A chitinase from the rice blast fungus (Magnaporthe oryzae) targets a rice (Oryza sativa) lectin to suppress plant immune response.
Abstract
The genome of rice blast fungus (Magnaporthe oryzae) encodes 15 glycoside hydrolase 18 family chitinases. In this study, we characterized the function of an M. oryzae extracellular chitinase, MoChi1, and its interaction with a host protein, OsMBL1, a jacalin-related Mannose-Binding Lectin (MBL) in rice (Oryza sativa). Deletion of MoChi1 resulted in reduced aerial hyphal formation and reduced virulence in rice by activating the expression of defense-responsive genes. We confirmed MoChi1 interaction with rice OsMBL1 in vitro and in vivo. OsMBL1 was induced by pathogen-associated molecular patterns and M. oryzae infection. Overexpression of OsMBL1 led to activation of rice defense-responsive genes and a chitin-induced reactive oxygen species burst, thereby enhancing resistance to M. oryzae. Knockdown of OsMBL1 enhances susceptibility of rice plants to M. oryzae. Furthermore, MoChi1 suppressed chitin-induced reactive oxygen species in rice cells and competed with OsMBL1 for chitin binding. Taken together, our study reveals a mechanism in which MoChi1 targets a host lectin to suppress rice immunity.
Plants are continuously exposed to pathogens and have evolved defense mechanisms to protect themselves against pathogen attacks. Early events in plant defense are initiated by pathogen-associated molecular patterns (PAMPs), such as bacterial flagellin or peptidoglycan, and fungal chitin-derived oligomers (Dangl and Jones, 2001; Desaki et al., 2006). Chitin, a linear β-1,4-linked homopolymer of N-acetylglucosamine (GlcNAc), is one of the key polysaccharide components of fungal cell walls and serves as a PAMP to induce immune responses in plant cells (Kuchitsu et al., 1993, 1997; Yamada et al., 1993; Kikuyama et al., 1997; Tanabe et al., 2006; Trouvelot et al., 2014).
Plants have evolved pattern recognition receptors (PRRs) to recognize PAMPs and initiate PAMP-triggered immunity (PTI), leading to the reinforcement of cell walls, the accumulation of reactive oxygen species (ROS), and the activation of defense-related genes (Dangl and Jones, 2001; Desaki et al., 2006). In rice (Oryza sativa), the soluble chitin (GlcNAc)8-receptor chitin elicitor binding protein (CEBiP) plays a major role by recruiting the chitin elicitor receptor kinase1 (OsCERK1) in chitin-mediated PTI defense (Ito et al., 1997; Shinya et al., 2010; Kouzai et al., 2014a, 2014b). Nevertheless, pathogens secret a series of effector proteins to target PRR or some key components in the PTI pathway to suppress plant immunity. The removal of PAMPs by pathogen effectors could also be part of a virulence mechanism, since this can diminish immunity responses. For example, the bacterium Pseudomonas syringae secrets an alkaline protease AprA to cleave flagellin to block defense signaling in plants (Pel et al., 2014). Fungal core effectors, such as effector extracellular protein 6, Secreted LysM Priotein 1 (Slp1), and Colletotrichum higginsianum extracellular LysM protein 1, which contain LysM motifs, can suppress chitin-mediated immunity (de Jonge et al., 2010; Mentlak et al., 2012; Takahara et al., 2016).
Lectins, a group of proteins that encode at least one noncatalytic carbohydrate-binding site, have diverse functions in plants and animals (Chrispeels and Raikhel, 1991; Peumans and Van Damme, 1995; Vandenborre et al., 2011). Plant jacalin-related lectins (JRLs) specifically bind to Man or Gal (Peumans et al., 2001), some of which are associated with host plant innate immunity. For example, the pepper (Capsicum annuum) Mannose-Binding Lectin (MBL) CaMBL1 is involved in defense responses to microbial pathogens (Hwang and Hwang, 2011). The rice JRL OsJAC1 confers resistance against pathogens by its dirigent and jacalin domain (Weidenbach et al., 2016). Wheat (Triticum aestivum) TaJA1 (an OsJAC1 ortholog) and TaJRLL1 (jacalin-related lectin-like) are both induced by pathogen infection and lead to resistance against fungal diseases (Xiang et al., 2011; Weidenbach et al., 2016; Han et al., 2018), demonstrating the importance of JRLs in plant immunity. However, it is unclear how JRLs interact with fungal pathogens.
Chitinases are hydrolytic enzymes that catalyze the degradation of the 1,4-β bond in chitin, which subsequently leads to the release of N-acetylglucosamine oligomers (Henrissat, 1991; Bussink et al., 2007; Gortari and Hours, 2013). Glycoside hydrolase 18 (GH_18) chitinases are found in fungi and are associated with fungal development and autolysis (Sasaki et al., 2002; Duo-Chuan, 2006; Seidl, 2008; Hartl et al., 2012). Disruption of the chitinase 1 in Saccharomyces cerevisiae leads to the failure of cell separation during the cell division cycle (Kuranda and Robbins, 1991). In Aspergillus nidulans, chitinase gene ChiB is induced by carbon/energy deprivation and plays a role in hyphal autolysis. ChiB deletion mutants were defective in germination and hyphal growth (Yamazaki et al., 2007).
Rice blast, caused by the filamentous fungus M. oryzae, is one of the most devastating diseases of rice (Ou, 1980; Dean et al., 2005). Rice blast disease has become a model pathosystem to study fungal pathogen-plant interactions because of the availability of genome sequences and genetic engineering of M. oryzae and rice (Valent, 1990; Ebbole, 2007). During the interaction between these species, M. oryzae delivers effector proteins to suppress host defense. To date, more than 10 different effectors have been identified, and at least five avirulence effector proteins are known to have direct rice target(s): AvrPik, AvrPita, Avr1-CO39, AvrPiz-t, and AVR-Pii (Kanzaki et al., 2012; Park et al., 2012; Cesari et al., 2013; Fujisaki et al., 2015; Singh et al., 2016). Secreted LysM Protein1 (Slp1), a LysM effector, does not attack rice proteins directly but competes with PRR protein CEBiP for chitin to block chitin signaling (Mentlak et al., 2012). In this study, we characterized the roles of an M. oryzae chitinase, MoChi1, and its interaction with OsMBL1, a rice jacalin-related MBL. Deletion of MoChi1 in M. oryzae leads to reduced infection and was correlated with increased expression of defense-related genes in rice.
Overexpression of OsMBL1 in rice conferred resistance against M. oryzae. This response was associated with elevated expression of defense-related genes in the PTI pathway. Furthermore, both MoChi1 and OsMBL1 show affinity toward chitin and compete for chitin. MoChi1 suppresses chitin-triggered ROS burst in rice, while OsMBL1 interferes with this process by altering the response time. Therefore, we hypothesized that the interaction of OsMBL1 and MoChi1 is responsible for the induction of defense responses by rice.
RESULTS
MoChi1 Is a Highly Conserved Chitinase from M. oryzae
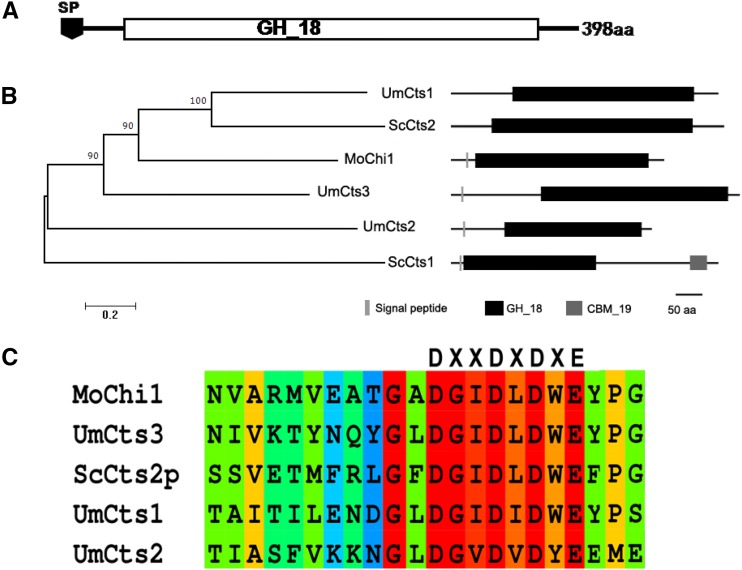
The genome of M. oryzae contains 15 genes annotated as GH_18 family chitinases (Supplemental Fig. S1). Previously, we learned that different chitinases in this family showed preferential expression in different cell types, such as vegetative hyphae, conidium, germ tube, and appressorium (Han et al., 2013). The deletion of each chitinase gene did not result in a pathogenic phenotype except for MoChi1 (Supplemental Fig. S2). MoChi1 (MGG_08054) encodes a protein with 389 amino acid residues with an N-terminal signal peptide and a GH_18 domain (Fig. 1A). Phylogenetic analysis showed that MoChi1 is orthologous to a Ustilago maydis chitinase protein, UmCts1 (Fig. 1B, top), which is known to degrade chitiooligosaccharides (GlcNAc)4 and (GlcNAc)6 into shorter chain oligomers (Langner et al., 2015). Conserved catalytic active residues DXXDXE are found in the MoChi1 coding region (Fig. 1C, bottom), suggesting that MoChi1 protein may have similar chitinolytic activity.
Figure 1.
Bioinformatic analysis of MoChi1. A, Schematic diagram of MoChi1 protein containing the GH_18 domain. B, Phylogeny of selective fungal chitinases. A bootstrap neighbor-joining phylogenetic tree was constructed based on full-length amino acid sequences of S. cerevisiae, U. maydis chitinase, and MoChi1 using MEGA6 with the default settings. The bootstrap values (%) with 1,000 repeats are indicated at the nodes. The protein organization on the right side was predicted by the Pfam database. aa, Amino acids. C, The conserved sequence with catalytic residues and active sites of S. cerevisiae chitinase 2 (ScCtS2p) and U. maydis chitinases Um10419 (UmCts1), Um06190 (UmCts2), and Um02758 (UmCts3) are provided for comparison.
MoChi1 Is an Extracellular Chitinase Released by M. oryzae
To test secretion characteristics, the open reading frame (ORF) of MoChi1 along with its 2-kb promoter fragment was amplified and fused with green fluorescent protein (GFP)-6*His tag. The MoChi1promoter:MoChi1ORF:GFP-6*His construct was transformed and expressed in M. oryzae strain Guy11. This fusion protein (70 kD) was harvested from supernatant fractions of MoChi1-GFP-6*His strain liquid cultures (Fig. 2A), indicating that MoChi1 is an extracellular protein. We also investigated the secretion feature of MoChi1 by using a yeast trap secretion assay following the method published previously (Lee et al., 2006). Two versions of plasmids were constructed by fusing with the yeast invertase (SUC2) gene lacking its own signal peptide, one containing full-length coding region (referred to as FL-MoChi1), the other containing the region without signal peptide but with start code ATG (referred to as NS-MoChi1). The secreted invertase catalyzes the hydrolysis of sucrose into glucose and fructose (Lee et al., 2006). Yeast colonies transformed with the constructs containing the signal peptide (FL-MoChi1-SUC2) grew and turned red on glucose and sucrose medium (Supplemental Fig. S3); however, the colonies carrying the nonsignal peptide fusion protein (NS-MoChi1-SUC2) did not grow well on the Suc medium, confirming that the predicted signal peptide of MoChi1 is functionally important for secretion of invertase SUC2.
