The necrotrophic pathogen Botrytis cinerea has a highly polygenic basis of virulence across genetically variable tomato, which should be considered when breeding for pathogen resistance.
Abstract
Although the impacts of crop domestication on specialist pathogens are well known, less is known about the interaction of crop variation and generalist pathogens. To study how genetic variation within a crop affects plant resistance to generalist pathogens, we infected a collection of wild and domesticated tomato accessions with a genetically diverse population of the generalist pathogen Botrytis cinerea. We quantified variation in lesion size of 97 B. cinerea genotypes (isolates) on six domesticated tomato genotypes (Solanum lycopersicum) and six wild tomato genotypes (Solanum pimpinellifolium). Lesion size was significantly affected by large effects of the host and pathogen’s genotype, with a much smaller contribution of domestication. This pathogen collection also enables genome-wide association mapping of B. cinerea. Genome-wide association mapping of the pathogen showed that virulence is highly polygenic and involves a diversity of mechanisms. Breeding against this pathogen would likely require the use of diverse isolates to capture all possible mechanisms. Critically, we identified a subset of B. cinerea genes where allelic variation was linked to altered virulence against wild versus domesticated tomato, as well as loci that could handle both groups. This generalist pathogen already has a large collection of allelic variation that must be considered when designing a breeding program.
INTRODUCTION
Plant disease is mediated by complex interactions among diverse host and pathogen molecular pathways. The disease outcome is the sum of host plant susceptibility/resistance and pathogen virulence/sensitivity mechanisms. The specific outcome of any interaction is highly dependent on the genetic variation within these pathways in both the host and pathogen. Over time, mutation and selection have led to distinct genetic architectures in the host and pathogen that are at least partly influenced by the host range of the pathogen. Specialist pathogens are a major focus in plant pathology. These pathogens are virulent on a narrow range of hosts, often a single plant species or genus. Most known plant genes for resistance to specialist pathogens confer qualitative resistance through innate immunity via large-effect loci that enable the recognition of the pathogen (Dangl and Jones, 2001; Jones and Dangl, 2006; Dodds and Rathjen, 2010; Pieterse et al., 2012). These recognition signals can be conserved pathogen patterns such as cell-wall polymers or flagellin, or alternatively, specific virulence factors that block perception of the pathogen, and in turn are detected by plant proteins that guard the signaling networks (Jones and Dangl, 2006; Bittel and Robatzek, 2007; Ferrari et al., 2007; Boller and He, 2009; Dodds and Rathjen, 2010). The evolution of large-effect qualitative loci has partly been driven by the narrow host range for the pathogen, which enhances co-evolution between host resistance genes and pathogen virulence mechanisms.
In contrast to specialist pathogens, generalist pathogens are virulent across a wide range of plant host species. Generalist pathogens potentially have less stringent co-evolution to specific hosts and their accompanying resistance mechanisms, because these pathogens can easily shift to new hosts in the environment. This allows generalist pathogens to evade the rapid evolution of new resistance mechanisms within specific hosts until they evolve to counter this new resistance. This niche-shifting ability may partially explain the observation that most natural resistance to generalist pathogens is highly polygenic and that the underlying plant genes for resistance are quantitative (Glazebrook, 2005; Nomura et al., 2005; Goss and Bergelson, 2006; Rowe and Kliebenstein, 2008; Barrett et al., 2009; Corwin et al., 2016a). Plant quantitative resistance genes to generalist pathogens include a broad array of direct defense genes, such as those involved in secondary metabolite production, cell wall formation, and defense proteins (Zhang et al., 2002; Denby et al., 2004; Zipfel et al., 2004; Ferrari et al., 2007; Rowe and Kliebenstein, 2008; Poland et al., 2009; Corwin et al., 2016a). Importantly, these quantitative plant resistance loci do not alter resistance to all genotypes (isolates) of a pathogen but instead interact with the infecting pathogen’s genotype. For example, the ability of the Arabidopsis (Arabidopsis thaliana) defense metabolite, camalexin, to provide resistance to Botrytis cinerea depends upon whether the specific isolate is sensitive or resistant to camalexin (Kliebenstein et al., 2005; Pedras and Ahiahonu, 2005; Stefanato et al., 2009; Pedras et al., 2011). Similarly, B. cinerea virulence on tomato varies with the isolate’s ability to detoxify tomatine (Quidde et al., 1998, 1999). In contrast to the polygenic nature of plant resistance to generalist pathogens, little is known about the genetic architecture of virulence within generalist pathogens and how this is affected by genetic variation in the plant (Bartoli and Roux, 2017). There are no reported naturally variable large-effect virulence loci in generalist pathogens, suggesting that virulence in generalist pathogens is largely quantitative and polygenic. This potential for interaction between polygenic virulence in generalist pathogens and equally polygenic resistance in host plants suggests that we need to work with genetic variation in both the host and pathogen to truly understand quantitative host-pathogen interactions.
Domestication of crop plants is a key evolutionary process in plants that has affected resistance to specialist pathogens. Domesticated plant varieties are typically more sensitive to specialist pathogens than their wild relatives (Smale, 1996; Rosenthal and Dirzo, 1997; Couch et al., 2005; Dwivedi et al., 2008), and pathogens may evolve higher virulence on domesticated hosts (Stukenbrock and McDonald, 2008). Furthermore, domestication typically imposes a genetic bottleneck that reduces genetic diversity in the crop germplasm, including decreased availability of resistance alleles against specialist pathogens (Tanksley and McCouch, 1997; Doebley et al., 2006; Chaudhary, 2013). These general evolutionary patterns of reduced resistance and allelic diversity found when studying the interaction of specialist pathogens with crop plants are assumed to hold for generalist pathogens and their domesticated hosts. However, there is less information about how crop host domestication affects disease caused by generalist pathogens, when the resistance to these pathogens is quantitative and polygenic rather than qualitative and monogenic. As such, there is a need to quantify the effect of domestication on a broad generalist pathogen in comparison with the rest of the crop’s standing variation to test how and whether domestication influences the pathogen.
B.cinerea provides a model generalist pathogen for studying quantitative interactions with plant hosts and underlying evolutionary processes. B. cinerea is a broad generalist pathogen that can infect most tested plants, from bryophytes to eudicots, and causes wide ranging pre- and post-harvest crop losses (Nicot and Baille, 1996; Elad et al., 2007; Fillinger and Elad, 2015). Individual isolates of B. cinerea show the same broad host range (Deighton et al., 2001; Finkers et al., 2007; Ten Have et al., 2007; Corwin et al., 2016b). This is in contrast to pathogens like Fusarium oxysporum where the species can infect diverse hosts, but each isolate is highly host specific (Katan, 1999; Ormond et al., 2010; Loxdale et al., 2011; Barrett and Heil, 2012). B. cinerea isolates display significant variation in virulence phenotypes, partly because genetic variation in specific virulence mechanisms, such as the production of the phytotoxins, botrydial and botcinic acid (Siewers et al., 2005; Dalmais et al., 2011). This genetic variation also influences cell wall degrading enzymes and key regulators of virulence such as VELVET, which quantitatively control virulence on multiple host plants (Rowe and Kliebenstein, 2007; Schumacher et al., 2012). This standing diversity in virulence mechanisms can contribute to the formation of quantitative differences in virulence between the isolates (Ten Have et al., 1998). This phenotypic variation is driven by a high level of sequence diversity spread across the genome (Rowe and Kliebenstein, 2007; Fekete et al., 2012). The polymorphism rate in B. cinerea, 37 single nucleotide polymorphisms (SNP)/kb, is much more variable than most previously studied plant pathogens (1-2 SNP/kb in Blumeria graminis, 1.5 SNP/kb in Melampsora larici-populina, 5.5 SNP/kb in the compact genome of the obligate biotroph Plasmodiophora brassicae, 12.3 SNP/kb in the wheat stem rust pathogen Puccinia graminis f. sp tritici) and human pathogens (3-6 SNP/kb in Mycobacterium tuberculosis). In addition to SNP diversity, genomic sequencing showed that B. cinerea has a high level of recombination and genomic admixture, as if it were a randomly intermating population (Supplemental Figure 1; Atwell et al., 2018). As such, a collection of B. cinerea isolates contain genetic variation in a wide range of virulence mechanisms, offering the potential to challenge the host with a blend of diverse virulence mechanisms to identify the pathogen variation controlling quantitative virulence.
A model pathosystem for studying quantitative host-pathogen interactions is the tomato–B. cinerea system, where the pathogen causes crop loss because of both pre- and post-harvest infection (Dean et al., 2012; Hahn, 2014; Romanazzi and Droby, 2016). Resistance to B. cinerea is a quantitative trait in tomato as with most other species, with identified tomato quantitative trait loci each explaining up to 15% of phenotypic variation for lesion size on stems (Díaz et al., 2002; Finkers et al., 2007; Ten Have et al., 2007; Rowe and Kliebenstein, 2008; Corwin et al., 2016a). Tomato is also a key model system to study how domestication influences plant physiology and resistance, including alterations in the circadian clock (Tanksley, 2004; Bai and Lindhout, 2007; Panthee and Chen, 2010; Bergougnoux, 2014; Müller et al., 2016), which can modulate resistance to B. cinerea (Sauerbrunn and Schlaich, 2004; Weyman et al., 2006; Bhardwaj et al., 2011; Hevia et al., 2015). This suggests that host plant diversity within tomato can alter traits known from other systems to influence B. cinerea resistance. Tomato domestication is typically considered a single event, followed by extensive crop improvement (Lin et al., 2014; Blanca et al., 2015). Thus, we are using the tomato–B. cinerea pathosystem to directly measure the interaction of domesticated crop variation with genetic variation in a generalist pathogen to better understand the evolution of this pathosystem.
In this study, we infected 97 genetically diverse B. cinerea isolates on a collection of domesticated tomato, Solanum lycopersicum, and wild tomato, Solanum pimpinellifolium, and quantified the interaction through lesion size in a detached leaf assay. Previous studies have examined B. cinerea resistance between domesticated and wild tomato species using single isolates of pathogens (Egashira et al., 2000; Nicot et al., 2002; Guimaraes et al., 2004; Ten Have et al., 2007; Finkers et al., 2008). These previous studies typically used individual wild and domesticated tomato accessions that were the founders of mapping populations and found a wide range of B. cinerea resistance. However, it is still unknown how domesticated and wild tomatoes compare in terms of B. cinerea resistance using multiple plant genotypes and a population of the pathogen. We selected accessions to sample major geographic origins of the progenitor species and focused the domesticated germplasm on diverse mid- to late- 20th century improved germplasm (Lin et al., 2014; Blanca et al., 2015). In this study, we asked whether B. cinerea virulence is controlled by host variation, pathogen variation, or the interaction between them. Lesion size in B. cinerea is a quantitative trait controlled by plant domestication status, plant genotype, and pathogen isolate. Finally, we aimed to identify the genetic basis of variation in B. cinerea virulence on S. lycopersicum and S. pimpinellifolium. We conducted genome-wide association (GWA) studies in B. cinerea to identify pathogen loci where genetic variation leads to altered virulence across the host genotypes, including a specific test for loci that influence responses to crop domestication. Few studies have conducted GWA analysis in plant pathogens for virulence phenotypes, and most of these were limited to a few variable loci or a few genetically distinct isolates (Dalman et al., 2013; Gao et al., 2016; Talas et al., 2016; Wu et al., 2017). Our previously sampled isolate collection includes genetic diversity across 272,672 SNPs (Supplemental Figure 1; Atwell et al., 2015, 2018; Zhang et al., 2017). We found that the genetic architecture of virulence of B. cinerea is highly quantitative, with hundreds of significant SNPs with small effect sizes associated with lesion area on each tomato genotype. Importantly, there is a subset of loci in the pathogen where allelic variation gives the isolates opposing responses to crop domestication. These pathogen loci could provide tools for understanding how domestication in tomato has influenced generalist pathogen resistance, which could facilitate breeding efforts.
