Abstract.
Dengue virus (DENV) causes asymptomatic to severe life-threatening infections and affects millions of people worldwide. Autophagy, a cellular degradative pathway, has both proviral and antiviral functions. Dengue virus triggers the autophagy pathway for the successful replication of its genome. However, the exact mechanism and the viral factors involved in activating this pathway remain unclear. This review summarizes the existing knowledge on the mechanism of autophagy induction and its significance during DENV infection.
DENGUE VIRUS
Dengue virus (DENV) is an RNA virus with lipid bilayer envelope embedded with virally encoded proteins. Inside the envelope is a capsid shell that contains the positive RNA genome. The size of the genome is around 11 kb that codes three structural (E, prM, and capsid) and seven nonstructural (NS1, NS2A, NS2B, NS3, NS4A, NS4B, and NS5) proteins.1 Biting of Aedes mosquitoes, mainly Aedes aegypti, transmits this virus to human and causes dengue. Dengue affects millions of people worldwide and is confined to hotter climates where the mosquito that transmits disease thrives.2,3 There are four dengue serotypes, that is, DENV-1, DENV-2, DENV-3, and DENV-4. The infection with any of the serotypes may be asymptomatic in majority of cases.4,5 However, in symptomatic cases, symptoms range from a mild flu-like syndrome known as dengue fever to most severe form dengue hemorrhagic fever (DHF). The latter is characterized by coagulopathy and increased vascular fragility and permeability, which may further progress to hypovolemic shock known as dengue shock syndrome (DSS).6,7 The antibodies generated after primary DENV infection provide lifelong immunity against the infecting serotypes. Rather than providing protection, these primary antibodies facilitate entry of the virus during secondary infection with different serotypes, leading to increased infectivity in the cells known as antibody-dependent enhancement (ADE).8,9 These cross-reactive antibodies are able to bind to Fc receptors present on the cell surface (monocytes, macrophages, mast cells, and basophils), and the Fc receptor-mediated endocytosis is faster and more efficient than the normal entry. Once inside the cell, the virus multiplies undetected, generating very high viral load, which causes serious diseases. Although the risk for DHF is higher during a secondary infection than during a primary infection, most cases with a secondary infection are either asymptomatic or symptomatic febrile illness. Only a minority of secondary infections develop DHF.9,10
AUTOPHAGY PATHWAY
Autophagy is a regulated cellular destructive pathway that is induced when the cell is under stress conditions such as hypoxia, nutrient depletion, and infection.11,12 During autophagy process, degradation of the substrates such as proteins, lipid droplets (LDs), and organelles occurs within the lysosome. This degradation is accountable for cellular waste clearance and recycling of the nutrients, which play a significant role in maintaining the cellular homeostasis process.12 In addition, autophagy plays an important role as a part of the innate and adaptive immune response in opposition to several bacterial and viral infections.11,13 This pathway has proviral and antiviral functions, depending on the type of infected virus. Viruses can either induce or inhibit the autophagy at initial or late stages of the pathway for their own benefit. Dengue virus and hepatitis C virus (HCV), hepatitis B virus (HBV), and varicella-zoster virus are the inducers, whereas HIV-1, herpes simplex virus-1, and poliovirus are the inhibitors at some stages of the pathway.14,15 Mostly, viruses activate autophagy during internalization via contact with surface receptors or by creating cellular stresses.15
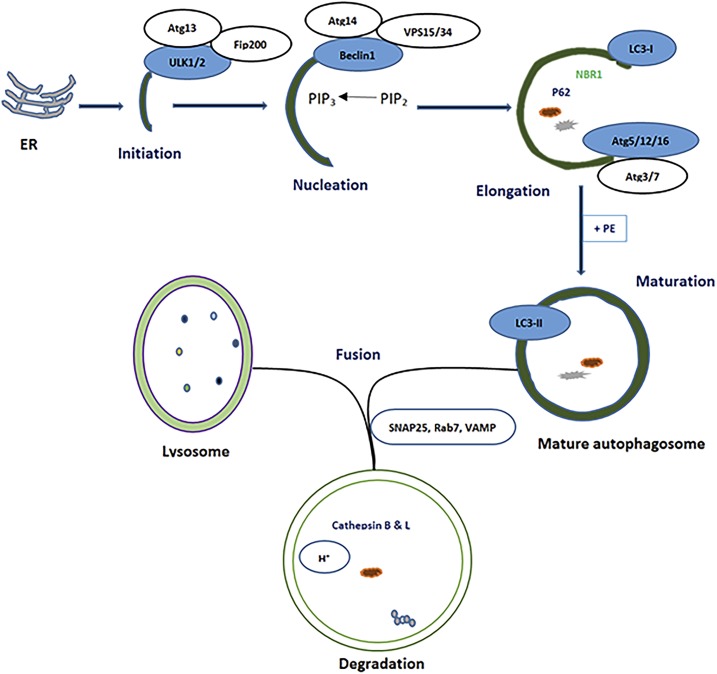
The general process of autophagy begins at the endoplasmic reticulum (ER) or trans-Golgi, with the formation of the budding membrane called pre-autophagosome or phagophore (initiation phase) through the involvement of proteins such as ULK1/2 (Unc-51–like autophagy-activating kinase) and Atg13 (autophagy protein).16 In the second step, the nucleation, ULK1 activates another protein called Beclin-1. The activated Beclin-1 forms the complex with other proteins called Vps15 and Vps34. This complex functions as a class III phosphoinositide 3-kinase (PI3K) to convert phosphatidylinositol diphosphate (PIP2) into phosphatidylinositol triphosphate (PIP3). In the third step, with the involvement of the various Atg proteins (Atg5, Atg12, Atg16, Atg4, Atg7, and Atg3), further elongation and lipidation of membrane—conversion of LC3-I (microtubule-associated protein light chain 3-I) to LC3-II—and recruitment of the cargo-binding proteins such as p62 and neighbor of BRCA1 gene (NBR1) occur.12,17 During the expansion stage, the phagophore engulfs intracellular-cargo-like organelles, LDs, and protein aggregates, thereby secluding the cargo in a double-membraned mature autophagosome.18,17 Finally, the mature autophagosome fuses and discharges its contents with the lysosome to form autolysosome. The degradation of the substrates occurs inside the acidic environment of the autolysosome with the involvement of the enzymes such as cathepsins B and L17 (Figure 1).
