Abstract
Objectives
Acute kidney injury (AKI) in hospitalized patients puts them at much higher risk for developing future health problems such as chronic kidney disease, stroke, and heart disease. Accurate AKI prediction would allow timely prevention and intervention. However, current AKI prediction researches pay less attention to model building strategies that meet complex clinical application scenario. This study aims to build and evaluate AKI prediction models from multiple perspectives that reflect different clinical applications.
Materials and Methods
A retrospective cohort of 76 957 encounters and relevant clinical variables were extracted from a tertiary care, academic hospital electronic medical record (EMR) system between November 2007 and December 2016. Five machine learning methods were used to build prediction models. Prediction tasks from 4 clinical perspectives with different modeling and evaluation strategies were designed to build and evaluate the models.
Results
Experimental analysis of the AKI prediction models built from 4 different clinical perspectives suggest a realistic prediction performance in cross-validated area under the curve ranging from 0.720 to 0.764.
Discussion
Results show that models built at admission is effective for predicting AKI events in the next day; models built using data with a fixed lead time to AKI onset is still effective in the dynamic clinical application scenario in which each patient’s lead time to AKI onset is different.
Conclusion
To our best knowledge, this is the first systematic study to explore multiple clinical perspectives in building predictive models for AKI in the general inpatient population to reflect real performance in clinical application.
Keywords: acute kidney injury, predictive modeling, prediction, machine learning, electronic medical record
INTRODUCTION
Acute kidney injury (AKI) is a sudden episode of kidney damage or failure occurring within a few hours or a few days, affecting 7–18% of hospitalized patients and more than 50% of patients in the intensive care unit (ICU).1 Despite preventive measures implemented in hospitals, the incidence rate of AKI is still increasing 11% annually in recent years and has increased by at least 20 times in the past 25 years.2 AKI has a mortality rate as high as 20% in general wards and can be up to 50% in the ICU and causes a significant increase in hospitalization costs that range from $5.4 to $24.0 billion in the United States.2,3 According to statistics, 2 million people die from AKI every year worldwide.4 Compared with patients without AKI, patients with AKI have an increased hospital length of stay of 3.2 days and increased hospitalization costs of $7933 per person.5 In addition, patients who recover from AKI still has an increased risk for developing chronic kidney disease and end-stage renal disease in the future.6
However, it is very difficult to recognize AKI risk early until too late because when AKI can be diagnosed by increases in serum creatinine (SCr) or decreases in urine output, kidney injury has already occurred. The pathogenesis underlying AKI is complex, determined by the interaction of multiple susceptible factors including advanced age and diabetes and exposure to insults, for example, sepsis and nephrotoxic medications. Recognizing patients at high AKI risk and treat them accordingly is more likely to result in better outcomes for patients than merely treating established AKI.
Current research in AKI prediction mainly focuses on patient in the ICU,7 or following certain diseases such as acute heart failure,8,9 burn,10 and so on, or certain surgeries such as cardiac surgery,11–13 lung transplantation,14 etc. There exists relatively less research on prediction models for AKI in the general hospitalized patients based on electronic medical record (EMR) data. Matheny et al15 presented one of the first risk stratification tools for predicting hospital acquired AKI (HA-AKI) utilizing EMR data and evaluated calibration drift in regression and machine learning models for AKI prediction over time.16 Cronin et al17 explored multiple machine learning methods for building risk stratification models for HA-AKI using National Veterans Health Administration data. Koyner et al18,19 adapted a discrete time survival analysis framework and Gradient Boosting Machine algorithm for AKI prediction and demonstrated excellent accuracy across different patient locations and admission SCr. In one of our previous work, we built machine learning models to predict AKI and evaluated the performance with a lead time from 1 to 3 days prior to the onset of AKI.20 Kate et al21 built machine learning models to predict who would develop AKI after 24 h of admission and detect AKI anytime during hospitalization in older adults, which is the first study that compared the performance between prediction and detection of AKI.
The existing studies discussed above have made important contributions to AKI prediction in the general hospital populations. The most commonly adopted model development and evaluation strategy is using AKI-onset as the anchor point for data extraction, which is an effective approach in retrospective data analysis to demonstrate the feasibility of predicting AKI early. However, how close the performance of the models built in such way reflects the actual prediction performance in clinical application where data is collected prospectively is an unknown. In effort to conduct a more comprehensive assessment of AKI prediction models, this study explores 4 evaluation perspectives designed to answer different AKI prediction questions faced in clinical practice: (1) predicting AKI from data before onset; (2) predicting AKI risk during hospital stay using only data at admission; (3) predicting if AKI will occur within a window of time from admission data and prior patient medical history; and (4) predicting if AKI will develop in the following day. The 4 evaluation perspectives imply different model building procedures with respect to data collection and prediction windows. The multi-perspective experiments offer a more comprehensive analysis of the current state of AKI prediction, which may provide useful guidance for future predictive modeling for clinical practice.
