Free fatty acids are toxic, but are stored in lipid droplets (LDs) that bud from the ER. Datta et al. show that cerebellar ataxia–linked protein Snx14 localizes to ER–droplet contacts and promotes LD growth. This implicates Snx14 in LD homeostasis which, when perturbed, contributes to disease.
Abstract
Lipid droplets (LDs) are nutrient reservoirs used by cells to maintain homeostasis. Nascent droplets form on the endoplasmic reticulum (ER) and grow following an influx of exogenous fatty acids (FAs). The budding of LDs requires extensive ER–LD crosstalk, but how this is regulated remains poorly understood. Here, we show that sorting nexin protein Snx14, an ER-resident protein associated with the cerebellar ataxia SCAR20, localizes to ER–LD contacts following FA treatment, where it promotes LD maturation. Using proximity-based APEX technology and topological dissection, we show that Snx14 accumulates specifically at ER–LD contacts independently of Seipin, where it remains ER-anchored and binds LDs in trans. SNX14KO cells exhibit perturbed LD morphology, whereas Snx14 overexpression promotes LD biogenesis and extends ER–LD contacts. Multi–time point imaging reveals that Snx14 is recruited to ER microdomains containing the fatty acyl-CoA ligase ACSL3, where nascent LDs bud. We propose that Snx14 is a novel marker for ER–LD contacts and regulates FA-stimulated LD growth.
Introduction
Fatty acids (FAs) are fundamental cellular metabolites used for membrane biosynthesis, cell signaling, and energy via their oxidation. Excess FAs are stored as triacylglyceride (TAG) housed within cytoplasmic organelles called lipid droplets (LDs). Defects in FA processing or the inability to store excess FAs in LDs lead to cellular lipotoxicity and are associated with metabolic syndromes such as diabetes, obesity, cardiovascular disease, and numerous neurological diseases (Listenberger et al., 2003).
LDs bud from the surface of the ER and receive TAG from the ER (Guo et al., 2009; Fujimoto and Parton, 2011). Although the mechanisms of LD biogenesis remain debated, it is generally accepted that neutral lipids accumulate at distinct microdomains within the ER membrane bilayer, leading to the formation of a lipid “lens” between the monolayer leaflets that gradually grows as neutral lipids coalesce (Athenstaedt and Daum, 2006). In fasted mammalian cells, ER microdomains containing nascent LDs designated preLDs have been observed and are marked by the enzyme acyl-CoA synthetase long chain family member 3 (ACSL3; Kassan et al., 2013). These small preLDs can grow in response to an influx of FAs such as oleic acid (OA), which is esterified by ACSL3 and combined with DAG via DAG O-acyltransferase (DGAT) enzymes into TAG. In Caenorhabditis elegans, the ER-localized fatty acid transport protein 1 (FATP1) interacts with the LD-localized DGAT2 to promote OA incorporation into TAG during LD growth (Xu et al., 2012). Furthermore, numerous studies implicate the protein Seipin in LD homeostasis, and Seipin localizes to ER–LD contacts in yeast and mammalian cells (Szymanski et al., 2007; Salo et al., 2016). Thus, LD homeostasis and growth requires extensive ER–LD interorganelle crosstalk, which ultimately governs the flux of lipids from the ER into the growing LD through either direct ER–LD contacts or recruitment of LDs to the ER surface (Wilfling et al., 2014). How this ER–LD crosstalk is coordinated remains poorly understood, and ER–LD contacts themselves remain poorly characterized, as they are difficult to observe by conventional microscopy.
Recent studies in yeast reveal that LD biogenesis can also be spatially restricted to distinct subregions of the ER surface. When yeast are deprived of a carbon source, LDs bud and accumulate on the surface of the nucleus (nuclear ER) which is in close apposition to the vacuole, a region known as the nuclear ER–vacuole junction (NVJ). NVJ-associated LD clustering is regulated by Mdm1, an ER-resident protein that interacts with the ACSL3 homologue Faa1 and promotes LD biogenesis (Hariri et al., 2018). Although mammalian cells lack NVJ contacts, Mdm1 is a member of the sorting nexin (Snx) protein family and is conserved in humans as four orthologues: Snx13, Snx14, Snx19, and Snx25. Snx14 loss-of-function mutations are associated with a distinct cerebellar ataxia termed spinocerebellar ataxia autosomal recessive 20 (SCAR20; OMIM 616354; Thomas et al., 2014; Shukla et al., 2017). This disease to date has been reported in 45 individuals from 24 families and is characterized by cerebellar hypertrophy, intellectual disability, and defects in speech. Recent studies reveal that human Snx14 localizes to the ER network, and its loss causes defects in neutral lipid homeostasis, although its function in lipid metabolism remains unclear (Bryant et al., 2018).
Here, we characterize Snx14 and mechanistically dissect how it regulates ER–LD crosstalk and LD maturation. Using proximity-based ascorbate peroxidase (APEX) technology combined with multi–time point imaging and biochemistry, we find that Snx14 localizes to ER microdomains containing preLDs following OA treatment, where it promotes LD maturation at ER–LD contacts.
Results
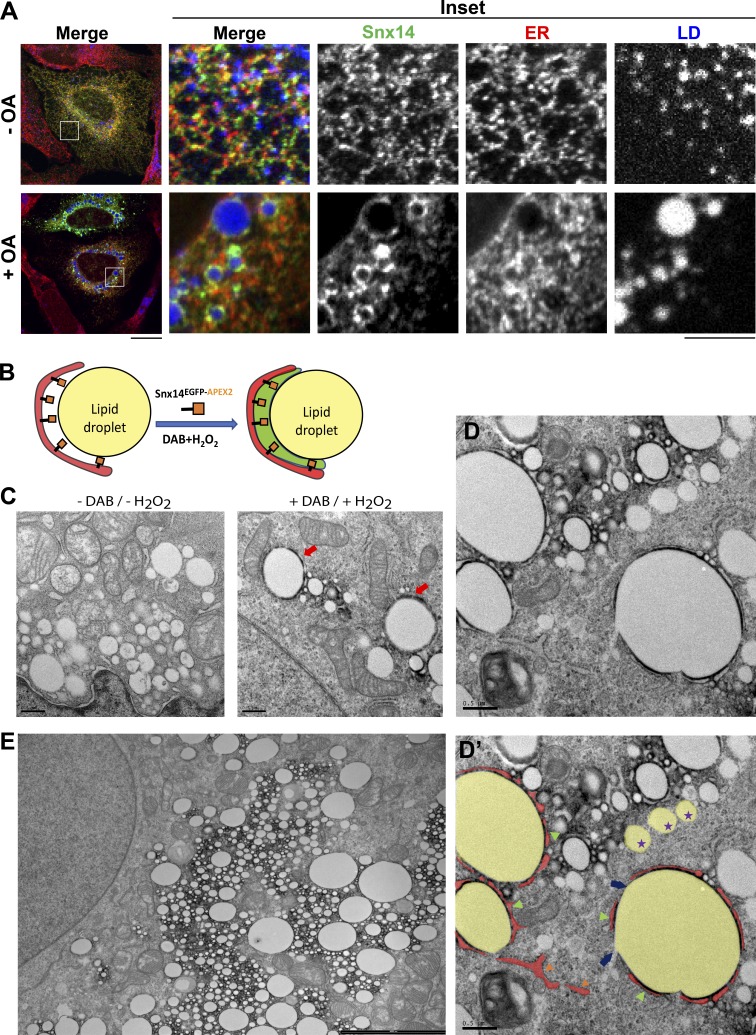
Snx14 localizes at ER–LD contacts after OA treatment
Previously, we demonstrated that Snx14 is an ER-resident protein whose loss in HEK293 cells perturbs ER-associated neutral lipid metabolism. Furthermore, the addition of OA, which is esterified in the ER before its incorporation into LDs as TAG, induced the accumulation of Snx14 near LDs (Bryant et al., 2018). To better understand the cellular function of Snx14, we investigated how its subcellular localization changed in response to OA treatment by culturing U2OS cells overnight with BSA-conjugated OA (denoted herein simply as OA treatment). In the absence of OA, confocal imaging of immunofluorescently labeled U2OS cells transiently expressing an untagged Snx14 revealed it was uniformly distributed across the ER. After 16 h of OA treatment to induce LD expansion, Snx14 redistributed and accumulated within a subdomain of the ER that formed ring-like structures surrounding LDs (Fig. 1 A). We observed the same result when we examined a cell line stably expressing a C-terminally EGFP-tagged Snx14 construct (Fig. S1 A). In both experiments, whether Snx14 remained ER associated or transferred to the LD surface following OA treatment could not be distinguished at this resolution. Nonetheless, as a consequence of this redistribution, quantification of the Snx14-ER colocalization between the two conditions revealed a significant loss of ER colocalization following OA treatment, changing from a Pearson’s coefficient (r) of 0.41 ± 0.07 in no OA to –0.06 ± 0.01 (Fig. S1 A).
Figure 1.
Snx14 localizes at ER–LD contacts after OA treatment. (A) Localization of Snx14 in U2OS cells in the absence and presence of OA, respectively. CoIF staining of the cells transiently expressing an untagged Snx14 were performed with α-Snx14 antibody (green), and α-HSP90B1 (ER marker, red) antibodies and LDs were stained with MDH (blue) and imaged by confocal microscopy. Inset of OA-treated cells displays Snx14 accumulating around LDs. Scale bar = 20 µm. Scale bar of insets = 5 µm. (B) Schematic diagram showing Snx14EGFP-APEX2 fusion at ER–LD contacts. (C) TEM micrographs showing untreated and DAB + H2O2–treated cells following OA treatment. The dark precipitate with DAB + H2O2 treatment indicates presence of Snx14EGFP-APEX2. Scale bar = 0.5 µm. (D) TEM micrograph showing SNX14EGFP-APEX2-expressing cell with DAB precipitate at ER–LD contacts. (D′) Image from D showing pseudocolored ER (red), LD (yellow), and green arrows pointing at DAB precipitate specifically at the junction of ER and LD. The blue arrows indicate a region of LD surface that lacks ER wrapping and also lacks DAB precipitate. Purple stars indicate LDs without any detectable ER association (and no DAB precipitate). The orange arrow indicates an ER membrane itself. Scale bar = 0.5 µm. (E) Lower-magnification TEM micrographs of a Snx14EGFP-APEX2-expressing cell following OA treatment and stained with DAB, showing precipitate surrounding various LDs entangled with the ER network. Scale bar = 0.5 µm.
To examine the accumulation of Snx14 near LDs at higher spatial resolution, we used the genetically encoded proximity-based labeling technology APEX (APEX2), which allows high-resolution subcellular localization of a protein of interest via thin-section transmission electron microscopy (TEM; Martell et al., 2012; Lam et al., 2015). We generated U2OS cells stably expressing Snx14EGFP-APEX2 and treated them with OA to stimulate LD production. These cells were then imaged in bright-field microscopy to confirm the function of the APEX2 enzyme tag. When treated with DAB and H2O2, but not in their absence, APEX2 catalyzes the formation of an electron-dense precipitate that is deposited within ∼10 nm of the enzyme (Figs. 1 B and S1 B).
