Bae et al. show that degradation of the mRNA encoding the adaptor Blos1 leads to the repositioning of late endosomes/lysosomes to the microtubule-organizing center in response to ER stress. This repositioning enhances cell survival during stress by promoting the clearance of protein aggregates.
Abstract
Cells respond to stress in the ER by initiating the widely conserved unfolded protein response. Activation of the ER transmembrane nuclease IRE1 leads to the degradation of specific mRNAs, but how this pathway affects the ability of cells to recover from stress is not known. Here, we show that degradation of the mRNA encoding biogenesis of lysosome-related organelles 1 subunit 1 (Blos1) leads to the repositioning of late endosomes (LEs)/lysosomes to the microtubule-organizing center in response to stress in mouse cells. Overriding Blos1 degradation led to ER stress sensitivity and the accumulation of ubiquitinated protein aggregates, whose efficient degradation required their independent trafficking to the cell center and the LE-associated endosomal sorting complexes required for transport. We propose that Blos1 regulation by IRE1 promotes LE-mediated microautophagy of protein aggregates and protects cells from their cytotoxic effects.
Introduction
Homeostasis in the ER is maintained through a conserved collection of mechanisms termed the unfolded protein response (UPR; Walter and Ron, 2011). The UPR in mammals is essential for development and is induced in many diseases, including cancer and neurodegenerative disorders (Hetz et al., 2013). Perhaps the least understood branch of the UPR, in terms of its biological function, is the degradation of mRNAs by the transmembrane nuclease inositol requiring enzyme 1 (IRE1; Hollien and Weissman, 2006; Hollien et al., 2009). IRE1 is activated by ER stress, defined as an imbalance between the load on the ER and its protein-processing capacity. IRE1’s cytosolic nuclease domain cleaves the mRNA encoding the transcription factor XBP1, initiating a splicing event that is required to produce the active XBP1 (Yoshida et al., 2001; Calfon et al., 2002; Lee et al., 2002), which then up-regulates genes involved in ER protein folding, processing, and degradation. IRE1 also cleaves and initiates the degradation of other mRNAs associated with the ER membrane. This pathway, termed regulated IRE1-dependent decay (RIDD), is independent of XBP1 and conserved across many species (Kimmig et al., 2012; Coelho et al., 2013; Levi-Ferber et al., 2015). In mammalian cells, IRE1 typically degrades only a few mRNAs that contain specific translationally stalled stem-loop structures (Moore and Hollien, 2015), making this an unlikely mechanism to reduce the protein folding load on the ER. In mice, degradation of particular RIDD targets has cell type–specific effects (So et al., 2012; Benhamron et al., 2014; Osorio et al., 2014). Whether there is a general function for RIDD, and how it affects the ability of mammalian cells to respond effectively to ER stress, are not known.
The most robust and consistently identified target of RIDD in mammalian cells (Bright et al., 2015) encodes biogenesis of lysosome-related organelles complex 1 (BLOC1) subunit 1, referred to here as BLOS1 and also known as general control of amino acid synthesis 5–like 1. BLOC1 mediates the formation of endosomal tubular structures and is important for sorting proteins to recycling endosomes and lysosome-related structures such as melanosomes (Delevoye et al., 2016; Dennis et al., 2016). Unlike some members of this complex, however, BLOS1 is essential for survival of mice (Scott et al., 2013; Zhang et al., 2014), perhaps due to its seemingly independent role in regulating mitochondrial protein acetylation, turnover, and metabolism (Scott et al., 2013, 2018; Wang et al., 2017).
BLOS1 also regulates lysosome trafficking in response to nutrient availability and growth factors (Pu et al., 2015; Filipek et al., 2017). BLOS1 and two other BLOC1 subunits form part of a second complex, the BLOC1-related complex (BORC; Pu et al., 2015). BORC couples late endosomes (LEs)/lysosomes to the small GTPase ARL8B and kinesin, thereby allowing for microtubule-based transport of lysosomes to the cell periphery (Pu et al., 2015; Guardia et al., 2016) and to the axon in neurons (Farías et al., 2017). Cells lacking BORC can traffic LEs/lysosomes to the cell center via dynein, and therefore display a characteristic clustering of LEs/lysosomes next to the nucleus (Pu et al., 2015). Similar lysosome clustering occurs in cultured cells deprived of serum (Korolchuk et al., 2011), which inhibits BORC function (Pu et al., 2017). This response is thought to enhance macroautophagy, the stress-regulated process by which cytosolic material is sequestered by double-membraned autophagosomes and degraded via fusion of these vesicles with lysosomes (Yin et al., 2016).
It has been unclear how the various functions reported for BLOS1 are related, and how regulation of BLOS1 by ER stress might affect trafficking in the endo-lysosomal system. Here, we show that degradation of the Blos1 mRNA by RIDD leads to LE/lysosomal repositioning and affects the clearance of protein aggregates during ER stress.
Results and discussion
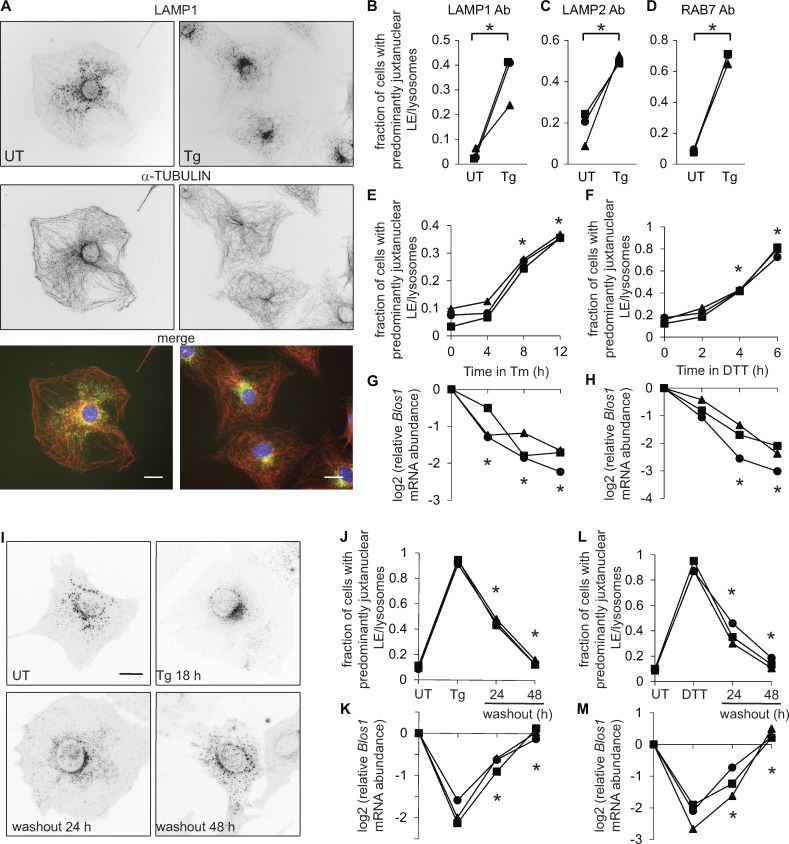
We predicted that during ER stress, when RIDD degrades the Blos1 mRNA, LEs and/or lysosomes would accumulate near the microtubule-organizing center (MTOC). To test this, we treated mouse MC3T3-E1 cells with thapsigargin (Tg), which induces ER stress by releasing calcium from the ER, then stained with antibodies for tubulin and the LE/lysosome marker LAMP1. In response to Tg, LAMP1 foci shifted from a disperse, cytosolic distribution to a condensed area on one side of the nucleus near the MTOC (Fig. 1, A and B), similar to the phenotype observed in BLOS1 knockdown or knockout cells (Pu et al., 2015).
