Abstract
PURPOSE:
Estrogen receptor-alpha (ER) is a therapeutic target of ER positive (ER+) breast cancers. Although ER signaling is complex, many mediators of this pathway have been identified. Specifically, phosphorylation of ER at serine 118 affects responses to estrogen and therapeutic ligands and has been correlated with clinical outcomes in ER+ breast cancer patients. We hypothesized that a newly described germline variant (S118P) at this residue would drive cellular changes consistent with breast cancer development and/or hormone resistance.
METHODS:
Isogenic human breast epithelial cell line models harboring ER S118P were developed via genome editing and characterized to determine the functional effects of this variant. We also examined the frequency of ER S118P in a case control study (N=536) of women with and without breast cancer with a familial risk.
RESULTS:
In heterozygous knock-in models, the S118P variant demonstrated no significant change in proliferation, migration, MAP Kinase pathway signaling, or response to the endocrine therapies tamoxifen and fulvestrant. Further, there was no difference in the prevalence of S118P between women with and without cancer relative to population registry databases.
CONCLUSIONS:
This study suggests that the ER S118P variant does not affect risk for breast cancer or hormone therapy resistance. Germline screening and modification of treatments for patients harboring this variant are likely not warranted.
Keywords: Breast cancer, ESR1, endocrine resistance
Introduction
In the United States of America, breast cancer is the most commonly diagnosed cancer among women with over 250,000 new cases diagnosed each year, and is the second leading cause of cancer death. Approximately 70% of these cancers express the estrogen receptor-alpha protein (ER) and are dependent on ER signaling for tumor growth and maintenance. ER is a steroid hormone receptor which, upon binding to estrogen, dimerizes to become an activated transcription factor. Activated ER can affect the expression of many downstream target genes and influence various aspects of the cell’s behavior, including proliferation. Additionally, ER participates in crosstalk with several other major signaling pathways in the cell, such as the MAP Kinase and PI3 Kinase pathways [1, 2]. A number of coactivators and corepressors can complex with ER to change its cellular location, responsiveness to ligand, and transcriptional activity [3, 4]. ER harbors a number of phosphorylation sites which are mediators of pathway interactions and transcriptional activities [5].
Due to ER’s functional role in the majority of breast cancers, research has led to targeted therapies against the estrogen signaling pathway. These therapies include selective ER modulators (SERMs; e.g. tamoxifen) and selective ER downregulators (SERDs; e.g. fulvestrant) both of which reduce the activity of ER in breast tissue. Aromatase inhibitors (AIs) constitute another class of endocrine therapies which decrease production of estrogen in non-ovarian tissues and thus are effective against ER+ disease in postmenopausal women or premenopausal women with ovarian suppression. However, ER-targeted hormone therapies are limited by a significant rate of drug resistance, both de novo and acquired [6]. Due to the importance of ER-targeted therapies for patient care, it is crucial to identify patients whose disease may be resistant to this class of drugs. While various studies have described resistance mechanisms [7], including mutations in the ER ligand binding domain (LBD) [8–12], other molecular mediators of resistance are unknown.
Review of next generation sequencing data from metastatic patient samples from the Johns Hopkins Molecular Tumor Board and other studies revealed several patients with an unusual ESR1 variant, rs200075329 (C/T) [13–15]. This single-nucleotide substitution results in a non-synonymous substitution of serine to proline at residue 118 of ER (ER S118P), in the region of the DNA-binding and transactivation function 1 domain of the protein. In silico analysis of the variant revealed conflicting and equivocal results, though only the PROVEAN program identified the variant as deleterious [16]. Serine 118 is an important phosphorylation site, affecting ER activity as well as recruitment of coregulators to ER at estrogen response elements [4]. S118 phosphorylation has been well-characterized and is mediated by several kinases including MAPK, mTOR, and CDK2 [17, 18]. Therefore, in addition to the potential change in protein structure resulting from this substitution, it also constitutes a potential loss of crucial regulation of ER signaling. Furthermore, phosphorylation at serine 118 has been shown to correlate with tamoxifen resistance [19]. Multiple laboratory studies have shown profound differences in ER function upon mutation of the S118 residue to an alanine or a glutamic acid, precluding or mimicking phosphorylation, respectively [4, 7, 20]. Due to the importance of the S118 residue in ER function, we hypothesized that S118P may be involved in ER+ breast cancer pathogenesis and/or drug resistance. In this study, we created genetically modified breast epithelial cell line models containing the S118P variant to determine the cellular phenotypes associated with this alteration. In addition, we queried germline DNA samples from women with a familial breast cancer risk to evaluate the allele frequency of ER S118P in the context of breast cancer incidence. These data were compared to publicly available general population data to assess enrichment of the variant in breast cancer.
Results
Isogenic targeting and overexpression of ER S118P in human breast epithelial cells
To study the functional consequences of ER S118P in human breast cells, three cell line models were developed. Isogenic knock-ins within the MCF7 and T47D human breast cancer cell line backgrounds were developed using AAV-mediated gene-targeting. The widely used MCF7 and T47D cell lines were each derived from metastatic breast adenocarcinomas and express endogenous ER. The S118P variant was incorporated into the host genome via adeno-associated viral gene targeting as previously described [21] producing targeted cells heterozygous for ER S118P (Figure 1A, B). These cell lines were designated MERSP (MCF7 ER S118P) and TERSP (T47D ER S118P) and two individually isolated clones were derived for each cell line. We also developed a “targeted wild-type” clone from each parental cell line, which underwent the same viral infection and subsequent isolation steps as the variant clones but does not carry the variant of interest. These were designated MWT and TWT, for MCF7 targeted wild-type and T47D targeted wild-type, respectively.
