Abstract.
American cutaneous leishmaniasis (ACL) is a common and important vector-borne parasitic zoonosis in Panamá. Here, we study Leishmania spp. infection rates and blood-feeding patterns among common sand flies in Trinidad de Las Minas, a rural community with hyperendemic ACL transmission, and where a deltamethrin fogging trial was performed. Sand flies were collected from April 2010 to June 2011 with light traps installed inside and in the peridomicile of 24 houses. We restricted our analysis to the most abundant species at the study site: Lutzomyia trapidoi, Lutzomyia gomezi, Lutzomyia panamensis, Lutzomyia triramula, and Lutzomyia dysponeta. We detected Leishmania spp. infection in sand flies by polymerase chain reaction (PCR) amplification of the internal transcribed spacer region 1 (ITS-1) in pooled females (1–10 females per pool). Host species of engorged sand flies were identified using a cytochrome b PCR. From 455 sand fly pools analyzed, 255 pools were positive for Leishmania spp., with an estimated infection rate (confidence interval) of 0.096 [0.080–0.115] before the deltamethrin fogging which slightly, but not significantly (P > 0.05), increased to 0.116 [0.098–0.136] after the deltamethrin fogging. Blood meal analysis suggested that pigs, goats, and birds were the most common sand fly blood sources, followed by humans and domestic dogs. DNA sequencing from a subsample of ITS-1 positive pools suggests that Leishmania panamensis, Leishmania naiffi, and other Leishmania spp. were the parasite species infecting the most common vectors at the study site. Our data confirm an association between sand fly species, humans, domestic dogs, and pigs and Leishmania spp. parasites in rural Panamá.
INTRODUCTION
American cutaneous leishmaniasis (ACL) is one of the most common parasitic zoonotic diseases in Panamá. Around 3,000 new cases are reported per year (60–100 new cases per 100,000 inhabitants), however, the true incidence is likely higher due to high underreporting of cases.1 According to epidemiological reports, from 1980 to 2012, 82% of cases come from rural, marginalized, and isolated geographical areas in the provinces of Bocas del Toro (29%), Panamá Oeste (17%), Coclé (16%), Colón (11%), Panamá Este (5%), and the Darién (4%).2 However, changing population dynamics of vectors and reservoirs, climate change, and land use have been identified as important factors driving ACL incidence dynamics.3–7
American cutaneous leishmaniasis is transmitted by sand flies (called “chitras” in Panamá), which are associated with heterogeneous environments with abundant vegetation and high humidity.8–10 A variety of mammal species are reservoirs of Leishmania spp.11–14 However, Choloepus hoffmanni (two-toed sloth) has been repeatedly identified as the main reservoir.12,14 The role of domestic animals in ACL transmission cycle remains unclear, however, canine infections are common in some Panamánian rural communities.15 Leishmania (Viannia) panamensis is the species most commonly isolated from human ACL lesion samples in Panamá.1,16 The recent implementation of molecular tools for epidemiological studies has allowed for better characterization of potential reservoirs and vectors involved in the transmission of parasites, including Leishmania spp. in ACL endemic regions.17–19 Similarly, these tools have allowed an analysis of sand fly host use and subsequent vector dispersion patterns; which is important in the design of vector surveillance, control, and prevention programs for endemic areas.20–24
This study uses molecular methods to quantify Leishmania spp. infection rate and blood-feeding patterns of sand flies collected inside and around houses in the community of Trinidad de Las Minas (TM), a rural community with a high incidence of ACL in Panamá Oeste Province.25 We specifically compare infection rates before and after a deltamethrin fogging control trial, to test whether concomitant with the observed 50–80% decrease in sand fly abundance there was a decrease in the infection rate in sand flies.26 Multivariate statistical methods were used to quantify the association between habitat use, blood sources, and infection in the five dominant sand fly species at the study site, Lutzomyia trapidoi, Lutzomyia gomezi, Lutzomyia panamensis, Lutzomyia triramula, and Lutzomyia dysponeta, which accounted for more than 80% of all the sand flies we collected during this study.27 The information presented here is then used to characterize ACL transmission dynamics in rural areas of Panamá.
