Abstract
The general transcription factor IID (TFIID) is a critical component of the eukaryotic transcription preinitiation complex (PIC) and is responsible for recognizing the core promoter DNA and initiating PIC assembly. We used cryo-electron microscopy (cryo-EM), chemical crosslinking-mass spectrometry (CX-MS) and biochemical reconstitution to determine the complete molecular architecture of TFIID and define the conformational landscape of TFIID in the process of TATA-box binding protein (TBP) loading onto promoter DNA. Our structural analysis revealed five structural states of TFIID in the presence of TFIIA and promoter DNA, showing that the initial binding of TFIID to the downstream promoter positions the upstream DNA and facilitates scanning of TBP for a TATA-box and the subsequent engagement of the promoter. Our findings provide a mechanistic model for the specific loading of TBP by TFIID onto the promoter.
The regulation of transcription initiation is arguably the primary way by which the expression of genes is controlled. The transcription pre-initiation complex (PIC), is responsible for the loading of RNA polymerase II (Pol II) onto DNA (1, 2). The assembly of the PIC begins with the recognition of the core promoter by TFIID, aided by TFIIA (3). The TATA-box binding protein (TBP), a component of TFIID, recruits TFIIB, which then loads Pol II-TFIIF (4). Lastly, the addition of TFIIE and TFIIH facilitates the opening of the transcription bubble (5). While the stepwise assembly of a TBP-based PIC has been well characterized structurally (6), the process by which TFIID loads TBP onto the promoter is not well understood.
TFIID is a ~1.3 MDa complex that contains, in addition to TBP, 13 TBP-associating factors (TAFs), with six of them (TAF4, −5, −6, −9, −10, −12) present in two copies (7–9) (fig. S1). At low resolution, human TFIID is composed of three lobes (lobes A, B, and C), with a fairly rigid connection between lobes B and C, and with lobe A more flexibly attached to this “BC core” (10). In previous work we showed that in a promoter bound complex (IIDAS, which we will refer to here as IIDA-SCP), containing TFIID, TFIIA and the super core promoter (SCP) (11), the promoter elements downstream of the transcription start site (TSS) are recognized by TAF1 and TAF2 in lobe C, while TBP binds the TATA box upstream of the TSS with the aid of TFIIA and lobe B (12).
Here we present cryo-electron microscopy (cryo-EM) structures of human TFIID, alone and in various stages of promoter binding. Together with chemical crosslinking mass spectrometry (CX-MS) data and biochemical reconstitution, we were able to determine the complete structure of TFIID and the functional conformational landscape of the complex. Our studies lead to a mechanistic model of TBP loading onto the promoter by TFIID and TFIIA, and provide insights into how TFIID may engage chromatin, respond to transcriptional activators, and serve as a scaffold for PIC assembly.
Overall structure of TFIID
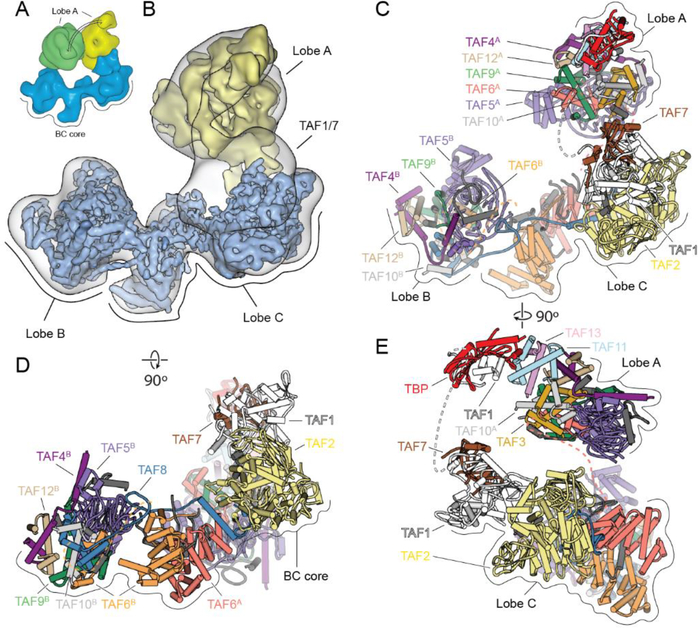
The flexible nature of TFIID has long hampered a high-resolution structural description of the intact complex (10). In previous work, we showed how the distribution of positions of the flexibly attached lobe A shifts upon binding of promoter DNA and TFIIA (10). Lobe A in apo-TFIID exists in a bimodal but continuous distribution of states, with roughly equal occupancy of two distinct, major states referred to as canonical and extended. While in the canonical state lobe A is near lobe C, in the extended state lobe A is between lobes B and C (Fig. 1A). The displacement of lobe A between these two states is ~100 Å. By sorting a large cryo-EM data set of free TFIID into two predominant states, refining them independently, and then combining the focused-refined regions, we were able to extend the resolution of the BC core to 4.5 Å (range of 4.2–6.5 Å) and to generate a three-dimensional (3D) reconstruction of lobe A at 9.5 Å (range of 8.5–15 Å) (Fig. 1B and figs. S2 and S3). We then used a combination cryo- EM, CX-MS, and structure prediction to generate a complete model of the complex.
Fig. 1. Cryo-EM Structure of TFIID.
(A) Cryo-EM reconstructions of TFIID, with the BC core in blue and Lobe A in yellow (canonical state) and green (extended state). (B) Transparent cryo-EM map of TFIID in the canonical state with fitted cryo-EM maps from focused refinements of the BC core and lobe A in solid blue and yellow, respectively. (C-E) TFIID structural model in front (C), top (D), and side views (E). See also Movie 1.
Compared to the IIDA-SCP structure (12), the density corresponding to the TAF1-TAF7 subcomplex within lobe C in apo-TFIID is poorly defined, indicating that this module is flexible in the unbound TFIID, but stabilized upon binding to promoter DNA (figs. S4 and S5). For the rest of lobe C, it was possible to dock into the density the model of the TAF6 HEAT repeat dimer, a segment from the C-terminal region of TAF8, and the TAF2 aminopeptidase-like domain (APD) from the previous IIDA-SCP structure (12), with adjustments and extensions made to fit the observed density (Fig. 1, C to E, and fig. S5).
Within lobe B, we were able to fit a homology model of the WD40 domain of TAF5, the crystal structures of TAF5 NTD2 domain and the histone-fold domain (HFD) heterodimers of TAF6-TAF9, TAF4-TAF12 and TAF8-TAF10, as well as to extend the models where additional densities were present in the cryo-EM map (Fig. 1, C and D, and fig. S5). The resulting atomic model for lobe B is consistent with our CX-MS data (fig. S6) and in agreement with previous biochemical studies (8, 13). To further validate our model, we heterologously co-expressed exclusively those segments of TAFs that we could directly model into the lobe B cryo-EM density, which comprised only 35% of the residues present in the full-length versions of the subunits (fig. S7). Three successive pull-downs using different affinity tags placed on TAF5, TAF4, and TAF8, followed by size exclusion chromatography, resulted in a pure, soluble complex containing stoichiometric amounts of all seven TAF fragments, supporting the formation of a stable complex from the components predicted by our structural model.
All the TAFs in lobe B, except for TAF8, have been proposed to exist in two copies within TFIID (8,14), suggesting that a similar architecture could exist within the flexible lobe A. We used a computational strategy, based on automated docking of different combinations of TFIID subunits into the lobe A cryo-EM density, to generate a complete model of lobe A (fig. S5). The core of the structure is equivalent to Lobe B, except for the replacement of TAF8 with TAF3 as the histone-fold partner of TAF10. Additionally, lobe A includes the TAF11/13 HFD pair and TBP (Fig. 1, C and E). Our placement of TAF11/13 adjacent to the TBP subunit is supported by the presence of chemical crosslinks between TAF11 and TBP (fig. S6) as well as in vivo and in vitro studies showing that the HFDs of TAF11/13 constitute the bridge between TBP and the rest of TFIID (15). Altogether, our structure defines the full architecture of human TFIID, revealing the complete evolutionarily-conserved regions of all the TAFs and TBP (fig. S1 and Movie 1).
