Abstract
Retinoid X receptor alpha (RXRα), a central member of the nuclear receptor superfamily and a key regulator of many signal transduction pathways, has been an attractive drug target. We previously discovered that an N-terminally truncated form of RXRα can be induced by specific ligands to form homotetramers, which, as a result of conformational selection, forms the basis for inhibiting the nongenomic activation of RXRα. Here, we report the identification and characterization of atorvastatin as a new RXRα tetramer stabilizer by using structure-based virtual screening and demonstrate that virtual library screening can be used to aid in identifying RXRα ligands that can induce its tetramerization. In this study, docking was applied to screen the FDA-approved small molecule drugs in the DrugBank 4.0 collection. Two compounds were selected and purchased for testing. We showed that the selected atorvastatin could bind to RXRα to promote RXRα-LBD tetramerization. We also showed that atorvastatin possessed RXRα-dependent apoptotic effects. In addition, we used a chemical approach to aid in the studies of the binding mode of atorvastatin.
Keywords: RXRα, RXRα tetramer, RXRα ligands, Virtual screening, Conformational selection
Graphical Abstract

1. Introduction
Retinoid X receptor alpha (RXRα), a unique member of the nuclear receptor superfamily, regulates a broad spectrum of physiological functions including cell differentiation, growth, and apoptosis, and is implicated in many diseases such as cancer, metabolic disorders and neurodegenerative diseases [1–5]. Thus, RXRα has been an attractive drug target, especially for anticancer therapy [1, 6, 7]. Similar to other nuclear receptors, structurally RXRα possesses three main functional domains: a disordered N-terminal region, a DNA-binding domain, and a ligand-binding domain (LBD). The LBD is characterized by a canonical ligand-binding pocket (LBP), a transactivation function domain 2, a coregulator-binding surface groove, and a dimerization surface. A well-accepted mechanism of RXRα action as a transcription factor is that RXRα acts as homodimers or as heterodimers partnering with many other nuclear receptors as ligand-mediated transcription factors through binding to specific DNA-response elements of the target genes [3, 8]. Ligand binding to the LBP induces a conformational change that triggers a cascade of events and lead to biological activities. Many natural and synthetic ligands have been discovered for this canonical LBP [9]. Targretin (bexarotene), a selective LBP ligand of RXRα, was approved for treating human cutaneous T-cell lymphoma [10, 11].
Besides functioning as a transcriptional factor, RXRα also play important extranuclear (nongenomic) roles through transcription-independent mechanisms [12–16]. RXRα migrates from the nucleus to the cytoplasm at different stages during development [17] and in response to differentiation [14], survival [18, 19], apoptosis [12], and inflammation [15, 16, 18, 19]. Studies from our laboratory show that RXRα can act in the cytoplasm to cross-talk with important biological processes such as mitochondria-dependent apoptotic pathway [12, 20], phosphatidylinositol 3-kinase (PI3K)/AKT-mediated cell survival pathway [19, 21], and NF-κB-mediated inflammatory pathway [22]. We have previously reported that RXRα is abnormally cleaved in various types of cancer cells, producing an N-terminally-truncated RXRα (tRXRα) protein. We showed that tRXRα resides in cytoplasm and is oncogenic in tumor cells. tRXRα acts to promotes phosphoinositide 3-kinase (PI3K)/AKT activation and enhance tumor cell growth via interacting with the p85α regulatory subunit of PI3K. Thus, molecules that can bind to RXRα to modulate its interaction with p85α may have therapeutic potential. Along this line, we have identified K-80003 (Fig. S1A), a designed analog of the non-steroidal anti-inflammatory drug (NSAID) Sulindac, as a promising anti-cancer agent. K-80003 induces apoptosis and inhibits the tRXRα-mediated PI3K/AKT survival pathway by binding to tRXRα and disrupting the interaction between tRXRα and p85α [19].F
Our recent studies of the molecular mechanism of K-80003 reveal that K-80003 utilizes conformational selection as a mechanism to inhibit the nongenomic function of tRXRα [23]. Besides functioning in the forms homodimers and heterodimers, RXRα can also form tetramers. We demonstrate that K-80003 binding promotes tRXRα tetramerization which results apoptosis via the inhibition of tRXRα interaction with p85α [23]. Crystal structure of the RXRα-LBD/K-80003 complex shows that the RXRα-LBD/K-80003 complex exists as a tetramer forming 2 large symmetric hydrophobic cavities where 3 molecules of K-80003 are bound per cavity. The fact that the cavity as a binding region can simultaneously accommodate 3 K-80003 molecules inspired us to ask if a molecule larger than K-80003 could be identified to mimic the binding of the 3 K-80003 molecules while retaining similar or better biological activities [21, 23].
Structure-based virtual screening is a powerful approach in drug discovery where the three-dimensional structure of the protein target is available [24]. In our previous work, we successfully discovered the first small molecule targeting the coregulator-binding site of RXRα using this approach [25]. Thus, we employed structure-based virtual screening to identify RXRα modulators that can bind and stabilize RXRα-LBD tetramers. As a pilot exercise we used an FDA-approved drug collection as the first screening library. Here we report the identification and characterization of drug atorvastatin as a non-canonical ligand of RXRα that stabilizes the RXRα tetrameric conformation to regulate RXRα nongenomic actions.
