Optimal activation of IRF3 is crucial for maintaining immune homeostasis. Huai et al. demonstrate that KAT8 acetylates IRF3 at lysine 359, inhibits IRF3 recruitment to promoters of type I interferon genes, and then decreases type I interferon production to attenuate antiviral innate immune responses.
Abstract
The transcription factor interferon regulatory factor 3 (IRF3) is essential for virus infection–triggered induction of type I interferons (IFN-I) and innate immune responses. IRF3 activity is tightly regulated by conventional posttranslational modifications (PTMs) such as phosphorylation and ubiquitination. Here, we identify an unconventional PTM of IRF3 that directly inhibits its transcriptional activity and attenuates antiviral immune response. We performed an RNA interference screen and found that lysine acetyltransferase 8 (KAT8), which is ubiquitously expressed in immune cells (particularly in macrophages), selectively inhibits RNA and DNA virus–triggered IFN-I production in macrophages and dendritic cells. KAT8 deficiency protects mice from viral challenge by enhancing IFN-I production. Mechanistically, KAT8 directly interacts with IRF3 and mediates IRF3 acetylation at lysine 359 via its MYST domain. KAT8 inhibits IRF3 recruitment to IFN-I gene promoters and decreases the transcriptional activity of IRF3. Our study reveals a critical role for KAT8 and IRF3 lysine acetylation in the suppression of antiviral innate immunity.
Introduction
Optimal activation of innate immune response is crucial for maintaining immune homeostasis and the elimination of invading pathogens, which involves diverse signaling pathway regulation and posttranslational modifications (PTMs; Deribe et al., 2010). PTMs are integral components of gene expression programs. To date, >200 different PTMs have been identified that influence diverse aspects of signaling regulation (Hirsch et al., 2017). PTMs also act as critical regulators of cellular signal transduction during innate immune responses (Deribe et al., 2010; Liu et al., 2016). In addition to conventional PTMs such as phosphorylation and ubiquitination, which have been extensively elucidated in cellular signaling pathways, other unconventional PTMs such as acetylation and methylation are increasingly being shown to control innate immune and inflammatory responses (Mowen and David, 2014; Cao, 2016; Li et al., 2016; Chen et al., 2017). For example, the methyltransferase Dnmt3a up-regulates expression of histone deacetylase HDAC9, which maintains the deacetylation status of the key pattern recognition receptor signaling molecule TBK1 and enhances its kinase activity (Li et al., 2016).
Lysine acetylation was first identified on histones >50 yr ago and has long been associated with gene activation (Phillips, 1963; Allfrey et al., 1964). Reversible lysine acetylation also occurs on nonhistone proteins outside of chromatin. In mammals, >8,000 acetyl-lysine sites are present on proteins that reside primarily in nuclear, cytoplasmic, and mitochondrial subcellular compartments (Choudhary et al., 2009; Schölz et al., 2015), and many of these modification sites are conserved across different species, implying functional significance (Weinert et al., 2011; Beltrao et al., 2012). Protein acetylation has a variety of effects, including regulating enzymatic activity, protein–protein interactions, nucleic acid binding, protein stability, and subcellular localization (Gu and Roeder, 1997; Ageta-Ishihara et al., 2013; Choudhary et al., 2014; Wang et al., 2016).
The MYST family proteins (including KAT5, KAT6A, KAT6B, KAT7, and KAT8), characterized by a highly conserved lysine acetyltransferase domain, are involved in a wide range of physiological processes in mammals (Thomas et al., 2000; Merson et al., 2006). KAT8 (also known as MOF or MYST1) is a major enzyme that catalyzes H4K16 acetylation in mammalian cells (Dou et al., 2005). KAT8 has an N-terminal chromodomain (reported to bind noncoding RNA) and a central MYST histone acetyltransferase domain (Akhtar et al., 2000). KAT8 participates in diverse biological processes, including embryonic development, DNA repair, autophagy, chromatin architecture, cellular lifespan, and mitochondrial DNA transcription, and has been implicated in cancer development and T cell differentiation (Shogren-Knaak et al., 2006; Thomas et al., 2008; Dang et al., 2009; Sharma et al., 2010; Füllgrabe et al., 2013; Gupta et al., 2013; Chatterjee et al., 2016; Luo et al., 2016). Ablation of KAT8 specifically in mouse T cells leads to defective cell differentiation and reduces T cell numbers and thymus size (Gupta et al., 2013). However, the role of KAT8 in innate immunity has not been reported.
The transcription factor IFN regulatory factor 3 (IRF3) is vital for the induction of IFN-I production in antiviral immunity (Sadler and Williams, 2008). The active state of IRF3 is tightly regulated by PTMs such as phosphorylation and ubiquitination (Taniguchi et al., 2001; Saitoh et al., 2006). Phosphorylation of IRF3 promotes IRF3 activity, and the importance of five critical serine or threonine residues of IRF3 (Ser396, Ser398, Ser402, Thr404, and Ser405) for its activation has been demonstrated (Lin et al., 1998, 1999). The prolyl isomerase Pin1, which is a negative regulator of IRF3, can interact with IRF3 and promote IRF3 degradation via polyubiquitination (Saitoh et al., 2006). Recently, unconventional PTMs have also been shown to regulate IRF3 activity. K366 monomethylation of IRF3, mediated by the methyltransferase NSD3, enhances the transcriptional activity of IRF3 and promotes IFN-I production (Wang et al., 2017). However, the role of acetylation in regulating IRF3 function remains unclear.
Here, we performed an RNA interference screen to investigate the potential roles of five MYST family members in inflammation and antiviral innate immunity. We identified KAT8 as a negative regulator in the production of IFN-I. Up to now, several reports have elucidated nonhistone acetylation by KAT8, but a regulatory function for KAT8 in host defense against infectious agents and the associated signal transduction pathways have not been reported. In this study, we report a novel role for KAT8 in down-regulating virus-induced IFN-I production by acetylating the transcription factor IRF3 at lysine 359, inhibiting the recruitment of IRF3 to IFN-I promoters, and decreasing its transcriptional activity.
Results
KAT8 deficiency selectively promotes IFN-I production
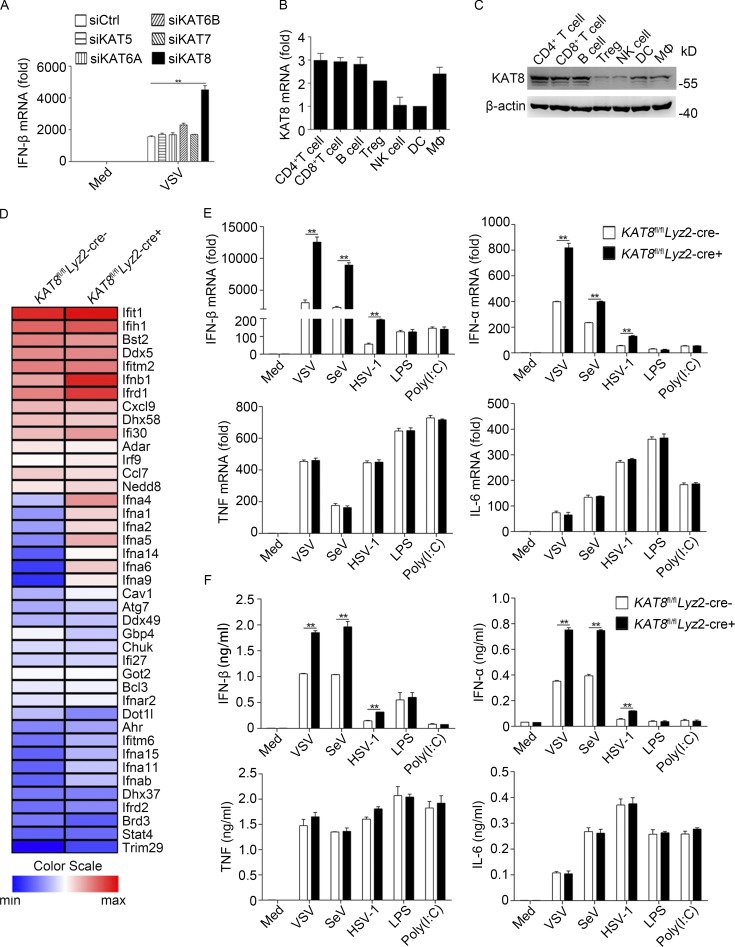
To identify the potential role for MYST family proteins in antiviral innate immunity, we performed a functional screen using siRNA targeting five MYST members (KAT5, KAT6A, KAT6B, KAT7, and KAT8) to observe the effect on IFN-β production in mouse peritoneal macrophages. Only knockdown of KAT8 was found to significantly enhance IFN-β production triggered by vesicular stomatitis virus (VSV; Fig. 1 A).
Figure 1.
Deficiency in KAT8 selectively promotes the production of IFN-I. (A) Q-PCR analysis of IFN-β mRNA in peritoneal macrophages transfected with control siRNA (siCtrl) or specific siRNA targeting KAT5 (siKAT5), KAT6A (siKAT6A), KAT6B (siKAT6B), KAT7 (siKAT7), or KAT8 (siKAT8) and then infected with VSV (1 MOI) for 8 h. (B) Q-PCR analysis of KAT8 mRNA in mouse immune cells; results were normalized to β-actin mRNA. (C) Immunoblot analysis of KAT8 and β-actin in mouse immune cells. (D) RNA sequencing analysis of KAT8fl/flLyz2-Cre− or KAT8fl/flLyz2-Cre+ peritoneal macrophages infected with VSV (1 MOI). (E) Q-PCR analysis of IFN-β, IFN-α, TNF, and IL-6 mRNA in KAT8fl/flLyz2-Cre− or KAT8fl/flLyz2-Cre+ peritoneal macrophages left untreated (Med), infected with VSV (1 MOI), SeV (1 MOI), or HSV-1 (10 MOI) for 8 h, or stimulated with LPS (100 ng/ml) or poly(I:C) (10 µg/ml) for 3 h. (F) ELISA of IFN-β, IFN-α, TNF, and IL-6 in supernatants of KAT8fl/flLyz2-Cre− or KAT8fl/flLyz2-Cre+ peritoneal macrophages infected with VSV (1 MOI), SeV (1 MOI), or HSV-1 (10 MOI) for 12 h or stimulated with LPS (100 ng/ml) or poly(I:C) (10 µg/ml) for 6 h. **, P < 0.01; two-tailed Student’s t test (A, B, E, and F). Data are representative of three independent experiments with similar results (C and D) or are from three independent experiments (A, B, E, and F; mean ± SEM).