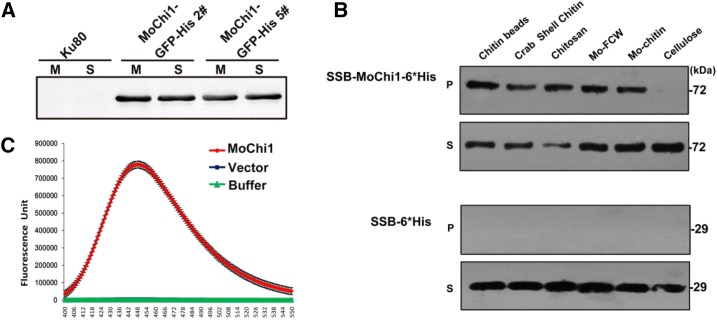
Figure 2.
Chitinolytic features of MoChi1. A, Secretion assay of MoChi1 fusion proteins from M. oryzae. M, Mycelia; S, supernatants. B, Polysaccharide affinity precipitation assay for recombinant SSB-MoChi1-6*His protein. At top, 10−3 μmol of MoChi1 coprecipitated with insoluble chitin magnetic beads, crab shell chitin, chitosan, fungal cell wall from M. oryzae (Mo-FCW), and colloidal chitin from M. oryzae (Mo-chitin) in insoluble pellet (P) and supernatant (S) fractions. At bottom, mock vector pSSBE1 coding SSB-6*His was used as the control. C, MoChi1 could degrade chitin in vitro. Short-chain fluorescent substrate MUC3 was used for chitiolytic activity. The 4-muthylumbelliferone released by MoChi1 was then detected with excitation wavelength of 360 nm and emission wavelength of 448 nm. Mock SSB-6*His protein and incubation buffer were used as controls. The enzymatic activity is presented as relative fluorescence units.
To verify whether MoChi1 has characteristics of a chitinase, we performed chitin-binding and chitinolytic activity assays. In the chitin-binding assay, after incubating with insoluble cell wall polysaccharides, SSB-MoChi1-6*His recombinant protein specifically coprecipitated with insoluble chitin beads (Chitin magnetic beads, New England Biolabs), crab shell chitin, chitosan, M. oryzae fungal cell wall, and M. oryzae chitin (Fig. 2B, top P and S). MoChi1 did not, however, precipitate with plant cell wall component cellulose and completely remained in the supernatant fraction (Fig. 2B, top, P and S). The control SSB-6*His protein was not detected in the insoluble pellets following affinity precipitation (Fig. 2B, bottom, P) and remained in the soluble fractions (Fig. 2B, bottom, S). These results demonstrated that MoChi1 specifically binds to chitin.
4-Methylumbelliferyl-β-d-N,N′,N′-triacetyl-chitotriose (MUC3) is a commercial fluorogenic substrate for chitinases. Chitinolytic activity of MoChi1 was measured as relative fluorescence units of the fluorescent 4-muthylumbelliferone enzymatically released from the nonfluorescent MUC3. Compared with the SSB-6*His protein and buffer that were used as controls, MoChi1 actively degraded MUC3 with much higher fluorescent intensity (Fig. 2C). Thus, MoChi1 is a chitinase secreted by M. oryzae, which can degrade MUC3 into shorter chains.
MoChi1 Is Involved in M. oryzae Aerial Growth
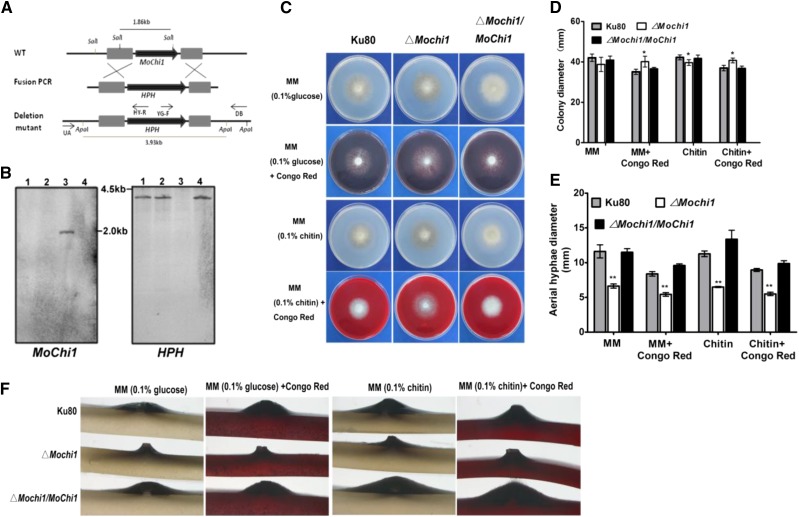
First, we checked the sublocalization of MoChi1 in M. oryzae with different stages. Most of the MoChi1 proteins accumulated around the vegetative hyphal septum (Supplemental Fig. S4). To examine the localization pattern of MoChi1 during fungal growth within rice cells, we engineered two strains that expressed the MoChi1:GFP fusion under a 1-kb promoter fragment from M. oryzae effectors Pathogenicity on Weeping Lovegrass 2 (PWL2) and Biotrophy-Associated Secreted protein 4 (BAS4; PWL2pro:MoChi1:GFP and BAS4pro:MoChi1:GFP, respectively), due to the poor fluorescence observed by its promoter. PWL2 is an identified biotrophic interfacial complex-localized effector and is known to be delivered into rice cells by M. oryzae during rice infection (Khang et al., 2010). BAS4 was confirmed to be a fungal biotrophy-associated secreted (BAS) protein and presumed to be an apoplastic effector (Khang et al., 2010). At 24 to 30 h post inoculation (hpi) in rice leaf sheath cells, fluorescence was observed to accumulate at the outline and the tip of invasive hyphae under the control of PWL2 and BAS4 promoters, as shown in Supplemental Figure S5. To investigate the potential roles of MoChi1 in M. oryzae, we replaced a 1.3-kb fragment of the MoCHI1 gene in strain Ku80 with the Hygromycin Phosphotransferase (HPH) gene (Fig. 3A). Three MoChi1 deletion mutants, ΔMochi1-1, ΔMochi1-3, and ΔMochi1-4, were verified by Southern blotting (Fig. 3B). We complemented the deletion mutants with the native promoter and ORF region of MoChi1 to generate strain ΔMochi1/MoChi1. Deletion of MoChi1 did not cause a strong defect in vegetative growth when measured by colony diameter. Growth of ΔMochi1 mutants appeared to be slightly slower (5%) on medium with glucose or chitin as the sole carbon source (Fig. 3, C and D). A discernible resistance to the cell wall perturbation agent Congo Red (Fig. 3, C and D) was observed with the ΔMochi1 deletion mutants. There was a more clear reduction in the amount of aerial hyphae produced in ΔMochi1 strains. The diameter of ΔMochi1 aerial hyphae around the plug was smaller than that of the control strains (Fig. 3, E and F). Taken together, deletion of MoChi1 in M. oryzae caused defects in aerial hyphal morphogenesis and conferred tolerance to the cell wall perturbation agent Congo Red.
Figure 3.
Cell integrity and aerial growth measurements in ΔMochi1. A, Schematic diagram of MoChi1 gene replacement. WT, Wild type. B, Southern-blot analysis of MoChi1 deletion mutants. SalI-digested genomic DNAs were hybridized with the ORF fragment of MoChi1 (left). ApaI-digested genomic DNAs were hybridized with HPH (right). Lanes 1, 2, and 4 present the MoChi1 deletion mutants ΔMochi1-1, ΔMochi1-3, and ΔMochi1-4, respectively; lane 3 presents wild-type strain Ku80. C and D, Colony growth (C) and colony diameters (D) of wild-type Ku80, ΔMochi1, and ΔMochi1/MoChi1 grown on mineral medium (MM) supplemented with the cell perturbation substance Congo Red. Glucose or colloidal chitin was added as the main carbon source. Congo Red (0.2 mg mL−1) was added to both media. The cultures were inoculated at 28°C for 10 d before being photographed or measured. Asterisks indicate significant differences against the wild type (*, P ≤ 0.05, one-way ANOVA). Means and ± sd were calculated from 10 independent replicates. E, Aerial hyphae diameter of wild-type Ku80, ΔMochi1, and ΔMochi1/MoChi1 on medium supplemented with MM-Glucose, MM-Glucose-Congo Red, MM-chitin, or MM-chitin-Congo Red. Asterisks indicate significant differences against the wild type (*, P ≤ 0.05 and **, P ≤ 0.01, one-way ANOVA). Means and ±sd were calculated from 10 independent replicates. F, The cross section for aerial appearance of Ku80, ΔMoChi1, and ΔMochi1/MoChi1 on agar plugs with different treatments.
MoChi1 Is Required for the Aggressiveness of M. oryzae
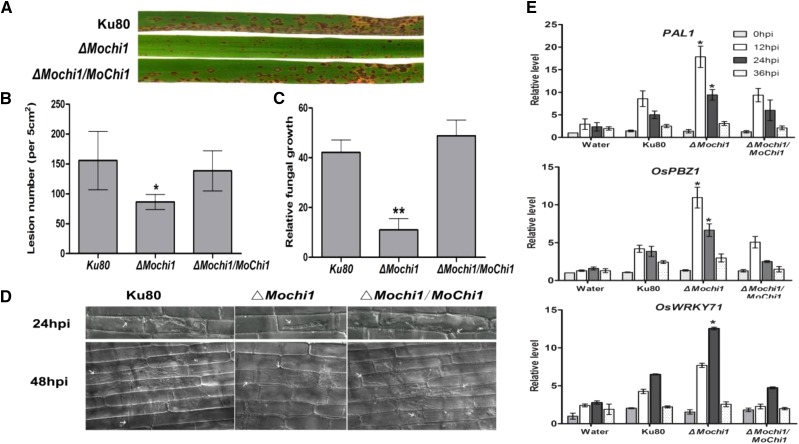
To determine whether MoChi1 contributes to pathogenicity in M. oryzae, conidia and agar plugs of wild-type Ku80, ΔMochi1, and ΔMochi1/MoChi1 strains were inoculated on susceptible rice (CO39) leaves and barley (Hordeum vulgare) leaves. The number and size of lesions on ΔMochi1-infected leaves were smaller at 5 d post inoculation than those observed in wild-type Ku80 and complementary strain infected leaves (Fig. 4, A and B; Supplemental Fig. S6). The amount of fungal biomass as determined by quantitative PCR of the M. oryzae transposable element gene Pot2 showed a 4- to 5-fold reduction in ΔMochi1 biomass relative to wild-type Ku80 and complementary strain ΔMochi1/MoChi1 inoculated rice leaves (Fig. 4C).
Figure 4.
Pathogenicity of M. oryzae is compromised in the ΔMochi1 mutant. A, Susceptible rice (CO39) was inoculated with suspension spores of M. oryzae wild type (Ku80), ΔMochi1, and complemented ΔMochi1/MoChi1 strains. B, Lesion number on rice leaves caused by M. oryzae wild type (Ku80), ΔMochi1, and ΔMochi1/MoChi1. C, Relative fungal growth as quantified by quantitative PCR using MoPot2 DNA as the target representing fungal biomass in infected rice leaves. For B and C, the ±sd of the measurements is indicated by the error bars. Statistical significance against the wild type is indicated by asterisks (*, P ≤ 0.05 and **, P ≤ 0.01, one-way ANOVA). D, Observations of infectious structures of the MoChi1 deletion mutant. The infectious hyphae were observed at 24 and 48 hpi for Ku80 wild type, ΔMochi1, and ΔMochi1/MoChi1 strains. The invasive hyphae are indicated by white arrows. E, Dynamic expression of PAL1, OsPBZ1, and OsWRKY71 in rice inoculated with M. oryzae. Leaves of 3-week-old CO39 seedlings were sprayed with the spores of Ku80, △Mochi1, and △Mochi1/MoChi1. A 0.02% (v/v) Tween 20 treatment was used as the control. The transcriptional profiles of selected genes were normalized to the Ct value of OsActin. Statistical differences against the wild-type strain at the same time points are indicated by asterisks (*, P ≤ 0.05, one-way ANOVA). The bar graphs show means ± sd from three replicates.
