and 24 h) to validate microarray results. Total RNA was extracted following Oñate-Sánchez and Vicente-Carbajosa (2008) and reverse transcribed using the Revert Aid H Minus First Strand cDNA Synthesis Kit (Fermentas). cDNAs from flowers, roots, siliques, and rosette leaves of 1-, 2-, and 3-week-old-Arabidopsis Col-0 plants were also prepared. RT-qPCR was performed for three biological samples as previously described (Santamaria et al., 2012a) using a SYBR Green Detection System (Roche) and the CFX Manager Software 2.0 (Bio-Rad). Similarly, gene expression levels involved in hormonal signaling pathways were analyzed in rosettes from Col-PP2-A5_2.3, pp2-a5, and wild-type plants in three biological samples from three independent experiments of noninfested and 1-, 6-, and 24-h infested plants (20 mites per plant). Gene expression was referred to as relative expression levels (2^-dCt) or fold change (2^-ddCt; Livak and Schmittgen, 2001). Ubiquitin was used for normalization. Specific primers were designed through the Salk Institute T-DNA primer design link (http://signal.salk.edu/tdnaprimers.2.html) or through the PRIMER 3 program (http://bioinfo.ut.ee/primer3-0.4.0/). Primer sequences are indicated in Supplemental Table S4. RT-qPCR was performed for three samples from three independent experiments. Six rosettes were pooled and frozen in liquid nitrogen for each experiment. Three biological replicates were analyzed per genotype and treatment (n = 3).
Structural and Evolutionary PP2-A5 Gene Analyses
PP2-A5 sequences were downloaded from the TAIR Web site (http://www.arabidopsis.org/). Amino acid sequence was subjected to a sequence search in the Pfam database v28.0 (http://pfam.xfam.org/) to identify possible domains. The SignalP 4.1 program (http://www.cbs.dtu.dk/Services/SignalP/) was used to search a signal peptide. PP2-A5 protein homologs from different plant species were obtained by BLASTP searches in the Phytozome v12.1 database (https://phytozome.jgi.doe.gov/) using an E-value lower than e−40 and a coverage higher than 50% of the query sequence (PP2-A5 protein). Additional searches were performed in the nonredundant National Center for Biotechnology Information protein database. Retrieved proteins were aligned by MUSCLE v3.8 (http://www.ebi.ac.uk/Tools/msa/muscle). A phylogenetic tree was constructed by the maximum likelihood PhyML v3.0 method (http://www.phylogeny.fr), using a BIONJ starting tree and applying the approximate likelihood-ratio test as a statistical test for nonparametric branch support. The displayed tree was visualized in the program MEGA 7.0 (http://www.megasoftware.net/).
Subcellular Localization of PP2-A5 in Transient and Stable Transgenic Plants
The open reading frame of the PP2-A5 gene translationally fused to the N terminus of the whole GFP reporter gene was cloned into the pGWB5 binary vector following Gateway technology instructions. As controls, the psmRS-GFP plasmid containing the GFP gene under the control of the cauliflower mosaic virus 35S promoter was used (Davis and Vierstra, 1998) as well as the pRTL2ΔNS/ss-RFP-HDEL plasmid containing the Arabidopsis chitinase signal sequence and the C-terminal HDEL ER retrieval signal, whose protein specifically localizes in the ER (Shockey et al., 2006). Transient transformation of onion (Allium cepa) epidermal cells was performed by particle cobombardment with a biolistic helium gun device (DuPont PDS-1000/Bio-Rad) as previously described (Diaz et al., 2005). Fluorescent images were acquired after 24 h of incubation at 22°C in the dark with the LEICA SP8 confocal microscope (Leica) using the appropriate filters. A. tumefaciens transient transformation assays were done as described by Voinnet et al. (2003) with modifications. A. tumefaciens strain GV3101 (C58C1, Rif) carrying the candidate vectors 35S:PP2-A5:GFP (pGWB5), 35S:GFP (pEAQ-HT-GFP), or 35S:P19 (pBIN61) were grown from single colonies for 24 h in 3 mL of Luria-Bertani medium with the appropriate antibiotics. After centrifugation, the bacteria pellets were resuspended in 3 mL of MA (10 mm MES, 10 mm MgCl2, and 150 µm acetosyringone) to a final OD of 0.3 and then incubated for at least 3 h at room temperature. Bacterial suspensions were infiltrated into the abaxial side of the third-youngest fully expanded Nicotiana benthamiana leaf using a needleless syringe. Fluorescent images were acquired after 3 d of infiltration as indicated above. For the subcellular localization of the fusion PP2-A5-GFP protein in Arabidopsis, seedlings from the Col-PP2-A5-GFP line were observed with the LEICA SP8 confocal microscope (Leica).