TFIID assembly around a dimeric subcomplex of TAFs
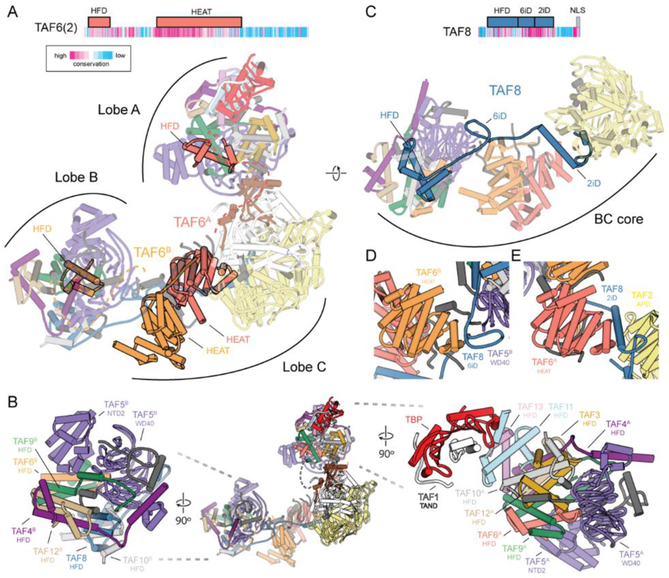
Our structure of human TFIID shows that the complex assembles around a dimeric, yet asymmetric arrangement of TAFs (fig. S6). Two copies of interacting TAF6 HEAT repeat domains are found at the center of the BC core, where they form a dimer with a 31 screw axis symmetry that bridges lobes B and C (Fig. 2A). The N-terminal HFDs of each copy of TAF6 are then separated between lobes A and B, and thus, TAF6, through the flexible connection between its HFD and HEAT repeat domain, tethers the entire complex together. This TAF6 connection is maintained throughout the various conformational states of TFIID (Fig. 2A). The HFD of TAF6 forms a heterodimer with the HFD of TAF9, which interacts with the WD40 and NTD2 regions of TAF5. The TAF6/9 HFD pair then forms a tetramer with the TAF4/12 HFD pair, and together these 5 subunits (TAF5, −6, −9, −4, −12) define the TAF subcomplex that is present in two copies within TFIID (Fig. 2B and figs. S7 and S8), one each in lobes A and B. The existence of a dimeric TAF-containing subcomplex has been previously proposed based on in vivo knockdown and in vitro biochemical studies (8,16). However, the structure within the native TFIID complex does not exhibit the symmetry previously proposed for a reconstituted subcomplex containing the same subunits, likely due to the presence of additional symmetry-breaking TAFs in the fully-formed, native complex (8) (fig. S7).
Fig. 2. Structural organization of human TFIID.
(A) Domain organization of TAF6, with sequence conservation colored based on ConSurf (69) scores (top). Model of TFIID with the TAF6 dimer highlighted (bottom). The dimer of TAF6 FIEAT repeats is centrally located within the complex. Dashed lines are shown connecting the TAF6 FIEAT domains with their corresponding FlFDs in lobes A and B. (B) Model of TFIID (center) and close ups of Lobe B (left) and Lobe A (right). (C) Domain organization of TAF8, with sequence conservation colored based on ConSurf (69) scores (top), and model of the BC core of TFIID with TAF8 highlighted (bottom). (D) The 6iD (TAF6 interacting domain) of TAF8 bridges the WD40 domain of TAF5 in Lobe B and the FIEAT repeat of TAF6 in Lobe C. (E) The 2iD (TAF2 interacting domain) of TAF8 bridges the FIEAT repeat of TAF6 and the APD of TAF2 within Lobe C. See also Movie 1.
The two sets of TAFs (4, 5, 6, 9,12) shared between lobes A and B act as a base for the assembly of the rest of each lobe. In lobe B, a hexamer of HFDs is formed by the TAF8/10, TAF6/9, and TAF4/12 HFD pairs. In lobe A, the TAF3/10 and TAF11/13 HFD pairs form an octamer-like structure with the TAF6/9 and TAF4/12 HFD pairs (Fig. 2B and Movie 1). Though the presence of higher-order histone-fold assemblies had been predicted to exist within TFIID, such a structure had not been visualized until now (fig. S8). It has been proposed that these nucleosome core-like structures may be involved in interaction with DNA and promoter binding (16–20). However, the surfaces of lobes A and B lack the large positively-charged patches observed in the nucleosomal histone octamer (fig. S8). The TAF6/9 HFD pair that was proposed to interact with the downstream DNA (20, 21) is actually located far from the DNA in the IIDA-SCP complex (fig. S8). We instead propose that HFDs serve as a structural scaffold within TFIID.
The difference in the flexibility of lobes A and B is likely due to the presence of TAF8 in lobe B, which stabilizes its connection with lobe C (Fig. 2C). In our model, the highly conserved middle region of TAF8 (residues 130–235) snakes through the BC core, interacting extensively with TAF2 and TAF6. Extending from its N-terminal HFD, the TAF6 interacting domain (6iD) of TAF8 forms a bridge between the WD40 of TAF5 in lobe B and the first of the HEAT repeats of TAF6 (Fig. 2D). The long helix of the TAF2-interacting domain (2iD) of TAF8 then bridges the second TAF6 HEAT repeat and the TAF2, and then TAF8 folds onto the surface of the TAF2 APD, effectively anchoring TAF2 to the rest of the complex. This network of interactions between TAF8, TAF6, and TAF2 (Fig. 2E) is consistent with previous biochemical studies (8,13).
Role of lobe B in the stabilization of upstream DNA binding
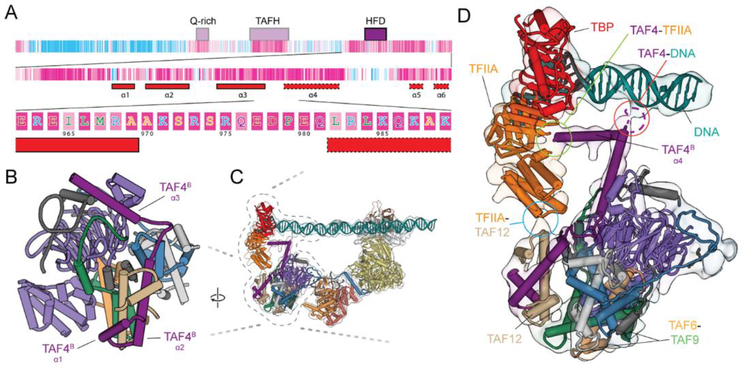
Our structural studies indicate that the function of lobe B is to stabilize the upstream DNA and bind TFIIA. Both of these functions involve the highly conserved C terminus of TAF4 (Fig. 3A). The HFD of TAF4, comprising helices α1 and α2, is followed by a large loop and a helix α3 that interacts with the WD40 of TAF5 (Fig. 3B). Docking of the lobe B structure into the IIDA-SCP map reveals that the highly conserved loop between α3 and a fourth helix in TAF4, α4, contacts the promoter DNA just downstream of the TATA-box (Fig. 3, C and D, and fig. S4). This loop has previously been shown to bind DNA in vitro (20), and in TAF4−/− human fibroblast cells stable expression of a TAF4 mutant lacking this loop results in the down-regulation of a subset of genes (22). From there, α4 continues toward the TBP/TFIIA density and is likely involved in TFIIA recruitment and the stabilization of the TFIIA-TBP-DNA module, in agreement with previous data (23) (Fig. 3D). The docking of lobe B into the IIDA-SCP map also revealed that the 4-helix bundle of TFIIA likely contacts the first helix-turn-helix of the TAF12 HFD (Fig. 3D). Thus, we propose that TAF4 and TAF12 within lobe B act to promote the binding of TBP to the upstream DNA by directly contacting both the DNA and TFIIA/TBP module. Therefore, the BC core of TFIID appears to act as a molecular ruler, placing TBP at a defined distance from the downstream promoter elements. This role suggests that maintaining a fairly rigid connection between lobes B and C is important for correctly positioning TBP with respect to the transcriptional start site (TSS), which in human core promoters are separated by ~30 base pairs (24, 25) (fig. S8).