METHODS
Study population
A retrospective cohort was built including 96 590 adult inpatients older than 18 years of age at a tertiary care, academic hospital (University of Kansas Health System – KUHS) from November 2007 to December 2016 with a length of stay of at least 2 days. The total number of encounters was179 370, considering there may be multiple admissions (encounters) of a patient. We excluded patient hospitalizations missing necessary data for outcome determination, that is, without enough SCr measurements (<2 times) for determining AKI. We also excluded patient admissions that had evidence of moderate or severe kidney dysfunction, that is, estimated glomerular filtration rate (eGFR) less than 60 mL/min/1.73 m2 or abnormal SCr level more than 1.3 mg/dL within 24 h of hospital admission. Although patients with reduced eGFR are at increased risk for AKI, we made the exclusion in this study because it is difficult to determine which of these patients had hospital-acquired versus community-acquired AKI without adequate longitudinal assessment of kidney function. The final analysis cohort consisted of 76 957 encounters among 96 590 inpatients.
AKI was defined using the Kidney Disease Improving Global Outcomes (KDIGO) SCr criteria. Baseline SCr level was defined as either the last measurement within 2-day time window prior to hospital admission or the first SCr measured after hospital admission. All SCr levels measured between admission and discharge were evaluated to determine the occurrence of HA-AKI. Out of total 76 957 encounters in the final analysis cohort, AKI events occurred in 7259 encounters and 69 698 encounters had no AKI events. The date distribution of those patients developing AKI in reference to their admission day is shown in the Supplementary Figure A1.
Data collection
KUHS’s de-identified clinical data repository HERON (Health Enterprise Repository for Ontological Narration)22,23 was queried to obtain clinical variables corresponding to each encounter in the final analysis cohort. De-identified data request was approved by the HERON Data Request Oversight Committee. Structured clinical variables extracted for each encounter included demographic information, vital signs, laboratory values, admission diagnosis, comorbidities, medications before admission, medication during hospitalization, and medical history, summing up to 1917 attributes. Among these, vital signs, laboratory values, medications, and medical history are associated with time stamps on when the values were recorded. Because of the longitudinal nature of the data, we could extract data according to different prediction windows and evaluation strategies for building predictive models.
A summary of clinical variables used to build the AKI prediction models is described in Table 1. SCr and eGFR were not included as predictive variables as they were used to determine AKI versus non-AKI encounters. For laboratory tests and vitals, only the last recorded value before a prediction point was used and their values were categorized as either “present and normal,” “present and abnormal,” or “unknown” according to standard reference ranges. Vitals were categorized into groups as described in our earlier work.20 Missing values in vitals and lab tests were captured as “unknowns” because information may be contained in the choice to not perform the measurement.
Table 1.
Clinical variables considered in building AKI predictive models
| Feature category | Number of variables | Details |
|---|---|---|
| Demographics | 3 | Age, gender, and race |
| Vitals | 5 | BMI, diastolic BP, systolic BP, pulse, and temperature |
| Lab tests | 14 | Albumin, ALT, AST, ammonia, blood bilirubin, BUN, Ca, CK-MB, CK, glucose, lipase, platelets, troponin, and WBC |
| Comorbidities | 29 | UHC comorbidity |
| Admission diagnosis | 315 | UHC APR-DRG |
| Medications | 1271 | All medications are mapped to RxNorm ingredient |
| Medical history | 280 | ICD9 codes mapped to CCS major diagnoses |
Abbreviations: AKI: acute kidney injury; BMI: body mass index; BP: blood pressure; ALT: alanine aminotransferase; AST: asparate aminotransferase; BUN: Blood Urea Nitrogen; CK-MB: Creatine Kinase-muscle/brain; WBC: white blood cell; UHC: University Healthsystem Consortium (http://www.vizientinc.com); APR-DRG: all patient refined diagnosis related group; CCS: Clinical Classifications Software; CK: Creatine Kinase.
All medication names were normalized by mapping to RxNorm ingredient. Only medication taken within 7 days before a prediction point was considered in the predictive models. Comorbidity and admission diagnosis, that is, all patient refined diagnosis related group variables, were collected from the University Health System Consortium (UHC), now known as Vizient (http://www.vizientinc.com), data source in HERON. Patient medical history was captured as major diagnoses (ICD-9 codes grouped according to the Clinical Classifications Software diagnosis categories by the Agency for Healthcare Research and Quality). Medical history, medication, comorbidity, and admission diagnosis variables took either “yes” or “no” values.
Vitals, labs, medical history, and medication variables were time-stamped relative to the admission date, referred here as time-dependent variables. Comorbidities, admission diagnosis, and demographics were presumed to be available at admission and not time dependent. AKI-onset was set as the day on which AKI can be diagnosed using the KDIGO criteria. Non-AKI onset was set as the day of the last normal SCr measurement for a patient during an encounter.
According to different prediction points and windows in various experimental settings, positive/negative examples were dynamically determined, while patients developing AKI before prediction point were excluded from a specific study cohort. Only those who have AKI occurred during a particular prediction window were regarded as positive examples. The specific process is described in the Supplementary Figure A2.
Experiment design
To comprehensively assess AKI prediction performance, we investigated the following 4 clinical perspectives that involve distinct prediction points and data collection windows. Each perspective has different model building and evaluation strategy to which we must pay attention, especially when we compare model performance across different perspectives and when we consider the effectiveness of utilizing these models in clinical practices.
