SEED CAROTENOID DEFICIENT functions in the methylerythritol phosphate pathway in maize and affects the accumulation of downstream isoprenoids.
Abstract
Plastid isoprenoids, a diverse group of compounds that includes carotenoids, chlorophylls, tocopherols, and multiple hormones, are essential for plant growth and development. Here, we identified and characterized SEED CAROTENOID DEFICIENT (SCD), which encodes an enzyme that functions in the biosynthesis of plastid isoprenoids in maize (Zea mays). SCD converts 2C-methyl-d-erytrithol 2,4-cyclodiphosphate to 1-hydroxy-2-methyl-2-(E)-butenyl 4-diphosphate in the penultimate step of the methylerythritol phosphate (MEP) pathway. In scd mutants, plant growth and development are impaired and the levels of MEP-derived isoprenoids, such as carotenoids, chlorophylls, and tocopherols, as well as abscisic and gibberellic acids, are reduced in leaves and seeds. This scd metabolic alteration varies among plant tissues and under different light conditions. RNA-sequencing of the scd mutant and wild type identified a limited number of differentially expressed genes in the MEP pathway, although isoprenoid levels were significantly reduced in scd seeds and dark-grown leaves. Furthermore, SCD-overexpressing transgenic lines showed little or no differences in isoprenoid levels, indicating that SCD may be subject to posttranslational regulation or not represent a rate-limiting step in the MEP pathway. These results enhance our understanding of the transcriptomic and metabolic regulatory roles of enzymes in the MEP pathway and of their effects on downstream isoprenoid pathways in various plant tissues and under different light conditions.
Isoprenoids (or terpenoids) are the most functionally and structurally diverse group of metabolites in living organisms, with over 55,000 natural molecules discovered to date (Thulasiram et al., 2007). Many isoprenoids play critical roles in plant growth and development and have important commercial applications. For example, carotenoids act as light-harvesting pigments that function in photoprotection against excess light during photosynthesis, with major impacts on plant growth and development (Demmig-Adams et al., 1996; Ruiz-Sola and Rodríguez-Concepción, 2012). In addition, carotenoids, such as provitamin A, are valuable for human health because of their antioxidant and nutritional qualities (Johnson, 2002; Goff and Klee, 2006).
All types of isoprenoids are derived from the common five-carbon precursors isopentenyl diphosphate (IPP) and its isomer dimethylallyl diphosphate (DMAPP; Bouvier et al., 2005). Condensation of IPP and DMAPP units results in the synthesis of short-chain prenyl diphosphates, which serve as branch points for the subsequent synthesis of all isoprenoid end products. Plants synthesize IPP and DMAPP via two independent pathways: the mevalonate (MVA) pathway for isoprenoid synthesis in the cytoplasm and the methylerythritol phosphate (MEP) pathway for isoprenoid synthesis in plastids (Kuzuyama and Seto, 2003; Hemmerlin et al., 2012; Vranová et al., 2013).
The MEP pathway has been well elucidated in the model plant Arabidopsis (Arabidopsis thaliana). This pathway starts with the production of 1-deoxy-d-xylulose-5-phosphate from glyceraldehyde 3-P and pyruvate, a process catalyzed by 1-deoxy-d-xylulose-5-phosphate synthase (DXS), and ends with the conversion of 1-hydroxy-2-methyl-2-(E)-butenyl 4-diphosphate (HMBPP) into a mixture of IPP and DMAPP by HMBPP reductase (HDR; Supplemental Fig. S1; Vranová et al., 2012, 2013). Almost all enzymes involved in the MEP pathway have been identified and characterized in Arabidopsis and are highly conserved with those in Escherichia coli (Estévez et al., 2000; Bauer et al., 2001; Carretero-Paulet et al., 2002; Querol et al., 2002; Hsieh and Goodman, 2005, 2006). Most mutants related to these genes have an albino seedling phenotype and impaired chloroplast development. By contrast, little is known about the MEP pathway in maize (Zea mays) beyond the isolation of DXS, 1-deoxy-d-xylulose-5-phosphate reductoisomerase (DXR), and HDR, which catalyze the first, second, and last steps of the MEP pathway, respectively (Hans et al., 2004; Cordoba et al., 2011; Lu et al., 2012).
HDS, also known as GcpE, IspG, CSB3, and CEH1, encodes 1-hydroxy-2-methyl-2-(E)-butenyl-4-diphosphate synthase, which catalyzes the conversion of 2C-methyl-d-erytrithol 2,4-cyclodiphosphate (MEcPP) to HMBPP, the penultimate step in the MEP pathway (Ostrovsky et al., 1998; Altincicek et al., 2001; Querol et al., 2002; Gutiérrez-Nava et al., 2004; Xiao et al., 2012). Consistent with other enzymes in the MEP pathway, HDS is encoded by a nuclear gene and is imported into plastids to synthesize the precursor of plastid isoprenoids. Knockout of HDS blocks the formation of plastid isoprenoids (Gutiérrez-Nava et al., 2004). For example, the complete loss of function of HDS in Arabidopsis and Nicotiana benthamiana inhibits chlorophyll and carotenoid biogenesis, resulting in a seedling-lethal albino phenotype (Gutiérrez-Nava et al., 2004; Page et al., 2004). By contrast, enhancing the level of active HDS protein in E. coli and Arabidopsis does not result in the increased accumulation of carotenoids, suggesting that enhanced flux through the MEP pathway during peak demand periods in bacteria and plastids does not require increased HDS activity (Flores-Pérez et al., 2008). Additional work is needed to understand the mechanism by which HDS regulates the metabolic flux from the MEP pathway to the downstream biosynthesis of isoprenoids.
In this study, we isolated the maize mutant seed carotenoid deficient (scd), which is characterized by albino plants with pale-yellow seeds, and determined that it is a spontaneous recessive mutant. We established that the causal mutation is located in SCD, which results in the loss of its function due to alternative transcript splicing. Phenotypic characterization of scd suggested that SCD plays an important role in controlling metabolic flux in the MEP pathway and indirectly affects the biosynthesis of a number of plastid isoprenoids, such as carotenoids, chlorophylls, tocopherols, abscisic acid (ABA), and gibberellic acids (GAs). This study provides molecular insights into the regulation of SCD in leaves and seeds and enhances our understanding of the mechanisms that control the MEP pathway during plant development.
RESULTS
The scd Mutant Has Carotenoid-Deficient Seeds and an Albino Seedling-Lethal Phenotype
We discovered the scd mutant, which has a visually distinctive phenotype consisting of pale-yellow seeds and white seedlings (Fig. 1, A–C), as a spontaneously occurring mutant in a recombinant inbred line population derived from a single cross between two elite inbred lines, Zong3 and Yu87-1 (Tang et al., 2007). The white seedlings generally die at the end of the V1 stage.
Figure 1.
Phenotypes of seeds and seedlings. A, A self-pollinated ear from an SCD/scd heterozygote. Homozygous mutant seeds exhibit carotenoid deficiency. B and C, Phenotypes of mature wild type (WT) and scd seeds (B) and V1-stage seedlings (C). D and E, HPLC analysis of carotenoid profiles in maize seeds (D) and V1-stage maize seedlings (E).
To obtain further insight into the nature of the scd mutant phenotype, we analyzed the carotenoid profiles of scd seeds and leaves and the chlorophyll profiles of scd leaves. The ability of the scd mutant to synthesize carotenoids and chlorophylls was greatly reduced in its leaves, and carotenoid levels were significantly reduced in scd seeds (Fig. 1, D and E; Supplemental Fig. S2). These results suggest that SCD likely functions upstream of carotenoid and chlorophyll biosynthesis and that its effects vary between seeds and leaves.
Carotenoids and chlorophylls play key roles in photoprotection and photosynthesis. To determine whether the effects of the scd mutation on carotenoid and chlorophyll levels in leaves are due to the pleiotropic effects of light damage, we grew scd and wild-type plants in the dark and analyzed the subsequent leaf carotenoid and chlorophyll profiles (Supplemental Fig. S3). The albino phenotype of scd did not revert to that of wild type, and little difference in carotenoid and chlorophyll levels was detected between scd mutants grown in either the light or the dark (Supplemental Figs. S2 and S3). These findings suggest that the albino phenotype of scd mutants is not because of the secondary effects of photo-oxidative stress.
The scd Mutation Affects Aleurone Cell Development and Results in Impaired Chloroplasts
To investigate the effect of the scd mutation on seed development, we analyzed wild type and scd seeds at 15 d after pollination (DAP) using transmission electron microscopy (TEM; Fig. 2, A and B). The aleurone and subaleurone cells showed substantial changes, especially with respect to the number of aleurone grains and vacuole shape. When compared with that in wild type, aleurone cells in scd contained a greater number of smaller aleurone grains and fewer larger vacuoles, whereas subaleurone cells contained fewer smaller aleurone grains and larger vacuoles (Fig. 2, C and D). It appears that the development of some organelles in the aleurone and subaleurone layers is blocked in the absence of functional SCD.
Figure 2.
Effects of the scd mutation on the morphology of developing seeds and seedling leaves of light- and dark-grown plants. A and B, Ultrastructures of cells in the aleurone and subaleurone layers of 15-DAP endosperms from wild type (WT; A) and scd (B). Bars = 5 μm. C, Relative number of aleurone grains and vacuoles. Error bars indicate the SE based on at least 10 biological replicates. D, Relative sizes of aleurone grains and vacuoles. Error bars indicate the SE based on at least 10 biological replicates. *P < 0.05, **P < 0.01, Student’s t test. E and F, Transverse sections of seedling leaves from wild type (E) and scd (F). Bars = 10 μm. G and H, Bundle sheath cells in seedling leaves from wild type (G) and scd (H) visualized by TEM. Bars = 5 μm. I and J, Ultrastructure of the chloroplasts in bundle sheath cells in seedling leaves from wild type (I) and scd (J). Bars = 1 μm. K and L, Transverse sections of dark-grown seedling leaves from wild type (K) and scd (L). Bars = 10 μm. M and N, Bundle sheath cells in dark-grown seedling leaves from wild type (M) and scd (N) visualized by TEM. Bars = 5 μm. O and P, Ultrastructures of chloroplasts in bundle sheath cells of dark-grown seedling leaves from wild type (O) and scd (P). Bars = 1 μm. BSc, bundle sheath cell; Mc, mesophyll cell; Ch, chloroplast; M, mitochondrion; AL, aleurone layer; AG, aleurone grain; SG, starch grain; PB, protein body; sER, smooth endoplasmic reticulum; rER, rough endoplasmic reticulum; N, nucleus; LB, lipid body; V, vacuole; VP, vacuolar precipitate; CW, cell wall.
