Abstract
Molecular exchanges between plants and biotrophic, hemibiotrophic, and necrotrophic oomycetes affect disease progression.
The microbial eukaryotes known as oomycetes comprise more than 1,500 species, including many important phytopathogens. Most exhibit filamentous growth and feed osmotrophically. Oomycetes appear fungus-like but are classified as stramenopiles along with brown algae and diatoms (Beakes et al., 2012). Unlike most fungi, oomycetes are diploid, have cell walls made primarily of cellulose and β-glucans instead of chitin, make aseptate hyphae, undergo oogamous reproduction, and produce few secondary metabolites (Fawke et al., 2015).
Oomycetes exhibit diverse lifestyles across terrestrial and aquatic niches. While best known as pathogens of leaves, stems, roots, and fruit, some oomycetes are endophytes, infect animals, or are saprophytes (Lamour and Kamoun, 2009; Ploch and Thines, 2011; Aram and Rizzo, 2018). Many are highly host adapted, unculturable on artificial media, and grow only on living plants as biotrophs. Examples include downy mildew pathogens such as Plasmopara viticola, which infects grapevine (Vitis vinifera), and Albugo candida, which causes white rust on crucifers (Kamoun et al., 2015). The obligate pathogens typically cause minimal damage to the plant but reduce yield and raise susceptibility to secondary infection or abiotic stress.
Many oomycetes are hemibiotrophs, which start infections like biotrophs but cause necrosis late in the disease cycle. Most belong to the genus Phytophthora, including Phytophthora cinnamomi, which infects hundreds of agricultural, forest, and ornamental hosts; Phytophthora infestans, which blights potato (Solanum tuberosum) and tomato (Solanum lycopersicum); and Phytophthora sojae, which colonizes soybean (Glycine max) and lupines. Some species, such as Ph. cinnamomi, shift to necrotrophy early in infection, while others, such as Ph. infestans, make the transition much later, reflecting differences in how the species balance the two trophic behaviors. Unlike many other oomycetes, Phytophthora spp. are culturable and amenable to transformation; thus, they have been the subject of many molecular studies.
The largest genus of necrotrophic oomycetes, which feed on nutrients from lysed cells, is Pythium. Most members of this group are opportunistic root pathogens with broad host ranges, such as Pythium ultimum, which infects vegetables, grains, and trees (Kamoun et al., 2015). Interestingly, some Pythium spp. also are mycoparasites (Benhamou et al., 2012). Also appearing to grow as a necrotroph is Aphanomyces euteiches, which causes root rot of legumes.
This review focuses on events at the plant-oomycete interface, where exchanges of host and pathogen molecules play critical roles in determining the outcome of the association (Fig. 1). Oomycete pathogens sense, bind, and absorb nutrients from their hosts and also interact with other microbes in the phyllosphere and rhizosphere. Meanwhile, plants detect and deliver defenses against infection. Plant-oomycete interfaces can be dynamic, varying with infection stage and as immune responses are deployed. Here, we discuss insights into these topics yielded by advances in cell biology, genome analysis, transcriptomics, and protein structure analysis.
Figure 1.
Interactions at plant-oomycete interfaces. Illustrated at center left is a biotrophic infection, starting from a sporangium and involving biflagellated zoospores, an appressorium formed from a germinated cyst, a primary infection vesicle (pv), an intercellular mycelium, and a haustorium. The effects of plant signals such as isoflavones, sucrose (suc), and amino acids (aa) on spore germination and/or homing are indicated. The bacterium at top right represents the effects of the microbiome on spores, as discussed in Box 2. The oval organelle marked “sequestered nutrients” represents a starch granule; this only releases significant carbohydrate to the pathogens during necrotrophy. The turquoise pentagon represents a nutrient such as sulfate that is located primarily in a plant vacuole. Yellow stars represent apoplastic effectors such as protease inhibitors (ae) and cytoplasmic effectors such as Crinklers (C) and RXLRs (R). The latter are shown inhibiting the delivery of defense materials, such as proteases and callose, to the apoplast and EHMx by secretory or autophagosomal vesicles of a mesophyll cell. These defense responses also occur in the epidermis. Shown at top right are the initial stages of infection initiated through a stomata (gray mycelium). Shown at right is an opportunistic necrotroph (spotted mycelium) entering through a wound, feeding from a lysed cell, and exiting into soil. Lysis of the host during infection by the necrotroph occurs due to the absence of defense-suppressing effectors, ROS generation, and early expression of NLPs, as discussed in Box 3.
PLANTS CAN ATTRACT UNWANTED GUESTS
Oomycetes employ several types of spores for dissemination and host infection (Box 1). These include both asexual and sexual spores (McCarren et al., 2005; Granke et al., 2009). Colonization by the majority of oomycetes begins when an asexual sporangium releases zoospores, which encyst and form a germ tube (Fig. 1). As discussed below, many aspects of spore behavior are influenced by plant signals. The microbiome also affects spores and can attenuate or worsen disease, as described in Box 2 (Lioussanne et al., 2008; Windstam and Nelson, 2008; Raaijmakers et al., 2010; Schlatter et al., 2017; Jack and Nelson, 2018).
Host signals can be sensed by the asexual sporangia since they are fully hydrated and metabolically active prior to germination, unlike most fungal spores, which are desiccated. While sporangia require only free water to germinate, this can be hastened by plant signals. Studies have shown that Pl. viticola releases zoospores faster on leaves than in a host-free system (Kiefer et al., 2002) and that Pythium spp. germination is accelerated by volatiles, sugars, and amino acids from seeds (Nelson, 1987). Root exudates, or sprouted potato tubers in the case of Ph. infestans, also stimulate the germination of sexual spores (oospores), which typically stay dormant in soil until a host is present (El-Hamalawi and Erwin, 1986; Pittis and Shattock, 1994). Studies with Ap. euteiches indicated that its oospores respond more to host than nonhost exudates (Shang et al., 2000). It is intriguing to consider that in the future, it may be possible to use plant signal mimics to cause oospores to undergo suicide germination before a crop is planted.
Zoospores exhibit several homing responses, including chemotaxis, electrotaxis, host-triggered encystment, and germ tube tropism (Deacon and Donaldson, 1993). These contribute to host specificity, especially with root pathogens. For instance, Ap. euteiches zoospores are attracted specifically to prunetin (Sekizaki et al., 1993), while Ph. sojae responds to daizein and genistein, which are produced by their respective hosts (Hosseini et al., 2014). These isoflavones also influence encystment and germ tube orientation (Morris et al., 1998). Recent data point to a role for G-proteins in these responses. Silencing of the Ph. sojae gene encoding its G-protein α-subunit interfered with zoospore motility and chemotaxis (Hua et al., 2008), and knockdowns of a G-protein α-subunit-interacting His triad protein inhibited chemotaxis (Zhang et al., 2016). In addition, encystment was stimulated and cyst germination was impaired by knocking down the expression of a protein that consists of a G-protein-coupled receptor domain coupled to a phosphatidylinositol phosphate kinase domain (Yang et al., 2013). Oomycetes express several novel G-protein-coupled receptor-like proteins with C-terminal accessory domains (van den Hoogen et al., 2018).
Pharmacological studies have shown that calcium influences most aspects of zoospore behavior. This explains the biology behind the strategy of reducing root diseases by adding gypsum (calcium sulfate) to soil, which impairs zoosporogenesis or causes encystment before a plant is reached (Mostowfizadeh-Ghalamfarsa et al., 2018). Many spore-specific calcium channels and calcium-regulated protein phosphatases and kinases have been identified, although none have been tested for function (Ah-Fong et al., 2017b).
Chemotaxis also occurs in foliar pathogens, where amino acids such as Gln attract zoospores, a process that also appears to involve G-proteins (Latijnhouwers et al., 2004). Amino acid signaling may explain why zoospores of Ph. infestans and many relatives concentrate near stomata (Dale and Irwin, 1991). Few Pl. viticola zoospores were drawn to stomata closed by exogenous abscisic acid, suggesting that the attractants are soluble or volatile substomatal chemicals. Such behavior is critical to Pl. viticola, which enters leaves only through stomata (Kiefer et al., 2002).
OOMYCETES ENTER PLANTS THROUGH MULTIPLE ROUTES
As water molds, most oomycetes prefer to grow in moist environments such as the apoplast. Entry into the plant may occur when zoospores or germ tubes pass through stomata or other natural openings, transit through wounds, or grow between root epidermal cells. Examples include Pl. viticola, which enters through stomata, Ph. cinnamomi, which can move through peridermal gaps, and Ph. infestans, which often enters tubers through lenticels. Ph. infestans, downy mildews, white rusts, and many Pythium spp. also penetrate tissue using appressoria. These swellings form when cyst germ tubes contact hydrophobic surfaces such as the cuticle, especially if epidermal cell boundaries or their topographic mimics are sensed (Bircher and Hohl, 1997).
Insight into the biology of oomycete appressoria has lagged behind that of fungi. However, a study in Ph. infestans using GFP-labeled F-actin identified an aster-like structure where appressoria contact the leaf, which may focus cargo transport to the penetration peg (Kots et al., 2017). Also, a basic leucine zipper domain transcription factor and mitogen-activated protein kinase were shown to regulate appressorium formation (Blanco and Judelson, 2005; Li et al., 2010). Genes induced in the appressorium stage by Phytophthora spp. include cell wall-degrading enzymes (CWDEs), defense-suppressing effectors, and potential adhesion proteins (Kebdani et al., 2010).
