Summary
Antibiotic resistance mediated by bacterial production of extended‐spectrum beta‐lactamase (ESBL) is a global threat to public health. ESBL resistance is most commonly hospital‐acquired; however, infections acquired outside of hospital settings have raised concerns over the role of livestock and wildlife in the zoonotic spread of ESBL‐producing bacteria. Only limited data are available on the circulation of ESBL‐producing bacteria in animals. Here, we report ESBL‐producing Escherichia coli in wild common vampire bats Desmodus rotundus and livestock near Lima, Peru. Molecular analyses revealed that most of this resistance resulted from the expression of bla CTX‐M‐15 genes carried by plasmids, which are disseminating worldwide in hospital settings and have also been observed in healthy children of Peru. Multilocus sequence typing showed a diverse pool of E. coli strains carrying this resistance that were not always host species‐specific, suggesting sharing of strains between species or infection from a common source. This study shows widespread ESBL resistance in wild and domestic animals, supporting animal communities as a potential source of resistance. Future work is needed to elucidate the role of bats in the dissemination of antibiotic‐resistant strains of public health importance and to understand the origin of the observed resistance.
Keywords: antibiotic resistance, Desmodus rotundus, ESBL‐producing Escherichia coli, multilocus sequence typing, Peru, plasmid typing
Impacts.
Extended‐spectrum beta‐lactamase (ESBL) resistance is a global threat to public health, and community‐acquired infections are increasingly reported.
There are limited data on the role of wildlife and livestock in the circulation of ESBL‐producing bacteria, particularly in Latin America.
The presence of ESBL‐producing Escherichia coli in both common vampire bats and livestock reveals a poorly understood and potentially zoonotic transmission cycle of ESBL resistance.
1. INTRODUCTION
Antimicrobial‐resistant (AMR) bacteria are responsible for hundreds of thousands of fatalities annually (World Health Organization, 2014). The majority of this problem is attributed to the spread of extended‐spectrum beta‐lactamase (ESBL)‐producing Escherichia coli and Klebsiella pneumonia (World Health Organization, 2014). Although most cases of ESBL‐producing bacteria are reported in clinical settings, increasing numbers of infections acquired outside of hospital settings (i.e., community‐acquired infections) have suggested that livestock and wildlife may form a zoonotic reservoir of ESBL for people living in close contact with animals (Guenther, Ewers, & Wieler, 2011). Supporting this hypothesis, ESBL‐producing E. coli have been found in several wild animals in Europe (Guenther et al., 2011); however, relatively few studies have been carried out on wild animals in low‐income countries (Carrillo‐Del Valle et al., 2016; Cristóbal‐Azkarate, Dunn, Day, & Amábile‐Cuevas, 2014; Guenther et al., 2011; Hasan et al., 2016; Liakopoulos et al., 2016), where the consequences of ESBL resistance can be exacerbated by a higher number of bacterial infections and more limited access to health facilities providing appropriate antibiotic treatment (Sosa et al., 2010).
While the role of bats as reservoirs of viruses is widely appreciated, bacterial infections have received less attention (Mühldorfer, 2013). Given their nightly feeding on antibiotic‐treated livestock and humans and widespread presence across Latin America, the common vampire bat, Desmodus rotundus, has a high risk of exposure to these bacteria and could potentially form a reservoir for transmission to other species. Although several recent studies have begun to characterize the diversity of pathogenic and non‐pathogenic bacteria in vampire bats (Adesiyun, Stewart‐Johnson, & Thompson, 2009; Bai et al., 2012; Carrillo‐Araujo et al., 2015; Chaverri, 2006; Zetun, Hoffmann, Silva, Souza, & Langoni, 2009), no study has evaluated this species for ESBL‐producing bacteria or investigated bacterial sharing with their livestock prey.
The aim of this study was to screen vampire bats and their surrounding livestock prey for ESBL‐producing Enterobacteriaceae in peri‐urban farms around Lima, and identify the bacterial strains and genes responsible for the observed phenotypic resistance to better understand the epidemiology of resistance in these communities. We report for the first time the presence of ESBL‐producing E. coli in vampire bats and livestock in Peru, suggesting a wide dissemination of AMR bacteria in the community.