We analyzed wild type and scd leaf sections from V1-stage seedlings by conventional optical microscopy and TEM to explore the cellular basis for the albino phenotype of the mutant (Fig. 2, E–J). We detected an extreme reduction in chloroplast numbers in mesophyll and vascular bundle sheath cells of scd compared with the wild type (Fig. 2, E and F). Moreover, plastids in scd lacked appressed internal membranes and contained short, linear, noncompact membranes (Fig. 2, G–J). When the plants were grown in the dark, chloroplasts were still present in both the mesophyll and vascular bundle sheath cells of V1-stage seedling leaves in scd, but their structures were disrupted (Fig. 2, K–P). Based on plastid morphology, it appears that chloroplast development in scd is arrested at an early stage of development. These results further demonstrate that the presence of impaired chloroplasts is caused by the scd mutation rather than the effect of photo-oxidative stress.
Map-Based Cloning and Functional Verification of SCD
Before isolating the SCD gene, we first asked whether the scd mutant is the result of a single gene disruption. We scored the number of seeds of each color (yellow and pale-yellow) in SCD/scd ears and found that the results fit a 3:1 ratio (yellow: pale-yellow seeds,  <
< 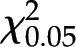 = 3.84; P > 0.05; Supplemental Table S1). Thus, scd is controlled by a single recessive gene.
= 3.84; P > 0.05; Supplemental Table S1). Thus, scd is controlled by a single recessive gene.
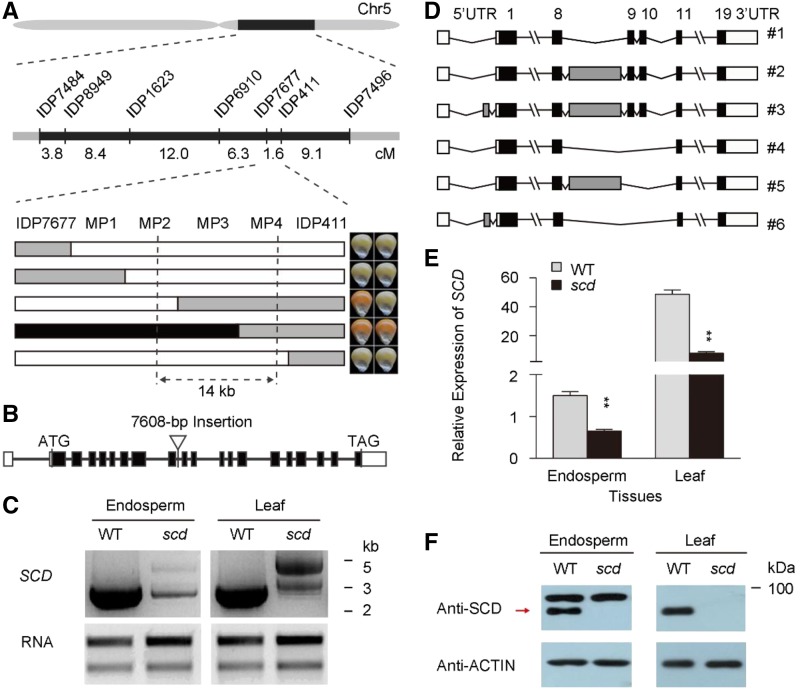
Subsequently, we carried out map-based cloning using an F2 population derived from the cross between B73 and SCD/scd heterozygotes. The SCD locus was initially mapped between the insertion-deletion (InDel) markers IDP7677 and IDP411 on chromosome 5 at a genetic distance of 1.6 cM and a physical distance of 1.86 Mb; it was subsequently fine-mapped within a 14-kb region between markers MP2 and MP4, based on five recombinants (Fig. 3A). This region contains two predicted genes, and genomic sequence analysis of the refined interval in scd and wild type revealed only a 7.6-kb insertion in the eighth intron of GRMZM2G137409 (Fig. 3B). BLAST analysis revealed that this inserted fragment shares 99% identity with the CRM centromeric retrotransposon in maize (AY129008.1), which has the typical structure of a long terminal repeat retrotransposon (Supplemental Fig. S4; Xu and Wang, 2007). The long terminal repeat retrotransposon insertion in GRMZM2G137409 in the scd mutant generates at least six alternatively spliced transcripts in V1-stage seedling leaves and 15-DAP endosperms, which leads to a sharp reduction in transcript levels and, consequently, a lack of protein from this transcript (Fig. 3, C–F). Therefore, we considered GRMZM2G137409 as a candidate gene for the SCD locus.
Figure 3.
Identification of the gene underlying the SCD locus. A, SCD was fine-mapped to a 14-kb region on chromosome 5. The markers for primary mapping, their genetic distances, and the genotypes and phenotypes of five key recombinants are shown. The black, white, and gray rectangles represent SCD, scd, and heterozygous genotypes, respectively. B, Gene structure of SCD and the position of the scd mutation. Introns are shown as solid lines, exons as black boxes, and untranslated regions as white boxes. The mutation site is represented by an inverted triangle. C, PCR product of the full-length SCD cDNA in 15-DAP endosperms and seedling leaves. D, Schematic representation of multiple SCD transcripts produced in scd 15-DAP endosperms and V1-stage seedling leaves. A 120-bp insertion in the 5′ untranslated region (5′UTR) and a footprint fragment with an insertion length ranging from 111 to 1593 bp between the eighth and ninth exons are shown as gray boxes; all other symbols are as in B. E, Relative expression of SCD in 15-DAP endosperms and seedling leaves. The RT-qPCR primers were designed based on exons 13 (forward) and 15 (reverse), which captures all transcript variants. Error bars indicate the SE based on at least three biological replicates. **P < 0.01, Student’s t test. F, Immunoblot analysis of SCD in 15-DAP endosperms and seedling leaves. Red arrow shows the target bands. WT, wild type.
We confirmed this result by identifying additional alleles with mutations in SCD. The classic maize mutant lemon white2 (lw2), which was also named vp12 and is referred to here as lw2-vp12, exhibits lemon-white endosperms and completely albino seedlings (Fig. 4, A and B; Maluf et al., 1997; Stinard, 2013). Genome sequence analysis suggested that GRMZM2G137409 is a prime candidate for the lw2-vp12 locus (Stinard, 2013). To investigate the genotype-phenotype relationship in this mutant, we sequenced GRMZM2G137409 in lw2-vp12 and the wild type parental line and found an 8-bp insertion in the first exon, resulting in a frameshift mutation (Fig. 4C). Expression analysis revealed severe reductions in the levels of GRMZM2G137409 mRNA and protein in 15-DAP endosperms and V1-stage seedling leaves of lw2-vp12 versus that in wild type (Fig. 4, D and E). Consistent with the phenotype of scd, lw2-vp12 had lower carotenoid and chlorophyll levels in seeds and seedling leaves under both light and dark conditions than the wild type (Fig. 4, F–J). These results clearly indicate that lw2-vp12 is another independent allele of scd.
Figure 4.
Molecular and genetic identification of lemon white 2 (lw2-vp12) as scd. A and B, Seed (A) and seedling (B) phenotypes of the lw2-vp12 mutant. C, The position of the causal mutation of SCD in the lw2-vp12 mutant. Introns are shown as solid lines, exons as black boxes, and untranslated regions as white boxes. The inverted triangles represent the insertions. D and E, Relative expression (D) and immunoblot analysis (E) of SCD in 15-DAP endosperms and seedling leaves of lw2-vp12. The RT-qPCR primers were designed based on exons 13 (forward) and 15 (reverse). Red arrow shows the target bands. F to H, Carotenoid levels in mature seeds (F) and light- (G) and dark-grown (H) seedling leaves. I and J, Chlorophyll levels in light- (I) and dark-grown (J) seedling leaves. Error bars indicate the SE based on at least three biological replicates. *P < 0.05, **P < 0.01, Student’s t test. WT, wild type; LUT, lutein; ZEA, zeaxanthin; BCRY, β-cryptoxanthin; AC, α-carotene; BC, β-carotene; VIO, violaxanthin; ANT, antheraxanthin; Chl a, chlorophyll a; Chl b, chlorophyll b.
We also identified two additional mutants in GRMZM2G137409, namely scd-1 and scd-2. These alleles have similar seed and seedling mutant phenotypes as scd (Supplemental Fig. S5A and B; Supplemental Table S1). Genomic sequence analysis of GRMZM2G137409 revealed that scd-1 carries a Mu transposon insertion in the first exon and scd-2 has a G-to-A substitution (Glu to stop codon, TAG) in the twelfth exon, introducing a premature stop codon (Supplemental Fig. S5C). These mutations also lead to decreased SCD protein levels (Supplemental Fig. S5D).
To confirm that the mutations in GRMZM2G137409 are responsible for the scd phenotype, we performed allelism tests using scd, lw2-vp12, scd-1, and scd-2 (Supplemental Fig. S5). The seed phenotypes displayed a 3:1 segregation ratio of normal color to pale-yellow color for all three F1 hybrids from crosses between SCD/scd plants and LW2-VP12/lw2-vp12, SCD-1/scd-1, or SCD-2/scd-2 plants (Supplemental Fig. S5E; Supplemental Table S1). Therefore, GRMZM2G137409 is the causal gene for the scd and lw2-vp12 mutants.
We engineered mutations in SCD in the inbred line B73-329 using clustered regularly interspaced short palindromic repeats (CRISPR)–Cas9 gene editing and obtained three null mutants, referred to as CR-scd-1, CR-scd-2, and CR-scd-3 (Supplemental Fig. S6). Genome sequencing identified multiple mutations in all mutants, and RT-quantitative PCR (RT-qPCR) analysis revealed a drastic reduction in GRMZM137409 mRNA expression in the leaves of all three CR-scd plants (Supplemental Fig. S6, B–D). Like the original scd mutant, these three CR-scd plants had albino phenotypes (Supplemental Fig. S6E). These findings suggest that the albino phenotype is caused by the loss of function of SCD, which is consistent with the original scd mutant and the results of allelism tests of the three independent scd mutants.
Characterization of SCD
We isolated the full-length complementary DNA (cDNA) of SCD from the leaves of wild type seedlings using 5′ and 3′ RACE (Supplemental Fig. S7). In addition to the PCR product covering the whole open reading frame region, the full-length SCD cDNA consists of a 203-bp 5′ untranslated region, a 2241-bp open reading frame (19 exons), and a 165-bp 3′ untranslated region (Fig. 3B). Sequence analysis using databases such as Ensemble Plants (http://plants.ensembl.org/) and NCBI (https://www.ncbi.nlm.nih.gov/) indicated that SCD is a single-copy gene in maize.
Sequence alignment with the predicted protein indicated that SCD encodes a putative HMBPP synthase (HDS) of 746 amino acids with an estimated molecular mass of 82 kD, sharing high protein sequence similarity with Arabidopsis HDS (Supplemental Fig. S8). The HDS enzyme catalyzes the conversion of MEcPP into HMBPP in the penultimate step of the MEP pathway (Ostrovsky et al., 1998; Altincicek et al., 2001; Querol et al., 2002; Gutiérrez-Nava et al., 2004; Page et al., 2004; Flores-Pérez et al., 2008). Arabidopsis plants with a knockout of HDS showed similar seed and leaf phenotypes to those of the maize scd mutants (Gutiérrez-Nava et al., 2004), confirming that SCD has conserved biological functions similar to those of its Arabidopsis putative ortholog.
