Excess fatty acids are toxic to cells but can be sequestered as triacylglycerides in lipid droplets. Hariri et al. show that the tethering protein Mdm1 spatially regulates this process at the junction between the endoplasmic reticulum and the yeast vacuole. These findings suggest that Mdm1 can drive spatially defined lipid droplet production to maintain cell homeostasis and protect against lipotoxicity.
Abstract
Lipid droplets (LDs) serve as cytoplasmic reservoirs for energy-rich fatty acids (FAs) stored in the form of triacylglycerides (TAGs). During nutrient stress, yeast LDs cluster adjacent to the vacuole/lysosome, but how this LD accumulation is coordinated remains poorly understood. The ER protein Mdm1 is a molecular tether that plays a role in clustering LDs during nutrient depletion, but its mechanism of function remains unknown. Here, we show that Mdm1 associates with LDs through its hydrophobic N-terminal region, which is sufficient to demarcate sites for LD budding. Mdm1 binds FAs via its Phox-associated domain and coenriches with fatty acyl–coenzyme A ligase Faa1 at LD bud sites. Consistent with this, loss of MDM1 perturbs free FA activation and Dga1-dependent synthesis of TAGs, elevating the cellular FA level, which perturbs ER morphology and sensitizes yeast to FA-induced lipotoxicity. We propose that Mdm1 coordinates FA activation adjacent to the vacuole to promote LD production in response to stress, thus maintaining ER homeostasis.
Introduction
Cells and organisms face constant fluctuations in nutrient levels; therefore, their ability to store energy may promote survival during starvation (Walther and Farese, 2012; Seo et al., 2017). At a cellular level, energy is stored in the form of neutral lipids packaged as lipid droplets (LDs) that form on the surface of the ER (Kassan et al., 2013; Choudhary et al., 2015). Remarkably little is known about how LDs form (Thiam et al., 2013). Current models suggest that neutral lipids, triacylglycerides (TAGs) and sterol esters, are synthesized and deposited between the ER monolayer leaflets (Walther and Farese, 2009). As neutral lipids reach a critical concentration, they coalesce into a “lens” that grows and eventually buds toward the cytoplasm, forming phospholipid monolayer–surrounded LDs (Walther et al., 2017).
LDs are increasingly considered ubiquitous organelles that function in maintaining cellular homeostasis beyond energy storage. Recent evidence suggests that LDs serve as depots for otherwise toxic lipids that perturb ER and mitochondrial functions (Nguyen et al., 2017). LDs also sequester lipotoxic fatty acids (FAs) through their incorporation into TAGs. FAs are esterified into fatty acyl–coenzyme A (FA-CoAs), which are subsequently used as building blocks in the stepwise assembly of TAGs. Consistent with this, mammalian cells lacking the TAG synthase DGAT1 display elevated sensitivity to FA-induced cytotoxicity and cell death (Listenberger et al., 2003; Chitraju et al., 2017). Similarly, LD-deficient yeast accumulates FAs within ER membranes, resulting in elevated ER stress and the induction of the unfolded protein response, as well as growth sensitivity when cultured in media containing lipotoxic FAs (Velázquez et al., 2016). Finally, inducing ER stress stimulates LD formation in yeast, underscoring the functional relationship between ER homeostasis and LD biogenesis (Fei et al., 2009; Basseri and Austin, 2012). However, the molecular mechanisms governing the interplay between ER stress responses and LD production remain poorly characterized.
What determines sites of LD biogenesis along the ER network is poorly understood. Stress-induced LD biogenesis appears to be highly spatially regulated and compartmentalized. Recent evidence suggests that, upon sensing a decline in nutrients, yeast exhibit a “bloom” of LDs that are spatially organized at a specific subregion of the ER surface adjacent to the vacuole (Barbosa and Siniossoglou, 2016; Hariri et al., 2018). This subregion constitutes an interorganelle contact site termed the nuclear ER–vacuole junction (NVJ; Pan et al., 2000). NVJ-associated LDs are a unique LD subpopulation decorated with specific proteins, including LD organizing proteins that regulate LD accumulation at the NVJ (Eisenberg-Bord et al., 2017; Teixeira et al., 2017). Another protein that clusters LDs at the NVJ is Mdm1, an integral ER membrane protein that binds to the vacuole in trans (Henne et al., 2015; Hariri et al., 2018). Mdm1 is sufficient to form ER–vacuole contact sites, and its overexpression promotes LD clustering at the NVJ, but its precise role in LD homeostasis remains unclear. Furthermore, Mdm1 is highly conserved in metazoans, and loss-of-function mutations in the human homologue Snx14 are associated with pediatric recessive cerebellar ataxia 20 disease (SCAR20; Thomas et al., 2014; Akizu et al., 2015). Recent studies have linked Snx14 function to neutral lipid metabolism, but its precise function also remains unknown (Bryant et al., 2018).
In the current study, we investigated the role of Mdm1 in NVJ-associated LD biogenesis. Our findings reveal that Mdm1 interacts with ER-associated LDs via its hydrophobic N-terminal region and regulates FA activation at LD bud sites. As such, loss of MDM1 perturbs FA metabolism and ER homeostasis, sensitizing yeast to FA-induced lipotoxicity. Our structural data provide insights into the molecular mechanism of this lipotoxicity, and of LD biogenesis in general. We propose that Mdm1 is uniquely positioned at three-way junctions connecting the ER, LDs, and vacuole to promote FA activation and incorporation into TAGs. Mdm1 thus promotes stress-induced LD biogenesis at the ER–vacuole interface where LDs can be efficiently delivered to the vacuole via lipophagy, thus helping maintain cellular homeostasis.
Results
Mdm1 associates with LD bud sites via its hydrophobic N-terminal region
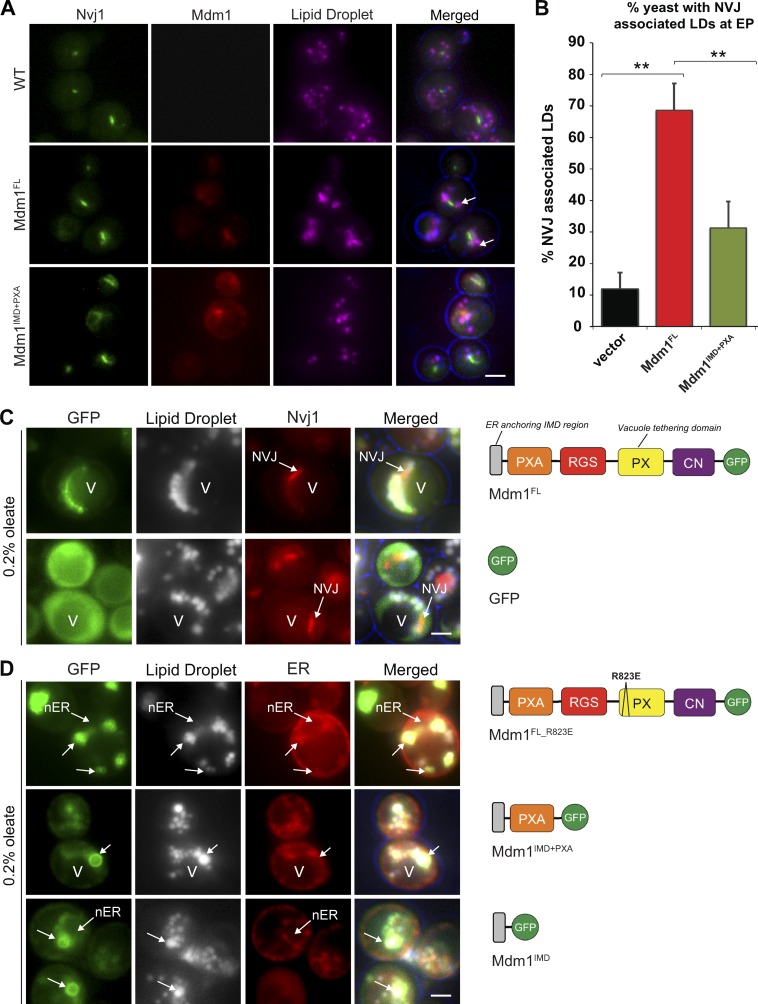
Previously, we demonstrated that yeast NVJs serve as sites for starvation-induced LD biogenesis (Hariri et al., 2018). The ER–vacuole tether Mdm1 coenriched with NVJ-associated LDs, suggesting that Mdm1 may directly interact with LDs and promote their clustering adjacent to the vacuole (Hariri et al., 2018). Overexpression of full-length Mdm1 (Mdm1FL), but not a truncated Mdm1 lacking its vacuole binding Phox (PX) homology domain, was sufficient to cluster LDs at the NVJ, suggesting that Mdm1 may directly associate with LDs at the NVJ (Fig. 1, A and B). To investigate how Mdm1 interacts with LDs and determine the structural features that are required for this interaction, we generated multiple GFP-tagged fragments of Mdm1 and examined their subcellular association with LDs using light microscopy.
Figure 1.
Mdm1 binds LDs via its N-terminal hydrophobic motif. (A) Light microscopy of yeast expressing chromosomally tagged Nvj1-GFP (green) and expressing either mCherry-tagged Mdm1FL or Mdm1IMD+PXA (red). LDs (magenta) visualized by MDH staining (arrows). Scale bar, 2 µm. (B) Quantification of images in A. Percentage of yeast cells with NVJ-associated LDs over the total number of cells counted; mean ± SD; n > 50 cells; **, P < 0.005; Student’s t test. EP, exponential phase. (C) Light microscopy of yeast expressing GFP-tagged Mdm1FL or soluble GFP in yeast expressing chromosomally tagged Nvj1-mCherry in the presence of 0.2% oleate. LDs (gray) visualized by MDH staining. V, vacuole. Diagrams depict Mdm1 fragments expressed. Scale bar, 2 µm. (D) Light microscopy of yeast expressing different GFP-tagged Mdm1 fragments in yeast expressing chromosomally tagged ER marker Ds-Red HDEL in the presence of 0.2% oleate. LDs (gray) visualized by MDH staining. Diagrams depict Mdm1 fragments expressed. RGS: regulator of G protein signaling, CN: C-terminal Nexin. Scale bar, 2 µm.
We cultured yeast in media containing oleate to stimulate LD production and growth. Upon oleate supplementation, Mdm1FL remained in the nuclear ER region surrounding the NVJ and associated with LDs adjacent to the vacuole, forming either foci at the base of LDs or crescent-shaped cups partially surrounding LDs (Fig. 1 C and Fig. S1, A–C). Mdm1 binds to the vacuole via its PX homology domain that binds PI3P with high affinity (Yu and Lemmon, 2001). To test whether vacuole binding was required for this Mdm1-LD association, we expressed an Mdm1FL containing a single amino acid mutation in the PX domain (Mdm1FL_R823E) that was previously shown to mitigate vacuole binding and NVJ localization (Henne et al., 2015). Interestingly, the association of Mdm1 with LDs was not affected by the PX mutation (Fig. 1 D and Fig. S1 D). Similarly, a construct encoding only the N-terminal integral membrane domain (IMD) and PX-associated (PXA) domain (Mdm1IMD+PXA) was sufficient to associate with LDs in oleate-treated yeast (Fig. 1 D and Fig. S1 E).