RESULTS
Experimental Design
To measure how genetic variation in tomato affects quantitative resistance to a population of a generalist pathogen, we infected a collection of 97 diverse B. cinerea isolates (genotypes) on wild and domesticated tomato genotypes. We selected 6 domesticated S. lycopersicum and 6 wild S. pimpinellifolium accessions, the closest wild relative of S. lycopersicum, to directly study how domestication has influenced resistance to B. cinerea (Supplemental Figure 2; Peralta et al., 2008; Müller et al., 2016). Our previously collected B. cinerea sample includes 97 isolates obtained from various eudicot plant hosts, including tomato stem tissue (2 isolates; T3, KT) and tomato fruit (3 isolates; KGB1, KGB2, Supersteak; Atwell et al., 2015, 2018 Zhang et al., 2017). We infected all 97 B. cinerea isolates onto each of the 12 plant genotypes in threefold replication across 2 independent experiments in a randomized complete block design, giving 6 measurements per plant-pathogen combination, for a total of 3276 lesions. Digital measurement of the area of the developing lesion provides a composite phenotype controlled by the interaction of host and pathogen genetics. This measurement of the plant–B. cinerea interaction has been used successfully in a number of molecular and quantitative genetic studies (Ferrari et al., 2003, 2007; Denby et al., 2004; Kliebenstein et al., 2005; Ten Have et al., 2007; Abuqamar et al., 2008; Rowe and Kliebenstein, 2008; Liu et al., 2014). It should be noted that we were not focusing on microbe- or pathogen-associated molecular pattern-specific host/pathogen interactions with this study; we were instead allowing for the identification of any mechanism that may influence the host/pathogen interaction including metabolism, development, or any other unknown component. If there is genetic variation affecting the trait, and the trait influences the interaction of host and pathogen, it would be a component of the experiment. This fits with the recently developing view that growth, development, and resistance in plants are highly integrated processes that may not be as distinct as once believed (Campos et al., 2016; Ballaré and Pierik, 2017; Züst and Agrawal, 2017; Izquierdo-Bueno et al., 2018).
Lesion Size (Phenotypic) Variation
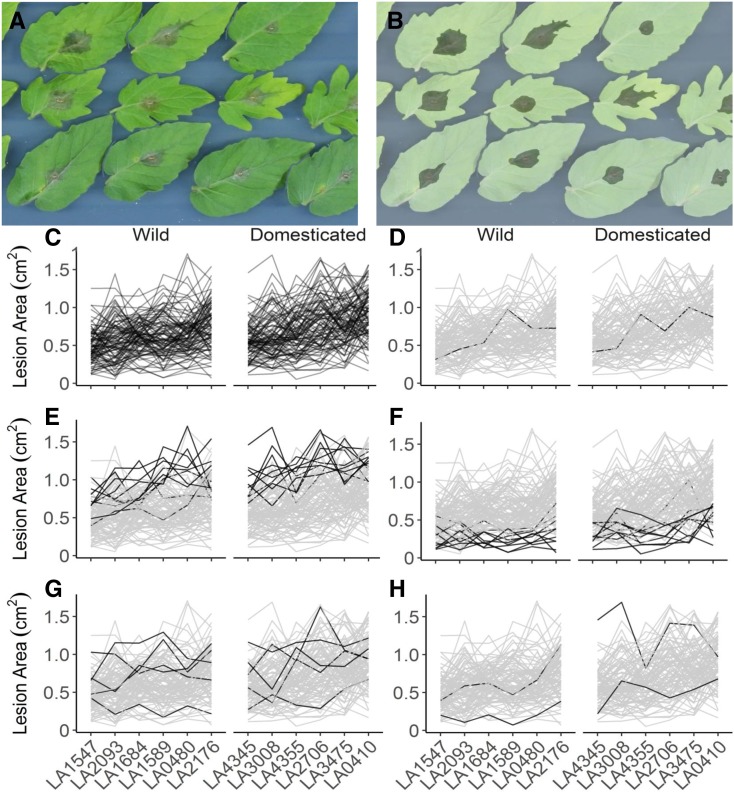
We collected images of all lesions at 24, 48, and 72 h after inoculation. At 24 h after inoculation, no visible lesions were present on the tomato leaves. At 48 h after inoculation, a thin ring of primary lesions became visible surrounding the location of the spore droplet, but no expansion was visible. At 72 h after inoculation, significant lesion growth was visible, but no lesions had spread to infect over half of the leaflet. We digitally measured the area of all developing lesions at 72 h after infection as a measure of virulence (Figure 1). We used the linear measurement of lesion area for several reasons. First, at 72 h after infection, B. cinerea lesion area appears to enter a relatively linear growth phase (Rowe et al., 2010). Second, this linear measurement behaves as a normally distributed trait (Kliebenstein et al., 2005; Corwin et al., 2016a; Atwell et al., 2018; Fordyce et al., 2018). Finally, B. cinerea isolates display large variation in their unit biomass per lesion area and, as such, growth in biomass is not the sole factor driving this measure (Corwin et al., 2016b). We observed a mean lesion size of 0.67 cm2 across the full experiment, with a coefficient of variation of 0.94 across the full isolate population on all tomato genotypes. Individual isolates were highly variable in their lesion size across tomato genotypes (Figures 1C to 1H and 2), with a mean lesion size per isolate of 0.14 cm2 to 1.29 cm2, and individual isolate coefficient of variation from 0.51 to 1.68 across all observations on all tomato genotypes (Supplemental Data Set 1). A subset of these isolates is highly virulent on tomato (mean lesion size > 1.05 cm2, Figure 1E), and a subset can be considered saprophytic (mean lesion size < 0.3 cm2, Figure 1F). Lesion size of B. cinerea on tomato showed a weak positive correlation with lesion size on A. thaliana from previous studies on both domesticated tomato (r = 0.247, P = 0.003) and wild tomato (r = 0.301, P = 0.016; Supplemental Figure 3; Zhang et al., 2017). This lack of correlation suggests the presence of both shared and unique mechanisms of quantitative virulence in the two species.
Figure 1.
Botrytis cinerea × Tomato Diversity, as Revealed by a Detached Leaf Assay and Digital Image Analysis.
(A) Individual tomato leaflets of 6 S. lycopersicum genotypes and 6 S. pimpinellifolium genotypes are in randomized rows, and spore droplets of individual B. cinerea isolates are in randomized columns. Digital images were collected 72 h after inoculation. Randomized leaflets were infected with single droplets of spore suspensions from 40 randomized B. cinerea isolates, and digital images were taken 72 h after inoculation.
(B) Digital masking of leaves and lesions was followed by automated measurement of the area of each lesion.
(C) to (H) Variation in lesion size resulting from the interaction of B. cinerea and diverse tomato genotypes.
(C) Average lesion sizes of single B. cinerea isolates (line traces) across tomato host genotypes grouped by domestication status.
(D) Highlight of the common reference B. cinerea isolate B05.10.
(E) Highlight of the ten highest-virulence isolates, as estimated based on mean virulence across all tomato genotypes.
(F) Highlight of the ten most saprophytic, or low virulence, isolates, as estimated based on mean virulence across all genotypes.
(G) Highlight of the five isolates collected from tomato tissue.
(H) Highlight of the two isolates with significant domestication sensitivity.
Contribution of Pathogen Genetics and Plant Genetics Effects to Resistance
To measure the relative contribution of genetic diversity in the plant and the pathogen to variation in the virulence/susceptibility phenotype, we used a general linear model (R lme4 package; Bates et al., 2015). This model directly tested the contribution of pathogen genotype (isolate), plant genotype, and plant domestication status to variation in lesion size. The final model showed that genetic variation within both the host plant and pathogen had significant effects on lesion growth, each explaining approximately the same portion of the variance (Table 1; Figure 1C). Interestingly, although tomato domestication status significantly affected B. cinerea virulence, it was to a much lower level than the other factors (Table 1). There was no evidence for significant interaction effects between pathogen isolate and plant genotype. Thus, the interaction between tomato and B. cinerea was significantly controlled by genetic diversity within the host plant and the pathogen, including a slight effect of domestication status.
Table 1. ANOVA Results of the Interaction between 12 Tomato Accessions and 95 B. cinerea Isolates Measured as Lesion Area.
| Effect | Type II Sum of Squares | F Value | Likelihood Ratio Test Statistic | Degrees of Freedom | P Value |
|---|---|---|---|---|---|
| Fixed Effect | |||||
| Isolate | 37.8 | 1.7 | 94 | 0.007 | |
| Domestication | 3.4 | 14.1 | 1 | 0.0006 | |
| Domestication/Plant | 39.3 | 16.2 | 10 | 5e-11 | |
| Isolate:Domestication | 15.8 | 0.7 | 94 | 0.99 | |
| Isolate:Domestication/Plant | 179.1 | 0.8 | 940 | 1 | |
| Random Effect | |||||
| Experiment | 136 | 1 | <2e-16 | ||
| Whole plant | 0.21 | 1 | 0.65 | ||
| Whole plant/leaf | 22.4 | 1 | 2e-06 | ||
| Whole plant/leaf/leaflet pair | 0 | 1 | 1 | ||
| Experiment:Isolate | 321 | 1 | <2e-16 |
Results of general linear modeling of lesion area for 12 tomato accessions by 95 B. cinerea isolates is shown (R lme4 package version 1.1-18-1; Bates et al., 2015). Two of the 97 isolates did not have replication across 2 experiments; therefore, they were dropped at this stage of analysis. The terms are as follows: Isolate is the 95 B. cinerea isolates; Domestication is wild tomato, S. pimpinellifolium, versus domesticated tomato, S. lycopersicum; Plant is 12 tomato genotypes nested within their respective domestication groupings. The experiment tests the random effect of 2 independent replicate experiments. The nested random effects of whole plant sampled, leaf sampled, and leaflet pair are included. In addition, interactions of these factors were tested (:). The degrees of freedom and p-value are shown. For fixed effects, the type II sum of squares and F-value are shown, and for random effects, the likelihood ratio test statistic is shown. P values in bold represent those crossing the significance threshold. Blank spaces within the table represent model terms for which that variance descriptor was not calculable due to it being a fixed or random effect.
Pathogen Specialization to the Source Host
One evolutionary model of plant–generalist pathogen interactions suggests that pathogen isolates within a generalist species may specialize for interaction with specific hosts. Alternatively, generalist isolates may show no host specialization or preference. Our collection of B. cinerea includes five isolates that may be adapted to tomato, because they were collected from S. lycopersicum. To test for evidence for specialization to the source host, we compared the virulence of the B. cinerea isolates obtained from tomato to the broader pathogen population. For B. cinerea genotypes isolated from tomato tissue versus other hosts, there was no significant difference in lesion size across all tomato genotypes (t test; n = 97, P = 0.14; Figure 1G). In fact, one isolate collected from tomato tissue (KGB1) was among the 10 least-virulent isolates and another (Triple3) was among the 10 most-virulent isolates (Figure 1G). This demonstrates that there is significant genetic variation in virulence across the B. cinerea isolates and that this collection of B. cinerea isolates from tomato does not display a strong host-specificity for tomato (Martinez et al., 2003; Ma and Michailides, 2005; Rowe and Kliebenstein, 2007; Samuel et al., 2012).
Pathogen Specialization to Host Genotype
Although we did not find evidence for B. cinerea preference for tomato based on isolate host source, the B. cinerea isolates may contain genetic variation at individual loci that allow them to better attack subsets of the tomato genotypes (Rowe and Kliebenstein, 2007; Kretschmer and Hahn, 2008; Corwin et al., 2016b). A visual analysis of the data suggested an interaction between the genomes of B. cinerea and tomato (Figure 1, C-H). However, when using the full model, we found no significant interaction between isolate and individual host genotype, even though there was a large fraction of variance within these terms (Table 1). This may indicate a lack of interaction between genetic variation in the host and pathogen. Interaction effects in large data sets can be difficult to identify using mixed models, so we used a second standard statistical approach, a Wilcoxon signed-rank test. We used model-adjusted lesion sizes as input to test whether the rank of B. cinerea isolate-induced lesion size significantly changes between pairs of tomato genotypes. When the full isolate population was used, the rank performance of the isolates significantly varied between host genotypes. When comparing mean lesion size between paired plant genotypes, 59% (39 out of 66) of tomato accession pairs had significantly different ranking of the isolates (Wilcoxon signed-rank test with Benjamini-Hochberg False Discovery Rate (FDR)-correction; Table 2; Supplemental Figure 4). A significant p-value indicates that the two host genotypes show evidence for different virulence interactions with the population of B. cinerea isolates, providing evidence for host × pathogen genotypic interactions. This pattern was consistent across domesticated host pairs, wild host pairs, or between-species host pairs (Wilcoxon signed-rank test with Benjamini-Hochberg FDR-correction; Table 2). This provides evidence that the population of B. cinerea does display differential responses to the genetic variation of tomato.
