Abstract
We report a facile route for fabricating a new class of nanomimics that overexpress hepatitis B virus (HBV) receptor via a natural biosynthetic procedure against HBV infection. We engineered a nine-transmembrane HBV-specific receptor, human sodium taurocholate co-transporting polypeptide (hNTCP), to naturally immobilize it onto the cellular surface and subsequently trigger the budding of hNTCP-anchoring membrane vesicles (hNTCP-MVs) that favor the HBV virion. We confirmed that hNTCP-MVs could rapidly block HBV infection in cell models. Furthermore, hNTCP-MVs treatment could effectively prevent viral infection, spreading, and replication in a human-liver-chimeric mouse model of HBV infection. Our findings demonstrate the receptor-mediated antiviral effect of hNTCP-MVs to trick HBV and offer novel opportunities for further development of antiviral strategies in nanomedicine.
Keywords: infection inhibition, hepatitis B virus, receptor, nanovesicles, nanomedicine
Graphical Abstract
A facile and bioinspired strategy for fabricating a new class of nanomimics that over-express Hepatitis B Virus (HBV) receptor via a natural biosynthetic procedure against HBV infection.
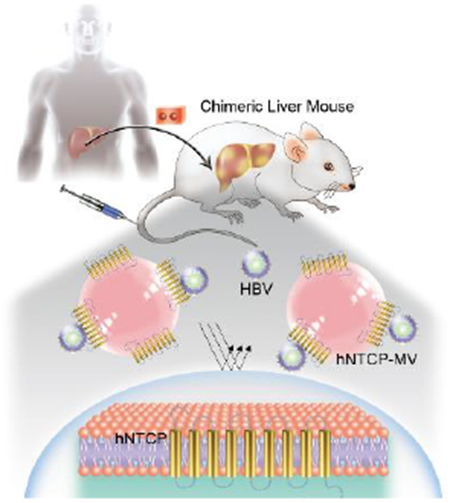
Chronic hepatitis B virus (HBV) infection places patients at high risk of death due to liver cirrhosis and hepatocellular carcinoma, and it remains a global public health issue.[1] Despite the use of preventive vaccines, it is estimated that there are still more than 290 million people who are persistently infected by HBV.[2] More importantly, a functional cure is seldomly achieved in patients with chronic HBV infection by treatment using currently approved anti-HBV drugs (usually <5% per year). Therefore, novel antiviral strategies are urgently needed.[3]
Cellular membrane vesicles (MVs) are directly formed from the natural cell membrane outward budding, they can reflect the antigenic content of the original cells, providing a new strategy for natural protein/vaccine delivery systems. Recently, MVs have been demonstrated as a novel targeted therapy with promising potential for the treatment of various diseases.[4] The receptors anchored on the cell membrane are generally key proteins for virus binding and entry, and they are also known as important therapeutic targets for various infectious diseases.[5] However, most MVs are surface engineered with functional motifs by chemical conjugation methods, which often suffer from poor biological stability and are limited in terms of in vivo delivery logistics and therapeutic activity.[6] It is clear that MVs engineered with highly stable “mimics” of receptor via a genetic modification procedure has the potential to address the limitations of receptor-mediated antiviral treatment with good specificity and safety profiles.[7]
Human sodium taurocholate co-transporting polypeptide (hNTCP) is a nine-transmembrane transporter consisting of 349 amino acids with a molecular mass of 56 kD. hNTCP has been demonstrated as a functional receptor of HBV and features essential high binding affinity for HBVpreS protein.[8] We thus hypothesized that MVs from NTCP-reconstituted cells with hNTCP overexpressed on the cell membrane could be an effective means of bestowing binding specificity towards HBV, which would lead to a receptor-mediated antiviral effect. To test this concept, we developed a facile route for fabricating hNTCP-MVs via a natural biosynthetic procedure, as shown in Figure 1. Specifically, we engineered MVs to display hNTCP receptors and validated the antiviral effect of hNTCP-MVs to trick HBV both in vitro and in vivo. We show that hNTCP-MVs can rapidly block HBV virion binding and reduce the expression of HBV DNA/surface antigen protein in HBV-replicating stable cells. Furthermore, hNTCP-MVs treatment can efficiently prevent HBV viral infection, spreading, and replication in a human-liver-chimeric mouse model of HBV infection. These findings highlight the novel roles of biomimetic binding strategies in which unique capabilities of viral bio-interfacing are employed against HBV infection.
Figure 1.
