Nguyen and Olzmann highlight new findings from the Henne laboratory that provide insights into the functions of lipid droplet tethers.
Abstract
Lipid droplets (LDs) are hubs for lipid metabolism that form membrane contact sites with multiple organelles. In this issue, Hariri et al. (2019. J. Cell Biol. https://doi.org/10.1083/jcb.201808119) reveal the functions of Mdm1-mediated endoplasmic reticulum (ER)–LD tethering in yeast and Datta et al. (2019. J. Cell Biol. https://doi.org/10.1083/jcb.201808133) identify a role for the Mdm1 orthologue, Snx14, as an ER–LD tether that regulates lipid metabolism in human cells.
Membrane-bound organelles mediate the segregation of numerous specialized biochemical reactions; however, this compartmentalization poses challenges because the exchange of materials between organelles is essential for cellular homeostasis. Membrane contact sites (MCSs) are regions where organelles are held in close apposition, thereby enabling the efficient transfer of lipids, metabolites, and ions (1). A hallmark of MCSs are proteins referred to as “tethers” that mediate organelle association by interacting with the two organelles directly or as part of a protein complex (1).
Lipid droplets (LDs) are ER–derived neutral lipid storage organelles (2). The biogenesis of LDs involves the deposition of neutral lipids between the leaflets of the ER, the phase separation of neutral lipids into a “lens” structure, and the emergence of the LD from the outer leaflet of the ER into the cytoplasm (2). LDs provide cells with a convenient lipid storage depot that can be accessed during periods of nutrient deprivation and that prevents cellular dysfunction by sequestering toxic lipids (2). LDs function as a nexus of cellular lipid metabolism and establish nutrient-regulated MCSs with the ER, mitochondria, Golgi, lysosomes, and peroxisomes (3). Although some LD tethers have been identified (2), their regulation and functions remain incompletely understood. In this issue, two studies provide new insights into ER–LD tethering, identifying Mdm1 (Hariri et al.) and its orthologue, Snx14 (Datta et al.), as ER–LD tethers that regulate lipid metabolism in yeast and human cells, respectively.
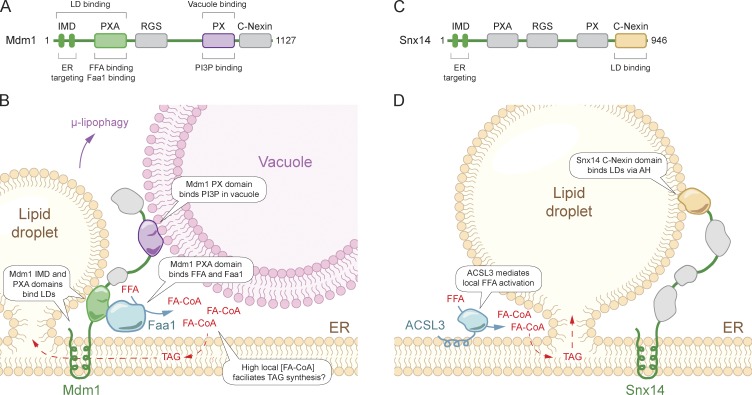
In yeast, Mdm1 is an ER-resident, multi-domain protein that functions as an ER–vacuole tether at the nuclear ER–vacuole junction (Fig. 1, A and B; 4) and facilitates the biogenesis of nuclear ER–vacuole junction–clustered LDs during periods of nutrient exhaustion (5). Mdm1 anchors to the ER via its N-terminal integral membrane domain (IMD) and associates with the vacuole via its phosphatidylinositol 3-phosphate–binding Phox (PX) domain (Fig. 1 B; 4). Through a series of elegant domain mapping experiments, Hariri et al. (6) find that the Mdm1 IMD is both necessary and sufficient to target Mdm1 to sites of LD biogenesis (Fig. 1 B). The IMD is a short stretch of amino acids that is predicted to form two transmembrane helices. How this region targets Mdm1 to sites of LD biogenesis and whether it presents a cytosolic LD-interacting interface is unclear. Mdm1 also contains an amphipathic helix in its PX-associated (PXA) domain that is sufficient to target LDs and may also contribute to Mdm1 LD binding (Fig. 1 B). Remarkably, replacing the vacuole-binding C terminus with a plasma membrane (PM)–targeting domain to recruit the IMD-PXA segment of Mdm1 to the PM is sufficient to promote LD biogenesis at the cortical ER proximal to the PM, demonstrating that this minimal segment of Mdm1 is capable of demarcating LD biogenesis sites.