MATERIALS AND METHODS
Study area.
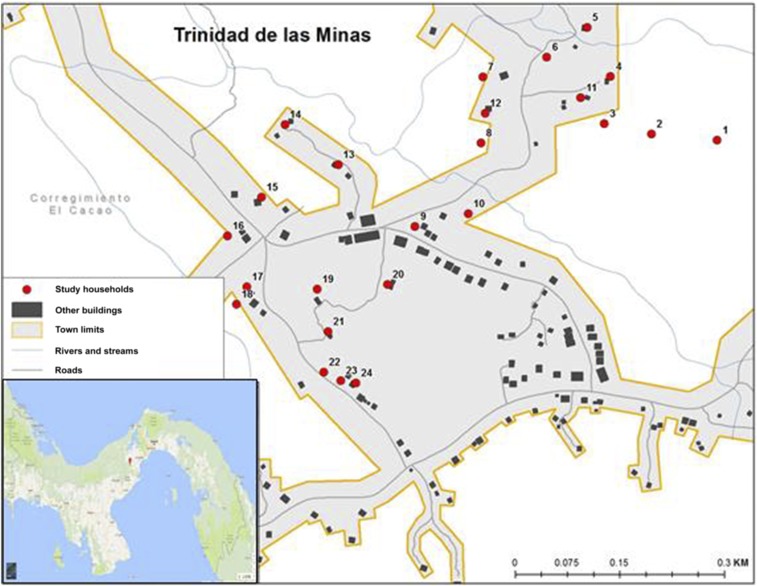
Sand fly collections were performed at TM (8°46′32″N; 79°59′45″W), a rural village located in the Capira District, Panamá Oeste Province. The village is located in a mountainous area with an altitude of 613 m; the vegetation is heterogeneous comprising secondary forest, farmland, grass, and herbaceous shrubs. The main economic activity in this area is subsistence agriculture. The climate is characterized by a dry season between the months of December and March and a rainy season from April to November. The annual average temperature is 25°C, with annual rainfall ranging from 2.101 to 2.200 mm, and an average relative humidity of 80%. The study was conducted in 24 of 128 houses present in the community (Figure 1), a sample large enough to test the impact of insecticide fogging on sand fly abundance.26 Houses were selected based on the presence of sand flies and confirmation of suspected wildlife reservoirs presence by residents. In addition, in 12 of these houses an intervention with deltamethrin thermal fogging was performed.26,27
Figure 1.
Geographical location of the studied houses at Trinidad de Las Minas, in Capira, West Panama, where sand flies collections were performed between April 2010 and June 2011. This figure appears in color at www.ajtmh.org.
Sand fly collection.
Sand flies were collected once a month for 15 months with light traps (HP Equipamentos e Instrumentos Biomédicos Indústria e Comércio LTDA, MG-Brazil). The traps were placed inside the main room and in the peridomicile (not exceeding 50 m from the house center) of each of the 24 selected houses (Figure 1). Sand fly sampling was carried out monthly between April 2010 and June 2011. In August and November 2010, and in January 2011 sampling at the study was not performed because of logistic and operational constraints. The houses selected for the intervention were fogged with deltamethrin in June 2010 and January 2011, and for the analysis, pre-fogging refers to samples collected between April and June 2010, and post-fogging to samples collected from July 2010 until June 2011. The traps were located at a height of 1.5 m and operated for 12 hours from 6:00 pm to 6:00 am. The collected specimens were placed in labeled plastic containers with 70% ethanol. Sand flies were transported to the Instituto Conmemorativo Gorgas de Estudios de la Salud where they were identified according to their morphological characteristics (genitalia, antenna and cibarium) using the taxonomic key by Young and Duncan.28 Specimens were kept at −4°C until final processing.