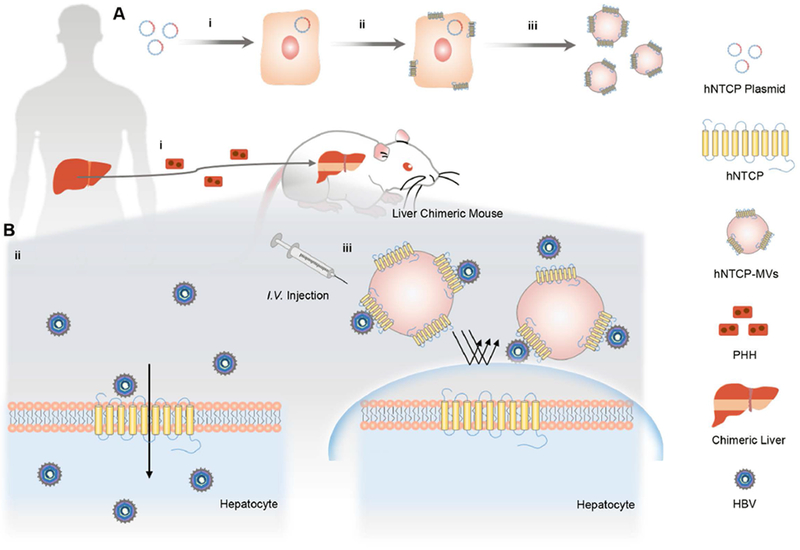
Schematic of the generation and antiviral treatment of hNTCP-MVs. A) Generation of hNTCP-MVs derived from hNTCP-overexpressing cells. i, ii) Engineering of hNTCP-overexpressing cell line stably expressing HBV receptor (hNTCP) on the cell membranes. iii) Harvesting of the cell membrane expressing hNTCP and preparation of hNTCP-MVs. B) Antiviral treatment of hNTCP-MVs. i) Construction of liver chimeric mouse model (Hu-FRGS) through transplantation of human primary hepatocytes (PHH), the natural host cells of HBV, to FRGS mouse. ii) PHH of Hu-FRGS infected with HBV in vivo. iii) The hNTCP-MVs bind to the dissociative HBV virion and prevent viral entry into PHH in Hu-FRGS.
hNTCP-overexpressing cell line[9] was generated by a well-characterized sleeping-beauty transposon-based system with the co-expression of red fluorescent protein mCherry and puromycin-resistant selective marker by plasmid transfection as we previously described (Figure 2A).[10] Immunofluorescence (IF) staining showed the cultures were positive for hNTCP, whereas the parental cells were not (Figure 2B). To evaluate their binding activity to the HBV virion, hNTCP-overexpressing cells were incubated with 0.2 μM FITC labeled HBV preS-peptides (HBVpreS/2-48myr-FITC peptide) in dark for 4 h and then washed with PBS.[11] The parental cells were used as a control (Figure 2C). Over 95% of the mCherry-positive hNTCP-overexpressing cells were bound to HBVpreS/2-48myr-FITC after 4 h of incubation, whereas the control cells showed negligible binding signal, manifesting the high biological activity of hNTCP protein expressed on the membrane of hNTCP-overexpressing cells (Figure 2D).
Figure 2.