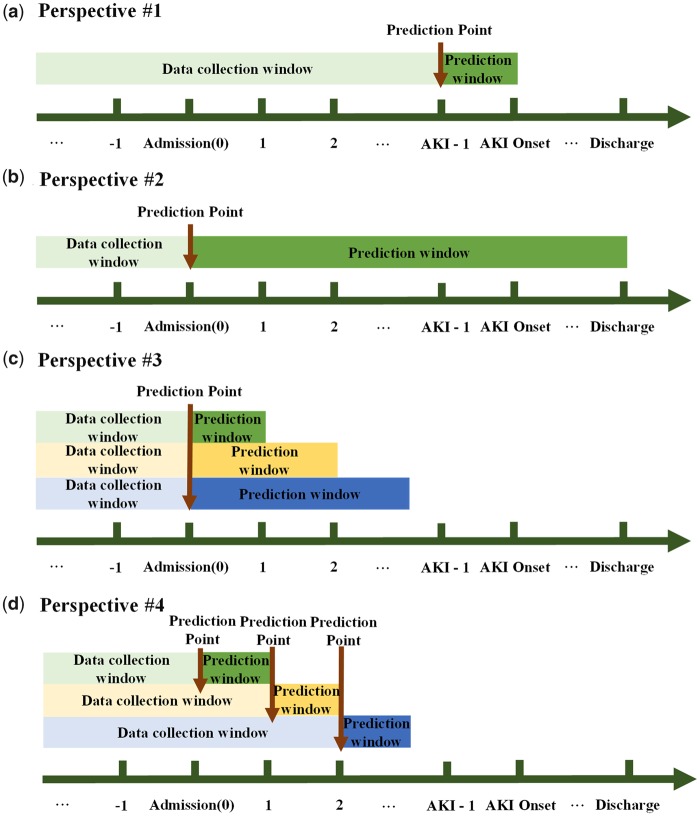
Perspective #1: Can we predict AKI before its onset using data before the onset time?
The perspective depicted in Figure 1(a) is the most commonly used approach in the medical informatics community to assess AKI prediction, that is, using AKI/non-AKI onset as the anchor points for prediction. Through this way, we can evaluate whether AKI can be predicted before its onset and how many days prior to onset accurate predictions can be made by changing the prediction window. From this perspective, we collected the latest data before AKI onset, excluding data on the day of AKI-onset, to evaluate the performance of prediction model, that is, data collection window is (past, AKI-onset—1 day), and prediction point is set at 1 day prior to AKI-onset.
Figure 1.
Different model building procedures in 4 perspectives. AKI: acute kidney injury. (a) Perspective #1 - Can we predict AKI before its onset using data before the onset time? (b) Perspective #2 - Can we predict at admission if AKI will occur for patients during their stay? (c) Perspective #3 - Can we predict at admission if AKI will occur within various numbers of days afterwards? (d) Perspective #4 - Can we predict if a patient will develop AKI within the next day in a clinical scenario?
Perspective #2: Can we predict at admission if AKI will occur for patients during their stay?
This perspective outlined in Figure 1(b) is to use admission day as the prediction point, predicting if a patient will develop AKI during their stay. This perspective is also used in current researches.17,21 Under this experiment, the data collection window becomes (past, admission) and the prediction window becomes (admission, discharge). This aligns with a clinical application scenario in which clinicians can use the models to make AKI risk prediction for everyone at admission and make subsequent treatment decisions.
Perspective #3: Can we predict at admission if AKI will occur within various numbers of days afterwards?
The purpose of this experiment is to explore whether there exists a validity period for predicting AKI following admission, as shown in Figure 1(c). Under this evaluation perspective, we used patient medical data before and include data collected on the admission day to predict a patient risk of developing HA-AKI in 1 day, 2 days, 3 days, 7 days, 15 days, and 30 days after admission. In this experiment, the data collection window is set at (past, admission) and upper bound of the prediction window varies with (admission, admission +1 day, 2 days, 3 days, 7 days, 15 days, and 30 days). The clinical application scenario for this experiment is using the models at admission time to predict patient AKI risk in the next 1 day, 2 days and etc., which can provide smaller granularity than that of Perspective #2. By comparing performance between these models, we can confirm how far ahead of time AKI can be effectively predicted.
Perspective #4: Can we predict if a patient will develop AKI within the next day in a clinical scenario?
This experiment allows us to assess whether the same performance for next-day AKI prediction made at admission carries through various number of days into the hospital stay as shown in Figure 1(d). This is a dynamic prediction perspective for clinical usage in which AKI risk prediction is made for patients at daily intervals after admission using the most recent data available before prediction point. The clinical application under this perspective requires a set of models built on dynamic data collection window, upper bound of which varies with (past, admission +0 day, 1 day, 2 days, 3 days, and 4 days), although only 1 model is required for clinical application in Perspectives #1 and #2.
Relations between perspectives
The relations between the above 4 perspectives are as follows. In Perspective #1, we use the data collected up to 1 day before AKI onset to evaluate whether we can predict AKI before its onset with a 1-day lead time. Although it is the widely used approach in the medical informatics community, the prerequisite is that we already know the date of AKI/non-AKI onset so that we could cut the data accordingly for model development and evaluation, but this information is not available in a real-life clinical application. Whether predictive models developed and evaluated on such data will generalize well to the real clinical application is not well understood. Perspective # 2 is designed to better reflect real clinical application, evaluating whether we can predict AKI event occurring on any day during hospitalization. Since Perspective #2 treats the entire hospital stay as a single long prediction window, prediction may not be granular enough, thus we designed Perspective #3 that builds a set of models with increasing length of the prediction windows to evaluate and compare the performance. Through Perspectives #2 and #3, we will be able to assess performance of the models built to predict possible AKI events at admission, but it is not enough. It is more clinical useful if a prediction model is available for predicting a patient’s next day AKI risk on a daily basis starting from admission to discharge using the most up to date data, thus we designed Perspective #4.