Table 2. Rank Order Shifts of 97 B. cinerea Isolates by Lesion Area across All of the Tomato Accessions.
| Wild | Domesticated | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| LA1547 | LA1589 | LA1684 | LA2093 | LA2176 | LA480 | LA2706 | LA3008 | LA3475 | LA410 | LA4345 | LA4355 | |
| Wild | ||||||||||||
| LA1547 | 2978 | 3988 | 2927 | 1865 | 3008 | 1710 | 3460 | 1597 | 1135 | 3928 | 2944 | |
| LA1589 | <0.001 | 5401 | 4699 | 3359 | 4662 | 3014 | 4918 | 2938 | 2340 | 5536 | 4454 | |
| LA1684 | NS | 0.029 | 3709 | 2552 | 3690 | 2296 | 4004 | 2205 | 1690 | 4537 | 3571 | |
| LA2093 | <0.001 | NS | 0.049 | 3013 | 4496 | 2732 | 4889 | 2588 | 1947 | 5534 | 4264 | |
| LA2176 | <0.001 | 0.004 | <0.001 | <0.001 | 5837 | 4029 | 6002 | 3963 | 3276 | 6706 | 5583 | |
| LA480 | <0.001 | NS | 0.044 | NS | 0.001 | 6143 | 4192 | 6286 | 6855 | 3575 | 4702 | |
| Domesticated | ||||||||||||
| LA2706 | <0.001 | <0.001 | <0.001 | <0.001 | NS | <0.001 | 6311 | 4523 | 3876 | 6917 | 5940 | |
| LA3008 | 0.009 | NS | NS | NS | <0.001 | NS | <0.001 | 2619 | 2082 | 5100 | 4049 | |
| LA3475 | <0.001 | <0.001 | <0.001 | <0.001 | NS | <0.001 | NS | <0.001 | 3815 | 7088 | 5984 | |
| LA410 | <0.001 | <0.001 | <0.001 | <0.001 | 0.002 | <0.001 | NS | <0.001 | NS | 7567 | 6602 | |
| LA4345 | 0.16 | 0.011 | NS | 0.011 | <0.001 | 0.021 | <0.001 | NS | <0.001 | <0.001 | 3439 | |
| LA4355 | <0.001 | NS | 0.02 | NS | 0.008 | NS | <0.001 | NS | <0.001 | <0.001 | <0.001 | |
Wilcoxon signed-rank test comparing model-corrected mean B. cinerea lesion areas on various tomato accessions. This tests for a change in the rank order of the 97 isolates between each pair of tomato accessions. A significant p value suggests that the relative performance of individual isolates is altered from one host to the other. The bottom left corner of the chart includes Benjamini-Hochberg FDR-corrected p values; the top right corner includes the test statistic (W). Bold text indicates significance at p < 0.01 after correction; italicized text indicates suggestive p values 0.01 < p < 0.1. NS, nonsignificant interactions. Blank spaces represent the self comparison diagonal and this was excluded as it is uninformative.
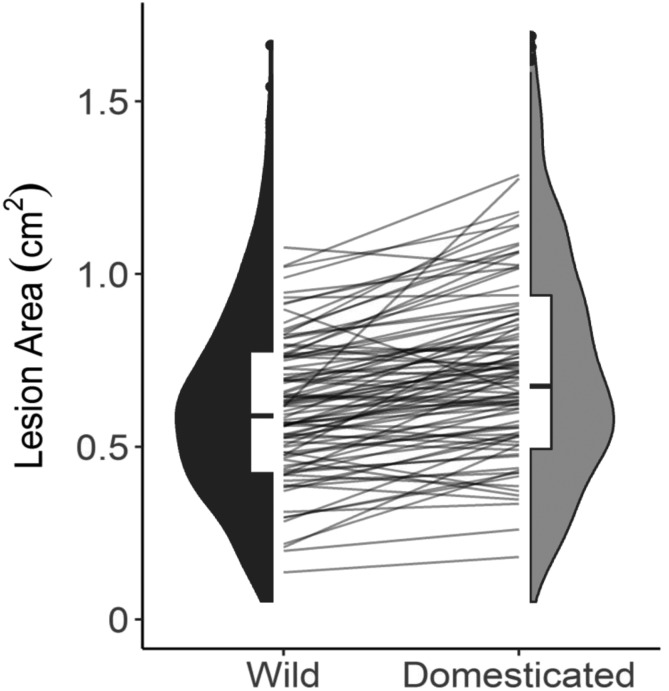
To focus on whether specific B. cinerea isolates may be sensitive to tomato domestication, we applied a Wilcoxon and Analysis of Variance (ANOVA) approach. Overall, most isolates (78/97, 80%) are more virulent on domesticated than wild tomato (Figure 3; Supplemental Data Set 1). Using a Wilcoxon signed-rank test to compare the rank of model-corrected mean lesion size of all the B. cinerea isolates on wild versus domesticated tomato, we found a significant difference (Wilcoxon signed-rank test, W = 5801, p-value = 0.0007; Figure 3). Although this shows a general population behavior, we used single-isolate ANOVAs to test whether any specific pathogen genotypes had a significant association with domestication. These general linear models included the fixed effects of plant, domestication, and the random effect of experiment. After adjustment for multiple testing, this identified two isolates (Fd2, Rose) with a significant effect of domestication on lesion size (P < 0.05, FDR corrected; Figure 1H), both of which are more virulent on domesticated tomato (Supplemental Data Set 2).
Figure 3.
Distribution of B. cinerea Virulence by Tomato Domestication Status.
The violin plots show the mean virulence of each B. cinerea isolate on the tomato genotypes, grouped as wild or domesticated germplasm. The domestication effect on lesion size is significant (ANOVA, P = 0.0006; Table 1). The interaction plot between the two violin plots connects the average lesion size of a single B. cinerea isolate between the wild and domesticated germplasm.
To assess whether isolates could appear domestication associated because random chance, we bootstrapped assignment of plant accessions to domestication groups. Ninety-six of the 100 bootstraps identified no isolates with domestication sensitivity, and the other four bootstraps identified only two isolates showing significant domestication association (FDR <0.01). Therefore, our individual isolate observations are in the 96th percentile. Although this is suggestive, a more precise estimate of isolate × domestication interactions would require larger experiments using either more replications or additional plant genotypes.
Domestication and Lesion Size Variation
Existing literature predominantly reports that crop domestication decreases plant resistance to pathogens (Smale, 1996; Rosenthal and Dirzo, 1997; Couch et al., 2005; Dwivedi et al., 2008; Stukenbrock and McDonald, 2008). Although we did observe the expected decreased resistance (by 18%) in domesticated tomato (Table 1; Figures 2 and 3;), domestication was a minor player in controlling lesion size variation, with most of the plant genetic signature coming from variation within both the wild and domesticated tomato species, contributing 12-fold more variation in resistance than domestication alone (Table 1). Removing the two domestication-associated isolates (Fd2, Rose) from our population did not eliminate the effect of tomato domestication on lesion size, because it was still significant and B. cinerea was still more virulent on domesticated tomato by 17% (Supplemental Table 1). To test how this mild domestication effect might be sensitive to shifts in the collection of tomato genotypes, we used the same bootstraps from above for the full model. Our observed domestication effect was in the top 80th percentile across all bootstraps, suggesting that although the domestication effect is small, it is relatively stable in response to shifts in the genotypes. However, a larger sample of S. lycopersicum and S. pimpinellifolium genotypes would be needed to develop a more precise estimate of any domestication effect on lesion size.
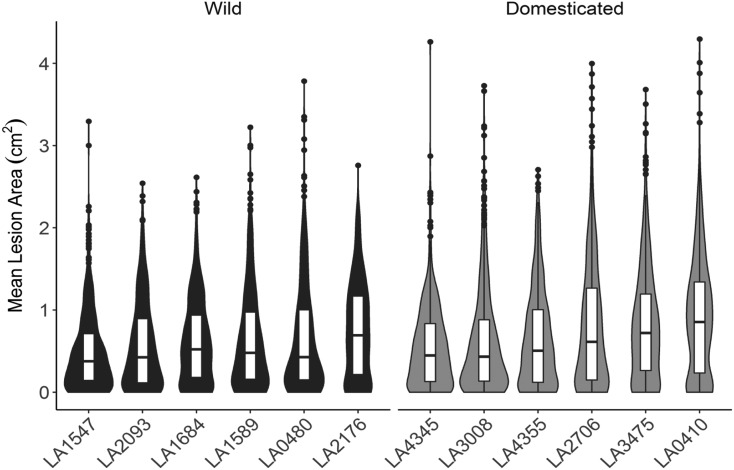
Figure 2.
Distribution of Tomato Genotype Susceptibility to Infection with 97 Genetically Diverse B. cinerea Isolates.
Violin plots show the distribution of lesion size caused by B. cinerea isolates on each tomato host genotype. Individual points are mean lesion size for each of the 97 different isolate-host pairs. The boxes show the 75th percentile distribution, and the horizontal line shows the mean resistance of the specific host genotype. The tomato genotypes are grouped based on their status as wild or domesticated germplasm.
In addition to altering trait means, domestication commonly decreases genetic variation in comparison with wild germplasm because of bottlenecks, including for tomato (Tanksley and McCouch, 1997; Doebley et al., 2006; Bai and Lindhout, 2007). We would expect this decreased genetic variation to limit phenotypic variation, including disease phenotypes. Interestingly, in this tomato population, we did not observe reduced variation in lesion size in the domesticated tomato. The wild and domesticated tomato genotypes showed similar variation in resistance (F-test, F96,96 = 1.39, P = 0.11; Figure 3; Supplemental Figure 2). Overall, there is a slight domestication impact on average resistance to B. cinerea, and no evidence of a phenotypic bottleneck because of domestication. This suggests that in the tomato–B. cinerea pathosystem, domestication is not a major part of the variation.
Quantitative Genetics of Pathogen Virulence on Tomato
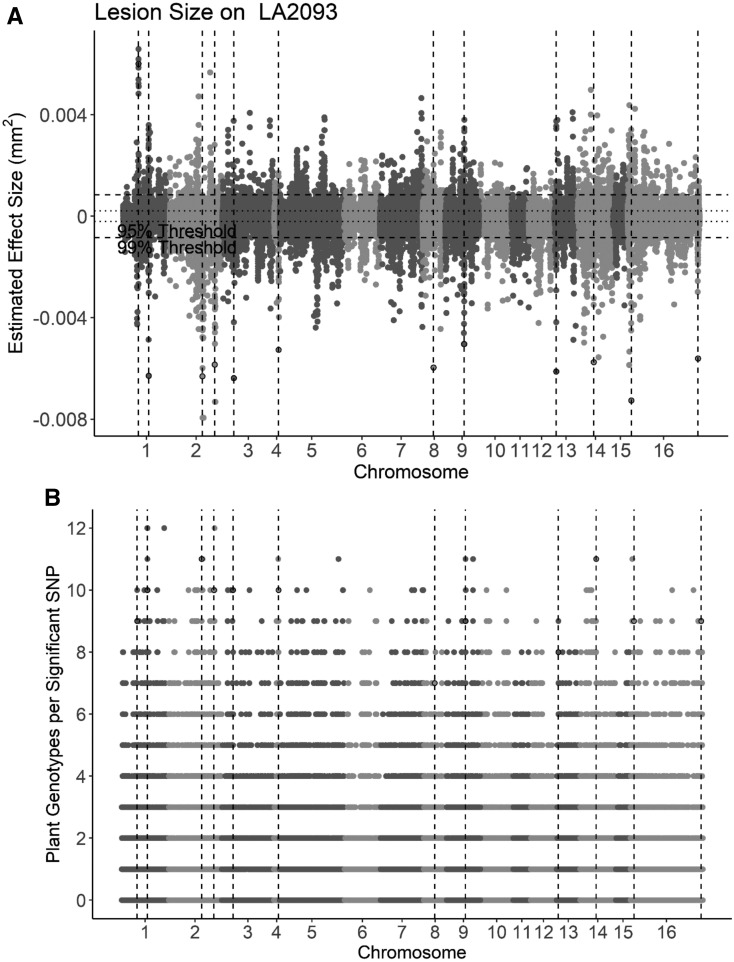
Genetic variation within B. cinerea had a large effect on virulence on tomato and showed some evidence for interaction with tomato domestication (Table 1). This suggests that there is genetic variation within the pathogen, in which some alleles enhance and other alleles decrease virulence, depending upon the plant’s genotype. To identify variable pathogen genes controlling differential virulence across plant genotypes, we conducted GWA mapping analysis within the pathogen using 272,672 SNPs compared with the B. cinerea T4 reference genome (Supplemental Figure 1; Atwell et al., 2018). Because of the large effect of plant genotype on resistance to B. cinerea, we performed GWA using model-corrected least-squared mean virulence measured on each tomato genotype as separate traits. We used a ridge-regression approach (bigRR) to estimate the phenotypic effects across the genome (Shen et al., 2013; Corwin et al., 2016a, 2016b; Francisco et al., 2016; Atwell et al., 2018). To determine significance of SNP effects under GWA, we permuted phenotypes 1000 times to calculate 95%, 99%, and 99.9% effect size thresholds within each plant host. At 1000 permutations, the 99.9% threshold is imprecise, but we included this approximate threshold to identify conservative SNP associations. GWA analysis showed that the genetic basis of B. cinerea virulence on tomato is highly polygenic. Consistent with a polygenic structure of this trait in the pathogen, GWA did not identify large-effect SNPs (Figure 4). The number of significant B. cinerea virulence SNPs identified by this ridge-regression approach (bigRR) varied by plant accession, from 1284 to 25,421 SNPs on the 12 different host genotypes (significance was determined by the SNP effect size estimate exceeding the 99% 1000-permutation threshold).