Fig. 3. Upstream promoter binding stabilized by Lobe B.
(A) Domain organization and sequence conservation of TAF4 based on ConSurf (69) scores. The first level shows the domain organization of TAF4. The second level zooms in on the C terminus and shows the secondary structure (solid outline corresponds to observed secondary structure and dashed the predicted secondary structure based on PSIPRED (70) results (α4 is not visible in the apo-TFIID structure, but becomes ordered upon interaction with the DNA). The third level shows the amino acid sequence of the loop between helices 3 and 4, which contain several conserved, positively charged residues that could be contacting the DNA. (B) Structure of lobe B. (C) Model of TFIID docked into the 11 DAS reconstruction. (D) Zoom up of part of c, highlighting the loop between helices 3 and 4 as it contacts the DNA (circled in red), helix 4 continuing on toward the TFIIA and TBP (circled in green), and the interaction between the TFIIA and TAF12 (circled in blue).
Our structure suggests a potential overlap between the contacts that TAF4 makes with the upstream promoter DNA in the IIDA-SCP complex and those established by the TFIIF winged-helix domain within the PIC (6, 26) (fig. S9). Additionally, the downstream promoter binding regions of TAF1 and TAF2 were also found to clash with Pol II in the closed PIC complex, and the path of the downstream promoter in the closed PIC is bent compared to the more linear path observed in the IIDA-SCP complex (12) (fig. S9). Thus, significant structural rearrangements in TFIID must occur during PIC assembly and transcription initiation, opening the question of whether TFIID can remain promoter-bound throughout the transcription initiation cycle.
Role of lobe A movement in TBP loading
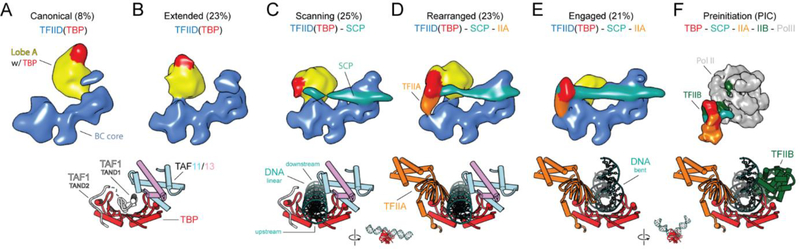
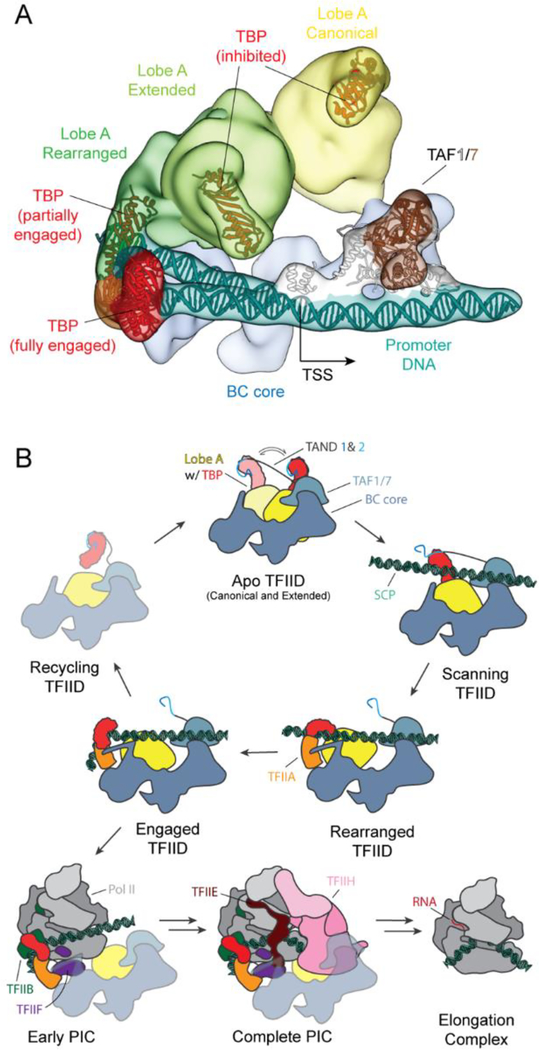
To gain insight into potential intermediate states in the process of TFIID binding to promoter DNA, we carried out cryo-EM analysis of a sample containing TFIID, TFIIA and SCP DNA that was a mixture of DNA-bound and unbound complexes. Extensive 3D sorting revealed the presence of five different states of TFIID that we propose correspond to different stages in its engagement with the promoter (Fig. 4, fig. S10, and Movie 2). We observed the same canonical and extended states that we saw in the free TFIID sample (Figs. 1A and 4, A and B), as well as the “engaged” state we previously described for the promoter-purified IIDA-SCP complex (12) (Fig. 3C and 4E). Additionally, we identified a state where lobe A is rotated toward lobe B, and TBP appears to occupy a conformation that would permit it to scan the DNA. We refer to this state, which includes only TFIID and DNA (no TFIIA) as the “scanning” state (Fig. 4C). The last state, called the rearranged state (described at low resolution in one of our previous studies ref. (10)), resembles the scanning state, but with the presence of TFIIA constraining the motion of lobe A and TBP within it. In both the scanning and rearranged states, the downstream DNA is stably bound to TFIID, while the upstream region remains flexible, as indicated by the poor density observed for the upstream DNA (Fig. 4D).
Fig. 4. Regulation of TBP DNA-binding activity by Lobe A.
Reconstructions of TFIID from the mixed dataset (which includes SCP and TFIIA), showing TFIID in the canonical (A), extended (B), scanning (C), rearranged (D), and engaged (E) states. (F) Human PIC cryo-EM map (EMD-2304) containing Pol II, TFIIA, TFIIB, TBP and promoter DNA (6). Models for TBP (PIC: PDB 5lYA) and its interacting partners are shown below each corresponding reconstructions. See also Movie 2.
By combining the mapping of TBP positions through the various states of promoter binding with previous biochemical and structural studies, we are able to propose a model of how TBP within TFIID would transition from being inhibited, to being DNA-engaged. We propose that in the canonical and extended states, TBP is bound by the TAND of TAF1 and by TAF11/13 within lobe A, both of which have been found to inhibit TBP from binding DNA (15, 27) (Fig. 4, A and B). The N terminus of the TAF1 TAND (TAND1) interacts with the DNA-binding cleft of TBP, whereas TAND2 binds the outer surface of TBP where a number of different TBP-interacting factors are known to interact, including TFIIA (28) (Fig. 4, A and B). In the scanning state, we propose that DNA displaces TAND1 and interacts with the cleft of TBP, but that the DNA remains in a linear, unbent form (Fig. 4C), due to a lack of defined DNA-TBP interaction, in contrast with what is seen for the bent DNA-TBP interaction (Fig. 4E). In the rearranged state, TFIIA would displace TAND2, releasing the connection between TAF1 and TBP and stabilizing the connection between lobes A and B (26, 28–30) (Fig. 4D). Finally, in the engaged state, TBP forms a stable complex with bent DNA, which causes the connection between TBP and TAF11 to break, and TBP to release from lobe A (Fig. 4E). This last step of lobe A release is essential for recruitment of TFIIB and for the assembly of the PIC, as it opens up the surface on TBP for TFIIB binding (31) (Fig. 4F).