To determine the specific region of the Mdm1 N-terminus that is required for LD binding, the IMD (amino acids 1–51) was fused to GFP (Mdm1IMD). In contrast to soluble GFP that distributed throughout the cytoplasm, Mdm1IMD was sufficient to associate with LDs (Fig. 1, C and D). Consistent with this, an Mdm1 construct lacking the IMD (Mdm1FL(−IMD)) lost LD binding, indicating the IMD is necessary and sufficient for LD association (Fig. S1 F). To determine whether LD association was a specific feature of the Mdm1 IMD, we replaced the IMD with the ER-anchored region of the tricalbin protein Tcb2 (Mdm1Tcb2+PXA), an ER protein that mediates ER–plasma membrane (PM) interorganelle tethering (Manford et al., 2012). This chimera localized throughout the ER network but failed to associate with LDs (Fig. S1 G). Altogether, these data demonstrate that the IMD of Mdm1 is required for LD association, whereas the C-terminal PX domain is required for vacuole binding and promotes Mdm1-mediated LD clustering at the NVJ.
The N-terminal region of Mdm1 defines sites of LD budding
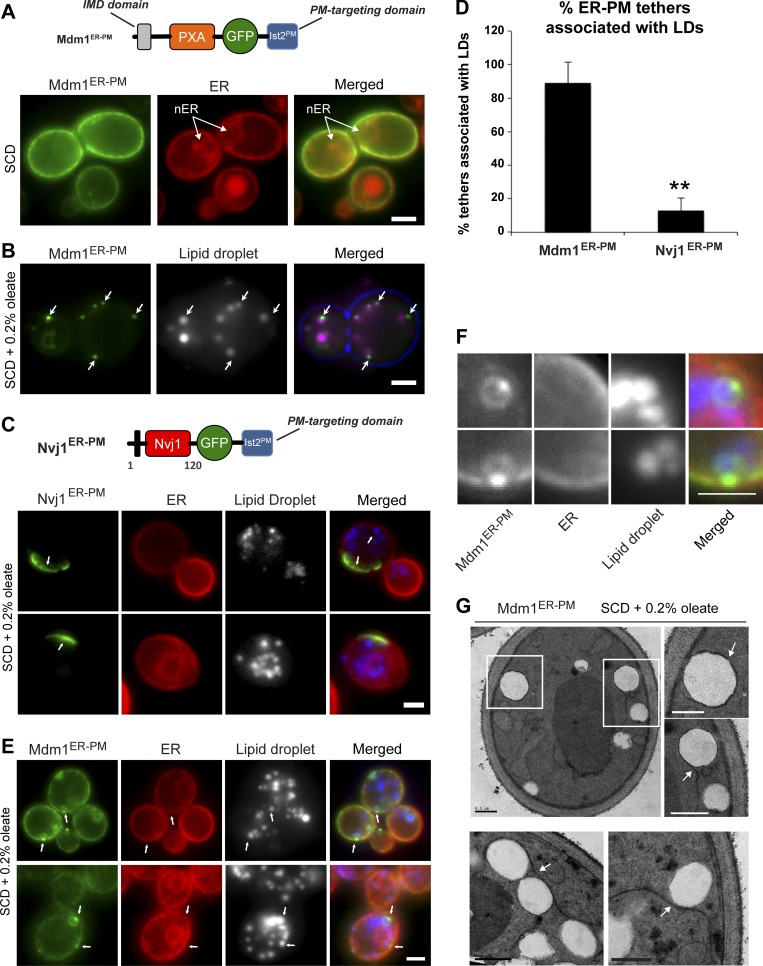
Since Mdm1 was sufficient to mediate LD clustering adjacent to the vacuole, we next investigated whether relocalizing Mdm1 to other regions of the cell would affect LD subcellular distribution. We generated an Mdm1 chimera that localized to ER–PM contact sites by replacing the Mdm1 vacuole-binding C-terminal region with the PM-binding region of Ist2, an ER–protein that functions as an interorganelle tether at ER–PM contact sites (Manford et al., 2012). This chimera (denoted Mdm1ER-PM) localized throughout the cortical ER (cER) immediately below the cell surface and was excluded from the nuclear ER where Mdm1 normally resides (Fig. 2 A). When yeast cells were cultured in media containing oleate, Mdm1ER-PM coalesced into distinct foci near the cell surface (Fig. 2 B). Strikingly, more than 80% of peripheral Mdm1ER-PM foci were associated with LDs near the cell surface, indicating that Mdm1ER-PM is demarcating LD bud sites along the cER network (Fig. 2, B and D).
Figure 2.
N-terminal region of Mdm1 is sufficient to define sites of LD budding. (A) Top: Diagram of GFP-tagged Mdm1ER-PM chimera with a C-terminal Ist2 PM-targeting domain. Bottom: Light microscopy of yeast expressing GFP-tagged Mdm1ER-PM chimera with Ds-Red HDEL marker to label the ER. Scale bar, 2 µm. (B) Light microscopy of yeast fed 0.2% oleate and expressing GFP-tagged Mdm1ER-PM chimera. LDs (gray) visualized by MDH staining (arrows). Scale bar, 2 µm. (C) Top: Diagram of GFP-tagged Nvj1ER-PM chimera with a C-terminal Ist2 PM-targeting domain. Bottom: Light microscopy of yeast fed 0.2% oleate and expressing GFP-tagged Nvj1ER-PM chimera with Ds-Red HDEL marker to label the ER. LDs (gray) visualized by MDH staining (arrows). Scale bar, 2 µm. (D) Quantification of yeast ER–PM tethers associated with LDs. Percentage of yeast cells over the total number of cells counted; mean ± SD; n > 50 cells; **, P < 0.005; Student’s t test. (E) Light microscopy of yeast fed 0.2% oleate and expressing GFP-tagged Mdm1ER-PM chimera with Ds-Red HDEL marker to label the ER. LDs (gray) visualized by MDH staining (arrows). Scale bar, 2 µm. (F) Higher magnification of images in E. Scale bar, 2 µm. (G) Thin-sectioning TEM of Mdm1ER-PM fed 0.2% oleate. Arrows indicate ER wrapping. Scale bars, 0.5 µm.
To test whether targeting of another ER–vacuole tether to the PM would similarly alter LD distribution, we generated a chimera that retargeted Nvj1 to the ER–PM contact sites (Nvj1ER-PM). This was similarly achieved by replacing the Vac8-binding C-terminus of Nvj1 with the PM-targeting domain of Ist2 (Fig. 2 C). Nvj1ER-PM formed distinct NVJ-like patches at the cell surface similar to those that normally form at the ER–vacuole interface (Fig. 2 C). However unlike Mdm1ER-PM, the distribution of Nvj1ER-PM did not change upon the addition of oleate, and only ∼10% of Nvj1ER-PM patches were in close proximity to LDs (Fig. 2 D). These data indicate that Mdm1ER-PM can specifically demarcate sites in the ER network where oleate-induced LDs form (Fig. 2 D). Additionally, multichannel labeling of Mdm1ER-PM, LDs, and the ER revealed that Mdm1ER-PM colocalized with the ER network adjacent to LDs, suggesting that LD-associated Mdm1 foci define the ER–LD interface (Fig. 2, E and F). Consistent with this, the association of Mdm1ER-PM with LDs required proper ER–LD contacts, as cells lacking the protein seipin (Sei1), an ER-resident protein proposed to enrich at ER–LD junctions, exhibited a significant reduction in Mdm1ER-PM accumulation adjacent to LDs (Fig. S2, A and B). Similarly, mCherry-tagged Sei1 colocalized with Mdm1ER-PM at LD bud sites, indicating that Mdm1ER-PM was coenriching with Sei1 at the ER–LD interface (Fig. S2, C and D).
To further investigate how Mdm1ER-PM associates with LDs and the ER, we generated a yeast construct encoding GFP-tagged LiveDrop, a well-characterized tool encoding a class I LD-targeting hairpin derived from the Drosophila melanogaster enzyme GPAT4 (Wang et al., 2016). This probe was previously reported to traffic between the ER bilayer and LD monolayer surface that is derived from the ER (Wang et al., 2016). GFP-LiveDrop localized primarily to the LD surface in yeast but could also be detected along the ER network and at the ER–LD interface of LDs connected to the ER (Fig. S2 E). Coexpression of Mdm1ER-PM-mCherry with GFP-LiveDrop revealed a co-enrichment of both proteins at the same LDs, with Mdm1ER-PM foci colocalizing with GFP-LiveDrop foci at the ER–LD interface (Fig. S2, F and G).
In addition to accumulating at ER–LD contact sites, Mdm1ER-PM also enriched as a ring across the surface of some LDs (Fig. 2 E and Fig. S2, F and G). To better resolve the topology of this Mdm1-LD association, we used thin-sectioning transmission electron microscopy (TEM) and examined cells expressing Mdm1ER-PM after oleate treatment. Interestingly, we observed wrapping of ER tubules around cortical LDs, suggesting Mdm1 may promote ER–LD contacts (Fig. 2 G). Taken together, these data suggest that Mdm1ER-PM enriches at the ER–LD interface and is capable of inducing ER-wrapping around LDs.
The PXA domain of Mdm1 binds FAs in vitro
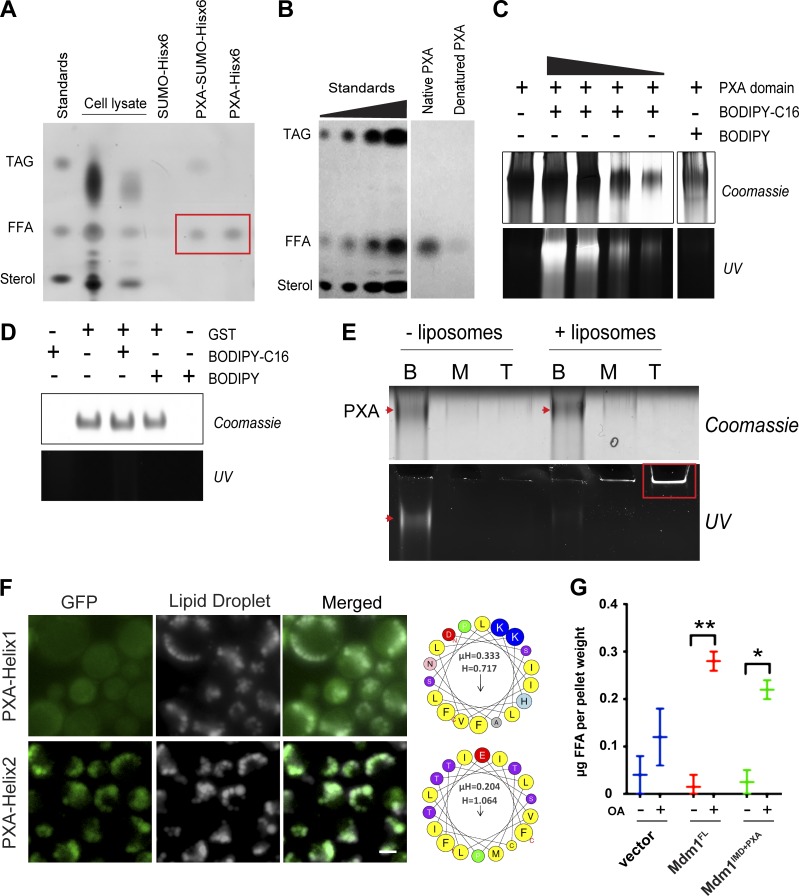
The observation that the Mdm1ER-PM chimera encoding only the IMD and PXA domain could demarcate sites of LD budding suggested that the PXA domain may play a role in LD formation and growth. The PXA domain is highly conserved in Mdm1 orthologues but has remained structurally and functionally uncharacterized. To investigate the function of the PXA domain, we first tested whether it could interact with lipids. We purified recombinant His6x-tagged and SUMO-His6x–tagged versions of Mdm1 PXA domain from Escherichia coli using a combination of affinity and size exclusion chromatography and validated that the protein domain was folded using circular dichroism spectroscopy (Fig. S3, A and B). Lipids were then extracted from purified yeast PXA and examined by using thin-layer chromatography (TLC). Both PXA-His6x and PXA-SUMO-His6x copurified with free FAs (FFAs) whereas His6x-SUMO alone did not (Fig. 3 A). This binding required protein folding, as denatured PXA domain failed to copurify with FFAs (Fig. 3 B).