We next analyzed the expression pattern of KAT8 and found it was universally expressed in different immune cells, including CD4+ T cells, CD8+ T cells, B cells, regulatory T cells, natural killer cells (NK cells), dendritic cells (DCs), and macrophages (Fig. 1, B and C). The expression of KAT8 showed limited change after VSV infection, and KAT8 exclusively localized in the nucleus of macrophages before or after viral infection (Fig. S1, A–C). Knockdown of KAT8 in macrophages also substantially enhanced the expression of IFN-α and IFN-β (both mRNA and protein), but not the proinflammatory cytokines TNF and IL-6, induced by infection with the RNA viruses VSV or Sendai virus (SeV) or the DNA virus HSV-1 (Fig. S1, D–F). However, stimulation by the TLR4 ligand LPS or the TLR3 ligand polyinosinic-polycytidylic acid (poly(I:C)) had no effect on the production of IFN-I and proinflammatory cytokines (Fig. S1, E and F), indicating that KAT8 was not involved in TLR4- and TLR3-triggered innate responses.
We next used CRISPR-Cas9 technology to knock out KAT8 in RAW264.7 cells to generate KAT8-KO cell lines (Fig. S2 A). Again, RNA and protein levels of IFN-α and IFN-β were significantly higher in KAT8-KO RAW264.7 cells than in control RAW264.7 cells in response to VSV infection (Fig. S2, B and C).
Next, we employed KAT8fl/flLyz2-Cre+ mice (Thomas et al., 2008), which undergo deletion of loxP-flanked KAT8 alleles (KAT8fl/fl) specifically in myeloid cells (mainly in macrophages) via Cre recombinase expressed from the myeloid cell–specific gene Lyz2 (Lyz2-Cre; Fig. S2 D). KAT8 deficiency in KAT8fl/flLyz2-Cre+ mice did not affect the development and differentiation of immune cells, including macrophages, NK cells, B cells, CD4+ T cells, and CD8+ T cells (Fig. S2, E and F). We then analyzed gene expression profiles in macrophages from KAT8-deficient (KAT8fl/flLyz2-Cre+) or KAT8-sufficient (KAT8fl/flLyz2-Cre−) mice upon VSV infection. We found that IFN and many IFN-stimulated genes (such as Ifi30, Ifrd1, and Ifitm6) were up-regulated in KAT8-deficient macrophages. Additionally, some non–IFN-stimulated genes (such as Ahr and Atg7) were also up-regulated (Fig. 1 D). Consistently, we found that mRNA and protein levels of IFN-α and IFN-β, but not proinflammatory cytokines TNF and IL-6, induced by infections with VSV, SeV, or HSV-1 were significantly higher in KAT8-deficient peritoneal macrophages than those in KAT8-sufficient counterparts (Fig. 1, E and F). However, the mRNA and protein levels of IFN-α, IFN-β, TNF, and IL-6 induced by LPS or poly(I:C) showed no difference between KAT8-deficient macrophages and control macrophages (Fig. 1, E and F). Furthermore, KAT8 deficiency in bone marrow–derived DCs increased mRNA and protein levels of IFN-α and IFN-β, but not TNF and IL-6, induced by VSV, SeV, or HSV-1 infection (Fig. S3, A–C). These data demonstrate that KAT8 selectively inhibits RNA and DNA virus infection–triggered production of IFN-I in macrophages and DCs.
KAT8 deficiency protects mice against viral infection by enhancing IFN-I production
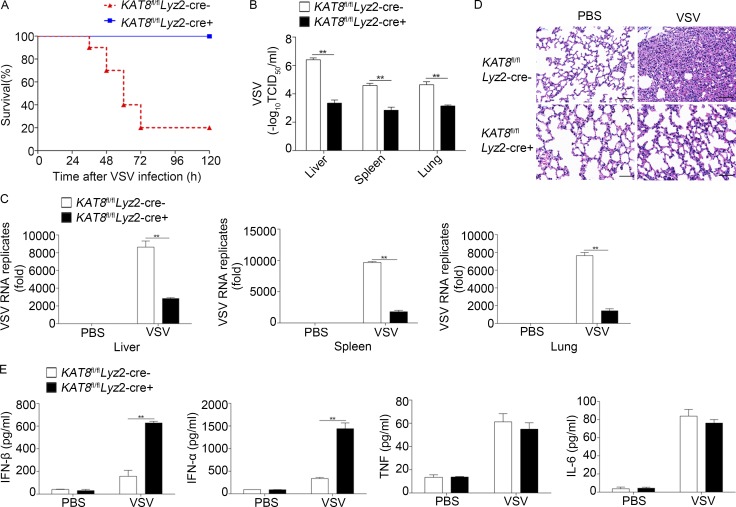
To further elucidate the importance of KAT8 in antiviral immunity, we challenged KAT8fl/flLyz2-Cre− and KAT8fl/flLyz2-Cre+ mice with VSV and found that the survival rate of KAT8-deficient mice was significantly higher than that of their KAT8-sufficient littermates (Fig. 2 A). VSV titers and replication in various organs were significantly lower in KAT8-deficient mice than in their KAT8-sufficient counterparts (Fig. 2, B and C). There was also less infiltration of inflammatory cells into lungs of KAT8-deficient mice after VSV infection (Fig. 2 D). KAT8-deficient mice also produced more IFN-α and IFN-β in serum in response to VSV infection (Fig. 2 E). Thus, KAT8 deficiency protects mice from virus challenge by selectively promoting IFN-I production.
Figure 2.
Deficiency of KAT8 protects mice against viral infection. (A) Survival of KAT8fl/flLyz2-Cre– or KAT8fl/flLyz2-Cre+ mice (n = 10 per group) after infection with VSV (5 × 107 plaque-forming units per gram body weight; Wilcoxon test). (B) VSV load in the liver, spleen, and lung of KAT8fl/flLyz2-Cre− or KAT8fl/flLyz2-Cre+ mice (n = 8 per group) 18 h after infection with VSV (as in A), assessed by endpoint-dilution assay and presented as 50% tissue culture infectious dose (TCID50). (C) Q-PCR analysis of VSV RNA in the liver, spleen, and lungs of KAT8fl/flLyz2-Cre− or KAT8fl/flLyz2-Cre+ mice 18 h after intraperitoneal injection of PBS or VSV (as in A). (D) Hematoxylin and eosin staining of sections of lungs from mice as in B. Bars, 50 µm. (E) ELISA of cytokines in serum from mice as in C. **, P < 0.01; two-tailed Student’s t test (B, C, and E). Data are from three independent experiments (B, C, and E; mean ± SEM) or are representative of three independent experiments with similar results (D).
KAT8 does not affect conventional innate signaling, including IRF3 phosphorylation, dimerization, and nuclear translocation
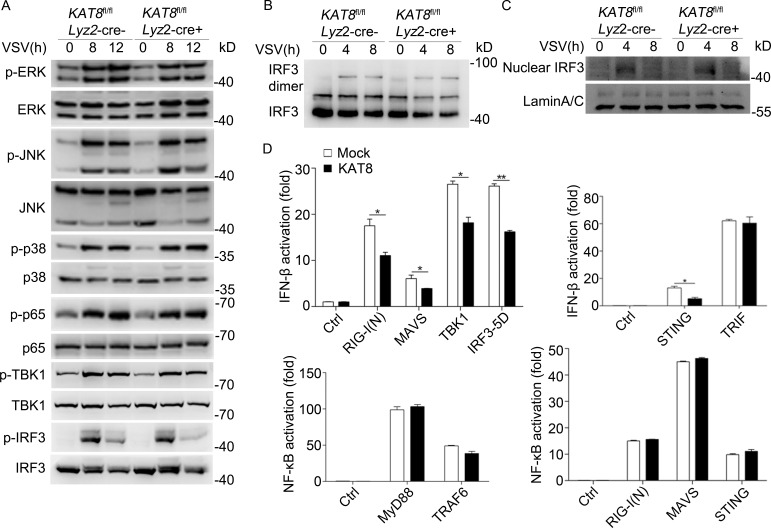
We next investigated the mechanism by which KAT8 suppressed virus infection–triggered IFN-I production by analyzing phosphorylation-induced activation of downstream signaling pathway components. There was no detectable difference in the phosphorylation of ERK, JNK, p38, and p65 between KAT8fl/flLyz2-Cre+ and KAT8fl/flLyz2-Cre− peritoneal macrophages after VSV infection (Fig. 3 A). Phosphorylation of the signaling molecule TBK1 and the transcription factor IRF3, which induce IFN-I, also showed no marked difference between KAT8fl/flLyz2-Cre+ and KAT8fl/flLyz2-Cre− peritoneal macrophages upon infection with VSV (Fig. 3 A). The dimerization and nuclear translocation of IRF3 also showed no change between KAT8fl/flLyz2-Cre+ and KAT8fl/flLyz2-Cre– peritoneal macrophages in response to VSV infection (Fig. 3, B and C). The same effects were also showed through knockdown of KAT8 via siRNA (Fig. S3, D–F). KAT8 overexpression did not influence activation of NF-κB luciferase reporter induced by MyD88, TRAF6, RIG-I(N), mitochondrial antiviral-signaling protein (MAVS), and stimulator ofinterferon genes (STING; Fig. 3 D). KAT8 overexpression suppressed activation of IFN-β luciferase reporter mediated by RIG-I(N), MAVS, TBK1, IRF3-5D, and STING, but not TIR domain–containing adaptor-inducing interferon-β (TRIF), in HEK293T cells (Fig. 3 D). Thus, KAT8 deficiency does not affect viral infection–induced activation of MAPK and NF-κB pathways. Since MAPK and NF-κB activation contribute to the production of inflammatory cytokines such as TNF and IL-6, these data further indicate that KAT8 deficiency has no effect on the production of inflammatory cytokines induced by TLR and virus.
Figure 3.
KAT8 targets IRF3. (A) Immunoblot analysis of phosphorylated (p-) or total ERK, JNK, p38, p65, TBK1, IRF3, or β-actin in KAT8fl/flLyz2-Cre− or KAT8fl/flLyz2-Cre+ peritoneal macrophages infected for the indicated times with VSV. (B) Immunoblot analysis of IRF3 dimerization in KAT8fl/flLyz2-Cre− or KAT8fl/flLyz2-Cre+ peritoneal macrophages infected for the indicated times with VSV. (C) Immunoblot analysis of IRF3 among nuclear proteins in KAT8fl/flLyz2-Cre− or KAT8fl/flLyz2-Cre+ peritoneal macrophages infected for the indicated times with VSV. (D) Luciferase activity assay of an IFN-β reporter in HEK293T cells transfected with KAT8 or Mock together with control vector, RIG-I(N), MAVS, TBK1, IRF3-5D, STING, or TRIF or luciferase activity assay of an NF-κB reporter in HEK293T cells transfected with KAT8 or Mock together with control vector, MyD88, TRAF6, RIG-I(N), MAVS, or STING assessed by dual-luciferase assay. *, P < 0.05; **, P < 0.01; two-tailed Student’s t test (D). Data are representative of three independent experiments with similar results (A–C) or are from three independent experiments (D; mean ± SEM).
