The highly conserved PROTEIN PHOSPHATASE4 complex coordinates the transcription and processing steps of microRNA biogenesis and plays broader roles in nuclear RNA metabolism in Arabidopsis (Arabidopsis thaliana).
Abstract
PROTEIN PHOSPHATASE4 (PP4) is a highly conserved Ser/Thr protein phosphatase found in yeast, plants, and animals. The composition and functions of PP4 in plants are poorly understood. Here, we uncovered the complexity of PP4 composition and function in Arabidopsis (Arabidopsis thaliana) and identified the composition of one form of PP4 containing the regulatory subunit PP4R3A. We show that PP4R3A, together with one of two redundant catalytic subunit genes, PROTEIN PHOSPHATASE X (PPX)1 and PPX2, promotes the biogenesis of microRNAs (miRNAs). PP4R3A is a chromatin-associated protein that interacts with RNA polymerase II and recruits it to the promoters of miRNA-encoding (MIR) genes to promote their transcription. PP4R3A likely also promotes the cotranscriptional processing of miRNA precursors, because it recruits the microprocessor component HYPONASTIC LEAVES1 to MIR genes and to nuclear dicing bodies. Finally, we show that hundreds of introns exhibit splicing defects in pp4r3a mutants. Together, this study reveals roles for Arabidopsis PP4 in transcription and nuclear RNA metabolism.
INTRODUCTION
PROTEIN PHOSPHATASE4 (PP4) is a highly conserved PP2A-type of Ser/Thr protein phosphatase found in eukaryotes that participates in a variety of cellular events (Cohen et al., 2005; Moorhead et al., 2009). The role of PP4 in DNA damage repair is particularly well documented, and its substrates involved in responses to DNA breaks and replication stress have been identified in animals (Chowdhury et al., 2008; Lee et al., 2010, 2012; Liu et al., 2012). These substrates include the chromatin condensation factor KAP1 (Kruppel-associated box-associated protein 1; Lee et al., 2012; Liu et al., 2012), Replication Protein A2, and the histone H2A variant γ-H2AX (Chowdhury et al., 2008; Lee et al., 2010). Arabidopsis (Arabidopsis thaliana) plants carrying mutations in the conserved PP4 regulatory subunit 3 (PP4R3) gene, PLATINUM SENSITIVITY (PSY) 2L/SUPPRESSOR OF MEK1 (SMEK)1, show enhanced sensitivity to the genotoxic drug cisplatin and exhibit activation of DNA repair signaling genes (Kataya et al., 2017). Thus, PP4 likely functions as a universal regulator of genome integrity.
Arabidopsis mutants in PSY2L/SMEK1 display pleiotropic phenotypes, strongly suggesting that PP4 plays a role in plant growth and development in addition to its role in DNA damage repair. PSY2L/SMEK1 was recently found to promote microRNA (miRNA) biogenesis in Arabidopsis (Su et al., 2017). Plant miRNAs are 20–24-nucleotide small RNAs that associate with ARGONAUTE (AGO) proteins (mainly AGO1) to mediate the post-transcriptional silencing of target genes by mRNA cleavage or translational repression. MiRNA biogenesis involves a series of coordinated steps (Rogers and Chen, 2013). DNA-dependent RNA polymerase II (Pol II) transcribes miRNA-encoding genes (MIR genes) into primary miRNAs (pri-miRNAs); this step is facilitated by Mediator, Elongator, and transcription factors such as NEGATIVE ON TATA LESS2 (NOT2) and CELL DIVISION CYCLE5 (CDC5; Xie et al., 2005; Zheng et al., 2009; Kim et al., 2011; Wang et al., 2013; Zhang et al., 2013; Fang et al., 2015). DICER-LIKE 1 (DCL1) processes pri-miRNAs into stem-loop precursors (pre-miRNAs) and then into miRNA/miRNA*duplexes in the nucleus (Kurihara and Watanabe, 2004). DCL1 associates with HYPONASTIC LEAVES 1 (HYL1) and SERRATE (SE) to form the microprocessor or Dicing complex, which is localized to subnuclear foci known as dicing bodies (D-bodies; Fang and Spector, 2007). HYL1 is regulated by phosphorylation/dephosphorylation: phosphorylation might be performed by MITOGEN-ACTIVATED PROTEIN KINASE3 and SUCROSE NONFERMENTING 1-RELATED PROTEIN KINASE2, whereas dephosphorylation by C-TERMINAL DOMAIN PHOSPHATASE-LIKE1, 2 (CPL1/2) is required for accurate miRNA processing (Manavella et al., 2012; Raghuram et al., 2015; Yan et al., 2017). PSY2L/SMEK1 is thought to promote miRNA biogenesis by dephosphorylating HYL1 to enhance its stability (Su et al., 2017).
Little is known about PP4 in Arabidopsis. Studies of the biological and molecular functions of PP4 have been restricted to mutants in PSY2L/SMEK1 (Kataya et al., 2017; Su et al., 2017). The two genes encoding the putative catalytic subunits, PROTEIN PHOSPHATASE X1 (PPX1) and PPX2, have not been characterized genetically. Thus, it is unclear whether the phenotypes of the psy2l/smek1 mutants reflect the functions of PP4. Biochemical characterization of the composition of PP4 has been limited to the demonstration that PSY2L/SMEK1 interacts with PPX1 and PPX2 in vivo (Su et al., 2017). Here, we report the potential existence of several forms of PP4 and the composition of one PP4 form. PSY2L/SMEK1 has an un-annotated and expressed homolog in the genome, and thus we named PSY2L/SMEK1 and this homolog PP4R3A and PP4R3B, respectively. Immunoprecipitation followed by mass spectrometry analysis with PP4R3A identified PPX1, PPX2, and PP4R2 LIKE (PP4R2L). Each ppx single mutant has no obvious phenotypes, but the double mutant resembles pp4r3a mutants, suggesting the presence of at least two PP4 forms with either PPX1 or PPX2 as the catalytic subunit. We show that not only PP4R3A, but also the catalytic subunit genes PPX1 and PPX2, are required for miRNA biogenesis. We show that PP4R3A is a chromatin-associated protein found at MIR genes and that it associates with and recruits Pol II to MIR genes to promote their transcription. Furthermore, we demonstrate that PP4 promotes the recruitment of HYL1 to MIR genes and the formation of D-bodies. Thus, PP4 coordinates the transcription and processing steps of miRNA biogenesis. Moreover, numerous protein-coding genes show intron retention in two pp4r3a mutants, indicating that PP4 plays broader roles in nuclear RNA metabolism.
RESULTS
Isolation of an Arabidopsis Mutant Defective in miRNA Biogenesis
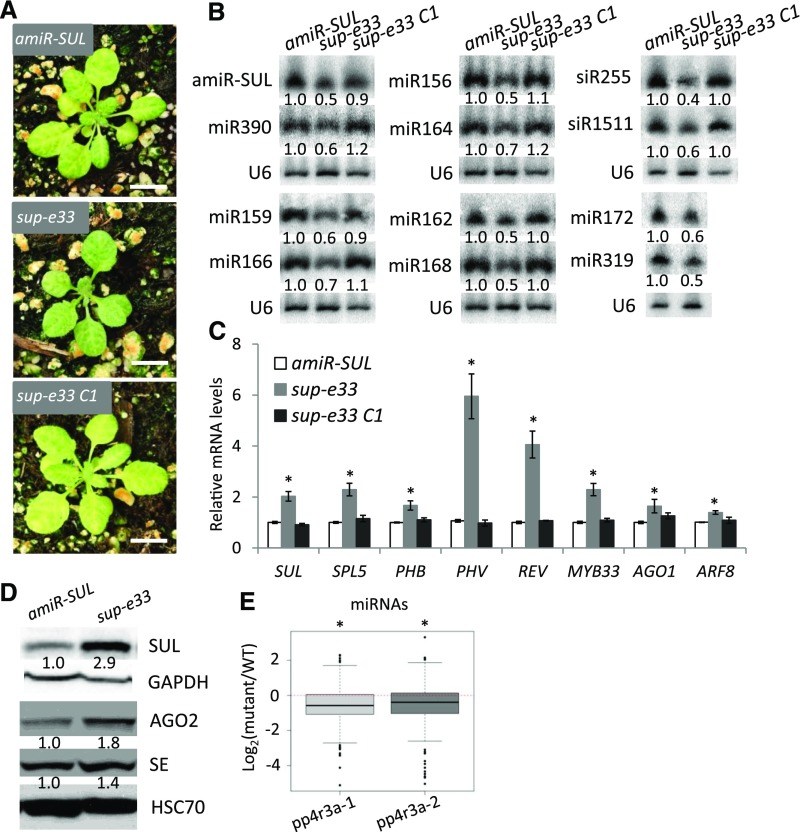
We took advantage of the visible vein-bleaching phenotype of the SUC2: amiR-SULFUR (amiR-SUL) transgenic line as a readout of miRNA activity to screen for mutants with potential defects in miRNA biogenesis or activity. In the amiR-SUL line, a phloem-specific promoter (SUC2) drives the expression of an artificial miRNA (amiR-SUL) that targets the SULFUR (SUL) gene, which is required for chlorophyll biosynthesis, leading to vein-centered leaf bleaching (de Felippes et al., 2011). We isolated a suppressor of amiR-SUL, sup-e33, with reduced leaf bleaching (Figure 1A; Supplemental Figure 1A and 1B). This phenotype suggested that sup-e33 is defective in amiR-SUL biogenesis or activity. Indeed, the levels of amiR-SUL were reduced in the mutant (Figure 1B); concomitantly, the expression of SUL was de-repressed, as reflected by both its transcript (Figure 1C) and protein levels (Figure 1D). The sup-e33 mutant exhibits pleiotropic phenotypes, including smaller plant size, early flowering, defective phyllotaxy and smaller silique-stem angles, and reduced fertility (Supplemental Figure 1). Because impaired miRNA biogenesis causes pleiotropic developmental defects (Park et al., 2002; Grigg et al., 2005; Zhang et al., 2013), we speculated that sup-e33 might be impaired in miRNA accumulation in general. We thus examined the abundance of endogenous miRNAs in 12-d-old amiR-SUL and sup-e33 seedlings. The accumulation of all miRNAs examined (miR166, miR164, miR159, miR390, miR319, miR172, miR156, and miR168) was reduced in sup-e33 compared with amiR-SUL (Figure 1B). We then examined the expression of seven known miRNA target genes (SQUAMOSA PROMOTER BINDING PROTEIN-LIKE5, PHABULOSA, PHAVOLUTA, REVOLUTA [REV], MYELOBLASTOSIS DOMAIN PROTEIN33 [MYB33], AGO1, and AUXIN RESPONSE FACTOR8) by reverse transcription quantitative PCR (RT-qPCR). As expected, the transcript levels of these genes were elevated in sup-e33 (Figure 1C). In addition, the protein levels of two miRNA targets (SE and AGO2) were elevated in sup-e33 (Figure 1D). The miRNAs that target these two genes, miR863-3p and miR403, were found to be at lower levels in the mutant based on small RNA-seq (Supplemental Data Set 1). Several miRNAs trigger the production of phased small interfering RNAs (siRNAs) from target genes (Fei et al., 2013; Komiya, 2017). We also detected the accumulation of miR173-triggered phased siRNAs known as trans-acting siRNAs (tasiRNAs; Allen et al., 2005). The levels of TRANS-ACTING SIRNA (TAS1)-siR255 and TAS2-siR1511 were lower in sup-e33 than in amiR-SUL (Figure 1B).