Figure 3.
PXA domain of Mdm1 binds FAs. (A) TLC on lipids extracted from proteins purified from E. coli: PXA-SUMO-hisx6 and PXA-hisx6. SUMO-hisx6 was used as a negative control. Red box: FFA bands. (B) TLC on lipids extracted from recombinant Mdm1PXA-hisx6 before and after denaturation by using guanidinium hydrochloride. (C) Native PAGE of the PXA domain with or without fluorescent palmitic acid (BODIPY-C16) or BODIPY. Coomassie staining (top) and fluorescence (UV excitation; bottom). (D) Native PAGE of the GST with or without fluorescent palmitic acid (BODIPY-C16) or BODIPY used as a negative control for B. Coomassie staining (top) and fluorescence (bottom). (E) Native PAGE of BODIPY-loaded PXA after flotation assay in the presence and absence of liposomes. Coomassie staining of the PXA protein (top; arrows) and fluorescence corresponding to bound lipids (bottom, arrow and box). (F) Left: Light microscopy of yeast expressing PXA-Helix1 or PXA-Helix2 N-terminally tagged with GFP. LDs (gray) visualized by MDH staining. Scale bar, 2 µm. Right: Helix1 and Helix2 helical wheel representations generated by using HeliQuest. (G) TLC quantification of FFAs (in micrograms) extracted from WT yeast overexpressing Mdm1FL or Mdm1IMD+PXA (with or without oleate; 4 h). Data represent mean ± SD normalized to cell pellet weight from TLC plate; n = 2. *, P < 0.01; **, P < 0.005; two-way ANOVA.
To further investigate whether the PXA domain could interact with FFAs, we co-incubated the recombinant protein with fluorescently tagged palmitic acid (BODIPY-C16) and analyzed the protein:lipid mix by native PAGE. This method was previously used to demonstrate lipid-binding abilities of the SMP domain of extended synaptotagmin 2 (Schauder et al., 2014). This revealed that the PXA domain comigrated with BODIPY-C16 but not with free BODIPY, suggesting a protein–lipid interaction (Fig. 3 C). In contrast, BODIPY-C16 failed to comigrate with purified GST, indicating that the BODIPY-C16 interacted specifically with the PXA domain (Fig. 3 D).
Next, we evaluated whether FA binding to the PXA domain was reversible by determining whether PXA domains preloaded with BODIPY-C16 could release bound lipids into a recipient membrane (Fig. S3, C and D). PXA domain was preloaded with BODIPY-C16 and, after removal of excess (unbound) lipids, incubated with nonfluorescent acceptor liposomes. The mixture was then placed at the bottom of a sucrose gradient and centrifuged to allow the liposomes and any associated protein or BODIPY-C16 to float to the top (Fig. S3 C). Whereas the BODIPY-C16–loaded PXA remained at the gradient bottom in the absence of liposomes, the addition of liposomes caused a redistribution of BODIPY-C16 to the top fraction (Fig. 3 E). Importantly, the PXA protein itself remained in the bottom fraction and did not float with the liposomes (Fig. 3 E). Therefore, the PXA domain exhibited negligible affinity for membranes but was able to reversibly bind and donate FAs to liposomes in vitro.
In addition to the IMD region, hydropathy plots revealed a highly hydrophobic region within the PXA domain of Mdm1 (Fig. S3 E). Moreover, primary amino acid sequence analysis of the PXA domain revealed that it contained two predicted hydrophobic helices (Helix1, 138–162; Helix2, 234–262), of which Helix2 was contained within the predicted hydrophobic region (Fig. S3 F). Previous studies indicated that cytosolic proteins that target to LDs often contain amphipathic helices that embed into the LD monolayer surface (Kory et al., 2016; Prévost et al., 2018). We tested whether PXA Helix1 and Helix2 targeted to LDs by expressing both helices fused to GFP and examining their localizations. While GFP-Helix1 remained diffuse in the cytoplasm, GFP-Helix2 colocalized with LDs, suggesting that this region may promote Mdm1-LD association (Fig. 3 F). Collectively, we conclude that the PXA domain binds to FFAs and potentially associates with LDs via a hydrophobic helix.
Given its ability to interact with FAs in vitro and its close proximity to LDs in vivo, we next interrogated whether the PXA domain could interact with FAs in vivo. We cultured WT yeast and yeast ectopically expressing either Mdm1FL or the Mdm1IMD+PXA fragment and exposed them overnight to oleate-containing media. We then extracted their intracellular lipids and determined whether the presence of Mdm1 fragments led to the accumulation of intracellular FFAs. Interestingly, ectopic expression of both Mdm1FL and Mdm1IMD+PXA caused an approximately twofold increase in intracellular FFAs compared with WT yeast (Fig. 3 G). Collectively, we conclude that the PXA domain binds to FFAs in vitro and is sufficient to promote intracellular FFA accumulation in vivo.
The PXA domain of Mdm1 interacts with FA-CoA ligase Faa1
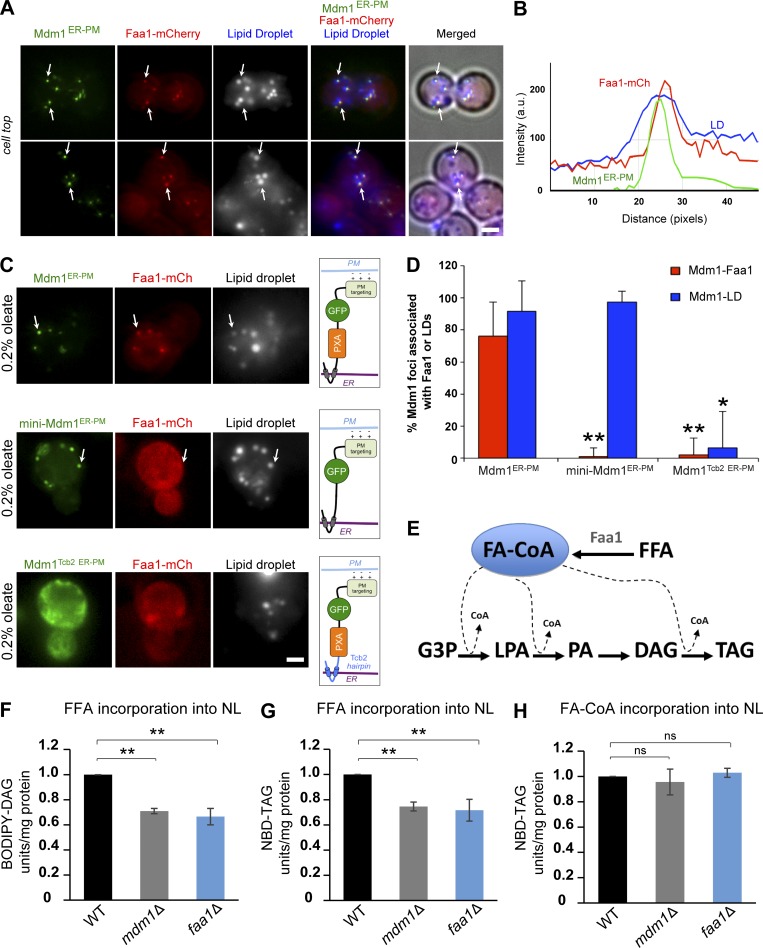
Using mass spectrometry-based proteomics, we previously showed that Mdm1 interacts with Faa1, a FA-CoA ligase that converts FFAs to FA-CoAs (Hariri et al., 2018). Faa1 normally resides in the cytosol but localizes to the LD surface in conditions when LD biogenesis is elevated, such as nutrient scarcity or following the addition of exogenous oleate (Currie et al., 2014). Given that relocalizing Mdm1 to cER–PM contacts also relocalized LD bud sites, we hypothesized that Mdm1ER-PM may be sufficient to recruit Faa1 to these sites. To test this hypothesis, Mdm1ER-PM was coexpressed in yeast stably expressing mCherry-tagged Faa1. Indeed, Faa1-mCherry formed foci at the cell periphery that colocalized with ∼78% of Mdm1ER-PM foci located at the base of LDs (Fig. 4, A, B and D).
Figure 4.
PXA domain of Mdm1 recruits FA-CoA ligase Faa1 to LD budding sites. (A) Left: Light microscopy of GFP-tagged Mdm1ER-PM in yeast stably expressing mCherry-tagged Faa1 in the presence of 0.2% oleate. Scale bar, 2 µm. LDs (gray) were visualized by using MDH. Arrows indicate co-enrichment of Mdm1ER-PM, Faa1, and LDs. Scale bar, 2 µm. (B) Line tracing of light microscopy images in (A). (C) Left: Light microscopy of different fragments of Mdm1ER-PM in yeast stably expressing mCherry-tagged Faa1 in the presence of 0.2% oleate. LDs (gray) were visualized by using MDH. Scale bar, 2 µm. Arrows indicate co-enrichment of Mdm1ER-PM, Faa1, and LDs. Right: Illustrated representation of the topology of different Mdm1ER-PM constructs being expressed. (D) Quantification of Mdm1ER-PM foci associated with Faa1 from images in A and C. Data represent the number of LD-associated foci over the total Mdm1ER-PM foci. Mean ± SD; n > 50 cells; *, P < 0.01; **, P < 0.005; Student’s t test. (E) Diagram depicting the pathway of neutral lipid synthesis. (F) Quantification of BODIPY-C16 incorporation into neutral lipid (NL) BODIPY-DAG in mdm1Δ and faa1Δ relative to WT yeast (1-h incubation). n = 3; **, P < 0.005; Student’s t test. Raw data are shown in Fig. S3G. (G) Quantification of BODIPY-C16 incorporation into neutral lipid (NL) BODIPY-TAG in mdm1Δ and faa1Δ relative to WT yeast (1-h incubation). n = 3; **, P < 0.005; Student’s t test. Raw data are shown in Fig. S3 G (high exposure). (H) Quantification of NBD-C16-CoA incorporation into NBD-TAG in mdm1Δ and faa1Δ relative to WT yeast (1-h incubation). n = 3; ns, nonsignificant; Student’s t test. Raw data are shown in Fig. S3 H.
To determine which regions encoded within Mdm1ER-PM were needed to recruit Faa1 to LDs, first we generated a smaller Mdm1ER-PM chimera (mini-Mdm1ER-PM) encoding only the IMD region of Mdm1 and lacking the PXA domain. Mini-Mdm1ER-PM was able to associate with cortical LD bud sites; however, it failed to recruit Faa1-mCherry to LDs (Fig. 4, C and D). Next, to investigate whether the PXA domain was sufficient for Faa1 recruitment, we generated another Mdm1 variant containing the PXA domain but replaced the Mdm1 IMD with the Tcb2 ER anchor (Mdm1Tcb2 ER-PM). As expected, this chimera distributed across the ER network and failed to associate with LD bud sites or recruit Faa1-mCherry (Fig. 4, C and D). Taken together, these data indicate that the PXA domain is necessary but not sufficient to recruit Faa1 to LD buds (Fig. 4, C and D). The IMD region is required to localize Mdm1 PXA domain to the LD bud site, which is sufficient to target Faa1 to these sites.