KAT8 directly interacts with IRF3 via its MYST domain
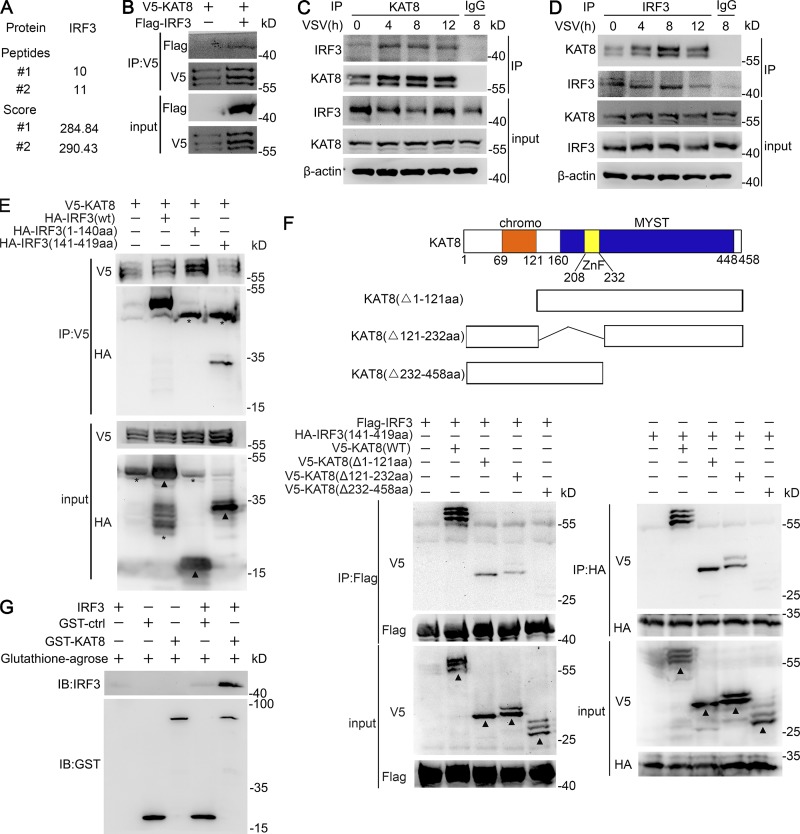
Next, KAT8-interacting proteins were identified by immunoprecipitation (IP) using anti-KAT8 antibody in mouse peritoneal macrophages infected with VSV followed by mass spectrometry (MS) analysis, which showed that IRF3 interacted with KAT8 (Fig. 4 A). We also confirmed the interaction in HEK293T cells transfected with V5-tagged KAT8 and Flag-tagged IRF3 (Fig. 4 B). Then, we sought to investigate whether KAT8 was able to interact with IRF3 endogenously. The endogenous KAT8–IRF3 complex was detected in peritoneal macrophages even in the absence of VSV infection; however, this interaction was enhanced after VSV infection (Fig. 4, C and D), consistent with the MS results.
Figure 4.
KAT8 interacts with IRF3 via the MYST domain. (A) MS analysis of KAT8-binding proteins in peritoneal macrophages infected with VSV (1 MOI). #1 and #2 indicates two independent experiments. (B) Immunoblot analysis of KAT8 or IRF3 in HEK293T cells transfected with V5-tagged KAT8 alone or together with Flag-tagged IRF3, assessed before (input) or after IP with antibody to V5 or Flag. (C) Immunoblot analysis of endogenous KAT8 or IRF3 in peritoneal macrophages infected with VSV for the indicated times, assessed before (input) or after IP with IgG (control) or antibody to KAT8. (D) Immunoblot analysis of endogenous KAT8 or IRF3 in peritoneal macrophages infected for the indicated times with VSV, assessed before (input) or after IP with IgG or antibody to IRF3. (E) Immunoblot analysis of HEK293T cells transiently transfected with HA-tagged WT or mutant IRF3 plus V5-tagged KAT8 and assessed 24 h later before (input) or after IP with antibody to V5. (F) Immunoblot analysis of HEK293T cells transiently transfected with V5-tagged WT or mutant KAT8 plus Flag-tagged IRF3 or HA-tagged mutant IRF3 (141–419 aa) and assessed 24 h later before (input) or after IP with antibody to Flag or HA. (G) Immunoblot (IB) analysis of IRF3 and KAT8 interaction in a GST pull-down assay. Data are representative of three independent experiments with similar results. *, nonspecific bands; arrowheads, specific bands.
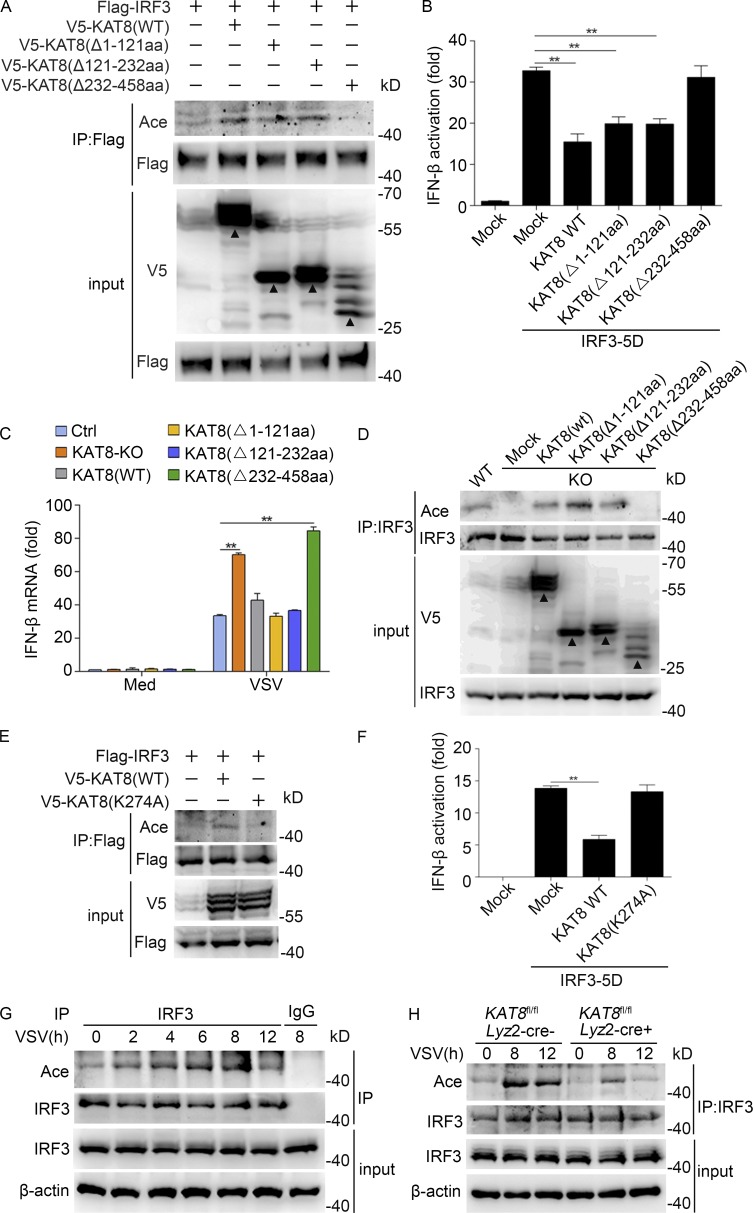
To map the regions of IRF3 responsible for the interaction with KAT8, we constructed two different hemagglutinin (HA)-tagged IRF3 truncations: N-terminal region 1–140 aa and C-terminal region 141–419 aa. The IRF3 truncation containing the C-terminal region (141–419 aa) was able to interact with KAT8, whereas the IRF3 truncation containing the N-terminal DNA-binding domain at 1–140 aa did not (Fig. 4 E). We further mapped the KAT8 domain required for the interaction with IRF3 and constructed three different V5-tagged KAT8 truncations, respectively lacking the chromodomain (Δ1–121 aa), C2HC zinc fingers (Δ121–232 aa), and the enzymatic MYST domain (Δ232–458 aa). Among the truncations, only the one lacking the MYST domain was unable to interact with the IRF3 C-terminal region (Fig. 4 F). Thus, the MYST domain of KAT8 was responsible for its interaction with the C-terminal region of IRF3. These data demonstrate that KAT8 interacts with the IRF3 C-terminal region through its MYST domain. Moreover, in vitro glutathione S-transferase (GST) pull-down assays confirmed the direct interaction of KAT8 and IRF3 (Fig. 4 G). Together, our findings demonstrate that KAT8 directly binds IRF3 through its MYST domain, suggesting a potential function of KAT8 in IRF3 acetylation.
KAT8 promotes IRF3 acetylation via its acetyltransferase activity
We then analyzed the acetylation of IRF3 in HEK293T cells transfected with V5-tagged KAT8 (WT) and the three different V5-tagged KAT8 truncations. KAT8 enhanced IRF3 acetylation, and this effect disappeared when the MYST domain of KAT8 was deleted (Fig. 5 A). Next, we detected IRF3-mediated Ifnb activation upon transfection with the three truncations of KAT8. Transfection of MYST domain–deficient KAT8 lacking acetyltransferase activity was unable to decrease Ifnb activation (Fig. 5 B).
Figure 5.
KAT8 promotes IRF3 acetylation via its MYST domain. (A) Immunoblot analysis of acetylation in HEK293T cells transiently transfected with V5-tagged WT or mutant KAT8 plus Flag-tagged IRF3 and assessed 24 h later before (input) or after IP with antibody to Flag. (B) Luciferase activity of an IFN-β reporter in HEK293T cells transfected with WT KAT8 or KAT8 truncations together with IRF3-5D. (C) KAT8-KO RAW264.7 cells were transfected with WT KAT8 or KAT8 truncations and then infected 24 h later with VSV (1 MOI) for 8 h. The expression of IFN-β at the mRNA level was measured by Q-PCR. (D) Immunoblot analysis of IRF3 acetylation and total IRF3 in KAT8-KO RAW264.7 cells transiently transfected with V5-tagged WT or mutant KAT8 and then infected 24 h later with VSV (1 MOI) for 8 h, assessed before (input) or after IP with antibody to IRF3. (E) Immunoblot analysis of acetylation in HEK293T cells transiently transfected with V5-tagged WT or mutant KAT8 plus Flag-tagged IRF3 and assessed 24 h later before (input) or after IP with antibody to Flag. (F) Luciferase activity of an IFN-β reporter in HEK293T cells transfected with WT KAT8 or KAT8 mutant (K274A) together with IRF3-5D. (G) Immunoblot analysis of endogenous acetylation of IRF3 in peritoneal macrophages infected for the indicated times with VSV (1 MOI), assessed before (input) or after IP with IgG or antibody to IRF3. (H) Immunoblot analysis of IRF3 acetylation in KAT8fl/flLyz2-Cre− or KAT8fl/flLyz2-Cre+ peritoneal macrophages infected for the indicated times with VSV (1 MOI), assessed before (input) or after IP with antibody to IRF3. **, P < 0.01 (one-way ANOVA; B and F); **, P < 0.01 (two-tailed Student’s t test; C). Data are representative of three independent experiments with similar results (A, D, E, G, and H) or are from three independent experiments (B, C, and F; mean ± SEM). Arrowheads, specific bands.