Figure 1.
Basal autophagy process showing different steps (Initiation, nucleation, elongation, maturation, and fusion). Atg = autophagy protein; Beclin-1 = mammalian ortholog of the yeast autophagy-related gene 6 (Atg6) and BEC-1 in the Caenorhabditis elegans nematode; ER = endoplasmic reticulum; Fip200 = focal adhesion kinase family interacting protein, a ULK interacting protein; LC3 = microtubule-associated protein light chain 3, marker of autophagy; NBR1 = neighbor of BRCA1 gene 1, cargo receptor protein; PE = phosphatidylethanolamine; PIP2 = phosphatidylinositol diphosphate; PIP3 = phosphatidylinositol triphosphate; p62 = a cargo receptor protein; Rab7 = Ras-related protein; SNAP25 = synaptosomal nerve–associated protein 25, a SNARE protein that mediates membrane fusion; ULK1/2 = Unc-51–like autophagy-activating kinase; VAMP = vesicle-associated membrane protein; VPS34 = phosphatidylinositol-3-kinase. This figure appears in color at www.ajtmh.org.
The autophagy process is regulated by different signaling pathways. One of them is the adenosine monophosphate–activated protein kinase (AMPK), a major controller of cellular energy homeostasis. This pathway is stimulated when the cell is deprived of energy. Adenosine monophosphate–activated protein kinase activation positively regulates the signaling events such as fatty acid oxidation and autophagy that refills the cellular adenosine triphosphate (ATP) and negatively regulates the energy-consuming biosynthetic processes.19 This kinase phosphorylates the ULK1/2 protein when the cell is deprived of energy to activate the autophagy process.20 Similarly, another signaling pathway called mammalian/mechanistic target of rapamycin (mTOR) is a central regulator of cell growth, proliferation and survival, protein synthesis, and autophagy process.21 Mammalian/mechanistic target of rapamycin functions by forming two different signaling complexes, that is, mTORC1 and mTORC2. The major function of the mTORC1 is to stimulate the cellular anabolic pathways such as protein, lipid, and nucleotide biosynthesis which are responsible for cell growth and proliferation. However, it inhibits the catabolic pathways such as lysosome biogenesis and autophagy. Nutrient and growth factor deprivation blocks the activity of the mTORC1.21,22 Blocking of the mTORC1 function results in increased ULK1/2 kinase activity which ultimately activates the autophagy pathway with the involvement of other autophagy (Atg) proteins.12,22 The detail process of autophagy activation and its regulation is reviewed elsewhere.23,24
DENGUE VIRUS AND AUTOPHAGY
Dengue virus infection has been shown to trigger the autophagy pathway. Inhibition of autophagy results in substantial drop in virus replication and the release of progeny virus from DENV-infected cells.25–29 The role of autophagy in DENV infection appears to be cell-type specific; it is nonproductive in monocytes30 but productive in Huh-7 cells.29 Dengue virus modulates host cell lipid metabolism to generate energy for its efficient replication.25 The mechanism and significance of the DENV-induced autophagy pathway are discussed in the following paragraph. The summary of the studies related to DENV-induced autophagy is mentioned in Table 1.
Table 1.