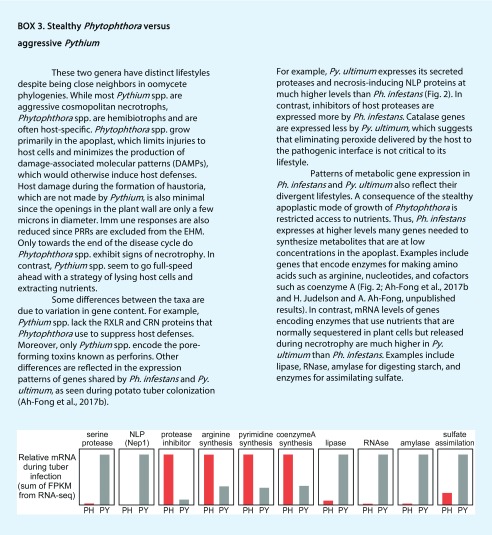
Mirroring the complexity of the plant cell wall, a typical oomycete expresses CWDEs belonging to as many as 28 glycosyl hydrolase groups (Blackman et al., 2015). A typical species of Phytophthora expresses about 200 genes encoding such proteins. Some of the (hemi)cellulases are predicted to bear glycophosphatidylinositol anchors and probably serve to expand the oomycete wall, which contains mostly cellulose plus β-1,3- and β-1,6-glucans (Mélida et al., 2013). Fewer types of CWDEs are expressed by biotrophs, as in the case of Albugo laibachii, which lacks pectate lyase and pectin esterase (Kemen et al., 2011). Studies in Ph. infestans and relatives show that CWDEs are expressed in stages during sporulation, germination, and in planta growth (Kebdani et al., 2010; Blackman et al., 2015). A less ordered pattern of expression was reported for Py. ultimum, which also expressed fewer CWDEs (Ah-Fong et al., 2017b). Other differences between Phytophthora and Pythium spp. are highlighted in Box 3. The pattern of CWDE expression in Py. ultimum suggests that the enzymes of this necrotroph may be used primarily to burst host cells rather than to digest plant walls for carbon. Indeed, cellulose is a poor carbon source for most oomycetes (Zerillo et al., 2013). Perhaps advanced imaging techniques such as superresolution confocal microscopy with specific organic fluorophores could be employed to obtain information about carbohydrate structure at penetration sites, the effects of CWDEs at different stages of infection, and polymer rearrangements resulting from plant defenses.
Most stages of infection require adherence of the pathogen to the host. Zoospores turn their ventral grooves toward the host prior to encystment, allowing vesicles to discharge a glue-like thrombospondin repeat protein toward the plant interface (Robold and Hardham, 2005). A protein containing a Sushi domain, which in animals mediates cell-cell adhesion, reaches the plant surface from other zoospore vesicles by kiss-and-run exocytosis (Zhang et al., 2013). Sticky substances also are released from germ tubes. The downy mildew Hyaloperonospora arabidopsidis was shown to secrete proteins and fibrillar β-1,3-glucans that bind its germ tubes to the substratum (Carzaniga et al., 2001). This may help resist detachment by wind or rain or protect against desiccation. Other potential adhesion proteins include mucin-like proteins (Larousse et al., 2014), jacalin-like and cellulose-binding elicitor (CBEL) lectins (Gaulin et al., 2002), and the ACWP family of acidic wall proteins (Resjö et al., 2017). Abnormal appressoria resulted from the knockdown of ACWP genes, suggesting that they contribute to adhesion or wall integrity. Knockdowns of CBEL showed that it helped hyphae bind to cellulosic substances but was not essential for pathogenicity (Gaulin et al., 2002). One of the few oomycete proteins known to concentrate in haustoria, Hmp1, is membrane anchored and weakly resembles lectins. Silencing Hmp1 in Ph. infestans impaired the formation of infection vesicles in epidermal cells and haustoria, suggesting that the protein helps the pathogen bind to host interfaces (Avrova et al., 2008).
PLANTS CAN DETECT OOMYCETES AND BRING DEFENSES TO THE INTERFACE
Plants have evolved sophisticated systems for detecting microbes. One involves the binding of pathogen-associated molecular patterns (PAMPs) to plasmalemma-spanning pattern recognition receptors (PRRs), which activates PAMP-triggered immunity (PTI; Saijo et al., 2018). The salicylic acid (SA) and jasmonic acid pathways both participate in PTI against necrotrophic and (hemi)biotrophic oomycetes (Halim et al., 2009). Disrupting jasmonate production in Arabidopsis (Arabidopsis thaliana) allowed Pythium irregulare, which typically infects wounded or otherwise compromised hosts, to become a more severe pathogen (Staswick et al., 1998).
Constituents of the cell wall or plasma membrane were among the first oomycete PAMPs to be identified. These include β-1,3- and β-1,6-glucans and arachidonic acid (Fawke et al., 2015; Robinson and Bostock, 2015). Medicago truncatula also responds to a chitosaccharide from Ap. euteiches; most other oomycete phytopathogens lack this PAMP, since they do not make chitin (Mélida et al., 2013; Nars et al., 2013). Proteinaceous PAMPs include a cell wall transglutaminase (Brunner et al., 2002), the glycosyl hydrolase domain of the oligopeptide elicitor (OPEL) protein (Chang et al., 2015), the cellulose-binding protein CBEL, the elicitin family of sterol-binding proteins (Takenaka et al., 2011; Derevnina et al., 2016), and the XEG1 endoglucanase (Wang et al., 2018). The latter are proposed to be used for sterol acquisition by Phytophthora and Pythium spp., which are sterol autotrophs. It is notable that Nep1-like proteins (NLPs) were classified recently as PAMPs by some researchers. Most NLPs in Phytophthora spp. are expressed late in infection and have been linked to necrotrophic growth (Feng et al., 2014). Analysis of crystal structures identified similarity with pore-forming cytotoxins of sea anemones, which suggests that NLPs destabilize the host plasmalemma (Lenarčič et al., 2017). Oomycetes are immune to NLPs, since the latter are specific for dicotyledonous sphingolipids. An NLP from Phytophthora parasitica was shown to elicit defenses in crucifers, which suggests that some NLPs affect plant cells both as pore-forming toxins and inducers of PTI (Böhm et al., 2014).
Receptors for three oomycete PAMPs are known. The infestin elicitin of Ph. infestans and related proteins are recognized in potato by elicitin response protein (ELR), a plasmalemma-associated factor that associates with SUPPRESSOR OF BIR1-1 (SOBIR1), which is a leucine-rich repeat (LRR) receptor kinase (Domazakis et al., 2018). This pairing is needed since ELR lacks an intracellular kinase domain. When infestin is detected, the ELR-SOBIR1 complex recruits the LRR receptor-like kinase BRI1-ASSOCIATED KINASE-1, which is a known hub in defense responses. SOBIR1 also participates in the reaction of Arabidopsis to NLPs, which are recognized by the LRR receptor RLP23 (Albert et al., 2015). Recently identified was Response to XEG1 (RXEG1), an LLR protein that recognizes XEG1, a glycoside hydrolase 12 endoglucanase that is made by Phytophthora spp. RXEG1also forms a complex with BRI1-ASSOCIATED KINASE-1 and SOBIR1 to transduce the defense signal (Wang et al., 2018). Interestingly, fungal glycoside hydrolase 12 proteins also have been shown to serve as PAMPS and act through the same signaling hub (Gui et al., 2017).
Once PTI is activated, defense molecules are delivered to plant-oomycete interfaces, including pathogenesis-related (PR) proteins, callose for thickening cell walls, and microbial toxins. Effector-triggered immunity reinforces and expands these responses and often leads to hypersensitive cell death. Since PTI and effector-triggered immunity are not oomycete specific, readers seeking more information are directed to other reviews (Kourelis and van der Hoorn, 2018; Saijo et al., 2018). However, oomycetes were used in many early studies of the cytoskeletal dynamics that occur during infection, which showed that plant actin microfilaments focused rapidly near penetration sites (Takemoto et al., 2003). This causes peroxisomes, nuclei, and the endomembrane transport network to move toward the infection, which may help deliver defenses (Li and Staiger, 2018). Some (hemi)biotrophic oomycetes have evolved counter defenses against these trafficking pathways and may have hijacked some to support haustoria.
While the delivery of proteases, glucanases, and callose to oomycete-plant interfaces through canonical secretory and exocytosis pathways is long established, autophagic vesicles were shown recently to surround Ph. infestans haustoria and also may convey defenses (Dagdas et al., 2018). It is unknown whether plants use exosomes against oomycetes, for example by transporting inhibitory small RNAs, as shown recently with fungi (Cai et al., 2018). Nevertheless, there are reports of lettuce (Lactuca sativa) and potato being engineered to resist Bremia lactucae and Ph. infestans by host-induced gene silencing using small RNAs targeting oomycete genes (Govindarajulu et al., 2015; Jahan et al., 2015).
Reactive oxygen species (ROS) are delivered to plant-oomycete interfaces through several pathways. ROS are derived from wall-bound peroxidases, respiratory burst oxidase homologs in plasmalemma, and glycolate oxidase in peroxisomes, which move to infection sites during cytoskeletal remodeling (Marino et al., 2012). ROS from tobacco (Nicotiana tabacum) roots have been implicated in blocking infection by Ph. parasitica zoospores, which interestingly die through programmed cell death (Galiana et al., 2005). Besides being antimicrobial, ROS strengthens cell walls by initiating lignin polymerization (Barros et al., 2015). Several other enzymes that fortify plant walls also are induced during PTI against oomycetes, including cinnamyl alcohol dehydrogenase and callose synthase (Wang et al., 2013; Hosseini et al., 2015).