In order to identify how MoChi1 contributes to virulence, we examined lesion development of ΔMochi1 mutants during growth in rice tissue. Penetration into rice cells typically occurs by 24 hpi. In the ΔMochi1 strain, the infection hyphae were generally restricted to one cell (Fig. 4D, middle). At 24 hpi, the wild type and complemented mutant had similar amounts of growth in planta (Fig. 4D, left and right). At 48 hpi, the colonization of the wild type continued into adjacent cells (Fig. 4D, bottom), but in contrast, colonization of the ΔMochi1 rarely progressed further. Thus, MoChi1 appears to be important for colonization after the initial infection of the rice cell.
Subsequently, to investigate if the reduced colonization is caused by the increased levels of defense responses in the host, we examined transcriptional changes of selected defense-responsive genes in rice plants when they were inoculated with ΔMochi1. The expression of PAL1, OsPBZ1, and OsWRKY71 (Fig. 4E) was up-regulated by 2- to 3-fold in ΔMochi1-challenged rice leaves at 24 hpi, compared with the Ku80 strain.
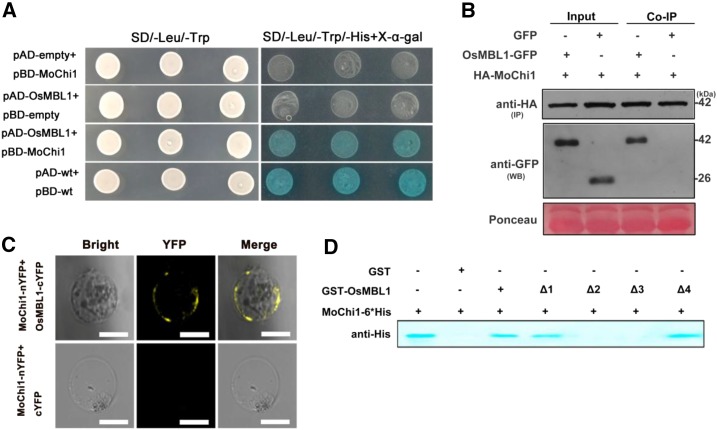
MoChi1 Interacts with the Rice JRL OsMBL1
To elucidate the molecular mechanism underlying MoChi1-mediated suppression of host immunity, we screened the potential rice targets of MoChi1 by using a yeast two-hybrid (Y2H) assay. With MoChi1 as the bait, we identified a total of 18 putative rice interacting proteins from the rice cDNA library (Supplemental Table S1). OsMBL1 showed the strongest interaction with MoChi1 in our Y2H screening; thus, we targeted this protein for further investigation. The interaction between MoChi1 and OsMBL1 was confirmed in yeast on synthetic drop-out (SD)-selective plates (SD/-Leu/-Trp/-His+X-α-gal; Fig. 5A) and validated by a coimmunoprecipitation (Co-IP) assay where Hemagglutinin tagged (HA) MoChi1 and OsMBL1-GFP were transiently expressed in Nicotiana benthamiana leaves using the agroinfiltration method. OsMBL1-GFP proteins were coimmunoprecipitated by HA-MoChi1 (Fig. 5B). We also employed the bimolecular fluorescence complementation (BiFC) assay using split yellow fluorescent protein fragments nYFP and cYFP, which were fused with MoChi1 (MoChi1-nYFP) and OsMBL1 (OsMBL1-cYFP), respectively, and transiently expressed in rice protoplast cells. We observed that the interacting MoChi1-OsMBL1 proteins appeared on cell membranes (Fig. 5C).
Figure 5.
OsMBL1 interacts with Mochi1 in vitro and in vivo. A, Y2H assay between pBD-MoChi1 and pAD-OsMBL1. Yeast cells were plated on SD/-Leu/-Trp and SD/-Leu/-Trp/-His + 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside acid (X-α-gal). pBD-wt and pAD-wt were used as positive controls by expressing wild-type fragment C of lambda cI repressor (aa 132-236). B, HA agarose bases Co-IP assay indicating interaction between HA-MoChi1 and OsMBL1-GFP in N. benthamiana leaves. Mock GFP was used as a negative control. HA-MoChi1 proteins were immunoprecipitated (IP) with anti-HA agarose beads. The GFP tagged OsMBL1 (OsMBL1-GFP) was detected by antibody anti GFP in western blotting (WB). Top and middle images represent immunoblot detection with anti-HA and anti-GFP antibodies, respectively; bottom image shows Ponceau S staining for equal loading. C, BiFC assay in rice protoplast cells. Top, Coexpression of MoChi1-nYFP with OsMBL1-cYFP; bottom, coexpression of MoChi1-nYFP with mock c-YFP. Bars = 50 μm. D, GST pull-down assay for the interaction of OsMBL1 and MoChi1. Full-length OsMBL1 (amino acids 1–145) and four truncated versions of OsMBL1 (Δ1, amino acids 20-145; Δ2, amino acids 1-80; Δ3, amino acids 20-80; and Δ4, amino acids 80-145) fusion proteins were precipitated with Glutathione-Sepharose beads and assayed for detecting the interaction with SSB-MoChi1-6*His proteins. GST was used as a negative control. An antibody against His was used to detect SSB-MoChi1-6*His fusion proteins.
To identify the region of OsMBL1 that is important for physical interaction with MoChi1, we expressed four truncated OsMBL1 proteins with a glutathione S-transferase (GST) tag and performed GST pull-down assay with MoChi1-His protein. As a positive control, we used full-length OsMBL1 to coimmunoprecipitate with MoChi1 (Fig. 5D, lane 3). The version of the OsMBL1 protein lacking 19 amino acids in the N terminus (Δ1) was able to bind to MoChi1 (Fig. 5D, lane 4); however, the version of the protein lacking the 66 amino acids in the C terminus (Δ2) failed to interact with MoChi1 (Fig. 5D, lane 5). We deleted both the N and C termini (Δ3), and no interaction was observed (Fig. 5D, lane 6). The version with only the 66-amino acid C terminus (Δ4) interacted with MoChi1 (Fig. 5D, lane 7), indicating that the C-terminal region of OsMBL1 is critical for the interaction with MoChi1.
Rice lectin Orysata, a homolog to OsMBL1, was predicted to contain four carbohydrate-binding sites (G134T135L136ID138) in its C terminus (Zhang et al., 2000), which are also found in OsMBL1 (G133T134L135ID137). When MoChi1 interacted with the C terminus of OsMBL1, we hypothesize that this motif has an important role in the MoChi1-OsMBL1 interaction. We conducted site-directed mutagenesis experiments by mutating these amino acid residues to Ala, and these mutant proteins were then detected by His tag pull-down assays. All mutants coimmunoprecipitated with MoChi1 (Supplemental Fig. S7), indicating that the interaction between MoChi1 and OsMBL1 was independent of the carbohydrate-binding motif.
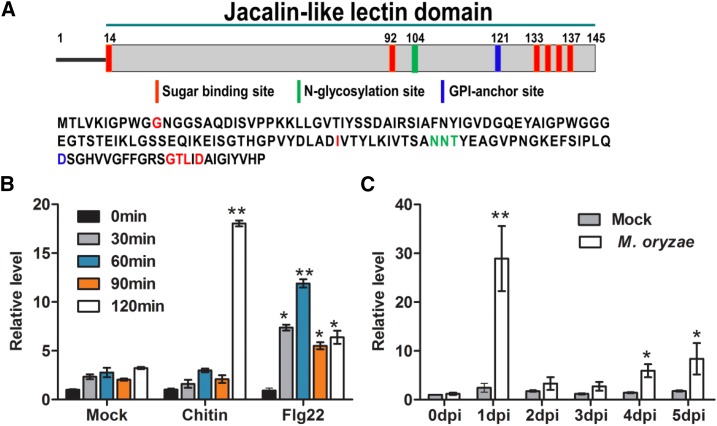
OsMBL1 Is Responsive to PAMPs and M. oryzae Infection
OsMBL1 is encoded with a jacalin-related Man-binding lectin domain with six carbohydrate sites, one glycosylation site, and one glycosylphosphatidylinositol-anchor site (Fig. 6A). To understand the roles of OsMBL1 in rice, we first studied the expression profiles of this gene against PAMPs and M. oryzae inoculation. The expression of OsMBL1 had a 6-fold increase at 2 h after chitin treatment when compared with the control (Fig. 6B). Flg22-activated OsMBL1 from as early as 30 min into treatment and as late as 2 h ranged from 2- to 5-fold (Fig. 6B). In addition, inoculation with an M. oryzae spore suspension induced a 15-fold increase in OsMBL1 expression in rice seedlings at 24 hpi (Fig. 6C).
Figure 6.
Induction of OsMBL1 toward PAMPs and M. oryzae. A, Domain schematic diagram of OsMBL1. B, Induction of OsMBL1 expression by PAMPs. Five-day-old rice seedlings were immersed in chitin (100 μg L−1) or flg22 (100 nm) solution for the indicated periods. C, Transcriptional profiles of OsMBL1 by inoculation with M. oryzae strain Guy11. The transcriptional expression of OsMBL1 was measured by quantitative PCR with normalizing to OsActin. dpi, Days post inoculation. In B and C, values represent means ± sd of three independent tests. Asterisks show statistical significance between treatment and mock at the same time point (*, P ≤ 0.05 and **, P ≤ 0.01, one-way ANOVA).
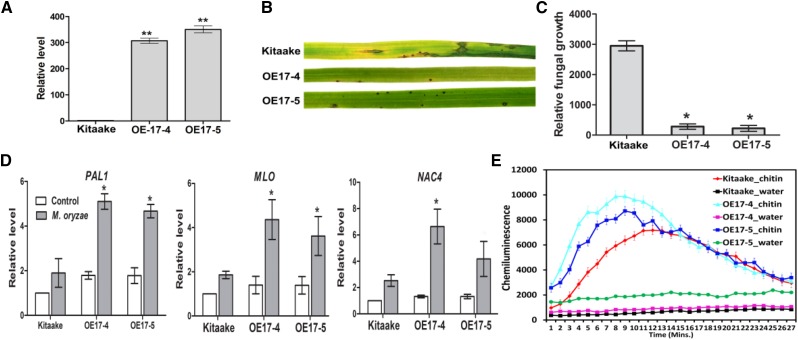
Overexpression of OsMBL1 Enhances Rice Resistance to M. oryzae
Two independent transgenic rice lines overexpressing OsMBL1 (OE17-4# and OE17-5#) were tested for OsMBL1 expression and found to have approximately 300- and 350-fold higher levels of OsMBL1 transcripts than the untransformed control line Kitaake (Fig. 7A). Overexpression of OsMBL1 activated some representative hormone-responsive defense-related genes. For example, compared with Kitaake, salicylic acid (SA) responsive genes PAL1 (2.5-fold), OsSGT1 (4.5-fold), OsPBZ1 (4-fold), and OsWRKY45 (12-fold) were transcriptionally up-regulated in OsMBL1-OE rice plants (Supplemental Fig. S8). Transcription of the jasmonic acid pathway gene OsAOS2 declined by 40% compared with the wild type (Supplemental Fig. S8). Transcription of the abscisic acid (ABA)-responsive gene OsABA45 was induced by more than 30-fold in OsMBL1-OE rice (Supplemental Fig. S8).
Figure 7.