Figure 1.
Model of Mdm1 and Snx14 in the formation and regulation of LD MCSs. (A) Yeast Mdm1 domain structure. (B) Mdm1 functions as a tri-organelle tether, physically linking the ER, vacuole, and LDs. Mdm1 binds FFA and recruits Faa1, providing high local concentrations of activated FA that promote efficient neutral lipid synthesis and prevent lipotoxicity. (C) Human Snx14 domain structure. (D) Snx14 acts as an ER–LD tether and promotes LD biogenesis. AH, amphipathic helix.
Hariri et al. (6) further find that Mdm1 facilitates fatty acid (FA) activation by directly binding to free fatty acids (FFAs) and by associating with the FA-activating enzyme Faa1 through its PXA domain (Fig. 1 B). Thus, the role of Mdm1 goes beyond simply tethering organelles together, providing a scaffold that facilitates the generation of a high local concentration of activated FAs and their subsequent incorporation into neutral lipids. This scaffolding function and promotion of lipid biosynthesis could contribute to a rudimentary triacylglycerol (TAG) biosynthesis metabolon. A metabolon refers to an assembly of consecutive enzymes, which provide a microenvironment that enhances metabolic flux and facilitates the sequestration of labile and/or toxic intermediates, such as in the biosynthetic pathways for coenzyme Q, FAs, purine nucleotides, and potentially TAG (7–10). Other TAG synthesis enzymes, such as Pah1 and Dga1, are present at sites of LD biogenesis and could be part of a TAG biosynthesis metabolon, though this remains to be tested. Consistent with a critical role for Mdm1 in correct lipid flux and storage, yeast lacking Mdm1 that were challenged with exogenous palmitoleate aberrantly accumulate LDs throughout the nuclear ER and exhibit altered ER morphologies, including tubular extensions (6). The ER morphological defects are further exacerbated in yeast lacking both Mdm1 and the enzymes required for neutral lipid synthesis, indicating that the LDs are generated to prevent lipotoxicity in the absence of Mdm1. Thus, Mdm1 promotes LD biogenesis at a tri-organelle junction by defining the site of biogenesis, organizing local FA metabolism, and acting as a tether to simultaneously bind the ER, vacuole, and LDs (Fig. 1 B). Mdm1 regulation of lipid metabolism is essential for ER homeostasis, and LD biogenesis proximal to the vacuole may facilitate LD autophagic breakdown via µ-lipophagy under nutrient-limiting conditions.
Mdm1 is conserved in humans as four sorting nexins (Snx), Snx13, Snx14, Snx19, and Snx25, which exhibit a similar domain structure to Mdm1 (Fig. 1, A and C; 4). Loss-of-function mutations in the gene encoding the ER-resident Snx14 are linked to autosomal recessive spinocerebellar ataxia 20 (SCAR20) disease (11). Snx14 has been observed in proximity to LDs and implicated in cholesterol ester metabolism (12), but the precise mechanisms connecting Snx14 to neutral lipid metabolism are unknown. Datta et al. (13) examined the possibility that the Mdm1 organelle tethering function is conserved in Snx14. Indeed, using proximity-based APEX labeling technology to visualize Snx14 by electron microscopy, Datta et al. (13) find that Snx14 is highly enriched at ER–LD contact sites. Consistent with a role for Snx14 as an ER–LD tether, knockout of Snx14 reduces ER–LD contacts and overexpression of Snx14 increases ER–LD contacts. Interestingly, several differences between the mechanism of Mdm1 and Snx14 ER–LD tethering were observed. In contrast to Mdm1 (Fig. 1 B), Snx14 interacts with LDs through an amphipathic helix present in the C-Nexin domain and does not associate with the lysosome, the human equivalent of the vacuole (Fig. 1 D). In addition, while Mdm1 binds and recruits Faa1 to sites of LD biogenesis, Snx14 is recruited downstream of the FA activating enzyme acyl CoA synthetase 3 (ACSL3). The presence of ACLS3 at sites of LD biogenesis suggests that a similar mechanism may still be present to generate high local concentrations of activated FAs, but whether Snx14 interacts with ACSL3 and whether the Snx14 PXA domain binds FFAs is not known.