Although most current studies adopting the same model building strategy as in Perspective #1 claim that the model can predict AKI at any time before AKI event, their evaluation result may be overestimated. In real-life clinical application, the testing data would contain samples with AKI onsets occurring on any day after admission whereas the training data extracted according to Perspective #1 uses AKI onset as the reference cut point so that the distance from prediction point to the AKI onset is the same among all samples. To assess whether model built from Perspective #1 can truly predict AKI in a real clinical situation, in this study we evaluated the model built from training data extracted according to Perspective #1 using testing data extracted according to Perspective #4.
Experimental methodology
Waikato Environment for Knowledge Analysis (Weka).24 was used to implement the following machine learning algorithms: Logistic Regression (batchSize = 100; numDecimalPlaces = 4; ridge = 1.0E−8), Naïve Bayes, Bayes Net (estimator = SimpleEstimator; searchAlgorithm = k2; batchSize = 100), Random Forest (bagSizePercent = 100; batchSize = 100; maxDepth = 0; numDecimalPlaces = 2; numIterations = 500), and an ensemble model with voting classifiers combining logistic regression and random forest (LR&RF_VotingEnsemble). The selected algorithms are wide-applied machine learning methods in AKI prediction research. We chose to use logistic regression and random forest to build the ensemble model because they showed superior performance on our data in the exploratory experiments. To reduce feature dimensionality for computational efficiency, we used the RemoveUseless node in Weka to remove attributes that do not vary at all or vary too much in the training set with a default threshold of 99%. Through this process, we discarded approximately 1000 variables. All machine learning models were evaluated using 10-fold cross-validation. Area under the receiver operating characteristic (ROC) curve (AUC), sensitivity, specificity, precision, and F-measure were reported to assess the prediction performance of our models and their 95% confidence interval (CI) was also reported for each model. We used the ThresholdSelector in Weka to obtain the best classification threshold for maximum F-measure.
RESULTS
Sample size change
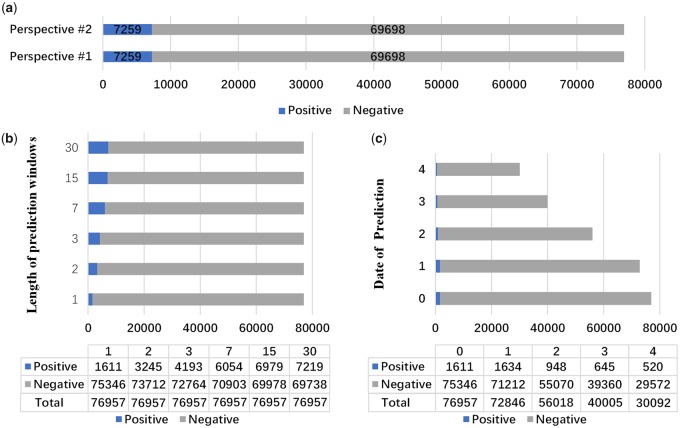
Different experimental design would result in very different sample size for model building and evaluation. In Perspectives #1 and #2, positive/negative samples do not change: there are 7259 positive samples and 69 698 negative samples as shown in Figure 2(a). The main difference is the data collection window.
Figure 2.
Sample size change in each perspective. AKI: acute kidney injury. (a) Perspective #1 vs #2; (b) Perspective #3; (c) Perspective #4.
In Perspective #3, the positive samples proportion increases with the increasing length of prediction window as shown in Figure 2(b).
In Perspective #4, the sample size becomes smaller over time as in Figure 2(c). Only patients who develop AKI 1 day after the prediction point would be considered as a positive sample. According to Supplementary Figure A2, those patients who have developed AKI or discharged before prediction point were excluded in the cohort. The number of patients in the study cohort were reduced to 30 092 (520 positive samples and 29 572 negative samples, 39.1% of total patients) at 4 days after admission.
Prediction performance
Table 2 summarizes the AUC and 95% CI obtained for AKI prediction models built and evaluated under clinical Perspectives #1 and #2. It is clear that the Ensemble method (the Voting classifier with Logistic Regression and Random Forest) achieved the best performance (AUC of 0.744 and 0.734 and F-measure of 0.330 and 0.318, respectively) in both evaluation strategies. Since the remaining Perspectives #3 and #4 are similar to Perspectives #1 and #2, the ensemble method is used as the base classifier in following experiments.
Table 2.