Toxic isoflavonoids, sesquiterpenes, polyacetylenes, and other molecules that are collectively named phytoalexins are believed to be delivered to the pathogen by ATP-binding cassette (ABC) transporters. The export of capsidiol during the elicitin-triggered defense response of Nicotiana benthamiana against Ph. infestans involves ABCG1 and ABCG2, which are up-regulated during PTI (Rin et al., 2017). Some phytoalexins, such as α-tomatine of tomato, are preformed in plants, while others are induced by infection, such as capsidiol of Nicotiana spp. and pepper (Capsicum annuum), glyceollin of soybean, and camalexin of crucifers (Hahn et al., 1985; Bednarek et al., 2005).
These defenses may combine to produce apoplastic (or intracellular) environments that are unfavorable to oomycetes. This may explain why necrotrophy begins earlier in some Phytophthora-plant pathosystems than others, although the water soaking that is often associated with plant cell death may keep the pathogen hydrated. The low level of free water resulting from silicon polymerization in the apoplast also was invoked to explain why soybean grown at high silicon concentrations was less susceptible to Ph. sojae (Rasoolizadeh et al., 2018).
MANY OOMYCETES HAVE EVOLVED ELABORATE COUNTER DEFENSES
Oomycetes exhibit stealthy behaviors during biotrophic growth that minimize the immune response and maintain host integrity, which helps these pathogens feed from living cells. This is not an issue for necrotrophs such as Pythium spp. (Box 3). The (hemi)biotrophs resist host defenses using cytoplasmic and apoplastic effector proteins that are secreted toward their interface with plants. Oomycetes also produce enzymes that may degrade phytoalexins or immune-response hormones. The existence of these enzymes and effectors highlights the power of selection in the pathogen and the importance of their plant targets to the host defense response.
One example involves the plant apoplastic Cys proteases Rcr3 and C14, which were shown by mutation and knockdowns in tomato and N. benthamiana to defend against Ph. infestans (Song et al., 2009; Kaschani et al., 2010). Ph. infestans and relatives antagonize these using effectors such as extracellular cystatin-like protease inhibitor 1 (EPIC1). Studies of EPIC1 from Ph. infestans and Phytophthora mirabilis (which infects Mirabilis jalapa) and the host proteases were performed, guided by the crystal structure of a related protease-inhibitor complex. Amino acid changes in EPIC1 were implicated in helping the pathogens jump to new host species (Dong et al., 2014). Orthologs of EPIC1 genes occur in downy mildew, white rust, and Pythium spp. genomes. Most oomycetes also can inhibit plant Ser proteases, and one from Phytophthora palmivora was shown to contribute to virulence against the rubber tree (Hevea brasiliensis; Ekchaweng et al., 2017).
Another example of host-pathogen coevolution in the apoplast comes from studies of the endoglucanase XEG1 from Ph. sojae. Soybean produces an inhibitor of this CWDE, which binds XEG1 and blocks its contribution to virulence. To counteract the plant, Ph. sojae secretes an inactive enzyme as a decoy (Ma et al., 2017). With the defense protein unproductively bound to this trap, Ph. sojae can invade soybean more easily. Orthologs of XEG1 and its decoy are conserved throughout the Phytophthora genus.
Another apoplastic effector that counteracts host defenses is the in planta-induced protein (IPI-O) of Ph. infestans. IPI-O contains an Arg-Gly-Asp motif that is believed to disrupt adhesion between the plant’s cell wall and plasmalemma by binding the lectin receptor kinase LecRK-I.9, thus promoting disease by reducing wall integrity (Bouwmeester et al., 2011). Intriguingly, IPI-O contains an RxLR motif (Arg-x-Leu-Arg) that is shared by many oomycete effectors that enter plant cells to interfere with plant defenses. In planta expression of IPI-O minus its signal peptide caused expanded lesions, suggesting that IPI-O acts both at the host plasmalemma and intracellularly (Chen and Halterman, 2017). The intracellular target, apparently, is resistance protein Rpi-blb1 (Champouret et al., 2009).
RxLRs along with CRN (Crinkler) proteins represent the known cytoplasmic effectors of oomycetes. These are absent from Pythium spp. (Box 3) but are encoded by large gene families in Phytophthora spp., downy mildews, and white rusts, albeit with divergent signature motifs in some species (Kemen et al., 2011). How these proteins move into plants is not fully clarified, but RxLR uptake may involve binding a receptor on lipid rafts, as shown for a host-targeted protein from the animal pathogenic oomycete Saprolegnia parasitica (Trusch et al., 2018). RxLRs and CRNs are known to defeat plant immune responses through many routes, which include reprogramming host gene expression, altering RNA metabolism, and binding to host proteins involved in signaling (Wang and Wang, 2018). In this review, mention will be made only of RxLRs that act at the oomycete-plant interface.
Many RxLRs affect the trafficking of defense molecules. AVRblb2 accumulates in plants near haustoria and blocks the secretion of C14 protease (Bozkurt et al., 2011). RxLR24 of Phytophthora brassicae interferes with the delivery of antimicrobial proteins such as PR-1 by attaching to a GTPase involved in exocytosis (Tomczynska et al., 2018). Trafficking also is blocked by Avr1 of Ph. infestans, which binds exocyst protein SEC5 (Du et al., 2015), REX3 of Ph. palmivora, which interferes with brefeldin-sensitive secretion (Evangelisti et al., 2017), and PexRD54 of Ph. infestans, which depletes the Joka2 cargo protein from the autophagosomal membrane-forming ATG8 complex (Dagdas et al., 2016). The latter is interesting since the pathogen may be hijacking autophagosomes to destroy defense molecules through selective autophagy.
Although their functions are unidentified, the RxLRs Avh241 of Ph. sojae and HaRxL77 of H. arabidopsidis localize to the plant plasmalemma and are hypothesized to bind PRRs at the plant-oomycete interface (Caillaud et al., 2012; Yu et al., 2012). Causing auxin levels to rise at the interface is Penetration Specific Effector1 of Ph. parasitica, which is made in appressoria and interferes with auxin carriers (Evangelisti et al., 2013). This may elevate plant susceptibility since auxin inhibits SA signaling.
Interestingly, some species of Pythium are known to produce the auxin indole-3-acetic acid (Gravel et al., 2007). While there is no proof that oomycetes make other plant hormones, the sunflower (Helianthus annuus) downy mildew Plasmopara halstedii apparently encodes all enzymes for synthesizing cytokinin, which some bacteria make to direct host nutrients to infection sites (Sharma et al., 2015). Pl. halstedii also seems capable of producing brassinolide from phytosterols, which would negatively regulate the immune response. Many oomycetes also encode a predicted isochorismatase, which may disrupt SA signaling by breaking down its precursor. Interestingly, the enzyme in Ph. sojae localizes to haustoria (Liu et al., 2014).
Unlike fungi, most oomycetes have a limited capacity to degrade phytoalexins. Perhaps to compensate, oomycetes encode many more ABC transporters, which may expel the toxins (Ah-Fong et al., 2017b). While many fungi can degrade α-tomatine, Phytophthora and Pythium spp. that are pathogenic on tomato fail to degrade this glycoalkaloid (Sandrock and Vanetten, 1998). Ph. sojae can break down some soybean phytoalexins but not the most bioactive, glyceollin (Stossel, 1983). Whether Pythium spp. have special mechanisms to counteract plant defenses is unknown. However, during tuber infection, mRNA levels of Py. ultimum ABC transporters were about twice those of their counterparts in Ph. infestans, suggesting that the transporters might help eliminate phytoalexins liberated from lysing cells (Ah-Fong et al., 2017b). Cytochrome P450 enzymes also were more highly expressed in Py. ultimum.
HAUSTORIA REPRESENT A SPECIALIZED INTERFACE
Biotrophic and hemibiotrophic oomycetes form intimate associations with their hosts using haustoria (Fig. 1). These specialized hyphae breach host cell walls and become enveloped by a host membrane called the extrahaustorial membrane (EHM). Between the haustorium and EHM is a carbohydrate-rich amorphous layer called the extrahaustorial matrix (EHMx), which likely is of plant and pathogen origin (Caillaud et al., 2014). Little is known about how haustoria form and function in oomycetes, including how the host machinery is coopted during their genesis and what limits their expansion; most haustoria are less than 25 µm long.