Figure 1.
The sup-e33 Mutant Is Defective in miRNA Biogenesis.
(A) Morphological phenotypes of 3-week-old SUC2:amiR-SUL (amiR-SUL), sup-e33, and rescued sup-e33 (sup-e33 C1) plants. Scale bars = 1 cm.
(B) RNA gel blot analysis to detect various miRNAs and tasiRNAs (siR155 and siR1511) in amiR-SUL, sup-e33, and rescued sup-e33 plants. U6 served as a loading control. The numbers represent relative abundance of small RNAs in the three genotypes.
(C) Transcript levels of miRNA targets, as determined by RT-qPCR. UBIQUITIN5 (UBQ5) served as an internal control. Error bars indicate sd from three independent experiments; asterisks indicate significant difference (t test, P < 0.05).
(D) Protein levels of miRNA targets (SUL/amiR-SUL, SE/miR863-3p, AGO2/miR403) in sup-e33, as determined by protein gel blot analysis. Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) and heat shock cognate (HSC)70 served as loading controls.
(E) Global abundance of miRNAs in Col and pp4r3a alleles, as determined by small RNA-seq. The small RNAs were normalized against total reads, abundance is expressed as RPM (reads per million mapped reads), and log2 fold changes (mutant/wild type) were plotted. Asterisk indicates that the mean log2 fold change is significantly below 0 (Wilcox. test, P < 2.3e-10).
A Mutation in PP4R3A, Encoding Regulatory Subunit 3 of PP4, Accounts for Compromised miRNA Accumulation
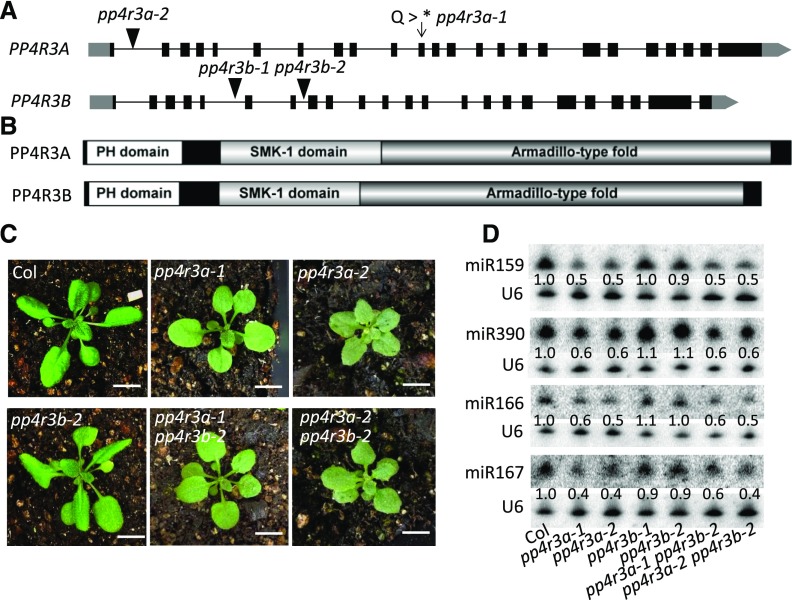
To identify the mutation in sup-e33, we backcrossed sup-e33 with the parental amiR-SUL line. More than 100 F2 segregants with the sup-33 phenotype were pooled for whole-genome re-sequencing. A C792 -to-T mutation in the open reading frame of AT3G06670 (this mutation is predicted to introduce a premature termination codon) was identified in sup-e33. In the F2 population of the backcross, this mutation was present in all 106 mutant plants tested, suggesting genetic linkage between the mutation and the phenotypes of sup-e33. We amplified genomic DNA encompassing the promoter and the coding region of AT3G06670 from wild-type plants and fused it to yellow fluorescent protein (YFP). This transgene, when introduced into sup-e33 plants, completely rescued the mutant phenotypes in terms of both leaf bleaching and miRNA defects (Figures 1A to 1C), confirming that the mutation in AT3G06670 leads to compromised miRNA accumulation. AT3G06670 encodes regulatory subunit 3 of PP4 (PP4R3, also known as PSY2L/SMEK1). We will refer to this gene as PP4R3A due to the presence of a homolog (see below).
To examine the phenotypes caused by the loss or reduced function of PP4R3A in an otherwise wild-type background, we crossed sup-e33 with wild type (Col) plants and obtained the pp4r3a-1 allele without the amiR-SUL transgene. We also acquired a T-DNA insertion allele, pp4r3a-2 (also known as smek1-2; Su et al., 2017). The pp4r3a-2 mutant displayed smaller plant size and darker green leaves than pp4r3a-1 (Figure 2C), suggesting that pp4r3a-2 has a stronger mutant phenotype than pp4r3a-1. In RNA-seq analysis, PP4R3A was found to be expressed at much lower levels in pp4r3a-2 than pp4r3a-1 and the wild type, suggesting that pp4r3a-2 is a strong knock-down allele (Supplemental Figure 2). The two mutants also had short roots (Supplemental Figure 3). We performed small RNA-sequencing (sRNA-seq) on 12-day-old Col, pp4r3a-1 and pp4r3a-2 seedlings (Figure 2A). Three sets of samples from each genotype were separately processed for library construction and sequencing, and the three independent experiments of the samples from each genotype were found to be reproducible (Supplemental Figure 4). A global reduction in miRNA accumulation was observed in the two pp4r3a mutants (Figure 1E; Supplemental Data Set 1), confirming that PP4R3A is required for miRNA biogenesis.
Figure 2.
Gene Cloning and Genetic Characterization of PP4R3A and PP4R3B.
(A) Gene structures of PP4R3A and PP4R3B (gray block, untranslated region; black blocks, exons; black lines, introns). The arrow indicates the point mutation site in sup-e33 (designated as pp4r3a-1), and the black triangles indicate the T-DNA insertion sites.
(B) Both PP4R3A and PP4R3B possess a pleckstrin homology (PH) domain, a suppressor of MEK-1 (SMK-1) domain, and an Armadillo fold.
(C) Morphological phenotypes of 3-week-old Col, pp4r3a-1 (sup-e33 in Col background), pp4r3a-2, pp4r3b-2, pp4r3a-1 pp4r3b-2, and pp44r3a-2 pp4r3b-2 plants. Scale bars = 1 cm.
(D) Determination of miRNA levels in pp4r3a, pp4r3b and pp4r3a pp4r3b by RNA gel blot analysis of small RNAs. The numbers represent relative abundance of small RNAs.
In the Arabidopsis genome, three clustered, annotated genes, AT5G49370, AT5G49380, and AT5G49390, share similarity to the N-terminal, middle, and C-terminal portions of PP4R3A, respectively. This observation suggested that the three “genes” could actually be one gene, which would be a homolog of PP4R3A. Indeed, a full-length transcript containing all three “genes” was obtained by RT-PCR and sequenced. The protein encoded by this transcript is highly similar in domain composition to the protein encoded by PP4R3A (Figures 2A and 2B). Thus, the three annotated genes actually represent one gene, which we named PP4R3B. We generated transgenic plants harboring the β-glucuronidase (GUS) reporter driven by the PP4R3A promoter and found that PP4R3A was mainly expressed in vascular tissue, with higher expression in roots than shoots (Supplemental Figure 5A). The promoter activity of PP4R3B was detectable in vascular tissue and mesophyll cells, but not in actively growing tissues such as young leaves or lateral root primordia (Supplemental Figure 5B).
To investigate whether PP4R3A and PP4R3B play redundant roles in morphogenesis and miRNA biogenesis in Arabidopsis, we obtained T-DNA insertion lines in PP4R3B; these alleles were named pp4r3b-1 and pp4r3b-2, respectively (Figure 2A). Neither pp4r3b allele had any obvious mutant phenotypes. The pp4r3a-1 pp4r3b-2 and pp4r3a-2 pp4r3b-2 double mutants resembled the corresponding pp4r3a single mutants in terms of plant morphology and in miRNA accumulation (Figures 2C and 2D), suggesting that PP4R3B does not play an overlapping role with PP4R3A in Arabidopsis development or miRNA biogenesis. We also introduced PP4R3B-YFP driven by the constitutive CaMV 35S promoter into pp4r3a-1 plants. The 11 transgenic T1 plants showed different levels of PP4R3B-YFP expression, but none of them exhibited morphological phenotypes indicative of transgene rescue of pp4r3a-1 (Supplemental Figure 6). These results suggest that PP4R3B cannot substitute for PP4R3A in terms of plant development or miRNA biogenesis. Because PP4R3B is expressed, it likely has an as-yet unknown function.
The PP4 Catalytic Subunits Promote miRNA Biogenesis
In the yeast Saccharomyces cerevisiae and mammals, PP4R3/Psy2 was identified as regulatory subunit 3 of the PP4 complex, which also contains the catalytic subunit (PP4C) and regulatory subunit 2 (PP4R2; Gingras et al., 2005; Hastie et al., 2006). To uncover the composition of the Arabidopsis PP4 complex, we introduced the PP4R3A:PP4R3A-YFP and PP4R3A:PP4R3A-HEMAGGLUTININ (HA) transgenes, respectively, into the pp4r3a-1 background. The transgenes were functional, because they fully rescued the morphological defects of the mutant (Supplemental Figure 7). We then immunoprecipitated (IP) PP4R3A-YFP from PP4R3A:PP4R3A-YFP/pp4r3a-1 plants and PP4R3A-HA from PP4R3A:PP4R3A-HA/pp4r3a-1 plants. In both experiments, Col plants were included as the negative control. Mass spectrometry (MS) analysis of the immunoprecipitates showed that PP4R3A associates with two catalytic subunits, PPX1 and PPX2, and the regulatory subunit PP4R2L (Supplemental Figure 8A and Supplemental Data Set 2). To determine whether PP4 acts in miRNA biogenesis, we searched for mutants in the catalytic subunit. However, T-DNA insertion mutants in PPX1 or PPX2 genes were not available. We therefore performed CRISPR/Cas9 (clustered regulatory interspaced short palindromic repeats/CRISPR-associated endonuclease9)-based gene editing (Yan et al., 2015) and successfully obtained ppx1 and ppx2 single mutants with base deletions (Supplemental Figure 8B). Neither single mutant showed any morphological defects (Figure 3A). In the F2 population of the cross between the two mutants, we found only three tiny seedlings displaying similar phenotypes to those of pp4r3a-2 among ∼150 seedlings (Figure 3A); these tiny plants indeed were ppx1 ppx2 double mutants, as revealed by genotyping. However, the proportion of double mutants in the F2 population was so low that we speculated that most of the double mutants were embryo lethal. Consistent with this hypothesis, we observed defective embryos in the siliques of ppx1/ppx1 PPX2/ppx2 plants (Supplemental Figure 8C). This finding suggests that the two PPX genes play largely redundant roles and together are essential, implying that either PPX protein assembles a PP4 complex. We harvested viable ppx1 ppx2 seedlings from the progeny of ppx1/ppx1 PPX2/ppx2 plants and measured the levels of mature miRNAs. Although both ppx single mutants resembled the wild type, the double mutant showed reduced levels of mature miRNAs and TAS1-siR255 (Figure 3B). Therefore, the biochemical and genetic data demonstrate that at least two subunits of the PP4 complex (PP4R3A and PPX) participate in miRNA biogenesis.