2. METHODS
In October 2015, we collected faecal swabs from 81 vampire bats in four colonies located in four districts (N = 29 in Mala district, 20 in Barranca, 20 in Huacho and 12 in Chancay) of the Lima Region of Peru (Figure 1a). We also collected fresh faecal swabs from 20 cows, eight pigs, five sheep, two horses and two donkeys from farms located <5 km from bat colonies, an area within the typical foraging range of vampire bats (Trajano, 1996). Samples were collected from six small‐scale farms that kept <30 animals each. Questionnaires to farmers and visual inspections of bite wounds confirmed that bats fed in all farms in the month prior to sampling and that farmers commonly treated their animals with tetracycline and penicillin. Twenty‐five per cent of samples came from livestock with fresh bites. Swabs from both bats and livestock were first screened for ESBL‐producing Enterobacteriaceae by direct incubation (on average, <1 hr after collection) at 37°C for 24 hr in ChromID ESBL (bioMérieux, Marcy l'Etoile, France). Samples exhibiting bacterial growth were subcultured to confirm growth after 24 and 48 hr of incubation. We used matrix‐assisted laser desorption/ionization–time‐of‐flight (MALDI‐TOF) mass spectrometry (Bruker Daltonics, Bremen, Germany) to identify the species of ESBL‐positive samples. From the 58 ESBL‐positive samples, 15 were E. coli (five from bats and 10 from livestock). The remaining 43 isolates included non‐Enterobacteriaceae Gram‐negative bacilli (e.g., Pseudomonas spp. and Acromobacter spp.). Susceptibility testing to individual β‐lactam antibiotics was performed using the disc diffusion method on Mueller–Hinton agar (Matuschek, Brown, & Kahlmeter, 2014) and was interpreted according to the European Committee on Antimicrobial Susceptibility Testing (EUCAST) clinical breakpoints (version 5.0). ESBL production was confirmed with the double‐disc synergy test.
Figure 1.
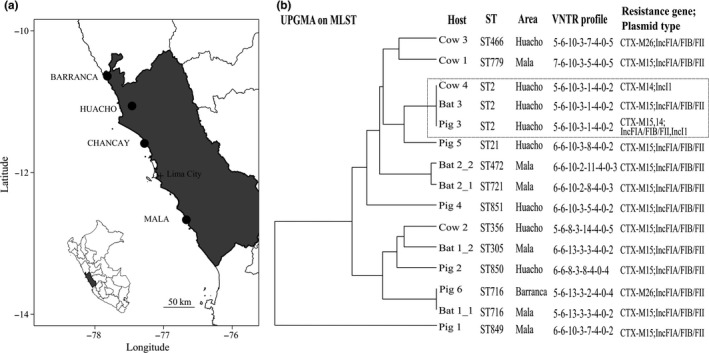
Map of the study area and phylogenetic relationships between ESBL‐producing bacteria from vampire bats and livestock in the Lima Region, Peru (a) Map with the location of the four vampire bat colonies sampled (b) Genetic relationship between isolates obtained by a Unweighted Pair Group Method with Arithmetic Mean (UPGMA) hierarchical clustering using the multilocus sequence typing (MLST) of loci dinB, icdA, pabB, polB, putP, trpA, trpB and uidA. The VNTR profile shown corresponds to the number of repeats from loci CVN001, CVN003, CVN004, CVN007, CVN014, CVN015, CVN016 and CCR001. The grey rectangle highlights isolates from three animals in Huacho showing the same genetic background. Isolates from the same individual are differentiated using an underscore
We assessed the genetic background of the ESBL‐producing E. coli by performing multilocus sequence typing (MLST) (http://bigsdb.web.pasteur.fr/) and variable number of tandem repeats (VNTR) (Lindstedt, Brandal, Aas, Vardund, & Kapperud, 2007). The genetic basis of the observed resistance was identified using multiplex PCR and DNA sequencing targeting the most prevalent ESBL‐encoding genes (e.g., bla CTX‐M, bla TEM , bla SHV and bla OXA‐1‐like genes) (Dallenne, Da Costa, Decré, Favier, & Arlet, 2010). Plasmids carrying resistant genes were identified and typed using the PCR‐based replicon typing method and the plasmid relaxase gene typing method (Carattoli et al., 2005; Compain et al., 2014).We also evaluated the ability of plasmids to conjugate using mating experiments with the azide‐resistant E. coli strain J53 as a recipient.