We next performed rescue experiments using WT KAT8 or KAT8 truncation to further elucidate KAT8-mediated regulation of IFN-I production and acetylation of IRF3 in KAT8-KO RAW264.7 cells. Overexpression of KAT8 lacking MYST domain, but not WT KAT8 or the other two KAT8 truncations, in KAT8-KO RAW264.7 cells up-regulated IFN-β mRNA expression (Fig. 5 C). Moreover, acetylation of IRF3 could not be detected in KAT8-KO RAW264.7 cells overexpressing KAT8 without MYST domain (Fig. 5 D). K274 acetylation of KAT8 is vital for the acetyltransferase activity of KAT8 (Kadlec et al., 2011). We then detected the acetylation of IRF3 in HEK293T cells transfected with V5-tagged WT KAT8 and KAT8 K274A mutant. WT KAT8 enhanced IRF3 acetylation, while KAT8 K274A mutant did not have this effect (Fig. 5 E). Furthermore, WT KAT8, but not K274A mutant, inhibited IRF3-mediated IFN-β luciferase reporter activation (Fig. 5 F). These data indicate that K274 is important for the acetyltransferase function of KAT8.
We also detected the acetylation status of IRF3 in RAW264.7 cells with stable overexpression of TAP-tagged IRF3 (TAP-IRF3 RAW264.7 cells) upon innate stimuli. The level of acetylation at lysine was significantly enhanced by infection with VSV, with no change upon stimulation with LPS or poly(I:C) (Fig. S4, A–C). The level of acetylation at lysine was also enhanced by VSV infection in peritoneal macrophages (Fig. 5 G). Furthermore, the IRF3 acetylation level was decreased in KAT8fl/flLyz2-Cre+ macrophages compared with that in KAT8fl/flLyz2-Cre− macrophages (Fig. 5 H). Similarly, knockdown of KAT8 down-regulated lysine acetylation of IRF3 in macrophages infected with VSV (Fig. S4 D). Additionally, the level of IRF3 acetylation was significantly reduced in KAT8-KO RAW264.7 cells compared with that in control RAW264.7 cells (Fig. S4 E). Moreover, the acetylation of KAT8 was increased in macrophages infected with VSV (Fig. S4 F). Considering that KAT8 acetylation is vital for the acetyltransferase activity of KAT8 (Kadlec et al., 2011), these data imply that the acetyltransferase activity of KAT8 may be enhanced upon viral infection. Collectively, these data show that KAT8 can enhance acetylation of IRF3 induced by virus infection via its acetyltransferase activity.
Identification of IRF3 acetylation at K359 mediated by KAT8 in antiviral innate immunity
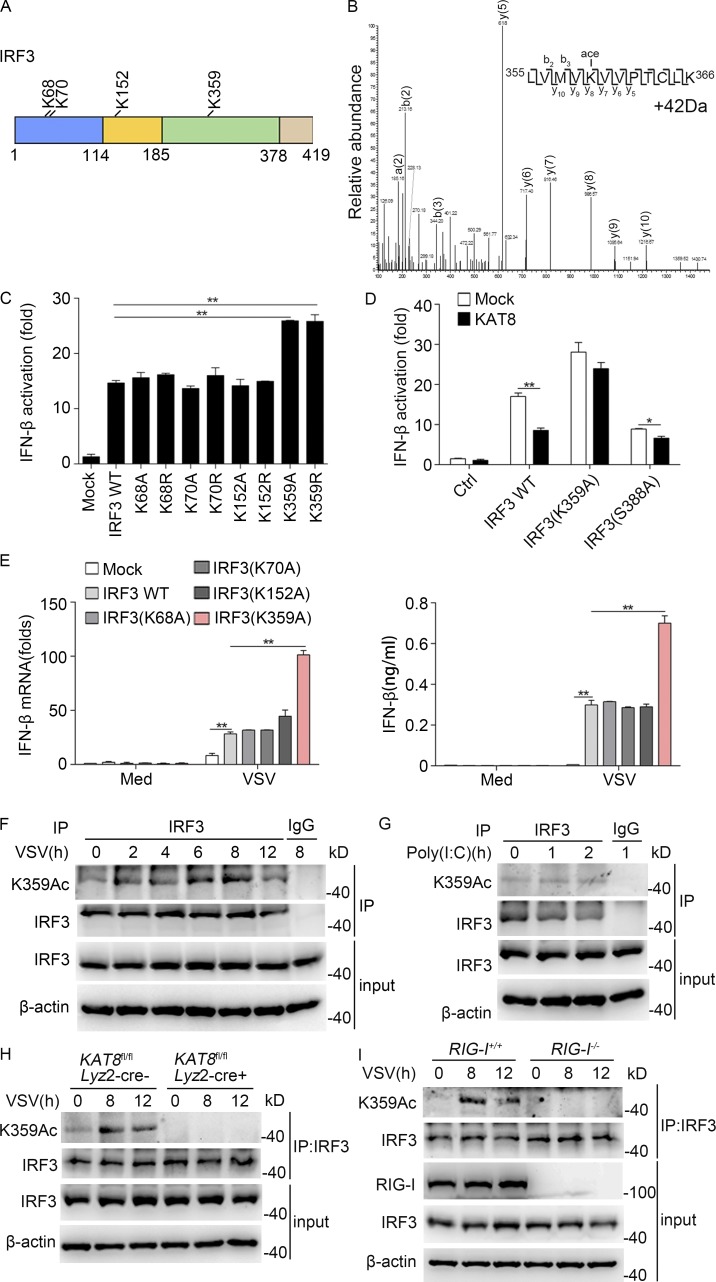
To further determine potential acetylated lysine residues of IRF3, we performed MS analysis in both untreated and VSV-infected TAP-IRF3 RAW264.7 cells. Four acetylated lysine residues, including K68, K70, K152, and K359, were identified (Fig. 6 A). K68 and K70 acetylation were present in untreated and VSV-infected macrophages, whereas K152 and K359 acetylation were only detected after VSV infection. In addition, a mass shift of 42 daltons was observed in VSV-infected macrophages for the IRF3 peptide LVMVKVVPTCLK, which is consistent with acetylation at the K359 residue (Fig. 6 B). To further investigate whether the acetylation of these sites on IRF3 could affect IRF3 transcriptional activity, we constructed acetylation-defective IRF3 mutants with each lysine residue substituted with either alanine (K68A, K70A, K152A, and K359A) or arginine (K68R, K70R, K152R, and K359R). Acetylation-defective substitution at K359 (K359A or K359R) significantly increased IRF3-driven IFN-β luciferase reporter activation, but those mutants at K68, K70, and K152 had no such effect (Fig. 6 C). Next, IRF3 mutants (IRF3-K152A and IRF3-K359A) were transfected along with KAT8 in 293T cells and assessed for their acetylation patterns. IRF3 acetylation level was markedly reduced upon transfection with IRF3-K359A (Fig. S4 G), suggesting that Lys359 may be the major acetylation site of IRF3 by KAT8.
Figure 6.
Acetylation of IRF3 at K359 in VSV-infected macrophages. (A) Illustration of acetylated lysine residues of IRF3 identified by an MS assay. (B) The tryptic peptide IRF3 (355–366 LVMVKVVPTCLK), consistent with acetylation, is characterized by a neutral addition of 42 D (neutral addition of H2O or NH3 omitted for clarity). (C) Luciferase activity of an IFN-β reporter in HEK293T cells transfected with control vector (Mock) or vectors encoding WT or mutant IRF3. (D) Luciferase activity of an IFN-β reporter in HEK293T cells transfected with control vector or vector encoding WT or mutant IRF3 (IRF3-K359A, and IRF3-S388A) along with vector encoding KAT8. (E) MEF cells from IRF3-deficient mice were transfected with WT or IRF3 mutants and then infected 24 h later with VSV (1 MOI) for 8 h. The mRNA level of IFN-β was measured by Q-PCR, and the production of IFN-β was measured by ELISA. (F) Immunoblot analysis of IRF3 acetylated at Lys359 (K359Ac) and total IRF3 in peritoneal macrophages infected for the indicated times with VSV (1 MOI), assessed before (input) or after IP with IgG or antibody to IRF3. (G) Immunoblot analysis of IRF3 acetylated at Lys359 (K359Ac) and total IRF3 in peritoneal macrophages stimulated for the indicated times with poly(I:C) (10 µg/ml), assessed before (input) or after IP with IgG or antibody to IRF3. (H) Immunoblot analysis of IRF3 acetylated at Lys359 (K359Ac) and total IRF3 in KAT8fl/flLyz2-Cre− or KAT8fl/flLyz2-Cre+ peritoneal macrophages infected with VSV (1 MOI), followed by IP with antibody to IRF3. (I) Immunoblot analysis of IRF3 acetylated at Lys359 (K359Ac) and total IRF3 in RIG-I–sufficient or RIG-I–deficient peritoneal macrophages infected with VSV (1 MOI), followed by IP with antibody to IRF3. **, P < 0.01 (one-way ANOVA; C); *, P < 0.05; **, P < 0.01 (two-tailed Student’s t test; D and E). Data are representative of three independent experiments with similar results (F–I) or are from three independent experiments (C–E; mean ± SEM).
Phosphorylation is considered the major PTM responsible for IRF3 activity (Sharma et al., 2003), which prompted us to analyze the interplay between acetylation and phosphorylation of IRF3. Ser396 (corresponding to Ser388 in mouse IRF3) is the main target of several kinases, such as TBK1, and a critical determinant of the activation status of IRF3 (Sharma et al., 2003). We thus constructed an IRF3-S388A mutant plasmid and transfected IRF3 and IRF3 mutants (IRF3-K359A or IRF3-S388A) along with KAT8 in 293T cells and detected the IFN-β luciferase reporter activity. In the presence of WT IRF3, KAT8 greatly reduced Ifnb activation. This effect was weakened by coexpression of IRF3 S388A mutant (Fig. 6 D), suggesting that acetylation of IRF3 may occur after its phosphorylation. This is in agreement with an earlier report that phosphorylation of IRF3 was crucial for the acetylation of IRF3 by p300 (Lin et al., 1999). Additionally, the inhibitory effect of KAT8 in IRF3-driven Ifnb activation disappeared with the expression of the acetylation-defective IRF3 mutant (IRF3-K359A; Fig. 6 D). This further indicates that KAT8 reduces IRF3-driven Ifnb activation via acetylation of IRF3 at K359. Next, we performed rescue experiments using IRF3- or IRF3 mutant–overexpressing plasmids to further elucidate the importance of K359 acetylation of IRF3 in regulating IFN-I production. Overexpression of IRF3 K359A in IRF3-deficient MEF cells significantly increased IFN-β production upon VSV infection (Fig. 6 E). These findings suggest that acetylation at the K359 site of IRF3 is most likely responsible for regulating IFN-I production.
