Abstract
Upstream open reading frames (uORFs) in 5’ UTRs of eukaryotic mRNAs are increasingly recognized as important elements that regulate cellular protein synthesis. Since uORFs can start from non-AUG codons, an enormous number of potential uORF initiation sites exists in 5’UTRs. However, only a subset of these sites is used and it has been unclear how actual start sites are selected. Studies of the DEAD-box helicase Ded1p from S. cerevisiae show that translation of uORFs with non-AUG initiation codons occurs upstream of mRNA structures that emerge with defective Ded1p. The data designate mRNA structure as important determinant for non-AUG initiation sites of uORFs. Ded1p can control this RNA structure and thereby regulate uORF translation.
Keywords: uORF, Ribosome profiling, RNA structure, Helicase, Translation, CLIP, DEAD-box, Riboswitch, Yeast, Meiosis, Near-cognate codon
Translation initiation is critical for the regulation of protein production in the cell (Shirokikh and Preiss, 2018). In eukaryotes, translation initiation involves more than a dozen protein factors (Hinnebusch, 2014). These translation initiation factors guide the small ribosomal subunit (SSU) to the start codon of the main ORF, by facilitating binding of the SSU to 5’ capped mRNAs, by aiding the SSU in the scanning of the mRNA region between the cap and the start codon of the main ORF, and by promoting recognition of the start codon and joining of the two ribosomal subunits (Hinnebusch, 2014).
The mRNA region between 5’ cap and start codon of the main ORF is commonly referred to as 5’ untranslated region (5’UTR). Notwithstanding the “untranslated” designation, translation in 5’UTRs is well known to regulate the synthesis of select proteins, including GCN4 and ATF4 (Hinnebusch et al., 2016). Over the last years it has become clear that translation in 5’UTRs is pervasive and exerts potentially broad impact on protein synthesis (Cabrera-Quio et al., 2016; Ingolia, 2016). Many 5’UTRs encode upstream ORFs (uORFs) that can comprise six to several hundreds of nucleotides (Hinnebusch et al., 2016). These uORFs can either be translated or skipped by the scanning pre-initiation complex (PIC) (Ingolia, 2016). Under several stress conditions such as nutrient deprivation, stress of the endoplasmatic reticulum, or hypoxia, translation and skipping of certain uORFs follows tightly regulated choreographies (Hinnebusch et al., 2016), (Starck et al., 2016). Defects in translation of uORFs and associated changes in the translation of main ORFs are also linked to diseases, including cancer, neurologic and metabolic disorders and heritable syndromes (Barbosa et al., 2013; Schulz et al., 2018).
Translation in 5’UTRs can create N-terminal extensions of main ORF products (Hinnebusch et al., 2016). However, most identified uORFs terminate before or shortly after the main ORF start codon, and are out of frame with the main ORF (Hinnebusch et al., 2016; Ingolia, 2016). Translation of these uORFs decreases protein synthesis from the main ORF in most studied examples (Johnstone et al., 2016). In some instances, peptides produced from uORFs have distinct functions (Andrews and Rothnagel, 2014). Yet, in most cases, the process of uORF translation, rather than the peptide product is thought to be critical for the regulatory function of the uORF (Hinnebusch et al., 2016).
Translation in 5’UTRs can initiate from canonical AUG start codons, but also from near cognate codons (Ingolia, 2016). These differ from canonical start codons by one nucleotide and can create alternative translation initiation sites (ATIS) (Hinnebusch et al., 2016). Near-cognate start codons represent roughly 14% of all codons (Kearse and Wilusz, 2017). While the efficiency of translation initiation from a near cognate codon is affected by its particular sequence and by the surrounding sequence (Kolitz et al., 2009; Spealman et al., 2018), there is still an enormous number of possible initiation sites 5’UTRs. Yet, only a comparably small subset of these near cognate initiation sites is typically used and it is not well understood how these sites in 5’UTRs are chosen.
