Rickman and Smogorzewska discuss recent studies that detail the cellular mechanisms protecting genome stability at sites of stalled DNA replication.
Abstract
The replisome, the molecular machine dedicated to copying DNA, encounters a variety of obstacles during S phase. Without a proper response to this replication stress, the genome becomes unstable, leading to disease, including cancer. The immediate response is localized to the stalled replisome and includes protection of the nascent DNA. A number of recent studies have provided insight into the factors recruited to and responsible for protecting stalled replication forks. In response to replication stress, the SNF2 family of DNA translocases has emerged as being responsible for remodeling replication forks in vivo. The protection of stalled replication forks requires the cooperation of RAD51, BRCA1, BRCA2, and many other DNA damage response proteins. In the absence of these fork protection factors, fork remodeling renders them vulnerable to degradation by nucleases and helicases, ultimately compromising genome integrity. In this review, we focus on the recent progress in understanding the protection, processing, and remodeling of stalled replication forks in mammalian cells.
Introduction
During DNA replication, the replisome encounters many obstacles that pose a risk to precisely copying the genetic material. The slowing or stalling of the progressing replication fork that results from such impediments is termed replication stress (Zeman and Cimprich, 2014). Endogenous sources of replication stress include a damaged DNA template, difficult-to-replicate regions such as repetitive sequences, active transcription machinery, RNA–DNA hybrids, DNA–protein adducts, and secondary DNA structures (Zeman and Cimprich, 2014). Alterations in the cell cycle associated with oncogene activation and rapid cell proliferation are also a source of replication stress due to insufficient deoxynucleotide triphosphate pools (Neelsen et al., 2013; Zeman and Cimprich, 2014; Ahuja et al., 2016). Cellular responses have evolved to manage replication stress in order to promote high-fidelity DNA replication to ensure cell viability. They protect against mutations and guard against tumorigenesis.
Replication stress is associated with the generation of single-stranded DNA (ssDNA) at the replication fork, which serves to recruit and activate the ataxia-telangiectasia and Rad3 related (ATR) kinase (Saldivar et al., 2017). The ATR kinase modulates the replication stress response by activating and recruiting DNA repair machinery, preventing new origin firing, promoting replication fork stability, and stimulating processing for replication restart (Saldivar et al., 2017). In the absence of ATR, replication stress leads to extensive ssDNA formation, which may result in replication protein A (RPA) exhaustion and DNA breakage (Toledo et al., 2013).
An improper response to replication stress can result in replication fork collapse. Replication fork collapse has often been used to describe the dissociation of the replication machinery or double-strand break (DSB) formation at stalled replication forks. In light of new data, replication fork collapse might be better defined as replication inactivation in which a fork is no longer able to resume replication (Cortez, 2015). Analysis of replication forks by iPOND (isolation of protein on nascent DNA) in mammalian cells has shown that in the absence of ATR activity, the core components of the replisome are stable. However, the proteome at the stalled fork is dynamically altered, reflecting the requirement for ATR activity to modulate the replication stress response in order to prevent fork collapse (Dungrawala et al., 2015).
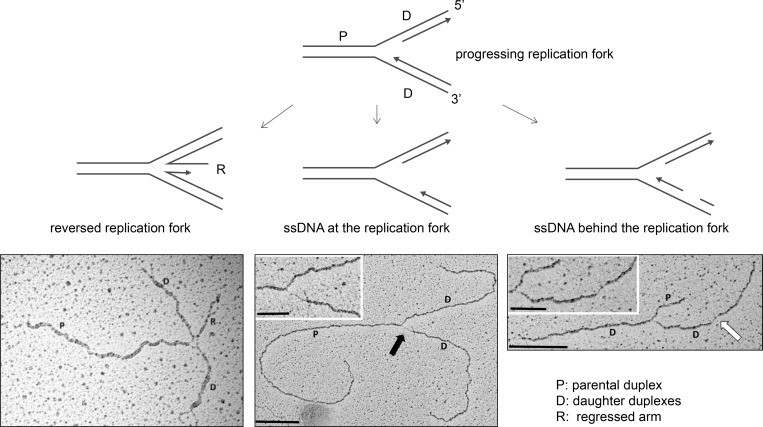
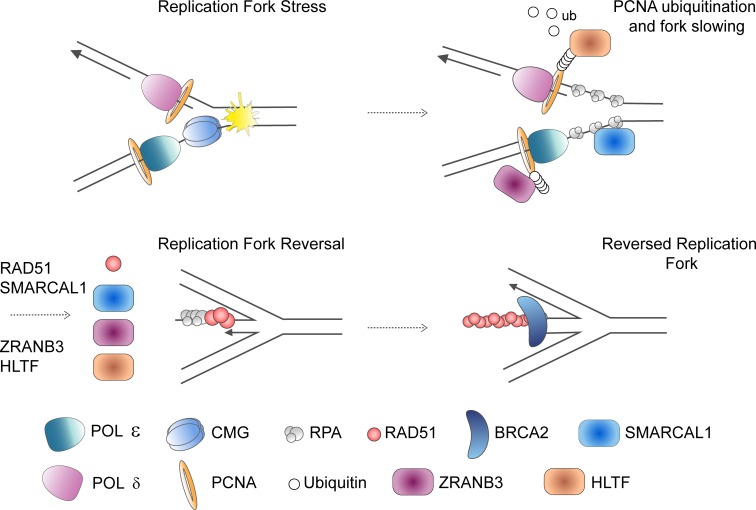
Replication forks that slow or stall can undergo remodeling into a reversed replication fork structure, which has been visualized by EM (Sogo et al., 2002; Ray Chaudhuri et al., 2012). Reversed forks are formed when the parental DNA strands reanneal and nascent DNA strands anneal, forming a “regressed arm,” a four-way joint molecule resembling a Holliday junction (Fig. 1). Replicating cells display a baseline level of reversed replication forks that is increased upon exogenous genotoxic stress, possibly as a result of ATR signaling (Ray Chaudhuri et al., 2012; Berti et al., 2013; Zellweger et al., 2015; Mutreja et al., 2018). A wide array of replication stress–producing agents, including topoisomerase inhibitors, DNA interstrand cross-linking agents, DNA synthesis inhibitors, alkylating agents, and UV radiation, increases replication fork reversal (Zellweger et al., 2015). Additionally, cells undergoing rapid proliferation use replication fork slowing and fork reversal as a means to protect against genomic instability produced by endogenous replication stress (Ahuja et al., 2016). Evidence to support fork reversal as a mechanism to protect against genomic instability is accumulating (Bétous et al., 2012; Ray Chaudhuri et al., 2012; Couch et al., 2013; Zellweger et al., 2015). Fork reversal may serve to protect against extensive ssDNA generation, provide DNA repair machinery access to the damaged template, or promote lesion bypass (Cortez, 2015). However, reversed replication forks are also liable to nuclease processing and DSB formation (Schlacher et al., 2011, 2012; Ying et al., 2012; Couch et al., 2013; Neelsen et al., 2013).
Figure 1.
Replication fork intermediates visualized by EM. To visualize replication fork intermediates, replicating cells are treated with the cross-linking agent psoralen, which cross-links DNA upon UVA exposure. The cross-linked duplex DNA is then visualized by EM, and replication intermediates are analyzed. ssDNA versus dsDNA is determined by measuring DNA fiber thickness. Progressing replication forks, reversed replication forks, and replication forks containing ssDNA gaps at the replication fork junction (thick black arrow) and behind the fork (thick white arrow) have been visualized. Scale bars indicate 0.5 kb in main images and 0.2 kb in insets. EM micrographs are reproduced with permission from Zellweger et al. (2015).
In this review, we focus on the processing that occurs at the replication fork when replication stress is encountered. Recently, a number of studies have provided insight into the dynamics of stalled replication forks and the proteins active in protecting replication forks to prevent genomic instability and permit resumption of replication. At the forefront of replication fork stability and protection are RAD51, BRCA1, and BRCA2, most well known for their role in homologous recombination. Until recently, it had been inferred that the role of these factors in the response to replication stress was the repair of DSBs at collapsed forks as necessary components of homology-directed repair (HDR). However, it is apparent their roles are much more pleiotropic. Single-molecule techniques such as DNA fiber analysis of replication tracks and EM of replication fork intermediates have greatly contributed to the understanding of these processes (Vindigni and Lopes, 2017). Armed with these techniques, the field is gaining insight into the activities of BRCA1, BRCA2, RAD51, other protection factors, and various nucleases and translocases at the stalled replication fork (Fig. 2), topics that will be discussed.
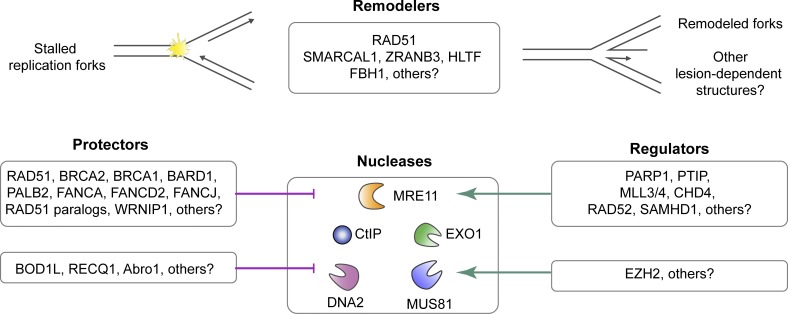
Figure 2.
