Cell wall porosity of fungi and plants could not be determined in vivo previously. Application of a novel method links dynamic changes in wall porosity to stress-induced cell elongation in plant roots and uptake of antifungal drugs in yeast cells.
Abstract
Even though cell walls have essential functions for bacteria, fungi, and plants, tools to investigate their dynamic structure in living cells have been missing. Here, it is shown that changes in the intensity of the plasma membrane dye FM4-64 in response to extracellular quenchers depend on the nano-scale porosity of cell walls. The correlation of quenching efficiency and cell wall porosity is supported by tests on various cell types, application of differently sized quenchers, and comparison of results with confocal, electron, and atomic force microscopy images. The quenching assay was used to investigate how changes in cell wall porosity affect the capability for extension growth in the model plant Arabidopsis thaliana. Results suggest that increased porosity is not a precondition but a result of cell extension, thereby providing new insight on the mechanism plant organ growth. Furthermore, it was shown that higher cell wall porosity can facilitate the action of antifungal drugs in Saccharomyces cerevisiae, presumably by facilitating uptake.
Introduction
Bacterial, fungal, and plant cells depend on cell walls for mechanical support, the determination of cell shape and size, and a multitude of additional functions that are essential for the life of these organisms (Cosgrove, 2000; Matteï et al., 2010; Winstel et al., 2013). Bacterial cell walls are a primary target for antibiotics (Brown and Wright, 2016; Young, 2016), just like the fungal cell wall is a target for the treatment of fungal infections (Burnham-Marusich et al., 2018). Plant cell walls form the starting material for many commercial products (Klemm et al., 2005) and are the main feedstock to produce second generation biofuels (Carroll and Somerville, 2009).
Cell walls vary in complexity, from the single polymer of Gram-positive bacteria, to the specialized network of different polysaccharides and proteins in plants (Burton et al., 2010; Meeske et al., 2015; Gow et al., 2017). The complexity, together with the nanometer-scale dimensions and the wall’s sensitivity to sample preparation processes, have limited the investigation of cell wall structure, especially in plant cell walls. The precise arrangement of the different cell wall components within a plant cell wall and the mechanism of dynamic changes in wall structure remain unknown (Cosgrove, 2016). In addition, the lack of simple tools to probe cell wall structure prevents a detailed functional characterization of the many genes that have been implicated in wall synthesis and remodeling (Somerville et al., 2004; Schneider and Persson, 2015; Taylor-Teeples et al., 2015).
One parameter of cell wall structure is the porosity, which describes the capacity for molecular movement within the wall and is related to the spacing between wall polysaccharides. Porosity correlates with cell wall digestibility and saccharification efficiency, as used for biofuel production (Himmel et al., 2007; Ding et al., 2012; Tavares et al., 2015). In rice, cell wall porosity was linked to mesophyll conductance for CO2, showing that it can even limit the photosynthetic capacity (Ellsworth et al., 2018). Furthermore, porosity influences the kinetics and capacity of a plant’s leaf water uptake (Boanares et al., 2018). In bacteria, porosity could be linked to cell growth (Huang et al., 2008; Turner et al., 2013). In plants, however, no consensus on the relationship of cell wall structure and cell wall extensibility could be reached so far (Bidhendi and Geitmann, 2016; Cosgrove, 2016), which reflects our inability to follow dynamic changes in wall structure.
Currently available methods for determining the porosity of cell walls have significant shortcomings (Adani et al., 2011). Transmission EM (TEM), which could be used to visualize areas of different density within cell walls at nanometer resolution, requires fixation and dehydration of the sample, potentially introducing artifacts. The less invasive cryo-EM has been used to visualize spaces between cellulose fibrils, but its resolution appears limited to 20 nm (Derksen et al., 2011; Zheng et al., 2017). A higher resolution and quantitative data of pore size distributions and pore surface area can be obtained by gas adsorption, but also requires harsh sample treatment (Adani et al., 2011). To assess the actual capacity for molecular movement within the wall, especially that of living cells, approaches based on fluorescence spectroscopy or microscopy have been developed. De Nobel et al. (1990) assessed the relative porosity of yeast cell walls by spectroscopically measuring the chemically induced release of UV-absorbing cellular compounds. This assay could show differences between fungal species, but impacts cell function and cannot be applied to complex tissues. Donaldson et al. (2015) used fluorescence quenching to quantify the porosity of dewatered wood, but their method is restricted to secondary cell walls, as it depends on lignin autofluorescence.
In the present study, we aimed to establish a method to quantify cell wall porosity in different cell types, including those with primary cell walls, in vivo. It was hypothesized that access of a small, freely diffusing extracellular molecule to the plasma membrane depends on the structure of the extracellular matrix. The hypothesis was tested and confirmed by measuring the quenching effect of an extracellular quencher on the membrane-specific dye FM4-64 in lipid vesicles, as well as mammalian, bacterial, fungal, and plant cells.
While this method is relevant for various applications, including assessing the saccharification potential of cellulosic biomass, its value was demonstrated here by investigating the relationship of cell wall structure and extension growth in the model plant Arabidopsis thaliana, as well as the effect of cell wall structure on the uptake of antifungal drugs in the yeast model Saccharomyces cerevisiae.
Results
Selection of quenchers for FM4-64
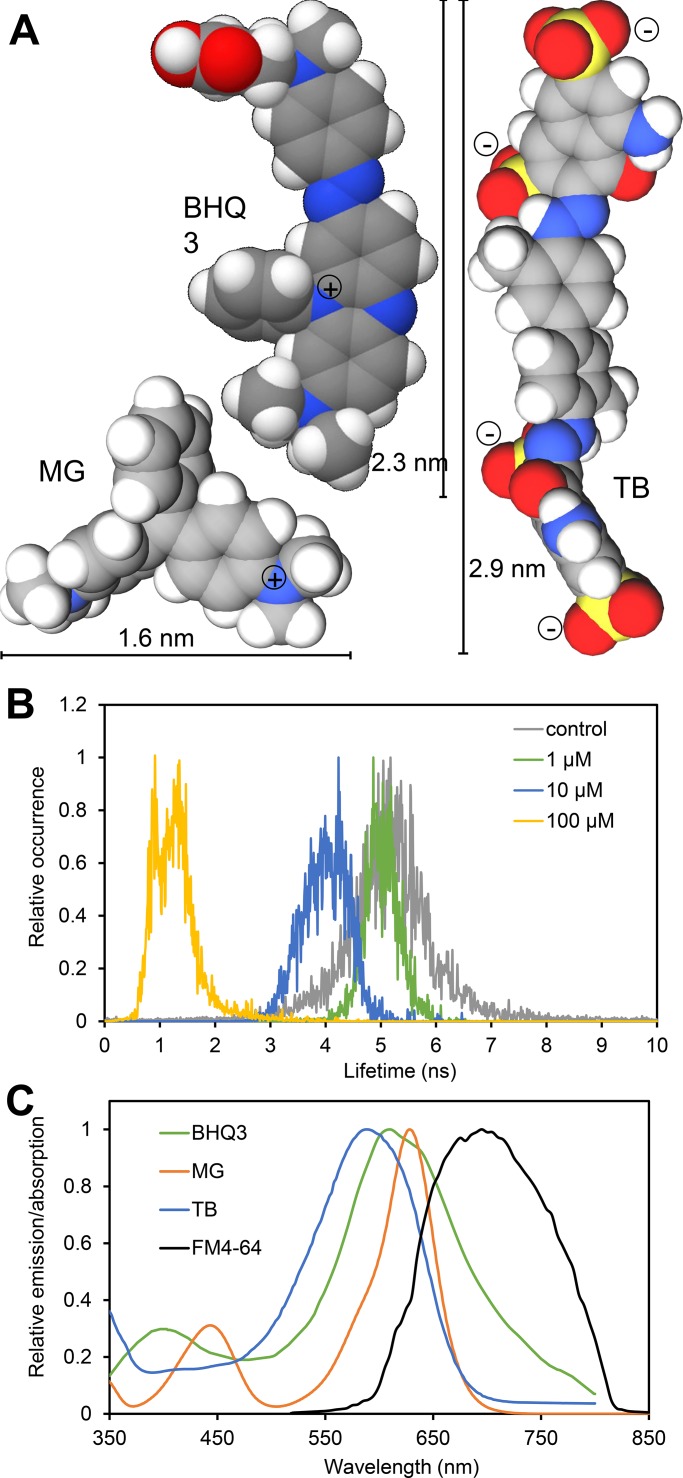
The requirements toward quenchers for the membrane-specific dye FM4-64 are (1) spectral overlap between dye emission and quencher absorption, (2) a dynamic quenching mechanism, and (3) membrane impermeability. Of the commonly used quenchers, Black Hole Quencher 3 (BHQ3), Malachite green (MG), and Trypan blue (TB) have adequate sizes to investigate the nano-scale structure of cell walls (Fig. 1 A). TB acts as a dynamic quencher, as demonstrated by the shortening of the fluorescence lifetime of the Bodipy FL fluorophore in the presence of TB (Fig. 1 B). A similar behavior has been shown previously for MG (Rolinski et al., 1999), while BHQ3 can act as dynamic as well as static quencher (Crisalli and Kool, 2011). Dynamic quenching capacity was furthermore indicated by quenching efficiency, correlating with the spectral overlap of quencher absorption and dye emission (Fig. S1). TB quenched the fluorescent dye Basic Fuchsin, whose emission spectrum overlaps the TB absorption spectrum almost completely, with much higher efficiency than Bodipy FL, whose emission spectrum only overlaps ∼20% (Fig. S1). TB did not show measurable quenching of Calcofluor white (CW), whose emission peak is completely separated from the peak of TB absorption (Fig. S1). The spectral overlap of quencher absorption with the emission of FM4-64 in the yeast plasma membrane was found to be between 20% for TB and 45% for BHQ3 (Fig. 1 C), yielding a useful degree of quenching (see below). The inability to cross membranes has been previously demonstrated for BHQ3 (Zhang et al., 2014), MG (Wilhelm et al., 2015), and TB (Strober, 2015). While all quenchers absorb excitation light at high concentrations, minimal absorption was observed at the concentrations used here (Fig. S1).
