CoQ lipids are built at the mitochondrial inner membrane by a multicomponent pathway. Subramanian et al. reveal that CoQ pathway components assemble via CoQ intermediates into domains at ER–mitochondria contacts, suggesting that CoQ domains function as metabolons that facilitate processive CoQ production and distribution.
Abstract
Coenzyme Q (CoQ) lipids are ancient electron carriers that, in eukaryotes, function in the mitochondrial respiratory chain. In mitochondria, CoQ lipids are built by an inner membrane–associated, multicomponent, biosynthetic pathway via successive steps of isoprenyl tail polymerization, 4-hydroxybenzoate head-to-tail attachment, and head modification, resulting in the production of CoQ. In yeast, we discovered that head-modifying CoQ pathway components selectively colocalize to multiple resolvable domains in vivo, representing supramolecular assemblies. In cells engineered with conditional ON or OFF CoQ pathways, domains were strictly correlated with CoQ production and substrate flux, respectively, indicating that CoQ lipid intermediates are required for domain formation. Mitochondrial CoQ domains were also observed in human cells, underscoring their conserved functional importance. CoQ domains within cells were highly enriched adjacent to ER–mitochondria contact sites. Together, our data suggest that CoQ domains function to facilitate substrate accessibility for processive and efficient CoQ production and distribution in cells.
Introduction
Mitochondria possess a protein-dense inner membrane organized into compositionally and morphologically distinct functional domains (Cogliati et al., 2016). The flat inner boundary membrane, parallel to the mitochondrial outer membrane, is enriched in mitochondrial protein import machinery (Vogel et al., 2006; Wurm and Jakobs, 2006). In contrast, the lamellar-like cristae membrane extends into the matrix and houses the electron transport chain complexes (Mannella et al., 1994). Within cristae, ATP synthase dimers bend and stabilize their highly curved edges, and respiratory chain complexes further assemble together into large megadalton supramolecular complexes, which may contribute to both oxidative phosphorylation efficiency and cristae compartment maintenance (Kühlbrandt, 2015). In addition, the MICOS complex assembles on the inner membrane into a scaffold structure that connects the outer and inner membranes (Rabl et al., 2009; Harner et al., 2011; Hoppins et al., 2011; von der Malsburg et al., 2011; Friedman et al., 2015). Together with the mitochondrial lipids cardiolipin and phosphatidylethanolamine, MICOS is thought to determine the copy number of cristae—likely by stabilizing the junctions that connect the boundary and cristae inner membrane domains (Friedman et al., 2015).
Metabolic pathways within mitochondria, such as the TCA cycle and the pyruvate dehydrogenase complex, also play roles in the higher-order organization of the mitochondrial compartment by assembling into large complexes or metabolons in a spatially defined manner within cells (Brandina et al., 2006; Chan et al., 2015; Webb et al., 2017). Proteins in the coenzyme Q (CoQ) biosynthetic pathway form a complex associated with the mitochondrial inner membrane and synthesize a highly hydrophobic prenylated benzoquinone lipid that is an essential component of the respiratory chain (Stefely and Pagliarini, 2017; Awad et al., 2018). The CoQ lipid functions to transfer electrons from respiratory complexes I and II to complex III and facilitates H+ pumping across the inner membrane to power ATP synthesis by Complex V. Essential components of the CoQ biosynthetic pathway are highly conserved across kingdoms and, in yeast, are required for growth on nonfermentable carbon sources, consistent with the essential role of CoQ in respiration (Tzagoloff et al., 1975; Tzagoloff and Dieckmann, 1990). In humans, mutations in genes required for CoQ synthesis cause heterogeneous phenotypes consistent with mitochondrial respiratory chain deficiency (Luna-Sánchez et al., 2015; Acosta et al., 2016). In addition, mouse models of a diverse array of mitochondrial diseases possess CoQ synthesis deficiencies, likely via secondary defects in the expression of CoQ pathway components, further highlighting the central role of CoQ biosynthesis in mitochondrial homeostasis (Montero et al., 2013; Kühl et al., 2017).
A majority of the CoQ biosynthetic pathway (∼13 components) is localized within mitochondria. However, the isoprene carbon units that create the CoQ tail and the 4-hydroxybenzoate quinone head precursor, produced from tyrosine, are synthesized by extramitochondrial pathways in the ER and cytosol (Fig. 1 A). In yeast, these building blocks are transported into mitochondria via unknown mechanisms. Within yeast mitochondria, Coq1 polymerizes highly charged isoprene units into a 6-unit tail, and Coq2 attaches the 4-hydroxybenzoate head to the isoprenyl tail to produce the 4-hydroxy-3-hexaprenyl benzoate (HHB) CoQ lipid intermediate (Ashby et al., 1992; Okada et al., 1996; Poon et al., 1997). Subsequently, the HHB head group is successively modified in a highly cooperative manner by the inner membrane–associated enzymes in the following order to produce CoQ6: Coq6, Coq3, Coq5, Coq7, and Coq3 (Barkovich et al., 1997; Poon et al., 1999; Jonassen and Clarke, 2000; He et al., 2014; Ozeir et al., 2015). Additional components play unclear roles in CoQ biosynthesis including Coq4, Coq8, Coq9, Coq10, and Coq11 (Barros et al., 2005; Xie et al., 2012; Lohman et al., 2014; Allan et al., 2015). Although their exact mechanism of action has not been resolved, bioinformatic, structural, and biochemical data indicate that a majority have lipid-binding properties, suggesting a chaperone-like role to promote substrate accessibility for the head-modifying enzymes (Rea et al., 2010; Lohman et al., 2014; Reidenbach et al., 2018). Consistent with this model, the ancient, atypical kinase Coq8 is an ATPase whose activity is stimulated by the head groups of CoQ intermediates and the inner membrane lipid cardiolipin, suggesting that Coq8 may function to extract highly hydrophobic CoQ intermediates (Stefely et al., 2015, 2016b; Reidenbach et al., 2018). Biochemical and proteomic analyses indicate that a subset of CoQ pathway components (Coq3–9 and Coq11) interact together in a highly interdependent manner to form a multi-subunit complex associated with the matrix side of the inner membrane in yeast and humans (Marbois et al., 2005; Tran and Clarke, 2007; Floyd et al., 2016; Stefely et al., 2016a). Consistent with this, cells lacking even late-acting enzymatic components of the CoQ pathway, such as Coq7, accumulate the earliest lipid precursor HHB (Poon et al., 1997). However, the mechanism underlying the cooperative assembly of the CoQ biosynthesis machinery is not understood.
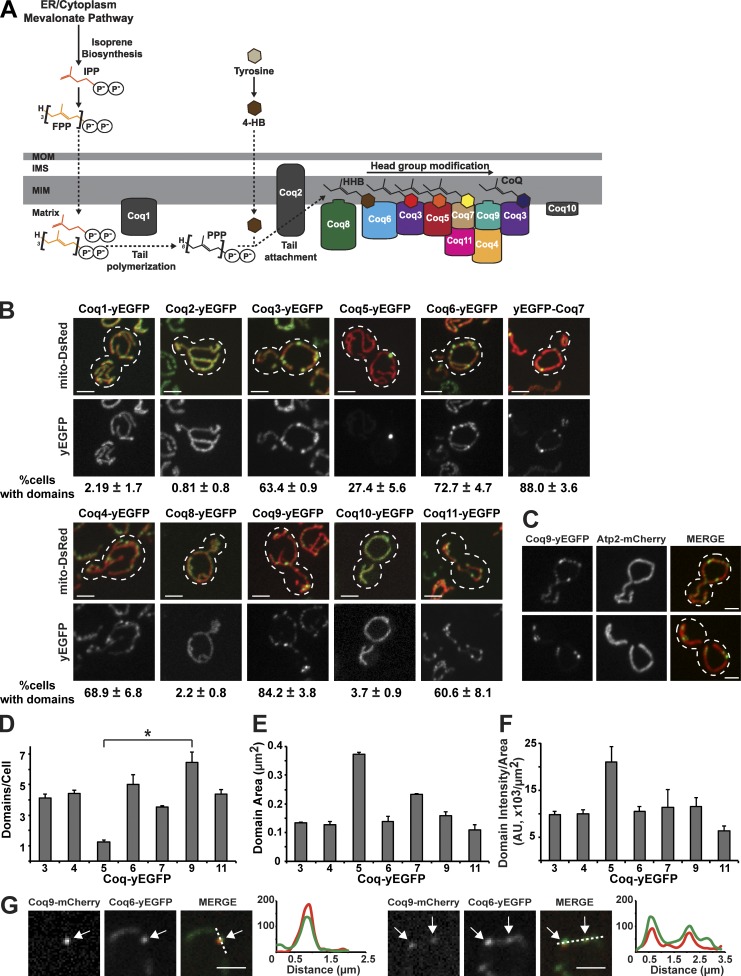
Figure 1.
Head-modifying Coq proteins localize to mitochondrial domains in vivo. (A) Working model of the yeast CoQ biosynthetic pathway. Coq1 polymerizes the tail (PPP) from IPP and/or FPP derived from the mevalonate pathway in the mitochondrial matrix. Coq2 attaches PPP to 4-HB, synthesized in the cytoplasm, to produce the intermediate CoQ lipid HHB. Coq8 functions to extract CoQ intermediate lipids to facilitate the nucleation of CoQ domains containing head-modifying components. CoQ6 is produced by known successive enzymatic modifications of the HHB head by Coq6, Coq3, Coq5, Coq7, and Coq3, as indicated by the head color of the associated CoQ lipid intermediates. Coq4, Coq8, Coq9, Coq10, and Coq11 play additional unclear roles in head modification. Coq1, Coq2, Coq8, and Coq10 do not localize to CoQ domains at steady state. (B) Representative max z-projection images of yeast cells expressing indicated Coq-yEGFP (green) from their endogenous loci and mito-DsRed (red). Percentage of cells with CoQ domains corresponding to respective yeast strains are shown below images as mean ± SEM; n > 70 cells from three independent experiments. Dotted lines represent cell boundary. (C) Representative max z-projection images of yeast cells expressing Coq9-yEGFP (green) and Atp2-mCherry (red) from their endogenous loci. Dotted lines represent cell boundary. (D–F) Quantification of average number of domains per cell (D), domain area in µm2 (E), and domain intensity per domain area in fluorescence intensity AU per µm2 (F) from cells imaged in B. Data represented as mean ± SEM; n > 70 cells from three independent experiments. (D) Kruskal–Wallis test, P = 0.01, followed by Dunn test, *, P < 0.02. (E) Kruskal–Wallis test, P = 0.02, followed by Dunn test, not significant. (F) Kruskal–Wallis test, P = 0.09, not significant. (G) Single-plane images of yeast cells expressing Coq9-mCherry (red) and Coq6-yEGFP (green). Arrows indicate CoQ domains analyzed by line scan of fluorescence intensity indicated by dashed lines. Y axis is fluorescence intensity (AU), and x axis is distance (µm). Scale bars = 2 µm.
Here, to gain insight into how CoQ pathway components assemble, we examined the spatial organization of Coq proteins in vivo in yeast. We discovered that after head-to-tail attachment, Coq head-modifying proteins form multiple discrete inner membrane domains in cells in a highly cooperative manner dependent on CoQ lipid intermediates. Within yeast cells, CoQ domains are spatially linked to ER–mitochondria contact sites. CoQ domains are also observed in human cells, indicating they possess a highly conserved function. Together, our data suggest that the cooperative assembly of CoQ domains at ER–mitochondria contact sites promotes access to CoQ lipid intermediates for efficient and processive CoQ production and distribution.
Results
Head-modifying Coq proteins localize to domains
To gain insight into the spatial organization of the CoQ biosynthetic complex in vivo, we directly examined the localization and distribution of fluorescent protein–tagged versions of Coq proteins using fluorescence microscopy in budding yeast (Fig. 1). Specifically, we imaged cells grown to log phase coexpressing yEGFP-tagged Coq proteins integrated at their endogenous chromosomal loci and mitochondrial-targeted DsRed (mito-DsRed) in media containing dextrose. CoQ is essential for growth on nonfermentable carbon sources (Fig. S1 A). Thus, the function of yEGFP-tagged Coq proteins was assessed by growth of cells harboring tagged versions on the nonfermentable carbon sources ethanol and glycerol (Fig. S1 A). In addition, CoQ abundance was directly measured using liquid chromatography–mass spectrometry (LC-MS) from cellular extracts (Fig. S1 B). Data from these assays show that in a majority of cases, growth rate and CoQ abundance in cells harboring yEGFP-tagged COQ genes were not significantly different from WT cells, indicating that tagged CoQ proteins were functional. In contrast, Coq5-yEGFP cells exhibited slow growth and possessed, in comparison to WT cells, the relatively lowest CoQ abundance and the highest levels of CoQ intermediates 3-polyprenyl-4-hydroxybenzoate (PPHB6) and aminated analogue of PPHB6 (PPAB6), which are early lipid CoQ intermediates in the pathway (Fig. S1, A–D). These data indicate that cells harboring Coq5-yEGFP possess only a partially functional CoQ pathway, which may be a consequence of the requirement of the RNA binding protein, Puf3, for native COQ5 mRNA targeting and expression (Lapointe et al., 2018).
