Abstract
Albomycins are peptidyl thionucleoside natural products that display antimicrobial activity against clinically important pathogens. Their structures are characterized by a thioheptose with atypical stereo-chemistry including a D-xylofuranose ring modified with a D-amino acid moiety. Herein it is demonstrated that AbmH is a pyridoxal 5′-phosphate (PLP)-dependent transaldolase that catalyzes a threo-selective aldol-type reaction to generate the thioheptose core with a D-ribofuranose ring and an L-amino acid moiety. The conversion of L-to D-amino acid configuration is catalyzed by the PLP-dependent epimerase AbmD. The D-ribo to D-xylo conversion of the thiofuranose ring appears according to gene deletion experiments to be mediated by AbmJ, which is annotated as a radical S-adenosyl-L-methionine (SAM) enzyme. These studies establish several key steps in the assembly of the thioheptose core during the biosynthesis of albomycins.
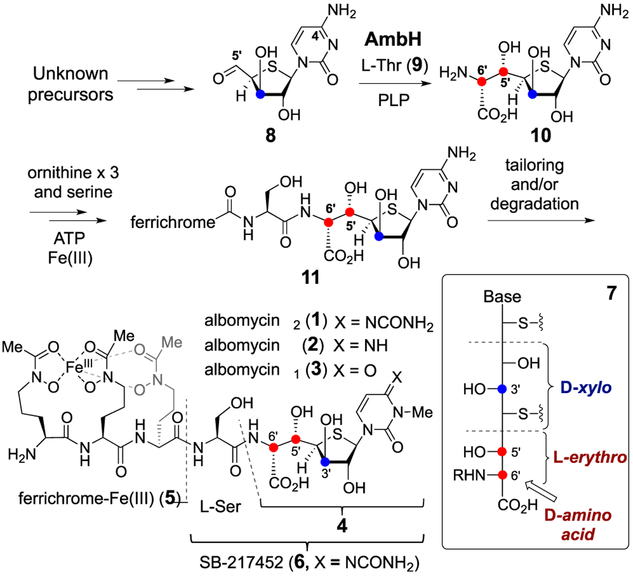
Albomycins (1–3) are sulfur-containing sideromycin antibiotics isolated from several species of Streptomy-cetes.1 The chemical structures of the albomycins consist of a 6′-amino-4′-thioheptose nucleoside (4) and an iron-chelating ferrichrome siderophore (5) that are connected via amide linkages to a serine residue (see Scheme 1).2 This allows albomycins to be actively transported via the bacterial iron uptake system into bacterial cells,3 where they are hydrolyzed by host peptidases liberating the thioheptose containing SB-217452 moiety (6) and the ferrichrome 5.4 SB-217452 (6) has been established as an inhibitor of bacterial seryl-tRNA synthetase due to its resemblance to seryl adenylate.4 Hence, albomycins have potent antimicrobial activities against many Gram-negative and Gram-positive bacteria.
Scheme 1.
Proposed Biosynthetic Pathway of Albomycins
The gene cluster responsible for albomycin production in Streptomyces sp. ATCC 700974 has been sequenced.5 Although several enzymes responsible for the tailoring modifications have been identified, little is known about how the 6′-amino-4′-thioheptose moiety (4) is biosynthesized. In particular, the D-configuration (6′R) of the α-amino acid and the D-xylo-configuration (3′R) of the furanose ring in the thioheptose core (see 4 and 7) are less commonly found in nature. Whether these unusual stereochemistries are inherited directly from the biosynthetic precursors or introduced after the core has been assembled is unknown.
The α-amino-β-hydroxy acid moiety at C5′-C6′ of 4 is found in several peptidyl nucleoside natural products such as the liposidomycins, caprazamycins, and muraymycins.6 It has been shown that this functional group is constructed in an aldol-type C─C bond forming reaction catalyzed by a PLP-dependent serine hydroxymethyl transferase homologue7 using L-threonine and an aldehyde precursor as substrates.8 A similar enzymatic conversion is also operative in the biosynthesis of obafluorin.9 On the basis of BLAST analysis, abmH in the albomycin gene cluster exhibits sequence similarity to genes encoding L-threonine:aldehyde transaldolases.5 Hence, the gene product AbmH was hypothesized to catalyze an aldol reaction between a 5′-oxo-4′-thionucleoside such as 8 and L-threonine (9) to form the C5′─C6′ bond (Scheme 1). The putative AbmH reaction product 10 may then be coupled with L-serine or a derivative thereof in the subsequent biosynthetic steps resulting in production of 11, which is the predicted precursor of 1.