Spider Mite Maintenance and Performance Analyses
A colony of Tetranychus urticae, London strain (Acari: Tetranychidae), provided by Dr. Miodrag Grbic (University of Western Ontario), was reared on beans (Phaseolus vulgaris) and maintained in growth chambers (Sanyo MLR-350-H) at 23°C ± 1°C, greater than 70% relative humidity, and a 16-h/8-h day/night photoperiod. Mites were synchronized as described by Santamaría et al. (2017). Spider mite development was studied on Col-PP2-A5_1.1, Col-PP2-A5_2.3, Col-PP2-A5_4.1, Col-pp2-a5, Kon-PP2-A5_1.2, Kon-PP2-A5 _4.2, and Kon-PP2-A5_5.3 transgenic lines and on nontransformed Col-0 and Kon controls. The T. urticae mortality and developmental assay was performed on detached leaves from 3-week-old plants. Small dishes (35 mm diameter) filled with some water and covered with Parafilm were used for these assays. The newest emerged leaf (about 1 cm long) from each plant was fit in the plate by introducing its petiole across the Parafilm to keep contact with water. Leaves were infested with 25 neonate larvae (24 h). After infestation, the plates were covered with a lid with ventilation and Parafilm to avoid possible escapes. Every day, the number of larvae becoming protonymph that died were counted in order to calculate developmental stages and mortality rates. Every 2 d, a new leaf from a new plant was added. Results were represented as percentages of mortality and the number of days that larva needed to become protonymph. Eight replicates from eight independent plants were used for each plant genotype.
Plant Damage Determination
Quantification of plant damage after arthropod feeding was done on Arabidopsis T2 entire plants from selected homozygous transgenic lines (Col-PP2-A5_1.1, Col-PP2-A5_2.3, Col-PP2-A5_4.1, Col-pp2-a5, Kon-PP2-A5_1.2, Kon-PP2-A5 _4.2, and Kon-PP2-A5_5.3) and from the nontransformed Col-0 and Kon controls. Three-week-old plants were infested with 20 T. urticae adults per plant. They were carefully transferred with a brush to the leaf surface. After 4 d of feeding, leaf damage was assessed by scanning the entire rosette using a scanner (HP Scanjet 5590 Digital Flatbed Scanner series) according to Cazaux et al. (2014). Leaf damage was calculated in mm2 using Adobe Photoshop CS software. Six replicates from six independent plants were used for each genotype.
RNA Sequencing Library Preparation, Sequencing, Alignment, and DEGs Analysis
Total RNA was extracted from 3-week-old Col-PP2-A5_2.3, Col-pp2-a5, and Col-0 wild-type seedlings as described by Oñate-Sánchez and Vicente-Carbajosa (2008). RNA integrity was analyzed using an Agilent 2100 Bioanalyzer with an RNA 6000 Chip and 2100 Expert software (Agilent Technologies). Total RNA was sent to the Beijing Genomics Institute. Double-stranded cDNA libraries obtained from purified mRNA were sequenced using Illumina HiSeq 2000 high-throughput sequencing technology. More than 10 million single-end reads of 50 nucleotides in length were obtained for each sample (n = 3). Three biological replicates from three independent experiments were used. For each biological replicate, six rosettes were pooled and frozen into liquid nitrogen. Raw reads with adaptor sequences and low-quality reads were removed. Clean reads were mapped to the Arabidopsis reference genome (http://www.arabidopsis.org/, TAIR version 10) using SOAPAligner/SOAP2 (Li et al., 2009). Alignment results were visualized by Integrative Genomics Viewer. Once all reads were assembled and annotated, transcript abundance was normalized using the reads per kilobase transcriptome per million mapped reads method (Mortazavi et al., 2008). DEGs between groups were obtained using the NOISeq method (Tarazona et al., 2011) with a log2 ratio (fold change) higher than 1 and a probability of differential expression higher than 0.8. Euler diagrams were made using the tools provided by the Venn Diagrams Web page (http://www.venndiagrams.net/). Gene enrichment analyses were performed with the Bonferroni step-down test using the ClueGO package (Bindea et al., 2009) in Cytoscape (Shannon et al., 2013). Heat maps were made by the ClustVis Web tool (http://biit.cs.ut.ee/clustvis/). Transcription factors were analyzed in AtTFDB (Yilmaz et al., 2011) and iTAK (Zheng et al., 2016) programs. Fisher’s exact tests were performed using GraphPad Prism 6 software.
Metabolite Analyses
Two-week-old-plants from Col-0, pp2-a5, and Col-PP2-A5 were infested with 25 female spider mites using a brush. Plant material from nontreated and infested plants was harvested after 24 h for glucosinolate and hormone analyses. For glucosinolate analysis, the whole rosette was collected from individual plants (n = 8) and glucosinolates were extracted and analyzed as desulfoglucosinolates as described previously (Kliebenstein et al., 2001; Crocoll et al., 2016). For hormone analysis, three rosettes were pooled and frozen in liquid nitrogen for each replicate. Four biological replicates were analyzed per genotype and treatment (n = 4). Frozen ground samples were extracted twice with methanol (1,250 mL, 80% [v/v]) followed by a 30-min incubation at 4°C. After centrifugation (15 min, 20,000g, 4°C), the pooled supernatants were passed over a C18 column (Phenomenex, Strata C18-E; 200 mg per 3 mL) using a vacuum. The eluate was evaporated to dryness, and samples were dissolved in 20% (v/v) methanol and filtered (0.22 µm) by centrifugation (5 min, 3,000 rpm). Phytohormones were analyzed by ultra-high-performance liquid chromatography coupled with triple quadrupolemass spectrometry on an AdvanceTM-UHPLC/EVOQTMElite-TQ-MS instrument (Bruker) equipped with a C-18 reverse-phase column (Kinetex 1.7 u XB-C18, 10 cm × 2.1 mm, 1.7 mm particle size; Phenomenex) by using a 0.05% (v/v) formic acid in water, pH 4 (solvent A)-methanol (solvent B) gradient at a flow rate of 0.4 mL min−1 at 40°C. The gradient applied was as follows: 10% to 50% B (15 min), 50% B (2 min), 50% to 100% B (0.1 min), 100% B (2.9 min), 100% to 10% B (0.1 min), and 10% B (5 min). Compounds were ionized by electrospray ionization with a spray voltage of +4,500 and −4,000 V in positive and negative mode, respectively. Heated probe temperature was 350°C, and cone temperature was 300°C. Quantification was based on response factors relative to (2H5)tZ (positive mode) and (2H6)ABA (negative mode). The individual hormones were monitored based on the following multiple reaction monitoring transitions: (2H5)tZ, (+) 225 > 137 [15 V]; (2H6)ABA, (−) 269 > 159 [7 V]; ABA, (−) 263 > 153 [7 V]; ACC, (+) 102 > 56 [15 V]; dihydrozeatin, (+) 222 > 136 [15 V]; dihydrozeatin riboside, (+) 354 > 222 [15 V]; IAA, (+) 176 > 130 [10 V]; 3-indoleacetamide, (+) 175 > 130 [15 V]; indole-3-aldehyde, (+) 146 > 118 [15 V]; indole-3-carboxy acid, (+) 162 > 118 [15 V]; iP, (+) 204 > 136 [10 V]; JA, (−) 209 > 59 [11 V]; JA-Ile, (−) 322 > 130 [17 V]; SA, (−) 137 > 93 [20 V]; tZ, (+) 220 > 136 [15 V]; tZ7G, (+) 382 > 220 [17 V]; and tZR, (+) 352 > 220 [15 V].