Performance of different methods on models in Perspectives #1 and #2
| Metrics | Naïve Bayes | Bayes net | Logistic regression | Random forest | LR&RF_VotingEnsemble |
|---|---|---|---|---|---|
| Models built in Perspective #1—data collection window (past, AKI-onset - 1 day) | |||||
| AUC | 0.687 (0.686–0.687) | 0.687 (0.687–0.687) | 0.726 (0.725–0.726) | 0.709 (0.708–0.710) | 0.744 (0.743–0.744) |
| F-measure | 0.261 (0.260–0.262) | 0.262 (0.261–0.262) | 0.317 (0.316–0.318) | 0.317 (0.316–0.318) | 0.330 (0.329–0.331) |
| Sensitivity (recall) | 47.6% (42.5–52.7%) | 47.5% (46.7–48.3%) | 40.6% (39.8–41.4%) | 40.7% (39.8–41.5%) | 40.3% (39.4–41.1%) |
| Specificity | 77.4% (76.8–77.9%) | 77.6% (77.0–78.1%) | 87.9% (87.5–88.4%) | 87.9% (87.4–88.4%) | 89.2% (88.8–89.6%) |
| Precision | 18.0% (17.8–18.1%) | 18.1% (17.9–18.2%) | 26.1% (25.9–26.4%) | 26.0% (25.6–26.4%) | 28.0% (27.7–28.3%) |
| Models built in Perspective #2–data collection window (past, admission) | |||||
| AUC | 0.676 (0.676–0.676) | 0.677 (0.677–0.677) | 0.719 (0.718–0.720) | 0.714 (0.713–0.715) | 0.734 (0.734–0.735) |
| F-measure | 0.253 (0.252–0.253) | 0.253 (0.252–0.254) | 0.308 (0.308–0.309) | 0.294 (0.293–0.295) | 0.318 (0.317–0.319 |
| Sensitivity (recall) | 45.4% (44.4–46.3%) | 45.7% (44.8–46.6%) | 40.3% (39.3–41.3%) | 40.4% (39.7–41.1%) | 40.6% (39.9–41.2%) |
| Specificity | 77.4% (76.8–77.9%) | 77.6% (77.0–78.1%) | 87.9% (87.5–88.4%) | 87.9% (87.4–88.4%) | 89.2% (88.8–89.6%) |
| Precision | 17.5% (17.4–17.6%) | 17.5% (17.4–17.7%) | 25.0% (24.6–25.3%) | 23.1% (22.8–23.4%) | 26.2% (25.8–26.5%) |
Abbreviations: AKI: acute kidney injury; AUC: area under the curve.
Evaluation results of the models built from Perspective #3 are shown in Table 3. It is apparent from Table 3 that the AUC of all models with different length of prediction window is higher than 0.72. The best length of prediction window is 1 day with an AUC of 0.764 (95% CI 0.762–0.766). The AUC reaches lowest level of 0.720 (95% CI 0.720–0.721) at prediction window of 3 days. As the length of prediction window increases, the F-measure increased from 0.184 to 0.316.
Table 3.
Performance of models in Perspective #3
| Metrics | 1 day | 2 days | 3 days | 7 days | 15 days | 30 days |
|---|---|---|---|---|---|---|
| Models built in Perspective #3—data collection window (past, admission) | ||||||
| AUC | 0.764 (0.762–0.766) | 0727 (0.726–0.728) | 0.720 (0.720–0.721) | 0.722 (0.722–0.722) | 0.730 (0.730–0.731) | 0.734 (0.734–0.734) |
| F-measure | 0.184 (0.182–0.186) | 0.213 (0.211–0.215) | 0.233 (0.231–0.234) | 0.278 (0.277–0.280) | 0.309 (0.308–0.310) | 0.316 (0.315–0.318) |
| Sensitivity (recall) | 18.1% (17.6–18.6%) | 23.8% (23.0–24.5%) | 28.1% (27.5–28.6%) | 37.0% (36.0–37.9%) | 38.6% (38.0–39.2%) | 40.8% (40.2–41.5%) |
| Specificity | 98.3% (98.2–98.4%) | 95.6% (95.4–95.9%) | 93.5% (93.2–93.7%) | 89.0% (88.6–89.5%) | 88.9% (88.6–89.3%) | 87.9% (87.4–88.3%) |
| Precision | 18.7% (18.1–19.3%) | 19.4% (18.9–19.8%) | 19.9% (19.6–20.2%) | 22.4% (22.0–22.7%) | 25.8% (25.5–26.1%) | 25.8% (25.5–26.2%) |
Abbreviation: AUC: area under the curve.
From Figure 2(b), we can see that the model to predict AKI in the next day (ie, prediction window length is 1 day) had least number of positive samples among all models in Perspective #3. Although the number of positive samples in training set is not the key to modeling performance, small number of positive examples for predictive modeling could still results in poor modeling performance. Since the model with the least number of samples performed the best as shown in Table 3, we want to further assess the validity of this model by following Kate et al21 to generate the model’s learning curve with respect to different sizes of training data as shown in Figure 3. As can be seen from Figure 3, the curve starts to flatten out as the percent of training data is over 50% and this model were little affected by a small amount of positive sample.
Figure 3.