We next generated an antibody specifically directed against IRF3 acetylated at K359. Dot-blot analysis indicated that this antibody specifically recognized IRF3 peptide acetylated at K359, but not IRF3 peptide acetylated at K68, K70, or K152 or a control peptide (Fig. S4 H). The level of IRF3 acetylation at K359 was significantly induced in TAP-IRF3 RAW264.7 cells by VSV infection, but not by stimulation with LPS or poly(I:C) (Fig. S4, I–K). We also used this antibody to analyze K359-acetylated IRF3 immunoprecipitated from whole-cell extracts of peritoneal macrophages infected with VSV. We detected K359-acetylated IRF3 in macrophages and found that the acetylation of IRF3 at K359 was enhanced after infection with VSV, but not stimulation with poly(I:C) (Fig. 6, F and G). Moreover, the acetylation of IRF3 at K359 markedly decreased in KAT8fl/flLyz2-Cre+ macrophages in response to infection with VSV (Fig. 6 H). Furthermore, VSV-induced K359 acetylation of IRF3 in macrophages was dependent on RIG-I–activated signaling (Fig. 6 I). Similarly, knockdown of KAT8 down-regulated the acetylation of IRF3 at K359 in macrophages infected with VSV (Fig. S4 L). Furthermore, the acetylation of IRF3 at K359 was significantly lower in KAT8-KO RAW264.7 cells than that in control cells in response to VSV infection (Fig. S4 M). Together, these results indicate that KAT8 enhances IRF3 acetylation at K359, and K359 acetylation of IRF3 plays a pivotal role in the regulation of IFN-I production.
KAT8 impairs IRF3 recruitment to IFN promoters by acetylating K359 of IRF3
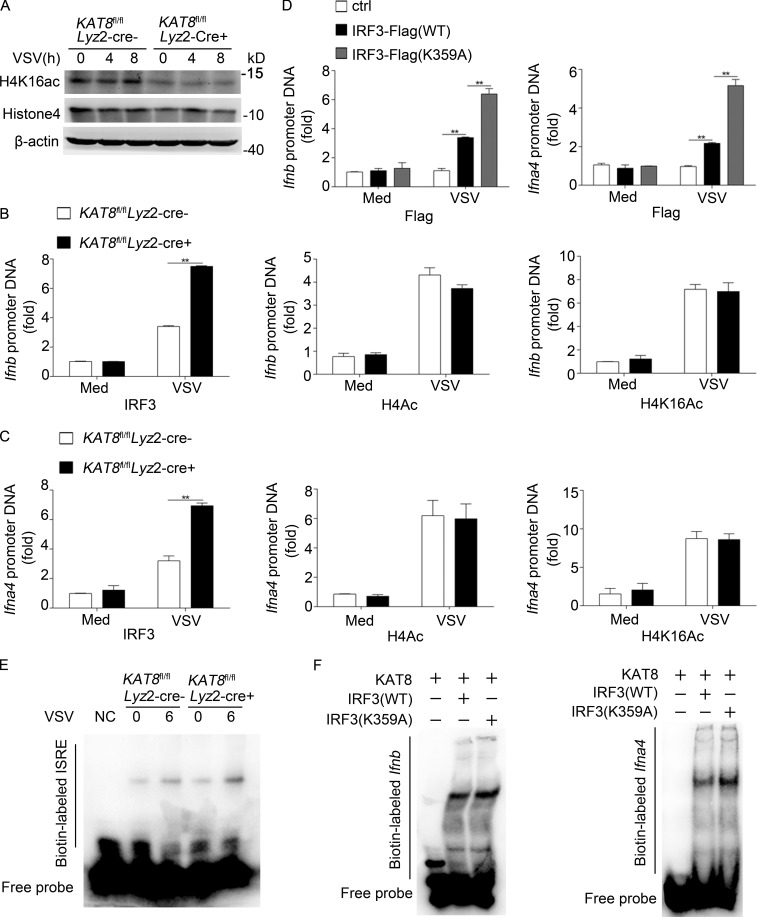
KAT8 regulates gene transcription mostly through acetylation of H4K16. We found lower levels of H4K16 acetylation in whole-cell lysates from KAT8fl/flLyz2-Cre+ macrophages compared with those in KAT8fl/flLyz2-Cre− macrophages (Fig. 7 A). We obtained similar results in KAT8-silenced macrophages (Fig. S5 A). Although KAT8 deficiency could decrease H4K16 acetylation, chromatin IP (ChIP) assays showed that there was no difference in abundance of H4ac and H4K16ac at promoters of Ifnb and Ifna genes between KAT8fl/flLyz2-Cre+ and KAT8fl/flLyz2-Cre− macrophages infected with VSV. However, the abundance of IRF3 at promoters of Ifnb and Ifna4 genes was significantly enhanced in KAT8fl/flLyz2-Cre+ macrophages compared with those in KAT8fl/flLyz2-Cre− macrophages with VSV infection (Fig. 7, B and C). Similarly, knockdown of KAT8 promoted IRF3 recruitment to promoters of Ifnb and Ifna4 genes but had no effect on the abundance of H4ac and H4K16ac at Ifn gene promoters (Fig. S5, B and C). To further confirm that K359 acetylation of IRF3 mediated by KAT8 is important for the recruitment of IRF3 to Ifn gene promoters, we transfected WT IRF3 or IRF3 K359A plasmids into IRF3-deficient MEFs. ChIP assays showed that the abundance of IRF3 K359A to promoters of Ifnb and Ifna genes markedly increased compared with the abundance of WT IRF3 (Fig. 7 D). An electrophoretic mobility shift assay (EMSA) with biotin-labeled interferon-sensitive response element (ISRE) showed the more binding of IRF3 to ISRE in KAT8fl/flLyz2-Cre+ macrophages as compared with that in KAT8fl/flLyz2-Cre− macrophages upon VSV infection (Fig. 7 E). Furthermore, an EMSA assay using recombinant WT IRF3 or mutant IRF3 (K359A) following treatment with KAT8 showed that mutant IRF3 (K359A) had more binding to the promoter of Ifna4 and Ifnb than WT IRF3 (Fig. 7 F). Collectively, these data indicate that KAT8 inhibits the recruitment of IRF3 to promoters of IFN-I genes by acetylating K359 of IRF3.
Figure 7.
KAT8 abrogates the abundance of IRF3 at IFN-I promoters. (A) Immunoblot analysis of H4K16ac, histone 4, or β-actin in KAT8fl/flLyz2-Cre− or KAT8fl/flLyz2-Cre+ peritoneal macrophages infected with VSV (1 MOI) for the indicated times. (B) ChIP analysis of IRF3, H4Ac, or H4K16Ac at the Ifnb promoter in KAT8fl/flLyz2-Cre− or KAT8fl/flLyz2-Cre+ peritoneal macrophages infected for 6 h with VSV (1 MOI) or left untreated. (C) ChIP analysis of IRF3, H4Ac, or H4K16Ac at Ifna4 promoter in KAT8fl/flLyz2-Cre− or KAT8fl/flLyz2-Cre+ peritoneal macrophages infected for 6 h with VSV (1 MOI) or left untreated. (D) ChIP analysis of IRF3 at Ifnb or Ifna4 promoter in MEF cells from IRF3-deficient mice overexpressed with WT or mutant IRF3 for 24 h and then infected with VSV (1 MOI) for 6 h. (E) EMSA of nuclear extract from KAT8fl/flLyz2-Cre− or KAT8fl/flLyz2-Cre+ peritoneal macrophages infected with VSV (1 MOI) for 6 h. The ISRE motif was biotin labeled. (F) Analysis of recombinant IRF3 WT or IRF3 mutant and IFN interaction in an EMSA assay. The Ifnb and Ifna motifs were biotin labeled. **, P < 0.01; two-tailed Student’s t test (B–D). Data are representative of three independent experiments with similar results (A, E, and F) or are from three independent experiments (B–D; mean ± SEM).
Discussion
PTMs of transcription factors such as IRF3 play crucial roles in the tight control of IFN-I production and innate immunity activation (Wang and Fish, 2012; Porritt and Hertzog, 2015; Huai et al., 2016). In this study, we identified KAT8 as a negative regulator that decreases virus-induced IFN-I by directly enhancing transcription factor IRF3 acetylation at K359 and preventing the recruitment of IRF3 to IFN-I promoters. Our findings report the crucial role of KAT8 and a specific acetylation residue of IRF3 in antiviral innate immunity.
IRF3 is a transcription factor and key inducer of IFN-I. IRF3 consists of an N-terminal DNA-binding domain and a C-terminal IRF association domain. The IRF association domain mediates phosphorylation-dependent homo-oligonucleotidemerization and hetero-oligonucleotidemerization with other IRF members and interacts with calmodulin-binding protein (CBP)/p300 (Lin et al., 1999). Under basal conditions, IRF3 is sequestered in the cytoplasm and is present as a monomer. Phosphorylation is considered the major PTM responsible for IRF3 activity. This phosphorylation induces IRF3 dimerization, translocation to the nucleus, and association with the coactivators CREB-binding protein p300, and the resultant complex activates the target genes in the nucleus. Ser396 is the target of the kinase such as TBK1 and a critical determinant for the activation of IRF3 (Sharma et al., 2003). Recently, the role of unconventional PTMs in the regulation of IRF3 activity is attracting more attention. For example, IRF3 methylation at K366 by NSD3 induced dissociation of IRF3 and protein phosphatase PP1cc, which maintains IRF3 phosphorylation and consequently promotes the production of IFN-I (Wang et al., 2017). SUMOylation of IRF3 represses IFN-I gene expression (Kubota et al., 2008). Although it is likely that IRF3 is subjected to a cascade of events responsible for regulating its biological activity, to date, no detailed mechanism or specific lysine residues of IRF3 acetylation have been reported to modulate IRF3 activity. Here, we demonstrate that K359 is a critical residue of IRF3, which is targeted by KAT8 for IRF3-mediated IFN-I production. In particular, acetylation of IRF3 at K359 was induced by viral infection, and such acetylation is essential for regulating virus-triggered induction of IFN-I. K359 of IRF3 is highly conserved among different kinds of mammals, suggesting the importance of this residue for the function of IRF3 in various kinds of mammals. In summary, IRF3 has different kinds of PTMs, which are involved in the synergetic tight control of IRF3 activation and IFN-I production to keep the immune balance.