Figure 1.
LEs/lysosomes cluster near the MTOC during ER stress. For all graphs throughout the figures: symbols represent independent experiments and are maintained among related panels; lines connect data from the same replicate experiment. (A) We treated MC3T3-E1 cells with Tg (2 µM, 18 h), fixed and stained with DAPI (blue) and antibodies for LAMP1 (green) and α-tubulin (red), and imaged on a wide-field fluorescence microscope. Nonmerged images are inverted for visualization. (B–D) We treated cells as in A; stained with antibodies for LAMP1, LAMP2, or RAB7; and scored cells for juxtanuclear LE/lysosome clustering. (E–H) We treated MC3T3-E1 cells stably expressing LAMP1-GFP with Tm (4 µg/ml) or DTT (2 mM), and then imaged live cells and collected RNA in parallel. We measured relative Blos1 mRNA levels by quantitative real-time RT-PCR. (I–M) We treated LAMP1-GFP cells with Tg (2 µM, 18 h) or DTT (2 mM, 4 h), then washed out the stressor to follow recovery. *P < 0.05, paired t test (B–D) or ANOVA followed by Tukey’s honestly significant difference test (E–H and J–M); n = 3. Only comparisons between stressed versus UT cells (E–H) or before versus after washout (J–M) are shown. Scale bars, 10 µm. Ab, antibody; UT, untreated.
We observed this clustering in fixed cells with an alternative LE/lysosome marker, LAMP2 (Fig. 1 C), with a more specific LE marker, RAB7 (Cheng et al., 2018; Fig. 1 D), and in living cells expressing LAMP1-GFP (Fig. 1, E and F). We also observed LE/lysosome clustering in response to other inducers of ER stress: tunicamycin (Tm), which inhibits N-linked glycosylation, and dithiothreitol (DTT), which reduces disulfide bonds (Fig. 1, E and F). The timing of LE/lysosome repositioning was similar to that of Blos1 mRNA degradation (Fig. 1, E–H). We did not detect any major changes in the distribution of mitochondria, microtubules, or actin (Fig. S1, A–C). The effect was conserved, although less dramatic, in HeLa cells (Fig. S1, D–H). Although we could not detect endogenous BLOS1 protein in mouse cells, we measured a significant decline in BLOS1 protein levels in HeLa cells in response to Tg (Fig. S1, G and H), consistent with the loss of BLOS1 leading to LE/lysosome repositioning. Finally, the effect was reversible in MC3T3-E1 cells that recovered from ER stress: washing out Tg or DTT led to a recovery of both peripheral LE/lysosome positioning and Blos1 mRNA levels (Fig. 1, J–M).
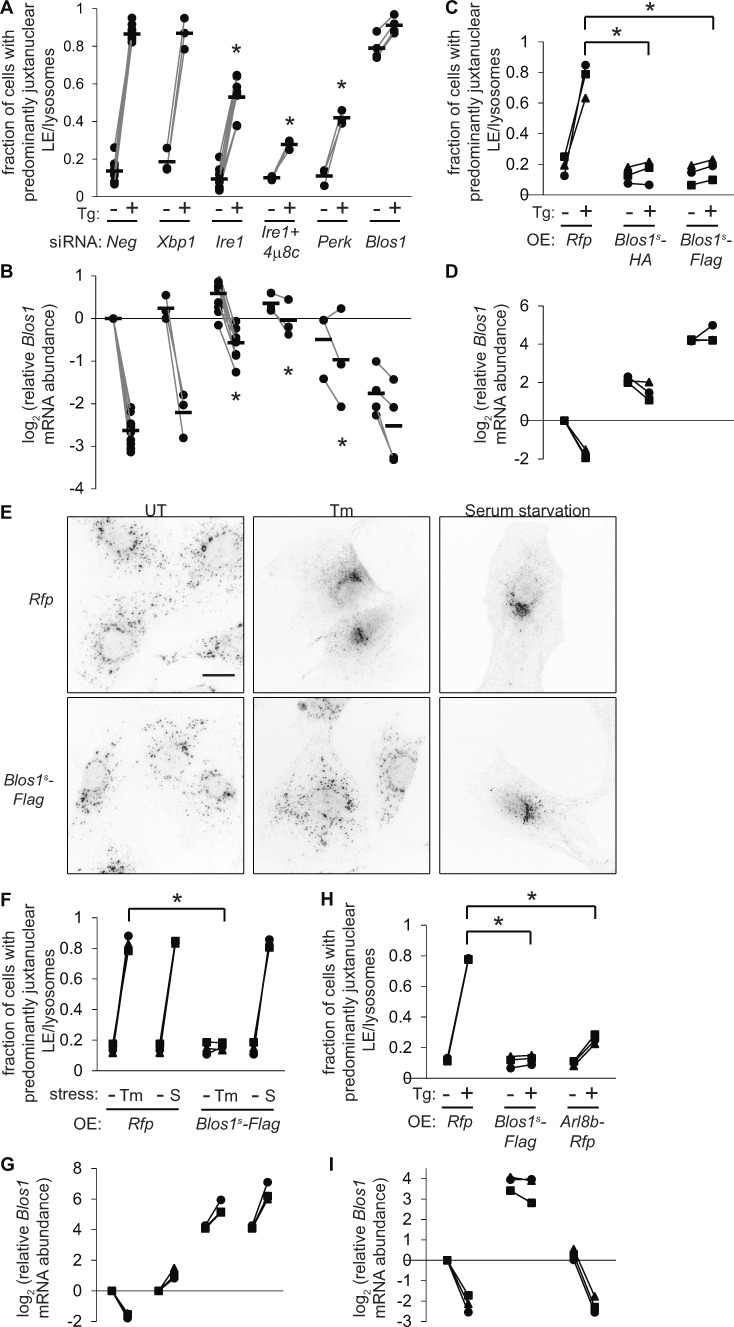
We depleted LAMP1-GFP cells of various UPR effectors and found that LE/lysosomal repositioning required IRE1 but not XBP1 (Fig. 2, A and B; and Fig. S2, A and B). Although the effect of IRE1 knockdown was incomplete, addition of the IRE1 nuclease inhibitor 4μ8c further prevented both Blos1 degradation and LE/lysosomal clustering (Fig. 2, A and B). Repositioning was also dependent on PKR-like ER kinase (PERK; Fig. 2, A and B), a second sensor of ER stress, which attenuates translation and is necessary for degradation of RIDD targets like Blos1 but is dispensable for Xbp1 splicing (Moore and Hollien, 2015). To test whether degradation of Blos1 specifically is required, we constructed stable cell lines overexpressing a stabilized variant of the Blos1 mRNA (Blos1s), which contains a silent point mutation (G360C in the coding sequence) that prevents its degradation by RIDD (Moore and Hollien, 2015). Expression of Blos1s completely blocked LE/lysosome repositioning during ER stress (Fig. 2, C–G) but did not affect Xbp1 splicing or degradation of other RIDD targets (Fig. S2, C–G). Blos1s expression also had no effect on LE/lysosome repositioning during serum starvation (Korolchuk et al., 2011; Pu et al., 2017; Fig. 2, E and F), suggesting that these cells are not generally compromised in their ability to traffic LEs/lysosomes and that RIDD of Blos1 is a novel mechanism controlling lysosome clustering.
Figure 2.