Faa1 is one of several yeast FA-CoA ligases that activate exogenous FFAs, such as oleate, converting them into FA-CoAs before their incorporation into downstream neutral lipids such as diacylglycerol (DAG) and TAG (Fig. 4 E). Since Mdm1 binds to FFAs in vitro, and Mdm1 and Faa1 coenrich at LD bud sites, we hypothesized that Mdm1 may influence Faa1’s ability to locally activate FFAs during oleate-induced LD biogenesis. To test this model, we performed in vitro FFA activation assays to quantitatively monitor FFA activation and incorporation into neutral lipids (McFie and Stone, 2011). Specifically, we monitored the incorporation of either fluorescently labeled FFA (BODIPY-C16) or FA-CoA (NBD-C16-CoA) into neutral lipids using TLC. After 1 h of incubation, we detected a similar reduction in the incorporation of BODIPY-C16 into DAG and TAG in both mdm1Δ and faa1Δ yeast lysates (Fig. 4, F and G; and Fig S3 G). In contrast, we observed no significant reduction in the incorporation of NBD-C16-CoA into TAG in these samples after 30 min or 1 h (Fig. 4 H and Fig. S3, H and I). This suggests that both Mdm1- and Faa1-deficient yeast lysates have similar defects in the activation and subsequent incorporation of FFAs into neutral lipids.
Loss of MDM1 perturbs ER morphology and sensitizes yeast to lipotoxicity
Having established that Mdm1 interacts with FFAs and colocalizes with Faa1 at LD bud sites, we next focused on dissecting the biological significance of this interaction. LD biogenesis has been suggested to promote ER homeostasis, as the aberrant accumulation of FAs can induce ER stress and lipotoxic cell death (Chitraju et al., 2017). Moreover, the addition of lipotoxic FAs induces LD biogenesis to sequester toxic lipids (Listenberger et al., 2003; Petschnigg et al., 2009). We previously reported that loss of MDM1 caused an increase in total cellular neutral lipids (Hariri et al., 2018). Consistent with this, yeast lacking Mdm1 accumulate approximately twofold more LDs that cluster on the surface of the nuclear envelope adjacent to the NVJ (Fig. S4, A and B). To better understand the cellular consequences of Mdm1 loss, we investigated the effect of MDM1 deletion on ER structure and homeostasis in the presence of exogenous FAs.
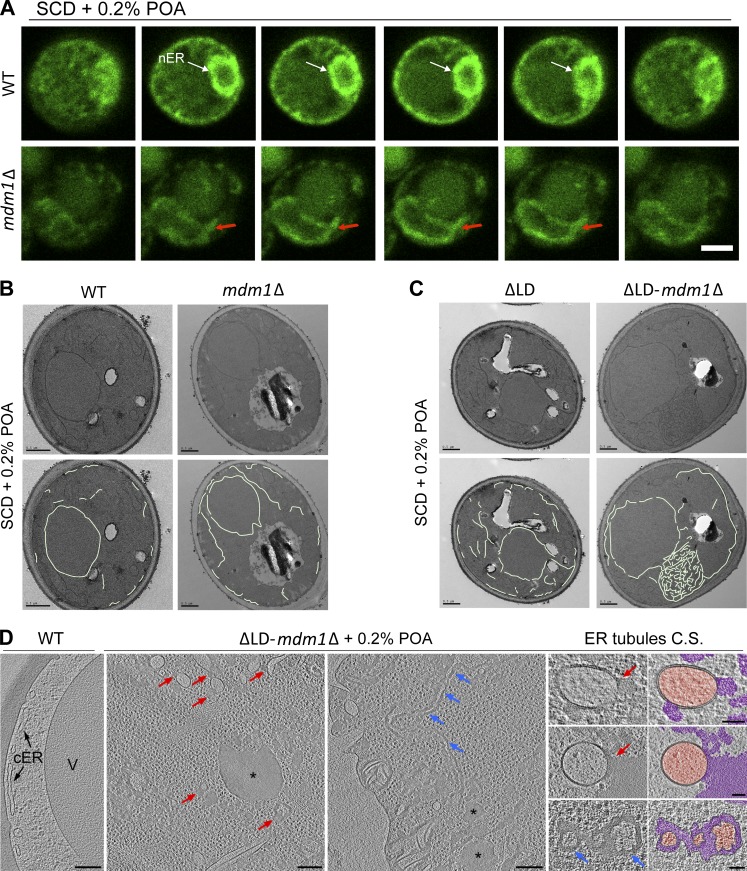
First, we examined the ER network of WT and mdm1Δ yeast expressing the ER marker GFP-HDEL using confocal microscopy. In contrast to WT yeast that exhibited regular ER and nuclear morphology, MDM1-deficient yeast displayed deformed and expanded nuclear envelopes when cultured in the presence of the exogenous FA palmitoleate, a free FA associated with lipotoxicity in yeast (POA; Fig. 5 A; Garbarino et al., 2009). Similarly, the cER showed numerous tubular extensions and puncta (Fig. S4 D). Using conventional plastic-section TEM, we confirmed that the ER network in mdm1Δ yeast contained atypical extensions (Fig. 5 B).
Figure 5.
Loss of MDM1 perturbs ER morphology and sensitizes yeast to lipotoxicty. (A) Slices from confocal microscopy of WT (top) and mdm1Δ (bottom) yeast fed 0.2% POA. ER (green) is marked by endogenous GFP-HDEL tagging. Arrows indicate deformed nuclear ER (nER) morphology in mdm1Δ yeast compared with WT. Scale bar, 2 µm. (B) Top: Thin-sectioning TEM of WT and mdm1Δ yeast fed 0.2% POA. Bottom: Manual tracing of the ER membrane. Scale bar 0.5 µm. (C) Top: Thin-sectioning TEM of LD-null (ΔLD) and ΔLD-mdm1Δ yeast fed 0.2% POA. Bottom: Manual tracing of the ER membrane. Scale bar, 0.5 µm. (D) Slices from cryotomographic reconstructions of WT and ΔLD-mdm1Δ yeast cryo-FIB sections (cultured in 0.2% POA for 6 h). Red arrows point at local bulges of FA deposits (asterisks) and blue arrows at large-range FA deposits between the membrane bilayer. Cross sections (C.S.) of ER tubules found in ΔLD-mdm1Δ yeast are shown both as raw tomographic slices and pseudocolored (orange, ER lumen; magenta, abnormal FA deposits). Scale bars, 200 nm (overviews) and 50 nm (C.S. zoom-ins in the right column).
We hypothesized that loss of MDM1 altered some aspect of ER homeostasis and that mdm1Δ yeast may elevate their LD stores to respond to this stress. As such, mdm1Δ yeast may be reliant on LD biogenesis to maintain ER homeostasis in the presence of exogenous FAs. To test this hypothesis, we generated a yeast strain that lacked MDM1 and in which we could controllably attenuate LD production (Fig. S4 C). This was achieved by ablating both sterol ester synthases (Are1 and Are2) and TAG synthase Lro1. The remaining TAG synthase Dga1 was placed under a galactose-inducible promoter, allowing the inhibition of LD biogenesis when the strain is cultured in glucose, and the production of TAG-containing LDs to be induced when cultured in galactose (Oelkers et al., 2002; Cartwright et al., 2015). For simplicity, we denote this strain as “ΔLD” when cultured in glucose. Strikingly, when grown in glucose the combined loss of MDM1 and LDs caused significant ER morphology defects and the formation of ER tangles in the cytoplasm as seen by using TEM (Fig. 5 C and Fig. S5 B). Remarkably, similar ER tangles have been reported in cells with elevated ER stress, as well as in yeast with altered ER lipid balance (Vevea et al., 2015). ER morphology defects could also be observed in ΔLD-mdm1Δ cells by confocal imaging as the appearance of bright ER foci in the cytoplasm (Fig. S4 E). Consistent with their perturbed ER morphology, both mdm1Δ and ΔLD-mdm1Δ strains grew poorly on solid media plates containing 0.2% POA, indicating a sensitivity to exogenous FAs (Fig. S4, F and G). A similar sensitivity was observed for ΔLDfaa1Δ yeast, indicating that, in the absence of proper LD biogenesis, defects in FA activation may cause sensitivity to excess FAs (Fig. S4 G). Consistent with this, deletion of FAA1 in the ΔLD yeast caused an ER morphology defect similar to the one caused by the absence of MDM1 (Fig. S5 C).
To examine the ER morphological defects in more detail in cells preserved in their near-to-native state, we rapidly froze WT and ΔLD-mdm1Δ yeast treated with 0.2% POA. We then used cryo-focused ion beam (cryo-FIB) milling to generate ∼200-nm-thick sections from the frozen cells, which were then imaged and 3D reconstructed by using cryo-electron tomography (cryo-ET). Strikingly, whereas the ER bilayer of WT yeast cells was clearly intact and regular (Fig. 5 D, left), the ΔLD-mdm1Δ cells and, in particular, their ER network displayed drastic structural defects. In the mutant, the ER membrane bilayer appeared to be irregular with the two membrane leaflets in some regions not clearly visible or separating from each other. In multiple locations, the phospholipid monolayer on the cytoplasmic side of the membrane bulged from the ER surface, surrounding an amorphous shaped, relatively electron-dense FA deposit (red arrows in Fig. 5 D and Video 1). Occasionally, the FA deposits were more widely spread, resulting in a large-range separation of the two ER leaflets (blue arrows in Fig. 5 D and Video 1), rather than formation of local bulges. Whereas regular spherically shaped LDs are not formed in the mutant, multiple irregular FA deposits of variable size were observed in the cytoplasm, as well as an unusually high amount of ribosomes and (protein and/or DNA) aggregates (Fig. 5 D and Video 1).
Consistent with this, TLC lipid analysis of ΔLD-mdm1Δ yeast cultured in 0.2% POA revealed that they contained elevated FFAs and reduced DAG compared with ΔLD yeast, consistent with a defect in FFA activation and subsequent incorporation into neutral lipids (Fig. S5 A). These observations collectively suggest that ΔLD-mdm1Δ yeast are unable to efficiently activate and process POA, which ultimately accumulates in the ER network and perturbs ER membrane integrity.
Loss of MDM1 perturbs LD biogenesis
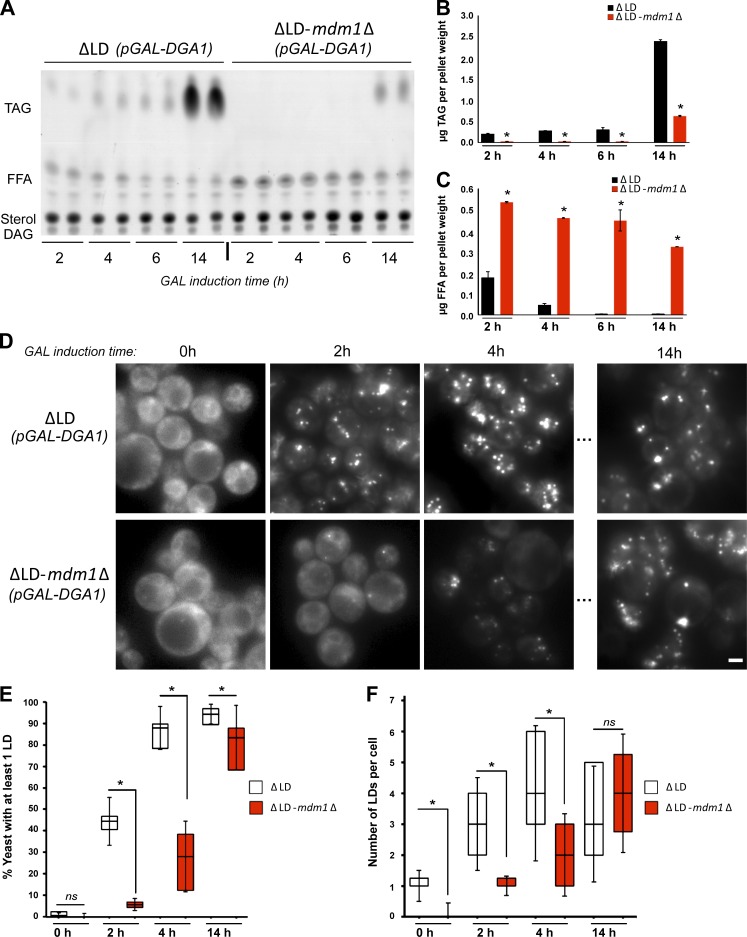
Since MDM1-deficient yeast exhibited FA-induced lipotoxicity in the presence of POA, we next investigated whether mdm1Δ yeast also displayed defects in FA incorporation into TAG during LD biogenesis. ΔLD yeast exhibit FA accumulation at steady-state due to the block in neutral lipid synthesis. However, FA accumulation was exacerbated in the ΔLD-mdm1Δ strain, suggesting a defect in FA homeostasis when MDM1 is also deleted (Fig. S5 D). To determine whether the increase in FFA was due to a defect in FA activation and incorporation into TAG, we conducted a kinetic assay that biochemically monitored FA conversion into TAG in living yeast. We cultured ΔLD and ΔLD-mdm1Δ strains in glucose, then switched them to galactose-containing media to induce pGAL-DGA1 expression and monitored Dga1-mediated production of TAG at different time points.