Summary of the studies related to DENV-induced autophagy pathways
| References | Cell lines/animals used | DENV serotype | Conclusion(s) |
|---|---|---|---|
| 29 | Huh-7, MEF, and BHK-2 | DENV-2 | DENV infection induces autophagy and causes autophagosome formation, which promotes virus replication in an ATG5-dependent manner |
| 26 | HepG2 | DENV-2 | DENV induces autophagy; viral NS1 and dsRNA can be detected on LC3-positive structures and inhibiting autophagosome/lysosome fusion increases virus yield. |
| 28 | HepG2 and LLC-MK2 | DENV-3 | Interactions between DENV and the host cell autophagy machinery are complex and may be determined in part by virus-encoded factors. |
| 25 | Huh-7.5, Huh7, HepG2, and BHK | DENV-2 | Regulation of lipid metabolism is the critical function of autophagy for DENV replication. Dengue virus-2–induced ER stress increases autophagy activity, DENV replication, and pathogenesis through two UPR-signaling pathways both in vitro and in vivo. |
| 50 | MDCK, 293T, Vero, HeLa, and Swiss Webster primary macrophage cells | DENV-2 (NS4A protein) | DENV NS4A–mediated upregulation of autophagy signaling is able to protect cell death and enhances viral replication. |
| 30 | U937 (human monocytic cell line), LLC-MK2 cells, and HEK293T/17 | DENV-2 | Autophagy is not a significant part of the DENV replication mechanism in monocytes, and there are distinct cell type–specific differences in the DENV-autophagy interaction |
| 54 | Six-day-old ICR suckling mice | DENV-2 | DENV induces amphisome and autophagosome formation as well as the autophagic flux in the brain of infected mice. Regulation of autophagy in vivo during DENV infection could influence physiopathological parameters, including disease symptoms, survival rate, and viral titer. |
| 51 | AG129 mice (129/Sv mice lacking IFN-α/β and IFN-γ receptors), BHK-21, HeLa, and Huh7.A.1 | DENV-2 | Inhibition of autophagy by spautin-1 generated heat-sensitive, noninfectious DENV particles, revealing a large effect of components of the autophagy pathway on viral maturation. |
| 53 | Human KU812 basophil precursor cells and HMC-1 immature mast cells | DENV-2 | Sub-neutralizing antibodies derived from dengue patient sera enhanced DENV infection and autophagy in basophils as well as mast cells. Autophagy plays an important role in DENV infection and replication in these cells. |
| 43 | Huh-7 and Vero | DENV-2 | In the course of DENV infection, autophagy shifts from a supporting to an antiviral role, which is countered by DENV. |
| 49 | Female BALB/c mice, HMEC-1, 293T, and Drosophila S2 cells | DENV-2 (NS1 protein) | DENV-NS1–induced vascular leakage through the secretion of MIF and the formation of autophagy. Inhibition of MIF or autophagy formation effectively reversed NS1-induced vascular leakage both in vitro and in mice. |
| 52 | Human myelogenous leukemia (K562) cells | DENV-3 | Both DENV and DENV-ADE infections induce an early interferon-stimulated gene (NOS2) expression through retinoic acid-inducible gene I–like receptor-MAVS signaling axis independent of the IFNs signaling. Dengue virus-ADE suppresses this early antiviral response through increased autophagy formation. The early induced autophagic proteins ATG5-ATG12 participate in the suppression of MAVS-mediated ISGs induction, thereby facilitating viral replication. |
| 27 | Huh-7 and BHK | DENV-1, DENV-2, DENV-3, DENV-4 | Infectious autophagy-associated dengue vesicles were released from the infected cells. The viral RNA is protected inside the vesicle, and neutralizing antibodies do not have the effect on the vesicle-mediated close contact transmission that is able to initiate the new round of infection in target cells. |
| 69 | A549 and THP-1 | DENV-2 | An autophagy-related miRNA, miR-146a, has been found to inhibit DENV-induced autophagy by targeting TRAF6 and potentially suppressing excessive inflammation in host cells to lessen the pathological damage of dengue infection. |
| 40 | HepG2 and HEK293T | DENV-2 | DENV infection activates AMPK signaling. Dengue virus induction of proviral lipophagy requires AMPK enzymatic activity. Inhibition of mTOR signaling during DENV infection is required to induce lipophagy. Dengue virus infection increases the accumulation of activated AMPK and produces a corresponding decrease in the mTORC1 activity. |
| 32 | MDCK, HELA, BHK, PERK (wild type and knockout), and MEF | DENV-2 | ER stress signaling, activated during infection, is required for virus-induced autophagy, protection of cells, and production of virus. Ataxia telangiectasia–mutated signaling is active in infected cells and affects ER stress response, dengue-induced protection, and autophagy. Endoplasmic reticulum stress pathway-dependent reactive oxygen species accumulation is important for induction of autophagy at later stages of infection. |
| 33 | Six-day-old ICR suckling mice, Huh-7, A549, BHK, and MEF-ATG5 wild-type and knockout cells | DENV-2 | DENV-2-induced ER stress increases the autophagy activity, DENV replication, and pathogenesis through two UPR signaling pathways both in vitro and in vivo. Targeting the molecules in ER stress and autophagy may become a potential treatment for DENV-infected patients. |
ADE = antibody-dependent enhancement; AMPK = adenosine monophosphate-activated protein kinase; BHK = baby hamster kidney; DENV = dengue virus; ER = endoplasmic reticulum; HELA = Henrietta Lacks; HMC-1 = human mast cell line-1; HMEC-1 = human microvascular endothelial cells-1; ICR = Institute of Cancer Research; LLC-MK2 = rhesus monkey kidney epithelial cells; MAVS = mitochondrial antiviral signaling; MDCK = Madin-Darby canine kidney; MEF = mouse embryonic fibroblast; MIF = migration inhibitory factor; mTOR = mammalian target of rapamycin; PERK = ER stress pathway; THP-1 = human leukemic monocyte-1; UPR = unfolded protein response.
Mechanism and significance of DENV-induced autophagy.
