Cotton Cys-rich repeat protein 1 participates in defense responses by protecting chitinase 28 from cleavage by a Ser protease of Verticillium dahliae.
Abstract
The apoplast serves as the first battlefield between the plant hosts and invading microbes; therefore, work on plant-pathogen interactions has increasingly focused on apoplastic immunity. In this study, we identified three proteins in the apoplast of cotton (Gossypium sp) root cells during interaction of the plant with the fungal pathogen Verticillium dahliae. Among these proteins, cotton host cells secrete chitinase 28 (Chi28) and the Cys-rich repeat protein 1 (CRR1), while the pathogen releases the protease VdSSEP1. Biochemical analysis demonstrated that VdSSEP1 hydrolyzed Chi28, but CRR1 protected Chi28 from cleavage by Verticillium dahliae secretory Ser protease 1 (VdSSEP1). In accordance with the in vitro results, CRR1 interacted with Chi28 in yeast and plant cells and attenuated the observed decrease in Chi28 level that occurred in the apoplast of plant cells upon pathogen attack. Knockdown of CRR1 or Chi28 in cotton plants resulted in higher susceptibility to V. dahliae infection, and overexpression of CRR1 increased plant resistance to V. dahliae, the fungus Botrytis cinerea, and the oomycete Phytophthora parasitica var nicotianae. By contrast, knockout of VdSSEP1 in V. dahliae destroyed the pathogenicity of this fungus. Together, our results provide compelling evidence for a multilayered interplay of factors in cotton apoplastic immunity.
INTRODUCTION
Microbial plant pathogens have developed various strategies to infect their plant hosts and cause diseases. In response, plants have developed a complex and multilayered immune system to recognize and combat the invading pathogens (Akira et al., 2006; Jones and Dangl, 2006; Spoel and Dong, 2012). Many microbials attack plant cells initially in the apoplast, an extracellular space in plant tissues that serves as the first battlefield between microbial invaders and their plant hosts (Doehlemann and Hemetsberger, 2013). Upon infection, pathogens secrete effectors, including proteases, hydrolytic enzymes, and protease inhibitors to facilitate the spread of disease (Jashni et al., 2015b; Li et al., 2016a; Ma et al., 2017). Conversely, plants export extracellular vesicles and compounds such as proteases and protease inhibitors to defend themselves against pathogens (Valueva and Mosolov, 2004; van der Hoorn, 2008; Kim et al., 2009; Rutter and Innes, 2017, 2018). Host proteases and hydrolytic enzymes can decompose microbial matrices to generate soluble pattern-recognition receptor ligands that initiate PTI responses (Liu et al., 2014). Therefore, apoplastic immunity is an important component of the plant immune responses to pathogens. Identifying the key players in apoplastic immunity will help us better understand the molecular mechanisms underlying plant-pathogen interactions.
Plant chitinases are a group of widely distributed and structurally heterogeneous enzymes. Accumulating evidence indicates that chitinase and its enzymatic products play a central role in plant-fungus interactions. In defending against fungal pathogen invasion, plants secrete chitinases into the apoplast to digest chitin in fungal cell wall, which produces pathogen-associated molecular patterns and ultimately leads to immune recognition (Collinge et al., 1993; van Loon et al., 2006; Cota et al., 2007; Miya et al., 2007; Wan et al., 2008; Sinha et al., 2014). Consistently, it has been reported that Cys-rich transmembrane chitin elicitor-binding protein (CEBiP) of rice (Oryza sativa) and chitin elicitor receptor kinase 1 (CERK1) of Arabidopsis (Arabidopsis thaliana) could bind the extracellular chitin and thus induced the defense response (Shinya et al., 2012, 2015; Kouzai et al., 2014). Also, transgenic plants overexpressing chitinase genes exhibit enhanced resistance to fungal attack (Schickler and Chet, 1997; de las Mercedes Dana et al., 2006; Maximova et al., 2006; Iqbal et al., 2012; Cletus et al., 2013).
A large gene family encodes proteins with one or more anti-fungal domains, which are composed of one or two Cys-rich domain of unknown function 26 motifs (DUF26) consisting of a C-X8-C-X2-C element. This motif was initially identified in the extracellular regions of Cys-rich receptor-like kinases (CRKs), plasmodesmata-located proteins (PDLP), and Cys-rich secretory DUF26 proteins (Chen, 2001; Bourdais and CRK Consortium et al., 2015). These proteins possess antifungal activity and meanwhile play a role in salt-stress responses (Zhang et al., 2009; Miyakawa et al., 2014). For example, overexpression of the salicylic acid-inducible genes CRK4, CRK5, CRK19, and CRK20 activates hypersensitive response-like cell death (Chen et al., 2003, 2004), and CRK4, CRK6, CRK13, and CRK36 increases tolerance to the bacterial pathogen Pseudomonas syringae pv tomato DC3000 (Pto DC3000; Acharya et al., 2007; Yeh et al., 2015). Arabidopsis PDLP1 and PDLP5 play important roles in the coordination of cell-to-cell communication in defense against the pathogens (Lee et al., 2011; Caillaud et al., 2014).
Compared with CRKs and PDLPs, our understanding of the physiological functions and molecular mechanisms used by Cys-rich secretory DUF26 proteins is limited. The rice root meander curling (OsRMC) protein, a secretory DUF26 protein, was implicated in the salt-stress response (Zhang et al., 2009). In addition, it was shown that the gymnosperm Ginkgo biloba secretes the DUF26 protein ginkbilobin2 (Gnk2), which contains a single DUF26 domain and is thought to function as an antimicrobial protein, binding mannose to inhibit the growth of fungi (Sawano et al., 2007; Miyakawa et al., 2014). These findings suggest that the secreted DUF26 proteins might function in biotic and abiotic stress responses in plants, but the molecular mechanisms of DUF26 proteins in plant-pathogen interactions are largely unknown.
Cotton Verticillium wilt is a severe vascular disease, which is caused by the soil-borne fungus Verticillium dahliae. Between the two major cotton cultivars, sea-island cotton (Gossypium barbadense) shows much higher resistance to V. dahliae infection than upland cotton (Gossypium hirsutum), the most widely grown cotton cultivar worldwide (Wilhelm et al., 1974). Exploring the key factors involved in the defense against V. dahliae invasion in G. barbadense can provide candidate genes for generating wilt-resistant G. hirsutum cultivars through molecular breeding (Li et al., 2014; Sun et al., 2014; Jun et al., 2015). Previously, we performed a comparative analysis of the apoplastic proteomes of G. barbadense upon V. dahliae infection to investigate the molecular mechanisms involved in innate immunity. We identified a set of apoplastic proteins from cotton root that are differentially expressed upon V. dahliae infection. For example, nucleoredoxin 1 (NRX1) functions as a reactive oxygen species scavenger that participates in apoplast immunity in response to V. dahliae infection (Li et al., 2016b). In this study, we characterized the physiological function of another cotton apoplastic protein, Cys-rich repeat 1 (CRR1), a Cys-rich secreted DUF26 protein, during the cotton-V. dahliae interaction. CRR1 expression was induced by V. dahliae infection. Overexpression of CRR1 in cotton conferred enhanced tolerance to V. dahliae, and downregulation of CRR1 increased susceptibility to the pathogen. We found that CRR1 interacted with the cotton chitinase Chi28, which is crucial for cotton defense against V. dahliae infection. Cotton Chi28 was degraded by the secretory Ser protease 1 (VdSSEP1) from V. dahliae. The interaction of CRR1 and Chi28 prevented cleavage of Chi28 by VdSSEP1. Our findings suggest that CRR1 stabilizes chitinase to counteract the protease-induced cleavage of Chi28. These findings shed light on the molecular role of DUF26 protein family members in plant defense against fungus pathogens and reveal a new player in plant apoplastic immunity.
RESULTS
Expression of Cotton CRR1 in Response to V. dahliae Infection
Previously, we identified a group of V. dahliae-responsive apoplastic proteins from the root of wilt-resistant sea-island cotton (G. barbadense) cv Hai 7124 (Li et al., 2016b). In this study, we focused on one of these proteins, a DUF26 motif-containing protein that is significantly induced in cotton roots after V. dahliae inoculation. Bioinformatics analysis suggested that this protein is secreted and has two anti-fungal domains, which are composed of two Cys-rich DUF26 motifs. SignalP (Emanuelsson et al., 2000) predicted that a 23-residue sequence at the N terminus functions as a signal peptide (Supplemental Figure 1A). We designated this protein as CRR1, based on the presence of a Cys-rich repeat.
To identify CRR1 homologs in cotton, we used its intact and conserved antifungal domain sequences to search the databases of G. hirsutum and G. barbadense (Li et al., 2015; Yuan et al., 2015). We chose the proteins with the highest E value as the closest homologs of CRR1 and identified three such proteins, which we designated CRR2, CRR3, and CRR4. CRR1 shares 86%, 85%, and 62% sequence identity with CRR2, CRR3, and CRR4 proteins, respectively (Supplemental Figure 1B). All four CRRs are present in G. hirsutum and G. barbadense and each gene shares nearly identical coding sequences in the two cotton species (Supplemental Figure 2).
To test whether expression of these cotton CRR genes responds to V. dahliae infection, we analyzed their transcript levels in the roots of G. barbadense and G. hirsutum. Expression of CRR1 and CRR2 was significantly up-regulated upon V. dahliae infection (Figure 1A) in both species. Moreover, CRR1 and CRR2 were expressed to higher levels in G. barbadense than in G. hirsutum. Compared with the mock-inoculated controls, expression of CRR1 was enhanced ∼15-fold in G. barbadense and 5-fold in G. hirsutum upon inoculation with the pathogen (Figure 1A). Among the four CRRs, as CRR1 showed the strongest induction of expression in response to pathogen (∼3-, 10-, and 7.5-folds higher than CRR2, CRR3, and CRR4, respectively), we chose CRR1 for further study.
Figure 1.
Expression of CRRs in Cotton.
(A) RT-qPCR analysis of CRR expression in cotton roots in response to V. dahliae infection. Total RNA was extracted from cotton roots inoculated with V. dahliae spores (1 × 106 conidia/mL) for 0, 6, 12, 24, 36, 48, 60, and 72 h, respectively, and 0 h indicates cotton plants without inoculation with the fungal spores. Error bars represent SD from three independent experiments (n = 3). Lowercase letters represent statistically significant differences (P < 0.05, nested ANOVA). The experiments were repeated three times with similar results.
(B) Histochemical GUS staining of wild-type (WT) and CRR1pro:GUS transgenic seedlings of G. hirsutum cotton without or with V. dahliae infection.
(C) Histochemical GUS staining of true leaves of wild-type and CRR1pro:GUS transgenic G. hirsutum plants without or with V. dahliae infection. Bar, 0.5 cm.
(D) Photographs of the lateral roots of non-inoculated and V. dahliae-infected transgenic cotton plants. Bar, 0.1 cm.
(E) Cross-section of the non-inoculated or V. dahliae-infected transgenic cotton root: epidermal cell (E), cortex cell (C), and vascular cell (V). Bar, 100 μm.
A promoter fusion to the β-glucuronidase (GUS) reporter gene was used to confirm the pathogen-induced expression of CRR1 in cotton plants, using a 1.5-kb upstream fragment of CRR1 from G. barbadense. Due to the lack of available techniques for transformation of G. barbadense, GUS expression driven by the CRR1 promoter sequence was tested in G. hirsutum. Staining for GUS activity showed that GUS was expressed at low levels in the root and leaf of cotton seedlings not infected by V. dahliae (Figure 1B). When the plant was treated with V. dahliae spores, GUS expression was strongly induced in the root and true leaf (Figures 1B and 1C), further confirming that CRR1 is a V. dahliae-responsive gene. CRR1 promoter-driven GUS expression was mainly detected in the epidermis, some cortex tissue, and vascular cells of the root as shown in Figures 1D and 1E.