3. RESULTS
We found 15 E. coli isolates that were intermediate or resistant to most β‐lactam antibiotics, except imipenem and ertapenem, in both vampire bats and livestock (Table S1). Molecular analysis revealed the presence of bla CTX‐M‐15 resistance gene alone in 11 isolates (all five bat and six livestock isolates including two cows and four pigs) and associated with bla CTX‐M‐14 in one isolate (“Pig 3,” Figure 1b). The bla CTX‐M‐26 and bla CTX‐M‐14 genes were found in two (“Cow 3” and “Pig 6”) and one (“Cow 4”) isolates of livestock, respectively. The bla CTX‐M15 and bla CTX‐M26 genes were associated with IncFIA/FIB/FII plasmid, and the bla CTX‐M14 was associated with IncI1 plasmid. Mating experiments showed that these plasmids were able to conjugate, suggesting their potential circulation between different bacterial strains.
Strain genotyping by MLST revealed high diversity among E. coli isolates across hosts and sampling locations: bat isolates belonged to sequence types (STs) ST2, ST305, ST472, ST716 and ST721, whereas livestock isolates belonged to ST2, ST21, ST356, ST466, ST479, ST716 and 3 novel STs registered as ST849, ST850 and ST851. All isolates had unique VNTR haplotypes, except for ST2, for which isolates from a bat, a cow and a pig in Huacho had the same haplotype.
4. DISCUSSION
Extended‐spectrum beta‐lactamase‐producing E. coli carrying bla CTX‐M‐15 genes have disseminated worldwide, causing a large number of nosocomial and community‐acquired urinary tract and bloodstream infections among humans (Guenther et al., 2011). bla CTX‐M‐15 and bla CTX‐M‐14 genes were reported in healthy children in Peru and Bolivia, and IncFIA/FIB/FII and IncI1 plasmids have been suggested to play a major role in the dissemination of these genes across humans in Latin America (Pallecchi et al., 2007). In our study, bla CTX‐M genes were carried by these same broad‐host‐range transmissible plasmids, suggesting that wildlife and livestock may also disseminate this ESBL resistance. The high diversity of E. coli STs found here, which do not overlap with clones found in wild birds of Chile or Argentina (Báez et al., 2015; Liakopoulos et al., 2016), suggests dissemination of these plasmids between different bacterial strains.
A pig and the bat carried the same bla CTX‐M‐15 gene within the same plasmid and the same E. coli ST, suggesting ESBL‐producing bacterial exchange between bats and livestock or contamination from a common source such as human sewage. Several routes of bacterial transmission between bats and livestock are possible. For example, faecal–oral transmission from livestock to bats could occur if bats ingest contaminated faecal material present on livestock skin or in the environment. Environmental contamination of bats with humans or livestock bacteria can increase if human or livestock sewage is used in farms. Alternatively, transmission is possible during blood feeding or contact with the wound, although this should be less frequent for E. coli, given that this bacterium is not commonly found in the blood of healthy animals. Finally, livestock and humans could also be exposed to bat bacteria through guano, as bat roosts in this area are mainly located in tunnels that cover parts of irrigation systems for crops.
The role of wildlife in the dissemination of resistant genes is supported by the recent isolation of ESBL‐producing bacteria carrying bla CTX‐M‐15 genes in wild birds of Nicaragua, Chile and Argentina, and raises public health concerns given the implications of these bacteria in both nosocomial and community‐acquired infections in humans (Guenther et al., 2011; Hasan et al., 2016; Liakopoulos et al., 2016). Our findings highlight the need to elucidate the role of bats and livestock in AMR bacterial transmission cycles and what risks these community sources may pose for zoonotic transmission to humans. Specifically, future work should investigate whether bat populations are capable of maintaining ESBL‐producing bacteria or whether these resistant bacteria result from frequent exchanges with livestock or humans through the environment (e.g., water and soil).