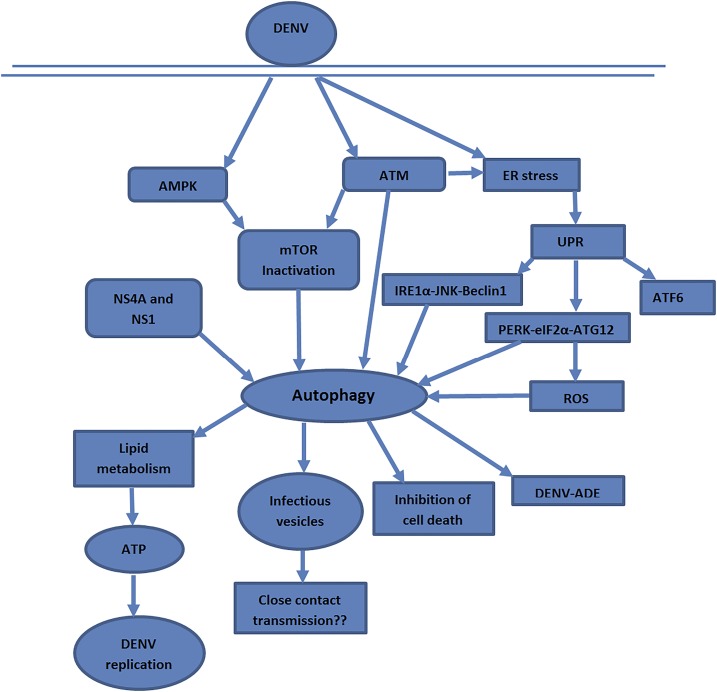
A diagram summarizing possible pathways of DENV-induced autophagy based on the information provided in the reviewed articles is shown in Figure 2. Three signaling pathways—AMPK, ER stress, and ataxia telangiectasia mutated (ATM) kinase—are associated with autophagy activation by DENV.
Figure 2.
Possible activation pathways and significance of autophagy. Dengue virus activates the autophagy pathway via AMPK, ATM, and ER stress pathway. NS4A and NS1 are the two nonstructural proteins involved in autophagy activation. The virus modulates the host cell lipid metabolism to derive the energy for replication and other processes. ADE = antibody-dependent enhancement; AMPK = adenosine monophosphate–activated protein kinase; ATF = activating transcription factor; ATM = ataxia telangiectasia mutated; ATP = adenosine triphosphate; DENV = dengue virus; eIF2α = eukaryotic initiation factor 2α; IRE1α = inositol-requiring protein-1α; JNK = c-Jun-N-terminal kinase; mTOR = mammalian target of rapamycin; PERK = protein kinase–like endoplasmic reticulum kinase; ROS = reactive oxygen species; UPR = unfolded protein response. This figure appears in color at www.ajtmh.org.
Dengue virus targets the ER and creates stress in the ER.31 The accumulation of the unfolded proteins in the ER during viral life cycle induces three hosts unfolded protein response (UPR) pathways, that is, inositol-requiring protein-1α (IRE1α), ER stress pathway (PERK), and activating transcription factor-6 (ATF6). The first two signaling events, that is, IRE1α with the involvement of c-Jun-N-terminal kinase and PERK with the involvement of eukaryotic initiation factor 2α, are the ultimate activators of the autophagy pathway, whereas ATF6 has no any significance on autophagy induction.32,33 In addition, the IRE1α catalyzes the splicing of mRNA that codes for a transcription factor called x-box binding protein (XBP-1)—a part of the ER stress response. The expression of XBP1 protein was found to elicit the autophagic activity in endothelial cells through transcriptional regulation of BECN-1 (a gene that encodes Beclin-1 protein). Knockdown of the XBP1 or IRE1α or BECN1 declines the LC3 expression and autophagosome formation.34 X-box binding protein1 was also found to play a role in the regulation of the immune response comprising CD8+ T-cell differentiation and many inflammatory processes.35,36 However, IRE1α–XBP1 signaling pathway has no impact on DENV-induced autophagy, but might be involved in the production of the cytokines IL-8 and TNF-α.33
The ATM kinase, a serine/threonine protein kinase signaling pathway, is a major regulator of DNA damage and oxidative stress response in eukaryotic cells.37 Knockdown of the ATM is associated with an elevated level of reactive oxygen species (ROS) and reactive nitrogen species. In response to elevated ROS, ATM stimulates the tumor suppressor tuberous sclerosis to suppress mTORC1 activity via the involvement of liver kinase B1/AMPK and induces autophagy.38,39 During DENV infection, the ATM activity is increased particularly in early stages. The increased activity of the ATM is reported to be interrelated with autophagy and ER stress signaling.32 Moreover, the activated ATM kinase stimulates the PERK pathway, which has a major role in the DENV-mediated autophagy induction. The ROS generated via the PERK pathway promotes autophagy at the late stage of the infection cycle.32
Apart from ATM and ER stress signaling pathways, DENV also activates the metabolic regulator AMPK to modulate the host lipid metabolism. The replication and production of infectious virions were considerably reduced in liver cells transfected with AMPKα1 siRNA.40 The activated AMPK blocks the activity of the mTORC1, thereby activating autophagy to be more accurate lipophagy by which the virus modulates the metabolic state of the infected cell. Silencing of the AMPK hindered the mobilization and depletion of LDs in cells infected with DENV. The virus enhances the breakdown of LDs, particularly triglycerides, in an autophagy-dependent manner to generate the ATP for efficient replication.40 On the other hand, besides altering the cellular metabolism, this selective autophagy directed toward LDs (lipophagy) may play some role in the secondary function of immune evasion. Targeting of autophagosomes to LDs can also switch autophagosomes from treating viral antigens for antigen presentation as an immune evasion approach. Moreover, the activated AMPK can promote autophagy in an mTOR-independent manner by directly phosphorylating ULK1/2 and phosphatidylinositol-3-kinase (VPS34-Beclin complex).41 However, the significance of this pathway in DENV has not been reported so far.