As expected, ΔLD yeast displayed a gradual decrease in FFAs as TAG levels increased when incubated in galactose-containing media (Fig. 6, A and B). In contrast, ΔLD-mdm1Δ yeast displayed elevated FFA levels and significantly delayed TAG synthesis, suggesting a defect in FA incorporation into TAG (Fig. 6, A and C). To determine whether the delayed TAG synthesis observed in ΔLD-mdm1Δ yeast also resulted in delayed LD biogenesis, we visualized LD production by staining LDs at set time internals following pGAL-DGA1 induction. As expected, ΔLD yeast produced many LDs within 2 h following addition of galactose whereas ΔLD-mdm1Δ yeast exhibited a significant delay in LD production (Fig. 6, D and E). After 14 h in galactose, the ΔLD-mdm1Δ yeast contained comparable numbers of LDs per yeast cell as ΔLD, indicating LD production was not blocked but delayed (Fig. 6 F).
Figure 6.
Loss of MDM1 perturbs LD biogenesis. (A) TLC of neutral lipids extracted at different time points after galactose (GAL) induction of Dga1 expression from ΔLD and ΔLD-mdm1Δ yeast. (B) Quantification of TAG levels from TLC in A (in micrograms). Data represent mean ± SD normalized to cell pellet weight; n = 2. *, P < 0.01; **, P < 0.005; two-way ANOVA. (C) Quantification of FFA levels from TLC in A (in micrograms). Data represent mean ± SD normalized to cell pellet weight; n = 2. *, P < 0.01; **, P < 0.005; two-way ANOVA. (D) Light microscopy ΔLD and ΔLD-mdm1Δ yeast at different time points after GAL induction of Dga1 expression. LDs (gray) visualized by MDH staining. Scale bar, 2 µm. (E) Quantification of the percentage of yeast cells with LDs at different time points after GAL induction of Dga1 expression. Data represent the number of cells over the total; mean ± SD; n > 50 cells; *, P < 0.01; Student’s t test. (F) Quantification of the number of LDs in ΔLD and ΔLD-mdm1Δ yeast at different time points after GAL induction of Dga1 expression. Data represent the number of LDs per cell over the total number of cells; mean ± SD; n > 50 cells; *, P < 0.01; Student’s t test.
Since Mdm1 and Faa1 colocalized at LD bud sites, we hypothesized that loss of Faa1 would similarly impact the ability to activate FFAs and incorporate them into TAG. Indeed, deletion of FAA1 in ΔLD yeast produced a similar delay in FA incorporation into TAG following overnight culturing in galactose (Fig. S5 E). Collectively, we conclude that Mdm1 and Faa1 promote FFA activation at LD bud sites.
Discussion
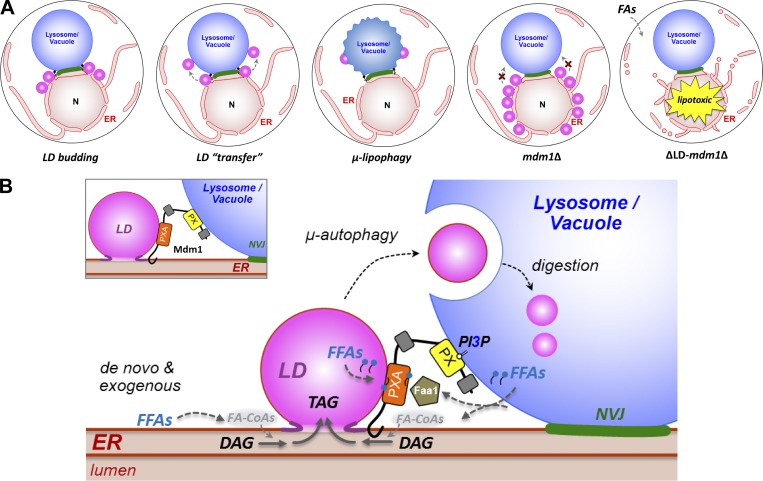
Recent studies reveal that yeast spatially clusters LDs adjacent to the vacuole during stress (Barbosa et al., 2015; Hariri et al., 2018). Specific proteins that localize with NVJ-associated LDs have been identified, but how LD clustering at the NVJ occurs and the physiological purpose of this process remain unclear (Eisenberg-Bord et al., 2017; Teixeira et al., 2017). One model posits that LD clustering near the NVJ facilitates their delivery into the vacuole lumen for lipophagic degradation, but the physiological relevance of this process remains largely uninvestigated (Wang et al., 2014; Barbosa and Siniossoglou, 2016; Seo et al., 2017). Here, we propose that Mdm1 regulates LD formation at the NVJ to promote ER homeostasis. By spatially regulating LD biogenesis adjacent to the vacuole, Mdm1 can promote the incorporation of lipids into LDs that can be efficiently delivered into the vacuole via μ-lipophagy (Fig. 7 A). Since LDs can sequester otherwise toxic lipids such as FAs, Mdm1-mediated LD biogenesis at the NVJ may represent an ER quality-control system to maintain lipid homeostasis and prevent FA-induced lipotoxicity by removing toxic lipids from the ER and delivering them to the vacuole (Fig. 7 A).
Figure 7.
Model for Mdm1-mediated LD formation and clustering. (A) Cartoon depicting NVJ-associated LD delivery to the vacuole via µ-lipopohagy. LDs initially form on the ER and accumulate near the NVJ, eventually translocating to the vacuole surface for lipophagy. In mdm1Δ yeast, LDs accumulate on the nuclear envelope. When exposed to exogenous FAs, ΔLD-mdm1Δ yeast exhibit ER morphological defects and lipotoxicity. (B) Illustration depicting a model for Mdm1’s role in localized FA processing and LD formation at the NVJ through interaction with Faa1.
How does Mdm1 associate with LDs? There appear to be at least two mechanisms. First, we find that Mdm1’s ER-anchored IMD is necessary and sufficient to associate with LDs. Consistent with this, we observe Mdm1 as foci or cups that partially surround LDs at ER–LD interfaces and colocalize with Seipin. Furthermore, we observe increased ER wrapping of cortical LDs in yeast expressing Mdm1ER-PM, consistent with the protein’s accumulation at ER–LD contact sites. Second, we identify a putative hydrophobic helical region in the PXA domain that is sufficient to target to LDs. This suggests that the PXA domain itself may associate with the LD monolayer surface (Fig. 7 B).
What is the physiological relevance of Mdm1-LD associations? We find that Mdm1 can spatially define sites of LD budding, as an ER–PM relocalized a minimal Mdm1 fragment containing only the IMD and PXA domains can ectopically redistribute LDs to the cell periphery. We also show that the PXA domain of Mdm1 binds FFAs in vitro and associates with the FA-CoA ligase Faa1 at LD bud sites. The precise nature of this Mdm1–Faa1 interaction is unclear, but we propose that the PXA domain mediates local enrichment of FA pools, which, in turn, promote the recruitment of Faa1. In the absence of MDM1, LD biogenesis is defective, which perturbs ER homeostasis, as MDM1-deficient yeast are sensitive to lipotoxic FAs. Altogether, we propose that Mdm1 regulates LD budding at three-way junctions between the ER, LDs, and vacuole to promote the spatially organized activation and incorporation of FFAs into LDs, which can subsequently be delivered to the vacuole (Fig. 7 B).
FAs are required for multiple steps in neutral lipid synthesis (Fig. 4 E). This suggests that a local pool of FA-CoAs may be required near LDs to promote efficient LD biogenesis. Indeed, the Caenorhabditis elegans FA-CoA ligase FATP1 binds to DGAT2 at the ER–LD interface and supplies local FA-CoAs for TAG synthesis (Xu et al., 2012). Similarly, in mammals ACSL5 interacts with DGAT2 and a ceramide synthase to promote ceramide storage in LDs (Senkal et al., 2017). These studies suggest a close spatial and functional coupling between FA activation and LD formation. To our knowledge, there is no established mechanism for localized FA processing at LDs in budding yeast. However, Dga1 enriches on the LD surface during active LD production, implying a need for localized FA activation during LD biogenesis (Jacquier et al., 2011). An intriguing model is that Mdm1 functions as an adaptor for spatially regulating Faa1 activity adjacent to LDs during times of elevated FA flux (Fig. 7 B).
In addition to its role in lipophagy, Mdm1-mediated LD clustering may play other roles in cellular homeostasis. During starvation, FAs generated within the vacuole from LD and membrane breakdown may use ER–vacuole contacts for efflux. How FAs exit the vacuole and how cells use FAs generated within the vacuole are currently unknown. We posit that Mdm1-mediated LD formation at the NVJ may provide a convenient escape route for vacuole-derived FAs that are packaged into LDs. Additionally, close LD–vacuole contact may provide a mechanism for lipid exchange between these organelles, which may promote vacuole surface remodeling that regulates signaling during starvation (Murley et al., 2017). Here, we show that LD formation at the ER–vacuole interface may protect the ER from stress due to the accumulation of lipid synthesis precursors such as FAs. Consistent with this, several studies reported that LDs are produced to mitigate ER stress by removing excess or toxic lipids from the ER (Ta et al., 2012; Vevea et al., 2015). By doing so, LDs provide a mechanism to maintain lipid balance and preserve organelle identity and cellular function. Mdm1 is highly conserved in humans as Snx14, and loss of this homologue contributes to progressive neuronal cell death observed in pediatric cerebellar ataxia disease SCAR20 (Akizu et al., 2015; Bryant et al., 2018). Although the mechanism for this disease remains unclear, an intriguing model is that Snx14 loss may cause defects in lipid homeostasis, leading to eventual neuronal death. New work reveals that Snx14 drives FA-stimulated LD growth at ER–LD contacts, indicating a conservation of function between Mdm1 and its human homologue (Datta et al., 2019). Further studies are needed to identify the cellular functions of Mdm1 homologues in human health and disease.