CRR1 Functions in Defense against V. dahliae Infection
We employed both G. hirsutum and G. barbadense to investigate the function of CRR1. Previously, it was reported that upland cotton (G. hirsutum) is susceptible to V. dahliae but sea-island cotton (G. barbadense) is resistant to V. dahliae infection (Wilhelm et al., 1974; Zhang et al., 2012; Chen et al., 2015). As an initial step, we confirmed the resistance of the two species side-by-side under our experimental conditions. Our results confirmed that G. barbadense cotton plants were much more resistant to V. dahliae infection than the G. hirsutum plants (Supplemental Figure 3).
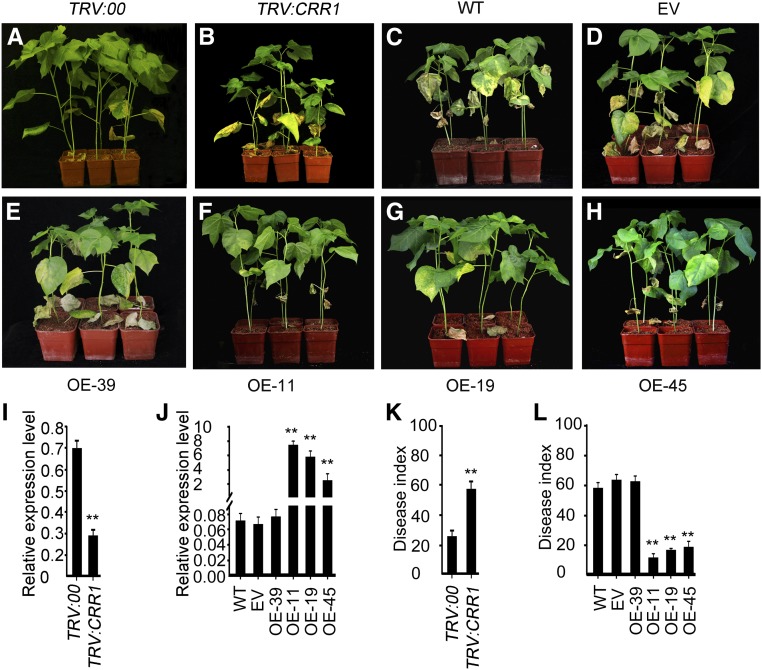
Then we evaluated the function of CRR1 in defense against V. dahliae infection in G. barbadense by examining plants with decreased CRR1 expression. We employed the vacuum infiltration method of virus-induced gene silencing (VIGS) to knock down the transcription of CRR1, using the gene-specific fragment. The cotton phytoene desaturase gene was used as an experimental marker for the silencing system (Supplemental Figure 4A). Two weeks after inoculation with the recombinant virus, the expression of CRR1 was strongly suppressed in the TRV:CRR1 G. barbadense plants compared with the TRV:00 control (Figure 2I). To see if off-target silencing of other CRR genes occurred in the VIGS plants, we examined the transcript levels of three other CRR genes and found that none of them was affected (Supplemental Figure 4B). The impact of CRR1 silencing on the wilt resistance of G. barbadense was then assessed, and we observed a significant decrease in resistance to V. dahliae infection (Figures 2A and 2B). Analysis of the disease indices further supported the results (Figure 2K).
Figure 2.
CRR1-Related Resistance to V. dahliae Infection in Cotton.
(A) and (B) Phenotypes of control and VIGS G. barbadense plants. Ten-day-old cotton plants were infiltrated with Agrobacterium cells carrying the VIGS-control vector (TRV:00) or TRV:CRR1 and inoculated with a suspension of V. dahliae spores at 2 weeks after infiltration. Photographs were taken at 2 weeks after V. dahliae inoculation.
(C) to (H) Phenotypes of the wild-type (WT) and empty vector (EV) transgenic and the CRR1-overexpressing (OE) G. hirsutum plants infected with fungal spores. Fifteen-day-old cotton plants were inoculated with a suspension of V. dahliae spores. Photographs were taken at 2 weeks after V. dahliae infection.
(I) and (J) RT-qPCR analysis of CRR1 expression in the VIGS G. barbadense roots (I), and wild-type, EV, CRR1-overexpressing G. hirsutum roots (J). Error bars indicate SD from three technical replicates of a single biological experiment. Asterisks indicate statistically significant differences, as determined by Student’s t test (**P < 0.01). The experiments were repeated three times with similar results.
(K) and (L) Disease index in the VIGS G. barbadense plants (K) and wild-type, EV, and CRR1-overexpressing G. hirsutum plants (L). Error bars represent SD of three biological replicates (n ≥ 36); asterisks indicate statistically significant differences, as determined by the Student’s t test (**P < 0.01).
In addition to using VIGS in G. barbadense, we also generated antisense transgenic G. hirsutum plants that can be transformed via Agrobacterium tumefaciens-mediated transformation. A recombinant plasmid harboring the full-length CRR1 complementary DNA (cDNA) in antisense orientation driven by the constitutive Cauliflower mosaic virus 35S promoter was introduced into the Verticillium wilt-susceptible G. hirsutum cultivar R15. Three CRR1-suppressed transgenic lines (An-4, An-7, and An-12) were used for further analysis. In these antisense plants, CRR1 expression was decreased to 20% of that of the wild type or empty vector (EV) control plants (Supplemental Figure 5B). Apart from CRR1, expression of CRR2 was also reduced, but to a lesser extent than CRR1 (75% of that of the wild-type or EV control plants), while CRR3 and CRR4 transcript levels were not significantly altered (Supplemental Figure 5C). V. dahliae challenge experiments showed that the antisense plants were more susceptible to V. dahliae, compared with the wild type and EV controls (Supplemental Figure 5A). The disease index values were higher in these plants as compared with the wild-type and EV control plants (Supplemental Figure 5D).
A gain-of-function approach was also used to characterize the function of CRR1 in defense against V. dahliae. Six independent CRR1-overexpressing transgenic lines were generated, and three (OE-11, OE-19, and OE-45) of them were chosen for phenotypic examination in the T2 generation. Transgenic G. hirsutum plants transformed with EV as well as a CRR1 transgenic line (OE-39) with unchanged CRR1 expression were used as controls (Figure 2J). The plants were inoculated with V. dahliae to test the defense-related role of CRR1. At 14 d after infection, overexpression of CRR1 led to a clear increase in disease resistance as compared with the wild type, EV, and OE-39 controls (Figures 2C to 2H). Further analysis showed that overexpression of CRR1 resulted in a reduced disease index (Figure 2L).
Collectively, these results demonstrated that expression level of CRR1 is tightly associated with resistance to V. dahliae infection in cotton.
CRR1 Interacts with a Class IV Chitinase in Cotton
As the secreted DUF26 protein ginkbilobin2 (Gnk2) from G. biloba was reported to inhibit the growth of fungi directly (Miyakawa et al., 2014), we attempted to see whether CRR1 had an inhibitory effect on V. dahliae by testing whether CRR1-soaked filter papers could inhibit V. dahliae growth on agar plates. However, these assays showed that CRR1 did not possess antifungal activity (Supplemental Figure 6).
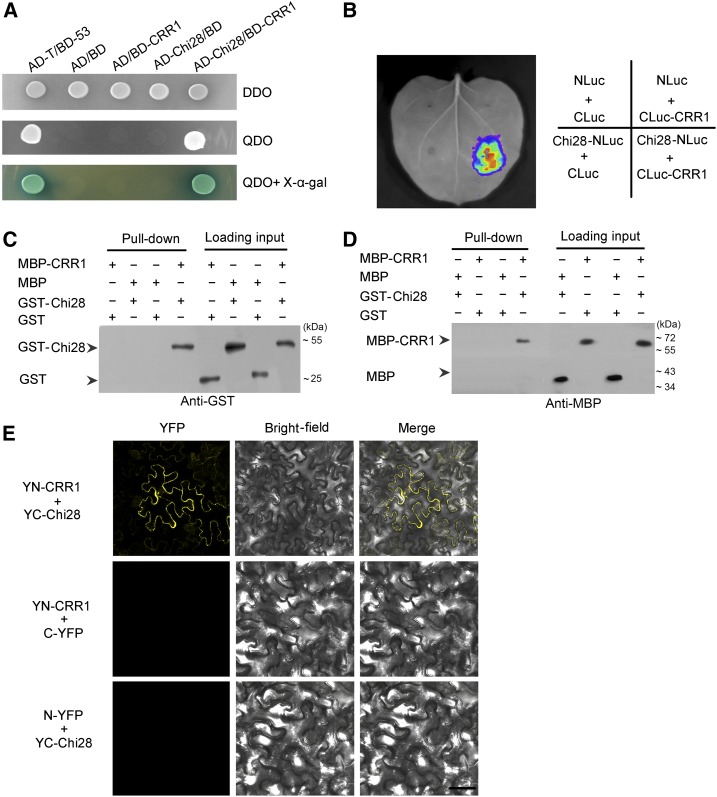
To explore the functional basis of CRR1 in the defense response, we screened for CRR1-interacting proteins in the yeast two-hybrid (Y2H) system. Screening of a cDNA library from G. barbadense roots infected by V. dahliae identified a class IV chitinase (Chi28) that strongly interacted with CRR1. Direct Y2H assays confirmed the interaction between the two proteins (Figure 3A). We performed a luciferase (Luc) complementation imaging assay to assess the interaction of CRR1 and Chi28 in plant cells. As shown in Figure 3B, Luc activity was detected in Nicotiana benthamiana leaf cells, but no Luc activity was detected in the negative controls.
Figure 3.
CRR1 Interacts with Class IV Chitinase 28 (Chi28).
(A) Yeast two-hybrid assay to detect interaction between CRR1 and Chi28. Yeast cells containing the indicated plasmids were grown on SD/-Leu/-Trp DO (DDO) plates and SD/-Leu/-Trp/-Ade/-His DO (QDO) plates (containing 40 mg/L X-α-gal) for 3 d. Interaction of AD/BD, AD/BD-CRR1, AD-Chi28/BD, and AD-T/BD-53 were used as the negative and positive controls, respectively.
(B) Luciferase (Luc) complementation imaging (LCI) analysis of the interaction between CRR1 and Chi28. The indicated plasmid pairs were transiently coexpressed in N. benthamiana. The luminescent signal was collected at 48 h after infiltration.
(C) and (D) In vitro pull-down assay. MBP-CRR1 fusion protein was used as bait, and GST-Chi28 fusion protein was used as prey. Anti-GST antibody was used to detect prey proteins (C); GST-Chi28 fusion protein was used as bait, and MBP-CRR1 fusion protein was used as prey. Anti-MBP antibody was used to detect prey proteins (D).
(E) Interaction test of CRR1 and Chi28 by BiFC assay in N. benthamiana. CRR1 was fused to the N-terminal fragment of YFP (YN-CRR1), and Chi28 was fused to the C-terminal fragment of YFP (YC-Chi28). YFP fluorescence indicates an interaction between the two proteins. Bar, 50 µm.
Next, we conducted a pull-down assay to further verify the interaction of the two proteins. We expressed and purified glutathione S-transferase (GST), GST-Chi28, Maltose binding protein (MBP), and MBP-CRR1 from the BL21 (DE3) strain of Escherichia coli and used these proteins in pull-down assays. Equal amounts of GST or GST-Chi28 proteins were incubated with immobilized MBP or MBP-CRR1 proteins. As expected, Chi28 bound to CRR1, but not to the control MBP proteins (Figure 3C). Likewise, CRR1 bound to Chi28, but not to the control GST proteins (Figure 3D). These results confirmed that CRR1 and Chi28 could interact. Furthermore, a bimolecular fluorescence complementation (BiFC) assay showed that the interaction of the two proteins occurred in the peripheral region of plant cells (Figure 3E).
