In this issue of JEM, Reinink et al. use comparative lipidomics to identify a new family of trehalose-containing cell wall lipids that are enriched in virulent Salmonella serovars. These lipids are structurally related to the important mycobacterial immunogen cord factor.
Abstract
In this issue of JEM, Reinink et al. (https://doi.org/10.1084/jem.20181812) use comparative lipidomics to identify a new family of trehalose-containing cell wall lipids that are enriched in virulent Salmonella serovars. These lipids are structurally related to the important mycobacterial immunogen cord factor.
Pathogens from the Gram-negative genus Salmonella infect around 1.3 billion people each year and are responsible for a spectrum of clinically significant human diseases (Coburn et al., 2007). Predominantly serovars of S. enterica, these pathogens can be loosely divided into two groups according to the type of disease they cause. Typhoidal serovars cause systemic illnesses such as typhoid fever, while nontyphoidal serovars cause localized gastroenteritis in otherwise healthy hosts (Makendi et al., 2016; Worley et al., 2018).
Genes encoding cell surface structures are major loci of genetic variation between Salmonella serovars. These structures include lipopolysaccharides and flagellin, both of which are used in Salmonella serotyping (Fierer and Guiney, 2001). Since our existing understanding of cell surface chemotypes does not fully explain the varied disease presentations caused by Salmonella serovars, it is reasonable to suggest that other cell wall lipids could have important roles to play in modulating the host response to Salmonella.
Insights from Stephanie R. Lovell-Read and Luiz Pedro Sorio de Carvalho.
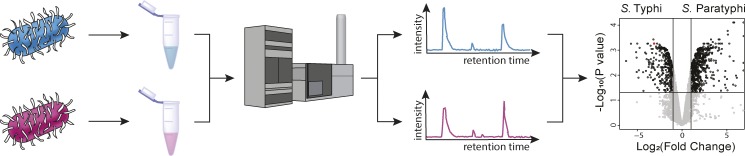
For this reason, a team composed of scientists from the Netherlands, the US, Japan, and the UK sought to identify lipids that could be linked to the pathogenicity of certain serovars of Salmonella. To accomplish this, Reinink et al. took advantage of the comprehensive comparative lipidomics methodology developed by the Moody laboratory to study Mycobacterium tuberculosis, as shown in the figure (Layre et al., 2011). By comparing the lipidome of the highly pathogenic serovar S. Typhi to that of the less pathogenic S. Paratyphi, the authors discovered a family of previously unknown trehalose phospholipids that were significantly enriched in S. Typhi. These lipids were conclusively identified as 6-phosphatidyltrehalose (PT) and 6,6′-diphosphatidyltrehalose (diPT) through a tour de force set of experiments, including extensive nontrivial mass spectrometry (MS), 1D and 2D nuclear magnetic resonance spectroscopy, and ultimately a nine-step chemical synthesis of diPT.
Overview of the comparative lipidomics method used in this study. Salmonella serovars with different pathogenicities were cultured under the same conditions. Their lipidomes were extracted and profiled using high-performance liquid chromatography (HPLC) MS. The HPLC retention times, mass-to-charge ratios, and peak intensities were combined to generate a list of unique species known as molecular events. The molecular events from the two serovars were then compared, allowing the identification of species that were more abundant in one serovar than in the other. In this study, the species of interest were those enriched in S. Typhi, the more pathogenic serovar, as they were more likely to be virulence associated. The volcano plot was adapted from Fig. 1 a of Reinink et al. (2019).
The newly discovered trehalose phospholipids are chemically related to the important mycobacterial immunogen trehalose dimycolate (TDM), or cord factor (Adam et al., 1967). This similarity prompted Reinink et al. (2019) to investigate the potential immunogenicity of trehalose phospholipids, specifically the ability of diPT to stimulate the Mincle receptor. Mincle, a myeloid cell C-type lectin pattern recognition receptor, is essential for the TDM-dependent inflammatory response (Ishikawa et al., 2009). Using reporter cell lines expressing Mincle and its signaling partner FcRγ, the authors confirmed that Mincle recognizes diPT in a dose-dependent manner.
Next, Reinink et al. (2019) investigated how trehalose phospholipids are synthesized in Salmonella. By considering PT and diPT as carbohydrate-substituted phosphatidylglycerols, bioinformatic analysis identified 12 genes that are potentially involved in the biosynthesis of these molecules. A systematic gene knockout experiment indicated that only one of these genes, clsB, was essential for trehalose phospholipid biosynthesis. Consequently, the authors propose that clsB, which was previously annotated as one of three cardiolipin synthases in Salmonella, is in fact a PT and diPT synthase.