Figure 1.
Characteristics of the quenchers BHQ3, MG, and TB. (A) Space filling model of hydrated quencher molecules in minimum energy configuration showing diameter and charges. (B) Fluorescence lifetime of Bodipy FL in PBS in the absence or presence of TB at the indicated concentrations showing a concentration-dependent reduction in lifetime. (C) Comparison of the absorption spectra of BHQ3, MG, and TB in PBS with the emission spectrum of FM4-64–labeled S. cerevisiae cells. For details see Materials and methods. Spectra were normalized to the respective maxima.
Quenching of plasma membrane–localized FM4-64 is influenced by the extracellular matrix
Despite the limited overlap with the excitation spectrum of TB (Fig. 1 C), fluorescence emission of FM4-64 incorporated in giant unilamellar vesicles (GUVs) gradually decreased in the presence of TB (Fig. 2, A and B), demonstrating its potential in this experimental system. To test how quenching is influenced by the extracellular matrix, we compared quenching efficiency and quenchable fraction in GUVs, human embryonic kidney 293 (HEK), Escherichia coli, and S. cerevisiae cells labeled with FM4-64. The amphiphilic nature of FM4-64 means that it has a high affinity to the nonpolar phospholipid bilayer, while its charged group prevents the dye molecule from crossing the membrane (Griffing, 2008; Wu et al., 2009). Importantly, FM4-64 does not bind to cell walls. Plasmolysis experiments on onion epidermis cells showed FM4-64 to be exclusively present in the plasma membrane (Fig. S2). Since FM4-64 can be internalized via endocytosis, measurements were restricted to 5 min after application of the dye, during which time only the plasma membrane is labeled (Vida and Emr, 1995; Bolte et al., 2004).
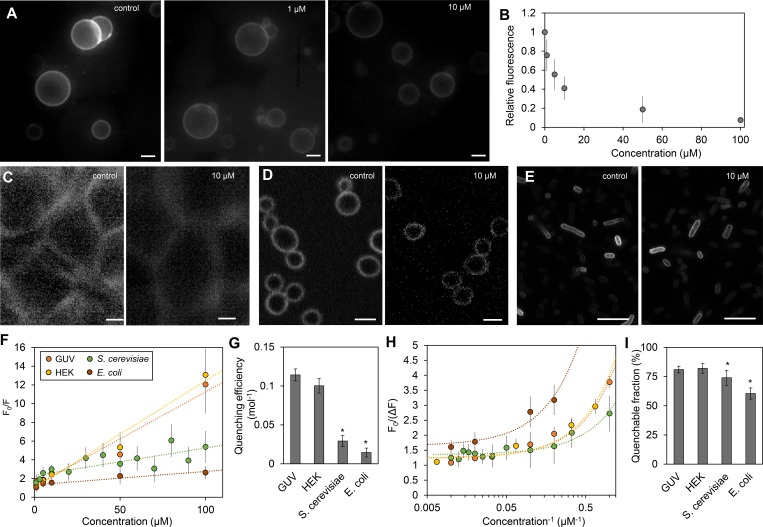
Figure 2.
Dependence of quenching of FM4-64 by TB on accessibility. (A and B) FM4-64–labeled GUVs imaged by fluorescence microscopy in the absence (control) or presence of TB at the indicated concentration (A), and the corresponding intensity plot (B). (C–E) FM4-64–labeled HEK cells (C), S. cerevisiae cells (D), and E. coli cells (E) were imaged in the absence and presence of TB by fluorescence microscopy. (F) Stern-Volmer plots of FM4-64 fluorescence quenching by TB in GUVs, HEK cells, S. cerevisiae cells, and E. coli cells. (G) The slope of the regression line indicates quenching efficiency shown in panel. (H and I) Corresponding modified Stern-Volmer (H) in which the intersection of the linear regression line corresponds to the fraction of quenchable fluorophores shown in panel I. Dotted lines depict linear regression. All error bars indicate standard deviation of the mean (n ≥ 6). Standard deviation of the quenchable fraction (I) was extrapolated from standard deviations of measurements at high quencher concentrations (H). Asterisks (*) in panels G and I indicate statistically significant (P < 0.05) difference to GUVs. Bars, 5 µm.
In all cases, addition of TB resulted in quenching of FM4-64 fluorescence (Fig. 2, C–E). To estimate the quenching efficiency and accessibility of FM4-64 to TB, fluorescence quenching data were analyzed by the Stern-Volmer equation (Eq. 1) and by the modified Stern-Volmer equation (Eq. 2; Lehrer, 1971). In HEK cells, quenching efficiency and quenchable fraction were similar to GUVs (Fig. 2, F–I). In the bacterial and yeast cells, efficiency and quenchable fraction were significantly lower (Fig. 2, F–I). The results demonstrate a lower accessibility of the plasma membrane–localized fluorophore in cells containing a cell wall.
Relationship of quenching efficiency and cell wall structure
After establishing that cell walls affect the quenching of plasma membrane–localized FM4-64, the relationship of cell wall structure and quenching efficiency was further investigated. Quenching experiments were performed on the root elongation zone of seedlings of Arabidopsis plants treated with chemicals known to affect cell wall structure as well as mutants with published cell wall phenotypes. The mutants have reduced amounts of or lack one or more polysaccharide component of the cell wall (Table 1). Chemicals included the cellulose synthesis inhibitors 2, 6-dichlorobenzonitrile (DCBN) and isoxaben, and the growth inducing polyethylene glycol (PEG; also used to simulate drought stress below). Experiments were conducted on epidermal cells, since these define organ morphology (Savaldi-Goldstein et al., 2007) and are accessible to dyes. In each experiment, FM4-64 staining was performed for <10 min to ensure that only the plasma membrane was labeled. It should be noted that the quenching assay is not only applicable to roots, but also works on other plant tissues, for example, maize leaves (Fig. S3).
Table 1. Information on Arabidopsis cell wall mutants used in this study.
| Mutant name | Gene | Description | Locus tag | Cell wall phenotype | Growth phenotype | References |
|---|---|---|---|---|---|---|
| cesa3S211A | CESA3 | Cellulose synthase family protein | AT5g05170 | ↓cellulose | Slightly dwarfed | Chen et al., 2016 |
| mur1 | MUR1 | GDP-d-mannose-4,6-dehydratase | At3g51160 | ↓ fucose, ↓RG II cross-linking | Slightly dwarfed | Reiter et al., 1997; O’Neill et al., 2001 |
| mur10 | CESA7 | Cellulose synthase family protein | AT5G17420 | ↓fucose ↓xylose ↑arabinose ↑mannose | Slightly dwarfed | Reiter et al., 1997; Bosca et al., 2006 |
| mur11 | SAC9 | sacI homology domain-containing protein | AT3G59770 | ↓fucose ↓xylose ↑arabinose | Dwarfed | Reiter et al., 1997; Austin et al., 2011 |
| rol1 | RHM1 | UDP-l-Rhamnose synthase | At1g78570 | ↓RG II modified RG I | Normal | Diet et al., 2006 |
| xxt1xxt2 | XXT1, XXT2 | Xyloglucan Xylosyltransferase 1, 2 | At4g02500 | ↓Xyloglucan | Dwarfed | Cavalier et al., 2008; Xiao et al., 2016 |
RG, rhamnogalacturonan.
Differences were observed between the three different quenchers (Fig. 3). In each plant, MG yielded the highest quenching efficiency, and BHQ3 yielded the lowest, with TB falling in between (Fig. 3 B). All mutants showed a reduction in quenching efficiency with TB and BHQ3 compared with wild-type plants, although the reduction was only significant for cesa3S211A, rol1, and xxt1xxt2 (Fig. 3 B). In contrast, quenching with MG did not show decreased efficiency in the xxt1xxt2 mutant and even increased in the cesa3S211A mutant (Fig. 3 B). Exposure of roots to the three chemicals increased quenching efficiency with all quenchers with the same order of efficiency, MG > TB > BHQ3, as observed in the cell wall mutants (Fig. 3 B). These results suggest that quenching efficiency depends on quencher size, as the smaller MG can quench more efficiently and is not hindered by the structural changes in the xxt1xxt2 and cesa3S211A mutants that affect penetration of the bigger BHQ3 and TB (see Fig. 1 for size comparison of quenchers).
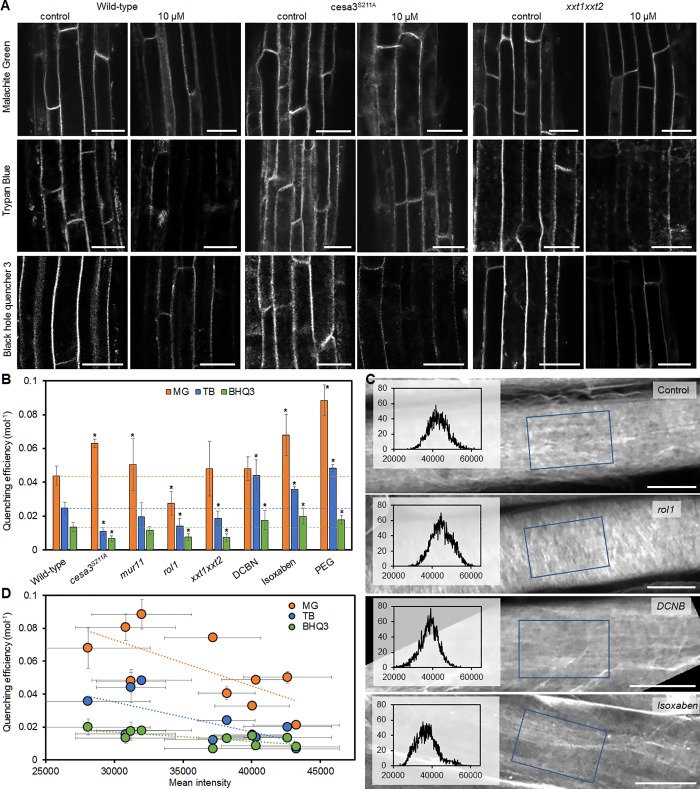
Figure 3.