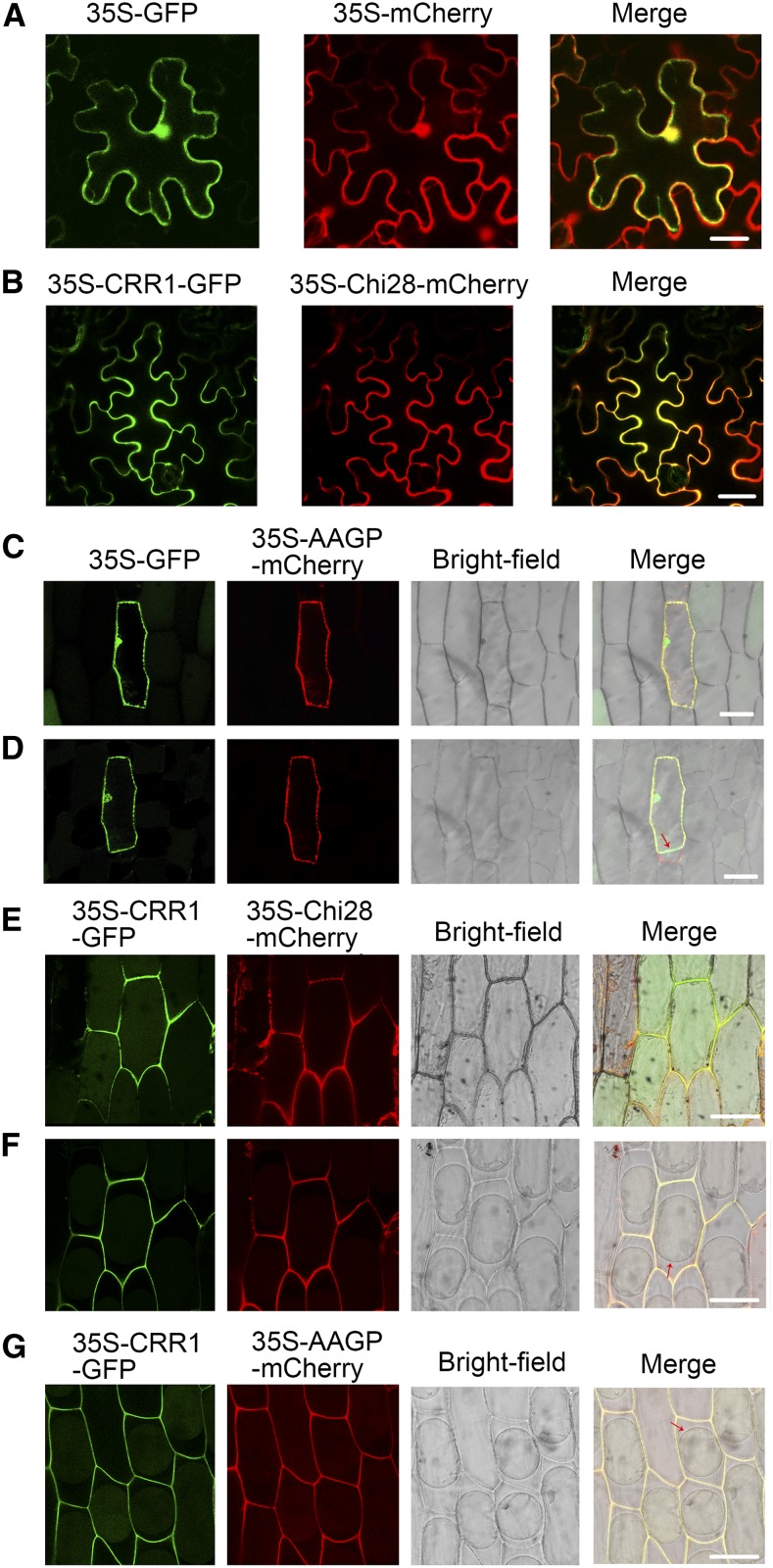
To obtain supporting evidence for the interaction of CRR1 and Chi28, subcellular localization of the two proteins was visualized. As shown in Figures 4A and 4B, CRR1 and Chi28 colocalized in the peripheral region in N. benthamiana leaf cells coexpressing CRR1-GFP and Chi28-mCherry. As CRR1 and Chi28 are putative apoplastic proteins, to observe the distribution more clearly, we examined their subcellular localization after transiently expressing these proteins fused to GFP or mCherry in onion epidermal cells. Fluorescence from the CRR1-GFP or Chi28-mCherry fusion proteins was detected in the apoplast (in the cell wall and in the compartment between the cell membrane and cell wall) after sucrose-induced plasmolysis (Figures 4E and 4F). However, the fluorescence from the GFP control was visualized only in the intracellular region and not in the extracellular region, in contrast to the cotton root apoplastic marker protein AAGP (apoplastic anion glutathione peroxidase; Figures 4C and 4D; Li et al., 2016b). The codistribution of CRR1-GFP with AAGP-mCherry further confirmed its apoplastic localization (Figure 4G).
Figure 4.
Subcellular Localization of CRR1 and Chi28.
(A) Localization of GFP and mCherry in N. benthamiana cells. Agrobacterium cells containing the indicated pair of GFP and mCherry plasmids were coinfiltrated into leaves of N. benthamiana. The signal was visualized by confocal microscopy. Bar, 20 μm.
(B) Colocalization of CRR1-GFP and Chi28-mCherry in the peripheral regions of N. benthamiana cells. Bar, 20 μm.
(C) Subcellular localization of GFP and mCherry-fused apoplastic marker protein AAGP in onion epidermal cells before plasmolysis. Bar, 30 μm.
(D) The same cells in (C) after plasmolysis. Arrow indicates the cell membrane. Bar, 30 μm.
(E) Colocalization of CRR1-GFP and Chi28-mCherry in onion epidermal cells before plasmolysis. Bar, 30 μm.
(F) The same cells in (E) after plasmolysis. Arrow indicates the cell membrane. Bar, 30 μm.
(G) Colocalization of CRR1-GFP and AAGP-mCherry in onion epidermal cells after plasmolysis. Arrow indicates the cell membrane. Bar, 30 μm.
Chi28 Participates in the Immune Response against V. dahliae
Chi28 belongs to the class IV chitinase subfamily, which has seven members in cotton (Chi28, 29, 30, 31, 34, 35, and 36) with protein sequence similarity ranging from 51% to 66% (Xu et al., 2016). Previously, it was reported that Chi28 was expressed more actively than other chitinase genes in G. barbadense root (Xu et al., 2016). We found that, in response to V. dahliae infection, expression of Chi28 was upregulated to much higher levels compared with the other six class IV chitinase genes in G. barbadense plants (Supplemental Figure 7). These results suggest that Chi28 may have an important role in the interaction between cotton and V. dahliae.
To specifically examine Chi28 protein, an antibody was generated using amino acids 146 to 156 of Chi28. To test the specificity of this antibody, all the class IV chitinase proteins of G. barbadense were expressed in E. coli and purified (Supplemental Figure 8A). Immunoblot analyses showed that this antibody recognized only Chi28 but not its homologs in class IV, including Chi29, 30, 31, 34, 35, and 36 (Supplemental Figure 8B).
To characterize the function of Chi28, expression of the gene was knocked down in G. barbadense through VIGS. In accordance with the reverse-transcription quantitative PCR (RT-qPCR) analysis, the immunoblot showed that Chi28 protein was down-regulated in the root apoplast of VIGS cotton plants (Figure 5B). Pathogen infection experiments showed that silencing of Chi28 significantly impaired the resistance of cotton plants to V. dahliae, indicating that Chi28 plays an important role in defense against V. dahliae (Figures 5A and 5C). The potential off-target silencing of other chitinase genes in class IV was checked in the Chi28 VIGS plants, and the transcript levels of the six genes were not significantly altered (Supplemental Figure 9).
Figure 5.
CRR1 Attenuates Chi28 Reduction in Cotton Root Apoplast.
(A) Reduced resistance to V. dahliae infection after silencing of Chi28 by VIGS in G. barbadense plants. Ten-day-old cotton plants were infiltrated with Agrobacterium cells carrying the VIGS-control vector (TRV:00) and TRV:Chi28. Photographs were taken at 2 weeks after V. dahliae inoculation.
(B) RT-qPCR and immunoblot analyses of Chi28 expression in TRV:00 and TRV:Chi28 G. barbadense plants. Cotton NRX1 was used as a control for apoplastic protein.
(C) Disease index of TRV:00 and TRV:Chi28 G. barbadense plants. Error bars represent SD of three biological replicates (n ≥ 36); asterisks indicate statistically significant differences, as determined by the Student’s t test (**P < 0.01).
(D) RT-qPCR analysis of Chi28 expression in wild-type (WT) and CRR1-transgenic G. hirsutum plants (OE-11, An-4) upon V. dahliae infection.
(E) Immunoblot analysis of Chi28 accumulation in the apoplastic compartment of wild-type and CRR1-transgenic G. hirsutum plants (OE-11, An-4) upon V. dahliae infection. Cotton NRX1 was used as a control for apoplastic protein.
(F) Immunoblot analysis of CRR1 accumulation in CRR1-overexpressing Arabidopsis plants during V. dahliae infection. β-tubulin was used as an internal control.
(G) Confocal observation of Chi28-mCherry in the leaves of wild-type and CRR1-overexpressing Arabidopsis plants during V. dahliae infection. All images are shown as micrographs projected from z-series stacks. Bar, 50 µm.
(H) Quantification of mCherry signals in the Arabidopsis leaf apoplast during V. dahliae infection. The intensities of the fluorescent signals from 30 cells were determined using Leica LAS AF Lite software. Data are means ± se (n = 30). Asterisks indicate statistically significant differences, as determined by Student’s t test (**P < 0.01); Similar results were obtained in three independent biological repeats.
CRR1 Stabilizes Chi28 in Cotton Root Apoplast
Since CRR1 interacts with Chi28, we hypothesized that CRR1 might associate with Chi28 to modulate immune responses against V. dahliae. To understand the functional link between these two proteins, we analyzed the expression of Chi28 in both wild-type and CRR1-transgenic G. hirsutum plants before and after V. dahliae infection. As shown in Figure 5D, Chi28 transcription was up-regulated in wild-type and CRR1-transgenic G. hirsutum plants treated with V. dahliae, and the increase of transcripts appeared comparable in these three types of plants.
We then detected Chi28 protein levels in wild-type and CRR1-transgenic G. hirsutum plants. We observed that unlike the sustained increase of Chi28 transcripts, Chi28 protein levels in the apoplast decreased in wild-type plants infected with V. dahliae. Interestingly, in the CRR1-overexpressing G. hirsutum plants, Chi28 protein levels did not significantly decrease upon V. dahliae infection and in the CRR1-suppressed G. hirsutum plants, Chi28 decreased much more than the wild-type and CRR1-overexpressing plants (Figure 5E).
In addition to cotton, we tested the functional link between CRR1 and Chi28 in Arabidopsis, which is susceptible to V. dahliae infection. Cotton Chi28-mCherry under the control of the 35S promoter was ectopically expressed in wild-type and CRR1-overexpressing (35S:CRR1-HA) Arabidopsis plants. The CRR1-, Chi28-mCherry-, and CRR1×Chi28-mCherry-overexpressing Arabidopsis plants showed increased resistance to V. dahliae infection. This was also indicated by the increased callose deposition in the transgenic leaves (Supplemental Figure 10). In Arabidopsis leaf epidermal cells, Chi28-mCherry was distributed in the peripheral regions of the cells. In Chi28-mCherry-overexpressing plants challenged with V. dahliae spores, the fluorescent signals from Chi28-mCherry decreased at the peripheral regions of the leaf epidermal cells, whereas in CRR1×Chi28-mCherry-overexpressing plants (Figure 5F), the decrease of Chi28-mCherry signals was significantly attenuated during infection (Figure 5G). This result was further supported by quantification of mCherry fluorescence signals in leaves of the Arabidopsis plants (Figure 5H).