Though TBP binds the TATA box sequence with the highest affinity of any DNA sequence, it has been observed to be a promiscuous DNA-binder (32,33). The mechanisms of TBP-inhibition within lobe A effectively represent an important role of TFIID as a TBP chaperone, stopping TBP from nonspecifically engaging with DNA outside of gene promoters, and therefore preventing aberrant PIC assembly and erroneous transcription initiation (33). We propose that, at the same time, the architecture and dynamics of TFIID facilitate the proper loading TBP at core promoters by progressively releasing those inhibitory interactions with TAFs, and, as explained below, strategically positioning TBP onto the upstream DNA.
Proposed mechanism of TBP loading by TFIID and consequent PIC recruitment
Superposition of the five conformational states of TFIID, canonical, extended, scanning, rearranged and engaged, illustrates the range of motion TBP experiences with respect to the BC core during the steps leading to full promoter engagement (Fig. 5A and Movie 2). The distance that TBP travels between these states is approximately 130, 40, 30 and 50 Ä respectively, and follows a curved path that directs TBP toward the upstream DNA. Taken together, these structures suggest a stepwise mechanism of TBP loading onto the promoter and the consequent recruitment of the rest of the PIC. In the first step, TAF1/7 and TAF2 in lobe C bind to downstream DNA. This initial DNA binding facilitates the positioning of the TATA-box where it can be reached by TBP as it travels with the mobile lobe A, thus helping the upstream DNA outcompete the inhibitory TAND1 from the cleft of TBP. In the second step, TFIIA displaces TAND2 from TBP and likely stabilizes the upstream DNA through its interaction with lobe B. In this way, the rearranged state constrains the position of lobe A and facilitates TBP binding to the upstream DNA. In the third step, TBP fully engages the promoter DNA, bending it and simultaneously causing a steric clash between the DNA and TAF11 that results in the release of TBP from the rest of lobe A (Fig. 5B and Movie 2).
Fig. 5. Mechanism of TBP loading by TFIID.
(A) Cryo-EM reconstructions of the canonical, extended, rearranged and engaged states of TFIID superimposed onto the BC core to show the range of motion of Lobe A and TBP. The TAF1/7 module is positioned based on the engaged state reconstruction, and the DNA models for both the engaged and rearranged states are shown. (B) Cartoon schematic for the process of TBP loading onto promoter DNA by TFIID, with subsequent PIC recruitment, assembly and progression to the elongation complex. See also Movie 2.
In the fourth step, TFIIB recognizes the fully engaged TBP-DNA complex and recruits with it Pol II/TFIIF. At this stage, the binding of the TFIIF winged-helix domain in Rap40 and Pol II would displace the TAF4 contact with upstream DNA and the interactions of lobe C with downstream core promoter sequences, respectively. This process could potentially result in the TAFs falling off the PIC, unless the interaction between TFIIA and TA F4 were sufficient to keep TFIID bound, or new contacts were to form between TFIID and the PIC at this stage of the assembly. While a number of interactions have been reported between TFIID and other GTFs in vitro (34–37), it has been shown that upon the addition of Pol II/TFIIB/TFIIF, TFIID remains associated with the promoter only in the presence of activators (38, 39). In this potential scenario, TFIID may not remain as part of the growing PIC, but could instead bind another TBP to be able to reform a new active complex once the previous clears the promoter (Fig. 5B). Additional experiments will be required to test this model and determine the precise role of TFIID in PIC assembly following TBP loading.
Approximately 80% of eukaryotic promoters lack a canonical TATA-box, yet loading of TBP is essential to initiate transcription for all protein genes (40). The mechanism of TBP loading by TFIID provides a way to promote TBP loading in the absence of a canonical TATA-box, and expands the potential for regulation through variation in the core promoter sequence. To structurally explore this concept, we assembled a promoter-bound complex using a mutant SCP (mSCP) that lacked a consensus TATA sequence (ACTGCCGT replacing TATAAAAG). The resulting IIDA-mSCP complex was purified via a DNA-pulldown and resulted in a sample that still bound the promoter DNA but appeared trapped in the rearranged state with TBP constrained onto the promoter (fig. S11). We did not observe any complexes in the engaged state, consistent with previous DNase foot-printing experiments that showed that TFIID is only able to weakly protect the TATA box using purified components (10). However, both in vitro transcription assays containing nuclear extracts and in vivo reporter assays showed transcription from mSCP templates (11, 41). Those results would indicate that other factors not present in the in DNase foot-printing experiments, but present in the nuclear extract, must be aiding TBP in the absence of a consensus TATA. Factors like transcriptional activators, chromatin marks or other coactivator complexes could play an essential role in allowing transcription from TATA-less promoters by facilitating the transition from the rearranged to the engaged states and thus the full engagement of TBP onto DNA.
TFIID as a coactivator and chromatin reader
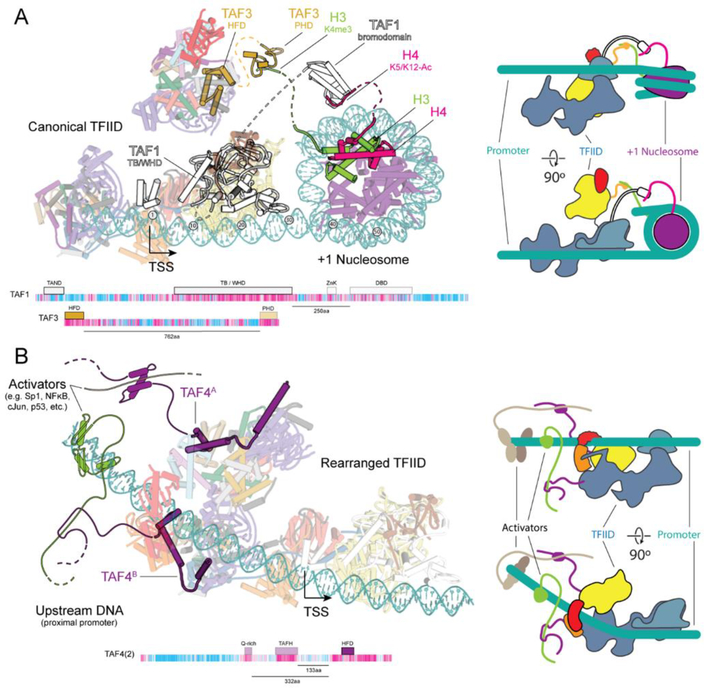
In vivo TFIID recruitment to the core promoter is aided by gene-specific activators and chromatin marks. Promoters are enriched in certain post-translational modifications of histones and in histone variants that distinguish them from the rest of the genome (42). Trimethylation of K4 on histone H3 (H3K4me3) and acetylation of H3 and H4 are especially enriched on the +1 nucleosome, located ~50 bp downstream of the TSS (43–46). TFIID recognizes H3K4me3 through the plant homeodomain (PHD) of TAF3, and the diacetylated H4 via the TAF1 double bromodomain (DBD) (47–49). A model of the downstream promoter extended with a +1 nucleosome shows how these domains, which our studies indicate are flexibly tethered to the core of TFIID, would be oriented toward the +1 nucleosome in the canonical state of TFIID, suggesting a mechanism of TFIID recruitment by the modified +1 nucleosomes of activated genes (Fig. 6A).
Fig. 6. Model of TFIID recruitment.