Figure 3.
Phenotypes of PP4 Catalytic Mutants.
(A) Morphologic phenotypes of the ppx1 and ppx2 single mutants and the ppx1 ppx2 double mutant. Scale bars = 1 cm.
(B) Accumulation of miRNAs and an siRNA in Col, ppx1, ppx2, and ppx1 ppx2 plants as detected by RNA gel blot analysis of small RNAs. U6 served as a loading control. The numbers indicate relative abundance of the small RNAs in the four genotypes.
PP4R3A Has Effects beyond Pri-miRNA Processing in miRNA Biogenesis
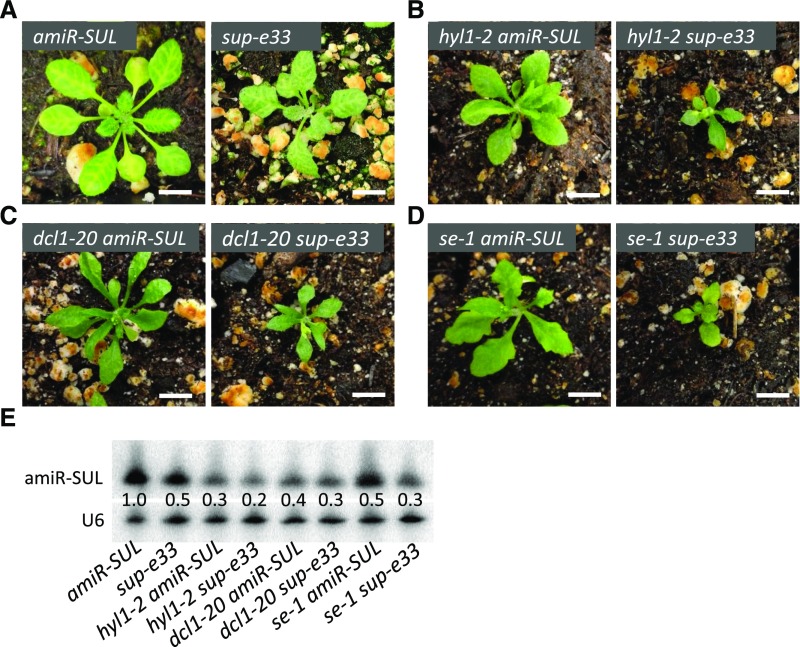
PP4R3A was previously reported to stabilize the DCL1 co-factor HYL1 and promote pri-miRNA processing; in two mutants in this gene, HYL1 protein levels were reduced to approximately half of wild-type levels (Su et al., 2017). However, we did not observe a reduction in HYL1 protein levels in pp4r3a-1 or pp4r3a-2 (this allele is the same as smek1-2; Su et al., 2017). In fact, an increase in HYL1 protein levels was observed at the normal growth temperature in the two alleles, although HYL1 protein levels were slightly reduced in pp4r3a-1 at 30°C (Supplemental Figure 9B). The lack of reduction in HYL1 protein levels in the pp4r3a mutants at 22°C suggested that the reduced miRNA abundance in the mutants is not solely attributable to defects in pri-miRNA processing. To examine this notion genetically, we crossed sup-e33 (pp4r3a-1 amiR-SUL) plants with mutants in the core dicing complex genes DCL1, HYL1, and SE in the amiR-SUL background. As expected, the dcl1-20, hyl1-2, and se-1 mutations compromised leaf bleaching in the amiR-SUL background (Figures 4A to 4D), which is consistent with the reduced levels of amiR-SUL in these backgrounds (Figure 4E). The double mutants had even lower levels of amiR-SUL compared with each single mutant (Figure 4E). The morphological phenotypes of the double mutants were also more severe than those of the corresponding single mutants (Figures 4A to 4D). The accumulation of endogenous miRNAs in the double mutants tended to further decrease compared with the corresponding single mutants (Supplemental Figure 10). Although the dcl1-20 and se-1 alleles are not null alleles (null alleles in these genes are embryo lethal; Golden et al., 2002; Grigg et al., 2005; Armenta-Medina et al., 2017), the hyl1-2 allele is likely a null allele, because no HYL1 protein was detected in this mutant (Cho et al., 2014). The finding that pp4r3a-1 enhanced the miRNA biogenesis defects of hyl1-2 indicates that PP4R3A plays a role in miRNA biogenesis beyond its reported role in stabilizing HYL1 protein.
Figure 4.
Genetic Interactions between sup-e33 and Pri-miRNA Processing Mutants.
(A) to (D) Morphological phenotypes of 3-week-old plants of the indicated genotypes. Scale bars = 1 cm.
(E) RNA gel blot analysis of small RNAs to determine the levels of amiR-SUL in various genotypes. U6 served as a loading control. The numbers represent relative abundance.
PP4R3A Promotes the Transcription of MIR Genes
To determine the potential reasons for reduced miRNA accumulation in the pp4r3a mutants, we examined the expression of known components in the miRNA biogenesis machinery. No changes in the transcript levels of these genes were found in the pp4r3a mutants (Supplemental Figure 9A). Moreover, the protein levels of some important factors in miRNA biogenesis, such as Pol II, HYL1, and SE, were not substantially reduced or were even increased (such as HYL1 and SE at 22°C) in the pp4r3a mutants compared with wild type (Supplemental Figure 9B). AGO1 protein levels were reduced in the two pp4r3a mutants (Supplemental Figure 9B), which is consistent with the global reduction in miRNA accumulation. We then examined the levels of HYL1, SE, and AGO1 at different growth temperatures. Intriguingly, the increase in HYL1 levels in pp4r3a-1 detected at 22°C and 16°C was not observed at 30°C, whereas the increase in SE levels detected at 22°C and 30°C was not observed at 16°C (Supplemental Figure 9B). These observations suggest that PP4R3A plays a role in the responses of plants to different temperatures, which deserves further investigation in the future.
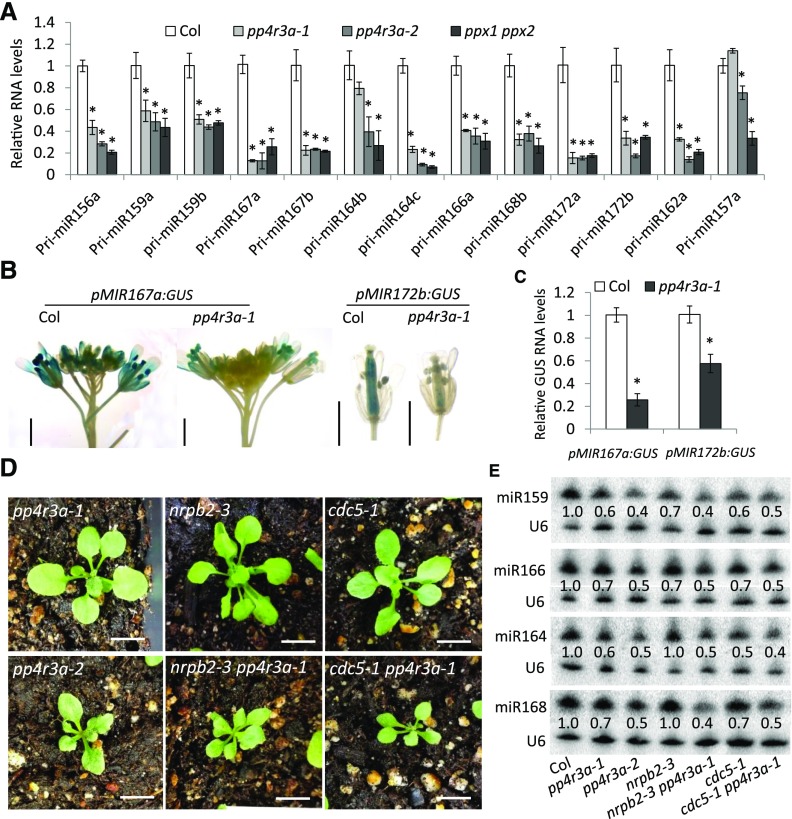
We then examined the transcript levels of 13 pri-miRNAs and found that the abundance of the tested pri-miRNAs, except for pri-miR164a and pri-miR157a, was drastically reduced in both pp4r3a alleles and ppx1 ppx2. The transcript levels of pri-miR164a and pri-miR157a were also reduced in pp4r3a-2 and ppx1 ppx2, although their levels were not obviously changed in pp4r3a-1 (Figure 5A). These results suggest that PP4 promotes the transcription of MIR genes. We examined the activities of MIR gene promoters in pp4r3a-1. We crossed two transgenic lines harboring the GUS reporter gene driven by the MIR167a or MIR172b promoter in the wild-type background with pp4r3a-1 plants and compared the expression of homozygous and isogenic transgenes between wild-type and pp4r3a-1 plants. GUS activity was greatly reduced in pp4r3a-1, as revealed by GUS staining. RT-qPCR analysis confirmed that GUS mRNA levels were reduced in pp4r3a-1 (Figures 5B and 5C), indicating that the transcription of MIR genes is impaired in pp4r3a-1. Consistent with the role of PP4R3A in MIR gene transcription, crosses with two other mutants with reduced transcription of MIR genes, nuclear rna polymerase II (nrpb)2-3 (Zheng et al., 2009) and cdc5-1 (Zhang et al., 2013), enhanced the weaker pp4r3a-1 mutant in terms of both plant morphology and miRNA levels (Figures 5D and 5E). In fact, the nrpb2-3 pp4r3a-1 and cdc5-1 pp4r3a-1 double mutants resembled the pp4r3a-2 allele, a strong knockdown allele (Figures 5D and 5E).
Figure 5.
Requirement of PP4R3A for MIR Transcription.
(A) Determination of pri-miRNA levels by RT-qPCR.
(B) GUS staining of representative samples harboring pMIR167a:GUS and pMIR172b:GUS in the Col and pp4r3a-1 backgrounds. Scale bars = 2 mm.
(C) GUS transcript levels in samples harboring pMIR167a:GUS and pMIR172b:GUS in the Col and pp4r3a-1 backgrounds, as determined by RT-qPCR.
(D) and (E) Genetic interactions between mutations in PP4R3A, NRPB2 and CDC5. (D) Additive phenotypes of nrpb2-3 pp4r3a-1 and cdc5-2 pp4r3a-1 double mutants. Scale bars = 1 cm. Note that the phenotypes of the double mutants were much stronger than the corresponding single mutants, but similar to that of pp4r3a-2. (E) The abundance of miRNAs in Col, pp4r3a-1, pp4r3a-2, nrpb2-3, cdc5-1, nrpb2-3 pp4r3a-1, and cdc5-1 pp4r3a-1, as detected by RNA gel blot analysis of small RNAs.
In (A) and (C), UBQ5 served as the internal control; error bars indicate sd from three independent experiments; asterisks indicate significant difference (t test, P < 0.05).