Together, these results indicate that the amount of Chi28 decreased in the apoplast of plants infected with V. dahliae and that CRR1 could help stabilize Chi28 to attenuate this decrease.
Chi28 Is a Target of the Protease VdSSEP1 from V. dahliae and CRR1 Protects It from Degradation by VdSSEP1
Fungal pathogens secrete proteases that target plant chitinases (Naumann and Price, 2012; Jashni et al., 2015a). As the amount of Chi28 proteins decreased during V. dahliae infection, we speculated that Chi28 might be a target of proteases from V. dahliae. To test this, we extracted secreted proteins from V. dahliae at 0, 1, and 2 d after incubation with cotton plants. We investigated the protein samples by mass spectrometry (MS), which identified 300 proteins; 13 of these V. dahliae proteins were annotated as proteases (Supplemental Figure 11A).
We then conducted a pull-down screen, in combination with MS, to identify proteins that might interact temporarily with Chi28. GST-tagged Chi28 protein was used as bait and secreted proteins extracted from the culture liquid of V. dahliae that had been incubated with cotton roots for 2 d were used as prey. The interacting proteins were collected and subjected to MS analysis. Six proteins were identified using this approach (Supplemental Figure 11B), including a secretory Ser protease (named VdSSEP1) whose content increased during infection (Supplemental Figure 11C). Therefore, we subjected VdSSEP1 to further study.
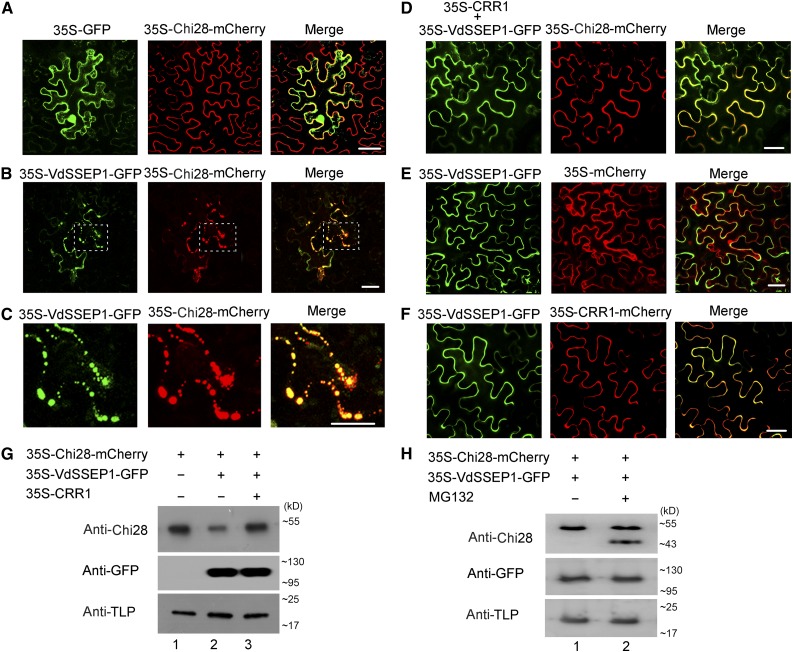
We examined whether VdSSEP1 has an effect on Chi28 in vivo. When GFP and Chi28-mCherry were coexpressed in N. benthamiana leaf cells, observation of fluorescent signals showed that Chi28 was distributed at the cell periphery (Figure 6A). However, in cells coexpressing VdSSEP1-GFP and Chi28-mCherry, aggregated fluorescent signals appeared, indicating that VdSSEP1 interacted with Chi28 and might affect its localization (Figures 6B and 6C). Moreover, when we expressed CRR1 along with VdSSEP1-GFP and Chi28-mCherry, the aggregated fluorescence signals from Chi28 disappeared, suggesting that CRR1 could perturb the interaction between VdSSEP1 and Chi28 (Figure 6D). However, when we coexpressed VdSSEP1-GFP with mCherry or CRR1-mCherry, the presence of VdSSEP1 did not affect the localization of mCherry or CRR1 (Figures 6E and 6F).
Figure 6.
CRR1 Perturbs the Interaction between VdSSEP1 and Chi28.
(A), (B), (D), (E), and (F) Agrobacterium strains containing the indicated plasmid pairs were coinfiltrated into the leaf cells of N. benthamiana. The fluorescence signal was collected at 48 h after infiltration. Bar, 20 µm.
(C) Enlarged images of the section outlined by white dashed lines in (B). Bar, 20 µm.
(G) and (H) Immunoblot analysis of Chi28 accumulation in the apoplastic compartment in the presence or absence of CRR1, VdSSEP1, or proteasome inhibitor, MG132. Tobacco apoplastic protein TLP was used as an internal control.
We also conducted immunoblot analyses to investigate the functional relevance among CRR1, Chi28, and VdSSEP1. The tobacco apoplastic thaumatin-like protein (TLP) was used as a control (Kim et al., 2013). As shown in Figure 6G, when Chi28 was expressed alone, a single Chi28 protein band could be revealed by the anti-Chi28 antibody (lane 1); when VdSSEP1 was coexpressed with Chi28, however, the band became thinner and weaker (lane 2); when CRR1 was expressed along with Chi28 and VdSSEP1 (lane 3), the band strength looked similar to that in lane 1. These results showed clearly that VdSSEP1 expression decreased the amount of Chi28 proteins and expression of CRR1 could eliminate this reduction.
Previous studies have showed that some Ser protease could cut the target protein into peptide fragments (Naumann and Price, 2012; Jashni et al., 2015a). However, in our experiment, we did not detect any band for the digested products on the gel though we observed a decrease of Chi28 abundance in the presence of VdSSEP1 (Figure 6G). We speculated that this was a result of successive degradation of the peptides, possibly by the proteasome system. To see if this holds true, we added the proteasome inhibitor MG132 in the test; a smaller band could be seen under this condition (Figure 6H), indicating that the Chi28 was digested by VdSSEP1 and the resulting fragments were further degraded by proteasome.
VdSSEP1 Is a Virulence Factor of V. dahliae
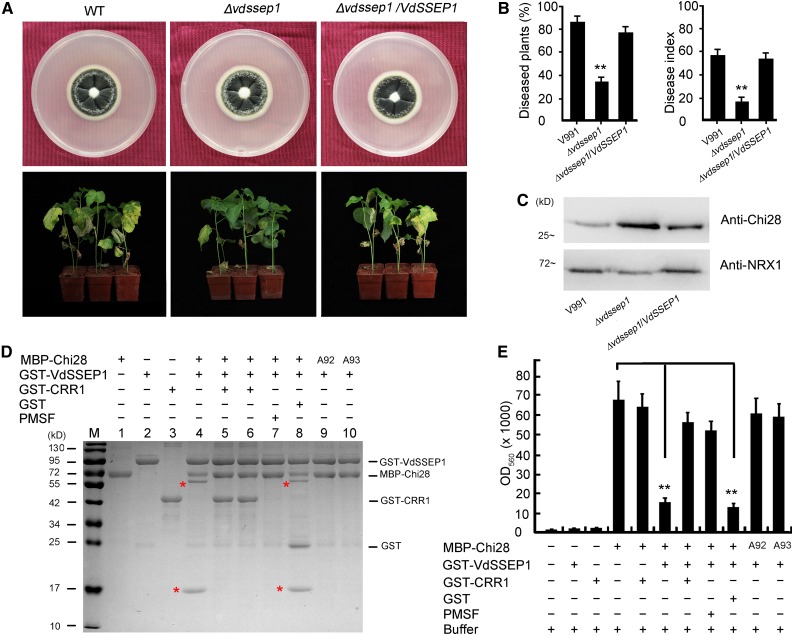
To examine the role of VdSSEP1 in V. dahliae pathogenesis and its action on Chi28, we generated the Δvdssep1 knockout mutant in V. dahliae (Supplemental Figure 12). Deletion of VdSSEP1 did not induce a notable change in hyphal growth (Figure 7A). However, the Δvdssep1 mutant showed much lower disease severity in cotton. Moreover, its pathogenicity was restored in the complemented strain (Figures 7A and 7B). These results suggest that VdSSEP1 is required for the full virulence of V. dahliae. We extracted secreted proteins from cotton plants treated with the wild type, Δvdssep1 mutant, and the complemented strains and subjected them to immunoblot analysis to measure Chi28 levels in the apoplast. As shown in Figure 7C, Chi28 protein levels were higher in the apoplast of cotton plants treated with Δvdssep1 versus the wild type and the VdSSEP1-complemented V. dahliae. These results indicated that the virulence of VdSSEP1 is related to the reduction of Chi28.
Figure 7.
CRR1 Helps Chi28 Counteract VdSSEP1-Mediated Cleavage.
(A) Colony morphology and virulence of Δvdssep1 and the complemented strains. The images in the top show strains grown on PDA plates at 2 weeks postincubation. The images in the bottom show cotton plants infected with wild type (WT) (V991), Δvdssep1, and the complemented strains for 2 weeks. Photographs were taken at 2 weeks after V. dahliae infection.
(B) Rate of diseased G. hirsutum plants and disease index of cotton plants infected with wild-type (V991), Δvdssep1, and the complemented strains. Error bars represent sd of three biological replicates (n ≥ 36); asterisks indicate statistically significant differences, as determined by Student’s t test (**P < 0.01).
(C) Immunoblot analysis of Chi28 protein levels in the apoplast of cotton roots infected with wild-type (V991), Δvdssep1, and the complemented strains. Cotton NRX1 was used as a control for apoplastic protein.
(D) Perturbation of VdSSEP1-mediated Chi28 cleavage by CRR1. The MBP-Chi28, GST-CRR1, and GST-VdSSEP1 fusion proteins were expressed in E. coli BL21 (DE3) and purified. The red asterisks indicated the newly generated peptides. A92 or A93 indicate mutations of Phe-92 or Tyr-93 to Ala.
(E) Chitinase activity of Chi28 in the presence of CRR1, VdSSEP1, or PMSF. Insoluble chitin azure was used as a substrate for Chi28, and the indicated proteins were added to the substrate. The OD560 value was then assessed, which represents the chitinase activity of the protein. Error bars represent sd of three biological replicates (n ≥ 10); asterisks indicate statistically significant differences, as determined by Student’s t test (**P < 0.01).
Biochemical Assessment of the Interplay among CRR1, Chi28, and VdSSEP1
To assess the functional relationships of CRR1, Chi28, and VdSSEP1 directly, we performed biochemical tests using purified proteins expressed in E. coli. In the presence of VdSSEP1, Chi28 was cleaved into two fragments, and the potential cleavage site was identified as Phe-92 and Tyr-93 by MS analysis (Supplemental Figure 13). When phenylmethanesulfonyl fluoride was included in the reaction, cleavage of Chi28 was blocked (Figure 7D), indicating that VdSSEP1 acted as a Ser protease and Chi28 was its substrate.
To test the specificity of cleavage of VdSSEP1 on Chi28, we tested VdSSEP2, another Ser protease identified in our secretome study. To this end, VdSSEP2 was expressed in E. coli and purified. Unlike VdSSEP1, VdSSEP2 was not able to digest Chi28 protein (Supplemental Figure 14). We also tested the specificity of VdSSEP1 on all the chitinases in class IV and found that VdSSEP1 degraded Chi28, but not the other chitinases (Supplemental Figure 15). These results demonstrated that VdSSEP1 specifically degraded Chi28.
