SME1 has a critical role in plant development and interaction with the environment by ensuring adequate splicing of specific precursor-messenger RNAs.
Abstract
The control of precursor-messenger RNA (pre-mRNA) splicing is emerging as an important layer of regulation in plant responses to endogenous and external cues. In eukaryotes, pre-mRNA splicing is governed by the activity of a large ribonucleoprotein machinery, the spliceosome, whose protein core is composed of the Sm ring and the related Sm-like 2-8 complex. Recently, the Arabidopsis (Arabidopsis thaliana) Sm-like 2-8 complex has been characterized. However, the role of plant Sm proteins in pre-mRNA splicing remains largely unknown. Here, we present the functional characterization of Sm protein E1 (SME1), an Arabidopsis homolog of the SME subunit of the eukaryotic Sm ring. Our results demonstrate that SME1 regulates the spliceosome activity and that this regulation is controlled by the environmental conditions. Indeed, depending on the conditions, SME1 ensures the efficiency of constitutive and alternative splicing of selected pre-mRNAs. Moreover, missplicing of most targeted pre-mRNAs leads to the generation of nonsense-mediated decay signatures, indicating that SME1 also guarantees adequate levels of the corresponding functional transcripts. In addition, we show that the selective function of SME1 in ensuring appropriate gene expression patterns through the regulation of specific pre-mRNA splicing is essential for adequate plant development and adaptation to freezing temperatures. These findings reveal that SME1 plays a critical role in plant development and interaction with the environment by providing spliceosome activity specificity.
INTRODUCTION
Plants are continuously challenged by changes in their environment, which frequently results in stressful situations. To cope with these scenarios, they have evolved different adaptive processes. Efforts aimed at identifying the molecular mechanisms underlying these processes revealed that they are strongly dependent on extensive transcriptional reprograming (Nakashima et al., 2014). Several studies have demonstrated that this reprograming, although tightly regulated at the transcriptional level, is also posttranscriptionally controlled (Guerra et al., 2015). Alternative precursor-messenger RNA (pre-mRNA) splicing, the differential recognition of introns and exons within a gene, constitutes one of the most significant posttranscriptional mechanisms controlling gene expression (Lee and Rio, 2015). In plants, the relevance of this mechanism in modulating gene expression is reflected in the fact that around 60% of all intron-containing genes from Arabidopsis (Arabidopsis thaliana) are subjected to alternative splicing (Zhang et al., 2017). Interestingly, this posttranscriptional mechanism is induced under abiotic stress conditions, and genes playing a key role in plant responses to such stresses have been shown to be prone to alternative splicing events (Deng et al., 2011; Seo et al., 2012, 2013; Liu et al., 2013; Calixto et al., 2018). These results clearly point out a crucial function of alternative splicing in controlling abiotic stress responses in plants. Yet, how the splicing machinery performs this function and how it is regulated remains poorly understood.
Pre-mRNA splicing is performed by the spliceosome, a highly evolutionary conserved multimegadalton ribonucleoprotein (RNP) complex composed of five small nuclear ribonucleoproteins (snRNPs) that constitute the core of this complex and hundreds of non-snRNPs (Fica and Nagai, 2017). Each snRNP consists of a snRNA (U1, U2, U4, U5, or U6) and an associated heptameric protein complex (Wilusz and Wilusz, 2013). In the case of U1, U2, U4, and U5 snRNPs, the corresponding snRNAs are associated with the Sm complex. The U6 snRNA, on the other hand, interacts with the related Sm-like (LSM) 2-8 complex. Recent genetic analyses have revealed the identity of several plant non-snRNPs involved in the control of responses to abiotic stresses, including high soil salinity, extreme temperature, drought and high light irradiation, by targeting pre-mRNAs corresponding to genes involved in those responses (Lee et al., 2006; Kim et al., 2008, 2017; Du et al., 2015; Feng et al., 2015). The Ser/Arg-rich proteins and heterogeneous nuclear ribonucleoproteins, for instance, which are important determinants of splice-site recognition (Black, 2003), have been shown to modulate pre-mRNA splicing, ensuring the correct plant responses to high salt, drought, and high temperature (Laloum et al., 2018). Moreover, the overexpression of certain non-snRNPs increases plant tolerance to adverse environmental situations (Guan et al., 2013). These data provide evidence that non-snRNPs have a pivotal role in pre-mRNA splicing of abiotic stress-related genes and, consequently, in plant adaptation to abiotic stresses.
In contrast to the non-snRNPs, little is known about the functions of the spliceosome core components (i.e., the snRNPs) in plant abiotic stress responses. Arabidopsis LSM5 and LSM4 proteins were characterized, both of them being related to abscisic acid and osmotic stress signaling (Xiong et al., 2001; Zhang et al., 2011). We previously reported that Arabidopsis has a functional LSM2-8 complex, the protein moiety of the U6 snRNP, that, as in yeast and animals, controls U6 snRNA stability (Perea-Resa et al., 2012). This function, furthermore, has relevant consequences in differentially modulating the constitutive and alternative pre-mRNA splicing of important regulators of Arabidopsis responses to abiotic stresses, depending on the environmental conditions (Carrasco-López et al., 2017). The Sm complex, the protein moiety of the U1, U2, U4, and U5 snRNPs, plays a central role in the assembly and nuclear shuttling of these snRNPs, ensuring adequate levels of the corresponding snRNAs (Gruss et al., 2017). The Sm proteins are characterized by having the Sm domain, which includes the Sm1 and Sm2 motifs joined by a variable linker region, and mediates their interaction with the snRNAs (Hermann et al., 1995). In eukaryotes, the Sm complex is formed by the seven Sm canonical proteins (SmB/B′, SmD1, SmD2, SmD3, SmE, SmF, and SmG) that are sequentially assembled as a heptameric ring around the snRNAs (Wilusz and Wilusz, 2013). It has been shown that human Sm proteins participate in the control of both constitutive and alternative pre-mRNA splicing (Saltzman et al., 2011). In plants, Sm proteins have been barely studied. As expected from their high level of conservation throughout evolution, in silico analyses of different plant genomes revealed that they contain numerous genes encoding presumed Sm proteins (Cao et al., 2011). To date, however, only two putative Arabidopsis Sms, SMD3 and SMD1, have been characterized. The analysis of smd3b and smd1b null mutants revealed that SMD3 and SMD1 are involved in plant development as well as in the processing of the few pre-mRNAs whose splicing patterns were examined (Swaraz et al., 2011; Elvira-Matelot et al., 2016). Nonetheless, whether they are functional Sm proteins that ensure precise levels of U snRNAs and what their relevance is in modulating pre-mRNA splicing at the genomic level is unknown. In addition, the involvement of SMD3 and SMD1 in plant responses to abiotic stresses has not been assessed.
Here, we report the molecular and functional characterization of SME1, an Arabidopsis homolog of the SmE subunit of the eukaryotic Sm ring, during plant development and in response to abiotic stress. Our results demonstrate that, according to the function of eukaryotic Sm proteins, SME1 promotes the accumulation of U1, U2, U4, and U5 snRNAs and is required for adequate constitutive and alternative splicing. Remarkably, the function of SME1 in controlling pre-mRNA splicing and, therefore, in maintaining adequate levels of functional transcripts, is modulated by environmental conditions at both qualitative and quantitative levels. Indeed, genome-wide transcriptomic analysis of sme1 plants revealed that, under control conditions, SME1 is essential for the correct splicing of a reduced number of selected pre-mRNAs, including various development-related ones. Under low temperature, however, SME1 ensures the appropriate splicing of a high number of pre-mRNAs, among which there are some cold-response regulators. Consistent with the activity of SME1 in pre-mRNA splicing, we show that SME1 is essential for Arabidopsis development from seedling establishment to floral transition and for adequate development of the cold acclimation process. We propose that SME1 plays a critical role in plant responses to endogenous and external signals by ensuring appropriate gene expression patterns through the regulation of specific pre-mRNA splicing.
RESULTS
SME1 Transcripts Accumulate in Response to Low Temperature Independently of Abscisic Acid and CBFs
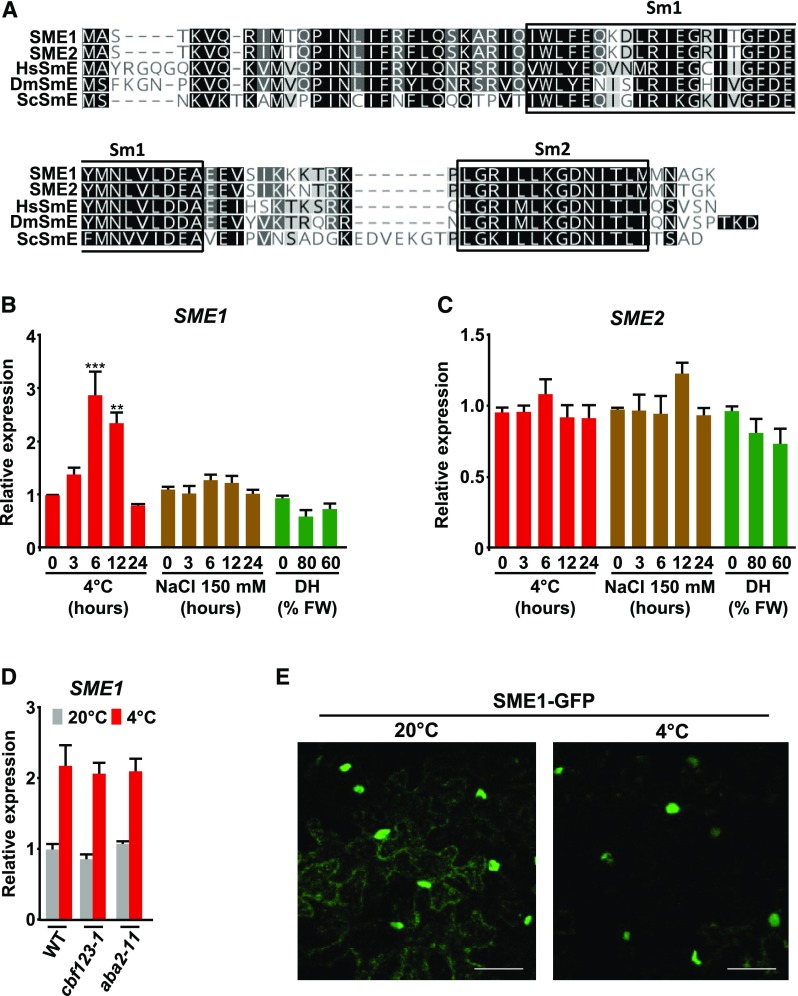
Previously, we reported that the LSM2-8 complex modulates spliceosome activity to differentially control the Arabidopsis response to abiotic stress (Carrasco-López et al., 2017). To further understand the mechanisms governing the spliceosome function under abiotic stress conditions, we decided to characterize the Arabidopsis SME, the only subunit of the Sm ring that has been described to interact with the LSM2-8 complex (Arabidopsis Interactome Mapping Consortium, 2011). The Arabidopsis genome contains two genes encoding proteins with the characteristic bipartite Sm-domain and high sequence identity (49 to 63%) to other eukaryotic SmEs (Figure 1A). We named these genes SME1 (At2g18740) and SME2 (At4g30330; Figure 1A). Expression data from the electronic Fluorescent Pictograph (eFP) browser database (bar.utoronto.ca) indicated that they are both actively expressed, and SME1 transcription might respond to abiotic stress, particularly to low temperature (Kilian et al., 2007). Confirming the eFP data, quantitative PCR (qPCR) experiments with RNA samples from Col-0 (wild-type) plants exposed to cold, high salt, or water stress showed a significant transient accumulation of SME1 transcripts only after cold treatment (Figure 1B). By contrast, the expression levels of SME2 did not vary in response to any treatment (Figure 1C). Consequently, we decided to focus on SME1 to proceed with our study. We investigated whether the cold accumulation of SME1 transcripts was dependent on the C-repeat Binding Factors (CBFs)and/or abscisic acid, which mediate the two main signaling pathways controlling cold-induced gene expression (Medina et al., 2011). Expression analyses in cold-treated CBF- and abscisic acid-deficient Arabidopsis mutants (cbf123-1 and aba2-11; González-Guzmán et al., 2002; Zhao et al., 2016) revealed that SME1 transcripts accumulate under low temperature conditions independently of CBFs and abscisic acid (Figure 1D).