LE/lysosome repositioning during ER stress is dependent on the degradation of Blos1 mRNA by RIDD. (A) We transfected MC3T3-E1 cells expressing LAMP1-GFP with siRNAs targeting UPR effectors or control siRNAs (Neg), then treated with Tg (2 µM, 12 h). Where indicated, we also added the IRE1 inhibitor 4μ8c (60 µM) 5 min before Tg. We then imaged and scored cells for juxtanuclear LE/lysosome clustering. (B) We collected RNA samples in parallel with A and measured Blos1 mRNA by qPCR. (C–G) We treated MC3T3-E1 cells stably expressing Rfp, Blos1s-HA, or Blos1s-Flag with Tg (2 µM), Tm (6 µg/ml), or serum-free media (S; 18 h). We then analyzed LE/lysosome clustering by LAMP1 immunostaining and Blos1 mRNA levels by qPCR. Panel E shows representative images, scale bar = 10 µm. (H and I) We incubated MC3T3-E1 cells stably expressing Rfp or Arl8b–Rfp with Tg (2 µM, 18 h), then analyzed as in C and D. *P < 0.05, stressed knockdown or overexpression cells versus controls, determined by ANOVA followed by Tukey’s honestly significant difference (A and B), or paired t tests corrected for multiple comparisons (C, F, and H), n = 3 except in A and B where the number of experiments is indicated by the symbols. UT, untreated; OE, overexpressed.
Control of LE/lysosome trafficking by BORC relies on its interaction with ARL8B. We therefore transfected MC3T3-E1 cells with ARL8B-RFP, whose overexpression can drive LEs/lysosomes to the periphery (Hofmann and Munro, 2006; Pu et al., 2015). ARL8B-overexpressing cells degraded Blos1 mRNA during ER stress (Fig. 2 I), but we observed LE/lysosome clustering in only 25% of cells (Fig. 2 H).
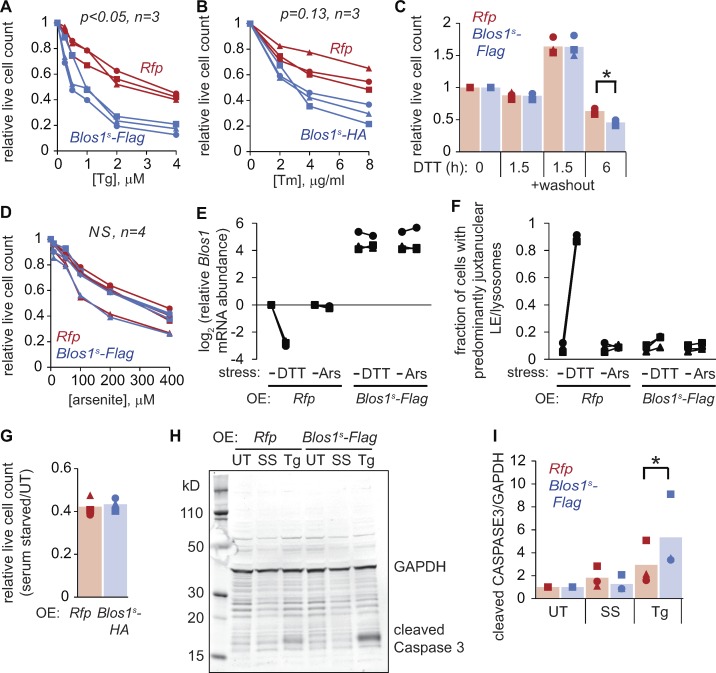
To understand the consequences of LE/lysosome repositioning during ER stress, we compared the stress sensitivity of cells expressing Blos1s with those expressing Rfp as a control. Blos1s cells died more readily after exposure to Tg, Tm, or 6 h DTT (Fig. 3, A–C) and accumulated cleaved caspase 3 to a higher degree than control cells (Fig. 3, H and I). The two cell lines were equally sensitive to short DTT treatments (1.5 h; Fig. 3 C), as reported previously (Bright et al., 2015) and consistent with the effect being dependent on LE/lysosome positioning. Blos1s expression did not sensitize cells to arsenite-induced stress (Fig. 3, D–F) or serum starvation (Fig. 3, G–I), suggesting a specific defect for Blos1s cells in the response to ER stress.
Figure 3.
Overriding RIDD of Blos1 sensitizes cells to ER stress. (A–D and G) We treated MC3T3-E1 cells stably expressing the indicated mRNAs with Tg or Tm (18 h), 2 mM DTT, arsenite (6 h), or serum-free media (18 h). We then counted live cells or washed out the DTT and counted cells after 24 h. (E and F) We treated cells with DTT (2 mM, 4 h) or arsenite (50 µM, 6 h) and measured Blos1 mRNA by qPCR and LE/lysosome clustering by LAMP1 immunostaining. (H and I) We incubated cells with Tg (4 µM) or serum-free media for 18 h, then measured cleaved caspase 3 levels by immunoblotting. For A, B, and D: P values were determined by two-factor ANOVA and compared Rfp with Blos1s cell lines. For C and I: *P < 0.05, paired t test for Rfp versus Blos1s cell lines, corrected for multiple comparisons. n = 3 except where indicated. UT, untreated; OE, overexpressed; Ars, arsenite; SS, serum starvation.
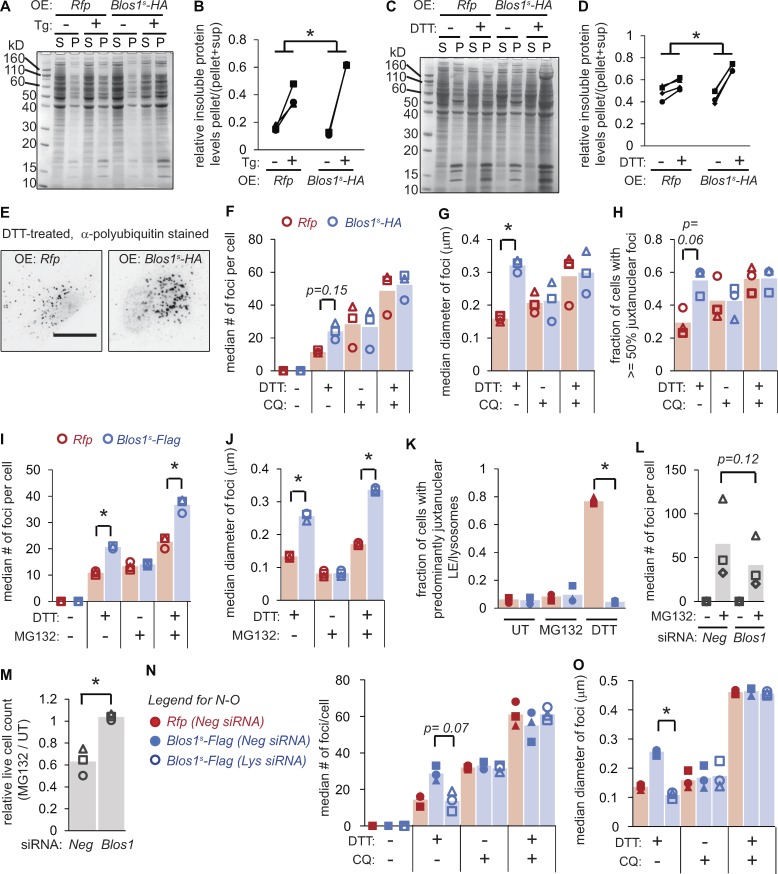
One way that LE/lysosome positioning could influence ER stress sensitivity is through the degradation of aggregated proteins. Cytosolic misfolded proteins are well-established substrates for lysosomes (Jackson and Hewitt, 2016), and certain aggregates are sequestered from the bulk cytosol by trafficking to the nuclear region in a manner reminiscent of the LE/lysosome clustering described here (Johnston et al., 1998). We therefore hypothesized that Blos1 degradation enhances the ability of lysosomes to degrade protein aggregates, by bringing them in close proximity. To test this, we first compared the relative amount of insoluble proteins in Blos1s- or Rfp-expressing cells exposed to ER stress. Blos1s cells accumulated higher levels of insoluble proteins in response to Tg or DTT (Fig. 4, A–D). We next labeled cells with an antibody for polyubiquitin chains. Cells treated with DTT or Tg (but not untreated cells) accumulated foci indicative of protein aggregation (Fig. 4 E and Fig. S3 A). Notably, the aggregates in Blos1s cells were larger and more numerous than those in control cells (Fig. 4, F–G, I, and J; and Fig. S3, A–C), and were predominantly localized to the one side of the nucleus (Fig. 4 H and Fig. S3 D), as predicted for aggregates that would otherwise be degraded by lysosomes near the MTOC.