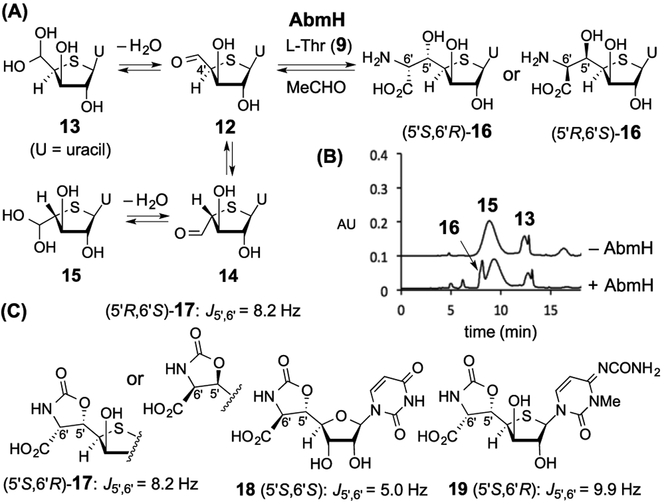
To test the proposed role of AbmH, N-His6-tagged AbmH was heterologously overexpressed and purified from Escherichia coli (Figure S2). Since attempts to synthesize 8 as a substrate for AbmH were unsuccessful due to complications involving the cross-coupling between the 4-amino and 5′-aldehyde groups, the uracil analogue 12 (existing as its hydrate form 13) was synthesized as an alternative substrate (Figure 1A and Supporting Information). When 12/13 (0.6 mM) was incubated with AbmH (3.2 μM) and L-threonine (9, 5 mM) in 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES) buffer (50 mM, pH 7.5), formation of several new compounds was detected by high-performance liquid chroma-tography (HPLC) (Figure 1B). On the basis of electrospray ionization-mass spectroscopy (ESI-MS) and nuclear Over-hauser effect spectroscopy (NOESY), the broad peak with a retention time at ~9 min was assigned as 15, which was the nonenzymatically generated 4′R-epimer of 13 (Figures S28). In contrast, a product peak at ~8 min displayed a mass consistent with an aldol adduct, such as 16 (calcd m/z for C11H16N3O7S+ [M + H]+ 334.0703; found 334.0686, see Figure S29). A similar ESI-MS result was also obtained for the minor peak at 6 min revealing the formation of another aldol adduct with the same chemical composition. Moreover, 1H NMR analysis indicated that the latter peak contained a mixture of two diastereomers of 16 (see Supporting Information).
Figure 1.
(A) AbmH reaction of 12/13. (B) HPLC-UV analysis. (C) Phosgene-derivatized AbmH-product 17 (5′S,6′R or its 5′R,6′S-isomer) and its analogues.
To establish the stereochemistry of the newly formed stereogenic centers C5′/C6′, the major AbmH product was isolated and derivatized with phosgene to afford a cyclic carbamate (see Supporting Information).10 1H NMR analysis of the carbamate (17) showed a value of 8.2 Hz for the J5′,6′ coupling that was significantly different from the reported J5′,6′ coupling (5.0 Hz) for 18, which has threo-stereochemistry at C5′/C6′ (Figure 1C).8a This outcome suggested an erythro-configuration at C5′/C6′ of 16 (5′S,6′R or its 5′R,6′S-isomer, see Figure 1A). This assignment is supported by comparison with the carbamate derivative 19 derived from 1, which has a J5′,6′ coupling of 9.9 Hz.