A recent study of the translation initiation factor Ded1p from S. cerevisiae has provided new insight into this question (Guenther et al., 2018). Ded1p is an RNA helicase of the DEAD-box family and can thus bind and remodel RNA and RNA-protein complexes in an ATP-dependent fashion (Sharma and Jankowsky, 2014). Ded1p is essential in S. cerevisiae and conserved in eukaryotes, and it is thought that the main functions of the enzyme are also conserved (Sharma and Jankowsky, 2014). The human Ded1p ortholog, DDX3X, has been linked to a multitude of cancers and to intellectual disability (Bol et al., 2015; Snijders Blok et al., 2015). Ded1p had been previously shown to function in translation initiation (Chuang et al., 1997; de la Cruz et al., 1997). As a helicase, Ded1p had been implicated in the remodeling of mRNA structure in 5’UTRs (Sen et al., 2015), although it was not known which mRNA structures are remodeled by the enzyme.
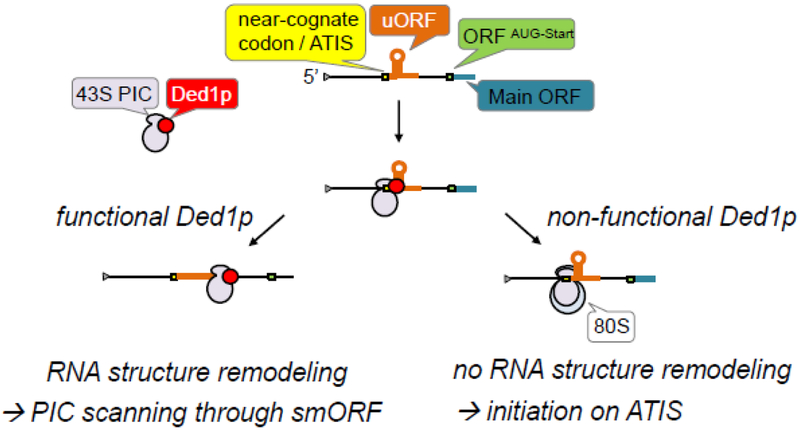
Our recent study aimed to systematically analyze the function of Ded1p in translation initiation in cells. We integrated transcriptome-wide analyses of translation, RNA structure, Ded1p-RNA binding and analysis of reporter constructs (Guenther et al., 2018). We found that Ded1p associates with the pre-initiation complex (PIC) at the mRNA entry channel, well positioned to remodel RNA structure ahead of the scanning PIC. Repression of Ded1p activity caused accumulation of mRNA structure in 5’UTRs and pervasive initiation of translation from near-cognate start codons immediately upstream of these structures (Fig.1). Translation from the corresponding main ORFs decreased in most cases, consistent with the notion that ribosomes on 5’UTRs form road-blocks for subsequent scanning PICs (Hinnebusch et al., 2016).
Figure 1. Defects in Ded1p cause translation initiation at near cognate codons that are proximal to mRNA structure.
Simplified schematics of the link between Ded1p function and activation of near cognate initiation condons in 5’UTRs.
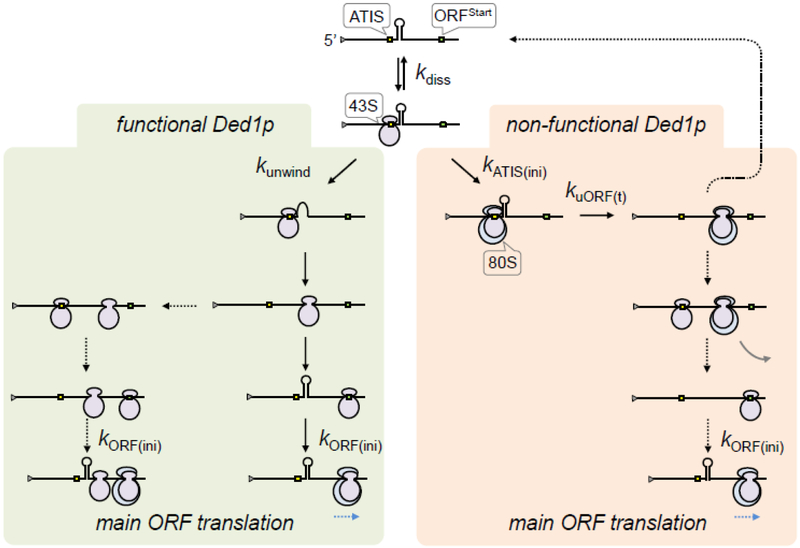
The data reveal that Ded1p couples remodeling of mRNA structure with activation of near cognate initiation codons, thereby impacting translation of the main ORF. The results suggest the model for Ded1p function shown in Fig.2. Ded1p associates with the PIC in the vicinity of the mRNA entry channel. The scanning PIC pauses before mRNA structures (indicated by Ded1p footprints on mRNA measured by iCLIP, Guenther et al., 2018). Functional Ded1p remodels the mRNA structures (indicated by DMS RNA structure probing in vivo), the PIC continues scanning and main ORF translation is not affected. If Ded1p is not functional or absent, the mRNA structures are not efficiently remodeled. The PIC either dissociates, or translation is initiated if a near cognate initiation codon (alternative translation initiation site -ATIS) or and AUG codon are located 5’ of the mRNA structure (indicated by ribosome profiling). Ribosomes initiating or translating in 5’UTRs impede subsequent scanning PICs (Hinnebusch et al., 2016), thereby decreasing translation efficiency for the main ORF. Scanning PICs following behind the translating ribosome can potentially continue scanning to the main ORF initiation site and thus initiate protein synthesis, although with reduced efficiency, compared to functional Ded1p.
