The alignment of chromosomes at the center of the mitotic spindle is a highly conserved step during cell division. Fonseca et al. find that mammalian cells with alignment defects due to lack of KIF18A function display interchromosomal compaction problems during anaphase and form abnormal nuclei after cell division.
Abstract
Chromosome alignment at the equator of the mitotic spindle is a highly conserved step during cell division; however, its importance to genomic stability and cellular fitness is not understood. Normal mammalian somatic cells lacking KIF18A function complete cell division without aligning chromosomes. These alignment-deficient cells display normal chromosome copy numbers in vitro and in vivo, suggesting that chromosome alignment is largely dispensable for maintenance of euploidy. However, we find that loss of chromosome alignment leads to interchromosomal compaction defects during anaphase, abnormal organization of chromosomes into a single nucleus at mitotic exit, and the formation of micronuclei in vitro and in vivo. These defects slow cell proliferation and are associated with impaired postnatal growth and survival in mice. Our studies support a model in which the alignment of mitotic chromosomes promotes proper organization of chromosomes into a single nucleus and continued proliferation by ensuring that chromosomes segregate as a compact mass during anaphase.
Introduction
Chromosome alignment at the mitotic spindle equator is a conserved feature of cell division in the majority of eukaryotic cells, suggesting that it has an essential function for accurate chromosome segregation. Possible functions of chromosome alignment include promoting attachments between chromosomes and spindle microtubules, preventing erroneous attachments, promoting equal chromosome segregation during anaphase, and coordinating anaphase and cytokinesis (Kops et al., 2010; Matos and Maiato, 2011; Pereira and Maiato, 2012; Maiato et al., 2017). Elucidating the importance of chromosome alignment has been technically difficult due to an inability to experimentally disrupt it without also altering attachments between kinetochores and spindle microtubules. Thus, it remains unclear how chromosome misalignment per se contributes to defects in chromosome copy number, development, and disease. New experimental models are, therefore, needed to address the functional importance of chromosome alignment to cellular and organismal physiology.
In mammalian cells, metaphase alignment requires the confinement of bioriented chromosome pairs to the spindle equator region. While the majority of chromosome pairs are located near the center of the spindle at the start of mitosis, some must be transported to the equator through a process called congression (Kapoor et al., 2006; Magidson et al., 2011). Paired chromosomes establish end-on attachments to microtubules emanating from opposite spindle poles via kinetochores, which assemble at the centromeric region of each chromosome. These bioriented chromosomes undergo microtubule-driven, oscillatory movements that initially permit excursions away from the equator (Skibbens et al., 1993). Therefore, the alignment of bioriented chromosomes requires mechanisms that regulate kinetochore-attached microtubules in a way that dampens these oscillations and limits them to an area around the spindle center.
Congression, biorientation, and chromosome confinement rely on kinesin-dependent mechanisms. CENP-E (kinesin-7) transports mono-oriented chromosomes to the spindle equator and works synergistically with KIF22 (kinesin-10) to promote the biorientation of chromosome pairs (Schaar et al., 1997; Kapoor et al., 2006; Barisic et al., 2014; Drpic et al., 2015). Loss of CENP-E or KIF22 function leads to chromosome segregation defects both in vitro and in vivo (Weaver et al., 2003; Ohsugi et al., 2008). However, the majority of chromosomes are able to align in cells lacking either CENP-E or KIF22 (Schaar et al., 1997; Levesque and Compton, 2001; Putkey et al., 2002), and the presence of attachment defects under these conditions complicates determination of the primary problem underlying chromosome segregation errors. Another kinesin motor, KIF18A (kinesin-8), is primarily responsible for the confinement of chromosome movements during metaphase (Zhu et al., 2005; Mayr et al., 2007). KIF18A concentrates at the plus ends of kinetochore microtubules and functions to reduce chromosome movements through direct suppression of kinetochore microtubule dynamics (Stumpff et al., 2008, 2012). Therefore, loss of KIF18A disrupts the alignment of all chromosomes.
Unlike CENP-E and KIF22, a role for KIF18A in promoting proper kinetochore microtubule attachments is cell type specific. Germ cells, as well as some genomically unstable tumor cell lines, require KIF18A function to satisfy the spindle assembly checkpoint and promote the metaphase to anaphase transition (Zhu et al., 2005; Mayr et al., 2007; Czechanski et al., 2015). These data suggest KIF18A has a role in establishing or maintaining kinetochore microtubule attachments. In contrast, primary mouse embryonic fibroblasts (MEFs) lacking KIF18A function progress through mitosis with normal timing, despite failing to align chromosomes (Czechanski et al., 2015). Thus, KIF18A’s alignment and attachment functions appear to be separable. Accordingly, Kif18a mutant mice survive to adulthood, although at slightly lower than the expected Mendelian ratio (Reinholdt et al., 2006; Czechanski et al., 2015). Collectively, these data implicate KIF18A-deficient somatic cells as a useful model system to determine the consequences of division with unaligned, but correctly attached, chromosomes.
Here we show that mitotic cell division in the absence of chromosome alignment does not significantly alter chromosome copy number. Instead, chromosome alignment is required for interchromosomal compaction during anaphase and the organization of chromosomes into a single nucleus at the completion of cell division. These defects reduce cellular proliferation and are associated with slow growth and subviability in mice. Our results define the physiological role of chromosome alignment independent of chromosome attachment, highlighting the importance of metaphase chromosome organization for proper nuclear envelope templating and proliferation in the next cellular generation.
Results
Human somatic cells deficient for KIF18A function divide in the absence of chromosome alignment
To determine if chromosome alignment is required for cell division in normal human cells, we tested the effects of KIF18A depletion on mitotic chromosome organization and progression through mitosis in a human retinal pigment epithelial cell line (RPE1) immortalized by human telomerase expression (hTERT). hTERT-RPE1 cells were derived from a female and are near diploid, containing a modal chromosome number of 46 with a single derivative X chromosome. These cells have a robust spindle assembly checkpoint response and display chromosome segregation errors under conditions that promote abnormal kinetochore microtubule attachments (Thompson and Compton, 2008; Uetake and Sluder, 2010).
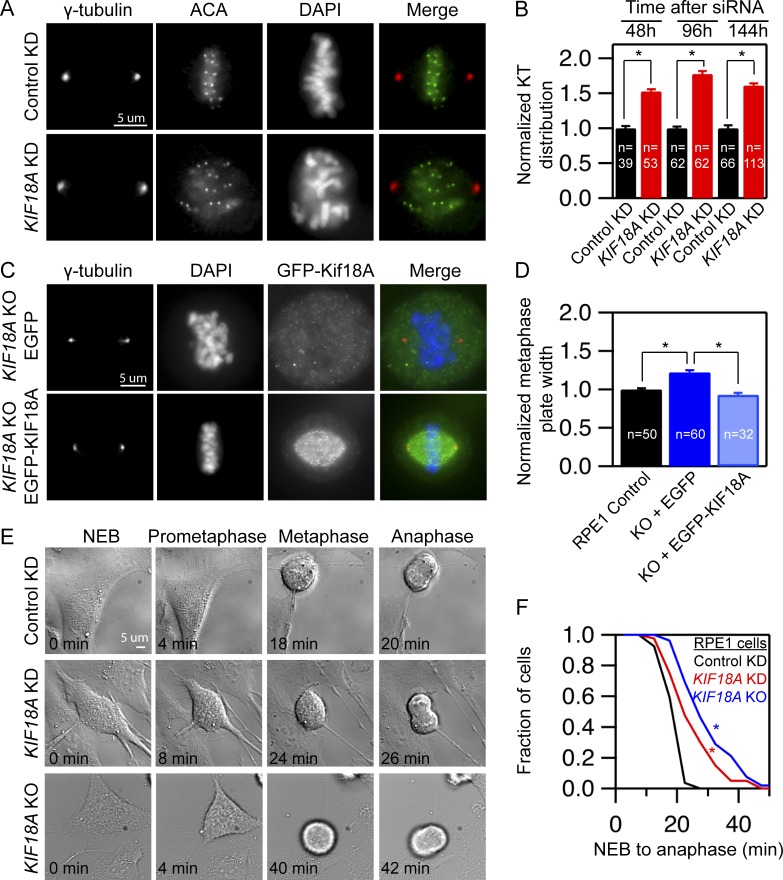
To determine the effects of KIF18A depletion on chromosome alignment in normal human somatic cells, we treated hTERT-RPE1 cells with control or KIF18A-specific siRNAs and then fixed and stained for kinetochores and centrosomes. Chromosome alignment was quantified by measuring the distribution of kinetochores along the spindle axis, as previously described (Stumpff et al., 2012; Fonseca and Stumpff, 2016). The distribution of kinetochores within the spindle was significantly increased in hTERT-RPE1 cells 48, 96, and 144 h after KIF18A knockdown (KD; Fig. 1, A and B). To achieve complete ablation of KIF18A function, we used CRISPR-Cas9 genomic editing to generate a homozygous KIF18A knockout (KO) hTERT-RPE1 line. Using antibodies that recognize either the C terminus or the motor domain of KIF18A, we confirmed that these KIF18A KO cells have undetectable KIF18A protein (Fig. S1). We found that the null cells also display chromosome alignment defects similar to those seen in KIF18A KD cells. Importantly, expression of EGFP-KIF18A, but not EGFP alone, rescues chromosome alignment in KIF18A KO cells (Fig. 1, C and D). Thus, the chromosome alignment defects in KIF18A KO cells are due specifically to the loss of KIF18A activity.
Figure 1.
Human retinal pigment epithelial cells lacking KIF18A function progress through mitosis with unaligned chromosomes. (A) Representative images of centrosomes and kinetochores in fixed hTERT-RPE1 cells treated with the indicated siRNAs. (B) Plot of kinetochore (KT) distribution at the indicated times following siRNA treatment measured using the FWHM of kinetochore fluorescence along the pole-to-pole axis. Data are from two (48 and 96 h) or three independent experiments (144 h). Mean ± SEM is shown. (C) Images of KIF18A KO cells transiently expressing EGFP or EGFP-KIF18A. Cells were fixed and stained for γ-tubulin and DNA. (D) Plot of metaphase plate width in control and KIF18A KO cells expressing GFP or GFP-KIF18A. Plate width was determined by measuring FWHM of DAPI fluorescence along the pole-to-pole axis. Data were collected from two independent experiments. Mean ± SEM is shown. (E) Stills from time-lapse DIC imaging of hTERT-RPE1 cells from the indicated treatment groups. (F) Cumulative frequency plot of time from nuclear envelope breakdown (NEB) to anaphase onset. n = 27 (control), n = 40 (KIF18A KD), and n = 52 (KIF18A KO). Data were from four independent experiments. All statistical comparisons were made using a Kruskal–Wallis one-way ANOVA with Dunn’s multiple comparisons test; *, P < 0.01.