Figure 1.
Low Temperature Promotes SME1 Expression and Nuclear Localization of the Corresponding Protein.
(A) Sequence alignment of SME proteins from Arabidopsis, Homo sapiens, Saccharomyces cerevisiae, and Drosophila melanogaster. Sm1 and Sm2 domains are shown. Black and gray shading indicates identical or similar residues, respectively, in at least half of the compared sequences. Dashes represent gaps inserted into the sequences for optimal alignment.
(B) and (C) Accumulation of SME1 (B) and SME2 (C) transcripts in 2-week-old wild-type (WT) plants exposed for the indicated number of hours to 4°C (red bars) or 150 mM NaCl (brown bars), or dehydrated (DH) until reaching the indicated percentage of fresh weight (FW) (green bars).
(D) Accumulation of SME1 transcripts in 2-week-old wild-type, cbf123-1, and aba2-11 plants grown under control conditions (20°C; gray bars) or exposed for 6 h to 4°C (red bars).
(E) Subcellular localization of SME1-GFP in N. benthamiana leaf cells under control (20°C) or cold (4°C, 24 h) conditions. Scale bars indicate 50 µm.
In (B) to (D), transcript levels, determined by qPCR, are represented as relative to values at 0 h or 0% DH (B and C) or to wild-type values (C). Data represent the mean of three independent experiments. Asterisks indicate significant differences (**P < 0.001, ***P < 0.0001) between treated and control (0 h) plants, as determined by one-way ANOVA (Dunnett’s post-hoc test). Error bars indicate the SD.
SME1 Preferentially Accumulates in the Nucleus in Response to Low Temperature
The subcellular distribution of SME1 was assessed by agroinfiltration of a SME1-GREEN FLUORESCENT PROTEIN (GFP) translational fusion (SME1PRO-SME1-GFP) in tobacco (Nicotiana benthamiana) leaves. Confocal microscopy examination of the transformed cells expressing the SME1PRO-SME1-GFP fusion allowed detection of green fluorescence in both nuclei and cytoplasm, indicating that, as previously described for other eukaryotic Sm proteins (Zieve, 1999; Gruss et al., 2017), SME1 simultaneously localizes to these subcellular compartments (Figure 1E). Since SME1 expression is regulated by low temperature, we also analyzed the subcellular localization of SME1 under cold conditions. Interestingly, after 24 h of exposure at 4°C, the SME1-GFP fusion protein was preferentially localized in nuclei (Figure 1E), indicating that SME1 accumulates in the nucleus in response to low temperature.
Arabidopsis SME1 Promotes the Accumulation of U1, U2, U4, and U5 snRNAs and Is Involved in pre-mRNA Splicing
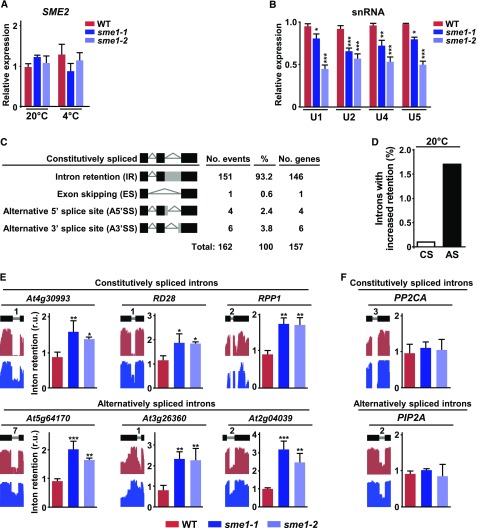
In metazoans and yeast, the Sm proteins are involved in pre-mRNA splicing by ensuring adequate levels of U1, U2, U4, and U5 snRNAs (Will and Lührmann, 2011). To determine whether SME1 has, indeed, a role in pre-mRNA splicing, we first identified two T-DNA lines each containing a single insertion in the fifth and second exons of SME1, respectively (Supplemental Figure 1A). qPCR assays showed that these lines, named sme1-1 and sme1-2, were severely affected in SME1 expression under control and low temperature conditions (Supplemental Figure 1B), indicating that they corresponded to null or severely hypomorphic alleles. SME2 transcripts were not affected in sme1 mutants in any case (Figure 2A), discarding a potential compensatory effect. Then, we used the sme1-1 and sme1-2 mutants to investigate if SME1 was required for the correct accumulation of U1, U2, U4, and U5 snRNAs in Arabidopsis. Results revealed a significant reduction in the levels of the four U snRNAs in the mutant plants (Figure 2B), demonstrating that SME1 is necessary for their proper accumulation in Arabidopsis.
Figure 2.
SME1 Promotes the Accumulation of U1, U2, U3, and U5 snRNAs and Controls Constitutive and Alternative Splicing in Arabidopsis.
(A) Accumulation of SME2 transcripts in 2-week-old wild-type (WT), sme1-1, and sme1-2 plants grown under control (20°C) or cold conditions (6 h, 4°C).
(B) Levels of U1, U2, U4, and U5 snRNAs in 2-week-old wild-type, sme1-1, and sme1-2 plants grown under control conditions.
(C) Quantification of altered splicing events (IR, ES, A5′SS, A3′SS) identified in sme1-1 with respect to wild-type plants grown under control conditions. Black bars represent exons, and gray bars represent intron regions retained in some events. The number of different genes affected in each case is also shown.
(D) Percentage of constitutively (CS) and alternatively (AS) spliced introns with increased retention in sme1-1 plants relative to the total number of the corresponding introns identified in wild-type plants.
(E) Validation of different IR events identified in sme1-1 mutants. In each case, the name of the gene containing the corresponding constitutively or alternatively spliced intron is indicated.
(F) Validation of constitutively and alternatively spliced introns not affected in sme1-1 mutants. The names of the genes containing the corresponding introns are indicated.
In (A) and (B), transcript levels, determined by qPCR, are represented as relative to wild-type values. In (E) and (F), a diagram of the pre-mRNA regions, including the considered introns (gray lines) with their relative positions in the representative gene model and the flanking exons (black bars), together with the captures of the corresponding read coverage tracks obtained from the IGV software, is shown in the left. The quantification of considered introns in wild-type, sme1-1, and sme1-2 plants by qPCR assays is displayed in the right. In (A), (B), (E), and (F), data represent the mean of three independent experiments. Asterisks indicate significant differences (*P < 0.01, **P < 0.001, ***P < 0.0001) between sme1 mutants and wild-type plants, as determined by one-way ANOVA (Dunnett’s post-hoc test). Error bars indicate the SD.
The implication of SME1 in pre-mRNA splicing was conclusively established by analyzing the impact of the sme1-1 mutation on the splicing events at the genome-wide level. We performed a high-coverage RNA sequencing (RNA-seq) analysis on 2-week-old wild-type and sme1-1 plants grown under control conditions. To identify and quantify splicing events, the resulting reads (around 50 million, 91 bp paired-end per genotype) were mapped to the Arabidopsis genome (TAIR10 version) using TopHat2 (Kim et al., 2013), and splicing events were counted with Regtools (https://github.com/griffithlab/regtools). When comparing data obtained from sme1-1 with those from wild-type plants, mutants showed alterations in 162 splicing events corresponding to 157 different genes (fold change ± 2; Fisher’s exact test, Q ≤ 0.05; Figure 2C; Supplemental Data Set 1). These alterations could be pooled into four main categories, namely intron retention (IR; 93.2%), exon skipping (ES; 0.6%), alternative 5′ splice site (A5′SS; 2.4%) and alternative 3′ splice site (A3′SS; 3.8%; Figure 2C). Intron retention was, by far, the most abundant category. Data revealed that SME1 controlled the adequate removal of 151 introns from 146 genes (Figure 2C; Supplemental Data Set 1), 84 corresponding to constitutively and 67 to alternatively spliced introns (Supplemental Data Set 1). These numbers represented around 0.1% and 1.7% of all constitutively (81,396) and alternatively (3,729) spliced introns identified in wild-type plants under our standard conditions, which were similar to those reported earlier (Figure 2D; Carrasco-López et al., 2017). Results from RNA-seq experiments were validated by analyzing, in independent RNA samples, the retention of a number of constitutively and alternatively spliced introns in sme1-1 and sme1-2 mutants by means of qPCR (Figure 2E). Constitutively and alternatively spliced introns not affected by the sme1 mutations were also validated (Figure 2F). In all cases, the IR were coincident with those inferred from RNA-seq experiments, indicating a very low false-positive rate.
Often, IR events originate transcripts with nonsense-mediated decay (NMD) signatures that are poorly translated (Yu et al., 2016). As a result, these events may influence the levels of functional transcripts and, thus, those of the corresponding proteins. The introns retained in sme1 plants generated 108 mRNAs (73.9% of all mRNAs with IR events) displaying one or more features of NMD targeted transcripts (Table 1; Supplemental Data Set 2). We concluded, therefore, that SME1 plays a role in regulating the spliceosome activity, by controlling the efficiency of both constitutive and alternative pre-mRNA splicing in Arabidopsis, this role being more significant on the latter, and the levels of functional transcripts.
Table 1. Introns Retained in sme1-1 Mutant under Control (20°C) or Cold (4°C) Conditions That Generate Transcripts with NMD Features.
| Environmental Condition | Intron Type | PTCa | 3′ UTR >350 ntb | SC→intron >55 ntc | 5′ uORFd | uORF Overlape | Genesf (%) | Non-NMD Genesg (%) |
|---|---|---|---|---|---|---|---|---|
| 20°C | Constitutive | 68 | 3 | 0 | 0 | 0 | 108 (73.9) | 38 (26.1) |
| Alternative | 35 | 4 | 0 | 0 | 1 | |||
| 4°C | Constitutive | 542 | 20 | 0 | 1 | 4 | 640 (77.2) | 189 (22.8) |
| Alternative | 129 | 10 | 0 | 2 | 2 |
Premature termination codon (PTC) and a 3′ UTR longer than 350 nucleotides (nt).
3′ UTR longer than 350 nt.
More than 55 nt between stop codon (SC) and a downstream intron.
Upstream open reading frame (uORF) longer than 35 amino acids.
uORF overlapping with the start codon of the main ORF.
Total number of genes containing at least one retained intron causing NMD features.
Total number of genes containing retained introns that do not generate NMD features.
SME1 Regulates Plant Development by Controlling the Splicing of Genes Involved in Different Developmental Stages from Seedling Establishment to Floral Transition and Development
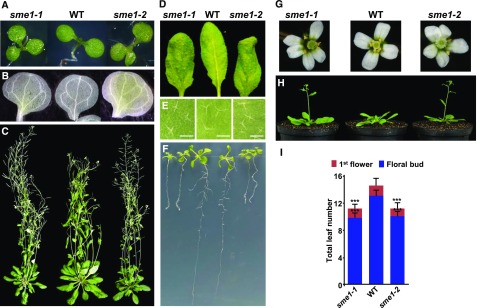
As other Arabidopsis mutants deficient in spliceosomal components (Swaraz et al., 2011; Perea-Resa et al., 2012; Elvira-Matelot et al., 2016; Mahrez et al., 2016), sme1 mutants exhibited substantial developmental defects. Both alleles identified showed a significant percentage of seedlings with alterations in the shape and number of cotyledons, and veins formed more closed loops than in wild-type cotyledons (Figures 3A and 3B; Supplemental Figures 2A and 2B). sme1-1 and sme1-2 rosettes were smaller than the wild-type ones (Figure 3C; Supplemental Figure 2C), and their leaves displayed a twisted shape (Figure 3D). Trichomes, in turn, had an atypical number of branches (Figure 3E; Supplemental Figure 2D). Regarding the radicular system, sme1 mutants exhibited notably shorter main roots, a complete absence of secondary roots and an evident increase in the number of root hairs (Figure 3F; Supplemental Figure 2E). Furthermore, primary and secondary inflorescences were longer in the mutants than in the wild type (Figure 3C), and mutant plants showed a higher number of flowers with an altered numbers of petals (Figure 3G; Supplemental Figure 2F). In addition, sme1 mutants flowered significantly earlier than wild-type plants (Figures 3H and 3I). All these data demonstrated that SME1 plays a key role in controlling plant development at different stages, from seedling establishment to floral transition and development.