(A) Model of TFIID bound to the promoter including a +1 nucleosome. The model Is compatible with the binding of flexible histone tails of H3 and H4 to the PHD (PDB ID 2K17, ref. (47)) of TAF3 and the bromodomain of BRD2 (PDB ID 2DVR (49)), a homolog of the DBD of TAF1, respectively. Dashed lines Indicate the connections between domains contained In the models of TFIID or the nucleosome, with the flexible domains that bridge the two. Domain architecture maps of TAF1 and TAF3 showing the distance between the structured domains modeled within TFIID and the domains that contact chromatin. A cartoon model of TFIID binding to the +1 nucleosome Is shown to the right. (B) Model of TFIID bound to the core promoter with bound activators at the upstream proximal promoter region. Activators are contacting the N terminus of TAF4 that contains activator Interacting regions, like the Q-rlch and TAFH domains. Domain maps of the highlighted TAFs Illustrate the distance between the domains that were part of the TFIID model (solid) and those domains that were not observed (transparent). Distances between the conserved C terminus and the domains that contact activators (TAFH and Q-rlch) are shown below the domain map. A cartoon model of TFIID binding to activators Is shown on the right.
Transcriptional activators determine cellular fate by directing the transcription of genes controlling development, differentiation, stimulus response, growth, and maintenance of homeostatic balance (50). While many activators have been shown to interact with different TAFs, the strongest evidence has been shown for binding of activators through the conserved Q-rich and TAFH domains of TAF4 within its long and flexible N terminus (51–53). A model generated by extending the upstream DNA in the TFIID rearranged state shows how both copies of TAF4 are positioned toward the upstream proximal promoter, which is known to remain cleared of nucleosomes and act as a binding site for transcriptional activators (45), so that they could interact with an activator via their flexible N-terminal domains. This model suggests that transcriptional activators may play a dual role in TFIID recruitment to the promoter, as well as in promoting TBP engagement by stabilizing the rearranged state of TFIID (Fig. 6B).
Implications for the structure and function of the SAGA transcription complex
The insights into the structure and mechanism of TFIID also shed light into the possible function of the large transcription factor SAGA, as the two complexes share a number of similar components (54) (fig. S12a). SAGA contains four main modules of different function: a TBP loading TAF-containing module, a histone acetyltransferase (HAT) module, a histone deubiquitinase (DUB) module, and an activator binding TRRAP module (55). In humans, the SAGA TAF module contains TAF9,10 and 12, which are shared with TFIID, as well as the SAGA-specific TAF5L and TAF6L, which are paralogs to TAF5 and TAF6 in TFIID. In addition, SAGA also contains TADA1, which substitutes for TAF4 in forming a HF-pair with TAF12, SUPT7, which can form a HF-pair with TAF10, and SUPT3H, which contains two HFDs homologous to those in TAF11 and 13. Therefore, SAGA contains homologous proteins for all the TAFs that make up the dimeric core of TFIID, but whether these exist in two copies within SAGA has not been determined. Using a model of lobe A, we aligned the common SAGA components with those in TFIID and were able to show that within the structurally modeled regions of TFIID, the homologous SAGA subunits are highly conserved (fig. S12b). We were also able to dock the TFIID-derived lobe A model containing only the SAGA homologous regions into the cryo-EM map of the Pichia pastoris SAGA complex (56), revealing its potential location within the complex (fig. S12c).
The TADA1 subunit of SAGA has a HFD similar to TAF4 but does not appear to retain the conserved C-terminal region that in TFIID interacts with DNA and TFIIA. The SUPT7L subunit of SAGA that could act as a replacement for TAF8 or TAF3, lacks strong sequence similarity to either of them outside of the HFD. The yeast ortholog of SUPT3H, Spt3, binds TBP but with much lower affinity than TAF11/13, as demonstrated by the fact that TBP does not immuno-purify with either human or yeast SAGA, but can still bind TBP (54, 57). The presence of SUPT3H in SAGA suggests that a lobe A-like module may exist within the complex, but whether such a module is involved in delivering TBP to promoters in vivo remains unclear. Existing models suggest that the activator binding components within SAGA bring it to the promoter to load TBP (58, 59).
Conclusions
Our studies provide a full structural description of human TFIID and its conformational landscape, and how these relate to core promoter engagement. The model we propose for TBP loading is likely conserved in eukaryotes as those regions that play critical roles in the process of TBP loading are all highly conserved (TAF1 and TAF2 downstream binding regions, TAF1 TAND, TAF4 C-terminal regions). Interestingly, even though the regions responsible for contacting the downstream promoter motifs in human TAF1 and TAF2 appear to be conserved in yeast, downstream promoter elements have not been identified in yeast despite a wealth of genomic data. Thus, it is likely that sequence-specific recognition plays a lesser role in downstream promoter binding in yeast TFIID, and that other factors, such as activators and chromatin marks, may play a more significant role in positioning TFIID. Our structures shed light on how TBP is regulated within TFIID to prevent it from non-specifically binding DNA and starting aberrant transcription events, while at the same time providing an explanation for how TFIID is able to load TBP onto both TATA and TATA-less promoters. Our structures also suggest how activators and chromatin marks may be directing TFIID recruitment and PIC assembly. Further studies will be needed to dissect the effects that these regulatory factors have on the mechanism of TBP loading and the details of TFIID dynamic rearrangements during PIC assembly.
Methods and materials summary
TFIID was immuno-purified from HeLa cells as described previously (10). For CL-MS 100 nM of TFIID was incubated with 150 nM TFIIA and 5 mM BS3 at room temperature for 2 hours and then quenched by the addition of 2.1μΜ ammonium bicarbonate. The crosslinked proteins were TCA precipitated and treated as described (60). Mass spectrometry and identification of BS3 crosslinked peptides was performed as described previously (60).
For the cryo-EM sample preparation of apo-TFIID, TFIID was crosslinked on ice for 5 min using 0.01% glutaraldehyde and then 4 μl was applied to a C-flat CF 2/2 holey carbon gird (Protochips) to which a thin continuous carbon film coated with polyethylenimine (PEI) had been applied to improve orientation distribution. For cryo-EM sample preparation of the mixed IIDA-SCP sample, TFIIA and SCP DNA were added at ~1.2x molar excess to TFIID and incubated for 3 min on ice followed by 2 min at 37°C and finally crosslinked on ice using 0.05% glutaraldehyde for 5 min before grid preparation. For cryo-EM sample preparation of the IIDA-mSCP complex was done are described in ref. (12) except that the promoter DNA contained a mutated TATA box, with the sequence TATAAAAG in the original SCP being replaced by ACTGCCGT.
The grids for apo-TFIID and IIDA-mSCP were loaded into a Titan Low-base electron microscope (FEI) and those for mixed IIDA-SCP were loaded into a Titan Krios electron microscope (FEI), both were operated at 300 keV acceleration voltage and equipped with a K2 direct electron detector (Ga-tan). Collected movies were motion corrected using MotionCor2 (61), CTF fitted were determined using Gctf (62), and particles were picked using Gautomatch (version 0.53, from K. Zhang, MRC-LMB, Cambridge). Data was processing was done using Relion (63, 64) and model building was done with O (65) and Coot (66) and model refinement was done using Phenix (67).
Depiction of molecular models were generated using PyMOL (The PyMOL Molecular Graphics System, version 1.8, Schrödinger) and the UCSF Chimera (68) package from the Computer Graphics Laboratory, University of California, San Francisco (supported by National Institutes of Health P41 RR-01081).
Supplementary Material
>Movie 1. Structural models for the canonical and engaged human TFIID. Structural models shown within the cryo-EM maps of the two estates aligned on the BC core (different regions of the maps were refined to different resolutions due to their different degrees of flexibility). Lobe A is shown only in the canonical state.
Movie 2. Functional states of TFIID during binding to core promoter DNA and TFIIA. Cryo-EM maps were obtained for apo-TFIID (canonical and extended), for purified mutant-SCP bound TFIID-TFIIA (rearranged), for purified SCP bound TFIID-TFIIA (engaged) and from a mixture of TFIID, TFIIA and DNA (all states). Atomic models were fit into the cryo-EM structures of the canonical, extended, rearrange and engaged state, and then used to generate a model of how TFIID transitions structurally through the process of core promoter binding.
