The extracellular space of Arabidopsis leaves contains diverse small RNAs, both inside and outside of extracellular vesicles, with the former being enriched in 10- to 17-nucleotide–long “tiny” RNAs.
Abstract
Small RNAs (sRNAs) that are 21 to 24 nucleotides (nt) in length are found in most eukaryotic organisms and regulate numerous biological functions, including transposon silencing, development, reproduction, and stress responses, typically via control of the stability and/or translation of target mRNAs. Major classes of sRNAs in plants include microRNAs (miRNAs) and small interfering RNAs (siRNAs); sRNAs are known to travel as a silencing signal from cell to cell, root to shoot, and even between host and pathogen. In mammals, sRNAs are transported inside extracellular vesicles (EVs), which are mobile membrane-bound compartments that participate in intercellular communication. In addition to sRNAs, EVs carry proteins, lipids, metabolites, and potentially other types of nucleic acids. Here we report that Arabidopsis (Arabidopsis thaliana) EVs also contain diverse species of sRNA. We found that specific miRNAs and siRNAs are preferentially loaded into plant EVs. We also report a previously overlooked class of “tiny RNAs” (10 to 17 nt) that are highly enriched in EVs. This RNA category of unknown function has a broad and very diverse genome origin and might correspond to degradation products.
INTRODUCTION
Small RNAs (sRNAs) are important 21- to 24-nucleotide (nt) noncoding signaling molecules involved in a wide variety of processes, including plant development, reproduction, and defense (Samad et al., 2017). sRNAs can be divided into two categories: microRNAs (miRNAs) and small interfering RNAs (siRNAs), based on the differences in their biogenesis and mode of action. miRNAs originate from a single-stranded, self-complementary, noncoding RNA that forms a hairpin structure. By contrast, siRNAs originate from a double-stranded RNA molecule synthesized by RNA-DEPENDENT RNA POLYMERASES. siRNAs can further be divided into two main categories: heterochromatic siRNAs (hc-siRNAs) and phased siRNAs (phasiRNAs), including trans-acting siRNAs (tasiRNAs). Both of these double-stranded RNA structures are recognized by Dicer-like proteins (DCL), which cleave these RNAs into defined length products. One strand of these products is then selectively loaded onto an ARGONAUTE protein and incorporated into the RNA-induced Silencing Complex (RISC). The RISC uses the sRNA in a sequence-homology–dependent manner to negatively regulate targets, typically mRNAs (Borges and Martienssen, 2015).
sRNAs are often mobile and function in non-cell–autonomous silencing, which can be either local or systemic. Local RNA silencing occurs among groups of adjacent cells and can gradually spread from cell to cell (Marín-González and Suárez-López, 2012; Dunoyer et al., 2013). Systemic silencing occurs over long distances and can spread throughout an entire plant. Although there are several documented cases of mobile sRNAs in plants, the mechanisms by which these RNAs move has yet to be clearly established. Local RNA silencing is thought to involve the transport of RNAs through plasmodesmata with the aid of plasmodesmata-interacting proteins. The same is thought to be true for systemic silencing, although this process also likely requires transport of RNA into and through the phloem. Systemic RNA silencing progresses in a vascular-like pattern reminiscent of the phloem and is graft-transmissible, moving bidirectionally up and down the plant in a manner that is heavily dependent on sink and source relationships (Lee and Frank, 2018).
Plant sRNAs are also known to move from plant cells into pathogens. Through a phenomenon known as Host-Induced Gene Silencing (HIGS), sRNAs produced in a plant cell can regulate the expression of genes in an invading pathogen or parasite. In recent years, HIGS has become a powerful tool for engineering resistance in crop plants to nematodes, insects, fungi, oomycetes, and parasitic weeds (Cai et al., 2018). Although multiple studies have developed artificial HIGS systems, the phenomenon can occur naturally as well, such as in the transfer of miRNAs miR166 and miR159 from cotton into the hemibiotrophic fungus, Verticillium dahliae (Zhang et al., 2016), and the transfer of siRNAs from Arabidopsis (Arabidopsis thaliana) into Botrytis cinerea (Cai et al., 2018). How this translocation is mediated is not well understood.
In mammals, extracellular vesicles (EVs) are known to mediate long-distance transport of both coding and noncoding RNAs. RNAs protected within the lumen of an EV are transported to distant cells, where they retain their function after delivery (Ratajczak et al., 2006; Valadi et al., 2007; Mittelbrunn et al., 2011). The RNA contents of mammalian EVs largely reflects that of their cell type of origin, but can show enrichment for certain mRNAs and miRNAs. The overrepresentation of specific sequences inside EVs suggests an active mechanism for loading RNAs into these compartments. Usually, miRNAs make up a sizable percentage of the RNAs within EVs. However, other classes of small noncoding RNAs are also present, including transfer RNAs (tRNAs), rRNAs, and fragments originating from both coding and noncoding RNAs. These other sRNAs are often highly enriched in EVs as compared with their cell of origin. The exact role of other noncoding RNAs and RNA fragments in EVs has yet to be determined. They may function in signaling or they may represent waste products that EVs remove from the cell (Bellingham et al., 2012; Nolte ’t Hoen et al., 2012; Huang et al., 2013; Chevillet et al., 2014; Van Balkom et al., 2015).
Although the RNA content of mammalian EVs has been extensively characterized, the RNA content of plant EVs has not been carefully assessed. To address this knowledge gap, we optimized methods for purifying EVs from plant apoplastic wash fluid (Rutter and Innes, 2017), performed sRNA-seq analysis on EV RNA, and compared the resulting data to sRNAs in the source leaf tissue and EV-depleted aplastic wash fluid. These analyses revealed that plant EVs are enriched in single-stranded sRNAs, especially a class of RNAs ranging in size from 10 to 17 nts. These RNAs seem to represent degradation products of sRNA production. Their abundance in plant EVs raises the question of whether they serve a function in either intercellular and/or inter-kingdom communication.