Relationship of quenching efficiency and cell wall structure in epidermis cells of Arabidopsis thaliana wild-type plants and cell wall mutants. (A) Wild-type, cesa3S211A, and xxt1xxt2 plant root epidermis cells were labeled with FM4-64 and imaged by fluorescence microscopy in the absence (control) and presence of the indicated quenchers (10 µM). (B) Quenching efficiency of different quenchers (MG, TB, and BHQ3) on wild-type, mutant plants, and wild-type plants treated with cell wall–modifying agents DCBN, isoxaben, and PEG. Lines mark the quenching efficiency in wild-type plants. (C) Root epidermis cells stained with the cellulose-specific dye S4B imaged by fluorescence microscopy. Blue rectangles indicate the area corresponding to the overlaid histograms. Lower intensity mean value indicates larger spacing between cellulose fibrils. (D) Quenching efficiency plotted against mean intensity showing a significant correlation for all three quenchers. Both parameters were measured on all samples shown in B, as well as other cell wall mutants listed in Table 1. Asterisks (*) in B indicate significant difference (P < 0.05) from wild type. All error bars indicate standard deviation of the mean. Regression lines in D indicate significant correlation (P < 0.05). Number of biological replicates n ≥ 4. Bars: 20 µm (A); 10 µm (C).
To further investigate how quenching efficiency relates to cell wall structure, we evaluated cellulose spacing of wild-type, mutant, and chemical-treated plants by staining the roots with the cellulose-specific dye, Pontamine Fast Scarlet 4B (S4B; Anderson et al., 2010; Liesche et al., 2013). With the intensity of cellulose fibrils normalized among all images, differences in cellulose fibril density become apparent (Fig. 3 C). These were quantified as histogram mean values of 2D projections of image stacks acquired throughout the cell wall. Lower mean values indicate a higher abundance of black pixels, i.e., spaces between cellulose fibrils. Plants treated with the cellulose synthesis inhibitors DCBN or isoxaben showed lower cellulose density compared with control plants and, accordingly, reduced mean values (Fig. 3 C). Cell wall mutants, such as rol1, showed an increased mean value and visible differences in the pattern of cellulose distribution (Fig. 3 C). Histogram mean value was found to significantly correlate with quenching efficiency for all three quenchers (Fig. 3 D).
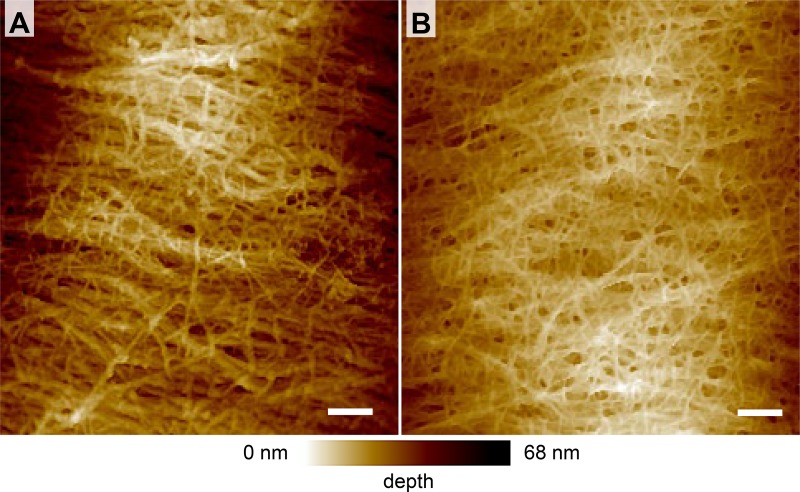
The correlation of quenching efficiency with cellulose spacing was further corroborated by atomic force microscopy (AFM) imaging of epidermal cells corresponding to those used in the quenching experiments. AFM images of the cesa3S211A mutant indicate a change in spacing of cellulose compared with wild-type (Fig. 4). Whereas in the wild-type wall, thick cellulose bundles with relatively large spaces in between were evident (Fig. 4 A), a tight network of thin cellulose fibrils was observed in the cesa3S211A mutant (Fig. 4 B). A similar difference has been observed in the epidermal cell walls of wild-type and the xxt1xxt2 mutant (Xiao et al., 2016). Whether a high number of small pores in the cesa3S211A mutant is responsible for the significantly increased MG quenching efficiency cannot be confirmed or excluded based on the AFM images.
Figure 4.
AFM images of the inner epidermal cell wall layer. (A and B) Spacing between wall polysaccharides appears larger in wild-type (A) than in the cesa3S211A mutant (B) plants. Bars, 100 nm.
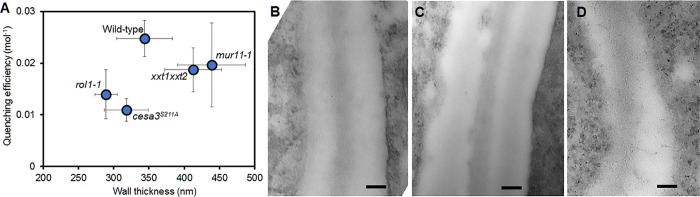
In contrast, no indication for a correlation of cell wall thicknesses, measured on TEM images and quenching efficiency, was found (Fig. 5). The walls of mutants mur11 and xxt1xxt2 were significantly thicker than those of wild-type plants, whereas walls of rol1 were significantly thinner (Fig. 5). For example, rol1 had a lower quenching efficiency but a thinner wall. The cesa3S211A mutant showed strong differences in quenching efficiency, but its wall thickness was not significantly different from wild type. The results strongly suggest that quenching efficiency depends on cell wall porosity and can, therefore, be used as indicator for this parameter.
Figure 5.
Cell wall thickness of root epidermis cells of Arabidopsis thaliana wild-type plants and cell wall mutants. (A) Cell wall thickness, as determined by TEM plotted against TB quenching efficiency, showing no correlation between the two parameters. (B–D) Representative electron micrographs of epidermal cell walls from wild type (B), mur11 (C), and rol1 (D). Error bars indicate standard deviation of the mean. Bars, 100 nm.
It should be noted that for TB quenching, changes in the quenchable fraction were also tested, but did not show significant differences (Fig. S4), which is why only quenching efficiencies are considered in the following. Furthermore, only TB was used in follow up experiments as it offered the highest dynamic range of the three quenchers, based on the experiments conducted on cell wall mutants.
Correlation of cell wall porosity and cell length in plants
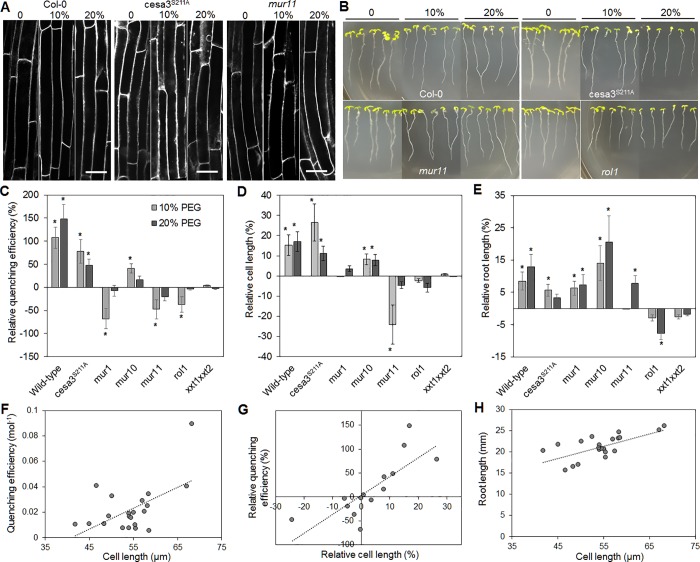
The mutant plants described above, as well as additional mutants that were tested (Table 1), all had significantly reduced root lengths (Fig. S4), indicating that reduced cell wall porosity might correlate with reduced wall extensibility and cell elongation, a connection that has been debated for a long time in the plant science community (Bidhendi and Geitmann, 2016; Cosgrove, 2016). To test this hypothesis, we induced cell extension by exposing wild-type and mutant plants to simulated drought stress, namely growth on 10 or 20% (wt/vol) PEG and determined quenching efficiencies (Fig. 6).
Figure 6.
Correlation analysis of quenching efficiency, cell length, and root length in Arabidopsis thaliana seedlings. TB quenching efficiency was determined in the FM4-64–labeled roots of wild-type and cell wall mutant plants exposed to different levels of drought stress, simulated by PEG treatment. (A and B) In addition to quenching efficiency, cell length (A) and root length (B) were measured in the absence (0%) and presence of PEG at moderate (10%) and high (20%) concentrations. Bars, 10 µm. (C–E) Changes relative to control conditions were quantified for quenching efficiency (C), cell length (D), and root length (E). (F–H) Quenching efficiency significantly correlated with cell length (F), as did relative quenching efficiency with relative cell length (G), and cell length with root length (H). Regression lines indicate significant (P < 0.05) correlation. All error bars indicate standard deviation of the mean. Asterisks (*) indicate significant difference (P < 0.05) compared with control conditions. Number of replicates n = 5 (C), n ≥ 25 (D), n ≥ 20 (E).