In another way, DENV interferes with the function of the cargo receptor that binds ubiquitin-tagged cargo and proceeds for the proteasomal degradation.42 One of the cargo receptors’ (p62, also known as SQSTM1) level is found to be reduced after the DENV infection. Stable overexpression of this protein suppresses the replication of DENV. This evidence suggests that p62 acts as a viral restriction factor, but later on, DENV mediates the proteasomal degradation of p62 for its own benefit. Therefore, during infection process, the virus initially stimulates and impedes general and selective autophagy, hindering autophagosome formation and degradation by lysosomes, and reducing autophagic receptor p62.43
DENV proteins’ role in autophagy.
Regarding the role of the DENV proteins, only two nonstructural proteins associated with autophagy pathways, that is, NS4A and NS1, were reported. During DENV infection, a pro-inflammatory cytokine called macrophage migration inhibitory factor (MIF) is secreted from different cells, such as liver cells, immune cells, and endothelial cells. It has been reported that MIF contributes to the pathogenesis of shock through autophagic degradation of the vascular endothelial cells, thereby causing vascular leakage.44,45 In addition, DENV NS1 protein is released and circulates in the bloodstream of the infected patient. The level of the NS1 protein triggers the endothelial permeability and has direct correlation with the severity of the disease.46 NS1 protein after interacting with Toll-like receptor 4 induces the vascular endothelial damage by stimulating the production of MIF and other inflammatory cytokines.47,48 Therefore, this NS1-mediated MIF production and autophagy might contribute to the pathogenesis of vascular leakage, and hence increase the severity of the dengue infection. Moreover, NS1 also brings about the conversion of LC3-I to LC3-II and degradation of p62, the markers for autophagy induction, in the endothelial cell line.49 Similarly, the DENV-2 NS4A has been shown to upregulate cellular autophagy in a PI3K-dependent manner in infected epithelial cells and consequently protects these cells from camptothecin- and staurosporine- induced apoptotic cell death. Inhibition of the autophagy on such cells restricts the replication of the virus. NS4A-mediated autophagy is, therefore, a key factor for the replication of viral genome.50
Autophagy and DENV-ADE.
During the autophagy, it has been shown that the infectious autophagy-associated dengue vesicles are released from the infected cells.51 These vesicles consist of viral proteins, namely, E, NS1, prM/M, viral RNA, LDs, and LC3-II, the markers of autophagy. These autophagy-associated infectious vesicles are also identified in the serum of patients infected with DENV. Neutralizing antibodies do not produce any effect on vesicle-enclosed viral proteins, and thus, these vesicles mediate close contact transmission in nearby cells to begin the new round of infection.27 It has been reported that both DENV and DENV-ADE infections induce early expression of interferon-stimulated genes in the interferon (IFN)-deficient monocytic cell line K562 by—retinoic acid–inducible gene I–like receptor—mitochondrial antiviral signaling axis independent of IFN signaling. In addition, DENV-ADE defeats this early antiviral response by increasing autophagy formation rather than by inducing IL-10 production.52 However, mast cells and basophils are also the Fc receptors bearing cells. The sub-neutralizing antibodies generated after primary infection bind on these cells and trigger the autophagy pathway, which plays an important role in DENV infection and replication in these cells.53 So the role of the autophagy during DENV-ADE infection is complex and needs further investigation.
AUTOPHAGY: A PROMISING THERAPEUTIC TARGET FOR DENV INFECTION
Autophagy is a cellular homeostatic pathway for the destruction and salvaging of the unnecessary cytoplasmic contents. Several viruses along with DENV, poliovirus, coxsackievirus, and HCV result in and require the machinery of the autophagy pathway for robust replication. Dengue virus–induced autophagy has the crucial role in viral RNA replication, maturation of viral particles, and pathogenesis of the disease.51,54 The UPR-related signaling pathways were responsible for autophagy induction.33 Similar to DENV, HCV and HBV also create an ER stress, and the UPR has been suggested to lead the autophagy induction.55,56 The autophagy proteins Atg5, Atg12, and Beclin-1 are the major proviral factors that are required to initiate the replication and translation of HCV RNA. Interference with the autophagy pathway repressed the replication and production of HCV in infected cells.57,58 Dengue virus induces autophagy at an early stage of infection to support RNA replication, but later on, it interferes with the lysosomal targeting of the autophagic vesicles to assist the production of infectious virus particles.43 The cleavage of the viral protein prM into pr and M by cellular furin is required for the viral maturation.59 Inhibition of the autophagy directly affects the assembly process by reducing the release of pr peptide, thereby producing noninfectious and heat-sensitive virus particles.51 Regulation of the autophagy in mice infected with DENV has been shown to affect physiopathological parameters, including disease symptoms, survival rate, and viral titer.54
The central metabolic regulator, AMPK, has different interactions depending on the species of the virus.60 In the viruses such as Rift valley fever virus and West Nile virus, the AMPKs are activated during infection and inhibit the replication of the virus by controlling the fatty acid synthesis through inactivation of acetyl-CoA carboxylase.61 Hepatitis C virus, a close member of the DENV, evades the antiviral activity of the AMPK by restricting its activation. Renovation of the AMPK not only inhibits the replication of the virus but also decreases the aggregation of lipid molecules in virus-infected cells.62 However, DENV activates the AMPK to trigger the autophagy pathway for the breakdown of the LDs to generate ATP, which is required for efficient DENV replication. Adenosine monophosphate–activated protein kinase signaling and AMPK-independent suppression of the mTORC1 activity are the major requirements for the stimulation of the lipophagy, but not for basal autophagy.40 The replication of the virus does not occur on cells lacking autophagic machinery. However, the external addition of the fatty acids promotes the viral multiplication of such cells. In addition, DENV also promotes the synthesis of the fatty acids at the viral replication site,63 similar to that of human cytomegalovirus. Human cytomegalovirus activates both the lipophagy and synthesis pathway simultaneously and the inhibition of the AMPK halts the viral replication.64 Adenosine monophosphate–activated protein kinase and fatty acid synthesis pathway inhibition might have a new therapeutic approach for DENV infection.