PP4R3A Associates with Chromatin and Is Required for Pol II Occupancy at MIR Promoters
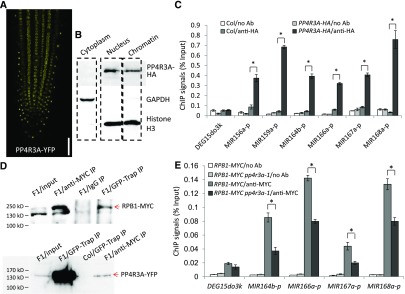
A classical bipartite-type nuclear localization signal is present in the Armadillo domain of PP4R3A. In PP4R3A:PP4R3A-YFP/pp4r3a-1 transgenic plants, YFP fluorescence was present in only the nucleus (Figure 6A). Given the finding that PP4R3A promotes MIR transcription and its known role in DNA damage repair, we wondered whether this protein is associated with chromatin. Using PP4R3A:PP4R3A-HA/pp4r3a-1 transgenic plants, we performed cytoplasmic/nuclear fractionation and extracted chromatin from the nuclear fraction. Protein gel blot analysis showed the presence of PP4R3A-HA in the nuclear rather than the cytoplasmic fraction, and also revealed its association with chromatin (Figure 6B). Because PP4R3A is required for MIR promoter activity, we examined whether PP4R3A binds MIR promoters. We performed chromatin immunoprecipitation (ChIP) assay with Col and pPP4R3A:PP4R3A-HA/pp4r3a-1 plants using an anti-HA antibody. Promoter regions of the tested MIRs were enriched in the immunoprecipitates from pPP4R3A:PP4R3A-HA/pp4r3a-1 plants (Figure 6C), indicating that PP4R3A is associated with MIR promoters.
Figure 6.
Analysis of PP4R3A Localization and Pol II Occupancy at MIR Promoters.
(A) YFP signals in the root of a 5-d-old PP4R3A:PP4R3A-YFP seedling. Scale bar = 200 μm.
(B) Protein gel blot analysis showing the cytoplasmic, nuclear, and chromatin distribution of the PP4R3A-HA protein from a PP4R3A:PP4R3A-HA/pp4r3a-1 transgenic line. GAPDH and histone H3 were used as protein markers for the cytoplasmic and nuclear/chromatin fractions, respectively.
(C) ChIP-qPCR analysis to determine the occupancy of PP4R3A at MIR promoters. ChIP was performed with no antibody (no Ab) or anti-HA antibody in Col and PP4R3A:PP4R3A-HA transgenic plants.
(D) Co-immunoprecipitation of PP4R3A with Pol II. F1 plants from a cross between PP4R3A:PP4R3A-YFP and RPB1:RPB1-4xMYC homozygous transgenic lines were used to perform IP, using either anti-MYC or GFP-Trap antibodies. Protein gel blot analysis to detect PP4R3A–YFP and RPB1-MYC was performed with anti-GFP and anti-MYC antibodies, respectively. The protein ladder is labeled on the left.
(E) ChIP-qPCR analysis to determine the occupancy of Pol II at MIR promoters using plants with a homozygous RPB1:RPB1-4xMYC transgene in the Col and pp4r3a-1 backgrounds. The intergenic region DEG15do3k between AT1G28310 and AT1G28320 was used as a negative control. The ChIP signal was normalized against input.
In (C) and (E), error bars indicate sd from three independent experiments; asterisks indicate significant difference (t test, P < 0.05).
MIR genes are transcribed by Pol II (Xie et al., 2005; Zheng et al., 2009). To determine whether PP4R3A associates with Pol II, we tagged the largest subunit of Pol II (RPB1) with the MYC epitope, crossed RPB1-MYC plants with PP4R3A-YFP/pp4r3a-1 plants, and extracted proteins from F1 plants expressing both RPB1-MYC and PP4R3A-YFP for co-immunoprecipitation (Co-IP). Reciprocal Co-IP was conducted and protein gel blot analysis showed that RPB1-MYC and PP4R3A-YFP was effectively enriched using anti-MYC antibody and green fluorescent protein (GFP)-Trap, respectively, and that RPB1-MYC was present in PP4R3A-YFP immunoprecipitates and vice versa (Figure 6D). Then, we analyzed the occupancy of Pol II at MIR promoters by ChIP, which was conducted on RPB1-MYC transgenic plants in Col (RPB1-MYC) and pp4r3a-1 (RPB1-MYC pp4r3a-1) backgrounds using an anti-MYC antibody. The promoter regions of all tested MIRs were enriched in RPB1-MYC and RPB1-MYC pp4r3a-1 immunoprecipitates; however, the enrichment fold in RPB1-MYC pp4r3a-1 was significantly reduced when compared with that in RPB1-MYC (Figure 6E).Taken together, these data show that PP4R3A promotes the transcription of MIR genes through recruitment of Pol II to MIR promoters.
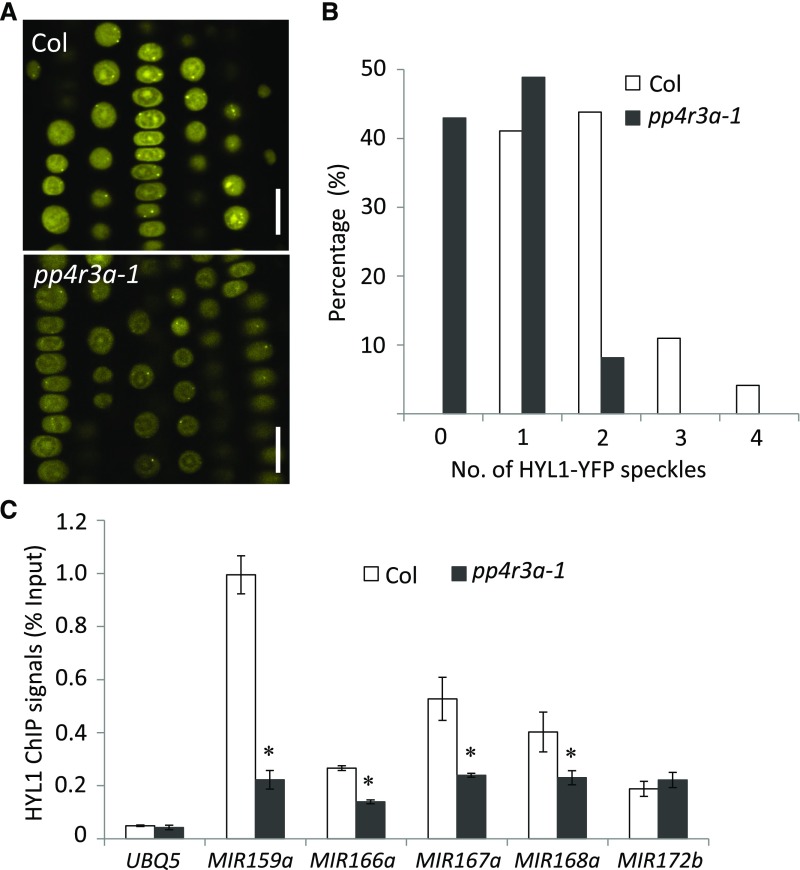
PP4R3A Promotes HYL1 Localization to D-Bodies and Its Association with MIR Loci
In mammals and plants, microprocessor and/or pri-miRNA transcripts are associated with chromatin and the processing of pri-miRNAs occurs co-transcriptionally (Morlando et al., 2008; Ballarino et al., 2009; Fang et al., 2015). In Arabidopsis, both DCL1 and HYL1 can be detected at MIR gene loci, suggesting co-transcriptional processing (Fang et al., 2015). We proposed that disruption of MIR transcription may affect co-transcriptional pri-miRNA processing. We crossed a HYL1-YFP line with pp4r3a-1 to generate HYL1-YFP pp4r3a-1 and compared the number of D-bodies in wild type and mutant backgrounds. The number of HYL1-YFP-containing D-bodies was dramatically reduced in pp4r3a-1 (Figures 7A and 7B), demonstrating that PP4R3A plays a role in the proper localization of HYL1 to D-bodies. To further confirm a defect in pri-miRNA co-transcriptional processing in pp4r3a-1, we performed HYL1 ChIP using anti-HYL1 antibodies. HYL1 was detected at the MIR loci, and its occupancy at four of five tested MIR loci was significantly reduced in pp4r3a-1 (Figure 7C). These results suggest that PP4R3A is required for the formation of D-bodies and the recruitment of HYL1 to MIR genes. Because both PP4R3A and HYL1 are found at MIR loci, we entertained the possibility that HYL1, by binding to nascent pri-miRNAs, recruits PP4R3A to MIR loci. We performed PP4R3A ChIP-qPCR at eight MIR promoters using PP4R3A:PP4R3A-HA transgenic plants in the wild type and hyl1-2 backgrounds and found no significant difference in PP4R3A occupancy at these promoters except for two (Supplemental Figure 11A). Thus, it is likely that PP4R3A recruits HYL1 to MIR loci but not vice versa. To determine whether PP4R3A recruits HYL1 to MIR loci through protein-protein interaction, we conducted Co-IP assays. PP4R3A-GFP was immunoprecipitated with GFP-Trap, and the immunoprecipitates were probed for HYL1 and SE by protein gel blot analysis. The two microprocessor components were not detected in the PP4R3A IP (Supplemental Figure 11B).
Figure 7.
HYL1 Localization and Chromatin Association are Impaired in pp4r3a-1.
(A) Representative confocal images of HYL1-YFP fluorescence in the meristematic zones of Col and pp4r3a-1 roots. Bars = 10 μm.
(B) Quantification of HYL1-YFP D-bodies. The x axis represents the number of HYL1-YFP D-bodies (speckles) per cell, and the y axis indicates the percentage of cells with the corresponding HYL1-YFP D-body numbers. The number of HYL1-YFP D-bodies was calculated from 146 root cells of Col and 135 root cells of pp4r3a-1.
(C) ChIP-qPCR assay to determine the association of HYL1 with MIR gene bodies in Col and pp4r3a-1. Error bars indicate sd from three independent experiments; asterisk indicates significant difference (t test, P < 0.05).
PP4R3A Promotes Transcription of Protein Coding Genes and Splicing of a Set of Introns
The association between PP4R3A and Pol II raised the possibility that PP4R3A affects the expression of other genes in addition to MIR genes. We therefore performed RNA-seq analysis of wild type, pp4r3a-1, and pp4r3-2 plants in three separate experiments. Clustering analysis showed that the replicates of each genotype were reproducible (Supplemental Figure 12A). First, we examined the abundance of pri-miRNAs. Among the 325 annotated pri-miRNAs in Araport11, those that were represented by reads of FPKM>2 were retained. These pri-miRNAs tended to accumulate to lower levels in both mutants compared with wild type (Supplemental Figure 12C). Because some pri-miRNAs overlapped with protein-coding genes, reads from the nearby genes could be mistakenly assigned to the pri-miRNAs. We therefore examined the pri-miRNAs without overlapping protein-coding genes. The levels of these pri-miRNAs were also reduced in both mutants compared with wild type (Supplemental Figure 12C), confirming that PP4R3A is a positive regulator of pri-miRNA accumulation. We then identified differentially expressed genes in each mutant relative to wild type. More than 1000 up- or downregulated genes were found in each pp4r3a mutant, and a large overlap was found for the up- or downregulated genes between the two mutant alleles (Figure 8A). Because the presence of allele-specific mis-regulated genes could be due to morphological differences between the two mutants, these genes were not investigated further. The overwhelming majority of genes that were upregulated in both alleles were enriched in genes that respond to various stimuli (Figure 8B), suggesting that PP4R3A suppresses plants' responses to various stimuli under normal conditions. This finding appears to be consistent with the observation that pp4r3a mutants are more sensitive to abscisic acid treatment than the wild type (Su et al., 2017). The genes that were downregulated in both alleles are involved in cellular oxidation-reduction processes, ion metabolism/transport, and signaling (Figure 8B). Surprisingly, despite the global reduction in miRNA levels in the mutants and the finding that several miRNA target transcripts were upregulated, as revealed by RT-qPCR (Figure 1C), a global trend of upregulation was not observed for known miRNA target genes (Supplemental Figure 12B). One possible explanation is that PP4R3A is also required for the transcription of these genes.