Figure 3.
Phenotypic Analysis of Arabidopsis sme1 Mutants.
(A) to (H) Morphological phenotypes of wild-type (WT), sme1-1, and sme1-2 plants. Three-day-old seedlings (A), cotyledon vein patterns (B), 7-week-old plants (C), rosette leaves of 4-week-old plants (D), trichomes on the leaves of 4-week-old plants (scale bars indicate 0.25 mm) (E), 14-d-old seedlings (F), flowers (G), and 4-week-old plants (H).
(I) Total number of leaves, after appearance of the floral bud (blue bars) or the first flower (red bars), in wild-type, sme1-1, and sme1-2 plants grown under control conditions. Data represent the mean of six independent experiments. Asterisks indicate significant differences (***P < 0.0001) between sme1 mutants and wild-type plants, as determined by one-way ANOVA (Dunnett’s post-hoc test). Error bars indicate the SD.
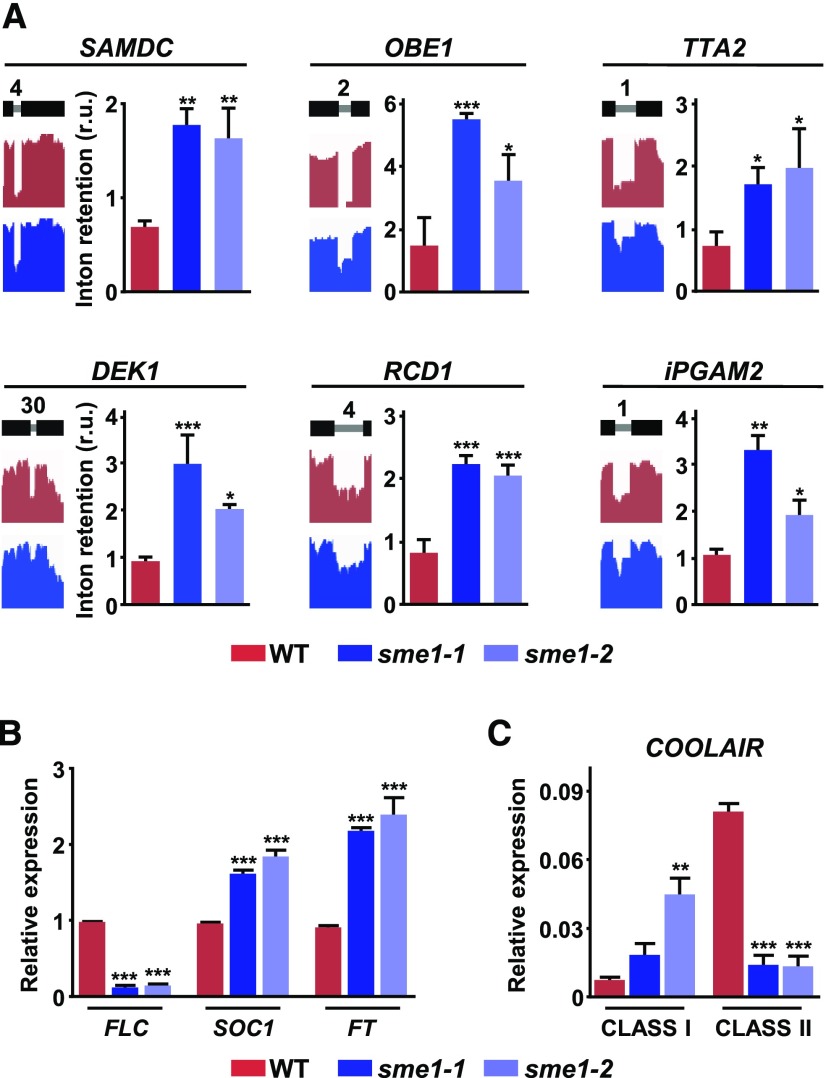
Interestingly, when examining the results of the RNA-seq analysis (Supplemental Data Set 1), 13 genes that had been implicated in regulating shoot and root architecture (Berardini et al., 2004) were affected in their patterns of spliced introns in the sme1-1 mutant. Furthermore, most introns retained in these genes led to pre-mRNAs (9) with one or more NMD signatures (Supplemental Data Set 3), suggesting that SME1 could control Arabidopsis development by ensuring the correct splicing of developmental genes and the appropriate levels of their functional transcripts. Supporting this assumption, the corresponding IR events in some of these genes, including those encoding the S-adenosylmethionine decarboxylase, which controls root and leaf growth (Ge et al., 2006), the plant homeodomain finger factors OBERON 1 and TITANIA 2, which are involved in embryonic root meristem initiation (Saiga et al., 2012), the calpain-type cysteine protease DEFECTIVE KERNEL 1, which controls cell fate in trichomes and flowers (Galletti et al., 2015), the WWE domain protein RADICAL-INDUCED CELL DEATH 1, which is related to cell division and differentiation (Teotia and Lamb, 2011), and the 2,3-BIPHOSPHOGLYCERATE-INDEPENDENT PHOSPHOGLYCERATE MUTASE 2, which modulates plant vegetative growth (Zhao and Assmann, 2011), were confirmed by qPCR experiments with appropriate primers in sme1-1 and sme1-2 mutant plants (Figure 4A).
Figure 4.
SME1 Regulates the Splicing and Accumulation of Development-Related Transcripts in Arabidopsis.
(A) Validation of different IR events identified in development-related genes in sme1-1 mutants. For each event, the name of the gene containing the corresponding retained intron is indicated. A diagram of the pre-mRNA regions, including the considered introns (gray lines) with their relative positions in the representative gene model and the flanking exons (black bars), together with the captures of the corresponding read coverage tracks obtained from the IGV software, is shown in the left. The quantification of retained introns in wild-type (WT), sme1-1, and sme1-2 plants by qPCR assays is displayed in the right.
(B) and (C) Accumulation of FLC, SOC1, and FT transcripts (B) and COOLAIR class I and class II splicing variants (C) in 9-day-old wild-type, sme1-1, and sme1-2 plants grown under control conditions.
In all cases, data represent the mean of three independent experiments. Asterisks indicate significant differences (*P < 0.01, **P < 0.001, ***P < 0.0001) between sme1 mutants and wild-type plants, as determined by one-way ANOVA (Dunnett’s post-hoc test). Error bars indicate the SD.
By contrast, we could not find any gene involved in flowering time whose splicing pattern was altered in the sme1-1 mutant. Nonetheless, the gene expression results generated from our RNA-seq data indicated that the transcript levels of FLOWERING LOCUS C (FLC), the major repressor of floral transition (Whittaker and Dean, 2017), were severely reduced in the mutant (Supplemental Data Set 4), which was consistent with its early flowering phenotype (Figures 3H and 3I). This result was corroborated by qPCR analysis with independent samples of sme1-1 and sme1-2 mutants (Figure 4B). Concomitantly with the low levels of FLC transcripts, the expression of the floral integrator genes SUPPRESSOR OF OVEREXPRESSION OF CO 1 and FLOWERING LOCUS T, the two main targets of FLC (Whittaker and Dean, 2017), were upregulated in sme1 mutants (Figure 4B). These data strongly suggested that SME1 would function as a flowering repressor by promoting FLC expression. It is well documented that FLC expression is tightly regulated by COOLAIR, a long noncoding antisense RNA expressed from the FLC locus (Liu et al., 2010), and that the modulation of the alternative splicing of this RNA is essential for its role in regulating FLC expression under warm temperatures (Marquardt et al., 2014). COOLAIR has two major splicing variants, the proximally polyadenylated class I and the distally polyadenylated class II (Supplemental Figure 3). While the accumulation of class I isoform results in the repression of FLC transcription and an early flowering, an increase of class II RNAs is accompanied by the activation of FLC expression with the corresponding delayed flowering (Marquardt et al., 2014). Given the role of SME1 in pre-mRNA splicing, we hypothesized that this protein could control FLC expression by regulating the splicing pattern of COOLAIR. qPCR experiments with samples from wild-type, sme1-1, and sme1-2 plants grown under control conditions revealed that, indeed, the absence of SME1 causes the accumulation of COOLAIR class I isoform and the reduction of the class II isoform (Figure 4C). These results were consistent with the reduced levels of FLC transcripts present in sme1 mutants and their early flowering phenotypes and provided evidence that SME1 modulates flowering time by ensuring the proper splicing pattern of COOLAIR. Together, our findings indicated that SME1 regulates plant development by controlling the splicing of genes involved in different developmental stages from seedling formation to floral transition and development.
SME1 Negatively Regulates the Capacity of Arabidopsis to Cold Acclimate
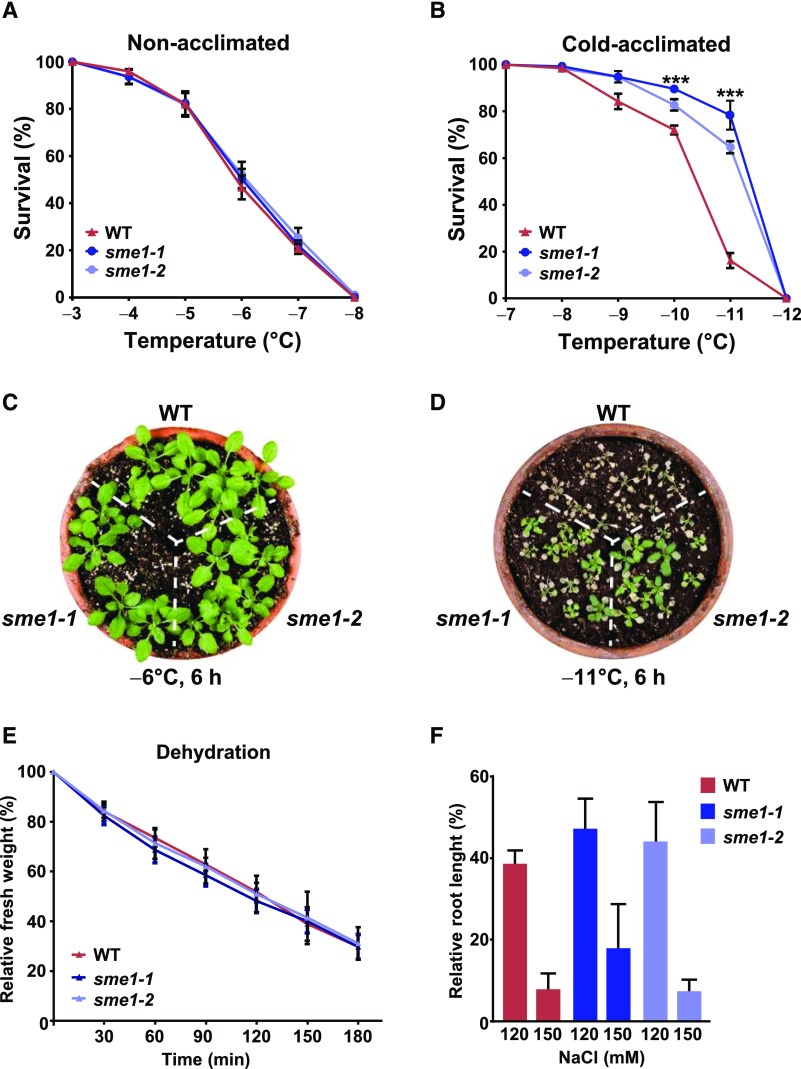
The expression pattern of SME1 (Figure 1B) suggested that it could be involved in the plant response to low temperature. This possibility was assessed by analyzing the tolerance of sme1 mutants to freezing temperatures before and after cold acclimation. The freezing tolerance of nonacclimated and cold-acclimated (4°C, 7 d) 2-week-old wild-type, sme1-1, and sme1-2 plants was estimated as the percentage of surviving plants after 6 h of exposure to different freezing temperatures. When not acclimated, all genotypes showed similar tolerance (Figures 5A and 5C). By contrast, after cold acclimation, sme1-1 and sme1-2 mutants displayed a significantly increased tolerance compared with wild-type plants, their LT50 (temperature that causes 50% lethality) being −11.4°C, −11.3°C, and −10.3°C, respectively (Figures 5B and 5D). We also investigated the potential implication of SME1 in plant tolerance to other abiotic stresses related to low temperature, such as water stress and high salt. Water stress was induced by maintaining rosettes from 2-week-old plants on dry filter paper for 3 h without watering. The rate of dehydration was established as the percentage of initial fresh weight that remained after different times of treatment. Salt tolerance was assayed in 5-day-old seedlings grown for 1 additional week on plates containing either 120 or 150 mM NaCl. Tolerance was determined by measuring the length of the main root of the seedlings after treatments. No significant differences between sme1 mutants and the wild type were found in any case (Figures 5E and 5F). These results demonstrated that SME1 functions as a negative regulator of the cold acclimation process but is not involved in the constitutive freezing tolerance of Arabidopsis nor in the Arabidopsis tolerance to water stress and high salt under our experimental conditions.