Statistical Analysis
The statistical analysis was performed using GraphPad Prism 6. One-way ANOVA was used for gene expression, damage analysis, and T. urticae performance. After one-way ANOVA, Tukey’s multiple comparisons test was applied. Two-way ANOVA was performed in the experiments in which genotype (G) and infestation (M) were simultaneously studied, in the gene expression analyses of hormone-related genes, and in the hormone and glucosinolate quantification assays. When the interaction (G × M) was significant, Tukey’s posthoc tests were applied to compare treatments. In the figures, significant differences (P < 0.05) among lines for different evaluated parameters are reported with different letters. A linear trend was drawn through mortality (%) and leaf damage (%) along the different plant genotypes. The R2 value indicates how well data fit the line. To test statistical significance of the correlation between mortality and leaf damage depending on the PP2-A5 gene expression, a Pearson product moment correlation test was performed. A negative correlation coefficient (r) and a P value lower than 0.05 described a negative correlation. A Kolmogorov-Smirnov test was used to test normality (P < 0.05). Brown-Forsythe and Bartlett’s tests were run during ANOVA to validate the homoscedasticity assumption (P > 0.05). The P values obtained from each statistical analysis are compiled in Supplemental Tables S5 and S6.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under these accession numbers: PP2-A5, AT1G65390; ERF11, AT1G28370; ORA47, AT1G74930; MYC2, AT1G32640; PDF1.2, AT5G44420; PDF1.2b, AT2G26020; VSP2, AT5G24770; PR1, AT2G14610; and DR22, AT5G25610.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Validation of splicing variant gene expression by RT-qPCR.
Supplemental Figure S2. Confocal stacks spanning N. benthamiana cells agroinfiltrated with 35S-PP2-A5-GFP and 35S-GFP control.
Supplemental Figure S3. Molecular characterization of Arabidopsis Col-0 transgenic lines.
Supplemental Figure S4. Quantification of auxin precursors and cytokinins in the three PP2-A5 Col-0 genotypes infested with T. urticae.
Supplemental Figure S5. Quantification of individual indole and benzenic glucosinolates in the three PP2-A5 Col-0 genotypes infested with T. urticae.
Supplemental Figure S6. Quantification of individual aliphatic glucosinolates in the three PP2-A5 Col-0 genotypes infested with T. urticae.
Supplemental Table S1. Development performance of T. urticae after feeding on the different PP2-A5 Col-0 and -Kon genotypes.
Supplemental Table S2. List of the significant results obtained from the enrichment GO analysis of the biological process assigned to the DEGs.
Supplemental Table S3. List of the significant results obtained from the enrichment GO analysis of the molecular functions assigned to the DEGs.
Supplemental Table S4. Primer sequences used in this work, indicating the purposed assay.
Supplemental Table S5. Statistical results obtained with GraphPad Prism 6 software.
Supplemental Table S6. Statistical results obtained with GraphPad Prism 6 software.
Supplemental Data Set S1. DEGs between Col-0 wild-type/PP2-A5 plants and Col-0 wild-type/pp2-a5 plants.
Footnotes
This work was supported by the Ministerio de Economía y Competitividad of Spain (BIO2017-83472-R and 618105-FACCE-Era Net Plus), the Government of Canada through the Ontario Research Fund (RE08-067) and the Natural Sciences and Engineering Research Council of Canada (NESR), the Danish National Research Foundation (DNRF grant 99), and The Danish Council for Independent Research (DFF-4181-00077).