Figure 4.
GWA of B. cinerea Lesion Size on Individual Tomato Genotypes.
B. cinerea chromosomes are differentiated by shading in alternating light and dark gray.
(A) Manhattan plot of estimated SNP effect sizes by bigRR (ridge-regression approach) for B. cinerea lesion size using a single tomato accession, LA2093. Permutation-derived thresholds are shown as horizontal dashed lines.
(B) The number of tomato accessions for which a B. cinerea SNP was significantly linked to lesion development by bigRR using the 99% permutation threshold. Frequency is the number of phenotypes in which the SNP exceeds the threshold. Vertical dotted lines identify regions with overlap between the top 100 large-effect SNPs for LA2093 and significance across the majority (≥6) of tomato genotypes tested.
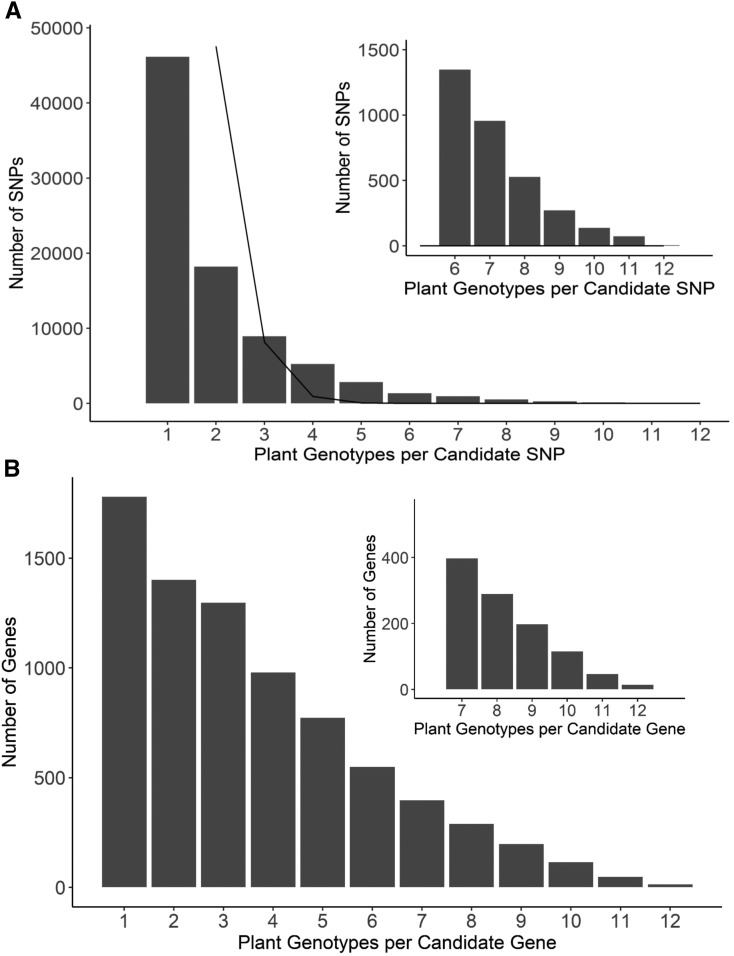
At the SNP level, fewer loci contribute to virulence across all host genotypes. We found five B. cinerea SNPs significantly linked to altered lesion size on all 12 tomato accessions (Figure 4B). In all, 215 SNPs were called in at least ten hosts, and 3300 SNPs were called in at least half of the hosts, whereas 27% (46,000) of the significant SNPs were linked to virulence on only a single host tomato genotype. These levels of overlap exceed the expected overlap due to random chance (Figure 5A). Although only a small subset of these B. cinerea SNPs were linked to virulence on all the tomato genotypes, we obtained better overlap across host genotypes by focusing on gene windows.
Figure 5.
Frequency of Overlap in B. cinerea GWA Significance across Tomato Accessions.
(A) The frequency with which the B. cinerea SNPs significantly associate with lesion size on the 12 tomato accessions using bigRR and the 99% permutation threshold. The black line indicates the expected frequency of random overlap, given the number of significant SNPs per plant genotype and the size of the total SNP set. The inset zooms in on the distribution for overlapping SNPs above 6 plant genotypes for easier visualization. There were no SNPs expected to overlap by random chance in the inset.
(B) The frequency with which B. cinerea genes significantly associated with lesion size on the 12 tomato accessions. Genes were called as significant if there was one significant SNP called at the 99% permutation threshold within the gene body, or within 2 kb of the gene body.
To focus on the small-effect genes linked to B. cinerea virulence, we classified a gene as significantly associated if there was one SNP linked to a trait using a 2-kbp window surrounding the start and stop codon for a given gene. This analysis identified 14 genes linked to differential virulence in all 12 tomato accessions by bigRR (Figure 5B; Supplemental Data Set 3A), as some SNPs within a gene had accession-specific phenotypes (significant in <12 tomato accessions). An additional 1045 genes were linked to differential virulence on 7 to 11 of the tomato accessions by bigRR (Figure 5B; Supplemental Data Set 3A).
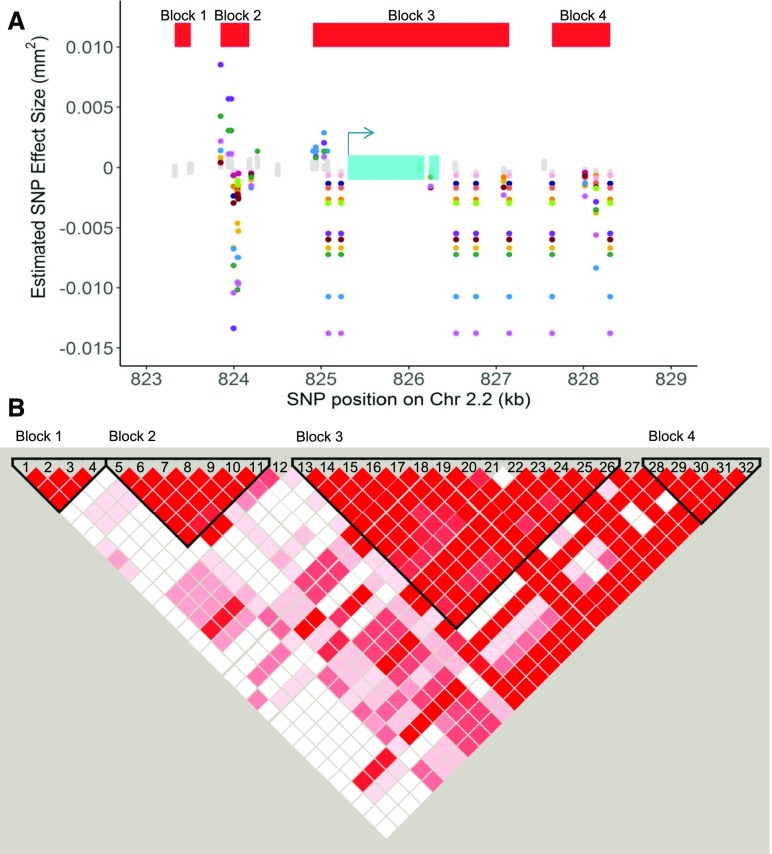
Of the 14 genes with SNPs significantly associated with B. cinerea virulence on all tomato genotypes by bigRR, most have not been formally linked to pathogen virulence. However, SNPs within a pectinesterase gene (BcT4_6001, Bcin14g00870) were associated with virulence across 11 tomato accessions. Pectinesterases are key enzymes for attacking the host cell wall, suggesting that variation in this pectinesterase locus and the other loci may influence pathogen virulence across all the tomato genotypes (Valette-Collet et al., 2003). Therefore, as an example of a virulence gene identified by our GWA methods, we looked for evidence of multiple haplotypes in this locus linked to virulence by visualizing the SNP effects across the pectinesterase gene. We plotted the effect sizes for all SNPs in this gene and investigated the linkage disequilibrium among these SNPs (Figure 6). This showed that the effect of SNPs across this gene vary in effect direction depending on tomato host genotype (Figure 6A). We identified two haplotype blocks contributing to the association of this gene to the virulence phenotype (Figure 6B). One block is associated with SNPs in the 5′ untranslated region in SNPs 5-11, and the second block is SNPs that span the entirety of the gene in SNPs 13-26. Interestingly, there are only two SNPs in the open reading frame of the associated gene (Figure 6). This suggests that the major variation surrounding this locus is controlling the regulatory motifs for this pectinesterase. Thus, there is significant genetic variation in B. cinerea virulence that is dependent upon the host’s genetic background. This suggests that the pathogen relies on polygenic small effect loci, potentially allowing selection to customize the level of virulence on different tomato hosts.
Figure 6.
Host Specificity of Significant SNPs Linked to the Gene BcT4_6001 (Bcin14g00870).
(A) SNPs with effects estimates above the 99% permutation threshold are colored by trait (plant accession in which the effect was estimated). Wild accessions are in the orange range (yellow to red shades), and domesticated accessions are in the blue range (green to purple shades). BcT4_6001 (Bcin14g00870) is a pectinesterase gene linked to at least one significant SNP on all 12 of the tested tomato accessions by bigRR. The annotated exons are depicted as turquoise rectangles, with the start codon marked with an arrow indicating the direction of transcription. Red rectangles indicate corresponding linkage disequilibrium blocks from Figure 6B.
(B) Linkage disequilibrium plot, including all pairwise comparisons of SNPs in the 2-kb region surrounding Bcin14g00870. The color scheme for each SNP pair is D'/LOD: white if LOD <2 and D’ <1, bright red for LOD ≥2 and D’=1, intermediate shades for LOD≥2 and D’<1.