Figure 4.
Overriding RIDD of Blos1 results in accumulation of protein aggregates. (A–D) We treated MC3T3-E1 cells stably expressing the indicated mRNAs with either Tg (2 µM, 18 h) or DTT (2 mM, 4 h), lysed in RIPA buffer, separated soluble (S) and insoluble (P) material by centrifugation, and analyzed by SDS-PAGE and Coomassie blue staining. A and C are representative gel images, and B and D show quantification of three experiments. *P < 0.05, paired t tests for Rfp compared with Blos1s cells (stressed/UT). (E–H) We treated cells as in C and D, except that we added CQ (120 µM) for the final 2 h of treatment, where indicated. We then fixed and stained cells with an antibody for polyubiquitin and analyzed foci as described in Materials and methods. E shows representative images (inverted, scale bar = 10 µm). (I–K) We treated cells with DTT (2 mM) and/or MG132 (10 µM) for 4 h, and then analyzed polyubiquitin foci as in F and G or LE/lysosome clustering by LAMP immunostaining. (L and M) We depleted MC3T3-E1 cells of Blos1 using siRNAs, then treated with MG132 (10 µM) and analyzed polyubiquitin foci (after 4 h) or counted live cells (after 18 h). (N and O) We depleted cells of the BORC component lyspersin (Lys) using siRNAs, then analyzed polyubiquitin foci as in F and G. Lys mRNA levels in depleted cells were 0.21 ± 0.09, relative to Neg controls, as measured by qPCR. *P < 0.05, Rfp versus Blos1s (F–K) or Neg versus Blos1 or Lys RNAi (L–O), from paired t tests with corrections for multiple comparisons; n = 3. All P values between 0.05 and 0.15 are shown. UT, untreated; OE, overexpressed.
To test whether the aggregate accumulation in Blos1s cells was a result of reduced lysosome-dependent degradation, rather than increased formation, we treated cells with chloroquine (CQ), which blocks the acidification of lysosomes. Including CQ for the final 2 h of the experiment led to increased aggregate accumulation, but we did not detect any differences in the aggregates between Blos1s- and Rfp-expressing cells (Fig. 4, F–H; and Fig. S3, B–D), suggesting that the difference between these cell lines is dependent on lysosomal function. In contrast, treatment of cells with MG132 to block proteasome function resulted in aggregate accumulation that remained exacerbated in Blos1s cells (Fig. 4, I and J), indicating that the enhanced aggregate clearance in control cells is not dependent on the proteasome.
Although MC3T3-E1 cells exposed to MG132 accumulated protein aggregates, they did not reposition their LEs/lysosomes (Fig. 4 K). To test whether clustering LEs/lysosomes at the MTOC would generally enhance aggregate degradation, we depleted cells of Blos1 and monitored their response to MG132. Knockdown of Blos1 reduced the accumulation of protein aggregates and protected cells from MG132 toxicity (Fig. 4, L and M).
To test whether protein aggregate accumulation in Blos1s cells was a consequence of loss of BORC function (rather than some other function of BLOS1), we attempted to reverse aggregate accumulation by depleting Blos1s cells of lyspersin, which is a component of BORC but not BLOC1 (Pu et al., 2015). Knocking down lyspersin resulted in fewer aggregates, similar to those in Rfp-expressing cells (Fig. 4, N and O).
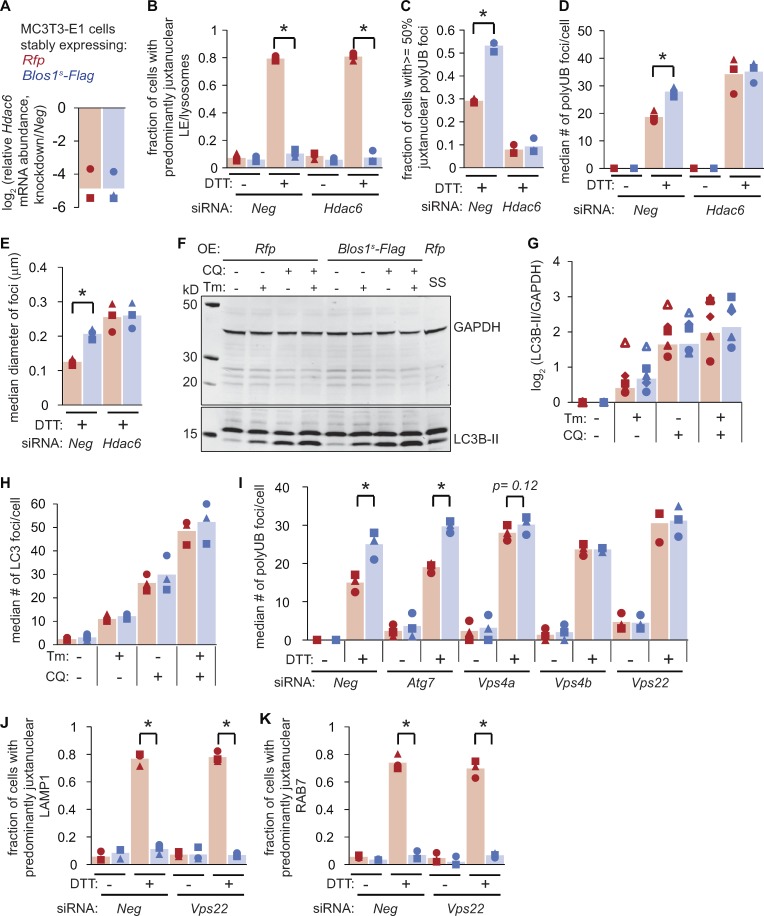
These results suggested that degradation of Blos1 during ER stress enhances the removal of protein aggregates by bringing together LEs/lysosomes with their protein substrates at the MTOC. To test whether the aggregates also need to traffic to the MTOC to be efficiently degraded, we depleted cells of histone deacetylase 6 (HDAC6), which is required for aggregate trafficking (Kawaguchi et al., 2003; Ouyang et al., 2012). HDAC6 knockdown did not affect the repositioning of LEs/lysosomes during ER stress (Fig. 5, A and B). However, while polyubiquitin foci were abundant in HDAC6-depleted, DTT-treated cells (Fig. 5 D), they were more distributed through the periphery of the cells and did not accumulate near the nucleus in either cell line (Fig. 5 C). Furthermore, Blos1s expression did not affect the number or size of these foci in HDAC6-depleted cells (Fig. 5, D and E). These data suggest that protein aggregates traffic independently of LEs/lysosomes, and that their ability to move to the cell center enhances their degradation during ER stress when Blos1 is degraded.