Materials and methods
Molecular biology, yeast genetics, and growth conditions
All yeast strains used in this study are detailed in Table S1. Yeast genetic manipulations were conducted using classical yeast knock-in/out protocols. For ectopic protein expression, genes were cloned into pBP73-C or -G vectors encoding either the GPD promoter or CPY promoter. All plasmids used in this study are detailed in Table S2. Yeast transformations were performed using the lithium acetate method. We used Ds-Red HDEL to label the ER. Strains were selected using antibiotics or dropout selection media. All chemicals used to make the yeast media were purchased from Sigma-Aldrich (succinic acid, sodium hydroxide, ammonium sulfate, yeast nitrogen base without amino acids, or ammonium sulfate). Yeast media was supplemented with a final concentration of 2% dextrose unless otherwise indicated. Oleate (Sigma-Aldrich; O1008) and POA (Sigma-Aldrich; P9417) were added to the culture media as indicated (0.2% oleate is equivalent to 6.32 mM, and 0.2% POA is equivalent to 7.04 mM).
LDs and vacuole staining
For LD staining, the fluorescent dye monodansylpentane (MDH/AutoDOT; Abgent) was added for 10 min. For vacuole staining, we incubated with CellTracker blue CMAC dye (Thermo Fisher Scientific) for 30 min to stain the vacuole lumen, and FM4-64 was added for 45 min to 1 h (or overnight) to stain the vacuole surface. Dyes were added to cell pellets that were washed and resuspended in a small volume of media with no added carbon source. Yeast cells were then pelleted, washed, and imaged.
Light microscopy
Imaging of live yeast cultures was performed by using EVOS FL Cell Imaging System (Thermo Fisher Scientific) at room temperature. Yeast cells were grown to the desired OD600 in the different growth condition. Before imaging, cells were pelleted (3,000 g for 5 min at room temperature), washed, and resuspended in a small volume of media without a carbon source. Then, 3 µl of the dense yeast suspension was transferred to a glass slide for imaging. Image analysis was performed using Fiji (ImageJ).
Confocal imaging
Imaging of live yeast cultures was performed by using a Zeiss LSM880 inverted laser scanning confocal microscope at room temperature. Yeast cells with HDEL-labeled ER were grown in SCD media overnight and the desired amount of POA (Sigma-Aldrich) was added for 4 h. Before imaging, cells were pelleted (3,000 g for 5 min at room temperature), washed, and resuspended in a small volume of media without a carbon source. Then, 3 µl of the dense yeast suspension was transferred to a glass slide for imaging. Optical sections (6×) of ∼0.5 µm thickness were obtained per condition. Image analysis was performed by using Fiji (ImageJ).
Neutral lipid analysis
Yeast cells were grown in the desired conditions and pelleted. Neutral lipids were extracted according to a modified protocol from Bligh and Dyer (1959). Neutral lipids were extracted from whole cells using chloroform/methanol method modified from the Bligh and Dyer method. After measuring the wet cell weight, pellets were lysed using glass bead–beating for 10 min in the presence of chloroform. All steps were performed in the cold room. Methanol was added, and the suspension was vortexed vigorously. Then, 500 mM NaCl prepared in 0.5% acetic acid was added to get the final concentration of chloroform:methanol:water to 2:2:1.8. Samples were spun at 4,000 rpm for 15 min in the cold room, and the bottom chloroform was recovered. The volume was recorded, and lipids were dried with Argon gas. Dried lipids were resuspended in chloroform to a final concentration normalized to the initial cell pellet weight (0.5 µl chloroform per 1 mg cell weight). One-dimensional TLC was used to separate the extracted lipids using hexane:diethyl ether:acetic acid (80:20:1) as a solvent to separate neutral lipids. TLC plates were spray-stained with 3% copper (II) acetate prepared in 8% phosphoric acid. Stained plates were incubated in the oven at 145°C for 1 h to overnight to develop the bands. Plates were resprayed and reheated as needed to visualize lipids. Where indicated, yeast cultures were diluted to OD600 of 0.2 and grown for 3 h in complete media supplemented with 2% glucose. After 3 h, 0.1% (∼3 mM) oleic acid was added overnight. After ∼14 h of treatment, cells were collected and prepared for lipid extraction.
TLC quantification
Stained TLC plates were scanned and then processed for quantification using Fiji (ImageJ). On each plate, we ran a serial dilution of standard neutral lipid mix of known concentration. The standard mixture was prepared in chloroform to a final concentration of 10 mg/ml. The standard mixture (1:10) was used to create a standard curve in which the x axis displayed the calculated lipid mass in micrograms, and the y-axis displayed the band intensity estimated by using Fiji (ImageJ). Statistical analysis was performed, and P values were calculated in GraphPad by using one-way ANOVA (NS > 0.05; *, P < 0.05; **, P < 0.005; ***, P < 0.0005; ****, P < 0.0001).
Yeast plating assays
Yeast precultures were grown overnight in SCD. The following day, cultures were diluted to OD600 0.5, and different concentrations of POA (Sigma-Aldrich; P9417) were added for 2 d. Prior to plating, cells were diluted to an OD600 of 0.3, and 5 µl was spotted on YPD agar plates. Cells were imaged at 2 d growth unless otherwise indicated.
Preparing liposomes by using extrusion
Lipids were purchased from Avanti Polar Lipids, Inc. A lipid mixture of 75 mol% DOPC and 25 mol% DOPS was prepared in chloroform and dried using argon gas, and rehydrating the lipid film in buffer (20 mM Hepes, pH 7.8, and 150 mM NaCl) to a final concentration of 2 mg/ml. An Avanti Mini-Extruder was used as suggested by the manufacturer. Liposomes were extruded (21×) through a Whatman membrane with 0.4-µm-pore size.
Recombinant protein expression and purification
Purification of recombinant PXA was done by using two different methods. (1) The PXA cloned from Saccharomyces cerevisiae was purified by nondenaturing solubilization of inclusion bodies (IBs) as described previously (Golovanov et al., 2004; Tsumoto et al., 2010). IBs were extracted from E. coli as described by Palmer and Wingfield (2004). Briefly, bacterial pellets were lysed by sonication on ice in lysis buffer (20 mM Hepes, pH 7.8, 150 mM NaCl, 1 mM BME, 0.5 mM PMSF, and protease inhibitor cocktail tablet) followed by centrifugation for 30 min at 8,000 g. Pellets were washed (2×) with 2% sodium deoxycholate in 1× TBS, then washed (2×) with distilled water. IB pellets can be stored at this point at −80°C until further need. For protein extraction, IB pellets were suspended in 0.5 M arginine solution (45 ml per 1-liter pellet) followed by centrifugation at 20,000 rpm. The supernatant was collected. The arginine buffer was dialyzed into the buffer of choice (20 mM Hepes, pH 7.8, 150 mM NaCl or 20 mM sodium phosphate 7.8, 50 mM NaCl plus, 1 mM BME, and 0.5 mM PMSF). The resultant protein was further purified by using liquid chromatography. The protein sample was first loaded on 5 ml Ni-Sepharose column (GE HisTrap; 20 mM Hepes, pH 7.8, 150 mM NaCl, and 40 mM imidazole). After washing, imidazole gradient buffer (20 mM Hepes, pH 7.8, 150 mM NaCl, and 500 mM imidazole) was used to elute the bound proteins. Fractions containing the PXA protein were pooled, concentrated, and further purified using Superdex 200 size exclusion chromatography. The resulting protein fold was validated using circular dichroism spectroscopy. (2) Purification of Oryzias latipes PXA was purified from the soluble fraction of E. coli lysates. Bacterial pellets were first resuspended in High Salt Lysis Buffer (20 mM Tris Base, 500 mM NaCl, protease inhibitors, and 10 mM BME or 1 mM DTT). DNase and 0.5 mg/ml lysozyme were added when the pellet was mostly resuspended followed by sonication on ice. Cell debris was then pelleted by centrifugation at 16,000 rpm for 20 min at 4°C. The cleared lysate was loaded into the 50-ml superloop and purified on Ni-Sepharose column (GE HisTrap 1 ml). The column was washed using High Salt Wash Buffer (20 mM Tris Base, 1 M NaCl, 20 mM imidazole, 10% vol/vol glycerol, and 2 mM octyl-β-glucoside, pH 7.5, with HCl). Bound proteins were eluted using Elution Buffer (20 mM Tris Base, 200 mM NaCl, 250 mM imidazole, 10% vol/vol glycerol, and 2 mM octyl-β-glucoside, pH 7.5, with HCl). Fractions containing the PXA domain were pooled and analyzed using size exclusion chromatography (GE Superdex200 Increase 10/300 GL). Fractions were analyzed by using SDS-PAGE. Fractions containing the PXA domain were pooled and concentrated, and proteins were frozen in liquid nitrogen and stored at −80°C if not immediately used.
Lipid loading
PXA loading with fluorescent lipids was performed as described previously by Schauder et al. (2014). In brief, purified S. cerevisiae PXA (19 µl at ∼3 mg/ml) was mixed with BODIPY-C16 (1 mg/ml in methanol). The mixture was incubated on ice for at least 1 h. PXA was separated from unbound BODIPY-C16 by using Ni-NTA spin columns (Qiagen). BODIPY 493/503 (4,4-Difluoro-1,3,5,7,8-Pentamethyl-4-Bora-3a,4a-Diaza-s-Indacene; catalog #D3922) and BODIPY FL C16 (4,4-Difluoro-5,7-Dimethyl-4-Bora-3a,4a-Diaza-s-Indacene-3-Hexadecanoic Acid; catalog #D3821) were purchased from Thermo Fisher Scientific.
Lipid extraction from recombinant protein
Lipids were extracted from recombinant PXA cloned from S. cerevisiae and O. latipes. The method for lipid extraction was described previously using the method of Bligh and Dyer (1959). Briefly, lipids were extracted as described by AhYoung et al. (2015) from 50 µl purified PXA protein (10 mg/ml) by the sequential addition of 3.75 volumes of chloroform:methanol (1:2 vol/vol), 1.25 volumes of chloroform, and 1.25 volumes of 0.5% acetic acid in 500 mM NaCl. Every step was followed by 1 min of vortexing. The samples were centrifuged at 14,000 g for 15 min. The bottom layer was gently recovered and dried. The resulting lipids were dissolved in 30 µl chloroform and resolved as mentioned in the section Neutral lipid analysis.
Flotation assay
This assay was originally developed by Bigay and Antonny (2005). Purified S. cerevisiae PXA protein was preloaded with BODIPY-C16 and purified as described above. Loaded PXA (25 µl at ∼1 mg/ml) was incubated with 55 µl liposomes (PC:PS; 75:25 mol%) and 75 µl 1× PBS. The mixture was placed in the bottom of a sucrose step gradient. Solutions containing 30% (100 µl), 25% (200 µl), or 0% (50 µl) sucrose were prepared in the buffer and layered carefully in centrifugation tubes. The assembled gradients were subject to ultracentrifugation for 90 min at 45,000 rpm in a TLS-55 rotor. Fractions were collected from the bottom to the top and analyzed by using native gel electrophoresis. Fluorescence was observed using Bio-Rad Laboratories ChemiDoc Touch imaging system, and protein was assessed using Coomassie staining.
FA activation and incorporation assay
This assay was modified from McFie and Stone (2011). Yeast lysate was extracted by vortexing cells in the presence of glass beads and lysis buffer (200 mM Tris-HCl, pH 8.4, 4 mM EDTA, 5 mM BME, 10% glycerol, 0.5 mM PMSF, 0.01% NP-40, and 1 PIC tablet per 50 ml buffer) for 10–12 min in the cold room. Cell debris was removed by spinning at 1,500 g for 5 min at 4°C, and the supernatant was collected and used for subsequent incubations. Yeast lysates (100 µg in 50 µl buffer) were mixed with 50 µM of BODIPY-C16 (Invitrogen; D3821) or NBD-C16-CoA (Avanti; 810705) in the presence of 20 µl 1 M Tris-HCl, pH 7.6, 4 µl 1 M MgCl2, 10 µl 4 mM 1–2-dioleoyl-sn-glycerol DOG (Sigma-Aldrich; D0138), 10 µl 12.5 mg/ml BSA, and 96 µl water per reaction and incubated at 30°C. Fluorescent neutral lipids from 150 µl reaction mixture were then extracted by adding 4 ml CHCl3/methanol (2:1, vol/vol) and 800 µl water then mixed by vortexing. Centrifuge at 3,000 rpm for 5 min separated aqueous and organic phases (lipids). Extracted lipids were separated on a TLC plate by using 1:2 (vol/vol) cyclohexane:ethyl acetate as a solvent. Fluorescent lipids were imaged by using Bio-Rad Laboratories ChemiDoc Touch imaging system and quantified by using imageJ. TopFluor DG (Avanti; 810294) and TopFluor TG (Avanti; 810298) were used as standards to validate fluorescent lipid bands.