RESULTS AND DISCUSSION
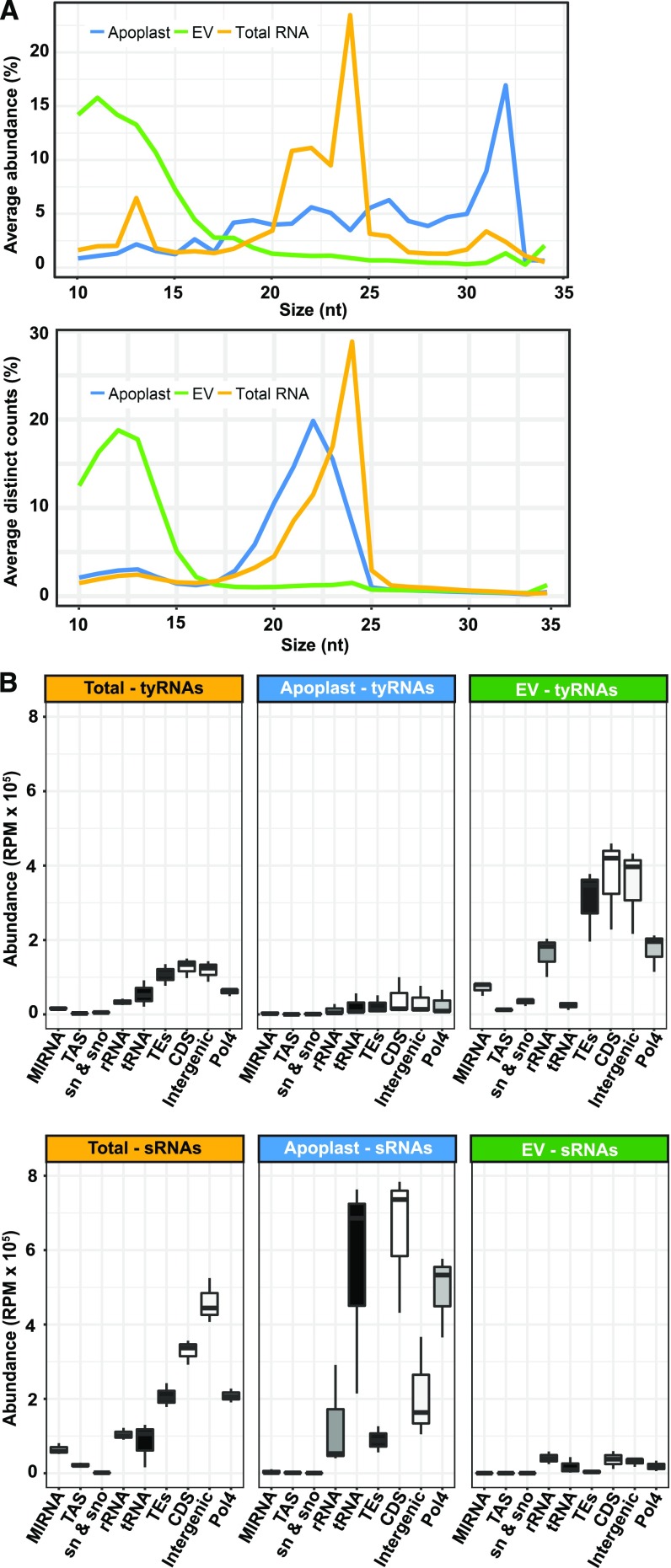
To analyze the sRNA content of EVs, we constructed sRNA libraries from three biological replicates of Arabidopsis rosette leaves (“total RNA,” henceforth), EVs, and apoplastic wash fluids depleted of EVs (“apoplast,” henceforth), as described in Rutter and Innes (2017). After an initial step of standard read trimming (adaptor removal and size selection from 18 to 34 nt), we observed that only 20% of the reads from EV samples were retained. This very low rate of retention was due to a large proportion of very short reads, from 10- to 17-nt long, present in the EV samples. We named these unusually short reads “tiny RNAs” (tyRNAs). Changing the trimming criteria to retain any read from 10 to 34 nt allowed us to achieve an average of ∼60% read retention (Supplemental Table 1). The size distribution of the EV and apoplast samples was unexpected (Figure 1A). Total RNA samples had the typical size distribution of Arabidopsis sRNAs, a peak at 24 nt followed by secondary peaks at 21 and 22 nt. However, EV samples had a high proportion of tyRNA reads between 10 and 15 nt and no peak in the 20-nt range, while apoplast samples had a high proportion of longer reads, between 30 and 32 nt (Figure 1A, left). When considering only distinct reads, where tags with the exact same sequence were counted only once (Figure 1A, right), EV samples continued to show an enrichment for tyRNAs. This suggests that the tyRNAs present in EVs were not only abundant, but also highly diverse in their sequence composition. Comparatively, apoplastic sRNAs were not enriched for tyRNA sequences. This strongly argues that tyRNA reads were not a result of degradation occurring during RNA isolation or library preparation, as these steps were performed in parallel for all RNA samples.
Figure 1.
EVs are Enriched in TyRNA Sequences Derived from Diverse Sources.
(A) Size distribution of sRNAs mapping to the genome; the percentage of each size class was calculated for each source of data, represented by the average of three replicate libraries. The x axis indicates the sRNA size and the y axis indicates its proportion. Left: sRNA sizes and proportions, as measured by total abundance of the reads; right: sRNA sizes and proportions, as calculated by distinct reads (unique sequences).
(B) The abundance of reads mapping to nine different features of the Arabidopsis genome. These include the following from left to right: miRNA precursors; TAS precursors; snRNA and snoRNAs; rRNAs; tRNAs; TEs; CDSs (including introns and UTRs); intergenic sequences; and siRNA precursors dependent on Pol4. RPM, reads per million.