The SCD protein was predicted to contain a chloroplast-targeting signal peptide, followed by a triose phosphate isomerase barrel domain (A) for dihydropteroate synthase, an inserted triose phosphate isomerase barrel domain (A*), and a 4Fe4S cluster-containing domain (B) for sulfite/nitrite reductase (Supplemental Fig. S8; Liu et al., 2012). The A* domain primarily plays a structural role and interacts with the A domain to form a structure that enables the A and B domains to interact in a “cup and ball” manner during catalysis (Liu et al., 2012). Among these domains, the A and B domains are highly conserved across species (Supplemental Fig. S8).
RT-qPCR analysis revealed that SCD was widely expressed in all maize tissues examined (Supplemental Fig. S9). SCD transcript levels were highest in leaves, followed by stems, roots, and other tissues. In developing seeds, SCD was expressed at high levels at 15 and 35 DAP but at relatively low levels at 20, 25, and 30 DAP; a similar pattern was observed in both endosperm and embryo tissues at the corresponding stages. When SCD was knocked out in scd, SCD expression was sharply downregulated, especially in tissues where SCD was normally expressed at high levels, such as in leaves, roots, stems, and 15-DAP seeds.
In addition, to examine the subcellular localization of SCD, we transiently expressed SCD-enhanced GFP (EGFP) fusions driven by the 35S promoter (35S::SCD-EGFP) in maize protoplasts (Supplemental Fig. S10). When SCD-EGFP was transiently expressed in protoplasts, EGFP fluorescence was observed in chloroplasts, in addition to red autofluorescence signals emitted from chlorophyll (Supplemental Fig. S10, E–G). This result indicates that SCD is imported into the plastid to carry out its function, which is in agreement with the movement of its Arabidopsis putative ortholog (Querol et al., 2002).
Given that SCD catalyzes the formation of HMBPP from MEcPP in the MEP pathway, we investigated whether the levels of metabolites in the MEP pathway were altered in scd (Fig. 5). The levels of the substrate metabolite MEcPP were uniformly higher in scd versus wild type seeds (∼20-fold increase), light-grown leaves (∼ 27-fold increase), and dark-grown leaves (∼230-fold increase), as was also the case for the upstream metabolite MEP (∼8-, 68-, and 280-fold increase in seeds, light-grown leaves, and dark-grown leaves, respectively). By contrast, the levels of the product HMBPP and the common isoprenoid precursors DMAPP and IPP were significantly reduced in both light- and dark-grown scd leaves, specifically 60.7% to 87.9% of those in wild type, whereas the levels of HMBPP, DMAPP, and IPP appeared to be reduced in scd seeds, but this decrease was not statistically significant. These findings suggest that the enzymatic activity of SCD is reduced in scd and that SCD encodes an HDS protein.
Figure 5.
Comparison of the levels of five metabolites of the MEP pathway in scd and wild type (WT) seeds and seedlings. A, Simplified diagram of the MEP pathway in plastids. The five metabolites shown in red were measured in this study. B–D, Content of the five metabolites in seeds (B) and light- (C) and dark-grown (D) seedling leaves. DMAPP/IPP represents the average of DMAPP and IPP. Error bars indicate the SE based on at least three biological replicates. *P < 0.05, **P < 0.01, Student’s t test. G3P, D-glyceraldehyde 3-phosphate; DXP, 1-deoxy-d-xylulose-5-phosphate; MCT, methylerythritol phosphate cytidyltransferase; CDP-ME, 4-diphosphocytidyl-2C-methyl-D-erythritol; CDP-MEP, 4-diphosphocytidyl-2C-methyl-D-erythritol 2-phosphate; CMK, 4-diphosphocytidyl-2C-methyl-D-erythritol kinase.
Metabolic Effects of the Loss of SCD Function
Due to the fundamental role of the MEP pathway in isoprenoid biosynthesis, we reasoned that the loss of function of SCD in this pathway might result in altered levels of numerous downstream isoprenoids. Indeed, analysis of lw2-vp12 indicated that vp12 embryos are deficient in ABA accumulation during kernel development (Maluf et al., 1997). Hence, in addition to carotenoids and chlorophylls, we analyzed 24 other metabolites, including three tocopherols and members of six phytohormone families, in seeds and light- and dark-grown leaves of scd seedlings. The ability of the scd and lw2-vp12 mutants to synthesize tocopherols was nearly lost in leaves but still remained in seeds, although at a significantly reduced level (Table 1; Supplemental Fig. S11). Light treatment had little effect on the tocopherol levels in scd.
Table 1. Comparison of tocopherol and phytohormone levels in scd and wild type seeds and seedling leaves.
Values are averages of at least three biological replicates ± se. Student’s t test compared with the corresponding wild type (*P < 0.05, **P < 0.01). DT, δ-tocopherol; GT, γ-tocopherol; AT, α-tocopherol; ABA, (+)-cis, trans-Abscisic acid; SA, salicylic acid; IAA, indole-3-acetic acid; ME-IAA, methyl indole-3-acetate; IBA, 3-indolebutyric acid; ICA, indole-3-carboxaldehyde; IP, N6-isopentenyladenine; tZ, trans-Zeatin; cZ, cis-Zeatin; DZ, dihydrozeatin; JA, (+)-jasmonic acid; H2JA, (+)-dihydrojasmonic acid; JA-ILE, N-[(-)-jasmonoyl]-(L)-Ile; GA1, gibberellin A1; GA3, gibberellin A3; GA9, gibberellin A9; GA15, gibberellin A15; GA19, gibberellin A19; GA20, gibberellin A20; GA24, gibberellin A24; GA53, gibberellin A53.
| Metabolites | Mature Seeds | Light-Grown Leaves | Dark-Grown Leaves | |||
|---|---|---|---|---|---|---|
| Wild Type | scd | Wild Type | scd | Wild Type | scd | |
| Tocopherols (µg/g) | ||||||
| DT | 1.11 ± 0.23 | 0.53 ± 0.13* | 11.47 ± 2.82 | 0.00** | 11.18 ± 0.70 | 0.00** |
| GT | 17.00 ± 1.00 | 5.52 ± 0.33** | 25.43 ± 1.62 | 0.00** | 27.21 ± 1.69 | 0.00** |
| AT | 10.34 ± 0.43 | 5.64 ± 0.39** | 20.98 ± 2.65 | 0.00** | 17.15 ± 1.03 | 0.00** |
| Abscisic acid (ng/g) | ||||||
| ABA | 0.00 | 0.00 | 2.39 ± 0.19 | 0.74 ± 0.06* | 9.16 ± 3.89 | 1.82 ± 0.17** |
| Salicylic acid (ng/g) | ||||||
| SA | 492.52 ± 46.60 | 313.44 ± 21.74** | 275.39 ± 56.91 | 415.13 ± 41.06 | 27.75 ± 2.09 | 28.05 ± 1.73 |
| Auxins (ng/g) | ||||||
| IAA | 264.49 ± 51.44 | 232.27 ± 6.16 | 0.87 ± 0.02 | 0.64 ± 0.01** | 2.75 ± 0.14 | 3.17 ± 0.17 |
| ME-IAA | 0.81 ± 0.32 | 0.68 ± 0.03 | 0.01 ± 0.00 | 0.03 ± 0.00** | 0.03 ± 0.00 | 0.12 ± 0.01* |
| IBA | 0.00 | 0.00 | 4.19 ± 0.08 | 1.49 ± 0.24** | 7.00 ± 0.75 | 14.4 ± 3.28 |
| ICA | 25.16 ± 3.73 | 33.66 ± 3.53 | 0.34 ± 0.08 | 0.61 ± 0.23 | 0.29 ± 0.02 | 0.67 ± 0.16 |
| Cytokinins (ng/g) | ||||||
| IP | 0.01 ± 0.00 | 0.01 ± 0.00 | 0.02 ± 0.01 | 0.03 ± 0.00 | 0.01 ± 0.00 | 0.01 ± 0.00 |
| tZ | 1.21 ± 0.31 | 1.65 ± 0.06 | 0.00 | 0.00 | 0.00 | 0.00 |
| cZ | 0.18 ± 0.05 | 0.19 ± 0.03 | 0.91 ± 0.08 | 1.20 ± 0.02 | 1.13 ± 0.01 | 1.00 ± 0.04 |
| DZ | 5.49 ± 1.08 | 5.37 ± 0.38 | 0.00 | 0.00 | 0.00 | 0.00 |
| Jasmonic acids (ng/g) | ||||||
| JA | 1.81 ± 0.24 | 1.69 ± 0.27 | 2.35 ± 0.16 | 0.97 ± 0.10** | 2.62 ± 0.93 | 1.83 ± 0.38 |
| H2JA | 1.94 ± 1.37 | 0.80 ± 0.23 | 0.08 ± 0.01 | 0.11 ± 0.03 | 0.10 ± 0.02 | 0.06 ± 0.01 |
| JA-ILE | 7.04 ± 0.44 | 5.27 ± 0.79 | 0.82 ± 0.09 | 0.72 ± 0.05 | 0.93 ± 0.18 | 1.38 ± 0.33 |
| Gibberellic acids (ng/g) | ||||||
| GA1 | 0.00 | 0.00 | 0.00 | 0.00 | 1.70 ± 0.15 | 1.55 ± 0.13 |
| GA3 | 0.00 | 0.00 | 0.30 ± 0.01 | 0.16 ± 0.02** | 0.37 ± 0.02 | 0.00** |
| GA9 | 0.40 ± 0.01 | 0.37 ± 0.03 | 0.26 ± 0.03 | 0.00** | 0.91 ± 0.10 | 0.31 ± 0.04* |
| GA15 | 0.11 ± 0.03 | 0.07 ± 0.00 | 0.10 ± 0.01 | 0.00** | 0.32 ± 0.04 | 0.00** |
| GA19 | 48.65 ± 0.12 | 48.48 ± 0.04 | 52.31 ± 0.35 | 48.11 ± 0.06** | 57.07 ± 0.60 | 0.00** |
| GA20 | 8.73 ± 0.62 | 6.52 ± 0.52 | 5.58 ± 1.11 | 0.29 ± 0.02* | 13.43 ± 1.58 | 0.83 ± 0.13* |
| GA24 | 24.44 ± 0.66 | 24.29 ± 0.09 | 0.00 | 0.00 | 0.00 | 0.00 |
| GA53 | 0.00 | 0.00 | 20.4 ± 0.10 | 0.00** | 21.7 ± 0.31 | 0.00** |
Among the 21 metabolites of the six phytohormone families identified, 16, 17, and 18 metabolites were identified in mature seeds, light-grown leaves, and dark-grown leaves in wild type, respectively (Table 1). Except for salicylic acid, no significant changes in the levels of these examined metabolites were detected in mature scd seeds. By contrast, the levels of ABA and six GAs, including GA3, GA9, GA15, GA19, GA20, and GA53, were significantly reduced in the seedling leaves of scd in both the light and dark, with the effects on GA19 levels highly sensitive to dark conditions. In addition, the levels of indole-3-acetic acid, 3-indolebutyric acid, and jasmonic acid were significantly reduced only in light-grown scd leaves. By contrast, the level of methyl indole-3-acetate consistently was higher in both light- and dark-grown scd leaves compared with the wild type. Collectively, these results indicate that the metabolic effects of SCD vary among plant tissues and growth conditions.