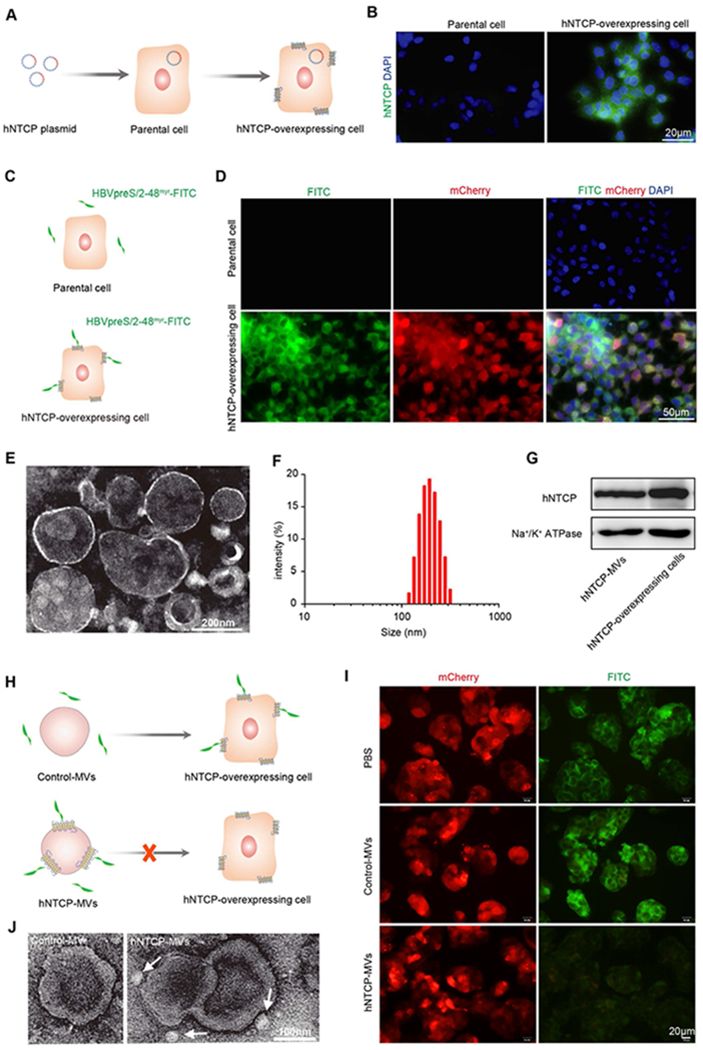
Generation and characterization of hNTCP-MVs. A) Establishment of hNTCP-overexpressing cell line stably expressing hNTCP on cell membranes. B) IF staining for hNTCP (green) expression in parental cells and hNTCP-overexpressing cells. Cell nuclei were stained with DAPI (blue). Scale bar: 20μm. C) Scheme of HBVpreS/2-48myr-FITC peptide binding. D) Fluorescence images of HepG2 and hNTCP-overexpressing cell lines at 4 hours after HBVpreS/2-48myr-FITC peptide (green) binding. Cell nuclei were stained with DAPI (blue). Scale bar: 50 μm. E) TEM images for hNTCP-MVs negatively stained with uranyl acetate. Scale bar: 200 nm. F) Size intensity curve of hNTCP-MVs measured by DLS. G) Western blot assay exhibited the expression of hNTCP on the hNTCP-MVs and whole cell lysate of hNTCP-overexpressing cell line. Na+/K+ ATPase was used as a loading control. H) Schematic of hNTCP-MVs competitively blocking HBVpreS/2-48myr-FITC peptide binding to hNTCP-overexpressing cells. I) Fluorescence images of PBS, hNTCP-MVs, and Control-MVs blocking HBVpreS/2-48myr-FITC binding to hNTCP-overexpressing cells. Scale bar: 20 μm. J) TEM images of HBV-hNTCP-MVs complexes negatively stained with uranyl acetate. Scale bar: 100 nm. HBV virion was indicated by white arrow.
The MVs derived from hNTCP-overexpressing cells (hNTCP-MVs) were generated using a previously reported procedure.[12] Briefly, cell membrane was collected by a previous reported freeze-thaw process. After sonication and centrifugation, major MVs were obtained. The vesicular structure of hNTCP-MVs was confirmed by transmission electron microscopy (TEM) (Figure 2E), and the vesicle size was approximately 200 nm, which matched the hydrodynamic diameter measured by dynamic light scattering (Figure 2F). The hNTCP-MVs maintained a stable size intensity within 10 days at 4 °C and after cryopreservation at −80 °C (Figure S1). Western blotting analysis demonstrated the presence of hNTCP on hNTCP-MVs (Figure 2G). To evaluate the biological activity, hNTCP-MVs and Control-MVs were used to competitively bind to the HBV virion and block virus-host binding. hNTCP-MVs showed significantly higher binding activity to HBVpreS/2-48myr-FITC peptide than the Control-MVs, which indicated the over-expression of hNTCP receptor mainly mediated the binding between the virion and hNTCP-MVs (Figure S2). Secondly, hNTCP-overexpressing cells in vitro cultured in 24-well plate were incubated with an excess of MVs (0.4 μg/μL) (Figure 2H). Fluorescence images showed over 90% of the binding between hNTCP-overexpressing cells and HBVpreS/2-48myr-FITC peptide was blocked by hNTCP-MVs, whereas the incubation of Control-MVs had no effect (Figure 2I). Notably, TEM images displayed the pattern of the HBV-hNTCP-MVs binding complex and indicated a size of 200-300 nm (Figure 2J and Figure S3). These findings demonstrate that hNTCP-MVs engineered with “mimics” of the multiple transmembrane HBV receptor are able to recognize and bind to the HBV virion with high specificity.