Summary of proteins and their roles in processing or protecting stalled replication forks. Recent observations suggest that many proteins promote the remodeling of DNA at stalled replication forks into reversed replication fork structures. The remodeled fork requires protection by BRCA1, BRCA2, RAD51, and several other factors that have been identified. In the absence of replication fork protection, the newly synthesized DNA is subject to degradation by nucleases. A number of proteins have also been identified as promoting the localization of these nucleases at the stalled fork. Further investigation is required to determine how these fork remodelers, nuclease regulators, and fork protectors may be operating to promote replication fork stability. It is possible that the various remodelers, protectors, regulators, and nucleases are operating in a coordinated fashion; however, it is also possible that their roles are DNA lesion and replication fork structure dependent.
Canonical HDR
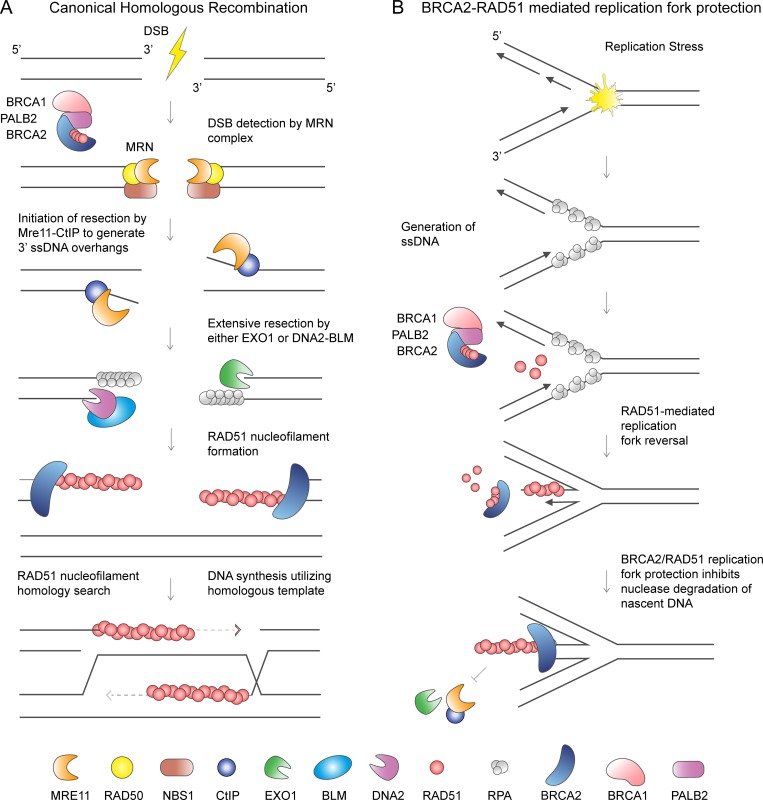
During HDR, BRCA1 localizes to DSBs to promote end resection that generates 3′ ssDNA overhangs (Schlegel et al., 2006; Chen et al., 2008). Resection is initiated by the MRE11–RAD50–NBS1 complex and the CtiP endonuclease to generate 3′ ssDNA tails that undergo more extensive resection by either the EXO1 exonuclease or the BLM–DNA2 helicase nuclease complex (Fig. 3 A; Sartori et al., 2007; Gravel et al., 2008; Mimitou and Symington, 2008; Zhu et al., 2008; Symington, 2014). BRCA2 then mediates the displacement of the ssDNA-binding protein RPA from the 3′ overhangs by loading the RAD51 recombinase. The RAD51 nucleofilament searches for homologous DNA in the sister chromatid, which is used as a template for precise DNA repair (Jasin and Rothstein, 2013). Following strand invasion and DNA synthesis, the four-stranded double Holliday junctions are dissolved by either the BLM–TOPOIIIa–RMI1-RMI2 complex or resolved by nucleolytic processing by GEN1 or MUS81–SLX1–SLX4 (Sarbajna and West, 2014).
Figure 3.
Distinct roles of BRCA2 and RAD51 in canonical homologous recombination and replication fork protection. (A) HDR of DSBs requires the formation of 3′ ssDNA overhangs. The MRE11–RAD50–NBS1 (MRN) complex senses DSB and with the CtiP endonuclease initiates DNA end resection. The EXO1 exonuclease or the BLM–DNA2 helicase nuclease complex is responsible for more extensive resection. BRCA2 loads and stabilizes RAD51 nucleofilaments on the ssDNA overhangs displacing the ssDNA binding protein RPA. RAD51 nucleofilament invades the sister chromatid to perform homology search. DNA synthesis proceeds using homologous DNA for precise repair. (B) Replication fork reversal is proposed to be a global response to replication stress that requires RAD51 and BRCA2 for fork reversal and fork protection. RAD51-mediated fork reversal entails the annealing of the newly replicated (nascent) strands of DNA and reannealing of the parental DNA strands. This function is proposed to require RAD51 independently of BRCA2. Subsequently, both RAD51 and BRCA2 are necessary to prevent nascent strand degradation.
BRCA2 and BRCA1 in replication fork protection
Besides the essential function of BRCA2 in HDR, its role in protecting stalled replication forks has now been extensively described. When BRCA2 is deficient, the newly duplicated (nascent) strand is degraded by MRE11 under conditions of prolonged hydroxyurea (HU) treatment that completely stalls replication fork progression (Fig. 3 B; Schlacher et al., 2011). The function of BRCA2 in replication fork protection was identified by studying the BRCA2 S3291A separation-of-function mutation (Schlacher et al., 2011). BRCA2 serine 3291 is a cyclin-dependent kinase phosphorylation site that regulates the C-terminal interaction of BRCA2, which is hypothesized to stabilize RAD51 nucleofilaments on ssDNA (Esashi et al., 2005; Davies and Pellegrini, 2007). The S3291A BRCA2 mutant is proficient for HDR activity but is unable to protect against nascent strand degradation by MRE11 (Schlacher et al., 2011; Feng and Jasin, 2017), which is presumed to be due to defective RAD51 nucleofilament formation at the stalled replication fork. Recent studies also suggest that the BRCA2 N-terminal domain and interaction with PALB2 is required for the recruitment and protection function of BRCA2 at stalled replication forks (Murphy et al., 2014; Hartford et al., 2016).
Just like during HDR, the ability of BRCA2 to deposit RAD51 onto ssDNA underlies its protective function at the stalled replication fork. This conclusion comes from experiments demonstrating that the disruption of RAD51 nucleofilaments by expression of the BRC4 peptide also results in nascent strand degradation. Conversely, overexpression of a RAD51 mutant, K133R, that forms stable nucleofilaments due to loss of ATPase activity required for its dissociation from DNA, renders replication forks resistant to degradation (Schlacher et al., 2011). Furthermore, depletion of RAD51 has been shown to result in nascent strand degradation at stalled replication forks and causes replication fork restart defects (Hashimoto et al., 2010; Petermann et al., 2010). However, not all studies agree on the exact role of RAD51 in nascent DNA resection at stalled forks (Thangavel et al., 2015; Feng and Jasin, 2017; Lemaçon et al., 2017; Mijic et al., 2017). A possible explanation for such a discrepancy might be different levels of RAD51 depletion (Bhat et al., 2018), resulting in distinctive cellular outcomes, as will be described when we discuss replication fork reversal.
The role of fork protection has been extended to include BRCA1 (Schlacher et al., 2012; Taglialatela et al., 2017) and the BRCA1-binding partner BARD1 (Billing et al., 2018). Mutations disrupting the BARD1 BRCT domain resulted in defective BARD1-BRCA1 recruitment to sites of replication stress and defective replication fork protection after HU (Billing et al., 2018). While both BRCA1 and BRCA2 are required in replication fork protection pathway, similar to HDR, it is likely their roles differ. Analysis of DNA replication and repair through the Tus/Ter replication fork barriers found that BRCA1, but not BRCA2, was required to suppress tandem duplications, providing evidence that these factors may operate differently during replication fork stalling (Willis et al., 2017). Another report suggests a role for BRCA1 in countering 53BP1 activity at the replication fork to promote cleavage-dependent processing (Xu et al., 2017b). However, these experiments have not tested if 53BP1 itself had an influence on nascent strand degradation upon complete replisome stalling. If it did, the additional downstream components recently identified as controlling the amount of ssDNA at a DSB, including REV7/MAD2L2, SHLD1, SHLD2, SHLD3, and DYNLL1 (Barazas et al., 2018; Dev et al., 2018; Ghezraoui et al., 2018; Gupta et al., 2018; He et al., 2018; Mirman et al., 2018; Noordermeer et al., 2018), should be tested as potential regulators of nascent strand degradation upon replication stress.
Since the discovery of the replication fork protection function of the canonical homologous recombination factors BRCA2 and BRCA1, multiple other DNA damage response proteins have also been found to have a role in this process including the RAD51 paralogs, the Fanconi anemia proteins FANCA, FANCD2, and FANCJ (Schlacher et al., 2012; Somyajit et al., 2015; Billing et al., 2018; Peng et al., 2018), as well as BOD1L, Abro1, RECQ1, and WRNIP1 (Higgs et al., 2015; Thangavel et al., 2015; Leuzzi et al., 2016; Xu et al., 2017a). These studies indicate that many factors influence replication fork protection and that further understanding is required to determine if they operate synergistically, act epistatically, or protect different replication fork intermediates produced by different types of replication stress.