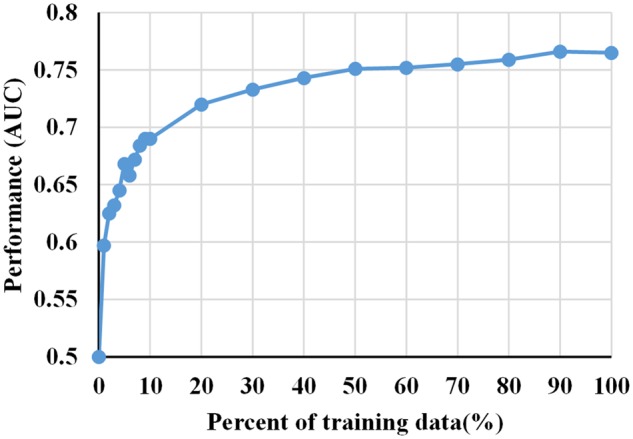
Learning curve of model built at admission to predict AKI in the next day in Perspective #3. AKI: acute kidney injury.
Table 4 shows the performance of different AKI prediction models used to predict AKI events within the next day from admission to 4 days after admission in Perspective #4. And we also used the evaluation strategy in Perspective #4 to evaluate the model built in Perspective #1 to confirm that we can use the model built from Perspective #1 to predict AKI in a real-life clinical application as Perspective #4. Figure 4 is a visual representation of AUC in Table 4. The most surprising aspect of the results is that the model built using Perspective #1 is better than models built from Perspective #4 in overall performance.
Table 4.
AUC of different AKI Prediction models at the first 5 days during hospitalization
| Metrics | Admission | Admission +1 | Admission +2 | Admission +3 | Admission +4 |
|---|---|---|---|---|---|
| Models built in Perspective #4 | |||||
| AUC | 0.764 (0.762–0.766) | 0.679 (0.677–0.681) | 0.652 (0.651–0.653) | 0.620 (0.616–0.624) | 0.600 (0.596–0.683) |
| F-measure | 0.184 (0.182–0.186) | 0.112 (0.111–0.114) | 0.066 (0.065–0.067) | 0.047 (0.045–0.049) | 0.049 (0.047–0.052) |
| Sensitivity (recall) | 18.1% (17.6–18.6%) | 16.4% (15.7–17.2%) | 15.2% (13.3–17.0%) | 10.0% (8.1–11.9%) | 12.7% (11.0–14.5%) |
| Specificity | 98.3% (98.2–98.4%) | 96.0% (95.7–96.2%) | 94.1% (93.2–94.9%) | 94.9% (93.8–96.0%) | 93.0% (91.8–94.1%) |
| Precision | 18.7% (18.1–19.3%) | 8.5% (8.3–8.7%) | 4.2% (4.1–4.4%) | 3.2% (2.9–3.4%) | 3.1% (2.9–3.3%) |
| Models built in Perspective #1 | |||||
| AUC | 0.736 (0.723–0.749) | 0.693 (0.679–0.706) | 0.667 (0.649–0.684) | 0.652 (0.622–0.682) | 0.648 (0.612–0.683) |
| F-measure | 0.108 (0.099–0.117) | 0.101 (0.094–0.107) | 0.067 (0.060–0.074) | 0.056 (0.048–0.064) | 0.060 (0.050–0.069) |
| Sensitivity (recall) | 46.6% (43.4–49.8%) | 39.8% (38.4–41.2%) | 37.5% (33.3–41.7%) | 37.4% (32.5–42.3%) | 41.8% (36.5–47.0%) |
| Specificity | 84.6% (83.5–85.8%) | 84.9% (83.6–86.2%) | 83.1% (81.7–84.6%) | 80.3% (78.7–82.0%) | 78.8% (77.0–80.6%) |
| Precision | 6.1% (5.6–6.6%) | 5.8% (5.3–6.2%) | 3.7% (3.3–4.1%) | 3.0% (2.6–3.5%) | 3.2% (2.7–3.8%) |
Abbreviations: AKI: acute kidney injury; AUC: area under the curve.
Figure 4.
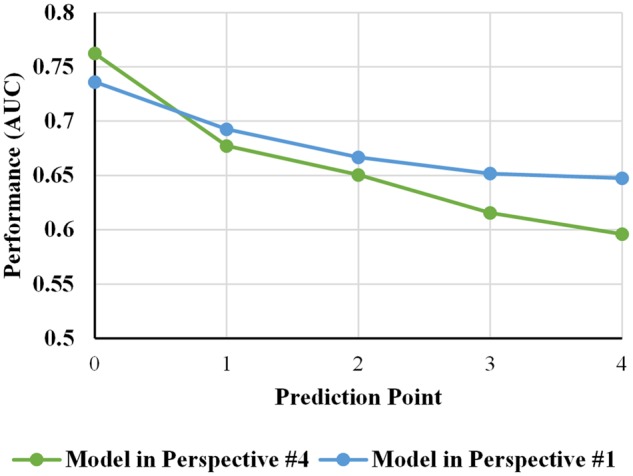
Comparison of performance of different AKI prediction models in Perspectives #1 and #4. AKI: acute kidney injury.