In wild-type plants, quenching efficiency increased upon PEG treatment (Fig. 4 C), indicating an increase in wall porosity. As expected cell length and root length increased with PEG concentration (Fig. 6, D and E). Regarding the cell wall mutants, a PEG-induced increase in quenching efficiency was observed in the cesa3S211A and the mur10 mutants, as well as an increase in cell and root length (Fig. 6, A–E). A significant reduction of quenching efficiency was observed in the mur1, mur11, and rol1 mutants (Fig. 6 C). For all plants that showed a reduction or no significant change in quenching efficiency, no significant PEG-induced increase in cell length was observed (Fig. 6, A and D). However, in case of mur1 and mur11, root length still increased (Fig. 6, B and E).
A correlation analysis was conducted using the data from wild-type and mutants under control and PEG-treatment conditions to test if quenching efficiency scales with cell length and root length. The analysis of absolute values (Fig. S4) showed a significant correlation of cell length and quenching efficiency (R = 0.45, P = 0.0272, n = 24; Fig. 6 F). Even higher significance was observed when testing correlation of relative quenching efficiency and relative cell length (R = 0.565, P = 0.0127, n = 16; Fig. 6 G). A correlation of quenching efficiency with root length was only found when comparing relative changes (R = 0.436, P = 0.035, n = 16). As expected, cell length showed a very strong correlation with root length (R = 0.677, P = 0.0003, n = 24; Fig. 6 H). The results support the notion of cell wall porosity being related to wall elasticity and, thereby, cell elongation. However, rather than being a precondition for cell elongation, increased porosity seems to be a consequence.
Cell wall porosity and drug uptake in yeast
To further illustrate the potential of the quenching assay, it was used to test if cell wall porosity influences the efficiency of antifungal drugs with a target inside the cell. Previously, lipid bilayers such as the plasma membrane in yeast or outer membrane of Gram-negative bacteria have been seen as the decisive barrier for the uptake of antibacterial or antifungal drugs (Lambert, 2002; Mishra et al., 2007). However, it was reported that disruption of the cell wall sensitizes the yeast model S. cerevisiae and the infectious Candida glabrata to the anti-malarial drug, chloroquine (Islahudin et al., 2013), indicating that cell wall structure can influence drug uptake. Here, experiments were performed on S. cerevisiae, which has a similar cell wall to the infectious Candida strains (Gow et al., 2017).
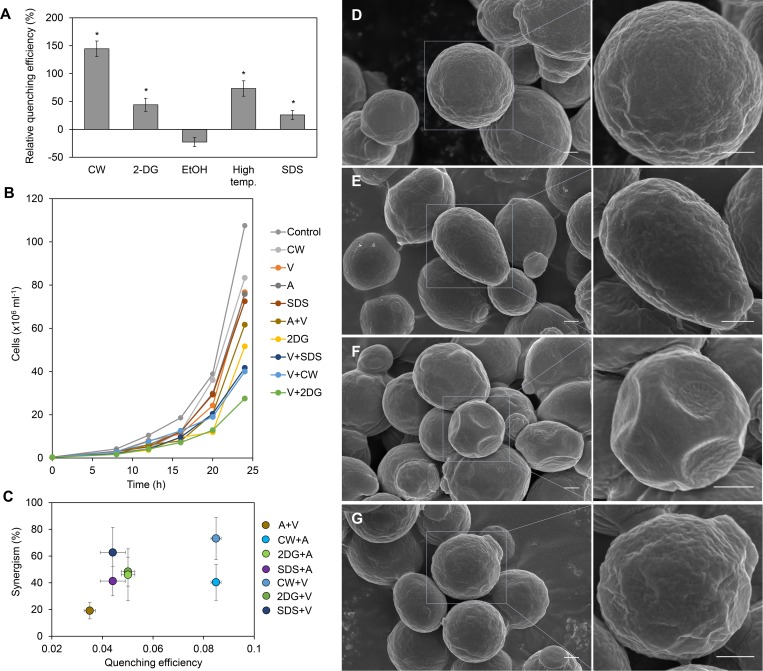
Several agents known to affect yeast cell wall structure (Okada et al., 2016) were tested for their influence on cell wall porosity using the quenching assay. Cells treated with CW, 2-Deoxyglucose (2-DG), a 42°C heat shock, and SDS were found to have a higher FM4-64 quenching efficiency compared with untreated cells, indicating an increased cell wall porosity (Fig. 7 A). The influence of CW, 2-DG, and SDS, as well as the influence of two known antifungal drugs with intracellular target, Amphotericin B and Voriconazole, on cell viability was tested. Amphotericin B and Voriconazole target the membrane integrity and ergosterol synthesis, respectively (ASDCD, 2018). At the minimal efficient concentrations, where separate application of these compounds reduced viability after 24 h by ∼50% (Fig. 7 B), combinations of wall-modifying agents with Amphotericin B or Voriconazole further reduced viability, indicating a synergistic effect (Fig. 7 B). Quantification of this effect showed that the efficiency of concomitant application of Amphotericin B and Voriconazole only increased by 15%, compared with separate applications of the two drugs (Fig. 7 C). All combinations of porosity-increasing chemicals with either Amphotericin B or Voriconazole led to an increase in treatment efficiency of at least 40% (Fig. 7 C). While there was no linear correlation between quenching efficiency and the synergistic effect (Fig. 7 C), these results do indicate a link between cell wall structure and efficiency of drug uptake.
Figure 7.
Effect of cell wall structure on antifungal drug efficiency in S. cerevisiae. (A) Relative FM4-64 quenching efficiency after treatment with cell wall–modifying factors compared with untreated cells (EtOH, ethanol). (B) Cell counts over 24 h culture period in the absence (control) or presence of cell wall–modifying chemicals and the antifungal drug Voriconazole (V). Combination of Voriconazole and cell wall–modifying chemicals led to strong reduction in cell number. Similar results for this viability were observed with another antifungal drug, Amphotericin B (A). (C) Synergism of cell wall–modifying chemicals and antifungal drugs plotted against quenching efficiency. (D–G) Field emission scanning electron micrographs of cells in the absence (D) or presence of the cell wall–modifying chemicals CW (E), 2-DG (F), and SDS (G) at two magnifications. Only 2-DG–treated cells showed visible signs of compromised cell wall integrity (F). Error bars indicate standard deviation of the mean (n = 4). Asterisks indicate significant difference (P < 0.05), compared with control conditions. Bars, 500 nm.
To check if the cell wall–modifying chemicals at the concentrations used for the quenching assay visibly compromise the cell wall or if they merely cause changes in the internal structure, cells were studied by field emission scanning EM (Fig. 7). Only the appearance of 2-DG–treated cells showed strong deviation from control cells (Fig. 7 F). The large indentations indicate that treatment led to major defects in the cell wall, whereas CW and SDS treatment did not affect appearance (Fig. 7, D–G).
Discussion
The quenching assay presented here is useful for quantifying the accessibility of the extracellular quencher molecule to the plasma membrane. Furthermore, the results demonstrate that this accessibility changes according to the structure of the extracellular matrix, i.e., the cell wall. Thereby, the assay constitutes a new quantitative method to probe the structure of cell walls in vivo.
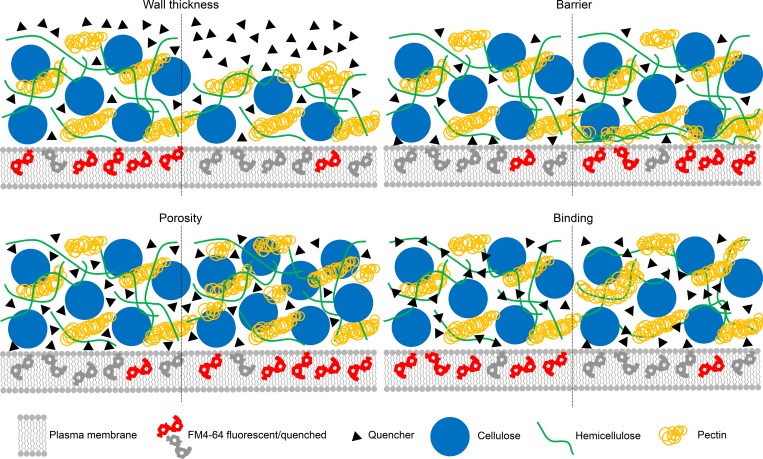
Which structural feature(s) of the cell wall determine the quenching efficiency? At least four parameters can be considered (Fig. 8). First, quenching efficiency could depend on the distance between the plasma membrane and the outer boundary of the extracellular matrix. No correlation of cell wall thickness and quenching efficiency was observed for Arabidopsis seedlings, although the relatively low sample number should be considered. Nevertheless, the result is in line with the fact that quenching is limited to ∼5-nm distance (Zu et al., 2017).
Figure 8.
Illustration of four parameters potentially affecting extracellular quenching: wall thickness, the barrierfunction of polysaccharides close the plasma membrane, wall porosity, and the binding capacity of quencher molecules to wall components. The experimental data indicate that wall porosity is the main parameter that determines quenching efficiency.
Second, quenching efficiency could depend on the contact area of plasma membrane and open apoplastic space. This is unlikely, because the quenching efficiency showed large differences between wild-type plants and some of the cell wall mutants, while the quenchable fraction, i.e., the amount of membrane dye that can be accessed by the quencher, did not significantly change. Furthermore, no visible differences were found between the plasma membrane–adjacent wall region of wild-type and xxt1xxt2 mutant cells (Xiao et al., 2016).