Figure 5.
Protein aggregate clearance during ER stress relies on HDAC6 and the ESCRTs. (A–E) We transfected cells with siRNAs targeting Neg or Hdac6, and then treated with DTT (2 mM, 4 h). We analyzed Hdac6 mRNA levels by qPCR (A), LE/lysosome repositioning by LAMP1 immunostaining (B), or polyubiquitin foci (C–E). (F) We treated cells with Tm (6 µg/ml) or serum-free media as a control (18 h). We added CQ (120 µM), where indicated for the final 2 h, and then analyzed LC3B processing by immunoblot. (G) Quantification of five independent experiments as in F. (H) We treated cells as in F and G, fixed and stained with an LC3B antibody, and counted LC3B foci. (I) We transfected cells with siRNAs targeting autophagy components or control siRNAs (Neg), then treated with DTT (2 mM, 4 h) and analyzed polyubiquitin foci. RNAi controls are shown in Fig. S3. (J and K) We depleted cells of Vps22, treated with DTT (2 mM, 4 h), and analyzed LE/lysosome repositioning by either LAMP1 (J) or RAB7 (K) immunostaining. All panels: *P < 0.05 for Rfp versus Blos1s cells, using paired t tests with corrections for multiple comparisons; P values between 0.05 and 0.15 are shown. n = 3 except for G. OE, overexpressed; SS, serum starvation.
Lysosome repositioning during starvation enhances macroautophagy (Korolchuk et al., 2011), which can degrade protein aggregates (Hyttinen et al., 2014), and BORC also influences autophagosome/lysosome fusion (Jia et al., 2017). We therefore examined macroautophagy by monitoring the processed, lipidated form of LC3B (LC3B-II), which is generated along with autophagosomes and is degraded upon their fusion with lysosomes. As reported previously (Ogata et al., 2006; Yorimitsu and Klionsky, 2007), LC3B-II levels increased during ER stress and increased further when we added CQ to block lysosome function. However, we did not detect any difference in LC3B-II levels between control and Blos1s cell lines in any of these conditions (Fig. 5, F and G). We obtained similar results by analyzing LC3B foci (Fig. 5 H). Finally, we depleted cells of the macroautophagy factor ATG7, and although this blocked LC3B processing (Fig. S3, E and F), polyubiquitin foci accumulated in Blos1s versus control cells, similar to cells with an intact macroautophagy pathway (Fig. 5 I). Therefore, these aggregates surprisingly do not appear to be degraded by macroautophagy.
To test whether Blos1 degradation affects other forms of autophagy (Galluzzi et al., 2017), we depleted Rfp and Blos1s cells of LAMP2, which is required for chaperone-mediated autophagy (Cuervo and Dice, 1996), or various VPS proteins, which are required for LE-mediated microautophagy (Sahu et al., 2011). Knockdown of LAMP2 did not affect protein aggregate accumulation in either cell line (Fig. S3, G–J). However, knockdown of VPS4a, VPS4b, or VPS22 increased the accumulation of aggregates and eliminated the effects of Blos1s overexpression (Fig. 5 I). These proteins are components of the endosomal sorting complexes required for transport (ESCRT) machinery and are essential for the inward budding of LEs/multivesicular bodies (Henne et al., 2011). We did not detect any effect of VPS22 knockdown on LC3B processing and degradation (Fig. S3, L and M) or on LE/lysosome repositioning (Fig. 5, J and K) during ER stress. We therefore propose that the ER stress–induced aggregates are engulfed by juxtanuclear LEs and subsequently degraded when LEs fuse or mature into lysosomes.
The pathway we describe here links the localized degradation of a single mRNA to the more global consequences of protein misfolding. Taken together, our results show that Blos1s-expressing cells are defective in ESCRT- and lysosome-dependent degradation of protein aggregates, likely due to their inability to down-regulate BORC and reposition LEs/lysosomes in response to stress. The source of these aggregates is not clear, but they may arise from overwhelming the proteasome-dependent degradation of misfolded ER proteins. For example, a disease-causing variant of the cystic fibrosis transmembrane conductance regulator, which is poorly folded and typically extracted from the ER for proteasomal degradation, forms juxtanuclear, ubiquitinated aggregates when overexpressed or when the proteasome is inhibited (Johnston et al., 1998).
Both the UPR and lysosomal function are thought to be key aspects of neurodegenerative diseases that involve protein misfolding and aggregation. Parkinson’s and Huntington’s disease are associated with large, juxtanuclear protein aggregates (Chin et al., 2008), and lysosome repositioning to the MTOC has also been observed in models of Huntington’s disease (Erie et al., 2015). Lysosome clustering may therefore be a general mechanism for combatting protein aggregates, and targeting BLOS1 or BORC may present a novel strategy for protecting cells from proteotoxicity.
Materials and methods
Cell culture, ER stress treatments, and RNAi
We cultured MC3T3-E1 cells (American Type Culture Collection) at 37°C and 5% CO2 in MEMα with nucleosides, l-glutamine, and no ascorbic acids (Life Technologies), supplemented with 10% FBS. We cultured HeLa cells in DMEM with 10% FBS. We obtained ER stressors and inhibitors from Sigma-Aldrich (DTT, Tg, arsenite, and CQ), Thermo Fisher Scientific (MG132), and EMD Millipore (Tm and 4μ8c). Long CQ treatments resulted in extensive cell death; thus, we included CQ for the final 2–4 h before sample collection. For cell viability measurements, we aspirated dead, floating cells, trypsinized attached cells, and counted on a hemocytometer. Counts were normalized to untreated conditions.
For knockdown experiments, we cultured cells in antibiotic-free media and used RNAiMAX (Invitrogen) to transfect multiple siRNAs for each target gene (from Qiagen or Sigma-Aldrich). We allowed cells to recover for 48–72 h before inducing ER stress.
Transfections and stable cell lines
We subcloned PCR products for Lamp1, Rfp, Blos1s-HA, Blos1s-Flag, or Arl8b-Rfp downstream of the human EF1a promoter in expression plasmids containing a hygromycin resistance gene. We transfected these plasmids into MC3T3-E1 cells using lipofectamine 2000 (Invitrogen), allowed cells to recover, and then selected with 100 µg/ml hygromycin B (Invitrogen) to generate stable polyclonal cell lines or monoclonal cell lines for Arl8b-Rfp. We maintained cells in hygromycin until the passage preceding each experiment.
Microscopy and immunostaining
We imaged cells using an Olympus IX-51 inverted microscope with a 60× (NA 1.25) oil objective at room temperature and a Q-imaging Qicam (SN Q25830) camera. For acquisition software we used QCapturePro 6.0. For live cell imaging of LAMP1-GFP cells and tracker dyes, we plated cells on glass-bottom dishes and allowed them to recover and adhere for ∼24 h before inducing ER stress. We imaged cells in PBS prewarmed to 37°C. We followed the manufacturer’s protocols for lysotracker red DND-99, mitotracker red CMXRos, and Alexa Fluor 555 phalloidin (all from Thermo Fisher Scientific).
For immunostaining, we grew cells on glass coverslips. For most antibodies, we fixed the cells with preheated (37°C) 4% paraformaldehyde and 1 mM MgCl in PBS for 15 min, and then permeabilized with 0.2% Triton X-100 (in PBS with MgCl, 20 min, room temperature). Antibodies are listed below. For staining of polyubiquitinated proteins, we pretreated the cells with digitonin (60 µg/ml, 15 min, 4°C) to allow access of the antibody to aggregated proteins, then fixed and permeabilized as described above. We incubated coverslips in blocking buffer (2% BSA, 0.02% Tween20, and 1 mM MgCl in PBS) for 5–10 min, and then with primary antibodies in blocking buffer for 1 h. After washing (0.02% Tween20, 1 mM MgCl, and PBS), we incubated with secondary antibodies in blocking buffer for 1 h, washed, and mounted on slides in ProLong Diamond Antifade mountant with Dapi (Invitrogen).