Tiny RNAs Mostly Originate from Coding Sequences, Transposable Elements, and Intergenic Regions
To understand the genomic origin of these tyRNAs, we mapped these reads to different features of the Arabidopsis genome, including genes encoding miRNA precursors (MIRNAs), genes giving rise to tasiRNAs (TAS genes), transposable elements (TEs), rRNAs, tRNAs, small nuclear and small nucleolar RNAs (snRNAs and snoRNAs), coding sequences (CDSs), potential RNA Polymerase IV products (Pol4; as defined in Blevins et al., 2015) and intergenic regions (Figure 1B). For reads that mapped to more than one feature, we counted each feature once. We performed the same analysis with the sRNA reads (18 to 34 nt) and observed that relative to sRNAs, tyRNAs were enriched in sequences that map to MIRNAs and TEs (Figure 1B). Although the EV samples contained a higher abundance of tyRNA reads, tyRNAs from both total RNA and EV samples mapped to a similar distribution of features. In both samples, the majority of tyRNAs mapped back to TEs, CDSs, and intergenic regions, which combined, comprise the largest proportion of the Arabidopsis genome (Figure 1B). We also detected high levels of tyRNAs derived from Pol4 precursors and rRNA regions, which represent 14% and 0.12% of the genome, respectively. Typically, rRNA represents ∼80% of total leaf RNA, so the relatively low frequency of rRNA reads in each library indicates that they were not significantly contaminated with degraded cellular RNA.
MIRNA-Derived tyRNAs Are Not Randomly Distributed
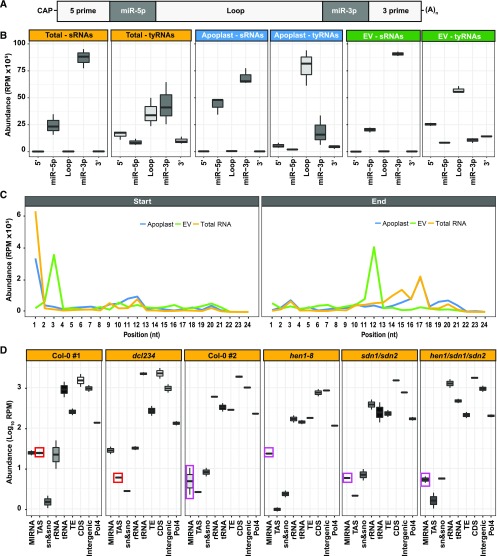
To determine whether tyRNAs are products of sRNA processing, we next analyzed their relative position in noncoding RNAs. The abundance of tyRNAs derived from miRNA precursors was almost twice that of snRNAs, snoRNAs, and tRNAs. Because this posits a clear preferential accumulation of MIRNA-derived tyRNAs in EVs, we analyzed the location of tyRNA sequences within miRNA precursors. In addition to a 5′ cap and a 3′ poly-A tail, miRNA precursors can be subdivided into the following segments: the 5′ region, miR-5p, the loop region, miR-3p, and the 3′ region (Figure 2A). sRNAs (18 to 34 nt) and tyRNAs (10 to 17 nt) mapped to these regions with different patterns of abundance (Figure 2B). In all of our samples, sRNAs originated almost exclusively from the miR-3p and miR-5p regions, which correspond to the mature miRNA/miRNA* duplex. tyRNAs, however, originated mostly from the loop region of the precursor, followed in abundance by the 5′ and 3′ regions (Figure 2B). These results suggest that MIRNA-derived tyRNAs are largely products of the remnant pieces after cleavage by DCL1. The terminal regions of miRNA precursors are known to undergo exonucleolytic decay via a process that includes RISC-INTERACTING CLEARING 3′-5′ EXORIBONUCLEASES to clear 5′ fragments (Yu et al., 2017b; Zhang et al., 2017) and EXORIBONUCLEASE4 to clear 3′ fragments (Souret et al., 2004). However, the process by which the leftover loop region is degraded is less well examined, but apparently results in the production of abundant tyRNAs.
Figure 2.
EV tyRNAs Are Derived from Specific Regions of Precursor RNAs.
(A) miRNA precursor structure, including from left to right, 5′ region, mature miRNA miR-5p, loop region, mature miRNA miR-3p, and 3′ region.
(B) EV tyRNAs are enriched in sequences that map to the loop region of miRNA precursors. The abundance of sRNAs and tyRNAs mapping to different regions of a miRNA precursor, as described in (A). The y axis represents abundance, in reads per million (RPM).
(C) EV tyRNAs mapping to mature miRNAs are cut out of the central region. Plots show the relative position of 5′ ends (left plot) or 3′ ends (right plot) of tyRNAs relative to nucleotide position within the parent miRNA. RPM, reads per million.
(D) Mutations that affect abundance of specific subclasses of sRNAs have a corresponding influence on tyRNA abundance. The abundance of total leaf tyRNAs mapping to different features of the Arabidopsis genome. These include the following from left to right: miRNA precursors, TAS precursors, snRNA and snoRNAs; rRNAs; tRNAs; TEs; CDSs (including introns and UTRs); intergenic sequences; and known Pol4 precursors. The y axis represents abundance, in reads per million (RPM), in a logarithmic (Log10) scale. Colored boxes highlight tyRNAs affected by mutations.
tyRNAs also mapped to the miRNA/miRNA* duplex, and we next checked the position of the tyRNAs within these ∼21- or 22-nt molecules (Figure 2C). Because the miRNA and miRNA* designations were removed from miRBase, we treated both sides of the duplex equally. The majority of EV tyRNAs that mapped to miRNAs originated from the center of the miRNA sequence (i.e. nucleotides 3 to 12; Figure 2C). By contrast, tyRNAs from total leaf libraries and apoplast mostly started at position 1 of the miRNA, followed by position 11, and they have an end position more widely distributed, from positions 13 to 21. These observations suggest that EV tyRNAs are not a random sampling of total cell tyRNAs, but are instead derived from a specific packaging process.
We also analyzed tyRNA abundances in a dcl234 triple mutant, which is deficient for the production of 22- and 24-nt siRNAs, including tasiRNAs (Henderson et al., 2006), but is unaffected in miRNA biogenesis. Consistent with expectations, MIRNA-derived tyRNA accumulation did not differ between the dcl234 triple mutant and wild-type Col-0. However, we observed a clear reduction in TAS-derived tyRNAs (Figure 2D), indicating that these tyRNAs are likely derived from tasiRNA precursors.