Role of BRCA2 and RAD51 in replication fork reversal
EM analysis of replication fork intermediates from BRCA2-depleted cells showed a decrease in the number of reversed replication forks. The reversed replication fork numbers were rescued upon inhibition of the MRE11 nuclease (Lemaçon et al., 2017; Mijic et al., 2017), which suggests that reversed forks form but are not protected from nucleolytic activity when BRCA2 levels are low. In agreement, reversed replication fork intermediates are detected at normal levels at early time points after replication stress but decrease over time (Lemaçon et al., 2017). Analysis of replication fork species by EM in RAD51-depleted cells also showed a decrease in reversed replication forks (Zellweger et al., 2015; Kolinjivadi et al., 2017; Mijic et al., 2017). However, these levels are not rescued by MRE11 inhibition (Mijic et al., 2017), which suggests that with inadequate RAD51 activity, replication forks are not reversed, precluding degradation.
Further insight into the role of RAD51 in reversed fork formation and fork protection came from studies of the RAD51-T131P mutant protein (Mijic et al., 2017). This dominant-negative RAD51 mutant was identified in an individual with Fanconi anemia–like syndrome. Due to hyperactive ATPase activity, the RAD51-T131P mutant interferes with normal RAD51 function (Wang et al., 2015). Cells expressing the RAD51-T131P mutant undergo MRE11-dependent nascent strand degradation at stalled replication forks, and similar results were observed in Xenopus laevis egg extracts reconstituted with the RAD51-T131P mutant (Mijic et al., 2017; Zadorozhny et al., 2017). Consistent with in vivo data, biochemical analysis showed that the T131P mutant cannot protect DNA substrates from MRE11-mediated degradation and the mutant forms abnormal nucleofilaments (Kolinjivadi et al., 2017; Zadorozhny et al., 2017). However, in contrast to RAD51-depleted cells, the decrease in reversed replication fork numbers in RAD51-T131P cells was rescued by MRE11 inhibition (Mijic et al., 2017). One interpretation of these results is that cells expressing RAD51-T131P mutant maintain enough RAD51 activity for replication fork reversal, but not enough function to protect the reversed replication fork from MRE11 degradation. It is possible that formation of stable RAD51 nucleofilaments may not be required for replication fork reversal activity but is required for protection of the regressed replication fork from nucleolytic activity (Mijic et al., 2017).
Taken together, the EM experiments suggest that RAD51, independently of BRCA2, is necessary to promote reversal of stalled replication forks, while both RAD51 and BRCA2 are required to protect the reversed fork from degradation (Fig. 3 B). However, all of the experiments addressing the fork protection function of BRCA2 were performed by RNAi depletion or the potentially hypomorphic BRCA2 ovarian carcinoma cell line, so it remains to be determined if BRCA2 is indeed not involved in replication fork reversal. A distinct possibility is that the decreased levels of BRCA2 achieved in those studies were enough for fork reversal, but not for fork protection. Identification of BRCA2 mutations that would affect one function, but not the other, will be necessary to fully understand the role of BRCA2 at the replication fork. Alternatively, it is possible that the function of RAD51 in replication fork reversal is dependent upon interaction with other factors such as the RAD51 paralogs (XRCC2, XRCC3, RAD51B, RAD51C, and RAD51D), RAD52, RAD54, (Sugawara et al., 2003; Suwaki et al., 2011), or the MMS22L-TONSL heterodimer (O’Donnell et al., 2010; Piwko et al., 2016).
In addition to the reversed replication fork structures identified by EM, other replication fork intermediates have also been observed. The depletion of RAD51 and BRCA2 in Xenopus egg extracts results in replication fork intermediates with increased ssDNA at and also behind the replication fork (Hashimoto et al., 2010; Kolinjivadi et al., 2017). Similar to unprotected reversed replication forks, internal ssDNA gaps behind the fork are substrates for MRE11, while ssDNA gaps at the replication fork junction are not (Hashimoto et al., 2010; Kolinjivadi et al., 2017). However, it was suggested that the ssDNA at the replication fork junction may be an intermediate that precedes replication fork reversal, which may then become a nuclease substrate after fork remodeling (Kolinjivadi et al., 2017). Further understanding of the roles of BRCA2 and RAD51 at the replication fork is needed in order to determine how they prevent ssDNA generation and if the mechanism of protection is distinct at different fork intermediates.
MRE11 recruitment to stalled forks
MRE11 travels with the replisome, and its recruitment to chromatin is enhanced by exogenous replication stress (Mirzoeva and Petrini, 2003; Robison et al., 2004; Dungrawala et al., 2015). While MRE11 is required for the processing of stalled replication forks, its aberrant activity at unprotected stalled replication forks in BRCA1/2-deficient cells contributes to increased genomic instability (Schlacher et al., 2011, 2012; Ying et al., 2012; Ray Chaudhuri et al., 2016; Taglialatela et al., 2017).
The presence of MRE11 at the replisome following replication stress is dependent on many factors, including PARP1, PTIP, and associated methyltransferases MLL3/MLL4 (Ding et al., 2016; Ray Chaudhuri et al., 2016; Mijic et al., 2017), chromatin remodeler CHD4 (Ray Chaudhuri et al., 2016), RAD52 (Mijic et al., 2017), and SAMHD1 (Coquel et al., 2018). Depletion or inhibition of these proteins results in decreased MRE11 association with the stressed replication fork and suppression of genomic instability in BRCA1- or BRCA2-deficient cells (Ding et al., 2016; Ray Chaudhuri et al., 2016; Mijic et al., 2017).
Contribution of other nucleases to nascent strand degradation
Recent studies also attribute the resection of unprotected nascent DNA at stalled forks to EXO1 and CTiP. Depletion of either of these nucleases rescues nascent strand degradation in BRCA2-deficient cells (Lemaçon et al., 2017). Similarly, knockdown of EXO1 rescues reversed fork levels that are decreased in BRCA1/2-deficient cells treated with replication stress–inducing drugs (Lemaçon et al., 2017). These data put forth a working model of resection at unprotected replication forks in the BRCA2-deficient setting that looks remarkably similar to the genetic requirements for resection at DSBs. In the absence of BRCA2, resection may be initiated by CTiP and MRE11 followed by more extensive processing by EXO1.
Current data on the involvement of DNA2 in the processing of stalled replication forks in BRCA2-deficient cells is still contradictory. One study found that DNA2 depletion does not rescue nascent strand degradation in BRCA2-deficient cells (Lemaçon et al., 2017), while another showed, in BRCA2-deficient B cells, that DNA2 inhibition is epistatic with MRE11 in the resection of nascent DNA (Ray Chaudhuri et al., 2016). It is unclear what accounts for these differences, but the studies use different cell types and assess the role of DNA2 using two different methods: siRNA depletion and a small-molecule inhibitor (Ray Chaudhuri et al., 2016; Lemaçon et al., 2017). Further investigation will be required to determine the dependency on DNA2 in fork processing in the BRCA2-deficient setting. It has been shown, however, that DNA2 does aberrantly process nascent DNA at stalled replication forks in cells defective for replication fork protection by RECQ1, BOD1L, and Abro1 (Higgs et al., 2015; Thangavel et al., 2015; Xu et al., 2017a). Under conditions of prolonged replication fork stalling, DNA2 and WRN are important for replication fork restart (Thangavel et al., 2015). DNA2 and WRN are also responsible for the hyperresection and ssDNA formation at DNA interstrand cross-links when DNA is not properly protected by RAD51 (Wang et al., 2015).
Role of MUS81 in creating DSBs at stalled replication forks
MUS81 is the nuclease responsible for DSB formation during replication stress leading to replication fork collapse (Hanada et al., 2007; Franchitto et al., 2008; Bétous et al., 2013b; Fugger et al., 2013). Increased MUS81 activity may lead to a high number of DSBs in the setting of replication stress associated with oncogene activation (Murfuni et al., 2013; Neelsen et al., 2013). However, it is now understood, that controlled DNA breakage by MUS81 is often a necessary compromise to promote genome stability and replication restart at forks challenged by replication stress (Hanada et al., 2007; Franchitto et al., 2008; Regairaz et al., 2011; Pepe and West, 2014). MUS81 activity is also required at late-replicating regions in the genome known as common fragile sites (CFSs), and without MUS81 processing, CFSs cause genomic instability during mitosis (Naim et al., 2013; Ying et al., 2013).
In the setting of BRCA2 deficiency, MUS81 depletion prevents DSB formation after fork stalling (Lemaçon et al., 2017). MUS81 depletion rescues reversed fork levels in BRCA2-deficient cells, but these replication fork intermediates appear to have an ssDNA flap, which may be a substrate for MUS81 cleavage that is generated by MRE11 or EXO1 resection. When the nascent strand degradation is inhibited by depletion of MRE11 or EXO1, the DSB formation also decreases, which is consistent with MUS81 working downstream of MRE11 and EXO1 processing (Lemaçon et al., 2017). Importantly, MUS81 is necessary for resistance to replication stress (HU) in BRCA2-deficient cells, and MUS81-dependent cleavage is necessary for fork restart (Lai et al., 2017; Lemaçon et al., 2017).