Next, cells were treated with OA and then exposed or not to DAB + H2O2 before processing for thin-section TEM. Both cell samples contained many LDs, but the DAB + H2O2–treated cells contained unambiguous electron dense DAB precipitates on the surface of distinct LDs (Fig. 1, C–E). High-magnification imaging revealed that the DAB stain accumulated specifically in spaces between the LD surface and ER tubular structures that were wrapped around the LD (Fig. 1, D and D′, green arrows). We also noted that regions of the LD surface not wrapped by ER contained no DAB stain (Fig. 1, D and D′, blue arrows). Clusters of small LDs also exhibited DAB stain around the LD surfaces, and ER tubular structures could be observed closely associated with these small LDs (Figs. 1 E and S1 D). Notably, there was no visible DAB stain associated with LDs that were not associated with ER (Fig. 1, D and D′, purple stars). We did not detect any obvious DAB stain at other interorganelle contact sites such as ER–mitochondria or mitochondria–LD contacts (Fig. S1 C, blue diamonds). Likewise, no obvious DAB stain was associated with the ER membrane network itself (Fig. 1, D and D′, orange arrows). These observations suggested that Snx14 may associate with the LD surface specifically at sites of ER–LD contact.
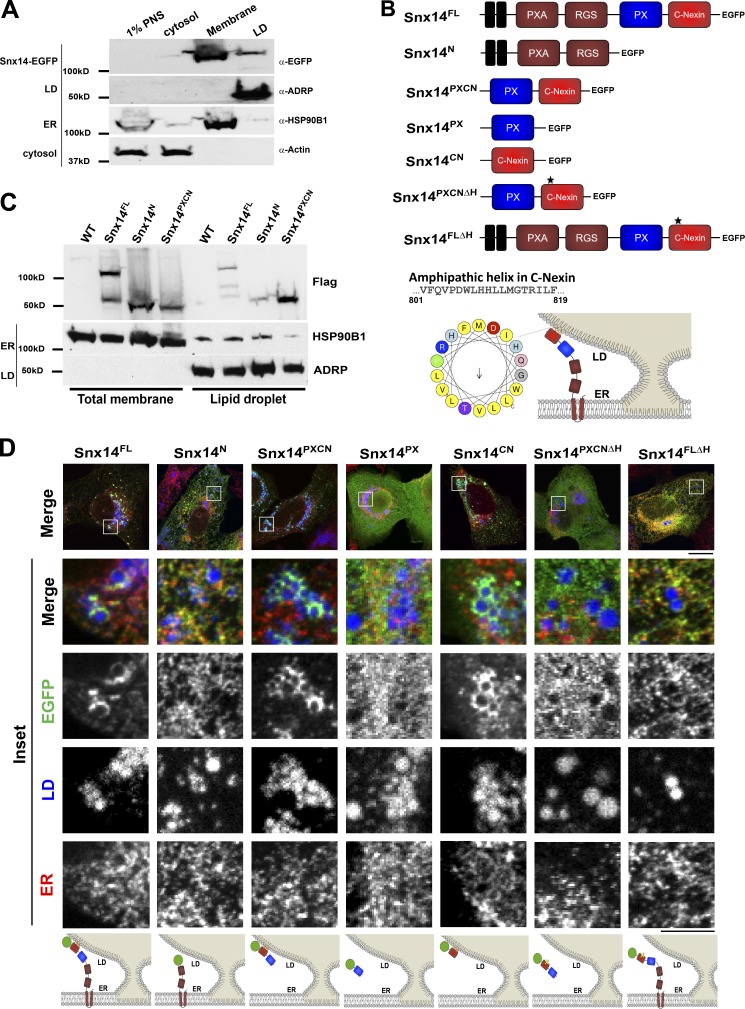
Snx14 interacts with LDs in trans from the ER surface
The enrichment of Snx14 specifically at ER–LD contacts following OA treatment suggested at least two possible models: (1) Snx14 translocates from the ER to the LD surface following OA treatment, or (2) Snx14 is retained in the ER and associates with LDs in trans at ER–LD contact sites. To test these models, we next conducted biochemical fractionation of OA-treated U2OS cells stably expressing Snx14EGFP to generate ER, LD, and cytoplasmic fractions and probed for Snx14 in each fraction using an anti-EGFP antibody. Snx14EGFP was primarily enriched in the ER membrane fraction, with negligible signal detected in the LD float fraction, suggesting that Snx14 remained associated with the ER following its association with the LD surface (Fig. 2 A). To confirm that endogenous Snx14 also remained in the ER following OA treatment, we also fractionated WT U2OS cells and probed for Snx14. We detected endogenous Snx14 in the membrane fraction similar to the Snx14EGFP stably expressing cell line (Fig. S2 A).
Figure 2.
Snx14 is topologically anchored in the ER and interacts with LDs in trans. (A) LD flotation assay of OA-treated U2OS cells expressing Snx14EGFP. Lanes indicate postnuclear supernatant (PNS), cytosol, total membrane, and LD float fractions. (B) Schematic diagram of Snx14 fragment constructs tagged with EGFP. Snx14FL depicts the full-length human Snx14. Snx14N is the N-terminal fragment from the start that includes PXA and RGS domains. Snx14PXCN includes the PX domain and C-Nexin domains. Snx14PX consists of PX and Snx14CN represents C-Nexin domain. An AH in the C-Nexin domain is identified as depicted in the schematic diagram. Snx14PXCNΔH indicates the PX and C-Nexin domain from which the AH is deleted. Snx14FLΔH depicts the full-length Snx14 with AH deletion. (C) Western blot showing distribution of Snx14FL, Snx14N, and Snx14PXCN tagged with 3XFlag among total membrane and LD fractions following OA treatment. (D) U2OS cells transfected with Snx14FL, Snx14N, Snx14PXCN, Snx14PX, Snx14CN, Snx14PXCNΔH, and Snx14FLΔH, respectively, and treated with OA for 16 h. CoIF staining with α−EGFP (green) and α-HSP90B1 (ER marker, red) and LDs stained with MDH (blue) and imaged by confocal microscopy. Scale bar = 20 µm. Inset scale bar = 5 µm. Cartoons represent the localization of the respective Snx14 fragments with respect to ER and LD.
To topologically dissect how Snx14 remained anchored at the ER but accumulated at ER–LD contact sites, we generated cell lines stably expressing 3XFlag-tagged fragments of Snx14: full length Snx14 (Snx14FL) or Snx14 fragments encoding its N-terminal transmembrane, PXA and RGS domains (Snx14N), or C-terminal PX and C-Nexin domains (Snx14PXCN). Whereas the Snx14N fragment contains predicted transmembrane helices, the Snx14PXCN fragment is predicted to be soluble and face the cytoplasm. To quantitatively assess the subcellular localization of these Snx14 constructs, we biochemically isolated membrane, LD, and cytosolic fractions following ∼16-h OA treatment and immunoblotted for each construct with anti-Flag antibody. Indeed, both Snx14FL and Snx14N were present primarily in the ER membrane fraction following overnight treatment with OA (Figs. 2 C and S2 B). However, Snx14PXCN was significantly associated with the LD fraction, suggesting it was sufficient to interact with LDs (Figs. 2 C and S2 B).
Next we evaluated the subcellular distribution of these constructs using EGFP-tagged versions and fluorescence confocal microscopy (Fig. 2 B). As previously observed, Snx14FL localized to ER–LD contacts following OA addition (Figs. 2 D and S2 C). In contrast, Snx14N distributed throughout the ER network following OA treatment, suggesting that removal of the C-terminal region attenuated LD association (Fig. 2 D). Consistent with this, Snx14PXCN accumulated around the surfaces of LDs, similar to its biochemical fractionation (Figs. 2 D and S2 C). We conclude that Snx14N remains anchored at the ER following OA treatment, but Snx14PXCN exhibits affinity for LDs and is sufficient for LD targeting.
To mechanistically dissect how the Snx14 C-terminal region interacts with LDs, we generated fragments encoding only the PX domain (Snx14PX) and C-Nexin domain (Snx14CN) and monitored their subcellular localizations. Interestingly, Snx14CN was sufficient to bind LD surfaces, whereas Snx14PX was cytosolic (Figs. 2 D and S2 C). Since hydrophobic and amphipathic helices (AHs) have been shown to mediate binding to LD surfaces in previous studies (Prevost et al., 2018), we analyzed the C-Nexin region and identified one putative AH region (Fig. 2 B). Strikingly, deletion of this AH region from both the C-terminal fragment (Snx14PXCNΔH) and full-length Snx14 (Snx14FLΔH) attenuated LD association following OA treatment (Figs. 2 D and S2 C). Taking these data collectively, we conclude that Snx14 localizes to ER–LD contacts by remaining anchored in the ER with its N-terminal region and binding the LD surface with a C-terminal AH region present in its C-Nexin domain.
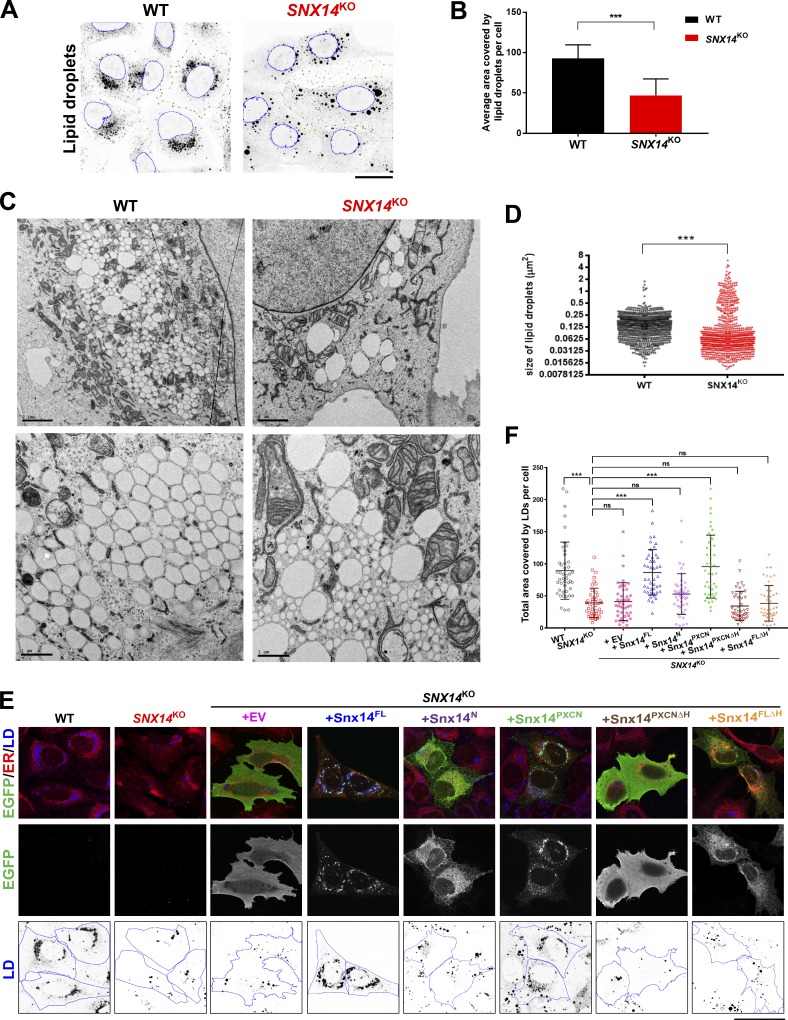
Loss of SNX14 alters LD morphology following OA treatment
Since ER–LD contacts have been proposed to regulate the flux of lipids into LDs during their expansion, we next evaluated whether loss of Snx14 would disrupt proper LD maturation following OA treatment. We generated SNX14 homozygous knockout (SNX14KO) U2OS cells using CRISPR/Cas9 technology with single guide RNA that targeted the first exon of the SNX14 ORF, resulting in deletion of several bases and an inserting base “A,” which led to the formation of a stop codon after 25 amino acids (Fig. S3 A). We confirmed that this CRISPR indel resulted in loss of Snx14 polypeptide by Western blot (Fig. S3 B). To determine whether loss of SNX14 affected OA-induced LD maturation, we treated WT and SNX14KO cells for ∼16 h with OA to induce LD expansion and imaged LDs by staining them with the fluorescent dye monodansylpentane (MDH). As expected, WT cells exhibited many LDs that were densely clustered near the nucleus. In contrast, SNX14KO cells displayed heterogeneous LDs of varying size and subcellular distribution (Fig. 3 A). Owing to the limited resolution of individual LDs by light microscopy, we calculated the total LD area per cell, which is indicative of both LD number and size. Indeed, quantification revealed that SNX14KO cells contained significantly less total LD area per cell, suggesting a defect in some aspect of LD homeostasis (Fig. 3 B).