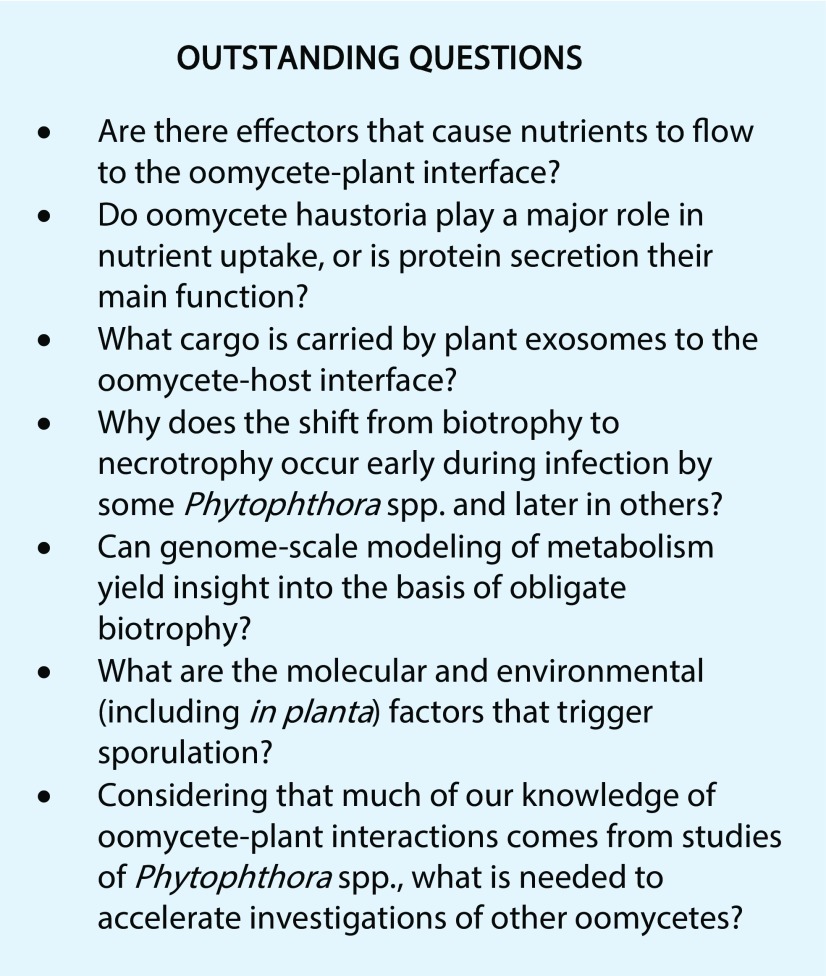
Recent studies with Ph. infestans and H. arabidopsidis indicated that the EHM is assembled de novo, as suggested for fungi (Lu et al., 2012; Bozkurt et al., 2015). Secretory vesicles are abundant near developing haustoria, along with trans-Golgi and late endosomal markers such as Rab5 and Rab7 GTPases (Caillaud et al., 2014; Inada et al., 2016). Within the EHM are plasmalemma proteins such as the Pen1 syntaxin, synaptotagmin, and remorin, which would be needed to deliver membrane material to growing haustoria. Some plant proteins are excluded from the EHM, including a calcium ATPase and at least some PRRs (Lu et al., 2012). Reduced ATPase activity could favor nutrient flow to the pathogen by reducing the plant’s capacity to retrieve nutrients from the EHMx, while PRR exclusion may minimize defense responses. Haustoria accommodation also causes host cells to reorganize their contents, with changes including endoplasmic reticulum aggregation, Golgi accumulation, and nuclear migration toward the haustoria (Lu et al., 2012). The nuclear shift might be part of a defense response or may be induced by the pathogen to facilitate the transport of CRN effectors, many of which act by reprogramming transcription (Song et al., 2015). Whether the reorganized endomembrane system delivers more transporters and/or nutrients to the EHM is an interesting question.
There are dissimilarities between haustoria of different species. While haustoria made by Phytophthora spp. are typically short and finger-like, those of downy mildews and white rusts are bulbous. Moreover, while Ph. infestans haustoria are anucleate and contain few mitochondria and endoplasmic reticulum, those of H. arabidopsidis have nuclei and many mitochondria and Golgi bodies (Mims et al., 2004). While the FLS2 PRR was excluded from the EHM with Ph. infestans, this was not the case with H. arabidopsidis (Lu et al., 2012). Downy mildew haustoria also are more likely to be surrounded by a callose collar than those of Phytophthora spp.
Several aspects of haustoria formation resemble plant defense responses. Deposition of the β-glucans that form callose collars involves secretory vesicles, multivesicular bodies, and Plasmodesmata-located-protein1 (PDLP1), which also is used to seal plasmodesmata during infection by other pathogens (Caillaud et al., 2014). Also possibly related to defense are haustorial encasements, which are double-layered membranes that often surround older haustoria (Lu et al., 2012). These are common with H. arabidopsidis but are seen less with Ph. infestans. Encasements might restrict the pathogen’s uptake of nutrients, impair effector translocation, or concentrate plant-derived antimicrobials. The EHM appears to have small invaginations, which also may promote its stability (Mims et al., 2004). The formation of these convolutions appears to involve PDLP1, since they increased when PDLP1 was overexpressed (Caillaud et al., 2014).
Although the contribution of oomycete haustoria to nutrient uptake is unclear, as discussed in the next section, the role of this structure in transporting proteins to the EHMx is demonstrated. Effectors are discharged from haustoria through at least two mechanisms. Secretion of the EPIC1 protease inhibitor was blocked by brefeldin A, indicating that it reaches the apoplast by the classic Golgi-mediated pathway (Wang et al., 2017). However, the delivery of RxLR Pi04314 was brefeldin A insensitive, suggesting that this cytoplasmic effector follows an alternative route even though it contains a classic signal peptide. Isochorismatases also are secreted but lack signal peptides, suggesting that they use the unconventional secretion pathway that has been documented in nonoomycetes (Liu et al., 2014).
NUTRIENT ACQUISITION AT THE PLANT-OOMYCETE INTERFACE
Although not proven, oomycete haustoria often are assumed to play a major role in nutrition. Nevertheless, they lack the neckband that encircles fungal haustoria, which is thought to help establish electrochemical gradients for nutrient transport by sealing the EHMx (Mims et al., 2004). Al. candida and H. arabidopsidis contain an electron-dense layer near their callose collars, which might function like a neckband, however (Soylu, 2004). In Ph. infestans, EHMx continuity with the apoplast was confirmed by studying the distribution of fluorescently tagged Avr3a (Whisson et al., 2007). Also unlike fungi, no haustoria-specific transporters are identified in oomycetes. While Ph. infestans and Py. ultimum both express ∼410 nutrient transporters, very few are specific to the haustoria-forming species (Ah-Fong et al., 2017b). Most nutrients may be drawn from the apoplast, considering that analyses of images of potato leaves infected by Ph. infestans indicate that its haustoria represent only about 2% of the total pathogen surface area (H. Judelson, unpublished data).
Regardless of where nutrients are acquired, plants contain myriad compounds to support pathogen growth. Metabolic models based on genome data indicate that most oomycetes can utilize the major plant hexoses, disaccharides, organic acids, starch, and sugar alcohols, although pentose utilization is restricted by the absence of arabinose isomerase (Rodenburg et al., 2018). Most oomycetes also can use the major nitrogen sources found in planta, including amino acids, ammonium, and nitrate. However, the biotrophs have reduced metabolic capabilities and, thus, a greater reliance on the host. While species of Phytophthora and Pythium each encode approximately 850 enzyme activities based on Enzyme Commission numbers, H. arabidopsidis and Al. candida encode only about 740 and 650, respectively (Judelson, 2017). These obligate biotrophs lack genes for nitrate assimilation and have impaired abilities to incorporate inorganic sulfur due to a lack of sulfite oxidase or reductase.
The metabolic deficiencies in the (hemi)biotrophs may help suppress immune responses, besides providing potential energy savings to the pathogen. The biotrophs are unable to make unsaturated fatty acids such as arachidonate, which are PAMPs in Phytophthora and Pythium spp. (Robinson and Bostock, 2015). The haustoria-forming oomycetes lack molybdopterin-utilizing pathways and consequently must acquire thiamine from the host. This may be beneficial, since this vitamin can up-regulate plant defenses, as demonstrated in the Pl. viticola-grape system (Boubakri et al., 2013).
It is important to differentiate the theoretical metabolism of oomycetes from what occurs in planta, since not all nutrients are at each plant-oomycete interface. While biotrophs are restricted to apoplastic nutrients, necrotrophs can access all compounds. Examples include starch and sulfate, which are sequestered within starch granules and vacuoles, respectively. Data from a study of Ph. infestans and Py. ultimum on potato tubers (Ah-Fong et al., 2017b) showed that while both encode α-amylase for starch utilization and adenylyl-sulfate kinase for incorporating sulfate, the Py. ultimum genes were expressed at greater than 10-fold higher levels (Box 3). This is the logical outcome of substrate-level induction. This situation changed during late infection when tissue colonized by Ph. infestans became necrotic, and mRNA levels for these enzymes equalized between the two pathogens. Similar patterns were observed for enzymes that act on other nutrients sequestered during biotrophic growth, such as phytate and lipids. This indicates that the terminal lifestyle of Ph. infestans is necrotrophic and not just necrogenic, thus addressing a debate in Phytophthora-host interactions. One position has been that plant necrosis does not benefit the pathogen and occurs simply because the pathogen no longer needs to suppress host defenses. The other viewpoint, which is supported by these results, is that necrosis is induced to liberate additional nutrients.
OOMYCETES HAVE AN EXIT STRATEGY
The final chapter in disease involves the pathogen’s egress from its host. Necrotrophs such as Pythium spp. can simply extend hyphae from macerated plant tissue into soil and transition to survival through saprophytic growth; sporulation is optional. In contrast, most (hemi)biotrophs must produce asexual spores. These are typically formed by root-rotting species at the crown, surface-exposed roots, or subterranean spaces adjoining roots. The task is more complicated for foliage-infecting (hemi)biotrophs, which usually sporulate from sporangiophores that pass through stomata (Farrell et al., 1969; Allègre et al., 2007). Most foliage-infecting oomycetes sporulate at night. This maximizes survival of the spores, which are prone to desiccation and lack UV-blocking pigments. Nocturnal sporulation is proposed to be regulated by cryptochromes in response to blue light (Xiang and Judelson, 2014) and requires modulating guard cell behavior, since stomata would normally be closed at night. Stomatal deregulation in the Pl. viticola-grape leaf interaction was proposed to be determined by a secreted glycoprotein, which caused stomata in colonized areas to remain open during darkness, water stress, and abscisic acid treatment (Allègre et al., 2007; Guillier et al., 2015). This effect resembles that caused by the bacterial toxin coronatine (Melotto et al., 2006).