References
- Agrawal AA, Vala F, Sabelis MW (2002) Induction of preference and performance after acclimation to novel hosts in a phytophagous spider mite: Adaptive plasticity? Am Nat 159: 553–565 [DOI] [PubMed] [Google Scholar]
- Agut B, Gamir J, Jacas JA, Hurtado M, Flors V (2014) Different metabolic and genetic responses in citrus may explain relative susceptibility to Tetranychus urticae. Pest Manag Sci 70: 1728–1741 [DOI] [PubMed] [Google Scholar]
- Agut B, Gamir J, Jaques JA, Flors V (2015) Tetranychus urticae-triggered responses promote genotype-dependent conspecific repellence or attractiveness in citrus. New Phytol 207: 790–804 [DOI] [PubMed] [Google Scholar]
- Agut B, Pastor V, Jaques JA, Flors V (2018) Can plant defence mechanisms provide new approaches for the sustainable control of the two-spotted spider mite Tetranychus urticae? Int J Mol Sci 19: 614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alba JM, Schimmel BCJ, Glas JJ, Ataide LMS, Pappas ML, Villarroel CA, Schuurink RC, Sabelis MW, Kant MR (2015) Spider mites suppress tomato defenses downstream of jasmonate and salicylate independently of hormonal crosstalk. New Phytol 205: 828–840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ament K, Kant MR, Sabelis MW, Haring MA, Schuurink RC (2004) Jasmonic acid is a key regulator of spider mite-induced volatile terpenoid and methyl salicylate emission in tomato. Plant Physiol 135: 2025–2037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arimura G, Tashiro K, Kuhara S, Nishioka T, Ozawa R, Takabayashi J (2000) Gene responses in bean leaves induced by herbivory and by herbivore-induced volatiles. Biochem Biophys Res Commun 277: 305–310 [DOI] [PubMed] [Google Scholar]
- Arimura G, Ozawa R, Nishioka T, Boland W, Koch T, Kühnemann F, Takabayashi J (2002) Herbivore-induced volatiles induce the emission of ethylene in neighboring lima bean plants. Plant J 29: 87–98 [DOI] [PubMed] [Google Scholar]
- Arnaiz A, Talavera-Mateo L, Gonzalez-Melendi P, Martinez M, Diaz I, Santamaria ME (2018) Involvement of Arabidopsis Kunitz trypsin inhibitors in defence against spider mites. Front Plant Sci 8: 9432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ataide LMS, Pappas ML, Schimmel BCJ, Lopez-Orenes A, Alba JM, Duarte MVA, Pallini A, Schuurink RC, Kant MR (2016) Induced plant-defenses suppress herbivore reproduction but also constrain predation of their offspring. Plant Sci 252: 300–310 [DOI] [PubMed] [Google Scholar]
- Balkema-Boomstra AG, Zijlstra S, Verstappen FW, Inggamer H, Mercke PE, Jongsma MA, Bouwmeester HJ (2003) Role of cucurbitacin C in resistance to spider mite (Tetranychus urticae) in cucumber (Cucumis sativus L.). J Chem Ecol 29: 225–235 [DOI] [PubMed] [Google Scholar]
- Barczak-Brzyżek A, Kiełkiewicz M, Górecka M, Kot K, Karpińska B, Filipecki M (2017) Abscisic Acid Insensitive 4 transcription factor is an important player in the response of Arabidopsis thaliana to two-spotted spider mite (Tetranychus urticae) feeding. Exp Appl Acarol 73: 317–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellande K, Bono JJ, Savelli B, Jamet E, Canut H (2017) Plant lectins and lectin receptor-like kinases: How do they sense the outside? Int J Mol Sci 18: E1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bencharki B, Boissinot S, Revollon S, Ziegler-Graff V, Erdinger M, Wiss L, Dinant S, Renard D, Beuve M, Lemaitre-Guillier C, et al. (2010) Phloem protein partners of Cucurbit aphid borne yellows virus: Possible involvement of phloem proteins in virus transmission by aphids. Mol Plant Microbe Interact 23: 799–810 [DOI] [PubMed] [Google Scholar]
- Bensoussan N, Santamaria ME, Zhurov V, Diaz I, Grbic M, Grbic V (2016) Plant-herbivore interaction: Dissection of the cellular pattern of Tetranychus urticae feeding on the host plant. Front Plant Sci 7: 1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernoux M, Ve T, Williams S, Warren C, Hatters D, Valkov E, Zhang X, Ellis JG, Kobe B, Dodds PN (2011) Structural and functional analysis of a plant resistance protein TIR domain reveals interfaces for self-association, signaling, and autoregulation. Cell Host Microbe 9: 200–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bindea G, Mlecnik B, Hackl H, Charoentong P, Tosolini M, Kirilovsky A, Fridman WH, Pagès F, Trajanoski Z, Galon J (2009) ClueGO: A Cytoscape plug-in to decipher functionally grouped Gene Ontology and pathway annotation networks. Bioinformatics 25: 1091–1093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cazaux M, Navarro M, Bruinsma KA, Zhurov V, Negrave T, Van Leeuwen T, Grbic V, Grbic M (2014) Application of two-spotted spider mite Tetranychus urticae for plant-pest interaction studies. J Vis Exp 89: e51738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen HY, Hsieh EJ, Cheng MC, Chen CY, Hwang SY, Lin TP (2016) ORA47 (octadecanoid-responsive AP2/ERF-domain transcription factor 47) regulates jasmonic acid and abscisic acid biosynthesis and signaling through binding to a novel cis-element. New Phytol 211: 599–613 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Crocoll C, Halkier BA, Burow M (2016) Analysis and quantification of glucosinolates. Curr Protoc Plant Biol 1: 385–409 [DOI] [PubMed] [Google Scholar]