As the in vivo analysis suggested that CRR1 could help protect Chi28 from degradation in the apoplast, we tested this function of CRR1 in vitro. CRR1 proteins were preincubated with Chi28 (Figure 7D, lane 5) or VdSSEP1 (Figure 7D, lane 6), and the enzymatic activity of VdSSEP1 on Chi28 was evaluated. As shown in Figure 7D, CRR1 could rescue VdSSEP1-mediated cleavage of Chi28.
We also addressed the question of whether CRR1 is a specific partner of Chi28. To this end, we assessed the ability of CRR2, CRR3, and CRR4 to protect Chi28 from cleavage by VdSSEP1. The three CRRs reduced the cleavage of Chi28 by VdSSEP1, but their efficiency was much lower than that of CRR1 (Supplemental Figure 16A). Quantitation of protein cleavage in the presence of CRR2, CRR3, and CRR4 showed that the amounts of the digested fragments were ∼40%, 60%, and 90% of that of the reaction without CRR protein (Supplemental Figure 16B). These data suggest that CRR1 plays a major role in protecting Chi28 from degradation by VdSSEP1 and indicate the possible minor involvement of other CRRs.
Additionally, we assayed the chitinase activity of Chi28 in the presence of VdSSEP1 or CRR1 using insoluble chitin azure as a substrate. Chi28 exhibited strong chitinase activity, which was not notably altered by coincubation with CRR1 but was clearly impaired in the presence of VdSSEP1. As expected, CRR1 prevented the reduction of Chi28 chitinase activity caused by VdSSEP1. Consistent with the results of cleavage site determination, the chitinase activity of Chi28 with mutations in Phe-92 or Tyr-93 was not significantly affected by VdSSEP1 (Figure 7E).
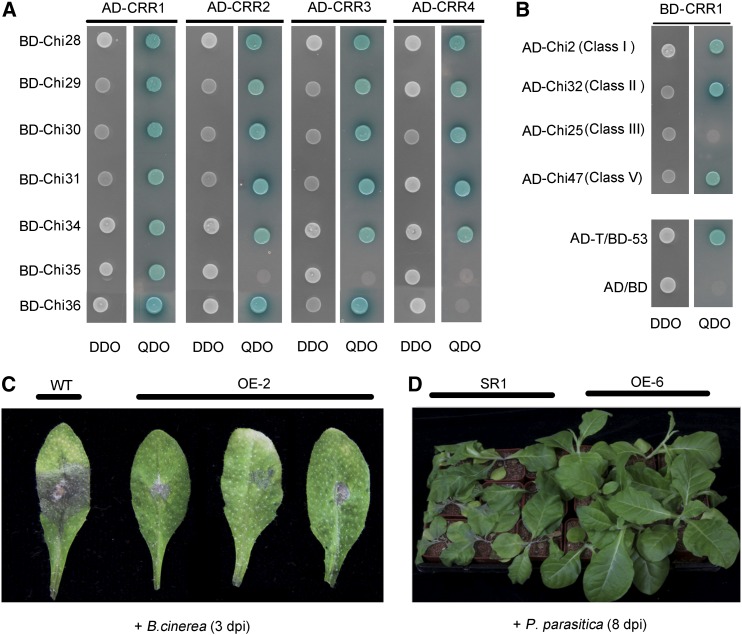
In cotton, chitinases are classified into five classes (Xu et al., 2016). We looked at the interaction between CRR1 and other chitinases. Y2H assays indicated that besides Chi28, CRR1 interacted with all of the other chitinases in class IV (Figure 8A). In addition, CRR1 could interact with Chi2 (class I), Chi32 (class II), and Chi47 (class V), but not with Chi25 (class III), which were randomly selected for the testing (Figure 8B). In addition to CRR1, we also examined the interactions between CRR2, CRR3, CRR4, and class IV chitinases. It appeared that most of the class IV chitinases could interact with these CRR proteins (Figure 8A).
Figure 8.
CRR1 Interacts with Various Types of Cotton Chitinases and Confers Fungal Resistance.
(A) Yeast two-hybrid assays to detect interactions between CRR proteins and class IV chitinases. Yeast cells containing the indicated plasmids were grown on SD/-Leu/-Trp DO (DDO) plates and SD/-Leu/-Trp/-Ade/-His DO (QDO) plates (containing 40 mg/L X-α-gal) for 3 d.
(B) Yeast two-hybrid assays to detect interactions between CRR1 and other subclasses of chitinases. AD/BD, negative control; AD-T/BD-53, positive control.
(C) CRR1 confers resistance to B. cinerea. Wild-type (WT) and CRR1-overexpressing (OE-2) Arabidopsis plants were infected with B. cinerea spores and imaged at 3 d postinoculation.
(D) CRR1 confers disease resistance to P. parasitica var nicotianae. Wild-type (SR1) and CRR1-overexpressing (OE-6) tobacco plants were infected with P. parasitica var nicotianae and imaged at 8 d post inoculation.
Together, these in vitro results further demonstrate that VdSSEP1 functions as a specific Ser protease that cleaves plant Chi28, and CRR1 protects Chi28, and possibly other chitinases, from cleavage by VdSSEP1.
CRR1 Confers Wide-Spectrum Resistance to Fungal Infections
In our study, in addition to testing V. dahliae, we inoculated CRR1-overexpressing Arabidopsis plants with another pathogen, the fungus Botrytis cinerea. Compared with wild-type, the transgenic plants were more tolerant to B. cinerea (Figure 8C). In addition to Arabidopsis, we generated CRR1-overexpressing transgenic tobacco (N. tabacum; SR1) and inoculated the plants with the fungus Phytophthora parasitica var nicotianae, which causes black shank disease in tobacco. Eight days after inoculation, the control plants wilted, whereas CRR1-overexpressing plants grew normally (Figure 8D). These results showed that overexpression of CRR1 could give plants broad-spectrum resistance, possibly by interactions between CRR1 and chitinases or other cofactors.
DISCUSSION
The apoplastic space is a battleground between plants and invading microorganisms. The pathogens secrete toxins and effectors, and the plants export toxic metabolites, hydrolytic enzymes, and defense-related proteins to the apoplastic compartment (Monaghan and Zipfel, 2012; El Kasmi et al., 2018). The battle in the apoplast determines the outcome of the attempted microbial colonization (Doehlemann and Hemetsberger, 2013). In this study, we identified three proteins in the apoplast of cotton root cells during interaction with the fungal pathogen V. dahliae and showed that cotton CRR1 safeguards Chi28 from cleavage by the V. dahliae virulence factor VdSSEP1. During the course of infection, cotton cells secrete Chi28 into the apoplast where they cleave fungal chitin, thereby producing pathogen-associated molecular patterns; however, the Chi28 proteins face cleavage by the fungal Ser protease VdSSEP1. Nevertheless, cotton CRR1 proteins counteract VdSSEP1 by protecting Chi28 from VdSSEP1-mediated degradation. Our findings may represent a new example of how plants have evolved a multilayered immune system to defend themselves against pathogen invasion.
DUF26 domains are present in CRKs, PDLPs, and Cys-rich secretory protein, and CRKs and PDLPs play crucial roles in signal transduction during plant-pathogen interactions (Chen, 2001; Bourdais and CRK Consortium et al., 2015). However, the precise physiological roles of DUF26-containing Cys-rich secretory proteins have not yet been elucidated. Proteome study has shown that Cys-rich secretory proteins undergo increased translation upon pathogen attack and osmotic stress (Delaunois et al., 2013). The secreted protein Gnk2 of G. biloba contains a DUF26 domain that was assumed to have antimicrobial function also acts in mannose binding to inhibit growth of Fusarium fungi (Sawano et al., 2007; Miyakawa et al., 2014). In this study, we found that cotton CRR1 belongs to the Cys-rich secretory protein family and that CRR1 expression is up-regulated in cotton plants challenged with V. dahliae, suggesting that it plays a role in cotton-pathogen interactions (Figure 1). Thus, in addition to the previously reported role of other Cys-rich secretory proteins in pathogen defense and osmotic stress responses, CRR1 also participates in plant defense responses to pathogens, providing additional evidence that DUF26-containing Cys-rich secretory proteins modulate immunity in the plant apoplast to counteract pathogen infection.
Unlike Gnk2, CRR1 did not directly inhibit the hyphal growth of V. dahliae (Supplemental Figure 6), suggesting that it might function by a different molecular mechanism from that of Gnk2. Both in vivo and in vitro experiments showed that CRR1 interacted with the apoplastic class IV chitinase Chi28, and CRR1-overexpressing cotton plants contained higher levels of Chi28 in the apoplast than the wild type when challenged with V. dahliae (Figure 5D). These results indicate that interaction of CRR1 and Chi28 in the apoplast helps to protect Chi28 from degradation. Moreover, as we observed widespread interactions among CRR1, CRR2, CRR3, CRR4, and various types of chitinases in the Y2H assays, we speculate that CRR-mediated stabilization of chitinases may make important contributions to chitin recognition in defense against different fungal pathogens in cotton, and the same could be true in other plants.
Crucial plant defense components such as pathogenesis-related proteins (PRs) are often attacked by pathogen effectors. For example, the wheat pathogen Parastagonospora nodorum secretes the protease SnTox3, which targets wheat PR-1 proteins to cause disease (Breen et al., 2016) and several fungal proteases modify the plant chitinase PR-3. The maize pathogens such as Fusarium verticillioides and Stenocarpella maydis could secrete virulence proteases to cleave the maize class IV chitin-binding domain (CBD) -chitinases (Naumann and Wicklow, 2010; Naumann and Price, 2012). In addition, the Ser protease and metalloprotease from Fusarium oxyspoyum were shown to be able to cut and remove CBD from two tomato CBD-chitinases (Jashni et al., 2015a). In line with these findings, in our study, we found that cotton Chi28 was truncated by the Ser protease VdSSEP1 from V. dahliae (Figure 7D). Incubation with recombinant VdSSEP1 led to the cleavage and removal of the CBD domain of Chi28 in vitro. MS analysis showed that VdSSEP1 cleaves Chi28 between Phe-92 and Tyr-93. Moreover, the Δvdssep1 mutant showed much lower disease severity in cotton, indicating that VdSSEP1 is required for the pathogenicity of V. dahliae. Therefore, the newly identified secreted protease VdSSEP1 is a virulence factor that targets plant chitinase to compromise chitin recognition of the host.
Upon pathogen infection, plants secrete protease inhibitors like wheat antimicrobial peptides (WAMP) to counter pathogen proteases. The WAMPs could protect class IV chitinases from degradation by a metalloprotease secreted from Fusarium verticillioides (Slavokhotova et al., 2014). In our study, we found that cotton CRR1 helped protect Chi28 from cleavage both in vivo and in vitro (Figures 5 and 7), suggesting that CRR1 may function as a protease inhibitor. However, whereas WAMPs bind to fungalysin, we did not detect an interaction between CRR1 and VdSSEP1. Instead, we detected strong interactions between CRR1 and chitinases. These observations indicate that CRR1 does not function like WAMP protease inhibitors. We speculate that CRR1 binds to cotton chitinases, thereby blocking the VdSSEP1 cleavage site. Further detailed experiments are needed to address this hypothesis.
Upon infection, plants secrete apoplastic chitinases to attack the fungal cell wall and generate soluble chitin fragments, thereby triggering immune responses. As a countermeasure, fungal-secreted apoplastic effectors such as proteases diminish chitin-mediated defense responses (Doehlemann and Hemetsberger, 2013). In this study, we show that cotton CRR1, a protector of chitinase, has evolved as an additional layer of the immune response, helping the plant withstand attack from pathogen proteases. Thus, the apoplastic protein CRR1 takes part in the plant-pathogen coevolutionary struggle.