Figure 5.
SME1 Negatively Regulates the Cold Acclimation Process in Arabidopsis.
(A) and (B) Freezing tolerance of nonacclimated (A) and cold-acclimated (7 d, 4°C) (B) 2-week-old wild-type (WT), sme1-1, and sme1-2 plants exposed for 6 h to the indicated freezing temperatures. Freezing tolerance was estimated as the percentage of plants surviving each specific temperature after 7 d of recovery under control conditions.
(C) and (D) Representative nonacclimated (C) and cold-acclimated (D) plants 7 d after being exposed to −6°C and −11°C, respectively, for 6 h.
(E) Tolerance to dehydration of 2-week-old wild-type, sme1-1, and sme1-2 plants. The rates of dehydration were established as the percentages of initial fresh weights that remained after allowing rosettes to dehydrate on filter papers for the indicated times.
(F) Tolerance to salt stress of 5-day-old wild-type, sme1-1, and sme1-2 seedlings. Tolerances were calculated as the relative main root lengths of seedlings exposed for 7 d to 120 or 150 mM NaCl with respect to seedlings grown under control conditions.
In (A), (B), (E), and (F), data represent the mean of three independent experiments. Asterisks indicate significant differences (***P < 0.0001) between sme1 mutants and wild-type plants, as determined by one-way ANOVA (Dunnett’s post-hoc test). Error bars indicate the SD.
SME1 Attenuates the Cold Acclimation Response by Ensuring the Adequate Splicing Patterns of Cold-Responsive Genes
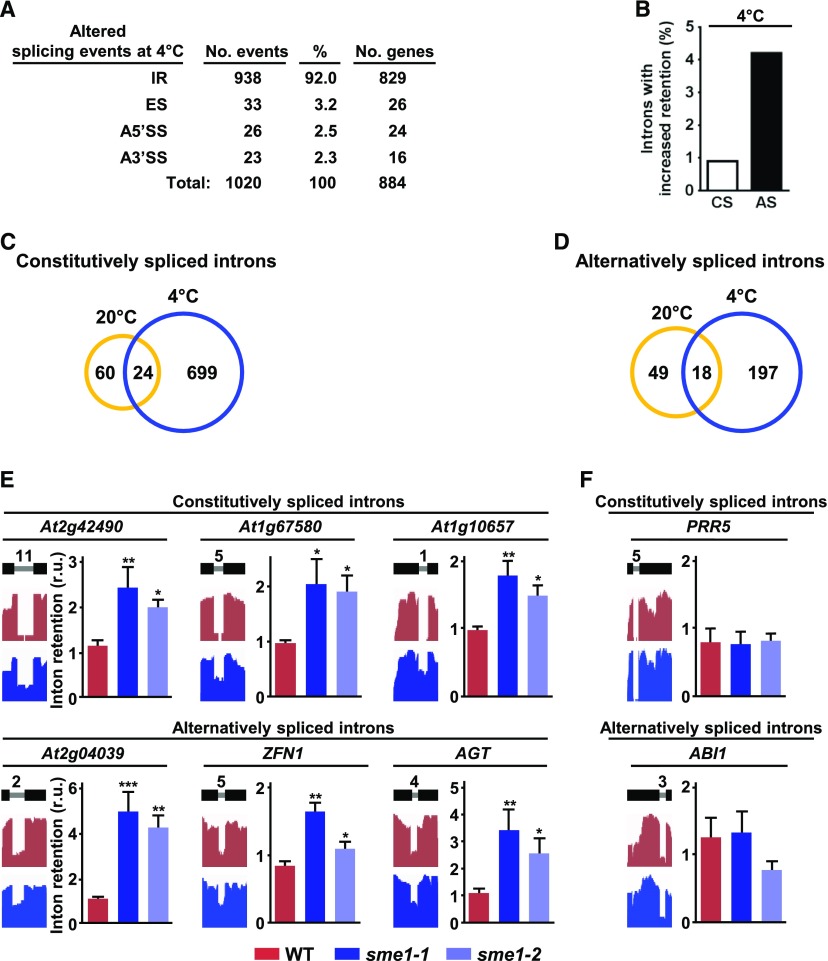
The arising question from the findings described above was what molecular mechanisms underlie SME1 function in attenuating cold acclimation. Considering the role of SME1 in controlling pre-mRNA splicing, to answer this question we performed an RNA-seq analysis on 2-week-old wild-type and sme1-1 plants exposed for 1 additional day to 4°C. As under control conditions , alternative splicing events were identified and quantified by mapping the resulting reads (around 50 million, 91 bp paired-end per genotype) to the Arabidopsis genome (TAIR10 version) using the TopHat2 software (Kim et al., 2013). Read alignments were also processed as described above. The comparison between the results obtained from the sme1-1 mutant and those from wild-type plants revealed alterations in 1020 splicing events corresponding to 884 different genes (fold change ± 2, Fisher’s exact test, Q ≤ 0.05; Figure 6A; Supplemental Data Set 5). Again, these events could be assembled into four main classes, i.e., IR, ES, A5′SS, and A3′SS. Although the number of events within each class was always higher than that observed at 20°C, the corresponding percentages with respect to the total number of events were, in general, quite similar (Figure 6A). Consequently, IR was also the most abundant splicing event alteration noticed in sme1-1 plants subjected to 4°C. In fact, in response to low temperature, SME1 controlled the correct splicing of 938 introns from 829 genes (Figure 6A; Supplemental Data Set 5), 723 corresponding to constitutively and 215 to alternatively spliced introns (Supplemental Data Set 5). These introns represented 0.9% and 4.2% of all constitutively (80,601) and alternatively (5,123) spliced introns, respectively, identified in wild-type plants exposed for 24 h to 4°C, which were similar to those previously reported (Figure 6B; Carrasco-López et al., 2017). When comparing the introns targeted by SME1 under cold conditions with those targeted at 20°C, 896 (95.5%) out of the 938 introns were found to be specifically regulated by SME1 in the cold, 699 corresponding to constitutively spliced (Figure 6C) and 197 to alternatively spliced ones (Figure 6D).
Figure 6.
Arabidopsis SME1 Controls the Splicing of Specific Constitutive and Alternative Spliced Introns in Response to Low Temperature.
(A) Quantification of altered splicing events (IR, ES, A5′SS, A3′SS) identified in sme1-1 with respect to wild-type (WT) plants in response to low temperature. The number of different genes affected in each case is also shown.
(B) Percentage of constitutively (CS) and alternatively (AS) spliced introns with increased retention in sme1-1 plants exposed to 4°C relative to the total number of the corresponding introns identified in wild-type plants exposed to the same conditions.
(C) and (D) Venn diagrams showing the overlap between the constitutively (C) and alternatively (D) spliced introns specifically retained in sme1-1 with respect to wild-type plants under control (20°C) or cold (4°C) conditions. The number of specific and common introns identified is indicated.
(E) Validation of different IR events identified in sme1-1 plants exposed to low temperature. In each case, the name of the gene containing the corresponding constitutively or alternatively spliced intron is indicated.
(F) Validation of constitutively and alternatively spliced introns not affected in sme1-1 plants exposed to low temperature. The names of the genes containing the corresponding introns are indicated.
In (E) and (F), a diagram of the pre-mRNA regions, including the considered introns (gray lines) with their relative positions in the representative gene model and the flanking exons (black bars), together with the captures of the corresponding read coverage tracks obtained from the IGV software, is shown in the left. The quantification of considered introns in wild-type, sme1-1, and sme1-2 plants by qPCR assays is displayed in the right. Data represent the mean of three independent experiments. Asterisks indicate significant differences (*P < 0.01, **P < 0.001, ***P < 0.0001) between sme1 mutants and wild-type plants, as determined by one-way ANOVA (Dunnett’s post-hoc test). Error bars indicate the SD.
RNA-seq data were validated by analyzing the retention of several constitutively and alternatively spliced introns in independent RNA samples from wild-type, sme1-1, and sme1-2 plants exposed to low temperature, by means of qPCR (Figure 6E). The events assayed corresponded to introns regulated specifically (At1g67580, At1g10657, ZINC FINGER PROTEIN 1, and ALANINE:GLYOXYLATE AMINOTRANSFERASE) and nonspecifically (At2g42490 and At2g04039) by SME1 in response to cold. We also validated constitutively and alternatively spliced introns not affected by the sme1-1 mutation (Figure 6F). In all cases, the retention patterns were coincident with those inferred from RNA-seq experiments, indicating that the rate of false positives was very low. Interestingly, most introns specifically retained in sme1-1 plants exposed to 4°C lead to mRNAs (640; 77.2% of all mRNAs with IR events) with one or more NMD signatures (Table 1; Supplemental Data Set 6), strongly suggesting a function for SME1 in maintaining the levels of selected functional transcripts when plants are subjected to cold. All these results provided genetic and molecular evidence that the Arabidopsis SME1 controls the correct splicing of selected pre-mRNAs in response to different environmental conditions and that this differential control is performed by targeting specific constitutively and alternatively spliced introns, depending on the condition to which plants are exposed. Furthermore, our results indicated that SME1 is essential to guarantee appropriate levels of functional transcripts and, in all likelihood, of proteins corresponding to the genes containing the targeted introns.