Figure 3.
Loss of Snx14 perturbs LD size and morphology but does not change neutral lipid levels. (A) Confocal micrographs of WT and SNX14KO cells treated with OA overnight. LDs visualized by MDH (black) and nucleus stained with Syto 85 orange fluorescent stain (blue outline). Images were processed so that LDs were converted to grayscale and inverted. Scale bar = 25 µm. (B) Quantification of average area covered by LDs per cell of representative images from A. Total LD area was derived from more than five fields of view, each consisting of approximately five cells or more of two different sets of experiments (total no. of cells >75; ***, P < 0.0001 unpaired t test with α = 0.05). (C) TEM micrographs of WT and SNX14KO cells treated with OA overnight to visualize LD distribution and morphology. The top panels are lower magnification (scale bar = 2 µm). The bottom panels are higher magnification (scale bar = 1 µm). (D) Scatter dot plot of cross-sectional areas of LDs in WT and SNX14KO cells as in C. Total LDs = 896; ***, P < 0.0001, Kolmogorov–Smirnov D test with α = 0.05. (E) Rescue of LD morphology in SNX14KO cells by readdition of empty vector (EV), Snx14FL, Snx14N, Snx14PXCN, Snx14PXCNΔH, and Snx14FLΔH, respectively, all tagged with EGFP. Cells were coIF stained with α-EGFP (green), α-HSP90B1 (ER, red), and LDs stained with MDH (blue) and imaged with confocal microscope. Scale bar = 50 µm. (F) Area covered by LDs in each cell from E analyzed and plotted. Total no. of cells quantified are 45 from two different sets of experiments (***, P < 0.0001, one-way ANOVA with α = 0.05). Line bars indicate mean ± SD.
To evaluate LD morphologies with better resolution, we next conducted thin-section TEM imaging of OA-treated cells. Similar to fluorescence imaging, we observed many LDs in WT cells of various sizes (Fig. 3 C). Quantification of the cross-sectional areas of ∼900 LDs revealed a broad distribution of LD sizes, although ∼55% of all LDs in WT cells ranged in size between 0.125 and 0.4 µm2 (Fig. 3, C and D; and Fig. S3, C and D′). In contrast, SNX14KO cells contained fewer and more sparsely distributed LDs that were also more heterogeneous in size and exhibited a more bimodal size distribution (Fig. 3, C and D; and Fig. S3 C). Indeed, only ∼10% of LDs in SNX14KO cells were between 0.125 and 0.4 µm2 in size (Fig. S3, C and D′). In fact, most LDs in SNX14KO cells were very small (∼78% <0.125 µm2 compared with ∼44% in WT cells), although the proportion of large LDs (>0.4 µm2) also increased in SNX14KO cells (∼14% compared with only ∼2% in WT; Fig. S3, C, D, and D″). We conclude that loss of SNX14 causes alterations in LD morphology following OA treatment, with a substantial increase in the proportion of very small and large LDs.
To confirm that the changes in LD morphologies were due to loss of Snx14, we reintroduced either full-length Snx14 or Snx14 fragments back into SNX14KO cells and monitored whether these constructs could rescue LD morphology. Whereas Snx14FL localized to ER–LD contacts and could restore the LD morphology defects, Snx14N remained in the ER network and did not rescue, suggesting that LD targeting is required for Snx14 function (Fig. 3, E and F). Similarly, a full-length version of Snx14 which lacked the AH region present in the C-Nexin domain also failed to localize to ER–LD contacts and could not functionally rescue LD morphology (Fig. 3, E and F). This indicates that Snx14’s localization to ER–LD contacts via this AH region is required for Snx14 function. Surprisingly, expression of Snx14PXCN was sufficient to restore LD morphology. However, Snx14PXCN required its LD targeting region to rescue, as a mutant fragment not containing the AH (Snx14PXCNΔH) could not rescue LD morphology (Fig. 3, E and F). Altogether, these results indicate that loss of Snx14 perturbs LD morphology, and Snx14 requires LD targeting via its C-terminal AH region for function.
Next, we examined whether loss of SNX14 altered the incorporation of OA into TAG. WT and SNX14KO cells were fed OA and collected at 2-h time intervals, and their lipids were extracted and evaluated by thin layer chromatography (TLC). As expected, TLC revealed a gradual increase in TAG and corresponding decrease in free FAs (FFAs) following OA treatment (Fig. S3 E). Surprisingly, loss of SNX14 did not significantly affect TAG levels at any time point (Fig. S3, E and F). Similarly, we did not detect significant alterations in cholesterol or cholesterol-ester levels in SNX14KO cells, collectively suggesting that loss of Snx14 primarily affects LD morphology (Fig. S3, F′ and F″). Given the clear LD morphological changes, it is possible that some neutral lipids are not properly transported from the ER into LDs during their formation. Alternatively, the increase in very small and very large LDs in SNX14KO cells may mean the LD population has become more bimodal in size but retained similar total lipid levels.
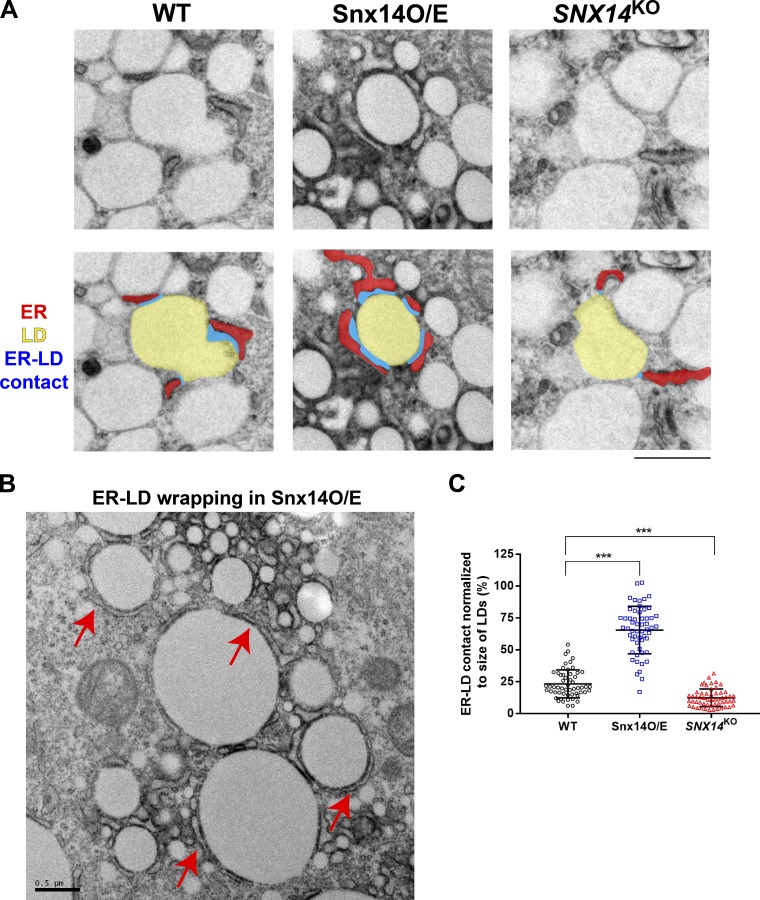
Snx14 expression modulates ER–LD contacts
LDs bud from the ER surface and establish and/or maintain contact with the ER to ensure proper LD growth. Since Snx14 localized at the interface between the ER and LDs, and its loss affected LD morphology, we next investigated how modulating Snx14 expression level would affect ER–LD contacts. We imaged WT, Snx14EGFP-APEX2-overexpressing (O/E), and SNX14KO cells treated with OA overnight by thin-section TEM. Strikingly, we observed significantly enhanced ER wrapping across the surfaces of LDs in Snx14 O/E samples (Fig. 4, A–C). In contrast, SNX14KO cells displayed mildly reduced ER–LD contacts that could be observed by TEM (Fig. 4, A and C). Indeed, after plotting the distribution of ER–LD contacts normalized to LD size, we observe a significant increase in the ER–LD contact size in Snx14 O/E cells, and a significant decrease in SNX14KO cells compared with the WTs (Fig. 4 C). These observations are consistent with a model where Snx14 localizes to ER–LD contacts and tethers these organelles together, so that increasing the Snx14 tether abundance enhances their contact.
Figure 4.
Snx14 expression modulates ER–LD contacts following OA addition. (A) TEM images showing the extent of ER–LD contacts in WT cells. Control WT cells, those overexpressing Snx14EGFP-APEX2, and SNX14KO cells were all treated with OA overnight. LDs are colored in yellow, ER in red, and the contact between ER–LD is depicted in blue. (B) ER–LD wrapping indicated by red arrows in Snx14EGFP-APEX2 cells. (C) Quantification of the length of ER–LD contact normalized to circumference of the LDs in the form of box plot in WT, Snx14EGFP-APEX2, and SNX14KO cells (n = 55; ***, P < 0.0001, Welch’s t test with α = 0.05).
Snx14 functions independently of Seipin at ER–LD contacts
The enrichment of Snx14 at ER–LD contacts, as well as the defects in LD morphology observed in SNX14KO cells, appeared reminiscent of recent studies on the protein Seipin. Seipin has been shown to localize to ER–LD contacts and is required for the proper maturation of nascent LDs into mature LDs following OA treatment (Szymanski et al., 2007; Fei et al., 2008; Salo et al., 2016; Wang et al., 2016). Furthermore, loss of Seipin leads to defects in LD morphology in yeast, insect, and mammalian cells. However, overall TAG levels are largely unaltered in SEIPINKO cells, similar to loss of Snx14. Since Seipin has also been proposed to be an ER–LD tether, we next investigated the functional relationship between Snx14 and Seipin.