Figure 2. Remodeling of mRNA by Ded1p and activation of near cognate initiation codons.
Ded1p associates with the PIC at the mRNA entry site of the small ribosomal subunit (Helix 16). Rate constants in the scheme are provided for illustrative purposes, they were not measured.
mRNA structures without proximal ATIS or AUG initiation sites can impact translation of the main ORF if these structures are only slowly or not at all remodeled without Ded1p. The balance of the various kinetic steps in the initiation mechanism is critical for the impact of Ded1p on main ORF translation. For example, if initiation at the main ORF is exceedingly slow, effects of non-functional Ded1p on main ORF translation diminish. A smaller impact of Ded1p would also be seen if PIC loading on the mRNA is slowed. Previously unrecognized steps might further affect the regulation of translation initiation by Ded1p, including the availability of large ribosomal subunits, and the rate of uORF elongation. Finally, regulation of initiation factors that promote association of Ded1p to the PIC could affect uORF and main ORF translation similar to nonfunctional Ded1p (Gao et al., 2016).
Besides providing new insight into the role of Ded1p in the regulation of translation initiation, the data reveal a remarkably simple mechanism for selecting initiation sites for upstream ORFs: mRNA structure that emerges when the helicase is inactive (Fig.1). This mechanism builds on the established observation that RNA structure promotes translation initiation on 5’ proximal cognate or near cognate codons (Hinnebusch, 2014). Examples include RNA structures following the initiation codon of main ORFs (Kozak, 1990), RAN translation, where RNA structures formed by aberrant expansion segments promote 5’ proximal translation initiation (Cleary and Ranum, 2017), and translation of the Histone H4 mRNA, which is initiated by a structure that, curiously, pairs with the small ribosomal subunit at Helix 16 (Martin et al., 2011), the region where Ded1p associates with the PIC (Guenther et al., 2018). Our observations suggest that the link between mRNA structure and uORF “activation” may be more prevalent than in the previously reported, isolated cases. Moreover, the data show that this mechanism can be controlled by a helicase and that mRNA structures with comparably low thermodynamic stability can elicit translation initiation from near cognate codons.
It might thus be instructive to view mRNA structures in the 5’ UTRs as Ded1p-sensitive riboswitches. Active Ded1p remodels the RNA structures, turns the switches off and suppresses uORF activation (Fig.1b). Inactive Ded1p lets the structures emerge, turns the switches on and activates the uORFs. Inactivation of Ded1p can be accomplished in multiple ways, including post-translational modifications, metabolites, such as AMP, low Ded1p levels, Ded1p sequestration in RNP granules, and regulation of factors involved in Ded1p recruitment to the PIC (Sharma and Jankowsky, 2014; Putnam and Jankowsky, 2013). Of note, our data indicate pervasive mRNA structures in 5’UTRs. In many cases, multiple structures likely overlap in a single 5’UTR, rendering 5’UTR structure far more complex then the often depicted isolated hairpins in unstructured RNA. These arrays of mRNA structure in 5’UTRs likely facilitate complex regulation, akin to riboswitches that respond to diverse inputs.