Figure 8.
Defects in Gene Expression and Splicing of Pre-mRNAs in pp4r3a Mutants.
(A) Venn diagrams showing the number of differentially expressed genes (DEGs) in each pp4r3a allele and the overlap of DEGs in the two alleles. Hyper- and hypo-DEGs are genes with increased and decreased expression, respectively.
(B) Gene Ontology enrichment analysis of common DEGs in the two pp4r3a alleles. In the pp4r3a mutants, upregulated genes are enriched in various stress responses Gene Ontology terms, whereas downregulated genes are involved in oxidation-reduction processes, cellular ion metabolism, and signaling.
(C) Scatterplots showing increased PI (percent of intron reads) in each pp4r3a mutant vs. wild type. Each dot represents an annotated intron in Araport11. The introns with a statistically significant increase in PI are shown in green.
(D) Venn diagram showing the number of introns with splicing defects in pp4r3a-1 and pp4r3a-2 and the overlap between the two alleles.
There is emerging evidence linking the processes of miRNA biogenesis and pre-mRNA splicing. On the one hand, introns or 5′ splice sites in some pri-miRNAs stimulate miRNA biogenesis (Bielewicz et al., 2013; Szweykowska-Kulińska et al., 2013). On the other hand, some proteins affect both miRNA biogenesis and the splicing of protein-coding genes, such as the microprocessor component SE, the Cap binding Complex components ABA HYPERSENSITIVE1/CAP-BINDING PROTEIN 80 and CAP-BINDING PROTEIN 20 (Laubinger et al., 2008), the splicing factors STABILIZED1, Arginine/serine (RS)-rich proteins RS40 and RS41 (Ben Chaabane et al., 2013; Chen et al., 2013, 2015), the K-homology domain RNA binding protein HOS5 (Chen et al., 2013, 2015), the proline-rich protein SICKLE (Zhan et al., 2012), and Modifier of snc1, 4-Associated Complex (MAC) components MAC3A, MAC3B, MAC7, PLEIOTROPIC REGULATORY LOCUS1 (PRL1), and PRL2 (Jia et al., 2017; Li et al., 2018). This prompted us to analyze whether the pp4r3a mutants show splicing defects. We analyzed splicing defects in the mutants based on RNA-seq data (see Methods for details). Each allele showed an increase in intron retention at more than 350 introns. This number is small considering that more than 70,000 introns were included in the analyses. Retention in both mutants was shown in 192 introns, and a large proportion of the genes with intron retention defects encode proteins that are targeted to organelles (Figures 8C and 8D; Supplemental Figure 13A and 13B). We then analyzed the expression of the genes with intron retention defects. No differential expression of these genes was observed between the wild type and pp4r3a mutants (Supplemental Figure 13C). Thus, there was no correlation between intron retention and gene expression levels at these genes. Because the MAC subunits MAC3A, MAC3B, PRL1, and PRL2 also promote intron splicing, we asked whether they affect the same introns as PP4R3A. We analyzed intron retention defects in the mac3 and prl mutants using the same method as for pp4r3a. As expected, a significant overlap (hypergeometric test, P < 9.93E-62) was found between the retained introns in mac3a mac3b and prl1 prl2 (Supplemental Figure 13D). There was also significant overlap in retained introns between the pp4r3a mutants and the mac mutants, but very few commonly affected introns were identified (Supplemental Figure 13D). We examined whether the introns with retention defects in the pp4r3a mutants had any common features. When compared with all introns, those with intron retention defects did not differ in terms of their positions along genes. The genes with intron retention defects did not differ from all Arabidopsis genes in terms of the number of introns. However, the introns with retention defects in the mutants tended to be longer as compared with the overall length distribution of introns (Supplemental Figure 14).
DISCUSSION
PP4, a Ser/Thr-specific phosphoprotein phosphatase (PP) conserved from yeast to mammals, is a heteromultimeric complex containing a catalytic subunit (PP4C) and two regulatory/scaffolding subunits, PP4R2 and PP4R3 (Gingras et al., 2005). Genes encoding homologs of all three subunits are present in plants, but our knowledge of the plant PP4 complex is rather limited. Little has been known about the composition of plant PP4 except that PP4R3 interacts in vivo with PPX1 and PPX2, two homologs of PP4C (Su et al., 2017). Reports of PP4 functions in plants were primarily based on studies of PP4R3, also known as PSY2L/SMEK1, in Arabidopsis (Kataya et al., 2017; Su et al., 2017). The current findings shed light on the composition and functions of Arabidopsis PP4. First, through IP of PP4R3 followed by mass spectrometry, we showed that PPX1, PPX2, and the PP4R2 homolog PP4R2L associate with PP4R3A in vivo. Second, through genetic analysis of the ppx1 and ppx2 single and double mutants, we showed that the two catalytic subunit genes are largely redundant and that the double mutant resembles the strong pp4r3a-2 mutant in terms of both morphological and molecular defects. This observation confirms that the PPX proteins function together with PP4R3A in vivo and suggests the presence of two PP4 forms, one containing PPX1 and PP4R3 and the other containing PPX2 and PP4R3. We also found that, in addition to PP4R3A, the Arabidopsis genome contains a second gene, which we named PP4R3B, which may encode regulatory subunit 3 of another form of PP4. We showed that only PP4R3A is critical for plant development and miRNA accumulation under normal growth conditions. PP4R3B is expressed and may exert functions under specific conditions.
Evolutionary analysis of PP catalytic subunits has shown that Arabidopsis PP4, like that of other eukaryotes, forms a distinct cluster with PP2A and PP6, which are members of the PP family (Moorhead et al., 2009; Lillo et al., 2014). The regulatory partners of PPs usually modulate the activity and substrate specificity of the catalytic subunits. However, it can be risky to study the functions of regulatory subunits as a substitute for PPs in plants. The Arabidopsis Tap46 (the homolog of yeast and mammalian Tap42/α4), a downstream effector of the target of the rapamycin signaling pathway, is a regulatory subunit that interacts with PP2A and PP2A-related phosphatases PP4 and PP6 with different affinities and plays essential roles in plant growth and responses to chilling (Harris et al., 1999; Ahn et al., 2011). Thus, the functions of Tap46 could reflect those of multiple PPs. In the case of PP4R3A, our IP MS analysis did not detect in vivo interactions with PP2A-type PPs. This observation, together with the similar phenotypes of pp4r3a-2 and ppx1 ppx2 mutants, suggests that PP4R3A serves as a specific regulatory subunit of PP4.
Several studies have revealed post-translational modifications of components of the miRNA biogenesis machinery, such as HYL1 and AGO1 (Cho et al., 2016). The double-stranded RNA binding protein, HYL1, forms a complex with DCL1 and SE to promote precise DCL1 cleavage as well as strand selection during AGO1 loading (Dong et al., 2008; Eamens et al., 2009). Manavella et al. (2012) showed that the dephosphorylation of HYL1 by a family of CPL phosphatases is required for its activity. Unlike the other components of the microprocessor, HYL1 is a short-lived protein that is degraded in the cytoplasm in the dark (Cho et al., 2014). The HYL1 phosphoisoforms (phosphorylated and nonphosphorylated) appear to affect the stability of the protein, as shown in two studies (Su et al., 2017; Achkar et al., 2018). However, these two studies led to different conclusions. One found that phosphorylated HYL1 forms a nuclear reserve of inactive protein protected from dark-induced degradation, whereas dephosphorylated HYL1 is unstable (Achkar et al., 2018). The other study (Su et al., 2017) found that PP4R3A stabilizes HYL1 by dephosphorylating it. Evidence supporting this claim includes the reduced HYL1 levels in the pp4r3a mutants and the finding that recombinant HYL1 is more unstable when incubated with smek1-1 lysates compared with wild-type lysates (Su et al., 2017). Under our growth conditions, we found an increase in HYL1 levels in two pp4r3a mutants (including one used in the Su et al. study) compared with wild type, but a small reduction in HYL1 levels was observed in the mutants at higher temperature. These conflicting observations suggest that HYL1 is regulated in an intricate manner in response to environmental conditions.
Emerging evidence suggests that pri-miRNAs are cotranscriptionally processed in plants. NOT2 (a core member of the CARBON CATABOLITE REPRESSION4 [CCR4]-NOT complex; Wang et al., 2013), PRL1, PRL2, CDC5, MAC3A, MAC3B and MAC7 (subunits of the Modifier of snc1, 4-associated complex; Zhang et al., 2013, 2014; Jia et al., 2017; Li et al., 2018), and ELP2 and ELP5 (subunits of the Elongator complex; Fang et al., 2015) interact with Pol II and/or the dicing complex to coordinate MIR transcription and pri-miRNA processing. We found that mutations in PP4R3A led to reduced occupancy of both Pol II and HYL1 at MIR genes, lower levels of pri-miRNAs, and fewer D-bodies compared with wild type. PP4R3A is a chromatin-associated protein that is present at the promoters of MIR genes and interacts with Pol II. One model is that PP4 recruits Pol II to MIR genes to promote their transcription, and at MIR loci, it dephosphorylates HYL1 to enable pri-miRNA processing (Su et al., 2017); hypo-phosphorylated HYL1 is active in pri-miRNA processing (Manavella et al., 2012). Alternatively, PP4 might dephosphorylate a chromatin protein to facilitate the access of both Pol II and hypo-phosphorylated HYL1 to chromatin.