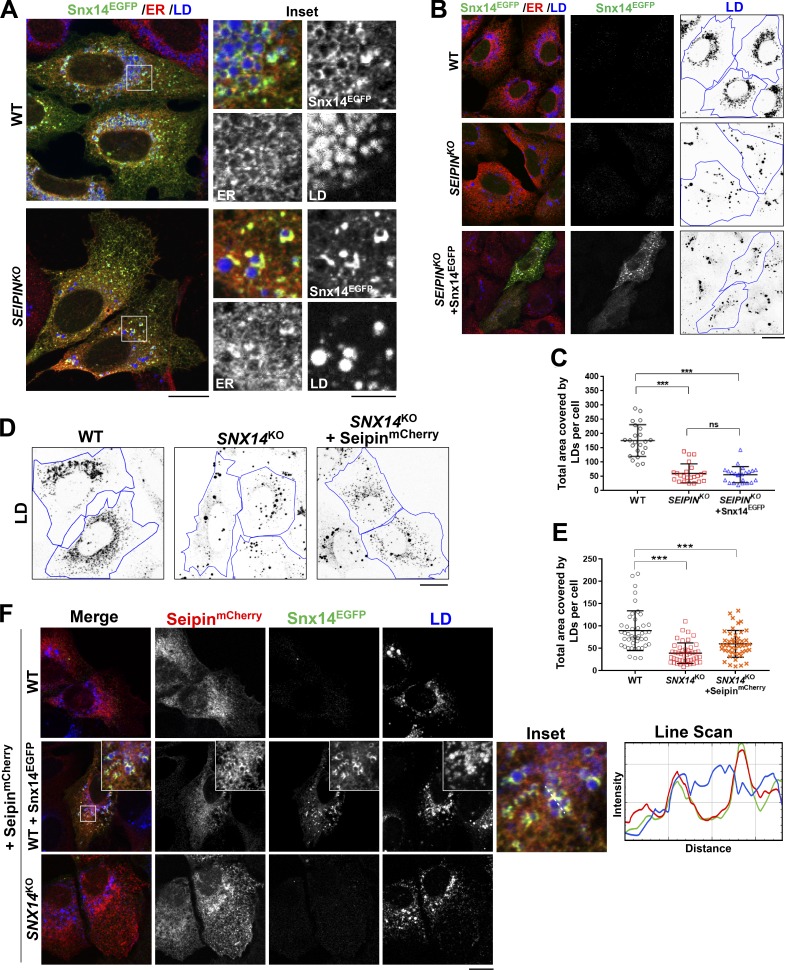
First, we evaluated whether Seipin is required for Snx14 to localize to ER–LD contacts. We ectopically expressed Snx14EGFP in SUM159 SEIPINKO cells, and observed that Snx14EGFP could still target to LDs following OA treatment (Figs. 5 A and S4 A). During these experiments, we noted that ectopic expression of Snx14EGFP was unable to rescue the LD morphology defects present in SEIPINKO cells (Fig. 5, B and C). To further investigate this functional relationship, we ectopically expressed mCherry-tagged Seipin in U2OS SNX14KO cells. Similarly, ectopic expression of SeipinmCherry was unable to rescue the LD morphology defects observed in SNX14KO cells, although LD area per cell was mildly different compared with SNX14KO alone (Fig. 5, D and E). We also depleted both proteins together but did not observe a cumulative effect on LD morphology when we siRNA depleted Snx14 in SEIPINKO cells (Fig. S4, B and C).
Figure 5.
Snx14 functions independently of ER–LD protein, Seipin. (A) Confocal micrographs showing localization of Snx14 in WT and SEIPINKO SUM159 cells. The cells were transfected with Snx14EGFP and treated with OA overnight. Labels were Snx14EGFP (α-EGFP), ER (α-HSP90B1), and LDs (MDH). Scale bar = 20 µm. Inset scale bar = 5 µm. (B) Test of rescue of LD morphology in SEIPINKO examined by ectopic expression of Snx14EGFP into SEIPINKO. Labels were Snx14EGFP (α-EGFP), ER (α-HSP90B1), and LDs (MDH stained, converted to grayscale, and inverted by ImageJ). Scale bar = 20 µm. (C) Area covered by LDs in each cell from B analyzed and plotted. Total no. of cells quantified is 23 from two different sets of experiments (***, P < 0.0001, one-way ANOVA with α = 0.05). Line bars indicate mean ± SD. (D) Test of rescue of LD morphology in SNX14KO examined by readdition of SeipinmCherry and comparing to that of WT and SNX14KO. Blue line depicts cell boundary. Black speckles represent LDs that are MDH-stained, converted to grayscale, and inverted by ImageJ. Scale bar = 20 µm. (E) Area covered by LDs in each cell from D analyzed and plotted. Total no. of cells quantified is 45 from two different sets of experiments (***, P < 0.0001, one-way ANOVA with α = 0.05). Line bars indicate mean ± SD. (F) Confocal micrographs of U2OS cells to examine localization of SeipinmCherry in WT cells, those transfected with Snx14EGFP, and in SNX14KO cells. Cells were transfected with Seipin-mCherry and treated with OA overnight. Labels were Snx14EGFP (α-EGFP), Seipin-mCherry (α-mCherry), and LDs (MDH). Scale bar = 20 µm.
Finally, we monitored the subcellular localization of both Snx14EGFP and SeipinmCherry when coexpressed. Overexpression of SeipinmCherry caused it to localize evenly across the ER network in WT cells, which was different than the punctate distribution shown near LDs in previous endogenous labeling studies (Salo et al., 2016; Wang et al., 2016). The distribution of SeipinmCherry was not altered in SNX14KO cells (Fig. 5 F). However, we observed an enrichment of SeipinmCherry at Snx14EGFP-positive ER–LD contacts when we coexpressed the two proteins together (Fig. 5 F). Collectively, we conclude that Seipin and Snx14 both accumulate at ER–LD contacts and influence LD growth and morphology, but are functionally independent of one another and play unique roles in LD maturation.
Snx14 localizes to ACSL3-associated preLDs during OA-induced LD expansion
The presence of Snx14 at ER–LD interfaces following OA treatment, as well as its requirement for proper LD morphology, suggested that Snx14 may be recruited to preexisting nascent LDs within the ER network to regulate LD growth following OA addition. To better dissect when and how Snx14 associates with LDs, we next monitored Snx14EGFP distribution in the ER during set time points following the addition of OA and its subsequent processing and incorporation into growing LDs.
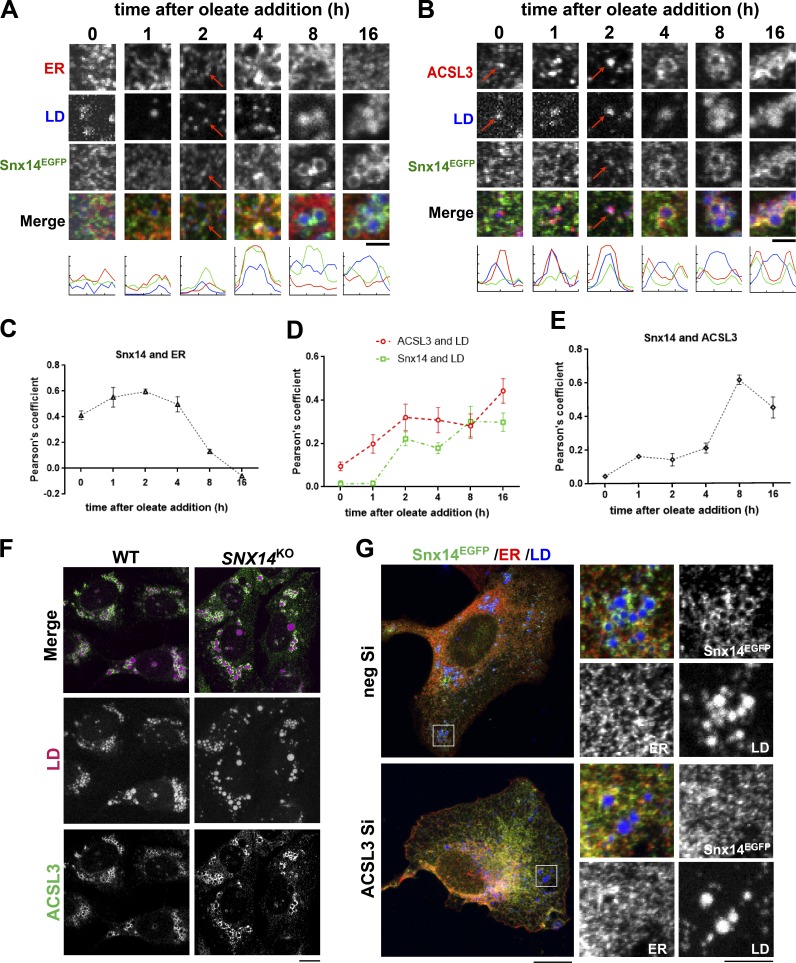
First, we fixed U2OS cells before the addition of OA and at 1, 2, 4, 8, and 16 h following OA treatment and performed coimmunofluorescence (coIF) labeling of Snx14EGFP and the ER network, as well as stained LDs with MDH. In cells receiving <2 h of OA, Snx14EGFP was distributed throughout the ER network with a Snx14-ER Pearson’s coefficient of r = ∼0.4–0.6 (Fig. 6, A and C; and Fig. S5 A). After 2 h of OA treatment, we observed a slight coalescence of Snx14EGFP into distinct ER-localized foci that were associated with small LDs (Figs. 6 A and S5 A). These Snx14EGFP foci became more prominent by 4 h of OA treatment, and by 8 h there was a significant decrease in Snx14-ER colocalization as Snx14EGFP accumulated around growing LDs (Fig. 6, A and C). This Snx14 accumulation around LDs corresponded with a significant decrease of the Snx14-ER Pearson’s coefficient, from r = ∼0.5 at 4 h to r = ∼0.13 at 8 h of OA treatment (Fig. 6, A and C). By 16 h of OA treatment, large LDs had appeared with Snx14 closely juxtaposed around them, and Snx14-ER colocalization was minimal.
Figure 6.
Snx14 localizes at ACSL3-positive preLDs following OA addition. (A) Confocal micrographs of immunofluorescently stained Snx14EGFP-expressing cells following OA treatment for t = 0, 1, 2, 4, 8, and 16 h. Labels were Snx14EGFP (α-EGFP), ER (α-HSP90B1), and LDs (MDH). Scale bar = 2 µm. Line scans show the spatial distribution of Snx14 (green) with respect to ER (red) and LDs (blue). (B) Confocal micrographs of immunofluorescently stained Snx14EGFP-expressing cells following OA treatment for t = 0, 1, 2, 4, 8, and 16 h. Labels were Snx14EGFP (α-EGFP), native ACSL3 (α-ACSL3), and LDs (MDH). Scale bar = 2 µm. Line scans show the spatial distribution of Snx14 (green) with respect to ACSL3 (red) and LDs (blue). Scale bar = 2 µm. (C) Quantification of Pearson’s coefficient between Snx14 and ER of n = ∼20 cells depicted in A. (D) Quantification of Pearson’s coefficient between ACSL3 and LDs, and Snx14 and LDs of n = ∼20 cells depicted in B. (E) Quantification of Pearson’s coefficient between Snx14 and ACSL3 of n = ∼20 cells depicted in B. (F) Confocal micrographs examining localization of ACSL3 in WT and SNX14KO cells treated with OA overnight. Labels were native ACSL3 (α-ACSL3, green) and LDs (MDH, magenta). Scale bar = 20 µm. (G) Confocal micrographs to examine localization of Snx14 in WT cells treated with negative scrambled siRNA (neg Si) or ACSL3-targeted siRNA (ACSL3 Si), respectively. The cells were transfected with Snx14EGFP along with the respective siRNAs and treated with OA overnight. Labels were Snx14EGFP (α-EGFP), ER (α-HSP90B1), and LDs (MDH). Scale bar = 20 µm. Inset scale bar = 5 µm.