The tail polymerase Coq1 and head-to-tail attachment protein Coq2 catalyze the first two steps in the CoQ biosynthetic pathway to produce the HHB lipid intermediate, whose head group is then processed by Coq6, Coq3, Coq5, Coq7, and Coq3 to produce CoQ6 (herein referred to as CoQ; Fig. 1 A). Coq1-yEGFP and Coq2-yEGFP localized to mitochondria, as previously described, with a uniform distribution similar to the mito-DsRed matrix marker (Fig. 1 B). In contrast, the yEGFP-tagged head-modifying Coq proteins localized to multiple discrete mitochondrial-associated foci in cells (Fig. 1 B, >25% of cell population, n > 200 cells), which we defined provisionally as domains (Fig. 1, B and C). Additionally, Coq proteins thought to function more indirectly in head modification were either uniformly localized to mitochondria (Coq8 and Coq10), similar to Coq1 and Coq2, or distributed within mitochondria in domains (Coq4, Coq9, and Coq11), similar to the head-modifying Coq enzymes within mitochondria in cells (Fig. 1 B). Previous work reported that ATP synthase components also localize to light resolvable regions within mitochondria, representing cristae (Jimenez et al., 2014). As shown in Fig. 1 C, under our experimental conditions, the distribution of Atp2-mCherry, an ATP synthase component and cristae marker, was relatively uniform within mitochondria compared with Coq9-yEGFP, which localized to discrete mitochondrial regions, indicating that the CoQ domains are unique and do not strictly correspond to cristae. Previous biochemical analyses of the CoQ biosynthetic pathway suggest that a subset of Coq proteins interact to form an inner membrane–associated complex (Marbois et al., 2005, 2009; Tran and Clarke, 2007; Floyd et al., 2016). Consistent with these data, domain-localized Coq proteins correspond to the subset of complex-associated Coq components, with the exception of Coq8. Our observations indicate that components that function in the head-group modification part of the CoQ biosynthetic pathway were specifically localized to domains (Fig. 1 A).
To gain insight into the properties and composition of CoQ domains, we analyzed the number of domains per cell (Fig. 1 D), the total domain area (Fig. 1 E), and domain intensity per domain area (Fig. 1 F) labeled by each of the domain-localized Coq-yEGFP proteins. For any given Coq-yEGFP, a comparable average number of four to six Coq-yEGFP–labeled domains per cell was observed (Fig. 1 C, >200 cells). The exception was the partially active Coq5-yEGFP, which labeled on average one domain per cell (Fig. 1 C, >80 cells). The average domain area and fluorescence intensity per domain area of Coq-yEGFP–labeled domains were also comparable among the Coq-yEGFP, suggesting that domains comprise all CoQ domain-forming components (Fig. 1, E and F). To directly determine if Coq proteins colocalized within domains, we examined the localization of a head modifier Coq6-yEGFP and a nonenzymatic CoQ component Coq9-mCherry coexpressed in cells from their endogenous loci by fluorescence microscopy. Line scan analysis revealed that Coq6-yEGFP and Coq9-mCherry localized together in domains (Fig. 1 G, n = 100 domains, >30 cells). Thus, with our characterization (Figs. 1, D–F), these data indicate that CoQ domains possess a similar composition within cells.
CoQ domains are conserved in human cells
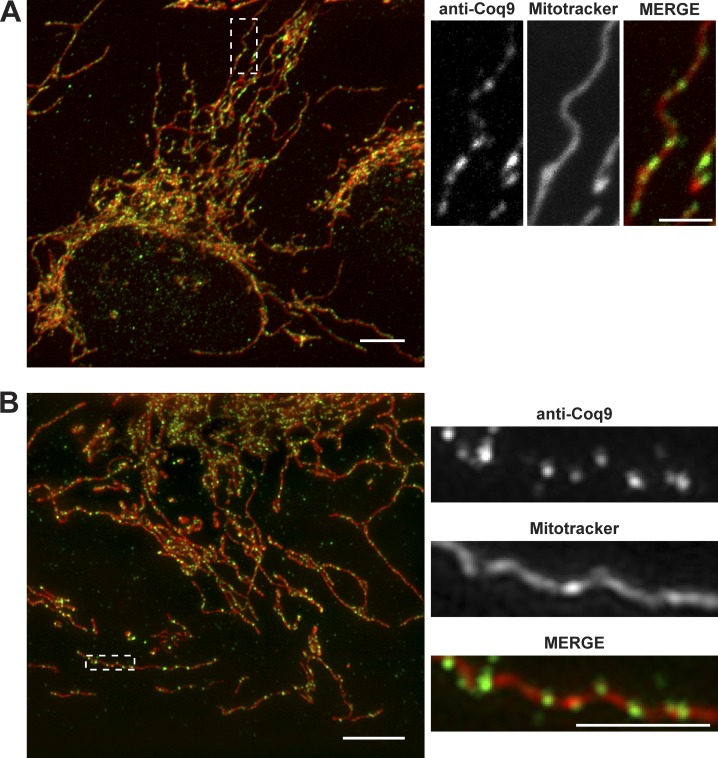
We asked whether mitochondrial CoQ domains are also found in human cells. Specifically, we examined the localization of the native human Coq9 orthologue using indirect immunofluorescence with Alexa Fluor 488–conjugated anti-human COQ9 antibodies in human osteosarcoma (U2OS) cells colabeled with mitochondrial vital dye, MitoTracker Red. Similar to our observations in yeast, endogenous human COQ9 localized to discrete foci that colocalized with MitoTracker Red (Fig. 2, A and B, 471 ± 52 domains per cell, n = 11 cells). To test the specificity of the anti-COQ9 antibody, we overexpressed COQ9-GFP in U2OS cells and observed the colocalization of GFP and anti-Coq9 fluorescence (Fig. S1 E). Thus, these data indicate that CoQ domains are observed within human mitochondria, consistent with the high degree of evolutionary conservation among Coq proteins and underscoring the functional importance of CoQ domains.
Figure 2.
CoQ domains form in human cells. (A and B) Representative max z-projection confocal (A) or structured illumination superresolution (B) images of fixed U2OS cells labeled with anti-COQ9 antibody conjugated to Alexa Fluor 488 (green) and MitoTracker Red (red). Dotted boxes indicate magnified regions in the inset (right). Scale bars = 10 µm (A) and 5 µm (B). Scale bars in the inset = 2 µm.
Functional status of CoQ pathway influences CoQ domain copy number
The hypomorphic Coq5-yEGFP mutant labeled fewer and larger domains in cells (Fig. 1, D and E), suggesting that the functional status of the CoQ pathway influences CoQ domain copy number. To test this, we compared Coq5-labeled domains in cells expressing endogenous Coq5-yEGFP to cells expressing a fully functional 13Myc-tagged version of Coq5, as assessed by cell growth on nonfermentable carbon sources and CoQ abundance in comparison to WT cells (Fig. S2, E–H). To detect Coq5-13Myc, we performed indirect immunofluorescence on yeast cells labeled with MitoTracker Red using anti-Myc Alexa Fluor 488–labeled antibodies (Fig. S2 A). As controls, we also examined the localization of functional 13Myc-tagged versions of Coq6, Coq8, or Coq9 in cells, expressed from their endogenous loci. Consistent with the behavior of the respective yEGFP-tagged versions, Coq6-13Myc and Coq9-13Myc localized to an average of four to six domains per cell, while Coq8-13Myc was uniformly distributed in mitochondria, compared with matrix labeled with MitoTracker Red (Fig. S2 B). However, in contrast to Coq5-yEGFP, which localized to an average of one to two domains per cell, Coq5-13Myc localized to a significantly greater number of domains per cell, similar to cells expressing other functional Coq-tagged proteins (Fig. S2, B and D). Taken together, these data suggest that the functional status of the CoQ pathway determines CoQ domain number and distribution in mitochondria. Alternatively, or in addition, these observations may suggest that Coq5 plays a more direct role in CoQ domain assembly.
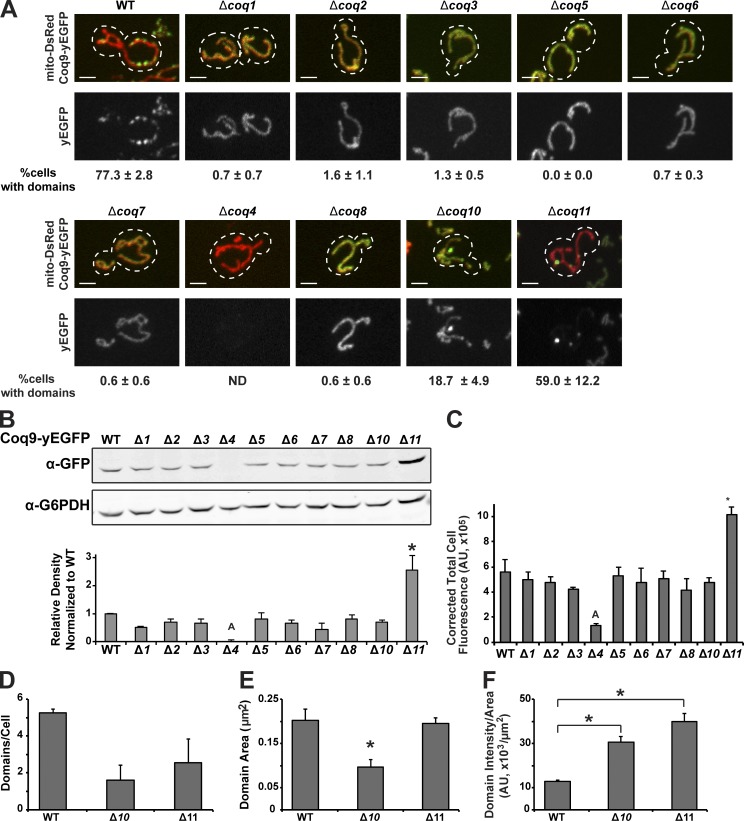
Essential CoQ biosynthetic components are required for CoQ domain formation
We asked which Coq proteins were required for CoQ domain formation by examining the localization of the CoQ domain marker Coq9-yEGFP in cells lacking individual COQ genes (Fig. 3). Although the tail polymerase and head-to-tail attachment proteins, Coq1 and Coq2, respectively, were not localized to domains, they and all of the other essential Coq proteins, Coq3, Coq4, Coq5, Coq6, Coq7, and Coq8, were required for formation of Coq9-yEGFP domains in cells (Fig. 3 A). In contrast, in cells lacking the nonessential CoQ components, Coq10 or Coq11, Coq9-labeled domains were observed, albeit in less of the Δcoq10 cell population compared with WT (Fig. 3, A and D). The apparent defect in domain assembly in Δcoq10 cells correlated with slower growth on nonfermentable carbon relative to WT, a phenotype that is tightly linked to CoQ production (Fig. S1 A). Coq9 expression was similar to WT in cells lacking Coq1, Coq2, Coq3, Coq5, Coq6, Coq7, Coq8, and Coq10, as assessed by Western blot analysis and by corrected total cell fluorescence, but was reduced in Δcoq4, consistent with published observations indicating that Coq4 is required for the stable expression of CoQ biosynthetic complex components (Fig. 3, B and C; Hsieh et al., 2007; Marbois et al., 2009). In addition, we observed increased expression of Coq9-yEGFP in Δcoq11 cells. Although the basis of this increase is not clear, it likely explains the relatively higher fluorescence intensity per domain area observed in Δcoq11 cells relative to WT (Fig. 3 F). Thus, with the possible exception of Δcoq4 cells, the absence or reduction of Coq-labeled domains in cells lacking COQ genes results from defective CoQ domain formation per se. The observation that all essential CoQ components are required for domain formation is consistent with biochemical analysis indicating that the loss of any single essential yeast Coq protein destabilizes the CoQ biosynthetic complex in cells (He et al., 2014; Stefely et al., 2016a).
Figure 3.