Next, we examined data previously generated from hen1, small DNA degrading nuclease (sdn1), sdn2, and hen1 sdn1 sdn2 mutants (Yu et al., 2017a; samples included in the PRJNA251351 Gene Expression Omnibus data set). HUA ENHANCER1 (HEN1) is a methyl transferase that stabilizes specific sRNAs by adding a methyl group at the 3′ end, which protects them from degradation (Li et al., 2005). When HEN1 is absent, sRNAs undergo an uridylation process that may promote the process of RNA decay. More typically, and in wild-type plants, sRNAs are degraded by exonucleases encoded by SMALL RNA DEGRADING NUCLEASE (SDN1) and SDN2. We observed that MIRNA-derived tyRNAs increased in hen1 mutants relative to wild-type Col-0, when miRNAs are destabilized, but remained at wild-type levels in sdn1 sdn2 and hen1 sdn1 sdn2 mutants, which lack the exonucleases responsible for miRNA degradation (Figure 2D). These results support our hypothesis that MIRNA-derived tyRNAs are degradation products.
1-Hit tyRNAs Map to Coding CDSs and Intergenic Regions
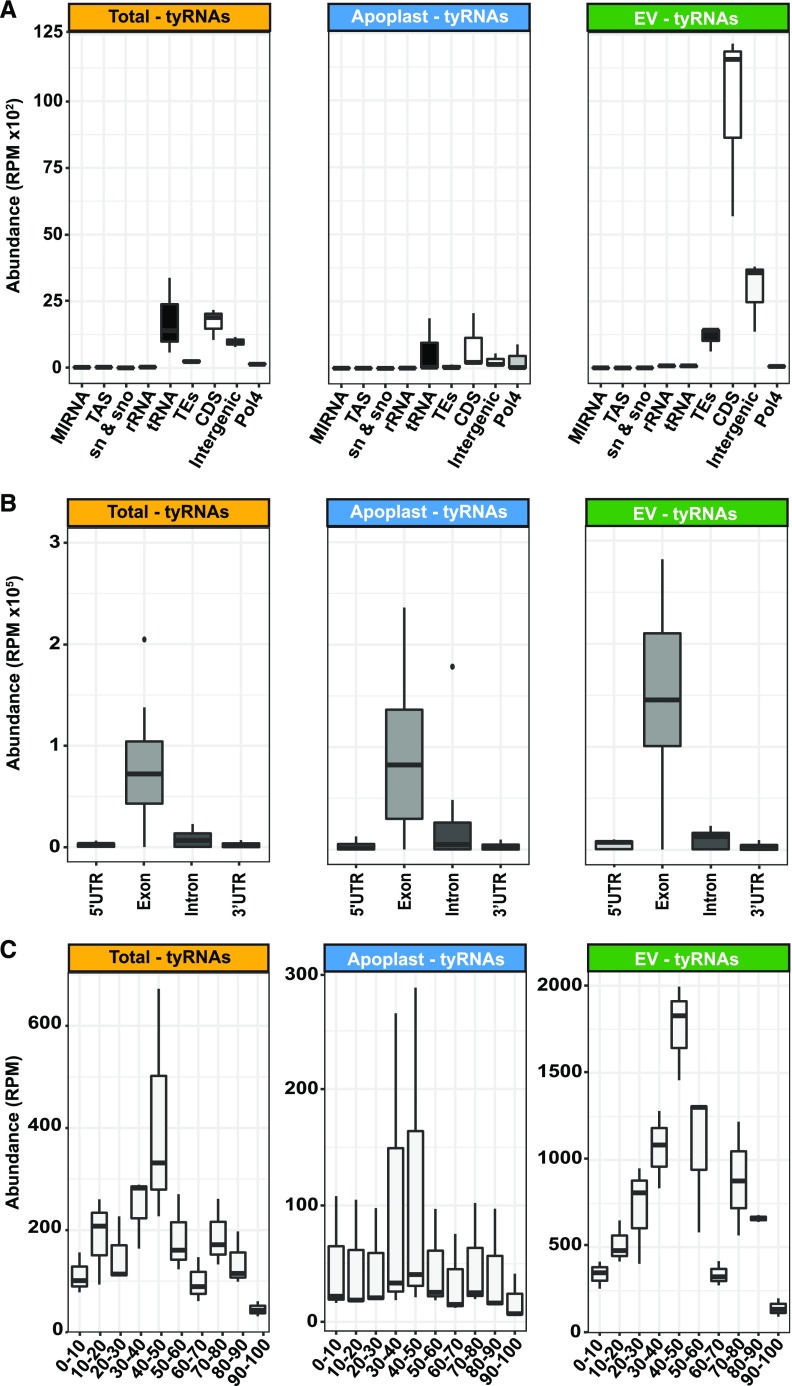
Due to the short length of tyRNAs, most tyRNAs map to multiple locations in the genome. To better determine their origin, we repeated the previously described analyses using only reads that map once to the genome (hereafter, “1-hit” reads). These 1-hit tyRNAs have a different size distribution, ranging from 13- to 17-nt long, with the majority being 16 to 17 nt (Supplemental Figure 1A).
The majority of the 1-hit tyRNAs mapped to coding (CDS) and intergenic regions (Figure 3A). We examined in more detail the exact origin of the CDS-derived tyRNAs by dividing each gene into the 5′ untranslated region (UTR), exons, introns and 3′ UTR. The 1-hit tyRNAs originated mostly from the sense strand of exons (Figure 3B), with no difference regarding the size of the tyRNAs (Supplemental Figure 1B). These results were independent of the sample type, suggesting that most of the CDS-derived tyRNAs originate from mature mRNAs.
Figure 3.
tyRNAs that Map to a Single Position in the Genome are Enriched in Exon-Derived Sense-Strand Sequences.
(A) The abundance of 1-hit tyRNAs mapping to different features of the Arabidopsis genome. RPM, reads per million.