MUS81-dependent processing of stalled replication forks is promoted by EZH2, a histone methyl transferase, but only in BRCA2-deficient and not in BRCA1-deficient cells (Rondinelli et al., 2017). The same study found that MUS81 depletion rescued replication fork degradation in BRCA2-deficient cells, which is in contrast to findings reported in a different study (Lemaçon et al., 2017). EZH2 and MRE11 codepletion further augments fork protection, suggesting a separate mechanism of MUS81 and MRE11 recruitment to stalled fork (Rondinelli et al., 2017).
Modulation of RAD51 activity by RADX and FBH1
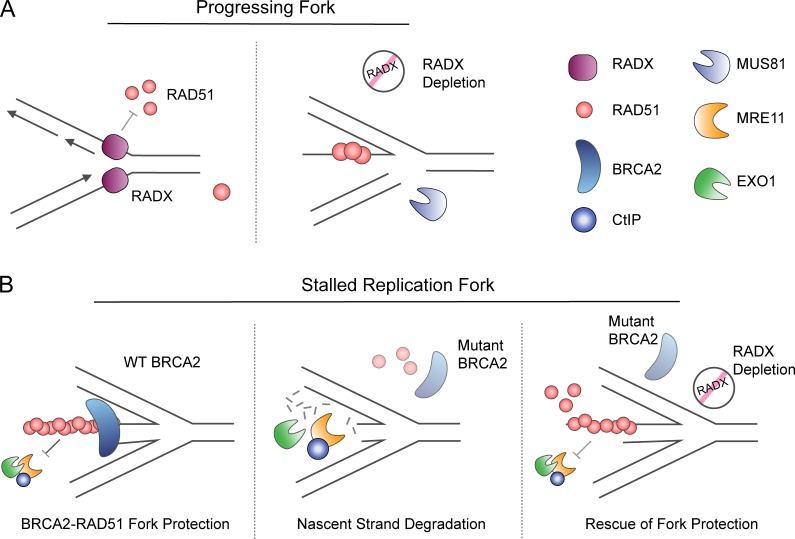
RADX is an ssDNA-binding protein recently identified as enriched at replication forks following replication stress (Dungrawala et al., 2017). It has sequence similarity to RPA and binds DNA using RPA-like oligonucleotide/oligosaccharide-binding folds. RADX is recruited to replication forks where it modulates RAD51 through a yet-to-be determined mechanism. RAD51, but not BRCA2 protein, accumulates at stalled replication forks in the absence of RADX. Conversely, RADX overexpression results in a reduction of RAD51 at stalled replication forks (Dungrawala et al., 2017), suggesting RADX antagonizes RAD51 function.
Although, RADX is not essential for survival in cells, where knockout has been tested, its absence slows replication forks and leads to DSB formation (Dungrawala et al., 2017; Schubert et al., 2017). These defects in RADX-deficient cells are rescued by depletion of RAD51, SMARCAL1, ZRANB3, and the MUS81 nuclease (Dungrawala et al., 2017). In light of the data described regarding the role of RAD51 in replication fork reversal, these results suggest that in the absence of RADX, hyperactivity of RAD51 interferes with normal replication and promotes inappropriate replication fork remodeling that results in increased processing by MUS81 (Dungrawala et al., 2017; Fig. 4 A).
Figure 4.
RADX modulates RAD51 activity at replication forks. (A) A proposed model of RADX function is to regulate RAD51 activity at the replication fork to prevent unnecessary RAD51 association and fork remodeling during unperturbed DNA replication. Upon RADX depletion, there is increased genomic instability and DSBs that may be the result of inappropriate replication fork remodeling, leading to increased fork cleavage by MUS81. (B) The depletion of RADX in BRCA2-deficient cells rescues nascent strand degradation at HU stalled replication forks without affecting homologous recombination. It is proposed that the removal of RADX results in increased RAD51 function, supporting improved RAD51 fork protection and the prevention of nascent strand degradation by nucleases.
RADX levels must be carefully controlled as overexpression of RADX also increases DNA damage due to enhanced nascent strand degradation at stalled replication forks. This may be the result of RADX antagonizing the RAD51 filament formation and fork reversal. Reciprocally, depletion of RADX and the concomitant increase in RAD51 at the stalled fork rescues nascent strand degradation, but not HDR, in BRCA2-deficient cells. Interestingly, RADX depletion also restores fork protection in cells deficient for BODL1 (Bhat et al., 2018). RAD51 nucleofilament formation in the absence of RADX may be significant enough to protect the regressed replication forks even in the absence of BRCA2 and BODL1 (Dungrawala et al., 2017; Bhat et al., 2018).
RADX depletion induces resistance to poly (ADP-ribose) polymerase (PARP) inhibitors in the BRCA2-deficient cells and increases resistance of RAD51-depleted cells to HU, camptothecin, and cisplatin (Dungrawala et al., 2017). Although still poorly understood, RADX appears to be a critical regulator of RAD51 at the stalled replication fork. How the cell balances RADX levels to enhance replication fork protection without induction of inappropriate fork reversal during unperturbed replication will be an important area of study (Fig. 4 B).
Another effector protein of RAD51 is FBH1, a 3′–5′ DNA helicase of the UvrD family that contains an F-Box domain, a proliferating cell nuclear antigen (PCNA) –interacting protein (PIP) box, and an AlkB homologue 2 PCNA interaction motif (APIM; Kim et al., 2002; Bacquin et al., 2013). FBH1 is recruited to sites of replication stress through interaction with PCNA (Fugger et al., 2009; Bacquin et al., 2013). In vitro, FBH1 has DNA unwinding activity and catalyzes fork regression (Kim et al., 2002; Fugger et al., 2015). FBH1 can form an SKP1–CUL1–F-box (SCF) complex with E3 ubiquitin ligase activity that targets K58/64 of RAD51 (Kim et al., 2002; Chu et al., 2015) and may negatively regulate RAD51 localization on chromatin (Chu et al., 2015). UCHL3 acts as a deubiquitinase that promotes the RAD51–BRCA2 interaction and positively regulates RAD51 chromatin localization (Luo et al., 2016).
Depletion of FBH1 increases RAD51 recruitment to chromatin but ultimately results in a reduced number of reversed fork species in response to HU, as the helicase activity of FBH1 is required for fork reversal (Fugger et al., 2015). FBH1 depletion decreases DSBs in response to replication stress, and codepletion with MUS81 does not further reduce DSBs (Fugger et al., 2013). Conversely, the overexpression of FBH1 impairs RAD51 foci formation at replication forks and results in increased ssDNA and DSBs (Fugger et al., 2009). Depletion of MUS81 in an FBH1 overexpression background rescues increased DSB formation. This places FBH1 activity upstream of MUS81 processing in promoting replication fork reversal (Fugger et al., 2013).
Unlike RADX depletion, FBH1 depletion in BRCA2-deficient cells does not rescue replication fork protection due to HU treatment (Higgs et al., 2015; Leuzzi et al., 2016). However, FBH1 depletion in cells deficient for the fork protection factors BOD1L or WRNIP1 does rescue nascent strand degradation of stalled replication forks (Higgs et al., 2015; Leuzzi et al., 2016). These differences provide insight into the layers of regulation of fork protection by these factors, placing BOD1L and WRNIP1 downstream of BRCA2. In this model, BRCA2 is required for stable RAD51 filament formation on the regressed replication fork, while BOD1L and WRNIP1 are important for protection of the loaded RAD51 from FBH1 and the nucleases. In cells deficient for BOD1L, nascent strand degradation is mediated by DNA2, whereas nascent strand degradation in WRNIP1-deficient cells is mediated by MRE11 (Higgs et al., 2015; Leuzzi et al., 2016). Understanding whether the ubiquitin ligase, helicase activity, or both functions of FBH1 is responsible for promoting nascent strand resection in cells deficient for replication protection requires further investigation.
DNA translocases in replication fork protection and processing
SMARCAL1, ZRANB3, and HLTF are ATPase-dependent DNA translocases of the SNF2 family of chromatin remodelers. These related proteins have been shown to have similar fork remodeling activity in vitro. SMARCAL1 demonstrates affinity for DNA fork structures and catalyzes strand annealing, fork regression, and branch migration (Yusufzai and Kadonaga, 2008; Bétous et al., 2012; Ciccia et al., 2012). ZRANB3 and HLTF also catalyze replication fork reversal in vitro (Blastyák et al., 2010; Yusufzai and Kadonaga, 2010; Ciccia et al., 2012; Yuan et al., 2012). Recent work has further expanded the role of SMARCAL1, ZRANB3, and HLTF to replication fork reversal in vivo (Fig. 5).
Figure 5.
Replication fork reversal mediated by translocases. Replication fork reversal is mediated in part by the SNF2 family chromatin remodelers SMARCAL1, ZRANB3, and HLTF. Whether the translocases work synergistically to reverse forks or operate on distinct fork structures is unclear and needs further investigation. HLTF is important for the polyubiquitination of PCNA that serves as a platform for recruitment of ZRANB3. SMARCAL1 is recruited to the replication fork through interaction with RPA.