Essential Coq proteins are required for CoQ domain formation in cells. (A) Representative max z-projection images of yeast cells expressing Coq9-yEGFP (green) from its endogenous loci and mito-DsRed (red) in indicated strains. Percentage of cells with CoQ domains corresponding to respective yeast strains are shown below images as mean ± SEM; n > 70 cells from three independent experiments. Dotted lines represent cell boundary. (B) Representative Western blot analysis (top) with indicated antibodies from cells expressing Coq9-yEGFP from its endogenous loci of whole-cell lysates from indicated strains. G6PDH antibody was used as a loading control. Quantification of the relative density of GFP/G6PDH normalized to WT (bottom) for three independent experiments. Data are represented as mean ± SEM. ANOVA, P = 2.8 × 10−6, followed by pairwise t test, *, P < 0.04, A = P < 0.01 for Δ4 compared with WT, Δ2, Δ3, Δ5, Δ6, Δ8, Δ11. (C) Corrected total cell fluorescence of indicated strains from A. ANOVA, P = 2.8 × 10−6, followed by pairwise t test, *, P < 0.002, A = P < 0.04 for Δ4 compared with WT, Δ1, Δ5, Δ7, Δ11. (D–F) Quantification of average number of domains per cell (D), domain area in µm2 (E), and domain intensity per domain area in fluorescence intensity AU per µm2 (F) from cells imaged in A. Data represented as mean ± SEM; n > 70 cells from three independent experiments. (D) Kruskal–Wallis test, P = 0.05, not significant. (E) ANOVA, P = 0.01, followed by pairwise t test, *, P < 0.04. (F) ANOVA, P = 0.0001, followed by pairwise t test, *, P < 0.001. Scale bars = 2 µm.
CoQ lipid substrates are required for domain formation
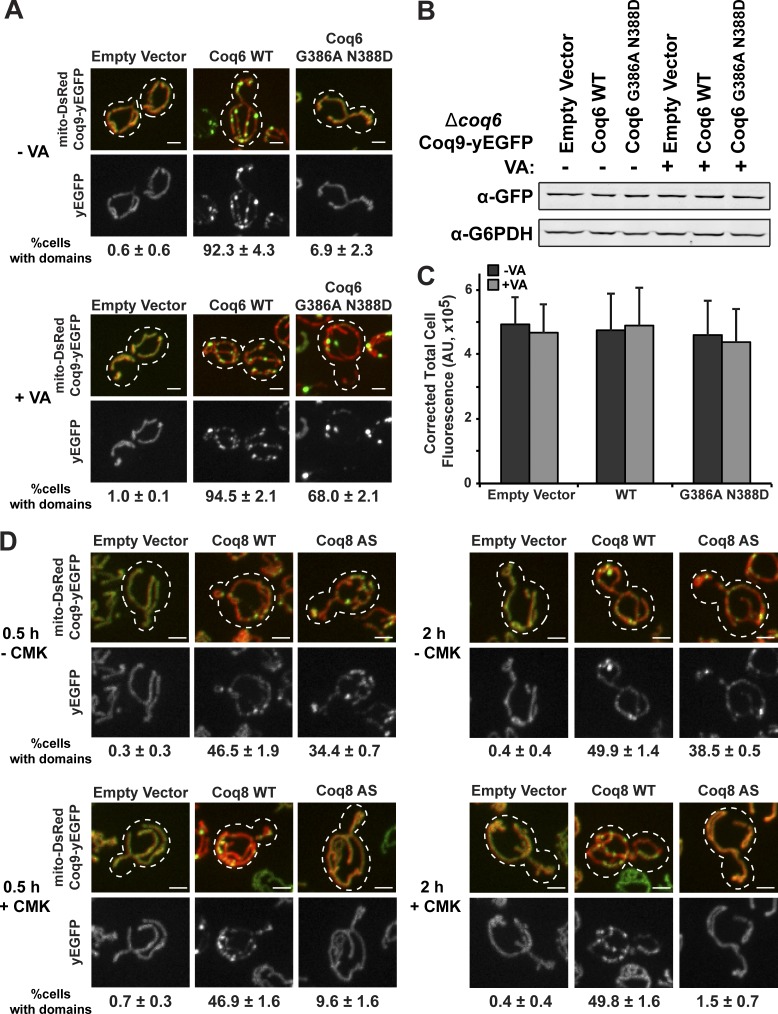
Our observations point to a role of CoQ lipid substrates in CoQ domain formation. To test this, we engineered a conditional ON CoQ pathway in cells and examined CoQ domains in the ON and OFF states using the CoQ domain marker, Coq9. Specifically, we used a hypomorphic allele of the essential head modifier flavoprotein monoxygenase, Coq6 G386A N388D, mutated in conserved residues implicated in NADPH and FAD binding (Palfey et al., 1994; Eppink et al., 1997; Ozeir et al., 2011). Exogenous addition of vanillic acid (VA), a dihydroxybenzoic acid derivative, bypasses the requirement for enzymatically active Coq6 in CoQ production in Coq6 G386A N388D mutant cells and restores CoQ biosynthesis and respiration (Ozeir et al., 2011). However, restoration of CoQ production in the Coq6 G386A N388D mutant requires the physical presence of Coq6 G386A N388D and the native biosynthetic pathway, suggesting that CoQ biosynthetic complex assembly must be preserved for the successful utilization of VA by the CoQ pathway (Ozeir et al., 2011).
We examined the distribution of Coq9-yEGFP in Δcoq6 cells and in cells expressing WT COQ6 or COQ6 G386A N388D. Coq9-yEGFP–labeled domains were observed only in Δcoq6 cells expressing WT Coq6, consistent with the essential role of active Coq6 in CoQ domain formation in vivo (Fig. 4 A). To determine whether CoQ lipid substrates are required for domain assembly, we examined Coq9-yEGFP distribution after addition of exogenous VA for >24 h. In the presence of VA, Coq9-yEGFP–labeled CoQ domains were observed in Δcoq6 cells expressing either WT Coq6 or the Coq6 G386A N388D mutant (Fig. 4 A). To test whether VA-conditional domain formation in Coq6 G386A N388D cells was correlated with CoQ biogenesis, we examined both CoQ domain formation and CoQ abundance at fixed time points 2, 10, and 26 h after VA addition. Consistently, we observed an increase in CoQ production over time (Fig. S3, A–C), with a corresponding increase in the percentage of cells with Coq9-labeled CoQ domains (Fig. S3 D). Both Western blot analysis and total cell fluorescence analysis (Fig. 4, B and C, respectively) indicated that Coq9-yEGFP expression was not significantly different in cells grown in either the presence or absence of VA. Thus, changes in Coq9-yEGFP expression did not contribute to VA-dependent domain formation. In addition, these data suggest that VA-dependent domains form in Coq6 G386A N388D cells by the concentration/assembly of the preexisting steady-state pool of Coq9-yEGFP. Consistent with this, Coq9-yEGFP intensity per domain area in Coq6 G386A N388D cells was threefold greater in VA-dependent domains compared with intensity per area for the more uniformly distributed Coq9-yEGFP observed within mitochondria in a majority of cells in the absence of VA (8 ± 1 vs. 25 ± 3 × 103 AU/µm2, respectively, n > 300 domains). These observations indicate that CoQ domains are supramolecular assemblies of Coq proteins.
Figure 4.
CoQ lipid intermediates are required for CoQ domain formation in cells. (A) Representative max z-projection images of Δcoq6 yeast cells expressing Coq9-yEGFP (green) from its endogenous loci, mito-DsRed (red), and empty vector, Coq6 WT, or Coq6 G386A N388D expressed from the TRP locus. Cells were grown in media without (top) or with (bottom) 1mM VA for >24 h. Percentage of cells with CoQ domains corresponding to respective yeast strains are shown below images as mean ± SEM; n > 70 cells from three independent experiments. Dotted lines represent cell boundary. (B) Representative Western blot analysis (top) with indicated antibodies from cells expressing Coq9-yEGFP from its endogenous loci of whole-cell lysates from indicated strains in A. G6PDH antibody was used as a loading control. (C) Corrected total cell fluorescence of indicated strains from A in the absence (gray) and presence (dark gray) of VA. n > 70 cells from three independent experiments. Data represented as mean ± SEM. Two-way ANOVA, P = 0.9, 0.9, 1.0, not significant. (D) Representative max z-projection images of Δcoq8 yeast cells expressing Coq9-yEGFP (green) from its endogenous locus, mito-DsRed (red), and empty vector, Coq8 WT, or Coq8 V202C M303C (Coq8 AS) expressed from a 2-µm plasmid. Cells were grown to log phase in media without (top) or with (bottom) 1 mM CMK for indicated times. Percentage of cells with CoQ domains corresponding to respective yeast strains are shown below images as mean ± SEM; n > 70 cells from three independent experiments. Dotted lines represent cell boundary. Scale bars = 2 µm.
We also examined CoQ domains by performing blue native–PAGE (BN-PAGE) analysis to examine detergent-soluble native Coq9-containing complexes from isolated mitochondria using anti-yeast Coq9 antibodies (Fig. S4 A). A >100-kD Coq9-containing complex was observed by BN-PAGE only in mitochondrial detergent extracts from Δcoq6 cells expressing WT Coq6 or from Coq6 G386A N388D cells grown in the presence of VA and not from either Δcoq6 cells with an empty vector or from Coq6 G386A N388D cells grown in the absence of VA (Fig. S4 A). Thus, these data support a model in which the recruitment of Coq proteins into Coq complexes, supramolecular assemblies, and/or domains is dependent on CoQ lipid intermediates formed at biosynthetic steps in the CoQ pathway subsequent to HHB synthesis.
To further test this model, we examined domain formation in cells engineered with a conditional OFF CoQ pathway. Specifically, we used an “irreversibly sensitized” version of the atypical kinase Coq8 (Coq8 AS), mutated in the ATP-binding pocket (V202C M303C) to allow binding of a halomethyl ketone (CMK; Rodríguez-Molina et al., 2016; Reidenbach et al., 2018). CMK specifically and covalently inhibits Coq8 AS enzymatic activity and CoQ production (Reidenbach et al., 2018). In Δcoq8 cells, Coq9-yEGFP–labeled domains were observed in a comparable population of cells expressing either WT Coq8 or Coq8 AS (Fig. 4 D). However, upon addition of CMK, a significant decrease in the population of cells with Coq9-yEGFP–labeled domains was observed only in cells expressing the Coq8 AS, with no domains detected in a majority of cells after a 2-h CMK incubation (Fig. 4 D). In addition, we measured CoQ abundance in conditional OFF cells in the presence and absence of CMK. A relatively small difference in CoQ abundance and CoQ intermediates was observed in the extracts from cells treated with CMK over the course of 2 h in Coq8 AS cell extracts relative to untreated cells (Fig. S3, E–G). These similar levels of CoQ are consistent with the reported long 49–125-h half-life of CoQ relative to the CMK treatment time (Thelin et al., 1992). Given that Coq9-yEGFP–labeled domains dispersed within 2 h of CMK treatment, these data suggest that under these conditions, CoQ domain formation is dependent on intermediate substrates, not CoQ abundance.
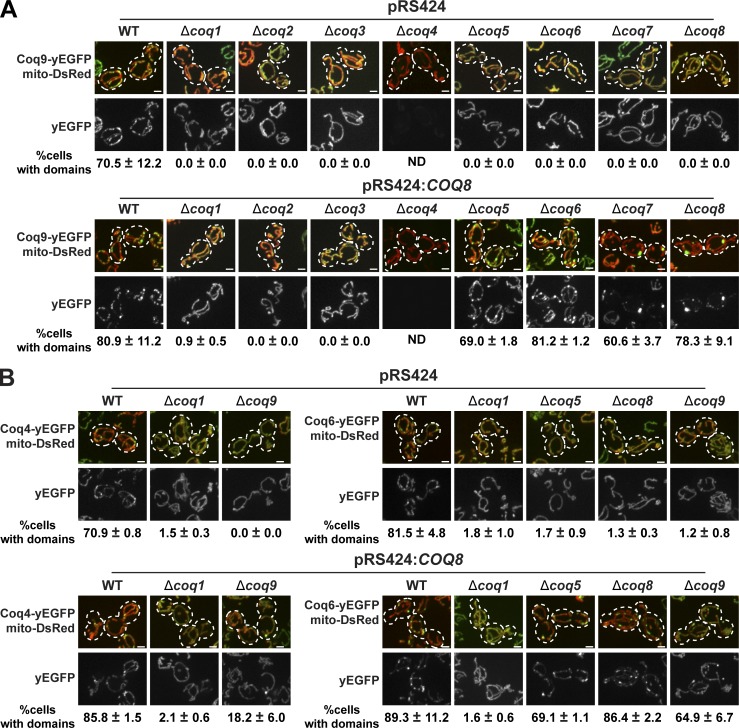
CoQ lipid intermediates are highly hydrophobic. Thus, we considered the possibility that CoQ domains function to promote the maintenance of substrate accessibility for efficient and processive head modification by essential head-modifying CoQ pathway components. Consistent with this idea, many of the “non-enzymatic” CoQ pathway components possess lipid-binding activities and may play lipid chaperone roles (Rea et al., 2010; Lohman et al., 2014; Reidenbach et al., 2018). One such component is Coq8, whose ATPase activity is stimulated by both cardiolipin and CoQ intermediate head groups, suggesting that Coq8 powers CoQ intermediate lipid extraction from the inner membrane to facilitate modification by head-modifying Coq enzymes (Reidenbach et al., 2018). Consistently, it was previously shown that overexpression of Coq8 suppressed CoQ biosynthetic complex assembly defects caused by mutation or absence of Coq5, Coq6, Coq7, or Coq9 and supported production of CoQ pathway intermediates, but not of terminal CoQ production (Xie et al., 2012; He et al., 2014). Thus, to further test the connection between domain formation and CoQ production and the role of CoQ lipid intermediates, we asked whether a multicopy plasmid containing COQ8 with its endogenous promoter (pRS424:COQ8) also suppressed defects in CoQ domain formation observed in cells lacking essential CoQ biosynthetic components (Fig. 5).