(B) The abundance of 1-hit tyRNAs mapping to 5′ UTRs, exons, introns, 3′ UTRs, and intergenic regions. The y axis represents abundance, in RPM.
(C) The abundance of 1-hit tyRNAs mapping to different relative positions in mRNAs. The x axis represents the relative position in full-length mRNA, expressed in percentage of the total length, and the y axis represents abundance, in RPM. Note that scales are different for each plot.
To further characterize the origin of the exon-derived tyRNAs, we analyzed their 5′ positions relative to the full-length mRNA, expressed as a percentage of the total length. In all samples, we observed a consistent peak originating at the center of the mRNA (Figure 3C). This persistent peak was more prominent in the EV samples than in whole leaf and apoplast samples. This pattern suggests that mRNAs may be first degraded by 5′ and 3′ exonucleases, with tyRNAs being derived from what is left over.
EVs Preferentially Load Specific miRNAs
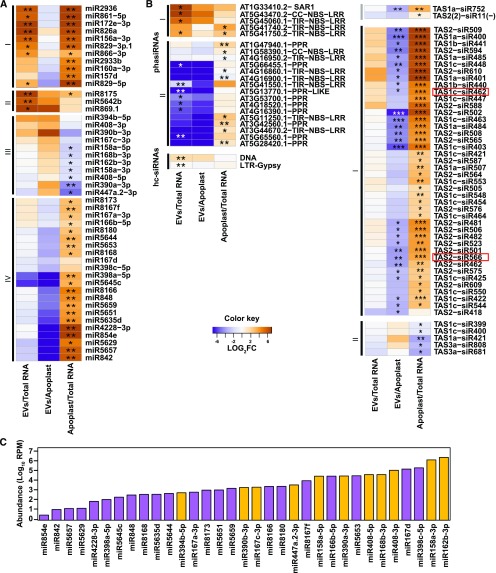
We next investigated whether sRNAs accumulate differentially in EVs relative to the apoplast and total leaf samples, focusing on four major classes of sRNAs (hc-siRNAs, phasiRNAs, tasiRNAs, and miRNAs; Figure 4). The most differentially represented sRNAs were miRNAs (Figure 4A). Group I consisted of miRNAs that were more abundant in EVs and apoplast samples as compared with total RNA. While the miRNAs within this group seem to be secreted from the cell, there is no specific accumulation in EVs. Group II miRNAs accumulated to high levels in EVs and were present at slightly lower levels in the apoplast samples, suggesting some accumulation in EVs. The miRNAs in group III were depleted in the apoplast samples relative to total RNA and EVs, suggesting that group III RNAs are preferentially secreted via EVs. The miRNAs in group IV had an opposite accumulation pattern, low in EVs as compared with apoplast and high in apoplast as compared with total RNA samples. These results indicate that a distinct subset of miRNAs are preferentially loaded into the EVs.
Figure 4.
A Subset of sRNAs Display Differential Abundance in EVs Versus Total Leaf RNA or Apoplastic RNA.
(A) Select miRNAs are enriched or depleted in EVs. Relative abundance of miRNAs in EVs and apoplast compared with abundance in total leaf RNA; the relative abundance is expressed as a heat map (see Key in center), with the samples being compared indicated below each heat map (*Q value ≤ 0.05 and **Q value ≤ 0.01).
(B) Select siRNAs are enriched in EVs. Heat map indicates relative abundance of siRNAs (including phasiRNAs, tasiRNAs, and hc-siRNAs) in the indicated samples. Boxed tasiRNAs indicate sRNAs taken up by B. cinerea (shown in Cai et al., 2018; *Q value ≤ 0.05, **Q value ≤ 0.01, and ***Q value ≤ 0.001).
(C) miRNA abundance does not correlate with EV enrichment. Absolute abundance in EVs of select miRNAs expressed in reads per million (RPM) on a logarithmic scale, including miRNAs enriched in EVs (orange) and those depleted in EVs (purple) relative to apoplastic RNA.
The four patterns of preferential miRNA secretion described above did not correlate with total miRNA abundance in leaves. miRNAs with similar abundances in the total leaf sample were represented in both groups III and IV (i.e. those that accumulated to both low and high levels in the EV samples; Figure 4C). For example, miRNAs miR390b-3p, miR167c-3p, miR8166, and miR8180 had a similar abundance in total RNA samples, but the first two were abundant in EV samples while the latter two displayed a low abundance in EVs.
Combined, our findings hint at two separate pathways for the secretion of miRNAs. While miRNAs present in groups I and II may be secreted through either pathway, with a slight preference for EV-dependent secretion in group II, miRNAs in group III are strongly dependent on EVs for secretion. In contrast, miRNAs in group IV are secreted primarily through an EV-independent pathway. There may be specific sorting mechanisms for loading miRNAs into these separate pathways. Whether such specificity might be directed by the miRNA sequence or duplex structure, or by something about its target or function, remains unknown.
siRNAs Are Poorly Represented in EVs
We next assessed the abundance of full-length siRNAs in EVs, including phasiRNAs, tasiRNAs, and hc-siRNAs. For phasiRNAs, we analyzed all members of families of genes encoding nucleotide-binding site leucine-rich repeat proteins, pentatricopeptide repeat proteins, and SUPPRESSOR OF AUXIN RESISTANCE proteins, which are the primary sources of phasiRNAs in Arabidopsis (Supplemental Data Set 1). We were able to identify 21 phasiRNA-producing loci that were differentially accumulated in one of our three comparisons (Figure 4B). These loci can be divided into two groups: Group I had high accumulation in EV and apoplast samples, as compared with total RNA; and group II had a low accumulation in EV samples as compared with apoplast and total RNA samples, and a high accumulation in apoplast as compared with total RNAs. Similarly, we analyzed the accumulation of the 24 different Arabidopsis tasiRNAs that have been identified (Zhang et al., 2014; Supplemental Data Set 1), and found only two that were differentially accumulated (Figure 4B, upper right), with low accumulation levels in EVs. We also analyzed 21- and 22-nt RNAs derived from the eight TAS genes identified in the Arabidopsis genome for their specific differential accumulation in our samples (Figure 4B, lower right). We observed that none of these TAS-derived sRNAs accumulated in EVs. Instead, all of the differentially accumulated TAS-derived sRNAs preferentially accumulated in the apoplast. This may indicate that phasiRNAs and tasiRNAs are preferentially directed to an apoplastic secretion pathway that is independent of EVs.