Substantial genomic resources are devoted to sporulation. In Ph. infestans, this involves the up-regulation of more than 3,000 genes (∼20% of the total), including those encoding storage, effector, and adhesion proteins, and several hundred components of flagella (Judelson et al., 2012; Ah-Fong et al., 2017a). Genes proven to regulate sporulation include MADS box and Myb transcription factors, a mitogen-activated protein kinase, and a cell cycle phosphatase (Ah-Fong and Judelson, 2011; Li et al., 2014; Xiang and Judelson, 2014). However, the primary trigger for sporulation is unknown. Nutrient limitation is thought to play a role, which is concordant with the finding that the nitrogen metabolite repression regulator NMRA is down-regulated near the onset of sporulation in Ph. infestans (Ah-Fong et al., 2017a). NMRA also was proposed to control the transcription of late-induced effectors in Phytophthora capsici (Pham et al., 2018). Spiking at the same time are mRNAs for genes used to assimilate nitrate, which is a nonpreferred nitrogen source compared with amino acids (Abrahamian et al., 2016). Since a study in Phytophthora cactorum found that adding amino acids or ammonium to media did not retard sporulation, the process also may be prompted by an accumulated metabolite (Elliott, 1989). The plant also may affect sporulation, since its metabolic pathways are linked to those of the pathogen during colonization. A dual-species systems approach to metabolism may help understand what influences sporulation, effector expression, phytohormone levels, and other aspects of plant-oomycete interactions.
CONCLUSION
Oomycetes have developed diverse lifestyles over their 400+ million years of evolution (Taylor et al., 2006). The (hemi)biotrophs have learned to coopt their hosts by suppressing defenses and coercing plants to form interfaces for effector and nutrient delivery. Such lifestyles may have evolved by exploiting pathways used by plants to harbor mutualists, since mutants of M. truncatula deficient in mycorrhizae formation were shown to have reduced susceptibility to Ph. palmivora (Rey et al., 2015). Many oomycetes have become host-adapted to the extent that they depend on plant metabolites for growth or reproduction. In contrast, necrotrophs have less-specialized lifestyles, as they can grow as saprophytes or pathogens, overpowering their hosts and perhaps even profiting from the plant immune response. Most oomycetes also have retained a flagellated life stage, which expands their access to new hosts, although this comes with a large genomic burden. Meanwhile, plants have evolved complex multilayered defenses that balance survival against the growth penalty that comes from activating the immune response (Ning et al., 2017).
Many of the defenses, counter defenses, spore behaviors, and interactions with other microbes that we have described have small individual effects on disease outcomes but are significant from an epidemiological perspective. The infection potential of an oomycete spore on plant tissue is usually much less than 100%, similar to the situation in fungi (Mellersh and Heath, 2002; Kong and Hong, 2016). The progress of an epidemic will be influenced by factors that raise or lower this infection potential or the time between penetration and sporulation (Willocquet et al., 2017). While many plant scientists aim to develop strong resistance against pathogens, natural defenses (as well as changes in pathogens that enhance fitness) need not have blockbuster effects to be retained during evolution.
Our knowledge of interactions involving Phytophthora spp. has grown dramatically due to the availability of genome sequences and tools for functional genomics, but research into other oomycetes has lagged. It is unfortunate that Pythium spp. have remained little studied despite their large agricultural impact, for example. Most Pythium spp. are easily cultured, so it should be possible for a new generation of researchers to improve our understanding of the genus. Studying the breadth of oomycetes is important since crop protection solutions developed for one group may not translate to others.
Footnotes
This work was supported by the National Science Foundation (grant no. IOS-1753749) and the National Institute of Food and Agriculture of the U.S. Department of Agriculture (grant no. 2016-67013-2481).
Articles can be viewed without a subscription.
References
- Abrahamian M, Ah-Fong AM, Davis C, Andreeva K, Judelson HS (2016) Gene expression and silencing studies in Phytophthora infestans reveal infection-specific nutrient transporters and a role for the nitrate reductase pathway in plant pathogenesis. PLoS Pathog 12: e1006097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ah-Fong AM, Judelson HS (2011) New role for Cdc14 phosphatase: Localization to basal bodies in the oomycete Phytophthora and its evolutionary coinheritance with eukaryotic flagella. PLoS ONE 6: e16725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ah-Fong AM, Kim KS, Judelson HS (2017a) RNA-seq of life stages of the oomycete Phytophthora infestans reveals dynamic changes in metabolic, signal transduction, and pathogenesis genes and a major role for calcium signaling in development. BMC Genomics 18: 198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ah-Fong AMV, Shrivastava J, Judelson HS (2017b) Lifestyle, gene gain and loss, and transcriptional remodeling cause divergence in the transcriptomes of Phytophthora infestans and Pythium ultimum during potato tuber colonization. BMC Genomics 18: 764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albert I, Böhm H, Albert M, Feiler CE, Imkampe J, Wallmeroth N, Brancato C, Raaymakers TM, Oome S, Zhang H, et al. (2015) An RLP23-SOBIR1-BAK1 complex mediates NLP-triggered immunity. Nat Plants 1: 15140. [DOI] [PubMed] [Google Scholar]
- Allègre M, Daire X, Héloir MC, Trouvelot S, Mercier L, Adrian M, Pugin A (2007) Stomatal deregulation in Plasmopara viticola-infected grapevine leaves. New Phytol 173: 832–840 [DOI] [PubMed] [Google Scholar]
- Aram K, Rizzo DM (2018) Distinct trophic specializations affect how Phytophthora ramorum and clade 6 Phytophthora spp. colonize and persist on Umbellularia californica leaves in streams. Phytopathology 108: 858–869 [DOI] [PubMed] [Google Scholar]
- Avrova AO, Boevink PC, Young V, Grenville-Briggs LJ, van West P, Birch PR, Whisson SC (2008) A novel Phytophthora infestans haustorium-specific membrane protein is required for infection of potato. Cell Microbiol 10: 2271–2284 [DOI] [PubMed] [Google Scholar]
- Barros J, Serk H, Granlund I, Pesquet E (2015) The cell biology of lignification in higher plants. Ann Bot 115: 1053–1074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beakes GW, Glockling SL, Sekimoto S (2012) The evolutionary phylogeny of the oomycete “fungi.” Protoplasma 249: 3–19 [DOI] [PubMed] [Google Scholar]
- Bednarek P, Schneider B, Svatos A, Oldham NJ, Hahlbrock K (2005) Structural complexity, differential response to infection, and tissue specificity of indolic and phenylpropanoid secondary metabolism in Arabidopsis roots. Plant Physiol 138: 1058–1070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benhamou N, le Floch G, Vallance J, Gerbore J, Grizard D, Rey P (2012) Pythium oligandrum: An example of opportunistic success. Microbiology 158: 2679–2694 [DOI] [PubMed] [Google Scholar]
- Bircher U, Hohl HR (1997) Environmental signalling during induction of appressorium formation in Phytophthora. Mycol Res 101: 395–402 [Google Scholar]
- Blackman LM, Cullerne DP, Torreña P, Taylor J, Hardham AR (2015) RNA-seq analysis of the expression of genes encoding cell wall degrading enzymes during infection of lupin (Lupinus angustifolius) by Phytophthora parasitica. PLoS ONE 10: e0136899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco FA, Judelson HS (2005) A bZIP transcription factor from Phytophthora interacts with a protein kinase and is required for zoospore motility and plant infection. Mol Microbiol 56: 638–648 [DOI] [PubMed] [Google Scholar]
- Böhm H, Albert I, Oome S, Raaymakers TM, Van den Ackerveken G, Nürnberger T (2014) A conserved peptide pattern from a widespread microbial virulence factor triggers pattern-induced immunity in Arabidopsis. PLoS Pathog 10: e1004491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boubakri H, Chong J, Poutaraud A, Schmitt C, Bertsch C, Mliki A, Masson JE, Soustre-Gacougnolle I (2013) Riboflavin (vitamin B-2) induces defence responses and resistance to Plasmopara viticola in grapevine. Eur J Plant Pathol 136: 837–855 [Google Scholar]
- Bouwmeester K, de Sain M, Weide R, Gouget A, Klamer S, Canut H, Govers F (2011) The lectin receptor kinase LecRK-I.9 is a novel Phytophthora resistance component and a potential host target for a RXLR effector. PLoS Pathog 7: e1001327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bozkurt TO, Schornack S, Win J, Shindo T, Ilyas M, Oliva R, Cano LM, Jones AME, Huitema E, van der Hoorn RAL, et al. (2011) Phytophthora infestans effector AVRblb2 prevents secretion of a plant immune protease at the haustorial interface. Proc Natl Acad Sci USA 108: 20832–20837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bozkurt TO, Belhaj K, Dagdas YF, Chaparro-Garcia A, Wu CH, Cano LM, Kamoun S (2015) Rerouting of plant late endocytic trafficking toward a pathogen interface. Traffic 16: 204–226 [DOI] [PubMed] [Google Scholar]