Quantitative Genetics of the Pathogen Response to Tomato Domestication
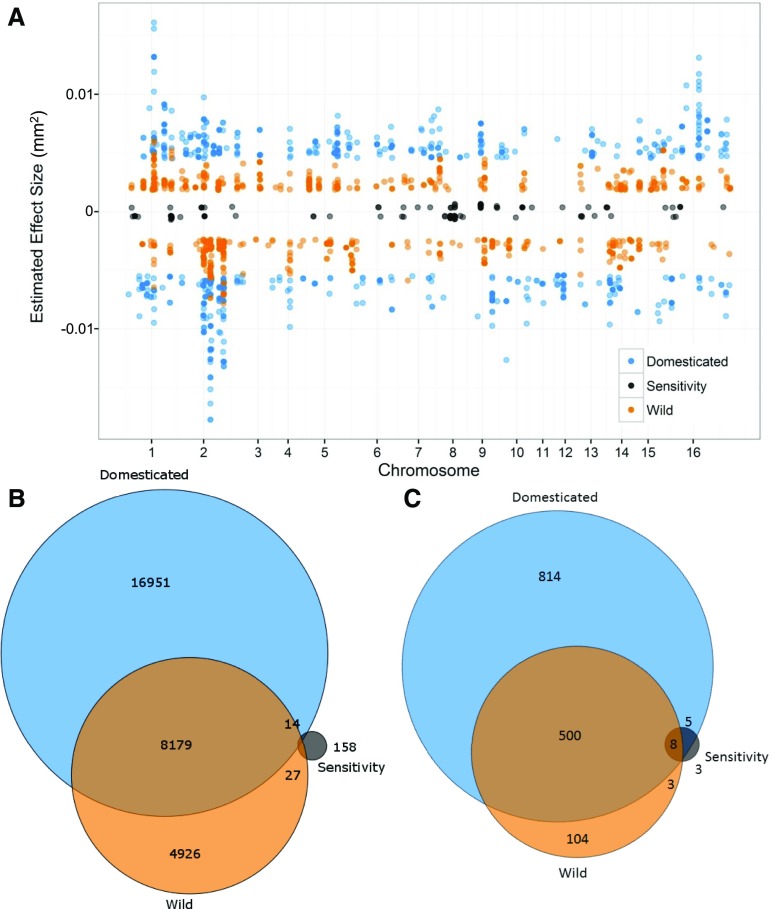
The identification of two isolates that differed on wild and domesticated tomato indicated that there may be some natural genetic variation in B. cinerea linked to this phenotypic variation. To directly map B. cinerea genes that control differential virulence on wild versus domesticated tomatoes, we used the least-squared mean virulence of each isolate across all wild and all domesticated tomato genotypes as two traits. We also calculated a domestication sensitivity trait: the relative difference in lesion size for each isolate between domesticated and wild hosts. Using these three traits, we conducted bigRR GWA within B. cinerea to map genes in the pathogen that respond to domestication shifts in the plant. The use of the mean lesion area of the B. cinerea isolates on the wild or domesticated tomato hosts uncovered a complex, highly polygenic pattern of significant SNPs similar to that of the individual tomato accessions (Figures 4 and 7). The significant SNP sets had a high degree of overlap between the wild phenotype and domesticated phenotype. By contrast, analysis of domestication sensitivity identified a more limited set of SNPs with less overlap to the mean lesion area on either domesticated or wild tomato (Figure 7). To query the underlying gene functions for these B. cinerea loci, we called genes as significant if there was one SNP within 2 kb of the gene (Figure 7C). The use of all 1251 genes linked to domestication traits by bigRR for functional enrichment analysis revealed only 22 significantly overrepresented biological functions (Fisher exact test, P < 0.05, Supplemental Data Set 3B) when compared with the whole-genome T4 gene annotation. The enrichments were largely surrounding enzyme and transport functions, which are known to be key components of how the pathogen produces toxic metabolites and conversely detoxifies plant defense compounds. Thus, there is an apparent subset of B. cinerea genes that may be specific to the genetic changes that occurred in tomato during domestication. Further work is needed to assess whether and how variation in these genes may be linked to altered virulence on domesticated and wild tomatoes.
Figure 7.
GWA Analysis of Domestication Sensitivity in B. cinerea.
Domestication sensitivity of each isolate was estimated as the difference between the average virulence on the wild and domesticated tomato germplasm. This value was then used for GWA mapping by bigRR.
(A) The top 1000 SNPs that significantly affect lesion size across domesticated tomato, wild tomato, or domestication sensitivity are shown. Significance is called as crossing the 99% permutation threshold.
(B) Venn diagram of overlapping SNPs identified as crossing the 99% permutation threshold for each trait.
(C) Venn diagram of overlapping genes identified as crossing the 99% permutation threshold for each trait. Genes were called as significant if there was one significant SNP within the gene body or within 2 kb of the gene body.
DISCUSSION
The genetics of plant resistance to generalist pathogens are mostly quantitative, depend upon pathogen isolate, and rely on genetic variation in both signal perception and direct defense genes (Kover and Schaal, 2002; Parlevliet, 2002; Glazebrook, 2005; Nomura et al., 2005; Goss and Bergelson, 2006; Tiffin and Moeller, 2006; Rowe and Kliebenstein, 2008; Barrett et al., 2009; Corwin et al., 2016a; Zhang et al., 2017). Previous studies of tomato resistance to B. cinerea have found a quantitative genetic architecture that varies between domesticated and wild tomato species, with higher resistance in the wild species (Egashira et al., 2000; Nicot et al., 2002; Guimaraes et al., 2004, Finkers, Heusden et al. 2007, Ten Have et al., 2007; Finkers et al., 2008). However, how the choice of B. cinerea isolate may alter the plant–pathogen interaction has been unclear. To address these questions, we used genetic variation in wild and domesticated tomato accessions in conjunction with a population of B. cinerea isolates. B. cinerea virulence on tomato, as measured by lesion size, was significantly affected by pathogen isolate, host genotype, and domestication status (Table 1). Pathogen isolate and tomato genotype were the strongest determinants of the interaction, with only a slight but significant decrease in resistance to the pathogen associated with domestication. Similarly, there was no evidence of a domestication bottleneck, with similar variance in resistance between the wild and domesticated tomato accessions (Table 1; Figure 2). There was also little evidence in this B. cinerea population for specialization to tomato, supporting the hypothesis that B. cinerea is a generalist at the isolate and species level (Figures 1C to 1H; Giraud et al., 1999; Martinez et al., 2003; Ma and Michailides, 2005). GWA mapping within the pathogen showed that the genetics underlying B. cinerea virulence on tomato are highly quantitative and vary across tomato genotypes and domestication status (Figures 5 and 7). This analysis identified a small subset of pathogen genes whose variation contributes to differential virulence on most of the hosts tested, and a set of pathogen genes whose variation is responsive to tomato domestication (Supplemental Data Set 3B).
Domestication and Altered Pathogen Virulence Genetics
In biotrophic pathogens, host domestication has decreased the diversity of resistance alleles because they are lost in the domestication bottleneck, as found for specialist pathogens (Tanksley and McCouch, 1997; Doebley et al., 2006; Hyten et al., 2006; Chaudhary, 2013). Surprisingly, we did not find evidence for a domestication bottleneck in the phenotypic resistance to B. cinerea (Figures 2 and 3). This is in contrast with genomic studies that explicitly show a genotypic bottleneck within tomato domestication (Miller and Tanksley, 1990; Koenig et al., 2013). Previous work in A. thaliana with these isolates has shown that if plant defenses such as jasmonic acid and salicylic acid signaling are nonfunctional, there is increased variation in B. cinerea virulence (Zhang et al., 2017). Thus, if these pathways had large-effect differences between wild and domesticated tomato, we would expect to see a wider range of B. cinerea virulence phenotypes in domesticated tomato (Zhang et al., 2017). The similarity in the variance suggests that any differences we are seeing are not caused by large-effect changes that abolish or greatly diminish specific defense signaling networks (Figures 2 and 3). These patterns, i.e., a mild decrease in resistance to B. cinerea because of plant domestication, and within-species plant variation exceeding the contribution of domestication itself, may be unique to interactions between B. cinerea and tomato, or more general. It is unclear whether this pattern is unique to tomato, or if each domestication event is unique.
Polygenic Quantitative Virulence and Breeding Complications
Our results indicate a highly polygenic basis of quantitative virulence of the generalist B. cinerea on tomato similar to the highly polygenic basis on the host side of the interaction (Zhang et al., 2017).The variation in lesion size is linked to numerous B. cinerea SNPs, each with small effect sizes (Figure 4A). Importantly, the tomato host accession greatly influenced which B. cinerea loci were significantly associated with lesion size (Figure 5). Thus, it is possible that there is specialization at the gene level, in which different alleles within the pathogen link to differential virulence on specific host genotypes (Giraud et al., 1999; Rowe and Kliebenstein, 2007; Blanco-Ulate et al., 2014). This polygenic architecture of virulence is different from virulence architecture in specialist pathogens that often have one or a few large effect genes that control virulence (Keen, 1992; De Feyter et al., 1993; Abramovitch and Martin, 2004; Boyd et al., 2013; Vleeshouwers and Oliver, 2014). Further studies are needed to compare how the host plant species may affect this image of genetic variation in virulence.
These results point to particular challenges for breeding durable resistance to B. cinerea, and possibly other generalist pathogens. The highly polygenic variation in virulence, combined with genomic sequencing showing that this pathogen is an inter-breeding population, suggests that the pathogen is actively blending a large collection of polymorphic virulence loci (Rowe and Kliebenstein, 2007; Fekete et al., 2012; Atwell et al., 2015,. 2018). Thus, it is insufficient to breed crop resistance against a single isolate of B. cinerea, because this resistance mechanism would likely be rapidly overcome by new genotypes within the field population of B. cinerea. By contrast, it is likely necessary to breed resistance using a population of the pathogen, and to focus on plant loci that target entire virulence pathways or mechanisms. The results in this study indicate that the specific genetics of the plant host, the host’s general domestication status, and the specific genetics of the pathogen isolate will all combine to affect how the estimated breeding value inferred from any experiment will translate to a field application (Table 1). As such, using a single or even a few pathogen isolates to guide resistance breeding in plants is unlikely to translate to durable resistance against B. cinerea as a species. Furthermore, the lack of evidence for a domestication bottleneck on tomato resistance to B. cinerea suggests that, at least for tomato, allelic variation in this generalist pathogen is sufficient to overcome introgression of wild resistance genes or alleles into the domesticated crop.
This study examined the contributions of host and pathogen natural genetic variation to the quantitative interaction in the tomato–B. cinerea pathosystem. B. cinerea has a highly quantitative genetic basis of virulence on tomato, which is dominated by pathogen effects but also sensitive to genetic variation linked to tomato domestication. Further studies are needed to test whether this pattern of domestication responses in tomato is similar to patterns in other crops. Because this population of B. cinerea can infect a wide range of hosts, it will be possible to directly conduct this study. By extending future work to additional domestication events, it may be possible to test whether independent crop domestication events have a consistent underlying genetic signal of the adaptation of B. cinerea to plant domestication.
METHODS
Tomato Genetic Resources
We obtained seeds for 12 selected tomato genotypes in consultation with the University of California Davis Tomato Genetics Resource Center. These include a diverse sample of 6 genotypes of domesticated tomato’s (Solanum lycopersicum) closest wild relative (Solanum pimpinellifolium) sampling across its major geographic regions (Peru, Ecuador) and 6 heritage and modern varieties of S. lycopersicum, focusing on mid- to late 20th century improved varieties (Lin et al., 2014; Blanca et al., 2015). Although genetic data are not available for all of our S. pimpinellifolium accessions, 9 of the 12 accessions have been genotyped and span the mappable diversity in domesticated tomato and its close relatives (Supplemental Figure 2; Sim et al., 2012). We bulked all genotypes and grew them under long-day (16-h photoperiod) greenhouse conditions at UC Davis in the Fall of 2014. We grew the plants under metal-halide lamps at 180-190 µEi using day/night temperatures at 25°C/18°C in 4″ pots filled with standard potting soil (Sunshine mix #1, Sun Gro Horticulture). The plants were watered once daily and pruned and staked to maintain upright growth. Fruits were collected at maturity and stored at 4°C in dry paper bags until seed cleaning. To clean the seeds, we incubated seeds and locule contents at 24°C in 1% protease solution (Rapidase C80 Max) for 2 h, then rinsed them in deionized water and air-dried. We stored the seeds in a cool, dry, dark location until use.
To grow plants for detached leaf assays, we bleach-sterilized all seeds and germinated them on paper in the growth chamber using flats covered with humidity domes. At 7 d, we transferred seedlings to soil (SunGro Horticulture) and grew all plants in growth chambers in 20°C, short-day (10-h photoperiod) conditions under metal-halide lamps at 180-190 µEi light intensity and 60% RH. We bottom-watered the plants with deionized water every 2 d for 2 weeks, and at week 3 watered every 2 d with added nutrient solution (0.5% Nitrogen-Phosphorus-Potassium fertilizer in a 2-1- 2 ratio; Grow More 4-18-38). The plants were used for detached leaf assays 6 weeks after transferring the seedlings to soil. Flowering in this system did not occur until at least 9 weeks of age for any accession, and as such we were sampling midway between the juvenile/adult transition and any flowering time decision. This window has been successfully used to minimize any major ontogenetic effects on the pathogen/host interaction in other systems (Corwin, Copeland et al. 2016).