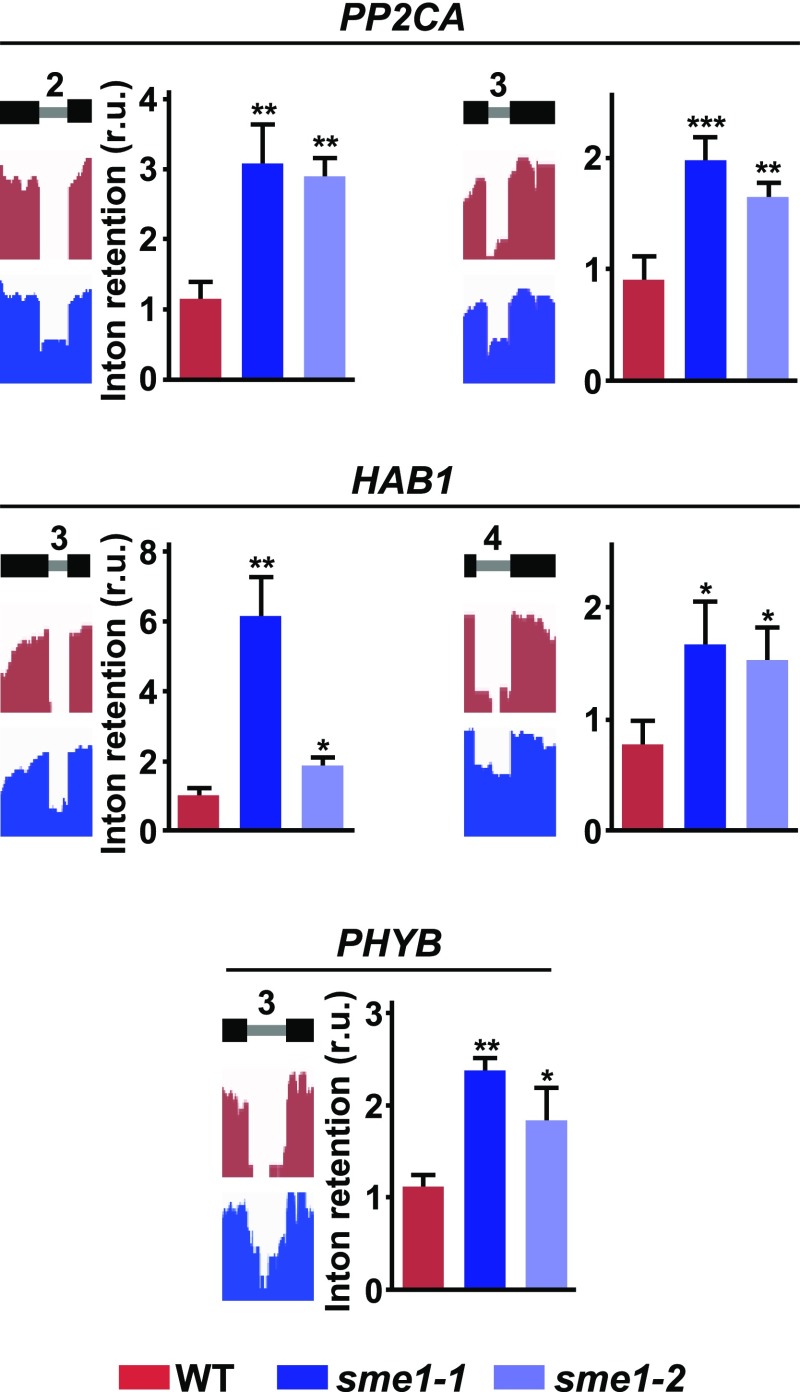
It is worth noting that a significant number of introns targeted by SME1 at 4°C (362; 43.7%) belonged to genes described to be regulated by low temperature (Kilian et al., 2007; Supplemental Data Set 7). More importantly, some of these genes, such as those encoding the protein phosphatases PROTEIN PHOSPHATASE 2CA (PP2CA) and HYPERSENSITIVE TO ABA1 (HAB1) or the light photoreceptor PHYTOCHROME B (PHYB), had been shown to act as negative regulators of cold acclimation in Arabidopsis (Tähtiharju and Palva, 2001; Lee and Thomashow, 2012; Ding et al., 2015). The IR events disclosed in these genes by our RNA-seq experiments, two in PP2CA and HAB1, and one in PHYB, were validated through qPCR assays using independent samples of wild-type, sme1-1, and sme1-2 plants exposed to 4°C (Figure 7). All these events lead to the generation of mRNA isoforms with features of NMD targets, which should account for the increased capacity to cold acclimate displayed by sme1 mutants. In addition, gene expression analysis from our RNA-seq experiments revealed that the levels of 532 transcripts corresponding to genes that had been reported to be regulated by low temperature (Kilian et al., 2007) were altered (± twofold; false discovery rate [FDR] < 0.001) in cold-exposed sme1-1 mutants compared with wild-type plants (Supplemental Data Set 8), which would also account for the freezing tolerant phenotype of cold-acclimated sme1 plants. This alteration in cold-related gene expression is, in all likelihood, an indirect consequence of the elevated number of introns retained in the mutant under low temperature conditions that contain NMD signatures (see above). Together, these findings pinpointed that SME1 would attenuate cold acclimation in Arabidopsis by ensuring the adequate levels of functional transcripts from selected genes having critical functions in this adaptive process through the accurate splicing of specific introns contained in them.
Figure 7.
SME1 Ensures the Accurate Splicing of Negative Regulators of the Arabidopsis Cold-Acclimation Process.
Validation of IR events identified in negative regulators of the cold acclimation process in sme1-1 plants exposed to low temperature. For each event, the name of the gene containing the retained intron is indicated. A diagram of the pre-mRNA regions, including the considered introns (gray lines) with their relative positions in the representative gene model and the flanking exons (black bars), together with the captures of the corresponding read coverage tracks obtained from the IGV software, is shown in the left. The quantification of retained introns in wild-type (WT), sme1-1, and sme1-2 plants by qPCR assays is displayed in the right. Data represent the mean of three independent experiments. Asterisks indicate significant differences (*P < 0.01, **P < 0.001, ***P < 0.0001) between sme1 mutants and wild-type plants, as determined by one-way ANOVA (Dunnett’s post-hoc test). Error bars indicate the SD.
DISCUSSION
The posttranscriptional control of gene expression is emerging as one of the most influential mechanisms regulating plant responses to both endogenous and external signals. In particular, the precise control of exon and intron rearrangement through the modulation of spliceosome activity has been shown to be essential for proper plant development and abiotic stress responses (Perea-Resa et al., 2012; Marquardt et al., 2014; Mahrez et al., 2016; Carrasco-López et al., 2017). The implication of the core components of the spliceosome in modulating its activity has been poorly studied. Here, we report the molecular and functional characterization of SME1, an Arabidopsis protein showing homology to the SmE subunit of the eukaryotic Sm complex, the protein moiety of the U1, U2, U4, and U5 snRNPs (Wilusz and Wilusz, 2013). Our data demonstrate that SME1 is, in fact, a functional Sm protein required for correct spliceosome activity. Moreover, SME1 has a critical role in controlling Arabidopsis development and interaction with its environment by ensuring the adequate processing of specific pre-mRNAs. While preparing this manuscript, the characterization of an Arabidopsis putative splicing regulator, PORCUPINE (PCP), which resulted to be identical to SME1, was reported (Capovilla et al., 2018). Conforming to our results, Capovilla and colleagues found that PCP expression is regulated by ambient temperature and that PCP controls the splicing of a number of pre-mRNAs under control and low-temperature conditions, its role being more relevant in the cold (16°C in this case). The implication of PCP in cold acclimation, however, was not explored. In contrast to our data, they did not observe extensive developmental alterations in pcp mutants grown at standard temperature, but after a long exposure to 16°C. Given that the same mutants have been analyzed in both studies, the differences existing between our results and those of Capovilla et al. (2018) must be due to the different experimental conditions used in each investigation.
The Arabidopsis genome contains two SME paralogs, which we have named SME1 and SME2, that encode proteins with high sequence identity (49 to 63%) to the eukaryotic SmE RNPs (Figure 1A). SME1 and SME2, in turn, only differ by two amino acids (Figure 1A), suggesting that they emerged from a gene duplication event, a common feature in the expansion of the Sm protein family in Arabidopsis (Cao et al., 2011). While SME1 transcripts accumulate in response to low temperature through an abscisic acid- and CBF-independent pathway, SME2 expression does not seem to be responsive to abiotic stress conditions. This functional diversity between the two paralogs might have contributed to the evolution of the regulatory role of SME1 in cold acclimation. The mechanisms underlying cold-induced accumulation of SME1 transcripts are still under investigation. Consistent with their role in U1, U2, U4, and U5 snRNP nuclear shuttling, yeast and animal Sm proteins have been described to be localized in both cytoplasm and nucleus (Zieve, 1999; Gruss et al., 2017). In concordance to these data, we found that SME1 is also distributed in those two subcellular compartments. Furthermore, our results reveal a substantial impact of low temperature on the subcellular distribution of SME1. Indeed, they indicate that in response to low temperature, SME1 mainly accumulates in the nucleus. This cold-induced nuclear accumulation of SME1 could be related to the prominent role of SME1 in controlling pre-mRNA splicing under cold conditions (see below).
Despite the elevated sequence conservation between SME1 and SME2, the morphological and molecular phenotypes exhibited by sme1 mutants indicate specific functions for SME1. We demonstrate that, consistent with the role of the Sm complex in yeast and metazoans (Wilusz and Wilusz, 2013), SME1 is involved in maintaining appropriate levels of U1, U2, U4, and U5 snRNAs and, consequently, in controlling the activity of the spliceosome in Arabidopsis. Indeed, our RNA-seq data reveal that SME1 regulates the efficiency of both constitutive and alternative pre-mRNA splicing. Although SME1 prevents the occurrence of some ES, A5′SS, and A3′SS events, its main function is to guarantee correct IR, its impact being higher on alternatively than constitutively spliced introns. A similar function has been described for other subunits of the Sm ring in animals (Saltzman et al., 2011). In plants, proteins showing homology to eukaryotic SMD1b and SMD3b subunits have been shown to control the splicing patterns of the small number of genes that were analyzed in each case (Swaraz et al., 2011; Elvira-Matelot et al., 2016). Yet, the role of these proteins in controlling the levels of U snRNAs and in pre-mRNA splicing at the genomic level remains to be determined. When comparing the introns targeted by SME1 with those targeted by the LSM2-8 complex (Carrasco-López et al., 2017), the protein moiety of the U6 snRNP, we found ∼27.1% overlap (Figure 8A), unveiling a reduced functional similarity of these spliceosome core components. Likewise, SME1 shares a low number (17%) of its targeted introns with those of the spliceosome snRNP assembly factor GEMIN2 (Schlaen et al., 2015). Remarkably, more than 70% of the introns that are retained in sme1 mutants give rise to mRNA isoforms with NMD features (Table 1). Since transcripts containing NMD signatures are barely translated (Yu et al., 2016), our results strongly suggest that SME1 guarantees adequate levels of functional transcripts and, consequently, of their corresponding proteins.
Figure 8.
SME1 and the LSM2-8 Complex Control the Splicing of Specific Introns in Arabidopsis Depending on the Environmental Conditions.
(A) and (B) Venn diagrams showing the introns targeted by SME1 and the LSM2-8 complex under control conditions (A) or in response to low temperature (B). The number of specific and common introns is indicated. The percentage of common genes with respect to the total introns targeted by SME1 is also displayed.
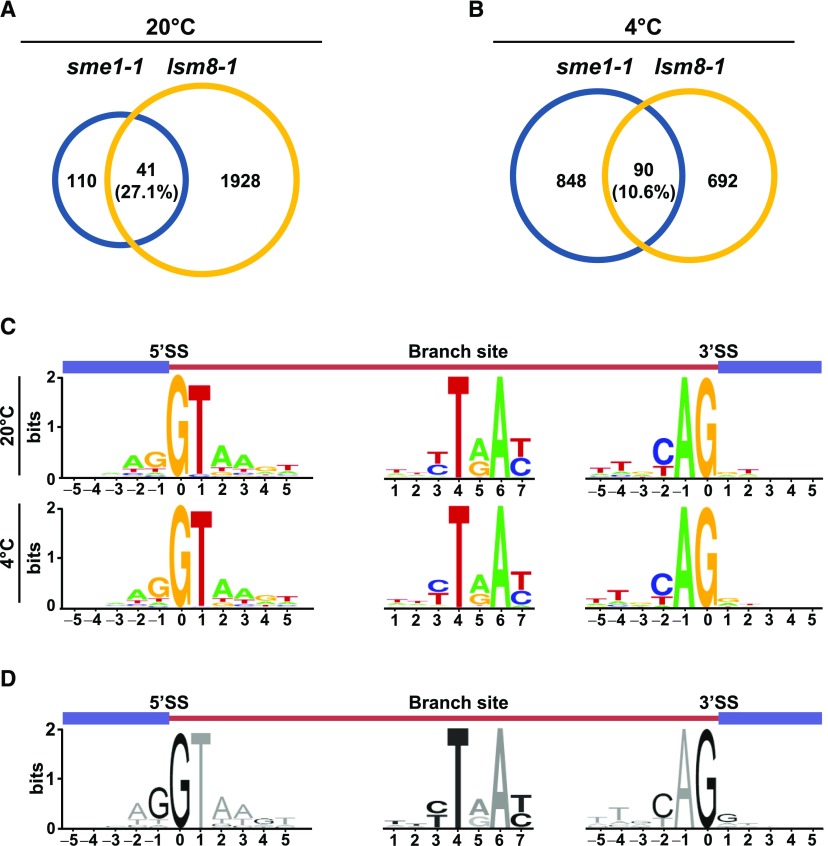
(C) Sequence LOGOs at the consensus 5′ and 3′ splice sites and the branch site of introns specifically retained in sme1-1 mutants when grown under control conditions (20°C) or exposed to low temperature (4°C).
(D) Sequence LOGOs at the consensus 5′ and 3′ splice sites and the branch site of all introns annotated in TAIR10.
In (C) and (D), a diagram of the pre-mRNA regions, including the retained introns (brown bars) and the flanking exons (blue bars) is shown in the top of each panel.