Identification of SCD-Associated Molecular Perturbation
To extend our knowledge of the changes in gene expression mediated by SCD, we generated transcriptome profiles for 15-DAP endosperms and embryos and light- and dark-grown V1-stage seedling leaves from scd and wild type. The abundance of reads for SCD exons downstream of the mutation site in scd was strongly reduced in endosperms, embryos, and light-grown leaves, but not in dark-grown leaves (Fig. 6A), which is highly consistent with the results of multiple transcript analysis and RT-qPCR (Fig. 3, D and E; Supplemental Fig. S12). Global transcriptome analysis identified 6,000 genes with significantly altered abundance in scd (>2.0-fold, P < 0.01), including 116 in endosperms, 175 in embryos, 735 in dark-grown leaves, and 4974 in light-grown leaves (Fig. 6B; Supplemental Data Set S1). Of these, no differentially expressed genes (DEGs) were common among the four tissues in the up- or down-regulated groups. The great differences in the altered gene expression between light- and dark-grown scd leaves demonstrate that transcriptional perturbation in scd leaves is greatly affected by light. The large differences among endosperms, embryos, and dark-grown leaves suggest that the downstream effects of SCD at the transcriptional level might be tissue specific to some extent.
Figure 6.
Overview of the transcriptomic changes caused by the loss of SCD in 15-DAP endosperms and embryos, and leaves from V1-stage seedlings grown in the light and dark. A, Read abundance across full-length SCD. The range of values of each read number is shown below the tissue name. B, Overlap of DEGs between the scd mutant and wild type (WT). C, GO enrichment analysis of functional categories of DEGs between scd and wild type. PS, photosynthesis; CHO, carbohydrate; OPP, oxidative phosphorylation; Misc, miscellaneous. D, Changes in the expression of genes in the major isoprenoid biosynthesis pathways. **False discovery rate (FDR) < 0.01. The names of all 60 genes are listed in Supplemental Table S2.
We then performed Gene Ontology (GO) enrichment analysis of the DEGs to better understand the biological processes affected by SCD (Fig. 6C). No biological process terms were commonly enriched in the four tissues, with 1, 5, 11, and 20 biological process terms enriched in endosperms, embryos, dark-grown leaves, and light-grown leaves, respectively. Among these biological process terms, photosynthesis was the most significantly enriched among down-regulated genes in light-grown scd leaves, most likely due to pleiotropic effects of light damage. Of the 1026 DEGs detected in endosperms, embryos, and dark-grown leaves, ∼4.09% (42/1026) of the genes were considerably enriched for the biological process categories involved in isoprenoid biosynthesis, photosynthesis, and the regulation of phytohormone levels. These results further confirm that SCD encodes a major enzyme in the MEP pathway, which produces the precursors of these isoprenoids.
We were particularly interested in biological processes associated with plastid isoprenoid biosynthesis, including carotenoid, chlorophyll, and tocopherol biosynthesis, and with the biosynthesis of their precursors in the MEP pathway in the plastid and isoprenoid precursor biosynthesis in the MVA pathway in the cytoplasm (Fig. 6D; Supplemental Table S2). Among the 60 genes identified in these metabolic pathways, 88% (53/60) were sharply down-regulated in light-grown scd leaves, especially genes in the MEP pathway and its downstream pathways. By contrast, no gene showed a significant change in expression in the endosperms, and 10% (6/60) and 21% (13/60) of genes were down- or up-regulated in embryos and dark-grown scd leaves, respectively.
Only two DEGs were commonly identified in embryos and dark-grown leaves. One of these genes is DXS2, encoding 1-deoxy-d-xylulose 5-phospate synthase, which catalyzes the first step in the MEP pathway (Walter et al., 2002; Cordoba et al., 2011); the other is CH1, encoding chlorophyllide a oxygenase, which functions in chlorophyll biosynthesis (Oster et al., 2000; Hoober and Eggink, 2001; Hirashima et al., 2006; Tanaka and Tanaka, 2007). Only DXS2 was down-regulated in both tissues of scd.
These results were confirmed by RT-qPCR analysis of the expression of 10 genes (Supplemental Fig. S12; Supplemental Table S3). These tissue-specific differences in gene expression are not consistent with the similar phenotypes of both scd leaves and seeds with respect to reduced carotenoid and tocopherol levels, suggesting that the nature of the transcriptional regulation of enzymes in the MEP pathway might vary among tissues. These results also indicate that this posttranscriptional regulation might be involved in the metabolic flux between the MEP pathway and the downstream isoprenoid biosynthesis pathway.
Overexpression of SCD Does Not Affect the Levels of Downstream Metabolites of the MEP Pathway
In contrast with the loss of SCD function, we reasoned that an increase in SCD expression might result in reduced substrate levels and enhanced metabolite production, consequently increasing the levels of downstream isoprenoids such as carotenoids, tocopherols, and chlorophylls. Thus, we generated transgenic lines expressing SCD cDNA under the control of the maize Ubiquitin1 promoter (pUbi) in maize inbred line B73-329. Two positive transgenic lines, OE1 and OE2, were obtained, and their enhanced expression of SCD was confirmed by both RT-qPCR and immunoblot analysis of T2 plants (Fig. 7, A–D). The overexpressing line OE1 was then backcrossed to SCD/scd plants and self-pollinated for two generations. The ears produced by selfing of SCDOE1/scd plants exhibited a 15:1 segregation ratio of yellow to pale-yellow seeds, indicating that scd was successfully complemented (Supplemental Fig. S13). In addition, no significant differences in the levels of downstream metabolites were detected between the complemented individuals and wild type (Supplemental Fig. S14). These results suggest that the scd mutant phenotype was fully rescued by overexpressing SCD.
Figure 7.
Characteristics of two SCD-overexpressing maize lines. A, Ears from OE1, OE2, and wild type (WT) plants. B, Seedling phenotypes of OE1, OE2, and wild type. C, Relative expression levels of SCD in 15-DAP endosperms, embryos, and seedling leaves from OE1, OE2, and wild type. D, Immunoblot analysis of SCD in 15-DAP endosperms and embryos and seedling leaves from OE1, OE2, and wild type. E and F, Levels of metabolites in the MEP pathway in seeds (E) and seedling leaves (F) of OE1, OE2, and wild type. DMAPP/IPP represents the average of DMAPP and IPP. Error bars indicate the SE based on at least three biological replicates. *P < 0.05, **P < 0.01, Student’s t test.
We subsequently measured five metabolites in the MEP pathway and the downstream isoprenoids in seeds and seedling leaves of the transgenic lines (Fig. 7; Supplemental Table S4). Only the levels of the substrate MEcPP were significantly reduced in the leaves of both transgenic lines. Unexpectedly, the levels of the isoprenoid precursors did not increase in the seeds and leaves of both transgenic lines, and only a few downstream isoprenoids increased in the seeds or leaves of one transgenic line.
We also examined the expression patterns of 24 genes in 15-DAP endosperms and embryos and V1-stage seedling leaves from the overexpressing transgenic lines OE1 and OE2 and from wild type B73-329 by RT-qPCR (Supplemental Fig. S15). These 24 genes included seven genes in the MEP pathway, two genes in the intermediate pathway between the MEP pathway and its downstream pathways, seven genes involved in carotenoid biosynthesis, four genes involved in tocopherol biosynthesis, and four genes involved in chlorophyll biosynthesis. Only a few genes were up-regulated in certain tissues, whereas SCD was up-regulated in all three tissues. These findings indicate that SCD overexpression does not induce the expression of genes in the MEP pathway or the downstream plastid isoprenoid pathway.
DISCUSSION
The MEP pathway plays a central role in plant growth and development, and several isoprenoids derived from this pathway are beneficial for human health and nutrition. Thus, identifying the major regulatory steps of the MEP pathway is important for potentially modulating the production of key isoprenoids. Here, through fine mapping, allelism tests, CRISPR/Cas9-mediated gene editing, and metabolite analysis, we determined that SCD functions as an HDS protein in the MEP pathway, consequently affecting the accumulation of a variety of isoprenoids, such as carotenoids, chlorophylls, tocopherols, ABA, and GAs. These findings provide important insights into the impact of the MEP pathway and its downstream metabolic and functional roles in maize.
SCD Is an Enzyme in the MEP Pathway
The MEP pathway produces the common metabolic precursors IPP and DMAPP for plastid isoprenoid biosynthesis, which involves the consecutive participation of eight enzymes, including an HDS enzyme that catalyzes the production of HMBPP from MEcPP in the plastid (Supplemental Fig. S1). Previous genome sequence analysis suggested that the lw2-vp12 locus harbors a candidate gene encoding an HDS protein in maize, which shares the highest sequence identity with AtHDS/AtGcpE in the maize genome (Stinard, 2013). Phenotypically, the lw2-vp12 mutant is a classical carotenoid mutant, with lemon-white endosperms and an albino seedling-lethal phenotype (Maluf et al., 1997), similar to that of the scd mutant identified in this study. Allelism tests performed with the lw2-vp12 and scd mutants demonstrated that the mutation in the lw2-vp12 mutation is allelic to the scd mutation, with both the lw2-vp12 and scd loci resolved to be a gene encoding an HDS protein. The enzymatic activity and function of AtGcpE/AtHDS as an HDS protein have been demonstrated in Arabidopsis (Seemann et al., 2005). Knockout mutations in CLB4/AtHDS result in white seeds and an albino seedling-lethal phenotype, with severely affected chloroplasts that are unable to synthesize photosynthetic pigments (Gutiérrez-Nava et al., 2004). The similar phenotypes observed in the scd/lw2-vp12 mutant point to the possibility that SCD functions as an HDS protein. This notion was further confirmed by the observation that this protein is localized to the plastid and that its substrate levels are elevated in scd, whereas the levels of its product are reduced.
Visibly, there is a slight difference between scd/lw2-vp12 and clb4 in terms of seed color, in that seeds are pale yellow in scd but white in clb4 (Gutiérrez-Nava et al., 2004). These phenotypes are indicators of carotenoid deficiency, as observed in other carotenoid-deficient mutants, such as y1, vp5, y9, vp9, ps1, and vp2 (Buckner et al., 1990; Li et al., 1996, 2007; Matthews et al., 2003; Singh et al., 2003). We found that scd seeds accumulated 13.6% of the total carotenoid levels of wild type seeds (Supplemental Fig. S2), suggesting that carotenoid biosynthesis still occurs in scd. Perhaps this is due to the remaining regulatory effects of low levels of SCD, because the multiple alternative transcript variants in the mutant include the normal SCD transcript (Figs. 3, C–E, and 6A). However, we cannot exclude the possibility that an active HDS enzyme other than SCD remains in the plastids of seeds, because immunoblot analysis revealed another visible band of the expected size that was not detected in leaves (Figs. 3F and 4D; Supplemental Fig. S5D), although the additional band might have been a nonspecific band detected by the SCD antibody in seeds. Further studies are needed to confirm the true nature of the carotenoid biosynthesis ability in scd seeds.