Third, the quenching efficiency might depend on the spacing between wall components, i.e., the molecular diffusion efficiency inside the cell wall. The comparison of quenching efficiency measurements with TEM and AFM images of the cell walls of wild-type and mutant plants supports this hypothesis. The walls of cesa3S211A and xxt1xxt2 mutants, which had a decreased quenching efficiency, both have a denser appearance than the respective walls wild-type plants (Xiao et al., 2016; Fig. 4). Importantly, quenching efficiency was influenced by the size of the quencher with MG showing better quenching than the larger BHQ3 and TB. TB is longer but narrower than BHQ3, which might cause the better quenching of TB compared with BHQ3 that was observed here. Charge is unlikely to influence quenching, as the negative charge of the cell wall (Crasnier et al., 1985) would be expected to facilitate access of the positively charged BHQ3, but not the negatively charged TB.
Fourth, quenching efficiency might depend on the molecular diffusion efficiency inside the wall, like in the third hypothesis, but this would depend on the binding of quencher molecules to cell wall components, instead of the availability of spaces between components. TB was previously proposed to have a moderate ability to bind β glucans and xyloglucan (Liesche et al., 2015). However, TB quenching efficiency in the different cell wall mutants does not support this observation. For example, in the xxt1xxt2 mutant, which lacks xyloglucans (Cavalier et al., 2008; Park and Cosgrove, 2012), quenching efficiency was lower than in wild-type plants. If TB diffusion was influenced by xyloglucan binding, then quenching efficiency should increase in the absence of these binding sites. Quenching efficiency was also significantly reduced in the rol1 mutant, even though this mutant has the same levels of xyloglucan and other glucans as wild-type plants (Diet et al., 2006).
In conclusion, it is most likely that the primary factor determining quenching efficiency is the spacing between cell wall components, especially between cellulose fibrils. That means that it would also be a good tool to test cell wall digestibility, which has previously been linked to cell wall porosity (Adani et al., 2011; Ding et al., 2012), in the same set of mutants analyzed here or in other plants for which the quenching measurements are made. This could be highly relevant for testing, maybe even as part of a breeding program, for biomass usability (Dixon, 2013). In this respect, the results also suggest that reducing certain cell wall components, at least in the primary wall, cannot be expected to be a good strategy for increasing digestibility, because decreased wall porosity might be a general response. This could explain why genetically modifying plants with increased expression of cell wall–modifying enzymes sometimes does not have a beneficial effect for digestibility (Tavares et al., 2015).
The potential of the quenching assay was illustrated by using it to investigate changes in wall porosity during drought-induced cell elongation in the model plant Arabidopsis. Root elongation is an important part of a plant’s drought response, as it enables access to residual water in the soil. It is achieved through a combination of higher cell division rates and cell elongation (Comas et al., 2013). Cell elongation depends on remodeling of the cell wall, and drought-induced wall remodeling is known to be enacted by enzymes that modify wall polysaccharides, especially expansins, xyloglucan endotransglucosylases/hydrolases, and pectin esterases (Tenhaken, 2015; Lampugnani et al., 2018). The question of how cell wall structure relates to wall mechanics and action of cell wall–loosening agents is seen as the “grand challenge” in the field of cell wall biology (Cosgrove, 2016). Results of the quenching assay provide some insight on this by demonstrating that cell wall porosity changes during wall remodeling. The occurrence of stress-induced cell elongation in mutants with strongly decreased wall porosity indicated that increased porosity is not a precondition for elongation but a consequence. This was supported by the higher significance of correlation of quenching efficiency and cell length relative to nonstressed conditions compared with the correlation of absolute values of the two parameters.
The results suggest the following to happen: stress-induced loosening of the cell wall is accompanied by an increased physical distance between wall components or the removal of certain materials between load-bearing components. In the cell wall mutants that are not able to remodel their cell walls in response to stress, cell wall–modifying enzymes can be present in the wall, but the lack/reduction of certain wall components prevents their action. These results could be extended by analyzing plants with altered amounts of cell wall–modifying enzymes, coupled with a detailed chemical analysis of cell wall composition and cross-linking. Facilitated by the noninvasiveness, the assay could thereby help to find the genetic basis of the adaptation of cell wall structure to environmental conditions. In addition, facilitated by the simplicity of the quenching assay, key genes controlling cell wall remodeling could be identified via forward genetics screening or through a genome wide association study.
The quenching assay presented here could also be used for chemical screens that aim to identify compounds that alter the cell walls of bacteria or fungi and thus have relevance for biomedical and biotechnology research. This was illustrated here by testing porosity of yeast cells in the presence of cell wall–modifying agents and linking these results to uptake of common antifungal drugs with intracellular targets. While results clearly demonstrate a synergistic effect due to modification of cell wall structure, it is not clear if there is a correlation with porosity or if the effect is due to another effect. For example, it might be possible that destabilizing the cell wall affects the plasma membrane and facilitates uptake or action through this. Investigation on a larger scale would be needed to determine the full potential of cell wall–modifying agents for antifungal drug treatment.
Materials and methods
Materials
The lipid 1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC) was purchased from Avanti Polar Lipids. N-(3-Triethylammoniumpropyl)-4-(6-(4-(Diethylamino) Phenyl) Hexatrienyl) Pyridinium Dibromide (FM4-64), Bodipy FL, and CW were purchased from Thermo Fisher Scientific. TB was purchased from Merck Millipore; Basic Fuchsin was purchased from Sigma-Aldrich; and SDS, MG, 2-DG, PEG (MW 8000), isoxaben, and DCNB was purchased from Solarbio. BHQ3 was purchased from LGC Biosearch. PBS contained 130 mM NaCl, 2.6 mM KCl, 7 mM Na2HPO4, and 1.2 mM KH2PO4 and was adjusted to pH 7.4. Phosphate buffer (PB) with pH 5.8 was obtained by mixing 0.2 M NaH2PO4 and 0.2 M Na2HPO4 with the volume ratio of 23:2 and then diluted to a concentration to 40 µM. PB with pH 6.8 obtained by mixing the two stock solutions with a ratio of 51:49. Tyrode’s balanced salt solution contained 136 mM NaCl, 2.6 mM KCl, 1.8 mM CaCl2, 1 mM MgCl2, 0.36 mM NaH2PO4, 5.56 mM d-Glucose, and 5 mM Hepes and was adjusted to pH 7.4.
Electroformation of GUVs
GUVs were produced by an electrophysical method (Angelova et al., 1992), as described in the following. DOPC was dissolved in chloroform to a final concentration of 0.5 mg/ml in a volume of 50 µl in pointy Schott glass tubes. 5 µl of the mixture was placed in small drops on both electrodes, and the chloroform was dissipated by vacuum application for 15 min. The electroformation chamber was filled with 300 µl sterile-filtered 300 mM sucrose solution. Vesicle formation was achieved by applying an AC voltage, 20 mV and 10 Hz, for 3 h, followed by 1 h at 2 V and 4 Hz. The GUVs (20 µl) were transferred on a microscope slide, and the same amount of PBS or quencher solution was added. GUVs were allowed to settle on the slide for 3 min. The quality of GUVs was checked using phase-contrast microscopy.
Cell cultures and plant cultivation
E. coli (DH5α) cells were grown at 37°C in Luria-Bertani liquid medium for 4 h. Bacteria were collected from l ml medium, washed with 1 ml PBS, and incubated with 100 µl PBS-based FM4-64 solution for 5 min. After washing with 1 ml PBS, cells were mixed with 10 µl quencher solutions on the slide and immediately analyzed under the microscope.
Yeast (S. cerevisiae) cells of the strain BY4742 were cultured in yeast extract peptone dextrose medium (1% yeast extract, 2% peptone, 2% glucose/dextrose, and water) at 30°C. For labeling with FM4-64, cells in 3 ml medium were grown to mid-logarithmic phase (OD600, ∼0.6–0.8), washed with PBS and suspended in 200 µl PBS-based FM4-64 solution for 3 min, followed by washing and imaging. Cells were used within 10 min. For the induction of changes in the yeast cell wall, 5 µg/ml CW, 0.02% (wt/vol) 2-DG, or 0.04 mg/ml SDS were added to the medium 2 h before quenching experiments were conducted.
HEK cells were maintained in Dulbecco’s modified Eagle’s medium (Invitrogen) supplemented with 10% fetal bovine serum at 37°C in 5% CO2. For labeling with FM4-64, cells were trypsinized using 0.25% trypsin-EDTA for 5–10 min and resuspended in Tyrode’s balanced salt solution. Cells were used for quenching assays within 1 h.
Arabidopsis ecotype Colombia (Col-0), and the homozygous mutants kor (N298), mur1 (N6244), mur10 (N8578), mur11 (N8579), xxt1xxt2 (N16349), rol1 (N16373; all obtained from the Nottingham Arabidopsis Stock Center), and CESA3S211A (Chen et al., 2016) were used in this study. Mutants were selected to represent different types of changes in cell wall composition and structure. After surface sterilization with 10% (vol/vol) sodium hypochlorite for 10 min, followed by vernalization for at least 3 d at 4°C, seedlings were grown on half-strength Murashige and Skoog (MS) plates containing 0.22% (wt/vol) MS salts (Phyto Technology Laboratories), 1% (wt/vol) sucrose, and 0.8% (wt/vol) agar powder (Solarbio), pH 5.8, under 16-h light/8-h dark in a growth chamber at 22°C. For drought treatment, 3-d-old seedlings were transplanted from normal half-strength MS plates to plates containing either 10 or 20% (wt/vol) PEG for 4 d. For treatment with the cellulose synthesis inhibitors DCBN (Desprez et al., 2002) and isoxaben (Tateno et al., 2016), 6-d-old Columbia seedlings grown on half-strength MS plates were transferred to plates containing 5 nM isoxaben or 0.5 µM DCBN. After 1-d incubation in the growth chamber with 16-h light/8-h dark cycle at 22°C, seedlings were used for the quenching assay and cell wall structure analysis.