This result highlights a major obstacle to understanding infection biology, namely the poor annotation of bacterial genomes and their metabolic products. In the era of high-throughput genome sequencing, functional annotation relies heavily on automated procedures, with the manually curated SwissProt database now comprising just 0.63% of the UniProt Knowledgebase (Gerlt, 2017). However, automated annotation pipelines are vulnerable to errors such as over-annotation. These errors propagate through sequence databases and can be difficult to identify and correct (Danchin et al., 2018).
Accurate annotation of multigene families (where multiple family members are present in the same genome) is particularly challenging since the enzymes in these families often undertake similar chemistry on closely related substrates (Zallot et al., 2016). With this in mind, it is perhaps unsurprising that the trehalose phospholipid synthase identified by Reinink et al. (2019) was originally annotated as a cardiolipin synthase. Nonetheless, the observation that S. Typhi clsB does not appear to encode a cardiolipin synthase should prompt a thorough analysis of the genes annotated as such, since this aspect of lipid biosynthesis is evidently more nuanced than its annotations suggest. Further experiments are clearly required to facilitate the unambiguous assignment of homologues of cardiolipin synthases in bacterial genomes, for example activity-based metabolomic profiling of recombinant enzymes or global metabolomic profiling of deletion mutants (Patti et al., 2012; Prosser et al., 2014).
Having identified that trehalose phospholipids are differentially expressed between Salmonella serovars, the authors evaluated whether PT and diPT are synthesized by other bacterial species possessing clsB orthologues. Of the species chemotyped, only Escherichia coli produced these compounds, and then only in four of the six strains tested. As noted by Reinink et al. (2019), further studies will be required to identify whether this is due to differences in clsB expression, enzyme functionality, or interactions with other enzymes.
Bacterial gene expression and cell wall composition can be extensively altered on host colonization. It would therefore be of interest to determine whether PT and diPT are core lipids in Salmonella or whether their levels change in response to the host environment. Future experiments should also seek to identify if receptors other than Mincle are involved in sensing trehalose-containing lipids. This would assist in the elucidation of the downstream effects of these molecules on the immune system and the organism as a whole, information that will be essential for a complete understanding of the role of trehalose phospholipids in host–pathogen interactions and infection.
Finally, the comparative lipidomics experiment identified hundreds of other statistically significant differences between the S. Typhi and S. Paratyphi lipidomes. Characterization of these hits, which will require the development of novel high-throughput approaches, will dramatically expand our understanding of cell envelope structure and function in S. Typhi.
In conclusion, the study by Reinink et al. (2019) highlights a quintessential fact: the vast majority of metabolites in bacteria remain to be discovered and structurally characterized, and a significant number of biosynthetic pathways have therefore yet to be elucidated. Several of these complex metabolites will exert important effects on the host and other bacteria and will consequently have translational applications.
Acknowledgments
The L.P.S. de Carvalho lab is funded by The Francis Crick Institute, which receives its core funding from Cancer Research UK (FC001060), the UK Medical Research Council (FC001060), and the Wellcome Trust (FC001060).
The authors declare no competing financial interests.
References
- Adam A., et al. 1967. Eur. J. Biochem. 2:460–468. 10.1111/j.1432-1033.1967.tb00160.x [DOI] [PubMed] [Google Scholar]
- Coburn B., et al. 2007. Immunol. Cell Biol. 85:112–118. 10.1038/sj.icb.7100007 [DOI] [PubMed] [Google Scholar]
- Danchin A., et al. 2018. Microb. Biotechnol. 11:588–605. 10.1111/1751-7915.13284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fierer J., and Guiney D.G.. 2001. J. Clin. Invest. 107:775–780. 10.1172/JCI12561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerlt J.A. 2017. Biochemistry. 56:4293–4308. 10.1021/acs.biochem.7b00614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa E., et al. 2009. J. Exp. Med. 206:2879–2888. 10.1084/jem.20091750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Layre E., et al. 2011. Chem. Biol. 18:1537–1549. 10.1016/j.chembiol.2011.10.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makendi C., et al. 2016. PLoS Negl. Trop. Dis. 10:e0004446 10.1371/journal.pntd.0004446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patti G.J., et al. 2012. Nat. Rev. Mol. Cell Biol. 13:263–269. 10.1038/nrm3314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prosser G.A., et al. 2014. EMBO Rep. 15:657–669. 10.15252/embr.201338283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinink P., et al. J. Exp. Med. 2019 doi: 10.1084/jem.20181812. [DOI] [Google Scholar]
- Worley J., et al. 2018. MBio. 9:e02303–e02318. 10.1128/mBio.02303-18 [DOI] [Google Scholar]
- Zallot R., et al. 2016. Life (Basel). 6:39 10.3390/life6030039 [DOI] [PMC free article] [PubMed] [Google Scholar]