METHODS
Plant Materials and Growth Conditions
Seeds of sea-island cotton (Gossypium barbadense cv Hai 7124) and upland cotton (G. hirsutum cv R15) were sterilized in 70% ethanol and followed by several rinses in sterile water. The cotton plants were grown in pots or hydroponically (Qu et al., 2005) at 28°C under 16-h/8-h light/dark photoperiod with 28-W fluorescent lamps. The roots of 10-d-old cotton plants were dip-inoculated with Verticillium dahliae spore suspension (1 × 106 conidia/mL) as previously described (Wang et al., 2011). The mock control indicates that cotton plants were treated with sterile water. For gene transcriptional analysis, roots were harvested at indicated time points in each experiment, frozen in liquid nitrogen, and stored at −80°C.
Arabidopsis (Arabidopsis thaliana, ecotype Columbia-1) and tobacco (Nicotiana benthamiana and N. tabacum cv SR1) plants were grown in a greenhouse under 16-h/8-h light/dark conditions at 23°C and 28°C, respectively.
Plasmid Construction and Plant Transformation
The open reading frame (ORF) of CRR1 (in sense or antisense orientation) was inserted into the binary expression vector pBIN438 to generate the overexpression or underexpression constructs. All constructs were confirmed by sequencing and then transformed into A. tumefaciens strain EHA105. Arabidopsis and tobacco transformation were performed by the Agrobacterium tumefaciens-mediated floral-dip method and leaf disc transformation, respectively. For transient expression in N. benthamiana, the ORFs of CRR1, VdSSEP1, and Chi28 were inserted into the binary expression vector pCAMBIA1300-GFP or pCAMBIA1300-mCherry to generate plasmids harboring 35S:CRR1-GFP, 35S:VdSSEP1-GFP, and 35S:Chi28-mCherry using homologous recombination cloning (ClonExpressMultiS One Step Cloning Kit, Vazyme Biotech, C112), respectively. Fully expanded N. benthamiana leaves were infiltrated with A. tumefaciens (strain GV3101) containing the recombinant plasmids. The infiltrated plants were incubated for 48 h at room temperature, and fluorescent signals were visualized under a confocal microscope (ZEISS LSM880 Airyscan) at 488 or 561 nm.
Pathogen Cultivation and Inoculation
The defoliating V. dahliae isolate V991 (Zhang et al., 2012), which was originally isolated from an infected upland cotton plant, was used in this study. For production of suspension spores, mycelia were grown on potato dextrose agar (PDA) medium, then collected and cultured in liquid Czapek’s medium at 150 rpm, 28°C for 3 to 5 d. Roots of 10-d-old cotton seedlings grown under hydroponic conditions were dip-inoculated with V. dahliae conidia suspension (1 × 106 conidia/mL) as previously described (Wang et al., 2011). To infect transgenic and VIGS cotton plants, plantlets in pots (250 mL) were inoculated by drenching soil with 30 mL conidial suspension (1 × 106 conidia/mL) or sterile water as mock. For Arabidopsis infection, 4-week-old plant roots were incubated with spore suspensions for 3 min, then transferred into fresh steam-sterilized water. The diseased seedlings were classified into five grades (grade 0, 1, 2, 3, and 4) based on the severity of disease after infection. And the disease index was calculated according to the formula: disease index = [(Σdisease grades × number of infected plants)/(total number of plants × 4)] × 100.
Spores of Botrytis cinerea strain BO5-10 were harvested from PDA media and adjusted to a concentration of 1 × 105 spores/mL with distilled water. Arabidopsis leaf was dropped with a 6-μL aliquot of spore suspension and the lesion size was measured at 3 d after inoculation.
The pathogen P. parasitica, was grown on millet medium plates at 24°C. The mycelia were ground and mixed with soil at a ratio of 1:1,000 (w/w). The mixture was used to inoculate soil-grown tobacco plants at the 5-leaf stage.
RNA Extraction and RT-qPCR Analysis
Total RNA was extracted using a Plant Total RNA Purification Kit (GeneMark) according to the manufacturer’s protocol. The RT-qPCR assay was performed using SYBR Green Real-Time PCR Master Mix (Toyobo, Japan), with cotton Histone3 as the internal control. All reactions were conducted in triplicate and primers used in this study are provided in Supplemental Table 1.
GUS Staining
The 1.5-kb upstream fragment of CRR1 from G. barbadense was amplified and cloned upstream of GUS in the pCAMBIA1301 vector. The recombinant plasmid was introduced into the G. hirsutum cultivar R15 to generate the transgenic cotton plants. The obtained T1 seedlings were used for GUS staining. Histochemical staining for GUS activity measurement in the transgenic plants was performed as previously described (Jefferson et al., 1987).
VIGS
For tobacco rattle virus (TRV)-based VIGS experiments, we employed a modified vacuum infiltration method (Liu et al., 2002; Qu et al., 2012). The gene-specific fragments for CRR1 and Chi28 were inserted into pTRV2. Agrobacterium cultures (OD600 = 1.5) harboring pTRV1 and pTRV2-CRR1 or pTRV2-Chi28 were mixed at a 1:1 ratio and agroinoculated into 10-d-old cotton roots by vacuum infiltration. G. barbadense phytoene desaturase was used as the experimental marker for this silencing system.
Protein Interaction Assays
The Y2H assay was performed as described in the Matchmaker Yeast Two-Hybrid System (Clontech) manual. A G. barbadense cDNA library was constructed in the yeast expression vector pGADT7 (Clontech) for screening prey proteins, and the coding region of CRR1 was cloned into the PGBKT7 vector (Clontech) to generate the construct BD-CRR1 as bait. After mating and prey protein screening, the BD-CRR1, AD-Chi28, and respective control vector constructs were cotransformed into yeast cells (strain AH109) and cultured on SD/-Leu/-Trp DO medium. After growth at 30°C for 72 h, independent colonies harboring the target constructs were verified by PCR amplification of both cotransformed genes. Verified colonies were selected on SD/-Leu/-Trp/-Ade/-His DO (QDO) medium, followed by the addition of 5 mm 3-AT (Sigma-Aldrich) and 40 mg/L X-α-Gal (Clontech) to determine galactosidase activity. The pGADT7-T/pGBKT7-53 (AD-T/BD-53) plasmid was used as a positive control, and pGADT7/pGBKT7 (AD/BD) was used as a negative control. For validating the interaction between CRRs and chitinases in Y2H, the cDNAs encoding CRR1-CRR4 and chitinases 28, 29, 30, 31, 34, 35, and 36 (without signal peptide) were cloned into AD and BD vectors, respectively. These recombinant plasmids were cotransformed in pairs into AH109 cells for examining on QDO medium.
For the pull-down assay, the MBP-CRR1 and GST-Chi28 fusion proteins were expressed in E. coli strain BL21 (DE3) and an in vitro protein-protein interaction assay was conducted according to the ProFound Pull-Down MBP Protein Kit. The eluted proteins were separated on an SDS-PAGE gel and detected by immunoblot using anti-GST or anti-MBP antibodies (1:4,000; Abcam, ab19256 and ab20216).
For the BiFC assay, the CRR1 and Chi28 cDNAs were amplified and cloned into the binary vector pSAT1 in frame with YFPN and YFPC to generate 35S:YN-CRR1 and 35S:YC-Chi28, respectively. The plasmids were introduced into Agrobacterium strain C58C1. Reconstitution of YFP fluorescence was analyzed by transient coexpression in cells of 4-week-old N. benthamiana leaves. YFP fluorescent signals were visualized under a confocal microscope activated at 514 nm.
For the firefly luciferase complementation imaging assay, the cDNAs of CRR1 and Chi28 were amplified and ligated to the DNA sequence encoding the C-terminal and N-terminal ends of split Luc to generate CLuc-CRR1 and NLuc-Chi28. The plasmids were subsequently introduced into Agrobacterium strain GV3101. Equal amounts of Agrobacterium cultures containing different CLuc and NLuc construct pairs were co-transformed into N. benthamiana. The plants were grown in the dark for 48 h and infiltrated leaves were visualized and sprayed with 1 mM luciferin (Promega). Luc signals were captured with a low-light cooled charge-coupled device camera (Night Owl LB985, Berthold Technologies), and relative Luc activity was measured. Quantitative analysis was performed using IndiGo software (Berthold Technologies).
Gene Knockout in V. dahliae
The upstream and downstream genomic sequences of VdSSEP1 and the hygromycin resistance cassette were amplified and fused in order into the pGKO vector via homologous recombination cloning (ClonExpressMultiS One Step Cloning Kit, Vazyme Biotech, C112; Wang et al., 2016; Zhou et al., 2017). A. tumefaciens-mediated transformation was performed as previously described with some modifications (Feng et al., 2018). A. tumefaciens cells (strain AGL1) carrying pGKO-VdSSEP1 were grown in YEP medium, the cultures were then transferred to minimal medium at a 1:100 ratio and grown at 28°C for 24 h. The A. tumefaciens cells were collected and diluted to OD600 = 0.2 in induction medium (1/2 minimal medium containing 5 mL/L glycerol) supplemented with 200 μm acetosyringone (AS). The cells were grown for an additional 6 h, then mixed with an equal volume of V991 conidial suspension (1 × 106 conidia/mL). The mixture (200 μL per plate) was plated onto induction medium in the presence of 200 μm acetosyringone and 40 mm MES (pH 5.3) overlaid with a nitrocellulose filter (0.22-μm pore, 60-mm diameter) and incubated for 3 d. The filter was transferred to PDA medium containing cefotaxime (200 mg/L), carbenicillin (200 mg/L), and hygromycin B (100 mg/L). Conidia of different transformants were suspended in sterile water and plated on PDA medium. To obtain the Δvdssep1 complemented strain, the ORF of VdSSEP1 was ligated into pSulPH binary vectors based on homologous recombination cloning as reported previously (Li et al., 2016b). The recombinant plasmids were transformed into V. dahliae as described above.
Preparation of Recombinant Proteins
The truncated cDNAs of Chi28 (without the sequence encoding the N-terminal signal peptide [SP], amino acids 1 to 23), VdSSEP1 (encoding the protease-like domain, amino acids 129 to 607), VdSSEP2 (encoding the protease-like domain, amino acids 23 to 428), CRR1-3 (without SP, amino acids 1 to 23), CRR4 (without SP, amino acids 1 to 20), Chi29 (without SP, amino acids 1 to 23), Chi30 (full length), Chi31 (without SP, amino acids 1 to 25), Chi34 (without SP, amino acids 1 to 22), Chi35 (without SP, amino acids 1 to 25), and Chi36 (without SP, amino acids 1 to 24) were fused to the pGEX6P-1 or pMAL-p2X vector to produce GST-Chi28, GST-VdSSEP1, GST-CRR1-4, MBP-CRR1, MBP-Chi28, MBP-Chi29, MBP-Chi30, MBP-Chi31, MBP-Chi34, MBP-Chi35, and MBP-Chi36 fusion proteins, respectively. All constructs were transformed into E. coli strain BL21 (DE3), and expression of recombinant protein were induced by addition of 0.5 mm isopropyl-β-D-thiogalactoside at 16°C overnight. Recombinant proteins were purified following the manufacturer’s instructions.
Preparation of Secreted Protein Extracts
For preparation of secreted proteins from the fungus, V. dahliae spores were cultured in liquid Czapek’s medium as described above. The roots of sterile cotton seedlings were inoculated with the V. dahliae conidia suspension (1 × 106 conidia/mL) for 24 or 48 h. The fungal mycelia were filtered out by Miracloth (Calbiochem), and the spores were removed by centrifugation at 3,700g for 15 min. The supernatant was concentrated in an ultra-filtration tube (3 kD, Millipore). To identify candidate Ser proteases, the purified GST-Chi28 proteins were bound to a GST affinity column and the extracted V. dahliae secreted proteins were incubated in the GST column for 3 h. The bound proteins were eluted in glutathione solution (10 mm in 20 mm Tris-HCl, pH 8.0) after five washes with phosphate buffered saline. The protein samples were analyzed by MS.