Live cell imaging was used to directly determine whether hTERT-RPE1 cells require KIF18A for progression through mitosis. In contrast to the long, spindle assembly checkpoint-dependent mitotic arrest displayed by KIF18A-depleted HeLa cells (Zhu et al., 2005; Mayr et al., 2007; Stumpff et al., 2008), the time from nuclear envelope breakdown to anaphase is moderately extended from a mean of 20.0 ± 3.3 min in control cells to 25.6 ± 8.2 and 31.0 ± 10.5 min in KIF18A KD and KO hTERT-RPE1 cells, respectively (Fig. 1, E and F). Consistent with these results, a larger fraction of asynchronously dividing KIF18A KD than control KD hTERT-RPE1 cells contained kinetochores positive for MAD1 (Fig. S2, A and B), a spindle assembly checkpoint protein that localizes to unattached or incompletely attached kinetochores (Magidson et al., 2016; Kuhn and Dumont, 2017). However, the number of MAD1-positive kinetochores per cell was similarly low in the control and KIF18A KD populations, and the fraction of cells with MAD1-positive kinetochores was dramatically reduced following metaphase arrest with MG132 (Fig. S2, B and C). Taken together, these data indicate that hTERT-RPE1 cells lacking KIF18A function are able to establish kinetochore microtubule attachments and complete cell division in the absence of chromosome alignment, albeit with reduced efficiency. Primary MEFs derived from Kif18Agcd2/gcd2 mutant mice, which carry a mutation that completely inactivates KIF18A, behave similarly but do not display a detectable mitotic delay compared with WT MEFs (Czechanski et al., 2015). These features permitted us to use KIF18A loss of function cells from mouse and human as tools to determine the consequences of undergoing cell division without mitotic chromosome alignment.
Chromosome alignment is largely dispensable for equal chromosome segregation
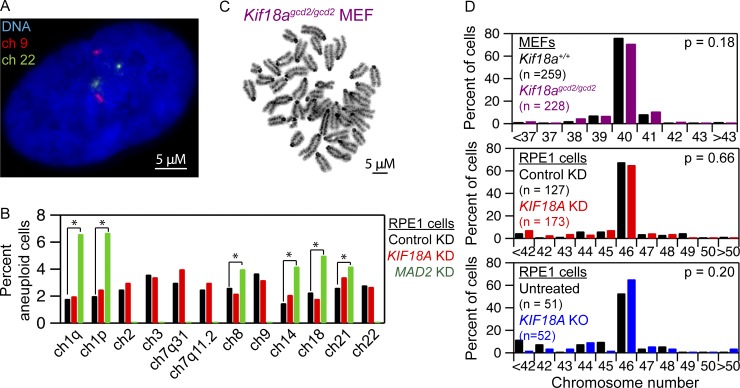
To determine whether chromosome alignment is required to maintain proper chromosome copy number, we analyzed hTERT-RPE1 cells treated with KIF18A siRNAs for 6 d using chromosome-specific fluorescence in situ hybridization (FISH) probes. hTERT-RPE1 cells display a doubling time of 14–24 h (Uetake and Sluder, 2004) and, therefore, are expected to complete approximately six divisions in the absence of chromosome alignment during the treatment period. KIF18A KD did not significantly alter the copy number of the 10 chromosomes we analyzed when compared with control siRNA-treated cells (Fig. 2, A and B; and Table S1). As a positive control, we analyzed chromosome copy number in hTERT-RPE1 cells treated with siRNAs targeting the spindle assembly checkpoint protein MAD2. Depletion of MAD2 promotes premature anaphase chromosome segregation before chromosome alignment in the presence of abnormal kinetochore microtubule attachments, which lead to chromosome segregation errors (Michel et al., 2001; Canman et al., 2002; Meraldi et al., 2004). In contrast to KIF18A KD cells, MAD2 KD cells were aneuploid for all chromosomes tested (Fig. 2 B and Table S1), consistent with previous reports (Michel et al., 2001). We also measured the total number of chromosomes present in mitotic KIF18A KD and KIF18A KO RPE1 cells, as well as primary embryonic fibroblasts from Kif18Agcd2/gcd2 mice. In all cases, chromosome numbers were comparable to those found in matched control cells (Fig. 2, C and D).
Figure 2.
Loss of KIF18A function does not alter chromosome copy number. (A) Image of an hTERT-RPE1 cell with chromosomes 9 (red) and 22 (green) labeled by FISH. (B) Plot of the percentage of cells aneuploid for the indicated chromosomes following treatment with control siRNA, KIF18A siRNA, or MAD2 siRNA. n = 1,500 cells for each condition from three independent experiments; *, P < 0.05 based on χ2 analyses. The effect of MAD2 KD on chromosomes 2, 3, 7, or 9 was not determined. (C) Image of Geimsa stained metaphase spread of an early passage (p0) MEF. (D) Quantification of metaphase chromosome numbers from WT and Kif18agcd2/gcd2, early passage (p0), pair-matched MEFs (top); hTERT-RPE1 cells treated with control or KIF18A siRNAs (middle); and control and KIF18A KO RPE1 cells (bottom). Indicated P values were calculated by χ2 analyses. Data were collected from three independent experiments for Kif18agcd2/gcd2 MEFs and siRNA-treated cells. Data were collected from two independent experiments for KIF18A KO hTERT-RPE1 cells.
To test the possibility that aneuploid KIF18A-deficient cells were being eliminated from the population via apoptosis, we measured the percentage of KIF18A KD cells positive for cleaved caspase-3, an apoptotic marker. KIF18A KD cells did not display an increase in cleaved caspase-3 relative to control treated cells 48 or 144 h after siRNA treatment (Fig. S3). Furthermore, our previous analyses of apoptosis in MEFs from Kif18Agcd2/gcd2 mice indicated no increase in TUNEL-positive cells compared with WT MEFs (Czechanski et al., 2015). Taken together, these data indicate that chromosome alignment per se is largely dispensable for the maintenance of euploidy and that abnormal kinetochore microtubule attachments likely underlie segregation errors in MAD2-depleted cells.
Loss of KIF18A leads to nuclear organization defects in interphase cells
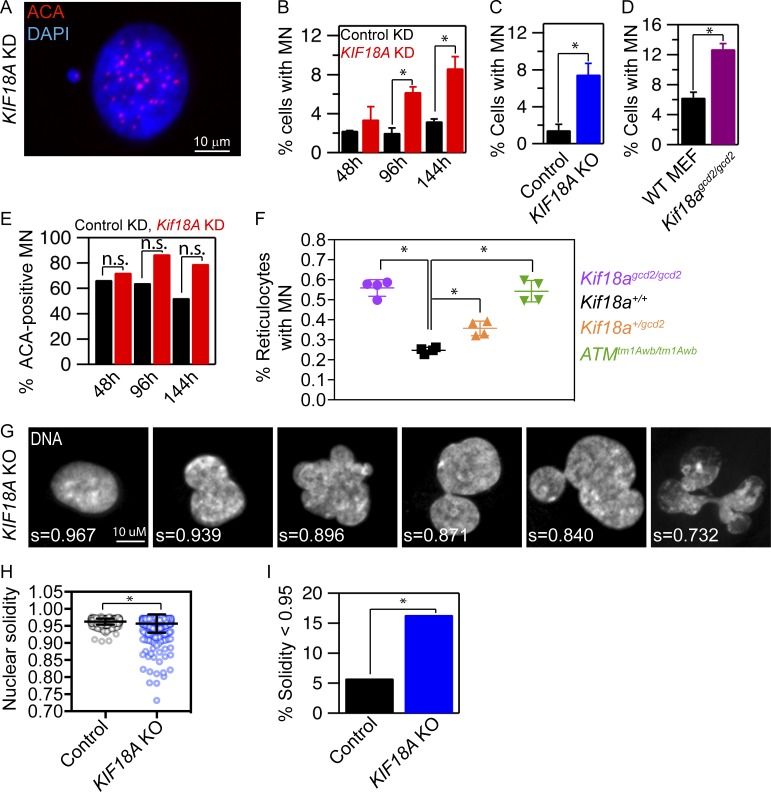
Despite the lack of evidence for chromosome copy number changes, we observed that KIF18A-deficient interphase cells display defects in nuclear organization. Loss of KIF18A function leads to an increase in cells with micronuclei (MN; Fig. 3 A). The percentage of KIF18A KD hTERT-RPE1 cells with MN increases with time following siRNA treatment (Fig. 3 B). Similarly, both KIF18A KO hTERT-RPE1 cells and Kif18Agcd2/gcd2 MEFs display a significant increase in MN compared with control cells (Fig. 3, C and D). The majority of these MN (∼70–85%) are positive for centromeres, consistent with most containing whole chromosomes (Fig. 3 E). KIF18A deficiency also leads to the formation of MN in vivo. The percentage of micronucleated reticulocytes is significantly increased in both Kif18Agcd2/gcd2 and in Kif18Agcd2/+ mice relative to controls in an additive manner (Fig. 3 F). The level of micronucleated reticulocytes in Kif18Agcd2/gcd2 mice is comparable to that of mice with genomic instability due to deficient DNA double strand break repair (ataxia telangiectasia-mutated ATMtm1Awb/tm1Awb; Fig. 3 F). These results indicate that KIF18A-dependent chromosome alignment is required to prevent MN formation and that a single functional allele of Kif18a is not sufficient for normal genetic stability.
Figure 3.
KIF18A-deficient cells form MN and abnormal nuclear shapes. (A) Representative image of a micronucleated hTERT-RPE1 cell labeled with DAPI and ACA to visualize DNA and centromeres, respectively. (B–D) Plots of the percentage of cells with MN in cells treated with the indicated siRNAs for 48, 96, or 144 h (B), control and KIF18A KO hTERT-RPE1 cells (C), and MEF from Kif18agcd2/gcd2 mice (D). n > 600 cells for each condition; data were compared via χ2 analyses. (E) Quantification of the percentage of MN containing centromeric DNA (ACA-positive) in control (black) and KIF18A siRNA (red) treated cells; data were compared via χ2 analyses. (F) Quantification of MN in mouse peripheral blood reticulocytes from Kif18a+/+, Kif18agcd2/gcd2, Kif18a+/gcd2, and Atmtm1Awb/tm1Awb. Data points represent the percentage of micronucleated cells from individual mice. Data were compared using a one-way ANOVA and Tukey’s multiple comparisons test. (G) Representative images of nuclear shapes and corresponding solidity values (s) observed in KIF18A KO hTERT-RPE1 cells. (H) Box and whisker plot of nuclear solidity values measured in control (n = 553) and KIF18A KO (n = 634) cells. Data distributions were compared using a Kolmogorov–Smirnov t test. (I) Plot of percentage of nuclei with solidity values two SDs below the average in control cells. Data were compared using a χ2 test. In all panels, *, P < 0.01. All data were collected from three independent experiments, and error bars indicate SD.
In addition to MN, KIF18A-deficient cells contain abnormally shaped primary nuclei with a lobed appearance (Fig. 3 G). To quantify this phenotype, we measured the solidity of DAPI-stained nuclei in control and KIF18A KO hTERT-RPE1 cells. KIF18A KO cells displayed a significant increase in the percentage of primary nuclei with solidity values that differed by more than two SDs from the mean solidity of control cells (Fig. 3, G–I). Thus, loss of KIF18A activity disrupts the normal convex shape of primary nuclei in hTERT-RPE1 cells.