- Davis SJ, Vierstra RD (1998) Soluble, highly fluorescent variants of green fluorescent protein (GFP) for use in higher plants. Plant Mol Biol 36: 521–528 [DOI] [PubMed] [Google Scholar]
- Delporte A, De Vos WH, Van Damme EJ (2014) In vivo interaction between the tobacco lectin and the core histone proteins. J Plant Physiol 171: 1149–1156 [DOI] [PubMed] [Google Scholar]
- Diaz I, Martinez M, Isabel-LaMoneda I, Rubio-Somoza I, Carbonero P (2005) The DOF protein, SAD, interacts with GAMYB in plant nuclei and activates transcription of endosperm-specific genes during barley seed development. Plant J 42: 652–662 [DOI] [PubMed] [Google Scholar]
- Díaz-Riquelme J, Zhurov V, Rioja C, Pérez-Moreno I, Torres-Pérez R, Grimplet J, Carbonell-Bejerano P, Bajda S, Van Leeuwen T, Martínez-Zapater JM, et al. (2016) Comparative genome-wide transcriptome analysis of Vitis vinifera responses to adapted and non-adapted strains of two-spotted spider mite, Tetranychus urticae. BMC Genomics 17: 74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinant S, Clark AM, Zhu Y, Vilaine F, Palauqui JC, Kusiak C, Thompson GA (2003) Diversity of the superfamily of phloem lectins (phloem protein 2) in angiosperms. Plant Physiol 131: 114–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dworak A, Nykiel M, Walczak B, Miazek A, Szworst-Łupina D, Zagdańska B, Kiełkiewicz M (2016) Maize proteomic responses to separate or overlapping soil drought and two-spotted spider mite stresses. Planta 244: 939–960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eggermont L, Stefanowicz K, Van Damme EJM (2018) Nictaba homologs from Arabidopsis thaliana are involved in plant stress responses. Front Plant Sci 8: 2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erb M, Meldau S, Howe GA (2012) Role of phytohormones in insect-specific plant reactions. Trends Plant Sci 17: 250–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glas JJ, Alba JM, Simoni S, Villarroel CA, Stoops M, Schimmel BC, Schuurink RC, Sabelis MW, Kant MR (2014) Defense suppression benefits herbivores that have a monopoly on their feeding site but can backfire within natural communities. BMC Biol 12: 98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grbić M, Van Leeuwen T, Clark RM, Rombauts S, Rouzé P, Grbić V, Osborne EJ, Dermauw W, Ngoc PCT, Ortego F, et al. (2011) The genome of Tetranychus urticae reveals herbivorous pest adaptations. Nature 479: 487–492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Q, Major IT, Howe GA (2018) Resolution of growth-defense conflict: Mechanistic insights from jasmonate signaling. Curr Opin Plant Biol 44: 72–81 [DOI] [PubMed] [Google Scholar]
- Jonckheere W, Dermauw W, Zhurov V, Wybouw N, Van den Bulcke J, Villarroel CA, Greenhalgh R, Grbić M, Schuurink RC, Tirry L, et al. (2016) The salivary protein repertoire of the polyphagous spider mite Tetranychus urticae: A quest for effectors. Mol Cell Proteomics 15: 3594–3613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonckheere W, Dermauw W, Khalighi M, Pavlidi N, Reubens W, Baggerman G, Tirry L, Menschaert G, Kant MR, Vanholme B, et al. (2018) A gene family coding for Salivary Proteins (SHOT) of the polyphagous spider mite Tetranychus urticae exhibits fast host-dependent transcriptional plasticity. Mol Plant Microbe Interact 31: 112–124 [DOI] [PubMed] [Google Scholar]
- Karley AJ, Mitchell C, Brookes C, McNicol J, O’Neill T, Roberts H, Graham J, Johnson SN (2016) Exploiting physical defence traits for crop protection: Leaf trichomes of Rubus idaeus have deterrent effects on spider mites but not aphids. Ann Appl Biol 168: 159–172 [Google Scholar]
- Kazan K, Manners JM (2013) MYC2: The master in action. Mol Plant 6: 686–703 [DOI] [PubMed] [Google Scholar]
- Kielkiewicz M, van de Vrie M (1990) Within-leaf differences in nutritive value and defence mechanism in chrysanthemum to the two spotted spider mite (Tetranychus urticae). Exp Appl Acarol 10: 33–43 [Google Scholar]
- Kliebenstein DJ, Kroymann J, Brown P, Figuth A, Pedersen D, Gershenzon J, Mitchell-Olds T (2001) Genetic control of natural variation in Arabidopsis glucosinolate accumulation. Plant Physiol 126: 811–825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kos SP, Klinkhamer PGL, Leiss KA (2014) Cross-resistance of chrysanthemum to western flower thrips, celery leafminer, and two spotted spider mite. Entomol Exp Appl 151: 198–208 [Google Scholar]
- Lannoo N, Van Damme EJ (2010) Nucleocytoplasmic plant lectins. Biochim Biophys Acta 1800: 190–201 [DOI] [PubMed] [Google Scholar]
- Lannoo N, Van Damme EJ (2014) Lectin domains at the frontiers of plant defense. Front Plant Sci 5: 397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JR, Boltz KA, Lee SY (2014) Molecular chaperone function of Arabidopsis thaliana phloem protein 2-A1, encodes a protein similar to phloem lectin. Biochem Biophys Res Commun 443: 18–21 [DOI] [PubMed] [Google Scholar]