Induction of defense-related genes in OsMBL1 overexpression rice. A, OsMBL1 transcription levels in wild-type Kitaake and OsMBL1-overexpressing rice lines. The values represent means ± sd of three independent tests. Asterisks show statistical significance between Kitaake and transgenic rice lines (**, P ≤ 0.01, one-way ANOVA). B, Inoculation of parental (Kitaake) and OsMBL1 overexpression lines with virulent M. oryzae strain Ku80. Images were taken 7 d after inoculation. C, Quantitative PCR was performed for quantification of relative fungal growth on inoculated leaves [2(Ct_OsUbiq-Ct_MoPot2) × 10,000]. The values represent means ± sd of five independent tests. Asterisk show statistical significance between Kitaake and transgenic rice lines (*, P ≤ 0.05, one-way ANOVA). D, Expression levels of selected defense-related genes in rice at 24 h after inoculation with M. oryzae. The transcriptional change was calculated by quantitative PCR and normalized to OsActin. The results represent means ± sd from three independent tests. The significant differences against Kitaake are indicated by asterisks (*, P ≤ 0.05, one-way ANOVA). E, Chitin-induced ROS burst in OsMBL1 overexpression and Kitaake plants. Rice leaf discs were treated with 8 nm chitin (GlcNAc)6 and water. ROS were detected with a luminol-chemiluminescence assay. Error bars represent the se (n = 3).
OsMBL1-OE plants exhibited enhanced resistance against M. oryzae (Fig. 7B). Measurement of fungal DNA by quantitative PCR was used to quantify fungal colonization of the host tissue. There was a 10-fold reduction in fungal colonization of the host in OsMBL1-OE plants (Fig. 7C). Three defense-related genes, PAL1 (3-fold), MLO (2-fold), and NAC4 (2- to 3-fold), were activated in OsMBL1 overexpressed rice at 24 hpi when inoculated with virulent M. oryzae (Fig. 7D). To determine the function of OsMBL1 in PTI, we measured the ROS burst in OsMBL1 transgenic rice and found that chitin-triggered ROS accumulation occurred earlier and stronger in OsMBL1 overexpression rice (OE-4 and OE-5) relative to control rice (Fig. 7E).
To validate the importance of OsMBL1 in rice innate immunity, we generated OsMBL1 RNA interference transgenic rice and used the punch inoculation method to conduct a pathogen growth assay. As expected, the lesion areas on OsMBL1-silenced rice leaves (osmbl1-3 and osmbl1-14) were larger than that on wild-type cv Nipponbare rice (Supplemental Fig. S9).
MoChi1 Competes with OsMBL1 for Chitin Binding and Suppresses Chitin-Induced ROS in Rice Cells
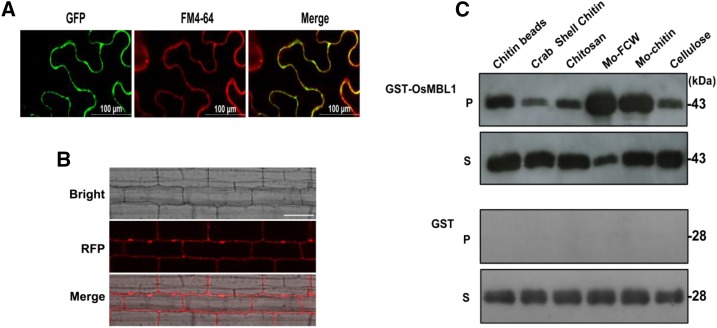
The oligosaccharides released by plant pathogens can be recognized by host PRR to initiate defense responses. The glycosylphosphatidylinositol anchor was predicted in the OsMBL1 C terminus, suggesting that this protein may associate with the plasma membrane. We hypothesized that OsMBL1 binds to oligosaccharides and plays a role in chitin recognition. To assess this idea, first we checked the subcellular localization of OsMBL1. Transient expression of OsMBL1 with GFP in N. benthamiana leaves showed that GFP-OsMBL1 accumulated at the plasma membrane (Fig. 8A). Similar results were observed with red fluorescent protein (RFP):OsMBL1 transgenic rice cells (Fig. 8B).
Figure 8.
OsMBL1 binds to chitin. A, Subcellular localization of OsMBL1-GFP transiently expressed in N. benthamiana leaves. Green fluorescence is shown in the left image (GFP). The middle image (FM4-64) shows plasma membrane stained with the membrane-binding lipophilic styryl dye FM4-64. The merged image of two fluorescence signals is shown at right. B, Subcellular localization of RFP-OsMBL1 in transgenic rice. Red fluorescence is shown in the middle image (RFP). The merged image of bright-field and red fluorescence signals is shown at bottom. C, OsMBL1 chitin-binding assay. At top, P represents GST-OsMBL1 coprecipitated with the insoluble polysaccharides chitin magnetic beads, crab shell chitin, chitosan, fungal cell wall from M. oryzae (Mo-FCW), chitin from M. oryzae (Mo-chitin), and cellulose, and S represents supernatant fractions. At bottom, for P and S, mock GST protein was used as a control and only stayed in supernatants. GST-tagged proteins were detected with immunoblotting by using anti-GST antibody.
To determine whether OsMBL1 binds to chitin, the purified GST-tagged OsMBL1 recombinant protein was incubated with different polysaccharides. OsMBL1 was pulled down with insoluble chitin beads, crab shell chitin, chitosan, fungal cell walls from M. oryzae, chitin from M. oryzae, and cellulose (Fig. 8C, top, P), showing that OsMBL1 could bind to M. oryzae cell walls and chitin as well as cellulose. The residual unbound proteins were found in the supernatants (Fig. 8C, top, S). Mock GST protein was used as a control and was only detected in the supernatant fraction after incubation (Fig. 8C, bottom, P and S).
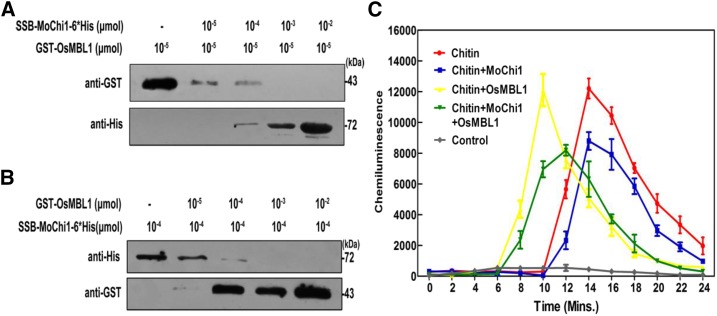
The LysM effector Slp1 from M. oryzae is a competitive inhibitor of the chitin receptor CEBiP and suppresses chitin-triggered immunity in rice (Mentlak et al., 2012). We reasoned that MoChi1 may have a similar effect toward OsMBL1 in chitin-binding activities. To test this hypothesis, a competitive binding assay was used to investigate whether MoChi1 competed with OsMBL1. Recombinant proteins SSB-MoChi1-6*His and GST-OsMBL1, 10−4 and 10−5 μmol, respectively (Supplemental Fig. S10), were used for binding to 100-μL chitin beads (Chitin magentic beads, New England Biolabs). Purified recombinant GST-OsMBL1 protein (10−5 μmol) was inoculated with titrating amounts of SSB-MoChi1-6*His proteins prior to binding to chitin beads. We observed a negative correlation between the amounts of MoChi1 added and the amount of OsMBL1 pulled down with the chitin beads (Fig. 9A), suggesting that MoChi1 competed with OsMBL1 to bind with the chitin bead. In parallel, we also introduced OsMBL1 as a competitor in the chitin pull-down assay and found that excess OsMBL1 disrupts the interaction between MoChi1 and chitin beads (Fig. 9B). These data demonstrated that MoChi1 and OsMBL1 compete with each other for chitin binding.
Figure 9.
Competition between OsMBL1 and MoChi1 in the binding of chitin. A, Purified recombinant OsMBL1 (10−5 μmol) was mixed with various amounts of MoChi1 (from 0, 10−5 to 10−2 μmol) prior to the addition of chitin beads (100 μL). B, Recombinant SSB-MoChi1-6*His proteins (10−4 μmol) were incubated with OsMBL1 in the presence of increasing amounts (from 0, 10−5 to 10−2 μmol) prior to the addition of chitin beads (100 μL for each). After further incubation, insoluble chitin beads were rinsed and collected for SDS-PAGE followed by immunoblots. Recombinant GST-OsMBL1 proteins (43 kDa) and SSB-MoChi1-6*His (73kDa) proteins were detected by anti-GST and anti-His antibodies, respectively. C, Influence of MoChi1 and OsMBL1 on chitin-triggered ROS burst in rice suspension cells. Production of ROS after induction with 8 nm (GlcNAc)6 was measured in the absence or presence of MoChi1 or OsMBL1. The experiment was performed three times with similar results. The values represent means ± se of three independent tests.
Given the ability of both MoChi1 and OsMBL1 to bind to chitin, we hypothesized that MoChi1 is involved in disrupting chitin-induced perception in the rice cell or suppresses OsMBL1 from binding chitin. We tested whether MoChi1 suppresses the chitin-induced ROS burst in rice suspension cells. In the presence of 8 nm chitin (GlcNAc)6, rice cells released ROS. Upon incubation with the SSB-MoChi1-6*His recombinant protein, we observed 28% suppression of ROS in the rice cells within 14 min after chitin treatment (Fig. 9C) compared with chitin alone. We also noticed that the GST-OsMBL1 recombinant protein caused a rapid induction of ROS in rice (Fig. 9C). Furthermore, coincubation of OsMBL1 and MoChi1 with chitin caused an earlier but lower level of induction of ROS burst (Fig. 9C), indicating that OsMBL1 may affect MoChi1-mediated suppression of ROS burst in rice cells by altering the response time.
DISCUSSION
Chitinases have been subjected to research for many decades, being involved in many biological processes. In this study, we set out to investigate the roles of chitinase in M. oryzae during the colonization in rice cells. Our results provide evidence that M. oryzae secrets the chitinase MoChi1 to suppress the chitin-induced ROS burst in rice cells and that this is an important strategy for disease development. MoChi1 is secreted by M. oryzae and accumulates on the outer layer of invasive hyphae at plant infection. The pattern of MoChi1 localization in living rice cells is different from the biotrophic interfacial complex-localized effectors (for example PWL2 and AvrPiz-t) delivered into plant cells (Khang et al., 2010), but rather is like the localization of apoplastic effectors BAS4 or Slp1, which have been reported to accumulate at the outline of invasive hyphae (Khang et al., 2010; Mentlak et al., 2012; Giraldo et al., 2013).
Being a chitinase, MoChi1 binds and hydrolyzes chitin into smaller pieces. So far, no clear evidence has shown what source of chitinase generates active chitin as a PAMP or hydrolyzes the active form of chitin into the inactive form. Some plant chitinases are reported to digest fungal cell walls for defense and act as pathogenesis-related (PR) proteins (Grover, 2012; Rawat et al., 2017). In contrast, fungal chitinase could degrade a certain degree of polymers of chitin to prevent it from being recognized by the plant. MoChi1 shares a conserved catalytic motif with U. maydis chitinases UmCts1 and UmCts2, both of which have activity toward short-chain chitooligosaccharide (GlcNAc)4 and (GlcNAc)6 (Langner et al., 2015).
Microbes produce chitinases to promote nutrient acquisition as well as growth and virulence. ChiB1, a class V chitinase from Penicillium chrysogenum, is associated with cell wall integrity and pellet formation (Kamerewerd et al., 2011). In this study, a lower density of aerial hyphae around an attached agar plug was observed for the ΔMochi1 mutant (Fig. 3F). The PcChiB1 deletion mutants and wild type strains can both grow with colloidal chitin as the only carbon source (Kamerewerd et al., 2011). Evidence reported here indicates that glycoside hydrolase_18 (GH_18) family chitinases in M. oryzae may have redundancy for nutrient acquisition, since ΔMochi1 mutants used colloidal chitin as well as Glc for growth without obvious differences.