To investigate the potential of hNTCP-MVs blocking the binding and entry into host cells of the HBV virion, hNTCP-overexpressing cells were incubated with hNTCP-MVs at various concentrations immediately after HBV infection (Figure 3A). We detected and analyzed HBV DNA and surface antigen (HBsAg) [13] to measure the viral load in this study. In contrast to the untreated controls (with an equal volume of PBS added), treatment with a 0.1 μg/μL dose of hNTCP-MVs significantly reduced HBsAg by 75.7 ± 2.5% (p < 0.001) and HBV DNA by 72.8 ± 1.6% (p < 0.001) in the supernatant 3 days post infection (3 dpi). The HBV suppression effects of hNTCP-MVs were dose-dependent and were prolonged to 6 dpi (Figure 3B, 3C, red column). In contrast to the hNTCP-MVs treated groups, the cells treated with equal doses of Control-MVs showed no significant HBV suppression effects (Figure 3B, 3C, blue column). Moreover, hNTCP-MVs treated cells, Control-MVs treated cells and untreated cells with HBV infection were detected by IF staining for intracellular expression of HBsAg. Notably, hNTCP-MVs treatment significant reduced intracellular HBsAg expression at 6 dpi (Figure 3D). The entecavir (ETV, a clinical nucleotide analogues used in the treatment of HBV infection) control group showed no HBsAg suppression but a significant and dose-dependent suppression of HBV DNA at 3 dpi (Figure S4A and S4B). These results imply that ETV can only suppress HBV repication, but has no effect on blocking HBV virion binding.
Figure 3.
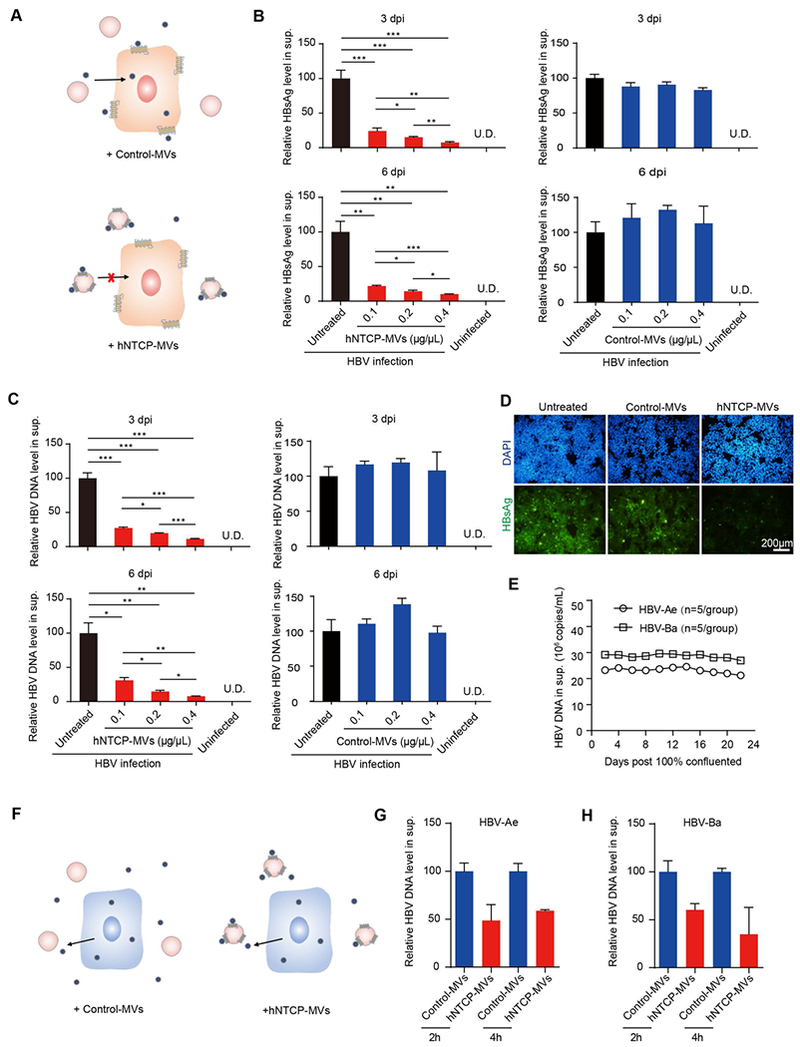
hNTCP-MVs prevent HBV infection and reduce viral load in vitro. A) Schematic of HBV-infected hNTCP-overexpressing cells that received hNTCP-MVs treatment. B) Relative HBsAg and C) HBV DNA levels in supernatant of uninfected hNTCP-overexpressing cells, untreated, hNTCP-MVs-treated and Control-MVs-treated hNTCP-overexpressing cells with HBV infection at 3 and 6 dpi (n = 3/group). D) IF staining for HBsAg (green) expression of untreated, hNTCP-MVs-treated and Control-MVs-treated hNTCP-overexpressing cells at 6 dpi. Scale bar: 200 μm. E) HBV DNA levels in cell culture supernatant of HBV-Ae/Ba cell lines from 0 to 24 days after 100% confluence (n=5/group). F) Schematic of HBV-Ae/Ba cells that received hNTCP-MVs treatment. G, H) Relative HBV DNA levels in supernatant of control and HBV-Ae/Ba cells treated with 0.4 μg/μL of hNTCP-MVs (n = 3/group).