Is this Ded1p-controlled uORF activation program used in biological processes? Several lines of evidence suggest use of this program. First, mRNAs whose translation is largely unaffected by Ded1p encode genes that belong to particular functional groups, including nucleosomes, proteins functioning in gluconeogenesis, and cell wall synthesis. Conversely, mRNAs whose translation is particularly sensitive to Ded1p encode proteins involved in mitotic cell cycle progression and RNA metabolism (Guenther et al., 2018). Second, sequence conservation around activated uORFs is higher than in other regions in 5’UTRs of fungi (Guenther et al., 2018). Third, a near cognate codons in the ALA1 transcript that is activated with non-functional Ded1p produces an N-terminal extension of Ala1p that targets the protein to the mitochondria (Tang et al., 2004). How the initiation site for this extension is chosen had been unknown.
Fourth, and perhaps most striking, our data indicate a role for the Ded1p-mediated mechanism of uORF activation in meiosis. A large number of uORFs starting from near cognate codons are utilized in meiosis (Brar et al., 2012). Many uORF initiation sites seen in meiosis overlap with those activated with non-functional Ded1p (Guenther et al., 2018). We find that Ded1p levels are markedly reduced during meiosis (Guenther et al., 2018). The link between Ded1p levels and uORF activation during meiosis suggests a role for Ded1p in this process. Collectively, these observations illustrate that the Ded1p-controlled uORF activation program is used in physiological cellular processes. Given the multitude of ways by which Ded1p can be regulated, it is possible that even more cellular processes utilize at least part of this mechanism.
In sum, the data indicate that intricate translation control and activation of uORFs can be based on simple, ubiquitous elements: a helicase, mRNA structure and near-cognate initiation codons. It is conceivable that the simplicity of the RNA elements - near cognate codons and proximal RNA structure - makes these regions particularly attractive to alter during evolution in order to modulate both uORF and main ORF translation (Spealman et al., 2018; Zhang et al., 2018).
Acknowledgements:
Work in our laboratory is supported by the NIH (GM118088 to E.J.)
REFERENCES
- Andrews SJ, and Rothnagel JA (2014). Emerging evidence for functional peptides encoded by short open reading frames. Nat Rev Genet 15, 193–204. [DOI] [PubMed] [Google Scholar]
- Barbosa C, Peixeiro I, and Romao L (2013). Gene expression regulation by upstream open reading frames and human disease. PLoS Genet 9, e1003529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bol GM, Xie M, and Raman V (2015). DDX3, a potential target for cancer treatment. Mol Cancer 14, 188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brar GA, Yassour M, Friedman N, Regev A, Ingolia NT, and Weissman JS (2012). High-resolution view of the yeast meiotic program revealed by ribosome profiling. Science 335, 552–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabrera-Quio LE, Herberg S, and Pauli A (2016). Decoding sORF translation - from small proteins to gene regulation. RNA Biol 13, 1051–1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang RY, Weaver PL, Liu Z, and Chang TH (1997). Requirement of the DEAD-Box protein ded1p for messenger RNA translation. Science 275, 1468–1471. [DOI] [PubMed] [Google Scholar]
- Cleary JD, and Ranum LP (2017). New developments in RAN translation: insights from multiple diseases. Curr Opin Genet Dev 44, 125–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Cruz J, Iost I, Kressler D, and Linder P (1997). The p20 and Ded1 proteins have antagonistic roles in eIF4E-dependent translation in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A 94, 5201–5206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Z, Putnam AA, Bowers HA, Guenther UP, Ye X, Kindsfather A, Hilliker AK, and Jankowsky E Coupling between the DEAD-box RNA helicases Ded1p and eIF4A. (2016) Elife 5, pii: e16408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guenther UP, Weinberg DE, Zubradt MM, Tedeschi FA, Stawicki BN, Zagore LL, Brar GA, Licatalosi DD, Bartel DP, Weissman JS, et al. (2018). The helicase Ded1p controls use of near-cognate translation initiation codons in 5’ UTRs. Nature 559, 130–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinnebusch AG (2014). The scanning mechanism of eukaryotic translation initiation. Annu Rev Biochem 83, 779–812. [DOI] [PubMed] [Google Scholar]