Fungal pathogens have strategies for protection against host innate immunity during infection. The main strategy is to release fungal proteins to target PRR or key components in the plant immunity signaling pathway. In M. oryzae, the well studied effectors and rice targets are mostly found with avirulence effectors (Avr). AvrPiz-t activates immunity mediated by resistance gene Piz-t in rice but without direct physical interaction. AvrPiz-t targets several rice proteins, including E3 ligases, transcription factors, and nucleoporin, to suppress rice immunity (Park et al., 2012, 2016; Wang et al., 2016; Tang et al., 2017).Another Avr effector from rice blast fungus AVR-Pii that indirectly recognizes rice resistance gene Pii specifically inhibits nicotinamide adenine dinucleotide phosphate (NADP)-malic enzyme2 to diminish the oxidative burst in rice and establish virulence of M. oryzae (Singh et al., 2016).
Evidence in this study shows that MoChi1 has a role in rice blast disease by suppressing defense gene expression (Fig. 4) and chitin-triggered ROS production in rice. Regarding its enzymatic activity, first, MoChi1 may digest chitin and prevent it from being recognized by PRRs. Second, MoChi1 interacts with rice proteins and compromises host innate immunity. We chose the second method for further study due to the availability of a rice cDNA library. A rice JRL, OsMBL1, was screened out and shown to interact with MoChi1 (Fig. 5, A–C). OsMBL1 expression was increased in response to PAMPs and M. oryzae treatments (Fig. 6, B and C). OsMBL1-OE plants show enhanced resistance to M. oryzae by induction of defense gene expression (Fig. 7). The ROS burst plays important roles in disease resistance (Bailey-Serres and Mittler, 2006; Pitzschke et al., 2006). We found that OsMBL1-OE plants accumulated ROS rapidly and highly after chitin treatment (Fig. 7). These results suggest that OsMBL1 may positively regulate rice disease resistance against M. oryzae.
Being a lectin, OsMBL1 binds to the cell wall of M. oryzae and chitin as well as to MoChi1 (Fig. 8C). The association of OsMBL1 with carbohydrates may play roles in recognition of pathogen cell wall fragments. Previous research shows that multiple PRR proteins in rice cells are implicated in recognition of chitin. CEBiP, a major receptor of chitin (GlcNAc)8, is essential for chitin defense in rice innate immunity (Ito et al., 1997; Shinya et al., 2010; Kouzai et al., 2014a). Gene silencing of CEBiP leads to decreases in resistance to fungal disease by weakening chitin-triggered defense responses in rice (Kaku et al., 2006; Kishimoto et al., 2010). CEBiP interacts with OsCERK1 to initiate chitin-mediated signaling pathways (Shimizu et al., 2010; Kouzai et al., 2014b). LysM proteins OsLYP4 and OsLYP6 also recognize chitin (GlcNAc)6 and act as an additional machinery for chitin perception (Liu et al., 2012). In our study, one role of OsMBL1 is likely to perceive and bind chitin as OsLYP4/6 does, and this perception complex should be further investigated.
Evidence reported here indicates that MoChi1 and OsMBL1 compete for chitin. These two proteins influence chitin-triggered ROS generation in different ways. SSB-MoChi1-6*His recombinant proteins were capable of suppressing chitin-triggered ROS production in rice cells. Application of GST-OsMBL1 recombinant proteins caused rapid induction of ROS (Fig. 9C), which is consistent with the results found in OsMBL1-OE rice plants. Therefore, it is likely that the interaction of OsMBL1 and MoChi1 leads to earlier defense response in rice.
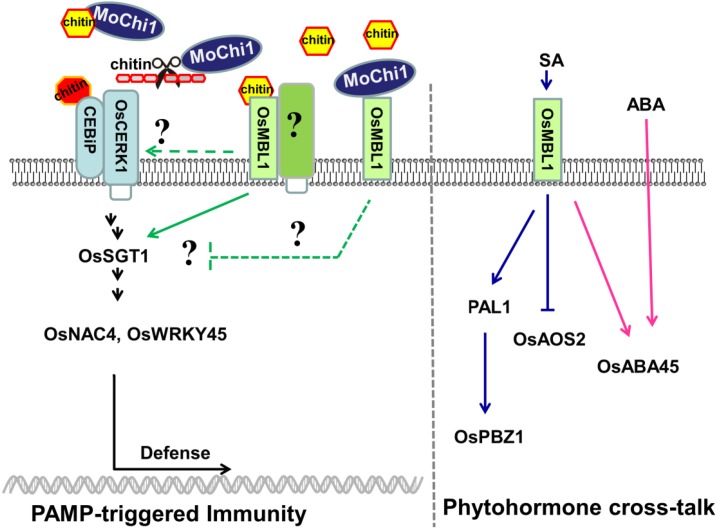
Based on the rice CEBiP-mediated PTI signaling pathway described by Kawano et al. (2014), we propose a potential model for MoChi1 and OsMBL1 during the M. oryzae-rice interaction (Fig. 10). OsMBL1, a JRL that perceives chitin, might be an additional receptor for chitin recognition. Elevated expression of OsMBL1 led to enhanced resistance against M. oryzae by activating defense-related genes and ROS burst in rice. The chitinase MoChi1 interacts with OsMBL1 and is required for full virulence by suppressing rice defense-related genes. MoChi1 could have two ways to disturb plant defense, by degrading chitin oligomers to suppress PTI or by sequestering chitin fragments from rice recognition. On the other hand, OsMBL1 interacts with MoChi1 to inhibit MoChi1 chitin binding and potentially prevents longer chitin oligomers from being hydrolyzed by MoChi1. Increasing amounts of OsMBL1 were accumulated on the plasma membrane and possibly bind more chitin oligomers to trigger PTI defense responses. Additionally, OsMBL1 was induced by plant hormones. OsMBL1 has roles in moderating SA and ABA pathways by positively regulating SA- and ABA-responsive genes, which may lead to enhanced resistance or tolerance to biotic and abiotic stresses.
Figure 10.
Roles of OsMBL1 in PTI and phytohormone pathways. MoChi1 hydrolyzes chitin and suppresses rice immunity by competing with OsMBL1 for chitin binding. OsMBL1 works upstream of the PTI pathway and could also participate in hormone cross talk in rice by positively regulating SA and ABA pathways. CEBiP, Chitin elicitor binding protein; OsABA45, ABA-responsive gene45; OsAOS2, allene oxide synthase2; OsCERK1, chitin elicitor receptor kinase1; OsNAC4, plant-specific NAC transcript factor4; OsPAL1, Phe ammonia-lyase1; OsPBZ1, probenazole-induced protein1; OsSTG1, suppressor of the G2 allele of skp1; OsWRKY45, WRKY transcript factor45.
While this article was being revised, another study was published identifying and characterizing MoChi1/MoChia1 (Yang et al., 2019). This work confirms our finding that MoChi1 is a catalytically active chitinase and that the cell wall is altered in the MoChi1 mutant. The defect in aggressiveness of MoChi1 mutants was also confirmed. The study by Yang et al. (2019) identified a rice tricopeptide-repeat protein, OsTPR1, as an interacting protein with MoChi1 protein and demonstrated that OsTRP1 was membrane localized and interacted with apoplastic MoChi1. In our model shown in Figure 10, we propose that a protein may interact with OsMBL1 to contribute to defense. It would be of some interest to determine if OsMBL1 interacts with OsTPR1. As noted, our preliminary Y2H analysis of OsMBL1 has identified some interacting partners, and additional partners, such as TPR1, may be found. Alternatively, OsMBL1 and OsTPR1 may act as independent sensors of MoChi1 to contribute to defense.
MATERIALS AND METHODS
Growth of Strains and Plants
The bacterial strains, Escherichia coli and Agrobacterium tumefaciens strain GV3101, were grown in Luria-Bertani medium at 37°C and 28°C, respectively. Magnaporthe oryzae isolates were grown in starch yeast extract medium for hyphal development at 28°C. Conidia were produced by growing cultures on rice bran medium (4% [w/v] rice polish and 1.5% [w/v] agar, pH 6) for 5 d in the dark followed by exposure to a 12-h light/dark cycle for a further 2 d at 25°C (Chen et al., 2008). To carry out plant inoculation assays, conidia were harvested from plate cultures and suspended at a concentration of 1 × 105 spores mL−1 in 0.2% (v/v) Tween 20.
Varieties of rice (Oryza sativa) were grown in a growth chamber at 28°C during the daytime and 26°C at night with 16 h of light and 8 h of darkness. The early-flowing japonica Kitaake seeds were used for transformation (Hiei and Komari, 2008) to generate transgenic plants overexpressing OsMBL1 under the control of the 35S promoter (Chen et al., 2009). Nicotiana benthamiana and barley (Hordeum vulgare ‘Golden Promise’) were grown in a growth chamber at 22°C with 16 h of light and 8 h of darkness.
Structure and Phylogenetic Analysis
Domain organization of M. oryzae GH_18 chitinases was predicted using NCBI conserved domains reserch (https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi?). The neighbor-joining tree was generated using MEGA6.0 (Tamura et al., 2013) with default parameters based on multiple alignments by ClustalW. The alignment of the chitinase proteins from M. oryzae, Saccharomyces cerevisiae, and Ustilago maydis was performed using the online alignment tool PRALINE (www.ibi.vu.nl/programs/pralinewww/). The FastTree phylogenic tree of chitinases was generated with default parameters based on multiple alignment by Mafft (Mafft-linsi-anysymbol; Liu et al., 2011).
Plasmid DNA Construction
For prokaryotic expression, the coding region of MoChi1 without the signal peptide was amplified from cDNA of M. oryzae strain Guy11 and then cloned into vector pSSBE1, which harbored a single-strain DNA-binding protein tag and a 6*His tag. OsMBL1 was constructed into the GST-tagged vector pEGX-KG as well as site-mutated fragments (A133G, A134T, A135L, A137D, 3Muts, and 4Muts). All these constructs were transformed and expressed in E. coli strain Rosetta (DE3).
For MoChi1 secretion from M. oryzae, the MoChi1 promoter and entire ORF region was amplified. The amplified fragment was fused with GFP-6*His by PCR and then cloned into the pKNT vector.
For MoChi1 cellular localization in M. oryzae vegetative hyphae, the promoter and coding region of MoChi1 was amplified and fused with GFP in the pKNTG vector. For localization at plant infection, the MoChi1 ORF-GFP region was fused with a 1-kb promoter fragment of PWL2 and BAS4.
For the yeast secretion assay, the full length of the MoChi1 coding region with or without the predicted signal peptide was cloned into the pYST2 vector.
For Co-IP assays, the coding sequences of MoChi1 and OsMBL1 were amplified and inserted into the pEarly Gate 201 and ImpGWB505 vectors, respectively, by Gateway cloning. The vectors were then transformed into A. tumefaciens GV3101 strain.
For the BiFC assay, the coding sequence of MoChi1 was cloned into pA7-nYFP, which harbors split N terminus of YFP (n-YFP). OsMBL1 was inserted into pA7-cYFP and fused with the C terminus of YFP (c-YFP).
To generate OsMBL1 overexpression rice, the coding region of OsMBL1 was cloned into the plant binary vector pCXDR (Chen et al., 2009). The OsMBL1 RNAi construct was made in pTCK303 (Wang et al., 2004). The region between 247 and 435 bp after the ATG start codon in the OsMBL1 coding sequence was cloned into the vector.