To extract apoplastic proteins from plants, a vacuum infiltration-centrifugation method was employed as described previously (Li et al., 2016b). For immunoblot analysis, 20 μg of the extracted protein was subjected to SDS-PAGE. Immunoblot experiment was performed using the antibodies (1:2,000 dilution) raised against Chi28, TLP, and GFP (Abcam, ab6556) as the primary antibodies, and horseradish peroxidase-conjugated goat anti-rabbit/mouse immunoglobulin G (1:3,000 dilution; Abmart, M21003S) as the secondary antibody. The antibody against Chi28 or tobacco TLP was designed and prepared using a Chi28- or TLP-specific peptide by the Abmart technology company.
Chi28 Cleavage and Chitinase Activity Assay
The chitinase cleavage assay was performed as previously described (Naumann and Price, 2012; Jashni et al., 2015a; Breen et al., 2016). Each reaction was performed in a total volume of 20 μL containing purified Chi28 (2 μm), VdSSEP1 (1 μm) or Chi28 (2 μm), VdSSEP1 (1 μm), and CRR1 (2 μm) in 50 mm sodium acetate buffer (pH 5.2). The reactions were incubated for 20 h at room temperature and stopped by the addition of SDS-PAGE loading buffer, and proteins were analyzed by SDS-PAGE. The peptide bands of cleaved proteins were excised from the gel and analyzed by MS.
The chitinase activity assay was based on the method reported previously (Jashni et al., 2015a). In brief, the insoluble chitin azure was washed with potassium phosphate buffer (100 mm, pH 6.0) and then re-suspended and mixed with purified Chi28, VdSSEP1, or CRR1 at a final concentration of 0.1 mg/mL and incubated for 20 h at 37°C. After centrifugation, the supernatants of these reactions were analyzed at 560 nm.
Accession Numbers
Sequence data for the genes described in this study can be found in the databases of G. barbadense and G. hirsutum (Li et al., 2015; Yuan et al., 2015; https://www.cottongen.org/) under the following accession numbers: CRR1 (Gbscaffold10424.22; CotAD_21912), CRR2 (Gbscaffold10424.17, Gbscaffold15923.5; CotAD_49962, CotAD_21909), CRR3 (Gbscaffold10424.16, Gbscaffold15923.4; CotAD_21908, CotAD_49965), CRR4 (Gbscaffold15923.8; CotAD_21911), Chi2 (Gbscaffold11871.2), Chi25 (Gbscaffold6583.1), Chi28 (Gbscaffold9972.13), Chi29 (Gbscaffold9972.16), Chi30 (Gbscaffold9972.18), Chi31 (Gbscaffold9972.19), Chi32 (Gbscaffold18847.8), Chi34 (Gbscaffold19845.1), Chi35 (Gbscaffold6967.1), Chi36 (Gbscaffold2259.17), Chi47 (Gbscaffold6881.5) and the GenBank/EMBL database under the accession numbers VdSSEP1 (VDAG-08100) and VdSSEP2 (VDAG-03905).
Supplemental Data
Supplemental Figure 1. Motif structures of cotton CRR1 and alignment of amino acid sequences of CRR proteins.
Supplemental Figure 2. Alignment of the nucleotide sequences of four cotton CRR genes in G. hirsutum and G. barbadense.
Supplemental Figure 3. Comparison of the verticillium wilt responses between G. hirsutum and G. barbadense cotton plants.
Supplemental Figure 4. Controls for cotton VIGS experiment and expression analysis of CRR2, CRR3, and CRR4 in CRR1-silenced G. barbadense plants.
Supplemental Figure 5. Increased susceptibility to V. dahliae infection of CRR1-suppressed G. hirsutum plants.
Supplemental Figure 6. Growth of V. dahliae without inhibition by the presence of CRR1 protein.
Supplemental Figure 7. Expression of G. barbadense chitinase genes in response to V. dahliae infection.
Supplemental Figure 8. SDS-PAGE analysis of the purified MBP-tagged class IV chitinases and immunoblot analysis using Chi28-specific antibody.
Supplemental Figure 9. Expression of class IV chitinase genes in Chi28-silenced G. barbadense plants.
Supplemental Figure 10. Increased V. dahliae resistance by ectopic overexpression of cotton CRR1 and Chi28-mCherry in Arabidopsis.
Supplemental Figure 11. Identification of VdSSEP1 interacting with Chi28.
Supplemental Figure 12. Construction of VdSSEP1 knockout mutant.
Supplemental Figure 13. Amino acid sequence of Chi28.
Supplemental Figure 14. SDS-PAGE analysis of cleavage of Chi28 by VdSSEP1 and VdSSEP2.
Supplemental Figure 15. SDS-PAGE analysis of cleavage of class IV chitinases by VdSSEP1.
Supplemental Figure 16. Cotton CRRs protect Chi28 from VdSSEP1-mediated cleavage.
Supplemental Table 1. Primers used in this study.
Dive Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
We thank Prof. Huishan Guo (Institute of Microbiology, Chinese Academy of Sciences) for kindly providing us with the pGKO vector. We thank Dr. Yule Liu (Tsinghua University) and Dr. Chaozu He (Hainan University) for kindly providing us with VIGS vector and pSulPH expression vector. This work was supported by the Strategic Priority Research Program of the Chinese Academy of Sciences (grant XDB11040600), the National Science Foundation of China (grants 31600978, 31772109, and 31401033), and the State Key Laboratory of Plant Genomics of China (grant 2016B0129-02).
AUTHOR CONTRIBUTIONS
G.-X.X. and L.-B.H conceived and designed the study; L.-B.H., Y.-B.L., and W.-Y.W. performed the experiments; Y.-B.L., L.-B.H., F.-X.W., J.L., J.-H.W., and N.-Q.Z. participated in data analysis and discussion; G.-L.J. and S.-J.W. generated the transgenic cotton plants. G.-X.X., H.-Y.W., and Y.-B.L. drafted the manuscript, and all authors read and approved the manuscript.
References
- Acharya B.R., Raina S., Maqbool S.B., Jagadeeswaran G., Mosher S.L., Appel H.M., Schultz J.C., Klessig D.F., Raina R. (2007). Overexpression of CRK13, an Arabidopsis cysteine-rich receptor-like kinase, results in enhanced resistance to Pseudomonas syringae. Plant J. 50: 488–499. [DOI] [PubMed] [Google Scholar]
- Akira S., Uematsu S., Takeuchi O. (2006). Pathogen recognition and innate immunity. Cell 124: 783–801. [DOI] [PubMed] [Google Scholar]
- Bourdais G., et al. ; CRK Consortium (2015). Large-scale phenomics identifies primary and fine-tuning roles for CRKs in responses related to oxidative stress. PLoS Genet. 11: e1005373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breen S., Williams S.J., Winterberg B., Kobe B., Solomon P.S. (2016). Wheat PR-1 proteins are targeted by necrotrophic pathogen effector proteins. Plant J. 88: 13–25. [DOI] [PubMed] [Google Scholar]
- Caillaud M.C., Wirthmueller L., Sklenar J., Findlay K., Piquerez S.J.M., Jones A.M.E., Robatzek S., Jones J.D.G., Faulkner C. (2014). The plasmodesmal protein PDLP1 localises to haustoria-associated membranes during downy mildew infection and regulates callose deposition. PLoS Pathog. 10: e1004496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z. (2001). A superfamily of proteins with novel cysteine-rich repeats. Plant Physiol. 126: 473–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J.Y., Huang J.Q., Li N.Y., Ma X.F., Wang J.L., Liu C., Liu Y.F., Liang Y., Bao Y.M., Dai X.F. (2015). Genome-wide analysis of the gene families of resistance gene analogues in cotton and their response to Verticillium wilt. BMC Plant Biol. 15: 148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K., Du L., Chen Z. (2003). Sensitization of defense responses and activation of programmed cell death by a pathogen-induced receptor-like protein kinase in Arabidopsis. Plant Mol. Biol. 53: 61–74. [DOI] [PubMed] [Google Scholar]
- Chen K., Fan B., Du L., Chen Z. (2004). Activation of hypersensitive cell death by pathogen-induced receptor-like protein kinases from Arabidopsis. Plant Mol. Biol. 56: 271–283. [DOI] [PubMed] [Google Scholar]
- Cletus J., Balasubramanian V., Vashisht D., Sakthivel N. (2013). Transgenic expression of plant chitinases to enhance disease resistance. Biotechnol. Lett. 35: 1719–1732. [DOI] [PubMed] [Google Scholar]
- Collinge D.B., Kragh K.M., Mikkelsen J.D., Nielsen K.K., Rasmussen U., Vad K. (1993). Plant chitinases. Plant J. 3: 31–40. [DOI] [PubMed] [Google Scholar]
- Cota I.E., Troncoso-Rojas R., Sotelo-Mundo R., Sánchez-Estrada A., Tiznado-Hernández M.E. (2007). Chitinase and β-1,3-glucanase enzymatic activities in response to infection by Alternaria alternata evaluated in two stages of development in different tomato fruit varieties. Sci. Hortic. (Amsterdam) 112: 42–50. [Google Scholar]
- de las Mercedes Dana M., Pintor-Toro J.A., Cubero B. (2006). Transgenic tobacco plants overexpressing chitinases of fungal origin show enhanced resistance to biotic and abiotic stress agents. Plant Physiol. 142: 722–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaunois B., Colby T., Belloy N., Conreux A., Harzen A., Baillieul F., Clément C., Schmidt J., Jeandet P., Cordelier S. (2013). Large-scale proteomic analysis of the grapevine leaf apoplastic fluid reveals mainly stress-related proteins and cell wall modifying enzymes. BMC Plant Biol. 13: 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doehlemann G., Hemetsberger C. (2013). Apoplastic immunity and its suppression by filamentous plant pathogens. New Phytol. 198: 1001–1016. [DOI] [PubMed] [Google Scholar]
- El Kasmi F., Horvath D., Lahaye T. (2018). Microbial effectors and the role of water and sugar in the infection battle ground. Curr. Opin. Plant Biol. 44: 98–107. [DOI] [PubMed] [Google Scholar]
- Emanuelsson O., Nielsen H., Brunak S., von Heijne G. (2000). Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J. Mol. Biol. 300: 1005–1016. [DOI] [PubMed] [Google Scholar]
- Feng Z., Tian J., Han L., Geng Y., Sun J., Kong Z. (2018). The Myosin5-mediated actomyosin motility system is required for Verticillium pathogenesis of cotton. Environ. Microbiol. 20: 1607–1621. [DOI] [PubMed] [Google Scholar]
- Iqbal M.M., Nazir F., Ali S., Asif M.A., Zafar Y., Iqbal J., Ali G.M. (2012). Over expression of rice chitinase gene in transgenic peanut (Arachis hypogaea L.) improves resistance against leaf spot. Mol. Biotechnol. 50: 129–136. [DOI] [PubMed] [Google Scholar]
- Jashni M.K., Dols I.H.M., Iida Y., Boeren S., Beenen H.G., Mehrabi R., Collemare J., de Wit P.J.G.M. (2015a). Synergistic action of a metalloprotease and a serine protease from Fusarium oxysporum f. sp lycopersici cleaves chitin-binding tomato chitinases, reduces their antifungal activity, and enhances fungal virulence. Mol. Plant Microbe Interact. 28: 996–1008. [DOI] [PubMed] [Google Scholar]
- Jashni M.K., Mehrabi R., Collemare J., Mesarich C.H., de Wit P.J.G.M. (2015b). The battle in the apoplast: further insights into the roles of proteases and their inhibitors in plant-pathogen interactions. Front. Plant Sci. 6: 584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson R.A., Kavanagh T.A., Bevan M.W. (1987). GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 6: 3901–3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones J.D., Dangl J.L. (2006). The plant immune system. Nature 444: 323–329. [DOI] [PubMed] [Google Scholar]