- Hinnebusch AG, Ivanov IP, and Sonenberg N (2016). Translational control by 5’-untranslated regions of eukaryotic mRNAs. Science 352, 1413–1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia NT (2016). Ribosome Footprint Profiling of Translation throughout the Genome. Cell 165, 22–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnstone TG, Bazzini AA, and Giraldez AJ (2016). Upstream ORFs are prevalent translational repressors in vertebrates. EMBO J 35, 706–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearse MG, and Wilusz JE (2017). Non-AUG translation: a new start for protein synthesis in eukaryotes. Genes Dev 31, 1717–1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolitz SE, Takacs JE and Lorsch JR (2009). Kinetic and thermodynamic analysis of the role of start codon/anticodon base pairing during eukaryotic translation initiation. RNA 15, 138–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M (1990). Downstream secondary structure facilitates recognition of initiator codons by eukaryotic ribosomes. Proc Natl Acad Sci U S A 87, 8301–8305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin F, Barends S, Jaeger S, Schaeffer L, Prongidi-Fix L, and Eriani G (2011). Cap-assisted internal initiation of translation of histone H4. Mol Cell 41, 197–209. [DOI] [PubMed] [Google Scholar]
- Putnam AA, and Jankowsky E AMP sensing by DEAD-box RNA helicases. (2013) .J Mol Biol 425, 3839–3845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz J, Mah N, Neuenschwander M, Kischka T, Ratei R, Schlag PM, Castanos-Velez E, Fichtner I, Tunn PU, Denkert C, et al. (2018). Loss-of-function uORF mutations in human malignancies. Sci Rep 8, 2395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen ND, Zhou F, Ingolia NT, and Hinnebusch AG (2015). Genome-wide analysis of translational efficiency reveals distinct but overlapping functions of yeast DEAD-box RNA helicases Ded1 and eIF4A. Genome Res 25, 1196–1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma D, and Jankowsky E (2014). The Ded1/DDX3 subfamily of DEAD-box RNA helicases. Crit Rev Biochem Mol Biol 49, 343–360. [DOI] [PubMed] [Google Scholar]
- Shirokikh NE, and Preiss T (2018). Translation initiation by cap-dependent ribosome recruitment: Recent insights and open questions. Wiley Interdiscip Rev RNA 9, e1473. [DOI] [PubMed] [Google Scholar]
- Snijders Blok L, Madsen E, Juusola J, Gilissen C, Baralle D, Reijnders MR, Venselaar H, Helsmoortel C, Cho MT, Hoischen A, et al. (2015). Mutations in DDX3X Are a Common Cause of Unexplained Intellectual Disability with Gender-Specific Effects on Wnt Signaling. Am J Hum Genet 97, 343–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spealman P, Naik AW, May GE, Kuersten S, Freeberg L, Murphy RF, and McManus J (2018). Conserved non-AUG uORFs revealed by a novel regression analysis of ribosome profiling data. Genome Res 28, 214–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starck SR, Tsai JC, Chen K, Shodiya M, Wang L, Yahiro K, Martins-Green M, Shastri N, and Walter P (2016). Translation from the 5’ untranslated region shapes the integrated stress response. Science 351, aad3867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang HL, Yeh LS, Chen NK, Ripmaster T, Schimmel P, and Wang CC (2004). Translation of a yeast mitochondrial tRNA synthetase initiated at redundant non-AUG codons. J Biol Chem 279, 49656–49663. [DOI] [PubMed] [Google Scholar]
- Zhang H, Dou S, He F, Luo J, Wei L, and Lu J (2018). Genome-wide maps of ribosomal occupancy provide insights into adaptive evolution and regulatory roles of uORFs during Drosophila development. PLoS Biol 16, e2003903. [DOI] [PMC free article] [PubMed] [Google Scholar]