B. cinerea Genetic Resources
We used a previously described collection of B. cinerea isolates that were isolated as single spores from natural infections of fruit and vegetable tissues collected in California and internationally (Atwell et al., 2015, 2018; Zhang et al., 2017). This included five isolates obtained from natural infections of tomato. We maintained B. cinerea isolates as conidial suspensions in 30% glycerol for long-term storage at −80°C. For regrowth, we diluted spore solutions to 10% concentration in filter-sterilized 50% grape juice and inoculated them onto 39 g/L potato dextrose agar medium. We grew the isolates at 25°C under a 12-h light/12-h dark cycle under incandescent lights at 100 µEi and propagated them every 2 weeks. Sequencing failed for 6 out of our 97 phenotyped isolates. For bigRR GWA mapping with the 91 isolates genotyped in this study, we used a total of 272,672 SNPs against the B. cinerea T4 genome with minor allele frequency 0.20 or greater and <10% missing calls across the isolates (SNP calls in at least 82/91 isolates; Atwell et al., 2018).
Detached Leaf Assay
To study the effect of genetic variation in the host and pathogen on lesion formation, we infected detached leaves of 12 diverse tomato varieties with the 97 B. cinerea isolates described above. We used a randomized complete block design for a total of 6 replicates across 2 experiments. In each experiment, this included a total of 10 plants per genotype randomized in 12 flats in 3 growth chambers. Each growth chamber block corresponded with a replicate of the detached leaf assay, such that growth chamber and replicate shared the same environmental block. At 6 weeks of age, we selected 5 leaves per plant (expanded leaves from the second true leaf or younger) and 2 leaflet pairs per leaf. We randomized the order of leaves from each plant, and the leaflets were placed on 1% Phytoagar in planting flats, with humidity domes. Our inoculation protocol followed previously described methods (Denby et al., 2004; Kliebenstein et al., 2005). Spores were collected from mature B. cinerea cultures grown on canned peach plates and diluted to 10 spores/ μL in filter-sterilized 50% organic grape juice. Spores in grape juice were maintained in a 4°C refrigerator or on ice from the time of collection to inhibit germination before inoculation. The diluted spore suspensions were homogenized by agitation continuously during the entire process of applying the spores to all samples to maintain the spores in the suspension and ensure even application across samples. For inoculation, 4 μL droplets were placed onto the detached leaflets at room temperature. The entire inoculation took ∼2 h per experiment. Mock-inoculated control leaves were treated with 4 μL of 50% organic grape juice without spores. Digital photos were taken of all leaflets at 24, 48, and 72 h after inoculation, and automated image analysis was used to measure lesion size.
Automated Image Analysis
Lesion area was digitally measured using the EBImage and CRImage packages (Pau et al., 2010; Failmezger et al., 2012) in the R statistical environment (R Development Core Team, 2008), as previously described (Corwin et al., 2016a, 2016b). Leaflets were identified as objects with green hue, and lesions were identified as low-saturation objects within leaves. Images masks were generated for both the leaf and lesion, then manually refined by a technician to ensure accurate object calling. The areas of these leaves and lesions were then automatically measured as pixels per lesion and converted to area using a 1-cm reference within each image.
Data Analysis
We analyzed lesion areas using general linear models for the full experiment to determine the contributions of plant and pathogen genotype (R lme4 package; Bates et al., 2015). Two of our 97 isolates that did not have replication across 2 experiments were dropped at this stage of analysis. We used the following linear models throughout our analyses.
Main mixed-effect model of lesion size variation
Within-plant accession mixed-effect model of lesion size
Within-isolate mixed-effect model of lesion size
where I represents fungal genotype (isolate), P represents plant genotype (accession), D represents domestication status, E represents experiment, W represents whole plant, L represents leaf, and A represents leaflet position. Factors with the subscript R are included in the analysis as random effects.
The within-plant accession model was used to calculate the significance of each factor and to obtain the least-squared means of lesion size for each B. cinerea isolate × tomato accession as well as for each B. cinerea isolate × domesticated/wild tomato. We also calculated a domestication sensitivity phenotype: Sensitivity = (Domesticated lesion size − Wild lesion size) / Domesticated lesion size.
We bootstrapped assignment of plant accessions to domestication groups to assess the robustness of our observed domestication effects. We randomly drew three genotypes from the domesticated and wild groupings and assigned them to a new pseudo-wild grouping. The other six genotypes were assigned as a pseudo-domesticated grouping, and the model was rerun. This bootstrapping was repeated 100 times with each representing a random draw. We used these to repeat the full model and to repeat the individual isolate models as a test of the robustness of the tomato domestication effect.
Using tomato sequence data from the SolCAP diversity panel that contained 9 of our 12 accessions, we determined pairwise genetic distances between our accessions (Sim et al., 2012). We calculated pairwise Euclidean distances between 426 wild and domesticated tomato accessions from Infinium SNP genotyping at 7720 loci using the R adegenet package (Jombart, 2008; Sim et al., 2012). Clustering was performed using R hclust (in the stats package) with the default UPGMA method (R Development Core Team, 2008).
We used several methods to examine host specialization to tomato within B. cinerea. First, we split our B. cinerea population into isolates collected from tomato tissue versus other hosts. We compared these groups by t test for virulence on domesticated tomato genotypes, wild tomato genotypes, or all tomato genotypes. Next, we used a Wilcoxon signed-rank test to compare the rank order distribution of model-adjusted lesion sizes across paired tomato genotypes. Also, to examine host specialization to tomato domestication within B. cinerea, we used a Wilcoxon signed-rank test to compare the rank order of model-adjusted lesion sizes across all domesticated versus all wild tomato genotypes. Finally, we conducted single-isolate ANOVAs with FDR correction on general linear models to identify isolates with a significant response to plant genotype or domestication status.
The model means and Domestication Sensitivity were used as the phenotypic input for GWA using bigRR, a heteroskedastic ridge regression method that incorporates SNP-specific shrinkage (Shen et al., 2013). This approach has previously had a high validation rate (Ober et al., 2015; Corwin et al., 2016a; Francisco et al., 2016; Kooke et al., 2016). The B. cinerea bigRR GWA was performed using 272,672 SNPs at minor allele frequency 0.20 or greater and <10% missing SNP calls as described above (Atwell et al., 2018). Because bigRR provides an estimated effect size, but not a p-value, significance was estimated using 1000 permutations to determine effect significance at 95%, 99%, and (approximately) 99.9% thresholds (Doerge and Churchill, 1996; Shen et al., 2013; Corwin et al., 2016a). SNPs were annotated by custom R scripts with gene transfer format file construction from the T4 gene models for genomic DNA by linking the SNP to genes within a 2-kbp window (http://www.broadinstitute.org, Staats and van Kan, 2012). Functional annotations are based on the T4 gene models for genomic DNA (http://www.broadinstitute.org, B. cinerea; Staats and van Kan, 2012). Additional genes of interest, based on a broad literature search of known virulence loci, were taken from National Center for Biotechnology Information (https://www.ncbi.nlm.nih.gov/) and included by mapping their sequences to the T4 reference using MUMmer v3.0 (Kurtz et al., 2004).
To predict expected overlap of significant SNPs across plant genotypes, we used the average number of significant SNPs per each of the 12 plant genotypes (14,000 SNPs) and calculated expected overlap between those 12 lists using binomial coefficients. Functional annotations of the gene lists are based on the T4 gene models for genomic DNA (http://www.broadinstitute.org, B. cinerea; Staats and van Kan, 2012).
Accession Numbers
The accession numbers of the major fungal genes described in this study are shown in Supplemental Table 2.
Supplemental Data
Supplemental Figure 1. Allele frequency spectrum of B. cinerea SNPs.
Supplemental Figure 2. Genetic distance between selected tomato accessions.
Supplemental Figure 3. Correlation between B. cinerea lesion size on tomato and on A. thaliana.
Supplemental Figure 4. Rank order plot of B. cinerea lesion size on two tomato genotypes.
Supplemental Table 1. Results of ANOVA following removal of domestication-associated isolates.
Supplemental Table 2. Sixty-three genes highlighted for high-level overlap of significant SNP hits across 11 to 12 tomato accessions.
Supplemental Data Set 1. Mean of B. cinerea lesion size of all isolates across all tomato accessions.
Supplemental Data Set 2. Results of single-isolate ANOVA on mixed effect model.
Supplemental Data Set 3. Gene and functional annotation based on T4 GWA results.
Acknowledgments
Financial support for this work was provided by the Danish National Research Foundation (Danmarks Grundforskningsfond) grant 99; National Science Foundation (NSF) grants IOS 1339125, MCB 1330337, and IOS 1021861; and the USDA National Institute of Food and Agriculture (NIFA), Hatch project number CA-D-PLS-7033-H.
AUTHOR CONTRIBUTIONS
N.E.S., S.A., G.S., R.G., and D.J.K. designed the research; N.E.S., G.S., R.F., R.G., D.G., and A.S. performed research; N.E.S., S.A., and R.F. contributed new analytic/computational/etc. tools; N.E.S. and D.J.K. analyzed data and wrote the paper.