Depletion of SMARCAL1, ZRANB3, or HLTF results in decreased detection of reversed replication fork intermediates by EM (Taglialatela et al., 2017; Vujanovic et al., 2017). Consistent with their function in fork reversal, depletion of any of the translocases rescues nascent strand degradation at unprotected stalled replication forks (Kolinjivadi et al., 2017; Taglialatela et al., 2017; Vujanovic et al., 2017). In the absence of any of the three translocases, cells become hypersensitive to replication stress–inducing agents and have increased genomic instability (Bansbach et al., 2009; Ciccia et al., 2009, 2012; Yuan et al., 2012; Taglialatela et al., 2017; Peng et al., 2018), but it remains unclear if the instability is secondary to the inability to reverse stalled replication forks.
All three translocases have been found to associate with the replication fork; however, how they do it is distinct. SMARCAL1 travels with the replication fork and becomes further enriched following replication stress through interaction with RPA (Bansbach et al., 2009; Ciccia et al., 2009; Postow et al., 2009; Yuan et al., 2009; Bétous et al., 2012; Dungrawala et al., 2015; Kolinjivadi et al., 2017). SMARCAL1’s interaction with RPA is important for providing substrate specificity to promote replication fork reversal and prevent activity during normal DNA replication (Bétous et al., 2013a). ATR phosphorylation of S652 of RPA-bound SMARCAL1 has been shown to be important for regulating its activity at the replication fork (Couch et al., 2013).
HLTF and ZRANB3 have been shown to interact with PCNA. HLTF contains a RING finger domain and a N-terminal HIRAN domain (Poole and Cortez, 2017). HLTF acts as ubiquitin ligase to polyubiquitinate PCNA in an MMS2-Ubc13–dependent manner (Motegi et al., 2008; Unk et al., 2008). In vitro studies indicate the HIRAN domain of HLTF recognizes the 3′ end of the leading strand to promote replication fork reversal (Kile et al., 2015). Recent work identified FANCJ as an HLTF interactor and suggests that HLTF activity may counter FANCJ activity at the fork to prevent unrestrained replication in the face of replication stress (Peng et al., 2018).
ZRANB3 localization to DNA is amplified upon induction of replication stress through PIP box– and APIM-dependent binding of PCNA (Ciccia et al., 2012; Weston et al., 2012; Yuan et al., 2012). ZRANB3 also contains an NPL4 zinc-finger motif that preferentially binds K63 polyubiquitinated PCNA and is required for its localization to sites of replication stress (Ciccia et al., 2012; Vujanovic et al., 2017).
Despite similar biochemical activity of the three translocases, it is clear that these proteins are not redundant and that dissection of their functions requires further characterization. They may have synergistic functions at the replication fork, nonoverlapping roles on different replication fork substrates, or additional roles outside of replication fork remodeling. The nonoverlapping function is highlighted by observations that SMARCAL1 activity is important for replication through difficult-to-replicate telomeric sequences, a function not attributed to ZRANB3 or HLTF (Poole et al., 2015). The distinct nature of SMARCAL1 activity is also emphasized by the disease Schimke immunosseous dysplasia (SIOD) that results from biallelic pathogenic mutations in SMARCAL1, but not HLTF or ZRANB3. Key features of SIOD are immunodeficiency, skeletal abnormalities, and renal failure (Boerkoel et al., 2002), and it remains to be determined if or how the lack of replication fork protection and genome instability contributes to this disease.
Concluding remarks
Investigations reviewed here strive to better elucidate the cellular response to replication stress and the role that replication fork reversal has in this response. These studies provided new insight about noncanonical roles and regulation of BRCA2 and RAD51 outside of HDR and shed light on how they protect replication fork intermediates from nucleolytic degradation. EM has permitted the observation that a number of factors, such as RAD51, FBH1, SMARCAL1, ZRANB3, and HLTF, have a role in promoting replication fork reversal in vivo. However, this work raises a number of questions. Alterations in the activity of a number of these factors causes increased genomic instability, so understanding the tradeoffs between protection against damage arising from replication and other sources of genome instability is of interest. The redundancy in roles of a number of factors, especially translocases and nucleases, leaves open the question of how these factors specifically operate in response to replication stress. As replication fork reversal has been shown to be a ubiquitous response to many types of replication stress, whether these factors, especially the translocases, act coordinately at the replication fork or have distinct roles depending on the type of DNA damage remains unknown.
The majority of the work discussed here was performed in the context of BRCA1 or BRCA2 deficiency, and it is unclear what the roles of these proteins are at the replication fork when BRCA proteins are present. The assumption is that a number of these factors recruited to the reversed replication forks have a role there even when the fork is properly protected. These roles need to be better understood and may differentiate between their diverse functions during replication in wild-type cells.
Acknowledgments
We thank Ryan White, Molly Kottemann, and Brooke Conti for comments on the manuscript.
The work on RAD51 and BRCA2 in the laboratory is supported by the National Institutes of Health (grant RO1 CA204127) and by the National Center for Advancing Translational Sciences, NIH Clinical and Translational Science Award program (grant UL1TR001866). K. Rickman was supported by the National Institute of General Medical Sciences of the National Institutes of Health (Medical Scientist Training Program award number T32GM007739 to the Weill Cornell/Rockefeller/Sloan-Kettering Tri-Institutional MD-PhD Program) and the William Randolph Hearst Foundation Fellowship at The Rockefeller University. A. Smogorzewska is a Howard Hughes Medical Institute Faculty Scholar.
The authors declare no competing financial interests.
Author contributions: Both authors co-wrote the manuscript and co-designed/drew the figures.
References
- Ahuja A.K., Jodkowska K., Teloni F., Bizard A.H., Zellweger R., Herrador R., Ortega S., Hickson I.D., Altmeyer M., Mendez J., and Lopes M.. 2016. A short G1 phase imposes constitutive replication stress and fork remodelling in mouse embryonic stem cells. Nat. Commun. 7:10660 10.1038/ncomms10660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacquin A., Pouvelle C., Siaud N., Perderiset M., Salomé-Desnoulez S., Tellier-Lebegue C., Lopez B., Charbonnier J.B., and Kannouche P.L.. 2013. The helicase FBH1 is tightly regulated by PCNA via CRL4(Cdt2)-mediated proteolysis in human cells. Nucleic Acids Res. 41:6501–6513. 10.1093/nar/gkt397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bansbach C.E., Bétous R., Lovejoy C.A., Glick G.G., and Cortez D.. 2009. The annealing helicase SMARCAL1 maintains genome integrity at stalled replication forks. Genes Dev. 23:2405–2414. 10.1101/gad.1839909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barazas M., Annunziato S., Pettitt S.J., de Krijger I., Ghezraoui H., Roobol S.J., Lutz C., Frankum J., Song F.F., Brough R., et al. 2018. The CST Complex Mediates End Protection at Double-Strand Breaks and Promotes PARP Inhibitor Sensitivity in BRCA1-Deficient Cells. Cell Reports. 23:2107–2118. 10.1016/j.celrep.2018.04.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berti M., Ray Chaudhuri A., Thangavel S., Gomathinayagam S., Kenig S., Vujanovic M., Odreman F., Glatter T., Graziano S., Mendoza-Maldonado R., et al. 2013. Human RECQ1 promotes restart of replication forks reversed by DNA topoisomerase I inhibition. Nat. Struct. Mol. Biol. 20:347–354. 10.1038/nsmb.2501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bétous R., Mason A.C., Rambo R.P., Bansbach C.E., Badu-Nkansah A., Sirbu B.M., Eichman B.F., and Cortez D.. 2012. SMARCAL1 catalyzes fork regression and Holliday junction migration to maintain genome stability during DNA replication. Genes Dev. 26:151–162. 10.1101/gad.178459.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bétous R., Couch F.B., Mason A.C., Eichman B.F., Manosas M., and Cortez D.. 2013a Substrate-selective repair and restart of replication forks by DNA translocases. Cell Reports. 3:1958–1969. 10.1016/j.celrep.2013.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bétous R., Glick G.G., Zhao R., and Cortez D.. 2013b Identification and characterization of SMARCAL1 protein complexes. PLoS One. 8:e63149 10.1371/journal.pone.0063149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhat K.P., Krishnamoorthy A., Dungrawala H., Garcin E.B., Modesti M., and Cortez D.. 2018. RADX Modulates RAD51 Activity to Control Replication Fork Protection. Cell Reports. 24:538–545. 10.1016/j.celrep.2018.06.061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billing D., Horiguchi M., Wu-Baer F., Taglialatela A., Leuzzi G., Nanez S.A., Jiang W., Zha S., Szabolcs M., Lin C.S., et al. 2018. The BRCT Domains of the BRCA1 and BARD1 Tumor Suppressors Differentially Regulate Homology-Directed Repair and Stalled Fork Protection. Mol. Cell. 72:127–139.e8. 10.1016/j.molcel.2018.08.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blastyák A., Hajdú I., Unk I., and Haracska L.. 2010. Role of double-stranded DNA translocase activity of human HLTF in replication of damaged DNA. Mol. Cell. Biol. 30:684–693. 10.1128/MCB.00863-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boerkoel C.F., Takashima H., John J., Yan J., Stankiewicz P., Rosenbarker L., André J.L., Bogdanovic R., Burguet A., Cockfield S., et al. 2002. Mutant chromatin remodeling protein SMARCAL1 causes Schimke immuno-osseous dysplasia. Nat. Genet. 30:215–220. 10.1038/ng821 [DOI] [PubMed] [Google Scholar]