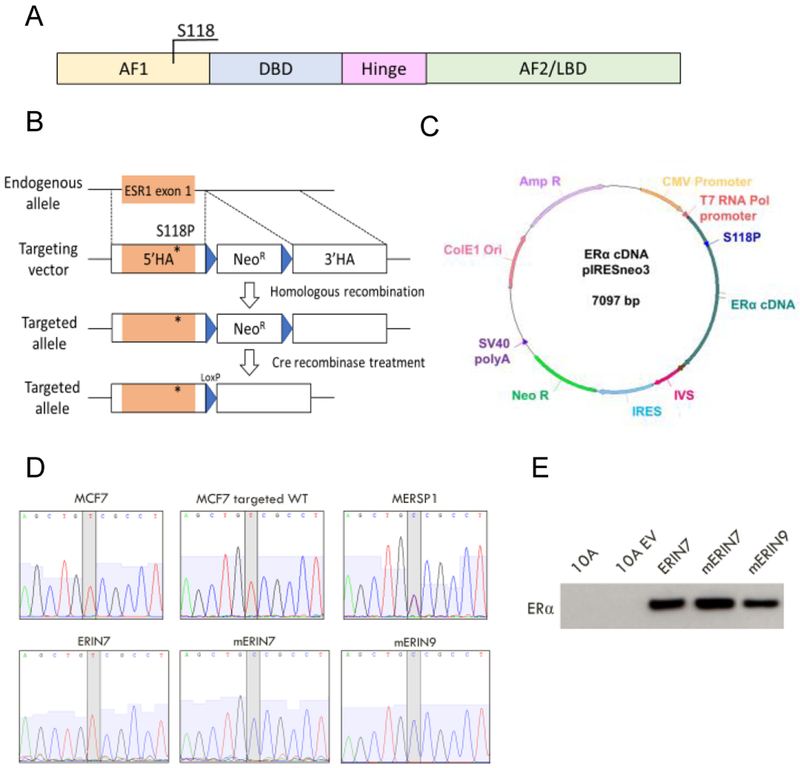
Figure 1: Generation of isogenic cell line models containing ER S118P.
A) Domains of the estrogen receptor-alpha protein and location of the serine 118 residue. AF1=Activation function 1; DBD=DNA-binding domain; AF2=Activation function 2; LBD=Ligand-binding domain. B) Recombinant AAV targeting strategy for knock-in of the variant S118P in ESR1 exon 1 in the ER-expressing cell lines MCF7 and T47D. 5’HA=5’ homology arm; 3’HA=3’ homology arm. C) pIRESneo3 plasmid containing mutated ER cDNA transcript for stable expression of variant ER in the non-ER-expressing cell line MCF10A. D) Sanger sequencing of complementary DNA from the entire panel of cell lines confirms expected heterozygous single base pair change in exon 1 of ESR1. E) Immunoblot analysis of exogenous ER expression for the MCF10A cell panel
In addition, the non-tumorigenic ER negative breast epithelial cell line, MCF10A, was used to develop an overexpression model of the ER S118P variant to evaluate homozygous ER S118P activity (Figure 1C). Our lab has previously established an overexpression model of wild-type ER in the MCF10A line, designated ERIN (ER in nontumorigenic) [22]. For this study we transfected in MCF10A an isogenic plasmid containing a copy of ER S118P cDNA to create two mERIN (mutant ER in nontumorigenic) cell line clones.
To confirm integration and stable expression of the variant protein in our isogenic knock-in models, we performed DNA and cDNA analysis by droplet digital PCR and Sanger sequencing (Figure 1D, Supp. Figure S1). All MERSP and TERSP clones demonstrated integration of the variant within the endogenous locus of a single allele. In the mERIN overexpression model, we confirmed high levels of expression of the variant mRNA and protein which were comparable to wild-type ER levels in the ERIN cell lines (Figure 1E).
Cells expressing ER S118P do not exhibit differential growth from parental controls
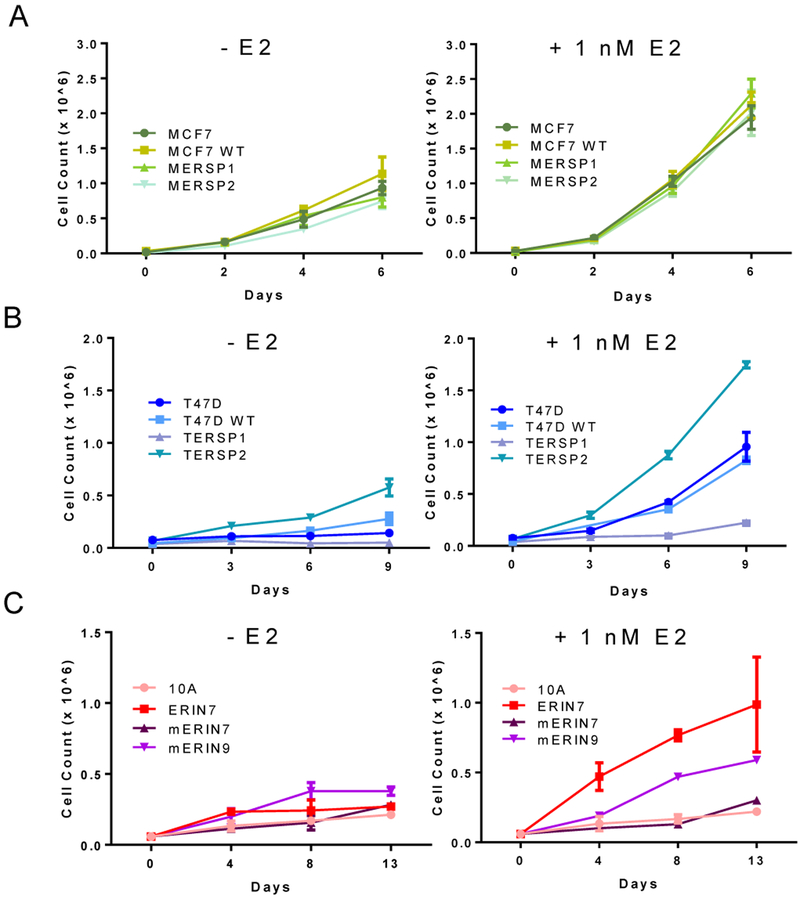
To determine any potential growth advantage of the ER S118P variant and varying responses to estrogen, one and two week cell proliferation assays were carried out in the presence and absence of estrogen. Among the MCF7 cell line panel, both MERSP1 and MERSP2 grew similarly to their parental and targeted wild-type counterparts, both in the presence of 1 nM 17-beta estradiol (E2) and vehicle control (Figure 2A). Interestingly, within the T47D cell line panel, the TERSP2 clone had a higher rate of proliferation than the parental and targeted wild-type cell lines, while the TERSP1 clone had a slower rate. Although both clones respond to 1 nM E2, the difference in their baseline growth rates was notable even in the presence of E2 (Figure 2B). This discrepancy in growth rates may be due to clonal variations among the T47D parental cell line population.