KAT8 is responsible for H4K16 acetylation and has been shown to acetylate nonhistone substrates such as p53 (Sykes et al., 2006). In this study, KAT8 plays an important role in attenuating antiviral immune response via directly mediating K359 acetylation of IRF3. It will be interesting to delineate the relationship between phosphorylation and acetylation of IRF3 in the context of antiviral innate immunity. Interestingly, KAT8 mainly locates in the nucleus in peritoneal macrophages with or without virus infection. Thus, we postulate that KAT8 acetylates IRF3 in the nucleus after the phosphorylation of IRF3. This is consistent with the prior finding that p300 is capable of acetylating IRF3 in vitro, but only when IRF3 is present as a dimer in the holocomplex (Suhara et al., 2002), which indicates that acetylation of IRF3 by p300 occurs in the nucleus and after IRF3 phosphorylation. It is reasonable to assume that KAT8 has no effect on the phosphorylation, dimerization, and nuclear translocation of IRF3. IRF3 acetylation by p300 promotes IRF3 DNA-binding ability and thus IFN-I production, while IRF3 acetylation by KAT8 has the opposite effect. We conclude that acetylation of IRF3 at discrete sites regulates a distinct function of IRF3.
Acetylation events have been shown to directly affect protein–protein interactions, protein–DNA interactions, and protein stability (Gu and Roeder, 1997; Ageta-Ishihara et al., 2013; Choudhary et al., 2014; Wang et al., 2016). Here, we discover that K359 acetylation of IRF3 by KAT8 reduces the recruitment of IRF3 to IFN-I promoters. The possible mechanisms responsible for this effect may be that K359 acetylation of IRF3 by KAT8 may change the structure of IRF3, resulting in the dissociation of IRF3 from IFN-I promoters. In addition, IRF3 acetylation may influence the interaction between cofactors and IRF3, resulting in less abundance of IRF3 at IFN-I promoters. Therefore, further investigation may be required to elucidate the more detailed mechanisms by which IRF3 acetylation by KAT8 affects the transcriptional activity of IRF3.
On the basis of our data, we propose that K359 acetylation of IRF3 by KAT8 abrogates its recruitment to IFN-I promoters, thereby contributing to down-regulation of IFN-I production in the antiviral innate response. Through the analysis of publically available gene-profiling data (Gene Expression Omnibus accession no. GDS4602), we found that KAT8 expression was significantly decreased in psoriasis (Nair et al., 2009). Since IFN-I is involved in the pathogenesis of psoriasis (Stockenhuber et al., 2018), it is possible that the alterative expression of KAT8 may lead to an imbalance in the production of IFN-I and IFN-inducible genes, which might be involved in psoriasis development. Thus, the roles of KAT8 in autoimmune disease need to be further investigated. In conclusion, our study provides a novel PTM layer of IRF3 that can attenuate antiviral innate immunity. Inhibition of KAT8 expression may be a promising intervention for treating viral diseases.
Materials and methods
Animal experiments
Male C57BL/6J mice (6–8 wk old) were from Joint Ventures Sipper BK Experimental Animal Company. KAT8fl/fl mice were kindly provided by Prof. T. Thomas (The Walter and Eliza Hall Institute of Medical Research, Parkville, Australia). IRF3-deficient mice were kindly provided by Prof. T. Taniguchi (University of Tokyo, Tokyo, Japan). RIG-I–deficient mice were generated and maintained as described previously (Wang et al., 2007; Hou et al., 2014). Mice were bred in pathogen-free conditions. All animal experiments were undertaken in accordance with the National Institutes of Health’s Guide for the Care and Use of Laboratory Animals with approval of the Scientific Investigation Board of Second Military Medical University, Shanghai, China.
Reagents and antibodies
LPS (Escherichia coli serotype 0111:B4) and poly(I:C) were from Sigma-Aldrich and have been previously described (Zhang et al., 2015). HSV-1 was a gift from Q. Li (Chinese Academy of Sciences, Beijing, China), VSV was a gift from W. Pan (Second Military Medical University, Shanghai, China), and SeV was a gift from B. Sun (Chinese Academy of Sciences, Shanghai, China). IRF3-his, IRF3 (K359A)-his, and KAT8-GST fusion proteins were custom produced by Detai Biologics. Protein A/G Plus-Agarose Immunoprecipitation Reagent (sc-2003) used for IP was from Santa Cruz Biotechnology. ChIP-grade protein G magnetic beads (9006) and cell lysis buffer (9803) were from Cell Signaling Technology. Antibodies against KAT8 (46862), TBK1 (3013), phospho-TBK1 (5483), IRF3 (4302), phospho-IRF3 (4947), p65 (8242), phospho-p65 (3033), ERK (9102), phospho-ERK (9106), JNK (9258), phospho-JNK (4668), p38 (9212), phospho-p38 (9211), β-Actin (4967), Lamin A/C (4777S), Flag-Tag (2044), HA-Tag (3724), and V5-Tag (13202) were from Cell Signaling Technology. Antibodies against acetyl-histone H4 (06-598), acetyl-histone H4 (Lys16; 07-329), and CBP epitope tag were from Millipore. Antibodies against acetyl (ab21623) and histone 4 (ab10158) were from Abcam. The polyclonal antibody against IRF3 K359 acetylation (IRF3 K359ac) was custom produced by Abmart. PE/cy7 anti-mouse CD3 (145-2C11; 100320), BV421 anti-mouse CD4 (GK1.5; 100437), BV605 anti-mouse CD8 (53–6.7; 100744), PE-Dazzle 594 anti-mouse NK1.1 (PK136; 108747), BV605 anti-mouse CD45 (30-F11; 103155), AF647 anti-mouse F4/80 (BM8; 123122), PE anti-mouse ly6c (H41.4; 128008), and FITC anti-mouse LY6G (1A8; 127606) were from BioLegend. FITC anti-mouse CD45 (30-F11) and anti-CD16/32 (2.4G2) were from BD Bioscience. Percp-cy5.5 anti-mouse CD11b (M1/70) and APC-cy7 anti-mouse CD19 (eBio103) were from eBioscience. Oregon Green 488 goat anti-rabbit IgG (H+L; REF011038) was from Life Technologies.
Cell culture
HEK293T and RAW264.7 cell lines were from ATCC. To obtain mouse primary peritoneal macrophages, mice (male or female, 6–8 wk old) were injected intraperitoneally with 3% fluid thioglycollate medium (Merck). 3 d later, peritoneal lavage fluids were collected and centrifuged. Cells were resuspended with DMEM containing 10% FBS (Gibco) and cultured in plates. 2 h later, nonadherent cells were removed, and the adherent monolayer cells were washed with DMEM and used as peritoneal macrophages. Primary MEF cells were isolated form IRF3-deficient embryos. Stable overexpression of TAP-tagged IRF3 cells (TAP-RAW264.7 cells) were established by using interplay TAP-expressing system (Merck) containing a CBP, and streptavidin-binding protein epitopes in TAP tag were established by our laboratory. The cells above were cultured in endotoxin-free DMEM supplemented with 10% FCS (Gibco). Bone marrow–derived DCs were generated by cultivating mouse bone marrow cells in RPMI 1640 medium containing 10% FBS (Gibco) supplemented with recombinant GM-CSF (20 ng/ml) and IL-4 (5 ng/ml; Peprotech). Fresh medium was added on day 3 and day 6, and the fully differentiated DCs were harvested on day 8 for the function assay. The generated DC population was analyzed by flow cytometry based on CD11c expression and further enriched using CD11c microbeads (Miltenyi Biotec).
Plasmid constructs and transfection
Expression vectors encoding V5-tagged KAT8 were constructed by PCR cloning into pcDNA3.1-V5 eukaryotic expression vector and FLAG-tagged IRF3 into pcDNA3.1-FLAG vector, respectively. Mutants and truncations of KAT8 and IRF3 were generated by PCR-based amplification. For transient transfection of plasmids in HEK293T cells, jetPEI reagent (PolyPlus) was used according to the manufacturer’s instructions. For transient transfection of plasmids in MEF cells and RAW264.7 cells, FuGENE HD transfection reagent (E2311; Promega) was used according to the manufacturer’s instructions.
RNA isolation and quantitative PCR (Q-PCR) assay
Total RNA was extracted from cells using TRIzol reagent (Invitrogen) following the instructions. Q-PCR analysis was performed using LightCycler 480 (Roche) and a SYBR RT-PCR kit (Takara). The mouse primer sequences used for Q-PCR were as follows: Ifnb forward, 5′-CAGCTCCAAGAAAGGACGAAC-3′; Ifnb reverse, 5′-GGCAGTGTAACTCTTCTGCAT-3′; Ifna forward, 5′-TACTCAGCAGACCTTGAACCT-3′; Ifna reverse, 5′-CAGTCTTGGCAGCAAGTTGAC-3′; Tnf forward, 5′-GACGTGGAACTGGCAGAAGAG-3′; Tnf reverse, 5′-TTGGTGGTTTGTGAGTGTGAG-3′; Il6 forward, 5′-TAGTCCTTCCTACCCCAATTTCC-3′; Il6 reverse, 5′-TTGGTCCTTAGCCACTCCTTC-3′; Kat8 forward, 5′-ACGAGGCGATCACCAAAGTG-3′; Kat8 reverse, 5′-AAGCGGTAGCTCTTCTCGAAC-3′; β-actin forward, 5′-AGTGTGACGT TGACATCCGT-3′; and β-actin reverse, 5′-GCAGCTCAGTAACAGTCCGC-3′. Data were normalized to β-actin expression.
ELISA
IFN-β, IFN-α, TNF, and IL-6 levels in the supernatants or sera were measured using a mouse IFN-β or IFN-α ELISA kit (PBL Biomedical Laboratories) as well as a mouse TNF or IL-6 ELISA kit (R&D Systems).
Immunoprecipitation and immunoblot analysis
Total proteins of cells were extracted with cell lysis buffer (Cell Signaling Technology) and additional protease inhibitor “cocktail” (Calbiochem) and 1 mM PMSF. Cytoplasm and nuclear protein were extracted with NE-PER extraction reagent (Thermo). Equal amounts of proteins were counted after concentrations measured with a bicinchoninic acid assay (Pierce) for Western blot or IP as described.