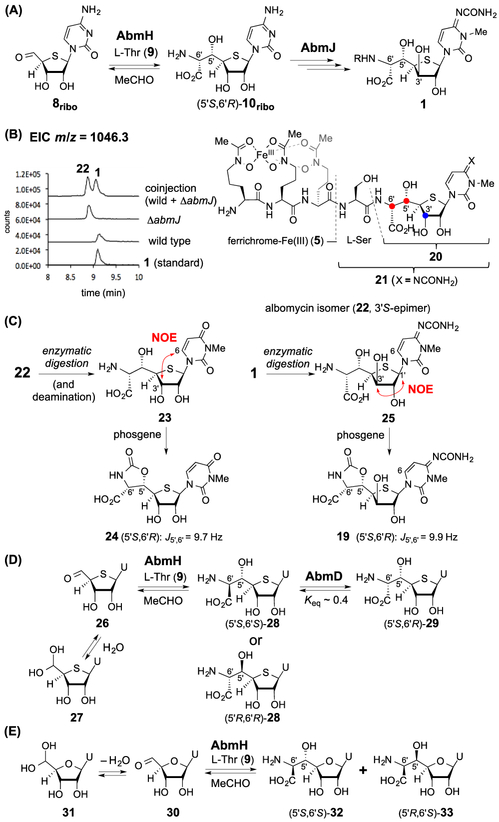
However, compound 8 represented by its synthetically accessible analogue 12/13 may not be the natural substrate of AbmH because it is a D-xylose-based nucleoside, which is unusual, and several stereoisomers of 16 were also observed during the incubation of 12/13 with AbmH. An alternative candidate is the 3′S-epimer of 8 (i.e., 8ribo, see Figure 2A), which has a D-ribose configuration. This would suggest that epimerization of C3′ is a necessary step prior to completion of the thioheptose core of 1. The abm cluster contains a gene, abmJ, which is annotated as encoding a radical SAM enzyme. It is well-known that certain radical SAM enzymes are capable of catalyzing epimerization reactions at unactivated centers.11,12 It was thus hypothesized that AbmH catalyzes the aldol-type reaction of 8ribo with L-threonine (9) to yield 10ribo prior to AbmJ-catalyzed C3′-epimerization to produce the thioheptose core in 1 (Figure 2A). To test this hypothesis, abmJ was in frame deleted in the producing strain (Figures 2B and S12). The ΔabmJ strain produced ferrichrome 5 as shown by HPLC and MS analysis; however, two new products were also observed. One had a mass consistent with that of 1 in its iron-chelated form (calcd m/z for C37H58FeN12O18S+ [M + H]+ 1046.3057; found 1046.2985, see Figure S29) and the other had a mass identical to that of SB-217452 (6, calcd m/z for C16H25N6O9S+ [M + H]+ 477.1398; found 477.1382, see Figure S29). However, neither of these two new products coeluted with 1 or 6 by HPLC, implying that they are epimers of 1 and 6 thereby implicating 22 and 21, respectively.
Figure 2.
(A) Alternative proposal of the albomycin biosynthesis. (B) LC–MS analysis of the metabolites from the ΔabmJ strain. Extracted ion chromatogram (EIC) traces corresponding to [M + H]+ signals from 1 or 22 are shown. (C) NOESY analysis and phosgene derivatization of the digested compounds. (D) Reaction of 26/27 with AbmH and AbmD to afford 28 and 29. (E) Reaction of 30/31 with AbmH.
Leucine amino peptidase was used to cleave the thionucleoside moiety from the iron-chelating ferrichrome to characterize the products isolated from the ΔabmJ strain by NMR (Figure 2C). When the resulting thionucleoside was derivatized using phosgene, a J5′,6′ coupling of 9.7 Hz in the corresponding carbamate was noted, indicating an erythro-configuration at C5′/C6′ consistent with 24. Moreover, 1H NMR NOESY analysis of the thionucleoside revealed a correlation between H-6 and H-3′ (Figures 2C and S28). These observations support assignment of the thionucleoside as 23. In contrast, a correlation between H-1′ and H-3′ was observed in 25 derived from hydrolysis of 1 (Figures 2C and S28). Collectively, these results indicate that the ΔabmJ metabolite is indeed 22, which has the S-configuration at C3′. Hence, the annotated radical SAM enzyme AbmJ appears to be necessary for C3′ epimerization from the S to R stereochemistry during the biosynthesis of 1. This hypothesis is also supported by the observation that complementation of the ΔabmJ mutant with abmJ partially restored production of 1 and 6 (Figure S13). While 1 showed antimicrobial activity against E. coli as previously reported,1b 22 exhibited significantly reduced bacterial growth inhibition based on disc-diffusion bioassays (Figure S26). These results imply the importance of the proposed C3′-epimerization for the biological activity of albomycins.