- Brunner F, Rosahl S, Lee J, Rudd JJ, Geiler C, Kauppinen S, Rasmussen G, Scheel D, Nürnberger T (2002) Pep-13, a plant defense-inducing pathogen-associated pattern from Phytophthora transglutaminases. EMBO J 21: 6681–6688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Q, Qiao L, Wang M, He B, Lin FM, Palmquist J, Huang SD, Jin H (2018) Plants send small RNAs in extracellular vesicles to fungal pathogen to silence virulence genes. Science 360: 1126–1129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caillaud MC, Piquerez SJM, Fabro G, Steinbrenner J, Ishaque N, Beynon J, Jones JDG (2012) Subcellular localization of the Hpa RxLR effector repertoire identifies a tonoplast-associated protein HaRxL17 that confers enhanced plant susceptibility. Plant J 69: 252–265 [DOI] [PubMed] [Google Scholar]
- Caillaud MC, Wirthmueller L, Sklenar J, Findlay K, Piquerez SJM, Jones AME, Robatzek S, Jones JDG, Faulkner C (2014) The plasmodesmal protein PDLP1 localises to haustoria-associated membranes during downy mildew infection and regulates callose deposition. PLoS Pathog 10: e1004496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carzaniga R, Bowyer P, O’Connell RJ (2001) Production of extracellular matrices during development of infection structures by the downy mildew Peronospora parasitica. New Phytol 149: 83–93 [DOI] [PubMed] [Google Scholar]
- Champouret N, Bouwmeester K, Rietman H, van der Lee T, Maliepaard C, Heupink A, van de Vondervoort PJ, Jacobsen E, Visser RG, van der Vossen EA, et al. (2009) Phytophthora infestans isolates lacking class I ipiO variants are virulent on Rpi-blb1 potato. Mol Plant Microbe Interact 22: 1535–1545 [DOI] [PubMed] [Google Scholar]
- Chang YH, Yan HZ, Liou RF (2015) A novel elicitor protein from Phytophthora parasitica induces plant basal immunity and systemic acquired resistance. Mol Plant Pathol 16: 123–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Halterman DA (2017) Determination of virulence contribution from Phytophthora infestans effector Ipi-O4 in a resistant potato host containing the RB gene. Physiol Mol Plant Pathol 100: 30–34 [Google Scholar]
- Dagdas YF, Belhaj K, Maqbool A, Chaparro-Garcia A, Pandey P, Petre B, Tabassum N, Cruz-Mireles N, Hughes RK, Sklenar J, et al. (2016) An effector of the Irish potato famine pathogen antagonizes a host autophagy cargo receptor. eLife 5: e10856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dagdas YF, Pandey P, Tumtas Y, Sanguankiattichai N, Belhaj K, Duggan C, Leary AY, Segretin ME, Contreras MP, Savage Z, et al. (2018) Host autophagy machinery is diverted to the pathogen interface to mediate focal defense responses against the Irish potato famine pathogen. eLife 7: e37476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dale ML, Irwin JAG (1991) Stomata as an infection court for Phytophthora megasperma f. sp. medicaginis in chickpea and a histological study of infection. Phytopathology 81: 375–379 [Google Scholar]
- Deacon JW, Donaldson SP (1993) Molecular recognition in the homing responses of zoosporic fungi, with special reference to Pythium and Phytophthora. Mycol Res 97: 1153–1171 [Google Scholar]
- Derevnina L, Dagdas YF, De la Concepcion JC, Bialas A, Kellner R, Petre B, Domazakis E, Du J, Wu CH, Lin X, et al. (2016) Nine things to know about elicitins. New Phytol 212: 888–895 [DOI] [PubMed] [Google Scholar]
- Domazakis E, Wouters D, Visser RGF, Kamoun S, Joosten MHAJ, Vleeshouwers VGAA (2018) The ELR-SOBIR1 complex functions as a two-component receptor-like kinase to mount defense against Phytophthora infestans. Mol Plant Microbe Interact 31: 795–802 [DOI] [PubMed] [Google Scholar]
- Dong S, Stam R, Cano LM, Song J, Sklenar J, Yoshida K, Bozkurt TO, Oliva R, Liu Z, Tian M, et al. (2014) Effector specialization in a lineage of the Irish potato famine pathogen. Science 343: 552–555 [DOI] [PubMed] [Google Scholar]
- Du Y, Mpina MH, Birch PRJ, Bouwmeester K, Govers F (2015) Phytophthora infestans RXLR effector AVR1 interacts with exocyst component Sec5 to manipulate plant immunity. Plant Physiol 169: 1975–1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekchaweng K, Evangelisti E, Schornack S, Tian M, Churngchow N (2017) The plant defense and pathogen counterdefense mediated by Hevea brasiliensis serine protease HbSPA and Phytophthora palmivora extracellular protease inhibitor PpEPI10. PLoS ONE 12: e0175795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Hamalawi ZA, Erwin DC (1986) Components in alfalfa Medicago sativa root extract and root exudate that increase oospore germination of Phytophthora megasperma f. sp. medicaginis. Phytopathology 76: 508–513 [Google Scholar]
- Elliott CG. (1989) Some aspects of nitrogen nutrition and reproduction in Phytophthora. Mycol Res 92: 34–44 [Google Scholar]
- Evangelisti E, Govetto B, Minet-Kebdani N, Kuhn ML, Attard A, Ponchet M, Panabières F, Gourgues M (2013) The Phytophthora parasitica RXLR effector penetration-specific effector 1 favours Arabidopsis thaliana infection by interfering with auxin physiology. New Phytol 199: 476–489 [DOI] [PubMed] [Google Scholar]
- Evangelisti E, Gogleva A, Hainaux T, Doumane M, Tulin F, Quan C, Yunusov T, Floch K, Schornack S (2017) Time-resolved dual transcriptomics reveal early induced Nicotiana benthamiana root genes and conserved infection-promoting Phytophthora palmivora effectors. BMC Biol 15: 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrell GM, Preece TF, Wren MJ (1969) Effects of infection by Phytophthora infestans on the stomata of potato leaves. Ann Appl Biol 63: 265–275 [Google Scholar]
- Fawke S, Doumane M, Schornack S (2015) Oomycete interactions with plants: Infection strategies and resistance principles. Microbiol Mol Biol Rev 79: 263–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng BZ, Zhu XP, Fu L, Lv RF, Storey D, Tooley P, Zhang XG (2014) Characterization of necrosis-inducing NLP proteins in Phytophthora capsici. BMC Plant Biol 14: 126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galiana E, Rivière MP, Pagnotta S, Baudouin E, Panabières F, Gounon P, Boudier L (2005) Plant-induced cell death in the oomycete pathogen Phytophthora parasitica. Cell Microbiol 7: 1365–1378 [DOI] [PubMed] [Google Scholar]
- Gaulin E, Jauneau A, Villalba F, Rickauer M, Esquerré-Tugayé MT, Bottin A (2002) The CBEL glycoprotein of Phytophthora parasitica var-nicotianae is involved in cell wall deposition and adhesion to cellulosic substrates. J Cell Sci 115: 4565–4575 [DOI] [PubMed] [Google Scholar]
- Govindarajulu M, Epstein L, Wroblewski T, Michelmore RW (2015) Host-induced gene silencing inhibits the biotrophic pathogen causing downy mildew of lettuce. Plant Biotechnol J 13: 875–883 [DOI] [PubMed] [Google Scholar]
- Granke LL, Windstam ST, Hoch HC, Smart CD, Hausbeck MK (2009) Dispersal and movement mechanisms of Phytophthora capsici sporangia. Phytopathology 99: 1258–1264 [DOI] [PubMed] [Google Scholar]
- Gravel V, Antoun H, Tweddell RJ (2007) Effect of indole-acetic acid (IAA) on the development of symptoms caused by Pythium ultimum on tomato plants. Eur J Plant Pathol 119: 457–462 [Google Scholar]
- Gui YJ, Chen JY, Zhang DD, Li NY, Li TG, Zhang WQ, Wang XY, Short DPG, Li L, Guo W, et al. (2017) Verticillium dahliae manipulates plant immunity by glycoside hydrolase 12 proteins in conjunction with carbohydrate-binding module 1. Environ Microbiol 19: 1914–1932 [DOI] [PubMed] [Google Scholar]
- Guillier C, Gamm M, Lucchi G, Truntzer C, Pecqueur D, Ducoroy P, Adrian M, Héloir MC (2015) Toward the identification of two glycoproteins involved in the stomatal deregulation of downy mildew-infected grapevine leaves. Mol Plant Microbe Interact 28: 1227–1236 [DOI] [PubMed] [Google Scholar]
- Hahn MG, Bonhoff A, Grisebach H (1985) Quantitative localization of the phytoalexin glyceollin I in relation to fungal hyphae in soybean roots infected with Phytophthora megasperma f. sp. glycinea. Plant Physiol 77: 591–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halim VA, Altmann S, Ellinger D, Eschen-Lippold L, Miersch O, Scheel D, Rosahl S (2009) PAMP-induced defense responses in potato require both salicylic acid and jasmonic acid. Plant J 57: 230–242 [DOI] [PubMed] [Google Scholar]
- Hosseini S, Heyman F, Olsson U, Broberg A, Jensen DF, Karlsson M (2014) Zoospore chemotaxis of closely related legume-root infecting Phytophthora species towards host isoflavones. Plant Pathol 63: 708–714 [Google Scholar]