2. Results and discussion
2.1. Structure-based virtual screening
In this study we screened an FDA-approved drug collection of 1908 compounds downloaded from DrugBank 4.0 [26]. The collection was first prepared by LigPrep [27] module in Maestro 10.5 and was then converted from 2D to 3D with conformation energy minimized using the OPLS3 force field. The crystal structure of RXRα-LBD tetramer retrieved from the Protein Data Bank (www.rcsb.org) (PDB code: 5TBP) was used for the docking study. The protein structure was prepared using Protein Preparation Wizard module [28] in Maestro 10.5, during which hydrogen atoms were added and crystallographic water molecules were removed. Missing side chains and loops were built using Prime [29] in Maestro 10.5. The binding site was defined based on the positions of all three bound K-80003 molecules (Fig.S1B), around which the 3D grid box was generated in a size of 20 Å per dimension for docking. Glide [30] in Maestro 10.5 was used for generating the grids and carrying out the docking studies. All compounds were docked using the standard precision (SP) mode for scoring. The virtual screening process is summarized in Fig.1A.
Fig.1.
Virtual Screening. (A) Structure-based virtual screening process. (B) Chemical structure of atorvastatin and lercanidipine.
2.2. Selection of compounds
There were 1736 molecules that could dock to the binding site. The first 10 compounds (Table S1) with the highest docking scores were selected for further evaluation using the following criteria: 1. How well the docked molecule interacts with the protein; 2. If the docked molecule occupies a reasonable size of the binding site; 3. If the compound is commercially available. As a result, 2 compounds, atorvastatin and lercanidipine were selected and purchased from commercial suppliers (Fig.1B). Both purchased compounds were confirmed with high-resolution mass spectrometry (HRMS).
2.3. Biological evaluation
2.3.1. Luciferase reporter assay evaluation
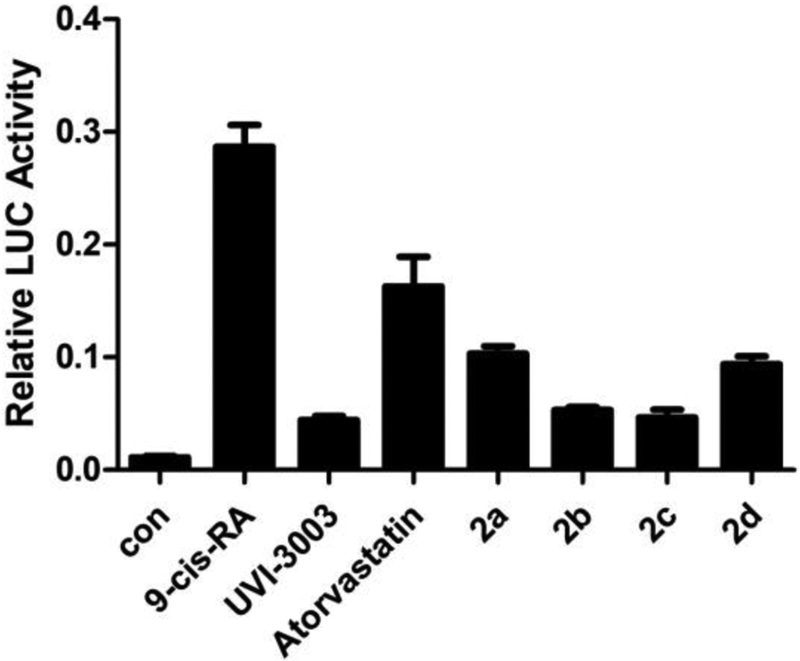
Luciferase reporter assay was first used to evaluate our docking results (Fig.2A). Gal4-RXRα-LBD strongly activates the Gal4 reporter in the presence of 9-cis-RA, which is inhibited by UVI-3003 [31], a known RXRα antagonist. We expected that atorvastatin and lercanidipine would act as antagonists in this assay if they could stabilize the tetrameric form of RXRα-LBD. Indeed, both atorvastatin and lercanidipine could inhibit the 9-cis-RA-induced Gal4 reporter activity (Fig. 2A) and consistently the compounds did not show any agonist activity at the concentrations used (Fig.S2). In addition, atorvastatin, but not lercanidipine, showed dose-dependent inhibitory effect. Thus, the reporter assay confirmed that atorvastatin could bind to RXRα-LBD.
Fig. 2.
Binding Evalution. (A) Inhibition of 9-cis-RA-induced Gal4 reporter activity. pBind-RXRα-LBD and pG5luc were transiently transfected into 293T cells. Cells were treated with 9-cis-RA (1 μM) in the presence of UVI-3003(1 μM), atorvastatin and lercanidipine (1 μM, 10 μM, 50 μM). Relative luciferase (LUC) activity was determined. (B) Fluorescence quenching effect of atorvastatin on RXRα-LBD. RXRα-LBD: 1 μM, atorvastatin: 0, 1, 3, 5 10, 15, 20, 30, 40 μM.
2.3.2. Binding evaluation by fluorescence quenching assay
To further confirm atorvastatin binds directly to RXRα, fluorescence quenching was conducted to analyze the binding affinity of atorvastatin to RXRα-LBD. Proteins possess intrinsic fluorescence mainly because of the aromatic amino acid residues, such as tryptophan and phenylalanine. The intrinsic fluorescence produced by these amino acid residues are highly sensitive to their local environment, that can be used to measure the binding ability of small molecules. In our work, there are Trp305, Phe439, Phe439 and Phe313 in the binding site of the RXRα tetramer, giving us the opportunity to use this method to evaluate the binding affinity of atorvastatin. Mixtures of RXRα-LBD and atorvastatin were analyzed by scanning fluorescence emission between 290 and 450 nm that were stimulated by an excitation wavelength of 280 nm. The results showed gradual fluorescence quenching when RXRα-LBD was exposed to different concentrations of atorvastatin (Fig.2B), suggesting that there were interactions between atorvastatin and RXRα-LBD. To calculate the Kd value, we used a total of 8 different compound concentrations from 1 μmol/L to 40 μmol/L to plot the fluorescence emission at 330 nm as a function of compound concentrations and then we performed a non-linear fitting of the curve [32] (Fig.S3). The results showed the Kd for the RXRα-LBD/atorvastatin complex is 10.34 ×10−6 M.
