The epigenetic regulator EZH2 has opposing roles during initiation and maintenance of acute myeloid leukemia.
Abstract
In this issue of JEM, Basheer et al. (https://doi.org/10.1084/jem.20181276) describe opposing roles of the epigenetic regulator Ezh2 during initiation and maintenance of acute myeloid leukemia (AML). Ezh2 was found to have tumor suppressive and oncogenic functions in different phases of the same malignancy.
The histone lysine N-methyltransferase EZH2 is the enzymatic component of the polycomb repressive complex 2 (PRC2) that controls stem cell maintenance and differentiation. Mutations in EZH2 exert context-specific and sometimes opposing effects on tumorigenesis. Oncogenic gain-of-function mutations found in patients with lymphoid malignancies (Morin et al., 2010) led to developing small molecule inhibitors of EZH2 that are currently being tested in clinical trials. Also, overexpression of the nonmutated EZH2 in breast, prostate, and renal cancers was associated with unfavorable prognosis (Kim and Roberts, 2016). In contrast, loss-of-function mutations in EZH2 were found in myeloproliferative neoplasms (MPNs; Ernst et al., 2010), and loss of Ezh2 accelerated progression in mouse models of MPN, indicating that in this context, EZH2 functions as a tumor suppressor (Sashida et al., 2016; Shimizu et al., 2016).

Insights from Radek C. Skoda and Juerg Schwaller.
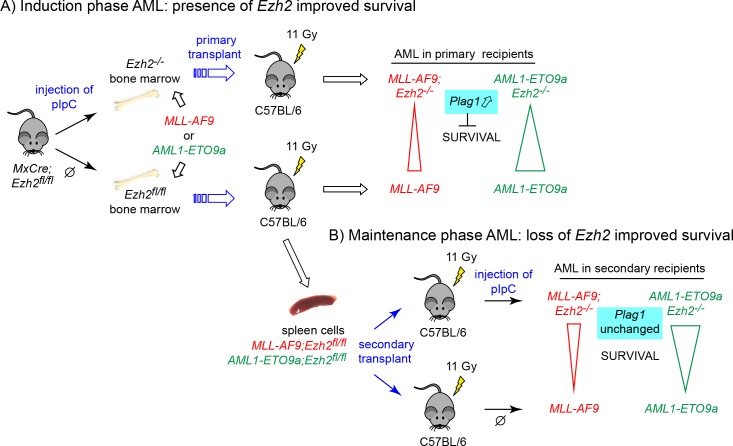
Basheer et al. add an additional layer of complexity to the already polymorphic actions of EZH2 in cancer. In mouse models of acute myeloid leukemia (AML), deletion of Ezh2 before retroviral transduction with oncogenic MLL-AF9 or AML1-ETO9a fusion genes accelerated disease and shortened survival, indicating that Ezh2 functions as a tumor suppressor. In contrast, when bone marrow cells were first transduced with MLL-AF9 or AML1-ETO9a and Ezh2 was deleted later in secondary recipients during the maintenance phase of AML, survival was prolonged, and disease severity was attenuated. Similar to the genetic ablation of Ezh2, treatment of secondary recipients with the Ezh2 inhibitor EPZ-6438 also resulted in prolonged survival of AML1-ETO9a mice. These data suggest that the nonmutated Ezh2 in this setting is required to unleash the full oncogenic effects of MLL-AF9 and AML1-ETO9a.
Experimental scheme of the in vivo experiments. (A) MxCre;Ezh2fl/fl mice were injected with pIpC to induce Cre-mediated deletion of the conditional Ezh2fl/fl alleles (upper panel) or injected with vehicle to leave Ezh2 intact (lower panel). Bone marrow cells were transduced with either MLL-AF9 or AML-ETO9a retroviruses and transplanted into lethally irradiated recipients. (B) Spleen cells from MxCre;Ezh2fl/fl mice were transplanted into secondary recipients and injected with pIpC to induce Cre-mediated deletion during the maintenance phase of the disease (upper panel). Noninduced recipients served as controls (lower panel).