ACKNOWLEDGMENTS
We thank S. Zheng for TAF4 monoclonal antibody, D. King for providing TAF4 antibody antigen peptide, A. lavarone for performing in-gel mass spectrometry data collection and analysis, S. Gradia and Berkeley Macrolab facility for 438 series plasmids, P. Grob, S. Howes, R. Zhang and L-A. Carlson for electron microscopy support, A. Chintangal and T. Houweling for computing support, C. Lopez and C. Yoshioka at the OSHU Cryo-EM Facility for help with collecting Krios data, and D. Herbst for discussion. We acknowledge the use of the LAWRENCIUM computing cluster at Lawrence Berkeley National Laboratory and the resources of the National Energy Research Scientific Computing Center, a Department of Energy Office of Science user facility supported by the Office of Science of the US Department of Energy under contract number DE-AC02-05CH11231.
Funding: This work was funded through NIGMS grants R01- GM63072 to E.N., R01-GM053451 to S.H., P50-GM076547 to J.R. and NCI grant R21-CA175849 to J.R. A.B.P.. R.K.L. was supported by the NIGMS Molecular Biophysics Training Grant (GM008295). B.J.G. was supported by fellowships from the Swiss National Science Foundation (projects P300PA_160983, P300PA_174355). E.N. is a Howard Hughes Medical Institute Investigator.
Footnotes
Competing interests: The authors declare no competing financial interests.
Data and materials availability: The cryo-EM maps and refined coordinate models reported here were deposited in the Electron Microscopy Data Bank with accession codes EMD-9298 (BC core), EMD-9299 (lobe B), EMD-9300 (lobe C), EMD-9302 (lobe A canonical), EMD- 9301 (lobe A extended), EMD-9305 (apo-TFIID canonical), and EMD-9306 (IIDA- SCP) and Protein Data Bank with accession codes PDB-6MZC (BC core), PDB- 6MZD (lobe A), PDB-6MZL (apo-TFIID canonical), and 6MZM (IIDA-SCP engaged).
SUPPLEMENTARY MATERIALS
www.sciencemag.org/cgi/content/full/science.aau8872/DC1
Materials and Methods
Figs. S1 to S12
Tables S1 to S3
References (71–95)
REFERENCES AND NOTES
- 1.Reinberg D, Horikoshi M, Roeder RG, Factors involved in specific transcription in mammalian RNA polymerase II. Functional analysis of initiation factors IIA and IID and identification of a new factor operating at sequences downstream of the initiation site. J. Biol. Chem 262,3322–3330 (1987). [PubMed] [Google Scholar]
- 2.Buratowski S, Hahn S, Guarente L, Sharp PA, Five Intermediate complexes in transcription Initiation by RNA polymerase II. Cell 56, 549–561 (1989). doi: 10.1016/0092-8674(89)90578-3 [DOI] [PubMed] [Google Scholar]
- 3.Cortes P, Flores O, Reinberg D, Factors Involved In specific transcription by mammalian RNA polymerase II: Purification and analysis of transcription factor IIA and Identification of transcription factor IIJ. Mol. Cell. Biol 12,413–421(1992). doi: 10.1128/MCB.13.1.413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Flores O, Lu H, Killeen M, Greenblatt J, Burton ZF, Reinberg D, The small subunit of transcription factor IIF recruits RNA polymerase II into the preinitiation complex. Proc. Natl. Acad. Sci. U.S.A 88, 9999–10003 (1991). doi: 10.1073/pnas.88.22.9999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Holstege FC, van der Vliet PC, Timmers ΗT, Opening of an RNA polymerase II promoter occurs in two distinct steps and requires the basal transcription factors IIE and IIH. EMB0 J. 15, 1666–1677 (1996). doi: 10.1002/j.1460-2075.1996.th00512.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.He Y, Fang J, Taatjes DJ, Nogales E, Structural visualization of key steps In human transcription Initiation. Nature 495, 481–486 (2013). doi: 10.1038/nature11991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dynlacht BD, Hoey T, Tjlan R, Isolation of Coactivators Associated with the TATA-Binding Protein That Mediate Transcriptional Activation. Cell 66,563–576 (1991). doi. 10.16/0092-8674(81)90019-2 [DOI] [PubMed] [Google Scholar]
- 8.Bienlossek C, Papai G, Schaffitzel C, Garzoni F, Chaillet M, Scheer E, Papadopoulos P, Tora L, Schultz P, Berger I, The architecture of human general transcription factor TFIID core complex. Nature 493, 699–702 (2013). doi: 10.1038/naturell791 [DOI] [PubMed] [Google Scholar]
- 9.Tora L, A unified nomenclature for TATA box binding protein (TBP)-associated factors (TAFs) Involved in RNA polymerase II transcription. Genes Dev. 16, 673–675 (2002). doi: 10.1101/gad.976402 [DOI] [PubMed] [Google Scholar]
- 10.Cianfrocco MA, Kassavetis GA, Grob P, Fang J, Juven-Gershon T, Kadonaga JT, Nogales E, Human TFIID binds to core promoter DNA In a reorganized structural state. Cell 152,120–131 (2013). doi: 10.1016/j.cell.3012.12.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Juven-Gershon T, Cheng S, Kadonaga JT, Rational design of a super core promoter that enhances gene expression. Nat. Methods 3, 917–922 (2006). doi: 10.1038/nmeth937 [DOI] [PubMed] [Google Scholar]
- 12.Louder RK, He Y, López-Blanco JR, Fang J, Chacón P, Nogales E, Structure of promoter-bound TFIID and model of human pre-initiation complex assembly. Nature 531, 604–609 (2016). doi: 10.1038/naturel7394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Trowltzsch S, Viola C, Scheer E, Conic S, Chavant V, Fournier M, Papai G, Ebong l.-O., Schaffitzel C, Zou J, Haffke M, Rappsilber J, Robinson CV, Schultz P, Tora L, Berger I, Cytoplasmic TAF2-TAF8-TAF10 complex provides evidence for nuclear holo-TFIID assembly from preformed submodules. Nat. Commun 6,6011 (2015). doi: 10.1038/ncomms7011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sanders SL, Garbett KA, Well PA, Molecular characterization of Saccharomyces cerevisiae TFIID. Mol. Cell. Biol 22, 6000–6013 (2002). doi: 10.1138/MCB.32.16.6000-6013.3002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gupta K, Watson AA, Baptista T, Scheer E, Chambers AL, Koehler C, Zou J, Obong-Ebong I, Kandiah E, Temblador A, Round A, Forest E, Man P, Bieniossek C, Laue ED, Lemke EA, Rappsilber J, Robinson CV, Devys D, Tora L, Berger I, Architecture of TAF11/TAF13/TBP complex suggests novel regulation properties of general transcription factor TFIID. eLife 6, e30395 (2017). doi: 10.7554/eLife.3Q395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wright KJ, Marr MT 2nd, Tjian R, TAF4 nucleates a core subcomplex of TFIID and mediates activated transcription from a TATA-less promoter. Proc. Natl. Acad. Sei. U.S.A 103, 12347–12352 (2006). doi: 10.1073/pnas.0605499103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hoffmann A, Chiang C-M, Oelgeschläger T, Xie X, Burley SK, Nakatani Y, Roeder RG, A histone octamer-like structure within TFIID. Nature 380, 356–359 (1996). doi: 10.1038/380356a0 [DOI] [PubMed] [Google Scholar]
- 18.Seileck W, Howley R, Fang Q, Podolny V, Fried MG, Buratowski S, Tan S, A histone fold TAF octamer within the yeast TFIID transcriptional coactivator. Nat. Struct. Biol 8, 695–700 (2001). doi: 10.1038/90408 [DOI] [PubMed] [Google Scholar]