hc-siRNAs largely originate from TEs and are thus repetitive in their nature, confounding analyses that require knowing their exact origin. Some hc-siRNAs may map to more than one TE with all the mapping locations corresponding to a single TE superfamily. Using hc-siRNAs that mapped within single superfamilies (Supplemental Data Set 1), we measured hc-siRNA levels and observed no clear patterns of accumulation in EV samples. Therefore, unlike miRNAs that demonstrated specificity in their EV accumulation, there was no evidence of specificity in EV-localized sRNAs from loci generating either hc-siRNA or any other siRNA analyzed here.
CONCLUSIONS
Here we have shown that EVs are highly enriched in sRNAs 10 to 17 nt in length, which we have named “tiny RNAs.” tyRNAs appear to be degradation products derived from multiple sources, including mRNAs, primary miRNAs, siRNAs, tasiRNAs, and hcRNAs. Whether this class of RNAs has a cellular function, or represent a waste- or by-product of cellular metabolism, remains unknown.
One potential function of tyRNAs could be as small activating RNAs (saRNAs). RNA activation is a mechanism by which sRNAs, usually 18- to 21-nt long, can activate gene transcription, and was first identified in human cells more than a decade ago (Li et al., 2006). More recently, several studies have identified animal saRNAs that are able to positively regulate gene expression by targeting UTRs (Vasudevan et al., 2007; Vasudevan and Steitz, 2007; Ørom et al., 2008). Since then, more than 15 examples of saRNAs have been described in animal model systems, such as rats and primates, in vivo and in vitro (Huang et al., 2010). saRNAs bind to promoter regions in a wide range of positions relative to their transcription starting site (Li et al., 2006; Janowski et al., 2007), as well as the 3′ UTR (Yue et al., 2010).
Here we also described that some miRNAs are specifically loaded into EVs, which reinforces the theory that sRNAs use EVs for long-distance movement through the plant, and possibly as a cross-kingdom delivery system. Recently it has been described that plants secrete EVs to transfer sRNAs into fungal pathogens (Cai et al., 2018). Specifically, it has been shown that two plant-derived tasiRNAs from TAS1c and TAS2 genes are transferred to B. cinerea via detergent-sensitive particles to target genes involved in vesicle trafficking. We also found that some TAS1- and TAS2-derived siRNAs are secreted out of the cell, but mainly through an apoplastic secretion mechanism. The discrepancies between our findings and those of Cai et al. (2018) may be due to differences in isolation methods, as Cai et al. (2018) used 100,000 × g as the relative centrifugal force rather than 40,000 × g to isolate EVs and they did not purify the vesicles away from any contaminating protein complexes. It may be that the tasiRNAs detected in Cai et al. (2018) were associated with other extracellular particles separate and distinct from EVs. This makes sense if, as our data suggests, plants employ multiple mechanisms for the secretion and long-distance transport of sRNAs.
METHODS
Plant Materials and Growth Conditions
Arabidopsis (Arabidopsis thaliana) seeds were germinated on 0.5× Murashige & Skoog medium containing 0.8% agar as described in Rutter and Innes (2017). The seeds were stored at 4°C for 2 d and then moved to short-day conditions illuminated using GE HI-LUMEN XL Starcoat 32 watt fluorescent bulbs (a 50:50 mixture of 3,500 K and 5,000 K spectrum bulbs; 9-h days, 22°C, 150 µEm−2 s−1). The seedlings were transferred after one week to Pro-Mix B Biofungicide potting mix supplemented with Osmocote slow-release fertilizer (14-14-14).
EV Isolation and Purification
Arabidopsis EVs were isolated from apoplastic wash and purified on an iodixanol density gradient, as described in Rutter and Innes (2017). Briefly, 6-week-old Arabidopsis rosettes were vacuum-infiltrated with Vesicle Isolation Buffer (VIB; 20 mM 2-[n-morpholino]ethanesulfonic acid, 2 mM CaCl2, and 0.01 M NaCl, pH 6.0). The rosettes were gently blotted to remove excess buffer and packaged inside needleless, 30-mL syringes. The syringes were centrifuged for 20 min at 700 × g (2°C, JA-14 rotor, Avanti J-20 XP Centrifuge; Beckman Coulter). The apoplastic wash collected from the rosettes was then filtered through a 0.22-μm membrane and centrifuged successively at 10,000 × g for 30 min to remove any large particles, 40,000 × g for 60 min to pellet the EVs and again at 40,000 × g to wash and re-pellet the EVs (2°C, SW41, Optima XPN-100 Ultracentrifuge; Beckman Coulter). After the first 40,000 × g step, 5 mL of the supernatant was retained on ice for RNA isolation. The washed EV pellet was resuspended in 1 mL of cold, sterile VIB and loaded on top of a discontinuous iodixanol gradient (5%, 10%, 20% and 40%; Optiprep; Sigma-Aldrich). The gradient underwent centrifugation at 100,000 × g for 17 h (2°C, SW41, Optima XPN-100 Ultracentrifuge; Beckman Coulter). After centrifugation, the first 5 mL of the gradient were discarded. The next three fractions of 0.8 mL were collected. The fractions were diluted in cold, sterile VIB and re-pelleted at 100,000 × g for 60 min (2°C, TLA100.3, Optima TLX Ultracentrifuge; Beckman Coulter). The pellets were resuspended in cold, sterile VIB, combined and brought up to a total volume of 100 μL using VIB.