The proposed activity of AbmJ also implies that the biosynthetic substrate for AbmH is 8ribo rather than 8. Therefore, to further test this hypothesis, the uracil analogue 26 (existing as its hydrate form 27) was prepared (see Supporting Information). Upon incubation of 26/27 and L-threonine (9) with AbmH, a single aldol product was observed (Figures 2D and S14) in addition to the nonenzymatically generated 4′R-epimer of 27 (structure not shown). 1H NMR analysis after phosgene derivatization of the aldol product demonstrated a coupling of J5′,6′ = 5.1 Hz indicating an unexpected threo-configuration at C5′/C6′ consistent with 28 (5′S,6′S or its 5′R,6′R-isomer). The formation of the threo-product from 26/27 by AbmH is difficult to reconcile with the results from the abmJ deletion experiments because in the latter case the isolated product 22 has an erythro-configuration at C5′/C6′. Hence, in albomycin biosynthesis, the nascent product of the AbmH reaction (the 28 equivalent) must undergo isomerization to give the 5′S,6′R-configuration as seen in 10ribo.
Epimerization of amino acids can be catalyzed by PLP-dependent enzymes.13 The gene abmD in the abm gene cluster encodes another PLP-dependent enzyme, although it does not show high sequence similarity to known racemases/epimerases. While it has been reported that overexpression of abmD increases the level of 1 production,14 the catalytic role of the gene product AbmD has not been characterized. To determine whether AbmD can catalyze the epimerization of the AbmH-product 28 (or its physiological equivalent), N-His6-tagged AbmD was heterologously overexpressed and purified from E. coli (Figure S2). The UV absorption at 413 nm indicated the presence of PLP as an internal aldimine in AbmD (Figure S3). When the AbmH product (28, 0.056 mM) was incubated with AbmD (4.6 μM) in HEPES buffer (50 mM, pH 7.5), a new product was detected by HPLC (Figures S15 and S16). ESI-MS analysis indicated that the product has the same mass as 28 (Figure S29). The product structure was characterized to be 29 by NMR and coelution with the synthetic 5′S,6′R-standard (Figures 2D, S18, and S66–S70).
To verify the position of epimerization (C5′ or C6′) resulting in 29, the AbmD reaction with 28 was run in D2O (92% D). Incorporation of a single deuterium into 29 (83% D) was observed, and no dideuteration was noted as would be expected for β-epimerization involving hydrogen exchange at C5′ (Figure S19). Likewise, when the reaction was conducted in H218O (90% 18O), no 18O-labeling was found in 29 as would be expected for β-epimerization involving hydroxyl-exchange at C5′ (Figure S19). These observations rule out β-epimerization (i.e., at C5′) as the process leading to 29 and thus indicate that AbmD-catalyzed epimerization only occurs at the α-carbon : (i.e., C6′) of the amino acid moiety (Figure S20). Hence, the AbmD-substrate, which is the AbmH-product, is (5′S,6′S)-28.
The demonstrated activities of AbmH and AbmD strongly suggest that 8ribo is the substrate in the AbmH-catalyzed C5′─C6′ formation. However, there remains the possibility that the natural substrates of AbmH and AbmD are actually 4′-oxynucleosides rather than 4′-thionucleosides such that the sulfur atom is inserted after the AbmH/AbmD-catalyzed reactions. To investigate this possibility, compound 30 (existing as its hydrate form 31) was chemically synthesized and incubated with AbmH under the standard assay conditions (Figure 2E). Two products were formed in ~1:1 ratio (Figure S21). One was identified as 32 with a threo-configuration (5′S,6′S) at C5′/C6′ (J5′,6′ = 5.2 Hz in the carbamate form) and the other was an erythro-heptose nucleoside (5′R,6′S)-33 (J5′,6′ = 9.7 Hz in the carbamate form) based on the results of phosgene derivatization and comparison with standards (Supporting Information and Figure S22). These observations showed that the 4′-oxynucleoside can also be converted by AbmH, but with much reduced diastereoselectivity and catalytic efficiency (a 14-fold difference between kcat/Km for 26/27 vs 30/31, see Table S1). Moreover, when the AbmH-products 32 and 33 were separately incubated with AbmD, no epimerization was observed in either case (Figure S23). These results indicate that AbmD can recognize the 4′-thiosugar 28 but not 4′-oxysugars 32/33. Accordingly, the sulfur atom must be incorporated into the sugar skeleton early in the biosynthesis of 1. However, the apparent kcat/Km of AbmD with 29 was estimated to be only (5.2 ± 0.3) × 10−4 min−1 μM−1 (Figure S25). While the poor catalytic efficiency of AbmD with 29 may be a consequence of 29 bearing a uracil rather than the biosynthetically more relevant cytosine nucleobase, the physiological substrate for the AbmD reaction ultimately remains ambiguous. Therefore, additional experiments will be needed to resolve this question and establish the timing between the AbmH and AbmD reactions.