- 19.Leurent C, Sanders S, Ruhlmann C, Mallouh V, Weil PA, Kirschner DB, Tora L, Schultz P, Mapping histone fold TAFs within yeast TFIID. EMBO J. 21,3424–3433 (2002). doi: 10.1093/emboj/cdf342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shao H, Revach M, Moshonov S, Tzuman Y, Gazit K, Albeck S, Unger T, Diksteln R, Core promoter binding by histone-like TAF complexes. Mol. Cell. Biol 25, 206–219 (2005). doi: 10.1138/MCR.35.1.306-319.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Burke TW, Kadonaga JT, The downstream core promoter element, DPE, Is conserved from Drosophila to humans and Is recognized by TAFII60 of Drosophila. Genes Dev. 11, 3020–3031 (1997). doi: 10.1101/gad.11.22.3020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gazit K, Moshonov S, Elfakess R, Sharon M, Mengus G, Davidson I, Dikstein R, TAF4/4b X TF12 displays a unique mode of DNA binding and is required for core promoter function of a subset of genes. J. Biol. Chem 284,26286–26296 (2009). doi: 10.1074/jbc.M109.011486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Layer JH, Weil PA, Direct TFIIA-TFIID protein contacts drive budding yeast ribosomal protein gene transcription. J. Biol. Chem. 288, 23273–23294 (2013). doi: 10.1074/jbc.M113.486829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.O’Shea-Greenfield A, Smale ST, Roles of TATA and Initiator elements In determining the start site location and direction of RNA polymerase II transcription. J. Biol. Chem 267,1391–1402 (1992). [PubMed] [Google Scholar]
- 25.Carnlnci P, Sandelin A, Lenhard B, Katayama S, Shlmokawa K, Ponjavlc J, Semple CAM, Taylor MS, Engström PG, Frith MC, Forrest ARR, Alkema WB, Tan SL, Plessy C, Kodzius R, Ravasi T, Kasukawa T, Fukuda S, Kanamori-Katayama M, Kltazume Y, Kawaji H, Kai C, Nakamura M, Konno H, Nakano K, Mottagui-Tabar S, Arner P, Chesi A, Gustincich S, Persichetti F, Suzuki H, Grimmond SM, Wells CA, Orlando V, Wahlestedt C, Liu ET, Harbers M, Kawai J, Bajic VB, Hume DA, Hayashizaki Y, Genome-wide analysis of mammalian promoter architecture and evolution. Nat. Genet. 38, 626–635 (2006). doi: 10.1038/ngl789 [DOI] [PubMed] [Google Scholar]
- 26.He Y, Yan C, Fang J, Inouye C, Tjian R, Ivanov I, Nogales E, Near-atomic resolution visualization of human transcription promoter opening. Nature 533, 359–365 (2016). doi: 10.1038/naturel7970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu D, Ishima R, Tong KI, Bagby S, Kokubo T, Muhandiram DR, Kay LE, Nakatani Y, Ikura M, Solution structure of a TBP-TAF(II)230 complex: Protein mimicry of the minor groove surface of the TATA box unwound by TBP. Cell 94, 573–583 (1998). doi: 10.1016/S0092-8674(00)81599-8 [DOI] [PubMed] [Google Scholar]
- 28.Anandapadamanaban M, Andresen C, Helander S, Ohyama Y, Siponen ΜI, Lundström P, Kokubo T, Ikura M, Moche M, Sunnerhagen M, HIgh-resolutlon structure of TBP with TAF1 reveals anchoring patterns In transcriptional regulation. Nat. Struct. Mol. Biol 20,1008–1014 (2013). doi: 10.1038/nsmb.2611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kokubo T, Swanson MJ, Nishikawa JI, Hinnebusch AG, Nakatani Y, The yeast TAF145 inhibitory domain and TFIIA competitively bind to TATA-blndlng protein. Mol. Cell. Biol 18,1003–1012 (1998). rioi: 10.1128/MGB.18.2.1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bagby S, Mal TK, Liu D, Raddatz E, Nakatani Y, Ikura M, TFIIA-TAF regulatory Interplay: NMR evidence for overlapping binding sites on TBP. FEBS Lett. 468, 149–154 (2000). doi: 10.1016/S0014-5793(00)01213-8 [DOI] [PubMed] [Google Scholar]
- 31.Nikolov DB, Chen H, Halay ED, Usheva AA, Hisatake K, Lee DK, Roeder RG, Burley SK, Crystal structure of a TFIIB-TBP-TATA-element ternary complex. Nature 377,119–128 (1995). doi: 10.1038/377119a0 [DOI] [PubMed] [Google Scholar]
- 32.Wong JM, Bateman E, TBP-DNA interactions In the minor groove discriminate between A:T and T:A base pairs. Nucleic Acids Res. 22, 1890–1896 (1994). doi: 10.1093/nar/22.10.1890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Coleman RA, Pugh BF, Evidence for functional binding and stable sliding of the TATA binding protein on nonspecific DNA. J. Biol. Chem. 270, 13850–13859 (1995). doi: 10.1074/jbc.270.23.13850 [DOI] [PubMed] [Google Scholar]
- 34.Hisatake K, Ohta T, Takada R, Guermah M, Horikoshi M, Nakatani Y, Roeder RG, Evolutionary conservation of human TATA-binding-polypeptide-associated factors TAFII31 and TAFII80 and Interactions of TAFII80 with other TAFs and with general transcription factors. Proc. Natl. Acad. Sel. U.S.A 92, 8195–8199 (1995). doi: 10.1073/pnas.92.18.8195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ruppert S, Tjian R, Human TAFII250 Interacts with RAP74: Implications for RNA polymerase II initiation. Genes Dev. 9, 2747–2755 (1995). doi: 10.1101/gad.9.22.2747 [DOI] [PubMed] [Google Scholar]
- 36.Dubrovskaya V, Lavigne AC, Davidson I, Acker J, Staub A, Tora L, Distinct domains of hTAFI100 are required for functional Interaction with transcription factor TFIIF beta (RAP30) and Incorporation into the TFIID complex. EMBOJ. 15, 3702–3712 (1996). doi: 10.1002/j.l460-2075.1996.tb00740.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gegonne A, Weissman JD, Lu H, Zhou M, Dasgupta A, Ribble R, Brady JN, Singer DS, TFIID component TAF7 functionally Interacts with both TFIIH and P-TEFb. Proc. Natl Acad. Sei. U.S.A. 105, 5367–5372 (2008). doi: 10.1073/pnas.08016,37105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yudkovsky N, Ranlsh JA, Hahn S, A transcription reinitiation Intermediate that is stabilized by activator. Nature 408, 225–229 (2000). doi: 10.1038/35041603 [DOI] [PubMed] [Google Scholar]
- 39.Yakovchuk P, Gilman B, Goodrich JA, Kugel JF, RNA polymerase II and TAFs undergo a slow isomerization after the polymerase Is recruited to promoter- bound TFIID. J. Mol. Biol 397, 57–68 (2010). doi: 10.1016/j.jmb.2010.01.025 [DOI] [PubMed] [Google Scholar]
- 40.FitzGerald PC, Shlyakhtenko A, Mir AA, Vinson C, Clustering of DNA sequences In human promoters. Genome Res. 14, 1562–1574 (2004). doi: 10.1101/gr.1953904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Um CY, Santoso B, Boulay T, Dong E, Ohler U, Kadonaga JT, The MTE, a new core promoter element for transcription by RNA polymerase II. Genes Dev. 18, 1606–1617 (2004). doi: 10.1101/gad.1193404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Barth TK, Imhof A, Fast signals and slow marks: The dynamics of histone modifications. Trends Biochem. Scl 35, 618–626 (2010). doi: 10.1016/j.tihs.2010.05.006 [DOI] [PubMed] [Google Scholar]
- 43.Kim TH, Barrera LO, Zheng M, Qu C, Singer MA, Richmond TA, Wu Y, Green RD, Ren B, A high-resolution map of active promoters In the human genome. Nature 436,876–880 (2005). doi: 10.1038/nature03877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Heintzman ND, Stuart RK, Hon G, Fu Y, Ching CW, Hawkins RD, Barrera LO, Van Calcar S, Qu C, Ching KA, Wang W, Weng Z, Green RD, Crawford GE, Ren B, Distinct and predictive chromatin signatures of transcriptional promoters and enhancers In the human genome. Nat. Genet 39,311–318 (2007). doi: 10.1038/ngl966 [DOI] [PubMed] [Google Scholar]
