Abstract
The pathway by which HIV-1 enters host cells is a prime target for novel drug discovery because of its critical role in the HIV-1 life cycle. The HIV-1 envelop glycoprotein gp120 plays an important role in initiating virus entry by targeting the primary cell receptor CD4. We explored the substitution of bulky molecular groups in region I in the NBD class of entry inhibitors. Previous attempts at bulky substitutions in that region abolished the antiviral activity, even though the binding site is hydrophobic. We synthesized a series of entry inhibitors containing 1,3-benzodioxolyl moiety or its bioisostere, 2,1,3-benzothiadiazole. The introduction of the bulkier groups was well tolerated, and despite only minor improvements in antiviral activity, the selectivity index (SI) improved significantly.
Keywords: HIV-1, ENV-pseudovirus, virus entry antagonist, structure-activity relationship (SAR), Reverse transcriptase (RT)
Graphical Abstract
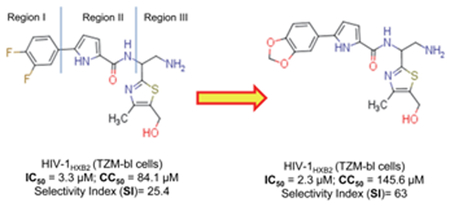
The 1,3-benzodioxolyl moiety and its bioisotere, 2,1,3-benzothiadiazole were well tolerated in the Region I, which site was traditionally known as not suitable for substitution by a bulky group. Three of the NBD inhibitors containing a 1,3-benzodioxolyl moiety showed some improvement in the antiviral activity, but more significantly the selectivity index (SI) increased by 2-3-fold compared to the control inhibitor NBD-14113.
Introduction
The development of anti-HIV-1 drugs with known and new targets, the reduction of the side effects of antiviral therapies, and, most critically, the optimization of antiretroviral combination therapies are helping many patients with HIV-1 infection to live a healthy life. Despite those successes, anti-HIV-1 drugs are still hampered by toxicities and drug resistance. Patient compliance with drug regimens also remains an issue. More research is needed to develop drugs with novel targets that block the HIV-1 replication cycle and work by different mechanisms than the currently available drugs.
Despite concerted efforts over the last two decades to develop a drug that blocks HIV-1 entry into host cells by preventing the HIV-1 envelop glycoprotein gp120 from binding to the cellular receptor CD4, there is no such drug yet available on the market. In 2015, the drug BMS-663068 (Fostemsavir) received the “Breakthrough Therapy” designation from the US FDA, representing a potential success in the long search for a drug that targets gp120. In 2017, ViiV Healthcare, the developer of Fostemsavir, announced positive phase 3 results from the BRIGHTE study of heavily treated patients with HIV infection.
Since 2005, our group has sought to develop HIV-1 host cell-entry inhibitors by targeting the Phe43 cavity in gp120. We have designed many potent gp120 antagonists since the discovery of NBD-556 (Figure 1), which acts as a CD4 mimic and gp120 agonist[1]. Because of the narrow space in the Phe43 cavity, the phenyl ring of NBD-556-type CD4 mimics will not tolerate a bulky substituent. For example, when we tried to introduce –C(=O)CH3 at the para- position of the phenyl ring, the antiviral activity was lost. Similarly, a bulkier group such as cycloheptyl in place of phenyl also completely abolished the antiviral activity[2]. We also attempted to introduce fluorine at the meta- position of the phenyl ring, but without succes[2–3]. Because none of the early attempts were promising, there has since been only limited exploration of substituent variations in the phenyl ring of NBD-556 and its analogs.
Figure 1.
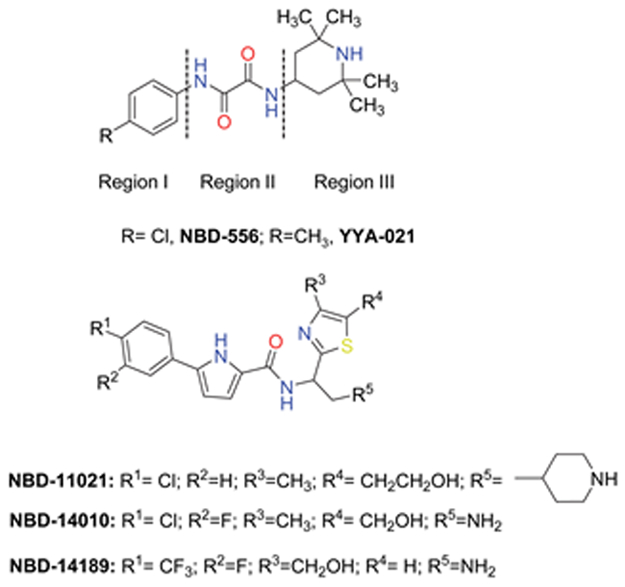
The chemical structure of NBD-556, YYA-021[6], and our next-generation lead compounds[1a, 4, 7].
We recently explored methoxy substitution in both the meta- and the para- positions of the phenyl ring of a new lead compound, NBD-14010, but both substitutions completely abolished the antiviral activity[4]. We hypothesize that the bulky and flexible nature of the methoxy substituent might be responsible for the loss of activity. To overcome the effect of the flexible substituent in region I, we decided to use 1,3-benzodioxole and its bioisostere 2,1,3-benzothiadiazole[5], which are more compact but bulkier than the methoxy substituent. Mizuguchi et al. in 2016 used a 1,3-benzodioxole moiety to replace a 4-chlorophenyl group of NBD-556. The resulting compound had about 10-fold less antiviral activity compared with NBD-556 and was also more toxic[8].
We have developed a set of new lead compounds (NBD-11021, NBD-14010 and NBD-14189) with markedly different structural profiles than NBD-556 and YYA-021[6] (Figure 1). We hypothesize that the structures of the new lead compounds might allow them to tolerate the introduction of 1,3-benzodioxole or its bioisostere 2,1,3-benzothiadiazole[5], resulting in retained antiviral activity, improved toxicity profile, and higher selectivity index (SI).
Here, we synthesized 22 entry inhibitors with substitutions of 1,3-benzodioxole or its bioisostere to generate a comprehensive understanding of the structure-activity relationship (SAR) of these bulky substituents in region I of the phenyl ring of NBD-type CD4 mimics.
Results and Discussion
Chemistry
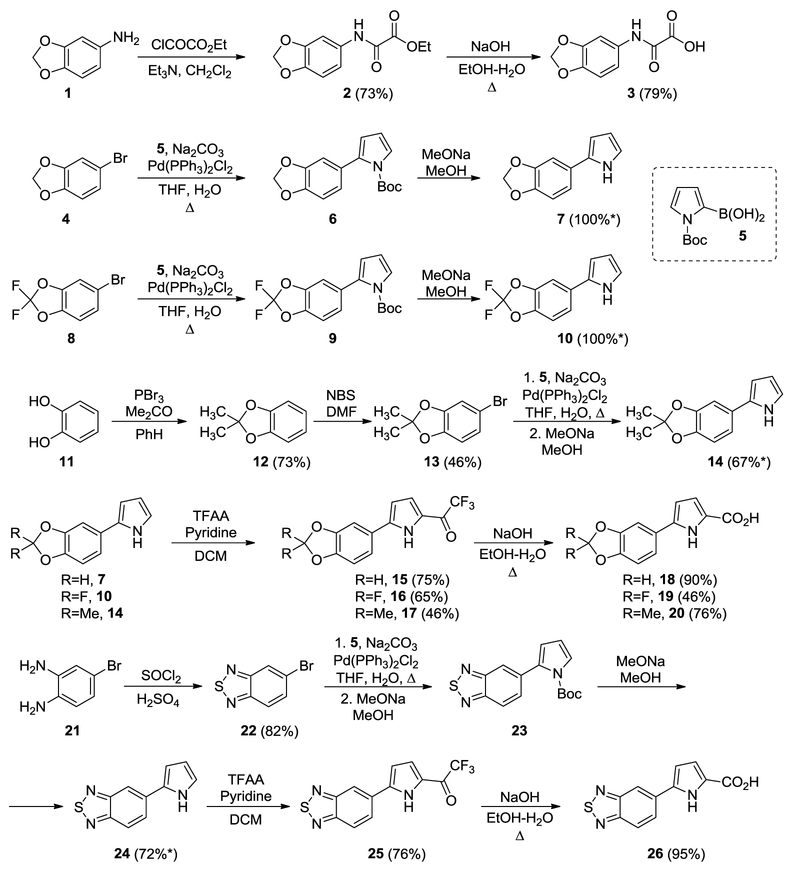
The synthesis of the novel NBD compounds is described in Schemes 1–3. First, we prepared a series of carboxylic acids with 1,3-benzodioxole or its bioisostere 2,1,3-benzothiadiazole (Scheme 1). Aniline 1 was acylated with ethyl 2-chloro-2-oxoacetate and after saponification yielded acid 3[2]. Acids 18-20, 26 were prepared using a general scheme involving Suzuki coupling of aryl bromides 4, 8, 13, and 22 with N-Boc-2-pyrrole boronic acid, followed by Boc cleavage, acylation, and semi-haloform reaction[9]. Aryl bromides 4 and 8 were commercially available. Aryl bromides 13 and 22 were prepared using procedures from the literature [10].
Scheme 1.
Synthesis of 1,3-benzodioxole and its bioisostere containing acids. *yields are over two steps.
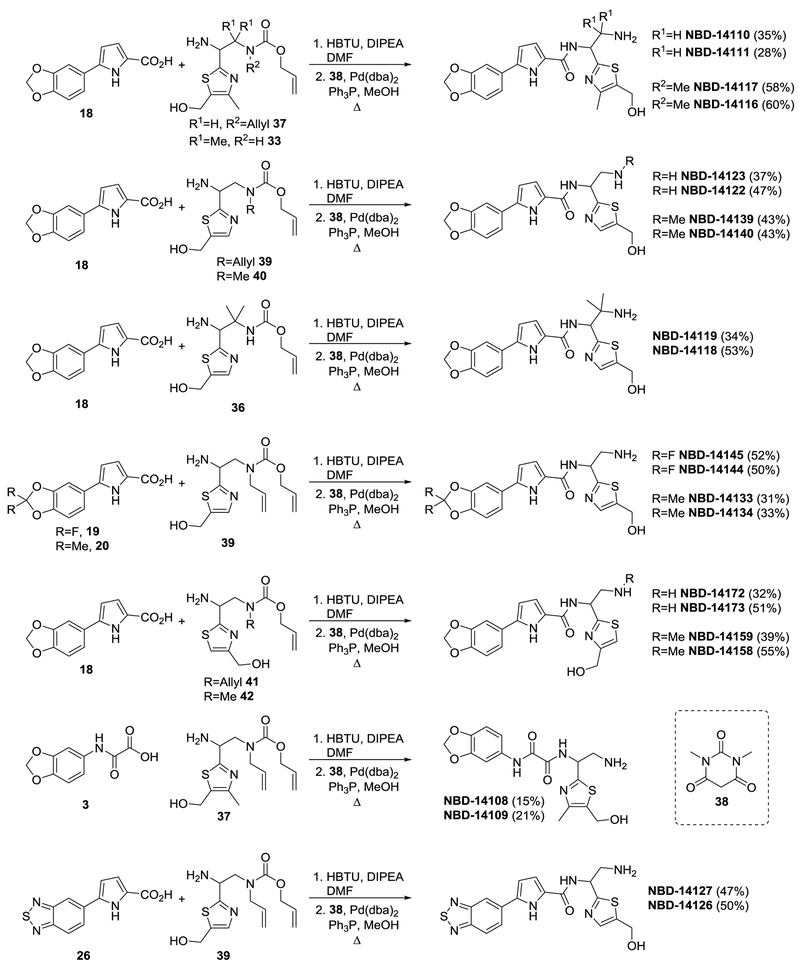
Scheme 3.
Synthesis of final NBD compounds.
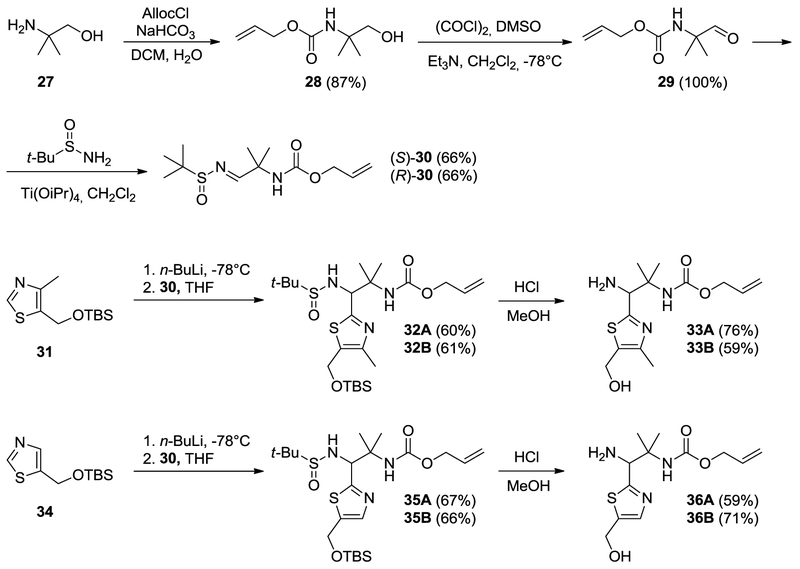
Protected amines 37, 39, 40, 41, and 42 and protected thiazoles 31 and 34 were prepared as described previously[4, 7b, 7c, 11]. Two protected amines, 33 and 36, with the gem-dimethyl moiety were prepared using 1,2-addition of thiazolyl anions of the two enantiopure imines 30, as per Scheme 2. Compounds 32 and 35 were obtained as inseparable mixtures of diastereomers with dr~4:1 – 3:1 (differs in configuration at C1, based on NMR). Because of the presence of acidic N-H amide, we used 2.5 equivalent of metallated thiazole. The absolute configuration of compounds 33 and 36 was not determined, and, as in prior works, compounds derived from (enantiomer 2)-30 were designated as A, and compounds derived from (enantiomer 1)-30 were designated as B. We prepared the final library of NBD compounds using peptide coupling, followed by protecting-group cleavage (Scheme 3)[12].
Scheme 2.
Synthesis of protected thiazole ring-containing amines.