Supporting information
ACKNOWLEDGEMENTS
DS and JB were funded by a Sir Henry Dale Fellowship, jointly funded by the Wellcome Trust and Royal Society (Grant 102507/Z/13/Z). CS, JB and DS were also funded by a CONCYTEC‐UK Embassy grant (No. 003‐2016‐FONDECYT). The Peruvian Government authorized sample collection and use of genetic resources for this study (RD‐273‐2012‐AG‐DGFFS‐DGEFFS, RD‐054‐2016‐SERFOR‐DGGSPFFS). Livestock sampling and bat sampling were approved by the MVLS College Ethics Committee of the University of Glasgow (200140112). We thank all farmers involved in this study for their cooperation and help with livestock sampling.
Benavides JA, Shiva C, Virhuez M, et al. Extended‐spectrum beta‐lactamase‐producing Escherichia coli in common vampire bats Desmodus rotundus and livestock in Peru. Zoonoses Public Health. 2018;65:454–458. 10.1111/zph.12456
REFERENCES
- Adesiyun, A. A. , Stewart‐Johnson, A. , & Thompson, N. N. (2009). Isolation of enteric pathogens from bats in Trinidad. Journal of wildlife diseases, 45, 952–961. 10.7589/0090-3558-45.4.952 [DOI] [PubMed] [Google Scholar]
- Báez, J. , Hernández‐García, M. , Guamparito, C. , Díaz, S. , Olave, A. , Guerrero, K. , … Valenzuela, N. (2015). Molecular characterization and genetic diversity of ESBL‐producing Escherichia coli colonizing the migratory Franklin's gulls (Leucophaeus pipixcan) in Antofagasta, North of Chile. Microbial Drug Resistance, 21, 111–116. 10.1089/mdr.2014.0158 [DOI] [PubMed] [Google Scholar]
- Bai, Y. , Recuenco, S. , Gilbert, A. T. , Osikowicz, L. M. , Gómez, J. , Rupprecht, C. , & Kosoy, M. Y. (2012). Prevalence and diversity of Bartonella spp. in bats in Peru. The American Journal of Tropical Medicine and Hygiene, 87, 518–523. 10.4269/ajtmh.2012.12-0097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carattoli, A. , Bertini, A. , Villa, L. , Falbo, V. , Hopkins, K. L. , & Threlfall, E. J. (2005). Identification of plasmids by PCR‐based replicon typing. Journal of Microbiological Methods, 63, 219–228. 10.1016/j.mimet.2005.03.018 [DOI] [PubMed] [Google Scholar]
- Carrillo‐Araujo, M. , Taş, N. , Alcántara‐Hernández, R. J. , Gaona, O. , Schondube, J. E. , Medellín, R. A. , … Falcón, L. I. (2015). Phyllostomid bat microbiome composition is associated to host phylogeny and feeding strategies. Frontiers in Microbiology, 6, 447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrillo‐Del Valle, M. D. , Jorge, A. , Díaz‐Aparicio, E. , Valdivia‐Flores, A. G. , Cisneros‐Guzmán, L. F. , Rosario, C. , … Maravilla, P. (2016). Characterization of Escherichia coli strains from red deer (Cervus elaphus) faeces in a Mexican protected natural area. European Journal of Wildlife Research, 62, 415–421. 10.1007/s10344-016-1015-z [DOI] [Google Scholar]
- Chaverri, G. (2006). Flora bacteriana aeróbica del tracto digestivo del vampiro común, Desmodus rotundus (Chiroptera: Phyllostomidae). Revista de Biología Tropical, 54, 717–724. [PubMed] [Google Scholar]
- Compain, F. , Poisson, A. , Le Hello, S. , Branger, C. , Weill, F.‐X. , Arlet, G. , & Decré, D. (2014). Targeting relaxase genes for classification of the predominant plasmids in Enterobacteriaceae. International Journal of Medical Microbiology, 304, 236–242. 10.1016/j.ijmm.2013.09.009 [DOI] [PubMed] [Google Scholar]
- Cristóbal‐Azkarate, J. , Dunn, J. C. , Day, J. M. , & Amábile‐Cuevas, C. F. (2014). Resistance to antibiotics of clinical relevance in the fecal microbiota of Mexican wildlife. PLoS ONE, 9, e107719 10.1371/journal.pone.0107719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dallenne, C. , Da Costa, A. , Decré, D. , Favier, C. , & Arlet, G. (2010). Development of a set of multiplex PCR assays for the detection of genes encoding important β‐lactamases in Enterobacteriaceae. Journal of Antimicrobial Chemotherapy, 65(3), 490–495. 10.1093/jac/dkp498 [DOI] [PubMed] [Google Scholar]