- Chen L., Nievera C.J., Lee A.Y., and Wu X.. 2008. Cell cycle-dependent complex formation of BRCA1.CtIP.MRN is important for DNA double-strand break repair. J. Biol. Chem. 283:7713–7720. 10.1074/jbc.M710245200 [DOI] [PubMed] [Google Scholar]
- Chu W.K., Payne M.J., Beli P., Hanada K., Choudhary C., and Hickson I.D.. 2015. FBH1 influences DNA replication fork stability and homologous recombination through ubiquitylation of RAD51. Nat. Commun. 6:5931 10.1038/ncomms6931 [DOI] [PubMed] [Google Scholar]
- Ciccia A., Bredemeyer A.L., Sowa M.E., Terret M.E., Jallepalli P.V., Harper J.W., and Elledge S.J.. 2009. The SIOD disorder protein SMARCAL1 is an RPA-interacting protein involved in replication fork restart. Genes Dev. 23:2415–2425. 10.1101/gad.1832309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciccia A., Nimonkar A.V., Hu Y., Hajdu I., Achar Y.J., Izhar L., Petit S.A., Adamson B., Yoon J.C., Kowalczykowski S.C., et al. 2012. Polyubiquitinated PCNA recruits the ZRANB3 translocase to maintain genomic integrity after replication stress. Mol. Cell. 47:396–409. 10.1016/j.molcel.2012.05.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coquel F., Silva M.J., Técher H., Zadorozhny K., Sharma S., Nieminuszczy J., Mettling C., Dardillac E., Barthe A., Schmitz A.L., et al. 2018. SAMHD1 acts at stalled replication forks to prevent interferon induction. Nature. 557:57–61. 10.1038/s41586-018-0050-1 [DOI] [PubMed] [Google Scholar]
- Cortez D. 2015. Preventing replication fork collapse to maintain genome integrity. DNA Repair (Amst.). 32:149–157. 10.1016/j.dnarep.2015.04.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couch F.B., Bansbach C.E., Driscoll R., Luzwick J.W., Glick G.G., Bétous R., Carroll C.M., Jung S.Y., Qin J., Cimprich K.A., and Cortez D.. 2013. ATR phosphorylates SMARCAL1 to prevent replication fork collapse. Genes Dev. 27:1610–1623. 10.1101/gad.214080.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies O.R., and Pellegrini L.. 2007. Interaction with the BRCA2 C terminus protects RAD51-DNA filaments from disassembly by BRC repeats. Nat. Struct. Mol. Biol. 14:475–483. 10.1038/nsmb1251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dev H., Chiang T.W., Lescale C., de Krijger I., Martin A.G., Pilger D., Coates J., Sczaniecka-Clift M., Wei W., Ostermaier M., et al. 2018. Shieldin complex promotes DNA end-joining and counters homologous recombination in BRCA1-null cells. Nat. Cell Biol. 20:954–965. 10.1038/s41556-018-0140-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding X., Ray Chaudhuri A., Callen E., Pang Y., Biswas K., Klarmann K.D., Martin B.K., Burkett S., Cleveland L., Stauffer S., et al. 2016. Synthetic viability by BRCA2 and PARP1/ARTD1 deficiencies. Nat. Commun. 7:12425 10.1038/ncomms12425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dungrawala H., Rose K.L., Bhat K.P., Mohni K.N., Glick G.G., Couch F.B., and Cortez D.. 2015. The Replication Checkpoint Prevents Two Types of Fork Collapse without Regulating Replisome Stability. Mol. Cell. 59:998–1010. 10.1016/j.molcel.2015.07.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dungrawala H., Bhat K.P., Le Meur R., Chazin W.J., Ding X., Sharan S.K., Wessel S.R., Sathe A.A., Zhao R., and Cortez D.. 2017. RADX Promotes Genome Stability and Modulates Chemosensitivity by Regulating RAD51 at Replication Forks. Mol. Cell. 67:374–386.e5. 10.1016/j.molcel.2017.06.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esashi F., Christ N., Gannon J., Liu Y., Hunt T., Jasin M., and West S.C.. 2005. CDK-dependent phosphorylation of BRCA2 as a regulatory mechanism for recombinational repair. Nature. 434:598–604. 10.1038/nature03404 [DOI] [PubMed] [Google Scholar]
- Feng W., and Jasin M.. 2017. BRCA2 suppresses replication stress-induced mitotic and G1 abnormalities through homologous recombination. Nat. Commun. 8:525 10.1038/s41467-017-00634-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franchitto A., Pirzio L.M., Prosperi E., Sapora O., Bignami M., and Pichierri P.. 2008. Replication fork stalling in WRN-deficient cells is overcome by prompt activation of a MUS81-dependent pathway. J. Cell Biol. 183:241–252. 10.1083/jcb.200803173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fugger K., Mistrik M., Danielsen J.R., Dinant C., Falck J., Bartek J., Lukas J., and Mailand N.. 2009. Human Fbh1 helicase contributes to genome maintenance via pro- and anti-recombinase activities. J. Cell Biol. 186:655–663. 10.1083/jcb.200812138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fugger K., Chu W.K., Haahr P., Kousholt A.N., Beck H., Payne M.J., Hanada K., Hickson I.D., and Sørensen C.S.. 2013. FBH1 co-operates with MUS81 in inducing DNA double-strand breaks and cell death following replication stress. Nat. Commun. 4:1423 10.1038/ncomms2395 [DOI] [PubMed] [Google Scholar]
- Fugger K., Mistrik M., Neelsen K.J., Yao Q., Zellweger R., Kousholt A.N., Haahr P., Chu W.K., Bartek J., Lopes M., et al. 2015. FBH1 Catalyzes Regression of Stalled Replication Forks. Cell Reports. 10:1749–P1757. 10.1016/j.celrep.2015.02.028 [DOI] [PubMed] [Google Scholar]
- Ghezraoui H., Oliveira C., Becker J.R., Bilham K., Moralli D., Anzilotti C., Fischer R., Deobagkar-Lele M., Sanchiz-Calvo M., Fueyo-Marcos E., et al. 2018. 53BP1 cooperation with the REV7-shieldin complex underpins DNA structure-specific NHEJ. Nature. 560:122–127. 10.1038/s41586-018-0362-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gravel S., Chapman J.R., Magill C., and Jackson S.P.. 2008. DNA helicases Sgs1 and BLM promote DNA double-strand break resection. Genes Dev. 22:2767–2772. 10.1101/gad.503108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta R., Somyajit K., Narita T., Maskey E., Stanlie A., Kremer M., Typas D., Lammers M., Mailand N., Nussenzweig A., et al. 2018. DNA Repair Network Analysis Reveals Shieldin as a Key Regulator of NHEJ and PARP Inhibitor Sensitivity. Cell. 173:972–988.e23. 10.1016/j.cell.2018.03.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanada K., Budzowska M., Davies S.L., van Drunen E., Onizawa H., Beverloo H.B., Maas A., Essers J., Hickson I.D., and Kanaar R.. 2007. The structure-specific endonuclease Mus81 contributes to replication restart by generating double-strand DNA breaks. Nat. Struct. Mol. Biol. 14:1096–1104. 10.1038/nsmb1313 [DOI] [PubMed] [Google Scholar]
- Hartford S.A., Chittela R., Ding X., Vyas A., Martin B., Burkett S., Haines D.C., Southon E., Tessarollo L., and Sharan S.K.. 2016. Interaction with PALB2 Is Essential for Maintenance of Genomic Integrity by BRCA2. PLoS Genet. 12:e1006236 10.1371/journal.pgen.1006236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto Y., Ray Chaudhuri A., Lopes M., and Costanzo V.. 2010. Rad51 protects nascent DNA from Mre11-dependent degradation and promotes continuous DNA synthesis. Nat. Struct. Mol. Biol. 17:1305–1311. 10.1038/nsmb.1927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y.J., Meghani K., Caron M.C., Yang C., Ronato D.A., Bian J., Sharma A., Moore J., Niraj J., Detappe A., et al. 2018. DYNLL1 binds to MRE11 to limit DNA end resection in BRCA1-deficient cells. Nature. 563:522–526. 10.1038/s41586-018-0670-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgs M.R., Reynolds J.J., Winczura A., Blackford A.N., Borel V., Miller E.S., Zlatanou A., Nieminuszczy J., Ryan E.L., Davies N.J., et al. 2015. BOD1L Is Required to Suppress Deleterious Resection of Stressed Replication Forks. Mol. Cell. 59:462–477. 10.1016/j.molcel.2015.06.007 [DOI] [PubMed] [Google Scholar]