The findings described in this work demonstrate that SME1 is essential for proper Arabidopsis development throughout the different phases of its life cycle. In fact, plants deficient in SME1 exhibit numerous alterations in both vegetative and reproductive developmental traits. We provide evidence that SME1 operates in plant development by ensuring the correct processing of numerous development-related genes. Thus, several constitutive and alternative introns targeted by this protein are located in genes encoding important regulators of Arabidopsis development, such as S-adenosylmethionine decarboxylase (leaf and root development), OBERON 1 and TITANIA 2 (embryo development), DEFECTIVE KERNEL 1 (trichome and flower development), RADICAL-INDUCED CELL DEATH 1 (cell division and differentiation), or 2,3-BIPHOSPHOGLYCERATE-INDEPENDENT PHOSPHOGLYCERATE MUTASE 2 (vegetative growth; Ge et al., 2006; Teotia and Lamb, 2011; Zhao and Assmann, 2011; Saiga et al., 2012; Galletti et al., 2015). Furthermore, missplicing of most of these introns (69.2%; Supplemental Data Set 3) generates mRNAs with characteristics of NMD targets, indicating that, at the end, SME1 plays a key role in regulating plant development by controlling the appropriate levels of the corresponding functional transcripts. Arabidopsis mutants deficient for SMD1b, SMD3b, and LSM8 genes also present severe developmental anomalies (Swaraz et al., 2011; Perea-Resa et al., 2012; Elvira-Matelot et al., 2016), emphasizing the essential role of the spliceosome core components in regulating plant development. Unfortunately, the absence of transcriptomic data for SMD1b and SMD3b does not allow determination of the overlapping degree between the introns targeted by SME1 and those targeted by these other Sm proteins. When compared with the available data for the LSM2-8 complex (Carrasco-López et al., 2017), only 38.5% of the introns targeted by SME1 that belong to development-related genes are shared, thus revealing that different spliceosome core components may confer specific catalytic activity to the splicing machinery and that the spliceosomal control of plant development is fine tuned by different components of this machinery. Interestingly, in addition to ensuring the correct splicing patterns of development-related genes, SME1 also operates in plant development by modulating the splicing of long noncoding RNAs. We demonstrate that SME1 controls the adequate splicing of COOLAIR, a long noncoding RNA that regulates the expression of FLC and, therefore, the timing of the switch to reproductive development in Arabidopsis (Liu et al., 2010). As mentioned, COOLAIR has two main splicing variants, the proximally polyadenylated class I and the distally polyadenylated class II (Marquardt et al., 2014). The accumulation of the proximally polyadenylated isoform results in repression of FLC expression and, consequently, early flowering. When the distally polyadenylated isoform accumulates, FLC expression is induced and concomitantly flowering is delayed (Marquardt et al., 2014). Our data unveil SME1 as a modulator of COOLAIR splicing with a potential role in vernalization. It would act promoting the accumulation of the class II isoform, which, in turn, would activate FLC expression. Similar to SME1, NUCLEAR SPECKLE RNA BINDING PROTEINS, which act as regulators of alternative splicing, have been recently reported to induce COOLAIR class II isoform accumulation delaying flowering (Bazin et al., 2018). In contrast to SME1, however, PRP8, a component of the U5 snRNP (Deng et al., 2016), has been described to enhance the accumulation of the class I isoform of COOLAIR (Marquardt et al., 2014), further supporting that different spliceosomal components may confer spliceosome activity specificity and constitute essential specific regulators of plant growth and development.
Consistent with the accumulation of SME1 transcripts in response to cold conditions, SME1 is involved in plant tolerance to low temperature. Arabidopsis plants deficient in this protein display increased freezing tolerance after cold acclimation, providing genetic evidence that SME1 negatively regulates this adaptive process. To our knowledge, the implication of a component of the Sm complex in abiotic stress responses has not been shown in any system and discloses an unanticipated potential of this core spliceosomal component to fine-tune the adaptation of a given organism to its environment. The RNA-seq analyses point out that, under low-temperature conditions, many introns targeted by SME1 (43.7%) are located in cold-responsive genes (Supplemental Data Set 7), some of them encoding important intermediates of freezing tolerance. Taking into account that missplicing of most of these introns (80.1%; Supplemental Data Set 9) ends up in the generation of pre-mRNA isoforms with features of NMD targets, our results indicate that SME1 guarantees adequate levels of functional transcripts and, consequently, proteins of the corresponding genes. Among the introns whose correct splicing is ensured by SME1 in response to low temperature, several belong to genes encoding proteins, such as PP2CA, HAB1, or PHYB, that are negative regulators of cold acclimation (Tähtiharju and Palva, 2001; Lee and Thomashow, 2012; Ding et al., 2015). Arabidopsis SME1, therefore, is required for the appropriate splicing of selected pre-mRNAs corresponding to proteins having an important function in the plant response to low temperature, which substantiates its major role in attenuating cold acclimation. Additionally, this function, as expected, is crucial to shape the correct cold transcriptomic profile. Indeed, our results show that SME1 regulates the expression levels of 532 cold-responsive genes (Supplemental Data Set 8), which represents 4.3% of the cold regulon and ultimately should contribute to the SME1 capacity to regulate Arabidopsis freezing tolerance. The LSM2-8 complex is the only spliceosome core component that, to date, has also been described to attenuate the Arabidopsis capacity to cold acclimate as a consequence of modulating pre-mRNA splicing (Carrasco-López et al., 2017). However, the comparison between the introns targeted by SME1 (938) and the introns targeted by the LSM2-8 complex (782) in the cold (Carrasco-López et al., 2017) revealed that only 90 (10.6%) are common (Figure 8B), which indicates that different core components of the spliceosome should also confer specific catalytic activities to this nuclear machinery under low-temperature conditions and that these activities determine the function of the spliceosome in cold acclimation.
It is remarkable that SME1, in addition to targeting selected introns depending on the environmental conditions, has a much more influential role under cold than control conditions. In fact, SME1 ensures the correct splicing of 938 introns when plants are exposed to 4°C while establishing the proper splicing of 151 introns at 20°C. This preferential function, moreover, comes about at the level of both constitutive (84/723) and alternative (67/215) spliced introns. However, this does not seem to be a common characteristic of the spliceosome core components. For instance, the LSM2-8 complex, which, as already mentioned, also negatively regulates the process of cold acclimation by targeting specific introns (Carrasco-López et al., 2017), controls the splicing of a similar number of introns under both control and low-temperature conditions (Carrasco-López et al., 2017), strongly suggesting that some core components of the spliceosome function mainly under particular environmental situations.
The substantial question emerging from the results discussed in this work is which molecular determinants underlie the specificity of SME1. An obvious possibility is that the selected introns targeted by this spliceosome core component belong to genes highly or exclusively transcribed under control conditions or in response to low temperature. The expression data from the eFP browser database (bar.utoronto.ca), however, reveals that this is not the case. Indeed, most genes containing specific introns targeted by SME1 at 20°C (65.7%) or 4°C (56.3%) show similar transcription levels under both conditions (Kilian et al., 2007). We also considered the possibility that the specific introns could contain singular sequence motifs. At both 20°C and at 4°C, the scanning of introns, using the MEME-suit to detect sequence motif enrichments, and the analysis of the frequencies of nucleotide sequences around their 5′ and 3′ splice sites or in their branch sites, using the WebLogo application, did not unveil substantial differences (Figure 8C). Moreover, we could not find important differences with the splice and branch site consensus sequences described for Arabidopsis (Szcześniak et al., 2013) in any case (Figure 8D). Recently, it has been proposed that intron selection by the Arabidopsis LSM2-8 complex could be determined by the size of introns (Carrasco-López et al., 2017). Our data suggest that this could also be the case for SME1. The comparison of the introns targeted by SME1 at 20°C and 4°C with the corresponding non-targeted ones evidenced that the former are significantly shorter (124.0 versus 164.1 bp [P < 0.0001] and 134.3 versus 163.8 bp [P < 0,0001] at 20°C and 4°C, respectively). Interestingly, it has been described that the demethylation of LSM4 is required for spliceosome assembly and adequate pre-mRNA splicing in plants and humans (Brahms et al., 2001; Zhang et al., 2011). Therefore, the possibility that posttranslational modifications of the SME1 protein could also contribute to its specificity remains to be determined. Finally, it is tempting to speculate that posttranscriptional chemical modifications and/or secondary structures of pre-mRNAs might also account for the functional specificity of SME1. Identifying the molecular mechanisms that determine this specificity constitutes an important goal for future studies that will provide insights into how plants respond and adapt to their changing environment.
METHODS
Plant Materials, Growth Conditions, Treatments, and Tolerance Assays
Arabidopsis (Arabidopsis thaliana) Columbia-0 (Col-0) ecotype was used in all experiments as wild-type plant. sme1-1 (Salk-119088), and sme1-2 (Salk-089521) mutant lines were obtained from the Nottingham Arabidopsis Stock Center and genotyped using the primers listed in Supplemental Data Set 10. The Arabidopsis cbf123-1 mutant was kindly provided by J.K. Zhu and aba2-11 mutant plants by P. Rodriguez. Plants were grown at 20°C under long-day photoperiods (16 h of cool-white fluorescent light, photon flux of 90 µmol m−2 s−1) in pots containing a mixture of organic substrate and vermiculite (3:1, v/v), or in Petri dishes containing 1/2 Murashige and Skoog medium supplemented with 1% (w/v) Suc and solidified with 0.9% (w/v) plant agar (germination medium [GM]).
Low temperature treatments for analyses of transcript accumulation were performed with 2-week-old plants exposed for the indicated times to 4°C in a growth chamber under long-day photoperiod (cool-white fluorescent light, photon flux of 40 µmol m−2 s−1). Salt application for transcript accumulation analyses was accomplished by transferring 2-week-old plants grown vertically on nylon meshes in Petri dishes under standard conditions to new plates containing GM medium supplemented with 150 mM NaCl for the indicated times. Dehydration for analyses of transcript accumulation was performed by allowing rosettes from 2-week-old Arabidopsis grown on soil to dehydrate on filter paper until attaining 80 or 60% of their fresh weight in a chamber set to 20°C and 60% humidity. Rosettes were detached from their roots immediately before being placed on filter papers. Tolerance to freezing temperatures was determined on 2-week-old plants grown on soil as described (Catalá et al., 2014). Tolerance to salt stress was analyzed by transferring 5-day-old seedlings grown vertically on nylon meshes in plates containing GM medium under control conditions to new plates supplemented with 120 or 150 mM NaCl.
Tolerances were quantified after 1 week of treatment, measuring the length of the main roots with respect to the roots of untreated plants. Tolerance to dehydration was assessed on rosettes from 2-week-old plants grown on soil, previously detached from their roots. Tolerances were estimated as the percentages of initial fresh weights that remained after allowing rosettes to dehydrate on filter papers for different times in a chamber set to 20°C and 60% humidity. In all cases, data reported are expressed as the SD of the mean of three independent experiments with 50 plants each.
Transcript Accumulation Analysis
For qPCR) experiments, total RNA was extracted using Purezol reagent (Bio-Rad) according to the manufacturer’s protocol. RNA samples were treated with DNase I (Roche) and quantified with a Nanodrop spectrophotometer (Thermo Fisher Scientific). Complementary DNA (cDNA) was synthesized with the iScript cDNA Synthesis Kit (Bio-Rad) following the manufacturer’s instructions and used as a template for qPCR assays employing the SsoFast EvaGreen Supermix (Bio-Rad) in an iQ2 thermal cycler machine (Bio-Rad) with the specific primers listed in Supplemental Data Set 10. The relative accumulation values were calculated using At4g26410 as a reference (Czechowski et al., 2005), and the ΔΔCT method to determine fold changes (Livak and Schmittgen, 2001).
To quantify the levels of the two major COOLAIR splicing variants, we basically followed the method previously described (Liu et al., 2010). Total RNA, extracted and quantified as described above, was reverse transcribed with the SuperScript III Kit (Invitrogen), according to the manufacturer’s instructions using oligo dT, and Coolair total_LP as primers (Supplemental Data Set 10). The cDNAs obtained were used as template for qPCR reactions in a LightCycler 480 System (Roche) using specific primers (Supplemental Data Set 10). The relative accumulation values were calculated using UBC21 (At5g25760) as a reference (Czechowski et al., 2005), and the ΔΔCT method to determine fold changes (Livak and Schmittgen, 2001).
Data disclosed are always expressed as means and the SD of three independent experiments with 50 plants each.