2.3.3. Atorvastatin induces RXRα-LBD tetramerization
We then asked if atorvastatin binding to RXRα can promote RXRα-LBD tetramerization as K-80003 does [23]. In non-denaturing polyacrylamide gel electrophoresis, purified RXRα-LBD protein in the absence of ligand existed as two distinct bands corresponding to homodimeric and homotetrameric RXRα-LBD respectively (Fig. 3). 9-cis-RA binding induces homodimerization and shifts the dimers/tetramers equilibrium to dimers (Fig. 3). As expected, atorvastatin induced RXRα-LBD tetramerization as K-80003 (Fig. 3). Furthermore, when incubated RXRα-LBD with 9-cis-RA and atorvastatin together, atorvastatin could competitively inhibit 9-cis-RA-induced dimerization to promote RXRα tetramerization (Fig. 3).
Fig.3.
Induction of RXRα-LBD tetramerization by atorvastatin. Equal amount of purified RXRα-LBD was incubated with K-80003, 9-cis-RA, DMSO, atorvastatin, 9-cis-RA/K-80003 or 9-cis-RA/atorvastatin and separated by non-denaturing polyacrylamide gel electrophoresis followed by Coomassie Bright Blue staining. The percentage of tetramer and dimer of RXRα-LBD was quantitated by densitometric analysis of the corresponding blots.
2.3.4. Atorvastatin displays apoptotic effects through RXRα binding
K-80003 is a promising anticancer agent, acting by inhibiting the RXRα-activated PI3K/AKT survival pathway and inducing apoptosis [19]. Therefore, we next determined whether atorvastatin had the same anticancer effect and if the effect was RXRα-dependent. As shown in Fig. 4, treatment of MCF-7 cancer cell lines with atorvastatin alone or in combination with TNF-α effectively induced PARP cleavage, an indication of apoptosis in cancer cells. To determine if the effect of atorvastatin on inducing PARP cleavage depended on the expression of RXRα, MCF-7 cells were transfected with RXRα siRNA and evaluated for the apoptotic effect of atorvastatin. The result showed that transfection of RXRα siRNA significantly reduced the level of RXRα in MCF-7 cells and weakened the atorvastatin-induced PARP cleavage (Fig. 4). These results demonstrated that RXRα played a crucial role in mediating the apoptotic effect of atorvastatin in cancer cells.
Fig.4.
Atorvastatin induces apoptosis of cancer cells in a RXRα-dependent manner. RXRα siRNA transfection inhibits the apoptotic effect of atorvastatin. MCF-7 cells transfected with control or RXRα siRNA for 48 hr were treated with atorvastatin (50 μM), K-80003 (30 μM) and/or TNF-α (20 ng/mL) for 12 hr and analyzed by immunoblotting.
2.4. Binding mode exploration
Besides the crystal structure of the tetrameric RXRα-LBD/K-80003 complex (PDB code: 5TBP), previously we also determined another crystal structure of RXRα-LBD tetramer in complex with a different ligand, K-8008 (PDB code: 4N8R). Although RXRα ligands K-8008 and K-80003 are structurally similar (K-8008 was designed based on the bioisostere concept by replacing the carboxylic acid in K-80003 with tetrazole) (Fig. S1A) and their complexes with RXRα-LBD adopt a tetrameric structure with each ligand-bound tetramer possessing 2 large symmetry-related interfacial cavities, the binding mode of K-80003 is different from the binding mode of K-8008 (Fig. S1B–C). In the complex structure of RXRα-LBD with K-80003, three molecules of K-80003 bind to the same large hydrophobic cavity, whereas only one molecule of K-8008 is bound in the crystal structure of RXRα-LBD/K-8008 complex (Fig. S1B–C) [21, 23]. Furthermore, we found that the orientations of Phe439 display large difference (Fig. S1D) in the area where ligand K-8008 or K-80003 binds. It is well known that docking results are influenced by many factors including docking methods and orientations of side chains in the binding site [33, 34]. Therefore, to explore the binding model, we docked atorvastatin to both crystal structures (4N8R and 5TBP). The results proposed 2 different binding models (Fig.5). In model A (Fig.5A), the docking score was −9.780 kcal mol−1, and atorvastatin interacted with the protein primarily via forming H-bonds with residues Arg316 and Ala327 and making van der Waals contacts with Trp305, Leu309, Leu326, Phe438, Phe439 and Leu436. The carboxylic acid group in atorvastatin appeared to be important for binding. In model B (Fig.5B), the docking score is −9.816 kcal mol−1, and the carboxylic acid group of atorvastatin formed a H-bond with Gln275 and the three benzene rings on atorvastatin made hydrophobic interactions with the surrounding hydrophobic amino acids including π-π interaction with Trp305.
Fig.5.
The possible binding modes of atorvastatin. (A) The predicted binding model A. (B) The predicted binding model B. Atorvastatin is shown in pink sticks, and the protein is shown in gray cartoon ribbon, key residues are shown in cyan sticks, and the potential H-bonds are shown in yellow dashes.
To help determine which model is more reasonable, four derivatives of atorvastatin with chemical modification of the carboxylic acid group were designed and synthesized. Methyl esterification, ethyl esterification, butyl esterification and hexyl esterification of the carboxylic acid were carried out to generate compounds 2a, 2b, 2c and 2d respectively using scheme 1. Model A showed that derivatization of the -COOH group not only would disrupt the H-bonds but also cause steric hindrance. Thus, we expected that esterification of the carboxylic acid group would weaken the binding of atorvastatin. In model B, there is still some unoccupied hydrophobic space around the carboxylic acid group, and alkylation modification of the carboxyl group would not necessarily lead to the weakening of the binding. Therefore, ester compounds would aid in assessing if model A or model B is the possible binding model for atorvastatin.