Next, we investigated whether Snx14EGFP was accumulating at ER microdomains containing nascent preLDs by costaining for Snx14EGFP and the acyl-CoA ligase ACSL3, an ER-resident protein and known preLD marker that promotes LD expansion following OA treatment (Kassan et al., 2013). As before, we performed colocalization analysis of IF-stained cells following OA treatment at 0, 1, 2, 4, 8, and 16 h. Even before OA treatment, we observed ACSL3 foci in the ER network, a subpopulation of which colocalized with MDH-stained LDs (Fig. 6 B, red arrows). However, MDH staining remained minimal, and several ACSL3 foci contained almost no detectable MDH, consistent with other studies where nascent LDs stain poorly with fluorescent dyes (Gao et al., 2017; Fig. 6, B and D; and Fig. S5 B). In contrast, Snx14EGFP displayed no detectable enrichment at ACSL3-positive preLD sites and exhibited no positive Pearson’s coefficient value for either ACSL3 or MDH-stained LDs before OA addition (Fig. 6, D and E). Following 2 h of OA treatment, we observed Snx14EGFP foci which colocalized with bright MDH-associated ACSL3 foci, along with an ∼2.5-fold increase in the Snx14-ACSL3 Pearson’s coefficient (Fig. 6 B, red arrows). This also coincided with a significant increase in the Snx14-LD Pearson’s coefficient to r = ∼0.2, which approached the ACSL3-LD coefficient for this time point (Fig. 6 D). By 4 h of OA treatment, Snx14 and ACSL3 colocalized, and this further increased at 8 and 16 h as Snx14EGFP accumulated around ACSL3-decorated LDs (Fig. 6, B, D, and E). These results indicate that Snx14EGFP does not detectably associate with ACSL3-positive preLDs under ambient conditions, but is recruited to the ER–LD interface following OA treatment during LD growth.
Since Snx14EGFP was accumulating at ACSL3-positive LD buds, we next asked whether loss of Snx14 or ACSL3 impacted the recruitment of one another to LDs. First, we stained for endogenous ACSL3 in WT and SNX14KO cells. Loss of Snx14 did not affect ACSL3 localization, and ACSL3 was detected on LD surfaces following OA treatment (Fig. 6 F). In contrast, siRNA depletion of ACSL3, which decreased LD abundance, also perturbed the association of Snx14EGFP with the remaining LDs following OA treatment, indicating that ACSL3 is necessary for Snx14EGFP accumulation at ER–LD contacts (Fig. 6 G). Collectively, we conclude that ACSL3 functions upstream of Snx14 and is required for the recruitment of Snx14 to preLDs during OA-induced LD growth.
Snx14 promotes LD biogenesis following OA addition
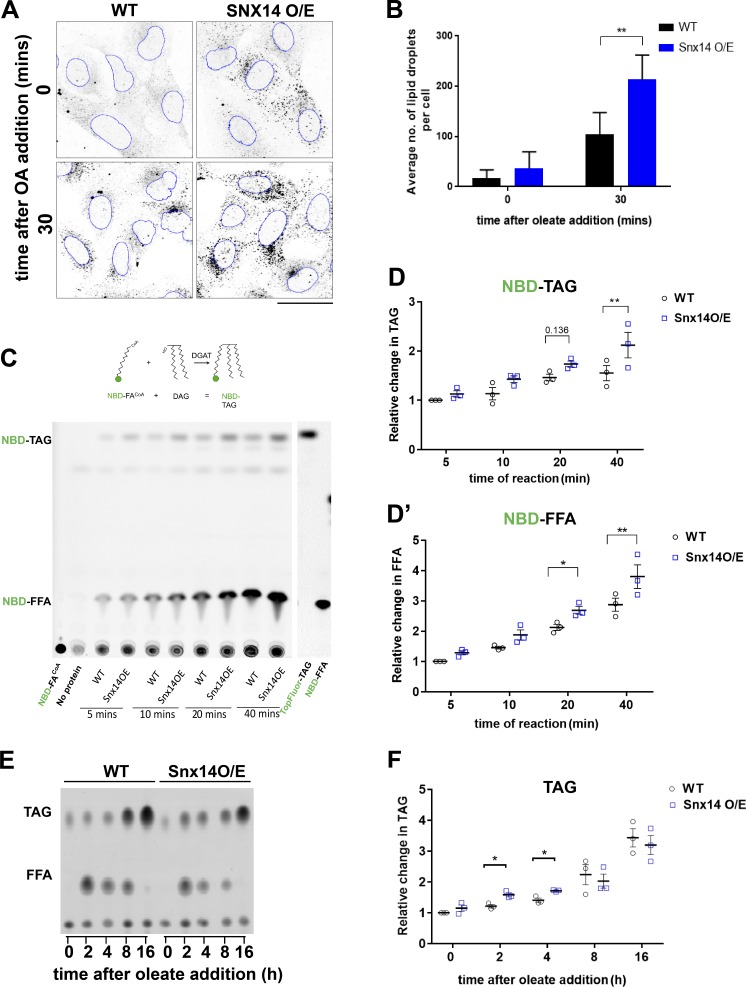
Since Snx14 accumulated at ACSL3-positive ER microdomains following OA treatment, we hypothesized that Snx14 may promote the incorporation of FAs into TAG during LD growth. To test this hypothesis, we ectopically overexpressed Flag-tagged Snx14 and examined its effect on OA-stimulated LD biogenesis in U2OS cells. We used confocal microscopy to quantify the number of MDH-stained LDs formed following a brief 30-min treatment with OA. Before OA treatment, Snx14 O/E cells exhibited no significant differences in MDH-stained LDs compared with WT cells (Fig. 7, A and B). However, following 30 min of OA treatment, Snx14 O/E cells exhibited a significant increase in the total number of MDH-stained LDs, suggesting that excess Snx14 promoted OA-induced LD biogenesis (Fig. 7, A and B). To eliminate the possibility that this was due to general ER stress caused by the overexpression of Snx14Flag, which might cause the production of LDs nonspecifically, we also monitored the unfolded protein response (UPR) by examining levels of spliced XBP1 transcripts, and found no significant differences (Fig. S6, A and B).
Figure 7.
Snx14 promotes LD biogenesis and enhances TAG synthesis. (A) Confocal micrographs of WT and Snx14Flag O/E U2OS cells either without OA or following 30 min OA treatment. LDs were labeled with Nile Red and converted to grayscale. ImageJ was used to mark the border of nuclei in blue. Scale bar = 50 µm. (B) Quantification of average number of LDs per cell of representative image from A. Each set of values was analyzed from five fields of view, each consisting of >10 cells of two different sets of experiments (total no. of cells >100; **, P < 0.001, multiple t test by Holm–Sidak method with α = 0.05). (C) Schematic diagram depicting fluorescent in vitro assay to measure NBD-FACoA incorporation into NBD-TAG. TLC of fluorescent NBD-FA-CoA incorporated into neutral lipids over time. WT and Snx14Flag O/E cells were pulsed with NBD-FA-CoA and collected at 5-, 10-, 20-, and 40-min time points. (D) Quantification of relative fold change in NBD-TAG (D) and NBD-FFA (D′) of TLC from C. Values are normalized to protein concentration and represent mean ± SEM (n = 3; *, P < 0.01; **, P < 0.001, multiple t test by Holm–Sidak method with α = 0.05). (E) TLC of neutral lipids in WT cells and cells overexpressing Snx14EGFP treated with OA for 0, 2, 4, 8, and 16 h. (F) Quantification of relative fold change in TAG (normalized to cell pellet weight) of TLC from E. Values represent mean ± SEM (n = 3; *, P < 0.01, multiple t test by Holm–Sidak method with α = 0.05).
To determine whether Snx14 influenced the incorporation of FAs into TAG during LD growth, we next monitored the incorporation of a fluorescently labeled fatty acyl-CoA (NBD-palmitoyl-CoA) into TAG when it was incubated with membranes isolated from WT and Snx14 O/E cells (Fig. 7 C). Using this in vitro assay, we observed that, over time, NBD-palmitoyl-CoA could be incorporated with exogenously added DAG into NBD-labeled TAG, which could be visibly detected by TLC (McFie and Stone, 2011). Notably, by 40 min of reaction time, there was a significant increase in the incorporation of NBD-palmitoyl-CoA into TAG in samples derived from Snx14 O/E cells compared with WT (Fig. 7, C and D). We also noted an increase in the level of NBD-labeled free palmitate (NBD-FFA; Fig. 7, C and D′).
Next, we determined whether Snx14 O/E also increased total TAG levels in intact cells following OA treatment. We monitored the change in TAG and FFAs by TLC in WT and Snx14 O/E cells following treatment with OA for 2, 4, 8, and 16 h (Fig. 7 E). Indeed, Snx14 O/E cells exhibited mild but significantly elevated TAG levels at early time points (2 and 4 h), as well as a significant decrease in FFAs at 2 h, coincident with the initial accumulation of Snx14 to ACSL3-positive preLDs observed by light microscopy (Figs. 7 F and S6 E). To confirm that these increases in TAG were due to increased TAG biogenesis and not decreased LD turnover, we monitored TAG mobilization by treating cells with the acyl-CoA synthesis inhibitor Triacsin C for 3 h to inhibit LD biogenesis and promote LD mobilization. We observed similar levels of TAG reduction in both WT and Snx14 O/E cell lines, suggesting that LD turnover is not blocked in Snx14 O/E cells (Fig. S6 F).
Finally, since ACSL3 and Snx14 colocalized at LDs following OA treatment, and Snx14 recruitment to LDs required ACSL3, we determined whether loss of ACSL3 would similarly affect TAG synthesis and LD biogenesis in WT and Snx14 O/E cells. Indeed, siRNA depletion of ACSL3 in both WT and Snx14 O/E cells similarly reduced LD numbers and TAG levels, suggesting that Snx14 O/E alterations were ACSL3 dependent (Fig. S6, C and D). In all, we conclude that Snx14 promotes the incorporation of OA into TAG during OA-induced LD growth in an ACSL3-dependent manner.