It has been demonstrated that several viruses trigger the autophagy pathway by their structural and nonstructural proteins. Hepatitis B virus X protein,65 Epstein-Barr virus (EBV) Rta transcription factor,66 SV40 T antigen,67 and HCV NS4B, NS5A, and NS5B68 were the inducers of autophagy. However, in the case of DENV, only two viral nonstructural proteins, NS1 and NS4A, have been linked with the autophagy pathway. The NS1-induced MIF secretion and autophagy may represent potential therapeutic targets for preventing vascular leakage in DHF/DSS.49 Cells infected with DENV are found to be resistant to apoptosis by external stimuli. This resistance to apoptosis is due to the NS4A protein which is dependent on autophagy. Inhibition of the autophagy by chemical treatment with wortmannin or 3-methyladenine, as well as by genetic treatment by the knockdown of Beclin-1 or Atg5, reverses the DENV-mediated protection against cell death.50 However, in vivo significance of the NS4A protein on this process requires further work.
The interaction of the viral factors with the host cell proteins to bring on autophagy is very complex and not fully known. The role and influences of autophagy on the pathogenesis of dengue infection need further exploration, which may lead to discover new therapeutic targets for the management of dengue.
Acknowledgments:
We are grateful to the Center of Excellence in Pharmacology and Molecular Biology of Malaria and Cholangiocarcinoma, Chulabhorn International College of Medicine at Thammasat University, Rangsit Center, Thailand, for the support in doing this review.
REFERENCES
- 1.Kuhn RJ, et al. 2002. Structure of dengue virus: implications for flavivirus organization, maturation, and fusion. Cell 108: 717–725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bhatt S, et al. 2013. The global distribution and burden of dengue. Nature 496: 504–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gyawali N, Bradbury RS, Taylor-Robinson AW, 2016. The epidemiology of dengue infection: harnessing past experience and current knowledge to support implementation of future control strategies. J Vector Borne Dis 53: 293–304. [PubMed] [Google Scholar]
- 4.Burke DS, Nisalak A, Johnson DE, Scott RM, 1988. A prospective study of dengue infections in Bangkok. Am J Trop Med Hyg 38: 172–180. [DOI] [PubMed] [Google Scholar]
- 5.Rafique I, Saqib MAN, Munir MA, Qureshi H, Taseer IUH, Iqbal R, Ahmed W, Akhtar T, Rizwanullah S, 2017. Asymptomatic dengue infection in adults of major cities of Pakistan. Asian Pac J Trop Med 10: 1002–1006. [DOI] [PubMed] [Google Scholar]
- 6.Halstead SB, 1988. Pathogenesis of dengue: challenges to molecular biology. Science 239: 476–481. [DOI] [PubMed] [Google Scholar]
- 7.Ranjit S, Kissoon N, 2011. Dengue hemorrhagic fever and shock syndromes. Pediatr Crit Care Med 12: 90–100. [DOI] [PubMed] [Google Scholar]
- 8.Acosta EG, Bartenschlager R, 2016. Paradoxical role of antibodies in dengue virus infections: considerations for prophylactic vaccine development. Expert Rev Vaccines 15: 467–482. [DOI] [PubMed] [Google Scholar]
- 9.Gan ES, Ting DH, Chan KR, 2017. The mechanistic role of antibodies to dengue virus in protection and disease pathogenesis. Expert Rev Anti Infect Ther 15: 111–119. [DOI] [PubMed] [Google Scholar]
- 10.Wang TT, et al. 2017. IgG antibodies to dengue enhanced for FcgammaRIIIA binding determine disease severity. Science 355: 395–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kudchodkar SB, Levine B, 2009. Viruses and autophagy. Rev Med Virol 19: 359–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mizushima N, Komatsu M, 2011. Autophagy: renovation of cells and tissues. Cell 147: 728–741. [DOI] [PubMed] [Google Scholar]
- 13.Deretic V, Levine B, 2018. Autophagy balances inflammation in innate immunity. Autophagy 14: 243–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dreux M, Chisari FV, 2010. Viruses and the autophagy machinery. Cell Cycle 9: 1295–1307. [DOI] [PubMed] [Google Scholar]
- 15.Jordan TX, Randall G, 2012. Manipulation or capitulation: virus interactions with autophagy. Microbes Infect 14: 126–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Axe EL, Walker SA, Manifava M, Chandra P, Roderick HL, Habermann A, Griffiths G, Ktistakis NT, 2008. Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum. J Cell Biol 182: 685–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mizushima N, 2007. Autophagy: process and function. Genes Dev 21: 2861–2873. [DOI] [PubMed] [Google Scholar]
- 18.Glick D, Barth S, Macleod KF, 2010. Autophagy: cellular and molecular mechanisms. J Pathol 221: 3–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Carling D, Mayer FV, Sanders MJ, Gamblin SJ, 2011. AMP-activated protein kinase: nature’s energy sensor. Nat Chem Biol 7: 512–518. [DOI] [PubMed] [Google Scholar]