Scheme 1.
Reagents and conditions: (a) ROH, H2SO4, r. t., 2.5h; (b) ROH, CH2Cl2, H2SO4, r. t., 2.5h.
Luciferase reporter assay was used to evaluate the binding of these four derivatives (Fig.6). Results showed that all 4 ester compounds (2a, 2b, 2c and 2d) inhibited the 9-cis-RA-induced Gal4 reporter activity more strongly than atorvastatin and compounds (2b and 2c) worked better than compounds (2a and 2d). These results implied that the aforementioned binding model B of atorvastatin was more reasonable. This is because the binding model A anticipated that derivatives 2a-d would display weaker inhibition of the 9-cis-RA-induced Gal4 reporter activity, whereas the binding model B supported that derivatives 2a-d could interact more strongly with the protein and display stronger inhibition of the 9-cis-RA-induced Gal4 reporter activity compared to atorvastatin.
Fig. 6.
Inhibition of the 9-cis-RA-induced Gal4 reporter activity. pBind-RXRα-LBD and pG5luc were transiently transfected into 293T cells. Cells were treated with 9-cis-RA (1 μM) in the presence of UVI-3003 (1 μM), atorvastatin, 2a, 2b, 2c and 2d (20 μM). Relative luciferase (LUC) activity was determined
Superimposition of the binding mode of atorvastatin in model B with the K-80003-bound RXRα-LBD structure showed that atorvastatin occupied the space taken by two of the three K-80003 molecules (Fig. 7A). Visual examination revealed that the unoccupied hydrophobic region near Gln275 (Fig.7B) could accommodate the alkyl group in compounds 2a, 2b, 2c and 2d, which would support the results that 2a, 2b, 2c and 2d displayed stronger inhibition of the 9-cis-RA-induced Gal4 reporter activity than atorvastatin. Consistently docking results showed that the alkoxy moiety in 2a, 2b, 2c or 2d bound to the unoccupied hydrophobic region next to Gln275 (Fig.7B), resulting in tighter binding. In summary, the data suggested that atorvastatin could bind to the RXRα tetramer via the binding model B shown in Fig. 5B.
Fig. 7.
Binding mode exploration. (A) The orthogonal view of RXRα tetramer showing one of the hydrophobic symmetric voids (semitransparent grey) at the interface between two dimers. 3 bound K-80003 molecules are shown as blue balls. Docked atorvastatin is shown as red balls. (B) Docking results of 2a, 2b, 2c and 2d. Atorvastatin is shown in red sticks, 2a is shown in wheat sticks, 2b is shown in yellow sticks, 2c is shown in green sticks and 2d is shown in purple sticks. Binding cavity is shown in semitransparent grey surface.
3. Conclusion
Atorvastatin, also known as Lipitor, is a synthetic HMG-CoA reductase inhibitor which has been widely used for lowering cholesterol. Here, we report that atorvastatin could target the binding site of the tetrameric RXRα-LBD via structure-based virtual screening. Further experiments confirmed that atorvastatin could induce RXRα-LBD tetramerization and had potential anticancer effects. In addition, we explored the binding model of atorvastatin through docking studies, chemical synthesis and biological evaluation. Our results demonstrated that molecules such as atorvastatin, larger than K-80003, can mimic 2 or more copies of K-80003 and occupy the large cavity in the RXRα-LBD tetramer to stabilize RXRα-LBD tetramer. Virtual screening is a feasible approach for discovering such molecules as novel RXRα ligands. Furthermore, out results imply that atorvastatin could be optimized as a lead compound targeting RXRα for anticancer therapy.
4. Experimental section
4.1. Virtual screening
Schrodinger suite (Schrödinger, LLC, New York, NY, 2016) was employed in our structure-based virtual screening. The 2D structures of the compounds were downloaded from DrugBank (www.drugbank.ca) and transformed to 3D by LigPrep module in Maestro 10.5. The crystal structure of RXRα tetramer was retrieved from the Protein Data Bank (www.rcsb.org) (PDB code: 5TBP) and was prepared by Protein Preparation Wizard module in Maestro 10.5. Molecular docking was performed by Glide module in Maestro 10.5. SP (standard precision) scoring was used. 5 poses per ligand were performed to post-docking minimization and the best of these 5 poses was collected. After docking, the top 10 compounds with the highest docking scores were selected for further evaluation using the following criteria: 1. How well the docked molecule interacts with the protein; 2. If the docked molecule occupies a reasonable size of the binding site; 3. If the compound is commercially available. Based on these 3 criteria, 2 compounds were selected and purchased for biological testing.
4.2. Biology
4.2.1. Cell Culture and Transfection
Human breast cancer cell line MCF-7 were cultured in MEM medium supplemented with 10% fetal bovine serum. These cells were maintained at 5% CO2 at 37 °C. Subconfluent cells with exponential growth were used throughout the experiments. Cell transfections were carried out by using Lipofectamine 2000 (Invitrogen) according to the instructions of the manufacturer.
4.2.2. Dual-Luciferase Reporter Assay
The experiments were performed as described previously [21]. Briefly, cells were seeded at a density of 3×103 cells/well in a 48-well plate. Cells were transfected with the corresponding plasmids for 24 hours and then treated with compounds for 18 hours. Cells were lysed and luciferase relative activity was tested by the Dual-Luciferase Reporter Assay System according to the manufacturer’s instructions. Transfection efficiency was normalized to Renilla luciferase activity.