M. orzyae Secretion Assay
M. oryzae strains were grown in liquid minimal medium (6g/L NaNO3, 0.52g/L KCl, 0.52g/L MgSO4, 1.52g/L KH2PO4, 0.5% [w/v] biotin pH6.5) that contained 0.1% (m/v) crab chitin as a carbon source and incubated at 28°C and 100 rpm for 4 d. The liquid was centrifuged at 4°C and 1,500g for 10 min, and the pellets were discarded. The harvested supernatants were filtered through a 0.45-μm membrane and then concentrated by using an Amicon ultracentrifugal filter 30k device. Total proteins were precipitated by 2 volumes of cold acetone at −20°C for 2 h and centrifuged at 4°C and 10,000g for 15 min. The pellets were washed by acetone and suspended in SDS-loading buffer for western blotting.
MoChi1 Chitinase Activity
Chitinase activity was assayed as previously described (Thompson et al., 2001) using the fluorogenic substrate MUC3 (Santa Cruz) with modifications. Briefly, 300 μL of sodium phosphate buffer (200 mm, pH 6.5) was mixed with substrate and incubated at 37°C for 15 min. The reaction was started by adding 70 μL of the protein extraction containing recombinant SSB-MoChi1-6*His protein. After overnight incubation at 37°C, the reaction was terminated by adding 100 μL of 3 m NaCO3. The fluorescence of 4-methylumbelliferone released by the chitinase activity was monitored using a Spectroflurometer FS5 (Edinburgh Instruments) with an excitation wavelength of 360 nm and an emission wavelength of 448 nm.
M. oryzae Cell Wall and Chitin Preparation
M. oryzae cell wall isolation was performed as previously described (Mitchell and Taylor, 1969). Colloidal chitin was prepared as described (Kang et al., 1999) with modifications. Briefly, 5 g of ground M. oryzae cell wall was dissolved in 50 mL of cold concentrated hydrochloric acid with stirring at 4°C overnight. The chitin solution was filtered through filter paper and precipitated by adding an ice-cold 15% (v/v) ethanol-water solution. The chitin colloidal suspension was stirred at 4°C overnight and collected by centrifugation (5,000g for 5 min at room temperature). Chitin pellets were then rinsed with distilled water until the pH of the suspension was 7.
Chitin-Binding Assay
This assay was performed as described (de Jonge et al., 2010; Liu et al., 2012) with modifications. In brief, insoluble chitin polysaccharides (commercial chitin beads, 20 mg of crab shell chitin, chitosan, M. oryzae cell wall, M. oryzae chitin, and cellulose) were incubated with MoChi1 or OsMBL1 recombinant proteins in binding buffer (20 mm Tris-HCl, pH 8, 1 mm EDTA, 500 mm NaCl, 0.05% [v/v] Triton X-100, 2 mm DTT, 1 mm phenylmethylsulfonyl fluoride [PMSF], and 50 mg mL−1 Roche complete protease inhibitor cocktail [Roche Diagnostics]) at 4°C overnight with end-over-end rotation. The protein tag SSB-His and GST were used as mock controls. For the competition assay, OsMBL1 or MoChi1 recombinant proteins were incubated with the competitor prior to chitin beads. The insoluble pellets were washed five times with binding buffer and then harvested for SDS-PAGE followed by western blotting. The presence of selective recombinant proteins in pellets or supernatant fractions was detected by anti-His or anti-GST antibodies (Abmart).
Y2H Assay
The HybriZAP-2.1 two-hybrid libraries system (Agilent) was used to screen MoChi1-interacting rice proteins. The coding region of MoChi1 without the signal peptide was cloned unframed into the bait vector pBD-GAL4 Cam by SacI/KpnI sites. The constructed pBD-MoChi1 plasmid DNA and pAD-GAL4 rice library were cotransformed into yeast strain YGR2. Candidate clones were grown on the SD/-His/-Leu/-Trp medium, which was supplemented with 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside acid (X-α-gal) for β-galactosidase assays for further confirmation according to the manufacturer’s protocol (Agilent).
Pull-Down Assays
Pull-down assays were performed following the instructions from the manufacturer. In the GST-pull down assay, E. coli strain Rosetta (DE3) carrying OsMBL1-GST was grown in a shaking incubator at 37°C and 200 rpm to OD600 = 0.6 before isopropyl β-d-thiogalactoside treatment. The proteins were then induced with 0.1 mm isopropyl β-d-thiogalactoside and shaken at 16°C and 200 rpm for 14 h. The cells were collected and resuspended with lysis buffer (20 mm KH2PO4-K2HPO4 and 300 mm NaCl supplemented with 0.1 mm PMSF and 50 mg mL−1 Roche complete protease inhibitor cocktail) and sonicated on ice. The mixture was centrifuged at 14,000g at 4°C for 45 min. The supernatant was then transferred and incubated with Glutathione-Sepharose beads (GE Life) at 4°C overnight and washed with lysis buffer. The supernatant containing expressed SSB-MoChi1-6*His protein was added to GST-OsMBL1-precipitated Glutathione-Sepharose 4B beads and incubated at 4°C for another 4 h with gentle end-over-end mixing. After five to six washes with lysis buffer, the beads were collected for SDS-PAGE and western-blotting analysis. SSB-MoChi1-6*His recombinant proteins were detected by anti-His antibody.
In the His tag pull-down assay, total proteins containing SSB-MoChi1-6*His were incubated with Ni-NTA beads (Qiagen) at 4°C with end-over-end mixing for 6 to 8 h. After five to six washes with lysis buffer, Ni-NTA beads bound with MoChi1-His were further incubated with GST-OsMBL1 extracts for another 6 to 8 h at 4°C. The beads were washed with lysis buffer and subsequently collected for SDS-PAGE and western blotting. MoChi1 and OsMBL1 recombinant proteins were detected with anti-His and anti-GST antibodies, respectively.
MoChi1 Gene Replacement and Complementation
The MoChi1 gene was replaced by split marker HPH as described (Chen et al., 2008). An 800-bp fragment upstream of the MoChi1 ORF was amplified and fused with an N-terminal HPH fragment. Then, a 927-bp fragment downstream of the MoChi1 ORF was amplified and fused with a C-terminal HPH fragment. These PCR fragments were then transformed into protoplast cells of wild-type strain Ku80. Hygromycin B-resistant colonies were picked up and replacement verified by PCR amplifications.
The complementation of MoChi1 was performed by constructing a 1-kb upstream fragment of the native promoter and MoChi1 ORF region into the pKNTG vector and then transformed into protoplast cells of the MoChi1 deletion strain. The neomycin-resistant transformants were screened and verified by PCR amplifications.
PAMPs and M. oryzae Inoculation
Rice seeds were surface sterilized and germinated on one-half-strength Murashige and Skoog medium at 25°C under 16-h-light/8-h-darkness conditions for 5 d. PAMP treatment was performed as previously described (Tanabe et al., 2006; Park et al., 2012) with modifications. Briefly, the 5-d-old rice seedlings were immersed in N-acetylchitohexaose (100 μg mL−1) and flg22 (100 nm). Sterilized water was used as a control. Rice leaves were collected at desired time points for quantitative PCR.
Conidia of M. oryzae strains were sprayed onto 3-week old rice seedlings. Rice lines (CO39 and Kitaake and OsMBL1 overexpression T2 lines were used for pathogenicity assays and for observation of infection with Guy11 (Ku80; Villalba et al., 2008) and ρMochi1 strains. The disease symptoms were allowed to develop for 6 or 7 d (Talbot et al., 1993). For each of the three assays, 30 leaves were harvested for DNA extraction. Three infection assays were performed, and DNA was isolated from infected leaves. The quantification of fungal biomass was determined by quantitative PCR (Park et al., 2012).
A. tumefaciens-Mediated Transient Expression in N. benthamiana
A. tumefaciens-mediated transient expression was performed as described (Ma et al., 2012) with modifications. Briefly, GV3101 strains carrying different constructs were grown in Luria-Bertani broth and collected by centrifugation at 2,500g and 25°C for 5 min. The pellets were then resuspended with MMA buffer (Murashige and Skoog medium without Gamborg B5 vitamins, 10 mm MES, 20% [w/v] sucrose, 10 mm MgCl2, and 200 μm acetosyringone, pH 5.7) to a final culture density of OD600 of 1. For coexpression, we used 1:1 for all combinations. The suspended culture was incubated for 2 to 4 h at 28°C before agroinfiltration into the leaves of N. benthamiana. The leaves of N. benthamiana were sprayed with water to keep moist prior to infiltration. The infiltrated plants were kept at 20°C to 25°C and shielded from direct light.
Co-IP Assay
Leaves of N. benthamiana were coinfiltrated with A. tumefaciens carrying MoChi1-HA and OsMBL1-GFP. The infiltrated leaves were harvested after 2 d and then ground to powder in liquid nitrogen. The powder was suspended in lysis buffer (50 mm Tris-MES, pH 8, 10 mm EDTA, 500 mm sucrose, 1 mm MgCl2, 5 mm DTT, 1 mm PMSF, and 50 mg mL−1 Roche complete protease inhibitor cocktail [Roche Diagnostics]). Anti-HA agarose bead suspension (Thermo) was added to the protein supernatants. The mixture was incubated at 4°C for 8 h with constant end-over-end rotation. Then the beads were rinsed with washing buffer (50 mm Tris-MES, pH 8, 10 mm EDTA, 500 mm sucrose, 1 mm MgCl2, and 5 mm DTT) until the A280 of the supernatant fraction was less than 0.01. Anti-HA agarose mixtures were collected for western blotting following SDS-PAGE. HA-MoChi1 and OsMBL1-GFP fusion proteins were detected by anti-HA and anti-GFP antibodies, respectively, according to the manufacturer’s instruction (Abmart).
BiFC Assay and Microscopy
Constructed vectors MoChi1-nYFP and OsMBL1-cYFP were cotransformed into rice protoplast cells as described (Chen et al., 2006). For OsMBL1-GFP subcellular localization, A. tumefaciens carrying ImpGWB505-OsMBL1 was infiltrated onto N. benthamiana leaves. Before microscopy, the infiltrated patches were stained with FM4-64 (4 μg mL−1). For localization in transgenic rice, the inner epidermal layer of leaf sheaths was peeled from RFP-OsMBL1 rice seedlings. Live cell imaging was performed by a Nikon A1 confocal microscope equipped with NIS-Elements Imaging Software. YFP was detected with excitation at 488 nm and emission at 515 nm. OsMBL1-GFP protein was visualized with excitation at 488 nm and emission at 525/40 nm. RFP-OsMBL1 and FM4-64 were detected with excitation at 561 nm and emission at 607/36 nm.
Quantitative PCR
Inoculated rice leaves were harvested at different time points and ground in liquid nitrogen for RNA extraction with TRIzol (Invitrogen). The first strand of cDNA was synthesized and used for quantitative PCR using an Eppendorf real-time PCR detection system (Eppendorf Master cycle rep Real plex). Primer sequences used in this study are listed in Supplemental Table S2.
ROS Measurements
For measurement of ROS burst in rice suspension cells, calli produced from mature seeds of rice cv Nipponbare were used to establish suspension cells as described (Ozawa and Komamine, 1989; Deshpande et al., 2012). Briefly, for callus induction, sterile seeds were cultivated on N6D solid medium containing 2 mg L−1 2,4-dichlorophenoxyacetic acid at 28°C under darkness for 30 d followed by their transfer into liquid N6D medium. The fragile calli were allowed to grow in a rotary shaker at 25°C and 150 rpm for 3 to 4 months with weekly subculture. Fine calli were filtered by medical gauze and harvested by centrifugation at a low speed for 10 min. The culture was diluted with a new fresh medium at 200 mg mL−1 and incubated at 150 rpm and 25°C for a further 2 to 3 h.