The coordination of splicing and miRNA biogenesis has been well documented (Stepien et al., 2017). Pri-miRNAs may contain introns or may reside in introns of host genes, such that miRNA biogenesis may entail intron removal from pri-miRNAs, and microprocessor-mediated pri-miRNA cropping from introns of host genes may affect the splicing of host pre-mRNAs. However, in Arabidopsis, it is estimated that only half of the known pri-miRNAs contain introns (Szweykowska-Kulińska et al., 2013; Stepien et al., 2017). Yet, many splicing-related proteins have been found to affect miRNA biogenesis in general. For example, the nuclear cap binding complex, known to promote pre-mRNA splicing and polyadenylation, is required for pri-miRNA processing (Laubinger et al., 2008; Raczynska et al., 2014). The K-homology domain protein REGULATOR OF CBF GENE EXPRESSION3 interacts with the CPL phosphatases and promotes dephosphorylation of HYL1 in specific plant tissues to promote miRNA biogenesis. REGULATOR OF CBF GENE EXPRESSION3 and CPL1 also interact with the Arg-Ser (RS)-rich splicing factors RS40 and RS41 to promote intron splicing in many stress-related genes under salt stress (Chen et al., 2013; Karlsson et al., 2015). The Arabidopsis MAC is an ortholog of NineTeen Complex/Precursor mRNA processing19 Complex, which is well known for its function in RNA splicing in yeast and mammals (Chanarat and Sträßer, 2013). Intriguingly, studies of MAC subunits such as MAC3A, MAC3B, MAC7, PRL1, and PRL2 revealed that MAC not only functions in pre-mRNA splicing, but it also functions in miRNA biogenesis through an association with Pol II and/or the microprocessor (Zhang et al., 2013, 2014; Jia et al., 2017; Li et al., 2018). PP4R3A is similar to MAC in many respects, such as its association with Pol II and promotion of pri-miRNA processing, as well as the splicing of hundreds of pre-mRNAs. Our analyses showed that MAC and PP4 act on a small number of common introns and larger numbers of different introns to promote their splicing. These complexes both also act in miRNA biogenesis in general. We suspect that the ever-growing list of proteins that affect both miRNA biogenesis and RNA splicing does not necessarily imply that the two processes are linked, but rather, the two processes may involve common factors.
METHODS
Plant Materials and Growth Conditions
All Arabidopsis (Arabidopsis thaliana) plants used in this study are in the Columbia accession background and were grown under long photoperiod conditions (16-h light/8-h dark; white light; light intensity, 120 μmol m−2 s−1) at 22°C. To assess the impact of temperature on the levels of HYL1, SE, and AGO1, 1-week-old seedlings were grown at 22°C and transferred to growth chambers at 16°C, 22°C, or 30°C for 1 more week. The T-DNA insertion mutants pp4r3b-1 (SALK_022912), pp4r3b-2 (SALK_138884), and pp4r3a-2 (SAIL_33_H01; this allele is the same as smek1-2; Su et al., 2017) were obtained from the ABRC; hyl1-2 (Han et al., 2004), nrpb2-3 (Zheng et al., 2009), cdc5-1 (Zhang et al., 2013), and dcl1-20 (Li et al., 2016) were previously described. The HYL1-YFP/pp4r3a-1, pMIR167a:GUS/pp4r3a-1, and pMIR172b:GUS/pp4r3a-1 lines were generated from crosses between pp4r3a-1 and the transgenic lines HYL1-YFP (Fang and Spector, 2007), pMIR167a:GUS (Zhang et al., 2014), and pMIR172b:GUS (Zhang et al., 2013), respectively.
Mutagenesis and Screening
The SUC2:amiR-SUL transgenic lines were previously described (de Felippes et al., 2011). A stable line with a moderate leaf bleaching phenotype and one copy of a T-DNA insertion (designated as amiR-SUL) was used as the parental line for ethyl methanesulfonate mutagenesis. The sup-e33 mutant was isolated based on its weaker leaf bleaching phenotype.
The method used for CRISPR/Cas9-mediated mutagenesis in the present study was reported previously (Yan et al., 2015). Genomic DNA was extracted from individual T1 or T2 transgenic plants, and the fragments flanking target sites were amplified with specific primer pairs and directly sequenced. The sequencing chromatograms were examined for exact patterns that indicate monoallelic or diallelic mutations.
DNA Constructs and Plant Transformation
To obtain the construct for the genetic complementation assay, a ∼9-kb genomic DNA fragment containing the PP4R3A coding region and promoter was amplified from the Col genome and cloned into pENTR/D-TOPO (Invitrogen). The fragment was subsequently integrated into pGWB40 and pEG301, generating pPP4R3A:PP4R3A-YFP and pPP4R3A:PP4R3A-HA, respectively. For the PP4R3A and PP4R3B promoter-GUS constructs, the DNA fragment containing either the PP4R3A or PP4R3B promoter was PCR amplified and cloned into the destination vector pMDC162. For the 35S:PP4R3B-YFP construct, the full-length CDS of PP4R3B without the stop codon was amplified and cloned into the pEG101 vector. CRISPR construction mainly involved two steps: first, the designed 20-bp guide RNA sequence targeting PPX1 (GCCCATCAACTAGTTATGGA) or PPX2 (GACCGTATAACTCTCATTAG) was cloned into the Bsa I-digested small guide RNA (sgRNA) scaffold where the sgRNA is driven by the AtU6-26 promoter, and then the AtU6-26:sgRNA cassette was introduced into the binary vector pYAO(Yan et al., 2015):Cas9 cassette-containing pCambia 1300 where the Cas9 gene is driven by the embryo-specific promoter of the YAOZHE (YAO) gene (Li et al., 2010).
The constructs were transferred into Agrobacterium tumefaciens strain GV3101 and introduced into plants by the floral dip method (Zhang et al., 2006). The primers used are listed in Supplemental Data Set 3.
GUS Staining and Microscopy
Plant tissues from homozygous transgenic lines were immersed in GUS staining buffer containing 50 mM sodium phosphate (pH 7.0), 0.5 mg/mL 5-Bromo-4-chloro-3-indolyl-β-D-glucuronic acid cyclohexylammonium salt (X-gluc), 0.1% Triton X-100, 2 mM potassium ferrocyanide, 2 mM potassium ferricyanide, and 10 mM EDTA and incubated at 37°C in the dark. The tissues were photographed under a stereomicroscope (Leica) after chlorophyll removal in 70% ethanol.
RT-qPCR and Small RNA Gel Blotting
Total RNAs were extracted from Arabidopsis tissues with TRIzol reagent (Medical Research Center) and treated with DNase I (Roche). First-strand complementary DNA was synthesized according to the manufacturer’s instructions (Thermo Fisher Scientific). RT-qPCR was performed in the CFX96 Real-time System (Bio-Rad) using SYBR Green I Supermix (Bio-Rad). UBQ5 served as an internal control. The procedure was previously described in detail (Jia et al., 2017). The primers are listed in Supplemental Data Set 3.
RNA gel blot analysis for small RNA detection was performed as described (Park et al., 2002). Total RNAs were extracted from 12-d-old seedlings. To detect miRNAs and tasiRNAs, 5′-end labeled ([γ-32P] ATP) antisense DNA oligonucleotides were used as probes. Signals were quantified with ImageQuant 5.2 software and normalized to that of small nuclear RNA U6. The sequences of probes are shown in Supplemental Data Set 3.
Construction and Data Analysis of Small RNA-seq and RNA-seq Libraries
To construct the small RNA libraries, 50 μg total RNAs from 12-d-old seedlings were separated on a 15% urea-PAGE gel from which RNAs in the desired size range (10-40 nucleotide) were recovered. Small RNA libraries were constructed using a New England BioLabs (NEB) Next Multiplex Small RNA Library Prep kit (NEB, E7300S) following the manufacturer’s instructions. Before RNA-seq library construction, poly(A)+ RNAs were isolated from 5 μg total RNAs from 12-d-old seedlings using a Magnetic mRNA Isolation kit (NEB, S1550S). RNA-seq libraries were constructed using a NEBNext mRNA Library Prep Reagent Set for Illumina (NEB, E6110). Both small RNA and RNA-seq libraries were pooled and sequenced on the Illumina HiSeq 2500 platform. The data analysis of small RNA seq and RNA-seq libraries was described (Jia et al., 2017). To analyze pri-miRNA levels, 325 pri-miRNAs annotated in Araport11 were interrogated, and those that were represented by reads of FPKM>2 were retained. Some pri-miRNA loci overlapped with protein-coding genes, whereas others did not (Lepe-Soltero et al., 2017); those not overlapping with protein-coding genes were separately analyzed. The 131 miRNA-target pairs validated from previous degradome or rapid-amplification of complementary DNA ends (RACE) experiments (Ma et al., 2018) were included in the analysis of the expression of miRNA target genes. The analysis of splicing defects was performed using Araport11 intron annotation and a previously developed pipeline known as SQUID (https://github.com/Xinglab/SQUID). In brief, the level of intron retention is calculated for each intron as the percentage of intron reads( ).
).
Co-immunoprecipitation and Protein Gel Blot Analysis
Approximately 0.5 g of tissue from 12-d-old F1 plants from a cross between PP4R3A-YFP and RPB1-MYC transgenic lines was ground in liquid nitrogen and homogenized in IP buffer containing 20 mM Tris·HCl (pH 7.5), 150 mM NaCl, 4 mM MgCl2, 75 μM ZnCl2, 0.1% Triton X-100, 1% glycerol, and EDTA-free protease inhibitor mixture (Roche). Cleared protein extracts were immunoprecipitated using anti-c-MYC Affinity Gel (Sigma, E6654) or GFP-Trap (ChromoTek, gtm-10). The bound proteins were washed twice with IP buffer and separated by SDS-PAGE. Protein gel blot analysis was performed using anti-MYC (1:1000, Millipore, 05-724) or GFP (1:1000, Roche, 11814460001) antibodies.
To determine protein levels, total cellular proteins from 12-d-old seedlings were resolved by SDS-PAGE, transferred to nitrocellulose membranes, and detected using antibodies against RPB1 (1:1000, Agrisera, AS111804), RPB2 (1:1000, Abcan, ab10338), HYL1 (1:2000, Agrisera, AS06136), AGO1 (1:2000, Agrisera, AS09527), AGO2 (1:2000, Agrisera, AS132682), and SE (1:1000, Agrisera, AS09532A).
Affinity Purification and Mass Spectrometric Analysis
Approximately 2 g of tissue from 12-d-old PP4R3A-YFP and PP4R3A-HA transgenic T3 plants, or Col plants as a negative control, were ground in liquid nitrogen. The powder was re-suspended in 10 mL of lysis buffer (50 mM Tris [pH 7.6], 150 mM NaCl, 5 mM MgCl2, 1% glycerol, 0.1% Nonidet P-40, 0.5 mM DTT, 1 mM PMSF, and protease inhibitor cocktail) and incubated on a rotator for 10 min at 4°C. The slurry was filtered through four layers of Miracloth and centrifuged at 13,000g for 15 min at 4°C. Before immunoprecipitation, the supernatant was incubated with 100 μL of Dynabeads (Invitrogen, 10002D) for 2 h at 4°C. The pre-cleared supernatant was immunoprecipitated using GFP-Trap (ChromoTek, gtm-10) or anti-HA magnetic beads (88836, Pierce) according to the manufacturer’s instructions. After incubation, the anti-GFP or anti-HA beads were washed three times with lysis buffer. The IP samples were resolved by SDS-PAGE and subjected to mass spectrometry as described (Sleat et al., 2006; Deng et al., 2016).
