Orr and Maiato discuss work from the Stumpff laboratory addressing the role of chromosome congression in mitotic fidelity and genomic stability.
Abstract
Chromosome alignment is a hallmark of mitosis in metazoans, but the physiological relevance of this orderly behavior has remained unclear. In this issue, Fonseca et al. (2019. J. Cell Biol. https://doi.org/10.1083/jcb.201807228) show that chromosome alignment ensures mitotic fidelity by promoting interchromosomal compaction during anaphase.
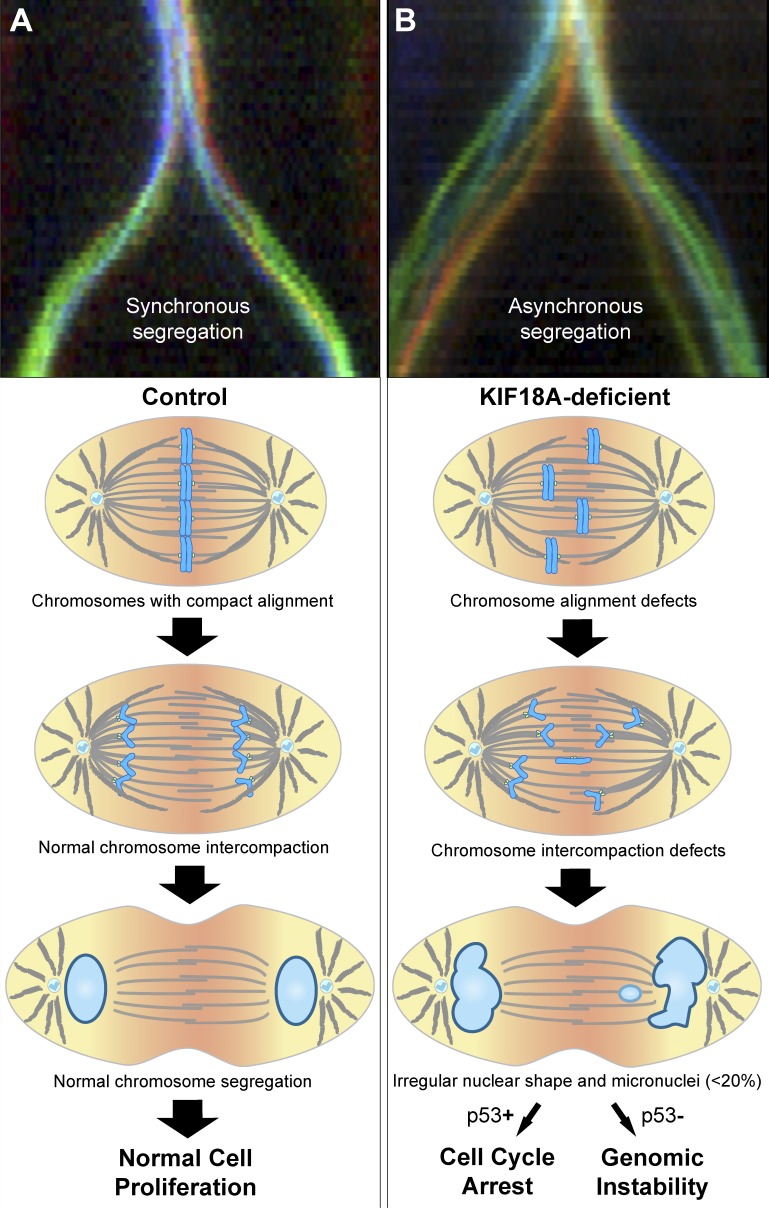
During mitosis, chromosomes align at the spindle equator to establish a metaphase plate. This process, termed chromosome congression, was speculated to promote mitotic fidelity by “forcing” the chromosomes to enter anaphase from the same starting line, followed by their synchronous poleward movement, thereby preventing chromosome dispersion (1; Fig. 1 A). However, appreciating the physiological relevance of chromosome congression for mitotic fidelity has proven difficult because it is technically challenging to specifically disrupt chromosome alignment without affecting microtubules, kinetochores, or both. KIF18A/Kinesin-8 is a microtubule motor that promotes chromosome congression by dampening kinetochore microtubule plus-end dynamics and reducing the oscillatory movements of bi-oriented chromosomes (2). Prior work established that KIF18A’s roles in chromosome alignment and kinetochore–microtubule attachments are cell type specific (3, 4), suggesting that the two functions are separable and dependent on cellular context. In this issue, Fonseca et al. use nontransformed in vitro and in vivo mammalian systems exploiting KIF18A’s dual functions to examine the physiological role of mitotic chromosome congression.