Figure 2: Proliferation of ER S118P cell lines is variable.
A) MCF7- and B) T47D-derived cell lines were seeded in 12 well plates in media supplemented with 5% serum (left) or 5% serum plus 1 nM estradiol (right). C) MCF10A-derived cell lines were seeded in 12 well plates in media supplemented with 5% serum (left) and 5% serum plus 100 nM estradiol (right). Data are shown as mean + SD of three replicates, and curves are representative of three independent experiments.
In the MCF10A cell line panel, overexpression of mutated ER (mERIN) clones grew similarly to the wild-type counterpart (ERIN7) in the absence of estrogen but showed a muted response to E2 when compared to WT (Figure 2C). Based on the observed proliferation rate across all cell line panels, ER S118P does not confer a measurable change in cell proliferation at baseline, and its presence as a single copy in our heterozygous models does not alter the response to estrogen. However, in the absence of wild-type ER as seen in our overexpression model, cells do not respond as strongly to estrogen indicating that the variant ER protein may not be as highly activated by its traditional ligand.
The ER signaling pathway is intact in ER S118P-expressing cells
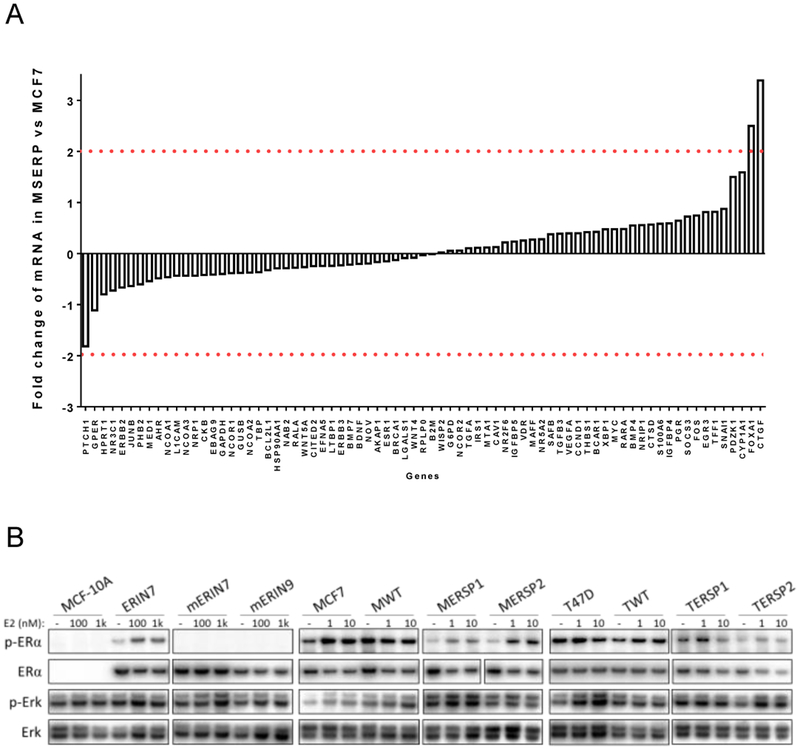
In light of ER’s classical role as a transcription factor and the phosphorylation of S118 as a dynamic effector of ER transcriptional regulation, we wanted to assess E2-induced expression of ER-regulated genes. Using a panel of 90 genes in the ER signaling pathway, we found that overall expression patterns with vehicle or 10 nM E2 treatment were relatively consistent between MCF7 and MERSP1 cells with small or negligible fold changes between cells (Figure 3A). This suggests there is no significant change in the ER-related gene expression patterns between the two cell lines and that the networks of ER transcriptional regulation are widely unaffected by the presence of the heterozygous ER S118P variant.
Figure 3: The S118P variant does not affect transcriptional regulation or downstream expression.
A) Gene expression analysis of genes regulated by ERα in MCF WT and MCF S118P cells as described in Methods. Fold change in gene expression between MCF7 S118P and MCF WT cells is plotted in the Y axis. Red lines indicate a change in expression ≥2. B) Immunoblot analysis of modified S118P panels as indicated. Cells were exposed to vehicle or stated E2 concentration for 24 hours and then lysed and analyzed by immunoblot analysis using antibodies listed in Materials and Methods.
ER signaling is known to interact with theMAP kinase signaling pathway and is crucial to the overall behavior of ER positive breast cells. Western blot analysis of phosphorylated Erk (p-Erk) activation was used to determine changes in the MAP kinase pathway in our isogenic cell lines. P-Erk levels were relatively consistent across all cell lines after E2 exposure suggesting no significant changes within the MAP kinase pathway (Figure 3B).
Cells expressing ER S118P remain sensitive to hormonal therapies and retain cancerous phenotypes similar to wild-type ER cells
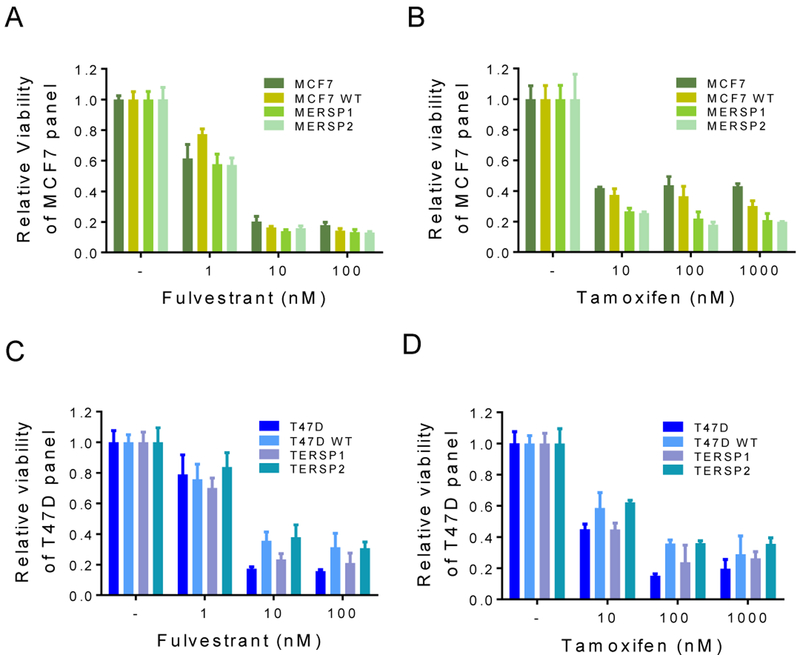
Given the importance of ER targeting in clinical breast oncology, we sought to determine whether our models would respond to hormone therapies commonly used in patients with ER+ disease, such as tamoxifen and fulvestrant. Tamoxifen and fulvestrant both interact directly with ER to reduce estrogen signaling in breast cancer cells, and other ER mutations have been shown to mediate resistance to these therapies [8, 10, 11]. Both the MCF7 and T47D ER S118P knock-in panels displayed dose dependent responses to tamoxifen and fulvestrant over nine days of drug treatment (Figure 4). There was no significant difference between the S118P variant and parental or WT suggesting that ER S118P does not interfere with ER protein-drug interaction.