We used the following antibodies for immunostaining: rat anti-mouse LAMP1 (DSHB 1D4B-s, 2 µg/ml) or LAMP2 (DSHB ABL-93, 2 µg/ml) with secondary donkey anti-rat IgG-Alexa Fluor 488 (Invitrogen A21208, 1:1,000); mouse anti-human LAMP1 (DSHB H4A3-s, 2 µg/ml) with secondary donkey anti-mouse IgG-Alexa Fluor 488 (Invitrogen A21202, 1:1,000); rabbit anti-RAB7 (Cell Signaling 9367, 1:100) or anti-tubulin (Cell Signaling 2144, 1:25) with secondary goat anti-rabbit IgG-Alexa Fluor 532 (Invitrogen A11009, 1:1,000); and mouse anti-polyubiquitin conjugate FK1 (Enzo BML-PW8805, 1:500) with secondary goat anti-mouse IgM μ chain Alexa Fluor 488 (Invitrogen A21042, 1:1,000).
For analyzing LEs/lysosomes or protein aggregates, we imaged cells in a systematic manner, ignoring only cells with insufficient signal or obvious morphological abnormalities. For most panels, we scored 24–81 cells per individual slide/dish, or 140–220 cells per condition. We then assigned random file names and had a researcher blinded to the conditions of the experiment score each cell. Cells with >50% of the LAMP1 (or RAB7) foci located on one side of the nucleus were counted as displaying “predominantly juxtanuclear LE/lysosomes.” We analyzed polyubiquitin foci in Figs. 4 and 5 by quantifying the size and number of foci using an ImageJ macro described previously (Dagda et al., 2008; Chu et al., 2009).
Quantitative real-time RT-PCR and Xbp1 splicing assay
We isolated total RNA using Quick RNA Miniprep kits (Zymo Research) and synthesized cDNA using 700 ng–2 µg total RNA as a template, a T18 primer, and Moloney murine leukemia virus reverse transcriptase (New England Biolabs). We measured the relative amount of specific mRNAs by quantitative PCR (qPCR) using a Mastercycler ep realplex (Eppendorf) or QuantStudio 3 (Life Technologies), with SYBR green as the fluorescent dye. We measured each sample in triplicate, quantified by comparison to serially diluted standard curve samples, and divided the target mRNA levels by those of ribosomal protein 19 (Rpl19) mRNA in the same sample. RNA levels were also normalized to those for control, unstressed cells from the same experiment.
We measured Xbp1 splicing by amplifying cDNA with primers encompassing Xbp1 splice site (5′-AGAAGAGAACCACAAACTCCAG-3′, 5′-GGGTCCAACTTGTCCAGAATGC-3′) and running the products on a 2% agarose gel. We then quantified the relative band intensities for the spliced and unspliced Xbp1 products.
We used the following primer pairs for qPCR: Blos1 (5′-CAAGGAGCTGCAGGAGAAGA-3′, 5′-CCAGGAGGGTGAAGTAAGAGG-3′), Scara3 (5′-TGCATGGATACTGACCCTGA-3′, 5′-GCCGTGTTACCAGCTTCTTC-3′), Col6a1 (5′-TGCTCAACATGAAGCAGACC-3′, 5′-TTGAGGGAGAAAGCTCTGGA-3′), Hgsnat (5′-TCTCCGCTTTCTCCATTTTG-3′, 5′-CGCATACACGTGGAAAGTCA-3′), BiP (5′-TCAGCATCAAGCAAGGATTG-3′, 5′-AAGCCGTGGAGAAGATCTGA-3′), Lys (5′-GTCCATGGGAATACAGGCCC-3′, 5′-AAGTTCCTAAAGCCCCTGGC-3′), Atg7 (5′-TCACTCACGGGGTTGCTCCT-3′, 5′-GATGGTAGGGCCGCTTGTTG-3′), Vps4A (5′-CTCCCTCATGCAGCCAGTCA-3′, 5′-TTCACTGTGGGCCGGGTAGT-3′), Vps4B (5′-GGCTTCATGGACAGCAGCTAGG-3′, 5′-TGCCACAAACCCCAAATTCA-3′), Vps22 (5′-GGATTCGGCATCATCCCTGT-3′, 5′-CCAGGCCAGTCCTTCCTTCA-3′), and Hdac6 (5′-ACCGCAGTCCGTACCTTGGA, 5′-CACTGCCCCGTTGTCTCCTT-3′). Housekeeping controls were Rpl19 from mouse (5′-CTGATCAAGGATGGGCTGAT-3′, 5′-GCCGCTATGTACAGACACGA-3′) or human (5′-ATGTATCACAGCCTGTACCTG-3′, 5′-TTCTTGGTCTCTTCCTCCTTG-3′).
Immunoblotting
We lysed cells in RIPA buffer (25 mM Tris, pH 7.6, 150 mM NaCl, 1% NP-40, 1% Na-deoxycholate, and 0.1% SDS) with protease inhibitors (Thermo Fisher Scientific) and phosphatase inhibitors (50 mM NaF and 0.2 mM Na-orthovanadate complexes). After incubating on ice for 30 min, we removed insoluble material by centrifugation (14,000 × g, 15 min, 4°C) and resolved the soluble proteins using 12% (for LC3B-II or cleaved caspase 3) or 4–12% (for LAMP2) polyacrylamide NuPage Bis-Tris gels (Invitrogen). We transferred proteins to nitrocellulose, incubated for 1 h in blocking buffer (5% BSA, 0.05% Tween20, 0.01% Triton X-100, and TBS) and probed using the antibodies listed below in blocking buffer (4°C, overnight). We washed the blots, incubated with secondary antibodies (in blocking buffer, 1 h, room temperature), and scanned using a Licor Odyssey Imager. We quantified bands using ImageJ and divided protein levels by GAPDH protein levels measured on the same blot.
We used the following antibodies for immunoblotting: mouse anti-human Bloc1s1 (Santa Cruz sc-515444, 1:250), rabbit anti-mouse cleaved caspase 3 (Cell Signaling 9664S, 1:1,000), rabbit anti-mouse LC3B (Sigma-Aldrich L7543, 1:1,000), rat anti-mouse LAMP2 (DSHB ABL-93, 0.5 µg/ml), and the loading control rabbit anti-mouse GAPDH (ProSci 3783, 1:20,000). For secondary antibodies, we used infrared dye-labeled goat anti-rabbit IgG (Licor 926-32210) or anti-rat IgG (Licor 925-68029), at 1:10,000.
SDS-PAGE analysis of protein aggregation
We collected identical numbers of cells for each sample, lysed cells, and separated soluble from insoluble proteins as described above (in the Immunoblotting section). After centrifugation, we added equal volumes of RIPA buffer to the pellets and sonicated using a Misonix XL2020 Ultrasonic Processor (Thermo Fisher Scientific, 10% power, 15 s). We then analyzed by SDS-PAGE using 12% polyacrylamide NuPage Bis-Tris gels (Invitrogen), followed by staining with Coomassie blue R250 (0.1% in 10% acetic acid and 50% methanol) and quantification of lane intensities using ImageJ.