Discussion
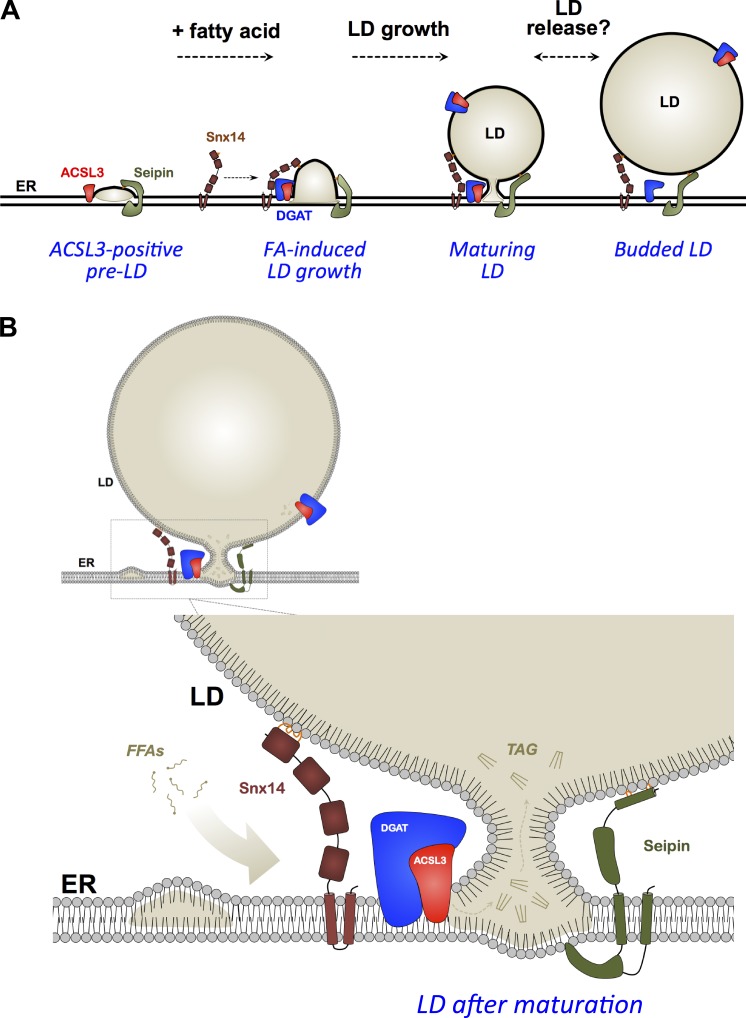
LD biogenesis and maturation involves distinct stages of lipid synthesis and ER membrane remodeling, and is executed by the coordinated effort of several proteins and enzymes that accumulate at ER microdomains from which LDs bud. During their initial maturation, LDs maintain contact with the ER, and this ER–LD bridge serves as a site for interorganelle lipid flux that drives LD growth. How, when, and whether an LD will be released from the ER surface remains poorly understood, and what factors regulate the dynamics of ER–LD crosstalk remain a major question in the LD field. Recent studies have highlighted proteins such as Seipin, Rab18, and others that localize at ER–LD contacts and promote LD maturation through interorganelle tethering (Salo et al., 2016; Wang et al., 2016; Xu et al., 2018). Here, we identify the SCAR20 disease-related protein Snx14 as a novel factor that contributes to FA-induced LD growth at ER–LD contacts. Using proximity-based APEX technology, we visualize Snx14 at ER–LD contact sites following OA treatment, the first use of APEX imaging technology at this interorganelle junction to our knowledge. Furthermore, we find that Snx14 remains topologically associated with the ER while at ER–LD contacts and binds LDs in trans. Consistent with its role as an ER–LD tether, Snx14 expression can modulate the extent of ER–LD contacts, and Snx14 O/E cells display extended ER–LD contact sites. We find with multi–time point imaging that Snx14 is not detectably present at nascent preLDs containing ACSL3, but is recruited to these ER sites following OA treatment as preLDs grow (Fig. 8 A).
Figure 8.
Working model for Snx14 in FA-induced LD maturation. (A) Stepwise cartoon of FA-induced LD biogenesis promoted by Snx14. (B) Model of Snx14 and other key factors at ER–LD contact site following LD maturation.
Intriguingly, the phenotypes observed in SNX14KO cells appear similar to loss of Seipin, another bridging protein that localizes to ER–LD contacts. Like Snx14, loss of Seipin perturbs LD morphology, but does not generally alter cellular TAG levels (Cartwright et al., 2015; Wang et al., 2016). Both proteins also appear to use similar mechanisms to target to ER–LD contacts (Sui et al., 2018; Yan et al., 2018). Seipin and Snx14 both contain transmembrane helices anchored in the ER. Snx14 contains an AH region on its C-Nexin domain that is necessary for LD binding and Snx14 function. Similarly, Drosophila melanogaster Seipin contains a hydrophobic helix in its lumenal domain and an N-terminal LD targeting region, both of which display affinity for LDs. Mutation of both these regions together perturbs Seipin function in LD formation, indicating that LD targeting by these regions is functionally important (Sui et al., 2018). Both proteins also play roles in LD maturation following OA addition. Seipin has also been reported to promote LD expansion in Drosophila S2 cells, as well as during adipocyte development (Pagac et al., 2016). Although further studies are required, an intriguing model is that both Seipin and Snx14 independently regulate LD maturation at the ER–LD interface by stabilizing the ER–LD bridge and helping to maintain this contact so that TAG can be efficiently produced in the ER and fluxed into the growing LD (Fig. 8 B). Consistent with this, Snx14 overexpression promoted FA incorporation into TAG. The precise mechanism of this is unclear, but one model is that Snx14 drives the stabilization and extension of ER–LD contacts, and these contacts are sites where TAG synthesis enzymes accumulate. In the absence of Snx14, compensatory mechanisms maintain neutral lipid synthesis, but proper LD growth is perturbed.
Human Snx14 displays functional conservation with yeast Mdm1, which also localizes to specific ER microdomains (yeast NVJs) and associates with LDs as they mature there. Furthermore, both proteins work in conjunction with fatty acyl-CoA ligases at sites of LD maturation. In yeast, Mdm1 functionally interacts with fatty acyl-CoA ligase Faa1 (Hariri et al., 2018). Similarly, Snx14 colocalizes with ACSL3 in human cells and requires ACSL3 for recruitment at ER–LD contacts, implying a conserved function in FA metabolism. However, whereas Mdm1 constitutively localizes to NVJ contact sites in yeast, human cells do not contain NVJ contacts, and Snx14 displays no detectable subcellular localization before its association with growing LDs. This difference may be attributed to the different phospholipid binding properties of the two proteins’ Phox homology (PX) domains. Mdm1 contains a PX domain that binds to vacuolar PtdIns(3)P, thus constitutively anchoring Mdm1 to ER microdomains adjacent to the vacuole surface. In contrast, structural and biochemical studies reveal that the Snx14 PX domain lacks residues necessary for PtdIns(3)P binding, and it displays negligible phospholipid binding affinity in vitro (Mas et al., 2014; Bryant et al., 2018). Instead, we find that Snx14 inducibly accumulates at distinct ER subdomains marked by ACSL3 following OA treatment. Once there, Snx14 remains associated with a growing LD via a C-terminal AH region in its C-Nexin domain. Further studies are needed to dissect these distinctions, which imply a functional divergence between Mdm1 and Snx14 regarding how they associate with different cellular organelles.
Recent studies reveal that many proteins retarget from the ER network to LDs during times of elevated TAG synthesis and LD growth. These generally include lipid synthesis enzymes that promote neutral lipid synthesis (GPAT4 and DGAT2) or phospholipid synthesis (CCT1) to promote proper LD maturation (Wilfling et al., 2013). These proteins also feature LD targeting mechanisms including either a hairpin membrane insertion motif that can interchangeably anchor in both the ER bilayer and LD monolayer (type I) or an AH that can insert into the LD monolayer surface (type II; Prevost et al., 2018). In contrast to type I LD-targeting proteins that leave the ER, Snx14 appears to maintain its ER localization throughout LD maturation. We propose that Snx14, like Seipin, constitutes its own class of LD-associated proteins that localize to ER–LD contacts during LD growth, but remain firmly anchored in the ER while binding the LD monolayer surface in trans. This allows them to act as interorganelle tethers firmly connecting the ER and LD during LD growth, and ensuring the two organelles maintain a connection as TAG produced in the ER is fluxed into the maturing LD (Fig. 8). Given its role in the pediatric cerebellar ataxia SCAR20, further analysis of Snx14 will no doubt yield more interesting and surprising discoveries in cell physiology. Indeed, further investigation of Mdm1 reveals its importance in maintaining ER homeostasis, as Mdm1-deficient yeast are sensitive to FA-induced lipotoxicity (Hariri et al., 2019). Taken together, this suggests that Snx14, like Mdm1, may play a functionally conserved role in FA homeostasis, which when perturbed contributes to neurological disease.
Materials and methods
Cell culture
U2OS cells were cultured in DMEM (Corning; 10-013-CV) supplemented with 10% Cosmic Calf Serum (Hyclone; SH30087.04) and 1% penicillin streptomycin solution (100×, Corning; 30-002-Cl). The cells were passaged when they reached 80–90% confluence with 0.25% trypsin-EDTA (Corning; 25-053-Cl). To promote LD biogenesis, cells were incubated with 600 µM of OA conjugated with 100 µM of FA-free BSA (Sigma-Aldrich; A3803) for the indicated period of time.
Cloning and transient transfection
All the Snx14 constructs, i.e., SNX14FL, SNX14N, SNX14PXCN, Snx14PX, Snx14CN, Snx14PXCNΔH, and Snx14FLΔH, tagged with EGFP at their C termini and untagged Snx14 were generated after PCR amplification of fragments of interest as described in Fig. 4 from Snx14 generated from cDNA library and cloning them into pEGFP-N2. A stop codon was inserted before EGFP in pEGFP-N2 vector to clone untagged Snx14. Seipinmcherry was generated after PCR amplification of Seipin from the cDNA library and was then cloned into pEGFP-N2 vector with an mCherry tag at the C-terminal and insertion of a stop codon before EGFP. All primers are available on request. The plasmids were transfected into U2OS cells using PolyFect Transfection reagent (Qiagen; 301105) and Optimem (Gibco; 31985-070) for 48 h before experiments.
Generation of stable cell line
All the C-terminally 3XFlag tagged constructs, i.e., SNX14FL, SNX14N, and SNX14PXCN, were generated following PCR amplification of Snx14 from cDNA library and cloning into the pMXs-3-FLAG vector. U2OS cells were transformed with retroviral vectors expressing puromycin-expressing plasmids. Cells expressing the plasmid of interest were selected via puromycin resistance and expanded to generate stable cell lines.
Generation of APEX2 tagged cell lines
Snx14EGFP-APEX2 was generated by first PCR amplifying full-length Snx14 from cDNA library, EGFP-APEX2 from pcDNA3-Connexin43EGFP-APEX2 (Addgene; plasmid 49385), and then cloning them both into one of the lentiviral vectors, plvx with puromycin resistance cassette. This cloned plasmid along with other lentiviral packaging vectors were transfected into 293T cells, followed by lentiviral production and transduction into U2OS cells to generate stable cell lines. All the generated stable cell lines were stored in liquid nitrogen before their use in experiments.