4.2.3. Western Blotting
The experiments were performed as described previously [21]. Briefly, cells were seeded at a density of 5×104 cells/well in a 12-well plate. Cells were transfected with siRNA control or siRXR for 48 hours and then treated with compounds for 12 hours. After treatment, cell lysates were prepared using NP-40 buffer. Cell lysates (determined by the Bradford protein assay) were boiled in sodium dodecyl sulfate (SDS) sample loading buffer, 20 μg of protein extracts from compounds-treated MCF-7 cells resolved by 10% SDS–polyacrylamide gel electrophoresis (SDS–PAGE) and transferred to nitrocellulose. The membranes were blocked in 5% milk in Tris-buffered saline and Tween 20 (TBST; 10 mM Tris–HCl [pH 8.0], 150 mM NaCl, 0.05% Tween 20) for 1 h at room temperature. After washing twice with TBST, the membranes were incubated with appropriate primary antibodies in TBST for 1 h and then washed thrice at room temperature, probed with horseradish peroxide-linked anti-immunoglobulin. After three washes with TBST, immunoreactive products were visualized using enhanced chemiluminescence reagents and autoradiography.
4.2.4. Antibody and reagents
PARP (Cat. 9542) was purchased from Cell Signal Technology (Beverly, MA, USA). RXRα (D-20) (Cat. sc- 553) and GAPDH (Cat. 47724) were purchased from Santa Cruz Biotechnology (Santa Cruz, CA, USA). SiRNA control and SiRXR were from Sigma.
4.2.5. Protein expression and purification
The human RXRα-LBD (223–462) was cloned as an N-terminal histidine-tagged fusion protein in pET15b expression vector and overproduced in Escherichia coli BL21 DE3 strain. Briefly, cells were harvested and sonicated, and the extract was incubated with the His60 Ni Superflow resin. The protein-resin complexes were washed and eluted with imidazole. The eluent was collected and concentrated to 5 mg/mL for subsequent trials.
4.2.6. Non-denaturing gel electrophoresis
Purified RXRα-LBD protein (0.2 mg mL−1) was incubated with DMSO, 9-cis-RA (0.5 mM), and/or K-80003, atorvastatin (20 mM) for 12 h at 4 °C in a total volume of 20 mL, and proteins were separated by 8% non-denaturing PAGE followed by Coomassie Bright Blue staining.
4.2.7. Fluorescence measurements
Fluorescence measurements were performed on Agilent Cary Eclipse Fluorescence Spectrophotometer using 10 mm quartz cuvette. The protein concentration of 1 mM was used for RXRα-LBD. Working solutions compounds were made in 1 mM DMSO, and used in a concentration range of to 40 μM. Protein was excited at 280 nm and the emission spectra were recorded between 290 and 450 nm at 25 °C using slits with a 5 nm band pass for excitation and emission, respectively. Fluorescence data were fitted to binding curves using the methods reported for dissociation constant (Kd) calculation [32]. All experiments were performed in triplicates and data were processed using the software Origin 2016.
4.3. Chemistry
4.3.1. General information
All commercially available starting materials and solvents were reagent grade and were purchased from Energy-chemical and used without further purification. Atorvastatin sodium and Lercanidipine hydrochloride were purchased from Hubei Jusheng Technology Co. Ltd. and its purity was 98%.
1H and 13C NMR were recorded on a Bruker Spectrospin DPX 600 MHz and Bruker Spectrospin DPX 151 MHz spectrometer, respectively using CDCl3 as a solvent and trimethylsilane (TMS) as the internal standard. Splitting patterns are designated as follows; s = singlet; d = doublet; t = triplet; m = multiplet; br = broad; J = coupling constant in hertz (Hz). Chemical shift values are given in ppm. HRMS were recorded by ESI-MS (Thermo Scientific Q Exactive).
4.3.2. General procedure for synthesis of 2a-2b
To a stirred solution of atorvastatin sodium (58 mg, 0.1 mmol) in ROH (2 mL) was added concentrated sulfuric acid (30 μL) in dropwise at 0 °C over 5 min. The resulting reaction mixture was stirred at room temperature for 2.5 h. Then 10% NaHCO3 aq. (5 mL) was added to quench the reaction. The resulting mixture was extracted with ethyl acetate. The organic phase was washed with saturated brine. The combined organic phase was dried over anhydrous MgSO4, and the solvent was removed under vacuum. The residue was purified by flash chromatography using gradient 12–66% ethyl acetate in hexanes to afford compound.
4.3.2.1. Methyl (3R,5R)-7-(2-(4-fluorophenyl)-5-isopropyl-3-phenyl-4-(phenylcarbamoyl)-1H-pyrrol-1-yl)-3,5-dihydroxyheptanoate (2a).
White solid, yield 88%. 1H NMR (Fig. S4, 600 MHz, CDCl3) δ 7.13 – 7.21 (m, 9H), 7.06 (d, J = 7.89 Hz, 2H), 6.97 – 7.02 (m, 3H), 6.86 (s, 1H), 4.08 – 4.19 (m, 2H), 3.91 – 3.97 (m, 1H), 3.73 – 3.77 (m, 1H), 3.71 (s, 3H), 3.69 (br. s., 1H), 3.63 (br. s., 1H), 3.57 (td, J = 7.13 and 14.17 Hz, 1H), 2.41 (d, J = 6.05 Hz, 2H), 1.61 – 1.71 (m, 2H), 1.54 (d, J = 6.97 Hz, 6H), 1.43 – 1.50 (m, 1H), 1.24 – 1.29 (m, 1H). 13C NMR (Fig. S4, 151 MHz, CDCl3) δ 173.2, 165.0, 162.4 (d, 1JC–F = 247.6 Hz), 141.6, 138.5, 134.7, 133.3(d, 3JC–F = 7.7 Hz), 130.6, 128.8, 128.8, 128.5, 128.4, 126.7, 123.7, 121.9, 119.7, 115.5(d, 2JC–F = 22 Hz), 115.4, 69.8, 69.1, 52.1, 41.8, 41.4, 41.2, 39.2, 26.3, 21.9, 21.8. HRMS (ESI): m/z [M + H]+ calcd for C34H38FN2O5, 573.2759; found, 573.2756.