Figure 5.
Coq8 overexpression suppresses defective CoQ domain formation in the absence of specific Coq proteins in cells. (A) Representative max z-projection images of yeast cells expressing Coq9-yEGFP (green) from its endogenous loci, mito-DsRed (red), and containing pRS424 or pRS424:COQ8 in indicated strains. Percentage of cells with CoQ domains corresponding to respective yeast strains are shown below images as mean ± SEM; n > 70 cells from three independent experiments. Dotted lines represent cell boundary. (B) Representative max z-projection images of yeast cells expressing Coq4-yEGFP (green) or Coq6-yEGFP (green) from their endogenous loci, mito-DsRed (red), and containing pRS424 or pRS424:COQ8 in indicated strains. Percentage of cells with domains corresponding to images above are represented as mean ± SEM; n > 70 cells from three independent experiments. Scale bars = 2 µm.
We imaged Coq9-yEGFP in Δcoq1, Δcoq2, Δcoq3, Δcoq4, Δcoq5, Δcoq6, Δcoq7, and Δcoq8 cells containing pRS424:COQ8. In the presence of pRS424:COQ8, Coq9-yEGFP–labeled domains were observed in Δcoq8 cells, as expected, and also in Δcoq5, Δcoq6, and Δcoq7 cells (Fig. 5 A). However, the addition of pRS424:COQ8 failed to suppress domain assembly defects in Δcoq1, Δcoq2, Δcoq3, and Δcoq4 cells (Fig. 5 A). These data correlate with previous biochemical data examining the effect of Coq8 overexpression in coq mutants on CoQ complex assembly and CoQ lipid intermediate production (Xie et al., 2012; He et al., 2014). pRS424:COQ8 also suppressed Coq4-yEGFP and Coq6-yEGFP domain formation defects in Δcoq5, Δcoq8, and Δcoq9, but not in Δcoq1, cells (Fig. 5 B), indicating that the effect of pRS424:COQ8 was not specific to Coq9-yEGFP domains. These observations are consistent with a role of CoQ lipid substrates in CoQ domain formation. Also consistent with this model is the failure of pRS424:COQ8 to suppress domain formation defects in Δcoq1 and Δcoq2 cells with pRS424:COQ8, as these cells lack CoQ intermediate lipids. For Δcoq3 and Δcoq4 cells, however, the failure to suppress the CoQ domain formation defect is likely a consequence of their respective distinctive functions in the CoQ pathway. Coq3 uniquely functions at both an early and a late step in the CoQ biosynthesis, which is arguably a more severe biosynthetic defect. However, it is also possible that the lack of suppression indicates that Coq3, like Coq4, functions in Coq protein complex assembly. Coq4 is important for CoQ biosynthetic component stability—a phenotype that is apparent from loss of Coq9-yEGFP in Δcoq4 cells (Figs. 3 B and 5 A) via a scaffolding role in complex assembly (Marbois et al., 2009). Together, these data are consistent with a role of CoQ domains in the maintenance of CoQ lipid intermediate accessibility from the inner membrane.
CoQ domains are spatially linked to ER–mitochondria contact sites
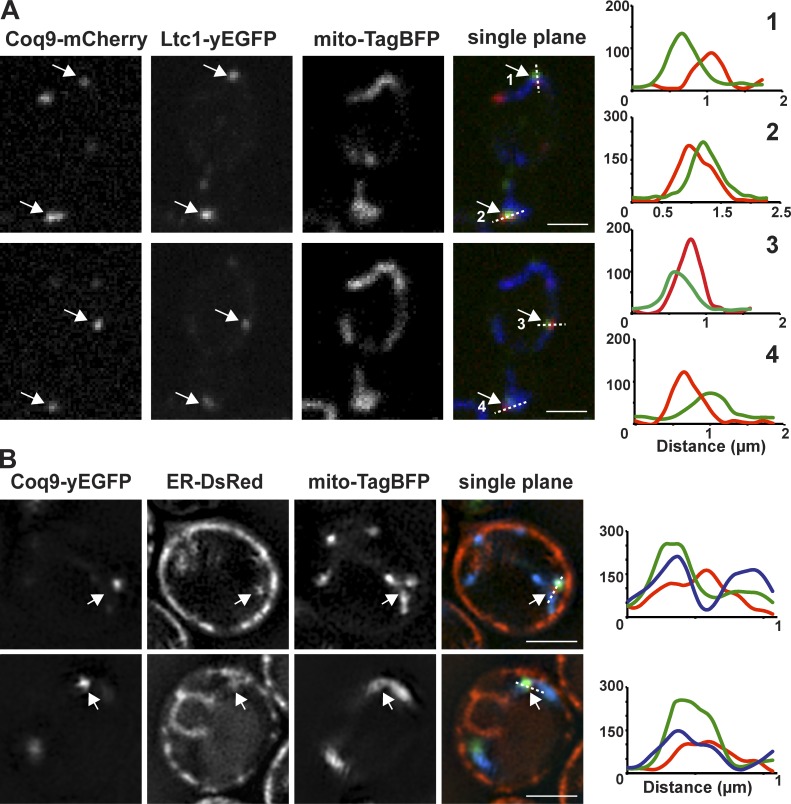
Our observations indicate that WT cells contain multiple mitochondrial CoQ domains. To examine whether CoQ domains are randomly distributed within mitochondria, we examined their spatial localization relative to ER–mitochondria contact sites, which we have previously shown to function as a nonrandom distribution mechanism for mitochondrial division machinery and replicating mtDNA nucleoids (Murley et al., 2013; Lewis et al., 2016). Two redundant ER–mitochondria tethers exist in yeast: Ltc1, a membrane sterol transporter protein localized to the ER, which partners with the mitochondrial outer membrane protein Tom71, and the multicomponent ERMES complex, which is thought to facilitate the transport of phospholipids between mitochondria and the ER (Kornmann et al., 2009; Elbaz-Alon et al., 2015; Gatta et al., 2015; Murley et al., 2015). Previous work has shown that both Ltc1 and ERMES components localize to punctate structures at regions of ER–mitochondria contact; however, ERMES marks only a subset of contacts (Murley et al., 2015). Thus, we labeled ER–mitochondria contacts using a functional yEGFP-tagged version of Ltc1 and observed that a vast majority of CoQ domains labeled with Coq9-yEcherry were spatially linked to Ltc1-yEGFP foci (Fig. 6 A, 76%, n = 467 domains and 243 cells). Consistent with this, Coq9-yEGFP–labeled domains were also spatially linked to regions of ER tubules, labeled with HDEL-DsRed, that were adjacent to mitochondria, labeled with the matrix marker mito-TagBFP (Fig. 6 B). These observations suggest that ER–mitochondria contacts may serve to specify regions of the mitochondrial inner membrane for CoQ domain formation and distribution.
Figure 6.
CoQ domains are spatially linked to ER–mitochondria contact sites in cells. (A) Single-plane images of yeast cells expressing Coq9-mCherry (red), Ltc1-yEGFP (green), and mito-TagBFP (blue). Arrows indicate regions analyzed by line scan of fluorescence intensity indicated by dashed lines. Y axis is fluorescence intensity (AU), and x axis is distance (µm). (B) Single-plane superresolution images of yeast cells expressing Coq9-yEGFP (green), ER-DsRed (ER, red), and mito-TagBFP (mitochondria, blue). Arrows indicate regions analyzed by line scan of fluorescence intensity indicated by dashed lines. Y axis is fluorescence intensity (AU), and x axis is distance (µm). Scale bars = 2 µm.
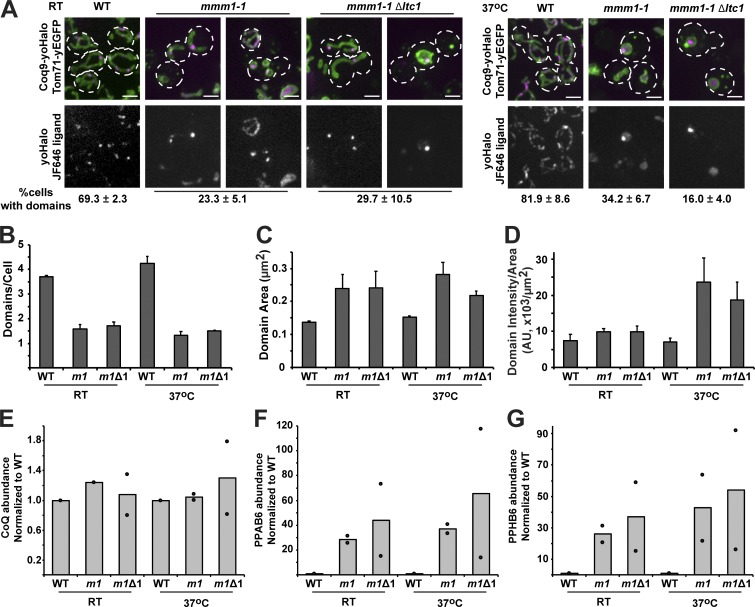
We examined the roles of ER–mitochondria contact site components, Ltc1 and the related Ltc family members, Ltc2–4, and Mmm1, which is an essential ERMES component, in CoQ domain formation and distribution and CoQ production. We did not observe any significant differences in the distribution or copy number of Coq9-yEGFP–labeled CoQ domains within the tubular mitochondrial network, as labeled by mito-dsRed, or CoQ production in Δltc1, Δltc1ltc2, or Δltc1ltc2ltc3ltc4 cells compared with WT, consistent with their WT growth rates on nonfermentable carbon (Elbaz-Alon et al., 2015; Murley et al., 2015; Fig. S5). ERMES deletion mutants possess abnormal large mitochondrial spheres, have severe growth defects on both fermentable and nonfermentable carbon sources, and lose mtDNA at a high frequency (Kornmann et al., 2009). In cells lacking ERMES components, these severe phenotypes rapidly select for dominant suppressor mutations, which map to the VPS13 gene (Lang et al., 2015). Thus, to avoid pleiotropic indirect effects and to examine more direct effects of the loss of Mmm1 function, we examined CoQ domains labeled by Coq9-yoHalo in the mmm1-1 temperature-sensitive mutant, expressing the mitochondrial outer membrane protein Tom71-yEGFP under permissive and acute (2 h) nonpermissive conditions. Under both permissive and nonpermissive conditions, we observed a decrease in the percentage of mmm1-1 cells containing CoQ domains relative to WT cells (Fig. 7 A). These data are consistent with the mmm1-1 allele being hypomorphic, as indicated by cells at permissive temperature (RT) containing large mitochondrial spheres characteristic of loss of ERMES function (Fig. 7 A, left panel at RT). Deletion of LTC1 enhanced the CoQ domain formation defect observed in mmm1-1 cells under the nonpermissive condition (Fig. 7 A). Additionally, under nonpermissive conditions, the number of domains per cell decreased, with an associated increase in domain intensity in mmm1-1 and mmm1-1Δltc1 cells (Fig. 7, B–D). As expected, CoQ levels were not significantly different in mutant cells under any condition, including the relatively short duration of nonpermissive treatment (Fig. 7 E). However, there was an increase in CoQ intermediates that correlated with the loss and alteration observed in CoQ domains in mmm1-1 and mmm1-1Δltc1 cells under permissive and nonpermissive conditions (Fig. 7, F and G). In addition, mmm1-1 and mmm1-1Δltc1 mutant cells that contained CoQ domains possessed an average of only one per cell, often localized within a single spherical mitochondria, in contrast to an average of four domains per WT cell (Fig. 7, A and B). These data suggest that ER–mitochondria contact sites are important for CoQ domain formation and CoQ production and may be important for normal CoQ domain distribution within cells. However, the exact mechanism underlying the role of ER–mitochondria contact sites in the CoQ pathway remains to be elucidated.
Figure 7.
ER–mitochondria contact site components are important for CoQ domain formation and distribution. (A) Representative max z-projection images of yeast cells expressing Coq9-yoHalo (magenta) and Tom71-yEGFP (green) from their endogenous loci in indicated strains. Cells were grown at RT or 37°C for 2 h. Percentage of cells with CoQ domains corresponding to respective yeast strains are shown below images as mean ± SEM; n > 70 cells from three independent experiments. Dotted lines represent cell boundary. (B–D) Quantification of average number of domains per cell (B), domain area in µm2 (C), and domain intensity per domain area in fluorescence intensity AU per µm2 (D) from cells imaged in A. Data represented as mean ± SEM; n > 70 cells from three independent experiments. (E–G) Relative CoQ (E), PPAB6 (F), and PPHB6 (G) abundance measurements of indicated strains. Abundance measurements were normalized to the corresponding WT at each temperature. Bars represent mean of two independent experiments, and data points (circles) represent the measurements for each experiment. Scale bars = 2 µm.