RNA Sample Preparation
RNA was isolated from three different sources: leaf tissue, EV-depleted supernatant, and purified EVs. Leaf RNA was isolated from 100 mg of leaf tissue harvested after collecting apoplastic wash, frozen, and ground to a powder using liquid nitrogen and suspended in 1 mL of TRIzol Reagent (Thermo Fisher Scientific). Supernatant was collected after the first 40,000 × g step, as described above, and combined with 0.1 volumes of 3 M sodium acetate and one volume of cold isopropyl alcohol. The supernatant sample was mixed briefly by vortexing and kept at −20°C for 1 h. After 1 h, the sample was centrifuged for 30 min at 12,000 × g and 4°C. The resulting pellet of RNA was resuspended in 1 mL of TRIzol Reagent. EV RNA was isolated from EVs purified on a discontinuous iodixanol gradient as described above. One-hundred microliters of EVs was added to 1 mL of TRIzol Reagent. Once all three samples were in TRIzol Reagent, RNA was extracted following the manufacturer’s instructions.
sRNA Libraries
sRNA libraries were constructed as described in Mathioni et al. (2017) using the NEBNext Small RNA Library Prep Set (no. E7330S; New England BioLabs). Due to the low abundance of RNA present in EVs and apoplast samples, 500 ng of total RNA was used as the starting material. Libraries were sequenced on a NextSeq instrument with single-end 75-bp reads at the Indiana University Center for Genomics and Bioinformatics.
Data Analysis
sRNA sequencing libraries were trimmed for adaptors using the software cutadapt v1.16 (Martin, 2011) with a minimum insert size of 10 nt and a maximum of 34 nt. Sequence quality was assessed using FastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/). Clean reads were aligned to the Arabidopsis genome (TAIR version 10), and all subsequent analyses were done using the software Bowtie2 (Langmead and Salzberg, 2012). For miRNA and tasiRNA analyses, the latest versions of miRBase (v22; Kozomara and Griffiths-Jones, 2014) and tasiRNA database (Zhang et al., 2014) were used, respectively. Mapping positions for miRNAs were assessed using the Bowtie2 output file. Differential accumulation analyses were performed using DEseq2 with default parameters, using not-normalized reads as input (Love et al., 2014) and graphical representations using the software ggplot2 (Wickham, 2009) in the R statistical environment. For box-and-whisker plots produced by ggplot2, the upper whisker represents the maximum value, or the top of the box plus 1.5 length of the box, whichever is less, while the lower whisker represents the minimum value, or the bottom of the box minus 1.5 length of the box whichever is greater.
Accession Numbers
All RNA sequencing data has been deposited into the Gene Expression Omnibus under the accession code GSE114696. Processed data can be accessed at https://mpss.danforthcenter.org/dbs/index.php?SITE=at_sRNA_EV.
Supplemental Data
Supplemental Figure 1. Characterization of 1-hit tyRNAs.
Supplemental Table 1. sRNA sequencing summary statistics.
Supplemental Data Set 1. Raw read count of full-length miRNAs and siRNAs sorted by sRNA class.
Dive Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
This work was supported by the United States National Science Foundation (grants IOS-1645745 and IOS-1842685 to R.W.I., and IOS-1842698 to B.C.M.). We thank Craig Pikaard for providing seed of dcl2/3/4 Arabidopsis triple mutant, and the Indiana University Physical Biochemistry Instrumentation Facility for access to ultracentrifuges and nanoparticle tracking equipment. We also thank the Indiana University Center for Genomics and Bioinformatics for assistance with the generation and analysis of sRNA-seq data.
AUTHOR CONTRIBUTIONS
P.B., B.D.R., R.W.I., and B.C.M. designed the research; P.B., B.D.R., and H.Z.K. performed the research; P.B. and R.P. analyzed the data; P.B., B.D.R., and R.W.I. wrote the article; all authors read and commented on the article.
Footnotes
Articles can be viewed without a subscription.
References
- Bellingham S.A., Coleman B.M., Hill A.F. (2012). Small RNA deep sequencing reveals a distinct miRNA signature released in exosomes from prion-infected neuronal cells. Nucleic Acids Res. 40: 10937–10949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blevins T., Podicheti R., Mishra V., Marasco M., Wang J., Rusch D., Tang H., Pikaard C.S. (2015). Identification of Pol IV and RDR2-dependent precursors of 24 nt siRNAs guiding de novo DNA methylation in Arabidopsis. eLife 4: e09591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borges F., Martienssen R.A. (2015). The expanding world of small RNAs in plants. Nat. Rev. Mol. Cell Biol. 16: 727–741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Q., Qiao L., Wang M., He B., Lin F.M., Palmquist J., Huang S.D., Jin H. (2018). Plants send small RNAs in extracellular vesicles to fungal pathogen to silence virulence genes. Science 360: 1126–1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevillet J.R., et al. (2014). Quantitative and stoichiometric analysis of the microRNA content of exosomes. Proc. Natl. Acad. Sci. USA 111: 14888–14893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunoyer P., Melnyk C., Molnar A., Slotkin R.K. (2013). Plant mobile small RNAs. Cold Spring Harb. Perspect. Biol. 5: 3–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson I.R., Zhang X., Lu C., Johnson L., Meyers B.C., Green P.J., Jacobsen S.E. (2006). Dissecting Arabidopsis thaliana DICER function in small RNA processing, gene silencing and DNA methylation patterning. Nat. Genet. 38: 721–725. [DOI] [PubMed] [Google Scholar]
- Huang X., et al. (2013). Characterization of human plasma-derived exosomal RNAs by deep sequencing. BMC Genomics 14: 319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang V., Qin Y., Wang J., Wang X., Place R.F., Lin G., Lue T.F., Li L.C. (2010). RNAa is conserved in mammalian cells. PLoS One 5: e8848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janowski B.A., Younger S.T., Hardy D.B., Ram R., Huffman K.E., Corey D.R. (2007). Activating gene expression in mammalian cells with promoter-targeted duplex RNAs. Nat. Chem. Biol. 3: 166–173. [DOI] [PubMed] [Google Scholar]