MN and abnormal nuclear shapes form as KIF18A-deficient cells exit mitosis
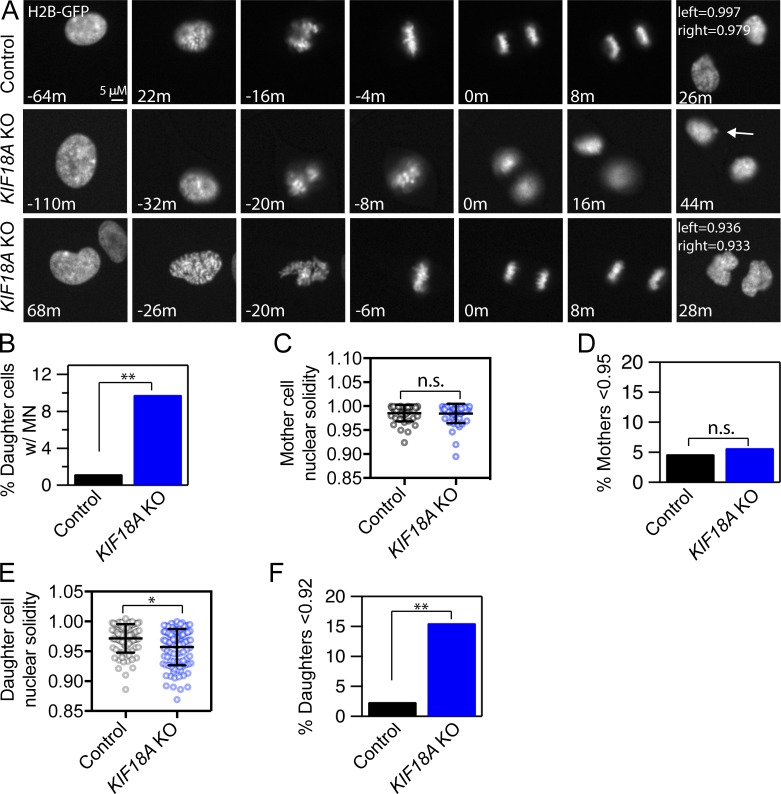
To determine if the interphase nuclear organization abnormalities observed in KIF18A-deficient cells result from mitotic defects, we analyzed when MN and lobed primary nuclei form relative to cell division in live KIF18A KO cells expressing H2B-GFP (Fig. 4; Fig. S4; and Videos 1, 2, and 3). We found that 9.8% of daughter cells formed MN as chromatin decondensation occurred at the end of mitosis in KIF18A KO cells compared with 1.2% in hTERT-RPE1 control cells (Fig. 4, A and B; and Table S2). This percentage is comparable to the fraction of cells with MN observed in fixed KIF18A KO cells (Fig. 3 C). Live KIF18A KO cells also exhibited abnormal nuclear shapes. Interestingly, the shape of primary nuclei in cells entering mitosis (mother cells) was comparable between KIF18A KO and control cells, suggesting that cells with abnormal nuclear shapes do not readily divide (Fig. 4, C and D; and Fig. S4 B). Despite entering mitosis with similar nuclear morphologies, the daughter cells resulting from KIF18A KO divisions formed a significantly higher fraction of abnormal nuclei than control cells during mitotic exit (Fig. 4, E and F). The percentage of daughters with abnormal nuclear shapes is strikingly similar to the percentage of cells with abnormal nuclear shapes observed in fixed samples (Fig. 3, H and I). These data strongly suggest that mitotic chromosome alignment defects underlie the majority of the nuclear organization abnormalities observed in interphase cells lacking KIF18A function.
Figure 4.
MN and abnormal nuclear shapes form as KIF18A-deficient cells exit mitosis. (A) Representative stills from time-lapse images of control and KIF18A KO hTERT-RPE1 cells expressing histone H2B-GFP. Note that MN (arrow, middle row) and lobed primary nuclei (bottom row) form as KIF18A KO cells exit mitosis. (B) Plot of the percentage of daughter cells that form MN during mitotic exit. Data were compared via χ2 test; **, P < 0.01. (C) Quantification of mother cell nuclear solidity measured 60 min before metaphase. Bars indicate mean and SD. Data were compared via Kolmogorov–Smirnov t test; P > 0.90. (D) Percentage of mother cell nuclear solidity values less than two SDs from the average control solidity. Data were compared via χ2 test; P > 0.90. (E) Quantification of daughter cell nuclear solidity 20 min after initial chromatin decondensation. Bars indicate mean and SD. Data were compared via Kolmogorov–Smirnov t test; *, P < 0.05. (F) Percentage of daughter cell nuclear solidity values two SDs below the average control solidity. Data were compared using χ2 tests; **, P < 0.01. All data were collected from three independent experiments.
Disruption of chromosome alignment leads to interchromosomal compaction defects in anaphase and abnormal lamin distribution in telophase
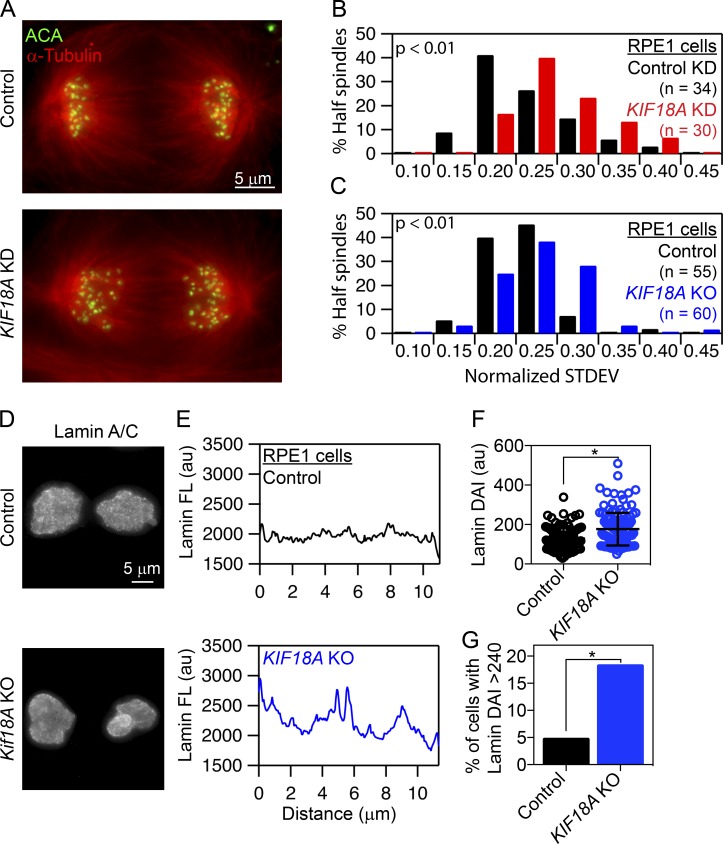
To gain an understanding of how nuclear organization problems result from mitotic defects in KIF18A-deficient cells, we compared chromosome segregation and nuclear envelope reformation in the presence and absence of KIF18A function. These analyses revealed that anaphase chromosomes are more broadly distributed in both KIF18A KO and KIF18A KD cells compared with controls (Fig. 5, A–C). Chromosome distributions during anaphase were quantified by measuring the SD of centrosome-to-kinetochore distances within each half spindle. KIF18A KD and KIF18A KO hTERT-RPE1 cells display a greater variance in pole-to-kinetochore distances than control cells (Fig. 5, B and C). These data indicate that KIF18A is required for interchromosomal compaction during anaphase.
Figure 5.
Loss of KIF18A function and chromosome alignment disrupts interchromosomal compaction during anaphase and Lamin A/C distribution during telophase. (A) Representative images of anaphase cells fixed and stained for α-tubulin (red) and centromeres (ACA; green). (B and C) Histograms of centromere to pole distance variance (calculated as SD) among all centromeres within a half spindle of control and KIF18A siRNA-treated hTERT-RPE1 cells (B) or control and KIF18A KO hTERT-RPE1 cells (C). Data were compared using a Kolmogorov–Smirnov t test; P < 0.01. (D) Representative images of control and KIF18A KO telophase cells labeled with Lamin A/C antibodies. (E) Plots of Lamin A/C fluorescence profiles along the long axis of telophase nuclei from control and KIF18A KO hTERT-RPE1 cells. (F) Box and whisker plot of Lamin A/C fluorescence variance in telophase nuclei calculated as the DAI in control and KIF18A KO cells. Bars indicate mean and SD. Distributions of DAI were compared using a Kolmogorov–Smirnov t test; P < 0.005. (G) Percentage of telophase cells with a DAI >2 SDs above the average control DAI. Data were compared using a χ2 test. *, P < 0.005 All data were collected from three independent experiments.
Defects in interchromosomal compaction are predicted to disrupt proper nuclear envelope templating during telophase (Mora-Bermúdez et al., 2007). To test this, we assayed the distribution of lamins in fixed telophase cells. The deviation from the average fluorescence intensity (DAI) along the long axis of each DNA mass in cells immunofluorescently labeled for Lamin A/C was determined (Fig. 5, D and E). KIF18A KO cells showed a significantly increased variance in Lamin A/C signal along the chromosome mass compared with controls, and the fraction of cells displaying a variance in lamin fluorescence more than two SDs higher than the mean in control cells was increased by threefold in the KIF18A KO population (Fig. 5, F and G). These data suggest that lamin is abnormally distributed during nuclear envelope reformation in the absence of KIF18A.
Lagging chromosomes in KIF18A-depleted cells travel longer distances in anaphase
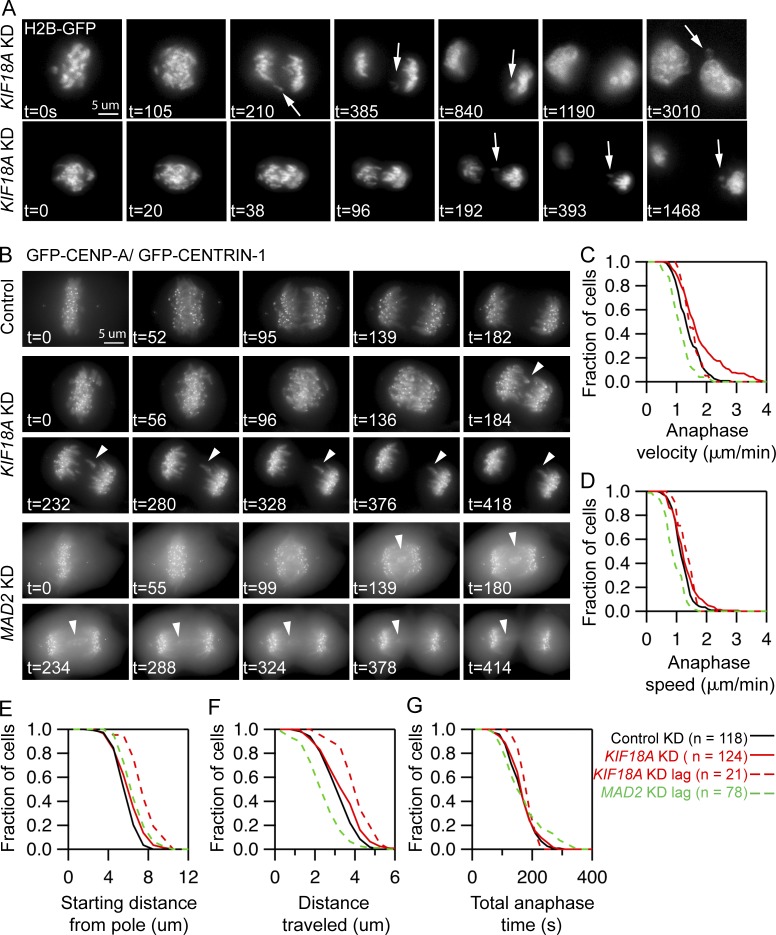
To determine the basis of interchromosomal compaction and nuclear organization defects in KIF18A-deficient cells, we imaged chromosome and kinetochore dynamics during mitosis with high temporal resolution. Analyses of H2B-GFP expressing anaphase cells confirmed interchromosomal compaction defects during anaphase after KIF18A KD (Fig. 6 A). Of 22 cells imaged from metaphase to telophase, 82% displayed obvious anaphase compaction defects (18 cells), and 68% had visible lagging chromosomes (28 total lagging chromosomes in 15 cells). In 11.4% of daughter cells formed from Kif18A KD cell divisions (5 of 44), we observed that a lagging chromosome was excluded from the primary nucleus and formed an MN, whereas lagging chromosomes integrated into the primary nucleus in all other cases (23 of 28 lagging chromosomes, 82%; Fig. 6 A). These data suggest that in the absence of KIF18A function, MN form around lagging mitotic chromosomes that arrive late to the poles during anaphase.