ROS detection was performed as described (Schwacke and Hager, 1992; Park et al., 2012) with modifications. Aliquots (200 μL) of the rice cell suspension were placed onto a 96-well ELISA PLATE (Costar 96 3590) and preincubated for 20 min before assays. Reaction mixtures (50 mm K3PO4, 17 mm luminol, and 10 mg mL−1 horseradish peroxidase) were mixed with the suspension cells for each treatment. Prior to the addition to rice cells, equivalent molar concentrations of each recombinant protein (6 μg of MoChi1, 3.528 μg of OsMBL1, and 2.184 μg of MBP) were incubated and equilibrated with 8 nm chitin (hexa-N-acetyl-chitohexaose, [GlcNAC]6) in buffer (50 mm Tris-HCl, pH 7, and 100 mm NaCl) for 2 h with constant shaking. The luminescence was measured continuously for 20 min with a Centro XS3 LB 960 (Berthold Technologies). Water was used as a control.
Measurement of the ROS burst in rice leaf discs was performed as described (Park et al., 2012) with modifications. Leaf discs from 4-week-old rice plants were cut with a mouse ear punch and preincubated in sterile distilled water overnight. Three leaf discs for each sample were placed on a 96-well ELISA PLATE (Costar 96 3590) containing 100 μm luminol, 5 μg mL−1 horseradish peroxidase, and 8 nm (GlcNAC)6, or water as a control. After treatment, ROS production in rice tissues was immediately monitored by luminescence assay with Thermo Scientific Varioskan Flash Microplate Readers. Three replications were performed for each sample and treatment. se values were calculated for each treatment.
Accession Numbers
The sequences of rice genes used in this article can be found in the Rice Genome Project Web site as follows: OsActin (LOC_Os03g50885), OsMBL1 (LOC_Os01g24710), PAL1 (LOC_Os02g41630), OsNAC4 (LOC_Os07g04560), OsWRKY45 (LOC_Os05g25770), OsWRKY71 (LOC_Os02g08440), OsSGT1 (LOC_Os01g43540), OsUbqi (LOC_Os03g13170), and OsPBZ1 (LOC_Os12g36880.1). The sequences of M. oryzae chitinases can be found in GenBank as follows: MoChi1 (MGG_08054), MGG_00086, MGG_07927, MGG_05533, MGG_01247, MGG_11231, MGG_03599, MGG_01876, MGG_04073, MGG_04534, MGG_10336, MGG_01336, MGG_17153, and MGG_08458.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Phylogeny of the M. oryzae GH_18 family.
Supplemental Figure S2. Pathogenicity of M. oryzae GH_18 chitinase mutants on plants.
Supplemental Figure S3. Secretory feature of MoChi1 by yeast trap analysis.
Supplemental Figure S4. Sublocalization of MoChi1 in M. oryzae.
Supplemental Figure S5. Sublocalization of MoChi1 in M. oryzae at plant infection.
Supplemental Figure S6. ΔMochi1 pathogenicity on barley.
Supplemental Figure S7. Carbohydrate-binding sites in the OsMBL1 C terminus are not required for interaction with MoChi1.
Supplemental Figure S8. Activation of representative defense-related genes in OsMBL1-OE rice.
Supplemental Figure S9. RNA silencing of OsMBL1 enhanced susceptibility to M. oryzae.
Supplemental Figure S10. Chitin-binding kinetic analysis of OsMBL1 and MoChi1.
Supplemental Table S1. List of the MoChi1-interacting rice proteins by Y2H assay.
Supplemental Table S2. Primer sequences used in this article.
Footnotes
This work was supported by the Ministry of Science and Technology of the People’s Republic of China (Chinese Ministry of Science and Technology) (2016YFD0100600) and the National Natural Science Foundation of China (NSFC) (31071654) to G.L.
References
- Bailey-Serres J, Mittler R (2006) The roles of reactive oxygen species in plant cells. Plant Physiol 141: 311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bussink AP, Speijer D, Aerts JM, Boot RG (2007) Evolution of mammalian chitinase(-like) members of family 18 glycosyl hydrolases. Genetics 177: 959–970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cesari S, Thilliez G, Ribot C, Chalvon V, Michel C, Jauneau A, Rivas S, Alaux L, Kanzaki H, Okuyama Y, et al. (2013) The rice resistance protein pair RGA4/RGA5 recognizes the Magnaporthe oryzae effectors AVR-Pia and AVR1-CO39 by direct binding. Plant Cell 25: 1463–1481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Zheng W, Zheng S, Zhang D, Sang W, Chen X, Li G, Lu G, Wang Z (2008) Rac1 is required for pathogenicity and Chm1-dependent conidiogenesis in rice fungal pathogen Magnaporthe grisea. PLoS Pathog 4: e1000202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Tao L, Zeng L, Vega-Sanchez ME, Umemura K, Wang GL (2006) A highly efficient transient protoplast system for analyzing defence gene expression and protein-protein interactions in rice. Mol Plant Pathol 7: 417–427 [DOI] [PubMed] [Google Scholar]
- Chen S, Songkumarn P, Liu J, Wang GL (2009) A versatile zero background T-vector system for gene cloning and functional genomics. Plant Physiol 150: 1111–1121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chrispeels MJ, Raikhel NV (1991) Lectins, lectin genes, and their role in plant defense. Plant Cell 3: 1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dangl JL, Jones JD (2001) Plant pathogens and integrated defence responses to infection. Nature 411: 826–833 [DOI] [PubMed] [Google Scholar]
- Dean RA, Talbot NJ, Ebbole DJ, Farman ML, Mitchell TK, Orbach MJ, Thon M, Kulkarni R, Xu JR, Pan H, et al. (2005) The genome sequence of the rice blast fungus Magnaporthe grisea. Nature 434: 980–986 [DOI] [PubMed] [Google Scholar]
- de Jonge R, van Esse HP, Kombrink A, Shinya T, Desaki Y, Bours R, van der Krol S, Shibuya N, Joosten MH, Thomma BP (2010) Conserved fungal LysM effector Ecp6 prevents chitin-triggered immunity in plants. Science 329: 953–955 [DOI] [PubMed] [Google Scholar]
- Desaki Y, Miya A, Venkatesh B, Tsuyumu S, Yamane H, Kaku H, Minami E, Shibuya N (2006) Bacterial lipopolysaccharides induce defense responses associated with programmed cell death in rice cells. Plant Cell Physiol 47: 1530–1540 [DOI] [PubMed] [Google Scholar]
- Deshpande A, Dhadi SR, Hager EJ, Ramakrishna W (2012) Anticancer activity of rice callus suspension culture. Phytother Res 26: 1075–1081 [DOI] [PubMed] [Google Scholar]
- Duo-Chuan L. (2006) Review of fungal chitinases. Mycopathologia 161: 345–360 [DOI] [PubMed] [Google Scholar]
- Ebbole DJ. (2007) Magnaporthe as a model for understanding host-pathogen interactions. Annu Rev Phytopathol 45: 437–456 [DOI] [PubMed] [Google Scholar]
- Fujisaki K, Abe Y, Ito A, Saitoh H, Yoshida K, Kanzaki H, Kanzaki E, Utsushi H, Yamashita T, Kamoun S, et al. (2015) Rice Exo70 interacts with a fungal effector, AVR-Pii, and is required for AVR-Pii-triggered immunity. Plant J 83: 875–887 [DOI] [PubMed] [Google Scholar]
- Giraldo MC, Dagdas YF, Gupta YK, Mentlak TA, Yi M, Martinez-Rocha AL, Saitoh H, Terauchi R, Talbot NJ, Valent B (2013) Two distinct secretion systems facilitate tissue invasion by the rice blast fungus Magnaporthe oryzae. Nat Commun 4: 1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gortari MC, Hours RA (2013) Biotechnological processes for chitin recovery out of crustacean waste: a Mini-review. Electron J Biotechnol 16: 14 [Google Scholar]
- Grover A. (2012) Plant chitinases: Genetic diversity and physiological roles. Crit Rev Plant Sci 31: 57–63 [Google Scholar]
- Han Y, Lin C, Wang Q, Lu G, Wang Z, Zhou J (2013) Expression investigation of the putative chitinase family genes in Magnaporthe oryzae. Redai Zuowu Xuebao 8: 1544–1551 [Google Scholar]
- Han Y, Zhong Z, Song L, Olsson S, Wang Z, Lu GD (2018) Evolutionary analysis of plant jacalin-related lectins (JRLs) family and expression of rice JRLs in response to Magnaporthe oryzae. J Integr Agric 17: 1252–1266 [Google Scholar]
- Hartl L, Zach S, Seidl-Seiboth V (2012) Fungal chitinases: Diversity, mechanistic properties and biotechnological potential. Appl Microbiol Biotechnol 93: 533–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henrissat B. (1991) A classification of glycosyl hydrolases based on amino acid sequence similarities. Biochem J 280: 309–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiei Y, Komari T (2008) Agrobacterium-mediated transformation of rice using immature embryos or calli induced from mature seed. Nat Protoc 3: 824–834 [DOI] [PubMed] [Google Scholar]
- Hwang IS, Hwang BK (2011) The pepper mannose-binding lectin gene CaMBL1 is required to regulate cell death and defense responses to microbial pathogens. Plant Physiol 155: 447–463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito Y, Kaku H, Shibuya N (1997) Identification of a high-affinity binding protein for N-acetylchitooligosaccharide elicitor in the plasma membrane of suspension-cultured rice cells by affinity labeling. Plant J 12: 347–356 [DOI] [PubMed] [Google Scholar]
- Kaku H, Nishizawa Y, Ishii-Minami N, Akimoto-Tomiyama C, Dohmae N, Takio K, Minami E, Shibuya N (2006) Plant cells recognize chitin fragments for defense signaling through a plasma membrane receptor. Proc Natl Acad Sci USA 103: 11086–11091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamerewerd J, Zadra I, Kürnsteiner H, Kück U (2011) PcchiB1, encoding a class V chitinase, is affected by PcVelA and PcLaeA, and is responsible for cell wall integrity in Penicillium chrysogenum. Microbiology 157: 3036–3048 [DOI] [PubMed] [Google Scholar]
- Kang SC, Park S, Lee DG (1999) Purification and characterization of a novel chitinase from the entomopathogenic fungus, Metarhizium anisopliae. J Invertebr Pathol 73: 276–281 [DOI] [PubMed] [Google Scholar]
- Kanzaki H, Yoshida K, Saitoh H, Fujisaki K, Hirabuchi A, Alaux L, Fournier E, Tharreau D, Terauchi R (2012) Arms race co-evolution of Magnaporthe oryzae AVR-Pik and rice Pik genes driven by their physical interactions. Plant J 72: 894–907 [DOI] [PubMed] [Google Scholar]
- Kawano Y, Kaneko-Kawano T, Shimamoto K (2014) Rho family GTPase-dependent immunity in plants and animals. Front Plant Sci 5: 522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khang CH, Berruyer R, Giraldo MC, Kankanala P, Park SY, Czymmek K, Kang S, Valent B (2010) Translocation of Magnaporthe oryzae effectors into rice cells and their subsequent cell-to-cell movement. Plant Cell 22: 1388–1403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuyama M, Kuchitsu K, Shibuya N (1997) Membrane depolarization induced by N-acetylchitooligosaccharide elicitor in suspension-cultured rice cells. Plant Cell Physiol 38: 902–909 [Google Scholar]
- Kishimoto K, Kouzai Y, Kaku H, Shibuya N, Minami E, Nishizawa Y (2010) Perception of the chitin oligosaccharides contributes to disease resistance to blast fungus Magnaporthe oryzae in rice. Plant J 64: 343–354 [DOI] [PubMed] [Google Scholar]