- Jasin M., and Rothstein R.. 2013. Repair of strand breaks by homologous recombination. Cold Spring Harb. Perspect. Biol. 5:a012740 10.1101/cshperspect.a012740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kile A.C., Chavez D.A., Bacal J., Eldirany S., Korzhnev D.M., Bezsonova I., Eichman B.F., and Cimprich K.A.. 2015. HLTF’s Ancient HIRAN Domain Binds 3′ DNA Ends to Drive Replication Fork Reversal. Mol. Cell. 58:1090–1100. 10.1016/j.molcel.2015.05.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., Kim J.H., Lee S.H., Kim D.H., Kang H.Y., Bae S.H., Pan Z.Q., and Seo Y.S.. 2002. The novel human DNA helicase hFBH1 is an F-box protein. J. Biol. Chem. 277:24530–24537. 10.1074/jbc.M201612200 [DOI] [PubMed] [Google Scholar]
- Kolinjivadi A.M., Sannino V., De Antoni A., Zadorozhny K., Kilkenny M., Techer H., Baldi G., Shen R., Ciccia A., Pellegrini L., et al. 2017. Smarcal1-Mediated Fork Reversal Triggers Mre11-Dependent Degradation of Nascent DNA in the Absence of Brca2 and Stable Rad51 Nucleofilaments. Mol. Cell. 67:867–881.e7. 10.1016/j.molcel.2017.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai X., Broderick R., Bergoglio V., Zimmer J., Badie S., Niedzwiedz W., Hoffmann J.S., and Tarsounas M.. 2017. MUS81 nuclease activity is essential for replication stress tolerance and chromosome segregation in BRCA2-deficient cells. Nat. Commun. 8:15983 10.1038/ncomms15983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemaçon D., Jackson J., Quinet A., Brickner J.R., Li S., Yazinski S., You Z., Ira G., Zou L., Mosammaparast N., and Vindigni A.. 2017. MRE11 and EXO1 nucleases degrade reversed forks and elicit MUS81-dependent fork rescue in BRCA2-deficient cells. Nat. Commun. 8:860 10.1038/s41467-017-01180-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leuzzi G., Marabitti V., Pichierri P., and Franchitto A.. 2016. WRNIP1 protects stalled forks from degradation and promotes fork restart after replication stress. EMBO J. 35:1437–1451. 10.15252/embj.201593265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo K., Li L., Li Y., Wu C., Yin Y., Chen Y., Deng M., Nowsheen S., Yuan J., and Lou Z.. 2016. A phosphorylation-deubiquitination cascade regulates the BRCA2-RAD51 axis in homologous recombination. Genes Dev. 30:2581–2595. 10.1101/gad.289439.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mijic S., Zellweger R., Chappidi N., Berti M., Jacobs K., Mutreja K., Ursich S., Ray Chaudhuri A., Nussenzweig A., Janscak P., and Lopes M.. 2017. Replication fork reversal triggers fork degradation in BRCA2-defective cells. Nat. Commun. 8:859 10.1038/s41467-017-01164-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimitou E.P., and Symington L.S.. 2008. Sae2, Exo1 and Sgs1 collaborate in DNA double-strand break processing. Nature. 455:770–774. 10.1038/nature07312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirman Z., Lottersberger F., Takai H., Kibe T., Gong Y., Takai K., Bianchi A., Zimmermann M., Durocher D., and de Lange T.. 2018. 53BP1-RIF1-shieldin counteracts DSB resection through CST- and Polα-dependent fill-in. Nature. 560:112–116. 10.1038/s41586-018-0324-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirzoeva O.K., and Petrini J.H.. 2003. DNA replication-dependent nuclear dynamics of the Mre11 complex. Mol. Cancer Res. 1:207–218. [PubMed] [Google Scholar]
- Motegi A., Liaw H.J., Lee K.Y., Roest H.P., Maas A., Wu X., Moinova H., Markowitz S.D., Ding H., Hoeijmakers J.H., and Myung K.. 2008. Polyubiquitination of proliferating cell nuclear antigen by HLTF and SHPRH prevents genomic instability from stalled replication forks. Proc. Natl. Acad. Sci. USA. 105:12411–12416. 10.1073/pnas.0805685105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murfuni I., Nicolai S., Baldari S., Crescenzi M., Bignami M., Franchitto A., and Pichierri P.. 2013. The WRN and MUS81 proteins limit cell death and genome instability following oncogene activation. Oncogene. 32:610–620. 10.1038/onc.2012.80 [DOI] [PubMed] [Google Scholar]
- Murphy A.K., Fitzgerald M., Ro T., Kim J.H., Rabinowitsch A.I., Chowdhury D., Schildkraut C.L., and Borowiec J.A.. 2014. Phosphorylated RPA recruits PALB2 to stalled DNA replication forks to facilitate fork recovery. J. Cell Biol. 206:493–507. 10.1083/jcb.201404111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutreja K., Krietsch J., Hess J., Ursich S., Berti M., Roessler F.K., Zellweger R., Patra M., Gasser G., and Lopes M.. 2018. ATR-Mediated Global Fork Slowing and Reversal Assist Fork Traverse and Prevent Chromosomal Breakage at DNA Interstrand Cross-Links. Cell Reports. 24:2629–2642.e5. 10.1016/j.celrep.2018.08.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naim V., Wilhelm T., Debatisse M., and Rosselli F.. 2013. ERCC1 and MUS81-EME1 promote sister chromatid separation by processing late replication intermediates at common fragile sites during mitosis. Nat. Cell Biol. 15:1008–1015. 10.1038/ncb2793 [DOI] [PubMed] [Google Scholar]
- Neelsen K.J., Zanini I.M., Herrador R., and Lopes M.. 2013. Oncogenes induce genotoxic stress by mitotic processing of unusual replication intermediates. J. Cell Biol. 200:699–708. 10.1083/jcb.201212058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noordermeer S.M., Adam S., Setiaputra D., Barazas M., Pettitt S.J., Ling A.K., Olivieri M., Álvarez-Quilón A., Moatti N., Zimmermann M., et al. 2018. The shieldin complex mediates 53BP1-dependent DNA repair. Nature. 560:117–121. 10.1038/s41586-018-0340-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Donnell L., Panier S., Wildenhain J., Tkach J.M., Al-Hakim A., Landry M.C., Escribano-Diaz C., Szilard R.K., Young J.T., Munro M., et al. 2010. The MMS22L-TONSL complex mediates recovery from replication stress and homologous recombination. Mol. Cell. 40:619–631. 10.1016/j.molcel.2010.10.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng M., Cong K., Panzarino N.J., Nayak S., Calvo J., Deng B., Zhu L.J., Morocz M., Hegedus L., Haracska L., and Cantor S.B.. 2018. Opposing Roles of FANCJ and HLTF Protect Forks and Restrain Replication during Stress. Cell Reports. 24:3251–3261. 10.1016/j.celrep.2018.08.065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pepe A., and West S.C.. 2014. MUS81-EME2 promotes replication fork restart. Cell Reports. 7:1048–1055. 10.1016/j.celrep.2014.04.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petermann E., Orta M.L., Issaeva N., Schultz N., and Helleday T.. 2010. Hydroxyurea-stalled replication forks become progressively inactivated and require two different RAD51-mediated pathways for restart and repair. Mol. Cell. 37:492–502. 10.1016/j.molcel.2010.01.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piwko W., Mlejnkova L.J., Mutreja K., Ranjha L., Stafa D., Smirnov A., Brodersen M.M., Zellweger R., Sturzenegger A., Janscak P., et al. 2016. The MMS22L-TONSL heterodimer directly promotes RAD51-dependent recombination upon replication stress. EMBO J. 35:2584–2601. 10.15252/embj.201593132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poole L.A., and Cortez D.. 2017. Functions of SMARCAL1, ZRANB3, and HLTF in maintaining genome stability. Crit. Rev. Biochem. Mol. Biol. 52:696–714. 10.1080/10409238.2017.1380597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poole L.A., Zhao R., Glick G.G., Lovejoy C.A., Eischen C.M., and Cortez D.. 2015. SMARCAL1 maintains telomere integrity during DNA replication. Proc. Natl. Acad. Sci. USA. 112:14864–14869. 10.1073/pnas.1510750112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postow L., Woo E.M., Chait B.T., and Funabiki H.. 2009. Identification of SMARCAL1 as a component of the DNA damage response. J. Biol. Chem. 284:35951–35961. 10.1074/jbc.M109.048330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray Chaudhuri A., Hashimoto Y., Herrador R., Neelsen K.J., Fachinetti D., Bermejo R., Cocito A., Costanzo V., and Lopes M.. 2012. Topoisomerase I poisoning results in PARP-mediated replication fork reversal. Nat. Struct. Mol. Biol. 19:417–423. 10.1038/nsmb.2258 [DOI] [PubMed] [Google Scholar]
- Ray Chaudhuri A., Callen E., Ding X., Gogola E., Duarte A.A., Lee J.E., Wong N., Lafarga V., Calvo J.A., Panzarino N.J., et al. 2016. Replication fork stability confers chemoresistance in BRCA-deficient cells. Nature. 535:382–387. 10.1038/nature18325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regairaz M., Zhang Y.W., Fu H., Agama K.K., Tata N., Agrawal S., Aladjem M.I., and Pommier Y.. 2011. Mus81-mediated DNA cleavage resolves replication forks stalled by topoisomerase I-DNA complexes. J. Cell Biol. 195:739–749. 10.1083/jcb.201104003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robison J.G., Elliott J., Dixon K., and Oakley G.G.. 2004. Replication protein A and the Mre11.Rad50.Nbs1 complex co-localize and interact at sites of stalled replication forks. J. Biol. Chem. 279:34802–34810. 10.1074/jbc.M404750200 [DOI] [PubMed] [Google Scholar]