Data presentation and statistics
At least three biological replicates were performed for every experiment. We displayed all replicate data in the figures, with symbols representing individual experiments and carried through multiple panels when samples were collected in parallel or when multiple measurements were made on the same sample. Lines connect data collected in a single replicate experiment. For statistical analysis of pairwise comparisons, we used Student’s two-tailed paired t tests and corrected for multiple comparisons in the same experiment/panel using the Holm-Bonferroni method. Where noted in the figure legends, we used ANOVA followed by Tukey’s honestly significant difference tests for multiple comparisons.
Online supplemental material
Fig. S1, which is related to Fig. 1, shows labeling of other cellular structures with and without ER stress and shows LE/lysosome repositioning, as well as the degradation of Blos1 mRNA and BLOS1 protein in human cultured cells. Fig. S2, which is related to Fig. 2, shows images for UPR-depleted cells and controls for the Blos1s cell lines. Fig. S3, which is related to Figs. 4 and 5, shows further elucidation of the mechanism of protein aggregate degradation as well as controls for various knockdown experiments.
Supplementary Material
Acknowledgments
We thank M. Babst and members of the Hollien laboratory for discussions.
This research was supported in part by the National Institutes of Health National Institute of General Medical Sciences (R35 GM119540).
The authors declare no competing financial interests.
Author contributions: J. Hollien, K.A. Moore, and D. Bae: conceptualization; D. Bae, K.A. Moore, J.M. Mella, S.Y. Hayashi, and J. Hollien: investigation, review, and editing; D. Bae and J. Hollien: original draft.
References
- Benhamron S., Hadar R., Iwawaky T., So J.S., Lee A.H., and Tirosh B.. 2014. Regulated IRE1-dependent decay participates in curtailing immunoglobulin secretion from plasma cells. Eur. J. Immunol. 44:867–876. 10.1002/eji.201343953 [DOI] [PubMed] [Google Scholar]
- Bright M.D., Itzhak D.N., Wardell C.P., Morgan G.J., and Davies F.E.. 2015. Cleavage of BLOC1S1 mRNA by IRE1 is sequence specific, temporally separate from XBP1 splicing, and dispensable for cell viability under acute endoplasmic reticulum stress. Mol. Cell. Biol. 35:2186–2202. 10.1128/MCB.00013-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calfon M., Zeng H., Urano F., Till J.H., Hubbard S.R., Harding H.P., Clark S.G., and Ron D.. 2002. IRE1 couples endoplasmic reticulum load to secretory capacity by processing the XBP-1 mRNA. Nature. 415:92–96. 10.1038/415092a [DOI] [PubMed] [Google Scholar]
- Cheng X.-T., Xie Y.-X., Zhou B., Huang N., Farfel-Becker T., and Sheng Z.-H.. 2018. Characterization of LAMP1-labeled nondegradative lysosomal and endocytic compartments in neurons. J. Cell Biol. 217:3127–3139. 10.1083/jcb.201711083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin J.A. Olzmann, and Li L.. 2008. Aggresome formation and neurodegenerative diseases: therapeutic implications. Curr. Med. Chem. 15:47–60. 10.2174/092986708783330692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu C.T., Plowey E.D., Dagda R.K., Hickey R.W., Cherra S.J. III, and Clark R.S.B.. 2009. Autophagy in neurite injury and neurodegeneration: in vitro and in vivo models. Methods Enzymol. 453:217–249. 10.1016/S0076-6879(08)04011-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coelho D.S., Cairrao F., Zeng X., Pires E., Coelho A.V., Ron D., Ryoo H.D., and Domingos P.M.. 2013. Xbp1-independent Ire1 signaling is required for photoreceptor differentiation and rhabdomere morphogenesis in Drosophila. Cell Reports. 5:791–801. 10.1016/j.celrep.2013.09.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuervo A.M., and Dice J.F.. 1996. A receptor for the selective uptake and degradation of proteins by lysosomes. Science. 273:501–503. 10.1126/science.273.5274.501 [DOI] [PubMed] [Google Scholar]
- Dagda R.K., Zhu J., Kulich S.M., and Chu C.T.. 2008. Mitochondrially localized ERK2 regulates mitophagy and autophagic cell stress: implications for Parkinson’s disease. Autophagy. 4:770–782. 10.4161/auto.6458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delevoye C., Heiligenstein X., Ripoll L., Gilles-Marsens F., Dennis M.K., Linares R.A., Derman L., Gokhale A., Morel E., Faundez V., et al. . 2016. BLOC-1 brings together the actin and microtubule cytoskeletons to generate recycling endosomes. Curr. Biol. 26:1–13. 10.1016/j.cub.2015.11.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis M.K., Delevoye C., Acosta-Ruiz A., Hurbain I., Romao M., Hesketh G.G., Goff P.S., Sviderskaya E.V., Bennett D.C., Luzio J.P., et al. . 2016. BLOC-1 and BLOC-3 regulate VAMP7 cycling to and from melanosomes via distinct tubular transport carriers. J. Cell Biol. 214:293–308. 10.1083/jcb.201605090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erie C., Sacino M., Houle L., Lu M.L., and Wei J.. 2015. Altered lysosomal positioning affects lysosomal functions in a cellular model of Huntington’s disease. Eur. J. Neurosci. 42:1941–1951. 10.1111/ejn.12957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farías G.G., Guardia C.M., De Pace R., Britt D.J., and Bonifacino J.S.. 2017. BORC/kinesin-1 ensemble drives polarized transport of lysosomes into the axon. Proc. Natl. Acad. Sci. USA. 114:E2955–E2964. 10.1073/pnas.1616363114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filipek P.A., de Araujo M.E.G., Vogel G.F., De Smet C.H., Eberharter D., Rebsamen M., Rudashevskaya E.L., Kremser L., Yordanov T., Tschaikner P., et al. . 2017. LAMTOR/Ragulator is a negative regulator of Arl8b- and BORC-dependent late endosomal positioning. J. Cell Biol. 216:4199–4215. 10.1083/jcb.201703061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galluzzi L., Baehrecke E.H., Ballabio A., Boya P., Bravo-San Pedro J.M., Cecconi F., Choi A.M., Chu C.T., Codogno P., Colombo M.I., et al. . 2017. Molecular definitions of autophagy and related processes. EMBO J. 36:1811–1836. 10.15252/embj.201796697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guardia C.M., Farías G.G., Jia R., Pu J., and Bonifacino J.S.. 2016. BORC functions upstream of kinesins 1 and 3 to coordinate regional movement of lysosomes along different microtubule tracks. Cell Reports. 17:1950–1961. 10.1016/j.celrep.2016.10.062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henne W.M., Buchkovich N.J., and Emr S.D.. 2011. The ESCRT pathway. Dev. Cell. 21:77–91. 10.1016/j.devcel.2011.05.015 [DOI] [PubMed] [Google Scholar]
- Hetz C., Chevet E., and Harding H.P.. 2013. Targeting the unfolded protein response in disease. Nat. Rev. Drug Discov. 12:703–719. 10.1038/nrd3976 [DOI] [PubMed] [Google Scholar]
- Hofmann I., and Munro S.. 2006. An N-terminally acetylated Arf-like GTPase is localised to lysosomes and affects their motility. J. Cell Sci. 119:1494–1503. 10.1242/jcs.02958 [DOI] [PubMed] [Google Scholar]
- Hollien J., and Weissman J.S.S.. 2006. Decay of endoplasmic reticulum-localized mRNAs during the unfolded protein response. Science. 313:104–107. 10.1126/science.1129631 [DOI] [PubMed] [Google Scholar]