- Kouzai Y, Mochizuki S, Nakajima K, Desaki Y, Hayafune M, Miyazaki H, Yokotani N, Ozawa K, Minami E, Kaku H, et al. (2014a) Targeted gene disruption of OsCERK1 reveals its indispensable role in chitin perception and involvement in the peptidoglycan response and immunity in rice. Mol Plant Microbe Interact 27: 975–982 [DOI] [PubMed] [Google Scholar]
- Kouzai Y, Nakajima K, Hayafune M, Ozawa K, Kaku H, Shibuya N, Minami E, Nishizawa Y (2014b) CEBiP is the major chitin oligomer-binding protein in rice and plays a main role in the perception of chitin oligomers. Plant Mol Biol 84: 519–528 [DOI] [PubMed] [Google Scholar]
- Kuchitsu K, Kikuyama M, Shibuya N (1993) N-Acetylchitooligosaccharides, biotic elicitor for phytoalexin production, induce transient membrane depolarization in suspension-cultured rice cells. Protoplasma 174: 79–81 [Google Scholar]
- Kuchitsu K, Yazaki Y, Sakano K, Shibuya N (1997) Transient cytoplasmic pH change and ion fluxes through the plasma membrane in suspension-cultured rice cells triggered by N-acetylchitooligosaccharide elicitor. Plant Cell Physiol 38: 1012–1018 [Google Scholar]
- Kuranda MJ, Robbins PW (1991) Chitinase is required for cell separation during growth of Saccharomyces cerevisiae. J Biol Chem 266: 19758–19767 [PubMed] [Google Scholar]
- Langner T, Öztürk M, Hartmann S, Cord-Landwehr S, Moerschbacher B, Walton JD, Göhre V (2015) Chitinases are essential for cell separation in Ustilago maydis. Eukaryot Cell 14: 846–857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SJ, Kelley BS, Damasceno CM, St John B, Kim BS, Kim BD, Rose JK (2006) A functional screen to characterize the secretomes of eukaryotic pathogens and their hosts in planta. Mol Plant Microbe Interact 19: 1368–1377 [DOI] [PubMed] [Google Scholar]
- Liu B, Li JF, Ao Y, Qu J, Li Z, Su J, Zhang Y, Liu J, Feng D, Qi K, et al. (2012) Lysin motif-containing proteins LYP4 and LYP6 play dual roles in peptidoglycan and chitin perception in rice innate immunity. Plant Cell 24: 3406–3419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu K, Linder CR, Warnow T (2011) RAxML and FastTree: Comparing two methods for large-scale maximum likelihood phylogeny estimation. PLoS ONE 6: e27731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L, Lukasik E, Gawehns F, Takken FL (2012) The use of agroinfiltration for transient expression of plant resistance and fungal effector proteins in Nicotiana benthamiana leaves. Methods Mol Biol 835: 61–74 [DOI] [PubMed] [Google Scholar]
- Mentlak TA, Kombrink A, Shinya T, Ryder LS, Otomo I, Saitoh H, Terauchi R, Nishizawa Y, Shibuya N, Thomma BP, et al. (2012) Effector-mediated suppression of chitin-triggered immunity by Magnaporthe oryzae is necessary for rice blast disease. Plant Cell 24: 322–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell AD, Taylor IE (1969) Cell-wall proteins of Aspergillus niger and Chaetomium globosum. J Gen Microbiol 59: 103–109 [DOI] [PubMed] [Google Scholar]
- Ou SH. (1980) Pathogen variability and host resistance in rice blast disease. Annu Rev Phytopathol 18: 167–187 [Google Scholar]
- Ozawa K, Komamine A (1989) Establishment of a system of high-frequency embryogenesis from long-term cell suspension cultures of rice (Oryza sativa L.). Theor Appl Genet 77: 205–211 [DOI] [PubMed] [Google Scholar]
- Park CH, Chen S, Shirsekar G, Zhou B, Khang CH, Songkumarn P, Afzal AJ, Ning Y, Wang R, Bellizzi M, et al. (2012) The Magnaporthe oryzae effector AvrPiz-t targets the RING E3 ubiquitin ligase APIP6 to suppress pathogen-associated molecular pattern-triggered immunity in rice. Plant Cell 24: 4748–4762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park CH, Shirsekar G, Bellizzi M, Chen S, Songkumarn P, Xie X, Shi X, Ning Y, Zhou B, Suttiviriya P, et al. (2016) The E3 ligase APIP10 connects the effector AvrPiz-t to the NLR receptor Piz-t in rice. PLoS Pathog 12: e1005529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pel MJ, van Dijken AJ, Bardoel BW, Seidl MF, van der Ent S, van Strijp JA, Pieterse CM (2014) Pseudomonas syringae evades host immunity by degrading flagellin monomers with alkaline protease AprA. Mol Plant Microbe Interact 27: 603–610 [DOI] [PubMed] [Google Scholar]
- Peumans WJ, Van Damme EJ (1995) Lectins as plant defense proteins. Plant Physiol 109: 347–352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peumans WJ, Van Damme EJ, Barre A, Rougé P (2001) Classification of plant lectins in families of structurally and evolutionary related proteins. Adv Exp Med Biol 491: 27–54 [DOI] [PubMed] [Google Scholar]
- Pitzschke A, Forzani C, Hirt H (2006) Reactive oxygen species signaling in plants. Antioxid Redox Signal 8: 1757–1764 [DOI] [PubMed] [Google Scholar]
- Rawat S, Ali S, Mittra B, Grover A (2017) Expression analysis of chitinase upon challenge inoculation to Alternaria wounding and defense inducers in Brassica juncea. Biotechnol Rep (Amst) 13: 72–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki C, Yokoyama A, Itoh Y, Hashimoto M, Watanabe T, Fukamizo T (2002) Comparative study of the reaction mechanism of family 18 chitinases from plants and microbes. J Biochem 131: 557–564 [DOI] [PubMed] [Google Scholar]
- Schwacke R, Hager A (1992) Fungal elicitors induce a transient release of active oxygen species from cultured spruce cells that is dependent on Ca2+ and protein-kinase activity. Planta 187: 136–141 [DOI] [PubMed] [Google Scholar]
- Seidl V. (2008) Chitinase of fiamententous fungi: A large group of diverse proteins with multiple physiological functions. Fungal Biol Rev 22: 36–42 [Google Scholar]
- Shimizu T, Nakano T, Takamizawa D, Desaki Y, Ishii-Minami N, Nishizawa Y, Minami E, Okada K, Yamane H, Kaku H, et al. (2010) Two LysM receptor molecules, CEBiP and OsCERK1, cooperatively regulate chitin elicitor signaling in rice. Plant J 64: 204–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinya T, Osada T, Desaki Y, Hatamoto M, Yamanaka Y, Hirano H, Takai R, Che FS, Kaku H, Shibuya N (2010) Characterization of receptor proteins using affinity cross-linking with biotinylated ligands. Plant Cell Physiol 51: 262–270 [DOI] [PubMed] [Google Scholar]
- Singh R, Dangol S, Chen Y, Choi J, Cho YS, Lee JE, Choi MO, Jwa NS (2016) Magnaporthe oryzae effector AVR-Pii helps to establish compatibility by inhibition of the rice NADP-malic enzyme resulting in disruption of oxidative burst and host innate immunity. Mol Cells 39: 426–438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahara H, Hacquard S, Kombrink A, Hughes HB, Halder V, Robin GP, Hiruma K, Neumann U, Shinya T, Kombrink E, et al. (2016) Colletotrichum higginsianum extracellular LysM proteins play dual roles in appressorial function and suppression of chitin-triggered plant immunity. New Phytol 211: 1323–1337 [DOI] [PubMed] [Google Scholar]
- Talbot NJ, Ebbole DJ, Hamer JE (1993) Identification and characterization of MPG1, a gene involved in pathogenicity from the rice blast fungus Magnaporthe grisea. Plant Cell 5: 1575–1590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol 30: 2725–2729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanabe S, Okada M, Jikumaru Y, Yamane H, Kaku H, Shibuya N, Minami E (2006) Induction of resistance against rice blast fungus in rice plants treated with a potent elicitor, N-acetylchitooligosaccharide. Biosci Biotechnol Biochem 70: 1599–1605 [DOI] [PubMed] [Google Scholar]
- Tang M, Ning Y, Shu X, Dong B, Zhang H, Wu D, Wang H, Wang GL, Zhou B (2017) The Nup98 homolog APIP12 targeted by the effector AvrPiz-t is involved in rice basal resistance against Magnaporthe oryzae. Rice (N Y) 10: 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson SE, Smith M, Wilkinson MC, Peek K (2001) Identification and characterization of a chitinase antigen from Pseudomonas aeruginosa strain 385. Appl Environ Microbiol 67: 4001–4008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trouvelot S, Héloir MC, Poinssot B, Gauthier A, Paris F, Guillier C, Combier M, Trdá L, Daire X, Adrian M (2014) Carbohydrates in plant immunity and plant protection: Roles and potential application as foliar sprays. Front Plant Sci 5: 592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valent B. (1990) Rice blast as a model system for plant pathology. Phytopathology 80: 33–36 [Google Scholar]
- Vandenborre G, Smagghe G, Van Damme EJ (2011) Plant lectins as defense proteins against phytophagous insects. Phytochemistry 72: 1538–1550 [DOI] [PubMed] [Google Scholar]
- Villalba F, Collemare J, Landraud P, Lambou K, Brozek V, Cirer B, Morin D, Bruel C, Beffa R, Lebrun MH (2008) Improved gene targeting in Magnaporthe grisea by inactivation of MgKU80 required for non-homologous end joining. Fungal Genet Biol 45: 68–75 [DOI] [PubMed] [Google Scholar]
- Wang R, Ning Y, Shi X, He F, Zhang C, Fan J, Jiang N, Zhang Y, Zhang T, Hu Y, et al. (2016) Immunity to rice blast disease by suppression of effector-triggered necrosis. Curr Biol 26: 2399–2411 [DOI] [PubMed] [Google Scholar]
- Wang Z, Chen C, Xu Y, Jiang R, Han Y, Xu Z, Chong K (2004) A practical vector for efficient knockdown of gene expression in rice (Oryza sativa L.). Plant Mol Biol Rep 22: 409–417 [Google Scholar]
- Weidenbach D, Esch L, Möller C, Hensel G, Kumlehn J, Höfle C, Hückelhoven R, Schaffrath U (2016) Polarized defense against fungal pathogens is mediated by the Jacalin-related lectin domain of modular Poaceae-specific proteins. Mol Plant 9: 514–527 [DOI] [PubMed] [Google Scholar]
- Xiang Y, Song M, Wei Z, Tong J, Zhang L, Xiao L, Ma Z, Wang Y (2011) A jacalin-related lectin-like gene in wheat is a component of the plant defence system. J Exp Bot 62: 5471–5483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada A, Shibuya N, Kodama O, Akatsuka T (1993) Induction of phytoalexin formation in suspension-cultured rice cells by N-acetylchitooligo-saccharides. Biosci Biotechnol Biochem 57: 405–409 [Google Scholar]
- Yamazaki H, Yamazaki D, Takaya N, Takagi M, Ohta A, Horiuchi H (2007) A chitinase gene, chiB, involved in the autolytic process of Aspergillus nidulans. Curr Genet 51: 89–98 [DOI] [PubMed] [Google Scholar]
- Yang C, Yu Y, Huang J, Meng F, Pang J, Zhao Q, Islam A, Xu N, Tian Y, Liu J (2019) Binding of the Magnaporthe oryzae chitinase MoChia1 by a rice tetratricopeptide repeat protein allows free chitin to trigger immune responses. Plant Cell 10.1105/tpc.18.00382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Peumans WJ, Barre A, Astoul CH, Rovira P, Rougé P, Proost P, Truffa-Bachi P, Jalali AA, Van Damme EJ (2000) Isolation and characterization of a jacalin-related mannose-binding lectin from salt-stressed rice (Oryza sativa) plants. Planta 210: 970–978 [DOI] [PubMed] [Google Scholar]