Quenching assay and imaging
FM4-64, from a 1 mM DMSO stock, was used at final a concentration of 50 µM for staining of GUVs, yeast, and HEK cells and at a concentration of 20 µM for staining plant cells. Stock solutions with different concentrations of BHQ3 and TB were made by dissolving the quenchers in PBS; MG was dissolved in PB. From these quencher solutions, 20 µl was added to the GUVs/cells, and 50 µl was added to the plant seedlings on the microscope slide to final concentrations ranging from 1 to 100 µM. After addition of quencher, samples were imaged immediately using a wide-field fluorescence microscope (DMi8, Leica Microsystems) for GUV experiments or a confocal microscope (Andor Revolution XD, Leica SP8 or a point-scanning confocal system equipped with CLSD-2SS Dual-Channel PMT Module [Thorlabs] on a Leica DMi8 microscope body) for experiments involving cells and plants. 100× oil immersion objectives with NA 1.3–1.49 were used for GUV, bacteria, yeast, and HEK cell imaging, and a 1.10 NA 40× water immersion objective was used for plant cells. Imaging parameters for FM4-64 were excitation at 530–550 nm and emission detection at 560–620 nm on the wide-field system and excitation at 514 nm, 532 nm, or 543 nm and emission detection at 570–616 nm or 550–610 nm on the confocal microscopes. Software supplied by the microscope manufacturers was used for image acquisition.
For each sample, the quenching assay was repeated at least three times, including the entire concentration range. As there was no significant variation between the data from different replicates, all data from each sample were pooled and analyzed together. While different intensity values were obtained from experiments performed using different confocal microscopes, there were no significant differences in quenching efficiency or quenchable fraction.
Absorption and emission spectrum measurements
Absorption scans were performed with an Infinite 200 Pro fluorescence spectrometer (Tecan) with the PBS-based solution at the concentration of 100 µM. CW, Basic Fuchsin, and Bodipy FL were all used at a concentration of 10 µg ml-1 and mixed with TB right before conducting the measurement. The optimal excitation and emission parameters used in these tests were determined in separate wavelength scans for each dye.
Image analysis and calculation of quenching efficiency
All image analysis was performed in ImageJ (Schindelin et al., 2012). To correct for background, signal in an area of each image where no GUVs or cells were present was measured. The black level of the image was assigned to this value and the remaining signal values were redistributed between 1 and 255 using the brightness and contrast tool. Then, fluorescence intensity was measured by determining the average intensity of a 3 × 5-µm region of interest (ROI) overlaid on the peripheral staining of each GUV or cell (Fig. S5). The ROI size was reduced to 1 × 2 µm for S. cerevisiae cells and 0.5 × 0.5 µm for bacteria. In plant cells, ROI width was kept constant, while ROI length varied in relation to the in-focus area of plasma membrane (Fig. S5).
Intensity values were transferred to Excel (Microsoft), and quenching efficiency and fraction of quenchable fluorophores were calculated according to the equations presented below. Quenching processes are generally described by the Stern-Volmer equation, which specifies the quenching efficiency
| (1) |
where F0 and F are the fluorescence intensity in the absence and presence of quencher at the concentration Q, respectively, and K is the quenching coefficient. In addition to quenching efficiency, the fraction of quencher-accessible fluorophores is used to characterize a quenching process. To determine the quenchable fraction of fluorophores, the Stern-Volmer equation (Eq. 1) can be modified to quantify the portion of fluorophores that are accessible for quenching, fa, according to Lehrer (1971),
| (2) |
where ΔF is the change in fluorescence intensity through quenching. Accordingly plotted, 1/fa of a quenching experiment is given as the point of intersection with the y axis.
Standard deviations were provided as error bars for all average values. The standard deviation of linear regressions was determined according to
with quenching efficiency b, R value of the fit R, and the number of experiments n. Welch’s t test was used to determine significance.
Relative quenching efficiencies were calculated by dividing values from experiments conducted under stress conditions with those conducted under control conditions.
Analysis of cellulose fibril spacing by confocal imaging
Roots of 7-d-old Arabidopsis seedlings were stained with 10 µg/ml S4B for 10 min and washed with PBS before imaging on a spinning-disc confocal system (Revolution WD; Andor) equipped with a CSU-W1 spinning-disc head (Yokogawa) and an iXon Ultra 888 EM charge-coupled device (Andor). Using the HCX PL Apo 100× NA 1.49 objective, 561-nm laser, and a TR-F607/36 bandpass filter (Semrock BrightLine), z stacks of the outer wall of epidermis cells were acquired. Image stacks were deconvolved using the Huygens software (Version 15.10; Scientific Volume Imaging) with the default settings using an automatically generated theoretical point spread function and the maximum likelihood estimator algorithm. Maximum projections were made from deconvolved image data in ImageJ. On these, the white level was adjusted so that the brightest cellulose fibrils of the outer wall are assigned the maximum value (65,535 for the 16-bit images). The black level was left unchanged. Thereby, differences in signal intensity between samples were eliminated, ensuring that darker areas indicate reduced presence of cellulose. Rectangular ROIs were drawn on the in-focus areas of the outer epidermal wall and mean values determined using ImageJ’s histogram function. At least six cells were measured for each sample.
Fluorescence lifetime imaging microscopy (FLIM)
FLIM was performed on a Microtime 200 laser-scanning confocal (PicoQuant). Bodipy FL was excited at 485 nm, and fluorescence emission was detected at 500–525 nm by a photon-counting hybrid photomultiplier detector. Electrical signals were processed by a time-correlated single photon-counting module (PicoQuant; HydraHarp 300). Analysis of FLIM images was performed using the SymPhoTime 64 software (PicoQuant), taking into account the instrument response function. Bodipy FL showed a single exponential decay. Time of photon arrival data are represented without fitting to a mathematical model. In these data, maximum peak height represents the fluorophore’s lifetime under the given conditions.
AFM
3-d-old dark-grown seedlings were bisected longitudinally and incubated in 2 M KOH at room temperature for 1 h and then in 1% Tween 20 for 30 min. After washing with double-distilled H2O (ddH2O) until pH 7.0 was reached, slices were placed between glass slides, and a load of 5 g was applied for 5 min. The innermost wall layer of primary cell walls was examined by scanning probe atomic force microscope (Bruker MultiMode V with NanoScope V Controller and SCANSYST-AIR probe). Contact AFM was performed in air at room temperature. Images of 2 µm2 size with 512 × 512-pixel resolution were recorded using the software NanoScope Analysis (Version 1.10). At least five areas per cell were scanned, and at least six cells from three samples were analyzed.
TEM
Whole, 7-d-old seedlings were vacuum infiltrated with 2.5% glutaraldehyde at 4°C for 6 h. After prefixing, the samples were gently rinsed with PBS solution (pH 6.8) for 4 times, 15 min each. Samples were fixed with 1% osmic acid at room temperature for 4 h, followed by three-times rinsing, 10 min each. The procedure of dehydration and infiltration with LR White resin was performed as described by Verhertbruggen et al. (2017). After complete polymerization in capsules in the absence of oxygen, at 55°C, the root elongation zone of LR White–embedded material were sectioned transversely with a diamond knife mounted on a Leica UC7 ultramicrotome to obtain ultra-thin sections of 70-nm thickness. After transfer to copper grids, samples were stained with uranyl acetate for 20 min and gently rinsed with ddH2O. Then, samples were counterstained with lead citrate for 10 min, rinsed with ddH2O and, after drying for 2 d, TEM images captured using a JEOL JEM-1230 under a voltage of 80 kV. Image files were analyzed by ImageJ to measure the thickness of the cell walls.
Yeast viability assay
Yeast cells of the strain BY4742 were cultured in yeast extract peptone dextrose medium at 30°C in the presence of either CW (0.25 µg/ml), 2-DG (0.01% [wt/vol]), SDS (2 µg/ml), Amphotericin B (0.5 µg/ml), or Voriconazole (0.25 µg/ml) or combinations of two of these substances. Every 2 h samples were taken, from which cell counts were determined using a hemocytometer. The data were analyzed using Microsoft Excel. Each assay was repeated three times. The synergistic effect was quantified as the average number of cells in culture after 24 h cultivation in the presence of two substances divided by the average number of cells of each treatment alone.
Field emission–scanning EM
Yeast cells were cultured as described above. For the different pretreatments 5 µg/ml CW, 0.04 mg/ml SDS, or 0.02% (wt/vol) 2-DG were added 2 h before fixation. Cells were fixed in 4% glutaraldehyde at 4°C for 4 h followed by gradient dehydration in eight steps. Critical-point drying was performed using a Leica EM CPD300 (Leica Microsystems) in automatic mode. Dried samples were immobilized on double-sided carbon tape on a SEM sample stage and coated with Pt using a Quorum Q150T sputter coater (Quorum) at 30 mA for 80 s. Images were acquired using a FEI Nova Nano SEM-450 (FEI) at a magnification of at least 10,000 times with an in-lens detector at 5 or 10 kV. Images were analyzed with Image J (Schindelin et al., 2012).
Statistical analysis
Welch’s two-sided t test was used to assess significance of difference between data points. A difference was considered significant when P < 0.05. Data distribution was assumed to be normal, but this was not formally tested. Pairwise Pearson Product Moment correlation was performed in SigmaPlot (Version 12.5; Systat Software). A correlation was considered significant when P < 0.05.
Online supplemental material
Fig. S1 complements Fig. 1 by providing further details on the properties of different quenchers. The correlation of quenching efficiency with spectral overlap of quencher absorption and dye emission (Fig. S1) supports the hypothesis that TB quenching is based on Förster resonance energy transfer. Fig. S2 provides an image from plasmolysis experiments performed on onion epidermis cells stained with FM4-64, indicating that the dye is associated with the plasma membrane and not the cell wall. Fig. S3 illustrates the applicability of the quenching assay to plant leaves. Fig. S4 provides quenchable fractions and quenching efficiencies in the various cell wall mutants used in this study, thereby complementing the information provided in Fig. 3 and Fig. 6. Fig. S5 shows how the measurement regions to quantify fluorescence intensity were defined on different types of samples.