- Hollien J., Lin J.H.J.H., Li H., Stevens N., Walter P., and Weissman J.S.. 2009. Regulated Ire1-dependent decay of messenger RNAs in mammalian cells. J. Cell Biol. 186:323–331. 10.1083/jcb.200903014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyttinen J.M.T., Amadio M., Viiri J., Pascale A., Salminen A., and Kaarniranta K.. 2014. Clearance of misfolded and aggregated proteins by aggrephagy and implications for aggregation diseases. Ageing Res. Rev. 18:16–28. 10.1016/j.arr.2014.07.002 [DOI] [PubMed] [Google Scholar]
- Jackson M.P., and Hewitt E.W.. 2016. Cellular proteostasis: degradation of misfolded proteins by lysosomes. Essays Biochem. 60:173–180. 10.1042/EBC20160005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia R., Guardia C.M., Pu J., Chen Y., and Bonifacino J.S.. 2017. BORC coordinates encounter and fusion of lysosomes with autophagosomes. Autophagy. 13:1648–1663. 10.1080/15548627.2017.1343768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston J.A., Ward C.L., and Kopito R.R.. 1998. Aggresomes: a cellular response to misfolded proteins. J. Cell Biol. 143:1883–1898. 10.1083/jcb.143.7.1883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaguchi Y., Kovacs J.J., McLaurin A., Vance J.M., Ito A., and Yao T.-P.. 2003. The deacetylase HDAC6 regulates aggresome formation and cell viability in response to misfolded protein stress. Cell. 115:727–738. 10.1016/S0092-8674(03)00939-5 [DOI] [PubMed] [Google Scholar]
- Kimmig P., Diaz M., Zheng J., Williams C.C., Lang A., Aragón T., Li H., and Walter P.. 2012. The unfolded protein response in fission yeast modulates stability of select mRNAs to maintain protein homeostasis. eLife. 1:e00048 10.7554/eLife.00048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korolchuk V.I., Saiki S., Lichtenberg M., Siddiqi F.H., Roberts E.A., Imarisio S., Jahreiss L., Sarkar S., Futter M., Menzies F.M., et al. . 2011. Lysosomal positioning coordinates cellular nutrient responses. Nat. Cell Biol. 13:453–460. 10.1038/ncb2204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K., Tirasophon W., Shen X., Michalak M., Prywes R., Okada T., Yoshida H., Mori K., and Kaufman R.J.. 2002. IRE1-mediated unconventional mRNA splicing and S2P-mediated ATF6 cleavage merge to regulate XBP1 in signaling the unfolded protein response. Genes Dev. 16:452–466. 10.1101/gad.964702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levi-Ferber M., Gian H., Dudkevich R., and Henis-Korenblit S.. 2015. Transdifferentiation mediated tumor suppression by the endoplasmic reticulum stress sensor IRE-1 in C. elegans. eLife. 4:e08005 10.7554/eLife.08005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore K., and Hollien J.. 2015. Ire1-mediated decay in mammalian cells relies on mRNA sequence, structure, and translational status. Mol. Biol. Cell. 26:2873–2884. 10.1091/mbc.E15-02-0074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogata M., Hino S., Saito A., Morikawa K., Kondo S., Kanemoto S., Murakami T., Taniguchi M., Tanii I., Yoshinaga K., et al. . 2006. Autophagy is activated for cell survival after endoplasmic reticulum stress. Mol. Cell. Biol. 26:9220–9231. 10.1128/MCB.01453-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osorio F., Tavernier S.J., Hoffmann E., Saeys Y., Martens L., Vetters J., Delrue I., De Rycke R., Parthoens E., Pouliot P., et al. . 2014. The unfolded-protein-response sensor IRE-1alpha regulates the function of CD8alpha+ dendritic cells. Nat. Immunol. 15:248–257. 10.1038/ni.2808 [DOI] [PubMed] [Google Scholar]
- Ouyang H., Ali Y.O., Ravichandran M., Dong A., Qiu W., MacKenzie F., Dhe-Paganon S., Arrowsmith C.H., and Zhai R.G.. 2012. Protein aggregates are recruited to aggresome by histone deacetylase 6 via unanchored ubiquitin C termini. J. Biol. Chem. 287:2317–2327. 10.1074/jbc.M111.273730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pu J., Schindler C., Jia R., Jarnik M., Backlund P., and Bonifacino J.S.. 2015. BORC, a multisubunit complex that regulates lysosome positioning. Dev. Cell. 33:176–188. 10.1016/j.devcel.2015.02.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pu J., Keren-Kaplan T., and Bonifacino J.S.. 2017. A Ragulator–BORC interaction controls lysosome positioning in response to amino acid availability. J. Cell Biol. 216:4183–4197. 10.1083/jcb.201703094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahu R., Kaushik S., Clement C.C., Cannizzo E.S., Scharf B., Follenzi A., Potolicchio I., Nieves E., Cuervo A.M., and Santambrogio L.. 2011. Microautophagy of cytosolic proteins by late endosomes. Dev. Cell. 20:131–139. 10.1016/j.devcel.2010.12.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott I., Webster B.R., Chan C.K., Okonkwo J.U., Han K., and Sack M.N.. 2013. GCN5-like protein 1 (GCN5L1) controls mitochondrial content through coordinated regulation of mitochondrial biogenesis and mitophagy. J. Biol. Chem. 289:2864–2872. 10.1074/jbc.M113.521641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott I., Wang L., Wu K., Thapa D., and Sack M.N.. 2018. GCN5L1/BLOS1 links acetylation, organelle remodeling, and metabolism. Trends Cell Biol. 28:346–355. 10.1016/j.tcb.2018.01.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- So J.S., Hur K.Y., Tarrio M., Ruda V., Frank-Kamenetsky M., Fitzgerald K., Koteliansky V., Lichtman A.H., Iwawaki T., Glimcher L.H., and Lee A.H.. 2012. Silencing of lipid metabolism genes through IRE1alpha-mediated mRNA decay lowers plasma lipids in mice. Cell Metab. 16:487–499. 10.1016/j.cmet.2012.09.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter P., and Ron D.. 2011. The unfolded protein response: from stress pathway to homeostatic regulation. Science. 334:1081–1086. 10.1126/science.1209038 [DOI] [PubMed] [Google Scholar]
- Wang L., Scott I., Zhu L., Wu K., Han K., Chen Y., Gucek M., and Sack M.N.. 2017. GCN5L1 modulates cross-talk between mitochondria and cell signaling to regulate FoxO1 stability and gluconeogenesis. Nat. Commun. 8:523 10.1038/s41467-017-00521-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Z., Pascual C., and Klionsky D.J.. 2016. Autophagy: machinery and regulation. Microb. Cell. 3:588–596. 10.15698/mic2016.12.546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yorimitsu T., and Klionsky D.J.. 2007. Endoplasmic reticulum stress: a new pathway to induce autophagy. Autophagy. 3:160–162. 10.4161/auto.3653 [DOI] [PubMed] [Google Scholar]
- Yoshida H., Matsui T., Yamamoto A., Okada T., and Mori K.. 2001. XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell. 107:881–891. 10.1016/S0092-8674(01)00611-0 [DOI] [PubMed] [Google Scholar]
- Zhang A., He X., Zhang L., Yang L., Woodman P., and Li W.. 2014. Biogenesis of lysosome-related organelles complex-1 subunit 1 (BLOS1) interacts with sorting nexin 2 and the endosomal sorting complex required for transport-I (ESCRT-I) component TSG101 to mediate the sorting of epidermal growth factor receptor into endo. J. Biol. Chem. 289:29180–29194. 10.1074/jbc.M114.576561 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.