- Hosseini S, Elfstrand M, Heyman F, Jensen DF, Karlsson M (2015) Deciphering common and specific transcriptional immune responses in pea towards the oomycete pathogens Aphanomyces euteiches and Phytophthora pisi. BMC Genomics 16: 627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua C, Wang Y, Zheng X, Dou D, Zhang Z, Govers F, Wang Y (2008) A Phytophthora sojae G-protein alpha subunit is involved in chemotaxis to soybean isoflavones. Eukaryot Cell 7: 2133–2140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inada N, Betsuyaku S, Shimada TL, Ebine K, Ito E, Kutsuna N, Hasezawa S, Takano Y, Fukuda H, Nakano A, et al. (2016) Modulation of plant RAB GTPase-mediated membrane trafficking pathway at the interface between plants and obligate biotrophic pathogens. Plant Cell Physiol 57: 1854–1864 [DOI] [PubMed] [Google Scholar]
- Jack ALH, Nelson EB (2018) A seed-recruited microbiome protects developing seedlings from disease by altering homing responses of Pythium aphanidermatum zoospores. Plant Soil 422: 209–222 [Google Scholar]
- Jahan SN, Åsman AK, Corcoran P, Fogelqvist J, Vetukuri RR, Dixelius C (2015) Plant-mediated gene silencing restricts growth of the potato late blight pathogen Phytophthora infestans. J Exp Bot 66: 2785–2794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Judelson HS. (2017) Metabolic diversity and novelties in the oomycetes. Annu Rev Microbiol 71: 21–39 [DOI] [PubMed] [Google Scholar]
- Judelson HS, Shrivastava J, Manson J (2012) Decay of genes encoding the oomycete flagellar proteome in the downy mildew Hyaloperonospora arabidopsidis. PLoS ONE 7: e47624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamoun S, Furzer O, Jones JD, Judelson HS, Ali GS, Dalio RJ, Roy SG, Schena L, Zambounis A, Panabières F, et al. (2015) The top 10 oomycete pathogens in molecular plant pathology. Mol Plant Pathol 16: 413–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaschani F, Shabab M, Bozkurt T, Shindo T, Schornack S, Gu C, Ilyas M, Win J, Kamoun S, van der Hoorn RA (2010) An effector-targeted protease contributes to defense against Phytophthora infestans and is under diversifying selection in natural hosts. Plant Physiol 154: 1794–1804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kebdani N, Pieuchot L, Deleury E, Panabières F, Le Berre JY, Gourgues M (2010) Cellular and molecular characterization of Phytophthora parasitica appressorium-mediated penetration. New Phytol 185: 248–257 [DOI] [PubMed] [Google Scholar]
- Kemen E, Gardiner A, Schultz-Larsen T, Kemen AC, Balmuth AL, Robert-Seilaniantz A, Bailey K, Holub E, Studholme DJ, Maclean D, et al. (2011) Gene gain and loss during evolution of obligate parasitism in the white rust pathogen of Arabidopsis thaliana. PLoS Biol 9: e1001094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiefer B, Riemann M, Büche C, Kassemeyer HH, Nick P (2002) The host guides morphogenesis and stomatal targeting in the grapevine pathogen Plasmopara viticola. Planta 215: 387–393 [DOI] [PubMed] [Google Scholar]
- Kong P, Hong C (2016) Soil bacteria as sources of virulence signal providers promoting plant infection by Phytophthora pathogens. Sci Rep 6: 33239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kots K, Meijer HJG, Bouwmeester K, Govers F, Ketelaar T (2017) Filamentous actin accumulates during plant cell penetration and cell wall plug formation in Phytophthora infestans. Cell Mol Life Sci 74: 909–920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kourelis J, van der Hoorn RAL (2018) Defended to the nines: 25 years of resistance gene cloning identifies nine mechanisms for R protein function. Plant Cell 30: 285–299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamour K, Kamoun S (2009) Oomycete Genetics and Genomics: Diversity, Interactions, and Research Tools. John Wiley and Sons, Hoboken, NJ [Google Scholar]
- Larousse M, Govetto B, Séassau A, Etienne C, Industri B, Theodorakopoulos N, Deleury E, Ponchet M, Panabières F, Galiana E (2014) Characterization of PPMUCL1/2/3, three members of a new oomycete-specific mucin-like protein family residing in Phytophthora parasitica biofilm. Protist 165: 275–292 [DOI] [PubMed] [Google Scholar]
- Latijnhouwers M, Ligterink W, Vleeshouwers VGAA, van West P, Govers F (2004) A Galpha subunit controls zoospore motility and virulence in the potato late blight pathogen Phytophthora infestans. Mol Microbiol 51: 925–936 [DOI] [PubMed] [Google Scholar]
- Lenarčič T, Albert I, Böhm H, Hodnik V, Pirc K, Zavec AB, Podobnik M, Pahovnik D, Žagar E, Pruitt R, et al. (2017) Eudicot plant-specific sphingolipids determine host selectivity of microbial NLP cytolysins. Science 358: 1431–1434 [DOI] [PubMed] [Google Scholar]
- Li J, Staiger CJ (2018) Understanding cytoskeletal dynamics during the plant immune response. Annu Rev Phytopathol 56: 513–533 [DOI] [PubMed] [Google Scholar]
- Li A, Wang Y, Tao K, Dong S, Huang Q, Dai T, Zheng X, Wang Y (2010) PsSAK1, a stress-activated MAP kinase of Phytophthora sojae, is required for zoospore viability and infection of soybean. Mol Plant Microbe Interact 23: 1022–1031 [DOI] [PubMed] [Google Scholar]
- Li A, Zhang M, Wang Y, Li D, Liu X, Tao K, Ye W, Wang Y (2014) PsMPK1, an SLT2-type mitogen-activated protein kinase, is required for hyphal growth, zoosporogenesis, cell wall integrity, and pathogenicity in Phytophthora sojae. Fungal Genet Biol 65: 14–24 [DOI] [PubMed] [Google Scholar]
- Lioussanne L, Jolicoeur M, St-Arnaud M (2008) Mycorrhizal colonization with Glomus intraradices and development stage of transformed tomato roots significantly modify the chemotactic response of zoospores of the pathogen Phytophthora nicotianae. Soil Biol Biochem 40: 2217–2224 [Google Scholar]
- Liu T, Song T, Zhang X, Yuan H, Su L, Li W, Xu J, Liu S, Chen L, Chen T, et al. (2014) Unconventionally secreted effectors of two filamentous pathogens target plant salicylate biosynthesis. Nat Commun 5: 4686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu YJ, Schornack S, Spallek T, Geldner N, Chory J, Schellmann S, Schumacher K, Kamoun S, Robatzek S (2012) Patterns of plant subcellular responses to successful oomycete infections reveal differences in host cell reprogramming and endocytic trafficking. Cell Microbiol 14: 682–697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Z, Zhu L, Song T, Wang Y, Zhang Q, Xia Y, Qiu M, Lin Y, Li H, Kong L, et al. (2017) A paralogous decoy protects Phytophthora sojae apoplastic effector PsXEG1 from a host inhibitor. Science 355: 710–714 [DOI] [PubMed] [Google Scholar]
- Marino D, Dunand C, Puppo A, Pauly N (2012) A burst of plant NADPH oxidases. Trends Plant Sci 17: 9–15 [DOI] [PubMed] [Google Scholar]
- McCarren KL, McComb JA, Shearer BL, Hardy GESJ (2005) The role of chlamydospores of Phytophthora cinnamomi: A review. Australasian Plant Pathology 34: 333–338 [Google Scholar]
- Mélida H, Sandoval-Sierra JV, Diéguez-Uribeondo J, Bulone V (2013) Analyses of extracellular carbohydrates in oomycetes unveil the existence of three different cell wall types. Eukaryot Cell 12: 194–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellersh DG, Heath MC (2002) Nonhost resistance to rust fungi in Arabidopsis. Plant Biol 2002: 127 [Google Scholar]
- Melotto M, Underwood W, Koczan J, Nomura K, He SY (2006) Plant stomata function in innate immunity against bacterial invasion. Cell 126: 969–980 [DOI] [PubMed] [Google Scholar]
- Mims CW, Richardson EA, Holt BF, Dangl JL (2004) Ultrastructure of the host-pathogen interface in Arabidopsis thaliana leaves infected by the downy mildew Hyaloperonospora parasitica. Can J Bot 82: 1001–1008 [Google Scholar]
- Morris PF, Bone E, Tyler BM (1998) Chemotropic and contact responses of Phytophthora sojae hyphae to soybean isoflavonoids and artificial substrates. Plant Physiol 117: 1171–1178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mostowfizadeh-Ghalamfarsa R, Hussaini K, Ghasemi-Fasaei R (2018) Effects of calcium salts in controlling Phytophthora pistaciae, the causal agent of pistachio gummosis. Eur J Plant Pathol 151: 475–485 [Google Scholar]
- Nars A, Lafitte C, Chabaud M, Drouillard S, Mélida H, Danoun S, Le Costaouëc T, Rey T, Benedetti J, Bulone V, et al. (2013) Aphanomyces euteiches cell wall fractions containing novel glucan-chitosaccharides induce defense genes and nuclear calcium oscillations in the plant host Medicago truncatula. PLoS ONE 8: e75039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson EB. (1987) Rapid germination of sporangia of Pythium spp. in response to volatiles from germinating seeds. Phytopathology 77: 1108–1112 [Google Scholar]
- Ning Y, Liu W, Wang GL (2017) Balancing immunity and yield in crop plants. Trends Plant Sci 22: 1069–1079 [DOI] [PubMed] [Google Scholar]
- Pham J, Stam R, Heredia VM, Csukai M, Huitema E (2018) An NMRA-like protein regulates gene expression in Phytophthora capsici to drive the infection cycle on tomato. Mol Plant Microbe Interact 31: 665–677 [DOI] [PubMed] [Google Scholar]
- Pittis JE, Shattock RC (1994) Viability, germination and infection potential of oospores of Phytophthora infestans. Plant Pathol 43: 387–396 [Google Scholar]
- Ploch S, Thines M (2011) Obligate biotrophic pathogens of the genus Albugo are widespread as asymptomatic endophytes in natural populations of Brassicaceae. Mol Ecol 20: 3692–3699 [DOI] [PubMed] [Google Scholar]