4.3.2.2. Ethyl (3R,5R)-7-(2-(4-fluorophenyl)-5-isopropyl-3-phenyl-4-(phenylcarbamoyl)-1H-pyrrol-1-yl)-3,5-dihydroxyheptanoate (2b).
White solid, yield 68%. 1H NMR (Fig. S5, 600 MHz, CDCl3) δ 7.14 – 7.21 (m, 9H), 7.06 (d, J = 7.89 Hz, 2H), 6.96 – 7.02 (m, 3H), 6.86 (s, 1H), 4.09 – 4.19 (m, 4H), 3.91 – 3.97 (m, 1H), 3.72 – 3.77 (m, 1H), 3.70 (br. s., 1H), 3.62 (s, 1H), 3.58 (td, J = 7.15 and 14.31 Hz, 1H), 2.40 (d, J = 6.05 Hz, 2H), 1.63 – 1.69 (m, 2H), 1.54 (d, J = 7.15 Hz, 6H), 1.44 – 1.50 (m, 1H), 1.25 – 1.28 (m, 4H). 13C NMR (Fig. S5, 151 MHz, CDCl3) δ 172.8, 165.0, 162.4(d, 1JC–F = 247.6 Hz), 141.7, 138.5, 134.8, 133.3(d, 3JC–F = 8.8 Hz), 130.6, 128.9, 128.8, 128.5, 128.5, 126.7, 123.6, 122.0, 119.7, 115.5(d, 2JC–F = 20.9 Hz), 69.8, 69.1, 61.1, 41.9, 41.4, 39.2, 26.3, 21.9, 21.8, 14.3. HRMS (ESI): m/z [M + H]+ calcd for C35H40FN2O5, 587.2916; found, 587.2915.
4.3.3. General procedure for synthesis of 2c-2d
To a stirred solution of atorvastatin sodium (58 mg, 0.1 mmol) in dichloromethane (2 mL) and ROH (1 mL) was added concentrated sulfuric acid (40 μL) in dropwise at 0 °C over 5 min. The resulting reaction mixture was stirred at room temperature for 2.5 h. Then 10% NaHCO3 aq. (5 mL) was added to quench the reaction. The resulting mixture was extracted with ethyl acetate. The organic phase was washed with saturated brine. The combined organic phase was dried over anhydrous MgSO4, and the solvent was removed under vacuum. The residue was purified by flash chromatography using gradient 12–66% ethyl acetate in hexanes to afford compound.
4.3.3.1. Butyl (3R,5R)-7-(2-(4-fluorophenyl)-5-isopropyl-3-phenyl-4-(phenylcarbamoyl)-1H-pyrrol-1-yl)-3,5-dihydroxyheptanoate (2c).
White solid, yield 59%. 1H NMR (Fig. S6, 600 MHz, CDCl3) δ 7.13 – 7.23 (m, 9H), 7.07 (d, J = 7.89 Hz, 2H), 6.96 – 7.03 (m, 3H), 6.86 (s, 1H), 4.08 – 4.18 (m, 4H), 3.90 – 3.98 (m, 1H), 3.72 – 3.77 (m, 1H), 3.68 (br. s., 1H), 3.60 (br. s., 1H), 3.54 – 3.59 (m, 1H), 2.40 (d, J = 6.05 Hz, 2H), 1.66 – 1.71 (m, 1H), 1.59 – 1.64 (m, 3H), 1.54 (d, J = 7.15 Hz, 6H), 1.43 – 1.51 (m, 1H), 1.33 – 1.41 (m, 2H), 1.25 – 1.28 (m, 1H), 0.93 (t, J = 7.43 Hz, 3H). 13C NMR (Fig. S6, 151 MHz, CDCl3) δ 172.9, 164.9, 162.4 (d, 1JC–F = 248.7 Hz), 141.7, 138.5, 134.8, 133.3 (d, 3JC–F = 7.7 Hz), 130.6, 128.9, 128.8, 128.5, 126.7, 123.6, 122.0, 119.7, 115.5 (d, 2JC–F = 22 Hz), 69.8, 69.2, 65.0, 41.9, 41.4, 41.4, 39.2, 30.6, 26.3, 21.9, 21.8, 19.2, 13.8. HRMS (ESI): m/z [M + H]+ calcd for C37H44FN2O5, 615.3229; found, 615.3227.
4.3.3.2. Hexyl (3R,5R)-7-(2-(4-fluorophenyl)-5-isopropyl-3-phenyl-4-(phenylcarbamoyl)-1H-pyrrol-1-yl)-3,5-dihydroxyheptanoate (2d).
Colorless oil, yield 70%. 1H NMR (Fig. S7, 600 MHz, CDCl3) δ 7.13 – 7.21 (m, 9H), 7.06 (d, J = 7.70 Hz, 2H), 6.96 – 7.02 (m, 3H), 6.86 (s, 1H), 4.08 – 4.18 (m, 4H), 3.91 – 3.97 (m, 1H), 3.72 – 3.77 (m, 1H), 3.69 (br. s., 1H), 3.61 (br. s., 1H), 3.55 – 3.59 (m, 1H), 2.40 (d, J = 6.05 Hz, 2H), 1.67 – 1.71 (m, 1H), 1.60 – 1.65 (m, 3H), 1.54 (d, J = 7.15 Hz, 6H), 1.44 – 1.51 (m, 1H), 1.26 – 1.35 (m, 7H), 0.89 (t, J = 6.69 Hz, 3H). 13C NMR (Fig. S7, 151 MHz, CDCl3) δ 172.9, 164.9, 162.4 (d, 1JC–F = 247.6 Hz), 141.7, 138.5, 134.8, 133.3 (d, 3JC–F = 8.8 Hz), 130.6, 128.9, 128.8, 128.5, 126.7, 123.6, 122.0, 119.7, 115.5 (d, 2JC–F = 20.9 Hz), 69.8, 69.1, 65.3, 41.9, 41.4, 41.4, 39.2, 31.5, 28.6, 26.3, 25.6, 22.6, 21.9, 21.8, 14.1. HRMS (ESI): m/z [M + H]+ calcd for C39H48FN2O5, 643.3542; found, 643.3539.