Figure 6.
In the absence of chromosome alignment, MN form around lagging chromosomes that travel long distances during anaphase. (A) Stills from time-lapse imaging of KIF18A-depleted cells expressing histone H2B-GFP. Arrows indicate lagging chromosomes that are excluded from the primary DNA mass and form MN. Data were collected from four independent experiments. (B) Representative images of live cells stably expressing GFP-CENP-A and GFP-CENTRIN-1 treated with control, KIF18A, or MAD2 siRNAs. Arrowheads indicate lagging chromosomes. (C–G) Survival plots of poleward anaphase velocity (µm/min; C), poleward anaphase speed (D), starting distance from the pole (E), distance traveled (F), and total anaphase time (G) for kinetochores in each experimental condition indicated. Dashed lines indicate the behavior of lagging chromosomes in KIF18A and MAD2 siRNA-treated cells. Data were compared using a Kruskal–Wallis test with post hoc Dunn’s multiple comparison tests. The starting distance from the pole and distance traveled for lagging chromosomes in KIF18A siRNA cells were significantly different than those in control siRNA cells or total kinetochores in KIF18A KD cells (P < 0.01). The anaphase velocity, speed, and distance traveled for lagging chromosomes in MAD2 KD cells are significantly different than those of kinetochores in control siRNA cells (P < 0.01). Data were collected from three independent experiments.
To understand the underlying defect causing lagging chromosomes in KIF18A-deficient cells, we tracked the movements of fluorescently labeled kinetochores in KIF18A and control siRNA-treated cells stably expressing EGFP-CENP-A and EGFP-Centrin-1 (Fig. 6 B; Magidson et al., 2011). These studies revealed that lagging chromosomes in KIF18A KD cells moved to the poles with normal speeds and velocities but began anaphase at significantly increased distances from the pole and, therefore, moved longer distances during anaphase (Fig. 6, C–F). However, the time that chromosomes were moving to the poles during anaphase was not changed in KIF18A KD cells relative to controls. In contrast, we found that lagging kinetochores in MAD2 KD cells move significantly slower but travel similar distances to kinetochores in control cells (Fig. 6, C–F). The slow movements of lagging chromosomes in MAD2 KD cells are likely due to merotelic attachments that are not corrected before the chromosomes prematurely segregate (Cimini et al., 2003). The differences between the movements of lagging chromosomes in KIF18A KD and MAD2 KD cells suggest that merotelic attachments do not significantly contribute to the defects in KIF18A KD cells. This conclusion is further supported by the low percentage of late anaphase KIF18A KD cells with midzone lagging chromosomes (2.4%, n = 286 cells), which is comparable to the number observed in late anaphase control cells (1.0%, n = 297 cells, P = 0.18). Late anaphase lagging chromosomes result from severe merotelic attachments (Cimini et al., 2004). Taken together, our data suggest that chromosome alignment defects in KIF18A KD cells abnormally position chromosomes at anaphase onset, in turn leading to interchromosomal compaction defects and lagging chromosomes during segregation.
A p53-dependent checkpoint reduces division of micronucleated KIF18A-deficient cells
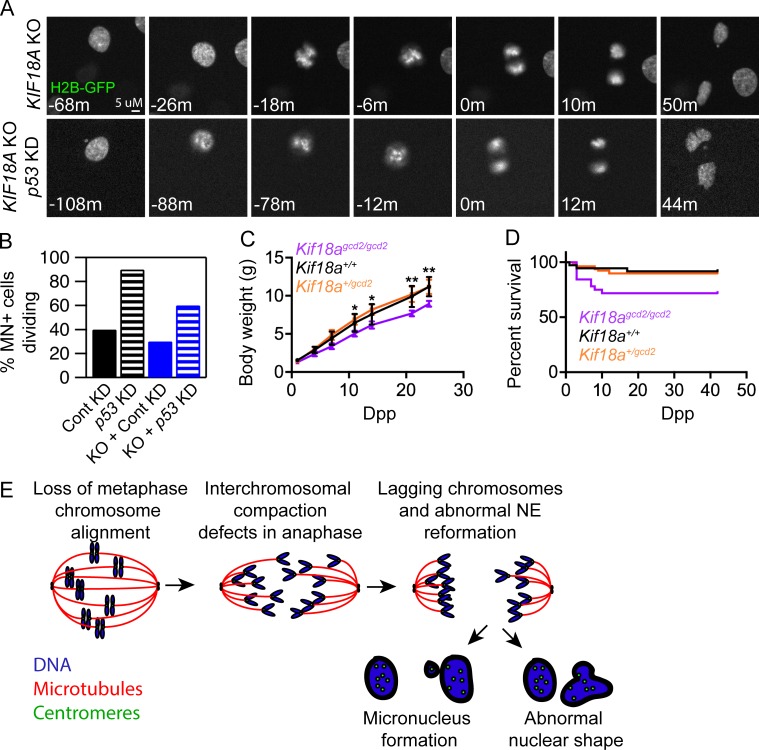
MN have been identified as markers of chromosome instability in tumors, and their presence can lead to chromothripsis, a phenomenon involving extensive structural rearrangements within a single chromosome (Crasta et al., 2012; Zhang et al., 2015). Interestingly, Kif18a mutant mice form MN in vivo (Fig. 3 F) but have been reported to be tumor resistant rather than predisposed to tumor formation (Zhang et al., 2010; Nagahara et al., 2011; Zhu et al., 2013). This raises the question of whether KIF18A-deficient cells with MN are able to continue to proliferate. We used long-term, live cell imaging to measure the rate of cell division for micronucleated cells in KIF18A KO and control hTERT-RPE1 populations. The percentage of micronucleated cell divisions was calculated as a function of the fraction of micronucleated cells in the population. These analyses revealed that micronucleated hTERT-RPE1 control and KIF18A KO cells display reduced division rates of 37.5% and 32.9%, respectively (Fig. 7, A and B; and Table S2). These division rates are consistent with those measured following induction of MN and chromosome segregation errors via treatments that do not cause mitotic arrest (Soto et al., 2017). Previous studies have indicated that MN formation can lead to cell cycle arrest via a p53-dependent checkpoint (Sablina et al., 1998; Thompson and Compton, 2010). Similarly, we found that the rate of division for micronucleated control and KIF18A KO cells increases twofold to threefold when p53 is depleted by siRNA treatment (Fig. 7 B and Table S2). These data indicate that MN limit proliferation in KIF18A KO cells, at least in part, through a p53-dependent mechanism.
Figure 7.
A p53-dependent mechanism limits the division of micronucleated KIF18A KO cells. (A) Still frames from time-lapse analyses of dividing histone 2B-GFP (H2B-GFP) expressing KIF18A KO cells and KIF18A KO cells treated with p53 siRNAs. (B) Plot of the percent of micronucleated cells that enter mitosis in control, p53 KD, KIF18A KO, or KIF18A KO + p53 KD hTERT-RPE1 cells. (C) Plot of body weights measured at the indicated dpp for each genotype listed. Error bars indicate SD. (D) Survival plot for mice of the indicated genotypes as a function of dpp. (E) Model for abnormal nuclear formation in the absence of chromosome alignment (see Discussion section for details). Data for all analyses were collected from three independent experiments.
Kif18A mutant mice display reduced growth rates and increased prewean mortality
Consistent with the reduction in proliferation observed in KIF18A KO hTERT-RPE1 cells, we previously found that MEFs derived from Kif18Agcd2/gcd2 mice grow more slowly than those from WT or Kif18Agcd2/+ mice, despite progressing through mitosis with normal timing (Czechanski et al., 2015). Furthermore, the percentage of homozygous mutant mice found in F2 litters (18%) was slightly below the expected Mendelian ratio, indicating lethality before 4 wk of age (Czechanski et al., 2015). To determine the timing of lethality and to assess growth, body weight was measured at regular intervals before 24 d postpartum (dpp). The body weights and growth rate of Kif18Agcd2/gcd2 mice were significantly reduced compared with either WT or Kif18Agcd2/+ littermates (Fig. 7 C). Approximately 30% of Kif18Agcd2/gcd2 mice also exhibited increased mortality during this time period, as predicted by our previous observations (Fig. 7 D; Czechanski et al., 2015). These phenotypes indicate that Kif18a is required for normal growth and viability during postnatal development.
Discussion
Chromosome compaction during anaphase is required to limit the frequency of lagging chromosomes and to promote formation of a single nucleus around all chromosomes at the end of cell division (Mora-Bermúdez et al., 2007; Ohsugi et al., 2008). This involves both the clustering of chromosomes together (interchromosomal compaction) and axial shortening of individual chromosome arms (intrachromosomal compaction). While mechanisms contributing to intrachromosomal compaction have been reported (Mora-Bermúdez et al., 2007; Ohsugi et al., 2008), the molecular control of interchromosomal compaction is less understood. The synchronous movement of anaphase chromosomes depends on mechanisms that equally distribute poleward forces across kinetochores (Matos et al., 2009). Our studies indicate anaphase chromosome clustering and the formation of a single nucleus in each daughter cell at mitotic exit also require the function of KIF18A. We propose that KIF18A-dependent metaphase chromosome alignment equalizes the distances that chromosomes must travel to reach the poles. This is consistent with the idea that the conserved equatorial positioning of metaphase chromosomes promotes synchronous anaphase segregation (Matos and Maiato, 2011; Pereira and Maiato, 2012; Maiato et al., 2017) and may preserve interphase chromosome organization (Gerlich et al., 2003). The model predicts that in the absence of metaphase chromosome alignment, chromosomes are not organized into a compact mass during anaphase, leading to improper templating of nuclear envelope components during telophase and the formation of abnormal nuclei at the completion of division (Fig. 7 E).
We observed that the vast majority of chromosome alignment–defective cells displayed lagging chromosomes and interchromosomal compaction defects, yet a much smaller fraction of daughter cells formed MN or abnormally shaped primary nuclei. The presence of mechanisms that spatially coordinate nuclear envelope formation with chromosome segregation can explain this discrepancy. For example, the assembly of noncore nuclear envelope components is prevented until chromosomes have segregated to the poles (Afonso et al., 2014; Liu et al., 2018). Most lagging chromosomes are able to integrate into the primary nucleus, reducing the frequency of MN formation compared with the frequency of lagging chromosomes. However, lagging chromosomes can still be excluded from the main nucleus even in the presence of late anaphase surveillance mechanisms, accounting for the low but consistent percentage of micronucleated cells we observed in the absence of chromosome alignment (Afonso et al., 2014).