- Raaijmakers JM, De Bruijn I, Nybroe O, Ongena M (2010) Natural functions of lipopeptides from Bacillus and Pseudomonas: More than surfactants and antibiotics. FEMS Microbiol Rev 34: 1037–1062 [DOI] [PubMed] [Google Scholar]
- Rasoolizadeh A, Labbé C, Sonah H, Deshmukh RK, Belzile F, Menzies JG, Bélanger RR (2018) Silicon protects soybean plants against Phytophthora sojae by interfering with effector-receptor expression. BMC Plant Biol 18: 97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resjö S, Brus M, Ali A, Meijer HJG, Sandin M, Govers F, Levander F, Grenville-Briggs L, Andreasson E (2017) Proteomic analysis of Phytophthora infestans reveals the importance of cell wall proteins in pathogenicity. Mol Cell Proteomics 16: 1958–1971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rey T, Chatterjee A, Buttay M, Toulotte J, Schornack S (2015) Medicago truncatula symbiosis mutants affected in the interaction with a biotrophic root pathogen. New Phytol 206: 497–500 [DOI] [PubMed] [Google Scholar]
- Rin S, Mizuno Y, Shibata Y, Fushimi M, Katou S, Sato I, Chiba S, Kawakita K, Takemoto D (2017) EIN2-mediated signaling is involved in pre-invasion defense in Nicotiana benthamiana against potato late blight pathogen, Phytophthora infestans. Plant Signal Behav 12: e1300733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson SM, Bostock RM (2015) β-Glucans and eicosapolyenoic acids as MAMPs in plant-oomycete interactions: Past and present. Front Plant Sci 5: 797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robold AV, Hardham AR (2005) During attachment Phytophthora spores secrete proteins containing thrombospondin type 1 repeats. Curr Genet 47: 307–315 [DOI] [PubMed] [Google Scholar]
- Rodenburg SYA, Seidl MF, de Ridder D, Govers F (2018) Genome-wide characterization of Phytophthora infestans metabolism: A systems biology approach. Mol Plant Pathol 19: 1403–1413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saijo Y, Loo EP, Yasuda S (2018) Pattern recognition receptors and signaling in plant-microbe interactions. Plant J 93: 592–613 [DOI] [PubMed] [Google Scholar]
- Sandrock RW, Vanetten HD (1998) Fungal sensitivity to and enzymatic degradation of the phytoanticipin alpha-tomatine. Phytopathology 88: 137–143 [DOI] [PubMed] [Google Scholar]
- Schlatter D, Kinkel L, Thomashow L, Weller D, Paulitz T (2017) Disease suppressive soils: New insights from the soil microbiome. Phytopathology 107: 1284–1297 [DOI] [PubMed] [Google Scholar]
- Sekizaki H, Yokosawa R, Chinen C, Adachi H, Yamane Y (1993) Studies on zoospore attracting activity. II. Synthesis of isoflavones and their attracting activity to Aphanomyces euteiches zoospore. Biol Pharm Bull 16: 698–701 [DOI] [PubMed] [Google Scholar]
- Shang H, Grau CR, Peters RD (2000) Oospore germination of Aphanomyces euteiches in root exudates and on the rhizoplanes of crop plants. Plant Dis 84: 994–998 [DOI] [PubMed] [Google Scholar]
- Sharma R, Xia X, Cano LM, Evangelisti E, Kemen E, Judelson H, Oome S, Sambles C, van den Hoogen DJ, Kitner M, et al. (2015) Genome analyses of the sunflower pathogen Plasmopara halstedii provide insights into effector evolution in downy mildews and Phytophthora. BMC Genomics 16: 741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J, Win J, Tian M, Schornack S, Kaschani F, Ilyas M, van der Hoorn RAL, Kamoun S (2009) Apoplastic effectors secreted by two unrelated eukaryotic plant pathogens target the tomato defense protease Rcr3. Proc Natl Acad Sci USA 106: 1654–1659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song T, Ma Z, Shen D, Li Q, Li W, Su L, Ye T, Zhang M, Wang Y, Dou D (2015) An oomycete CRN effector reprograms expression of plant HSP genes by targeting their promoters. PLoS Pathog 11: e1005348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soylu S. (2004) Ultrastructural characterisation of the host-pathogen interface in white blister-infected Arabidopsis leaves. Mycopathologia 158: 457–464 [DOI] [PubMed] [Google Scholar]
- Staswick PE, Yuen GY, Lehman CC (1998) Jasmonate signaling mutants of Arabidopsis are susceptible to the soil fungus Pythium irregulare. Plant J 15: 747–754 [DOI] [PubMed] [Google Scholar]
- Stossel P. (1983) In vitro effects of glyceollin on Phytophthora megasperma f. sp. glycinea. Phytopathology 73: 1603–1607 [Google Scholar]
- Takemoto D, Jones DA, Hardham AR (2003) GFP-tagging of cell components reveals the dynamics of subcellular re-organization in response to infection of Arabidopsis by oomycete pathogens. Plant J 33: 775–792 [DOI] [PubMed] [Google Scholar]
- Takenaka S, Yamaguchi K, Masunaka A, Hase S, Inoue T, Takahashi H (2011) Implications of oligomeric forms of POD-1 and POD-2 proteins isolated from cell walls of the biocontrol agent Pythium oligandrum in relation to their ability to induce defense reactions in tomato. J Plant Physiol 168: 1972–1979 [DOI] [PubMed] [Google Scholar]
- Taylor TN, Krings M, Kerp H (2006) Hassiella monospora gen. et sp. nov., a microfungus from the 400 million year old Rhynie chert. Mycol Res 110: 628–632 [DOI] [PubMed] [Google Scholar]
- Tomczynska I, Stumpe M, Mauch F (2018) A conserved RxLR effector interacts with host RABA-type GTPases to inhibit vesicle-mediated secretion of antimicrobial proteins. Plant J 95: 187–203 [DOI] [PubMed] [Google Scholar]
- Trusch F, Loebach L, Wawra S, Durward E, Wuensch A, Iberahim NA, de Bruijn I, MacKenzie K, Willems A, Toloczko A, et al. (2018) Cell entry of a host-targeting protein of oomycetes requires gp96. Nat Commun 9: 2347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Hoogen DJ, Meijer HJG, Seidl MF, Govers F (2018) The ancient link between G-protein-coupled receptors and C-terminal phospholipid kinase domains. MBio 9: e02119-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Wang Y (2018) Phytophthora sojae effectors orchestrate warfare with host immunity. Curr Opin Microbiol 46: 7–13 [DOI] [PubMed] [Google Scholar]
- Wang S, Boevink PC, Welsh L, Zhang R, Whisson SC, Birch PRJ (2017) Delivery of cytoplasmic and apoplastic effectors from Phytophthora infestans haustoria by distinct secretion pathways. New Phytol 216: 205–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Bouwmeester K, van de Mortel JE, Shan W, Govers F (2013) A novel Arabidopsis-oomycete pathosystem: Differential interactions with Phytophthora capsici reveal a role for camalexin, indole glucosinolates and salicylic acid in defence. Plant Cell Environ 36: 1192–1203 [DOI] [PubMed] [Google Scholar]
- Wang Y, Xu Y, Sun Y, Wang H, Qi J, Wan B, Ye W, Lin Y, Shao Y, Dong S, et al. (2018) Leucine-rich repeat receptor-like gene screen reveals that Nicotiana RXEG1 regulates glycoside hydrolase 12 MAMP detection. Nat Commun 9: 594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whisson SC, Boevink PC, Moleleki L, Avrova AO, Morales JG, Gilroy EM, Armstrong MR, Grouffaud S, van West P, Chapman S, et al. (2007) A translocation signal for delivery of oomycete effector proteins into host plant cells. Nature 450: 115–118 [DOI] [PubMed] [Google Scholar]
- Willocquet L, Savary S, Yuen J (2017) Multiscale phenotyping and decision strategies in breeding for resistance. Trends Plant Sci 22: 420–432 [DOI] [PubMed] [Google Scholar]
- Windstam S, Nelson EB (2008) Differential interference with Pythium ultimum sporangial activation and germination by Enterobacter cloacae in the corn and cucumber spermospheres. Appl Environ Microbiol 74: 4285–4291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang Q, Judelson HS (2014) Myb transcription factors and light regulate sporulation in the oomycete Phytophthora infestans. PLoS ONE 9: e92086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Zhao W, Hua C, Zheng X, Jing M, Li D, Govers F, Meijer HJG, Wang Y (2013) Chemotaxis and oospore formation in Phytophthora sojae are controlled by G-protein-coupled receptors with a phosphatidylinositol phosphate kinase domain. Mol Microbiol 88: 382–394 [DOI] [PubMed] [Google Scholar]
- Yu X, Tang J, Wang Q, Ye W, Tao K, Duan S, Lu C, Yang X, Dong S, Zheng X, et al. (2012) The RxLR effector Avh241 from Phytophthora sojae requires plasma membrane localization to induce plant cell death. New Phytol 196: 247–260 [DOI] [PubMed] [Google Scholar]
- Zerillo MM, Adhikari BN, Hamilton JP, Buell CR, Lévesque CA, Tisserat N (2013) Carbohydrate-active enzymes in Pythium and their role in plant cell wall and storage polysaccharide degradation. PLoS ONE 8: e72572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Blackman LM, Hardham AR (2013) Transient fusion and selective secretion of vesicle proteins in Phytophthora nicotianae zoospores. PeerJ 1: e221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Zhai C, Hua C, Qiu M, Hao Y, Nie P, Ye W, Wang Y (2016) PsHint1, associated with the G-protein α subunit PsGPA1, is required for the chemotaxis and pathogenicity of Phytophthora sojae. Mol Plant Pathol 17: 272–285 [DOI] [PMC free article] [PubMed] [Google Scholar]