Extraction of Chromatin-Associated Proteins
Twelve-day-old seedlings were subjected to nuclear-cytoplasmic fractionation as previously described (Wang et al., 2011). The nuclei pellet was re-suspended in low-salt buffer (10 mM Tris-HCl [pH 7.4], 0.2 mM MgCl2, and 1% Triton-X 100) supplemented with protease inhibitor cocktail (Roche), incubated on a rotator at 4°C for 30 min, and centrifuged at 6,500g for 10 min at 4°C. The chromatin pellet was washed twice with 5 volumes of no-salt buffer (3 mM EDTA and 0.2 mM EGTA), re-suspended in 2-5 volumes of 0.2 N HCl, and incubated on a rotator at 4°C for 4 h. The solution was centrifuged at 14,000 rpm for 10 min at 4°C, and the supernatant (consisting of acid-soluble proteins from the chromatin fraction) was collected and neutralized with the same volume of 1M Tris-HCl (pH 8.0). GAPDH (Santa Cruz, sc-47724) and histone H3 (Abcam, ab10799) serve as the cytoplasmic and the nuclear marker, respectively.
ChIP
The ChIP assays of PP4R3A, Pol II, and HYL1 were performed as described previously (Yamaguchi et al., 2014). For ChIP of HYL1, 12-d-old Col and pp4r3a-1 seedlings were processed and anti-HYL1 antibodies (Agrisera, AS06136) were used for immunoprecipitation; for PP4R3A, 12-d-old seedlings harboring PP4R3A:PP4R3A-HA and the negative control (wild type) were processed, and HA-probe (Santa Cruz, sc-7392) was used to pull down the protein–DNA complexes. Transgenic lines harboring isogenic RPB1-MYC in the Col and pp4r3a-1 backgrounds were used for Pol II ChIP. Quantitative PCR was performed using immunoprecipitated DNA samples; relative enrichment was calculated by normalizing the amount of ChIP-ed DNA to the corresponding amount in the input. The primers used are listed in Supplemental Data Set 3.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: PP4R3A (AT3G06670), PP4R3B (AT5G49370, AT5G49380, AT5G49390), PP4R2L (AT5G17070), PPX1 (AT4G26720), PPX2 (AT5G55260), SUL (AT4G18480), SQUAMOSA PROMOTER BINDING PROTEIN-LIKE5 (AT3G15270), PHABULOSA (AT2G34710), PHAVOLUTA (AT1G30490), REV (AT5G60690), MYB33 (AT5G06100), AGO1 (AT1G48410), AUXIN RESPONSE FACTOR8 (AT5G37020), AGO2 (AT1G31280), SE (AT2G27100), DCL1 (AT1G01040), HYL1 (AT1G09700), CDC5 (AT1G09770), RPB1 (AT4G35800), RPB2/NRPB2 (AT4G21710), CPL1 (AT4G21670), HEN1 (AT4G20910), MOS2 (AT1G33520), NOT2A (AT1G07705), NOT2B (AT5G18230), PRL1 (AT4G15900), DDL (AT3G20550), TGH (AT5G23080), CPB20 (AT5G44200), CPB80 (AT2G13540), and HST (AT3G05040). The small RNA- and RNA-seq data were deposited in the Gene Expression Omnibus database under accession number GSE121456.
Supplemental Data
Supplemental Figure 1. Phenotypes of sup-e33 mutant plants.
Supplemental Figure 2. PP4R3A expression in wide type and two pp44r3 mutants, as revealed by RNA-seq.
Supplemental Figure 3. Mutations in PP4R3A lead to shortened roots.
Supplemental Figure 4. Hierarchical clustering analysis showing the degree of similarity among the sRNA-seq libraries.
Supplemental Figure 5. Tissue-specific expression patterns of pPP4R3A:GUS and pPP4R3B:GUS.
Supplemental Figure 6. The 35S:PP4R3B transgene does not rescue the phenotypes of pp4r3a-1.
Supplemental Figure 7. Phenotypes of pp4r3a-1 rescued with YFP- or HA-tagged PP4R3A transgenes.
Supplemental Figure 8. Composition of PP4, and mutants of PP4 catalytic subunit genes.
Supplemental Figure 9. Mutations in PP4R3A do not affect the expression of genes involved in miRNA biogenesis.
Supplemental Figure 10. Accumulation of endogenous miRNAs in microprocessor mutants and in double mutants between sup-e33 and microprocessor mutants.
Supplemental Figure 11. PP4R3A does not associate with the microprocessor subunits HYL1 and SE in vivo, and HYL1 is not required for the occupancy of PP4R3A at MIR promoters.
Supplemental Figure 12. RNA-seq of wild type and two pp4r3a mutants reveals global changes of the expression of miRNA targets and pri-miRNAs in the mutants.
Supplemental Figure 13. RNA-seq reveals changes in gene expression and defects in pre-mRNA splicing in the mutants.
Supplemental Figure 14. Features of retained introns and genes with intron retention in two pp4r3a alleles.
Supplemental Data Set 1. Levels of miRNAs in wild type (Col), pp4r3a-1, and pp4r3-2, as determined by small RNA sequencing.
Supplemental Data Set 2. List of proteins identified by IP-MS.
Supplemental Data Set 3. Sequences of primers and probes used in this study.
Dive Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
We are grateful to Dr. B. Yu (University of Nebraska–Lincoln) for sharing Arabidopsis mutants and transgenic lines. We thank Dr. H. Zheng (Rutgers University) for technical support in mass spectrometry analysis. This project was funded by Guangdong Innovation Research Team Fund (2014ZT05S078), National Science Foundation of China (91740202, 31571332), National Institutes of Health (GM129373) and China Postdoctoral Science Foundation (2017M612717).
AUTHOR CONTRIBUTIONS
S.W., B.M., and X.C. conceived the project; S.W., L.Q., L.L., and Y.Z. designed and performed the experiments; S.L., C.Y., L.G., L.Z., and Y.Q. analyzed the data; S.W. and X.C. wrote the manuscript.
References
- Achkar N.P., et al. (2018). A quick HYL1-dependent reactivation of microRNA production is required for a proper developmental response after extended periods of light deprivation. Dev. Cell 46: 236–247.e6. [DOI] [PubMed] [Google Scholar]
- Ahn C.S., Han J.A., Lee H.S., Lee S., Pai H.S. (2011). The PP2A regulatory subunit Tap46, a component of the TOR signaling pathway, modulates growth and metabolism in plants. Plant Cell 23: 185–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen E., Xie Z., Gustafson A.M., Carrington J.C. (2005). microRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell 121: 207–221. [DOI] [PubMed] [Google Scholar]
- Armenta-Medina A., Lepe-Soltero D., Xiang D., Datla R., Abreu-Goodger C., Gillmor C.S. (2017). Arabidopsis thaliana miRNAs promote embryo pattern formation beginning in the zygote. Dev. Biol. 431: 145–151. [DOI] [PubMed] [Google Scholar]
- Ballarino M., Pagano F., Girardi E., Morlando M., Cacchiarelli D., Marchioni M., Proudfoot N.J., Bozzoni I. (2009). Coupled RNA processing and transcription of intergenic primary microRNAs. Mol. Cell. Biol. 29: 5632–5638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben Chaabane S., Liu R., Chinnusamy V., Kwon Y., Park J.H., Kim S.Y., Zhu J.K., Yang S.W., Lee B.H. (2013). STA1, an Arabidopsis pre-mRNA processing factor 6 homolog, is a new player involved in miRNA biogenesis. Nucleic Acids Res. 41: 1984–1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bielewicz D., Kalak M., Kalyna M., Windels D., Barta A., Vazquez F., Szweykowska-Kulinska Z., Jarmolowski A. (2013). Introns of plant pri-miRNAs enhance miRNA biogenesis. EMBO Rep. 14: 622–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chanarat S., Sträßer K. (2013). Splicing and beyond: The many faces of the Prp19 complex. Biochim. Biophys. Acta 1833: 2126–2134. [DOI] [PubMed] [Google Scholar]
- Chen T., Cui P., Chen H., Ali S., Zhang S., Xiong L. (2013). A KH-domain RNA-binding protein interacts with FIERY2/CTD phosphatase-like 1 and splicing factors and is important for pre-mRNA splicing in Arabidopsis. PLoS Genet. 9: e1003875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T., Cui P., Xiong L. (2015). The RNA-binding protein HOS5 and serine/arginine-rich proteins RS40 and RS41 participate in miRNA biogenesis in Arabidopsis. Nucleic Acids Res. 43: 8283–8298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho S.K., Ben Chaabane S., Shah P., Poulsen C.P., Yang S.W. (2014). COP1 E3 ligase protects HYL1 to retain microRNA biogenesis. Nat. Commun. 5: 5867. [DOI] [PubMed] [Google Scholar]
- Cho S.K., Ryu M.Y., Shah P., Poulsen C.P., Yang S.W. (2016). Post-translational regulation of miRNA pathway components, AGO1 and HYL1, in plants. Mol. Cells 39: 581–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhury D., Xu X., Zhong X., Ahmed F., Zhong J., Liao J., Dykxhoorn D.M., Weinstock D.M., Pfeifer G.P., Lieberman J. (2008). A PP4-phosphatase complex dephosphorylates gamma-H2AX generated during DNA replication. Mol. Cell 31: 33–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen P.T., Philp A., Vázquez-Martin C. (2005). Protein phosphatase 4--from obscurity to vital functions. FEBS Lett. 579: 3278–3286. [DOI] [PubMed] [Google Scholar]
- de Felippes F.F., Ott F., Weigel D. (2011). Comparative analysis of non-autonomous effects of tasiRNAs and miRNAs in Arabidopsis thaliana. Nucleic Acids Res. 39: 2880–2889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng X., Lu T., Wang L., Gu L., Sun J., Kong X., Liu C., Cao X. (2016). Recruitment of the NineTeen Complex to the activated spliceosome requires AtPRMT5. Proc. Natl. Acad. Sci. USA 113: 5447–5452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong Z., Han M.H., Fedoroff N. (2008). The RNA-binding proteins HYL1 and SE promote accurate in vitro processing of pri-miRNA by DCL1. Proc. Natl. Acad. Sci. USA 105: 9970–9975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eamens A.L., Smith N.A., Curtin S.J., Wang M.B., Waterhouse P.M. (2009). The Arabidopsis thaliana double-stranded RNA binding protein DRB1 directs guide strand selection from microRNA duplexes. RNA 15: 2219–2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang Y., Spector D.L. (2007). Identification of nuclear dicing bodies containing proteins for microRNA biogenesis in living Arabidopsis plants. Curr. Biol. 17: 818–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang X., Cui Y., Li Y., Qi Y. (2015). Transcription and processing of primary microRNAs are coupled by Elongator complex in Arabidopsis. Nat. Plants 1: 15075. [DOI] [PubMed] [Google Scholar]
- Fei Q., Xia R., Meyers B.C. (2013). Phased, secondary, small interfering RNAs in posttranscriptional regulatory networks. Plant Cell 25: 2400–2415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gingras A.C., Caballero M., Zarske M., Sanchez A., Hazbun T.R., Fields S., Sonenberg N., Hafen E., Raught B., Aebersold R. (2005). A novel, evolutionarily conserved protein phosphatase complex involved in cisplatin sensitivity. Mol. Cell. Proteomics 4: 1725–1740. [DOI] [PubMed] [Google Scholar]