Chromosome alignment per se does not appear to be required for proper kinetochore microtubule attachments in normal mammalian somatic cells. Despite severe disruption of chromosome alignment, KIF18A-deficient somatic cells from mouse and human progress through mitosis with relatively normal timing compared with the long mitotic delays observed in germ cells and HeLa cells lacking KIF18A function (Zhu et al., 2005; Mayr et al., 2007; Czechanski et al., 2015). These data suggest that chromosome alignment–defective MEFs and hTERT-RPE1 cells establish attachments that satisfy the spindle assembly checkpoint with kinetics similar to or slightly slower than control cells, respectively. Furthermore, the lagging chromosomes resulting from chromosome alignment defects displayed distinct behaviors compared with those that result from merotelic attachments, which typically segregate with reduced velocities and often stall at the midzone during anaphase (Cimini et al., 2004). These data strongly support the conclusion that chromosome alignment defects, rather than abnormal attachments, lead to nuclear formation problems in the absence of KIF18A function. Furthermore, while we cannot completely rule out that a small, stable population of KIF18A contributes directly to late mitotic functions that affect the organization of chromosomes into a single nucleus, the majority of KIF18A protein is rapidly degraded at anaphase onset (Sedgwick et al., 2013).
Our analyses of chromosome copy number also suggest that metaphase alignment may not be necessary for the equal segregation of chromosomes. However, it is surprising that a population of cells where ∼10% are micronucleated display no change in chromosome copy number. We believe this apparent discrepancy can be explained by sensitivity limitations of standard assays for chromosome copy number. If it is assumed that there is no bias in selecting which chromosomes form MN, any particular chromosome would be micronucleated in ∼0.4% of KIF18A-deficient cells (10% of cells with MN/23 chromosomes). This level of copy number change would not be detectable in our FISH analyses of primary nuclei. Furthermore, because micronucleated KIF18A-deficient cells display a low rate of division, any copy number defects in these cells are unlikely to impact our analyses of mitotic chromosome spreads. Thus, we acknowledge that there may be a low level of aneuploidy induced by loss of KIF18A function and chromosome alignment that was not detected in our assays. However, our data indicate that chromosomes are equally segregated in the majority of chromosome alignment–defective cells, supporting the conclusion that alignment is largely dispensable for maintaining chromosome copy number.
We found that the cellular defects resulting from loss of chromosome alignment are correlated with long-term consequences that negatively impact cell proliferation, early mammalian growth, and survival. At least a portion of micronucleated cells resulting from chromosome alignment defects undergoes a p53-dependent cell cycle arrest. This response is comparable to that of cells containing MN formed as a result of genomic instability, and since we did not observe a significant mitotic delay associated with loss of KIF18A function, this is suggestive of DNA damage (Sablina et al., 1998; Thompson and Compton, 2010; Santaguida et al., 2017; Soto et al., 2017). Cell cycle arrest could account for the reduced proliferation of Kif18agcd2/gcd2 MEFs and reduced growth of Kif18agcd2/gcd2 mice (Czechanski et al., 2015). Survival is also reduced in some Kif18agcd2/gcd2 mice, suggesting that loss of chromosome alignment may compromise viability. However, we cannot exclude that KIF18A is required to satisfy the spindle assembly checkpoint in essential cell lineages in developing mice, as seen in murine germ cells and some human cell lines (Mayr et al., 2007; Czechanski et al., 2015; Janssen et al., 2018), or that the motor has a role in microtubule-driven processes outside of mitosis that are required for mammalian development. For example, kinesin-binding protein has been shown to interact with and regulate the activity of a variety of kinesins, including KIF18A, in the context of neuronal microtubule dynamics (Kevenaar et al., 2016). While KIF18A-deficient mice do not display the overt phenotypes that are typically found in mice with disrupted axonal microtubule dynamics, such as progressive peripheral neuropathy, microcephaly, or ataxia, more work is needed to understand the underlying causes of death for the ∼30% of Kif18a mutant animals that are lost before wean (Hirst et al., 2017; Wilson et al., 2017).
We speculate that mice harboring defects in chromosome alignment may be sensitized to additional genetic or environmental factors that are detrimental to the mitotic/meiotic spindle or to genome integrity. In fact, the intermediate phenotypes we observed in mice and in cell lines heterozygous for Kif18a mutations show that even subtle defects in chromosome alignment contribute to semidominant phenotypes. Therefore, it will also be important to determine if Kif18a mutant mice are predisposed to aneuploidy and to explore long-term consequences of this in the context of stem cell exhaustion, tumorigenesis, and overall population fitness. Interestingly, Kif18a mutant mice are resistant to induced colorectal cancer, suggesting that like aneuploidy, MN may have complex effects on oncogenesis (Weaver et al., 2007; Silk et al., 2013; Sheltzer et al., 2017).
In summary, our data suggest that the organization of chromosomes at the spindle equator during metaphase primarily functions to cluster chromosomes for segregation and ensure the proper formation of a single nucleus in each daughter cell. In the absence of this function, nuclear organization defects, such as MN and abnormal nuclear shape, may lead to reduced postnatal mammalian growth and survival.
Materials and methods
Animal ethics statement
All procedures involving mice were approved by The Jackson Laboratory’s Institutional Animal Care and Use Committee and performed in accordance with the National Institutes of Health guidelines for the care and use of animals in research. A cryorecovery of CAST.129S1(B6)-Kif18agcd2/Jcs (RRID: MMRRC_034325-JAX; Czechanski et al., 2015) × C57BL/6J (RRID: IMSR_JAX:000664) was used to establish a breeding colony, which was maintained by sibling intercrossing.
Cell culture and transfections
hTERT-RPE-1 cells (ATCC) were maintained at 37°C with 5% CO2 in MEM-α (Life Technologies) containing 10% FBS (Life Technologies) and antibiotics. For time course experiments, cells were plated in a 6-well dish and transfected with 150 pmol of siRNAs using RNAiMAX (Life Technologies) following the manufacturer’s instructions. Fresh siRNAs were added every 48 h. Cells were then seeded on 25-mm coverslips (Electron Microscopy Sciences) for imaging. For other fixed cell assays, cells were seeded on 12-mm acid-washed coverslips and transfected with 30 pmol siRNA complexed with RNAiMAX. For live cell imaging, cells were seeded in 35-mm poly-L-lysine–coated glass bottom dishes (MatTek) 24 h before addition of siRNA. Plasmid transfections were performed using a Nucleofector 4D system (Lonza).
MEF derivation and culture
For derivation of MEFs, F3 embryos were harvested and euthanized at 12.5–14.5 d postcoitum (copulation plug = 0.5 d postcoitum). Tissue samples were collected for genotyping, and each embryo was processed individually as previously described to avoid cross-contamination between genotypes (Czechanski et al., 2014, 2015). Individual, eviscerated, decapitated embryos were washed in cold PBS, transferred to clean 100-mm dishes, macerated with forceps in 3–5 ml of 0.05% Trypsin/EDTA (Thermo Fisher, cat. no. 25300120), and then dissociated through 18G needles. Tissue suspensions were transferred to 15-ml conical tubes, and an equal volume of prewarmed 37°C, MEF medium (DMEM [Thermo Fisher, cat. no. 11960044], 10% FBS [Lonza, cat. no. 14-501F, lot no. 0000217266], 100 U/ml penicillin/streptomycin [penicillin-streptomycin 1,000 U/ml, Thermo Fisher, cat. no. 15140122], 1× Glutamax [Thermo Fisher, cat. no. 35050061], 0.2 µM filtered) was added. Large fragments were allowed to settle, and cell suspensions were transferred to clean 15-ml conical tubes and centrifuged for 5 min at 200 xg at room temperature. Cell pellets were resuspended in 5 ml of prewarmed, 37°C MEF media and plated onto clean 60-mm tissue culture dishes (P0). Cultures were incubated at 37°C, 5% CO2 for 2–4 d until 80% confluent. Cultures were then harvested in 0.05% trypsin/EDTA, aliquots were removed for genotype confirmation, and cultures were resuspended in freeze medium (10% DMSO/10% FBS/80%MEF) at 0.5–1 × 106 cells/ml, dispensed into 1-ml aliquots in cryovials, and then gradually frozen (−1°C/min) at −80°C overnight in CoolCell freezing containers. Frozen vials were then immersed in liquid nitrogen for storage. Tissue and cell samples were genotyped by The Jackson Laboratory Transgenic Genotyping Service using an endpoint assay designed to detect the R308K missense gcd2 mutation at Chr2: 109,908,059 G/T, GRCm38, mm10.
CRISPR targeting of KIF18A
A single guide RNA (sgRNA) guide sequence was designed against the 15th exon in the C terminus of Kif18A with the sequence 5′-CTAATGCCATCTCCCTTGAA(AGG)-3′. The PAM sequence is in parentheses. Primers were designed with this sequence and cloned into pCR-Blunt with gBLOCK through site-directed mutagenesis and confirmed by sequencing.
For transfection of the complex, ∼2 × 106 hTERT-RPE1 cells plated into a 60-mm dish with 3 ml MEM-α media (Thermo Fisher), supplemented with 10% FBS and 1% Penicillin/Streptomycin (Thermo Fisher), were treated with 1 µg pCR-Blunt sgRNA and 1 µg pX458 Cas9-GFP with 16 µl Lipofectamine LTX (Thermo Fisher), incubated for 20 min at room temperature in 250 µl OptiMeM (Thermo Fisher). Similar amounts of hTERT-RPE1 cells were also transfected as negative and positive controls for FACS: 2 µg pCR-Blunt sgRNA (neg) and 2 µg pMAX GFP (Lonza) each with 16 µl Lipofectamine LTX incubated for 20 min at room temperature in 250 µl OptiMeM. The treated hTERT-RPE1 cells were incubated at 37°C for 48 h.
The Cas9 + sgRNA–treated cells were then strained and sorted for positive GFP signal using a BD FACSAria cell sorter equipped with 488-nm Coherent Sapphire laser. Positive and negative controls were used to place bins for determining the cell population desired for sorting. 1 µg/ml DAPI was also included in the sample as a live/dead marker. Cells with high GFP signal and low DAPI signal were selected and automatically placed one cell per well into 96-well plates containing 100 µl MEM-α media supplemented with 20% FBS. Cell colonies that grew were subsequently tracked and replated into larger well sizes periodically over a period of ∼3 wk. From a 24-well size dish, a colony was divided equally into two wells of a 6-well dish. One well was used to continue growing the cell population; the other, at confluency, was lysed and the genome extracted using a Blood Mini Kit (Qiagen) following the manufacturer’s Appendix B protocol for cultured cells.
Cell genomes were screened by PCR amplification of the exon region of interest using genomic-specific primers designed with the BLAT bioinformatics design tool (University of California, Santa Cruz, Genome Bioinformatics). Forward primer is 5′-GTAATAAACTGGTCACTGACACCCAAACCC-3′; reverse primer is 5′-GGGTAATTTACACTTCGAGCTCTTGATGTCTTC-3′. The guide was designed with a unique restriction enzyme (BslI; New England Biolabs) cut site overlapping the Cas9 cleavage site, such that mutations to the genome would negate BslI cutting. PCR amplified genomes that did not cut were then sequenced using the forward primer above. The KIF18A-deficient CRISPR line used in this study has frame shifts in each allele, resulting in premature truncations at amino acid residues 675 and 673.