Antiviral screening and SAR analysis
We evaluated the HIV-1-inhibitory activity and cytotoxicity of the NBD compounds. We assessed the anti-HIV-1 activity in a single-cycle assay by infecting TZM-bl indicator cells with the pseudovirus HIV-1HXB2. We also assessed the anti-HIV-1 activity in a multi-cycle assay by infecting MT-2 human T-cells with the full-length, lab-adapted virus HIV-1IIIB, as described previously[4]. As a control, we used NBD-14113, which contains the sterically smallest substituent fluorine (F) at the meta- and para- positions. NBD-14110, which has the 1,3-benzodioxole moiety in region I, showed a slight improvement in antiviral activity in both the single-cycle and the multi-cycle assays and also a ~1.7-fold improvement in cytotoxicity (CC50) compared with the control. NBD-14111 lost its antiviral activity, although it still had reduced cytotoxicity compared with the control. We substituted the hydrogens of the α-carbon of the ethylamine group in NBD-14110 and NBD-14111 with two methyl groups to understand the SAR effect of the bulk in that region. The resulting compounds (NBD-14116 and NBD-14117) showed similar antiviral activity but became more cytotoxic. When we removed the methyl group from the thiazole of NBD-14110 to convert it to NBD-14122, we observed a ~1.9-fold reduction in antiviral activity. However, the antiviral activity of NBD-14123 (converted from NBD-14111) remained similar to that of NBD-14111, and there was no noticeable change in cytotoxicity. The addition of a methyl group in the primary amine of NBD-14139 and NBD-14140 resulted in marginal improvement of antiviral activity; however, the cytotoxicity remained the same. Interestingly, when we replaced the hydrogens of the α-carbon of the ethylamine of NBD-14123 with two methyl groups, the antiviral activity of NBD-14119 was improved by ~2.3-fold, while that of NBD-14118 remained similar to that of NBD-14116; the cytotoxicity of both NBD-14118 and NBD-14119 was higher, as was observed with NBD-14117 and NBD-14118.
We further explored the SAR by adding di-fluoro at position 2 of the 1,3-benzodioxole moiety. NBD-14144 showed ~4.5-fold higher antiviral activity compared with NBD-14122. NBD-14145 showed less improvement (~1.7-fold) in antiviral activity. It is worth noting, however, that the cytotoxicity of both compounds became much higher (~3.9-fold). When we added two methyl groups at position 2 of the moiety, NBD-14133 and NBD-14134 lost their antiviral potency. The positional switching of CH2OH in NBD-14172 improved the antiviral potency by about 1.5-fold compared with that of NBD-14123, whereas similar switching in NBD-14122 reduced the antiviral activity of the resultant (enantiomer 1)enantiomer NBD-14173 by ~1.9 fold; the cytotoxicity was higher for both compounds, however. Addition of a methyl group in the primary amine of NBD-14158 and NBD-14159 did not have any effect on antiviral potency or cytotoxicity.
When we attempted to introduce the 1,3-benzodioxole moiety in the oxalamide-containing NBD-14108 and NBD-14109, the antiviral activity was completely lost, unlike what was observed by Ohashi et al. [7a]. We attempted to replace the 1,3-benzodioxole moiety with its bioisostere, 2,1,3-benzothiadiazole[5]. The resulting NBD-14127 showed ~1.6-fold improvement in antiviral potency compared with NBD-14123, and NBD-14126 showed about 2.8-fold improvement in antiviral activity compared with NBD-14122. The cytotoxicity of both NBD-14126 and NBD-14127 was higher by about 1.5-fold compared with that of the parent compounds.
The SAR analysis indicated that bulkier groups such as 1,3-benzodioxole or its bioisostere 2,1,3-benzothiadiazole[5] are well tolerated to replace the phenyl moiety in region I. This was also validated by docking one of the best leads, NBD-14110, in the Phe43 cavity of HIV-1 gp120 (Figure 2) using Glide software from Schrodinger (Cambridge, USA). When we examined the SI [SI = CC50/IC50)], NBD-14110, NBD-14123, and NBD-14159 turned out to be the best leads for further exploration.
Figure 2.
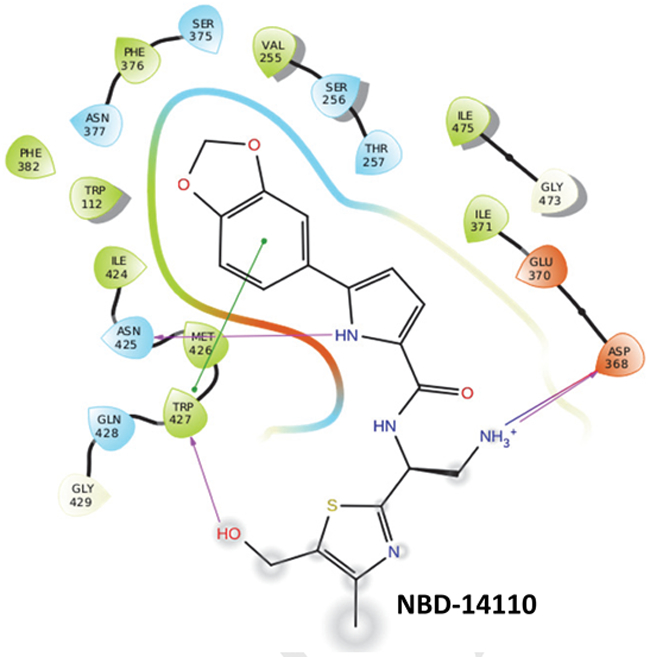
2D-interaction of NBD-14110 with HIV-1 gp120 in the Phe43 cavity based on Glide-based docking.
Antiviral activity of the polycyclic NBD compounds against a large panel of HIV-1 Env-pseudotyped reference viruses.
We selected the three new NBD compounds (NBD-14110, NBD-14123, and NBD-14159) based on their anti-HIV-1 activity and the SI after comparing data from the single-cycle and multi-cycle cell-based assays. We evaluated those compounds against a selection of 34 HIV-1 clones of clinical isolates of subtypes A, B, C, and D and of recombinant subtype A (Arec), including A1/D, A2/D, and AG. We compared the three selected compounds with the previously described compound NBD-14113[13]. We found that the three polycyclic compounds exhibited anti-HIV-1 activity at low micromolar concentrations, with overall mean IC50 values very similar to that of NBD-14113 (Table 2). When we analyzed the data obtained from the different viral subtypes, the mean IC50 for NBD-14113 across the subtype Arec and subtype C viruses was slightly higher than that across the subtype A, B, and D viruses. NBD-14110 was equally active against all the viral subtypes, as indicated by the similar mean IC50 values obtained for that compound. NBD-14123 and NBD-14159 were slightly more active against the subtype A, Arec, B and D viruses than against subtype C viruses, as shown by the mean IC50 values. Moreover, NBD-14110, NBD-14123, and NBD-14159 were weak inhibitors of the control pseudovirus VSV-G, indicating that the inhibitory activity of these compounds is specific to HIV-1. We observed similar activity of these three molecules against all the clinical isolated tested. We reason that despite possible sequence difference in the Phe43 cavity of the isolates tested, the variation in substituents in the molecule are loated in the α-carbon atom of the primary amine and the thiazole ring (CH3 vs. H) which have least impact in the binding of the molecules in the cavity as demonstrated in Figure 2.
Table 2.
Neutralization activity of NBD compounds against a panel of HIV-1 Env pseudoviruses
| IC50 (µM)a | ||||||
|---|---|---|---|---|---|---|
| Subtype | NIH # | ENV | NBD-14113 | NBD-14110 | NBD-14123 | NBD-14159 |
| A | 11887 | Q259ENV.W6 | 2.1±0.3 | 1.9±0.2 | 2.3±0.2 | 1.3±0.2 |
| 11888 | QB726.70M.ENV.C4 | 4±0.2 | 3.7±0.3 | 3.5±0.1 | 2.7±0.2 | |
| 11891 | QF495.23M.ENV.A3 | 4±0.2 | 3±0.1 | 3.3±0.2 | 3.7±0.1 | |
| - | BG505-T332N | 3.3±0.2 | 4.2±0.5 | 4±0.5 | 3.3±0.2 | |
| - | KNH1144 | 3.8±0.1 | 5.9±0.4 | 4.1±0.4 | 4±0.3 | |
| A1/D | 11901 | QA790.204I.ENV.A4 | 5.5±0.3 | 2.1±0.6 | 1.6±0.1 | 4±0.4 |
| 11903 | QA790.204I.ENV.C8 | 4.3±0.1 | 2.4±0.2 | 1.4±0.2 | 2.1±0.1 | |
| 11904 | QA790.204I.ENV.E2 | 3±0.1 | 1.5±0.2 | 2.5±0.3 | 3.2±0.2 | |
| A2/D | 11906 | QG393.60M.ENV.B7 | 5.9±0.1 | 5.2±0.3 | 5.9±0.2 | 3.8±0.4 |
| AG | 11595 | CRF02_AG, clone 251 | 3±0.2 | 2.5±0.1 | 3.1±0.2 | 3.5±0.1 |
| 11597 | CRF02_AG, clone 253 | 4.8±0.3 | 3.2±0.2 | 3.2±0.2 | 3.1±0.1 | |
| 11599 | CRF02_AG, clone 257 | 4.6±0.5 | 5.4±0.2 | 4.4±0.1 | 4.8±0.2 | |
| 11607 | CRF02_AG, clone 33 | 3.5±0.2 | 5±0.3 | 3.5±0.4 | 2.7±0.2 | |
| B | 11018 | QH0692, clone 42 | 4.3±0.4 | 8.2±0.2 | 6.4±0.1 | 5.8±0.7 |
| 11022 | PVO, clone 4 | 4±0.3 | 1.4±0.3 | 1.9±0.2 | 4.3±0.7 | |
| 11023 | TRO, clone 11 | 5.1±0.1 | 2.4±0.4 | 3.6±0.2 | 4.4±0.6 | |
| 11035 | REJO4541, clone 67 | 3.1±1 | 3.4±0.3 | 3.4±0.2 | 3.1±0.4 | |
| 11036 | RHPA4259, clone 7 | 3.7±0.4 | 2.8±0.3 | 3.1±0.7 | 3.7±0.4 | |
| 11037 | THRO4156, clone 18 | 3.8±0.1 | 2.7±0.2 | 3.1±0.1 | 2.7±0.2 | |
| 11561 | 1054.TC4.1499 | 3.6±0.2 | 4.2±0.4 | 2.4±0.3 | 4.1±0.1 | |
| 11572 | 9021_14.B2.4571 | 3.2±0.2 | 4.5±0.5 | 2.1±0.6 | 3.7±0.3 | |
| 11578 | WEAUd15.410.5017b | 3.7±0.3 | 1.6±0.1 | 3.4±0.2 | 3.3±0.2 | |
| - | B41 | 3.4±0.1 | 2±0.1 | 2.8±0.3 | 1.7±0.1 | |
| C | 11308 | Du422, clone 1 | 3±0.1 | 1.8±0.2 | 2.5±0.3 | 3.1±0.3 |
| 11314 | ZM109F.PB4 | 1.9±0.1 | 4.9±0.1 | 3±0.2 | 3.3±0.1 | |
| 11317 | CAP210.2.00.E8 | 3.7±0.1 | 5.5±0.4 | 5.3±0.2 | 3.5±0.2 | |
| 11500 | HIV-00836-2, clone 5 | 4.3±0.4 | 1.7±0.1 | 3.2±0.2 | 4.9±0.3 | |
| 11504 | HIV-16936-2, clone 21 | 5.9±0.2 | 3.5±0.1 | 4.4±0.5 | 5.3±0.2 | |
| 11506 | HIV-25711-2, clone 4 | 4.7±0.4 | 4.4±0.4 | 5.7±0.7 | 5.8±0.3 | |
| 11507 | HIV-25925-2, clone 22 | 4.8±0.3 | 4.7±0.1 | 5.6±0.3 | 4.2±0.5 | |
| 11909 | QB099.391M.ENV.C8 | 4±0.3 | 4.3±0.5 | 4.4±0.6 | 5±0.1 | |
| D | 11911 | QA013.70I.ENV.H1 | 3.5±0.1 | 2.9±0.3 | 2.5±0.4 | 4.3±0.7 |
| 11916 | QD435.100M.ENV.B5 | 2.2±0.3 | 2.4±0.2 | 3.5±0.3 | 3.3±0.2 | |
| 11918 | QD435.100M.ENV.E1 | 4.8±0.6 | 5.9±0.4 | 5.3±0.4 | 3.5±0.2 | |
| Control | VSV-Gc | 26.3±1.4 | 51.6±4.4 | 45.3±2.3 | 41.3±3.4 | |
| Mean ± SEM (µM): Overall (n=34) | 3.9±0.17 | 3.56±0.27 | 3.54±0.22 | 3.68±0.18 | ||
| Subtype A (n=5) | 3.44±0.36 | 3.74±0.66 | 3.44±0.32 | 3±0.48 | ||
| Subtype Arec (n=8) | 4.33±0.39 | 3.4±0.55 | 3.2±0.52 | 3.4±0.29 | ||
| Subtype B (n=10) | 3.79±0.18 | 3.32±0.63 | 3.22±0.4 | 3.68±0.35 | ||
| Subtype C (n=7) | 4.04±0.43 | 3.85±0.5 | 4.26±0.44 | 4.39±0.36 | ||
| Subtype D (n=3) | 3.5±0.75 | 3.7±1.1 | 3.77±0.82 | 3.7±0.31 | ||
The reported IC50 values represent the mean ± standard deviation (n = 3).
R5X4-tropic virus; all the rest are CCR5-tropic viruses.
VSV-G was tested in U87-CD4-CCR5 cells. The CC50 for NBD-14113 was ~75 µM, and the CC50 for NBD-14110, NBD-14123 and NBD-14159 was >100 µM in these cells.
Additionally, we tested the anti-HIV-1 activity of the new polycyclic NBD compounds against a set of paired infant and maternal HIV-1 Env molecular clones of subtype A and C/D, which were isolated from infants infected between birth and 6 weeks postpartum and from the respective chronically infected mothers [14]. The infant variants were poorly neutralized by combinations of Mab 2G12, biz, 2F5, and 4E10 [14]. We detected no differences in the activity of the NBD compounds against the vertically transmitted viral types, indicating that all the polycyclic compounds neutralized the infant and maternal HIV-1 variants equally (Table 3). NBD-14159 was the most effective compound with an overall mean IC50 of 3.9±0.3 µM. NBD-14110 was the least effective with an overall mean IC50 of 5±0.5 µM.
Table 3.
Neutralization activity of NBD compounds against an HIV-1 panel of “Paired Infant (B) and Maternal (M) Env Molecular Clones.”
| IC50 (µM)a | ||||||
|---|---|---|---|---|---|---|
| Subtype | NIH # | ENV | NBD-14113 | NBD-14110 | NBD-14123 | NBD-14159 |
| A | 11518-B | BG505.W6M.ENV.C2 | 4.4±0.3 | 4.8±0.3 | 4.4±0.1 | 4.7±0.5 |
| A | 11528-M | MG505.W0M.ENV.A2 | 4.7±0.8 | 6.1±0.5 | 3.7±0.2 | 4.6±0.1 |
| A | 11519-B | B1206.W6P.ENV.A1A | 4.8±0.4 | 5.6±0.2 | 4.9±0.3 | 4±0.3 |
| A | 11531-M | MI206.W0M.ENV.D1 | 3.4±0.2 | 4.7±0.2 | 3.4±0.3 | 3.4±0.2 |
| A | 11521-B | BJ613.W6M.ENV.E1 | 2.8±0.4 | 2.8±0.1 | 2.5±0.1 | 3±0.1 |
| A | 11534-M | MJ613.W0M.ENV.A2 | 4.9±0.2 | 4.9±0.1 | 4±0.1 | 3.1±0.3 |
| A | 11525-B | BL274.W6M.ENV.A3 | 5.7±0.3 | 7.2±0.1 | 5.7±0.3 | 4.2±0.5 |
| A | 11540-M | ML274.W0M.ENV.B1 | 4.5±0.6 | 7.1±0.3 | 5.6±0.4 | 5.4±0.5 |
| C/D | 11522-B | BK184.W6M.ENV.D2 | 3.6±0.2 | 3.7±0.6 | 3.6±0.1 | 4.2±0.1 |
| C/D | 11536-M | MK184.W0M.ENV.E4 | 2.6±0.3 | 3.4±0.2 | 3.7±0.1 | 2.6±0.1 |
| Mean ± SEM (µM): Overall (n=10) | 4.1±0.3 | 5±0.5 | 4.2±0.3 | 3.9±0.3 | ||
| Infant (B) | 4.3±0.5 | 4.8±0.8 | 4.2±0.6 | 4±0.3 | ||
| Mother (M) | 4±0.4 | 5.2±0.6 | 4.1±0.4 | 3.8±0.5 | ||
The reported IC50 values represent the mean ± standard deviation (n = 3).