Discussion
Our data show that head-modifying proteins in the CoQ biosynthetic pathway selectively form light-resolvable, supramolecular assemblies or domains in cells. In this manner, CoQ domain assembly is analogous to other multicomponent metabolic pathways, such as the de novo purine nucleotide biosynthetic pathway, the TCA cycle, and the glycolytic pathway, that dynamically assemble into metabolons in response to substrates and other regulatory inputs (Chan et al., 2015; Webb et al., 2017). Metabolons are thought to function as microenvironments that sequester pathway intermediates for processive modification and prevent intermediates from participating in other metabolic pathways (Sweetlove and Fernie, 2018). Given that CoQ lipid intermediates are extremely hydrophobic, their accessibility to enzymes operating at the aqueous/lipid bilayer interface is likely to be rate limiting at every step in CoQ production. Thus, we postulate that one primary function of CoQ domains is to create and maintain a relatively high concentration of accessible CoQ lipid intermediates for the efficient and processive production of the quinone head group—an idea supported by the strict correlation we observe between domain formation and CoQ production. Our data indicate that CoQ domain formation requires CoQ lipid intermediates formed after HHB, the earliest lipid substrate in the pathway, consistent with published work indicating that cells accumulate HHB in the absence of acting enzymes in the pathway (Poon et al., 1997; He et al., 2014). The functional importance of CoQ domains is underscored by our observation that they are conserved from yeast to human cells. Given the link between defects in CoQ production and divergent mammalian mitochondrial diseases, our findings raise the possibility that CoQ domains also may be a robust biomarker for functional CoQ production in human cells, which might enable a deeper understanding of the role of CoQ biogenesis defects in human diseases.
Our data provide mechanistic insights into CoQ domain formation and a working model for CoQ biosynthesis (Fig. 1 A). We speculate that the highly cooperative assembly of CoQ domains requires the coordinate binding of CoQ lipid intermediates by Coq proteins and Coq protein scaffolds that function to concentrate and organize Coq enzymes responsible for modifying the lipid head group. Published work suggests that Coq8 facilitates head group modification by mediating the extraction of CoQ intermediate lipids from the inner membrane: it is an ATPase whose activity is stimulated by both CoQ head intermediates and the mitochondrial inner membrane lipid, cardiolipin (Reidenbach et al., 2018). In our analysis, Coq8 behaved as an outlier relative to other components that function in the CoQ head modification part of the pathway, as it did not concentrate in CoQ domains. As such, our data suggest that Coq8 is not a steady-state component of the CoQ complex and are consistent with a role as a CoQ lipid “chaperone” whose activity is permissive for CoQ domain nucleation and maintenance. Published work on Coq4 suggests that it may assist with complex scaffolding: it interacts with Coq3, Coq6, and Coq9 (Marbois et al., 2005; Allan et al., 2015). Consistent with a Coq4 protein scaffolding role, overexpression of Coq8 failed to suppress CoQ domain formation defects in Δcoq4 cells. In addition, an x-ray structure of a related Coq4 domain in the cyanobacteria protein Alr8543 revealed a hydrophobic binding pocket containing a geranylgeranyl monophosphate lipid, suggesting that Coq4 may coordinately bind the long isoprenyl tail of CoQ intermediates and head modification enzymes (Rea et al., 2010). Indeed, higher molecular species containing Coq9 were only detected by BN-PAGE in the presence of CoQ lipid intermediates, further indicating that CoQ lipid production and CoQ domain assembly may be obligatorily coupled.
Our data indicate that within mitochondria in cells, CoQ domains are positioned nonrandomly adjacent to ER–mitochondria contacts. Cells defective in ER–mitochondria contact site components displayed defects in CoQ domain copy number and distribution and accumulated CoQ intermediates. Although the molecular basis of the link between CoQ domains and the ER is not known, these observations suggest a role for ER–mitochondria contacts in CoQ domain formation and CoQ production and distribution. Active positioning of other metabolons at defined subcellular localizations has been observed, such as the localization of the cytosolic de novo purine nucleotide biosynthetic and glycolytic metabolons adjacent to mitochondria (French et al., 2016; Kohnhorst et al., 2017; Pedley and Benkovic, 2017). In these cases, the colocalization is thought to fuel and coordinate metabolic pathways via the exchange of substrates, cofactors, and products. The spatial organization of the mevalonate pathway, which produces the isoprene units used inside mitochondria for Coq1-dependent tail polymerization, has not been fully established. However, many components of this pathway, including HMG-CoA reductase, are localized to the ER. Thus, it is possible that CoQ domains are positioned at the contacts as a consequence of substrate flux; however, Coq1 and Coq2, which directly use the isoprene units, are not enriched at CoQ domains. Alternatively, ER–mitochondria contacts may enrich regions of associated inner membrane in specific proteins and lipids, such as cardiolipin, that modulate the activity of Coq8 and recruit additional Coq proteins (Lohman et al., 2019). Our previous work has demonstrated that ER–mitochondria contacts link the distribution of mitochondria and mtDNA (Murley et al., 2013; Lewis et al., 2016). In this manner, ER–mitochondria contacts may also function to spatially organize and integrate essential mitochondrial structures. Thus, they could similarly function to control the spatial organization of CoQ domains to distribute the CoQ lipid within mitochondria to drive oxidative phosphorylation and within cells to extramitochondrial compartments, where it may play additional redox-related roles (Zhang et al., 1996; Fernández-Ayala et al., 2005).
Materials and methods
Plasmid and strain construction
Saccharomyces cerevisiae strains constructed in this study are based on the W303 (ade2-1; leu2-3; his3-11, 15; trp 1-1; ura3-1; can1-100) genetic background. All deletions were derived using PCR-based homologous recombination replacing the entire ORF of targeted genes with NatMX6 cassettes (Longtine et al., 1998; Sheff and Thorn, 2004; Hoppins et al., 2011). All C-terminal tags were generated using PCR-based targeted homologous recombination using the following cassettes: yEGFP::SpHIS5, mCherry::SpHIS5, and yoHalo::CaUra3. Strains were constructed by mating and/or by direct transformation using the lithium acetate method. Correct targeting of chromosomal integrations was confirmed by PCR. The N-terminal fluorescent protein fusion allele of Coq7 was generated using the Delitto Perfecto system previously described (Storici and Resnick, 2006; Stuckey et al., 2011; Stuckey and Storici, 2013). The GSKU cassette was PCR amplified from pGSKU, integrated into Coq7 locus by homologous recombination between amino acids 36 and 37, and verified by PCR. yEGFP was PCR amplified and integrated by homologous recombination to replace the GSKU cassette. Loss of the GSKU cassette was selected for by growth on 5-fluoroorotic acid. The resulting strain was mated back to W303, sporulated, and selected by fluorescence microscopy. Haploid double-tagged or deletion strains expressing Coq9-yEGFP::His were constructed by crossing followed by sporulation or PCR-based targeted homologous recombination. The temperature-sensitive allele mmm1-1 expressing Coq9-yoHalo::CaUra3 and Tom71-yeGFP::His was constructed by mating mmm1-1 Tom71-yeGFP::His or mmm1-1 Δltc1::Nat Tom71-yeGFP::His to Coq9-yoHalo::CaUra3, followed by sporulation.
To generate pRS304-COQ6, genomic DNA from W303A yeast strain was used to amplify a PCR product containing COQ6 plus its native promoter (∼500 bp 5′ of start codon) and ∼500 bp after its stop codon and was then cloned into linearized pRS304 using a Gibson isothermal assembly reagent according to the manufacturer’s instructions. This plasmid was used to generate pRS304-COQ6 G386A N388D by isothermal assembly of PCR products that when recombined, included the two nucleotide changes. pRS304-COQ6 plasmids were integrated at the TRP1 locus and introduced by linearization using Bsu36I and lithium acetate transformation, followed by selection on SD media-Trp agar plates. To generate pRS424-COQ8, genomic DNA from W303A yeast strain was used to amplify a PCR product containing COQ8 plus its native promoter (∼500 bp 5′ of start codon) and ∼500 bp after its stop codon and was then cloned into linearized pRS424 using a Gibson isothermal assembly reagent according to the manufacturer’s instructions. pRS426-GPD, pRS426-GPD-COQ8, and pRS426-GPD-COQ8 M202C were previously described in more detail (Reidenbach et al., 2018). To generate pFA6a-yoHalo::CaUra3, yoHalo-Tag plasmid was a gift of B. Glick (University of Chicago; Day et al., 2018) and was used to amplify a PCR product containing yoHalo. yEGFP was cut out of pFA6a-yEGFP::CaUra3 using PacI and AscI and replaced with yoHalo using a Gibson isothermal assembly reagent according to the manufacturer’s instructions.
pVT100U-mtTagBFP, a 2-µm plasmid driving mitochondrial matrix-targeted TagBFP (mt-TagBFP) expression (Friedman et al., 2018); pYX142-mtDsRed and pYX142-mtGFP, a CEN/ARS plasmid driving mitochondrial matrix-targeted DsRed (mt-dsRed) or GFP (mtGFP) expression from a TDI promoter (Westermann and Neupert, 2000); and pK1803 (DsRed-HDEL), which expresses a fusion protein containing the Kar2 signal sequence, DsRed, and an HDEL retention sequence (Madrid et al., 2006) are all previously described in more detail.
Human CoQ9 was cloned from HCT116 cDNA using Platinum SuperFi DNA polymerase (Thermo Fisher Scientific). The CoQ9 gene was then PCR amplified with the primers 5′-CGACGGTACCGCGGGCCCGGATGGCGGCGGCGGCGGTATC-3′ and 5′-CACCATGGTGGCGACCGGTGAGCTTCCTCCGCCCCGACGCTGGTTTAGACCTGTCAAGTTCT-3′ and inserted into a BamHI-digested pEGFP-N1 backbone using Gibson isothermal assembly reagent according to the manufacturer’s instructions.
Yeast growth assays
Cells were grown to log phase in yeast extract peptone dextrose (YPD), pelleted, and resuspended in water at a concentration of 0.5 OD600/ml. 4 µl of 10-fold serial dilutions were plated on YPD and yeast extract, peptone, ethanol, glycerol (YPEG) plates. Cells were grown for ∼48 h (YPD) and ∼48–60 h (YPEG) at 30°C.
Fluorescence microscopy
For all fluorescence microscopy experiments with yeast, cells were grown at 30°C to log phase in the appropriate synthetic medium to select for plasmids. Cells were mounted and imaged on a glass-bottom SensoPlate (Greiner 655892) pretreated with concanavalin A (C2010, 0.25 mg/ml; Sigma-Aldrich) for 1 h followed by drying. Cells were imaged at 25°C.
For VA treatments in Fig. 4, cells were grown in synthetic media with Dex–Leu to mid-log phase and incubated in the presence or absence of 1 mM VA (Sigma-Aldrich) for the entire growth in liquid media. For fixed time points in Fig. S3, mid-log phase cells were grown in the absence or presence of 1 mM VA for 0, 2, 10, or 26 h. For CMK treatments in Fig. 4, cells were grown overnight and to mid-log phase in synthetic media with Dex–Leu–Ura and 10–12 h in synthetic media without pABA and folate with Dex–Ura. For fixed time points, cells were grown to mid-log phase and incubated for 0.5 or 2 h in the absence or presence of 1 mM CMK (Reidenbach et al., 2018).
For temperature-sensitive experiments in Fig. 7, cells were grown in synthetic media with Dex Complete to mid-log phase at RT and incubated at RT or 37°C for 2 or 24 h. To visualize Coq9-yoHalo, JF646 ligand, provided by L. Lavis (Janelia Research Campus; Grimm et al., 2015), was added to cells to a final concentration of 1 µM, incubated at RT or 37°C for 30 min, and rinsed in synthetic media with Dex Complete, similar to previous description (Day et al., 2018).
For confocal microscopy, images of Z-series (step size of 0.2 µm) of cells were collected using the spinning-disk module of a Marianas SDC Real Time 3D Confocal-TIRF microscope (Intelligent Imaging Innovations) fitted with a 100× 1.46 NA objective and a Photometrics QuantEM EMCCD camera. Images were captured with SlideBook (Intelligent Imaging Innovations), and linear adjustments were made using ImageJ (National Institutes of Health).
For confocal microscopy for VA time-course experiments in Fig. S3, images of Z-series (step size of 0.2 µm) of cells were collected using the Nikon High Content Analysis microscope with spinning-disk confocal (Nikon Instruments) fitted with a 100× 1.46 NA objective and an ORCA-Flash 4.0 V2 Digital CMOS camera. Images were captured with NIS-Elements (Nikon).