Plasmids and siRNAs
H2B-GFP was a gift from Geoff Wahl (The Salk Institute, La Jolla, CA; Addgene plasmid no. 11680). Cells were transfected with pools of siRNAs targeting the KIF18A sequences 5′-UCUCGAUUCUGGAACAAGCAG-3′ and 5′-CCACUUUAUGAAAUCCAGCTG-3′, the MAD2 sequences 5′-UAUUUUCCUCAUGUCAUCCTT-3′ and 5′-AGAUGGAUAUAUGCCACGCTT-3′, or scrambled negative control Silencer siRNA no. 1 (Life Technologies).
Generation of anti-KIF18A motor domain antibodies
To generate antibodies against the motor domain of KIF18A, nucleotides 1–1089 of the coding region (GenBank accession no. BC048347) were PCR amplified and inserted into the BamHI/EcoRI sites of pGEX6P-1 (GE Healthcare). GST-KIF18A-NT was expressed in and purified from BL-21(DE3) cells using glutathione agarose (Sigma-Aldrich) and used to immunize rabbits (Cocalico). KIF18A-specific antibodies were affinity purified by passing anti-GST-depleted serum over Affi-Gel 10 (Bio-Rad) covalently coupled to GST-KIF18A-NT. Affinity-purified antibodies were dialyzed into PBS, frozen in liquid nitrogen, and stored at −80°C.
Cell fixation and immunofluorescence
hTERT-RPE1 cells were fixed in −20°C methanol (Thermo Fisher), 1% paraformaldehyde (Electron Microscopy Sciences) in −20°C Methanol, 4% Paraformaldehyde in 1× TBS (2.6 mM potassium chloride, 24.7 mM Tris base, and 136.8 mM NaCl at pH 7.4), or 0.5% Glutaraldehyde (Electron Microscopy Sciences) in Microtubule Stabilization Buffer (1× BRB80, 4 mM EGTA, and 0.5% Triton X-100). Cells were then washed in 1× TBS and blocked in antibody dilution buffer (Abdil; tris buffered saline, pH 7.4, 1% bovine serum albumin, 0.1% Triton X-100, and 0.1% sodium azide) containing 20% goat serum. Cells were incubated with the following primary antibodies for 1 h at room temperature in Abdil: mouse anti-α tubulin at 1µg/ml (Sigma-Aldrich), mouse anti-γ-tubulin at 1 µg/ml (Sigma-Aldrich), rat anti-YL at 2 µg/ml (Millipore), mouse anti-human Lamin A/C at 1 µg/ml (Millipore), rabbit anti-cleaved caspase-3 at 1:100 (Cell Signaling), mouse anti-p53BP at 0.8 µg/ml (Santa Cruz Biotechnology), or rabbit anti-GFP at 8 µg/ml (Molecular Probes by Life Technologies). Cells were incubated with the following antibodies overnight at 4°C: human anti-centromere antibodies (ACA; Antibodies Inc.) at 2 µg/ml, rabbit anti-KIF18A (C-terminal) at 2 µg/ml (Bethyl Antibodies), and rabbit anti-KIF18A (N-terminal) at 3 µg/ml. Cells were incubated for 1 h at room temperature with goat secondary antibodies against mouse, rabbit, or human IgG conjugated to Alex Fluor 488, 594, or 647 (Molecular Probes by Life Technologies). Coverslips were mounted on glass slides with Prolong Gold antifade reagent plus DAPI (Molecular Probes by Life Technologies).
Fluorescent in situ hybridization and chromosome spreads
For fluorescent in situ hybridization and chromosome spread experiments, hTERT-RPE1 cells and MEFs were dissociated and placed directly in hypotonic solution (0.8% sodium citrate and 0.4% potassium chloride) for 20 min, prefixed in Carnoy’s fixative (3:1 Methanol:Glacial Acetic Acid), pelleted, resuspended in Carnoy’s fixative, and washed four times with Carnoy’s fixative. For chromosome spreads, cells were dropped onto a glass slide inside a Thermotron Drying Chamber set at 25°C and 37% humidity for optimal spreading. Slides were dried and heated to 65°C for 30 min (or overnight for chromosome banding) and either mounted with Prolong Gold containing DAPI or trypsin banded and Giemsa stained. For fluorescent in situ hybridization, slides were immersed in 2× SSC at room temperature for 2 min, then dehydrated in 70%, 85%, and 100% ethanol for 1 min each at room temperature and air dried. Commercial probes for locus specific regions 1p36, 1qter, 2 centromere, 3 centromere, 9q34, and 22q11.2 (Cytocell) and for locus specific regions 7q11.2, 7q31, 8q21, 21q22, 14q32, and 18q21(Abbott Molecular) were used. Probes were applied to samples, coverslipped, and rubber cemented. Slides and probes were codenatured at 73°C for 2 min and hybridized at 37°C overnight in a hybridization chamber (Thermo Fisher). Slides were then washed in 0.4× SSC/0.3%NP-40 for 2 min at 73°C, rinsed in 2× SSC/0.1% NP-40 for 1 min at room temperature, counterstained with DAPI (Abbott Molecular), and coverslipped.
Microscopy
Cells were imaged on a Nikon Ti-E inverted microscope (Nikon Instruments) controlled by NIS Elements software (Nikon Instruments) with a Spectra-X light engine (Lumencore), Clara cooled-CCD camera (Andor), 37°C environmental chamber, and the following Nikon objectives: Plan Apo 20× differential interference contrast (DIC) M N2 0.75 NA, Plan Apo 40× DIC M N2 0.95NA, Plan Apo λ 60× 1.42 NA, and APO 100× 1.49 NA.
Live cell imaging
Cells were transferred into CO2-independent media with 10% FBS and 1% penicillin and streptomycin (Gibco by Life Technologies) for imaging via fluorescence or DIC microscopy. For DIC imaging and long-term imaging of H2B-GFP expressing cells, single focal plane images were acquired at 2-min intervals with a 20× or 40× objective. For high–temporal resolution imaging of chromosomes, optical sections were collected with 1.2 µm spacing through the entire H2B-GFP mass every 10 s using a 60× objective. For imaging kinetochore movements in hTERT-RPE-1 cells stably expressing EGFP-CENP-A/EGFP-Centrin-1 (graciously gifted by Alexey Khodjakov, Department of Health, Wadsworth Center, Albany, NY) optical sections were collected at 1 µm spacing every 5 s using a 60× objective (Magidson et al., 2011).
Live kinetochore tracking
Maximum intensity projections of 3.6–6 µm were made from optical sections of siRNA-treated hTERT-RPE-1 cells stably expressing EGFP-CENP-A/EGFP-Centrin-1. Images were analyzed using MtrackJ in ImageJ. Centrin-1 was tracked throughout the movie from the end of metaphase through the end of anaphase B. CENP-A foci positions were tracked during poleward movement. Kinetochore positions were normalized relative to the Centrin-1 track. Each kinetochore track was opened in Igor, and kinetochore velocities were measured by averaging the slopes of regression lines fit to track data using a five-point sliding window from the beginning to the end of anaphase. Speed measurements include pauses and reversals, whereas velocity measurements do not. Pauses were defined as a stop in poleward kinetochore movement that occurred for 30 s or longer. Reversals were characterized by away-from-pole movement that lasted for at least 30 s.
MN counts
For fixed cells, MN counts were made using single focal plane images of DAPI stained cells. Image acquisition was started at a random site at the top edge of the coverslip, and images were acquired every two fields of view using a 40× objective. For live cell studies, the percentage of micronucleated cells was determined for each population by counting the number of cells with a preexisting MN in the first frame of each time-lapse data set. MN were defined as H2B-GFP foci spatially separated from the primary nucleus that migrated similarly to the primary nucleus of the cell. NIS Elements software (Nikon) was used to review the images and track MN dynamics, as well as nuclear lobing. The percentage of cells dividing with a preexisting MN was calculated as the percentage of divisions wherein a cell with a preexisting MN entered mitosis divided by the percentage of preexisting MN in that population (as estimated from the percentage of MN-containing cells present in the first frame of each movie).
Peripheral blood MN assay
MN assays of peripheral blood were conducted as previously described (Dertinger et al., 1996; Reinholdt et al., 2004). Peripheral blood was collected from the retro-orbital sinus of male and female laboratory mice, Mus musculus aged 12–18 wk, n = 5 for each Kif18a genotype, and n = 4 for ATM. Strains used were CAST.129S1(B6)-Kif18agcd2/Jcs (RRID: MMRRC_034325-JAX) × C57BL/6J (RRID: IMSR_JAX:000664) F2 and F3 homozygous mutant and littermate WT controls, as well as 129S6/SvEvTac-Atmtm1Awb/J (RRID: IMSR_JAX:002753; Barlow et al., 1996). 75 µl of blood was immediately mixed with 100 µl of heparin, and the mixture was then pipetted into 2 ml of ice-cold (−80°C) 100% methanol with vigorous agitation to prevent clumping. Samples were stored at −80°C overnight before processing for flow cytometry.
Sample preparation and flow cytometry
Each blood sample was removed from −80°C storage and washed with 12 ml of autoclaved, ice-cold bicarbonate buffer (0.9% NaCl and 5.3 mM sodium bicarbonate, pH 7.5), centrifuged at 500 xg for 5 min and resuspended in a minimum of carryover buffer (∼100 µl). 20 µl of each sample was added to a 5-ml polystyrene round-bottomed tube, and to each sample an 80-µl solution of CD71-FITC and RNaseA (1 mg/ml) was added. Additional control samples were CD71-FITC alone and an additional sample with bicarbonate buffer alone to which propidium iodide (PI) would be later added (see below). Cells were incubated at 4°C for 45 min, washed with 2 ml cold bicarbonate buffer, and centrifuged as above. Cell pellets were stored on ice and then, immediately before flow cytometric analysis, resuspended in 1 ml of ice-cold PI solution (1.25 mg/ml) to stain DNA.
Flow cytometry
Samples were processed on a BD Bioscience LSRII fluorescence-activated cell sorter gated for FITC and PI and set to collect 20,000 CD71-positive events at 5,000 events per second. The CD71-FITC and PI control samples were used to calibrate for autofluorescence. Reticulocytes (Retic, CD71+, PI− [in the presence of RNase A]), mature red blood cells (RBC, CD71−, PI−), micronucleated normochromatic erythrocytes (NCE-MN, CD71−, PI+), and micronucleated reticulocytes (Ret-MN, CD71+, PI+) were measured using FlowJo software. The total percent of spontaneous MN in NCE was NCE-MN/(NCE-MN + RBC) × 100. Genotyping for Kif18agcd2 was as described above. Genotyping for Atmtm1Awb mice was performed by The Jackson Laboratory Transgenic Genotyping service using standard PCR.
Anaphase compaction measurements
To measure anaphase compaction, optical sections were collected with 200-nm spacing through the entire spindle of fixed cells labeled with gamma-tubulin and ACA. The centroid of the gamma-tubulin focus in each half spindle was used as a reference point, and 3D distance measurements to each ACA-labeled kinetochore were made using ImageJ (National Institutes of Health). The SD of these distances was computed for each half spindle and used for comparison.