IF staining
Cells were fixed with 4% PFA solution in PBS for 15 min at RT. For IF staining, fixed cells were washed with PBS, permeabilized with 0.2% NP-40 in PBS at RT for 3 min, and then blocked in IF buffer (PBS containing 3% BSA, 0.1% NP-40, and 0.02% sodium azide) for 40 min. The cells were then incubated with primary antibody in IF buffer for 1 h, washed thrice with PBS, incubated with secondary antibody in IF buffer for 30 min, given three washes with PBS, and then mounted, before being used for microscopic studies. The primary antibodies used are mouse anti-Hsp90B1 (1:300; Sigma-Aldrich; AMAb91019), rabbit anti-Snx14 (HPA017639), rabbit anti-EGFP (1:300), rabbit anti-Flag (1:200; Sigma-Aldrich; F7425), mouse anti-ACSL3 (1:200; Novus Biologicals; H00002181-B01P). The secondary antibodies are donkey anti-mouse AF488 (Thermo Fisher Scientific; A21202), and donkey anti-rabbit AF594 (Thermo Fisher Scientific; A21207) used at a dilution of 1:1,000. LDs were visualized by staining the cells with MDH (1:1,000; Abgent; SM1000a) for 15 min. Nucleus was stained with 5 μM Syto 85 orange fluorescent stain (Molecular Probes; S-11366).
Confocal microscopy and image analysis
The images of the cells were taken using a 63× oil-immersion objective in a Zeiss LSM 780 confocal microscope. Approximately four Z-sections of each image were taken. ImageJ was used for the representation and quantification of images. The merged images were generated by max z-projection. For others, one plane was shown. The MDH-stained blue LDs were converted to grayscale and inverted, and the cell boundary was drawn with a freehand line tool.
Analysis of area covered by LDs
The area covered by LDs was calculated using ImageJ (Fiji), first by thresholding with “convert to mask,” then “analyze particles” was used. Upon treatment of WT and SNX14KO cells overnight with OA, the LDs in WT showed perinuclear clustering and were hard to distinguish one from the other. So we quantified the average area covered by LDs, which is an estimation of both size and numbers of LDs. For the rescue experiments, the transfected cells were selected, and then the area covered by LDs in each cell was calculated and plotted.
Analysis of LDs numbers
The quantification of number of LDs was adapted from Kassan et al. (2013). Again, the “analyze particles” plugin of ImageJ was used to calculate number of LDs <5 µm2. Owing to the limitation of resolution and clustering effect of LDs, the number of LDs whose area was >5 µm2 was calculated from the sum total area covered by all the LDs divided by the mean area of LDs (i.e., 0.5 µm2).
Fluorescence colocalization analysis
Pearson’s coefficient (r) was quantified for all the z-stacks of the image after choosing a threshold value to exclude the background signal using Imaris Image Analysis software.
Analysis of ER–LD contacts from TEM images
The length of each detected contact made by an ER membrane with the LD (marked in blue in Fig. 4) was calculated and normalized to the perimeter of the LD. These lengths were then plotted as dot plots.
DAB labeling of Snx14EGFP-APEX2
Cells stably expressing Snx14EGFP-APEX2 were fixed with 2.5% glutaraldehyde in cacodylate buffer (100 mM sodium cacodylate with 2 mM CaCl2, pH 7.4) for 30 min. DAB tablets (Sigma-Aldrich; D5905) were dissolved in PBS to a final concentration of 0.5 mg/ml DAB, and 10 mM of 30% H2O2 was added. The fixed cells were incubated with the DAB solution for 30 min at RT. The cells were then washed thrice with PBS, and then they were imaged by bright-field microscope to test expression and activity of APEX2. The cells were then processed to be imaged by EM.
EM
The cell lines of interest were cultured under desired conditions and processed in the University of Texas Southwestern Electron Microscopy Core Facility. Cells were fixed on MatTek dishes with 2.5% (vol/vol) glutaraldehyde in 0.1 M sodium cacodylate buffer. After three rinses in 0.1 M sodium cacodylate buffer, they were postfixed in 1% osmium tetroxide and 0.8% K3[Fe(CN6)] in 0.1 M sodium cacodylate buffer for 1 h at room temperature. Cells were rinsed with water and en bloc stained with 2% aqueous uranyl acetate overnight. Next, they were rinsed in buffer and dehydrated with increasing concentrations of ethanol, infiltrated with Embed-812 resin, and polymerized in a 60°C oven overnight. Blocks were sectioned with a diamond knife (Diatome) on an Ultracut UCT (7) ultramicrotome (Leica Microsystems) and collected onto copper grids, poststained with 2% aqueous uranyl acetate and lead citrate. Images were acquired on a Tecnai G2 spirit transmission EM (FEI) equipped with an LaB6 source using a voltage of 120 kV. The APEX-tagged samples were processed for EM as described above except that they were not poststained, and the methods used were based on the protocol from Martell et al. (2012).
Generation of SNX14KO U2OS cell lines using CRISPR-Cas9
In U2OS cells, the Snx14 gene was targeted with the gRNA 5′-GAAGCAGAACAGCGGGTACTGG-3′ using CRISPR-Cas9 system. Single cells were then seeded in one well of a 96-well plate, colonies were expanded, and loss of Snx14 was validated using Western blot. These clones were next sequenced, and the one that generates a very short peptide of 25 residues was selected for experiments.
Neutral lipid analysis by TLC
Lipids were extracted from cultured cells using a protocol adapted from Bligh and Dyer (1959). Cells cultured in complete media were washed twice in PBS, scraped, and collected, and their weights were measured. The cell pellets were lysed by vortexing in the presence of chloroform, followed by methanol and 500 mM NaCl in 0.5% acetic acid to the final concentration such that the ratio of chloroform:methanol:water was 2:2:1.8. The suspension was centrifuged at 4,000 rpm for 15 min at 4°C and the bottom chloroform layer, comprising the lipids, was collected. This lipid layer was dried and then again resuspended in chloroform to a final concentration normalized to the initial cell pellet weight. TLC was used to separate the extracted lipids. Hexane:diethyl ether:acetic acid (80:20:1) was used as a solvent to separate neutral lipids extracted from the cells. The bands in the TLC plates were developed by spray-staining with 3% copper acetate prepared in 8% phosphoric acid and incubating in the oven at 135°C for ∼1 h. Plates were resprayed and reheated as needed to visualize lipids. Developed TLC plates were scanned and then processed for quantification using Fiji (ImageJ). On each plate we ran a serial dilution of standard neutral lipid mixture of known concentrations to create a standard curve between the lipid mass versus the band intensity to estimate the lipid mass from the samples of interest.
Fluorescent acyltransferase activity assay
This fluorescent-based assay to measure DGAT activity in vitro was based on the protocol adapted from McFie and Stone (2011). In short, total membranes were isolated from cells, 80 µg of which was used with NBD-palmitoyl CoA as substrate to react with DAG added externally to produce NBD-TAG. The fluorescent neutral lipids were then detected by TLC where cyclohexane:ethylacetate (1:2) was used as solvent and quantified using a Bio-Rad fluorescent imaging system.
Isolation of total LDs
Total LDs were isolated from U2OS cells treated with OA overnight according to the protocol adapted from Ding et al. (2013). In brief, cells from eight 150-mm dishes were washed twice with ice-cold PBS and then collected by scraping them into ice-cold PBS. The cells were pelleted by centrifugation at 1,000 g for 5 min at 4°C, resuspended in buffer A (20 mM tricine and 250 mM sucrose, pH 7.8) containing 0.2 µM PMSF and protease inhibitor cocktail (Thermo Fisher Scientific; A32965) and kept on ice for 20 min. The cells were disrupted using Nitrogen bomb at a pressure of 35 bar N2 on ice for 15 min. The homogenate was centrifuged at 3,000 g for 10 min at 4°C, and the supernatant was transferred into SW 40 Ti tubes. Then 2 ml of buffer B (20 mM Hepes, 100 mM KCl, and 2 mM MgCl2, pH 7.4) was loaded onto the top of the supernatant, and the gradient was centrifuged at 182,000 g for 1 h at 4°C. The topmost white layer, which consists of LDs, was collected to a 1.5-ml microcentrifuge tube. The middle part, which is the cytosol, and the pellet, which is the membrane fraction, were also collected.
Western blot
The LD and membrane fractions collected were diluted respectively in 200 and 500 µl of buffer B containing protease inhibitor cocktail (Thermo Fisher Scientific; 78430) and 0.2 µM PMSF. Samples were prepared in 2× Laemmli Sample Buffer (Bio-Rad; 161-0737) supplemented with 5% β-mercaptoethanol, heated at 95°C for 10 min, and then run on a polyacrylamide gel. The proteins were then transferred onto a PVDF membrane. Next, the membrane-containing proteins were blocked with 5% milk in TBS-0.1%Tween (TBST) buffer for 1 h at RT, incubated overnight with primary antibodies, which were Rab anti-EGFP (1:2,000), mouse anti-Hsp90B1 (1:1,000), Rab anti-actin (1:1,000), mouse anti-ADRP (1:50; Fitzgerald), mouse anti-Flag (1:1,000), and Rab anti-Snx14 (1:400; Sigma-Aldrich HPA017639), washed thrice with TBST, incubated with HRP-conjugated secondary antibodies (1:5,000), and developed with Clarity Western ECL blotting substrate (Bio-Rad; 1705061) and imaging with x-ray film. For company sources of antibodies, please refer to above IF staining section.
For quantitative densitometry, relative protein expression was quantified by analyzing band intensities using ImageJ. For determining LD:membrane protein ratios (as in Fig. S2 B), equal volumes of total membrane and LD fractions were loaded onto the gel. To correct for their dilutions, the membrane fraction signals were multiplied by 2.5.
siRNA transfection
Cells were transfected with Neg siRNA (Ambion; AM4611), Snx14 siRNA (Ambion; 4392420, ID s32927), and ACSL3 siRNA (Ambion; 4390824, ID s4999, s4997) using Lipofectamine 2000 (Invitrogen; 11668-019) in Optimem (Gibco; 31985-070). After 6 h, the transfected media was changed to complete media for another 48 h before any experiments.
Quantifying ER stress
ER stress was quantified by analyzing the transcript level of spliced xbp1. For this, total RNA was extracted from cells, followed by qRT-PCR reaction. Cells treated with 5 µg/ml of tunicamycin, which causes UPR stress, was used a control.
For RNA extraction, cells from the dishes were directly dissolved in TRIzol, then treated with chloroform, followed by centrifugation at 12,000 g for 15 min. The colorless upper aqueous layer was collected and isopropanol added to precipitate RNA, which was then washed by ethanol, air-dried, and resuspended in RNase-free water. RNA concentration was measured with Nano Spectrophotometer and was used to make cDNA with commercial kit from Bio-Rad (iScript cDNA Synthesis kit 1708891) according to the manufacturer’s instructions. The cDNA was used to run quantitative PCR to detect signals for xbp1 mRNA relative to β-actin (actb) used as a control using predesigned primers KiCq (Sigma-Aldrich).
Online supplemental material
Fig. S1 shows that Snx14 localizes to the ER–LD interface. Fig. S2 shows topological dissection of Snx14 LD association. Fig. S3 shows that loss of SNX14 affects LD morphology but not TAG synthesis. Fig. S4 shows that Snx14 localizes to ER–LD contacts independently of Seipin. Fig. S5 shows that Snx14 localizes to ACSL3-positive preLDs. Fig. S6 shows that Snx14 promotes LD biogenesis in an ACSL3-dependent manner.