Figure 4: ER S118P cells remain sensitive to tamoxifen and fulvestrant.
MCF7 cells and all targeted cell line derivatives were treated with 1 nM E2 and indicated concentrations of fulvestrant (A) or tamoxifen (B) over nine days. T47D cells and all targeted cell line derivatives were treated with 1 nM E2 and indicated concentrations of fulvestrant (C) or tamoxifen (D) over nine days. Data are shown as mean + SD of three replicates relative to cells treated with 1 nM E2 + vehicle, and graphs are representative of three independent experiments.
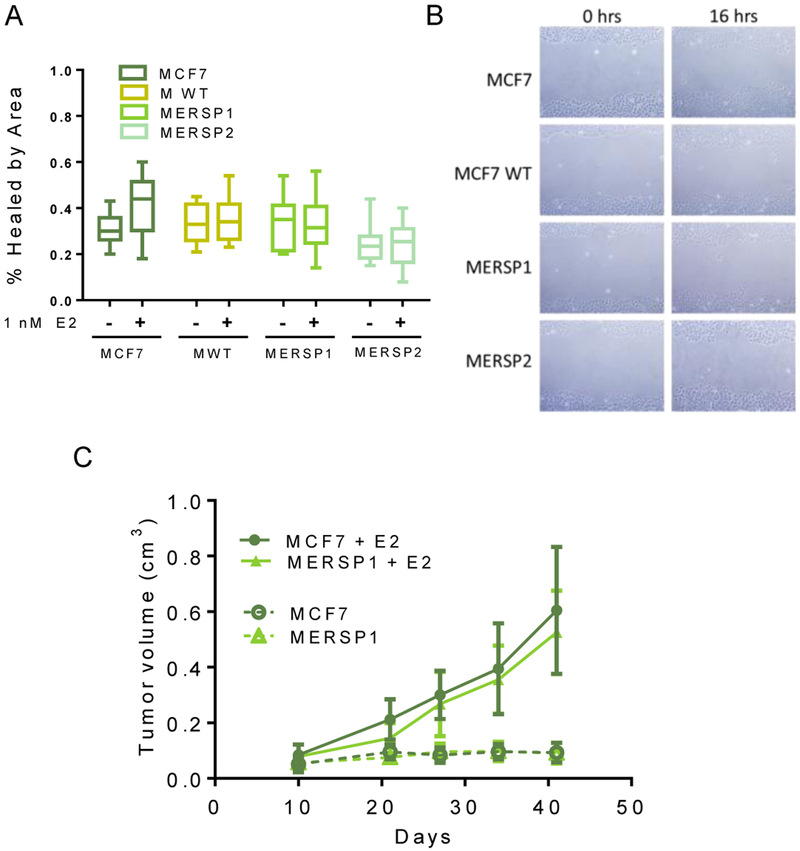
Due to recent studies which suggest that ER LBD mutations at Y537 and D538 can increase the metastatic potential of cells, we were also interested in the migratory abilities of ER S118P cells [23]. Changes in the migratory properties of cells can be attributed to changes in metastatic potential. In order to determine if the S118P variant demonstrated these changes, scratch wound healing assays were carried out on the MCF7 S118P panel. Regardless of the presence of E2, there were no measurable differences in wound closure between cells expressing ER S118P and the wild-type counterparts (Figure 5 A, B).
Figure 5: ER S118P does not exhibit increased metastatic or tumorigenic potential.
A) MCF7 and targeted cell line derivatives were seeded in 6 well plates with and without 1 nM E2. Once confluent, wells were scratched with a pipet tip and monitored over 16 hours for wound closure. Percent healing of the wound by area was determined by subtracting cell coverage at 0 hours from cell coverage at 16 hours. B) Representative images of scratch wound healing. Data are representative of four independent experiments. C) MCF7 and MERSP1 xenograft formation is estrogen-dependent. MCF7 and MERSP1 cells were injected into nude mice with and without estrogen supplementation implants and mice were monitored for 45 days. Both cell lines formed tumors only in the presence of estrogen. Data are shown as mean + SD of five mice per group.
In addition to changes in metastatic potential, the LBD ER mutations have been shown to constitutively activate ER, leading to E2 independent growth in vitro and in vivo [8, 10, 11]. Accordingly, we assessed the ability of the S118P variant cells to form tumors in vivo. Parental MCF7 and MERSP1 cells were injected into the mammary fat pad of athymic nude mice and tumor growth was monitored over seven weeks. As expected, MCF7 cells were only able to form tumors with exogenous estrogen supplementation. Similarly, MERSP1 cells were non-tumorigenic in the absence of estrogen but formed large, rapidly-progressing tumors in the presence of estrogen (Figure 5C). The size and growth rate of resulting tumors were similar between the parental and variant cell lines. T47D and MCF-10A cell lines were not assessed due to their inability to grow in athymic nude mice.