For structured illumination superresolution microscopy, cells were grown to log phase, concentrated by centrifugation, and directly mounted and imaged on MICRO slides (VWR 48300-025). Images of Z-series (step size of 0.5 µM) of cells were collected using the Nikon Structured Illumination Super-Resolution Microscope (N-SIM) fitted with a 100× 1.46 NA objective. Images were captured in the 3D-SIM mode using NIS-Elements (Nikon), and SIM processing was performed using the SIM module in the Nikon software package. Linear adjustments were made using ImageJ.
Indirect immunofluorescence in yeast cells
In Fig. S2, yeast cells were grown in YPD media to log phase and incubated with 500 nM MitoTracker Red CMX Ros (Thermo Fisher Scientific) according to the manufacturer’s instructions for 30 min, protected from light. Fixed as previously described (Swayne et al., 2009), cells were washed in YPD media and grown for 1 h in 3.7% PFA in YPD media at 30°C. Cells were centrifuged and resuspended in Tris/DTT (0.1 M Tris-SO4, pH 9.4, and 0.01 M DTT) and incubated for 20 min at 30°C. Cells were centrifuged, resuspended in zymolyase 20T (5 mg/ml) in wash solution (0.025 M potassium phosphate, pH 7.5, and 0.8 M KCl), and incubated for 20 min at 30°C. Cells were centrifuged, washed two times in NS+ (0.02 M Tris-HCl, pH 7.5, 0.04 g/ml sucrose, 1 mM EDTA, pH 8.0, 1 mM MgCl2, 0.1 mM ZnCl2, 0.05 mM CaCl2, 1 mM PMSF, and 1% wt/vol NaN3), and resuspended in 2 vol of NS+. Spheroplasts were used for indirect immunofluorescence. Cells were mounted and imaged on glass-bottom SensoPlate (Greiner 655892) pretreated with Polylysine solution (C2010, 0.25 mg/ml; Sigma-Aldrich) for 1 h followed by drying. Cells were incubated with primary anti-Myc antibody (Thermo Fisher Scientific) at 1:200 for 2 h and with Alexa Fluor–conjugated secondary antibody (Thermo Fisher Scientific) at 1:1,000. Cells were washed with PBT (PBS containing 1% wt/vol BSA, 0.1% Triton X-100, and 0.1% wt/vol sodium azide) between antibody incubations. Before imaging, mounting solution (0.1% wt/vol p-phenylenediamine and 90% vol/vol glycerol in PBS) was added to cells.
Mammalian cell growth, transfection, and indirect immunofluorescence
For Fig. 2, U2OS cells were grown in high-glucose DMEM supplemented with 10% FBS and 1% penicillin/streptomycin. Cells were seeded at ∼0.25 × 105 cells per ml in 35-mm glass-bottom dishes (Mattek) and allowed to attach for 24 h before mitochondrial staining with 500 nM MitoTracker Red CMX Ros (Thermo Fisher Scientific) according to manufacturer instructions. Cells were then directly fixed in 4% PFA in PBS, pH 7.4, for 20 min at RT, protected from light. Dishes were washed twice in PBS, permeabilized in 0.1% Triton X-100 for 20 min, and blocked in 3% BSA PBS solution for 1 h at RT. Primary anti-Coq9 antibodies (ProteinTech) were added at 1:10,000 in PBST (PBS, pH 7.4, 1% BSA, and 0.1% Tween-20) overnight at 4°C, rinsed three times in PBS and incubated with Alexa Fluor–conjugated secondary antibodies (Thermo Fisher Scientific) at 1:2,000 dilution in PBST for 1 h. Dishes were washed at least three times before imaging. Imaging was performed as previously described above using the spinning-disk module of an inverted objective fluorescence microscope or Nikon Structured Illumination Super-Resolution Microscope.
For Fig. S1 E, the day before transfection, U2OS cells were plated on 12-mm coverslips (#1.5) in a 24-well plate at a density of 75,000 cells per well. Transfection was performed with Lipofectamine 2000 (Thermo Fisher Scientific) according to the manufacturer instructions using 25 ng of FUGW-CoQ9-eGFP plasmid, 175 ng of pGEM 3Z f(+) plasmid as carrier DNA, and 1.25 µl of Lipofectamine 2000 per well. Cells were fixed and imaged as previously described above using the spinning-disk module of an inverted objective fluorescence microscope.
Lipid extraction
Cells were grown to log phase in YPD or appropriate synthetic media with Dex to select for plasmids, centrifuged, and flash-frozen dropwise in liquid N2. Frozen pellets of yeast (108 cells) were thawed on ice and mixed with glass beads (0.5 mm diameter, 100 µl). CHCl3/MeOH (2:1, vol/vol, 4°C, 900 ml) and CoQ10 (10 µl, 10 µM, 0.1 nmol) were added and vortexed (2 × 30 s). HCl (1 M, 200 µl, 4°C) was added and vortexed (2 × 30 s). The samples were centrifuged (5,000 g, 2 min, 4°C) to complete phase separation. 400 µl of the organic phase was transferred to a clean tube and dried under Ar(g). The organic residue was reconstituted in ACN/IPA/H2O (65:30:5, vol/vol/vol, 100 µl) for LC-MS analysis.
LC-MS lipid analysis
LC-MS analysis was performed on an Acquity CSH C18 column held at 50°C (100 mm × 2.1 mm × 1.7 µm particle size; Waters) using a Vanquish Binary Pump (400 µl/min flow rate; Thermo Fisher Scientific). Mobile phase A consisted of 10 mM ammonium acetate in ACN:H2O (70:30, vol/vol) containing 250 µl/l acetic acid. Mobile phase B consisted of 10 mM ammonium acetate in IPA:ACN (90:10, vol/vol) with the same additives. Mobile phase B was initially held at 50% for 1.5 min and then increased to 99% over 7.5 min and held there for 2 min. The column was reequilibrated for 2.5 min before the next injection. 10 µl of samples were injected by a Vanquish Split Sampler HT autosampler (Thermo Fisher Scientific).
The LC system was coupled to a Q Exactive mass spectrometer by a HESI II heated ESI source maintained at 300°C (Thermo Fisher Scientific). The inlet capillary was kept at 300°C, sheath gas was set to 25 units and auxiliary gas to 10 units, and the spray voltage was set to 5,000 V. For identification of CoQ6, the MS was operated in positive mode, and a mass of 591.44 was targeted for fragmentation. Automatic gain control (AGC) target was set to 5 × 105, and resolving power was set to 140,000. Quantification was performed in Xcalibur (Thermo Fisher Scientific) by monitoring the product ion 197.08 Th, corresponding to the Q headgroup, for each targeted mass. The MS was operated in negative mode, and masses (545.40 and 544.42, respectively) were targeted to quantify CoQ intermediates PPHB6 and PPAB6. AGC target was set to 5 × 105, and resolving power was set to 140,000.
Whole-cell extracts and mitochondrial isolation
For whole-cell extracts, cells were grown to log phase in YPD or synthetic complete media with Dex or Leu to select for plasmids. Whole-cell lysates of 0.25 OD600 cells were obtained by alkaline extraction (0.255 M NaOH and 1% β-mercaptoethanol), precipitated with 9% trichloroacetic acid, washed with acetone, dried, and resuspended in 50 µl MURB protein sample buffer (100 mM MES, pH 7.0, 1% SDS, 3 M urea, and 10% β-mercaptoethanol). 10 µl of sample was used for Western blot analysis.
For isolation of mitochondria, cells were grown to log phase in synthetic complete media with Dex or Leu to select for plasmids. Cells were grown to log phase, and 500 OD600 cells were centrifuged at 2,700 rpm. Cell pellets were washed with water, resuspended in NMIB (0.6 M sorbitol, 20 mM Hepes, 100 mM KOAc, 5 mM MgCl2, 50 mM KCl, and 1 mM PMSF) and 1× Protease Inhibitor Mixture I (Calbiochem), flash-frozen dropwise in liquid N2, and lysed using a Freezer/Mill (SPEX). Cell lysates were centrifuged at 4,500 rpm for 5 min, and supernatant was centrifuged at 10,500 rpm for 15 min. Pellets were resuspended in 400 µl NMIB with Inhibitor Mixture I. 25 µg protein, determined by Bradford or BCA assay, was added to MURB sample buffer and used for Western blot analysis.
Western blot analysis
Samples were boiled for 5 min, analyzed by SDS-PAGE, transferred to nitrocellulose, and immunoblotted with primary antibodies at indicated concentrations: anti-GFP antibody (1:2,000, University of California, Davis, NeuroMab clone N86/8), anti-Coq4 antibody (Belogrudov et al., 2001), anti-Coq9 antibody (Hsieh et al., 2007), anti-GP6DH antibody (1:2,000, Sigma-Aldrich), and anti-Porin antibody (1:5,000, Thermo Fisher Scientific). The appropriate secondary antibodies conjugated to DyLight680 or DyLight800 (1:10,000, Thermo Fisher Scientific) were used and visualized with the Odyssey Infrared Imaging System (LI-COR). Linear adjustments to images were made using ImageJ. Relative density from Western blot analysis was determined using Imaging Studio Lite.
BN-PAGE analysis
BN-PAGE was performed as previously described (Wittig et al., 2006). Briefly, 200 µg mitochondria were suspended in 20 µl solubilization buffer (50 mM NaCl, 50 mM imidazole, 2 mM 6-aminohexanoic acid, and 1 mM EDTA, pH 7.0), and membrane proteins were solubilized on ice for 10 min with Triton X-100 to a detergent/protein ratio of 2 g/g. After centrifugation (20 min at 20,000 g), the supernatant was collected, and protein concentrations were quantified by BCA. Glycerol was added to a final concentration of 5%, and Coomassie dye from a 5% Coomassie blue G-250 stock suspension was added to a final detergent/dye ratio of 8 g/g. An equal amount of protein (5 or 25 µg) was loaded and run on a 3.5–16% or 5–16% gel with 25 mM imidazole (pH 7.0) as anode buffer and 50 mM Tricine, 7.5 mM imidazole, and 0.02% Coomassie blue G-250 as cathode buffer. The cathode buffer was changed to 50 mM Tricine, 7.5 mM imidazole, and 0.002% Coomassie blue G-250 after the protein had migrated through one-third of the resolving gel. Gels were run at 4°C at an initial voltage of 100 V. When the sample had reached the resolving gel, the voltage was increased to 200–250 V until the Coomassie dye approached the gel front. Native gels were incubated with agitation for 20 min in 300 mM Tris, 100 mM acetic acid, and 1% SDS, pH 8.6. The gels were then placed between two glass plates for 1 h at RT to allow the SDS to spread evenly within the gel and denature the protein. Proteins were then electroblotted on PVDF membrane at 4°C for 12 h using SDS-free transfer buffer (150 mM Tris and 50 mM acetic acid, pH 8.6). Proteins were transferred at 90 mA and voltage limited to 20 V.
Quantification and statistical analysis
To quantify domain area (µm2) and domain intensity/area (AU/µm2), background subtraction, threshold, and watershed were performed using max z-projections in ImageJ of images taken under all the same imaging conditions for each experiment. Particles were analyzed from threshold and watershed 488-channel images with size (0.05–2.0, 0.4–2.0) and circularity (0.4–1.0) for yeast and mammalian cells, respectively. Regions of interest identified from particle analysis were selected on the original image and measured for particle area (µm2) and integrated density (AU). Particle area was averaged for each experiment. The integrated density with the background subtracted was divided by the corresponding particle area to determine the intensity/area and averaged for each experiment. At least 70 cells were quantified and imaged for three separate experiments.
To quantify the corrected total cell fluorescence, cells were outlined from single-plane bright-field images, and the integrated density was determined using the sum of z-projections using ImageJ. The integrated density – (area of selected cell × mean fluorescence of background readings) was determined and averaged for at least 70 cells per experiment.
All statistical analyses were performed using R version 3.4.4. One-way ANOVA analyses were followed by post hoc pairwise t test with Bonferroni correction. Normality of residuals was tested by visual inspection of Q-Q plots followed by Shapiro–Wilk tests. Homoscedasticity was tested with a Levene test. When ANOVA assumptions were violated, a nonparametric Kruskal–Wallis test was used followed by a post-hoc Dunn test. Analyses with P values smaller than 0.05 were considered significant.
Online supplemental material
Fig. S1 shows functional analysis of Coq-GFP strains as assessed by serial dilutions on fermentable carbon source containing media and CoQ lipid abundance measurements and also shows the specificity of anti-Coq9 antibody in human cells. Fig. S2 shows that CoQ production and CoQ domain distribution were affected in yeast cells expressing hypomorphic Coq5-yEGFP and also shows indirect immunofluorescence of fixed yeast cells of select Coq proteins. Fig. S3 shows CoQ lipid measurements and the percentage of cells with CoQ domains as a function of time in COQ6 G386A N388D cells after VA addition and in COQ8 AS cells following addition of CMK. Fig. S4 contains BN-PAGE analysis showing the presence of large molecular Coq9-containing species correlated with CoQ domains in cells. Fig. S5 shows that CoQ domains were present in LTC-deficient cells and also shows CoQ lipid abundance measurements for LTC-deficient cells.