- Kozomara A., Griffiths-Jones S. (2014). miRBase: Annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 42: D68–D73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B., Salzberg S.L. (2012). Fast gapped-read alignment with Bowtie 2. Nat. Methods 9: 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.Y., Frank M. (2018). Plasmodesmata in phloem: Different gateways for different cargoes. Curr. Opin. Plant Biol. 43: 119–124. [DOI] [PubMed] [Google Scholar]
- Li J., Yang Z., Yu B., Liu J., Chen X. (2005). Methylation protects miRNAs and siRNAs from a 3′-end uridylation activity in Arabidopsis. Curr. Biol. 15: 1501–1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L.-C., Okino S.T., Zhao H., Pookot D., Place R.F., Urakami S., Enokida H., Dahiya R. (2006). Small dsRNAs induce transcriptional activation in human cells. Proc. Natl. Acad. Sci. USA 103: 17337–17342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love M.I., Huber W., Anders S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15: 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marín-González E., Suárez-López P. (2012). “And yet it moves”: Cell-to-cell and long-distance signaling by plant microRNAs. Plant Sci. 196: 18–30. [DOI] [PubMed] [Google Scholar]
- Martin M. (2011). Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 17: 10–12. [Google Scholar]
- Mathioni S.M., Kakrana A., Meyers B.C. (2017). Characterization of plant small RNAs by next generation sequencing. Curr. Protoc. Plant Biol. 2: 39–63. [DOI] [PubMed] [Google Scholar]
- Mittelbrunn M., Gutiérrez-Vázquez C., Villarroya-Beltri C., González S., Sánchez-Cabo F., González M.Á., Bernad A., Sánchez-Madrid F. (2011). Unidirectional transfer of microRNA-loaded exosomes from T cells to antigen-presenting cells. Nat. Commun. 2: 282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolte ’t Hoen E.N., Buermans H.P., Waasdorp M., Stoorvogel W., Wauben M.H., ’t Hoen P.A. (2012). Deep sequencing of RNA from immune cell-derived vesicles uncovers the selective incorporation of small non-coding RNA biotypes with potential regulatory functions. Nucleic Acids Res. 40: 9272–9285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ørom U.A., Nielsen F.C., Lund A.H. (2008). MicroRNA-10a binds the 5' UTR of ribosomal protein mRNAs and enhances their translation. Mol. Cell 30: 460–471. [DOI] [PubMed] [Google Scholar]
- Ratajczak J., Miekus K., Kucia M., Zhang J., Reca R., Dvorak P., Ratajczak M.Z. (2006). Embryonic stem cell-derived microvesicles reprogram hematopoietic progenitors: Evidence for horizontal transfer of mRNA and protein delivery. Leukemia 20: 847–856. [DOI] [PubMed] [Google Scholar]
- Rutter B.D., Innes R.W. (2017). Extracellular vesicles isolated from the leaf apoplast carry stress-response proteins. Plant Physiol. 173: 728–741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samad A.F.A., Sajad M., Nazaruddin N., Fauzi I.A., Murad A.M.A., Zainal Z., Ismail I. (2017). MicroRNA and transcription factor: Key players in plant regulatory network. Front. Plant Sci. 8: 565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souret F.F., Kastenmayer J.P., Green P.J. (2004). AtXRN4 degrades mRNA in Arabidopsis and its substrates include selected miRNA targets. Mol. Cell 15: 173–183. [DOI] [PubMed] [Google Scholar]
- Valadi H., Ekström K., Bossios A., Sjöstrand M., Lee J.J., Lötvall J.O. (2007). Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 9: 654–659. [DOI] [PubMed] [Google Scholar]
- Van Balkom B.W.M., Eisele A.S., Pegtel D.M., Bervoets S., Verhaar M.C. (2015). Quantitative and qualitative analysis of small RNAs in human endothelial cells and exosomes provides insights into localized RNA processing, degradation and sorting. J. Extracell. Vesicles 4: 26760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasudevan S., Steitz J.A. (2007). AU-rich-element-mediated upregulation of translation by FXR1 and Argonaute 2. Cell 128: 1105–1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasudevan S., Tong Y., Steitz J.A. (2007). Switching from repression to activation: microRNAs can up-regulate translation. Science 318: 1931–1934. [DOI] [PubMed] [Google Scholar]
- Wickham H. (2009). ggplot2: Elegant Graphics for Data Analysis. Springer, New York. [Google Scholar]
- Yu Y., et al. (2017a). ARGONAUTE10 promotes the degradation of miR165/6 through the SDN1 and SDN2 exonucleases in Arabidopsis. PLoS Biol. 15: e2001272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y., Jia T., Chen X. (2017b). The “how” and “where” of plant microRNAs. New Phytol. 216: 1002–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue X., Schwartz J.C., Chu Y., Younger S.T., Gagnon K.T., Elbashir S., Janowski B.A., Corey D.R. (2010). Transcriptional regulation by small RNAs at sequences downstream from 3′ gene termini. Nat. Chem. Biol. 6: 621–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C., Li G., Zhu S., Zhang S., Fang J. (2014). tasiRNAdb: A database of ta-siRNA regulatory pathways. Bioinformatics 30: 1045–1046. [DOI] [PubMed] [Google Scholar]
- Zhang T., Zhao Y.L., Zhao J.H., Wang S., Jin Y., Chen Z.Q., Fang Y.Y., Hua C.L., Ding S.W., Guo H.S. (2016). Cotton plants export microRNAs to inhibit virulence gene expression in a fungal pathogen. Nat. Plants 2: 16153. [DOI] [PubMed] [Google Scholar]
- Zhang Z., Hu F., Sung M.W., Shu C., Castillo-González C., Koiwa H., Tang G., Dickman M., Li P., Zhang X. (2017). RISC-interacting clearing 3′-5′ exoribonucleases (RICEs) degrade uridylated cleavage fragments to maintain functional RISC in Arabidopsis thaliana. eLife 6: 1–29. [DOI] [PMC free article] [PubMed] [Google Scholar]