The ER S118P allele frequency is comparable between the general population and families at high risk for breast cancer
The ER S118P variant was first noted due to the rise in commercial NGS assays that did not employ germline filtering. Although it was assumed that this variant may be somatic, further analysis determined that ER S118P was a germline variant in at least one patient. During the course of our study, the variant was added to the Single Nucleotide Polymorphism Database (dbSNP; identifier rs200075329) and identified as a variant of unknown clinical significance (NA). To determine the frequency of this variant in a population of women with familial breast cancer risk. To avoid confounding statistical analysis, all samples containing germline BRCA1 and BRCA2 variants, including pathogenic and variants of unknown significance (VUS), were excluded. We hypothesized that ER S118P may be associated with either a higher or lower breast cancer incidence, due to potential changes and dysregulation in the ER signaling pathways.
The minor allele frequency of rs200075329, corresponding to ER S118P, varies from 0.0016 to 0.0098 in the general population according to various exome sequencing studies (Table 1, Supplemental Table 1). In a cohort of 536 individuals with a family history of breast cancer (Breast and Ovarian Surveillance Service (BOSS) cohort), consisting of 268 breast cancer patients and 268 age-matched controls (Supplemental Table 2), we identified four heterozygous carriers of the ER S118P variant (Table 2). In our cohort, the minor allele frequency of this variant was 0.004. Based on these data, the frequency of the variant was also not different in breast cancer patients and age-matched controls.
Table 1: ER S118P variant allele frequency in the general population.
Publicly available datasets establish a minor allele frequency for this variant ranging from 0.0016 to 0.0098.
| Source | Variant Allele Count | Total Allele Count | Allele Frequency | |
|---|---|---|---|---|
| Exome Aggregation Consortium (ExAC) | 312 | 31964 | .0098 | *2 homozygotes |
| Exome Variant Server (GO-ESP) | 31 | 12278 | .0025 | *0 homozygotes |
| Atherosclerosis Risk in Communities | 44 | 15726 | .0028 | |
| TOPMed | 78 | .0027 | ||
| 1000 Genomes | 8 | .0016 |
Table 2: Breast and Ovarian Surveillance Service (BOSS) ER S118P carrier characteristics.
S118P carrier characteristics within a cohort of individuals with a family history of breast cancer.
| Variable | ER S118 WT (N=532) | ER S118P (N=4) |
|---|---|---|
| Breast cancer survivor, % | 50.0 | 50.0 |
| Age at baseline, years, mean (SD) | 50.3 (11.2) | 52.2 (13.0) |
| Postmenopausal, % | 52.3 | 75.0 |
| Age at menopause, years, mean (SD) | 49.2 (5.8) | 52.3 (9.1) |
| BMI, kg/m2, mean(SD) | 26.6 (6.1) | 26.0 (4.9) |
| Physical activity, MET-h/week mean (SD)a | 27.1 (31.1) | 19.3 (22.8) |
| Alcohol intake, mean (SD) | 6.3 (10.1) | 4.1 (3.9) |
| Smoking, % | ||
| Never | 57.1 | 0 |
| Former | 37.8 | 100.0 |
| Current | 4.5 | 0 |
| Missing | <1 | 0 |
metabolic equivalents from recreational and occupational activity
Discussion
The importance of phosphorylation at the serine 118 residue of ER in cellular dynamics and hormone response has been described in numerous studies [24]. ER is involved in complex transcriptional networks and S118 phosphorylation has effects on the binding of coregulatory partners, activation time course, and response to ligand binding [5]. Phosphorylation of the S118 residue has also been shown to affect levels of ER in the cell via changes in degradation dynamics as well as binding to the ESR1 promoter [25].
Previous studies demonstrated cells expressing an alanine at residue 118 (S118A) of ER displayed significant changes in transcriptional regulation activities, estrogen-induced growth stimulation, and response to tamoxifen [4, 7, 26]. Disruption of ER phosphorylation at both the S118 and S167 site have also been shown to confer marked phenotypic changes in a breast cancer cell model [27]. To our knowledge, ER S118 mutants have only been studied in overexpression constructs and do not accurately represent variants observed in tumors. Through genome editing, mutations can be expressed under the endogenous promotor and have the capacity to interact with wild-type ER protein present on the remaining unaltered allele, as it occurs in the germline. We hypothesized that heterozygous knock-in of a naturally-occurring ER variant, ER S118P, would affect ER signaling and therefore response to ER ligands and possibly risk of breast cancer occurrence. However, the present studies have shown that a single copy of ER S118P in several different human breast cancer and non-cancerous cell line models is not sufficient to alter proliferation, migration, or sensitivity to endocrine therapies in vitro. In addition, we did not see significant changes in expression of ER-regulated genes or activation of key signaling pathways, all of which are known to be affected by ER S118 phosphorylation dynamics. Although a slight upregulation of Erk phosphorylation in response to E2 was observed, it was not specific to the heterozygous ER S118P cells and did not lead to a detectable cellular phenotype [28]. Finally, these cells behave similarly in vivo to their wild-type counterparts as estrogen-dependent xenografts in athymic nude mice.
We did not find any evidence that this ER variant contributes to a significant cellular or molecular phenotype in breast cancer cells according to the heterozygous models we have characterized. We hypothesize that the remaining wild-type ER protein in heterozygous models is sufficient to carry out normal estrogen signaling in the cell. Although significant crystallographic studies would be required to determine how often mutant ER is binding with WT, we hypothesize that occurrence of these heterodimers is not sufficient to mitigate or ablate ER activity. Furthermore, if the protein structure is not significantly altered by this variant, phosphorylation of only one subunit may be sufficient for normal transcriptional regulation. Therefore, we speculate that the majority of ER dimers function normally without obvious phenotypic changes.
Finally, we have analyzed a population of individuals with familial risk of breast cancer . We hypothesized that this variant may be involved in cellular signaling changes associated with the development of breast cancer and may have a higher prevalence than the general population in women with a familial risk However, the variant was present at a minor allele frequency of 0.004, consistent with the range of minor allele frequencies identified in the general population. This result suggests that ER S118P is unlikely to be a common modifier of breast cancer development.
Despite negative results, our study provides important functional information about a germline variant identified in breast cancer patients. Germline variants have led to confusion and uncertainty regarding how to best manage patients who are prescribed endocrine therapies, notably CYP2D6 variants that were thought to be poor metabolizers of tamoxifen [29]. Our current study strongly suggests that the S118P variant, despite the importance of this amino acid in wild-type ER signaling, is not increased in women with a familial risk and may not be associated with breast cancer incidence nor therapeutic response to ER targeted therapies in pre-clinical models. As such, we would advocate that screening for S118P is not warranted and should not be further pursued as a predictive or prognostic marker.