- 20.Kim J, Kundu M, Viollet B, Guan KL, 2011. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat Cell Biol 13: 132–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jung CH, Ro S-H, Cao J, Otto NM, Kim D-H, 2010. mTOR regulation of autophagy. FEBS Lett 584: 1287–1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim YC, Guan K-L, 2015. mTOR: a pharmacologic target for autophagy regulation. J Clin Invest 125: 25–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.He C, Klionsky DJ, 2009. Regulation mechanisms and signaling pathways of autophagy. Annu Rev Genet 43: 67–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang Z, Klionsky DJ, 2010. Mammalian autophagy: core molecular machinery and signaling regulation. Curr Opin Cell Biol 22: 124–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Heaton NS, Randall G, 2010. Dengue virus-induced autophagy regulates lipid metabolism. Cell Host Microbe 8: 422–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Panyasrivanit M, Khakpoor A, Wikan N, Smith DR, 2009. Co-localization of constituents of the dengue virus translation and replication machinery with amphisomes. J Gen Virol 90: 448–456. [DOI] [PubMed] [Google Scholar]
- 27.Wu YW, Mettling C, Wu SR, Yu CY, Perng GC, Lin YS, Lin YL, 2016. Autophagy-associated dengue vesicles promote viral transmission avoiding antibody neutralization. Sci Rep 6: 32243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Khakpoor A, Panyasrivanit M, Wikan N, Smith DR, 2009. A role for autophagolysosomes in dengue virus 3 production in HepG2 cells. J Gen Virol 90: 1093–1103. [DOI] [PubMed] [Google Scholar]
- 29.Lee YR, Lei HY, Liu MT, Wang JR, Chen SH, Jiang-Shieh YF, Lin YS, Yeh TM, Liu CC, Liu HS, 2008. Autophagic machinery activated by dengue virus enhances virus replication. Virology 374: 240–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Panyasrivanit M, Greenwood MP, Murphy D, Isidoro C, Auewarakul P, Smith DR, 2011. Induced autophagy reduces virus output in dengue infected monocytic cells. Virology 418: 74–84. [DOI] [PubMed] [Google Scholar]
- 31.Pena J, Harris E, 2011. Dengue virus modulates the unfolded protein response in a time-dependent manner. J Biol Chem 286: 14226–14236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Datan E, Roy SG, Germain G, Zali N, McLean JE, Golshan G, Harbajan S, Lockshin RA, Zakeri Z, 2016. Dengue-induced autophagy, virus replication and protection from cell death require ER stress (PERK) pathway activation. Cell Death Dis 7: e2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee YR, Kuo SH, Lin CY, Fu PJ, Lin YS, Yeh TM, Liu HS, 2018. Dengue virus-induced ER stress is required for autophagy activation, viral replication, and pathogenesis both in vitro and in vivo. Sci Rep 8: 489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Margariti A, et al. 2013. XBP1 mRNA splicing triggers an autophagic response in endothelial cells through BECLIN-1 transcriptional activation. J Biol Chem 288: 859–872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kamimura D, Bevan MJ, 2008. Endoplasmic reticulum stress regulator XBP-1 contributes to effector CD8+ T cell differentiation during acute infection. J Immunol 181: 5433–5441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kaser A, et al. 2008. XBP1 links ER stress to intestinal inflammation and confers genetic risk for human inflammatory bowel disease. Cell 134: 743–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shiloh Y, Ziv Y, 2013. The ATM protein kinase: regulating the cellular response to genotoxic stress, and more. Nat Rev Mol Cell Biol 14: 197. [PubMed] [Google Scholar]
- 38.Alexander A, et al. 2010. ATM signals to TSC2 in the cytoplasm to regulate mTORC1 in response to ROS. Proc Natl Acad Sci USA 107: 4153–4158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Alexander A, Kim J, Walker CL, 2010. ATM engages the TSC2/mTORC1 signaling node to regulate autophagy. Autophagy 6: 672–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jordan TX, Randall G, 2017. Dengue virus activates the AMP kinase-mTOR axis to stimulate a proviral lipophagy. J Virol 91: e02020–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Russell RC, Tian Y, Yuan H, Park HW, Chang Y-Y, Kim J, Kim H, Neufeld TP, Dillin A, Guan K-L, 2013. ULK1 induces autophagy by phosphorylating Beclin-1 and activating Vps34 lipid kinase. Nat Cell Biol 15: 741–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zaffagnini G, Martens S, 2016. Mechanisms of selective autophagy. J Mol Biol 428: 1714–1724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Metz P, Chiramel A, Chatel-Chaix L, Alvisi G, Bankhead P, Mora-Rodriguez R, Long G, Hamacher-Brady A, Brady NR, Bartenschlager R, 2015. Dengue virus inhibition of autophagic flux and dependency of viral replication on proteasomal degradation of the autophagy receptor p62. J Virol 89: 8026–8041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen H-R, Chuang Y-C, Chao C-H, Yeh T-M, 2015. Macrophage migration inhibitory factor induces vascular leakage via autophagy. Biol Open 4: 244–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chuang Y-C, Lei H-Y, Liu H-S, Lin Y-S, Fu T-F, Yeh T-M, 2011. Macrophage migration inhibitory factor induced by dengue virus infection increases vascular permeability. Cytokine 54: 222–231. [DOI] [PubMed] [Google Scholar]
- 46.Beatty PR, Puerta-Guardo H, Killingbeck SS, Glasner DR, Hopkins K, Harris E, 2015. Dengue virus NS1 triggers endothelial permeability and vascular leak that is prevented by NS1 vaccination. Sci Transl Med 7: 304ra141. [DOI] [PubMed] [Google Scholar]