Figure 1.
Chromosome alignment ensures anaphase synchrony to promote mitotic fidelity. Top panels (courtesy of Irina Matos and António Pereira) show illustrative chromokymographs of synchronous (A) and asynchronous (B) Drosophila melanogaster S2 cells undergoing anaphase (colors represent the tracking of individual kinetochores). Models illustrate the progression and fate for normal (A) and Kif18A-deficient (B) cells. Kif18A-deficient cells show impaired chromosome alignment and interchromosomal compaction during anaphase, leading to the formation of irregular daughter nuclei and/or micronuclei. Only a minor fraction of lagging chromosomes results in micronuclei, consistent with anaphase surveillance mechanisms (e.g., dependent on a midzone Aurora B phosphorylation gradient) that spatially control nuclear envelope reassembly. Cell proliferation in the presence of micronuclei is limited by p53, thereby ensuring genomic stability.
Fonseca et al. (5) combined the use of embryonic fibroblasts and peripheral blood cells from KIF18A knockout mice, with the inactivation of Kif18A function by siRNA and CRISPR/Cas9 editing in immortalized (but not transformed) human retinal pigment epithelial cells (hTERT-RPE1 cells). Live-cell imaging of mitotic progression and quantifications of MAD1-positive kinetochores revealed that KIF18A inactivation in nontransformed cells did not significantly compromise kinetochore–microtubule attachments or timely anaphase onset, in contrast with previous observations in cancer cells (3). KIF18A-deficient cells entered anaphase without chromosome alignment but quantification of chromosome distribution revealed interchromosomal compaction defects as they exited mitosis (5). These defects led to the formation of lagging chromosomes, micronuclei, and irregular daughter nuclei, as well as decreased cell proliferation, impaired postnatal growth, and reduced overall survival in mice (5). Intriguingly, FISH with chromosome-specific probes showed that, despite the defects resulting from KIF18A inactivation, there were no detectable alterations in chromosome copy number (5). Overall, these data suggest that although chromosome alignment prevents the formation of lagging chromosomes and micronuclei in anaphase, mechanisms exist that limit the prevalence of chromosome copy number alterations in nontransformed cells.
These findings are in line with previous ones proposing that flux-dependent force equalization on kinetochores promotes mitotic fidelity by ensuring the formation of a tight metaphase plate and the subsequent synchronous poleward segregation of chromosomes during anaphase (6; Fig. 1, A and B). More recently, it was shown that late-aligning chromosomes are more prone to lag behind in anaphase (7). This led to the proposal that delayed chromosome alignment compromises bi-orientation and the establishment of functional/correct kinetochore–microtubule attachments that might account for chromosomal instability in cancer cells (7). However, the observation by Fonseca et al. that nontransformed KIF18A-deficient cells did not show detectable alterations in kinetochore–microtubule attachments or chromosome copy number (5) suggests that chromosome alignment and maintenance of nuclear shape are largely dispensable for chromosomal stability in mammals. Nevertheless, technical limitations in the detection of chromosome copy number alterations by FISH and/or mild kinetochore–microtubule attachment defects that satisfy the spindle assembly checkpoint could not be excluded. Alternative approaches such as single-cell sequencing and correlative light and electron microscopy might prove useful in the future to rule out these possibilities.
Fonseca et al. also found that lagging chromosomes originating from anaphase asynchrony in KIF18A-deficient cells traveled at normal speed but needed to cover longer distances to reach the spindle pole (5). In contrast, lagging chromosomes generated by disrupting spindle assembly checkpoint function (MAD2 depletion) traveled equal distances as control cells (albeit at a slower speed), leading to detectable alterations in chromosome copy number (5). These data suggest that the origin of lagging chromosomes (due to merotelic kinetochore–microtubule attachments or defects in interchromosomal compaction) is an important determinant of chromosome copy number in human cells. Despite these differences, the authors show via p53 inactivation studies that the proliferation of the few micronucleated cells (≤8%) resulting from KIF18A inactivation was limited by a p53-dependent mechanism (5), in agreement with previous reports (8). This explains, at least in part, how nontransformed cells avoid the perpetuation of chromosome missegregation events to ensure genomic stability (Fig. 1 B).