SCD Has Pleiotropic Effects on Metabolite Biosynthesis and Plant Development
The MEP pathway produces metabolic precursors for plastid isoprenoids, suggesting that disturbing MEP pathway activity alters the accumulation of the downstream isoprenoids, consequently leading to pleiotropic phenotypes. Most MEP pathway genes identified to date are associated with carotenoid accumulation (Lois et al., 2000; Estévez et al., 2001; Hsieh and Goodman, 2005; Xing et al., 2010; Lu et al., 2012), as is SCD, which was identified in this study. In addition to carotenoid deficiency caused by the knockout of SCD, SCD/HDS colocalized with a quantitative trait locus cluster for zeaxanthin, xanthophylls, and total carotenoids (Venado et al., 2017), pointing to the possible association between SCD and carotenoids, although no significant association was observed in a genome-wide association study of 281 maize inbred lines (Owens et al., 2014). Consistent with the reduction in carotenoid levels, we also observed a reduction in tocopherol levels among the three tissues examined.
Furthermore, the loss of SCD function weakened the ability of the scd mutant to synthesize chlorophylls, ABA, and several GAs in light- and dark-grown leaves and indole-3-acetic acid, indole-3-carboxaldehyde, and jasmonic acid in light-grown leaves. Among the phytohormones whose levels were altered in scd, only ABA levels were previously shown to be reduced in developing embryos of the lw2-vp12 mutant (Maluf et al., 1997). However, ABA was not detected in scd seeds, perhaps because dried mature seeds were used in our study (McCarty and Carson, 1991). Nonetheless, the finding that ABA levels were reduced in both light- and dark-grown leaves confirms the notion that SCD has a metabolic effect on the accumulation of ABA.
In addition to the metabolic effects of SCD, the scd mutant exhibited severely impaired seedling development, a phenotype similar to that of other albino mutants in Arabidopsis and maize harboring mutations that affect the enzymes involved in MEP-related biosynthetic pathways (Mandel et al., 1996; Araki et al., 2000; Estévez et al., 2000; Budziszewski et al., 2001; Gutiérrez-Nava et al., 2004; Guevara-García et al., 2005; Hsieh et al., 2008; Cordoba et al., 2011; Lu et al., 2012). It is conceivable that the inability to carry out photosynthesis, as reflected by severely disrupted chloroplasts, leads to seedling lethality in the scd mutant at early stages of development. Because chlorophylls are required for the formation of stacked thylakoids (Von Wettstein et al., 1995), it is reasonable to link the early arrest of chloroplast differentiation in scd plants with the absence of chlorophyll. However, other factors could also lead to the formation of albino plants, such as reduced levels of some phytohormones such as ABA, GAs, and so on (Buckner et al., 1990; Li et al., 1996; Matthews et al., 2003; Singh et al., 2003; Myers et al., 2011). In addition, distinct changes were observed in the aleurone and subaleurone cells of developing scd seeds, including the number and size or shape of aleurone grains and vacuoles. How the development of the aleurone and subaleurone layers in seeds is associated with SCD in the MEP pathway remains unknown.
The metabolic and developmental effects of SCD varied among tissues and light conditions. A similar situation was observed in the maize w3 mutant, whose phenotypes result from a plastoquinone-9 deficiency that in turn limits the contributions of plastoquinone-9 to the formation of downstream metabolites such as carotenoids, chlorophylls, and tocopherols (Hunter et al., 2018). The ability of the scd mutant to continue to accumulate isoprenoids in seeds reflects the fact that these seeds are not subject to severe photo-oxidative stress. The light-mediated metabolic differences in leaves under light and dark growth conditions can be explained primarily by the different levels of damage to the photosynthesis system under different light conditions. Previous studies have shown that dark treatment weakens the activity of the MEP pathway (Botella-Pavía et al., 2004; Hans et al., 2004; Rodríguez-Concepción et al., 2004; Rodríguez-Villalón et al., 2009), which is consistent with our findings (Fig. 6). Therefore, the reduction in levels of the isoprenoid precursor in the dark might be weakened, which results in the limited reduction in the levels of downstream products.
Post-Transcriptional Regulation of SCD in the MEP Pathway
Although the function of SCD in maize was confirmed in this study, it remains unclear how SCD is regulated during the complex metabolic flux to biosynthesize MEP-derived isoprenoids. The MEP pathway is thought to branch to produce many families of metabolites, and the common core reactions of this pathway are thought to be tightly regulated to assure the coordination of metabolic flux. Due to the pleiotropic effects of light damage, we only compared the DEGs identified in the developing endosperms and embryos and dark-grown leaves of scd versus the wild type. Unexpectedly, the number of down-regulated DEGs involved in a known biological process related to isoprenoid biosynthesis was far smaller in the dark than in the light, with none identified in the endosperms and only two and three DEGs from the carotenoid and chlorophyll pathways identified in dark-grown leaves and embryos, respectively (Fig. 7). One possible explanation for this finding involves the existence of a posttranscriptional or posttranslational regulatory mechanism, as has been found in other species (Hemmerlin et al., 2012; Hemmerlin, 2013). This molecular regulatory mechanism operates for other enzymes in the MEP pathway, e.g. DXS in Arabidopsis (Guevara-García et al., 2005).
Theoretically, up-regulating the genes in the MEP pathway will force metabolic flux to the downstream isoprenoids, consequently leading to the enhanced accumulation of isoprenoids. For example, overexpressing genes for certain upstream precursor pathway enzymes (e.g. DXS, DXR, and HDR) was shown to force metabolic flux to the downstream pathway (Estévez et al., 2001; Botella-Pavía et al., 2004; Carretero-Paulet et al., 2006). By contrast, no obvious coordinated changes were detected for the levels of isoprenoids in the leaves or seeds of SCD-overexpressing transgenic lines, and the up-regulation of SCD expression was not coordinated with that of other genes controlling plastid isoprenoid biosynthesis. Indeed, the expression levels of SCD/HDS are not significantly correlated with carotenoid contents in maize endosperms (Vallabhaneni and Wurtzel, 2009). Similar results were also obtained for SCD in E. coli and Arabidopsis (Flores-Pérez et al., 2008) and DXR, another gene in the MEP pathway, in Arabidopsis (Xing et al., 2010). Perhaps the expression levels of SCD are normally higher than needed and thus metabolic changes will occur only after changes in the enzyme levels reach a certain threshold, e.g. knockout of SCD. In addition, as suggested by the transcriptional changes detected in scd, SCD might undergo posttranscriptional or posttranslational regulation. For example, the mislocalization of SCD protein might have a regulatory effect when the protein levels in SCD-overexpressing transgenic lines are elevated. However, the likelihood of such an effect is reduced in scd when it is rescued by SCD overexpression in transgenic plants, pointing to the correct localization of SCD. Thus, it appears that metabolic balance is more sensitive to the down-regulation of the MEP pathway than its up-regulation.
In summary, we demonstrated that SCD affects the production of plastid isoprenoid precursors in the MEP pathway in maize, subsequently affecting the accumulation of its downstream isoprenoids, thereby affecting plant development. The metabolic and transcriptional changes caused by the loss of SCD function varied among tissues and light conditions. The identification of a few DEGs related to MEP-derived isoprenoid biosynthesis suggests that posttranscriptional regulation might be a major regulatory mechanism of SCD. These findings advance our understanding of the roles of SCD in plants.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
The scd mutant in maize originated from a natural mutation that occurred in recombinant inbred line RIL105 derived from a cross between two elite inbred lines, Zong3 and Yu87-1 (Tang et al., 2007). Three additional scd mutants were obtained from the Maize Genetics Cooperation Stock Center. lw2-vp12 (stock number 512I lw2-vp12) was derived from an unknown tropical variety. scd-1 (stock number 523B lw2-UFMu-07766) was isolated from the UniformMu line UFMu-07766 in the W22 background. scd-2 (stock number 518D lw2) is a spontaneous mutant in the maize variety ‘Golden Glory’ background. The mutation sites of these alleles were determined by sequencing with the primers listed in Supplemental Table S5. The plants were cultivated in a greenhouse under light or dark conditions or in an experimental field at the China Agricultural University in Beijing and Hainan, China. Seedlings were grown in growth chambers under a 16-h-light/8-h-dark cycle for light conditions or a 24-h-dark cycle for dark conditions.
Measurement of the Chemical Composition of Seeds and Leaves
To analyze the chemical composition of seeds, 50 seeds were randomly chosen from each well-grown ear. To analyze the chemical composition of leaves, 0.1–0.2 g of tissue was collected from fresh seedling leaves. The chlorophyll pigments were extracted in 95% (v/v) ethanol, and the levels of chlorophyll a and chlorophyll b were determined with a UV/visible spectroscopy spectrophotometer to measuring absorbance at 665 and 649 nm, respectively (Lichtenthaler, 1987). To measure carotenoid and tocopherol contents in both seeds and leaves, HPLC was performed (Chander et al., 2008). All measurements were replicated at least three times.
Microscopy
To visualize leaf and seed development, leaves sampled from V1-stage wild type and scd seedlings and seeds dissected from self-pollinated wild type and SCD/scd ears at 15 DAP were fixed for 2 h in 2.5% (v/v) glutaraldehyde buffered with 0.1 M, pH 7.2, phosphate buffer, washed three times in 0.1 M, pH 7.2, phosphate buffer, postfixed for 2 h in 1% (w/v) osmic acid, washed in phosphate buffer, dehydrated in an acetone gradient up to 100% (v/v), and slowly embedded in Spurr epoxy resin.
For light microscopy analysis of leaf tissue, 0.75-μm semithin sections were cut with an OM-U3 microtome (Reichert-Jung) and photographed under an Olympus BX-60 microscope using a Jenoptik C5 camera system. For TEM of leaves and seeds, samples were sectioned at 80 nm with a Leica EM UC6 ultramicrotome, affixed to grids, and stained with uranyl acetate and lead citrate. Digital images were acquired using JEOL 2100 200 kV scanning and JEM-1230 transmission electron microscopes.
Map-Based Cloning of SCD
To map the SCD locus, an SCD/scd plant was crossed with B73 plants. Primary mapping was performed with 356 simple sequence repeat or InDel markers using 509 individuals from the F2 mapping population. Subsequently, 828 recessive individual plants with pale-yellow seeds and white leaves were used for fine mapping of SCD with 11 markers (Supplemental Table S5). To identify the mutation site, the corresponding genomic fragments in the refined interval were sequenced in scd and wild type using 27 primer pairs (Supplemental Table S5).