In summary, the in vitro investigation of the substrate specificity and stereochemistry of the AbmH/AbmD-catalyzed reactions in conjunction with the abmJ deletion study establishes several key steps in the assembly of the thioheptose core during biosynthesis of albomycins. Although epimerizations catalyzed by AbmD and AbmJ do not result in major structural alterations, they appear to be essential for conferring the antimicrobial activity of 1. The present study not only reveals the intricacy of the construction of a thionucleoside-containing natural product, but also enhances our knowledge regarding the correlation of structural variation and the biological activity important for the survival of the producing microorganisms.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by grants from the National Institutes of Health (GM035906 and 1 S10 OD021508-01 for NMR) and the Welch Foundation (F-1511). We thank Jessi Cai for the assistance in the preparation of AbmH.
Footnotes
Supporting Information
The Supporting Information is available free of charge on the ACSPublicationswebsite at DOI:10.1021/jacs.8b12565.
Details regarding experimental procedures and spectroscopic data (PDF)
Crystallographic data (CIF)
Notes
The authors declare no competing financial interest.
REFERENCES
- (1) (a).Gause GF Recent studies on albomycin, a new antibiotic. Br. Med. J 1955, 2, 1177–1179. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Stapley EO; Ormond RE Similarity of albomycin and grisein. Science 1957, 125, 587–589. [DOI] [PubMed] [Google Scholar]; (c) Lin Z; Xu X; Zhao S; Yang X; Guo J; Zhang Q; Jing C; Chen S; He Y Total synthesis and antimicrobial evaluation of natural albomycins against clinical pathogens. Nat. Commun 2018, 9, 3445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2) (a).Benz G Enzymatische spaltung der desferriform der albomycine δ1, δ2. Liebigs Ann. Chem 1984, 1984, 1399–1407. [Google Scholar]; (b) Benz G; Born L; Brieden M; Grosser R; Kurz J; Paulsen H; Sinnwell V; Webber B Absolute konfiguration der desferriform der albomycine. Liebigs Ann. Chem 1984, 1984, 1408–1423. [Google Scholar]; (c) Paulsen H; Brieden M; Benz G Synthese des sauersstoffanalogons der desferriform von δ1-Albomycin. Liebigs Ann. Chem 1987, 1987, 565–575. [Google Scholar]
- (3) (a).Pramanik A; Braun V Albomycin uptake via a Ferric hydroxamate transport system of Streptococcus pneumoniae R6. J. Bacteriol 2006, 188, 3878–3886. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Ferguson AD; Braun V; Fiedler HP; Coulton JW; Diederichs K; Welte W Crystal structure of the antibiotic albomycin in complex with the outer membrane transporter FhuA. Protein Sci. 2000, 9, 956–963. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Clarke TE; Braun V; Winkelmann G; Tari LW; Vogel HJ X-ray crystallographic structures of the Escherichia coli periplasmic protein FhuD bound to hydroxamate-type siderophores and the antibiotic albomycin. J. Biol. Chem 2002, 277, 13966–13972. [DOI] [PubMed] [Google Scholar]
- (4).Stefanska AL; Fulston M; Houge-Frydrych CSV; Jones JJ; Warr SR A potent seryl tRNA synthetase inhibitor SB-217452 isolated from a Streptomyces species. J. Antibiot 2000, 53, 1346–1353. [DOI] [PubMed] [Google Scholar]