Clinically, suppressing serum HBV DNA in patients with sustained viremia is critical to achieving a functional cure.[3] Thus, we investigated whether hNTCP-MVs could suppress HBV DNA in a cell model with stable HBV DNA expression. Generally, HBV-replicating stable cell lines were used to model host cells with a sustained infection with HBV.[10] In this study, HBV-Ae/Ba stable cells (genotype A/B with replication-competent HBV genome stably integrated (Figure 3E) )were incubated with hNTCP-MVs to evaluate the antiviral effect of hNTCP-MVs (Figure 3F). In contrast to the treatment with the Control-MVs, treatment with hNTCP-MVs rapidly reduced the HBV DNA level in the supernatant by approximately 50% after incubation in both cell lines (Figure 3G, 3H). However, ETV showed no suppression of HBV DNA in HBV-Ae/Ba stable cells after treatment (Figure S4C). To rule out the possibility that the decreased HBV expression was caused by nonspecific cytotoxicity, we performed CCK8 assay with those MVs. The CCK8 results showed over 90% cell viability, which indicated a very low cytotoxicity of hNTCP-MVs (Figure S5). Taken together, these in vitro results suggest the antiviral effect of hNTCP-MVs via efficient and competitive binding to the dissociative HBV virion and blocking virus-host binding.
Next, we evaluated the antiviral effect of hNTCP-MVs in vivo in human liver chimeric mice (Hu-FRGS), a classic mouse model of HBV infection. Firstly, to obtain Hu-FRGS mice modeling HBV-induced viremia in human patients, immunodeficient FRGS mice were engrafted with primary human hepatocytes by splenic injection as we previously described.[14] Hu-FRGS mice received a caudal vein injection of 500 μL of hNTCP-MVs suspension (1 μg/μL) immediately post infection (Figure 4A). All the Hu-FRGS mice retained serum human albumin (hALB) levels of 1-3 mg/mL (Figure S6A), which indicated a considerable and stable human liver chimerism supporting HBV infection. In contrast to the Control-MVs treatment, the hNTCP-MVs treatment reduced serum HBsAg and HBV DNA levels by over 90% at 20 dpi. This indicated that hNTCP-MVs can effectively prevent HBV infection and the progression of viremia (Figure 4B, 4C). Importantly, immunohistochemistry (IHC) staining showed that hNTCP-MVs treatment prevented the widespread expression of HBsAg and HBV core antigen (HBcAg) in chimeric human liver lobes (Figure 4D). In addition, the treated mice showed normal physiological characteristics and liver function, suggesting the safety of the hNTCP-MVs regimen (Figure S6B and Figure S7). As expected, the ETV control group showed no effect on prevention of HBV spreading in chimeric human liver lobes (Figure S8D). Over all, these results strongly demonstrate that hNTCP-MVs treatment prolonged the suppression effect of HBV infection, spreading, and replication in vivo. The curative therapeutic effect also indicated promising potential for future clinical translation, upon further optimization and investigation in large animal models.
Figure 4.