Supplementary Material
Acknowledgments
We thank Michael Paddy at the University of California, Davis, Molecular and Cellular Biology Imaging Facility for advice with fluorescence microscopy. We thank members of the Nunnari and Pagliarini laboratories for stimulating discussions. K. Gov and M. Wong provided technical help.
The 3iMarianas spinning-disk confocal and Nikon High Content Analysis spinning-disk confocal used in this study were purchased using National Institutes of Health (NIH) Shared Instrumentation grants 1S10RR024543-01 and 1S10OD019980-01A1, respectively. K. Subramanian was supported by NIH training grant 5T32GM007377-34. S. Lewis was supported by NIH grant F32GM113388 and a Burroughs Wellcome Postdoctoral Enrichment Award. J. Nunnari is supported by NIH grants R37GM097432 and R01GM126081. D.J. Pagliarini is supported by NIH grants R01GM112057 and R01GM115591, and J.J. Coon and D.J. Pagliarini are supported by GM108538.
J. Nunnari is on the scientific advisory board of Mitobridge, an Astellas Company. The authors declare no further conflict of interest.
Author contributions: K. Subramanian: conceptualization, data curation, formal analysis, methodology, and writing–original draft and review and editing. J. Nunnari: conceptualization, formal analysis, funding acquisition, supervision, and writing–original draft and review and editing. A. Jochem, S. Lewis, and M. Le Vasseur: data curation, formal analysis, methodology, and writing–review and editing. B.R. Paulson, T.R. Reddy, and J.D. Russell: data curation. J.J. Coon and D.J. Pagliarini: conceptualization, funding acquisition, supervision, and writing–review and editing.
References
- Acosta M.J., Vazquez Fonseca L., Desbats M.A., Cerqua C., Zordan R., Trevisson E., and Salviati L.. 2016. Coenzyme Q biosynthesis in health and disease. Biochim. Biophys. Acta. 1857:1079–1085. 10.1016/j.bbabio.2016.03.036 [DOI] [PubMed] [Google Scholar]
- Allan C.M., Awad A.M., Johnson J.S., Shirasaki D.I., Wang C., Blaby-Haas C.E., Merchant S.S., Loo J.A., and Clarke C.F.. 2015. Identification of Coq11, a new coenzyme Q biosynthetic protein in the CoQ-synthome in Saccharomyces cerevisiae. J. Biol. Chem. 290:7517–7534. 10.1074/jbc.M114.633131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashby M.N., Kutsunai S.Y., Ackerman S., Tzagoloff A., and Edwards P.A.. 1992. COQ2 is a candidate for the structural gene encoding para-hydroxybenzoate:polyprenyltransferase. J. Biol. Chem. 267:4128–4136. [PubMed] [Google Scholar]
- Awad A.M., Bradley M.C., Fernández-Del-Río L., Nag A., Tsui H.S., and Clarke C.F.. 2018. Coenzyme Q10 deficiencies: pathways in yeast and humans. Essays Biochem. 62:361–376. 10.1042/EBC20170106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkovich R.J., Shtanko A., Shepherd J.A., Lee P.T., Myles D.C., Tzagoloff A., and Clarke C.F.. 1997. Characterization of the COQ5 gene from Saccharomyces cerevisiae. Evidence for a C-methyltransferase in ubiquinone biosynthesis. J. Biol. Chem. 272:9182–9188. 10.1074/jbc.272.14.9182 [DOI] [PubMed] [Google Scholar]
- Barros M.H., Johnson A., Gin P., Marbois B.N., Clarke C.F., and Tzagoloff A.. 2005. The Saccharomyces cerevisiae COQ10 gene encodes a START domain protein required for function of coenzyme Q in respiration. J. Biol. Chem. 280:42627–42635. 10.1074/jbc.M510768200 [DOI] [PubMed] [Google Scholar]
- Belogrudov G.I., Lee P.T., Jonassen T., Hsu A.Y., Gin P., and Clarke C.F.. 2001. Yeast COQ4 encodes a mitochondrial protein required for coenzyme Q synthesis. Arch. Biochem. Biophys. 392:48–58. 10.1006/abbi.2001.2448 [DOI] [PubMed] [Google Scholar]
- Brandina I., Graham J., Lemaitre-Guillier C., Entelis N., Krasheninnikov I., Sweetlove L., Tarassov I., and Martin R.P.. 2006. Enolase takes part in a macromolecular complex associated to mitochondria in yeast. Biochim. Biophys. Acta. 1757:1217–1228. 10.1016/j.bbabio.2006.07.001 [DOI] [PubMed] [Google Scholar]
- Chan C.Y., Zhao H., Pugh R.J., Pedley A.M., French J., Jones S.A., Zhuang X., Jinnah H., Huang T.J., and Benkovic S.J.. 2015. Purinosome formation as a function of the cell cycle. Proc. Natl. Acad. Sci. USA. 112:1368–1373. 10.1073/pnas.1423009112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cogliati S., Enriquez J.A., and Scorrano L.. 2016. Mitochondrial Cristae: Where Beauty Meets Functionality. Trends Biochem. Sci. 41:261–273. 10.1016/j.tibs.2016.01.001 [DOI] [PubMed] [Google Scholar]
- Day K.J., Casler J.C., and Glick B.S.. 2018. Budding yeast has a minimal endomembrane system. Dev. Cell. 44:56–72.e4. 10.1016/j.devcel.2017.12.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbaz-Alon Y., Eisenberg-Bord M., Shinder V., Stiller S.B., Shimoni E., Wiedemann N., Geiger T., and Schuldiner M.. 2015. Lam6 Regulates the Extent of Contacts between Organelles. Cell Reports. 12:7–14. 10.1016/j.celrep.2015.06.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eppink M.H., Schreuder H.A., and Van Berkel W.J.. 1997. Identification of a novel conserved sequence motif in flavoprotein hydroxylases with a putative dual function in FAD/NAD(P)H binding. Protein Sci. 6:2454–2458. 10.1002/pro.5560061119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández-Ayala D.J., Brea-Calvo G., López-Lluch G., and Navas P.. 2005. Coenzyme Q distribution in HL-60 human cells depends on the endomembrane system. Biochim. Biophys. Acta. 1713:129–137. 10.1016/j.bbamem.2005.05.010 [DOI] [PubMed] [Google Scholar]
- Floyd B.J., Wilkerson E.M., Veling M.T., Minogue C.E., Xia C., Beebe E.T., Wrobel R.L., Cho H., Kremer L.S., Alston C.L., et al. 2016. Mitochondrial Protein Interaction Mapping Identifies Regulators of Respiratory Chain Function. Mol. Cell. 63:621–632. 10.1016/j.molcel.2016.06.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- French J.B., Jones S.A., Deng H., Pedley A.M., Kim D., Chan C.Y., Hu H., Pugh R.J., Zhao H., Zhang Y., et al. 2016. Spatial colocalization and functional link of purinosomes with mitochondria. Science. 351:733–737. 10.1126/science.aac6054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman J.R., Mourier A., Yamada J., McCaffery J.M., and Nunnari J.. 2015. MICOS coordinates with respiratory complexes and lipids to establish mitochondrial inner membrane architecture. eLife. 4:e07739 10.7554/eLife.07739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman J.R., Kannan M., Toulmay A., Jan C.H., Weissman J.S., Prinz W.A., and Nunnari J.. 2018. Lipid Homeostasis Is Maintained by Dual Targeting of the Mitochondrial PE Biosynthesis Enzyme to the ER. Dev. Cell. 44:261–270.e6. 10.1016/j.devcel.2017.11.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatta A.T., Wong L.H., Sere Y.Y., Calderón-Noreña D.M., Cockcroft S., Menon A.K., and Levine T.P.. 2015. A new family of StART domain proteins at membrane contact sites has a role in ER-PM sterol transport. eLife. 4:e07253 10.7554/eLife.07253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimm J.B., English B.P., Chen J., Slaughter J.P., Zhang Z., Revyakin A., Patel R., Macklin J.J., Normanno D., Singer R.H., et al. 2015. A general method to improve fluorophores for live-cell and single-molecule microscopy. Nat. Methods. 12:244–250. 10.1038/nmeth.3256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harner M., Körner C., Walther D., Mokranjac D., Kaesmacher J., Welsch U., Griffith J., Mann M., Reggiori F., and Neupert W.. 2011. The mitochondrial contact site complex, a determinant of mitochondrial architecture. EMBO J. 30:4356–4370. 10.1038/emboj.2011.379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He C.H., Xie L.X., Allan C.M., Tran U.C., and Clarke C.F.. 2014. Coenzyme Q supplementation or over-expression of the yeast Coq8 putative kinase stabilizes multi-subunit Coq polypeptide complexes in yeast coq null mutants. Biochim. Biophys. Acta. 1841:630–644. 10.1016/j.bbalip.2013.12.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoppins S., Collins S.R., Cassidy-Stone A., Hummel E., Devay R.M., Lackner L.L., Westermann B., Schuldiner M., Weissman J.S., and Nunnari J.. 2011. A mitochondrial-focused genetic interaction map reveals a scaffold-like complex required for inner membrane organization in mitochondria. J. Cell Biol. 195:323–340. 10.1083/jcb.201107053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh E.J., Gin P., Gulmezian M., Tran U.C., Saiki R., Marbois B.N., and Clarke C.F.. 2007. Saccharomyces cerevisiae Coq9 polypeptide is a subunit of the mitochondrial coenzyme Q biosynthetic complex. Arch. Biochem. Biophys. 463:19–26. 10.1016/j.abb.2007.02.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimenez L., Laporte D., Duvezin-Caubet S., Courtout F., and Sagot I.. 2014. Mitochondrial ATP synthases cluster as discrete domains that reorganize with the cellular demand for oxidative phosphorylation. J. Cell Sci. 127:719–726. 10.1242/jcs.137141 [DOI] [PubMed] [Google Scholar]
- Jonassen T., and Clarke C.F.. 2000. Isolation and functional expression of human COQ3, a gene encoding a methyltransferase required for ubiquinone biosynthesis. J. Biol. Chem. 275:12381–12387. 10.1074/jbc.275.17.12381 [DOI] [PubMed] [Google Scholar]
- Kohnhorst C.L., Kyoung M., Jeon M., Schmitt D.L., Kennedy E.L., Ramirez J., Bracey S.M., Luu B.T., Russell S.J., and An S.. 2017. Identification of a multienzyme complex for glucose metabolism in living cells. J. Biol. Chem. 292:9191–9203. 10.1074/jbc.M117.783050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornmann B., Currie E., Collins S.R., Schuldiner M., Nunnari J., Weissman J.S., and Walter P.. 2009. An ER-mitochondria tethering complex revealed by a synthetic biology screen. Science. 325:477–481. 10.1126/science.1175088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kühl I., Miranda M., Atanassov I., Kuznetsova I., Hinze Y., Mourier A., Filipovska A., and Larsson N.G.. 2017. Transcriptomic and proteomic landscape of mitochondrial dysfunction reveals secondary coenzyme Q deficiency in mammals. eLife. 6:e30952 10.7554/eLife.30952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kühlbrandt W. 2015. Structure and function of mitochondrial membrane protein complexes. BMC Biol. 13:89 10.1186/s12915-015-0201-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang A.B., John Peter A.T., Walter P., and Kornmann B.. 2015. ER-mitochondrial junctions can be bypassed by dominant mutations in the endosomal protein Vps13. J. Cell Biol. 210:883–890. 10.1083/jcb.201502105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapointe C.P., Stefely J.A., Jochem A., Hutchins P.D., Wilson G.M., Kwiecien N.W., Coon J.J., Wickens M., and Pagliarini D.J.. 2018. Multi-omics reveal specific targets of the RNA-binding protein Puf3p and its orchestration of mitochondrial biogenesis. Cell Syst. 6:125–135.e6. 10.1016/j.cels.2017.11.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis S.C., Uchiyama L.F., and Nunnari J.. 2016. ER-mitochondria contacts couple mtDNA synthesis with mitochondrial division in human cells. Science. 353:aaf5549 10.1126/science.aaf5549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohman D.C., Forouhar F., Beebe E.T., Stefely M.S., Minogue C.E., Ulbrich A., Stefely J.A., Sukumar S., Luna-Sánchez M., Jochem A., et al. 2014. Mitochondrial COQ9 is a lipid-binding protein that associates with COQ7 to enable coenzyme Q biosynthesis. Proc. Natl. Acad. Sci. USA. 111:E4697–E4705. 10.1073/pnas.1413128111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohman D.C., Aydin D., Von Bank H., Smith R., Linke V., Weisenhorn E., McDevitt M.T., Hutchins P., Wilkerson E.M., Wancewicz B., et al. 2019. An isoprene lipid-binding protein promotes eukaryotic coenzyme Q biosynthesis. Mol. Cell. 73:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine M.S., McKenzie A. III, Demarini D.J., Shah N.G., Wach A., Brachat A., Philippsen P., and Pringle J.R.. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 14:953–961. [DOI] [PubMed] [Google Scholar]