- Rondinelli B., Gogola E., Yücel H., Duarte A.A., van de Ven M., van der Sluijs R., Konstantinopoulos P.A., Jonkers J., Ceccaldi R., Rottenberg S., and D’Andrea A.D.. 2017. EZH2 promotes degradation of stalled replication forks by recruiting MUS81 through histone H3 trimethylation. Nat. Cell Biol. 19:1371–1378. 10.1038/ncb3626 [DOI] [PubMed] [Google Scholar]
- Saldivar J.C., Cortez D., and Cimprich K.A.. 2017. The essential kinase ATR: ensuring faithful duplication of a challenging genome. Nat. Rev. Mol. Cell Biol. 18:622–636. 10.1038/nrm.2017.67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarbajna S., and West S.C.. 2014. Holliday junction processing enzymes as guardians of genome stability. Trends Biochem. Sci. 39:409–419. 10.1016/j.tibs.2014.07.003 [DOI] [PubMed] [Google Scholar]
- Sartori A.A., Lukas C., Coates J., Mistrik M., Fu S., Bartek J., Baer R., Lukas J., and Jackson S.P.. 2007. Human CtIP promotes DNA end resection. Nature. 450:509–514. 10.1038/nature06337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlacher K., Christ N., Siaud N., Egashira A., Wu H., and Jasin M.. 2011. Double-strand break repair-independent role for BRCA2 in blocking stalled replication fork degradation by MRE11. Cell. 145:529–542. 10.1016/j.cell.2011.03.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlacher K., Wu H., and Jasin M.. 2012. A distinct replication fork protection pathway connects Fanconi anemia tumor suppressors to RAD51-BRCA1/2. Cancer Cell. 22:106–116. 10.1016/j.ccr.2012.05.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlegel B.P., Jodelka F.M., and Nunez R.. 2006. BRCA1 promotes induction of ssDNA by ionizing radiation. Cancer Res. 66:5181–5189. 10.1158/0008-5472.CAN-05-3209 [DOI] [PubMed] [Google Scholar]
- Schubert L., Ho T., Hoffmann S., Haahr P., Guérillon C., and Mailand N.. 2017. RADX interacts with single-stranded DNA to promote replication fork stability. EMBO Rep. 18:1991–2003. 10.15252/embr.201744877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sogo J.M., Lopes M., and Foiani M.. 2002. Fork reversal and ssDNA accumulation at stalled replication forks owing to checkpoint defects. Science. 297:599–602. 10.1126/science.1074023 [DOI] [PubMed] [Google Scholar]
- Somyajit K., Saxena S., Babu S., Mishra A., and Nagaraju G.. 2015. Mammalian RAD51 paralogs protect nascent DNA at stalled forks and mediate replication restart. Nucleic Acids Res. 43:9835–9855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugawara N., Wang X., and Haber J.E.. 2003. In vivo roles of Rad52, Rad54, and Rad55 proteins in Rad51-mediated recombination. Mol. Cell. 12:209–219. 10.1016/S1097-2765(03)00269-7 [DOI] [PubMed] [Google Scholar]
- Suwaki N., Klare K., and Tarsounas M.. 2011. RAD51 paralogs: roles in DNA damage signalling, recombinational repair and tumorigenesis. Semin. Cell Dev. Biol. 22:898–905. 10.1016/j.semcdb.2011.07.019 [DOI] [PubMed] [Google Scholar]
- Symington L.S. 2014. End resection at double-strand breaks: mechanism and regulation. Cold Spring Harb. Perspect. Biol. 6:6 10.1101/cshperspect.a016436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taglialatela A., Alvarez S., Leuzzi G., Sannino V., Ranjha L., Huang J.W., Madubata C., Anand R., Levy B., Rabadan R., et al. 2017. Restoration of Replication Fork Stability in BRCA1- and BRCA2-Deficient Cells by Inactivation of SNF2-Family Fork Remodelers. Mol. Cell. 68:414–430.e8. 10.1016/j.molcel.2017.09.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thangavel S., Berti M., Levikova M., Pinto C., Gomathinayagam S., Vujanovic M., Zellweger R., Moore H., Lee E.H., Hendrickson E.A., et al. 2015. DNA2 drives processing and restart of reversed replication forks in human cells. J. Cell Biol. 208:545–562. 10.1083/jcb.201406100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toledo L.I., Altmeyer M., Rask M.B., Lukas C., Larsen D.H., Povlsen L.K., Bekker-Jensen S., Mailand N., Bartek J., and Lukas J.. 2013. ATR prohibits replication catastrophe by preventing global exhaustion of RPA. Cell. 155:1088–1103. 10.1016/j.cell.2013.10.043 [DOI] [PubMed] [Google Scholar]
- Unk I., Hajdú I., Fátyol K., Hurwitz J., Yoon J.H., Prakash L., Prakash S., and Haracska L.. 2008. Human HLTF functions as a ubiquitin ligase for proliferating cell nuclear antigen polyubiquitination. Proc. Natl. Acad. Sci. USA. 105:3768–3773. 10.1073/pnas.0800563105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vindigni A., and Lopes M.. 2017. Combining electron microscopy with single molecule DNA fiber approaches to study DNA replication dynamics. Biophys. Chem. 225:3–9. 10.1016/j.bpc.2016.11.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vujanovic M., Krietsch J., Raso M.C., Terraneo N., Zellweger R., Schmid J.A., Taglialatela A., Huang J.W., Holland C.L., Zwicky K., et al. 2017. Replication Fork Slowing and Reversal upon DNA Damage Require PCNA Polyubiquitination and ZRANB3 DNA Translocase Activity. Mol. Cell. 67:882–890.e5. 10.1016/j.molcel.2017.08.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang A.T., Kim T., Wagner J.E., Conti B.A., Lach F.P., Huang A.L., Molina H., Sanborn E.M., Zierhut H., Cornes B.K., et al. 2015. A Dominant Mutation in Human RAD51 Reveals Its Function in DNA Interstrand Crosslink Repair Independent of Homologous Recombination. Mol. Cell. 59:478–490. 10.1016/j.molcel.2015.07.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weston R., Peeters H., and Ahel D.. 2012. ZRANB3 is a structure-specific ATP-dependent endonuclease involved in replication stress response. Genes Dev. 26:1558–1572. 10.1101/gad.193516.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willis N.A., Frock R.L., Menghi F., Duffey E.E., Panday A., Camacho V., Hasty E.P., Liu E.T., Alt F.W., and Scully R.. 2017. Mechanism of tandem duplication formation in BRCA1-mutant cells. Nature. 551:590–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu S., Wu X., Wu L., Castillo A., Liu J., Atkinson E., Paul A., Su D., Schlacher K., Komatsu Y., et al. 2017a Abro1 maintains genome stability and limits replication stress by protecting replication fork stability. Genes Dev. 31:1469–1482. 10.1101/gad.299172.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y., Ning S., Wei Z., Xu R., Xu X., Xing M., Guo R., and Xu D.. 2017b 53BP1 and BRCA1 control pathway choice for stalled replication restart. eLife. 6:e30523 10.7554/eLife.30523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying S., Hamdy F.C., and Helleday T.. 2012. Mre11-dependent degradation of stalled DNA replication forks is prevented by BRCA2 and PARP1. Cancer Res. 72:2814–2821. 10.1158/0008-5472.CAN-11-3417 [DOI] [PubMed] [Google Scholar]
- Ying S., Minocherhomji S., Chan K.L., Palmai-Pallag T., Chu W.K., Wass T., Mankouri H.W., Liu Y., and Hickson I.D.. 2013. MUS81 promotes common fragile site expression. Nat. Cell Biol. 15:1001–1007. 10.1038/ncb2773 [DOI] [PubMed] [Google Scholar]
- Yuan J., Ghosal G., and Chen J.. 2009. The annealing helicase HARP protects stalled replication forks. Genes Dev. 23:2394–2399. 10.1101/gad.1836409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan J., Ghosal G., and Chen J.. 2012. The HARP-like domain-containing protein AH2/ZRANB3 binds to PCNA and participates in cellular response to replication stress. Mol. Cell. 47:410–421. 10.1016/j.molcel.2012.05.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yusufzai T., and Kadonaga J.T.. 2008. HARP is an ATP-driven annealing helicase. Science. 322:748–750. 10.1126/science.1161233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yusufzai T., and Kadonaga J.T.. 2010. Annealing helicase 2 (AH2), a DNA-rewinding motor with an HNH motif. Proc. Natl. Acad. Sci. USA. 107:20970–20973. 10.1073/pnas.1011196107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zadorozhny K., Sannino V., Beláň O., Mlčoušková J., Špírek M., Costanzo V., and Krejčí L.. 2017. Fanconi-Anemia-Associated Mutations Destabilize RAD51 Filaments and Impair Replication Fork Protection. Cell Reports. 21:333–340. 10.1016/j.celrep.2017.09.062 [DOI] [PubMed] [Google Scholar]
- Zellweger R., Dalcher D., Mutreja K., Berti M., Schmid J.A., Herrador R., Vindigni A., and Lopes M.. 2015. Rad51-mediated replication fork reversal is a global response to genotoxic treatments in human cells. J. Cell Biol. 208:563–579. 10.1083/jcb.201406099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeman M.K., and Cimprich K.A.. 2014. Causes and consequences of replication stress. Nat. Cell Biol. 16:2–9. 10.1038/ncb2897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Z., Chung W.H., Shim E.Y., Lee S.E., and Ira G.. 2008. Sgs1 helicase and two nucleases Dna2 and Exo1 resect DNA double-strand break ends. Cell. 134:981–994. 10.1016/j.cell.2008.08.037 [DOI] [PMC free article] [PubMed] [Google Scholar]