- Jun Z., Zhang Z., Gao Y., Zhou L., Fang L., Chen X., Ning Z., Chen T., Guo W., Zhang T. (2015). Overexpression of GbRLK, a putative receptor-like kinase gene, improved cotton tolerance to Verticillium wilt. Sci. Rep. 5: 15048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J.Y., Park S.C., Hwang I., Cheong H., Nah J.W., Hahm K.S., Park Y. (2009). Protease inhibitors from plants with antimicrobial activity. Int. J. Mol. Sci. 10: 2860–2872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S.G., Wang Y., Lee K.H., Park Z.Y., Park J., Wu J., Kwon S.J., Lee Y.H., Agrawal G.K., Rakwal R., Kim S.T., Kang K.Y. (2013). In-depth insight into in vivo apoplastic secretome of rice-Magnaporthe oryzae interaction. J. Proteomics 78: 58–71. [DOI] [PubMed] [Google Scholar]
- Kouzai Y., Nakajima K., Hayafune M., Ozawa K., Kaku H., Shibuya N., Minami E., Nishizawa Y. (2014). CEBiP is the major chitin oligomer-binding protein in rice and plays a main role in the perception of chitin oligomers. Plant Mol. Biol. 84: 519–528. [DOI] [PubMed] [Google Scholar]
- Lee J.Y., Wang X., Cui W., Sager R., Modla S., Czymmek K., Zybaliov B., van Wijk K., Zhang C., Lu H., Lakshmanan V. (2011). A plasmodesmata-localized protein mediates crosstalk between cell-to-cell communication and innate immunity in Arabidopsis. Plant Cell 23: 3353–3373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F., et al. (2015). Genome sequence of cultivated Upland cotton (Gossypium hirsutum TM-1) provides insights into genome evolution. Nat. Biotechnol. 33: 524–530. [DOI] [PubMed] [Google Scholar]
- Li C., He X., Luo X., Xu L., Liu L., Min L., Jin L., Zhu L., Zhang X. (2014). Cotton WRKY1 mediates the plant defense-to-development transition during infection of cotton by Verticillium dahliae by activating JASMONATE ZIM-DOMAIN1 expression. Plant Physiol. 166: 2179–2194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Kim P., Yu L., Cai G., Chen S., Alfano J.R., Zhou J.M. (2016a). Activation-dependent destruction of a co-receptor by a Pseudomonas syringae effector dampens plant immunity. Cell Host Microbe 20: 504–514. [DOI] [PubMed] [Google Scholar]
- Li Y.B., Han L.B., Wang H.Y., Zhang J., Sun S.T., Feng D.Q., Yang C.L., Sun Y.D., Zhong N.Q., Xia G.X. (2016b). The thioredoxin GbNRX1 plays a crucial role in homeostasis of apoplastic reactive oxygen species in response to Verticillium dahliae infection in cotton. Plant Physiol. 170: 2392–2406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., et al. (2014). Host-induced bacterial cell wall decomposition mediates pattern-triggered immunity in Arabidopsis. eLife 3: 01990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Schiff M., Dinesh-Kumar S.P. (2002). Virus-induced gene silencing in tomato. Plant J. 31: 777–786. [DOI] [PubMed] [Google Scholar]
- Ma Z., et al. (2017). A paralogous decoy protects Phytophthora sojae apoplastic effector PsXEG1 from a host inhibitor. Science 355: 710–714. [DOI] [PubMed] [Google Scholar]
- Maximova S.N., Marelli J.P., Young A., Pishak S., Verica J.A., Guiltinan M.J. (2006). Over-expression of a cacao class I chitinase gene in Theobroma cacao L. enhances resistance against the pathogen, Colletotrichum gloeosporioides. Planta 224: 740–749. [DOI] [PubMed] [Google Scholar]
- Miya A., Albert P., Shinya T., Desaki Y., Ichimura K., Shirasu K., Narusaka Y., Kawakami N., Kaku H., Shibuya N. (2007). CERK1, a LysM receptor kinase, is essential for chitin elicitor signaling in Arabidopsis. Proc. Natl. Acad. Sci. USA 104: 19613–19618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyakawa T., Hatano K., Miyauchi Y., Suwa Y., Sawano Y., Tanokura M. (2014). A secreted protein with plant-specific cysteine-rich motif functions as a mannose-binding lectin that exhibits antifungal activity. Plant Physiol. 166: 766–778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monaghan J., Zipfel C. (2012). Plant pattern recognition receptor complexes at the plasma membrane. Curr. Opin. Plant Biol. 15: 349–357. [DOI] [PubMed] [Google Scholar]
- Naumann T.A., Price N.P.J. (2012). Truncation of class IV chitinases from Arabidopsis by secreted fungal proteases. Mol. Plant Pathol. 13: 1135–1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naumann T.A., Wicklow D.T. (2010). Allozyme-specific modification of a maize seed chitinase by a protein secreted by the fungal pathogen Stenocarpella maydis. Phytopathology 100: 645–654. [DOI] [PubMed] [Google Scholar]
- Qu J., Ye J., Geng Y.F., Sun Y.W., Gao S.Q., Zhang B.P., Chen W., Chua N.H. (2012). Dissecting functions of KATANIN and WRINKLED1 in cotton fiber development by virus-induced gene silencing. Plant Physiol. 160: 738–748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu Z.L., Wang H.Y., Xia G.X. (2005). GhHb1: a nonsymbiotic hemoglobin gene of cotton responsive to infection by Verticillium dahliae. Biochim. Biophys. Acta 1730: 103–113. [DOI] [PubMed] [Google Scholar]
- Rutter B.D., Innes R.W. (2017). Extracellular vesicles isolated from the leaf apoplast carry stress-response proteins. Plant Physiol. 173: 728–741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutter B.D., Innes R.W. (2018). Extracellular vesicles as key mediators of plant-microbe interactions. Curr. Opin. Plant Biol. 44: 16–22. [DOI] [PubMed] [Google Scholar]
- Sawano Y., Miyakawa T., Yamazaki H., Tanokura M., Hatano K. (2007). Purification, characterization, and molecular gene cloning of an antifungal protein from Ginkgo biloba seeds. Biol. Chem. 388: 273–280. [DOI] [PubMed] [Google Scholar]
- Schickler H., Chet I. (1997). Heterologous chitinase gene expression to improve plant defense against phytopathogenic fungi. J. Ind. Microbiol. Biotechnol. 19: 196–201. [Google Scholar]
- Shinya T., Motoyama N., Ikeda A., Wada M., Kamiya K., Hayafune M., Kaku H., Shibuya N. (2012). Functional characterization of CEBiP and CERK1 homologs in arabidopsis and rice reveals the presence of different chitin receptor systems in plants. Plant Cell Physiol. 53: 1696–1706. [DOI] [PubMed] [Google Scholar]
- Shinya T., Nakagawa T., Kaku H., Shibuya N. (2015). Chitin-mediated plant-fungal interactions: catching, hiding and handshaking. Curr. Opin. Plant Biol. 26: 64–71. [DOI] [PubMed] [Google Scholar]
- Sinha M., Singh R.P., Kushwaha G.S., Iqbal N., Singh A., Kaushik S., Kaur P., Sharma S., Singh T.P. (2014). Current overview of allergens of plant pathogenesis related protein families. Sci. World J. 2014: 543195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slavokhotova A.A., Naumann T.A., Price N.P.J., Rogozhin E.A., Andreev Y.A., Vassilevski A.A., Odintsova T.I. (2014). Novel mode of action of plant defense peptides: hevein-like antimicrobial peptides from wheat inhibit fungal metalloproteases. FEBS J. 281: 4754–4764. [DOI] [PubMed] [Google Scholar]
- Spoel S.H., Dong X. (2012). How do plants achieve immunity? Defence without specialized immune cells. Nat. Rev. Immunol. 12: 89–100. [DOI] [PubMed] [Google Scholar]
- Sun L., Zhu L., Xu L., Yuan D., Min L., Zhang X. (2014). Cotton cytochrome P450 CYP82D regulates systemic cell death by modulating the octadecanoid pathway. Nat. Commun. 5: 5372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valueva T.A., Mosolov V.V. (2004). Role of inhibitors of proteolytic enzymes in plant defense against phytopathogenic microorganisms. Biochemistry (Mosc.) 69: 1305–1309. [DOI] [PubMed] [Google Scholar]
- van der Hoorn R.A.L. (2008). Plant proteases: From phenotypes to molecular mechanisms. Annu. Rev. Plant Biol. 59: 191–223. [DOI] [PubMed] [Google Scholar]
- van Loon L.C., Rep M., Pieterse C.M. (2006). Significance of inducible defense-related proteins in infected plants. Annu. Rev. Phytopathol. 44: 135–162. [DOI] [PubMed] [Google Scholar]
- Wan J., Zhang X.C., Neece D., Ramonell K.M., Clough S., Kim S.Y., Stacey M.G., Stacey G. (2008). A LysM receptor-like kinase plays a critical role in chitin signaling and fungal resistance in Arabidopsis. Plant Cell 20: 471–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F.X., Ma Y.P., Yang C.L., Zhao P.M., Yao Y., Jian G.L., Luo Y.M., Xia G.X. (2011). Proteomic analysis of the sea-island cotton roots infected by wilt pathogen Verticillium dahliae. Proteomics 11: 4296–4309. [DOI] [PubMed] [Google Scholar]
- Wang S., Xing H., Hua C., Guo H.S., Zhang J. (2016). An improved single-step cloning strategy simplifies the Agrobacterium tumefaciens-mediated transformation (ATMT)-based gene-disruption method for Verticillium dahliae. Phytopathology 106: 645–652. [DOI] [PubMed] [Google Scholar]
- Wilhelm S., Sagen J.E., Tietz H. (1974). Resistance to Verticillium wilt in cotton: sources, techniques of identification, inheritance trends, and the resistance potential of multiline cultivars. Phytopathology 64: 924–931. [Google Scholar]
- Xu J., Xu X., Tian L., Wang G., Zhang X., Wang X., Guo W. (2016). Discovery and identification of candidate genes from the chitinase gene family for Verticillium dahliae resistance in cotton. Sci. Rep. 6: 29022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh Y.H., Chang Y.H., Huang P.Y., Huang J.B., Zimmerli L. (2015). Enhanced Arabidopsis pattern-triggered immunity by overexpression of cysteine-rich receptor-like kinases. Front. Plant Sci. 6: 322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan D., et al. (2015). The genome sequence of Sea-Island cotton (Gossypium barbadense) provides insights into the allopolyploidization and development of superior spinnable fibres. Sci. Rep. 5: 17662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Tian L.H., Zhao J.F., Song Y., Zhang C.J., Guo Y. (2009). Identification of an apoplastic protein involved in the initial phase of salt stress response in rice root by two-dimensional electrophoresis. Plant Physiol. 149: 916–928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W.W., Jian G.L., Jiang T.F., Wang S.Z., Qi F.J., Xu S.C. (2012). Cotton gene expression profiles in resistant Gossypium hirsutum cv. Zhongzhimian KV1 responding to Verticillium dahliae strain V991 infection. Mol. Biol. Rep. 39: 9765–9774. [DOI] [PubMed] [Google Scholar]
- Zhou T.T., Zhao Y.L., Guo H.S. (2017). Secretory proteins are delivered to the septin-organized penetration interface during root infection by Verticillium dahliae. PLoS Pathog. 13: e1006275. [DOI] [PMC free article] [PubMed] [Google Scholar]