References
- Abramovitch R.B., Martin G.B. (2004). Strategies used by bacterial pathogens to suppress plant defenses. Curr. Opin. Plant Biol. 7: 356–364. [DOI] [PubMed] [Google Scholar]
- Abuqamar S., Chai M.-F., Luo H., Song F., Mengiste T. (2008). Tomato protein kinase 1b mediates signaling of plant responses to necrotrophic fungi and insect herbivory. Plant Cell 20: 1964–1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atwell S., Corwin J.A., Soltis N.E., Subedy A., Denby K.J., Kliebenstein D.J. (2015). Whole genome resequencing of Botrytis cinerea isolates identifies high levels of standing diversity. Front. Microbiol. 6: 996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atwell S., Corwin J., Soltis N., Kliebenstein D. (2018). Resequencing and association mapping of the generalist pathogen Botrytis cinerea. bioRxiv. In press. [Google Scholar]
- Bai Y., Lindhout P. (2007). Domestication and breeding of tomatoes: What have we gained and what can we gain in the future? Ann. Bot. 100: 1085–1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballaré C.L., Pierik R. (2017). The shade-avoidance syndrome: Multiple signals and ecological consequences. Plant Cell Environ. 40: 2530–2543. [DOI] [PubMed] [Google Scholar]
- Barrett L.G., Heil M. (2012). Unifying concepts and mechanisms in the specificity of plant-enemy interactions. Trends Plant Sci. 17: 282–292. [DOI] [PubMed] [Google Scholar]
- Barrett L.G., Kniskern J.M., Bodenhausen N., Zhang W., Bergelson J. (2009). Continua of specificity and virulence in plant host-pathogen interactions: causes and consequences. New Phytol. 183: 513–529. [DOI] [PubMed] [Google Scholar]
- Bartoli C., Roux F. (2017). Genome-wide association studies in plant pathosystems: Toward an ecological genomics approach. Front. Plant Sci. 8: 763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates D., Maechler M., Bolker B., Walker S. (2015). Fitting linear mixed-effects models using lme4. J. Stat. Softw. 67: 1–48. [Google Scholar]
- Bergougnoux V. (2014). The history of tomato: From domestication to biopharming. Biotechnol. Adv. 32: 170–189. [DOI] [PubMed] [Google Scholar]
- Bhardwaj V., Meier S., Petersen L.N., Ingle R.A., Roden L.C. (2011). Defence responses of Arabidopsis thaliana to infection by Pseudomonas syringae are regulated by the circadian clock. PLoS One 6: e26968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bittel P., Robatzek S. (2007). Microbe-associated molecular patterns (MAMPs) probe plant immunity. Curr. Opin. Plant Biol. 10: 335–341. [DOI] [PubMed] [Google Scholar]
- Blanca J., Montero-Pau J., Sauvage C., Bauchet G., Illa E., Díez M.J., Francis D., Causse M., van der Knaap E., Cañizares J. (2015). Genomic variation in tomato, from wild ancestors to contemporary breeding accessions. BMC Genomics 16: 257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco-Ulate B., Morales-Cruz A., Amrine K.C., Labavitch J.M., Powell A.L., Cantu D. (2014). Genome-wide transcriptional profiling of Botrytis cinerea genes targeting plant cell walls during infections of different hosts. Front. Plant Sci. 5: 435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boller T., He S.Y. (2009). Innate immunity in plants: An arms race between pattern recognition receptors in plants and effectors in microbial pathogens. Science 324: 742–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd L.A., Ridout C., O’Sullivan D.M., Leach J.E., Leung H. (2013). Plant-pathogen interactions: Disease resistance in modern agriculture. Trends Genet. 29: 233–240. [DOI] [PubMed] [Google Scholar]
- Campos M.L., Yoshida Y., Major I.T., de Oliveira Ferreira D., Weraduwage S.M., Froehlich J.E., Johnson B.F., Kramer D.M., Jander G., Sharkey T.D., Howe G.A. (2016). Rewiring of jasmonate and phytochrome B signalling uncouples plant growth-defense tradeoffs. Nat. Commun. 7: 12570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhary B. (2013). Plant domestication and resistance to herbivory. Int. J. Plant Genomics 2013: 572784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corwin J.A., Copeland D., Feusier J., Subedy A., Eshbaugh R., Palmer C., Maloof J., Kliebenstein D.J. (2016a). The quantitative basis of the Arabidopsis innate immune system to endemic pathogens depends on pathogen genetics. PLoS Genet. 12: e1005789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corwin J.A., Subedy A., Eshbaugh R., Kliebenstein D.J. (2016b). Expansive phenotypic landscape of Botrytis cinerea shows differential contribution of genetic diversity and plasticity. Mol. Plant Microbe Interact. 29: 287–298. [DOI] [PubMed] [Google Scholar]
- Couch B.C., Fudal I., Lebrun M.-H., Tharreau D., Valent B., van Kim P., Nottéghem J.-L., Kohn L.M. (2005). Origins of host-specific populations of the blast pathogen Magnaporthe oryzae in crop domestication with subsequent expansion of pandemic clones on rice and weeds of rice. Genetics 170: 613–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalmais B., Schumacher J., Moraga J., LE Pêcheur P., Tudzynski B., Collado I.G., Viaud M. (2011). The Botrytis cinerea phytotoxin botcinic acid requires two polyketide synthases for production and has a redundant role in virulence with botrydial. Mol. Plant Pathol. 12: 564–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalman K., Himmelstrand K., Olson Å., Lind M., Brandström-Durling M., Stenlid J. (2013). A genome-wide association study identifies genomic regions for virulence in the non-model organism Heterobasidion annosum s.s. PLoS One 8: e53525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dangl J. L., Jones J. D. (2001). Plant pathogens and integrated defence responses to infection. Nature 411: 826–833. [DOI] [PubMed] [Google Scholar]
- Dean R., Van Kan J.A., Pretorius Z.A., Hammond-Kosack K.E., Di Pietro A., Spanu P.D., Rudd J.J., Dickman M., Kahmann R., Ellis J., Foster G.D. (2012). The Top 10 fungal pathogens in molecular plant pathology. Mol. Plant Pathol. 13: 414–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Feyter R., Yang Y., Gabriel D.W. (1993). Gene-for-genes interactions between cotton R genes and Xanthomonas campestris pv. malvacearum avr genes. Mol. Plant Microbe Interact. 6: 225–237. [DOI] [PubMed] [Google Scholar]
- Deighton N., Muckenschnabel I., Colmenares A.J., Collado I.G., Williamson B. (2001). Botrydial is produced in plant tissues infected by Botrytis cinerea. Phytochemistry 57: 689–692. [DOI] [PubMed] [Google Scholar]
- Denby K.J., Kumar P., Kliebenstein D.J. (2004). Identification of Botrytis cinerea susceptibility loci in Arabidopsis thaliana. Plant J. 38: 473–486. [DOI] [PubMed] [Google Scholar]
- Díaz J., Ten Have A., van Kan J.A. (2002). The role of ethylene and wound signaling in resistance of tomato to Botrytis cinerea. Plant Physiol. 129: 1341–1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodds P.N., Rathjen J.P. (2010). Plant immunity: Towards an integrated view of plant-pathogen interactions. Nat. Rev. Genet. 11: 539–548. [DOI] [PubMed] [Google Scholar]
- Doebley J.F., Gaut B.S., Smith B.D. (2006). The molecular genetics of crop domestication. Cell 127: 1309–1321. [DOI] [PubMed] [Google Scholar]
- Doerge R.W., Churchill G.A. (1996). Permutation tests for multiple loci affecting a quantitative character. Genetics 142: 285–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dwivedi S.L., Upadhyaya H.D., Stalker H.T., Blair M.W., Bertioli D.J., Nielen S., Ortiz R. (2008). Enhancing crop gene pools with beneficial traits using wild relatives. In Janick J, ed., Plant Breeding Reviews, John Wiley & Sons, New Jersey, 179. [Google Scholar]
- Egashira H., Kuwashima A., Ishiguro H., Fukushima K., Kaya T., Imanishi S. (2000). Screening of wild accessions resistant to gray mold (Botrytis cinerea Pers.) in Lycopersicon. Acta Physiol. Plant. 22: 324–326. [Google Scholar]
- Elad Y., Williamson B., Tudzynski P., Delen N. (2007). Botrytis spp. and diseases they cause in agricultural systems–an introduction. In Elad Y, Williamson B, Tudzynski P, Delen N, eds, Botrytis: Biology, pathology and control. Springer: Dordrecht, pp 1–8. [Google Scholar]
- Failmezger H., Yuan Y., Rueda O., Markowetz F., Failmezger M. H. (2012). CRImage: CRImage a package to classify cells and calculate tumour cellularity. R package version 1.24.0. https://rdrr.io/bioc/CRImage/
- Fekete É., Fekete E., Irinyi L., Karaffa L., Árnyasi M., Asadollahi M., Sándor E. (2012). Genetic diversity of a Botrytis cinerea cryptic species complex in Hungary. Microbiol. Res. 167: 283–291. [DOI] [PubMed] [Google Scholar]
- Ferrari S., Plotnikova J.M., De Lorenzo G., Ausubel F.M. (2003). Arabidopsis local resistance to Botrytis cinerea involves salicylic acid and camalexin and requires EDS4 and PAD2, but not SID2, EDS5 or PAD4. Plant J. 35: 193–205. [DOI] [PubMed] [Google Scholar]
- Ferrari S., Galletti R., Denoux C., De Lorenzo G., Ausubel F.M., Dewdney J. (2007). Resistance to Botrytis cinerea induced in Arabidopsis by elicitors is independent of salicylic acid, ethylene, or jasmonate signaling but requires PHYTOALEXIN DEFICIENT3. Plant Physiol. 144: 367–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fillinger S., Elad Y. (2015). Botrytis-the Fungus, the Pathogen and Its Management in Agricultural Systems. Springer International Publishing, Switzerland [Google Scholar]
- Finkers R., van Heusden A.W., Meijer-Dekens F., van Kan J.A., Maris P., Lindhout P. (2007). The construction of a Solanum habrochaites LYC4 introgression line population and the identification of QTLs for resistance to Botrytis cinerea. Theor. Appl. Genet. 114: 1071–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkers R., Bai Y., van den Berg P., van Berloo R., Meijer-Dekens F., Ten Have A., van Kan J., Lindhout P., van Heusden A.W. (2008). Quantitative resistance to Botrytis cinerea from Solanum neorickii. Euphytica 159: 83–92. [Google Scholar]
- Fordyce R.F., Soltis N.E., Caseys C., Gwinner R., Corwin J.A., Atwell S., Copeland D., Feusier J., Subedy A., Eshbaugh R., Kliebenstein D.J. (2018). Digital imaging combined with genome-wide association mapping links loci to plant-pathogen interaction traits. Plant Physiol. 178: 1406–1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francisco M., Joseph B., Caligagan H., Li B., Corwin J.A., Lin C., Kerwin R.E., Burow M., Kliebenstein D.J. (2016). Genome wide association mapping in Arabidopsis thaliana identifies novel genes involved in linking allyl glucosinolate to altered biomass and defense. Front. Plant Sci. 7: 1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y., Liu Z., Faris J.D., Richards J., Brueggeman R.S., Li X., Oliver R.P., McDonald B.A., Friesen T.L. (2016). Validation of genome-wide association studies as a tool to identify virulence factors in Parastagonospora nodorum. Phytopathology 106: 1177–1185. [DOI] [PubMed] [Google Scholar]
- Giraud T., Fortini D., Levis C., Lamarque C., Leroux P., Lobuglio K., Brygoo Y. (1999). Two sibling species of the Botrytis cinerea complex, transposa and vacuma, are found in sympatry on numerous host plants. Phytopathology 89: 967–973. [DOI] [PubMed] [Google Scholar]
- Glazebrook J. (2005). Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu. Rev. Phytopathol. 43: 205–227. [DOI] [PubMed] [Google Scholar]
- Goss E.M., Bergelson J. (2006). Variation in resistance and virulence in the interaction between Arabidopsis thaliana and a bacterial pathogen. Evolution 60: 1562–1573. [PubMed] [Google Scholar]
- Guimaraes R.L., Chetelat R.T., Stotz H.U. (2004). Resistance to Botrytis cinerea in Solanum lycopersicoides is dominant in hybrids with tomato, and involves induced hyphal death. Eur. J. Plant Pathol. 110: 13–23. [Google Scholar]
- Hahn M. (2014). The rising threat of fungicide resistance in plant pathogenic fungi: Botrytis as a case study. J. Chem. Biol. 7: 133–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hevia M.A., Canessa P., Müller-Esparza H., Larrondo L.F. (2015). A circadian oscillator in the fungus Botrytis cinerea regulates virulence when infecting Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 112: 8744–8749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyten D.L., Song Q., Zhu Y., Choi I.-Y., Nelson R.L., Costa J.M., Specht J.E., Shoemaker R.C., Cregan P.B. (2006). Impacts of genetic bottlenecks on soybean genome diversity. Proc. Natl. Acad. Sci. USA 103: 16666–16671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izquierdo-Bueno I., González-Rodríguez V.E., Simon A., Dalmais B., Pradier J.M., Le Pêcheur P., Mercier A., Walker A.S., Garrido C., Collado I.G., Viaud M. (2018). Biosynthesis of abscisic acid in fungi: Identification of a sesquiterpene cyclase as the key enzyme in Botrytis cinerea. Environ. Microbiol. 20: 2469–2482. [DOI] [PubMed] [Google Scholar]
- Jombart T. (2008). adegenet: A R package for the multivariate analysis of genetic markers. Bioinformatics 24: 1403–1405. [DOI] [PubMed] [Google Scholar]
- Jones J.D., Dangl J.L. (2006). The plant immune system. Nature 444: 323–329. [DOI] [PubMed] [Google Scholar]
- Katan T. (1999). Current status of vegetative compatibility groups in Fusarium oxysporum. Phytoparasitica 27: 51–64. [Google Scholar]