ChIP assay
ChIP was performed following the instructions of the Chromatin Immunoprecipitation Assay Kit (Millipore). Target gene promoter sequences in both input DNA and recovered DNA immunocomplexes were assessed by Q-PCR analysis. The primers were as follows: Ifnb1 promoter forward, 5′-CCAGGAGCTTGAATAAAATGAA-3′; Ifnb1 promoter reverse, 5′-TGCAGTGAGAATGATCTTCCTT-3′. Ifna4 promoter forward, 5′-ATCCCAGACACACAAGCAGAGAG-3′; and Ifna4 promoter reverse, 5′-GGCTGTGGGTTTGAGTCTTCT-3′. Data were normalized by input DNA for each sample.
Luciferase assay
Cells were transiently transfected with the indicated combinations of plasmids. The total amount of transfected DNA in each dish was kept constant by the addition of empty vector wherever necessary. Cell extracts were prepared 24 h after transfection, and luciferase activity was measured using the Dual-Glo Luciferase Assay System (Promega). Luciferase activity was measured and normalized to Renilla.
CRISPR-Cas9–mediated gene knockout
Cas9/green fluorescent protein and KAT8 guide RNA plasmids (Shanghai Biomodel Organism Science & Technology Development Co.) were transiently transfected into RAW 264.7 cells using FuGENE HD transfection reagent according to the manufacturer’s instructions. Single transfected cells were sorted into individual wells in a 96-well plate using the MoFlo XDP and then expanded and screened by immunoblot.
RNA interference
siRNAs against KAT5, KAT6A, KAT6B, KAT7, KAT8, and negative control were from Dharmacon. Macrophages were transfected with siRNA by using RNAi MAX reagent (Invitrogen) according to a transfection procedure.
GST pull-down assay
GST pull-down assays were performed using a GST Protein Interaction Pull-Down Kit (Pierce) following the manufacturer’s instruction. In brief, 500 µg purified IRF3 protein was incubated with 100 µg GST-tagged KAT8 proteins or GST control protein in Tris-buffered saline buffer with glutathione agarose beads overnight at 4°C. The incubated proteins were then washed and immunoblotted using anti-IRF3 antibody.
EMSA
Peritoneal macrophages were infected with VSV (1 multiplicity of infection [MOI]) for 6 h, and then nuclear extracts from cells were prepared following the manufacturer’s instructions using NE-PER Nuclear and Cytoplasmic Extraction Reagents (Thermo Fisher) supplemented with protease inhibitor cocktail (Calbiochem) and 1 mM PMSF. The nuclear extracts were incubated with a biotin-labeled ISRE (forward, 5′-GGGAAAGGGAAACCGAAACTGA-3′; reverse, 5′-TCAGTTTCGGTTTCCCTTTCCC-3′) probe. Recombinant WT IRF3 or IRF3 mutation (K359A), together with recombinant KAT8, were incubated with biotin-labeled IFN-β probe (forward, 5′-GAAAACTGAAAGGGAGAACTGAAAGT-3′; reverse, 5′-CTTTTGACTTTCCCTCTTGACTTTCA-3′) or a biotin-labeled IFN-α probe (forward, 5′-GAAAGTGAAAAGAGAATTGGAAAGC-3′; reverse, 5′-CTTTCACTTTTCTCTTAACCTTTCG-3′).
RNA sequencing analysis
Total RNA was isolated using an RNeasy mini kit (Qiagen). Total RNA was quantified using a NanoDrop ND-2000 spectrophotometer (Thermo Fisher Scientific), and RNA integrity was determined using an Agilent 2100 system and an RNA 6000 Nano kit (Agilent Technologies). The paired-end libraries were constructed using a TruSeq Stranded mRNA LTSample Prep Kit (Illumina) according to the manufacturer’s instructions. The libraries were then sequenced on an Illumina platform (HiSeq X Ten; Illumina, Shanghai OE Biotech Co., Ltd.), and 150-bp paired-end reads were generated. Raw data (raw reads) were processed using Trimmomatic. The reads containing poly-N and low-quality reads were removed to obtain the clean reads. Then, the clean reads were mapped to the reference genome using hisat2. The fragments per kilobase of transcript per million mapped reads value of each gene was calculated using cufflinks, and the read counts of each gene were obtained by htseq-count. Differentially expressed genes were identified using the DESeq (2012) R package functions estimate Size Factors and nbinomTest. Gene ontology enrichment and Kyoto Encyclopedia of Genes and Genomes pathway enrichment analysis of differentially expressed genes were respectively performed using R based on the hypergeometric distribution. The RNA sequencing data have been deposited in the Gene Expression Omnibus with accession no. GSE124551.
Confocal microscopy
Macrophages were fixed with 4% paraformaldehyde for 15 min and permeabilized with 0.2% Triton X-100 for 5 min. After blocking with 5% BSA, cells were labeled with anti-KAT8 antibody followed by staining with corresponding secondary antibodies. Cells were observed with a Leica TCS SP2 confocal laser microscope.
Flow cytometric analysis
For flow cytometry, the 2.4G2 antibody was used as a pretreatment to minimize nonspecific binding. Cell suspensions were stained with fluorescence-conjugated primary antibodies for 30 min at 4°C. Flow cytometry data were acquired using the Fortessa flow cytometer (Becton Dickinson) and analyzed by FlowJo software (Becton Dickinson).
Statistical analysis
Statistical significance between two groups was determined by unpaired two-tailed Student’s t test or ANOVA. The statistical significance of survival curves was estimated according to the method of Kaplan and Meier, and the curves were compared with the generalized Wilcoxon’s test. Differences were considered to be significant when P < 0.05.
Online supplemental material
Fig. S1 analyzes the production of IFN-I in KAT8 knockdown peritoneal macrophages. Fig. S2 shows that KAT8 KO selectively promotes the production of IFN-I in RAW264.7 cells and that KAT8 deficiency has no influence on cell development and differentiation in vivo. Fig. S3 shows that KAT8 KO selectively promotes virus-induced IFN-I production in DCs and that KAT8 knockdown has no effect on virus-induced phosphorylation, dimerization, and nuclear translocation of IRF3 in macrophages. Fig. S4 detects IRF3 acetylation at K359 in VSV-infected macrophages. Fig. S5 analyzes the abundance of IRF3 at IFN-I promoters in KAT8 knockdown peritoneal macrophages.
Acknowledgments
We thank X. Sun and M. Jin for technical assistance.
This work was supported by the National Natural Science Foundation of China (81788101, 31570871, and 31770970), the Chinese Academy of Medical Sciences Innovation Fund for Medical Sciences (2016-12M-1-003), and the Shuguang Program of the Shanghai Education Development Foundation and the Shanghai Municipal Education Commission (18SG33).
All authors declare no competing financial interests.
Author contributions: W. Huai, X. Liu, C. Wang, Y. Zhang, Xi Chen, Xiang Chen, S. Xu, and N. Li performed the experiments; T. Thomas provided KAT8fl/fl mice; X. Cao, W. Huai, and X. Liu analyzed the data and wrote the paper; and X. Cao designed and supervised the study.
References
- Ageta-Ishihara N., Miyata T., Ohshima C., Watanabe M., Sato Y., Hamamura Y., Higashiyama T., Mazitschek R., Bito H., and Kinoshita M.. 2013. Septins promote dendrite and axon development by negatively regulating microtubule stability via HDAC6-mediated deacetylation. Nat. Commun. 4:2532 10.1038/ncomms3532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akhtar A., Zink D., and Becker P.B.. 2000. Chromodomains are protein-RNA interaction modules. Nature. 407:405–409. 10.1038/35030169 [DOI] [PubMed] [Google Scholar]
- Allfrey V.G., Faulkner R., and Mirsky A.E.. 1964. Acetylation and methylation of histones and their possible role in the regulation of rna synthesis. Proc. Natl. Acad. Sci. USA. 51:786–794. 10.1073/pnas.51.5.786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beltrao P., Albanèse V., Kenner L.R., Swaney D.L., Burlingame A., Villén J., Lim W.A., Fraser J.S., Frydman J., and Krogan N.J.. 2012. Systematic functional prioritization of protein posttranslational modifications. Cell. 150:413–425. 10.1016/j.cell.2012.05.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao X. 2016. Self-regulation and cross-regulation of pattern-recognition receptor signalling in health and disease. Nat. Rev. Immunol. 16:35–50. 10.1038/nri.2015.8 [DOI] [PubMed] [Google Scholar]
- Chatterjee A., Seyfferth J., Lucci J., Gilsbach R., Preissl S., Böttinger L., Mårtensson C.U., Panhale A., Stehle T., Kretz O., et al. . 2016. MOF Acetyl Transferase Regulates Transcription and Respiration in Mitochondria. Cell. 167:722–738.e23. 10.1016/j.cell.2016.09.052 [DOI] [PubMed] [Google Scholar]
- Chen K., Liu J., Liu S., Xia M., Zhang X., Han D., Jiang Y., Wang C., and Cao X.. 2017. Methyltransferase SETD2-Mediated Methylation of STAT1 Is Critical for Interferon Antiviral Activity. Cell. 170:492–506.e14. 10.1016/j.cell.2017.06.042 [DOI] [PubMed] [Google Scholar]
- Choudhary C., Kumar C., Gnad F., Nielsen M.L., Rehman M., Walther T.C., Olsen J.V., and Mann M.. 2009. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science. 325:834–840. 10.1126/science.1175371 [DOI] [PubMed] [Google Scholar]
- Choudhary C., Weinert B.T., Nishida Y., Verdin E., and Mann M.. 2014. The growing landscape of lysine acetylation links metabolism and cell signalling. Nat. Rev. Mol. Cell Biol. 15:536–550. 10.1038/nrm3841 [DOI] [PubMed] [Google Scholar]
- Dang W., Steffen K.K., Perry R., Dorsey J.A., Johnson F.B., Shilatifard A., Kaeberlein M., Kennedy B.K., and Berger S.L.. 2009. Histone H4 lysine 16 acetylation regulates cellular lifespan. Nature. 459:802–807. 10.1038/nature08085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deribe Y.L., Pawson T., and Dikic I.. 2010. Post-translational modifications in signal integration. Nat. Struct. Mol. Biol. 17:666–672. 10.1038/nsmb.1842 [DOI] [PubMed] [Google Scholar]