DISCUSSION
In predicting AKI events 1 day before its onset, the ensemble prediction model developed and evaluated on data extracted according to Perspective #1 achieved the best AUC of 0.744. However, as discussed above in the experiment design, the testing data corresponding to this evaluation result does not conform to the real-life clinical application, which suggests that the model may not achieve the same performance in clinical application. To investigate prediction performance in more realistic clinical application scenarios, we made AKI predictions at hospital admission for forecasting patient risk of developing AKI on any day during their stay and found that the ensemble prediction model trained and tested on data from Perspective #2 yielded an AUC of 0.734. To further analyze, how the performance of AKI prediction at hospital admission varies with the prediction window, that is, AKI occurring on a specific number of days after admission, the ensemble model developed and evaluated on Perspective #3 demonstrated the best AUC of 0.764 for AKI occurring within 1 day after admission despite the fact that it had the fewest number of positive samples; and AUC for longer prediction windows (2–30 days) is significantly lower ranged from 0.720 to 0.734. Finally, to explore whether we can dynamically predict if a patient will develop AKI within the next day at daily intervals starting from admission to discharge (Perspective #4), the best performance achieved by the ensemble model is AUC of 0.764 and it decreases as time goes on which may be due to the shrinking number of positive samples. It is important to know that good prediction (AUC = 0.764) can be made at hospital admission for forecasting patient AKI risk within the next day as it best reflects real clinical usage of the prediction model. The comprehensive evaluation of AKI prediction models from 4 different clinical perspectives provides us a more realistic performance range with AUC from 0.720 to 0.764.
As noted above that models built and evaluated on data extracted according to Perspective #1 may not effectively reflect the performance in clinical application, we cross tested model developed using Perspective #1 on data extracted according to Perspective #4 and observed that performance of the model built at 1 day before the AKI onset is worse than previous evaluation on the data extracted using Perspective #1. However, the overall performance of the cross testing is surprisingly better than models built and tested using the same strategy, that is, Perspective #4. There may be multiple explanations for this phenomenon. A major factor may be related to sample size. As shown in Figure 2, AKI prevalence rate in Perspective #1 is 9.4% whereas the AKI prevalence rate in Perspective #4 ranged from 2.1% to 1.4% in decreasing order as the prediction point moved. In other words, there are much more positive samples for training the predictive model using model building strategy Perspective #1 versus #4. This is interesting because it may suggest that modeling strategy in Perspective #1 could be sufficiently good for building accurate AKI prediction model when there are abundant samples in which it only requires 1 model being built rather than dynamically building models on a daily basis. Furthermore, AKI prediction is an imbalanced classification problem with an overall cohort in this study containing only 9.4% of AKI cases (10:1 positive to negative ratio) and model building strategy Perspective #4 exacerbated the imbalanced problem. The effect of imbalanced dataset is clearly reflected in the reported F-measure. For instance, under Perspective #3 the AKI prevalence rate increased from 2.1% to 9.4% across different prediction window, the AUC did not vary greatly (0.764–0.734) but F-measure varied from 0.184 to 0.316.
Compared with current research, models presented in this study achieved a relatively good performance. Model built in this study for predicting AKI at 1 day prior to onset from Perspective #1 achieved a comparable AUC of 0.744 as our previous study (AUC of 0.765).19 using the same experimental set up but different cohort and clinical variables. Additionally, though not directly comparable due to different study population, our model built from Perspective #2 achieved a higher AUC of 0.734 than the model described by Kate et al,21 which achieved an AUC of 0.664 under similar experimental set up except they were predicting risk of AKI occurring during entire hospital stay after 24 h of admission rather than at admission as in our design.
A limitation of this study is that there is no comprehensive index to measure the overall performance of models built from Perspective #1 and Perspective #4 to dynamically predict AKI events. Similar to the relationship between AUC and ROC curve, an index depicting the overall performance can help clinicians compare the performance between different models directly. Further work needs to be done to establish such comprehensive index.
CONCLUSION
In this study, we explored different strategies for evaluating the discriminative performance of EMR-based machine learning models for predicting hospital acquired AKI. The multi-perspective experimental analysis showed that the EMR-based AKI prediction performance ranges from 0.720 to 0.764 in AUC in different real-world clinical application scenario. There are 2 interest findings in our experiments. First, we can build an effective model at admission to predict AKI risk in the next day on a training set with small positive samples. Second, predictive models built before onset with a fixed lead time, that is, the most commonly used modeling strategy in current research, is demonstrated through our comprehensive analysis as an effective and robust way to obtain dynamic risk predictions if there are abundant samples. In summary, the important take away message is that we need to focus on designing rational modeling and evaluation strategies that best reflect real-world application based on how the model will be used in clinic. It may also be useful to develop online learning algorithms that address the streaming nature of the EMR data.
SUPPLEMENTARY MATERIAL
Supplementary material is available at Journal of the American Medical Informatics Association online.
FUNDING
This research was partially supported by the Major Research Plan of the National Natural Science Foundation of China (Key Program, Grant No. 91746204), the Science and Technology Development in Guangdong Province (Major Projects of Advanced and Key Techniques Innovation, Grant No. 2017B030308008), and Guangdong Engineering Technology Research Center for Big Data Precision Healthcare (Grant No. 603141789047). The dataset used for analysis described in this study was obtained from KUMC’s HERON clinical data repository which is supported by institutional funding and by the KUMC CTSA grant UL1TR002366 from NCRR/NIH.
CONTRIBUTORS
Y.H. and M.L. designed and conceptualized the overall study. M.L. and L.R.W. contributed in EMR data extraction. J.H. carried out data post-processing to generate the final analysis datasets, developed and evaluated the predictive models. X.Z. and L.W. provided guidance on experiment implementation and manuscript writing. All authors reviewed the manuscript critically for scientific content, and all authors gave final approval of the manuscript for publication.