One critical finding in the study by Fonseca et al. was that most lagging chromosomes (>80%) induced through KIF18A disruption in nontransformed human cells reintegrated the main daughter nuclei and only rarely resulted in micronuclei (5). At first sight, this is at odds with previous reports in human transformed U2OS and HCT116 cells in which micronuclei were detected in nearly 80% of anaphase cells with lagging chromosomes (9). However, these differences might reflect the type of defect that gives rise to lagging chromosomes, the possibility that only a small fraction of (multiple) lagging chromosomes during anaphase result in micronuclei, the normal versus transformed cellular state, or a combination of these factors. It will be important to distinguish between these possibilities by developing sensitive live-cell assays that allow the unequivocal tracking of individual lagging chromosomes in transformed versus nontransformed cells, while assessing respective causes and consequences.
In addition to alterations in copy number, other chromosome rearrangements reported in some human cancers such as chromothripsis were recently proposed to arise from irreversible nuclear envelope defects on micronuclei (10). The observation by Fonseca et al. that most lagging chromosomes are able to reintegrate the main nuclei (5), together with the existence of mechanisms that limit the proliferation of nontransformed cells with micronuclei, suggest that chromothripsis originating from mitotic errors is a rare event whose significance for the etiology of human cancers remains unclear. In agreement, KIF18A knockout mice form micronuclei in vivo (5) but were reported to be tumor resistant, rather than predisposed to tumor formation (11). The clear bias for the resolution and reintegration of lagging chromosomes in nontransformed cells (5) is consistent with the existence of surveillance mechanisms operating during anaphase that actively promote mitotic fidelity by spatially regulating nuclear envelope reassembly (12; Fig. 1, A and B). Therefore, micronuclei formation is not necessarily a pathological condition that inevitably links mitotic errors to chromothripsis and tumorigenesis. Nevertheless, it remains possible that, when combined with faulty surveillance mechanisms (including p53 inactivation, a common feature in human cancers), mitotic errors leading to micronuclei formation promote genomic instability and rapid tumor evolution in mammals (Fig. 1 B). Further work will be necessary to explore these possibilities.
Acknowledgments
We would like to thank Irina Matos and António Pereira for providing chromokymographs of normal and asynchronous anaphases after perturbation of spindle microtubule flux in Drosophila S2 cells.
Work in the Maiato Lab is supported by the European Research Council under the European Union’s Horizon 2020 research and innovation program (grant agreement number 681443) and FLAD Life Science 2020.
The authors declare no competing financial interests.
References
- 1.Maiato H., et al. Biology (Basel). 2017 doi: 10.3390/biology6010013. [DOI] [Google Scholar]
- 2.Stumpff J., et al. 2008. Dev. Cell. 14:252–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mayr M.I., et al. 2007. Curr. Biol. 17:488–498. [DOI] [PubMed] [Google Scholar]
- 4.Czechanski A., et al. 2015. Dev. Biol. 402:253–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fonseca C.L., et al. J. Cell Biol. 2019 doi: 10.1083/jcb.201807228. [DOI] [Google Scholar]
- 6.Matos I., et al. 2009. J. Cell Biol. 186:11–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kuniyasu K., et al. Biomolecules. 2018 doi: 10.3390/biom9010010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thompson S.L., and Compton D.A.. 2010. J. Cell Biol. 188:369–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thompson S.L., and Compton D.A.. 2011. Proc. Natl. Acad. Sci. USA. 108:17974–17978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu S., et al. 2018. Nature. 561:551–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhu H., et al. 2013. Biochem. Biophys. Res. Commun. 438:97–102. [DOI] [PubMed] [Google Scholar]
- 12.Afonso O., et al. 2014. Science. 345:332–336. [DOI] [PMC free article] [PubMed] [Google Scholar]