Materials and Methods
Cell culture
MCF7, T47D, and MCF10A parental cell lines were obtained from ATCC and verified via short tandem repeat profiling by the Johns Hopkins Genetic Resource Core Facility. All cells were grown in 5% CO2 at 37° C. MCF7 and T47D lines and their derivatives were maintained in DMEM containing 5% fetal bovine serum and 1% penicillin/streptomycin. The MCF10A line and its derivatives were maintained in DMEM:F12 containing 5% horse serum, 1% pen/strep, epidermal growth factor at 20 ng/ml, insulin at 10 μg/ml, hydrocortisone at 0.5 μg/ml, and cholera toxin at 0.1 μg/ml. For assays, cells were arrested in clear DMEM:F12 with 1% charcoal dextran treated fetal bovine serum and 1% pen/strep and assayed in clear DMEM:F12 with 10% charcoal dextran treated fetal bovine serum and 1% pen/strep (all MCF10A media contained insulin, cholera toxin, and hydrocortisone at the concentrations stated above, but EGF was omitted in arrest and assay formulations).
Gene targeting
MCF7 and T47D parental cell lines were genetically altered using a recombinant AAV vector containing the T>C single base-pair substitution in ESR1 exon 1 resulting in ER S118P. Cells were targeted, screened, and validated as previously described [28]. Two independently derived clones containing the variant and one targeted wild-type clone were isolated from each parental cell line and confirmed via gDNA and cDNA analysis by Sanger sequencing and droplet digital PCR. Targeted cells were maintained in DMEM media as described above.
Overexpression
MCF10A cells were transfected using Fugene 6 (Promega) with the pIRESneo3 vector containing a copy of ER cDNA modified with the S118P variant. Cells with stable expression of the neomycin resistance gene were selected with Geneticin (Life Technologies) at 120 ug/ml. Two independently derived clones were validated for stable expression of ER S118P via cDNA ddPCR. Proper expression was confirmed via immunoblotting for the presence of total ER but absence of phosphorylated ER S118. ERIN cell lines were previously established in the lab to overexpress wild-type ER from the same parent vector and were used as the wild-type control for these cells [22]. ERIN and mERIN lines were maintained in DMEM:F12 media as stated above with the addition of Geneticin at 120 μg/ml.
Cell proliferation and drug response assays
17-beta estradiol (E2), 4-hydroxytamoxifen (tamoxifen), and fulvestrant were obtained from Sigma-Aldrich and diluted according to manufacturer recommendations. Cells were seeded at low confluency (3,000 MCF7 cells/well, 50,000 MCF10A cells/well, 60,000 T47D cells/well) in 12 well cell culture plates in arrest media for 24 hours. Cells were visualized on day 0 and estrogen/drug/vehicle were added, at the stated concentrations, to assay media. Media containing the indicated additives was replaced every three days. Cells were trypsinized, uniformly resuspended, and counted using the Vi-Cell XR (Beckman Coulter) on indicated days. All results are an average of cell counts from triplicate wells.
Scratch assays
Cells were seeded at 50% confluency in 6 well cell culture plates in assay media. Once cells reached 90% confluency, assay media containing 1 nM 17-beta estradiol (E2) or vehicle were added to indicated plates. After 8 hours, a 200 uL pipet tip was used to created two perpendicular scratches in the lawn of cells; cells were then rinsed twice with clear HBSS to remove floating cell debris and previously indicated media was replaced. Two visual fields per well were imaged at time 0 and 16 hours. Images were then analyzed in ImageJ using the MiToBo plugin for percent of field containing cells, and timepoints were compared to determine migration of cells into scratched area.
Immunoblotting
Cells were seeded in arrest media for 24 hours followed by indicated treatment. Cells were collected and protein lysates were prepared using Laemmli buffer. Lysates were analyzed by immunoblot as described previously [21]. Primary antibodies included: phospho-ERα S118 (Cell Signaling 2511), ERα (Cell Signaling 8644), phospho-Erk (Cell Signaling 4370), Erk (Cell Signaling 9102), GAPDH (Cell Signaling 5174).
Quantitative PCR
Cells were seeded in arrest media for 24 hours, followed by 10% CD assay media for 24 hours. Cells were exposed to 17-beta estradiol at 1 nM for 45 minutes and then processed for RNA and cDNA. PrimePCR pathway plates for estrogen receptor signaling (Bio-Rad) with SsoAdvanced Universal Sybr Green Supermix (Bio-Rad) were used to process and analyze the samples according to the Bio-Rad protocol.
Xenografts
1 × 106 cells MCF7 cells suspended in 200 μL of Geltrex LDEV-Free Reduced Growth Factor Basement Membrane Matrix (Life Technologies) were injected subcutaneously into the mammary fat pad of 8- to 10-week-old female athymic nude mice (Harlan Laboratories) with and without slow-release estrogen implants as previously described [21]. Tumor volumes were calculated as a pseudosphere from length, width, and height measurements. All animal experiments were performed in accordance with institutional guidelines and The National Institutes of Health Guide for the Care and Use of Laboratory Animals guidelines.
Statistics
Statistical analyses were performed using GraphPad Prism 6 software. Significance levels are indicated by an asterisk: *P ≤ 0.05. Error bars represent ± SD. Relative proliferation rates were analyzed by two-way ANOVA.
Cohort genotyping
Genomic DNA extracted from lymphocytes was obtained from Kala Visvanathan at the Johns Hopkins Center for Clinical Cancer Genetics and Prevention. Samples were analyzed via droplet digital PCR on the QX200 platform (Bio-Rad) per the manufacturer’s protocol for probe-based ddPCR. Primers and probes used were as follows: forward primer (5’-CACTCAACAGCGTGTCT-3’), reverse primer (5’-CTCGTTCTCCAGGTAGTAG-3’), wild-type probe (5’-AGCTGTCGCCTTTCCTGCAG-3’), variant probe (5’-AGCTGCCGCCTTTCCTGCAG-3’).
Supplementary Material
Acknowledgments
The authors would like to thank the Exome Aggregation Consortium and the groups that provided exome variant data for comparison. A full list of contributing groups can be found at http://exac.broadinstitute.org/about.