RNA Extraction and RT-qPCR
Total RNA was extracted from various organs using an RNAprep Pure Plant Kit (Tiangen). First-strand cDNA was synthesized using a PrimeScript II first strand cDNA synthesis kit (TAKARA). RT-qPCR was carried out in triplicate for each sample using a SYBR Green I Kit (TAKARA) on a 7500 Real Time PCR System (ABI). Maize TUBBLIN was used as a control for normalization between samples (Supplemental Table S2). Relative transcript levels were calculated using the comparative threshold cycle method (Livak and Schmittgen, 2001).
RACE and Identification of Various Transcripts in the scd Mutant
Total RNA was extracted from V1-stage leaves and 15-DAP endosperms from the scd mutant and wild type. For RACE, first-strand cDNA from wild type leaf tissue was synthesized using a 5′-Full RACE Kit and 3′-Full RACE Core Set (TAKARA). SCD-specific primers were designed to amplify 5′-and 3′-RACE-ready cDNAs (Supplemental Table S5). Sequences from the 5′-and 3′-RACE products were assembled to obtain full-length SCD cDNA. To isolate various transcripts in scd leaves and endosperms, the synthesized first-strand cDNA was amplified using primer pair SCD_FL (Supplemental Table S5). The PCR products from wild type were directly sequenced, whereas the PCR products from the scd mutant were cloned into the pMDTM20 T-Vector (TAKARA), and at least 30 clones were sequenced.
Antibody Preparation, Protein Extraction, and Immunoblot Analysis
A polypeptide containing SCD amino acid residues 733–745 was synthesized and used to raise polyclonal antibodies in rabbits, which were then affinity purified by the Beijing ComWin Biotech Company (China). A 1:100 dilution of the resulting antibody was used for protein blotting. The specificity of SCD antibodies was confirmed by immunoblot analysis of wild type. Total proteins were isolated from various organs as described (Bernard et al., 1994). An equal amount of protein from each sample (30 μg) was loaded onto an 8% SDS-polyacrylamide gel and blotted onto a 0.2-µm nitrocellulose membrane (Schleicher and Schuell). The membranes were blocked with 5% (w/v) nonfat dry milk in Tris-buffered saline with 0.1% (v/v) Tween 20 and incubated with primary antigen-specific antiserum at a 1:200 dilution and secondary alkaline phosphatase–conjugated goat antirabbit IgG (Bio-Rad) at 1:3000 dilution. Immunoblot analysis was performed using the SCD antibody at 1:100 dilution, with Actin antibody (ABclonal) used as the control at 1:1000 dilution.
CRISPR/Cas9 Gene Editing and Genotyping
CRISPR/Cas9 mutagenesis and the generation of transgenic maize plants were performed as described previously (Xing et al., 2014). Briefly, the CRISPR/Cas9 binary vector pBUE-2gRNA-SCD was constructed to produce defined deletions in the coding region of SCD using two single guide RNAs (sgRNAs) alongside the Cas9 endonuclease gene (Supplemental Table S5). The construct was transformed into Agrobacterium tumefaciens strain EHA105 and used to transform immature embryos of maize inbred line B73-329 via Agrobacterium-mediated transformation at China Agricultural University Transgenic Facility Center. To genotype the first-generation (T0) transgenic lines, the white and green leaves of three regenerated plants were separately collected. DNA and RNA were simultaneously extracted using an RNAprep Pure Plant Kit (Tiangen). Each sample was genotyped using a forward primer based on the sequence to the left of sgRNA1 and a reverse primer to the right of sgRNA2 (Supplemental Table S5). PCR products were cloned into the pMDTM20 T-Vector (TAKARA), and at least six clones per PCR product were sequenced. Three positive transgenic plants were subjected to RT-qPCR using the SCD primer pair (Supplemental Table S3).
Subcellular Localization
The coding sequence of SCD was amplified using cDNA from V1-stage wild type seedlings as the template. The resulting PCR product was inserted into the pEZS-NL vector to generate the SCD-EGFP fusion protein driven by the 35S promoter. Maize protoplasts were isolated as described (Yoo et al., 2007), transformed with the 35S::SCD-EGFP construct, incubated for 16–20 h in the dark at 25°C, and analyzed for GFP expression under a Zeiss 710 confocal microscope.
Measurement of Metabolites in the MEP Pathway
The relative levels of the targeted metabolites in the MEP pathway in maize seed and leaf samples were quantified by liquid chromatography–electrospray ionization–tandem mass spectrometry (Li and Sharkey, 2013). The metabolites were extracted from 50 mg of powdered tissue using 1 mL of 50% (v/v) methanol and separated and quantified on a C18 5u column (150 mm × 4.6 mm; Phenomenex) fitted to a 5500 triple quadrupole-linear ion trap (QTRAP) mass spectrometer (AB Sciex). Standards for MEP pathway metabolites, including MEP, MEcPP, HMBPP, IPP, and DMAPP, were acquired from Echelon Biosciences. The metabolites were quantified using the scheduled multiple reaction monitoring method (Liu et al., 2017). Among the detected metabolites, DMAPP and IPP could not be differentiated from each other; thus, the metabolite content was calculated as the average of DMAPP and IPP.
Analysis of Phytohormone Levels by Ultra-HPLC Tandem Electrospray Mass Spectrometry
Phytohormones were extracted and purified in three biological replicates using a protocol modified from Cai et al. (2016). Fresh plant materials were harvested, immediately frozen in liquid nitrogen, and stored at −80°C until use. The samples were ground in liquid nitrogen, and 120 mg (fresh weight) of material per sample was transferred into a 1.5-mL centrifuge tube. Following the addition of solution containing 80% (v/v) methanol (1.2 mL), the mixture was vortexed six times every 0.5 h. After incubation at 4°C for 12 h, the supernatant was collected following centrifugation at 12,000 g for 15 mins at 4°C. The supernatant was evaporated to dryness under a mild nitrogen stream at 35°C and redissolved in 0.1 mL solution containing 30% (v/v) methanol. The solution was centrifuged, and the supernatant was collected for phytohormone analysis. Twenty-one standards (Supplemental Table S6) were purchased from Olchemim Ltd. and Sigma. Stock solutions of all standards were prepared at a concentration of 10 mg/mL in acetonitrile and stored at −18°C until use. To construct the standard curve, the stock solutions were diluted with acetonitrile into working solutions, and 18 gradient-mixed standard solutions ranging from 0.02 to 2000 ng/mL (0.02, 0.05, 0.1, 0.2, 0.5, 1, 2, 5, 10, 20, 30, 50, 100, 200, 400, 800,1600, and 2000 ng/mL) were prepared.
The levels of auxins, GAs, cytokinins, jasmonic acids, salicylic acid, and ABA in each sample and mixed standard were detected by ultra-HPLC tandem electrospray mass spectrometry (UPLC-MS/MS; AB Sciex). Separation was performed on an ACQUITY UPLC HSS T3 C18 column (1.8 µm, 2.1 mm × 100 mm) at 40°C, with a mobile phase flow rate of 0.35 mL/min. The mobile phase consisted of water (A) and acetonitrile (B), both containing 0.04% (v/v) acetic acid. A linear gradient elution program was applied as follows: initial 5% B for 11 min, 95% B at 11.1 min for 1 min, 5% B at 12.1 min for 3 min, and 5% B at 15 min. The injection volume was 5.0 µL. The effluent was alternatively connected to an electrospray ionization (ESI)-QTRAP mass spectrometer. The API 6500 QTRAP LC/MS/MS system, equipped with an ESI Turbo Ion-Spray interface, was controlled by Analyst 1.6 software (AB Sciex). The ESI source operation parameters were as follows: ion source, turbo spray; source temperature, 500°C; ion spray voltage, 5500 V; curtain gas, 35.0 psl; and collision gas set to medium. Detailed information about each measured metabolite is provided in Supplemental Table S6. Data acquisition and processing were performed using Analyst 1.6 software (AB Sciex).
RNA-Sequence Analysis
Eighteen samples, including 15-DAP endosperms and embryos and V1-stage leaves from scd and wild type plants, were subjected to RNA-sequence (RNA-seq). Three independent biological replicates were analyzed per tissue. RNA-seq libraries were constructed according to the standard Illumina RNA-seq protocol, followed by paired-end sequencing on the Illumina HiSeq 2500 platform. Reads were sequenced, demultiplexed, and filtered using the default pipeline and parameters. On average, 126.1 million 125-bp paired-end reads were generated per sample (Supplemental Data Set S1). The reads were aligned to the maize B73 RefGen_V3 reference genome using Tophat2 (Kim et al., 2013), and the number of uniquely mapped reads covering each gene model (AGPv3) was determined using HTSeq software (Anders et al., 2015). The counts were normalized according to library size, and genes with less than one count per million were filtered out. DEGs between scd and wild type plants were identified using the software package edgeR (Robinson et al., 2010), allowing a false discovery rate of 0.01. The potential functions and enriched GO terms of the DEGs were analyzed using MapMan software (http://mapman.gabipd.org; Thimm et al., 2004). To investigate the enrichment of GO categories, Fisher’s exact test was used to analyze the number of genes per bin. P-values were adjusted for multiple tests based on the false discovery rate using the Benjamini and Hochberg method (Benjamini and Hochberg, 1995). Bins for which the false discovery rate was < 0.05 were considered to be enriched.
Overexpression of SCD and Genotyping and Phenotyping of the Resulting Lines
The full-length coding sequence of SCD was amplified from cDNA with primer pair PBCXUN_SCD (Supplemental Table S6) and cloned into the binary vector PBCXUN under the control of the maize ubiquitin-1 promoter, using XcmI as the restriction enzyme. The recombinant plasmid PBCXUN-SCD was introduced into Agrobacterium strain EHA105, which was then transformed into B73-329 at the China Agricultural University Transgenic Facility Center. The SCD-specific primer pair ZmSCD was used to screen for transgene-positive plants, which were further confirmed by RT-qPCR using the same primers (Supplemental Table S3). To evaluate the phenotypic effects of overexpressing SCD, the levels of metabolites in the MEP pathway and downstream isoprenoids, i.e. carotenoids and tocopherols, were measured in seedlings and seeds as described above, as were chlorophyll levels in seedlings.
Statistical Analysis
Two-tailed Student’s t test was used to test the significance of expression and metabolite differences between mutants or overexpressing transgenic lines and wild types.
Accession Numbers
Sequence data from this study can be found in the GenBank/EMBL data libraries under accession number MF784564 for SCD, MG181953 for scd, and SRP116562 and SRP159627 for the RNA-seq data.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Simplified diagram of the MVA and MEP pathways and the biosynthesis of their downstream products in plant cells.
Supplemental Figure S2. Comparison of carotenoid and chlorophyll contents in scd vs. WT.
Supplemental Figure S3. Phenotypes of seedlings grown in the dark.
Supplemental Figure S4. Schematic representation of the 7.6-kb terminal repeat retrotransposon insertion in scd.
Supplemental Figure S5. Allelism tests using three additional scd alleles.
Supplemental Figure S6. CRISPR/Cas9-induced mutations in SCD result in lethal seedlings.