- Luna-Sánchez M., Díaz-Casado E., Barca E., Tejada M.A., Montilla-García Á., Cobos E.J., Escames G., Acuña-Castroviejo D., Quinzii C.M., and López L.C.. 2015. The clinical heterogeneity of coenzyme Q10 deficiency results from genotypic differences in the Coq9 gene. EMBO Mol. Med. 7:670–687. 10.15252/emmm.201404632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madrid A.S., Mancuso J., Cande W.Z., and Weis K.. 2006. The role of the integral membrane nucleoporins Ndc1p and Pom152p in nuclear pore complex assembly and function. J. Cell Biol. 173:361–371. 10.1083/jcb.200506199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mannella C.A., Marko M., Penczek P., Barnard D., and Frank J.. 1994. The internal compartmentation of rat-liver mitochondria: tomographic study using the high-voltage transmission electron microscope. Microsc. Res. Tech. 27:278–283. 10.1002/jemt.1070270403 [DOI] [PubMed] [Google Scholar]
- Marbois B., Gin P., Faull K.F., Poon W.W., Lee P.T., Strahan J., Shepherd J.N., and Clarke C.F.. 2005. Coq3 and Coq4 define a polypeptide complex in yeast mitochondria for the biosynthesis of coenzyme Q. J. Biol. Chem. 280:20231–20238. 10.1074/jbc.M501315200 [DOI] [PubMed] [Google Scholar]
- Marbois B., Gin P., Gulmezian M., and Clarke C.F.. 2009. The yeast Coq4 polypeptide organizes a mitochondrial protein complex essential for coenzyme Q biosynthesis. Biochim. Biophys. Acta. 1791:69–75. 10.1016/j.bbalip.2008.10.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montero R., Grazina M., López-Gallardo E., Montoya J., Briones P., Navarro-Sastre A., Land J.M., Hargreaves I.P., and Artuch R.. Coenzyme Q10 Deficiency Study Group . 2013. Coenzyme Q10 deficiency in mitochondrial DNA depletion syndromes. Mitochondrion. 13:337–341. 10.1016/j.mito.2013.04.001 [DOI] [PubMed] [Google Scholar]
- Murley A., Lackner L.L., Osman C., West M., Voeltz G.K., Walter P., and Nunnari J.. 2013. ER-associated mitochondrial division links the distribution of mitochondria and mitochondrial DNA in yeast. eLife. 2:e00422 10.7554/eLife.00422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murley A., Sarsam R.D., Toulmay A., Yamada J., Prinz W.A., and Nunnari J.. 2015. Ltc1 is an ER-localized sterol transporter and a component of ER-mitochondria and ER-vacuole contacts. J. Cell Biol. 209:539–548. 10.1083/jcb.201502033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada K., Suzuki K., Kamiya Y., Zhu X., Fujisaki S., Nishimura Y., Nishino T., Nakagawa T., Kawamukai M., and Matsuda H.. 1996. Polyprenyl diphosphate synthase essentially defines the length of the side chain of ubiquinone. Biochim. Biophys. Acta. 1302:217–223. 10.1016/0005-2760(96)00064-1 [DOI] [PubMed] [Google Scholar]
- Ozeir M., Mühlenhoff U., Webert H., Lill R., Fontecave M., and Pierrel F.. 2011. Coenzyme Q biosynthesis: Coq6 is required for the C5-hydroxylation reaction and substrate analogs rescue Coq6 deficiency. Chem. Biol. 18:1134–1142. 10.1016/j.chembiol.2011.07.008 [DOI] [PubMed] [Google Scholar]
- Ozeir M., Pelosi L., Ismail A., Mellot-Draznieks C., Fontecave M., and Pierrel F.. 2015. Coq6 is responsible for the C4-deamination reaction in coenzyme Q biosynthesis in Saccharomyces cerevisiae. J. Biol. Chem. 290:24140–24151. 10.1074/jbc.M115.675744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palfey B.A., Entsch B., Ballou D.P., and Massey V.. 1994. Changes in the catalytic properties of p-hydroxybenzoate hydroxylase caused by the mutation Asn300Asp. Biochemistry. 33:1545–1554. 10.1021/bi00172a035 [DOI] [PubMed] [Google Scholar]
- Pedley A.M., and Benkovic S.J.. 2017. A New View into the Regulation of Purine Metabolism: The Purinosome. Trends Biochem. Sci. 42:141–154. 10.1016/j.tibs.2016.09.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poon W.W., Do T.Q., Marbois B.N., and Clarke C.F.. 1997. Sensitivity to treatment with polyunsaturated fatty acids is a general characteristic of the ubiquinone-deficient yeast coq mutants. Mol. Aspects Med. 18:S121–S127. 10.1016/S0098-2997(97)00004-6 [DOI] [PubMed] [Google Scholar]
- Poon W.W., Barkovich R.J., Hsu A.Y., Frankel A., Lee P.T., Shepherd J.N., Myles D.C., and Clarke C.F.. 1999. Yeast and rat Coq3 and Escherichia coli UbiG polypeptides catalyze both O-methyltransferase steps in coenzyme Q biosynthesis. J. Biol. Chem. 274:21665–21672. 10.1074/jbc.274.31.21665 [DOI] [PubMed] [Google Scholar]
- Rabl R., Soubannier V., Scholz R., Vogel F., Mendl N., Vasiljev-Neumeyer A., Körner C., Jagasia R., Keil T., Baumeister W., et al. 2009. Formation of cristae and crista junctions in mitochondria depends on antagonism between Fcj1 and Su e/g. J. Cell Biol. 185:1047–1063. 10.1083/jcb.200811099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rea S.L., Graham B.H., Nakamaru-Ogiso E., Kar A., and Falk M.J.. 2010. Bacteria, yeast, worms, and flies: exploiting simple model organisms to investigate human mitochondrial diseases. Dev. Disabil. Res. Rev. 16:200–218. 10.1002/ddrr.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reidenbach A.G., Kemmerer Z.A., Aydin D., Jochem A., McDevitt M.T., Hutchins P.D., Stark J.L., Stefely J.A., Reddy T., Hebert A.S., et al. 2018. Conserved lipid and small-molecule modulation of COQ8 reveals regulation of the ancient kinase-like UbiB family. Cell Chem. Biol. 25:154–165.e11. 10.1016/j.chembiol.2017.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-Molina J.B., Tseng S.C., Simonett S.P., Taunton J., and Ansari A.Z.. 2016. Engineered Covalent Inactivation of TFIIH-Kinase Reveals an Elongation Checkpoint and Results in Widespread mRNA Stabilization. Mol. Cell. 63:433–444. 10.1016/j.molcel.2016.06.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheff M.A., and Thorn K.S.. 2004. Optimized cassettes for fluorescent protein tagging in Saccharomyces cerevisiae. Yeast. 21:661–670. 10.1002/yea.1130 [DOI] [PubMed] [Google Scholar]
- Stefely J.A., and Pagliarini D.J.. 2017. Biochemistry of Mitochondrial Coenzyme Q Biosynthesis. Trends Biochem. Sci. 42:824–843. 10.1016/j.tibs.2017.06.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefely J.A., Reidenbach A.G., Ulbrich A., Oruganty K., Floyd B.J., Jochem A., Saunders J.M., Johnson I.E., Minogue C.E., Wrobel R.L., et al. 2015. Mitochondrial ADCK3 employs an atypical protein kinase-like fold to enable coenzyme Q biosynthesis. Mol. Cell. 57:83–94. 10.1016/j.molcel.2014.11.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefely J.A., Kwiecien N.W., Freiberger E.C., Richards A.L., Jochem A., Rush M.J.P., Ulbrich A., Robinson K.P., Hutchins P.D., Veling M.T., et al. 2016a Mitochondrial protein functions elucidated by multi-omic mass spectrometry profiling. Nat. Biotechnol. 34:1191–1197. 10.1038/nbt.3683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefely J.A., Licitra F., Laredj L., Reidenbach A.G., Kemmerer Z.A., Grangeray A., Jaeg-Ehret T., Minogue C.E., Ulbrich A., Hutchins P.D., et al. 2016b Cerebellar Ataxia and Coenzyme Q Deficiency through Loss of Unorthodox Kinase Activity. Mol. Cell. 63:608–620. 10.1016/j.molcel.2016.06.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storici F., and Resnick M.A.. 2006. The delitto perfetto approach to in vivo site-directed mutagenesis and chromosome rearrangements with synthetic oligonucleotides in yeast. Methods Enzymol. 409:329–345. 10.1016/S0076-6879(05)09019-1 [DOI] [PubMed] [Google Scholar]
- Stuckey S., and Storici F.. 2013. Gene knockouts, in vivo site-directed mutagenesis and other modifications using the delitto perfetto system in Saccharomyces cerevisiae. Methods Enzymol. 533:103–131. 10.1016/B978-0-12-420067-8.00008-8 [DOI] [PubMed] [Google Scholar]
- Stuckey S., Mukherjee K., and Storici F.. 2011. In vivo site-specific mutagenesis and gene collage using the delitto perfetto system in yeast Saccharomyces cerevisiae. Methods Mol. Biol. 745:173–191. 10.1007/978-1-61779-129-1_11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swayne T.C., Boldogh I.R., and Pon L.A.. 2009. Imaging of the cytoskeleton and mitochondria in fixed budding yeast cells. Methods Mol. Biol. 586:171–184. 10.1007/978-1-60761-376-3_9 [DOI] [PubMed] [Google Scholar]
- Sweetlove L.J., and Fernie A.R.. 2018. The role of dynamic enzyme assemblies and substrate channelling in metabolic regulation. Nat. Commun. 9:2136 10.1038/s41467-018-04543-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thelin A., Schedin S., and Dallner G.. 1992. Half-life of ubiquinone-9 in rat tissues. FEBS Lett. 313:118–120. 10.1016/0014-5793(92)81425-L [DOI] [PubMed] [Google Scholar]
- Tran U.C., and Clarke C.F.. 2007. Endogenous synthesis of coenzyme Q in eukaryotes. Mitochondrion. 7(Suppl):S62–S71. 10.1016/j.mito.2007.03.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzagoloff A., and Dieckmann C.L.. 1990. PET genes of Saccharomyces cerevisiae. Microbiol. Rev. 54:211–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzagoloff A., Akai A., and Needleman R.B.. 1975. Assembly of the mitochondrial membrane system. Characterization of nuclear mutants of Saccharomyces cerevisiae with defects in mitochondrial ATPase and respiratory enzymes. J. Biol. Chem. 250:8228–8235. [PubMed] [Google Scholar]
- Vogel F., Bornhövd C., Neupert W., and Reichert A.S.. 2006. Dynamic subcompartmentalization of the mitochondrial inner membrane. J. Cell Biol. 175:237–247. 10.1083/jcb.200605138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von der Malsburg K., Müller J.M., Bohnert M., Oeljeklaus S., Kwiatkowska P., Becker T., Loniewska-Lwowska A., Wiese S., Rao S., Milenkovic D., et al. 2011. Dual role of mitofilin in mitochondrial membrane organization and protein biogenesis. Dev. Cell. 21:694–707. 10.1016/j.devcel.2011.08.026 [DOI] [PubMed] [Google Scholar]
- Webb B.A., Dosey A.M., Wittmann T., Kollman J.M., and Barber D.L.. 2017. The glycolytic enzyme phosphofructokinase-1 assembles into filaments. J. Cell Biol. 216:2305–2313. 10.1083/jcb.201701084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westermann B., and Neupert W.. 2000. Mitochondria-targeted green fluorescent proteins: convenient tools for the study of organelle biogenesis in Saccharomyces cerevisiae. Yeast. 16:1421–1427. [DOI] [PubMed] [Google Scholar]
- Wittig I., Braun H.P., and Schägger H.. 2006. Blue native PAGE. Nat. Protoc. 1:418–428. 10.1038/nprot.2006.62 [DOI] [PubMed] [Google Scholar]
- Wurm C.A., and Jakobs S.. 2006. Differential protein distributions define two sub-compartments of the mitochondrial inner membrane in yeast. FEBS Lett. 580:5628–5634. 10.1016/j.febslet.2006.09.012 [DOI] [PubMed] [Google Scholar]
- Xie L.X., Ozeir M., Tang J.Y., Chen J.Y., Jaquinod S.K., Fontecave M., Clarke C.F., and Pierrel F.. 2012. Overexpression of the Coq8 kinase in Saccharomyces cerevisiae coq null mutants allows for accumulation of diagnostic intermediates of the coenzyme Q6 biosynthetic pathway. J. Biol. Chem. 287:23571–23581. 10.1074/jbc.M112.360354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Turunen M., and Appelkvist E.L.. 1996. Restricted uptake of dietary coenzyme Q is in contrast to the unrestricted uptake of alpha-tocopherol into rat organs and cells. J. Nutr. 126:2089–2097. 10.1093/jn/126.9.2089 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.