Conventional TEM
Yeast cells were grown in the desired conditions and processed in the University of Texas Southwestern Electron Microscopy Core Facility using a protocol adapted from Wright (2000). In brief, cells were fixed in potassium permanganate, dehydrated, and stained in uranyl acetate and embedded in Spurr Resin. Specimen blocks were polymerized at 60°C overnight and sectioned at 70 nm with a diamond knife (Diatome) on a Leica Ultracut UCT 6 ultramicrotome (Leica Microsystems). Sections were poststained with 2% uranyl acetate in water and lead citrate. Sections were placed on copper grids (Thermo Fisher Scientific). Images were acquired on a Tecnai G2 spirit TEM (FEI) equipped with a LaB6 source at 120 kV by using a Gatan Ultrascan charge-coupled device camera.
Cryo-sample preparation for cryo-EM
Yeast cells were grown as described above and added to a glow-discharged (30 s at −35 mA) holey carbon copper grid (R2/2; Quantifoil Micro Tools GmbH). After blotting excess medium from the grid backside for ∼3.0 s with Whatman filter paper, the grid was plunge-frozen in liquid ethane by using a homemade plunge-freezer. Grids were mounted in notched cryo-FIB Autogrids (Thermo Fisher Scientific) and stored in liquid nitrogen until used.
Cryo-FIB milling
Autogrids with vitrified cells were loaded into a shuttle under cryogenic conditions and transferred into a Scios dual-beam instrument (FIB/SEM; Thermo Fisher Scientific) equipped with a cryo-stage that was precooled to −185°C. The sample surface was coated with layers of platinum (sputter-coater: 1 keV and 30 mA for 10 s, and gas injection system: 27°C preheating temperature for 10 s and condensed with the gallium-ion beam for 5 min at 30 kV and 0.5 nA) for sample protection during milling and increased conductivity (Schaffer et al., 2017). Suitable cells for cryo-FIB–milling were identified on an overview image of the grid generated by using the Maps software in SEM mode. By using the cryo-stage tilt, the target region of interest was properly oriented for milling with a shallow angle of 8–14° between gallium-ion beam and EM grid. To generate a “cell section” (self-supporting lamella) that is thin enough for cryo-ET, the sample was milled with a 30 keV gallium ion beam by using a beam current of 0.1 nA for initial bulk milling, 50–30 pA for the thinning steps, and 10 pA for final polishing. The milling progress was observed by using SEM imaging at 5 keV and 25 pA. Frozen-hydrated material was ablated until the thickness of the lamellae was 150–200 nm.
Cryo-ET and image processing
The grids with lamellae were imaged by using a Titan Krios transmission electron microscope (Thermo Fisher Scientific) at 300 keV and under low-dose conditions. Cryo-ET was performed as previously described (Fu et al., 2018) using the microscope control software SerialEM (Mastronarde, 2005). Briefly, tilt series were recorded using a dose-symmetric tilting scheme (Hagen et al., 2017) and a tilt range from −60° to +60° with an increment of 2°. Images were recorded on a K2 Summit direct electron detector (Gatan) in counting mode (15 frames, 0.4-s exposure time per frame, and a dose rate of 8 electrons/pixel/s for each tilt image) at 26,000 magnification with an effective pixel size of 5.5 Å. The total electron dose per tilt series was limited to ∼100 e/Å2. The defocus was set to −0.5 µm while using a Volta-Phase-Plate (Danev et al., 2014), and the postcolumn energy filter (Gatan) was operated in zero-loss mode (20-eV slit width). Using the IMOD software package (Kremer et al., 1996), we first motion-corrected and averaged the K2 frames for each tilt image, then the tilt series images were aligned fiducial-less by using patch tracking (800 × 800-pixel size), before calculating the 3D reconstruction by weighted back-projection. To reduce noise, the cryo-tomograms were slightly filtered by using a weighted median filter, and in selected figure panels (Fig. 5 D, right) the gray values were false-colored by using Photoshop.
Online supplemental material
Fig. S1 shows that Mdm1 binds lipid dropets via its N-terminal region. Fig. S2 shows that Mdm1 colocalizes with LDs at ER–LD interfaces. Fig. S3 shows that the Mdm1 PXA domain is hydrophobic and binds FAs, and loss of Mdm1 perturbs FA activation. Fig. S4 shows that loss of Mdm1 sensitizes yeast to lipotoxic stress. Fig. S5 shows that loss of Mdm1 perturbs TAG synthesis. Table S1 lists yeast strains used in this study. Table S2 lists yeast expression plasmids used in this study. Video 1 shows tomographic reconstruction of ΔLD-mdm1Δ cells exposed to lipotoxic stress.
Supplementary Material
Acknowledgments
We thank Sandra Schmid and Joel Goodman for helpful discussions and critical reading of the manuscript. We thank Jonathan Friedman for helpful discussions. We thank Maya Schuldiner for sharing the yeast GFP collection. We thank Kate Luby-Phelps and the University of Texas Southwestern Electron Microscopy Core Facility for expert technical assistance. We thank Daniel Stoddard for training and management of the University of Texas Southwestern Medical Center Cryo-Electron Microscopy Facility that is supported in part by the Cancer Prevention and Research Institute of Texas Core Facility Support Award RP170644.
D. Nicastro is supported by Cancer Prevention and Research Institute of Texas grant RP140082. W.M. Henne is supported by grants from the Welch Foundation (I-1873), the Searle Foundation (SSP-2016-1482), the National Institutes of Health National Institute of General Medical Sciences (GM119768), American Federation for Aging Research (A15198), and the University of Texas Southwestern Endowed Scholars Program.
The authors declare no competing financial interests.
Author contributions: H. Hariri and W.M. Henne, conception and design of the study, data acquisition, analysis and interpretation, drafting and revising the article, and preparing the figures. H. Hariri, conventional electron microscopy and biochemical assays. H. Hariri and W.M. Henne, multichannel imaging and light microscopy and quantitative image analysis. H. Hariri and N. Speer, thin-layer chromatography. H. Hariri and S. Datta, confocal microscopy. J. Bowerman and S. Roger, generating yeast strains. G. Fu, E. Reetz, and D. Nicastro, cryo-sample preparation, data acquisition, and analysis. J.R. Feathers and R. Ugrankar, technical assistance.
References
- AhYoung A.P., Jiang J., Zhang J., Khoi Dang X., Loo J.A., Zhou Z.H., and Egea P.F.. 2015. Conserved SMP domains of the ERMES complex bind phospholipids and mediate tether assembly. Proc. Natl. Acad. Sci. USA. 112:E3179–E3188. 10.1073/pnas.1422363112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akizu N., Cantagrel V., Zaki M.S., Al-Gazali L., Wang X., Rosti R.O., Dikoglu E., Gelot A.B., Rosti B., Vaux K.K., et al. . 2015. Biallelic mutations in SNX14 cause a syndromic form of cerebellar atrophy and lysosome-autophagosome dysfunction. Nat. Genet. 47:528–534. 10.1038/ng.3256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbosa A.D., and Siniossoglou S.. 2016. Spatial distribution of lipid droplets during starvation: Implications for lipophagy. Commun. Integr. Biol. 9:e1183854 10.1080/19420889.2016.1183854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbosa A.D., Sembongi H., Su W.-M., Abreu S., Reggiori F., Carman G.M., and Siniossoglou S.. 2015. Lipid partitioning at the nuclear envelope controls membrane biogenesis. Mol. Biol. Cell. 26:3641–3657. 10.1091/mbc.E15-03-0173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basseri S., and Austin R.C.. 2012. Endoplasmic Reticulum Stress and Lipid Metabolism: Mechanisms and Therapeutic Potential. Biochem. Res. Int. 2012:841362 10.1155/2012/841362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bigay J., and Antonny B.. 2005. Real-time assays for the assembly-disassembly cycle of COP coats on liposomes of defined size. Methods Enzymol. 404:95–107. 10.1016/S0076-6879(05)04010-3 [DOI] [PubMed] [Google Scholar]
- Bligh E.G., and Dyer W.J.. 1959. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 37:911–917. 10.1139/y59-099 [DOI] [PubMed] [Google Scholar]
- Bryant D., Liu Y., Datta S., Hariri H., Seda M., Anderson G., Peskett E., Demetriou C., Sousa S., Jenkins D., et al. . 2018. SNX14 mutations affect endoplasmic reticulum-associated neutral lipid metabolism in autosomal recessive spinocerebellar ataxia 20. Hum. Mol. Genet. 27:1927–1940. 10.1093/hmg/ddy101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cartwright B.R., Binns D.D., Hilton C.L., Han S., Gao Q., and Goodman J.M.. 2015. Seipin performs dissectible functions in promoting lipid droplet biogenesis and regulating droplet morphology. Mol. Biol. Cell. 26:726–739. 10.1091/mbc.E14-08-1303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chitraju C., Mejhert N., Haas J.T., Diaz-Ramirez L.G., Grueter C.A., Imbriglio J.E., Pinto S., Koliwad S.K., Walther T.C., and Farese R.V. Jr. 2017. Triglyceride Synthesis by DGAT1 Protects Adipocytes from Lipid-Induced ER Stress during Lipolysis. Cell Metab. 26:407–418.e3. 10.1016/j.cmet.2017.07.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhary V., Ojha N., Golden A., and Prinz W.A.. 2015. A conserved family of proteins facilitates nascent lipid droplet budding from the ER. J. Cell Biol. 211:261–271. 10.1083/jcb.201505067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Currie E., Guo X., Christiano R., Chitraju C., Kory N., Harrison K., Haas J., Walther T.C., and Farese R.V. Jr. 2014. High confidence proteomic analysis of yeast LDs identifies additional droplet proteins and reveals connections to dolichol synthesis and sterol acetylation. J. Lipid Res. 55:1465–1477. 10.1194/jlr.M050229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danev R., Buijsse B., Khoshouei M., Plitzko J.M., and Baumeister W.. 2014. Volta potential phase plate for in-focus phase contrast transmission electron microscopy. Proc. Natl. Acad. Sci. USA. 111:15635–15640. 10.1073/pnas.1418377111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta S., Liu Y., Hariri H., Bowerman J., and Henne W.M.. 2019. Cerebellar ataxia disease-associated Snx14 promotes lipid droplet growth at ER-droplet contacts. J. Cell Biol. jcb.201808133 10.1083/jcb.201808133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenberg-Bord M., Mari M., Weill U., Rosenfeld-Gur E., Moldavski O., Castro I.G., Soni K.G., Harpaz N., Levine T.P., Futerman A.H., et al. . 2017. Identification of seipin-linked factors that act as determinants of a lipid droplet subpopulation. J. Cell Biol. 217:269–282. 10.1083/jcb.201704122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fei W., Wang H., Fu X., Bielby C., and Yang H.. 2009. Conditions of endoplasmic reticulum stress stimulate lipid droplet formation in Saccharomyces cerevisiae. Biochem. J. 424:61–67. 10.1042/BJ20090785 [DOI] [PubMed] [Google Scholar]