Nuclear shape analyses
For fixed cell analyses, single focal plane images of DAPI-stained nuclei were acquired with a 20× objective. The shape of each nucleus was measured using the solidity measurement function in ImageJ. Nuclei were first thresholded and selected with the magic wand tool before shape measurements. For live cells expressing H2B-GFP, mother cell and daughter cell nuclear solidity were quantified from single focal plane images at the indicated time points relative to anaphase onset. Mother cell nuclear solidity measurements were made 1 h before metaphase. Daughter cell nuclear solidity was measured 20 min after the second frame of chromosome decondensation in telophase.
Telophase lamin distribution measurements
Single focal plane images of WT and mutant KIF18A KO hTERT-RPE1 cells labeled with Lamin A/C were acquired through the central plane of the nucleus and opened in ImageJ. A line was drawn across the Lamin A/C signal along the long axis of the nucleus, and the intensity of each pixel was imported into an Igor Pro (Wavemetrics) macro. The fluorescence intensity of each pixel along the line scan was subtracted from a linear regression line fit to the fluorescence distribution. The SD of these values was determined for each nucleus and defined as the DAI.
Quantification of chromosome alignment
Chromosome alignment was quantified by calculating the full width at half maximum (FWHM) of ACA or DAPI fluorescence distribution along the pole-to-pole axis in ImageJ as described previously (Kim et al., 2014).
Cleaved caspase-3
Cleaved caspase-3 immunofluorescence intensity was quantified using ImageJ. Cells expressing greater than two times the average fluorescence intensity measured in control cells were considered positive for cleaved caspase-3.
MAD1-positive kinetochore quantification
Control or Kif18A KD cells were treated with MG132 for 2 h, 48 h after addition of siRNAs. MAD1-positive kinetochores were scored in ImageJ based on fluorescence intensity. A kinetochore was considered to be MAD1-positive if the MAD1 immunofluorescence intensity overlapping with the ACA-labeled kinetochore was at least 1.2× higher than the cytoplasimic MAD1 signal.
Viability and growth, Kif18agcd2 mice
A cohort of 91 B6;CAST-Kif18agcd2 F3 mice representing all three Kif18agcd2 genotypes were monitored for growth. Animals were weighed every 3 d through wean and then monitored twice weekly through 8 wk of age. Survival data were plotted using Prism 7, and survival curves were compared using log-rank (Mantel–Cox) and Gehan–Breslow–Wilcoxon tests. Growth data were also plotted using Prism 7, and multiple t tests at each time point were used to compare the mean weights across genotypes (Holm–Sidak, α = 0.05).
Online supplemental material
Fig. S1 shows analyses of KIF18A protein in KIF18A KD and KO hTERT-RPE1. Fig. S2 shows analyses of MAD1 at kinetochores in KIF18A KD hTERT-RPE1 cells. Fig. S3 shows results of cleaved caspase-3 assays in KIF18A KD hTERT-RPE1 cells. Fig. S4 displays results from live imaging assays of H2B-GFP expressing KIF18A KO hTERT-RPE1 cells. Table S1 summarizes data from FISH experiments, and Table S2 summarizes measurements from live cell analyses of H2B-GFP expressing hTERT-RPE1 cells. Videos include time-lapse fluorescence videos of control (Video 1) and KIF18A KO (Videos 2 and 3) hTERT-RPE1 cells expressing H2B-GFP.
Supplementary Material
Acknowledgments
The authors thank Catherine Buck (University of Vermont Medical Center) for technical assistance with FISH and chromosome spread analyses and the University of Vermont Flow Cytometry and Cell Sorting Facility for assistance with sorting GFP-Cas9–expressing cells.
This work was supported by National Institutes of Health grants GM121491 (to J. Stumpff), R03 HD078485 (to L.G. Reinholdt), and R01 GM086610 (to R. Ohi); by a Lake Champlain Cancer Research Organization grant (to J. Stumpff); by Susan G. Komen grant CCR16377648 (to J. Stumpff); by a Leukemia and Lymphoma Career Development Award (to R. Ohi); by U.S. Department of Defense Peer Reviewed Cancer Research Program Horizon Award W81XWH-17-1-0371 (to H.L.H. Malaby); and by a Vermont Space Grant Consortium fellowship (to L.A. Sepaniac).
The authors declare no competing financial interests.
Author contributions: Conceptualization, L.G. Reinholdt and J. Stumpff; methodology, C.L. Fonseca, H.L.H. Malaby, L.A. Sepaniac, W. Martin, C. Byers, A. Czechanski, M. Tang, R. Ohi, L.G. Reinholdt, and J. Stumpff; investigation, C.L. Fonseca, H.L.H. Malaby, L.A. Sepaniac, W. Martin, C. Byers, A. Czechanski, D. Messinger, M. Tang, L.G. Reinholdt, and J. Stumpff; resources, H.L.H. Malaby, W. Martin, C. Byers, A. Czechanski, R. Ohi, L.G. Reinholdt, and J. Stumpff; writing, original draft, C.L. Fonseca, H.L.H. Malaby, L.A. Sepaniac, L.G. Reinholdt, and J. Stumpff; writing, review and editing, C.L. Fonseca, H.L.H. Malaby, L.A. Sepaniac, R. Ohi, L.G. Reinholdt, and J. Stumpff; visualization, C.L. Fonseca, H.L.H. Malaby, L.A. Sepaniac, L.G. Reinholdt, and J. Stumpff; supervision, L.G. Reinholdt and J. Stumpff.
References
- Afonso O., Matos I., Pereira A.J., Aguiar P., Lampson M.A., and Maiato H.. 2014. Feedback control of chromosome separation by a midzone Aurora B gradient. Science. 345:332–336. 10.1126/science.1251121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barisic M., Aguiar P., Geley S., and Maiato H.. 2014. Kinetochore motors drive congression of peripheral polar chromosomes by overcoming random arm-ejection forces. Nat. Cell Biol. 16:1249–1256. 10.1038/ncb3060 [DOI] [PubMed] [Google Scholar]
- Barlow C., Hirotsune S., Paylor R., Liyanage M., Eckhaus M., Collins F., Shiloh Y., Crawley J.N., Ried T., Tagle D., and Wynshaw-Boris A.. 1996. Atm-deficient mice: a paradigm of ataxia telangiectasia. Cell. 86:159–171. 10.1016/S0092-8674(00)80086-0 [DOI] [PubMed] [Google Scholar]
- Canman J.C., Salmon E.D., and Fang G.. 2002. Inducing precocious anaphase in cultured mammalian cells. Cell Motil. Cytoskeleton. 52:61–65. 10.1002/cm.10032 [DOI] [PubMed] [Google Scholar]
- Cimini D., Moree B., Canman J.C., and Salmon E.D.. 2003. Merotelic kinetochore orientation occurs frequently during early mitosis in mammalian tissue cells and error correction is achieved by two different mechanisms. J. Cell Sci. 116:4213–4225. 10.1242/jcs.00716 [DOI] [PubMed] [Google Scholar]
- Cimini D., Cameron L.A., and Salmon E.D.. 2004. Anaphase spindle mechanics prevent mis-segregation of merotelically oriented chromosomes. Curr. Biol. 14:2149–2155. 10.1016/j.cub.2004.11.029 [DOI] [PubMed] [Google Scholar]
- Crasta K., Ganem N.J., Dagher R., Lantermann A.B., Ivanova E.V., Pan Y., Nezi L., Protopopov A., Chowdhury D., and Pellman D.. 2012. DNA breaks and chromosome pulverization from errors in mitosis. Nature. 482:53–58. 10.1038/nature10802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czechanski A., Byers C., Greenstein I., Schrode N., Donahue L.R., Hadjantonakis A.-K., and Reinholdt L.G.. 2014. Derivation and characterization of mouse embryonic stem cells from permissive and nonpermissive strains. Nat. Protoc. 9:559–574. 10.1038/nprot.2014.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czechanski A., Kim H., Byers C., Greenstein I., Stumpff J., and Reinholdt L.G.. 2015. Kif18a is specifically required for mitotic progression during germ line development. Dev. Biol. 402:253–262. 10.1016/j.ydbio.2015.03.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dertinger S.D., Torous D.K., and Tometsko K.R.. 1996. Simple and reliable enumeration of micronucleated reticulocytes with a single-laser flow cytometer. Mutat. Res. 371:283–292. 10.1016/S0165-1218(96)90117-2 [DOI] [PubMed] [Google Scholar]
- Drpic D., Pereira A.J., Barisic M., Maresca T.J., and Maiato H.. 2015. Polar ejection forces promote the conversion from lateral to end-on kinetochore-microtubule attachments on mono-oriented chromosomes. Cell Reports. 13:460–468. 10.1016/j.celrep.2015.08.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonseca C., and Stumpff J.. 2016. Quantification of mitotic chromosome alignment. In The Mitotic Spindle. P. Chang and R. Ohi, editors. Springer, New York. 253–262. 10.1007/978-1-4939-3542-0_16 [DOI] [PubMed] [Google Scholar]
- Gerlich D., Beaudouin J., Kalbfuss B., Daigle N., Eils R., and Ellenberg J.. 2003. Global chromosome positions are transmitted through mitosis in mammalian cells. Cell. 112:751–764. 10.1016/S0092-8674(03)00189-2 [DOI] [PubMed] [Google Scholar]
- Hirst C.S., Stamp L.A., Bergner A.J., Hao M.M., Tran M.X., Morgan J.M., Dutschmann M., Allen A.M., Paxinos G., Furlong T.M., et al. 2017. Kif1bp loss in mice leads to defects in the peripheral and central nervous system and perinatal death. Sci. Rep. 7:16676 10.1038/s41598-017-16965-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssen L.M.E., Averink T.V., Blomen V.A., Brummelkamp T.R., Medema R.H., and Raaijmakers J.A.. 2018. Loss of Kif18A results in spindle assembly checkpoint activation at microtubule-attached kinetochores. Curr. Biol. 28:2685–2696.e4. 10.1016/j.cub.2018.06.026 [DOI] [PubMed] [Google Scholar]
- Kapoor T.M., Lampson M.A., Hergert P., Cameron L., Cimini D., Salmon E.D., McEwen B.F., and Khodjakov A.. 2006. Chromosomes can congress to the metaphase plate before biorientation. Science. 311:388–391. 10.1126/science.1122142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kevenaar J.T., Bianchi S., van Spronsen M., Olieric N., Lipka J., Frias C.P., Mikhaylova M., Harterink M., Keijzer N., Wulf P.S., et al. 2016. Kinesin-binding protein controls microtubule dynamics and cargo trafficking by regulating kinesin motor activity. Curr. Biol. 26:849–861. 10.1016/j.cub.2016.01.048 [DOI] [PubMed] [Google Scholar]