- Le Roux C, Huet G, Jauneau A, Camborde L, Trémousaygue D, Kraut A, Zhou B, Levaillant M, Adachi H, Yoshioka H, et al. (2015) A receptor pair with an integrated decoy converts pathogen disabling of transcription factors to immunity. Cell 161: 1074–1088 [DOI] [PubMed] [Google Scholar]
- Li R, Yu C, Li Y, Lam TW, Yiu SM, Kristiansen K, Wang J (2009) SOAP2: An improved ultrafast tool for short read alignment. Bioinformatics 25: 1966–1967 [DOI] [PubMed] [Google Scholar]
- Li Z, Zhang L, Yu Y, Quan R, Zhang Z, Zhang H, Huang R (2011) The ethylene response factor AtERF11 that is transcriptionally modulated by the bZIP transcription factor HY5 is a crucial repressor for ethylene biosynthesis in Arabidopsis. Plant J 68: 88–99 [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Llorente F, Muskett P, Sánchez-Vallet A, López G, Ramos B, Sánchez-Rodríguez C, Jordá L, Parker J, Molina A (2008) Repression of the auxin response pathway increases Arabidopsis susceptibility to necrotrophic fungi. Mol Plant 1: 496–509 [DOI] [PubMed] [Google Scholar]
- Lumba S, Toh S, Handfield LF, Swan M, Liu R, Youn JY, Cutler SR, Subramaniam R, Provart N, Moses A, et al. (2014) A mesoscale abscisic acid hormone interactome reveals a dynamic signaling landscape in Arabidopsis. Dev Cell 29: 360–372 [DOI] [PubMed] [Google Scholar]
- Martel C, Zhurov V, Navarro M, Martinez M, Cazaux M, Auger P, Migeon A, Santamaria ME, Wybouw N, Diaz I, et al. (2015) Tomato whole genome transcriptional response to Tetranychus urticae identifies divergence of spider mite-induced responses between tomato and Arabidopsis. Mol Plant Microbe Interact 28: 343–361 [DOI] [PubMed] [Google Scholar]
- Maserti BE, Del Carratore R, Croce CM, Podda A, Migheli Q, Froelicher Y, Luro F, Morillon R, Ollitrault P, Talon M, et al. (2011) Comparative analysis of proteome changes induced by the two spotted spider mite Tetranychus urticae and methyl jasmonate in citrus leaves. J Plant Physiol 168: 392–402 [DOI] [PubMed] [Google Scholar]
- Michiels K, Van Damme EJ, Smagghe G (2010) Plant-insect interactions: What can we learn from plant lectins? Arch Insect Biochem Physiol 73: 193–212 [DOI] [PubMed] [Google Scholar]
- Migeon A, Dorkeld F (2018) Spider Mites Web: A Comprehensive Database for the Tetranychidae. Institute for Agronomy Research, Center for Biology and Management of Populations, Montpellier, France.www1.montpellier.inra.fr/CBGP/spmweb. (February 25, 2019)
- Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B (2008) Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods 5: 621–628 [DOI] [PubMed] [Google Scholar]
- Nakagawa T, Kurose T, Hino T, Tanaka K, Kawamukai M, Niwa Y, Toyooka K, Matsuoka K, Jinbo T, Kimura T (2007) Development of series of gateway binary vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. J Biosci Bioeng 104: 34–41 [DOI] [PubMed] [Google Scholar]
- Nandety RS, Caplan JL, Cavanaugh K, Perroud B, Wroblewski T, Michelmore RW, Meyers BC (2013) The role of TIR-NBS and TIR-X proteins in plant basal defense responses. Plant Physiol 162: 1459–1472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naseem M, Philippi N, Hussain A, Wangorsch G, Ahmed N, Dandekar T (2012) Integrated systems view on networking by hormones in Arabidopsis immunity reveals multiple crosstalk for cytokinin. Plant Cell 24: 1793–1814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimma S, Ve T, Williams SJ, Kobe B (2017) Towards the structure of the TIR-domain signalosome. Curr Opin Struct Biol 43: 122–130 [DOI] [PubMed] [Google Scholar]
- Nishimura MT, Anderson RG, Cherkis KA, Law TF, Liu QL, Machius M, Nimchuk ZL, Yang L, Chung EH, El Kasmi F, et al. (2017) TIR-only protein RBA1 recognizes a pathogen effector to regulate cell death in Arabidopsis. Proc Natl Acad Sci USA 114: E2053–E2062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oñate-Sánchez L, Vicente-Carbajosa J (2008) DNA-free RNA isolation protocols for Arabidopsis thaliana, including seeds and siliques. BMC Res Notes 1: 93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park YL, Lee JH (2002) Leaf cell and tissue damage of cucumber caused by twospotted spider mite (Acari: Tetranychidae). J Econ Entomol 95: 952–957 [DOI] [PubMed] [Google Scholar]
- Qi L, Yan J, Li Y, Jiang H, Sun J, Chen Q, Li H, Chu J, Yan C, Sun X, et al. (2012) Arabidopsis thaliana plants differentially modulate auxin biosynthesis and transport during defense responses to the necrotrophic pathogen Alternaria brassicicola. New Phytol 195: 872–882 [DOI] [PubMed] [Google Scholar]
- Rehrig EM, Appel HM, Jones AD, Schultz JC (2014) Roles for jasmonate- and ethylene-induced transcription factors in the ability of Arabidopsis to respond differentially to damage caused by two insect herbivores. Front Plant Sci 5: 407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rioja C, Zhurov V, Bruinsma K, Grbic M, Grbic V (2017) Plant-herbivore interactions: A case of an extreme generalist, the two-spotted spider mite Tetranychus urticae. Mol Plant Microbe Interact 30: 935–945 [DOI] [PubMed] [Google Scholar]