Ezh2 is the enzymatic component of PRC2. The SET domain of Ezh2 catalyzes the addition of repressive methyl marks on lysine 27 on histone 3 (H3K27) that are preferentially deposited near gene promoters and enhancers, and thereby contribute to silencing gene expression. Basheer et al. (2019) performed detailed RNA sequencing and chromatin modifications analyses in c-kit+ bone marrow cells isolated during the AML initiation phase. In addition to Lin28b, an oncogene previously shown to be up-regulated upon Ezh2 deletion, they also found elevated expression of Plag1, a zinc-finger transcription factor, previously identified as a cooperating oncogene to the leukemogenic CBFβ-SMMHC fusion (Landrette et al., 2005). Basheer et al. (2019) found that forced expression of Plag1 in bone marrow cells transduced with MLL-AF9 accelerated the development of AML. Interestingly, Plag1 was not up-regulated following Ezh2 ablation in a published dataset during the maintenance of MLL-AF9 AML, and treatment with an Ezh2 inhibitor GSK343 did not increase expression of Plag1, suggesting that this effect is selective for the early stage of AML.
Indeed, the expression profiles observed by Basheer et al. (2019) following excision of Ezh2 during the induction phase of AML showed little overlap with their datasets obtained following loss in the maintenance phase of MLL-AF9 AML (60/496 genes, ∼12%), suggesting that the contrasting phenotypes may be due to derepression of different genes during AML induction and maintenance. Chromatin immunoprecipitation sequencing analysis revealed that the loss of Ezh2-mediated methyltransferase activity decreased H3K27me3 marks preferentially at bivalent promoters and less frequently in gene enhancer regions. In contrast to MPN, where loss of H3K27me3 marks was frequently accompanied by increase in the reciprocal histone activation mark, H3K27Ac, no such increase in H3K27Ac was noted in the context of AML.
The study by Basheer et al. (2019) confirms earlier observations of slower progression of AML in secondary recipients of MLL-AF9–expressing bone marrow stem and progenitor cells, when Ezh2 was genetically ablated (Neff et al., 2012). The current study adds new data by showing that during disease initiation, loss of Ezh2 had the opposite effect and shortened survival. The basis for these stage-specific differences in expression profiles induced by loss of Ezh2 remains to be determined. The order in which Ezh2 is deleted in respect to introducing the MLL-AF9 and AML1-ETO9a fusion genes could be a contributing factor to the differences in the expression profiles and AML kinetics. Slower progression and prolonged survival in both studies was observed when deletion of Ezh2 was induced in cells already transformed by MLL-AF9. In the study by Neff et al. (2012), the Cre-mediated deletion of the loxP conditional Ezh2fl/fl alleles was induced 3 wk after transplantation of the MLL-AF9 transduced bone marrow cells, while in the study by Basheer et al. (2019), deletion of Ezh2fl/fl alleles was further delayed and induced only in secondary recipients. A third study by Fujita et al. (2018) used two other fusion genes (MLL-AF10 and MOZ-TIF2) and also observed improved survival when Ezh2fl/fl was deleted in already transduced bone marrow cells 3 d after transplantation.
Additional factors contributing to the complexities could also stem from the partially redundant actions of Ezh1 and Ezh2. Indeed, survival of MLL-AF10 or MOZ-TIF2–driven AML was further improved when both Ezh1 and Ezh2 were genetically deleted (Fujita et al., 2018). Similarly, genetic ablation of Eed1, a component of the PRC2 that is essential for both Ezh1 and Ezh2 activity, substantially prolonged survival compared with Ezh2 loss alone (Neff et al., 2012). Thus, the shortened survival due to loss of Ezh2 during AML initiation appears to be linked to the experimental setting, where Ezh2 was deleted before retroviral transduction with the oncogenic fusion gene. Loss of Ezh2 is known to alter the composition of the hematopoietic progenitor and stem cell pool (Xie et al., 2014), and thus the prior loss of Ezh2 may alter the target cell population transduced and clonally selected by MLL-AF9 and AML1-ETO9a oncogenes. The stem cell versus progenitor origin of AML can affect AML kinetics and phenotypes (Stavropoulou et al., 2016). Similarly, differences in the phenotypes of MPNs were observed when JAK2-V617F was acquired before a mutation in the epigenetic regulator TET2 compared with patients in whom a TET2 mutation occurred before JAK2-V617F (Ortmann et al., 2015).