- Kim H., Fonseca C., and Stumpff J.. 2014. A unique kinesin-8 surface loop provides specificity for chromosome alignment. Mol. Biol. Cell. 25:3319–3329. 10.1091/mbc.e14-06-1132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kops G.J.P.L., Saurin A.T., and Meraldi P.. 2010. Finding the middle ground: How kinetochores power chromosome congression. Cell. Mol. Life Sci. 67:2145–2161. 10.1007/s00018-010-0321-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn J., and Dumont S.. 2017. Spindle assembly checkpoint satisfaction occurs via end-on but not lateral attachments under tension. J. Cell Biol. 216:1533–1542. 10.1083/jcb.201611104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levesque A.A., and Compton D.A.. 2001. The chromokinesin Kid is necessary for chromosome arm orientation and oscillation, but not congression, on mitotic spindles. J. Cell Biol. 154:1135–1146. 10.1083/jcb.200106093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S., Kwon M., Mannino M., Yang N., Renda F., Khodjakov A., and Pellman D.. 2018. Nuclear envelope assembly defects link mitotic errors to chromothripsis. Nature. 561:551–555. 10.1038/s41586-018-0534-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magidson V., O’Connell C.B., Lončarek J., Paul R., Mogilner A., and Khodjakov A.. 2011. The spatial arrangement of chromosomes during prometaphase facilitates spindle assembly. Cell. 146:555–567. 10.1016/j.cell.2011.07.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magidson V., He J., Ault J.G., O’Connell C.B., Yang N., Tikhonenko I., McEwen B.F., Sui H., and Khodjakov A.. 2016. Unattached kinetochores rather than intrakinetochore tension arrest mitosis in taxol-treated cells. J. Cell Biol. 212:307–319. 10.1083/jcb.201412139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiato H., Gomes A., Sousa F., and Barisic M.. 2017. Mechanisms of chromosome congression during mitosis. Biology (Basel). 6:E13 10.3390/biology6010013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matos I., and Maiato H.. 2011. Prevention and correction mechanisms behind anaphase synchrony: Implications for the genesis of aneuploidy. Cytogenet. Genome Res. 133:243–253. 10.1159/000323803 [DOI] [PubMed] [Google Scholar]
- Matos I., Pereira A.J., Lince-Faria M., Cameron L.A., Salmon E.D., and Maiato H.. 2009. Synchronizing chromosome segregation by flux-dependent force equalization at kinetochores. J. Cell Biol. 186:11–26. 10.1083/jcb.200904153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayr M.I., Hümmer S., Bormann J., Grüner T., Adio S., Woehlke G., and Mayer T.U.. 2007. The human kinesin Kif18A is a motile microtubule depolymerase essential for chromosome congression. Curr. Biol. 17:488–498. 10.1016/j.cub.2007.02.036 [DOI] [PubMed] [Google Scholar]
- Meraldi P., Draviam V.M., and Sorger P.K.. 2004. Timing and checkpoints in the regulation of mitotic progression. Dev. Cell. 7:45–60. 10.1016/j.devcel.2004.06.006 [DOI] [PubMed] [Google Scholar]
- Michel L.S., Liberal V., Chatterjee A., Kirchwegger R., Pasche B., Gerald W., Dobles M., Sorger P.K., Murty V.V.V.S., and Benezra R.. 2001. MAD2 haplo-insufficiency causes premature anaphase and chromosome instability in mammalian cells. Nature. 409:355–359. 10.1038/35053094 [DOI] [PubMed] [Google Scholar]
- Mora-Bermúdez F., Gerlich D., and Ellenberg J.. 2007. Maximal chromosome compaction occurs by axial shortening in anaphase and depends on Aurora kinase. Nat. Cell Biol. 9:822–831. 10.1038/ncb1606 [DOI] [PubMed] [Google Scholar]
- Nagahara M., Nishida N., Iwatsuki M., Ishimaru S., Mimori K., Tanaka F., Nakagawa T., Sato T., Sugihara K., Hoon D.S.B., and Mori M.. 2011. Kinesin 18A expression: Clinical relevance to colorectal cancer progression. Int. J. Cancer. 129:2543–2552. 10.1002/ijc.25916 [DOI] [PubMed] [Google Scholar]
- Ohsugi M., Adachi K., Horai R., Kakuta S., Sudo K., Kotaki H., Tokai-Nishizumi N., Sagara H., Iwakura Y., and Yamamoto T.. 2008. Kid-mediated chromosome compaction ensures proper nuclear envelope formation. Cell. 132:771–782. 10.1016/j.cell.2008.01.029 [DOI] [PubMed] [Google Scholar]
- Pereira A.J., and Maiato H.. 2012. Maturation of the kinetochore-microtubule interface and the meaning of metaphase. Chromosome Res. 20:563–577. 10.1007/s10577-012-9298-8 [DOI] [PubMed] [Google Scholar]
- Putkey F.R., Cramer T., Morphew M.K., Silk A.D., Johnson R.S., McIntosh J.R., and Cleveland D.W.. 2002. Unstable kinetochore-microtubule capture and chromosomal instability following deletion of CENP-E. Dev. Cell. 3:351–365. 10.1016/S1534-5807(02)00255-1 [DOI] [PubMed] [Google Scholar]
- Reinholdt L., Ashley T., Schimenti J., and Shima N.. 2004. Forward genetic screens for meiotic and mitotic recombination-defective mutants in mice. Methods Mol. Biol. 262:87–107. 10.1385/1-59259-761-0:087 [DOI] [PubMed] [Google Scholar]
- Reinholdt L.G., Munroe R.J., Kamdar S., and Schimenti J.C.. 2006. The mouse gcd2 mutation causes primordial germ cell depletion. Mech. Dev. 123:559–569. 10.1016/j.mod.2006.05.003 [DOI] [PubMed] [Google Scholar]
- Sablina A.A., Ilyinskaya G.V., Rubtsova S.N., Agapova L.S., Chumakov P.M., and Kopnin B.P.. 1998. Activation of p53-mediated cell cycle checkpoint in response to micronuclei formation. J. Cell Sci. 111:977–984. 10.1016/S0165-1218(96)90021-X [DOI] [PubMed] [Google Scholar]
- Santaguida S., Richardson A., Iyer D.R., M’Saad O., Zasadil L., Knouse K.A., Wong Y.L., Rhind N., Desai A., and Amon A.. 2017. Chromosome mis-segregation generates cell-cycle-arrested cells with complex karyotypes that are eliminated by the immune system. Dev. Cell. 41:638–651.e5. 10.1016/j.devcel.2017.05.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaar B.T., Chan G.K., Maddox P., Salmon E.D., and Yen T.J.. 1997. CENP-E function at kinetochores is essential for chromosome alignment. J. Cell Biol. 139:1373–1382. 10.1083/jcb.139.6.1373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedgwick G.G., Hayward D.G., Di Fiore B., Pardo M., Yu L., Pines J., and Nilsson J.. 2013. Mechanisms controlling the temporal degradation of Nek2A and Kif18A by the APC/C-Cdc20 complex. EMBO J. 32:303–314. 10.1038/emboj.2012.335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheltzer J.M., Ko J.H., Replogle J.M., Habibe Burgos N.C., Chung E.S., Meehl C.M., Sayles N.M., Passerini V., Storchova Z., and Amon A.. 2017. Single-chromosome gains commonly function as tumor suppressors. Cancer Cell. 31:240–255. 10.1016/j.ccell.2016.12.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silk A.D., Zasadil L.M., Holland A.J., Vitre B., Cleveland D.W., and Weaver B.A.A.. 2013. Chromosome missegregation rate predicts whether aneuploidy will promote or suppress tumors. Proc. Natl. Acad. Sci. USA. 110:E4134–E4141. 10.1073/pnas.1317042110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skibbens R.V., Skeen V.P., and Salmon E.D.. 1993. Directional instability of kinetochore motility during chromosome congression and segregation in mitotic newt lung cells: A push-pull mechanism. J. Cell Biol. 122:859–875. 10.1083/jcb.122.4.859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soto M., Raaijmakers J.A., Bakker B., Spierings D.C.J., Lansdorp P.M., Foijer F., and Medema R.H.. 2017. p53 prohibits propagation of chromosome segregation errors that produce structural aneuploidies. Cell Reports. 19:2423–2431. 10.1016/j.celrep.2017.05.055 [DOI] [PubMed] [Google Scholar]
- Stumpff J., von Dassow G., Wagenbach M., Asbury C., and Wordeman L.. 2008. The kinesin-8 motor Kif18A suppresses kinetochore movements to control mitotic chromosome alignment. Dev. Cell. 14:252–262. 10.1016/j.devcel.2007.11.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stumpff J., Wagenbach M., Franck A., Asbury C.L., and Wordeman L.. 2012. Kif18A and chromokinesins confine centromere movements via microtubule growth suppression and spatial control of kinetochore tension. Dev. Cell. 22:1017–1029. 10.1016/j.devcel.2012.02.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson S.L., and Compton D.A.. 2008. Examining the link between chromosomal instability and aneuploidy in human cells. J. Cell Biol. 180:665–672. 10.1083/jcb.200712029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson S.L., and Compton D.A.. 2010. Proliferation of aneuploid human cells is limited by a p53-dependent mechanism. J. Cell Biol. 188:369–381. 10.1083/jcb.200905057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uetake Y., and Sluder G.. 2004. Cell cycle progression after cleavage failure: Mammalian somatic cells do not possess a “tetraploidy checkpoint”. J. Cell Biol. 165:609–615. 10.1083/jcb.200403014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uetake Y., and Sluder G.. 2010. Prolonged prometaphase blocks daughter cell proliferation despite normal completion of mitosis. Curr. Biol. 20:1666–1671. 10.1016/j.cub.2010.08.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver B.A.A., Bonday Z.Q., Putkey F.R., Kops G.J.P.L., Silk A.D., and Cleveland D.W.. 2003. Centromere-associated protein-E is essential for the mammalian mitotic checkpoint to prevent aneuploidy due to single chromosome loss. J. Cell Biol. 162:551–563. 10.1083/jcb.200303167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver B.A.A., Silk A.D., Montagna C., Verdier-Pinard P., and Cleveland D.W.. 2007. Aneuploidy acts both oncogenically and as a tumor suppressor. Cancer Cell. 11:25–36. 10.1016/j.ccr.2006.12.003 [DOI] [PubMed] [Google Scholar]
- Wilson R., Geyer S.H., Reissig L., Rose J., Szumska D., Hardman E., Prin F., McGuire C., Ramirez-Solis R., White J., et al. 2017. Highly variable penetrance of abnormal phenotypes in embryonic lethal knockout mice. Wellcome Open Res. 1:1 10.12688/wellcomeopenres.9899.2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C., Zhu C., Chen H., Li L., Guo L., Jiang W., and Lu S.H.. 2010. Kif18A is involved in human breast carcinogenesis. Carcinogenesis. 31:1676–1684. 10.1093/carcin/bgq134 [DOI] [PubMed] [Google Scholar]
- Zhang C.-Z., Spektor A., Cornils H., Francis J.M., Jackson E.K., Liu S., Meyerson M., and Pellman D.. 2015. Chromothripsis from DNA damage in micronuclei. Nature. 522:179–184. 10.1038/nature14493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu C., Zhao J., Bibikova M., Leverson J.D., Bossy-Wetzel E., Fan J.-B., Abraham R.T., and Jiang W.. 2005. Functional analysis of human microtubule-based motor proteins, the kinesins and dyneins, in mitosis/cytokinesis using RNA interference. Mol. Biol. Cell. 16:3187–3199. 10.1091/mbc.e05-02-0167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H., Xu W., Zhang H., Liu J., Xu H., Lu S., Dang S., Kuang Y., Jin X., and Wang Z.. 2013. Targeted deletion of Kif18a protects from colitis-associated colorectal (CAC) tumors in mice through impairing Akt phosphorylation. Biochem. Biophys. Res. Commun. 438:97–102. 10.1016/j.bbrc.2013.07.032 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.