Supplementary Material
Highlights:
A pilot virtual screening was performed to identify RXRα-LBD tetramer stablizers.
Atorvastatin was identified to bind to RXRα and promote the RXRα-LBD tetramer formation
Atorvastatin induces PARP cleavage in RXRα-dependent manner.
Acknowledgements
This work was supported by the grants from the National Nature Science Fund of China (NSFC-31271453, NSFC-31471318, NSFC-91129302 and NSFC-81301888), the Fundamental Research Funds for the Central Universities (2013121038, 20720180052), the Natural Science Foundation of Fujian Province of China (No. 2018J01133), and the US National Institutes of Health (CA198982)
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Altucci L, Leibowitz MD, Ogilvie KM, de Lera AR, Gronemeyer H, RAR and RXR modulation in cancer and metabolic disease, Nat Rev Drug Discov 6(10) (2007) 793–810. [DOI] [PubMed] [Google Scholar]
- [2].Evans RM, Mangelsdorf DJ, Nuclear Receptors, RXR, and the Big Bang, Cell 157(1) (2014) 255–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Lefebvre P, Benomar Y, Staels B, Retinoid X receptors: common heterodimerization partners with distinct functions, Trends Endocrin Met 21(11) (2010) 676–683. [DOI] [PubMed] [Google Scholar]
- [4].Szanto A, Narkar V, Shen Q, Uray IP, Davies PJA, Nagy L, Retinoid X receptors: X-ploring their (patho)physiological functions, Cell Death Differ 11 (2004) S126–S143. [DOI] [PubMed] [Google Scholar]
- [5].Cramer PE, Cirrito JR, Wesson DW, Lee CYD, Karlo JC, Zinn AE, Casali BT, Restivo JL, Goebel WD, James MJ, Brunden KR, Wilson DA, Landreth GE, ApoE-Directed Therapeutics Rapidly Clear beta-Amyloid and Reverse Deficits in AD Mouse Models, Science 335(6075) (2012) 1503–1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Su Y, Zeng ZP, Chen ZW, Xu D, Zhang WD, Zhang XK, Recent Progress in the Design and Discovery of RXR Modulators Targeting Alternate Binding Sites of the Receptor, Curr Top Med Chem 17(6) (2017) 663–675. [DOI] [PubMed] [Google Scholar]
- [7].Zhang XK, Zhou H, Su Y, Targeting truncated RXR alpha for cancer therapy, Acta Bioch. Bioph. Sin 48(1) (2016) 49–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Egea PF, Mitschler A, Rochel N, Ruff M, Chambon P, Moras D, Crystal structure of the human RXR alpha ligand-binding domain bound to its natural ligand: 9-cis retinoic acid, Embo J 19(11) (2000) 2592–2601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Dominguez M, Alvarez S, de Lera AR, Natural and Structure-based RXR Ligand Scaffolds and Their Functions, Curr Top Med Chem 17(6) (2017) 631–662. [DOI] [PubMed] [Google Scholar]
- [10].Dawson MI, Zhang XK, Discovery and design of retinoic acid receptor and retinoid X receptor class- and subtype-selective synthetic analogs of all-trans-retinoic acid and 9-cis-retinoic acid, Curr Med Chem 9(6) (2002) 623–637. [DOI] [PubMed] [Google Scholar]
- [11].Querfeldt C, Nagelli LV, Rosen ST, Kuzel TM, Guitart J, Bexarotene in the treatment of cutaneous T-cell lymphoma, Expert Opin Pharmaco 7(7) (2006) 907–915. [DOI] [PubMed] [Google Scholar]
- [12].Cao X, Liu W, Lin F, Li H, Kolluri SK, Lin B, Han Y-H, Dawson MI, Zhang X.-k., Retinoid X receptor regulates Nur77/TR3-dependent apoptosis by modulating its nuclear export and mitochondrial targeting., Mol Cell Biol 24(22) (2004) 9705–9725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Casas F, Daury L, Grandemange S, Busson M, Seyer P, Hatier R, Carazo A, Cabello G, Wrutniak-Cabello C, Endocrine regulation of mitochondrial activity: involvement of truncated RXRalpha and c-Erb Aalpha1 proteins, FASEB J 17(3) (2003) 426–36. [DOI] [PubMed] [Google Scholar]
- [14].Katagiri Y, Takeda K, Yu ZX, Ferrans VJ, Ozato K, Guroff G, Modulation of retinoid signalling through NGF-induced nuclear export of NGFI-B, Nat Cell Biol 2(7) (2000) 435–40. [DOI] [PubMed] [Google Scholar]
- [15].Ghose R, Zimmerman TL, Thevananther S, Karpen SJ, Endotoxin leads to rapid subcellular re-localization of hepatic RXRalpha: A novel mechanism for reduced hepatic gene expression in inflammation, Nucl Recept 2(1) (2004) 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Zimmerman TL, Thevananther S, Ghose R, Burns AR, Karpen SJ, Nuclear export of retinoid X receptor alpha in response to interleukin-1beta-mediated cell signaling: roles for JNK and SER260, J Biol Chem 281(22) (2006) 15434–40. [DOI] [PubMed] [Google Scholar]
- [17].Fukunaka K, Saito T, Wataba K, Ashihara K, Ito E, Kudo R, Changes in expression and subcellular localization of nuclear retinoic acid receptors in human endometrial epithelium during the menstrual cycle, Mol Hum Reprod 7(5) (2001) 437–46. [DOI] [PubMed] [Google Scholar]