Conflict of interest statement. None declared.
Supplementary Material
REFERENCES
- 1. Chawla LS, Bellomo R, Bihorac A, et al. Acute kidney disease and renal recovery: consensus report of the Acute Disease Quality Initiative (ADQI) 16 Workgroup. Nat Rev Nephrol 2017; 134: 241–57. [DOI] [PubMed] [Google Scholar]
- 2. Luo M, Yang Y, Xu J, et al. A new scoring model for the prediction of mortality in patients with acute kidney injury. Sci Rep 2017; 71: 7862.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Brown JR, Rezaee ME, Marshall EJ, et al. Hospital mortality in the United States following acute kidney injury. Biomed Res Int 2016; 2016: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Li PK, Burdmann EA, Mehta RL.. Acute kidney injury: global health alert. Kidney Int 2013; 833: 372–6. [DOI] [PubMed] [Google Scholar]
- 5. Silver SA, Long J, Zheng Y, et al. Cost of acute kidney injury in hospitalized patients. J Hosp Med 2017; 122: 70–6. [DOI] [PubMed] [Google Scholar]
- 6. Chawla LS, Eggers PW, Star RA, et al. Acute kidney injury and chronic kidney disease as interconnected syndromes. N Engl J Med 2014; 3711: 58–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Laszczynska O, Severo M, Azevedo A.. Electronic medical record-based predictive model for acute kidney injury in an acute care hospital. Stud Health Technol Inform 2016; 228: 810–2. [PubMed] [Google Scholar]
- 8. Wang Y-N, Cheng H, Yue T, et al. Derivation and validation of a prediction score for acute kidney injury in patients hospitalized with acute heart failure in a Chinese cohort. Nephrology 2013; 187: 489–96. [DOI] [PubMed] [Google Scholar]
- 9. Zhou LZ, Yang XB, Guan Y, et al. Development and validation of a risk score for prediction of acute kidney injury in patients with acute decompensated heart failure: a prospective cohort study in China. J Am Heart Assoc 2016; 511: e004035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Schneider DF, Dobrowolsky A, Shakir IA, et al. Predicting acute kidney injury among burn patients in the 21st century. J Burn Care Res 2012; 332: 242–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Palomba H, de Castro I, Neto ALC, et al. Acute kidney injury prediction following elective cardiac surgery: AKICS Score. Kidney Int 2007; 725: 624–31. [DOI] [PubMed] [Google Scholar]
- 12. Ng SY, Sanagou M, Wolfe R, et al. Prediction of acute kidney injury within 30 days of cardiac surgery. J Thorac Cardiovasc Surg 2014; 1476: 1875–83, 1883.e1. [DOI] [PubMed] [Google Scholar]
- 13. Demirjian S, Schold JD, Navia J, et al. Predictive models for acute kidney injury following cardiac surgery. Am J Kidney Dis 2012; 593: 382–9. [DOI] [PubMed] [Google Scholar]
- 14. Grimm JC, Lui C, Kilic A, et al. A risk score to predict acute renal failure in adult patients after lung transplantation. Ann Thorac Surg 2015; 991: 251–7. [DOI] [PubMed] [Google Scholar]
- 15. Matheny ME, Miller RA, Ikizler TA, et al. Development of inpatient risk stratification models of acute kidney injury for use in electronic health records. Med Decis Making 2010; 306: 639–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Davis SE, Lasko TA, Chen G, et al. Calibration drift in regression and machine learning models for acute kidney injury. J Am Med Inform Assoc 2017; 246: 1052–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Cronin RM, VanHouten JP, Siew ED, et al. National veterans health administration inpatient risk stratification models for hospital-acquired acute kidney injury. J Am Med Inform Assoc 2015; 225: 1054–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Koyner JL, Carey KA, Edelson DP, et al. The development of a machine learning inpatient acute kidney injury prediction model. Crit Care Med 2018; 467: 1070–7. [DOI] [PubMed] [Google Scholar]
- 19. Koyner JL, Adhikari R, Edelson DP, et al. Development of a multicenter ward-based AKI prediction model. Clin J Am Soc Nephrol 2016; 1111: 1935–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Cheng P, Waitman LR, Hu Y, et al. Predicting inpatient acute kidney injury over different time horizons: how early and accurate? AMIA Annu Symp Proc 2017; 2017: 565–74. [PMC free article] [PubMed] [Google Scholar]
- 21. Kate RJ, Perez RM, Mazumdar D, et al. Prediction and detection models for acute kidney injury in hospitalized older adults. BMC Med Inform Decis Mak 2016; 16: 39.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Waitman LR, Warren JJ, Manos EL, et al. expressing observations from electronic medical record flowsheets in an i2b2 based clinical data repository to support research and quality improvement. AMIA Ann Symp Proc 2011; 2011: 1454–63. [PMC free article] [PubMed] [Google Scholar]
- 23. Murphy SN, Weber G, Mendis M, et al. Serving the enterprise and beyond with informatics for integrating biology and the bedside (i2b2). J Am Med Inform Assoc 2010; 172: 124–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Witten IH, Frank E, Hall MA, et al. Data Mining: Practical Machine Learning Tools and Techniques Burlington, MA, USA: Morgan Kaufmann; 2016.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.