- Dou Y., Milne T.A., Tackett A.J., Smith E.R., Fukuda A., Wysocka J., Allis C.D., Chait B.T., Hess J.L., and Roeder R.G.. 2005. Physical association and coordinate function of the H3 K4 methyltransferase MLL1 and the H4 K16 acetyltransferase MOF. Cell. 121:873–885. 10.1016/j.cell.2005.04.031 [DOI] [PubMed] [Google Scholar]
- Füllgrabe J., Lynch-Day M.A., Heldring N., Li W., Struijk R.B., Ma Q., Hermanson O., Rosenfeld M.G., Klionsky D.J., and Joseph B.. 2013. The histone H4 lysine 16 acetyltransferase hMOF regulates the outcome of autophagy. Nature. 500:468–471. 10.1038/nature12313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu W., and Roeder R.G.. 1997. Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell. 90:595–606. 10.1016/S0092-8674(00)80521-8 [DOI] [PubMed] [Google Scholar]
- Gupta A., Hunt C.R., Pandita R.K., Pae J., Komal K., Singh M., Shay J.W., Kumar R., Ariizumi K., Horikoshi N., et al. . 2013. T-cell-specific deletion of Mof blocks their differentiation and results in genomic instability in mice. Mutagenesis. 28:263–270. 10.1093/mutage/ges080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch C.L., Wrana J.L., and Dent S.Y.R.. 2017. KATapulting toward pluripotency and cancer. J. Mol. Biol. 429:1958–1977. 10.1016/j.jmb.2016.09.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou J., Zhou Y., Zheng Y., Fan J., Zhou W., Ng I.O., Sun H., Qin L., Qiu S., Lee J.M., et al. . 2014. Hepatic RIG-I predicts survival and interferon-α therapeutic response in hepatocellular carcinoma. Cancer Cell. 25:49–63. 10.1016/j.ccr.2013.11.011 [DOI] [PubMed] [Google Scholar]
- Huai W., Song H., Yu Z., Wang W., Han L., Sakamoto T., Seiki M., Zhang L., Zhang Q., and Zhao W.. 2016. Mint3 potentiates TLR3/4- and RIG-I-induced IFN-β expression and antiviral immune responses. Proc. Natl. Acad. Sci. USA. 113:11925–11930. 10.1073/pnas.1601556113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadlec J., Hallacli E., Lipp M., Holz H., Sanchez-Weatherby J., Cusack S., and Akhtar A.. 2011. Structural basis for MOF and MSL3 recruitment into the dosage compensation complex by MSL1. Nat. Struct. Mol. Biol. 18:142–149. 10.1038/nsmb.1960 [DOI] [PubMed] [Google Scholar]
- Kubota T., Matsuoka M., Chang T.H., Tailor P., Sasaki T., Tashiro M., Kato A., and Ozato K.. 2008. Virus infection triggers SUMOylation of IRF3 and IRF7, leading to the negative regulation of type I interferon gene expression. J. Biol. Chem. 283:25660–25670. 10.1074/jbc.M804479200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Zhang Q., Ding Y., Liu Y., Zhao D., Zhao K., Shen Q., Liu X., Zhu X., Li N., et al. . 2016. Methyltransferase Dnmt3a upregulates HDAC9 to deacetylate the kinase TBK1 for activation of antiviral innate immunity. Nat. Immunol. 17:806–815. 10.1038/ni.3464 [DOI] [PubMed] [Google Scholar]
- Lin R., Heylbroeck C., Pitha P.M., and Hiscott J.. 1998. Virus-dependent phosphorylation of the IRF-3 transcription factor regulates nuclear translocation, transactivation potential, and proteasome-mediated degradation. Mol. Cell. Biol. 18:2986–2996. 10.1128/MCB.18.5.2986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin R., Mamane Y., and Hiscott J.. 1999. Structural and functional analysis of interferon regulatory factor 3: localization of the transactivation and autoinhibitory domains. Mol. Cell. Biol. 19:2465–2474. 10.1128/MCB.19.4.2465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Qian C., and Cao X.. 2016. Post-translational modification control of innate immunity. Immunity. 45:15–30. 10.1016/j.immuni.2016.06.020 [DOI] [PubMed] [Google Scholar]
- Luo H., Shenoy A.K., Li X., Jin Y., Jin L., Cai Q., Tang M., Liu Y., Chen H., Reisman D., et al. . 2016. MOF Acetylates the Histone Demethylase LSD1 to Suppress Epithelial-to-Mesenchymal Transition. Cell Reports. 15:2665–2678. 10.1016/j.celrep.2016.05.050 [DOI] [PubMed] [Google Scholar]
- Merson T.D., Dixon M.P., Collin C., Rietze R.L., Bartlett P.F., Thomas T., and Voss A.K.. 2006. The transcriptional coactivator Querkopf controls adult neurogenesis. J. Neurosci. 26:11359–11370. 10.1523/JNEUROSCI.2247-06.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mowen K.A., and David M.. 2014. Unconventional post-translational modifications in immunological signaling. Nat. Immunol. 15:512–520. 10.1038/ni.2873 [DOI] [PubMed] [Google Scholar]
- Nair R.P., Duffin K.C., Helms C., Ding J., Stuart P.E., Goldgar D., Gudjonsson J.E., Li Y., Tejasvi T., Feng B.J., et al. Collaborative Association Study of Psoriasis . 2009. Genome-wide scan reveals association of psoriasis with IL-23 and NF-kappaB pathways. Nat. Genet. 41:199–204. 10.1038/ng.311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips D.M. 1963. The presence of acetyl groups of histones. Biochem. J. 87:258–263. 10.1042/bj0870258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porritt R.A., and Hertzog P.J.. 2015. Dynamic control of type I IFN signalling by an integrated network of negative regulators. Trends Immunol. 36:150–160. 10.1016/j.it.2015.02.002 [DOI] [PubMed] [Google Scholar]
- Sadler A.J., and Williams B.R.. 2008. Interferon-inducible antiviral effectors. Nat. Rev. Immunol. 8:559–568. 10.1038/nri2314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitoh T., Tun-Kyi A., Ryo A., Yamamoto M., Finn G., Fujita T., Akira S., Yamamoto N., Lu K.P., and Yamaoka S.. 2006. Negative regulation of interferon-regulatory factor 3-dependent innate antiviral response by the prolyl isomerase Pin1. Nat. Immunol. 7:598–605. 10.1038/ni1347 [DOI] [PubMed] [Google Scholar]
- Schölz C., Weinert B.T., Wagner S.A., Beli P., Miyake Y., Qi J., Jensen L.J., Streicher W., McCarthy A.R., Westwood N.J., et al. . 2015. Acetylation site specificities of lysine deacetylase inhibitors in human cells. Nat. Biotechnol. 33:415–423. 10.1038/nbt.3130 [DOI] [PubMed] [Google Scholar]
- Sharma G.G., So S., Gupta A., Kumar R., Cayrou C., Avvakumov N., Bhadra U., Pandita R.K., Porteus M.H., Chen D.J., et al. . 2010. MOF and histone H4 acetylation at lysine 16 are critical for DNA damage response and double-strand break repair. Mol. Cell. Biol. 30:3582–3595. 10.1128/MCB.01476-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma S., tenOever B.R., Grandvaux N., Zhou G.P., Lin R., and Hiscott J.. 2003. Triggering the interferon antiviral response through an IKK-related pathway. Science. 300:1148–1151. 10.1126/science.1081315 [DOI] [PubMed] [Google Scholar]
- Shogren-Knaak M., Ishii H., Sun J.M., Pazin M.J., Davie J.R., and Peterson C.L.. 2006. Histone H4-K16 acetylation controls chromatin structure and protein interactions. Science. 311:844–847. 10.1126/science.1124000 [DOI] [PubMed] [Google Scholar]
- Stockenhuber K., Hegazy A.N., West N.R., Ilott N.E., Stockenhuber A., Bullers S.J., Thornton E.E., Arnold I.C., Tucci A., Waldmann H., et al. . 2018. Foxp3+ T reg cells control psoriasiform inflammation by restraining an IFN-I-driven CD8+ T cell response. J. Exp. Med. 215:1987–1998. 10.1084/jem.20172094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suhara W., Yoneyama M., Kitabayashi I., and Fujita T.. 2002. Direct involvement of CREB-binding protein/p300 in sequence-specific DNA binding of virus-activated interferon regulatory factor-3 holocomplex. J. Biol. Chem. 277:22304–22313. 10.1074/jbc.M200192200 [DOI] [PubMed] [Google Scholar]
- Sykes S.M., Mellert H.S., Holbert M.A., Li K., Marmorstein R., Lane W.S., and McMahon S.B.. 2006. Acetylation of the p53 DNA-binding domain regulates apoptosis induction. Mol. Cell. 24:841–851. 10.1016/j.molcel.2006.11.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniguchi T., Ogasawara K., Takaoka A., and Tanaka N.. 2001. IRF family of transcription factors as regulators of host defense. Annu. Rev. Immunol. 19:623–655. 10.1146/annurev.immunol.19.1.623 [DOI] [PubMed] [Google Scholar]
- Thomas T., Voss A.K., Chowdhury K., and Gruss P.. 2000. Querkopf, a MYST family histone acetyltransferase, is required for normal cerebral cortex development. Development. 127:2537–2548. [DOI] [PubMed] [Google Scholar]
- Thomas T., Dixon M.P., Kueh A.J., and Voss A.K.. 2008. Mof (MYST1 or KAT8) is essential for progression of embryonic development past the blastocyst stage and required for normal chromatin architecture. Mol. Cell. Biol. 28:5093–5105. 10.1128/MCB.02202-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B.X., and Fish E.N.. 2012. The yin and yang of viruses and interferons. Trends Immunol. 33:190–197. 10.1016/j.it.2012.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C., Wang Q., Xu X., Xie B., Zhao Y., Li N., and Cao X.. 2017. The methyltransferase NSD3 promotes antiviral innate immunity via direct lysine methylation of IRF3. J. Exp. Med. 214:3597–3610. 10.1084/jem.20170856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D., Kon N., Lasso G., Jiang L., Leng W., Zhu W.G., Qin J., Honig B., and Gu W.. 2016. Acetylation-regulated interaction between p53 and SET reveals a widespread regulatory mode. Nature. 538:118–122. 10.1038/nature19759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Zhang H.X., Sun Y.P., Liu Z.X., Liu X.S., Wang L., Lu S.Y., Kong H., Liu Q.L., Li X.H., et al. . 2007. Rig-I-/- mice develop colitis associated with downregulation of G alpha i2. Cell Res. 17:858–868. 10.1038/cr.2007.81 [DOI] [PubMed] [Google Scholar]
- Weinert B.T., Wagner S.A., Horn H., Henriksen P., Liu W.R., Olsen J.V., Jensen L.J., and Choudhary C.. 2011. Proteome-wide mapping of the Drosophila acetylome demonstrates a high degree of conservation of lysine acetylation. Sci. Signal. 4:ra48 10.1126/scisignal.2001902 [DOI] [PubMed] [Google Scholar]
- Zhang Q., Zhao K., Shen Q., Han Y., Gu Y., Li X., Zhao D., Liu Y., Wang C., Zhang X., et al. . 2015. Tet2 is required to resolve inflammation by recruiting Hdac2 to specifically repress IL-6. Nature. 525:389–393. 10.1038/nature15252 [DOI] [PMC free article] [PubMed] [Google Scholar]