Supplemental Figure S7. Full-length SCD cDNA isolated using 5′ and 3′ RACE.
Supplemental Figure S8. Amino acid sequence alignment of SCD in maize and its putative orthologs in Arabidopsis thaliana, Cyanidioschyzon merolae, and Escherichia coli.
Supplemental Figure S9. Expression patterns of SCD in various tissues at different developmental stages in WT and scd.
Supplemental Figure S10. Subcellular localization of SCD.
Supplemental Figure S11. Comparison of tocopherol contents between the lw2-vp2 mutant and WT.
Supplemental Figure S12. Expression patterns of 10 genes in 15-DAP endosperms and embryos and leaves from light-grown and dark-grown V1-stage scd and WT seedlings, as estimated by RT-qPCR.
Supplemental Figure S13. Schematic diagram of functional complementation of scd via crossing with OE1, an SCD-overexpressing transgenic line.
Supplemental Figure S14. Comparison of carotenoid, chlorophyll, and tocopherol contents between the complemented individuals, SCD overexpressors, and wild type.
Supplemental Figure S15. Expression patterns of 24 genes in 15-DAP endosperms and embryos and V1-stage seedling leaves from overexpressing transgenic lines OE1, OE2, and WT, as estimated by RT-qPCR.
Supplemental Table S1. χ2 test of the ratio of yellow seeds to pale-yellow seeds (3:1) derived from heterozygotes of four scd mutant alleles and from the F1 hybrids between the heterozygotes for scd, lw2-vp12, and two additional alleles, scd-1 and scd-2.
Supplemental Table S2. List of the 60 genes related to carotenoid biosynthesis, providing an overview of the transcriptome profiles.
Supplemental Table S3. Primers used for RT-qPCR in this study.
Supplemental Table S4. Comparison of metabolite levels downstream of the MEP pathway in seeds and seedlings between the two SCD-overexpressing lines and wild type.
Supplemental Table S5. Primers used for mapping, sequencing, and vector construction in this study.
Supplemental Table S6. ESI+-MS/MS parameters used for phytohormone measurements.
Supplemental Data Set S1. List of DEGs in 15-DAP endosperms, 15-DAP embryos, and light- and dark-grown seedling leaves.
Acknowledgments
We greatly appreciate Dr. Philip Stinard from the Maize Genetics Cooperation Stock Center for providing maize genetic stocks, Dr. Byung-Ho Kang at University of Florida for help in identifying plastids in maize seeds, and the Maize Functional Genomic Project of China Agricultural University for generating the CRISPR/Cas9-mediated mutants and overexpressing seeds. The authors declare that they have no competing interests.
Footnotes
This work was supported by the National Natural Science Foundation of China (NSFC) (31671697) and the National Key Research and Development Program of China (2016YFD0100503).
References
- Altincicek B, Kollas AK, Sanderbrand S, Wiesner J, Hintz M, Beck E, Jomaa H (2001) GcpE is involved in the 2-C-methyl-D-erythritol 4-phosphate pathway of isoprenoid biosynthesis in Escherichia coli. J Bacteriol 183: 2411–2416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S, Pyl PT, Huber W (2015) HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics 31: 166–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araki N, Kusumi K, Masamoto K, Niwa Y, Iba K (2000) Temperature-sensitive Arabidopsis mutant defective in 1-deoxy-D-xylulose 5-phosphate synthase within the plastid non-mevalonate pathway of isoprenoid biosynthesis. Physiol Plant 108: 19–24 [Google Scholar]
- Bauer J, Hiltbrunner A, Kessler F (2001) Molecular biology of chloroplast biogenesis: Gene expression, protein import and intraorganellar sorting. Cell Mol Life Sci 58: 420–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y (1995) Controlling the false discovery rate: A practical and powerful approach to multiple testing. J R Stat Soc Series B Stat Methodol 57: 289–300 [Google Scholar]
- Bernard L, Ciceri P, Viotti A (1994) Molecular analysis of wild-type and mutant alleles at the Opaque-2 regulatory locus of maize reveals different mutations and types of O2 products. Plant Mol Biol 24: 949–959 [DOI] [PubMed] [Google Scholar]
- Botella-Pavía P, Besumbes O, Phillips MA, Carretero-Paulet L, Boronat A, Rodríguez-Concepción M (2004) Regulation of carotenoid biosynthesis in plants: Evidence for a key role of hydroxymethylbutenyl diphosphate reductase in controlling the supply of plastidial isoprenoid precursors. Plant J 40: 188–199 [DOI] [PubMed] [Google Scholar]
- Bouvier F, Rahier A, Camara B (2005) Biogenesis, molecular regulation and function of plant isoprenoids. Prog Lipid Res 44: 357–429 [DOI] [PubMed] [Google Scholar]
- Buckner B, Kelson TL, Robertson DS (1990) Cloning of the y1 Locus of maize, a gene involved in the biosynthesis of carotenoids. Plant Cell 2: 867–876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budziszewski GJ, Lewis SP, Glover LW, Reineke J, Jones G, Ziemnik LS, Lonowski J, Nyfeler B, Aux G, Zhou Q, et al. (2001) Arabidopsis genes essential for seedling viability: Isolation of insertional mutants and molecular cloning. Genetics 159: 1765–1778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai WJ, Ye TT, Wang Q, Cai BD, Feng YQ (2016) A rapid approach to investigate spatiotemporal distribution of phytohormones in rice. Plant Methods 12: 47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carretero-Paulet L, Ahumada I, Cunillera N, Rodríguez-Concepción M, Ferrer A, Boronat A, Campos N (2002) Expression and molecular analysis of the Arabidopsis DXR gene encoding 1-deoxy-D-xylulose 5-phosphate reductoisomerase, the first committed enzyme of the 2-C-methyl-D-erythritol 4-phosphate pathway. Plant Physiol 129: 1581–1591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carretero-Paulet L, Cairó A, Botella-Pavía P, Besumbes O, Campos N, Boronat A, Rodríguez-Concepción M (2006) Enhanced flux through the methylerythritol 4-phosphate pathway in Arabidopsis plants overexpressing deoxyxylulose 5-phosphate reductoisomerase. Plant Mol Biol 62: 683–695 [DOI] [PubMed] [Google Scholar]
- Chander S, Guo YQ, Yang XH, Zhang J, Lu XQ, Yan JB, Song TM, Rocheford TR, Li JS (2008) Using molecular markers to identify two major loci controlling carotenoid contents in maize grain. Theor Appl Genet 116: 223–233 [DOI] [PubMed] [Google Scholar]
- Cordoba E, Porta H, Arroyo A, San Román C, Medina L, Rodríguez-Concepción M, León P (2011) Functional characterization of the three genes encoding 1-deoxy-D-xylulose 5-phosphate synthase in maize. J Exp Bot 62: 2023–2038 [DOI] [PubMed] [Google Scholar]
- Demmig-Adams B, Gilmore AM, Adams WW III (1996) Carotenoids 3: In vivo function of carotenoids in higher plants. FASEB J 10: 403–412 [DOI] [PubMed] [Google Scholar]
- Estévez JM, Cantero A, Romero C, Kawaide H, Jiménez LF, Kuzuyama T, Seto H, Kamiya Y, León P (2000) Analysis of the expression of CLA1, a gene that encodes the 1-deoxyxylulose 5-phosphate synthase of the 2-C-methyl-D-erythritol-4-phosphate pathway in Arabidopsis. Plant Physiol 124: 95–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estévez JM, Cantero A, Reindl A, Reichler S, León P (2001) 1-Deoxy-D-xylulose-5-phosphate synthase, a limiting enzyme for plastidic isoprenoid biosynthesis in plants. J Biol Chem 276: 22901–22909 [DOI] [PubMed] [Google Scholar]
- Flores-Pérez U, Pérez-Gil J, Rodríguez-Villalón A, Gil MJ, Vera P, Rodríguez-Concepción M (2008) Contribution of hydroxymethylbutenyl diphosphate synthase to carotenoid biosynthesis in bacteria and plants. Biochem Biophys Res Commun 371: 510–514 [DOI] [PubMed] [Google Scholar]
- Goff SA, Klee HJ (2006) Plant volatile compounds: Sensory cues for health and nutritional value? Science 311: 815–819 [DOI] [PubMed] [Google Scholar]
- Guevara-García A, San Román C, Arroyo A, Cortés ME, de la Luz Gutiérrez-Nava M, León P (2005) Characterization of the Arabidopsis clb6 mutant illustrates the importance of posttranscriptional regulation of the methyl-D-erythritol 4-phosphate pathway. Plant Cell 17: 628–643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutiérrez-Nava MdeL, Gillmor CS, Jiménez LF, Guevara-García A, León P (2004) CHLOROPLAST BIOGENESIS genes act cell and noncell autonomously in early chloroplast development. Plant Physiol 135: 471–482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hans J, Hause B, Strack D, Walter MH (2004) Cloning, characterization, and immunolocalization of a mycorrhiza-inducible 1-deoxy-d-xylulose 5-phosphate reductoisomerase in arbuscule-containing cells of maize. Plant Physiol 134: 614–624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemmerlin A. (2013) Post-translational events and modifications regulating plant enzymes involved in isoprenoid precursor biosynthesis. Plant Sci 203-204: 41–54 [DOI] [PubMed] [Google Scholar]
- Hemmerlin A, Harwood JL, Bach TJ (2012) A raison d’être for two distinct pathways in the early steps of plant isoprenoid biosynthesis? Prog Lipid Res 51: 95–148 [DOI] [PubMed] [Google Scholar]
- Hirashima M, Satoh S, Tanaka R, Tanaka A (2006) Pigment shuffling in antenna systems achieved by expressing prokaryotic chlorophyllide a oxygenase in Arabidopsis. J Biol Chem 281: 15385–15393 [DOI] [PubMed] [Google Scholar]
- Hoober JK, Eggink LL (2001) A potential role of chlorophylls b and c in assembly of light-harvesting complexes. FEBS Lett 489: 1–3 [DOI] [PubMed] [Google Scholar]
- Hsieh MH, Goodman HM (2005) The Arabidopsis IspH homolog is involved in the plastid nonmevalonate pathway of isoprenoid biosynthesis. Plant Physiol 138: 641–653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh MH, Goodman HM (2006) Functional evidence for the involvement of Arabidopsis IspF homolog in the nonmevalonate pathway of plastid isoprenoid biosynthesis. Planta 223: 779–784 [DOI] [PubMed] [Google Scholar]
- Hsieh MH, Chang CY, Hsu SJ, Chen JJ (2008) Chloroplast localization of methylerythritol 4-phosphate pathway enzymes and regulation of mitochondrial genes in ispD and ispE albino mutants in Arabidopsis. Plant Mol Biol 66: 663–673 [DOI] [PubMed] [Google Scholar]