Supplementary Material
Acknowledgments
We thank Daniel Cosgrove and Charles T. Anderson (Penn State University) for critical discussion and helpful comments on the manuscript. Imaging was performed at the Center for Advanced Bioimaging Denmark and Life Science Research Core Services of Northwest A&F University.
T.G. Pomorski acknowledges support from the Villum Foundation (project grant number 022868). J. Liesche acknowledges support from the National Science Foundation China (project grant number 31700313) and the Science Foundation of Shaanxi Province (100 Talent Program).
The authors declare no competing financial interests.
Author contributions: T.G. Pomorski and J. Liesche devised the quenching assay and control experiments. X. Liu and J. Liesche devised the experimental plan for plant cell wall analysis. J. Liesche performed FLIM and conducted the quenching experiments on vesicles and mammalian cells. X. Liu conducted quenching experiments on bacteria, yeast, and plants, and measured cellulose fibril spacing and yeast viability. X. Liu and J. Li performed the EM. S.Chen, H. Zhao, and B. Liu performed the AFM. Johannes Liesche wrote the manuscript with support of all authors.
References
- Adani F., Papa G., Schievano A., Cardinale G., D’Imporzano G., and Tambone F.. 2011. Nanoscale structure of the cell wall protecting cellulose from enzyme attack. Environ. Sci. Technol. 45:1107–1113. 10.1021/es1020263 [DOI] [PubMed] [Google Scholar]
- Anderson C.T., Carroll A., Akhmetova L., and Somerville C.. 2010. Real-time imaging of cellulose reorientation during cell wall expansion in Arabidopsis roots. Plant Physiol. 152:787–796. 10.1104/pp.109.150128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angelova M., Soleau S., Meleard P., Faucon J.F., and Bothorel P.. 1992. Preparation of giant vesicles by external AC electric fields. Kinetics and applications. Prog. Colloid Polym. Sci. 89:127–131. 10.1007/BFb0116295 [DOI] [Google Scholar]
- ASDCD Synergistic drug combination DataBase. http://asdcd.amss.ac.cn/; last accessed 20. October 2018.
- Austin R.S., Vidaurre D., Stamatiou G., Breit R., Provart N.J., Bonetta D., Zhang J., Fung P., Gong Y., Wang P.W., et al. 2011. Next-generation mapping of Arabidopsis genes. Plant J. 67:715–725. 10.1111/j.1365-313X.2011.04619.x [DOI] [PubMed] [Google Scholar]
- Bidhendi A.J., and Geitmann A.. 2016. Relating the mechanics of the primary plant cell wall to morphogenesis. J. Exp. Bot. 67:449–461. 10.1093/jxb/erv535 [DOI] [PubMed] [Google Scholar]
- Boanares D., Ferreira B.G., Kozovits A.R., Sousa H.C., Isaias R.M.S., and França M.G.C.. 2018. Pectin and cellulose cell wall composition enables different strategies to leaf water uptake in plants from tropical fog mountain. Plant Physiol. Biochem. 122:57–64. 10.1016/j.plaphy.2017.11.005 [DOI] [PubMed] [Google Scholar]
- Bolte S., Talbot C., Boutte Y., Catrice O., Read N.D., and Satiat-Jeunemaitre B.. 2004. FM-dyes as experimental probes for dissecting vesicle trafficking in living plant cells. J. Microsc. 214:159–173. 10.1111/j.0022-2720.2004.01348.x [DOI] [PubMed] [Google Scholar]
- Bosca S., Barton C.J., Taylor N.G., Ryden P., Neumetzler L., Pauly M., Roberts K., and Seifert G.J.. 2006. Interactions between MUR10/CesA7-dependent secondary cellulose biosynthesis and primary cell wall structure. Plant Physiol. 142:1353–1363. 10.1104/pp.106.087700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown E.D., and Wright G.D.. 2016. Antibacterial drug discovery in the resistance era. Nature. 529:336–343. 10.1038/nature17042 [DOI] [PubMed] [Google Scholar]
- Burnham-Marusich A.R., Hubbard B., Kvam A.J., Gates-Hollingsworth M., Green H.R., Soukup E., Limper A.H., and Kozel T.R.. 2018. Conservation of Mannan Synthesis in Fungi of the Zygomycota and Ascomycota Reveals a Broad Diagnostic Target. MSphere. 3:3 10.1128/mSphere.00094-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton R.A., Gidley M.J., and Fincher G.B.. 2010. Heterogeneity in the chemistry, structure and function of plant cell walls. Nat. Chem. Biol. 6:724–732. 10.1038/nchembio.439 [DOI] [PubMed] [Google Scholar]
- Carroll A., and Somerville C.. 2009. Cellulosic Biofuels Annual Review of Plant Biology. Annual Review of Plant Biology. Vol. 60 Annual Reviews, Palo Alto: 165–182. [DOI] [PubMed] [Google Scholar]
- Cavalier D.M., Lerouxel O., Neumetzler L., Yamauchi K., Reinecke A., Freshour G., Zabotina O.A., Hahn M.G., Burgert I., Pauly M., et al. 2008. Disrupting two Arabidopsis thaliana xylosyltransferase genes results in plants deficient in xyloglucan, a major primary cell wall component. Plant Cell. 20:1519–1537. 10.1105/tpc.108.059873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S., Jia H., Zhao H., Liu D., Liu Y., Liu B., Bauer S., and Somerville C.R.. 2016. Anisotropic Cell Expansion Is Affected through the Bidirectional Mobility of Cellulose Synthase Complexes and Phosphorylation at Two Critical Residues on CESA3. Plant Physiol. 171:242–250. 10.1104/pp.15.01874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comas L.H., Becker S.R., Cruz V.M., Byrne P.F., and Dierig D.A.. 2013. Root traits contributing to plant productivity under drought. Front. Plant Sci. 4:442 10.3389/fpls.2013.00442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosgrove D.J. 2000. Loosening of plant cell walls by expansins. Nature. 407:321–326. 10.1038/35030000 [DOI] [PubMed] [Google Scholar]
- Cosgrove D.J. 2016. Plant cell wall extensibility: connecting plant cell growth with cell wall structure, mechanics, and the action of wall-modifying enzymes. J. Exp. Bot. 67:463–476. 10.1093/jxb/erv511 [DOI] [PubMed] [Google Scholar]
- Crasnier M., Moustacas A.-M., and Ricard J.. 1985. Electrostatic effects and calcium ion concentration as modulators of acid phosphatase bound to plant cell walls. Eur. J. Biochem. 151:187–190. 10.1111/j.1432-1033.1985.tb09084.x [DOI] [PubMed] [Google Scholar]
- Crisalli P., and Kool E.T.. 2011. Multi-path quenchers: efficient quenching of common fluorophores. Bioconjug. Chem. 22:2345–2354. 10.1021/bc200424r [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Nobel J.G., Klis F.M., Munnik T., Priem J., and van den Ende H.. 1990. An assay of relative cell wall porosity in Saccharomyces cerevisiae, Kluyveromyces lactis and Schizosaccharomyces pombe. Yeast. 6:483–490. 10.1002/yea.320060605 [DOI] [PubMed] [Google Scholar]
- Derksen J., Janssen G.J., Wolters-Arts M., Lichtscheidl I., Adlassnig W., Ovecka M., Doris F., and Steer M.. 2011. Wall architecture with high porosity is established at the tip and maintained in growing pollen tubes of Nicotiana tabacum. Plant J. 68:495–506. 10.1111/j.1365-313X.2011.04703.x [DOI] [PubMed] [Google Scholar]
- Desprez T., Vernhettes S., Fagard M., Refrégier G., Desnos T., Aletti E., Py N., Pelletier S., and Höfte H.. 2002. Resistance against herbicide isoxaben and cellulose deficiency caused by distinct mutations in same cellulose synthase isoform CESA6. Plant Physiol. 128:482–490. 10.1104/pp.010822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diet A., Link B., Seifert G.J., Schellenberg B., Wagner U., Pauly M., Reiter W.D., and Ringli C.. 2006. The Arabidopsis root hair cell wall formation mutant lrx1 is suppressed by mutations in the RHM1 gene encoding a UDP-L-rhamnose synthase. Plant Cell. 18:1630–1641. 10.1105/tpc.105.038653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding S.Y., Liu Y.S., Zeng Y., Himmel M.E., Baker J.O., and Bayer E.A.. 2012. How does plant cell wall nanoscale architecture correlate with enzymatic digestibility? Science. 338:1055–1060. 10.1126/science.1227491 [DOI] [PubMed] [Google Scholar]
- Dixon R.A. 2013. Microbiology: Break down the walls. Nature. 493:36–37. 10.1038/493036a [DOI] [PubMed] [Google Scholar]
- Donaldson L.A., Kroese H.W., Hill S.J., and Franich R.A.. 2015. Detection of wood cell wall porosity using small carbohydrate molecules and confocal fluorescence microscopy. J. Microsc. 259:228–236. 10.1111/jmi.12257 [DOI] [PubMed] [Google Scholar]
- Ellsworth P.V., Ellsworth P.Z., Koteyeva N.K., and Cousins A.B.. 2018. Cell wall properties in Oryza sativa influence mesophyll CO2 conductance. New Phytol. 219:66–76. 10.1111/nph.15173 [DOI] [PubMed] [Google Scholar]
- Gow N.A.R., Latge J.P., and Munro C.A.. 2017. The Fungal Cell Wall: Structure, Biosynthesis, and Function. Microbiol. Spectr. 5:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffing L.R. 2008. FRET analysis of transmembrane flipping of FM4-64 in plant cells: is FM4-64 a robust marker for endocytosis? J. Microsc. 231:291–298. 10.1111/j.1365-2818.2008.02042.x [DOI] [PubMed] [Google Scholar]