The authors would like to thank the NHLBI GO Exome Sequencing Project and its ongoing studies which produced and provided exome variant calls for comparison: the Lung GO Sequencing Project (HL-102923), the WHI Sequencing Project (HL-102924), the Broad GO Sequencing Project (HL-102925), the Seattle GO Sequencing Project (HL-102926) and the Heart GO Sequencing Project (HL-103010).
The Atherosclerosis Risk in Communities study has been funded in whole or in part with Federal funds from the National Heart, Lung, and Blood Institute, National Institutes of Health, Department of Health and Human Services, under Contract nos. (HHSN268201700001I, HHSN268201700002I, HHSN268201700003I, HHSN268201700005I, HHSN268201700004I). The authors thank the staff and participants of the ARIC study for their important contributions
The authors would like to thank the BOSS cohort participants.
Whole genome sequencing (WGS) for the Trans-Omics in Precision Medicine (TOPMed) program was supported by the National Heart, Lung and Blood Institute (NHLBI). Centralized read mapping and genotype calling, along with variant quality metrics and filtering were provided by the TOPMed Informatics Research Center (3R01HL-117626–02S1). Phenotype harmonization, data management, sample-identity QC, and general study coordination, were provided by the TOPMed Data Coordinating Center (3R01HL-120393–02S1). We gratefully acknowledge the studies and participants who provided biological samples and data for TOPMed.
Funding:
This work was supported by: The Komen Foundation (B.H.P.), NIH CA214494 (B.H.P.), NIH CA088843 and CA194024 (K.C. and B.H.P.), GM008752 (B.B.), NIH CA009314 (CR), and NIH CA009071 (W.B.D., E.C. and J.D.). We would also like to thank and acknowledge the support of the Commonwealth Foundation, the Breast Cancer Research Foundation, The Canney Foundation, the M&E Foundation, Avon Breast Cancer Research Program Network, and the Johns Hopkins Fetting Fund.
Footnotes
Disclosure of Potential Conflicts of Interest:
B.H.P. has ownership interest and is a paid member of the scientific advisory board of Loxo Oncology and is a paid consultant for Foundation Medicine, Inc, Jackson Labs, Roche, Casdin Capital and H3 Biomedicine. Under separate licensing agreements between Horizon Discovery, LTD and The Johns Hopkins University, B.H.P. is entitled to a share of royalties received by the University on sales of products. The terms of this arrangement are being managed by the Johns Hopkins University in accordance with its conflict of interest policies. No other authors declare potential conflicts of interest.
None of the authors have a financial relationship with the organizations that sponsored the research.
Ethical Statement
All studies were performed in compliance with institutional ethical standards.
Ethical Approval:
All animal experiments were performed in accordance with institutional and The National Institutes of Health Guide for the Care and Use of Laboratory Animals guidelines.
Informed Consent:
All study participants provided informed consent under an IRB approved protocol.
References
- 1.Campbell RA, Bhat-Nakshatri P, Patel NM, Constantinidou D, Ali S, Nakshatri H: Phosphatidylinositol 3-kinase/AKT-mediated activation of estrogen receptor alpha: a new model for anti-estrogen resistance. The Journal of biological chemistry 2001, 276(13):9817–9824. [DOI] [PubMed] [Google Scholar]
- 2.Migliaccio A, Piccolo D, Castoria G, Di Domenico M, Bilancio A, Lombardi M, Gong W, Beato M, Auricchio F: Activation of the Src/p21ras/Erk pathway by progesterone receptor via cross-talk with estrogen receptor. The EMBO journal 1998, 17(7):2008–2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McKenna NJ, Nawaz Z, Tsai SY, Tsai MJ, O’Malley BW: Distinct steady-state nuclear receptor coregulator complexes exist in vivo. Proceedings of the National Academy of Sciences of the United States of America 1998, 95(20):11697–11702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Duplessis TT, Williams CC, Hill SM, Rowan BG: Phosphorylation of Estrogen Receptor alpha at serine 118 directs recruitment of promoter complexes and gene-specific transcription. Endocrinology 2011, 152(6):2517–2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Anbalagan M, Rowan BG: Estrogen receptor alpha phosphorylation and its functional impact in human breast cancer. Molecular and cellular endocrinology 2015. [DOI] [PubMed] [Google Scholar]
- 6.Clarke R, Liu MC, Bouker KB, Gu Z, Lee RY, Zhu Y, Skaar TC, Gomez B, O’Brien K, Wang Y et al. : Antiestrogen resistance in breast cancer and the role of estrogen receptor signaling. Oncogene 2003, 22(47):7316–7339. [DOI] [PubMed] [Google Scholar]
- 7.Abukhdeir AM, Vitolo MI, Argani P, De Marzo AM, Karakas B, Konishi H, Gustin JP, Lauring J, Garay JP, Pendleton C et al. : Tamoxifen-stimulated growth of breast cancer due to p21 loss. Proceedings of the National Academy of Sciences of the United States of America 2008, 105(1):288–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Robinson DR, Wu YM, Vats P, Su F, Lonigro RJ, Cao X, Kalyana-Sundaram S, Wang R, Ning Y, Hodges L et al. : Activating ESR1 mutations in hormone-resistant metastatic breast cancer. Nature genetics 2013, 45(12):1446–1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Toy W, Shen Y, Won H, Green B, Sakr RA, Will M, Li Z, Gala K, Fanning S, King TA et al. : ESR1 ligand-binding domain mutations in hormone-resistant breast cancer. Nat Genet 2013, 45(12):1439–1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jeselsohn R, Yelensky R, Buchwalter G, Frampton G, Meric-Bernstam F, Gonzalez-Angulo AM, Ferrer-Lozano J, Perez-Fidalgo JA, Cristofanilli M, Gomez H et al. : Emergence of constitutively active estrogen receptor-alpha mutations in pretreated advanced estrogen receptor-positive breast cancer. Clinical cancer research : an official journal of the American Association for Cancer Research 2014, 20(7):1757–1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li S, Shen D, Shao J, Crowder R, Liu W, Prat A, He X, Liu S, Hoog J, Lu C et al. : Endocrine-therapy-resistant ESR1 variants revealed by genomic characterization of breast-cancer-derived xenografts. Cell reports 2013, 4(6):1116–1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Merenbakh-Lamin K, Ben-Baruch N, Yeheskel A, Dvir A, Soussan-Gutman L, Jeselsohn R, Yelensky R, Brown M, Miller VA, Sarid D et al. : D538G mutation in estrogen receptor-alpha: A novel mechanism for acquired endocrine resistance in breast cancer. Cancer research 2013, 73(23):6856–6864. [DOI] [PubMed] [Google Scholar]