- Keen N.T. (1992). The molecular biology of disease resistance. Plant Mol. Biol. 19: 109–122. [DOI] [PubMed] [Google Scholar]
- Kliebenstein D.J., Rowe H.C., Denby K.J. (2005). Secondary metabolites influence Arabidopsis/Botrytis interactions: variation in host production and pathogen sensitivity. Plant J. 44: 25–36. [DOI] [PubMed] [Google Scholar]
- Koenig D., et al. (2013). Comparative transcriptomics reveals patterns of selection in domesticated and wild tomato. Proc. Natl. Acad. Sci. USA 110: E2655–E2662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kooke R., Kruijer W., Bours R., Becker F.F., Kuhn A., Buntjer J., Doeswijk T., Guerra J., Bouwmeester H.J., Vreugdenhil D. (2016). Genome-wide association mapping and genomic prediction elucidate the genetic architecture of morphological traits in Arabidopsis thaliana. Plant Physiol 170:2187-1203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kover P.X., Schaal B.A. (2002). Genetic variation for disease resistance and tolerance among Arabidopsis thaliana accessions. Proc. Natl. Acad. Sci. USA 99: 11270–11274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kretschmer M., Hahn M. (2008). Fungicide resistance and genetic diversity of Botrytis cinerea isolates from a vineyard in Germany. J. Plant Dis. Prot. 115: 214–219. [Google Scholar]
- Kurtz S., Phillippy A., Delcher A.L., Smoot M., Shumway M., Antonescu C., Salzberg S.L. (2004). Versatile and open software for comparing large genomes. Genome Biol. 5: R12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin T., et al. (2014). Genomic analyses provide insights into the history of tomato breeding. Nat. Genet. 46: 1220–1226. [DOI] [PubMed] [Google Scholar]
- Liu B., Hong Y.-B., Zhang Y.-F., Li X.-H., Huang L., Zhang H.-J., Li D.-Y., Song F.-M. (2014). Tomato WRKY transcriptional factor SlDRW1 is required for disease resistance against Botrytis cinerea and tolerance to oxidative stress. Plant Sci. 227: 145–156. [DOI] [PubMed] [Google Scholar]
- Loxdale H.D., Lushai G., Harvey J.A. (2011). The evolutionary improbability of ‘generalism’ in nature, with special reference to insects. Biol. J. Linn. Soc. Lond. 103: 1–18. [Google Scholar]
- Ma Z., Michailides T.J. (2005). Genetic structure of Botrytis cinerea populations from different host plants in California. Plant Dis. 89: 1083–1089. [DOI] [PubMed] [Google Scholar]
- Martinez F., Blancard D., Lecomte P., Levis C., Dubos B., Fermaud M. (2003). Phenotypic differences between vacuma and transposa subpopulations of Botrytis cinerea. Eur. J. Plant Pathol. 109: 479–488. [Google Scholar]
- Miller J.C., Tanksley S.D. (1990). RFLP analysis of phylogenetic relationships and genetic variation in the genus Lycopersicon. Theor. Appl. Genet. 80: 437–448. [DOI] [PubMed] [Google Scholar]
- Müller N.A., et al. (2016). Domestication selected for deceleration of the circadian clock in cultivated tomato. Nat. Genet. 48: 89–93. [DOI] [PubMed] [Google Scholar]
- Nicot P.C., Baille A. (1996). Integrated control of Botrytis cinerea on greenhouse tomatoes. In Nicot PC, Nguyen C, eds, Aerial Plant Surface Microbiology. Plenum Press, New York, pp. 169–189. [Google Scholar]
- Nicot P., Moretti A., Romiti C., Bardin M., Caranta C., Ferriere H. (2002). Differences in susceptibility of pruning wounds and leaves to infection by Botrytis cinerea among wild tomato accessions. TGC Report 52: 24–26. [Google Scholar]
- Nomura K., Melotto M., He S.-Y. (2005). Suppression of host defense in compatible plant-Pseudomonas syringae interactions. Curr. Opin. Plant Biol. 8: 361–368. [DOI] [PubMed] [Google Scholar]
- Ober U., Huang W., Magwire M., Schlather M., Simianer H., Mackay T.F. (2015). Accounting for genetic architecture improves sequence based genomic prediction for a Drosophila fitness trait. PLoS One 10: e0126880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ormond E.L., Thomas A.P., Pugh P.J., Pell J.K., Roy H.E. (2010). A fungal pathogen in time and space: the population dynamics of Beauveria bassiana in a conifer forest. FEMS Microbiol. Ecol. 74: 146–154. [DOI] [PubMed] [Google Scholar]
- Panthee D.R., Chen F. (2010). Genomics of fungal disease resistance in tomato. Curr. Genomics 11: 30–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parlevliet J.E. (2002). Durability of resistance against fungal, bacterial and viral pathogens; present situation. Euphytica 124: 147–156. [Google Scholar]
- Pau G., Fuchs F., Sklyar O., Boutros M., Huber W. (2010). EBImage--an R package for image processing with applications to cellular phenotypes. Bioinformatics 26: 979–981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedras M.S.C., Ahiahonu P.W. (2005). Metabolism and detoxification of phytoalexins and analogs by phytopathogenic fungi. Phytochemistry 66: 391–411. [DOI] [PubMed] [Google Scholar]
- Pedras M.S.C., Hossain S., Snitynsky R.B. (2011). Detoxification of cruciferous phytoalexins in Botrytis cinerea: spontaneous dimerization of a camalexin metabolite. Phytochemistry 72: 199–206. [DOI] [PubMed] [Google Scholar]
- Peralta I., Spooner D., Knapp S. (2008). The taxonomy of tomatoes: A revision of wild tomatoes (Solanum section Lycopersicon) and their outgroup relatives in sections Juglandifolium and Lycopersicoides. Syst. Bot. Monogr. 84: 1–186. [Google Scholar]
- Pieterse C.M., Van der Does D., Zamioudis C., Leon-Reyes A., Van Wees S.C. (2012). Hormonal modulation of plant immunity. Annu. Rev. Cell Dev. Biol. 28: 489–521. [DOI] [PubMed] [Google Scholar]
- Poland J.A., Balint-Kurti P.J., Wisser R.J., Pratt R.C., Nelson R.J. (2009). Shades of gray: The world of quantitative disease resistance. Trends Plant Sci. 14: 21–29. [DOI] [PubMed] [Google Scholar]
- Quidde T., Osbourn A., Tudzynski P. (1998). Detoxification of α-tomatine by Botrytis cinerea. Physiol. Mol. Plant Pathol. 52: 151–165. [Google Scholar]
- Quidde T., Büttner P., Tudzynski P. (1999). Evidence for three different specific saponin-detoxifying activities in Botrytis cinerea and cloning and functional analysis of a gene coding for a putative avenacinase. Eur. J. Plant Pathol. 105: 273–283. [Google Scholar]
- R Development Core Team (2008). R: A language and environment for statistical computing. (Vienna, Austria: R Foundation for Statistical Computing; ). [Google Scholar]
- Romanazzi G., Droby S. (2016). Control Strategies for Postharvest Grey Mould on Fruit Crops. In Fillinger S, Elad Y, eds, Botrytis–the Fungus, the Pathogen and its Management in Agricultural Systems. Springer, Cham, pp 217–228. [Google Scholar]
- Rosenthal J.P., Dirzo R. (1997). Effects of life history, domestication and agronomic selection on plant defence against insects: Evidence from maizes and wild relatives. Evol. Ecol. 11: 337–355. [Google Scholar]
- Rowe H.C., Kliebenstein D.J. (2007). Elevated genetic variation within virulence-associated Botrytis cinerea polygalacturonase loci. Mol. Plant Microbe Interact. 20: 1126–1137. [DOI] [PubMed] [Google Scholar]
- Rowe H.C., Kliebenstein D.J. (2008). Complex genetics control natural variation in Arabidopsis thaliana resistance to Botrytis cinerea. Genetics 180: 2237–2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowe H.C., Walley J.W., Corwin J., Chan E.K.-F., Dehesh K., Kliebenstein D.J. (2010). Deficiencies in jasmonate-mediated plant defense reveal quantitative variation in Botrytis cinerea pathogenesis. PLoS Pathog. 6: e1000861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuel S., Veloukas T., Papavasileiou A., Karaoglanidis G.S. (2012). Differences in frequency of transposable elements presence in Botrytis cinerea populations from several hosts in Greece. Plant Dis. 96: 1286–1290. [DOI] [PubMed] [Google Scholar]
- Sauerbrunn N., Schlaich N.L. (2004). PCC1: A merging point for pathogen defence and circadian signalling in Arabidopsis. Planta 218: 552–561. [DOI] [PubMed] [Google Scholar]
- Schumacher J., Pradier J.-M., Simon A., Traeger S., Moraga J., Collado I.G., Viaud M., Tudzynski B. (2012). Natural variation in the VELVET gene bcvel1 affects virulence and light-dependent differentiation in Botrytis cinerea. PLoS One 7: e47840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen X., Alam M., Fikse F., Rönnegård L. (2013). A novel generalized ridge regression method for quantitative genetics. Genetics 193: 1255–1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siewers V., Viaud M., Jimenez-Teja D., Collado I.G., Gronover C.S., Pradier J.-M., Tudzynski B., Tudzynski P. (2005). Functional analysis of the cytochrome P450 monooxygenase gene bcbot1 of Botrytis cinerea indicates that botrydial is a strain-specific virulence factor. Mol. Plant Microbe Interact. 18: 602–612. [DOI] [PubMed] [Google Scholar]
- Sim S.-C., Durstewitz G., Plieske J., Wieseke R., Ganal M.W., Van Deynze A., Hamilton J.P., Buell C.R., Causse M., Wijeratne S., Francis D.M. (2012). Development of a large SNP genotyping array and generation of high-density genetic maps in tomato. PLoS One 7: e40563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smale M. (1996). Understanding global trends in the use of wheat diversity and international flows of wheat genetic resources. http://ageconsearch.umn.edu/record/7670
- Staats M., van Kan J.A. (2012). Genome update of Botrytis cinerea strains B05.10 and T4. Eukaryot. Cell 11: 1413–1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefanato F.L., Abou-Mansour E., Buchala A., Kretschmer M., Mosbach A., Hahn M., Bochet C.G., Métraux J.P., Schoonbeek H.J. (2009). The ABC transporter BcatrB from Botrytis cinerea exports camalexin and is a virulence factor on Arabidopsis thaliana. Plant J. 58: 499–510. [DOI] [PubMed] [Google Scholar]
- Stukenbrock E.H., McDonald B.A. (2008). The origins of plant pathogens in agro-ecosystems. Annu. Rev. Phytopathol. 46: 75–100. [DOI] [PubMed] [Google Scholar]
- Talas F., Kalih R., Miedaner T., McDonald B.A. (2016). Genome-wide association study identifies novel candidate genes for aggressiveness, deoxynivalenol production, and azole sensitivity in natural field populations of Fusarium graminearum. Mol. Plant Microbe Interact. 29: 417–430. [DOI] [PubMed] [Google Scholar]
- Tanksley S.D. (2004). The genetic, developmental, and molecular bases of fruit size and shape variation in tomato. Plant Cell 16 (Suppl): S181–S189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanksley S.D., McCouch S.R. (1997). Seed banks and molecular maps: Unlocking genetic potential from the wild. Science 277: 1063–1066. [DOI] [PubMed] [Google Scholar]
- Ten Have A., Mulder W., Visser J., van Kan J.A. (1998). The endopolygalacturonase gene Bcpg1 is required for full virulence of Botrytis cinerea. Mol. Plant Microbe Interact. 11: 1009–1016. [DOI] [PubMed] [Google Scholar]
- Ten Have A., van Berloo R., Lindhout P., van Kan J.A. (2007). Partial stem and leaf resistance against the fungal pathogen Botrytis cinerea in wild relatives of tomato. Eur. J. Plant Pathol. 117: 153–166. [Google Scholar]
- Tiffin P., Moeller D.A. (2006). Molecular evolution of plant immune system genes. Trends Genet. 22: 662–670. [DOI] [PubMed] [Google Scholar]
- Valette-Collet O., Cimerman A., Reignault P., Levis C., Boccara M. (2003). Disruption of Botrytis cinerea pectin methylesterase gene Bcpme1 reduces virulence on several host plants. Mol. Plant Microbe Interact. 16: 360–367. [DOI] [PubMed] [Google Scholar]
- Vleeshouwers V.G., Oliver R.P. (2014). Effectors as tools in disease resistance breeding against biotrophic, hemibiotrophic, and necrotrophic plant pathogens. Mol. Plant Microbe Interact. 27: 196–206. [DOI] [PubMed] [Google Scholar]
- Weyman P.D., Pan Z., Feng Q., Gilchrist D.G., Bostock R.M. (2006). A circadian rhythm-regulated tomato gene is induced by Arachidonic acid and Phythophthora infestans infection. Plant Physiol. 140: 235–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J.Q., Sakthikumar S., Dong C., Zhang P., Cuomo C.A., Park R.F. (2017). Comparative genomics integrated with association analysis identifies candidate effector genes corresponding to Lr20 in phenotype-paired Puccinia triticina isolates from Australia. Front. Plant Sci. 8: 148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L.P., Khan A., Niño-Liu D., Foolad M.R. (2002). A molecular linkage map of tomato displaying chromosomal locations of resistance gene analogs based on a Lycopersicon esculentum x Lycopersicon hirsutum cross. Genome 45: 133–146. [DOI] [PubMed] [Google Scholar]
- Zhang W., Corwin J. A., Copeland D., Feusier J., Eshbaugh R., Chen F., Atwell S., Kliebenstein D. J. (2017). Plastic transcriptomes stabilize immunity to pathogen diversity: The jasmonic acid and salicylic acid networks within the Arabidopsis/Botrytis pathosystem. Plant Cell 29: 2727–2752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zipfel C., Robatzek S., Navarro L., Oakeley E.J., Jones J.D., Felix G., Boller T. (2004). Bacterial disease resistance in Arabidopsis through flagellin perception. Nature 428: 764–767. [DOI] [PubMed] [Google Scholar]
- Züst T., Agrawal A.A. (2017). Trade-offs between plant growth and defense against insect herbivory: An emerging mechanistic synthesis. Annu. Rev. Plant Biol. 68: 513–534. [DOI] [PubMed] [Google Scholar]