Subcellular Localization
The SME1PRO-SME1-GFP fusion was generated by amplifying the genomic region of the SME1 gene, including 1380 bp of its promoter region, with specific primers (Supplemental Data Set 10). The resulting PCR product was then introduced into the pMDC107 binary vector (Curtis and Grossniklaus, 2003) using the In-Fusion HD cloning kit (Takara Clontech). For subcellular localization of SME1, transient expression of the SME1PRO-SME1-GFP fusion protein was analyzed by confocal microscopy 3 d after agroinfiltration in leaves of 3-week-old Nicotiana benthamiana plants exposed to 20°C or 4°C for 24 h, as reported (English et al., 1997). Microscopy images were collected using a TCS SP2 confocal laser spectral microscope (Leica Microsystems). The excitation line for imaging GFP was 488 nm. Microscopy analyses were performed in triplicate with independent samples.
RNA-seq Experiments
Total RNAs for RNA-seq experiments were extracted with Purezol reagent (Bio-Rad) and purified with the RNeasy Plant Mini Kit (Qiagen). cDNA libraries were generated from three independent RNA preparations each. RNA quality determination, library preparation, and subsequent sequencing in an Illumina HiSeq 2000 platform were performed by the staff of the Beijing Genome Institute. Approximately 50 million 91-bp paired-end reads per sample were generated and > 90% reads were aligned to the TAIR10 Col-0 reference genome using SOAP2 (Li et al., 2009) with default parameters. Transcript levels were calculated using the reads per kilobase per million reads method (Mortazavi et al., 2008). Differentially expressed genes were identified using the algorithm developed by Audic and Claverie (1997) to obtain a P-value for each gene between any pair of samples. Then, an FDR analysis was performed to determine the threshold of P values in multiple tests. We established an FDR ≤ 0.001 and a fold change ± 2 as cutoffs for any given differentially expressed gene.
The Expression Browser tool of The Bio-Analytic Resource for Plant Biology (http://bar.utoronto.ca) was used to determine the genes from our RNA-seq data that, in addition of displaying IR events or altered expression in the sme1-1 mutant, were cold induced. In this analysis, the selected settings were “AtGenExpressstress series” as data set and “cold stress” as research area (Kilian et al., 2007). The total number of cold-regulated genes in Arabidopsis (12,184) was estimated from the data described by Kilian et al. (2007), considering those genes showing a fold change ± 2 in at least one time point or tissue analyzed.
Detection of Differential Alternative Splicing Events
The identification and quantification of alternative splicing events was mainly performed as previously described (Carrasco-López et al., 2017). In brief, reads obtained from the RNA-seq experiments were aligned to the TAIR10 Col-0 reference genome using TopHat2 with default parameters (Kim et al., 2013). Then, splicing events on each alignment file were identified through Regtools (https://github.com/griffithlab/regtools) with an anchor length of 0. Reads whose spliced region contained an exon annotated in TAIR10 were marked as ES events. Spliced reads that shared an identical 3′ splicing site but different 5′ splicing sites in at least one of the samples analyzed were annotated as A5′SS. Similarly, spliced reads that shared 5′ splicing sites but different 3′ splicing sites were annotated as A3′SS. To reduce the false positive rate, only events with more than five reads in at least one of the samples analyzed were considered. For the identification of IR events, a list of nonoverlapping introns per gene in the TAIR10 annotation were constructed. Introns that were covered in > 90% of their length with an average depth of more than five reads per base in at least one sample were considered for differential retention analysis. Intron retention levels were always calculated as a ratio between the expression levels of the introns and those of the corresponding genes. All introns annotated as retained in the wild type sample were classified as alternatively spliced. On the other hand, introns not annotated as retained were classified as constitutively spliced. The Q value for differential splicing events between two samples was determined using the DESeq2 package in R (Love et al., 2014). Alternatively spliced events with a fold change ± 2 and a Q value < 0.05, after correction for multiple testing using the Benjamini-Hochberg method, were annotated as significant. Events exclusively observed in sme1-1 mutants and with a mean of at least five reads per base throughout their sequences, were also considered significant, and their assigned Q value was zero.
Validation of IR Events
Validation of IR events was performed by qPCR analysis using specific primers (Supplemental Data Set 10) and three independent RNA samples from those utilized for the RNA-seq experiments. The expression of At4g26410 was used as reference to normalize the data obtained (Czechowski et al., 2005), and the ΔΔCT method was used to determine fold changes (Livak and Schmittgen, 2001). In all cases, IR levels were calculated as a ratio between the expression levels of the introns and those of the corresponding genes. Data presented are expressed as the SD of the mean of three independent experiments with 50 plants each.
Sequence Analysis
The size and GC content of introns and exons were calculated from the TAIR10 reference genome. Introns annotated as retained were classified based on their NMD pathway characteristics. The identification was based on the TAIR10 reference genome and the criteria previously described (Yu et al., 2016; i.e., premature termination codon and a 3′ UTR longer than 350 nt, 3′ UTR longer than 350 nt, more than 55 nt between the stop codon and a downstream intron, upstream open reading frame (uORF) longer than 35 amino acids, uORF overlapping with the start codon of the main ORF). WebLogo application (http://weblogo.threeplusone.com) was used to calculate and represent the nucleotide frequency in the 5′ and 3′ splice sites (five nucleotides of both the intron and the adjacent exon) and the branch site. The sequences of the 5′ and 3′ splice sites were obtained from the TAIR10 genome, and those of the branch sites were obtained from the database of plant splice sites and splicing signals ERISdb (Szcześniak et al., 2013). The identification of nucleotide motifs in the sequences of introns was performed by scanning the whole introns with MEME (www.meme-suit.org).
Sequence alignment of SME proteins from Arabidopsis (At2g18740.1 and At4g30330.1), Saccharomyces cerevisiae (YOR159C), Homo sapiens (NP_003085), and Drosophila melanogaster (NP_609162) was generated using ClustalW2 software (Larkin et al., 2007) and edited with Geneious software.
Statistical Analysis
Data sets were statistically analyzed with Prism 6 software (GraphPad Software). Comparisons between multiple groups were made by one-way ANOVA followed by Dunnett’s post-hoc test. ANOVA results are shown in Supplemental Data Set 11.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL data libraries under the accession numbers described in Supplemental Data Set 12. The RNA-seq data were submitted to the Gene Expression Omnibus database (www.ncbi.nlm.nih.gov/geo/) under accession number GSE116964.
Supplemental Data
The following materials are available in the online version of this article:
Supplemental Figure 1. Arabidopsis sme1-1 and sme1-2 mutants are null or severely hypomorphic.
Supplemental Figure 2. Quantification of developmental phenotypes shown by Arabidopsis sme1 mutants.
Supplemental Figure 3. Schematic representation of COOLAIR splicing variants.
Supplemental Data Set 1. Altered splicing events identified in sme1-1 plants grown at 20°C.
Supplemental Data Set 2. Genes with IR events in sme1-1 plants grown at 20°C that originate NMD signatures.
Supplemental Data Set 3. Genes showing IR events in sme1-1 plants grown at 20°C that are involved in Arabidopsis development.
Supplemental Data Set 4. Genes with altered expression in sme1-1 plants grown at 20°C.
Supplemental Data Set 5. Altered splicing events identified in sme1-1 plants exposed to 4°C.
Supplemental Data Set 6. Genes with IR events in sme1-1 plants exposed to 4°C that originate NMD signatures.
Supplemental Data Set 7. Genes with IR in sme1-1 plants exposed to 4°C whose expression has been described to be regulated by low temperature.
Supplemental Data Set 8. Cold-regulated genes with altered expression in sme1-1 plants exposed to 4°C.
Supplemental Data Set 9. Genes showing IR events in sme1-1 plants exposed to 4°C whose expression has been described to be regulated by low temperature and display NMD signatures.
Supplemental Data Set 10. Specific primers used in this article.
Supplemental Data Set 11. ANOVA results.
Supplemental Data Set 12. Accession numbers and full names of the genes mentioned in this article.
Dive Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
NMD Gramene: nonsense-mediated mRNA decay
NMD Araport: nonsense-mediated mRNA decay
Acknowledgments
We thank Jian-Kang Zhu (PSC/Purdue University) for the cbf123-1 mutant and Pedro Rodríguez (IBMCP/UPV) for the aba2-11 mutant. Furthermore, we thank J.J. Sanchez-Serrano and C. Carrasco for helpful discussions and comments. This research was supported by grants from AEI/FEDER, UE (BIO2016-79187-R to J.S., BIO2016-77559-R to J.A.J., and RYC-2013-14689 to P.C.).
AUTHOR CONTRIBUTIONS
R.H., R.C., M.M.C., P.C., and J.S. designed the research. R.H., R.C., M.M.C., and P.C. performed the research. R.H., R.C., J.M.J.-G., M.M.C., P.C., M.P., J.A.J., and J.S. analyzed the data. R.C. and J.S. wrote the paper.