- Roth C, Lüdke D, Klenke M, Quathamer A, Valerius O, Braus GH, Wiermer M (2017) The truncated NLR protein TIR-NBS13 is a MOS6/IMPORTIN-α3 interaction partner required for plant immunity. Plant J 92: 808–821 [DOI] [PubMed] [Google Scholar]
- Santamaria ME, Cambra I, Martinez M, Pozancos C, González-Melendi P, Grbic V, Castañera P, Ortego F, Diaz I (2012a) Gene pyramiding of peptidase inhibitors enhances plant resistance to the spider mite Tetranychus urticae. PLoS ONE 7: e43011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santamaria ME, Arnaiz A, Gonzalez-Melendi P, Martinez M, Diaz I (2018a) Plant perception and short-term responses to phytophagous insects and mites. Int J Mol Sci 19: 1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santamaria ME, Diaz I, Martinez M (2018c) Dehydration stress contributes to the enhancement of plant defense response and mite performance on barley. Front Plant Sci 9: 458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santamaría ME, Hernández-Crespo P, Ortego F, Grbic V, Grbic M, Diaz I, Martinez M (2012b) Cysteine peptidases and their inhibitors in Tetranychus urticae: A comparative genomic approach. BMC Genomics 13: 307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santamaría ME, González-Cabrera J, Martínez M, Grbic V, Castañera P, Díaz L, Ortego F (2015) Digestive proteases in bodies and faeces of the two-spotted spider mite, Tetranychus urticae. J Insect Physiol 78: 69–77 [DOI] [PubMed] [Google Scholar]
- Santamaría ME, Martinez M, Arnaiz A, Ortego F, Grbic V, Diaz I (2017) MATI, a novel protein involved in the regulation of herbivore-associated signaling pathways. Front Plant Sci 8: 975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santamaría ME, Arnaiz A, Velasco-Arroyo B, Grbic V, Diaz I, Martinez M (2018b) Arabidopsis response to the spider mite Tetranychus urticae depends on the regulation of reactive oxygen species homeostasis. Sci Rep 8: 9432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarmento RA, Lemos F, Bleeker PM, Schuurink RC, Pallini A, Oliveira MG, Lima ER, Kant M, Sabelis MW, Janssen A (2011) A herbivore that manipulates plant defence. Ecol Lett 14: 229–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarris PF, Duxbury Z, Huh SU, Ma Y, Segonzac C, Sklenar J, Derbyshire P, Cevik V, Rallapalli G, Saucet SB, et al. (2015) A plant immune receptor detects pathogen effectors that target WRKY transcription factors. Cell 161: 1089–1100 [DOI] [PubMed] [Google Scholar]
- Schäfer M, Meza-Canales ID, Navarro-Quezada A, Brütting C, Vanková R, Baldwin IT, Meldau S (2015) Cytokinin levels and signaling respond to wounding and the perception of herbivore elicitors in Nicotiana attenuata. J Integr Plant Biol 57: 198–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schouppe D, Ghesquière B, Menschaert G, De Vos WH, Bourque S, Trooskens G, Proost P, Gevaert K, Van Damme EJ (2011) Interaction of the tobacco lectin with histone proteins. Plant Physiol 155: 1091–1102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweighofer A, Kazanaviciute V, Scheikl E, Teige M, Doczi R, Hirt H, Schwanninger M, Kant M, Schuurink R, Mauch F, et al. (2007) The PP2C-type phosphatase AP2C1, which negatively regulates MPK4 and MPK6, modulates innate immunity, jasmonic acid, and ethylene levels in Arabidopsis. Plant Cell 19: 2213–2224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon PT, Grimes M, Kutlu B, Bot JJ, Galas DJ (2013) RCytoscape: Tools for exploratory network analysis. BMC Bioinformatics 14: 217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shigenaga AM, Argueso CT (2016) No hormone to rule them all: Interactions of plant hormones during the responses of plants to pathogens. Semin Cell Dev Biol 56: 174–189 [DOI] [PubMed] [Google Scholar]
- Shockey JM, Gidda SK, Chapital DC, Kuan JC, Dhanoa PK, Bland JM, Rothstein SJ, Mullen RT, Dyer JM (2006) Tung tree DGAT1 and DGAT2 have nonredundant functions in triacylglycerol biosynthesis and are localized to different subdomains of the endoplasmic reticulum. Plant Cell 18: 2294–2313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staudacher H, Schimmel BCJ, Lamers MM, Wybouw N, Groot AT, Kant MR (2017) Independent effects of a herbivore’s bacterial symbionts on its performance and induced plant defences. Int J Mol Sci 18: 182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki T, España MU, Nunes MA, Zhurov V, Dermauw W, Osakabe M, Van Leeuwen T, Grbic M, Grbic V (2017a) Protocols for the delivery of small molecules to the two-spotted spider mite, Tetranychus urticae. PLoS ONE 12: e0180658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki T, Nunes MA, España MU, Namin HH, Jin P, Bensoussan N, Zhurov V, Rahman T, De Clercq R, Hilson P, et al. (2017b) RNAi-based reverse genetics in the chelicerate model Tetranychus urticae: A comparative analysis of five methods for gene silencing. PLoS ONE 12: e0180654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarazona S, García-Alcalde F, Dopazo J, Ferrer A, Conesa A (2011) Differential expression in RNA-seq: A matter of depth. Genome Res 21: 2213–2223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vacante V. (2016) The Handbook of Mites of Economic Plants: Identification, Bio-ecology and Control. CABI International, Wallingford, UK [Google Scholar]