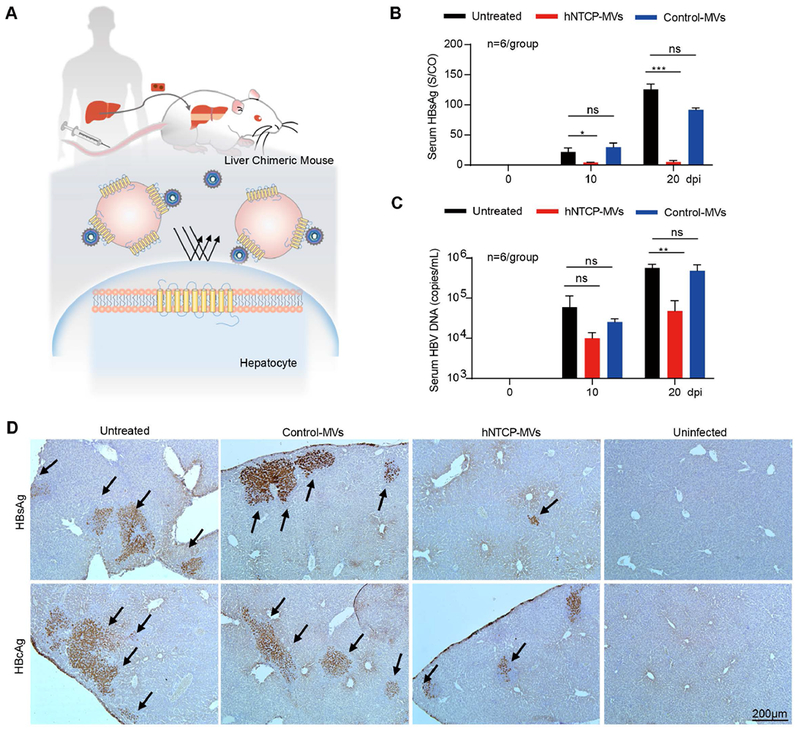
hNTCP-MVs prevent HBV infection in liver humanized mouse model. A) Schematic design of HBV-infected Hu-FRGS mice that received hNTCP-MVs treatment. B) Serum HBsAg and C) HBV DNA levels of untreated, Control-MVs-treated, and hNTCP-MVs-treated Hu-FRGS mice with HBV infection from 0 to 20 dpi (n = 6/group). D) IHC staining for HBsAg and HBcAg expression in chimeric human liver lobes of untreated, Control-MVs-treated, and hNTCP MVs-treated Hu-FRGS mice with HBV infection. Hu-FRGS mice without HBV infection were used as a control. Scale bar: 200 μm.
In summary, we successfully developed a new class of bioengineered nanodecoy anchoring natural HBV receptor (hNTCP) for use against HBV infection. We confirmed that the nanomimics are able to recognize and bind to the dissociative HBV virion with high specificity and sensitivity. The results show promise for inhibiting viral-host cell attack, suppressing viral load, and preventing viral spreading both in vitro and in vivo by mimicking a natural HBV infection process. Compared to soluble receptor proteins, MVs bestow natural properties of the source cell membrane in a straightforward manner, including unique virus bio-interfacing capabilities and lipid-based structures. The laterally mobile, two-dimensional lipid-bilayer environment and specific molecular compositions modulate the conformational equilibria of receptors and coordinate the bioactivity of membrane constituents. [15] Furthermore, the approach avoids the potential risk of inhibitors and antagonists, such as CsA, which might subvert the normal cellular functions of receptor proteins by inactivation of host receptors. [16] NTCP also plays an important role in the enterohepatic circulation of bile salts. Prolonged inactivation of hNTCP might lead to a harmful high level of serum conjugated bile salts.[17] Most importantly, to target different kinds of pathogens, the receptor can be changed, modified, and even further co-expressed by genetic engineering methods. Thus, the flexibility of bioinspired nanotechnology could potentially allow the prevention and treatment of multiple infectious diseases in future clinical translations.
Supplementary Material
Acknowledgments
This work was supported by the Major State Basic Research Development Program of China (2017YFA0205201, 2014CB744503,and 2013CB733802), the National Natural Science Foundation of China (81422023, 81371596, 81672023, U1705281, and U1505221), The National Scientific and Technological Major Project (2018ZX100302), the Fundamental Research Funds for the Central Universities (20720160065 and 20720150141), and the Program for New Century Excellent Talents in University, China (NCET-13-0502). The authors also thank Dr. Qingbing Zheng, Dr. Xiaoyong Wang and Dr. Qingliang Zhao for technical surpport. All animal experiments were approved by the Animal Management and Ethics Committee of Xiamen University.
Footnotes
Supporting information for this article is given via a link at the end of the document.
Contributor Information
Dr. Xuan Liu, State Key Laboratory of Molecular Vaccinology and Molecular Diagnostics & Center for Molecular Imaging and Translational Medicine, School of Public Health, Xiamen University, 361102, China
Dr. Lunzhi Yuan, State Key Laboratory of Molecular Vaccinology and Molecular Diagnostics & National Institute of Diagnostics and Vaccine Development in Infectious Disease, School of Public Health and School of Life Science, Xiamen University, 361102, China
Liang Zhang, State Key Laboratory of Molecular Vaccinology and Molecular Diagnostics & National Institute of Diagnostics and Vaccine Development in Infectious Disease, School of Public Health and School of Life Science, Xiamen University, 361102, China.