Antiviral activity of the polycyclic NBD compounds against three HIV-1 isolates in human PBMC.
The new polycyclic NBD compounds were tested in human PBMC cells against 3 competent HIV-1 isolates. We used two lab-adapted viruses, the CXCR4-tropic HIV-1LAI, and the CCR5-tropic HIV-1BaL and the CCR5-tropic HIV-197USSN54, primary clinical isolate of subtype A. We found that all the NBD compounds neutralized the lab-adapted viruses with similar antiviral potency as shown by the IC50 in range of 1.9–3.8 µM (Table 4). NBD-14110 showed a slightly better activity against the primary isolate HIV-197USSN54. The cytotoxicity assay indicated that the CC50 for these compounds tested in the human PBMC was ≥ 47 µM.
Table 4.
Anti-HIV-1 activity (IC50) and cytotoxicity (CC50) of Polycyclic NBD compounds tested in human PBMC
| Inhibitors | IC50 (μM)a | CC50 (μM)a |
||
|---|---|---|---|---|
| HIV- 1LAI |
HIV-1BaL | HIV- 197USSN54 |
||
| NBD-14113 | 3.8±1.1 | 3.8±0.4 | 3.1±0.5 | 47.1±2.6 |
| NBD-14110 | 3.5±0.9 | 2.8±0.6 | 1.1±0.3 | 62.8±2.9 |
| NBD-14123 | 2.9±0.9 | 1.9±0.2 | 1.8±0.6 | 61.3±4 |
| NBD-14159 | 2.6±0.7 | 2.8±0.4 | 3.6±0.4 | 68.3±5 |
The reported IC50 and CC50 values represent the mean ± standard deviation (n = 3).
NBD compounds do not enhance HIV-1 entry into CD4-negative cells and showed moderate activity against HIV-1 reverse transcriptase (RT)
We evaluated whether the selected polycyclic compounds NBD-14110, NBD-14123, and NBD-14159 support HIV-1 entry into CD4-negative cells. We used NBD-556 (an HIV-1 entry agonist [13, 15]) as a control. Although NBD-556 enhanced the infection of the Cf2Th-CCR5 cells, we found that none of the new polycyclic NBD compounds enhanced HIV-1 infectivity in those cells. To exclude false negative results we performed a cytotoxicity assay by treating these cells with escalating concentrations of the above NBD compounds and we found that the CC50 detected for NBD-14113 was ~90 µM while NBD-556, NBD-14110, NBD-14123 and NBD-14159 had a CC50 >100 µM. Taken together, these results suggest that the HIV-1 entry antagonist property is maintained in these new generation NBD compounds as well (Figure 3).
Figure 3.
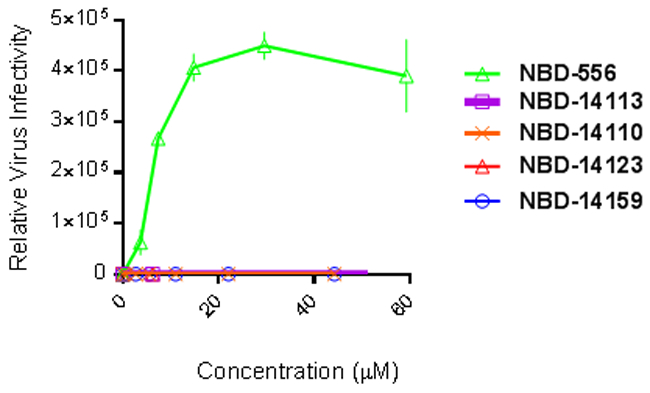
Infectivity of CD4 negative Cf2Th–CCR5 cells by HIV-1ADA (CD4-dependent virus). Cf2Th–CCR5 cells were infected with HIV-1ADA in the presence of the NBD entry inhibitors. The relative virus infectivity indicates the ratio between the amount of infection detected in the presence of the compounds and the amount of infection detected in the absence of the compounds. Three independent experiments were performed in triplicate, and the graph is representative of one experiment; the values represent the mean ± standard deviation.
Because some NBD entry antagonists have shown moderate activity against HIV-1 reverse transcriptase (RT)[4], we evaluated the activity of the polycyclic NBD compounds against that enzyme. We used NBD-556 and Nevirapine as controls. As previously reported, 300 μM NBD-556 had no activity against HIV-1 RT[13, 15], whereas Nevirapine was extremely potent against HIV-1 RT, with an IC50 of 0.20 μM (Table 4). All of the polycyclic NBD compounds inhibited HIV-1 RT with an IC50 in the range of 28–45 μM. Those IC50 values are more comparable to the values obtained by testing the NBD compounds against the control pseudovirus VSV-G than to those obtained in the HIV-1 neutralization assays, suggesting that the HIV-1 RT is not the primary target of the polycyclic NBD compounds.
S375Y and S375W mutants are not resistant to NBD compounds
Previously, we reported that some specific amino acid substitutions in the CD4-binding site of the ENV gp120 rendered the mutant virus resistant to the NBD compounds, but S375H, S375W, and S375Y mutant viruses were not. In this study, we tested NBD-14110 against the mutant pseudovirus HIV-1HXB2 carrying amino acid substitution S375Y and S375W in the ENV gp120 region. This compound showed similar potency against this mutant as was observed against HIV-1HXB2-WT (data not shown). The data indicate that the S375Y and S375W mutant viruses are sensitive to NBD-14110.
Conclusions
We explored the effect of substituting a phenyl ring in region I of the phenyl ring of NBD CD4 mimics with a bulkier moiety such as 1,3-benzodioxole or its bioisostere. Three new NBD compounds (NBD-14110, NBD-14123, and NBD-14159) with a 1,3-benzodioxole substituent exhibited the best improvement in SI compared with the control, NBD-14113. Furthermore, those inhibitors inhibited all of the tested clinical HIV-1 isolates with similar potency. The 1,3-benzodioxole series compounds showed poor activity against HIV-1 RT, indicating that they primarily target gp120, thereby preventing HIV-1 entry into host cells. We anticipate that these new lead compounds can be further optimized to develop more potent and HIV-1 entry-specific inhibitors.
Experimental Section
Synthesis
ethyl 2-(benzo[d][1,3]dioxol-5-ylamino)-2-oxoacetate (2)
Aniline 1 (10.10 g, 73.6 mmol) was dissolved in CH2Cl2 (74 mL), Et3N (11.30 mL, 73.6 mmol) was added. To the resulted solution ethyl chlorooxoacetate (9.05 mL, 73.6 mmol) was added dropwise as a solution in CH2Cl2 (74 mL) with cooling on a water bath. The reaction mixture was stirred for 1 hour and washed with 5% HCl (~200 mL). The layers were separated, and the organic layer was extracted with CH2Cl2 (2 × 100 mL). The combined organic layers were dried over Na2SO4, filtered and evaporated. The residue was recrystallized from ethanol (50 mL) to give 12.75 g (73%) of dirty-white solid.
1H NMR (CDCl3, 400 MHz) δ = 1.29 (t, J=7.0 Hz, 3 H), 4.28 (q, J=7.0 Hz, 2 H), 5.99 (s, 2 H), 6.87 (d, J=8.4 Hz, 1 H), 7.22 (d, J=7.5 Hz, 1 H), 7.36 (s, 1 H), 10.66 (s, 1 H).
13C NMR (CDCl3, 100 MHz) δ = 13.8, 62.4, 101.2, 102.3, 108.0, 113.7, 131.7, 144.1, 147.1, 155.1, 160.7.
2-(benzo[d][1,3]dioxol-5-ylamino)-2-oxoacetic acid (3)
The ester 2 (12.75 g, 53.80 mmol) was suspended in ethanol (108 mL), and a solution of NaOH (4.30 g, 108 mmol, 2.0 equiv) in water (108 mL) was added. The resulting mixture was heated at reflux for 2 days, cooled to room temperature and acidified with conc. HCl (~10 mL). To the resulted homogeneous solution, water was added until the product precipitates (~200 mL), and the acid was filtered off. M = 8.84 g. Yield = 79%.
1H NMR: (DMSO-d6, 400 MHz) δ = 5.99 (s, 2 H), 6.87 (d, J=8.4 Hz, 1 H), 7.24 (dd, J=8.4, 1.7 Hz, 1 H), 7.39 (d, J=1.7 Hz, 1 H), 10.60 (s, 1 H), 12.37 – 14.87 (br. s, 1 H).
13C NMR (DMSO-d6, 100 MHz): δ = 101.2, 102.2, 108.0, 113.5, 132.0, 144.0, 147.1, 156.5, 162.2.
General procedure for cross-coupling reaction and Boc cleavage:
Aryl bromide (1 equiv) was dissolved in THF (1 M), Boc-pyrrole boronic acid (1.2 equiv) was added to the solution followed by aqueous Na2CO3 (2 equiv) solution (1 M). Under a constant flow of nitrogen Pd(PPh3)2Cl2 (1 mol %) was added. The reaction mixture was vigorously stirred and heated under reflux for 4–5 hours. The reaction mixture was cooled to room temperature diluted with water and extracted with DCM (3×100 mL). The organic layer was dried over Na2SO4 and evaporated. To the residue was added freshly prepared (from Na and MeOH) NaOMe solution (~1M in MeOH, 2–3 equiv) and left overnight. The reaction mixture was diluted with water and extracted with DCM (3×100 mL). The organic layer was dried over Na2SO4 and evaporated. The residue was used without further purification.
2-(benzo[d][1,3]dioxol-5-yl)-1H-pyrrole (7)
Compound 7 was prepared according to the general procedure for cross-coupling reaction and Boc cleavage from 4 (10.00 g). M = 9.80 g. Yield >100% (over two steps).
1H NMR (CDCl3, 400 MHz) δ = 5.98 (s, 2 H), 6.29 (q, J=2.7 Hz, 1 H), 6.41 (br. s, 1 H), 6.81 – 6.85 (m, 2 H), 6.94 (dd, J=9.3, 1.2 Hz, 1 H), 6.98 (s, 1 H), 8.32 (br. s, 1 H).
13C NMR (CDCl3, 100 MHz) δ = 101.2, 105.3, 105.5, 108.8, 110.1, 117.4, 118.5, 127.6, 132.3, 146.3, 148.3.
2-(2,2-difluorobenzo[d][1,3]dioxol-5-yl)-1H-pyrrole (10)
Compound 10 was prepared according to the general procedure for cross-coupling reaction and Boc cleavage from 8 (8.43 g). M = 8.15 g. Yield >100% (over two steps).
1H NMR (CDCl3, 400 MHz) δ = 6.33 (q, J=2.7 Hz, 1 H), 6.46 – 6.50 (m, 1 H), 6.85 – 6.89 (m, 1 H), 7.04 (d, J=8.9 Hz, 1 H), 7.12 – 7.17 (m, 2 H), 8.39 (br. s, 1 H).
13C NMR (CDCl3, 100 MHz) δ = 105.6, 106.5, 109.8, 110.3, 119.0, 119.5, 129.5, 131.0, 131.8 (t, J = 255.1 Hz), 142.2, 144.4.
2,2-dimethylbenzo[d][1,3]dioxole (12)
PBr3 (8.54 mL, 91 mmol, 0.4 equiv) was added dropwise to a stirred solution of pyrocatechol (25 g, 227 mmol) and acetone (50 mL, 681 mmol, 3 equiv) in benzene (230 mL). The reaction mixture was stirred until HBr ceased to evolve (~4 hours) and then poured into NaOH aqueous solution (100 g in 0.5 L). The organic layer was separated, and the aqueous layer was extracted with DCM (3×100 mL). The organic layer was dried over Na2SO4 and evaporated. The residue was dissolved in hexane and filtered through a small plug of silica (~1 cm). The filtrate was evaporated to give a pure 12 as a colorless liquid. M = 24.86 g. Yield = 73%.
1H NMR (CDCl3, 400 MHz) δ = 1.70 (s, 6 H), 6.73 – 6.84 (m, 4 H).
13C NMR (CDCl3, 100 MHz) δ = 26.0 (2C), 108.6 (2C), 117.5, 121.1 (2C), 147.4 (2C).
5-bromo-2,2-dimethylbenzo[d][1,3]dioxole (13)
12 (14.64 g, 97.6 mmol) was dissolved in DMF (98 mL), and NBS (17.40 g, 97.7 mmol) was added. The reaction mixture was stirred for 1 day, diluted with water (~1L) and extracted with hexane (3×100 mL). The combined organic layers were dried over Na2SO4 and evaporated. The residue was distilled at reduced pressure (bp=80 ºC, 1 torr). M = 10.35 g. Yield = 46%.
1H NMR (CDCl3, 400 MHz) δ = 1.68 (s, 6 H), 6.60 (d, J=8.2 Hz, 1 H), 6.87 (d, J=2.0 Hz, 1 H), 6.90 (dd, J=8.2, 1.8 Hz, 1 H).
13C NMR (CDCl3, 100 MHz) δ = 25.9 (2C), 109.5, 112.2, 112.5, 119.1, 123.8, 146.9, 148.5.
2-(2,2-dimethylbenzo[d][1,3]dioxol-5-yl)-1H-pyrrole (14)
Compound 14 was prepared according to the general procedure for cross-coupling reaction and Boc cleavage from 13. M = 2.46 g. Yield = 67% (over two steps).
1H NMR (CDCl3, 400 MHz) δ = 1.70 (s, 6 H), 6.27 (dd, J=5.7, 2.7 Hz, 1 H), 6.35 – 6.39 (m, 1 H), 6.74 (d, J=8.6 Hz, 1 H), 6.79 – 6.83 (m, 1 H), 6.85 – 6.93 (m, 2 H), 8.33 (br. s, 1 H).
13C NMR (CDCl3, 100 MHz) δ = 26.0 (2C), 105.2, 108.6, 110.0, 116.9, 118.2, 118.3, 127.0, 132.5, 146.3, 148.1.