- Hunter CT, Saunders JW, Magallanes-Lundback M, Christensen SA, Willett D, Stinard PS, Li QB, Lee K, DellaPenna D, Koch KE (2018) Maize w3 disrupts homogentisate solanesyl transferase (ZmHst) and reveals a plastoquinone-9 independent path for phytoene desaturation and tocopherol accumulation in kernels. Plant J 93: 799–813 [DOI] [PubMed] [Google Scholar]
- Johnson EJ. (2002) The role of carotenoids in human health. Nutr Clin Care 5: 56–65 [DOI] [PubMed] [Google Scholar]
- Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL (2013) TopHat2: Accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol 14: R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzuyama T, Seto H (2003) Diversity of the biosynthesis of the isoprene units. Nat Prod Rep 20: 171–183 [DOI] [PubMed] [Google Scholar]
- Li Z, Sharkey TD (2013) Metabolic profiling of the methylerythritol phosphate pathway reveals the source of post-illumination isoprene burst from leaves. Plant Cell Environ 36: 429–437 [DOI] [PubMed] [Google Scholar]
- Li F, Murillo C, Wurtzel ET (2007) Maize Y9 encodes a product essential for 15-cis-zeta-carotene isomerization. Plant Physiol 144: 1181–1189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li ZH, Matthews PD, Burr B, Wurtzel ET (1996) Cloning and characterization of a maize cDNA encoding phytoene desaturase, an enzyme of the carotenoid biosynthetic pathway. Plant Mol Biol 30: 269–279 [DOI] [PubMed] [Google Scholar]
- Lichtenthaler HK. (1987) Chlorophylls and carotenoids: Pigments of photosynthetic biomembranes. Methods Enzymol 148: 350–382 [Google Scholar]
- Liu R, Hong J, Xu X, Feng Q, Zhang D, Gu Y, Shi J, Zhao S, Liu W, Wang X, et al. (2017) Gut microbiome and serum metabolome alterations in obesity and after weight-loss intervention. Nat Med 23: 859–868 [DOI] [PubMed] [Google Scholar]
- Liu YL, Guerra F, Wang K, Wang W, Li J, Huang C, Zhu W, Houlihan K, Li Z, Zhang Y, Nair SK, Oldfield E (2012) Structure, function and inhibition of the two- and three-domain 4Fe-4S IspG proteins. Proc Natl Acad Sci USA 109: 8558–8563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Lois LM, Rodríguez-Concepción M, Gallego F, Campos N, Boronat A (2000) Carotenoid biosynthesis during tomato fruit development: Regulatory role of 1-deoxy-D-xylulose 5-phosphate synthase. Plant J 22: 503–513 [DOI] [PubMed] [Google Scholar]
- Lu XM, Hu XJ, Zhao YZ, Song WB, Zhang M, Chen ZL, Chen W, Dong YB, Wang ZH, Lai JS (2012) Map-based cloning of zb7 encoding an IPP and DMAPP synthase in the MEP pathway of maize. Mol Plant 5: 1100–1112 [DOI] [PubMed] [Google Scholar]
- Maluf MP, Saab IN, Wurtzel ET, Sachs MM (1997) The viviparous12 maize mutant is deficient in abscisic acid, carotenoids, and chlorophyll synthesis. J Exp Bot 48: 1259–1268 [Google Scholar]
- Mandel MA, Feldmann KA, Herrera-Estrella L, Rocha-Sosa M, León P (1996) CLA1, a novel gene required for chloroplast development, is highly conserved in evolution. Plant J 9: 649–658 [DOI] [PubMed] [Google Scholar]
- Matthews PD, Luo R, Wurtzel ET (2003) Maize phytoene desaturase and zeta-carotene desaturase catalyse a poly-Z desaturation pathway: Implications for genetic engineering of carotenoid content among cereal crops. J Exp Bot 54: 2215–2230 [DOI] [PubMed] [Google Scholar]
- McCarty DR, Carson CB (1991) The molecular genetics of seed maturation in maize. Physiol Plant 81: 267–272 [Google Scholar]
- Myers AM, James MG, Lin Q, Yi G, Stinard PS, Hennen-Bierwagen TA, Becraft PW (2011) Maize opaque5 encodes monogalactosyldiacylglycerol synthase and specifically affects galactolipids necessary for amyloplast and chloroplast function. Plant Cell 23: 2331–2347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oster U, Tanaka R, Tanaka A, Rüdiger W (2000) Cloning and functional expression of the gene encoding the key enzyme for chlorophyll b biosynthesis (CAO) from Arabidopsis thaliana. Plant J 21: 305–310 [DOI] [PubMed] [Google Scholar]
- Ostrovsky D, Diomina G, Lysak E, Matveeva E, Ogrel O, Trutko S (1998) Effect of oxidative stress on the biosynthesis of 2-C-methyl-D-erythritol-2,4-cyclopyrophosphate and isoprenoids by several bacterial strains. Arch Microbiol 171: 69–72 [DOI] [PubMed] [Google Scholar]
- Owens BF, Lipka AE, Magallanes-Lundback M, Tiede T, Diepenbrock CH, Kandianis CB, Kim E, Cepela J, Mateos-Hernandez M, Buell CR, et al. (2014) A foundation for provitamin A biofortification of maize: Genome-wide association and genomic prediction models of carotenoid levels. Genetics 198: 1699–1716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page JE, Hause G, Raschke M, Gao W, Schmidt J, Zenk MH, Kutchan TM (2004) Functional analysis of the final steps of the 1-deoxy-D-xylulose 5-phosphate (DXP) pathway to isoprenoids in plants using virus-induced gene silencing. Plant Physiol 134: 1401–1413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Querol J, Campos N, Imperial S, Boronat A, Rodríguez-Concepción M (2002) Functional analysis of the Arabidopsis thaliana GCPE protein involved in plastid isoprenoid biosynthesis. FEBS Lett 514: 343–346 [DOI] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK (2010) edgeR: A bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26: 139–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-Concepción M, Forés O, Martinez-García JF, González V, Phillips MA, Ferrer A, Boronat A (2004) Distinct light-mediated pathways regulate the biosynthesis and exchange of isoprenoid precursors during Arabidopsis seedling development. Plant Cell 16: 144–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-Villalón A, Gas E, Rodríguez-Concepción M (2009) Phytoene synthase activity controls the biosynthesis of carotenoids and the supply of their metabolic precursors in dark-grown Arabidopsis seedlings. Plant J 60: 424–435 [DOI] [PubMed] [Google Scholar]
- Ruiz-Sola MA, Rodríguez-Concepción M (2012) Carotenoid biosynthesis in Arabidopsis: A colorful pathway. The Arabidopsis Book 10: e0158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seemann M, Wegner P, Schünemann V, Bui BTS, Wolff M, Marquet A, Trautwein AX, Rohmer M (2005) Isoprenoid biosynthesis in chloroplasts via the methylerythritol phosphate pathway: The (E)-4-hydroxy-3-methylbut-2-enyl diphosphate synthase (GcpE) from Arabidopsis thaliana is a [4Fe-4S] protein. J Biol Inorg Chem 10: 131–137 [DOI] [PubMed] [Google Scholar]
- Singh M, Lewis PE, Hardeman K, Bai L, Rose JK, Mazourek M, Chomet P, Brutnell TP (2003) Activator mutagenesis of the pink scutellum1/viviparous7 locus of maize. Plant Cell 15: 874–884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stinard P. (2013) Data-mining the B73 genome sequence for carotenoid biosynthesis gene candidates. Maize Genet Coop Newsl 86: 29–31 [Google Scholar]
- Tanaka R, Tanaka A (2007) Tetrapyrrole biosynthesis in higher plants. Annu Rev Plant Biol 58: 321–346 [DOI] [PubMed] [Google Scholar]
- Tang JH, Teng WT, Yan JB, Ma XQ, Meng YJ, Dai JR, Li JS (2007) Genetic dissection of plant height by molecular markers using a population of recombinant inbred lines in maize. Euphytica 155: 117–124 [Google Scholar]
- Thimm O, Bläsing O, Gibon Y, Nagel A, Meyer S, Krüger P, Selbig J, Müller LA, Rhee SY, Stitt M (2004) MAPMAN: A user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J 37: 914–939 [DOI] [PubMed] [Google Scholar]
- Thulasiram HV, Erickson HK, Poulter CD (2007) Chimeras of two isoprenoid synthases catalyze all four coupling reactions in isoprenoid biosynthesis. Science 316: 73–76 [DOI] [PubMed] [Google Scholar]
- Vallabhaneni R, Wurtzel ET (2009) Timing and biosynthetic potential for carotenoid accumulation in genetically diverse germplasm of maize. Plant Physiol 150: 562–572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venado RE, Owens BF, Ortiz D, Lawson T, Mateos-Hernandez M, Ferruzzi MG, Rocheford TR (2017) Genetic analysis of provitamin A carotenoid β-cryptoxanthin concentration and relationship with other carotenoids in maize grain (Zea mays L.). Mol Breed 37: 127 [Google Scholar]
- Von Wettstein D, Gough S, Kannangara CG (1995) Chlorophyll biosynthesis. Plant Cell 7: 1039–1057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vranová E, Coman D, Gruissem W (2012) Structure and dynamics of the isoprenoid pathway network. Mol Plant 5: 318–333 [DOI] [PubMed] [Google Scholar]
- Vranová E, Coman D, Gruissem W (2013) Network analysis of the MVA and MEP pathways for isoprenoid synthesis. Annu Rev Plant Biol 64: 665–700 [DOI] [PubMed] [Google Scholar]
- Walter MH, Hans J, Strack D (2002) Two distantly related genes encoding 1-deoxy-d-xylulose 5-phosphate synthases: differential regulation in shoots and apocarotenoid-accumulating mycorrhizal roots. Plant J 31: 243–254 [DOI] [PubMed] [Google Scholar]
- Xiao Y, Savchenko T, Baidoo EEK, Chehab WE, Hayden DM, Tolstikov V, Corwin JA, Kliebenstein DJ, Keasling JD, Dehesh K (2012) Retrograde signaling by the plastidial metabolite MEcPP regulates expression of nuclear stress-response genes. Cell 149: 1525–1535 [DOI] [PubMed] [Google Scholar]
- Xing HL, Dong L, Wang ZP, Zhang HY, Han CY, Liu B, Wang XC, Chen QJ (2014) A CRISPR/Cas9 toolkit for multiplex genome editing in plants. BMC Plant Biol 14: 327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing S, Miao J, Li S, Qin G, Tang S, Li H, Gu H, Qu LJ (2010) Disruption of the 1-deoxy-D-xylulose-5-phosphate reductoisomerase (DXR) gene results in albino, dwarf and defects in trichome initiation and stomata closure in Arabidopsis. Cell Res 20: 688–700 [DOI] [PubMed] [Google Scholar]
- Xu Z, Wang H (2007) LTR_FINDER: An efficient tool for the prediction of full-length LTR retrotransposons. Nucleic Acids Res 35: W265-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo SD, Cho YH, Sheen J (2007) Arabidopsis mesophyll protoplasts: A versatile cell system for transient gene expression analysis. Nat Protoc 2: 1565–1572 [DOI] [PubMed] [Google Scholar]