Supplementary Material
Acknowledgments
The authors sincerely thank Drs. Joel Goodman, Sandra Schmid, and Russell De-Bose Boyd for their critical assistance throughout the course of this study. We also thank Dr. Kate Luby-Phelps and Anza Darehshouri for technical assistance with the confocal and electron microscopy. We also acknowledge Drs. Tobias Walther and Robert Farese (Harvard Medical School, Boston, MA) and Dr. Rob Yang (University of New South Wales, Sydney, Australia) for sending SEIPINKO cells. We also thank Jade Bowerman for technical help and other members of the Henne laboratory for helpful discussions and feedback. We also thank Drs. Xiaoyu Hu and Michael Reese for assistance with fluorescent FA TLCs, and Dr. Rupali Ugrankar for quantitative PCR assistance.
W.M. Henne, S. Datta, Y. Liu, and H. Hariri are supported by grants from the Welch Foundation (I-1873), the Searle Foundation (SSP-2016-1482), the National Institutes of Health National Institute of General Medical Sciences (GM119768), and the University of Texas Southwestern Endowed Scholars Program.
The authors declare no competing financial interests.
Author contributions: W.M. Henne and S. Datta conceptualized and designed the experiments. S. Datta conducted imaging, biochemical and other experiments, and analysis. W.M. Henne and S. Datta drafted and revised the article. Y. Liu generated SNX14KO cell lines and overexpressed stable cell lines and provided early experimental assistance. H. Hariri provided conceptual assistance in the design of the study and biochemical assistance in the lipid extraction (TLC). J. Bowerman provided technical assistance.
References
- Athenstaedt, K., and Daum G.. 2006. The life cycle of neutral lipids: synthesis, storage and degradation. Cell. Mol. Life Sci. 63:1355–1369. 10.1007/s00018-006-6016-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bligh, E.G., and Dyer W.J.. 1959. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 37:911–917. 10.1139/y59-099 [DOI] [PubMed] [Google Scholar]
- Bryant, D., Liu Y., Datta S., Hariri H., Seda M., Anderson G., Peskett E., Demetriou C., Sousa S., Jenkins D., et al. 2018. SNX14 mutations affect endoplasmic reticulum-associated neutral lipid metabolism in autosomal recessive spinocerebellar ataxia 20. Hum. Mol. Genet. 27:1927–1940. 10.1093/hmg/ddy101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cartwright, B.R., Binns D.D., Hilton C.L., Han S., Gao Q., and Goodman J.M.. 2015. Seipin performs dissectible functions in promoting lipid droplet biogenesis and regulating droplet morphology. Mol. Biol. Cell. 26:726–739. 10.1091/mbc.E14-08-1303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding, Y., Zhang S., Yang L., Na H., Zhang P., Zhang H., Wang Y., Chen Y., Yu J., Huo C., et al. 2013. Isolating lipid droplets from multiple species. Nat. Protoc. 8:43–51. 10.1038/nprot.2012.142 [DOI] [PubMed] [Google Scholar]
- Fei, W., Shui G., Gaeta B., Du X., Kuerschner L., Li P., Brown A.J., Wenk M.R., Parton R.G., and Yang H.. 2008. Fld1p, a functional homologue of human seipin, regulates the size of lipid droplets in yeast. J. Cell Biol. 180:473–482. 10.1083/jcb.200711136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimoto, T., and Parton R.G.. 2011. Not just fat: the structure and function of the lipid droplet. Cold Spring Harb. Perspect. Biol. 3:a004838. 10.1101/cshperspect.a004838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao, Q., Binns D.D., Kinch L.N., Grishin N.V., Ortiz N., Chen X., and Goodman J.M.. 2017. Pet10p is a yeast perilipin that stabilizes lipid droplets and promotes their assembly. J. Cell Biol. 216:3199–3217. 10.1083/jcb.201610013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, Y., Cordes K.R., Farese R.V. Jr., and Walther T.C.. 2009. Lipid droplets at a glance. J. Cell Sci. 122:749–752. 10.1242/jcs.037630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hariri, H., Rogers S., Ugrankar R., Liu Y.L., Feathers J.R., and Henne W.M.. 2018. Lipid droplet biogenesis is spatially coordinated at ER-vacuole contacts under nutritional stress. EMBO Rep. 19:57–72. 10.15252/embr.201744815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hariri, H., Speer N., Bowerman J., Roger S., Fu G., Reetz E., Datta S., Feathers J.R., Ugrankar R., Nicastro D., and Henne M.. 2019. Mdm1 maintains endoplasmic reticulum homeostasis by spatially regulating lipid droplet biogenesis. J. Cell Biol. 10.1083/jcb.201808119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassan, A., Herms A., Fernández-Vidal A., Bosch M., Schieber N.L., Reddy B.J., Fajardo A., Gelabert-Baldrich M., Tebar F., Enrich C., et al. 2013. Acyl-CoA synthetase 3 promotes lipid droplet biogenesis in ER microdomains. J. Cell Biol. 203:985–1001. 10.1083/jcb.201305142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam, S.S., Martell J.D., Kamer K.J., Deerinck T.J., Ellisman M.H., Mootha V.K., and Ting A.Y.. 2015. Directed evolution of APEX2 for electron microscopy and proximity labeling. Nat. Methods. 12:51–54. 10.1038/nmeth.3179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Listenberger, L.L., Han X., Lewis S.E., Cases S., Farese R.V. Jr., Ory D.S., and Schaffer J.E.. 2003. Triglyceride accumulation protects against fatty acid-induced lipotoxicity. Proc. Natl. Acad. Sci. USA. 100:3077–3082. 10.1073/pnas.0630588100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martell, J.D., Deerinck T.J., Sancak Y., Poulos T.L., Mootha V.K., Sosinsky G.E., Ellisman M.H., and Ting A.Y.. 2012. Engineered ascorbate peroxidase as a genetically encoded reporter for electron microscopy. Nat. Biotechnol. 30:1143–1148. 10.1038/nbt.2375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mas, C., Norwood S.J., Bugarcic A., Kinna G., Leneva N., Kovtun O., Ghai R., Ona Yanez L.E., Davis J.L., Teasdale R.D., and Collins B.M.. 2014. Structural basis for different phosphoinositide specificities of the PX domains of sorting nexins regulating G-protein signaling. J. Biol. Chem. 289:28554–28568. 10.1074/jbc.M114.595959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFie, P.J., and Stone S.J.. 2011. A fluorescent assay to quantitatively measure in vitro acyl CoA:diacylglycerol acyltransferase activity. J. Lipid Res. 52:1760–1764. 10.1194/jlr.D016626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagac, M., Cooper D.E., Qi Y., Lukmantara I.E., Mak H.Y., Wu Z., Tian Y., Liu Z., Lei M., Du X., et al. 2016. SEIPIN Regulates Lipid Droplet Expansion and Adipocyte Development by Modulating the Activity of Glycerol-3-phosphate Acyltransferase. Cell Reports. 17:1546–1559. 10.1016/j.celrep.2016.10.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prevost, C., Sharp M.E., Kory N., Lin Q., Voth G.A., Farese R.V. Jr., and Walther T.C.. 2018. Mechanism and determinants of amphipathic helix-containing protein targeting to lipid droplets. Dev. Cell. 44:73–86.e4. 10.1016/j.devcel.2017.12.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salo, V.T., Belevich I., Li S., Karhinen L., Vihinen H., Vigouroux C., Magré J., Thiele C., Hölttä-Vuori M., Jokitalo E., and Ikonen E.. 2016. Seipin regulates ER-lipid droplet contacts and cargo delivery. EMBO J. 35:2699–2716. 10.15252/embj.201695170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla, A., Upadhyai P., Shah J., Neethukrishna K., Bielas S., and Girisha K.M.. 2017. Autosomal recessive spinocerebellar ataxia 20: Report of a new patient and review of literature. Eur. J. Med. Genet. 60:118–123. 10.1016/j.ejmg.2016.11.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sui, X., Arlt H., Brock K.P., Lai Z.W., DiMaio F., Marks D.S., Liao M., Farese R.V. Jr., and Walther T.C.. 2018. Cryo-electron microscopy structure of the lipid droplet-formation protein seipin. J. Cell Biol. 217:4080–4091. 10.1083/jcb.201809067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szymanski, K.M., Binns D., Bartz R., Grishin N.V., Li W.P., Agarwal A.K., Garg A., Anderson R.G., and Goodman J.M.. 2007. The lipodystrophy protein seipin is found at endoplasmic reticulum lipid droplet junctions and is important for droplet morphology. Proc. Natl. Acad. Sci. USA. 104:20890–20895. 10.1073/pnas.0704154104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas, A.C., Williams H., Setó-Salvia N., Bacchelli C., Jenkins D., O’Sullivan M., Mengrelis K., Ishida M., Ocaka L., Chanudet E., et al. 2014. Mutations in SNX14 cause a distinctive autosomal-recessive cerebellar ataxia and intellectual disability syndrome. Am. J. Hum. Genet. 95:611–621. 10.1016/j.ajhg.2014.10.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, H., Becuwe M., Housden B.E., Chitraju C., Porras A.J., Graham M.M., Liu X.N., Thiam A.R., Savage D.B., Agarwal A.K., et al. 2016. Seipin is required for converting nascent to mature lipid droplets. eLife. 5:e16582. 10.7554/eLife.16582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilfling, F., Wang H., Haas J.T., Krahmer N., Gould T.J., Uchida A., Cheng J.X., Graham M., Christiano R., Fröhlich F., et al. 2013. Triacylglycerol synthesis enzymes mediate lipid droplet growth by relocalizing from the ER to lipid droplets. Dev. Cell. 24:384–399. 10.1016/j.devcel.2013.01.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilfling, F., Thiam A.R., Olarte M.J., Wang J., Beck R., Gould T.J., Allgeyer E.S., Pincet F., Bewersdorf J., Farese R.V. Jr., and Walther T.C.. 2014. Arf1/COPI machinery acts directly on lipid droplets and enables their connection to the ER for protein targeting. eLife. 3:e01607. 10.7554/eLife.01607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, D., Li Y., Wu L., Li Y., Zhao D., Yu J., Huang T., Ferguson C., Parton R.G., Yang H., and Li P.. 2018. Rab18 promotes lipid droplet (LD) growth by tethering the ER to LDs through SNARE and NRZ interactions. J. Cell Biol. 217:975–995. 10.1083/jcb.201704184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, N., Zhang S.O., Cole R.A., McKinney S.A., Guo F., Haas J.T., Bobba S., Farese R.V. Jr., and Mak H.Y.. 2012. The FATP1-DGAT2 complex facilitates lipid droplet expansion at the ER-lipid droplet interface. J. Cell Biol. 198:895–911. 10.1083/jcb.201201139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan, R., Qian H., Lukmantara I., Gao M., Du X., Yan N., and Yang H.. 2018. Human SEIPIN binds anionic phospholipids. Dev. Cell. 47:248–256.e4. 10.1016/j.devcel.2018.09.010 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.