- (5).Zeng Y; Kulkarni A; Yang Z; Patil PB; Zhou W; Chi X; Van Lanen S; Chen S Biosynthesis of albomycin δ2 provides a template for assembling siderophore and aminoacyl-tRNA synthetase inhibitor conjugates. ACS Chem. Biol 2012, 7, 1565–1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6) (a).Winn M; Goss RJ; Kimura K; Bugg TD Antimicrobial nucleoside antibiotics targeting cell wall assemblyrecent advances in structure-function studies and nucleoside biosynthesis. Nat. Prod. Rep 2010, 27, 279–304. [DOI] [PubMed] [Google Scholar]; (b) Walsh CT; Zhang W Chemical logic and enzymatic machinery for biological assembly of peptidyl nucleoside antibiotics. ACS Chem. Biol 2011, 6, 1000–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7) (a).Schirch L; Gross T Serine transhydroxymethylase. Identification as the threonine and allothreonine aldolases. J. Biol. Chem 1968, 243, 5651–5655. [PubMed] [Google Scholar]; (b) Gutierrez ML; Garrabou X; Agosta E; Servi S; Parella T; Joglar J; Clapeś P Serine hydroxymethyl transferase from Streptococcus thermophilus and l-threonine aldolase from Escherichia coli as stereocomplementary biocatalysts for the synthesis of β-hydroxy-α,ω-diamino acid derivatives. Chem. - Eur. J 2008, 14, 4647–4656. [DOI] [PubMed] [Google Scholar]
- (8) (a).Barnard-Britson S; Chi X; Nonaka K; Spork AP; Tibrewal N; Goswami A; Pahari P; Ducho C; Rohr J; Van Lanen SG Amalgamation of nucleosides and amino acids in antibiotic biosynthesisdiscovery of an l-threonine:uridine-5′-aldehyde transaldolase. J. Am. Chem. Soc 2012, 134, 18514–18517. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Cai W; Goswami A; Yang Z; Liu X; Green KD; Barnard-Britson S; Baba S; Funabashi M; Nonaka K; Sunkara M; Morris AJ; Spork AP; Ducho C; Garneau-Tsodikova S; Thorson JS; Van Lanen SG The biosynthesis of capuramycin-type antibioticsidentification of the A-102395 biosynthetic gene cluster, mechanism of self-resistance, and formation of uridine-5′-carboxamide. J. Biol. Chem 2015, 290, 13710–13724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9) (a).Scott TA; Heine D; Qin Z; Wilkinson B An L-threonine transaldolase is required for l-threo-β-hydroxy-α-amino acid assembly during obafluorin biosynthesis. Nat. Commun 2017, 8, 15935. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Schaffer JE; Reck MR; Prasad NK; Wencewicz TA β-Lactone formation during product release from a nonribosomal peptide synthetase. Nat. Chem. Biol 2017, 13, 737–744. [DOI] [PubMed] [Google Scholar]
- (10).Futagawa S; Inui T; Shiba T Nuclear magnetic resonance study of the stereoisomeric 2-oxazolidone and 2-phenyl-2-oxazoline derivatives of α-amino-β-hydroxy acids. Bull. Chem. Soc. Jpn 1973, 46, 3308–3310. [Google Scholar]
- (11) (a).Freeman MF; Gurgui C; Helf MJ; Morinaka BI; Uria AR; Oldham NJ; Sahl HG; Matsunaga S; Piel J Metagenome mining reveals polytheonamides as posttranslationally modified ribosomal peptides. Science 2012, 338, 387–390. [DOI] [PubMed] [Google Scholar]; (b) Benjdia A; Guillot A; Ruffie P; Leprince J; Berteau O Post-translational modification of ribosomally synthesized peptides by a radical SAM epimerase in Bacillus subtilis. Nat. Chem 2017, 9, 698–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Kudo F; Hoshi S; Kawashima T; Kamachi T; Eguchi T Characterization of a radical S-adenosyl-l-methionine epimerase, NeoN, in the last step of neomycin B biosynthesis. J. Am. Chem. Soc 2014, 136, 13909–13915. [DOI] [PubMed] [Google Scholar]
- (13) (a).Yoshimura T; Esak N Amino acid racemasesfunctions and mechanisms. J. Biosci. Bioeng 2003, 96, 103–109. [DOI] [PubMed] [Google Scholar]; (b) Du Y-L; Ryan KS Pyridoxal phosphate-dependent reactions in the biosynthesis of natural products. Nat. Prod. Rep 2019, DOI: 10.1039/C8NP00049B. [DOI] [PubMed] [Google Scholar]
- (14).Kulkarni A; Zeng Y; Zhou W; Van Lanen S; Zhang W; Chen S A branch point of Streptomyces sulfur amino acid metabolism controls the production of albomycin. Appl. Environ. Microbiol 2016, 82, 467–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.