- 47.Calandra T, Bernhagen J, Mitchell RA, Bucala R, 1994. The macrophage is an important and previously unrecognized source of macrophage migration inhibitory factor. J Exp Med 179: 1895–1902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Modhiran N, Watterson D, Muller DA, Panetta AK, Sester DP, Liu L, Hume DA, Stacey KJ, Young PR, 2015. Dengue virus NS1 protein activates cells via Toll-like receptor 4 and disrupts endothelial cell monolayer integrity. Sci Transl Med 7: 304ra142. [DOI] [PubMed] [Google Scholar]
- 49.Chen HR, Chuang YC, Lin YS, Liu HS, Liu CC, Perng GC, Yeh TM, 2016. Dengue virus nonstructural protein 1 induces vascular leakage through macrophage migration inhibitory factor and autophagy. PLoS Negl Trop Dis 10: e0004828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.McLean JE, Wudzinska A, Datan E, Quaglino D, Zakeri Z, 2011. Flavivirus NS4A-induced autophagy protects cells against death and enhances virus replication. J Biol Chem 286: 22147–22159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mateo R, Nagamine CM, Spagnolo J, Mendez E, Rahe M, Gale M, Jr., Yuan J, Kirkegaard K, 2013. Inhibition of cellular autophagy deranges dengue virion maturation. J Virol 87: 1312–1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Huang X, et al. 2016. Antibody-dependent enhancement of dengue virus infection inhibits RLR-mediated Type-I IFN-independent signalling through upregulation of cellular autophagy. Sci Rep 6: 22303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fang YT, Wan SW, Lu YT, Yao JH, Lin CF, Hsu LJ, Brown MG, Marshall JS, Anderson R, Lin YS, 2014. Autophagy facilitates antibody-enhanced dengue virus infection in human pre-basophil/mast cells. PLoS One 9: e110655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lee YR, Hu HY, Kuo SH, Lei HY, Lin YS, Yeh TM, Liu CC, Liu HS, 2013. Dengue virus infection induces autophagy: an in vivo study. J Biomed Sci 20: 65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dash S, Chava S, Aydin Y, Chandra PK, Ferraris P, Chen W, Balart LA, Wu T, Garry RF, 2016. Hepatitis C virus infection induces autophagy as a prosurvival mechanism to alleviate hepatic ER-stress response. Viruses 8: 150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lazar C, Uta M, Branza-Nichita N, 2014. Modulation of the unfolded protein response by the human hepatitis B virus. Front Microbiol 5: 433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ke PY, Chen SS, 2011. Activation of the unfolded protein response and autophagy after hepatitis C virus infection suppresses innate antiviral immunity in vitro. J Clin Invest 121: 37–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dreux M, Chisari FV, 2009. Autophagy proteins promote hepatitis C virus replication. Autophagy 5: 1224–1225. [DOI] [PubMed] [Google Scholar]
- 59.Hsieh SC, Wu YC, Zou G, Nerurkar VR, Shi PY, Wang WK, 2014. Highly conserved residues in the helical domain of dengue virus type 1 precursor membrane protein are involved in assembly, precursor membrane (prM) protein cleavage, and entry. J Biol Chem 289: 33149–33160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chukkapalli V, Heaton NS, Randall G, 2012. Lipids at the interface of virus–host interactions. Curr Opin Microbiol 15: 512–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Moser TS, Schieffer D, Cherry S, 2012. AMP-activated kinase restricts Rift Valley fever virus infection by inhibiting fatty acid synthesis. PLoS Pathog 8: e1002661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mankouri J, Tedbury PR, Gretton S, Hughes ME, Griffin SDC, Dallas ML, Green KA, Hardie DG, Peers C, Harris M, 2010. Enhanced hepatitis C virus genome replication and lipid accumulation mediated by inhibition of AMP-activated protein kinase. Proc Natl Acad Sci USA 107: 11549–11554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Heaton NS, Perera R, Berger KL, Khadka S, Lacount DJ, Kuhn RJ, Randall G, 2010. Dengue virus nonstructural protein 3 redistributes fatty acid synthase to sites of viral replication and increases cellular fatty acid synthesis. Proc Natl Acad Sci USA 107: 17345–17350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.McArdle J, Moorman NJ, Munger J, 2012. HCMV targets the metabolic stress response through activation of AMPK whose activity is important for viral replication. PLoS Patho 8: e1002502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhang HT, Chen GG, Hu BG, Zhang ZY, Yun JP, He ML, Lai PB, 2014. Hepatitis B virus x protein induces autophagy via activating death-associated protein kinase. J Viral Hepat 21: 642–649. [DOI] [PubMed] [Google Scholar]
- 66.Hung CH, et al. 2014. Regulation of autophagic activation by Rta of Epstein-Barr virus via the extracellular signal-regulated kinase pathway. J Virol 88: 12133–12145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kumar SH, Rangarajan A, 2009. Simian virus 40 small T antigen activates AMPK and triggers autophagy to protect cancer cells from nutrient deprivation. J Virol 83: 8565–8574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wang L, Ou J-hJ, 2015. Hepatitis C virus and autophagy. Biol Chemistry 396: 1215–1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pu J, Wu S, Xie H, Li Y, Yang Z, Wu X, Huang X, 2017. miR-146a inhibits dengue-virus-induced autophagy by targeting TRAF6. Arch Virol 162: 3645–3659. [DOI] [PMC free article] [PubMed] [Google Scholar]