- 45.Schönes DE, Cui K, Cuddapah S, Roh T-Y, Barski A, Wang Z, Wei G, Zhao K, Dynamic regulation of nucleosome positioning In the human genome. Cell 132, 887–898 (2008). dni: 10.1016/j.cfill.2008.02.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mavrich TN, Jiang C, loshlkhes IP, LI X, Venters BJ, Zanton SJ, Tomsho LP, Ql J, Glaser RL, Schuster SC, Gllmour DS, Albert I, Pugh BF, Nucleosome organization In the Drosophila genome. Nature 453, 358–362 (2008). doi: 10.1038/natnre06929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.van Ingen H, van Schalk FMA, Wienk H, Ballering J, Rehmann H, Dechesne AC, Kruljzer JAW, Liskamp RMJ, Timmers HTM, Boelens R, Structural Insight Into the recognition of the H3K4me3 mark by the TFIID subunit TAF3. Structure 16,1245–1256 (2008). doi: 10.1016/j.str.20Q8.04.015 [DOI] [PubMed] [Google Scholar]
- 48.Jacobson RH, Ladurner AG, King DS, Tjian R, Structure and function of a human TAFII250 double bromodomaln module. Science 288,1422–1425 (2000). doi: 10.1176/science.788.5470.14?7 [DOI] [PubMed] [Google Scholar]
- 49.Umehara T, Nakamura Y, Jang MK, Nakano K, Tanaka A, Ozato K, Padmanabhan B, Yokoyama S, Structural basis for acetylated histone H4 recognition by the human BRD2 bromodomaln. J. Biol. Chem. 285, 7610–7618 (2010). doi: 10.1074/jbc.M109.062422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Levine M, Cattoglio C, Tjian R, Looping back to leap forward: Transcription enters a new era. Cell 157,13–25 (2014). doi: 10.1016/j.cell.2014.02.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liu W-L, Coleman RA, Ma E, Grob P, Yang JL, Zhang Y, Dailey G, Nogales E, Tjian R, Structures of three distinct activator-TFIID complexes. Genes Dev. 23, 1510–1521 (2009). doi: 10.1101/gad.1790709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hiblno E, Inoue R, Suglyama M, Kuwahara J, Matsuzaki K, Hoshino M, Identification of heteromolecular binding sites In transcription factors Spl and TAF4 using high-resolutlon nuclear magnetic resonance spectroscopy. Protein Sei. 26, 2280–2290 (2017). doi: 10.1002/prn.3287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang X, Truckses DM, Takada S, Matsumura T, Tañese N, Jacobson RH, Conserved region I of human coactivator TAF4 binds to a short hydrophobic motif present In transcriptional regulators. Proc. Natl. Acad. Sci. U.S.A. 104, 7839–7844 (2007). doi: 10.1073/pnas.0608570104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Martinez E, Palhan VB, Tjernberg A, Lymar ES, Gamper AM, Kundu TK, Chait BT, Roeder RG, Human STAGA complex Is a chromatin-acetylating transcription coactivator that Interacts with pre-mRN A splicing and DNA damagebinding factors In vivo. Mol. Cell. Biol 21, 6782–6795 (2001). doi: 10.1128/M0B.21.20.6782-6795.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Helmlinger D, Tora L, Sharing the SAGA. Trends Biochem. Sci 42, 850–861 (2017). doi: 10.1016/j.tibs.2017.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sharov G, Voltz K, Durand A, Kolesnikova O, Papal G, Myasnikov AG, Dejaegere A, Shem A. Ben, Schultz P, Structure of the transcription activator target Tral within the chromatin modifying complex SAGA. Nat. Commun 8, 1556 (2017). doi: 10.1038/s41467-017-01564-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Han Y, Luo J, Ranish J, Hahn S, Architecture of the Saccharomyces cerevisiae SAGA transcription coactivator complex. EMBO J. 33, 2534–2546 (2014). doi: 10.15252/embj.201488638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Belotserkovskaya R, Sterner DE, Deng M, Sayre ΜH, Lieberman PM, Berger SL, Inhibition of TATA-binding protein function by SAGA subunits Spt3 and Spt8 at Gcn4-actlvated promoters. Mol. Cell. Biol. 20, 634–647 (2000). doi: 10.1128/M0B.20.2.634-647.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Larschan E, Winston F, The S cerevisiae SAGA complex functions In vivo as a coactivator for transcriptional activation by Gal4. Genes Dev. 15, 1946–1956 (2001). doi: 10.1101/gad.911501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tuttle LM, Pacheco D, Warfield L, Luo J, Ranlsh J, Hahn S, Klevuit RE, Gcn4- Mediator Specificity Is Mediated by a Large and Dynamic Fuzzy Protein-Protein Complex. Cell Rep. 22, 3251–3264 (2018). doi: 10.1016/j.celrep.2018.02.097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zheng SQ, Palovcak E, Armache J-P, Verba KA, Cheng Y, Agard DA, MotlonCor2: Anisotropic correction of beam-induced motion for improved cryo- electron microscopy. Nat. Methods 14,331–332 (2017). doi: 10.1038/nmeth.4193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang K, Gctf: Real-time CTF determination and correction. J. Struct. Biol 193, 1–12 (2016). doi: 10.1016/j.jsh.2015.11.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Scheres SHW, RELION: Implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol 180,519–530 (2012). doi: 10.1016/j.jsb.2012.09.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kimanius D, Forsberg BO, Scheres SH, Lindahl E, Accelerated cryo-EM structure determination with parallelisation using GPUs in RELION-2. eLife 5, el8722 (2016). doi: 10.7554/eLife.18722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jones TA, Interactive electron-density map interpretation: From INTER to 0. Acta Crystallogr. D 60, 2115–2125 (2004). doi: 10.1107/S0907444904023509 [DOI] [PubMed] [Google Scholar]
- 66.Emsley P, Lohkamp B, Scott WG, Cowtan K, Features and development of Coot. Acta Crystallogr. D 66,486–501 (2010). doi: 10.1107/S0907444910007493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, Headd JJ, Flung L-W, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH, PHENIX-. A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D 66, 213–221 (2010). doi: 10.1107/S0907444909052925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE, UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004). doi: 10.1002/jcc.20084 [DOI] [PubMed] [Google Scholar]
- 69.Ashkenazy H, Erez E, Martz E, Pupko T, Ben-Tal N, ConSurf 2010: Calculating evolutionary conservation in sequence and structure of proteins and nucleic acids. Nucleic Acids Res. 38, W529–W533 (2010). doi: 10.1093/nar/gkq399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Buchan DWA, Minneci F, Nugent TCO, Bryson K, Jones DT, Scalable web services for the PSIPRED Protein Analysis Workbench. Nucleic Acids Res. 41, W349–W357 (2013). doi: 10.1093/nar/gkt381 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
>Movie 1. Structural models for the canonical and engaged human TFIID. Structural models shown within the cryo-EM maps of the two estates aligned on the BC core (different regions of the maps were refined to different resolutions due to their different degrees of flexibility). Lobe A is shown only in the canonical state.
Movie 2. Functional states of TFIID during binding to core promoter DNA and TFIIA. Cryo-EM maps were obtained for apo-TFIID (canonical and extended), for purified mutant-SCP bound TFIID-TFIIA (rearranged), for purified SCP bound TFIID-TFIIA (engaged) and from a mixture of TFIID, TFIIA and DNA (all states). Atomic models were fit into the cryo-EM structures of the canonical, extended, rearrange and engaged state, and then used to generate a model of how TFIID transitions structurally through the process of core promoter binding.