Yalin Mu, State Key Laboratory of Molecular Vaccinology and Molecular Diagnostics & Center for Molecular Imaging and Translational Medicine, School of Public Health, Xiamen University, 361102, China.
Xiaoling Li, State Key Laboratory of Molecular Vaccinology and Molecular Diagnostics & National Institute of Diagnostics and Vaccine Development in Infectious Disease, School of Public Health and School of Life Science, Xiamen University, 361102, China.
Dr. Chao Liu, State Key Laboratory of Molecular Vaccinology and Molecular Diagnostics & Center for Molecular Imaging and Translational Medicine, School of Public Health, Xiamen University, 361102, China
Dr. Peng Lv, State Key Laboratory of Molecular Vaccinology and Molecular Diagnostics & Center for Molecular Imaging and Translational Medicine, School of Public Health, Xiamen University, 361102, China
Dr. Yali Zhang, State Key Laboratory of Molecular Vaccinology and Molecular Diagnostics & National Institute of Diagnostics and Vaccine Development in Infectious Disease, School of Public Health and School of Life Science, Xiamen University, 361102, China
Prof. Tong Cheng, State Key Laboratory of Molecular Vaccinology and Molecular Diagnostics & National Institute of Diagnostics and Vaccine Development in Infectious Disease, School of Public Health and School of Life Science, Xiamen University, 361102, China
Prof. Quan Yuan, State Key Laboratory of Molecular Vaccinology and Molecular Diagnostics & National Institute of Diagnostics and Vaccine Development in Infectious Disease, School of Public Health and School of Life Science, Xiamen University, 361102, China.
Prof. Ningshao Xia, State Key Laboratory of Molecular Vaccinology and Molecular Diagnostics & National Institute of Diagnostics and Vaccine Development in Infectious Disease, School of Public Health and School of Life Science, Xiamen University, 361102, China
Dr. Xiaoyuan Chen, Laboratory of Molecular Imaging and Nanomedicine, National Institute of Biomedical Imaging and Bioengineering (NIBIB), National Institutes of Health (NIH), Bethesda, MD 20892, USA
Prof. Gang Liu, State Key Laboratory of Molecular Vaccinology and Molecular Diagnostics & Center for Molecular Imaging and Translational Medicine, School of Public Health, Xiamen University, 361102, China; State Key Laboratory of Cellular Stress Biology, Innovation Center for Cell Biology, Xiamen University, 361102, China.
References
- [1].a) Trepo C, Chan HL, Lok A, Lancet 2014, 384, 2053–2063 [DOI] [PubMed] [Google Scholar]; b) Dienstag JL, N. Engl. J. Med 2008, 359, 1486–1500. [DOI] [PubMed] [Google Scholar]
- [2].a) C. Polaris Observatory, Lancet Gastroenterol. Hepatol. 2018, 3, 383–40329599078 [Google Scholar]; b) Loomba R, Liang TJ, Gastroenterology 2017, 152, 1297–1309 [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Nayagam S, Thursz M, Sicuri E, Conteh L, Wiktor S, Low-Beer D, Hallett TB, Lancet Infect. Dis. 2016, 16, 1399–1408. [DOI] [PubMed] [Google Scholar]
- [3].Durantel D, Zoulim F, J. Hepatol 2016, 64, S117–S131. [DOI] [PubMed] [Google Scholar]
- [4].a) Hu CM, Fang RH, Wang KC, Luk BT, Thamphiwatana S, Dehaini D, Nguyen P, Angsantikul P, Wen CH, Kroll AV, Carpenter C, Ramesh M, Qu V, Patel SH, Zhu J, Shi W, Hofman FM, Chen TC, Gao W, Zhang K, Chien S, Zhang L, Nature 2015, 526, 118–121 [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Farokhzad OC, Nature 2015, 526, 47–48 [DOI] [PubMed] [Google Scholar]; c) Hu Q, Sun W, Qian C, Wang C, Bomba HN, Gu Z, Adv. Mater 2015, 27, 7043–7050 [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Zhang X, Wang C, Wang J, Hu Q, Langworthy B, Ye Y, Sun W, Lin J, Wang T, Fine J, Cheng H, Dotti G, Huang P, Gu Z, Adv. Mater 2018, 30, e1707112. [DOI] [PubMed] [Google Scholar]