- [18].Wang GH, Jiang FQ, Duan YH, Zeng ZP, Chen F, Dai Y, Chen JB, Liu JX, Liu J, Zhou H, Chen HF, Zeng JZ, Su Y, Yao XS, Zhang XK, Targeting truncated retinoid X receptor-alpha by CF31 induces TNF-alpha-dependent apoptosis, Cancer research 73(1) (2013) 307–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Zhou H, Liu W, Su Y, Wei Z, Liu J, Kolluri SK, Wu H, Cao Y, Chen J, Wu Y, Yan T, Cao X, Gao W, Molotkov A, Jiang F, Li WG, Lin B, Zhang HP, Yu J, Luo SP, Zeng JZ, Duester G, Huang PQ, Zhang XK, NSAID sulindac and its analog bind RXR alpha and inhibit RXR alpha-dependent AKT signaling, Cancer Cell 17(6) (2010) 560–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Lin B, Kolluri S, Lin F, Liu W, Han YH, Cao X, Dawson MI, Reed JC, and Zhang XK, Conversion of Bcl-2 from Protector to Killer by Interaction with Nuclear Orphan Receptor Nur77/TR3, Cell 116 (2004) 527–540. [DOI] [PubMed] [Google Scholar]
- [21].Chen L, Wang ZG, Aleshin AE, Chen F, Chen J, Jiang F, Alitongbieke G, Zeng Z, Ma Y, Huang M, Zhou H, Cadwell G, Zheng JF, Huang PQ, Liddington RC, Zhang XK, Su Y, Sulindac-derived RXRalpha modulators inhibit cancer cell growth by binding to a novel site, Chem Biol 21(5) (2014) 596–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Zeng Z, Sun Z, Huang M, Zhang W, Liu J, Chen L, Chen F, Zhou Y, Lin J, Huang F, Xu L, Zhuang Z, Guo S, Alitongbieke G, Xie G, Xu Y, Lin B, Cao X, Su Y, Zhang X.-k., Zhou H, Nitrostyrene Derivatives Act as RXR alpha Ligands to Inhibit TNF alpha Activation of NF-kappa B, Cancer research 75(10) (2015) 2049–2060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Chen L, Aleshin AE, Alitongbieke G, Zhou Y, Zhang X, Ye X, Hu M, Ren G, Chen Z, Ma Y, Zhang D, Liu S, Gao W, Cai L, Wu L, Zeng Z, Jiang F, Liu J, Zhou H, Cadwell G, Liddington RC, Su Y, Zhang XK, Modulation of nongenomic activation of PI3K signalling by tetramerization of N-terminally-cleaved RXRalpha, Nat Commun 8 (2017) 16066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Lionta E, Spyrou G, Vassilatis DK, Cournia Z, Structure-Based Virtual Screening for Drug Discovery: Principles, Applications and Recent Advances, Curr Top Med Chem 14(16) (2014) 1923–1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Chen F, Liu J, Huang M, Hu M, Su Y, Zhang XK, Identification of a New RXRalpha Antagonist Targeting the Coregulator-Binding Site, ACS Med Chem Lett 5(7) (2014) 736–741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Law V, Knox C, Djoumbou Y, Jewison T, Guo AC, Liu Y, Maciejewski A, Arndt D, Wilson M, Neveu V, Tang A, Gabriel G, Ly C, Adamjee S, Dame ZT, Han B, Zhou Y, Wishart DS, DrugBank 4.0: shedding new light on drug metabolism, Nucleic Acids Res 42(Database issue) (2014) D1091–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Schrödinger, Release 2016–1: LigPrep, Version 3.7, Schrödinger, LLC, New York, NY, 2016. [Google Scholar]
- [28].Sastry GM, Adzhigirey M, Day T, Annabhimoju R, Sherman W, Protein and ligand preparation: parameters, protocols, and influence on virtual screening enrichments, J Comput Aided Mol Des 27(3) (2013) 221–34. [DOI] [PubMed] [Google Scholar]
- [29].Schrödinger, Release 2016–1: Prime, Version 4.3, Schrödinger, LLC, New York, NY, 2016. [Google Scholar]
- [30].Schrödinger, Release 2016–1: Glide, Version 7.0, Schrödinger, LLC, New York, NY, 2016. [Google Scholar]
- [31].Nahoum V, Perez E, Germain P, Rodriguez-Barrios F, Manzo F, Kammerer S, Lemaire G, Hirsch O, Royer CA, Gronemeyer H, de Lera AR, Bourguet W, Modulators of the structural dynamics of the retinoid X receptor to reveal receptor function, P Natl Acad Sci USA 104(44) (2007) 17323–17328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Wang ZX, Kumar NR, Srivastava DK, A Novel Spectroscopic Titration Method for Determining the Dissociation-Constant and Stoichiometry of Protein Ligand Complex, Anal Biochem 206(2) (1992) 376–381. [DOI] [PubMed] [Google Scholar]
- [33].Zhao H, Caflisch A, Molecular dynamics in drug design, Eur J Med Chem 91 (2015) 4–14. [DOI] [PubMed] [Google Scholar]
- [34].Conda-Sheridan M, Park EJ, Beck DE, Reddy PV, Nguyen TX, Hu B, Chen L, White JJ, van Breemen RB, Pezzuto JM, Cushman M, Design, synthesis, and biological evaluation of indenoisoquinoline rexinoids with chemopreventive potential, Journal of medicinal chemistry 56(6) (2013) 2581–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.