General procedure for aryl pyrrole trifluoroacetylation:
To the solution appropriate 2-aryl-pyrrole and pyridine (1.2 equiv) in dichloromethane (1 M) trifluoroacetic anhydride (1.2 equiv) was added dropwise with an external cold water bath cooling. After the addition is complete, the mixture was stirred for 1 hour, and the yellow-greenish precipitate was filtered and washed with DCM. If no precipitation occurred, the reaction mixture was evaporated and triturated with water. The precipitate was filtered and recrystallized from ethanol.
1-(5-(benzo[d][1,3]dioxol-5-yl)-1H-pyrrol-2-yl)-2,2,2-trifluoroethanone (15)
Compound 15 was prepared according to the general procedure for aryl pyrrole trifluoroacetylation from 7. M = 7.14 g. Yield = 75%.
1H NMR: (DMSO, 400 MHz) δ = 6.06 (s, 2 H), 6.82 (s, 1 H), 6.97 (d, J=8.1 Hz, 1 H), 7.22 (d, J=1.3 Hz, 1 H), 7.50 (d, J=8.1 Hz, 1 H), 7.61 (s, 1 H), 12.68 (s, 1 H).
13C NMR: (DMSO, 100 MHz) δ = 101.5, 106.5, 108.7, 110.4, 117.2 (q, J = 289.8 Hz), 120.9, 123.5 (q, J = 2.9 Hz), 123.8, 125.5, 143.7, 147.9, 148.1, 167.1 (q, J = 34.4 Hz).
1-(5-(2,2-difluorobenzo[d][1,3]dioxol-5-yl)-1H-pyrrol-2-yl)-2,2,2-trifluoroethanone (16)
Compound 16 was prepared according to the general procedure for aryl pyrrole trifluoroacetylation from 10. M = 7.52 g. Yield = 65%.
1H NMR: (DMSO, 400 MHz) δ = 6.95 (dd, J=4.2, 2.4 Hz, 1 H), 7.23 – 7.29 (m, 1 H), 7.48 (d, J=8.4 Hz, 1 H), 7.85 (dd, J=8.5, 1.7 Hz, 1 H), 8.06 (d, J=1.5 Hz, 1 H), 12.89 (br. s, 1 H).
13C NMR: (DMSO, 100 MHz) δ = 107.9, 110.3, 110.9, 117.2 (q, J = 289.8 Hz), 122.9, 123.0 (q, J = 3.5 Hz), 126.3, 126.8, 131.4 (t, J = 253.8 Hz), 142.1, 143.1, 143.5, 168.0 (q, J = 289.8 Hz).
1-(5-(2,2-dimethylbenzo[d][1,3]dioxol-5-yl)-1H-pyrrol-2-yl)-2,2,2-trifluoroethanone (17)
Compound 17 was prepared according to the general procedure for aryl pyrrole trifluoroacetylation from 14. 17 was purified by means of column chromatography, eluent hexane/EtOAc, 1:1. M =2.46 g. Yield = 46%.
1H NMR: (DMSO, 400 MHz) δ = 1.65 (s, 6 H), 6.80 (dd, J=3.9, 2.3 Hz, 1 H), 6.89 (d, J=8.2 Hz, 1 H), 7.17 – 7.26 (m, 1 H), 7.47 (dd, J=8.2, 1.2 Hz, 1 H), 7.51 (s, 1 H), 12.67 (s, 1 H).
13C NMR: (DMSO, 100 MHz) δ = 25.5 (2C), 106.4, 108.6, 110.4, 117.3 (q, J = 289.8 Hz), 118.8, 120.6, 123.4, 123.6 (q, J = 2.9 Hz), 125.5, 144.1, 147.5, 147.8, 167.0 (q, J = 34.4 Hz).
General procedure for haloform reaction:
The solution of NaOH (3 equiv) in water-ethanol mixture (0.33 M, 1:1) was added 2,2,2-trifluoroethanone (1 equiv). The resulting reaction mixture was refluxed for 12 hours and cooled to room temperature. Concentrated aqueous HCl solution (~12 M, 3 equiv) was added dropwise. The resulting precipitate is filtered off and washed with water. If no precipitation occurs, the reaction mixture was extracted with Et2O or EtOAc (3×100 mL). The combined organic layer was washed with brine, dried over MgSO4 and evaporated. If necessary, the acids can be purified using chromatography. Eluent: hexanes/EtOAc, 1:1.
5-(benzo[d][1,3]dioxol-5-yl)-1H-pyrrole-2-carboxylic acid (18)
Compound 18 was prepared according to the general procedure for haloform reaction from 15. M = 5.24 g. Yield = 90%.
1H NMR: (DMSO, 400 MHz) δ = 6.01 (s, 2 H), 6.50 (dd, J=3.5, 2.6 Hz, 1 H), 6.76 (dd, J=3.6, 2.3 Hz, 1 H), 6.90 (d, J=8.1 Hz, 1 H), 7.32 (dd, J=8.1, 1.7 Hz, 1 H), 7.45 (d, J=1.5 Hz, 1 H), 11.78 (s, 1 H), 12.27 (br. s, 1H).
13C NMR: (DMSO, 100 MHz) δ = 101.2, 105.7, 107.2, 108.6, 116.6, 119.0, 123.6, 126.0, 136.7, 146.5, 147.8, 162.0.
5-(2,2-difluorobenzo[d][1,3]dioxol-5-yl)-1H-pyrrole-2-carboxylic acid (19)
Compound 19 was prepared according to the general procedure for haloform reaction from 16. M = 389 mg. Yield = 46%.
1H NMR: (DMSO, 400 MHz) δ = 6.65 (s, 1 H), 6.79 (s, 1 H), 7.40 (d, J=8.6 Hz, 1 H), 7.69 (d, J=8.4 Hz, 1 H), 7.93 (s, 1 H), 12.02 (s, 1 H), 12.40 (br. s, 1 H).
13C NMR: (DMSO, 100 MHz) δ = 106.9, 108.3, 110.3, 116.5, 121.3, 124.7, 128.7, 131.5 (t, J = 251.9 Hz), 135.3, 141.8, 143.5, 162.0.
5-(2,2-dimethylbenzo[d][1,3]dioxol-5-yl)-1H-pyrrole-2-carboxylic acid (20)
Compound 20 was prepared according to the general procedure for haloform reaction from 17. M = 1.55 g. Yield = 76%.
1H NMR: (DMSO, 400 MHz) δ = 1.63 (s, 6 H), 6.43 – 6.50 (m, 1 H), 6.76 (dd, J=3.4, 2.3 Hz, 1 H), 6.80 (d, J=8.1 Hz, 1 H), 7.28 (dd, J=8.2, 1.3 Hz, 1 H), 7.35 (d, J=1.2 Hz, 1 H), 11.77 (s, 1 H), 12.22 (br. s, 1 H).
13C NMR: (DMSO, 100 MHz) δ = 25.6 (2C), 105.6, 107.0, 108.4, 116.5, 118.2, 118.5, 123.4, 125.4, 136.9, 146.2, 147.3, 161.9.
5-bromobenzo[c][1,2,5]thiadiazole (22)
Phenylendiamine 21 (12.46 g, 67.0 mmol) was added to SOCl2 (44 mL, 603 mmol, 9 equiv) followed by H2SO4 (1.93 mL, 36.2 mmol, 0.54 equiv). The reaction mixture was heated at reflux for 2 hours, cooled and evaporated. The residue was suspended in water (~0.5 L), and the product was extracted with DCM (3×100 mL). The combined organic layers were dried over Na2SO4 and evaporated. The residue was purified using column chromatography, eluent hexane/EtOAc, 10:1. M = 11.80 g. Yield = 82%.
1H NMR (CDCl3, 400 MHz) δ = 7.62 (dd, J=9.2, 1.7 Hz 1 H), 7.83 (d, J=9.2 Hz, 1 H), 8.18 (d, J=1.6 Hz, 1 H).
13C NMR (CDCl3, 100 MHz) δ = 122.2, 123.9, 124.6, 133.2, 153.4, 155.3.
5-(1H-pyrrol-2-yl)benzo[c][1,2,5]thiadiazole (24)
Compound 24 was prepared according to the general procedure for cross-coupling reaction and Boc cleavage from 22. The substance was triturated with heхane/EtOAc 10:1. M = 7.27 g. Yield = 72% (over two steps).
1H NMR: (DMSO, 400 MHz) δ = 6.20 (s, 1 H), 6.84 (d, J=2.4 Hz, 1 H), 7.02 (s, 1 H), 8.00 (d, J=9.2 Hz, 1 H), 8.08 (d, J=9.2 Hz, 1 H), 8.21 (s, 1 H), 11.65 (br. s, 1 H).
13C NMR: (DMSO, 100 MHz) δ = 109.2, 109.9, 111.3, 121.2, 121.8, 128.7, 129.6, 134.0, 153.1, 155.4.
1-(5-(benzo[c][1,2,5]thiadiazol-5-yl)-1H-pyrrol-2-yl)-2,2,2-trifluoroethanone (25)
Compound 25 was prepared according to the general procedure for aryl pyrrole trifluoroacetylation from 24. M = 8.20 g. Yield = 76%.
1H NMR: (DMSO, 400 MHz) δ = 7.12 (s, 1 H), 7.26 (s, 1 H), 8.05 (d, J=9.2 Hz, 1 H), 8.21 (d, J=9.2 Hz, 1 H), 8.72 (s, 1 H), 13.06 (s, 1 H).
13C NMR: (DMSO, 100 MHz) δ = 112.6, 115.5, 117.0 (q, J = 289.1 Hz), 117.6, 118.4, 121.4, 123.0, 126.9, 128.6, 130.9, 141.4, 154.0, 154.5, 167.9, 168.1 (q, J = 35.3 Hz).
5-(benzo[c][1,2,5]thiadiazol-5-yl)-1H-pyrrole-2-carboxylic acid (26)
Compound 26 was prepared according to the general procedure for haloform reaction from 25. M = 6.75 g. Yield = 95%.
1H NMR: (DMSO, 400 MHz) δ = 6.86 (s, 1 H), 6.93 (s, 1 H), 8.04 (d, J=9.3 Hz, 1 H), 8.20 (d, J=9.2 Hz, 1 H), 8.62 (s, 1 H), 12.46 (br. s., 1 H), 12.31 (s, 1 H).
13C NMR: (DMSO, 100 MHz) δ = 110.4, 114.9, 116.5, 121.2, 126.0, 128.9, 132.6, 134.7, 153.5, 155.0, 161.7.
allyl (1-hydroxy-2-methylpropan-2-yl)carbamate (28)
2-amino-2-methylpropan-1-ol 27 (46 mL, 482 mmol, 1.5 equiv) was dissolved in dichloromethane (470 mL), and water (470 mL) and sodium bicarbonate (80.6 g, 960 mmol, 3 equiv) were then added. To this vigorously stirred solution, allyl chloroformate (34 mL, 320 mmol) was added dropwise, and the mixture was stirred at room temperature for 16 hours. The organic layer was separated, and the aqueous layer was extracted with dichloromethane (100×2 mL), the extract was dried over anhydrous sodium sulfate. After filtration, the filtrate was concentrated to give the pure compound. M = 47.93 g. Yield = 87%.
1H NMR (CDCl3, 400 MHz): δ = 1.29 (s, 6H), 3.39 (br. s, 1H), 3.61 (s, 2H), 4.53 (d, J=5.4 Hz, 2H), 4.93 (br. s, 1H), 5.23 (dd, J=10.5, 1.0 Hz, 1H), 5.31 (dd, J=17.2, 1.6 Hz, 1H), 5.92 (dddd, J=16.8, 10.9, 5.7, 5.4 Hz, 1H).
13C NMR (CDCl3, 100 MHz): δ = 24.3 (2C), 54.4, 65.4, 70.1, 117.7, 132.8, 156.0.
allyl (2-methyl-1-oxopropan-2-yl)carbamate (29)
A solution of oxalyl chloride (35 mL, 0.413 mol, 1.5 equiv) in CH2Cl2 (100 mL) was added dropwise to a solution of DMSO (49 mL, 0.690 mol, 2.5 equiv) in CH2Cl2 (415 mL) at – 70 to – 80 °C. The resulting solution was stirred for 10 minutes, and a solution of alcohol 28 (47.93 g, 277 mmol) in CH2Cl2 (415 mL) was added dropwise at the same temperature. After 15 minutes Et3N (193 mL, 1.385 mol, 5.0 equiv) was added, and 5 minutes later the reaction mixture was allowed to warm to r.t. The reaction mixture was quenched with water (1 L), and the layers were separated. The organic layer was dried over Na2SO4 and evaporated. The residue was used without purification. M = 47.40 g. Yield = 100 %.
1H NMR (CDCl3, 400 MHz): δ = 1.39 (s, 6H), 4.56 (d, J=5.4 Hz, 2H), 5.23 (dd, J=10.2, 1.0 Hz, 1H), 5.31 (d, J=17.2 Hz, 1H), 5.41 (br. s, 1 H), 5.91 (dddd, J=16.8, 11.0, 5.6, 5.4 Hz, 1H), 9.44 (s, 1H).
13C NMR (CDCl3, 100 MHz): δ = 21.8 (2C), 59.5, 65.7, 118.0, 132.6, 155.2, 200.6.
(E)-allyl (1-((tert-butylsulfinyl)imino)-2-methylpropan-2-yl)carbamate (30)
To a solution of aldehyde 29 (10.00 g, 58.5 mmol) in CH2Cl2 (100 mL), R or S tert-butylsulfonamide (7.80 g, 64.4 mmol, 1.1 equiv) was added in one portion, followed by the addition of Ti(OiPr)4 (17.3 mL, 58.4 mmol, 1 equiv). The resulting solution was stirred overnight (TLC indicated consumption of the SM), and silica (50 mL) was added to the reaction mixture followed by water (3 mL, ~3 equiv). The resulted suspension was stirred for 10 minutes and evaporated. Chromatography with 10:1 hexanes/EtOAc, then pure EtOAc eluent gave an oil.
M = 10.67 g. Yield = 66% (S-isomer).
M = 10.53 g. Yield = 66% (R-isomer).
1H NMR (CDCl3, 400 MHz): δ = 1.17 (s, 9H), 1.47 (d, J=2.4 Hz, 6H), 4.45 – 4.51 (m, 2H), 5.16 (dd, J=10.5, 1.1 Hz, 1H), 5.25 (dd, J=17.2, 1.4 Hz, 1H), 5.63 (br. s, 1H), 5.79 – 5.93 (m, 1 H), 7.88 (s, 1 H).