- 13.Parsons HA, Beaver JA, Cimino-Mathews A, Ali SM, Axilbund J, Chu D, Connolly RM, Cochran RL, Croessmann S, Clark TA et al. : Individualized Molecular Analyses Guide Efforts (IMAGE): A Prospective Study of Molecular Profiling of Tissue and Blood in Metastatic Triple-Negative Breast Cancer. Clinical cancer research : an official journal of the American Association for Cancer Research 2017, 23(2):379–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hortobagyi GN, Chen D, Piccart M, Rugo HS, Burris HA 3rd, Pritchard KI, Campone M, Noguchi S, Perez AT, Deleu I et al. : Correlative Analysis of Genetic Alterations and Everolimus Benefit in Hormone Receptor-Positive, Human Epidermal Growth Factor Receptor 2-Negative Advanced Breast Cancer: Results From BOLERO-2. Journal of clinical oncology : official journal of the American Society of Clinical Oncology 2016, 34(5):419–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sfakianos JP, Cha EK, Iyer G, Scott SN, Zabor EC, Shah RH, Ren Q, Bagrodia A, Kim PH, Hakimi AA et al. : Genomic Characterization of Upper Tract Urothelial Carcinoma. European urology 2015, 68(6):970–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rebai M, Rebai A: In silico characterization of functional SNP within the oestrogen receptor gene. J Genet 2016, 95(4):865–874. [DOI] [PubMed] [Google Scholar]
- 17.Bunone G, Briand PA, Miksicek RJ, Picard D: Activation of the unliganded estrogen receptor by EGF involves the MAP kinase pathway and direct phosphorylation. The EMBO journal 1996, 15(9):2174–2183. [PMC free article] [PubMed] [Google Scholar]
- 18.Chen D, Washbrook E, Sarwar N, Bates GJ, Pace PE, Thirunuvakkarasu V, Taylor J, Epstein RJ, Fuller-Pace FV, Egly JM et al. : Phosphorylation of human estrogen receptor alpha at serine 118 by two distinct signal transduction pathways revealed by phosphorylation-specific antisera. Oncogene 2002, 21(32):4921–4931. [DOI] [PubMed] [Google Scholar]
- 19.Chen M, Cui YK, Huang WH, Man K, Zhang GJ: Phosphorylation of estrogen receptor alpha at serine 118 is correlated with breast cancer resistance to tamoxifen. Oncology letters 2013, 6(1):118–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cheng J, Zhang C, Shapiro DJ: A functional serine 118 phosphorylation site in estrogen receptor-alpha is required for down-regulation of gene expression by 17beta-estradiol and 4-hydroxytamoxifen. Endocrinology 2007, 148(10):4634–4641. [DOI] [PubMed] [Google Scholar]
- 21.Zabransky DJ, Yankaskas CL, Cochran RL, Wong HY, Croessmann S, Chu D, Kavuri SM, Red Brewer M, Rosen DM, Dalton WB et al. : HER2 missense mutations have distinct effects on oncogenic signaling and migration. Proc Natl Acad Sci U S A 2015, 112(45):E6205–6214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Abukhdeir AM, Blair BG, Brenner K, Karakas B, Konishi H, Lim J, Sahasranaman V, Huang Y, Keen J, Davidson N et al. : Physiologic estrogen receptor alpha signaling in non-tumorigenic human mammary epithelial cells. Breast cancer research and treatment 2006, 99(1):23–33. [DOI] [PubMed] [Google Scholar]
- 23.Jeselsohn R, Bergholz JS, Pun M, Cornwell M, Liu W, Nardone A, Xiao T, Li W, Qiu X, Buchwalter G et al. : Allele-Specific Chromatin Recruitment and Therapeutic Vulnerabilities of ESR1 Activating Mutations. Cancer cell 2018, 33(2):173–186 e175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Anbalagan M, Rowan BG: Estrogen receptor alpha phosphorylation and its functional impact in human breast cancer. Mol Cell Endocrinol 2015, 418 Pt 3:264–272. [DOI] [PubMed] [Google Scholar]
- 25.Tian D, Solodin NM, Rajbhandari P, Bjorklund K, Alarid ET, Kreeger PK: A kinetic model identifies phosphorylated estrogen receptor-alpha (ERalpha) as a critical regulator of ERalpha dynamics in breast cancer. FASEB journal : official publication of the Federation of American Societies for Experimental Biology 2015, 29(5):2022–2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Murphy LC, Weitsman GE, Skliris GP, Teh EM, Li L, Peng B, Davie JR, Ung K, Niu YL, Troup S et al. : Potential role of estrogen receptor alpha (ERalpha) phosphorylated at Serine118 in human breast cancer in vivo. The Journal of steroid biochemistry and molecular biology 2006, 102(1–5):139–146. [DOI] [PubMed] [Google Scholar]
- 27.Huderson BP, Duplessis TT, Williams CC, Seger HC, Marsden CG, Pouey KJ, Hill SM, Rowan BG: Stable inhibition of spedfic estrogen receptor alpha (ERalpha) phosphorylation confers increased growth, migration/invasion, and disruption of estradiol signaling in MCF-7 breast cancer cells. Endocrinology 2012,153(9):4144–4159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gustin JP, Karakas B, Weiss MB, Abukhdeir AM, Lauring J, Garay JP, Cosgrove D, Tamaki A, Konishi H, Konishi Y et al. : Knockin of mutant PIK3CA activates multiple oncogenic pathways. Proc Natl Acad Sci U S A 2009, 106(8):2835–2840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dean L: Tamoxifen Therapy and CYP2D6 Genotype In: Medical Genetics Summaries. edn. Edited by Pratt V, McLeod H, Rubinstein W, Dean L, Kattman B, Malheiro A. Bethesda (MD); 2012. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.