References
- Arabidopsis Interactome Mapping Consortium (2011). Evidence for network evolution in an Arabidopsis interactome map. Science 333: 601–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Audic S., Claverie J.M. (1997). The significance of digital gene expression profiles. Genome Res. 7: 986–995. [DOI] [PubMed] [Google Scholar]
- Bazin J., Romero N., Rigo R., Charon C., Blein T., Ariel F., Crespi M. (2018). Nuclear speckle RNA binding proteins remodel alternative splicing and the non-coding Arabidopsis transcriptome to regulate a cross-talk between auxin and immune responses. Front. Plant Sci. 9: 1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berardini T.Z., et al. (2004). Functional annotation of the Arabidopsis genome using controlled vocabularies. Plant Physiol. 135: 745–755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black D.L. (2003). Mechanisms of alternative pre-messenger RNA splicing. Annu. Rev. Biochem. 72: 291–336. [DOI] [PubMed] [Google Scholar]
- Brahms H., Meheus L., de Brabandere V., Fischer U., Lührmann R. (2001). Symmetrical dimethylation of arginine residues in spliceosomal Sm protein B/B′ and the Sm-like protein LSm4, and their interaction with the SMN protein. RNA 7: 1531–1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calixto C.P.G., Guo W., James A.B., Tzioutziou N.A., Entizne J.C., Panter P.E., Knight H., Nimmo H., Zhang R., and Brown J.W.S. (2018). Rapid and dynamic alternative splicing impacts the Arabidopsis cold response transcriptome. Plant Cell 30: 1424–1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao J., Shi F., Liu X., Jia J., Zeng J., Huang G. (2011). Genome-wide identification and evolutionary analysis of Arabidopsis sm genes family. J. Biomol. Struct. Dyn. 28: 535–544. [DOI] [PubMed] [Google Scholar]
- Capovilla G., Delhomme N., Collani S., Shutava I., Bezrukov I., Symeonidi E., de Francisco Amorim M., Laubinger S., Schmid M. (2018). PORCUPINE regulates development in response to temperature through alternative splicing. Nat. Plants 4: 534–539. [DOI] [PubMed] [Google Scholar]
- Carrasco-López C., Hernández-Verdeja T., Perea-Resa C., Abia D., Catalá R., Salinas J. (2017). Environment-dependent regulation of spliceosome activity by the LSM2-8 complex in Arabidopsis. Nucleic Acids Res. 45: 7416–7431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catalá R., López-Cobollo R., Mar Castellano M., Angosto T., Alonso J.M., Ecker J.R., Salinas J. (2014). The Arabidopsis 14-3-3 protein RARE COLD INDUCIBLE 1A links low-temperature response and ethylene biosynthesis to regulate freezing tolerance and cold acclimation. Plant Cell 26: 3326–3342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis M.D., Grossniklaus U. (2003). A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol. 133: 462–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czechowski T., Stitt M., Altmann T., Udvardi M.K., Scheible W.R. (2005). Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol. 139: 5–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng X., Lu T., Wang L., Gu L., Sun J., Kong X., Liu C., Cao X. (2016). Recruitment of the NineTeen Complex to the activated spliceosome requires AtPRMT5. Proc. Natl. Acad. Sci. USA 113: 5447–5452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Y., Humbert S., Liu J.X., Srivastava R., Rothstein S.J., Howell S.H. (2011). Heat induces the splicing by IRE1 of a mRNA encoding a transcription factor involved in the unfolded protein response in Arabidopsis. Proc. Natl. Acad. Sci. USA 108: 7247–7252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Y., Li H., Zhang X., Xie Q., Gong Z., Yang S. (2015). OST1 kinase modulates freezing tolerance by enhancing ICE1 stability in Arabidopsis. Dev. Cell 32: 278–289. [DOI] [PubMed] [Google Scholar]
- Du J.L., Zhang S.W., Huang H.W., Cai T., Li L., Chen S., He X.J. (2015). The splicing factor PRP31 is involved in transcriptional gene silencing and stress response in Arabidopsis. Mol. Plant 8: 1053–1068. [DOI] [PubMed] [Google Scholar]
- Elvira-Matelot E., Bardou F., Ariel F., Jauvion V., Bouteiller N., Le Masson I., Cao J., Crespi M.D., Vaucheret H. (2016). The nuclear ribonucleoprotein SmD1 interplays with splicing, RNA quality control and posttranscriptional gene silencing in Arabidopsis. Plant Cell 28: 426–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- English J.J., Davenport G.F., Elmayan T., Vaucheret H., Baulcombe D.C. (1997). Requirement of sense transcription for homology-dependent virus resistance and trans-inactivation. Plant J. 12: 597–603. [Google Scholar]
- Feng J., Li J., Gao Z., Lu Y., Yu J., Zheng Q., Yan S., Zhang W., He H., Ma L., Zhu Z. (2015). SKIP confers osmotic tolerance during salt stress by controlling alternative gene splicing in Arabidopsis. Mol. Plant 8: 1038–1052. [DOI] [PubMed] [Google Scholar]
- Fica S.M. and Nagai K. (2017). Cryo-electron microscopy snapshots of the spliceosome: Structural insights into a dynamic ribonucleoprotein machine. Nat. Struct. Mol. Biol. 24: 791–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galletti R., Johnson K.L., Scofield S., San-Bento R., Watt A.M., Murray J.A.H., Ingram G.C. (2015). DEFECTIVE KERNEL 1 promotes and maintains plant epidermal differentiation. Development 142: 1978–1983. [DOI] [PubMed] [Google Scholar]
- Ge C., Cui X., Wang Y., Hu Y., Fu Z., Zhang D., Cheng Z., Li J. (2006). BUD2, encoding an S-adenosylmethionine decarboxylase, is required for Arabidopsis growth and development. Cell Res. 16: 446–456. [DOI] [PubMed] [Google Scholar]
- González-Guzmán M., Apostolova N., Bellés J.M., Barrero J.M., Piqueras P., Ponce M.R., Micol J.L., Serrano R., Rodríguez P.L. (2002). The short-chain alcohol dehydrogenase ABA2 catalyzes the conversion of xanthoxin to abscisic aldehyde. Plant Cell 14: 1833–1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruss O.J., Meduri R., Schilling M., Fischer U. (2017). UsnRNP biogenesis: Mechanisms and regulation. Chromosoma 126: 577–593. [DOI] [PubMed] [Google Scholar]
- Guan Q., Wu J., Zhang Y., Jiang C., Liu R., Chai C., Zhu J. (2013). A DEAD box RNA helicase is critical for pre-mRNA splicing, cold-responsive gene regulation, and cold tolerance in Arabidopsis. Plant Cell 25: 342–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerra D., Crosatti C., Khoshro H.H., Mastrangelo A.M., Mica E., Mazzucotelli E. (2015). Post-transcriptional and post-translational regulations of drought and heat response in plants: A spider’s web of mechanisms. Front. Plant Sci. 6: 57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermann H., Fabrizio P., Raker V.A., Foulaki K., Hornig H., Brahms H., Lührmann R. (1995). snRNP Sm proteins share two evolutionarily conserved sequence motifs which are involved in Sm protein-protein interactions. EMBO J. 14: 2076–2088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilian J., Whitehead D., Horak J., Wanke D., Weinl S., Batistic O., D’Angelo C., Bornberg-Bauer E., Kudla J., Harter K. (2007). The AtGenExpress global stress expression data set: Protocols, evaluation and model data analysis of UV-B light, drought and cold stress responses. Plant J. 50: 347–363. [DOI] [PubMed] [Google Scholar]
- Kim D., Pertea G., Trapnell C., Pimentel H., Kelley R., Salzberg S.L. (2013). TopHat2: Accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14: R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim G.-D., Cho Y.-H., Lee B.H., Yoo S.-D. (2017). STABILIZED1 modulates pre-mRNA splicing for thermotolerance. Plant Physiol. 173: 2370–2382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J.S., Jung H.J., Lee H.J., Kim K.A., Goh C.H., Woo Y., Oh S.H., Han Y.S., Kang H. (2008). Glycine-rich RNA-binding protein 7 affects abiotic stress responses by regulating stomata opening and closing in Arabidopsis thaliana. Plant J. 55: 455–466. [DOI] [PubMed] [Google Scholar]
- Laloum T., Martín G., Duque P. (2018). Alternative splicing control of abiotic stress responses. Trends Plant Sci. 23: 140–150. [DOI] [PubMed] [Google Scholar]
- Larkin M.A., et al. (2007). Clustal W and Clustal X version 2.0. Bioinformatics 23: 2947–2948. [DOI] [PubMed] [Google Scholar]
- Lee C.M., Thomashow M.F. (2012). Photoperiodic regulation of the C-repeat binding factor (CBF) cold acclimation pathway and freezing tolerance in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 109: 15054–15059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y., Rio D.C. (2015). Mechanisms and regulation of alternative pre-mRNA splicing. Annu. Rev. Biochem. 84: 291–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee B.H., Kapoor A., Zhu J., Zhu J.K. (2006). STABILIZED1, a stress-upregulated nuclear protein, is required for pre-mRNA splicing, mRNA turnover, and stress tolerance in Arabidopsis. Plant Cell 18: 1736–1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R., Yu C., Li Y., Lam T.-W., Yiu S.M., Kristiansen K., Wang J. (2009). SOAP2: An improved ultrafast tool for short read alignment. Bioinformatics 25: 1966–1967. [DOI] [PubMed] [Google Scholar]
- Liu F., Marquardt S., Lister C., Swiezewski S., Dean C. (2010). Targeted 3′ processing of antisense transcripts triggers Arabidopsis FLC chromatin silencing. Science 327: 94–97. [DOI] [PubMed] [Google Scholar]
- Liu J., Sun N., Liu M., Liu J., Du B., Wang X., Qi X. (2013). An autoregulatory loop controlling Arabidopsis HsfA2 expression: Role of heat shock-induced alternative splicing. Plant Physiol. 162: 512–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K.J., Schmittgen T.D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Δ Δ C(T)) method. Methods 25: 402–408. [DOI] [PubMed] [Google Scholar]
- Love M.I., Huber W., Anders S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15: 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahrez W., Shin J., Muñoz-Viana R., Figueiredo D.D., Trejo-Arellano M.S., Exner V., Siretskiy A., Gruissem W., Köhler C., Hennig L. (2016). BRR2a affects flowering time via FLC splicing. PLoS Genet. 12: e1005924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marquardt S., Raitskin O., Wu Z., Liu F., Sun Q., Dean C. (2014). Functional consequences of splicing of the antisense transcript COOLAIR on FLC transcription. Mol. Cell 54: 156–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medina J., Catalá R., Salinas J. (2011). The CBFs: Three arabidopsis transcription factors to cold acclimate. Plant Sci. 180: 3–11. [DOI] [PubMed] [Google Scholar]
- Mortazavi A., Williams B.A., McCue K., Schaeffer L., Wold B. (2008). Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 5: 621–628. [DOI] [PubMed] [Google Scholar]
- Nakashima K., Yamaguchi-Shinozaki K., Shinozaki K. (2014). The transcriptional regulatory network in the drought response and its crosstalk in abiotic stress responses including drought, cold, and heat. Front. Plant Sci. 5: 170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perea-Resa C., Hernández-Verdeja T., López-Cobollo R., del Mar Castellano M., Salinas J. (2012). LSM proteins provide accurate splicing and decay of selected transcripts to ensure normal Arabidopsis development. Plant Cell 24: 4930–4947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saiga S., Möller B., Watanabe-Taneda A., Abe M., Weijers D., Komeda Y. (2012). Control of embryonic meristem initiation in Arabidopsis by PHD-finger protein complexes. Development 139: 1391–1398. [DOI] [PubMed] [Google Scholar]
- Saltzman A.L., Pan Q., Blencowe B.J. (2011). Regulation of alternative splicing by the core spliceosomal machinery. Genes Dev. 25: 373–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlaen R.G., Mancini E., Sanchez S.E., Perez-Santángelo S., Rugnone M.L., Simpson C.G., Brown J.W.S., Zhang X., Chernomoretz A., Yanovsky M.J. (2015). The spliceosome assembly factor GEMIN2 attenuates the effects of temperature on alternative splicing and circadian rhythms. Proc. Natl. Acad. Sci. USA 112: 9382–9387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo P.J., Park M.J., Lim M.H., Kim S.G., Lee M., Baldwin I.T., Park C.M. (2012). A self-regulatory circuit of CIRCADIAN CLOCK-ASSOCIATED1 underlies the circadian clock regulation of temperature responses in Arabidopsis. Plant Cell 24: 2427–2442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo P.J., Park M.J., Park C.M. (2013). Alternative splicing of transcription factors in plant responses to low temperature stress: Mechanisms and functions. Planta 237: 1415–1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swaraz A.M., Park Y.D., Hur Y. (2011). Knock-out mutations of Arabidopsis SmD3-b induce pleotropic phenotypes through altered transcript splicing. Plant Sci. 180: 661–671. [DOI] [PubMed] [Google Scholar]
- Szcześniak M.W., Kabza M., Pokrzywa R., Gudyś A., Makałowska I. (2013). ERISdb: A database of plant splice sites and splicing signals. Plant Cell Physiol. 54: e10. [DOI] [PubMed] [Google Scholar]
- Tähtiharju S., Palva T. (2001). Antisense inhibition of protein phosphatase 2C accelerates cold acclimation in Arabidopsis thaliana. Plant J. 26: 461–470. [DOI] [PubMed] [Google Scholar]
- Teotia S., Lamb R.S. (2011). RCD1 and SRO1 are necessary to maintain meristematic fate in Arabidopsis thaliana. J. Exp. Bot. 62: 1271–1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittaker C., Dean C. (2017). The FLC locus: A platform for discoveries in epigenetics and adaptation. Annu. Rev. Cell Dev. Biol. 33: 555–575. [DOI] [PubMed] [Google Scholar]
- Will C.L., Lührmann R. (2011). Spliceosome structure and function. Cold Spring Harb. Perspect. Biol. 3: a003707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilusz C.J., Wilusz J. (2013). Lsm proteins and Hfq: Life at the 3′ end. RNA Biol. 10: 592–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong L., Gong Z., Rock C.D., Subramanian S., Guo Y., Xu W., Galbraith D., Zhu J.K. (2001). Modulation of abscisic acid signal transduction and biosynthesis by an Sm-like protein in Arabidopsis. Dev. Cell 1: 771–781. [DOI] [PubMed] [Google Scholar]
- Yu H., Tian C., Yu Y., Jiao Y. (2016). Transcriptome survey of the contribution of alternative splicing to proteome diversity in Arabidopsis thaliana. Mol. Plant 9: 749–752. [DOI] [PubMed] [Google Scholar]
- Zhang R., et al. (2017). A high quality Arabidopsis transcriptome for accurate transcript-level analysis of alternative splicing. Nucleic Acids Res. 45: 5061–5073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., et al. (2011). Arabidopsis floral initiator SKB1 confers high salt tolerance by regulating transcription and pre-mRNA splicing through altering histone H4R3 and small nuclear ribonucleoprotein LSM4 methylation. Plant Cell 23: 396–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Z., Assmann S.M. (2011). The glycolytic enzyme, phosphoglycerate mutase, has critical roles in stomatal movement, vegetative growth, and pollen production in Arabidopsis thaliana. J. Exp. Bot. 62: 5179–5189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C., Zhang Z., Xie S., Si T., Li Y., Zhu J.K. (2016). Mutational evidence for the critical role of CBF transcription factors in cold acclimation in Arabidopsis. Plant Physiol. 171: 2744–2759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zieve G.W. (1999). The cytoplasmic sites of the snRNP protein complexes are punctate structures that are responsive to changes in metabolism and intracellular architecture. Exp. Cell Res. 247: 249–256. [DOI] [PubMed] [Google Scholar]