- Golden T.A., Schauer S.E., Lang J.D., Pien S., Mushegian A.R., Grossniklaus U., Meinke D.W., Ray A. (2002). SHORT INTEGUMENTS1/SUSPENSOR1/CARPEL FACTORY, a Dicer homolog, is a maternal effect gene required for embryo development in Arabidopsis. Plant Physiol. 130: 808–822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grigg S.P., Canales C., Hay A., Tsiantis M. (2005). SERRATE coordinates shoot meristem function and leaf axial patterning in Arabidopsis. Nature 437: 1022–1026. [DOI] [PubMed] [Google Scholar]
- Han M.H., Goud S., Song L., Fedoroff N. (2004). The Arabidopsis double-stranded RNA-binding protein HYL1 plays a role in microRNA-mediated gene regulation. Proc. Natl. Acad. Sci. USA 101: 1093–1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris D.M., Myrick T.L., Rundle S.J. (1999). The Arabidopsis homolog of yeast TAP42 and mammalian alpha4 binds to the catalytic subunit of protein phosphatase 2A and is induced by chilling. Plant Physiol. 121: 609–617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastie C.J., Vázquez-Martin C., Philp A., Stark M.J., Cohen P.T. (2006). The Saccharomyces cerevisiae orthologue of the human protein phosphatase 4 core regulatory subunit R2 confers resistance to the anticancer drug cisplatin. FEBS J. 273: 3322–3334. [DOI] [PubMed] [Google Scholar]
- Jia T., Zhang B., You C., Zhang Y., Zeng L., Li S., Johnson K.C.M., Yu B., Li X., Chen X. (2017). The Arabidopsis MOS4-associated complex promotes microRNA biogenesis and precursor messenger RNA splicing. Plant Cell 29: 2626–2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlsson P., Christie M.D., Seymour D.K., Wang H., Wang X., Hagmann J., Kulcheski F., Manavella P.A. (2015). KH domain protein RCF3 is a tissue-biased regulator of the plant miRNA biogenesis cofactor HYL1. Proc. Natl. Acad. Sci. USA 112: 14096–14101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kataya A.R.A., Creighton M.T., Napitupulu T.P., Sætre C., Heidari B., Ruoff P., Lillo C. (2017). PLATINUM SENSITIVE 2 LIKE impacts growth, root morphology, seed set, and stress responses. PLoS One 12: e0180478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y.J., Zheng B., Yu Y., Won S.Y., Mo B., Chen X. (2011). The role of Mediator in small and long noncoding RNA production in Arabidopsis thaliana. EMBO J. 30: 814–822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komiya R. (2017). Biogenesis of diverse plant phasiRNAs involves an miRNA-trigger and Dicer-processing. J. Plant Res. 130: 17–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurihara Y., Watanabe Y. (2004). Arabidopsis micro-RNA biogenesis through Dicer-like 1 protein functions. Proc. Natl. Acad. Sci. USA 101: 12753–12758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laubinger S., Sachsenberg T., Zeller G., Busch W., Lohmann J.U., Rätsch G., Weigel D. (2008). Dual roles of the nuclear cap-binding complex and SERRATE in pre-mRNA splicing and microRNA processing in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 105: 8795–8800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D.H., Pan Y., Kanner S., Sung P., Borowiec J.A., Chowdhury D. (2010). A PP4 phosphatase complex dephosphorylates RPA2 to facilitate DNA repair via homologous recombination. Nat. Struct. Mol. Biol. 17: 365–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D.H., Goodarzi A.A., Adelmant G.O., Pan Y., Jeggo P.A., Marto J.A., Chowdhury D. (2012). Phosphoproteomic analysis reveals that PP4 dephosphorylates KAP-1 impacting the DNA damage response. EMBO J. 31: 2403–2415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lepe-Soltero D., Armenta-Medina A., Xiang D., Datla R., Gillmor C.S., Abreu-Goodger C. (2017). Annotating and quantifying pri-miRNA transcripts using RNA-Seq data of wild type and serrate-1 globular stage embryos of Arabidopsis thaliana. Data Brief 15: 642–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H.J., Liu N.Y., Shi D.Q., Liu J., Yang W.C. (2010). YAO is a nucleolar WD40-repeat protein critical for embryogenesis and gametogenesis in Arabidopsis. BMC Plant Biol. 10: 169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S., Le B., Ma X., Li S., You C., Yu Y., Zhang B., Liu L., Gao L., Shi T., Zhao Y., Mo B., et al. (2016). Biogenesis of phased siRNAs on membrane-bound polysomes in Arabidopsis. eLife 5: e22750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S., Liu K., Zhou B., Li M., Zhang S., Zeng L., Zhang C., Yu B. (2018). MAC3A and MAC3B, two core subunits of the MOS4-associated complex, positively influence miRNA biogenesis. Plant Cell 30: 481–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lillo C., Kataya A.R., Heidari B., Creighton M.T., Nemie-Feyissa D., Ginbot Z., Jonassen E.M. (2014). Protein phosphatases PP2A, PP4 and PP6: Mediators and regulators in development and responses to environmental cues. Plant Cell Environ. 37: 2631–2648. [DOI] [PubMed] [Google Scholar]
- Liu J., Xu L., Zhong J., Liao J., Li J., Xu X. (2012). Protein phosphatase PP4 is involved in NHEJ-mediated repair of DNA double-strand breaks. Cell Cycle 11: 2643–2649. [DOI] [PubMed] [Google Scholar]
- Ma X., Liu C., Gu L., Mo B., Cao X., Chen X. (2018). TarHunter, a tool for predicting conserved microRNA targets and target mimics in plants. Bioinformatics 34: 1574–1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manavella P.A., Hagmann J., Ott F., Laubinger S., Franz M., Macek B., Weigel D. (2012). Fast-forward genetics identifies plant CPL phosphatases as regulators of miRNA processing factor HYL1. Cell 151: 859–870. [DOI] [PubMed] [Google Scholar]
- Moorhead G.B., De Wever V., Templeton G., Kerk D. (2009). Evolution of protein phosphatases in plants and animals. Biochem. J. 417: 401–409. [DOI] [PubMed] [Google Scholar]
- Morlando M., Ballarino M., Gromak N., Pagano F., Bozzoni I., Proudfoot N.J. (2008). Primary microRNA transcripts are processed co-transcriptionally. Nat. Struct. Mol. Biol. 15: 902–909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park W., Li J., Song R., Messing J., Chen X. (2002). CARPEL FACTORY, a Dicer homolog, and HEN1, a novel protein, act in microRNA metabolism in Arabidopsis thaliana. Curr. Biol. 12: 1484–1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raczynska K.D., Stepien A., Kierzkowski D., Kalak M., Bajczyk M., McNicol J., Simpson C.G., Szweykowska-Kulinska Z., Brown J.W., Jarmolowski A. (2014). The SERRATE protein is involved in alternative splicing in Arabidopsis thaliana. Nucleic Acids Res. 42: 1224–1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghuram B., Sheikh A.H., Rustagi Y., Sinha A.K. (2015). MicroRNA biogenesis factor DRB1 is a phosphorylation target of mitogen activated protein kinase MPK3 in both rice and Arabidopsis. FEBS J. 282: 521–536. [DOI] [PubMed] [Google Scholar]
- Rogers K., Chen X. (2013). Biogenesis, turnover, and mode of action of plant microRNAs. Plant Cell 25: 2383–2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sleat D.E., Zheng H., Qian M., Lobel P. (2006). Identification of sites of mannose 6-phosphorylation on lysosomal proteins. Mol. Cell. Proteomics 5: 686–701. [DOI] [PubMed] [Google Scholar]
- Stepien A., Knop K., Dolata J., Taube M., Bajczyk M., Barciszewska-Pacak M., Pacak A., Jarmolowski A., Szweykowska-Kulinska Z. (2017). Posttranscriptional coordination of splicing and miRNA biogenesis in plants. Wiley Interdiscip. Rev. RNA 8: e1403. [DOI] [PubMed] [Google Scholar]
- Su C., Li Z., Cheng J., Li L., Zhong S., Liu L., Zheng Y., Zheng B. (2017). The protein phosphatase 4 and SMEK1 complex dephosphorylates HYL1 to promote miRNA biogenesis by antagonizing the MAPK cascade in Arabidopsis. Dev. Cell 41: 527–539.e5. [DOI] [PubMed] [Google Scholar]
- Szweykowska-Kulińska Z., Jarmolowski A., Vazquez F. (2013). The crosstalk between plant microRNA biogenesis factors and the spliceosome. Plant Signal. Behav. 8: e26955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Song X., Gu L., Li X., Cao S., Chu C., Cui X., Chen X., Cao X. (2013). NOT2 proteins promote polymerase II-dependent transcription and interact with multiple MicroRNA biogenesis factors in Arabidopsis. Plant Cell 25: 715–727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Ye R., Xin Y., Fang X., Li C., Shi H., Zhou X., Qi Y. (2011). An importin β protein negatively regulates MicroRNA activity in Arabidopsis. Plant Cell 23: 3565–3576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z., Allen E., Fahlgren N., Calamar A., Givan S.A., Carrington J.C. (2005). Expression of Arabidopsis MIRNA genes. Plant Physiol. 138: 2145–2154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi N., Winter C.M., Wu M.F., Kwon C.S., William D.A., Wagner D. (2014). PROTOCOLS: Chromatin immunoprecipitation from Arabidopsis tissues. The Arabidopsis Book 12: e0170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan J., et al. (2017). The SnRK2 kinases modulate miRNA accumulation in Arabidopsis. PLoS Genet. 13: e1006753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan L., Wei S., Wu Y., Hu R., Li H., Yang W., Xie Q. (2015). High efficiency genome editing in Arabidopsis using Yao promoter-driven CRISPR/Cas9 system. Mol. Plant 8: 1820–1823. [DOI] [PubMed] [Google Scholar]
- Yan L., Wei S., Wu Y., Hu R., Li H., Yang W., Xie Q. (2015). High-Efficiency Genome Editing in Arabidopsis Using YAO Promoter-Driven CRISPR/Cas9 System. Mol. Plant 8: 1820–1823. [DOI] [PubMed] [Google Scholar]
- Zhan X., Wang B., Li H., Liu R., Kalia R.K., Zhu J.K., Chinnusamy V. (2012). Arabidopsis proline-rich protein important for development and abiotic stress tolerance is involved in microRNA biogenesis. Proc. Natl. Acad. Sci. USA 109: 18198–18203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S., Xie M., Ren G., Yu B. (2013). CDC5, a DNA binding protein, positively regulates posttranscriptional processing and/or transcription of primary microRNA transcripts. Proc. Natl. Acad. Sci. USA 110: 17588–17593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S., Liu Y., Yu B. (2014). PRL1, an RNA-binding protein, positively regulates the accumulation of miRNAs and siRNAs in Arabidopsis. PLoS Genet. 10: e1004841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Henriques R., Lin S.S., Niu Q.W., Chua N.H. (2006). Agrobacterium-mediated transformation of Arabidopsis thaliana using the floral dip method. Nat. Protoc. 1: 641–646. [DOI] [PubMed] [Google Scholar]
- Zheng B., Wang Z., Li S., Yu B., Liu J.Y., Chen X. (2009). Intergenic transcription by RNA polymerase II coordinates Pol IV and Pol V in siRNA-directed transcriptional gene silencing in Arabidopsis. Genes Dev. 23: 2850–2860. [DOI] [PMC free article] [PubMed] [Google Scholar]