13C NMR (CDCl3, 100 MHz): δ = 22.4 (3C), 24.7, 24.9, 56.5, 57.2, 65.4, 117.8, 132.8, 154.6, 171.3.
allyl (1-(5-(((tert-butyldimethylsilyl)oxy)methyl)-4-methylthiazol-2-yl)-1-(1,1-dimethylethylsulfinamido)-2-methylpropan-2-yl)carbamate (32)
Thiazole 31 (23.66 g 97.4 mmol, 2.5 equiv) was dissolved in THF (97 mL) and cooled to −78 °C. At this temperature, n-BuLi (2.5 M, 39 mL, 97.5 mmol, 2.5 equiv) was added dropwise under the nitrogen atmosphere. The reaction mixture was stirred for 30 min at −78 °C, and (enantiomer 2)-30 (10.67 g, 38.9 mmol) was added dropwise as a solution in THF (40 mL). The reaction mixture was slowly (∼1 h) warmed to 0 °C, and poured into water (200 mL). The biphasic mixture was extracted with CH2Cl2 (3×100 mL). The combined organic phases were dried over anhydrous Na2SO4 and evaporated. The residue was purified by column chromatography. Eluent, hexanes/EtOAc 10:1, 3:1, 1:1 then pure EtOAc. M = 12.07 g. Yield = 60%. Compounds that were prepared from S-30 were designated as A, compounds that were prepared from R-30 were designated as B. 32B was prepared using the same conditions from (enantiomer 1)-30. M = 12.11 g. Yield = 61%.
1H NMR (CDCl3, 400 MHz): δ = 0.09 (s, 6H), 0.83 – 0.95 (m, 9H), 1.23 (s, 9H), 1.32 (s, 3H), 1.42 (s, 3H), 1.54 (s, 3H), 2.33 (s, 1H), 4.47 – 4.63 (m, 2H), 4.78 (s, 2H), 5.11 (s, 1H), 5.21 (dd, J=10.3, 1.1 Hz, 1H), 5.30 (dd, J=17.2, 1.6 Hz, 1H), 5.77 (br. s, 1H), 5.85 – 5.97 (m, 1H).
allyl (1-amino-1-(5-(hydroxymethyl)-4-methylthiazol-2-yl)-2-methylpropan-2-yl)carbamate (33)
The 1 M HCl-MeOH solution was prepared by dropwise addition of AcCl (7.10 mL, 0.10 mol) to methanol (100 mL) with cooling on a water bath. The resulting solution was cooled to ambient temperature and added to the flask containing 32A (12.07 g, 23.3 mmol). After the dissolution, the reaction mixture was stirred for 1 h, evaporated, dissolved in CH2Cl2 (100 mL), and washed with 10% aqueous K2CO3 solution (100 mL). The layers were separated, and the aqueous layer was extracted with CH2Cl2 (2×100 mL). The combined organic phases were dried over anhydrous Na2SO4 and evaporated. The residue was separated by column chromatography. Eluent CH2Cl2/MeOH (50:1, 10:1). M = 5.28 g. Yield = 76%.
The second enantiomer (33B) was prepared according to the same procedure from 32B. M = 4.11 g. Yield = 59%.
1H NMR (CDCl3, 400 MHz): δ = 1.33 (d, 6 H), 2.32 (s, 3 H), 2.76 (br. s, 3 H), 4.35 (s, 1H), 4.50 (d, J=5.3 Hz, 2H), 4.70 (s, 2H), 5.18 (d, J=10.4 Hz, 1H), 5.28 (d, J=17.1 Hz, 1H), 5.72 (s, 1H), 5.90 (dddd, J=16.8, 11.0, 5.6, 5.4 Hz, 1H).
13C NMR (CDCl3, 100 MHz): δ = 15.1, 23.7, 23.9, 56.0, 56.3, 60.1, 65.2, 117.6, 131.6, 133.0, 148.4, 155.3, 169.4.
Allyl (1-(5-(((tert-butyldimethylsilyl)oxy)methyl)thiazol-2-yl)-1-(1,1-dimethylethylsulfinamido)-2-methylpropan-2-yl)carbamate (35)
Thiazole 34 (36.30 g, 158.5 mmol, 2.5 equiv) was dissolved in THF (158 mL) and cooled to −78 °C. At this temperature, n-BuLi (2.5 M, 63 mL, 157.5 mmol, 2.5 equiv) was added dropwise under the nitrogen atmosphere. The reaction mixture was stirred for 10 min at −78 °C, and (enantiomer 2)-30 (17.35 g, 63.3 mmol) was added dropwise as a solution in THF (63 mL). The reaction mixture was slowly (∼1 h) warmed to 0 °C, and poured into water (0.5 L). The biphasic mixture was extracted with CH2Cl2 (3×100 mL). The combined organic phases were dried over anhydrous Na2SO4 and evaporated. The residue was purified by column chromatography. Eluent, hexanes/EtOAc 10:1, then pure EtOAc. M = 21.45 g. Yield = 67%.
35B was prepared using the same conditions from (enantiomer 1)-30. M = 20.08 g. Yield = 66%.
35 was obtained as an inseparable mixture of diastereomers (dr~3:1).
1H NMR: (CDCl3, 400 MHz) δ = 0.07 (s, 6 H), 0.88 (s, 9 H), 1.21 (s, 6 H), 1.30 (s, 3 H), 1.40 (s, 3 H), 1.52 (s, 3 H), 4.45 – 4.59 (m, 2 H), 4.80 – 4.86 (m, 3 H), 5.10 (s, 1 H), 5.18 (dd, J=10.5, 1.0 Hz, 1 H), 5.28 (dd, J=17.2, 1.4 Hz, 1 H), 5.66 – 5.95 (m, 2 H), 7.51 (s, 1 H).
13C NMR (CDCl3, 100 MHz): δ = −5.2, 22.7, 22.8 (3C), 25.4, 25.8 (3C), 25.8, 25.9, 56.5, 56.5, 58.5, 65.6, 117.8, 132.8, 138.8, 140.3, 156.1, 170.8.
Аllyl (1-amino-1-(5-(hydroxymethyl)thiazol-2-yl)-2-methylpropan-2-yl)carbamate (36)
The 1 M HCl-MeOH solution was prepared by dropwise addition of AcCl to methanol with cooling on a water bath. The resulting solution (~200 mL) was cooled to an ambient temperature and added to the flask containing 35A (21.45 g, 42.6 mmol). After the dissolution, the reaction mixture was stirred for 1 h, evaporated, dissolved in CH2Cl2 (200 mL), and washed with 10% aqueous K2CO3 solution (200 mL). The layers were separated, and the aqueous layer was extracted with CH2Cl2 (2×100 mL). The combined organic phases were dried over anhydrous Na2SO4 and evaporated. The residue was separated by column chromatography. Eluent CH2Cl2/MeOH (50:1, 10:1). M = 7.18 g. Yield = 59%.
The second enantiomer (36B) was prepared according to the same procedure from 35B. M = 8.02 g. Yield = 71%.
1H NMR: (CDCl3, 400 MHz) δ = 1.29 (s, 6 H), 2.73 (br. s, 3 H), 4.41 (s, 1 H), 4.47 (d, J=5.0 Hz, 2 H), 4.72 (s, 2 H), 5.15 (d, J=10.5 Hz, 1 H), 5.25 (d, J=17.1 Hz, 1 H), 5.66 (s, 1 H), 5.78 – 5.93 (m, J=16.8, 11.0, 5.5, 5.5 Hz, 1 H), 7.44 (s, 1 H).
13C NMR (CDCl3, 100 MHz): δ = 23.6, 23.8, 56.1, 57.0, 59.9, 65.2, 117.6, 132.9, 139.2, 139.4, 155.2, 172.4.
General procedure for amide coupling:
DIPEA (1 equiv) was added to an appropriate acid (3, 18, 19, 20, 26; 1 equiv) followed by DMF (10 mL per 1 g of acid) and then HBTU (1 equiv). The resulting solution was stirred for 5 min and added to a solution of appropriate amine (33, 36, 37, 39-42 1 equiv) in DMF (10 mL per 1 g of amine) in several portions. The reaction mixture was stirred overnight; DMF was evaporated, and the residue was dissolved in DCM (50 mL) and successively washed with 5% aqueous NaOH and 10% tartaric acid or citric acid aqueous solutions (50 mL). The organic layer was dried over Na2SO4, filtered, evaporated, and dry loaded on silica. Eluting with hexanes/EtOAc (1:1, then pure EtOAc) gave the target compounds. The products were used in the next step without analysis.
General procedure for deprotection:
To a solution containing protected compound (5 mmol) and N,N′-dimethyl barbituric acid (NDMBA, 15 mmol, 3 equiv) in MeOH (50 mL), PPh3 (10 mol %) was added under a nitrogen atmosphere followed by Pd(dba)2 (5 mol %). The mixture was stirred for 1 day under reflux. After cooling, 50 mL of DCM was added, and the organic phase was shaken with 10% aqueous K2CO3 (50 mL) to remove the unreacted NDMBA. The organic layer was separated, and the aqueous layer was extracted with DCM/EtOH (∼4:1, (2−4) × 50 mL). The combined organic layers were dried over Na2SO4, filtered, and concentrated. Purification by flash chromatography afforded amine as a slightly brown or yellowish solid. Eluent CHCl3/MeOH saturated with NH3 (10:1). When yields are listed the first one is for compounds prepared from ‘A’ amine, the second one is for compounds prepared from ‘B’ amine.
N-(2-amino-1-(5-(hydroxymethyl)-4-methylthiazol-2-yl)ethyl)-5-(benzo[d][1,3]dioxol-5-yl)-1H-pyrrole-2-carboxamide (NBD-14110 and NBD-14111)
Compounds NBD-14110 and NBD-14111 were obtained following the general procedure for amide coupling and then the general procedure for deprotection from amine 37 and acid 18.
NBD-14110: M = 406 mg. Yield = 35% (over two steps).
rt = 1.108 min. Purity = 100%. LC–MS: m/z [M+H]+ = 401 Da.
NBD-14111: M = 212 mg. Yield = 28% (over two steps).
rt = 1.074 min. Purity = 100%. LC–MS: m/z [M+H]+ = 401 Da.
m.p. = 110–115°C.
1H NMR: (DMSO, 400 MHz) δ = 1.69 (br. s, 2 H), 2.25 (s, 3 H), 2.96 (dd, J=13.3, 8.1 Hz, 1 H), 3.08 (dd, J=13.3, 5.2 Hz, 1 H), 4.52 (s, 2 H), 5.12 (dd, J=13.2, 7.6 Hz, 1 H), 5.36 (br. s, 1 H), 6.01 (s, 2 H), 6.49 (d, J=3.5 Hz, 1 H), 6.91 (d, J=8.2 Hz, 1 H), 6.93 (d, J=3.8 Hz, 1 H), 7.29 (dd, J=8.1, 1.0 Hz, 1 H), 7.42 (s, 1 H), 8.42 (d, J=7.9 Hz, 1 H), 11.58 (br. s, 1 H).
13C NMR: (DMSO, 100 MHz) δ = 14.8, 45.7, 54.3, 55.0, 101.0, 105.4, 106.4, 108.5, 112.9, 118.4, 126.2, 126.5, 132.4, 134.9, 146.1, 146.9, 147.6, 160.4, 169.4
HRMS (ESI): m/z calcd for C19H21N4O4S [M+H]+ 401.1278, found 401.1278.
N-(2-amino-1-(5-(hydroxymethyl)-4-methylthiazol-2-yl)-2-methylpropyl)-5-(benzo[d][1,3]dioxol-5-yl)-1H-pyrrole-2-carboxamide (NBD-14117 and NBD-14116)
Compounds NBD-14117 and NBD-14116 were obtained following the general procedure for amide coupling and then the general procedure for deprotection from amine 33 and acid 18.
NBD-14117: M = 502 mg. Yield = 58% (over two steps).
rt = 1.260 min. Purity = 100%. LC–MS: m/z [M+H]+ = 429 Da.
NBD-14116: M = 494 mg. Yield = 60% (over two steps).
rt = 1.247 min. Purity = 100%. LC–MS: m/z [M+H]+ = 429 Da.
mp= 115–120°C (decomp.).
1H NMR: (DMSO-d6, 400 MHz) δ = 1.07 (s, 3 H), 1.11 (s, 3 H), 1.98 (br. s, 2 H), 2.27 (s, 3 H), 4.54 (d, J=3.4 Hz, 2 H), 5.18 (d, J=8.9 Hz, 1 H), 5.34 – 5.43 (m, 1 H), 6.02 (s, 2 H), 6.49 (d, J=3.5 Hz, 1 H), 6.92 (d, J=8.1 Hz, 1 H), 6.95 (d, J=3.5 Hz, 1 H), 7.27 (dd, J=8.1, 1.7 Hz, 1 H), 7.40 (d, J=1.6 Hz, 1 H), 8.07 (d, J=9.0 Hz, 1 H), 11.62 (br. s, 1 H).
13C NMR (DMSO-d6, 100 MHz) δ = 14.9, 27.6, 28.4, 52.6, 55.0, 58.9, 101.0, 105.4, 106.4, 108.5, 113.4, 118.4, 126.2, 126.3, 132.6, 135.0, 146.2, 146.5, 147.7, 159.9, 167.5.
HRMS (ESI): m/z calcd for C21H25N4O4S [M+H]+ 429.1591, found 429.1588.
N-(2-amino-1-(5-(hydroxymethyl)thiazol-2-yl)ethyl)-5-(benzo[d][1,3]dioxol-5-yl)-1H-pyrrole-2-carboxamide (NBD-14123 and NBD-14122)
Compounds NBD-14123 and NBD-14122 were obtained following the general procedure for amide coupling and then the general procedure for deprotection from amine 39 and acid 18.
NBD-14123: M = 337 mg. Yield = 37% (over two steps).
rt = 1.193 min. Purity = 100%. LC–MS: m/z [M+H]+ = 387 Da.
NBD-14122: M = 512 mg. Yield = 47% (over two steps).
rt = 1.136 min. Purity = 100%. LC–MS: m/z [M+H]+ = 387 Da.
m.p. = 200–205°C.
1H NMR: (DMSO, 400 MHz) δ = 1.68 (br. s, 2 H), 2.99 (dd, J=13.1, 7.8 Hz, 1 H), 3.12 (dd, J=13.2, 5.4 Hz, 1 H), 4.59 (s, 2 H), 5.18 (dd, J=13.2, 7.7 Hz, 1 H), 5.40 – 5.50 (m, 1 H), 6.01 (s, 2 H), 6.50 (d, J=3.7 Hz, 1 H), 6.91 (d, J=8.2 Hz, 1 H), 6.94 (d, J=3.7 Hz, 1 H), 7.25 – 7.33 (m, 1 H), 7.42 (s, 1 H), 7.53 (s, 1 H), 8.45 (d, J=7.9 Hz, 1 H), 11.58 (br. s, 1 H).
13C NMR: (DMSO, 100 MHz) δ = 45.6, 54.4, 55.7, 101.0, 105.4, 106.4, 108.5, 112.9, 118.4, 126.2, 126.5, 135.0, 139.0, 140.0, 146.1, 147.7, 160.5, 172.2.
HRMS (ESI): m/z calcd for C18H19N4O4S [M+H]+ 387.1122, found 387.1126.
5-(benzo[d][1,3]dioxol-5-yl)-N-(1-(5-(hydroxymethyl)thiazol-2-yl)-2-(methylamino)ethyl)-1H-pyrrole-2-carboxamide (NBD-14139 and NBD-14140)
Compounds NBD-14139 and NBD-14140 were obtained following the general procedure for amide coupling and then the general procedure for deprotection from amine 40 and acid 18.