The work by Basheer et al. (2019) suggests that targeting the enzymatic activity of EZH2 by small molecules might be beneficial in AML patients. Are trials of EZH2 inhibitors warranted in patients with AML? If so, how can the “maintenance” phase of AML be defined in patients? Analysis of patient data revealed that survival of AML patients with EZH2 mutations (in most cases heterozygotes for mutations predicted to be loss-of-function) was inferior compared with patients without EZH2 mutations. This result is not encouraging, as inhibiting EZH2 protein might also have unfavorable effects. However, pharmacological inhibition of Ehz2 activity differed from genetic ablation of Ezh2, as, e.g., GSK343 did not increase expression of Plag1, and pharmacological EZH2 inhibition and did not up-regulate Hox expression during maintenance (Khan et al., 2013). The latter finding somehow contrasts with observations by Göllner et al. (2017) that suppression of EZH2 protein expression resulted in derepression of HOX genes in AML cell lines and primary cells in vitro and in vivo, and that knockdown of HOXB7 and HOXA9 increased sensitivity to cytotoxic drugs or tyrosine kinase inhibitors.
Dual small-molecule inhibitors that suppress H3K27 methylation by blocking the enzymatic activity of both EZH2 and EZH1 show some promise. A dual EZH1/2 inhibitor (UNC1999) showed significant antileukemic activity by promoting differentiation, suppressing clonogenic growth, and inducing apoptosis of MLL-AF9 and MLL-ENL transformed cells (Xu et al., 2015). Similarly, another EZH1/EZH2 dual inhibitor (OR-S1) suppressed proliferation of MLL-AF10 or MOZ-TIF2 transformed cells in vitro and reduced the number of leukemic stem cells, resulting in significantly delayed propagation of the disease by transplantation (Fujita et al., 2018). Will blocking the catalytic activity of EZH2 be sufficient? Observations in other cancers suggest PRC2-independent and methylation-independent roles of EZH2 may provide a route to escape enzymatic inhibition (Kim and Roberts, 2016). Proteolysis-targeting chimera (PROTAC)–mediated protein degradation of epigenetic regulators, such as Brd4, PCAF, or TRIM24, has recently been shown to exert potent antitumor effects (Scheepstra et al., 2019). Therefore, PROTAC-mediated degradation of EZH2 protein could be an attractive alternative to inhibiting EZH2 methyltransferase activity. Overall, the work by Basheer et al. (2019) provides an exciting basis for further studies into the context-specific roles of EZH2 in AML that may better inform therapeutic strategies.
References
- Basheer F., et al. J. Exp. Med. 2019 doi: 10.1084/jem.20181276. [DOI] [Google Scholar]
- Ernst T., et al. 2010. Nat. Genet. 42:722–726. 10.1038/ng.621 [DOI] [PubMed] [Google Scholar]
- Fujita S., et al. 2018. Leukemia. 32:855–864. 10.1038/leu.2017.300 [DOI] [PubMed] [Google Scholar]
- Göllner S., et al. 2017. Nat. Med. 23:69–78. 10.1038/nm.4247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan S.N., et al. 2013. Leukemia. 27:1301–1309. 10.1038/leu.2013.80 [DOI] [PubMed] [Google Scholar]
- Kim K.H., and Roberts C.W.. 2016. Nat. Med. 22:128–134. 10.1038/nm.4036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landrette S.F., et al. 2005. Blood. 105:2900–2907. 10.1182/blood-2004-09-3630 [DOI] [PubMed] [Google Scholar]
- Morin R.D., et al. 2010. Nat. Genet. 42:181–185. 10.1038/ng.518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neff T., et al. 2012. Proc. Natl. Acad. Sci. USA. 109:5028–5033. 10.1073/pnas.1202258109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortmann C.A., et al. 2015. N. Engl. J. Med. 372:601–612. 10.1056/NEJMoa1412098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sashida G., et al. 2016. J. Exp. Med. 213:1459–1477. 10.1084/jem.20151121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheepstra M., et al. 2019. Comput. Struct. Biotechnol. J. 17:160–176. 10.1016/j.csbj.2019.01.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu T., et al. 2016. J. Exp. Med. 213:1479–1496. 10.1084/jem.20151136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stavropoulou V., et al. 2016. Cancer Cell. 30:43–58. 10.1016/j.ccell.2016.05.011 [DOI] [PubMed] [Google Scholar]
- Xie H., et al. 2014. Cell Stem Cell. 14:68–80. 10.1016/j.stem.2013.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu B., et al. 2015. Blood. 125:346–357. 10.1182/blood-2014-06-581082 [DOI] [PMC free article] [PubMed] [Google Scholar]