- Fu G., Wang Q., Phan N., Urbanska P., Joachimiak E., Lin J., Wloga D., and Nicastro D.. 2018. The I1 dynein-associated tether and tether head complex is a conserved regulator of ciliary motility. Mol. Biol. Cell. 29:1048–1059. 10.1091/mbc.E18-02-0142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garbarino J., Padamsee M., Wilcox L., Oelkers P.M., D’Ambrosio D., Ruggles K.V., Ramsey N., Jabado O., Turkish A., and Sturley S.L.. 2009. Sterol and diacylglycerol acyltransferase deficiency triggers fatty acid-mediated cell death. J. Biol. Chem. 284:30994–31005. 10.1074/jbc.M109.050443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golovanov A.P., Hautbergue G.M., Wilson S.A., and Lian L.-Y.. 2004. A simple method for improving protein solubility and long-term stability. J. Am. Chem. Soc. 126:8933–8939. 10.1021/ja049297h [DOI] [PubMed] [Google Scholar]
- Hagen W.J.H., Wan W., and Briggs J.A.G.. 2017. Implementation of a cryo-electron tomography tilt-scheme optimized for high resolution subtomogram averaging. J. Struct. Biol. 197:191–198. 10.1016/j.jsb.2016.06.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hariri H., Rogers S., Ugrankar R., Liu Y.L., Feathers J.R., and Henne W.M.. 2018. Lipid droplet biogenesis is spatially coordinated at ER-vacuole contacts under nutritional stress. EMBO Rep. 19:57–72. 10.15252/embr.201744815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henne W.M., Zhu L., Balogi Z., Stefan C., Pleiss J.A., and Emr S.D.. 2015. Mdm1/Snx13 is a novel ER-endolysosomal interorganelle tethering protein. J. Cell Biol. 210:541–551. 10.1083/jcb.201503088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacquier N., Choudhary V., Mari M., Toulmay A., Reggiori F., and Schneiter R.. 2011. Lipid droplets are functionally connected to the endoplasmic reticulum in Saccharomyces cerevisiae. J. Cell Sci. 124:2424–2437. 10.1242/jcs.076836 [DOI] [PubMed] [Google Scholar]
- Kassan A., Herms A., Fernández-Vidal A., Bosch M., Schieber N.L., Reddy B.J.N., Fajardo A., Gelabert-Baldrich M., Tebar F., Enrich C., et al. . 2013. Acyl-CoA synthetase 3 promotes lipid droplet biogenesis in ER microdomains. J. Cell Biol. 203:985–1001. 10.1083/jcb.201305142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kory N., Farese R.V. Jr., and Walther T.C.. 2016. Targeting Fat: Mechanisms of Protein Localization to Lipid Droplets. Trends Cell Biol. 26:535–546. 10.1016/j.tcb.2016.02.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kremer J.R., Mastronarde D.N., and McIntosh J.R.. 1996. Computer visualization of three-dimensional image data using IMOD. J. Struct. Biol. 116:71–76. 10.1006/jsbi.1996.0013 [DOI] [PubMed] [Google Scholar]
- Listenberger L.L., Han X., Lewis S.E., Cases S., Farese R.V. Jr., Ory D.S., and Schaffer J.E.. 2003. Triglyceride accumulation protects against fatty acid-induced lipotoxicity. Proc. Natl. Acad. Sci. USA. 100:3077–3082. 10.1073/pnas.0630588100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manford A.G., Stefan C.J., Yuan H.L., Macgurn J.A., and Emr S.D.. 2012. ER-to-plasma membrane tethering proteins regulate cell signaling and ER morphology. Dev. Cell. 23:1129–1140. 10.1016/j.devcel.2012.11.004 [DOI] [PubMed] [Google Scholar]
- Mastronarde D.N. 2005. Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol. 152:36–51. 10.1016/j.jsb.2005.07.007 [DOI] [PubMed] [Google Scholar]
- McFie P.J., and Stone S.J.. 2011. A fluorescent assay to quantitatively measure in vitro acyl CoA:diacylglycerol acyltransferase activity. J. Lipid Res. 52:1760–1764. 10.1194/jlr.D016626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murley A., Yamada J., Niles B.J., Toulmay A., Prinz W.A., Powers T., and Nunnari J.. 2017. Sterol transporters at membrane contact sites regulate TORC1 and TORC2 signaling. J. Cell Biol. 216:2679–2689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen T.B., Louie S.M., Daniele J.R., Tran Q., Dillin A., Zoncu R., Nomura D.K., and Olzmann J.A.. 2017. DGAT1-Dependent Lipid Droplet Biogenesis Protects Mitochondrial Function during Starvation-Induced Autophagy. Dev. Cell. 42:9–21.e5. 10.1016/j.devcel.2017.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oelkers P., Cromley D., Padamsee M., Billheimer J.T., and Sturley S.L.. 2002. The DGA1 gene determines a second triglyceride synthetic pathway in yeast. J. Biol. Chem. 277:8877–8881. 10.1074/jbc.M111646200 [DOI] [PubMed] [Google Scholar]
- Palmer I., and Wingfield P.T.. 2004. Preparation and extraction of insoluble (inclusion-body) proteins from Escherichia coli. Curr. Protoc. Protein Sci. 6:Unit 6.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan X., Roberts P., Chen Y., Kvam E., Shulga N., Huang K., Lemmon S., and Goldfarb D.S.. 2000. Nucleus-vacuole junctions in Saccharomyces cerevisiae are formed through the direct interaction of Vac8p with Nvj1p. Mol. Biol. Cell. 11:2445–2457. 10.1091/mbc.11.7.2445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petschnigg J., Wolinski H., Kolb D., Zellnig G., Kurat C.F., Natter K., and Kohlwein S.D.. 2009. Good fat, essential cellular requirements for triacylglycerol synthesis to maintain membrane homeostasis in yeast. J. Biol. Chem. 284:30981–30993. 10.1074/jbc.M109.024752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prévost C., Sharp M.E., Kory N., Lin Q., Voth G.A., Farese R.V. Jr., and Walther T.C.. 2018. Mechanism and Determinants of Amphipathic Helix-Containing Protein Targeting to Lipid Droplets. Dev. Cell. 44:73–86.e4. 10.1016/j.devcel.2017.12.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaffer M., Mahamid J., Engel B.D., Laugks T., Baumeister W., and Plitzko J.M.. 2017. Optimized cryo-focused ion beam sample preparation aimed at in situ structural studies of membrane proteins. J. Struct. Biol. 197:73–82. 10.1016/j.jsb.2016.07.010 [DOI] [PubMed] [Google Scholar]
- Schauder C.M., Wu X., Saheki Y., Narayanaswamy P., Torta F., Wenk M.R., De Camilli P., and Reinisch K.M.. 2014. Structure of a lipid-bound extended synaptotagmin indicates a role in lipid transfer. Nature. 510:552–555. 10.1038/nature13269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senkal C.E., Salama M.F., Snider A.J., Allopenna J.J., Rana N.A., Koller A., Hannun Y.A., and Obeid L.M.. 2017. Ceramide Is Metabolized to Acylceramide and Stored in Lipid Droplets. Cell Metab. 25:686–697. 10.1016/j.cmet.2017.02.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo A.Y., Lau P.-W., Feliciano D., Sengupta P., Gros M.A.L., Cinquin B., Larabell C.A., and Lippincott-Schwartz J.. 2017. AMPK and vacuole-associated Atg14p orchestrate μ-lipophagy for energy production and long-term survival under glucose starvation. eLife. 6:e21690 10.7554/eLife.21690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ta M.T., Kapterian T.S., Fei W., Du X., Brown A.J., Dawes I.W., and Yang H.. 2012. Accumulation of squalene is associated with the clustering of lipid droplets. FEBS J. 279:4231–4244. 10.1111/febs.12015 [DOI] [PubMed] [Google Scholar]
- Teixeira V., Johnsen L., Martínez-Montañés F., Grippa A., Buxó L., Idrissi F.-Z., Ejsing C.S., and Carvalho P.. 2017. Regulation of lipid droplets by metabolically controlled Ldo isoforms. J. Cell Biol. 217:127–138. 10.1083/jcb.201704115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiam A.R., Farese R.V. Jr., and Walther T.C.. 2013. The biophysics and cell biology of lipid droplets. Nat. Rev. Mol. Cell Biol. 14:775–786. 10.1038/nrm3699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas A.C., Williams H., Setó-Salvia N., Bacchelli C., Jenkins D., O’Sullivan M., Mengrelis K., Ishida M., Ocaka L., Chanudet E., et al. . 2014. Mutations in SNX14 cause a distinctive autosomal-recessive cerebellar ataxia and intellectual disability syndrome. Am. J. Hum. Genet. 95:611–621. 10.1016/j.ajhg.2014.10.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsumoto K., Abe R., Ejima D., and Arakawa T.. 2010. Non-denaturing solubilization of inclusion bodies. Curr. Pharm. Biotechnol. 11:309–312. 10.2174/138920110791111924 [DOI] [PubMed] [Google Scholar]
- Velázquez A.P., Tatsuta T., Ghillebert R., Drescher I., and Graef M.. 2016. Lipid droplet-mediated ER homeostasis regulates autophagy and cell survival during starvation. J. Cell Biol. 212:621–631. 10.1083/jcb.201508102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vevea J.D., Garcia E.J., Chan R.B., Zhou B., Schultz M., Di Paolo G., McCaffery J.M., and Pon L.A.. 2015. Role for Lipid Droplet Biogenesis and Microlipophagy in Adaptation to Lipid Imbalance in Yeast. Dev. Cell. 35:584–599. 10.1016/j.devcel.2015.11.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walther T.C., and Farese R.V. Jr. 2009. The life of lipid droplets. Biochim. Biophys. Acta. 1791:459–466. 10.1016/j.bbalip.2008.10.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walther T.C., and Farese R.V. Jr. 2012. Lipid droplets and cellular lipid metabolism. Annu. Rev. Biochem. 81:687–714. 10.1146/annurev-biochem-061009-102430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walther T.C., Chung J., and Farese R.V. Jr. 2017. Lipid Droplet Biogenesis. Annu. Rev. Cell Dev. Biol. 33:491–510. 10.1146/annurev-cellbio-100616-060608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C.-W., Miao Y.-H., and Chang Y.-S.. 2014. A sterol-enriched vacuolar microdomain mediates stationary phase lipophagy in budding yeast. J. Cell Biol. 206:357–366. 10.1083/jcb.201404115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Becuwe M., Housden B.E., Chitraju C., Porras A.J., Graham M.M., Liu X.N., Thiam A.R., Savage D.B., Agarwal A.K., et al. . 2016. Seipin is required for converting nascent to mature lipid droplets. eLife. 5:e16582 10.7554/eLife.16582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright R. 2000. Transmission electron microscopy of yeast. Microsc. Res. Tech. 51:496–510. [DOI] [PubMed] [Google Scholar]
- Xu N., Zhang S.O., Cole R.A., McKinney S.A., Guo F., Haas J.T., Bobba S., Farese R.V. Jr., and Mak H.Y.. 2012. The FATP1-DGAT2 complex facilitates lipid droplet expansion at the ER-lipid droplet interface. J. Cell Biol. 198:895–911. 10.1083/jcb.201201139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J.W., and Lemmon M.A.. 2001. All phox homology (PX) domains from Saccharomyces cerevisiae specifically recognize phosphatidylinositol 3-phosphate. J. Biol. Chem. 276:44179–44184. 10.1074/jbc.M108811200 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.