- Vandenborre G, Miersch O, Hause B, Smagghe G, Wasternack C, Van Damme EJ (2009) Spodoptera littoralis-induced lectin expression in tobacco. Plant Cell Physiol 50: 1142–1155 [DOI] [PubMed] [Google Scholar]
- Vandenborre G, Groten K, Smagghe G, Lannoo N, Baldwin IT, Van Damme EJ (2010) Nicotiana tabacum agglutinin is active against lepidopteran pest insects. J Exp Bot 61: 1003–1014 [DOI] [PubMed] [Google Scholar]
- Van Holle S, Smagghe G, Van Damme EJ (2016) Overexpression of Nictaba-like lectin genes from Glycine max confers tolerance toward Pseudomonas syringae infection, aphid infestation and salt stress in transgenic Arabidopsis plants. Front Plant Sci 7: 1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Leeuwen T, Dermauw W (2016) The molecular evolution of xenobiotic metabolism and resistance in Chelicerate mites. Annu Rev Entomol 61: 475–498 [DOI] [PubMed] [Google Scholar]
- Villarroel CA, Jonckheere W, Alba JM, Glas JJ, Dermauw W, Haring MA, Van Leeuwen T, Schuurink RC, Kant MR (2016) Salivary proteins of spider mites suppress defenses in Nicotiana benthamiana and promote mite reproduction. Plant J 86: 119–131 [DOI] [PubMed] [Google Scholar]
- Voinnet O, Rivas S, Mestre P, Baulcombe D (2003) An enhanced transient expression system in plants based on suppression of gene silencing by the p19 protein of tomato bushy stunt virus. Plant J 33: 949–956 [DOI] [PubMed] [Google Scholar]
- Vos IA, Verhage A, Schuurink RC, Watt LG, Pieterse CM, Van Wees SC (2013) Onset of herbivore-induced resistance in systemic tissue primed for jasmonate-dependent defenses is activated by abscisic acid. Front Plant Sci 4: 539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams SJ, Sohn KH, Wan L, Bernoux M, Sarris PF, Segonzac C, Ve T, Ma Y, Saucet SB, Ericsson DJ, et al. (2014) Structural basis for assembly and function of a heterodimeric plant immune receptor. Science 344: 299–303 [DOI] [PubMed] [Google Scholar]
- Wittstock U, Burow M (2010) Glucosinolate breakdown in Arabidopsis: Mechanism, regulation and biological significance. The Arabidopsis Book 8: e0134, [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wybouw N, Zhurov V, Martel C, Bruinsma KA, Hendrickx F, Grbić V, Van Leeuwen T (2015) Adaptation of a polyphagous herbivore to a novel host plant extensively shapes the transcriptome of herbivore and host. Mol Ecol 24: 4647–4663 [DOI] [PubMed] [Google Scholar]
- Ximénez-Embún MG, Castañera P, Ortego F (2017) Drought stress in tomato increases the performance of adapted and non-adapted strains of Tetranychus urticae. J Insect Physiol 96: 73–81 [DOI] [PubMed] [Google Scholar]
- Xu F, Kapos P, Cheng YT, Li M, Zhang Y, Li X (2014) NLR-associating transcription factor bHLH84 and its paralogs function redundantly in plant immunity. PLoS Pathog 10: e1004312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yilmaz A, Mejia-Guerra MK, Kurz K, Liang X, Welch L, Grotewold E (2011) AGRIS: The Arabidopsis Gene Regulatory Information Server, an update. Nucleic Acids Res 39: D1118–D1122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Shi H, Chen L, Wang X, Lü B, Zhang S, Liang Y, Liu R, Qian J, Sun W, et al. (2011) Harpin-induced expression and transgenic overexpression of the phloem protein gene AtPP2-A1 in Arabidopsis repress phloem feeding of the green peach aphid Myzus persicae. BMC Plant Biol 11: 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Bernoux M, Bentham AR, Newman TE, Ve T, Casey LW, Raaymakers TM, Hu J, Croll TI, Schreiber KJ, et al. (2017) Multiple functional self-association interfaces in plant TIR domains. Proc Natl Acad Sci USA 114: E2046–E2052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y, Jiao C, Sun H, Rosli HG, Pombo MA, Zhang P, Banf M, Dai X, Martin GB, Giovannoni JJ, et al. (2016) iTAK: A program for genome-wide prediction and classification of plant transcription factors, transcriptional regulators, and protein kinases. Mol Plant 9: 1667–1670 [DOI] [PubMed] [Google Scholar]
- Zhu Z, Xu F, Zhang Y, Cheng YT, Wiermer M, Li X, Zhang Y (2010) Arabidopsis resistance protein SNC1 activates immune responses through association with a transcriptional corepressor. Proc Natl Acad Sci USA 107: 13960–13965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhurov V, Navarro M, Bruinsma KA, Arbona V, Santamaria ME, Cazaux M, Wybouw N, Osborne EJ, Ens C, Rioja C, et al. (2014) Reciprocal responses in the interaction between Arabidopsis and the cell-content-feeding chelicerate herbivore spider mite. Plant Physiol 164: 384–399 [DOI] [PMC free article] [PubMed] [Google Scholar]