NBD-14139: M = 385 mg. Yield = 43% (over two steps).
rt = 0.746 min. Purity = 100%. LC–MS: m/z [M+H]+ = 401 Da.
NBD-14140: M = 400 mg. Yield = 43% (over two steps).
rt = 0.941 min. Purity = 100%. LC–MS: m/z [M+H]+ = 401 Da.
m.p. = 115–120°C.
1H NMR: (DMSO, 400 MHz) δ = 1.91 (br. s, 1 H), 2.32 (s, 3 H), 2.99 (dd, J=12.3, 8.3 Hz, 1 H), 3.05 (dd, J=12.5, 5.6 Hz, 1 H), 4.59 (d, J=5.3 Hz, 2 H), 5.39 (td, J=7.9, 6.0 Hz, 1 H), 5.46 (t, J=5.6 Hz, 1 H), 6.01 (s, 2 H), 6.50 (dd, J=3.4, 2.1 Hz, 1 H), 6.89 – 6.94 (m, 2 H), 7.29 (dd, J=8.1, 1.5 Hz, 1 H), 7.42 (d, J=1.5 Hz, 1 H), 7.53 (s, 1 H), 8.47 (d, J=8.1 Hz, 1 H), 11.59 (s, 1 H).
13C NMR: (DMSO, 100 MHz) δ = 35.5, 50.3, 54.5, 55.8, 101.0, 105.4, 106.5, 108.5, 113.0, 118.4, 126.2, 126.4, 135.0, 138.9, 140.1, 146.2, 147.7, 160.3, 172.2.
HRMS (ESI): m/z calcd for C19H21N4O4S [M+H]+ 401.1278, found 401.1283.
N-(2-amino-1-(5-(hydroxymethyl)thiazol-2-yl)-2-methylpropyl)-5-(benzo[d][1,3]dioxol-5-yl)-1H-pyrrole-2-carboxamide (NBD-14119 and NBD-14118)
Compounds NBD-14119 and NBD-14118 were obtained following the general procedure for amide coupling and then the general procedure for deprotection from amine 36 and acid 18.
NBD-14119: M = 331 mg. Yield = 34% (over two steps).
rt = 1.119 min. Purity = 98%. LC–MS: m/z [M+H]+ = 415 Da.
NBD-14118: M = 475 mg. Yield = 53% (over two steps).
rt = 1.121 min. Purity = 96%. LC–MS: m/z [M+H]+ = 415 Da.
mp= 105–110°C (decomp.).
1H NMR: (DMSO-d6, 400 MHz) δ = 1.09 (s, 3 H), 1.12 (s, 3 H), 2.01 (br. s, 2 H), 4.62 (s, 2 H), 5.24 (d, J=8.8 Hz, 1 H), 5.48 (br. s, 1 H), 6.02 (s, 2 H), 6.49 (d, J=3.8 Hz, 1 H), 6.92 (d, J=8.1 Hz, 1 H), 6.95 (d, J=3.7 Hz, 1 H), 7.28 (dd, J=8.1, 1.7 Hz, 1 H), 7.40 (d, J=1.7 Hz, 1 H), 7.56 (s, 1 H), 8.10 (d, J=9.0 Hz, 1 H), 11.62 (br. s, 1 H).
13C NMR (DMSO-d6, 100 MHz) δ = 27.5, 28.5, 52.7, 55.8, 59.2, 101.0, 105.4, 106.5, 108.6, 113.5, 118.5, 126.2, 126.3, 135.1, 138.7, 140.2, 146.2, 147.7, 160.0, 170.3.
HRMS (ESI): m/z calcd for C20H23N4O4S [M+H]+ 415.1435, found 415.1439.
N-(2-amino-1-(5-(hydroxymethyl)thiazol-2-yl)ethyl)-5-(2,2-difluorobenzo[d][1,3]dioxol-5-yl)-1H-pyrrole-2-carboxamide (NBD-14145 and NBD-14144)
Compounds NBD-14145 and NBD-14144 were obtained following the general procedure for amide coupling and then the general procedure for deprotection from amine 39 and acid 19.
NBD-14145: M = 124 mg. Yield = 52% (over two steps).
rt = 1.143 min. Purity = 100%. LC–MS: m/z [M+H]+ = 423 Da.
NBD-14144: M = 290 mg. Yield = 50% (over two steps).
rt = 1.196 min. Purity = 100%. LC–MS: m/z [M+H]+ = 423 Da.
m.p. = 145–150°C.
1H NMR: (DMSO, 400 MHz) δ = 1.81 (br. s, 2 H), 2.99 (dd, J=13.1, 7.9 Hz, 1 H), 3.12 (dd, J=13.3, 5.3 Hz, 1 H), 4.59 (s, 2 H), 5.18 (dd, J=13.1, 7.6 Hz, 1 H), 5.46 (br. s., 1 H), 6.64 (d, J=3.8 Hz, 1 H), 6.99 (d, J=3.8 Hz, 1 H), 7.40 (d, J=8.4 Hz, 1 H), 7.53 (s, 1 H), 7.65 (dd, J=8.5, 1.7 Hz, 1 H), 7.90 (d, J=1.5 Hz, 1 H), 8.55 (d, J=7.8 Hz, 1 H), 11.83 (br. s, 1 H).
13C NMR: (DMSO, 100 MHz) δ = 45.5, 54.4, 55.8, 106.5, 107.7, 110.3, 112.8, 120.8, 127.4, 128.9, 131.2 (t, J = 252.5 Hz), 133.6, 139.0, 140.0, 141.4, 143.3, 160.5, 172.0.
HRMS (ESI): m/z calcd for C18H17F2N4O4S [M+H]+ 423.0933, found 423.0935.
N-(2-amino-1-(5-(hydroxymethyl)thiazol-2-yl)ethyl)-5-(2,2-dimethylbenzo[d][1,3]dioxol-5-yl)-1H-pyrrole-2-carboxamide (NBD-14133 and NBD-14134)
Compounds NBD-14133 and NBD-14134 were obtained following the general procedure for amide coupling and then the general procedure for deprotection from amine 39 and acid 20.
NBD-14133: M = 360 mg. Yield = 31% (over two steps).
rt = 1.279 min. Purity = 97%. LC–MS: m/z [M+H]+ = 415 Da.
NBD-14134: M = 426 mg. Yield = 33% (over two steps).
rt = 1.289 min. Purity = 99%. LC–MS: m/z [M+H]+ = 415 Da.
m.p. = 100–105°C (decomp.).
1H NMR: (DMSO, 400 MHz) δ = 1.64 (s, 6 H), 2.99 (dd, J=13.1, 7.8 Hz, 1 H), 3.12 (dd, J=13.2, 5.3 Hz, 1 H), 4.60 (s, 2 H), 5.18 (dd, J=13.0, 7.6 Hz, 1 H), 5.48 (br. s., 1 H), 6.47 (d, J=3.8 Hz, 1 H), 6.82 (d, J=8.1 Hz, 1 H), 6.95 (d, J=3.7 Hz, 1 H), 7.24 (dd, J=8.2, 1.6 Hz, 1 H), 7.31 (d, J=1.5 Hz, 1 H), 7.54 (s, 1 H), 8.46 (d, J=7.8 Hz, 1 H), 11.56 (br. s, 1 H). Two exchangeable protons are missing
13C NMR: (DMSO, 100 MHz) δ = 25.6 (2C), 45.7, 54.4, 55.8, 105.3, 106.3, 108.3, 113.0, 118.05, 118.08, 125.7, 126.4, 135.3, 139.0, 140.0, 145.9, 147.3, 160.5, 172.2.
HRMS (ESI): m/z calcd for C20H23N4O4S [M+H]+ 415.1435, found 415.1440.
N-(2-amino-1-(4-(hydroxymethyl)thiazol-2-yl)ethyl)-5-(benzo[d][1,3]dioxol-5-yl)-1H-pyrrole-2-carboxamide (NBD-14172 and NBD-14173)
Compounds NBD-14172 and NBD-14173 were obtained following the general procedure for amide coupling and then the general procedure for deprotection from amine 41 and acid 18.
NBD-14172: M = 323 mg. Yield = 32% (over two steps).
rt = 1.174 min. Purity = 100%. LC–MS: m/z [M+H]+ = 387 Da.
NBD-14173: M = 503 mg. Yield = 51% (over two steps).
rt = 1.136 min. Purity = 100%. LC–MS: m/z [M+H]+ = 387 Da.
m.p. = 100–105°C.
1H NMR: (DMSO-d6, 400 MHz) δ = 1.73 (br. s, 2 H), 2.98 (dd, J=13.1, 7.8 Hz, 1 H), 3.12 (dd, J=13.2, 5.3 Hz, 1 H), 4.52 (d, J=3.2 Hz, 2 H), 5.14 – 5.23 (m, 1 H), 5.29 (t, J=5.1 Hz, 1 H), 6.01 (s, 2 H), 6.50 (d, J=3.7 Hz, 1 H), 6.91 (d, J=8.2 Hz, 1 H), 6.94 (d, J=3.7 Hz, 1 H), 7.27 (s, 1 H), 7.29 (dd, J=8.2, 1.7 Hz, 1 H), 7.42 (d, J=1.6 Hz, 1 H), 8.47 (d, J=8.1 Hz, 1 H), 11.60 (br. s, 1 H).
13C NMR (DMSO-d6, 100 MHz): δ = 45.8, 54.3, 59.8, 101.0, 105.4, 106.5, 108.5, 113.0, 114.0, 118.4, 126.2, 126.5, 135.0, 146.2, 147.7, 157.6, 160.5, 172.5.
HRMS (ESI): m/z calcd for C18H19N4O4S [M+H]+ 387.1122, found 387.1121.
5-(benzo[d][1,3]dioxol-5-yl)-N-(1-(4-(hydroxymethyl)thiazol-2-yl)-2-(methylamino)ethyl)-1H-pyrrole-2-carboxamide (NBD-14159 and NBD-14158)
Compounds NBD-14159 and NBD-14158 were obtained following the general procedure for amide coupling and then the general procedure for deprotection from amine 42 and acid 18.
NBD-14159: M = 611 mg. Yield = 39% (over two steps).
rt = 1.071 min. Purity = 100%. LC–MS: m/z [M+H]+ = 401 Da.
NBD-14158: M = 769 mg. Yield = 55% (over two steps).
rt = 1.056 min. Purity = 100%. LC–MS: m/z [M+H]+ = 401 Da.
m.p. = 150–155°C.
1H NMR: (DMSO-d6, 400 MHz) δ = 1.90 (br. s, 1 H), 2.33 (s, 3 H), 3.01 (dd, J=12.5, 8.4 Hz, 1 H), 3.08 (dd, J=12.5, 5.4 Hz, 1 H), 4.54 (s, 2 H), 5.29 (br. s, 1 H), 5.43 (td, J=8.1, 5.6 Hz, 1 H), 6.01 (s, 2 H), 6.50 (d, J=3.5 Hz, 1 H), 6.91 (d, J=8.2 Hz, 1 H), 6.94 (d, J=3.5 Hz, 1 H), 7.26 – 7.31 (m, 2 H), 7.42 (d, J=1.7 Hz, 1 H), 8.48 (d, J=8.2 Hz, 1 H), 11.59 (br. s, 1 H).
13C NMR (DMSO-d6, 100 MHz) δ = 35.5, 50.3, 54.7, 59.8, 101.0, 105.4, 106.5, 108.5, 113.1, 114.1, 118.4, 126.2, 126.4, 135.1, 146.2, 147.7, 157.6, 160.3, 172.5.
HRMS (ESI): m/z calcd for C19H21N4O4S [M+H]+ 401.1278, found 401.1278.
N1-(2-amino-1-(5-(hydroxymethyl)-4-methylthiazol-2-yl)ethyl)-N2-(benzo[d][1,3]dioxol-5-yl)oxalamide (NBD-14108 and NBD-14109)
Compounds NBD-14108 and NBD-14109 were obtained following the general procedure for amide coupling and then the general procedure for deprotection from amine 37 and acid 3.
NBD-14108: M = 106 mg. Yield = 15% (over two steps).
rt = 0.950 min. Purity = 100%. LC–MS: m/z [M+H]+ = 379 Da.
NBD-14109: M = 151 mg. Yield = 21% (over two steps).
rt = 0.947 min. Purity = 100%. LC–MS: m/z [M+H]+ = 379 Da.
m.p. = 155–160°C.
1H NMR: (DMSO, 400 MHz) δ = 1.75 (br. s, 2 H), 2.24 (s, 3 H), 3.04 (d, J=6.1 Hz, 2 H), 4.52 (d, J=4.3 Hz, 2 H), 4.98 (t, J=5.8 Hz, 1 H), 5.40 (t, J=4.9 Hz, 1 H), 6.00 (s, 2 H), 6.89 (d, J=8.4 Hz, 1 H), 7.32 (d, J=8.6 Hz, 1 H), 7.45 (s, 1 H), 9.39 (br. s, 1 H), 10.64 (br. s, 1 H).
13C NMR: (DMSO, 100 MHz) δ = 14.9, 45.2, 54.9, 55.0, 101.2, 102.2, 108.0, 113.6, 131.9, 132.8, 143.9, 146.9, 147.0, 157.9, 160.4, 167.4.
HRMS (ESI): m/z calcd for C16H19N4O5S [M+H]+ 379.1071, found 379.1066.
N-(2-amino-1-(5-(hydroxymethyl)thiazol-2-yl)ethyl)-5-(benzo[c][1,2,5]thiadiazol-5-yl)-1H-pyrrole-2-carboxamide (NBD-14127 and NBD-14126)
Compounds NBD-14127 and NBD-14126 were obtained following the general procedure for amide coupling and then the general procedure for deprotection from amine 39 and acid 26.
NBD-14127: M = 419 mg. Yield = 47% (over two steps).
rt = 1.001 min. Purity = 100%. LC–MS: m/z [M+H]+ = 401 Da.
NBD-14126: M = 475 mg. Yield = 50% (over two steps).
rt = 1.134 min. Purity = 100%. LC–MS: m/z [M+H]+ = 401 Da.
m.p. = 140–145°C.
1H NMR: (DMSO, 400 MHz) δ = 1.78 (br. s, 2 H), 3.02 (dd, J=13.1, 7.8 Hz, 1 H), 3.15 (dd, J=13.2, 5.3 Hz, 1 H), 4.60 (s, 2 H), 5.22 (dd, J=12.6, 7.0 Hz, 1 H), 5.46 (br. s, 1 H), 6.94 (d, J=3.8 Hz, 1 H), 7.07 (d, J=3.8 Hz, 1 H), 7.55 (s, 1 H), 8.05 (d, J=9.2 Hz, 1 H), 8.19 (d, J=9.3 Hz, 1 H), 8.57 (s, 1 H), 8.64 (d, J=7.6 Hz, 1 H), 12.02 (br. s, 1 H).
13C NMR: (DMSO, 100 MHz) δ = 45.6, 54.5, 55.8, 110.0, 113.1, 114.2, 121.2, 128.9, 132.9, 133.2, 139.0, 140.1, 153.5, 155.1, 160.3, 171.9.
HRMS (ESI): m/z calcd for C17H17N6O2S2 [M+H]+ 401.0849, found 401.0852.
Cells and viruses
MT-2 cells [16], TZM-bl cells [17] and U87-CD4+-CXCR5+ cells and U87-CD4+-CCR5+ cells [18] were obtained through the NIH ARP. HEK 293T cells were purchased from ATCC. CD4-negative Cf2Th-CCR5+ cells and Env expression vector pSVIIIenv-ADA were kindly provided by Dr. J. G. Sodroski [19]. The human PBMC (Peripheral blood mononuclear cells) were isolated from buffy coats of healthy HIV-1 negative donor obtained from the New York Blood Center (New York, NY) and grown in RPMI 1640 medium supplemented with fetal bovine serum (FBS) penicillin and streptomycin. The PBMC were stimulated with 5 mg/mL phytohemagglutinin (PHA) and 20 U/mL interleukin 2 (IL-2). HIV-1 Env molecular clone expression vector pHXB2-env (X4) DNA was obtained through the NIH ARP [20]. HIV-1 Env molecular clones of gp160 genes for HIV-1 Env pseudovirus production were obtained as follows: clones representing the standard panels A, A1/D, A2/D, C (QB099.391M.Env.C8) and D were obtained through the NIH ARP from Dr. J. Overbaugh [21]. The HIV-1 Env molecular clones of subtype A/G were obtained through the NIH ARP, from Drs. D. Ellenberger, B. Li, M. Callahan and S. Butera [22]. The HIV-1 Env panel of standard reference subtype B Env clones were obtained through the NIH ARP from Drs. D. Montefiori, F. Gao and M. Li (PVO, clone 4 (SVPB11), TRO, clone 11 (SVPB12) and QH0692, clone 42 (SVPB6)); from Drs. B. H. Hahn and J. F. Salazar-Gonzalez (pREJO4541, clone 67 (SVPB16) and pRHPA4259, clone 7 (SVPB14)); from Drs. B. H. Hahn and D. L. Kothe (pTHRO4156 clone 18 (SVPB15)) [23]. The subtype B clones pWEAUd15.410.5017, p1054.TC4.1499 and p9021_14.B2.4571 were obtained through the NIH ARP from Drs. B. H. Hahn, B. F. Keele and G. M. Shaw [24]. The subtype C HIV-1 reference panel of Env clones were also obtained through the NIH ARP from Drs. D. Montefiori, F. Gao, C. Williamson and S. A. Karim (Du422.1, clone 1); from Drs. E. Hunter and C. Derdeyn (ZM109F.PB4); from Drs. L. Morris, K. Mlisana, and D. Montefiori, (CAP210.2.00.E8) [25]. The HIV-1 Subtype C Panel of Indian gp160 Env Clones HIV-00836–2 clone 5, HIV-16936–2 clone 21, HIV-25711–2 clone 4 and HIV-225925–2 clone 22 were obtained through the NIH ARP from Drs. R. Paranjape, S. Kulkarni and D. Montefiori [22]. The HIV-1 Env “Panel of Paired Infant and Maternal Env Molecular Clones” of subtype A and C/D, were obtained through the NIH ARP from Dr. J. Overbaugh [14]. The Env pseudotyped genes of BG505.T332N, KNH1144, and B41 were kindly provided by Dr. J. P. Moore of the Weil Cornell Medical College, NY.
The Env-deleted proviral backbone plasmids pSG3Δenv DNA (from Drs. J. C. Kappes and X. Wu) [17, 23b] and pNL4–3.Luc.R-.E- DNA (from Dr. N. Landau) [26] were obtained through the NIH ARP. MLV gag-pol-expressing vector pVPack-GP, Env-expressing vector pVPack-VSV-G and a pFB-luc vector were obtained from Stratagene (La Jolla, CA).
HIV-1IIIB laboratory-adapted strain was obtained through the NIH ARP.
Pseudovirus preparation
Pseudoviruses capable of single cycle infection were prepared as previously described [27]. Briefly, 5 × 106 HEK293T cells were transfected with a solution containing the same amounts of an HIV-1 Env-deleted pro-viral backbone plasmid pSG3Δenv DNA or pNL4–3.Luc.R-.E- DNA and an HIV-1 Env-expression plasmid with FuGENE HD (Promega). VSV-G pseudovirus was prepared by transfecting the HEK 293T cells with a combination of the Env-expressing plasmid pVPack-VSV-G, the MLV gag-pol-expressing plasmid pVPack-GP, the pFB-luc plasmid and FuGENE HD. Pseudovirus-containing supernatants were collected two days after transfection, filtered, tittered and stored.
Measurement of antiviral activity
Single-cycle infection assay in TZM-bl cells.
The new generation of polycyclic NBD compounds was evaluated in single-cycle infection assay for their anti-HIV-1 activity by infecting TZM-bl cells with an HIV-1 pseudovirus expressing the Env from the lab-adapted HIV-1HXB-2 (X4). Also, NBD-14113, NBD-14110, NBD-14123, and NBD-14159 were tested against a large number of HIV-1 pseudotyped viruses expressing the Env from a panel of diverse clinical isolates as previously described [27]. To this end, TZM-bl cells were platted at 1×104 / well in a 96-well tissue culture plate and cultured. Following overnight incubation, aliquots of HIV-1 pseudoviruses were pre-treated with graded concentrations of the small molecules for 30 min and added to the cells. Following 3 days of incubation, the cells were washed and lysed. 20 µl of the lysates were transferred to a white plate and mixed with the luciferase assay reagent (Promega). The luciferase activity was immediately measured with a Tecan infinite M1000 reader, and the percent inhibition by the compounds and IC50 (the half maximal inhibitory concentration) values were calculated by the GraphPad Prism software.
Multi-cycle infection assay in MT-2 cells.
The antiviral activity of the small polycyclic molecules was also evaluated against the full-length laboratory-adapted HIV-1IIIB as previously described [28]. In short, aliquots of HIV-1IIIB at 100 TCID50 were pre-incubated with an equal volume of graded concentrations of compounds for 30 min then added to the MT-2 cells at 1×104/well in a 96-well tissue culture plate and incubated overnight. The culture supernatants were then replaced with fresh media and cultured for 4 days. Finally, the supernatants were collected and mixed with an equal volume of 5% Triton X-100 and tested for p24 antigen by sandwich-ELISA. The percent inhibition of p24 production and IC50 values were calculated by the GraphPad Prism software.
Multi-cycle infection assay in PBMC.
The inhibitory activity of the NBD compounds on infection of PBMC by 3 HIV-1 isolates was determined as previously described [15]. The PHA-stimulated PBMC (5 × 104 cells/well) were infected with lab-adapted (HIV-1LAI and HIV-1BaL) and primary (HIV-197USSN54 [29]) HIV-1 isolates at 500 TCID50 in the absence or presence of graded concentrations of compounds. 100 µl of culture media were replaced every 3 days with fresh media. The supernatants were collected 7 days post-infection and tested for p24 antigen by ELISA. The percent inhibition of p24 production and IC50 values were calculated by using the GraphPad Prism software.
Single-cycle infection assay in U87-CD4-CCR5 cells.
The activity of the small polycyclic molecules was also tested against control pseudovirus VSV-G, obtained as described above. Briefly, U87-CD4-CCR5 cells were platted in a 96-well tissue culture plate at 1 × 104 / well and cultured overnight. The following day, aliquots of VSV-G pseudovirus pre-treated with graded concentrations of the polycyclic compounds for 30 min, were added to the cells and incubated for 3 days. Cells were washed and lysed with 40 µl of lysis buffer. The lysates were then transferred to a white plate and mixed with the luciferase assay reagent. The luciferase activity was immediately measured to calculate the percent of inhibition and IC50 values by using the GraphPad Prism software.
Evaluation of cytotoxicity
TZM-bl, U87-CD4-CCR5 and U87-CD4-CXCR4 cells.
The cytotoxicity of the small polycyclic molecules in these cells was determined by using the colorimetric method CellTiter 96® AQueous One Solution Cell Proliferation Assay (MTS) (Promega) following the manufacturer’s instructions. Briefly, the cells were platted in a 96-well tissue culture plate at 1 × 104 / well and cultured at 37 °C. Following overnight incubation, the cells were incubated with 100 µl of the compounds at graded concentrations and cultured for 3 days. The MTS reagent was added to the cells and incubated for 4 h at 37 ºC. The absorbance was recorded at 490 nm. The percent of cytotoxicity and the CC50 (the concentration for 50 % cytotoxicity) values were calculated as above.
MT-2 cells.
The cytotoxicity of the NBD small molecules was measured in MT-2 cells with the colorimetric method CellTiter 96® AQueous One Solution Cell Proliferation Assay (MTS). Briefly, 100 µl of a small molecule at graded concentrations was added to an equal volume of cells (105 cells / ml) in 96-well plates. The following day, the culture supernatants were replaced with fresh media and incubated for 4 days. The MTS reagent was added to the cells and incubated for 4 h at 37 ºC. The absorbance was recorded and the percent of cytotoxicity and the CC50 values were calculated as above.
Uninfected PBMC cells.
The toxicity of the small molecules in freshly isolated and stimulated PBMC cells was measured with the colorimetric method CellTiter 96® AQueous One Solution Cell Proliferation Assay. Briefly, 100 µl of escalating concentrations of compounds was added to an equal volume of cells (5 × 105 / ml) in a 96-well plate and incubated overnight. 100 µl of culture media were replaced every 3 days with fresh media and incubated for a total of 7 days. The MTS reagent was added to the cells and incubated for 4 h at 37 ºC. The absorbance was recorded and the percent of cytotoxicity and the CC50 values were calculated as above.
Cf2Th-CCR5 cells.
The Cf2Th-CCR5 cells were platted in a 96-well tissue culture plate at 6 × 103 / well and cultured at 37 °C. Following overnight incubation, the cells were incubated with 100 µl of the compounds at graded concentrations and cultured for 48 h. As described above, the MTS reagent was added to the cells and incubated for 4 h at 37 ºC. The absorbance was recorded and the percent of cytotoxicity and the CC50 values were calculated as above.
Single-cycle infection assay in U87-CD4-CXCR5 cells.
The activity of the small polycyclic molecules was also tested against control pseudovirus VSV-G, obtained as described above. Briefly, U87-CD4-CXCR5 cells were platted in a 96-well tissue culture plate at 1 × 104 / well and cultured overnight. The following day, aliquots of VSV-G pseudovirus pre-treated with graded concentrations of the polycyclic compounds for 30 min, were added to the cells and incubated for 3 days. Cells were washed and lysed with 40 µl of lysis buffer. The lysates were then transferred to a white plate and mixed with the luciferase assay reagent. The luciferase activity was immediately measured to calculate the percent of inhibition and IC50 values by using the GraphPad Prism software.
Evaluation of cytotoxicity
TZM-bl cells.
The cytotoxicity of the small polycyclic molecules in TZM-bl cells was determined by using the colorimetric method CellTiter 96® AQueous One Solution Cell Proliferation Assay (MTS) (Promega) following the manufacturer’s instructions. Briefly, TZM-bl cells were platted in a 96-well tissue culture plate at 1×104 / well and cultured at 37 °C. Following overnight incubation, the cells were incubated with 100 µl of the compounds at graded concentrations and cultured for 3 days. The MTS reagent was added to the cells and incubated for 4 h at 37 ºC. The absorbance was recorded at 490 nm. The percent of cytotoxicity and the CC50 (the concentration for 50 % cytotoxicity) values were calculated as above.
MT-2 cells.
The cytotoxicity of the NBD small molecules was measured in MT-2 cells with the colorimetric method CellTiter 96® AQueous One Solution Cell Proliferation Assay (MTS). Briefly, 100 µl of a small molecule at graded concentrations was added to an equal volume of cells (105 cells/ml) in 96-well plates. The following day, the culture supernatants were replaced with fresh media and incubated for 4 days. Four hours after the addition of MTS reagent the soluble intracellular formazan was quantitated and the percent of cytotoxicity and the CC50 values were calculated as above.
Assay in Cf2Th-CCR5 cells
CD4-negative Cf2Th-CCR5 cells were infected with the luciferase-expressing recombinant CD4-dependent pseudovirus HIV-1ADA as previously described[4]. Briefly, the Cf2Th-CCR5 cells were plated at 6×103 cells/well in a 96-well tissue culture plate. Following overnight incubation, aliquots of the pseudovirus HIV-1ADA were pre-treated with graded concentrations of the small polycyclic molecules for 30 min then, added to the cells and cultured for 48 hours. Cells were washed with PBS and lysed with 40 µl of cell lysis reagent. Lysates were transferred to a white 96-well plate and mixed with 100 µl of luciferase assay reagent. The luciferase activity was immediately measured to obtain the relative infection concerning the untreated control. The Relative virus infectivity indicates the amount of infection detected in the presence of the compounds divided by the amount of infection detected in the absence of the compounds.
Quantitative Determination of HIV-1 Reverse Transcriptase activity.
The polycyclic NBD compounds were also evaluated for their activity against the HIV-1 Reverse Transcriptase (RT) by using the Colorimetric Reverse Transcriptase Assay (Roche) and following the manufacturer’s instructions. NBD-556 and Nevirapine (non-nucleoside reverse transcriptase inhibitor (NNRTI)) were used as controls.
ENV-mutated pseudovirus assay
We introduced the amino acid substitutions S375Y and S375W into the pHXB2-Env expression vector by site-directed mutagenesis (Stratagene) using mutagenic oligonucleotides as previously described[7c]. The ENV plasmids carrying the above amino acid substitutions were sequenced and analyzed with the Geneious R8 software (Biomatters, New Zealand). The pseudoviruses were obtained by using the Env-deleted proviral backbone plasmids pNL4–3.Luc.R-.E- DNA as described above. U87-CD4-CXCR4 cells were infected with the ENV-mutated pseudoviruses pretreated for 30 min with escalating concentrations of the NBD compounds and incubated for 3 days. Cells were washed with PBS and lysed with 40 ml of cell culture lysis reagent. Lysates were transferred to a white 96-well plate and mixed with 100 ml of luciferase assay reagent. We immediately measured the luciferase activity to calculate the IC50 as described above.
Supplementary Material
Table 1.
Anti-HIV-1 activity (IC50) and cytotoxicity (CC50) of NBD compounds in single-cycle (TZM-bl cells) and multi-cycle (MT-2 cells) assays
| Code (enantiomer) | Structure | TZMb-l | MT−2 | ||||
|---|---|---|---|---|---|---|---|
| IC50 (µM) | CC50 (µM) | SI | IC50 (µM) | CC50 (µM) | SI | ||
| NBD-14113 (enantiomer 1) |  |
3.3±0.3a | 84.1±1.3 | 25.5 | 4.8±0.8a | 96.8±1.8 | 20.2 |
| NBD-14110 (enantiomer 2) | 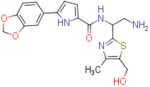 |
2.3±0.1 | 145.6±7.6 | 63.3 | 3.8±1.4 | 193±6.3 | 50.8 |
| NBD-14111 (enantiomer 1) | 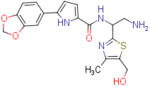 |
7±0.4 | 145.8±5.2 | 20.8 | 9.8±4.8 | 170±9 | 17.3 |
| NBD-14116 (enantiomer 1) | 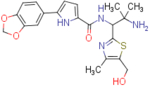 |
5.7±0.4 | 55.2±5.4 | 9.7 | 2.3±0.3 | 98±2.2 | 42.6 |
| NBD-14117 (enantiomer 2) | 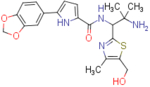 |
2.9±0.7 | 66.9±1.4 | 23.1 | 2.5±1 | 98±1.5 | 39.2 |
| NBD-14122 (enantiomer 1) | 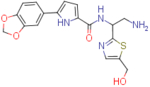 |
6.8±0.7 | 154.4±1.5 | 22.7 | 5.1±0.4 | 182.8±3 | 35.8 |
| NBD-14123 (enantiomer 2) | 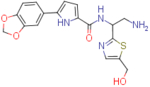 |
4.3±0.8 | 142.3±2.6 | 33.1 | 4.5±0.9 | 191.5±2.6 | 42.5 |
| NBD-14139 (enantiomer 2) | 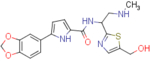 |
4.6±0.6 | 149±11.3 | 32.4 | 7.5±1.7 | 178.1±8 | 23.7 |
| NBD-14140 (enantiomer 1) | 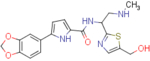 |
3.7±1.3 | 153±11.5 | 41.3 | 9.32±2.8 | 173.2±8 | 18.5 |
| NBD-14118 (enantiomer 1) | 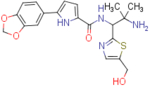 |
5.4±0.2 | 64.3±3.7 | 11.9 | 4.3±2 | 123±10.5 | 28.6 |
| NBD-14119 (enantiomer 2) | 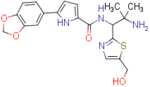 |
1.9±0.4 | 69.2±3.8 | 36.4 | 2.8±0.9 | 106±4.4 | 37.8 |
| NBD-14144 (enantiomer 1) | 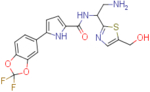 |
1.5±0.1 | 37.9±2.4 | 25.2 | 2.7±0.2 | 43.4±3.6 | 16.1 |
| NBD-14145 (enantiomer 2) | 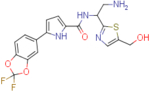 |
2.5±0.2 | 38.7±1.3 | 15.5 | 3.1±0.5 | 41±2.7 | 13.2 |
| NBD-14133 (enantiomer 2) | >24 | 64±1.2 | - | 16.2±0.4 | 96.5±1.2 | 5.9 | |
| NBD-14134 (enantiomer 1) | >24 | 61.1±1.4 | - | 15.3±5 | 89.3±1.3 | 5.8 | |
| NBD-14172 (enantiomer 2) | 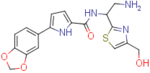 |
2.8±1.3 | 92.3±2.9 | 32.9 | 2.4±0.3 | 156.1±14.3 | 65.0 |
| NBD-14173 (enantiomer 1) | 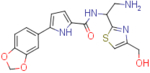 |
8.4±2 | 105.2±7.9 | 12.5 | 8.2±2.7 | 176.1±4.3 | 21.5 |
| NBD-14158 (enantiomer 1) | 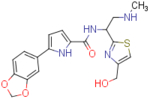 |
8.1±1.1 | 157.3±7.8 | 19.4 | 9.5±0.5 | 192.4±4.2 | 20.2 |
| NBD-14159 (enantiomer 2) | 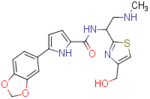 |
2.7±0.3 | 95.8±1.4 | 35.5 | 2.98±0.1 | 180±2 | 60.4 |
| NBD-14108 (enantiomer 2) | 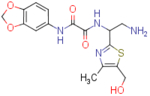 |
>53 | ≥750 | - | >79 | ≥650 | - |
| NBD-14109 (enantiomer 1) | 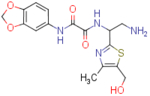 |
>53 | ≥750 | - | >79 | ≥650 | - |
| NBD-14126 (enantiomer 1) | 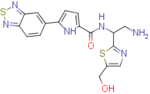 |
2.4±0.5 | 79.9±1 | 33.3 | 4±0.7 | 117.4±4.3 | 29.3 |
| NBD-14127 (enantiomer 2) | 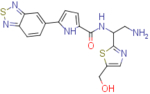 |
2.7±0.3 | 96.7±2.9 | 35.8 | 7.3±1.5 | 137±1.5 | 18.8 |
Data previously reported [13]; the reported IC50 and CC50 values represent the means ± standard deviation (SD), n=3.
Table 5.
Antiviral Activity of NBD Compounds against HIV-1 Reverse Transcriptase
| Inhibitors | IC50 (μM)a |
|---|---|
| NBD-556 | >300 |
| NBD-14110 | 30.6±2.7 |
| NBD-14113 | 45.9±3.6 |
| NBD-14123 | 39.5±2.8 |
| NBD-14159 | 28.1±7.9 |
| Nevirapine | 0.2 |
The reported IC50 values represent the means ± standard deviations (n = 3).
Acknowledgments
This study was supported by funds from NIH Grant R01 AI104416 (AKD), the New York Blood Center (AKD.
References:
- [1] a ).Zhao Q, Ma L, Jiang S, Lu H, Liu S, He Y, Strick N, Neamati N, Debnath AK, Virology 2005, 339, 213–225; [DOI] [PubMed] [Google Scholar]; b ) Schon A, Madani N, Klein JC, Hubicki A, Ng D, Yang X, Smith AB III, Sodroski J, Freire E, Biochemistry 2006, 45, 10973–10980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Curreli F, Choudhury S, Pyatkin I, Zagorodnikov VP, Bulay AK, Altieri A, Kwon YD, Kwong PD, Debnath AK, J. Med. Chem 2012, 55, 4764–4775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Lalonde JM, Elban MA, Courter JR, Sugawara A, Soeta T, Madani N, Princiotto AM, Kwon YD, Kwong PD, Schon A, Freire E, Sodroski J, Smith AB III, Bioorg. Med. Chem 2011, 19, 91–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Curreli F, Kwon YD, Belov DS, Ramesh RR, Kurkin AV, Altieri A, Kwong PD, Debnath AK, J Med Chem 2017, 60, 3124–3153. [DOI] [PubMed] [Google Scholar]
- [5].Mederski WWKR, Osswald M, Dorsch D, Anzali S, Christadler M, Schmitges C-J, Wilm C, Bioorganic & Medicinal Chemistry Letters 1998, 8, 17–22. [DOI] [PubMed] [Google Scholar]
- [6].Yamada Y, Ochiai C, Yoshimura K, Tanaka T, Ohashi N, Narumi T, Nomura W, Harada S, Matsushita S, Tamamura H, Bioorg. Med. Chem. Lett 2010, 20, 354–358. [DOI] [PubMed] [Google Scholar]
- [7] a ).Ohashi N, Harada S, Mizuguchi T, Irahara Y, Yamada Y, Kotani M, Nomura W, Matsushita S, Yoshimura K, Tamamura H, ChemMedChem 2016, 11, 940–946; [DOI] [PubMed] [Google Scholar]; b ) Curreli F, Kwon YD, Zhang H, Scacalossi D, Belov DS, Tikhonov AA, Andreev IA, Altieri A, Kurkin AV, Kwong PD, Debnath AK, J. Med. Chem 2015, 58, 6909–6927; [DOI] [PMC free article] [PubMed] [Google Scholar]; c ) F. B. Curreli DS; Kwon YD; Ramesh R; Furimsky AM; O’Loughlin K; Byrge PC; Iyer LV; Mirsalis JC; Kurkin AV; Altieri A Debnath AK, Eur J Med Chem 2018. 154, 367–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Mizuguchi T, Harada S, Miura T, Ohashi N, Narumi T, Mori H, Irahara Y, Yamada Y, Nomura W, Matsushita S, Yoshimura K, Tamamura H, Bioorg Med Chem Lett 2016, 26, 397–400. [DOI] [PubMed] [Google Scholar]
- [9].Belov DS, Ivanov VN, Curreli F, Kurkin AV, Altieri A, Debnath AK, Synthesis 2017, 49, 3692–3699. [Google Scholar]
- [10].Ivanov AV, Svinareva PA, Tomilova LG, Zefirov NS, Russian Chemical Bulletin 2001, 50, 919–920. [Google Scholar]
- [11].Curreli F, Belov DS, Ramesh RR, Patel N, Altieri A, Kurkin AV, Debnath AK, Bioorg. Med. Chem 2016, 24, 5988–6003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Belov DS, Curreli F, Kurkin AV, Altieri A, Debnath AK ChemistrySelect 2018, 3, 6450–6453. [Google Scholar]
- [13].Curreli F, Kwon YD, Belov DS, Ramesh RR, Kurkin AV, Altieri A, Kwong PD, Debnath AK, J Med Chem 2017. [DOI] [PubMed] [Google Scholar]
- [14].Wu X, Parast AB, Richardson BA, Nduati R, John-Stewart G, Mbori-Ngacha D, Rainwater SM, Overbaugh J, J.Virol 2006, 80, 835–844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Curreli F, Kwon YD, Zhang H, Scacalossi D, Belov DS, Tikhonov AA, Andreev IA, Altieri A, Kurkin AV, Kwong PD, Debnath AK, J Med Chem 2015, 58, 6909–6927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Haertle T, Carrera CJ, Wasson DB, Sowers LC, Richman DD, Carson DA, J.Biol.Chem 1988, 263, 5870–5875. [PubMed] [Google Scholar]
- [17].Wei X, Decker JM, Liu H, Zhang Z, Arani RB, Kilby JM, Saag MS, Wu X, Shaw GM, Kappes JC, Antimicrob.Agents Chemother. 2002, 46, 1896–1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Bjorndal A, Deng H, Jansson M, Fiore JR, Colognesi C, Karlsson A, Albert J, Scarlatti G, Littman DR, Fenyo EM, J.Virol 1997, 71, 7478–7487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Kolchinsky P, Mirzabekov T, Farzan M, Kiprilov E, Cayabyab M, Mooney LJ, Choe H, Sodroski J, J.Virol 1999, 73, 8120–8126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Page KA, Landau NR, Littman DR, J.Virol 1990, 64, 5270–5276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21] a ).Blish CA, Jalalian-Lechak Z, Rainwater S, Nguyen MA, Dogan OC, Overbaugh J, J.Virol 2009, 83, 7783–7788; [DOI] [PMC free article] [PubMed] [Google Scholar]; b ) Long EM, Rainwater SM, Lavreys L, Mandaliya K, Overbaugh J, AIDS RH 2002, 18, 567–576. [DOI] [PubMed] [Google Scholar]
- [22].Kulkarni SS, Lapedes A, Tang H, Gnanakaran S, Daniels MG, Zhang M, Bhattacharya T, Li M, Polonis VR, McCutchan FE, Morris L, Ellenberger D, Butera ST, Bollinger RC, Korber BT, Paranjape RS, Montefiori DC, Virology 2009, 385, 505–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23] a ).Li M, Gao F, Mascola JR, Stamatatos L, Polonis VR, Koutsoukos M, Voss G, Goepfert P, Gilbert P, Greene KM, Bilska M, Kothe DL, Salazar-Gonzalez JF, Wei X, Decker JM, Hahn BH, Montefiori DC, J.Virol 2005, 79, 10108–10125; [DOI] [PMC free article] [PubMed] [Google Scholar]; b ) Wei X, Decker JM, Wang S, Hui H, Kappes JC, Wu X, Salazar-Gonzalez JF, Salazar MG, Kilby JM, Saag MS, Komarova NL, Nowak MA, Hahn BH, Kwong PD, Shaw GM, Nature 2003, 422, 307–312. [DOI] [PubMed] [Google Scholar]
- [24].Keele BF, Giorgi EE, Salazar-Gonzalez JF, Decker JM, Pham KT, Salazar MG, Sun C, Grayson T, Wang S, Li H, Wei X, Jiang C, Kirchherr JL, Gao F, Anderson JA, Ping LH, Swanstrom R, Tomaras GD, Blattner WA, Goepfert PA, Kilby JM, Saag MS, Delwart EL, Busch MP, Cohen MS, Montefiori DC, Haynes BF, Gaschen B, Athreya GS, Lee HY, Wood N, Seoighe C, Perelson AS, Bhattacharya T, Korber BT, Hahn BH, Shaw GM, Proc.Natl.Acad.Sci.U.S.A 2008, 105, 7552–7557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25] a ).Derdeyn CA, Decker JM, Bibollet-Ruche F, Mokili JL, Muldoon M, Denham SA, Heil ML, Kasolo F, Musonda R, Hahn BH, Shaw GM, Korber BT, Allen S, Hunter E, Science 2004, 303, 2019–2022; [DOI] [PubMed] [Google Scholar]; b ) Li M, Salazar-Gonzalez JF, Derdeyn CA, Morris L, Williamson C, Robinson JE, Decker JM, Li Y, Salazar MG, Polonis VR, Mlisana K, Karim SA, Hong K, Greene KM, Bilska M, Zhou J, Allen S, Chomba E, Mulenga J, Vwalika C, Gao F, Zhang M, Korber BT, Hunter E, Hahn BH, Montefiori DC, J.Virol 2006, 80, 11776–11790; [DOI] [PMC free article] [PubMed] [Google Scholar]; c ) Williamson C, Morris L, Maughan MF, Ping LH, Dryga SA, Thomas R, Reap EA, Cilliers T, van HJ, Pascual A, Ramjee G, Gray G, Johnston R, Karim SA, Swanstrom R, AIDS RH 2003, 19, 133–144. [DOI] [PubMed] [Google Scholar]
- [26] a ).Connor RI, Chen BK, Choe S, Landau NR, Virol. 1995, 206, 935–944; [DOI] [PubMed] [Google Scholar]; b ) He J, Choe S, Walker R, di MP, Morgan DO, Landau NR, J.Virol 1995, 69, 6705–6711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27] a ).Curreli F, Belov DS, Ramesh RR, Patel N, Altieri A, Kurkin AV, Debnath AK, Bioorg.Med.Chem 2016; [DOI] [PMC free article] [PubMed] [Google Scholar]; b ) Curreli F, Kwon YD, Zhang H, Yang Y, Scacalossi D, Kwong PD, Debnath AK, Antimicrob.Agents Chemother 2014, 58, 5478–5491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Zhang H, Zhao Q, Bhattacharya S, Waheed AA, Tong X, Hong A, Heck S, Curreli F, Goger M, Cowburn D, Freed EO, Debnath AK, J.Mol.Biol 2008, 378, 565–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Sullivan PS, Do AN, Ellenberger D, Pau CP, Paul S, Robbins K, Kalish M, Storck C, Schable CA, Wise H, Tetteh C, Jones JL, McFarland J, Yang C, Lal RB, Ward JW, J Infect Dis 2000, 181, 463–469. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.