Snx14 ER–LD tethering is necessary for correct LD biogenesis as loss of Snx14 results in numerous small LDs and a few very large LDs, and Snx14 lacking the LD-binding amphipathic helix in the C-Nexin domain is unable to rescue the phenotype. The loss of seipin, a key LD biogenesis factor that may also function as an ER–LD tether (2), results in a very similar phenotype to loss of Snx14 though seipin and Snx14 do not appear to be functionally redundant. Whether the ER–LD tethering function of Snx14 plays a role in SCAR20 disease pathogenesis remains to be determined. The traditional view has been that neurons do not contain LDs, but recent findings identify LDs in neurons under a variety of conditions, such as following disruption of the spastic paraplegia-associated TAG hydrolase DDHD2 (14). Emerging studies also highlight the importance of glial LDs and neuron-to-glial lipid transfer in preventing neuronal lipotoxicity (14). Thus, LDs serve important functions in the central nervous system.
The studies from Hariri et al. (6) and Datta et al. (13) add to the growing inventory of LD tethers and beautifully illustrate the ability of tethering proteins to not only mediate organelle association but also to function as scaffolds that spatially organize cellular processes such as lipid metabolism. It is still early days for our understanding of the molecular basis and functions of LD MCSs. Certainly, additional LD tethers remain undiscovered. As new LD tethers are identified, an important challenge that arises is to understand their regulation, structure, function, and coordination as well as their roles under different metabolic conditions and in different cell types.
Acknowledgments
This work was supported by grants from the National Institutes of Health (R01GM112948 to J.A. Olzmann) and from the American Heart Association (16GRNT30870005 to J.A. Olzmann). J.A. Olzmann is a Chan Zuckerberg Biohub investigator.
The authors declare no competing financial interests.
References
- 1.Prinz W.A. 2014. J. Cell Biol. 205:759–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Olzmann J.A., and Carvalho P. Nat. Rev. Mol. Cell Biol. 2018 doi: 10.1038/s41580-018-0085-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Valm A.M., et al. 2017. Nature. 546:162–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Henne W.M., et al. 2015. J. Cell Biol. 210:541–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hariri H., et al. 2018. EMBO Rep. 19:57–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hariri H., et al. J. Cell Biol. 2019 doi: 10.1083/jcb.201808119. [DOI] [Google Scholar]
- 7.Subramanian K., et al. J. Cell Biol. 2019 doi: 10.1083/jcb.201808044. [DOI] [Google Scholar]
- 8.Sweetlove L.J., and Fernie A.R.. 2018. Nat. Commun. 9:2136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Coleman R.A. J. Lipid Res. 2019 doi: 10.1194/jlr.S091843. [DOI] [Google Scholar]
- 10.Srere P.A. 1985. Trends Biochem. Sci. 10:109–110. [Google Scholar]
- 11.Thomas A.C., et al. 2014. Am. J. Hum. Genet. 95:611–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bryant D., et al. 2018. Hum. Mol. Genet. 27:1927–1940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Datta S., et al. J. Cell Biol. 2019 doi: 10.1083/jcb.201808133. [DOI] [Google Scholar]
- 14.Pennetta G., and Welte M.A.. 2018. Dev. Cell. 45:427–432. [DOI] [PMC free article] [PubMed] [Google Scholar]