- Himmel M.E., Ding S.Y., Johnson D.K., Adney W.S., Nimlos M.R., Brady J.W., and Foust T.D.. 2007. Biomass recalcitrance: engineering plants and enzymes for biofuels production. Science. 315:804–807. 10.1126/science.1137016 [DOI] [PubMed] [Google Scholar]
- Huang K.C., Mukhopadhyay R., Wen B., Gitai Z., and Wingreen N.S.. 2008. Cell shape and cell-wall organization in Gram-negative bacteria. Proc. Natl. Acad. Sci. USA. 105:19282–19287. 10.1073/pnas.0805309105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islahudin F., Khozoie C., Bates S., Ting K.-N., Pleass R.J., and Avery S.V.. 2013. Cell wall perturbation sensitizes fungi to the antimalarial drug chloroquine. Antimicrob. Agents Chemother. 57:3889–3896. 10.1128/AAC.00478-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klemm D., Heublein B., Fink H.P., and Bohn A.. 2005. Cellulose: fascinating biopolymer and sustainable raw material. Angew. Chem. Int. Ed. Engl. 44:3358–3393. 10.1002/anie.200460587 [DOI] [PubMed] [Google Scholar]
- Lambert P.A. 2002. Cellular impermeability and uptake of biocides and antibiotics in Gram-positive bacteria and mycobacteria. J. Appl. Microbiol. 92(s1, Suppl):46S–54S. 10.1046/j.1365-2672.92.5s1.7.x [DOI] [PubMed] [Google Scholar]
- Lampugnani E.R., Khan G.A., Somssich M., and Persson S.. 2018. Building a plant cell wall at a glance. J. Cell Sci. 131:2017373 10.1242/jcs.207373 [DOI] [PubMed] [Google Scholar]
- Lehrer S.S. 1971. Solute perturbation of protein fluorescence. The quenching of the tryptophyl fluorescence of model compounds and of lysozyme by iodide ion. Biochemistry. 10:3254–3263. 10.1021/bi00793a015 [DOI] [PubMed] [Google Scholar]
- Liesche J., Ziomkiewicz I., and Schulz A.. 2013. Super-resolution imaging with Pontamine Fast Scarlet 4BS enables direct visualization of cellulose orientation and cell connection architecture in onion epidermis cells. BMC Plant Biol. 13:226 10.1186/1471-2229-13-226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liesche J., Marek M., and Günther-Pomorski T.. 2015. Cell wall staining with Trypan blue enables quantitative analysis of morphological changes in yeast cells. Front. Microbiol. 6:107 10.3389/fmicb.2015.00107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matteï P.J., Neves D., and Dessen A.. 2010. Bridging cell wall biosynthesis and bacterial morphogenesis. Curr. Opin. Struct. Biol. 20:749–755. 10.1016/j.sbi.2010.09.014 [DOI] [PubMed] [Google Scholar]
- Meeske A.J., Sham L.T., Kimsey H., Koo B.M., Gross C.A., Bernhardt T.G., and Rudner D.Z.. 2015. MurJ and a novel lipid II flippase are required for cell wall biogenesis in Bacillus subtilis. Proc. Natl. Acad. Sci. USA. 112:6437–6442. 10.1073/pnas.1504967112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra N.N., Prasad T., Sharma N., Payasi A., Prasad R., Gupta D.K., and Singh R.. 2007. Pathogenicity and drug resistance in Candida albicans and other yeast species. A review. Acta Microbiol. Immunol. Hung. 54:201–235. 10.1556/AMicr.54.2007.3.1 [DOI] [PubMed] [Google Scholar]
- O’Neill M.A., Eberhard S., Albersheim P., and Darvill A.G.. 2001. Requirement of borate cross-linking of cell wall rhamnogalacturonan II for Arabidopsis growth. Science. 294:846–849. 10.1126/science.1062319 [DOI] [PubMed] [Google Scholar]
- Okada H., Kono K., Neiman A.M., and Ohya Y.. 2016. Examination and Disruption of the Yeast Cell Wall. Cold Spring Harb. Protoc. 2016:pdb.top078659 10.1101/pdb.top078659 [DOI] [PubMed] [Google Scholar]
- Park Y.B., and Cosgrove D.J.. 2012. A revised architecture of primary cell walls based on biomechanical changes induced by substrate-specific endoglucanases. Plant Physiol. 158:1933–1943. 10.1104/pp.111.192880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiter W.D., Chapple C., and Somerville C.R.. 1997. Mutants of Arabidopsis thaliana with altered cell wall polysaccharide composition. Plant J. 12:335–345. 10.1046/j.1365-313X.1997.12020335.x [DOI] [PubMed] [Google Scholar]
- Rolinski O.J., Birch D.J.S., McCartney L.J., and Pickup J.C.. 1999. Fluorescence resonance energy transfer from allophycocyanin to malachite green. Chem. Phys. Lett. 309:395–401. 10.1016/S0009-2614(99)00707-1 [DOI] [Google Scholar]
- Savaldi-Goldstein S., Peto C., and Chory J.. 2007. The epidermis both drives and restricts plant shoot growth. Nature. 446:199–202. 10.1038/nature05618 [DOI] [PubMed] [Google Scholar]
- Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., et al. 2012. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 9:676–682. 10.1038/nmeth.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider R., and Persson S.. 2015. Another brick in the wall. Science. 350:156–157. 10.1126/science.aad3200 [DOI] [PubMed] [Google Scholar]
- Somerville C., Bauer S., Brininstool G., Facette M., Hamann T., Milne J., Osborne E., Paredez A., Persson S., Raab T., et al. 2004. Toward a systems approach to understanding plant cell walls. Science. 306:2206–2211. 10.1126/science.1102765 [DOI] [PubMed] [Google Scholar]
- Strober W. 2015. Trypan Blue Exclusion Test of Cell Viability. Curr. Protoc. Immunol. 111:1–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tateno M., Brabham C., and DeBolt S.. 2016. Cellulose biosynthesis inhibitors - a multifunctional toolbox. J. Exp. Bot. 67:533–542. 10.1093/jxb/erv489 [DOI] [PubMed] [Google Scholar]
- Tavares E.Q.P., De Souza A.P., and Buckeridge M.S.. 2015. How endogenous plant cell-wall degradation mechanisms can help achieve higher efficiency in saccharification of biomass. J. Exp. Bot. 66:4133–4143. 10.1093/jxb/erv171 [DOI] [PubMed] [Google Scholar]
- Taylor-Teeples M., Lin L., de Lucas M., Turco G., Toal T.W., Gaudinier A., Young N.F., Trabucco G.M., Veling M.T., Lamothe R., et al. 2015. An Arabidopsis gene regulatory network for secondary cell wall synthesis. Nature. 517:571–575. 10.1038/nature14099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenhaken R. 2015. Cell wall remodeling under abiotic stress. Front. Plant Sci. 5:771 10.3389/fpls.2014.00771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner R.D., Hurd A.F., Cadby A., Hobbs J.K., and Foster S.J.. 2013. Cell wall elongation mode in Gram-negative bacteria is determined by peptidoglycan architecture. Nat. Commun. 4:1496 10.1038/ncomms2503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhertbruggen Y., Walker J.L., Guillon F., and Scheller H.V.. 2017. A Comparative Study of Sample Preparation for Staining and Immunodetection of Plant Cell Walls by Light Microscopy. Front. Plant Sci. 8:1505 10.3389/fpls.2017.01505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vida T.A., and Emr S.D.. 1995. A new vital stain for visualizing vacuolar membrane dynamics and endocytosis in yeast. J. Cell Biol. 128:779–792. 10.1083/jcb.128.5.779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilhelm M.J., Sharifian Gh M., and Dai H.L.. 2015. Chemically Induced Changes to Membrane Permeability in Living Cells Probed with Nonlinear Light Scattering. Biochemistry. 54:4427–4430. 10.1021/acs.biochem.5b00600 [DOI] [PubMed] [Google Scholar]
- Winstel V., Liang C., Sanchez-Carballo P., Steglich M., Munar M., Bröker B.M., Penadés J.R., Nübel U., Holst O., Dandekar T., et al. 2013. Wall teichoic acid structure governs horizontal gene transfer between major bacterial pathogens. Nat. Commun. 4:2345 10.1038/ncomms3345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y., Yeh F.L., Mao F., and Chapman E.R.. 2009. Biophysical characterization of styryl dye-membrane interactions. Biophys. J. 97:101–109. 10.1016/j.bpj.2009.04.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao C., Zhang T., Zheng Y., Cosgrove D.J., and Anderson C.T.. 2016. Xyloglucan Deficiency Disrupts Microtubule Stability and Cellulose Biosynthesis in Arabidopsis, Altering Cell Growth and Morphogenesis. Plant Physiol. 170:234–249. 10.1104/pp.15.01395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young K.D. 2016. Microbiology: The bacterial cell wall takes centre stage. Nature. 537:622–624. 10.1038/537622a [DOI] [PubMed] [Google Scholar]
- Zhang P., Lock L.L., Cheetham A.G., and Cui H.. 2014. Enhanced cellular entry and efficacy of tat conjugates by rational design of the auxiliary segment. Mol. Pharm. 11:964–973. 10.1021/mp400619v [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y., Cosgrove D.J., and Ning G.. 2017. High-Resolution Scanning Electron Microscopy (FESEM) Imaging of Cellulose Microfibril Organization in Plant Primary Cell Walls. Microsc. Microanal. 23:1048–1054. 10.1017/S143192761701251X [DOI] [PubMed] [Google Scholar]
- Zu F.L., Yan F.Y., Bai Z.J., Xu J.X., Wang Y.Y., Huang Y.C., and Zhou X.G.. 2017. The quenching of the fluorescence of carbon dots: A review on mechanisms and applications. Mikrochim. Acta. 184:1899–1914. 10.1007/s00604-017-2318-9 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.