- Guenther, S. , Ewers, C. , & Wieler, L. (2011). Extended‐spectrum beta‐lactamases producing E. coli in wildlife, yet another form of environmental pollution? Frontiers in Microbiology, 2, 246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasan, B. , Laurell, K. , Rakib, M. M. , Ahlstedt, E. , Hernandez, J. , Caceres, M. , & Järhult, J. D. (2016). Fecal carriage of extended‐spectrum β‐lactamases in healthy humans, poultry, and wild birds in León, Nicaragua—a shared pool of bla CTX‐M genes and possible interspecies clonal spread of extended‐spectrum β‐lactamases‐producing Escherichia coli . Microbial Drug Resistance, 22, 682–687. 10.1089/mdr.2015.0323 [DOI] [PubMed] [Google Scholar]
- Liakopoulos, A. , Olsen, B. , Geurts, Y. , Artursson, K. , Berg, C. , Mevius, D. J. , & Bonnedahl, J. (2016). Molecular characterization of extended‐spectrum cephalosporin‐resistant Enterobacteriaceae from wild birds (Kelp gulls) in South America. Antimicrobial Agents and Chemotherapy, 60, 6924–6927. 10.1128/AAC.01120-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindstedt, B.‐A. , Brandal, L. T. , Aas, L. , Vardund, T. , & Kapperud, G. (2007). Study of polymorphic variable‐number of tandem repeats loci in the ECOR collection and in a set of pathogenic Escherichia coli and Shigella isolates for use in a genotyping assay. Journal of Microbiological Methods, 69, 197–205. 10.1016/j.mimet.2007.01.001 [DOI] [PubMed] [Google Scholar]
- Matuschek, E. , Brown, D. , & Kahlmeter, G. (2014). Development of the EUCAST disk diffusion antimicrobial susceptibility testing method and its implementation in routine microbiology laboratories. Clinical Microbiology and Infection, 20, O255–O266. 10.1111/1469-0691.12373 [DOI] [PubMed] [Google Scholar]
- Mühldorfer, K. (2013). Bats and bacterial pathogens: A review. Zoonoses and Public Health, 60, 93–103. 10.1111/j.1863-2378.2012.01536.x [DOI] [PubMed] [Google Scholar]
- Pallecchi, L. , Bartoloni, A. , Fiorelli, C. , Mantella, A. , Di Maggio, T. , Gamboa, H. , … Rossolini, G. M. (2007). Rapid dissemination and diversity of CTX‐M extended‐spectrum β‐lactamase genes in commensal Escherichia coli isolates from healthy children from low‐resource settings in Latin America. Antimicrobial Agents and Chemotherapy, 51, 2720–2725. 10.1128/AAC.00026-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sosa, A. D. J. , Amábile‐Cuevas, C. F. , Byarugaba, D. K. , Hsueh, P.‐R. , Kariuki, S. , & Okeke, I. N. (2010). Antimicrobial resistance in developing countries. New York, NY: Springer; 10.1007/978-0-387-89370-9 [DOI] [Google Scholar]
- Trajano, E. (1996). Movements of cave bats in Southeastern Brazil, with emphasis on the population ecology of the Common Vampire Bat, Desmodus Rotundus (Chiroptera). Biotropica, 28(1), 121–129. 10.2307/2388777 [DOI] [Google Scholar]
- World Health Organization . (2014). Antimicrobial resistance: 2014 global report on surveillance. Geneva, Switzerland: World Health Organization. [Google Scholar]
- Zetun, C. , Hoffmann, J. , Silva, R. , Souza, L. C. D. , & Langoni, H. (2009). Leptospira spp. and Toxoplasma gondii antibodies in vampire bats (Desmodus rotundus) in Botucatu region, SP, Brazil. Journal of Venomous Animals and Toxins including Tropical Diseases, 15, 546–552. 10.1590/S1678-91992009000300014 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials


