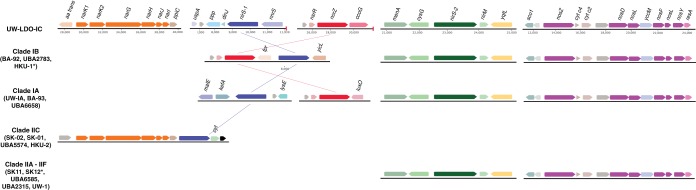
FIG 3.
Schematic illustration of denitrifying gene loci in different genomes of “Ca. Accumulibacter phosphatis.” Changes in the positions of homologous genes are indicated by lines; the arrows show the direction of transcription. Genes are drawn to scale. Proteins with unknown function are colored gray and transposases black. The end of each scaffold is indicated with a red line. aa trans, amino acid carrier family; nar, nitrate reductase; ppiC, peptidyl-prolyl cis-trans isomerase C; uspA, universal stress protein A; ppp, protein phosphatase; dinJ, DNA-damage-inducible protein J; nir, nitrite reductase; nsrR, nitric oxide-sensitive repressor; nor, nitric oxide reductase; ccoG, type cbb3 cytochrome oxidase biogenesis protein; menA, 1,4-dihydroxy-2-naphthoate octaprenyltransferase; cysG, siroheme synthase; yqfl, ATP/GTP-binding protein; sco1, cytochrome c oxidase biogenesis protein; nos, nitrous oxide reductase; cyt, cytochrome; resA, cytochrome c-type biogenesis protein; fpr, ferredoxin-NADP reductase; yicL, carboxylate/amino acid/amine transporter; malE, carboxylate/amino acid/amine transporter; kefA, potassium efflux system protein; lysE, l-lysine exporter family protein; luxO, two-component signal response regulator. *, genome lacking one of the scaffolds.