- [5].a) Marsh M, Helenius A, Cell 2006, 124, 729–740 [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Glebe D, Urban S, World J. Gastroenterol. 2007, 13, 22–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].a) Zhang P, Chen Y, Zeng Y, Shen C, Li R, Guo Z, Li S, Zheng Q, Chu C, Wang Z, Zheng Z, Tian R, Ge S, Zhang X, Xia NS, Liu G, Chen X, Proc. Natl. Acad. Sci. U. S. A 2015, 112, E6129–6138 [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Zhang P, Liu G, Chen X, Nano today 2017, 13, 7–9 [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Zhang P, Zhang L, Qin Z, Hua S, Guo Z, Chu C, Lin H, Zhang Y, Li W, Zhang X, Chen X, Liu G, Adv. Mater 2018, 30, e1705350. [DOI] [PubMed] [Google Scholar]
- [7].a) Lauster D, Glanz M, Bardua M, Ludwig K, Hellmund M, Hoffmann U, Hamann A, Bottcher C, Haag R, Hackenberger CPR, Herrmann A, Angew. Chem. Int. Ed 2017, 56, 5931–5936 [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Cagno V, Andreozzi P, D’Alicarnasso M, Jacob Silva P, Mueller M, Galloux M, Le Goffic R, Jones ST, Vallino M, Hodek J, Weber J, Sen S, Janecek ER, Bekdemir A, Sanavio B, Martinelli C, Donalisio M, Rameix Welti MA, Eleouet JF, Han Y, Kaiser L, Vukovic L, Tapparel C, Kral P, Krol S, Lembo D, Stellacci F, Nat. Mater 2018, 17, 195–203 [DOI] [PubMed] [Google Scholar]; c) Najer A, Wu D, Bieri A, Brand F, Palivan CG, Beck HP, Meier W, ACS nano 2014, 8, 12560–12571. [DOI] [PubMed] [Google Scholar]
- [8].Yan H, Zhong G, Xu G, He W, Jing Z, Gao Z, Huang Y, Qi Y, Peng B, Wang H, Fu L, Song M, Chen P, Gao W, Ren B, Sun Y, Cai T, Feng X, Sui J, Li W, eLife 2012, 1, e00049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Sun Y, Qi Y, Peng B, Li W, Methods Mol. Biol. 2017, 1540, 1–14. [DOI] [PubMed] [Google Scholar]
- [10].Wu Y, Zhang TY, Fang LL, Chen ZX, Song LW, Cao JL, Yang L, Yuan Q, Xia NS, Virol J. Methods 2016, 234, 96–100. [DOI] [PubMed] [Google Scholar]
- [11].Meier A, Mehrle S, Weiss TS, Mier W, Urban S, Hepatology 2013, 58, 31–42. [DOI] [PubMed] [Google Scholar]
- [12].Hu CM, Zhang L, Aryal S, Cheung C, Fang RH, Zhang L, Proc. Natl. Acad. Sci. U. S. A 2011, 108, 10980–10985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Yuan L, Liu X, Zhang L, Li X, Zhang Y, Wu K, Chen Y, Cao J, Hou W, Zhang J, Zhu H, Yuan Q, Tang Q, Cheng T, Xia N, Front. Microbiol 2018, 9, 908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Zhang TY, Yuan Q, Zhao JH, Zhang YL, Yuan LZ, Lan Y, Lo YC, Sun CP, Wu CR, Zhang JF, Zhang Y, Cao JL, Guo XR, Liu X, Mo XB, Luo WX, Cheng T, Chen YX, Tao MH, Shih JW, Zhao QJ, Zhang J, Chen PJ, Yuan YA, Xia NS, Gut 2016, 65, 658–671. [DOI] [PubMed] [Google Scholar]
- [15].Lingwood D, Simons K, Science 2010, 327, 46–50. [DOI] [PubMed] [Google Scholar]
- [16].Watashi K, Sluder A, Daito T, Matsunaga S, Ryo A, Nagamori S, Iwamoto M, Nakajima S, Tsukuda S, Borroto-Esoda K, Sugiyama M, Tanaka Y, Kanai Y, Kusuhara H, Mizokami M, Wakita T, Hepatology 2014, 59, 1726–1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Nkongolo S, Ni Y, Lempp FA, Kaufman C, Lindner T, Esser-Nobis K, Lohmann V, Mier W, Mehrle S, Urban S, J. Hepatol 2014, 60, 723–731. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