DNA extraction.
Female sand flies, engorged and non-engorged, were grouped in pools of 1–10 specimens. Pools were made by species and according to collection month and sampling environment (domicile and peridomicile). Some of the pools exclusively included non-engorded sand flies, whereas the remaining pools were a mixture of engorged and non-engorged sand flies. Each sand fly pool was placed in an Eppendorf tube, where they were first macerated in a solution of 180 μL of 1× phosphate-buffered saline. Next, a DNA extraction was performed following the protocol from Qiagen DNeasy Blood & Tissue (Qiagen, Germantown, MD). The isolated DNA was stored at −20°C until further use.
Detection of Leishmania spp. infection in sand flies.
Leishmania spp. detection in sand flies was performed by amplifying the Internal transcribed spacer region 1 (ITS-1) with LITSR (5′CTGGATCATTTTCCGATG3) and L5.8S (5′TGATACCACTTATCGCACTT3′) primers, following the PCR conditions described by Schönian et al.23 and Kuhls et al.29 As positive control, DNA from a promastigote culture of L. (Viannia) panamensis MHOM/PA/98/WR 2306 reference strain was used. As negative control, we used the mix of all reagents without a DNA template.
Blood meal analysis.
Sand fly pools found positive for Leishmania spp. infection (ITS-1 PCR) were analyzed to detect the blood meal source. A multiplex PCR that amplifies the cytochrome b (Cyt b) gene from the mitochondrial DNA of the following mammals: pig (453 pb), human (334 pb), goat (132 pb), dog (680 pb), and cow (561 pb).30 Negative samples for blood meals with this protocol were further analyzed using universal primers of Cyt b for mammals (623 pb) and birds (300 pb), following conditions described by Fornadel and Norris.31 Amplified products by PCR were analyzed by gel electrophoresis in 1.5% agarose in 0.5× Tris/Borate/EDTA buffer and visualized by ethidium bromide staining.
We verified Leishmania spp. DNA amplification using between 1 and 4 ITS-1 PCR positive pools for each of the studied species, excluding pools that were positive for blood ingestion. We only considered sand fly pools without blood contents to ensure that any Leishmania spp. DNA amplification did not come from infected blood from vertebrate hosts but from parasites in the sand flies. The PCR-amplified products from these pools were purified using a QIAquick PCR purification kit (Qiagen) and then sequenced using ITS-1 forward and reverse primers,23,29 with an ABI Prism 3130 × sequencer (Applied Biosystems, Foster City, CA). Resulting sequences were then edited and aligned using MEGA 7.0.18 (Pennsylvania State University, State College, PA).32 Then, each DNA sequence was compared with ITS-1 DNA sequences available in the GenBank by performing a Basic Local Alignment Search Tool (BLAST) search of the National Center for Biotechnology Information Database (http://www.ncbi.nlm.nih.gov/BLAST/). The nucleotide sequences generated from the analyzed pools were deposited in the GenBank database. Our criterion to identify DNA sequences as belonging to a given Leishmania spp. was based on greater than 95% sequence identity in the BLAST analysis, otherwise DNA sequences were identified to the genus level.
Statistical analysis.
Leishmania spp. prevalence for the five dominant sand fly species at our study site was estimated for pools collected before and after the insecticide thermal fogging applications. This was performed to assess changes in the infection rate by Leishmania spp. in the dominant sand fly species before and after the insecticide thermal fogging. We used the maximum likelihood estimation method developed for unequal pool size by Farrington et al.,33 which can be easily fit with generalized linear models with a cloglog link, with confidence intervals estimated by inverting the likelihood ratio test.34 We used this method assuming the sensitivity and specificity of the L. panamensis diagnostic test were nearly 1, given that our diagnostic PCR is a common gold standard to determine Leishmania spp. infections.1 The association between the presence of a blood meal, Leishmania spp. infection, trap location (peridomicile or domicile), intervention status (fogged or control houses), the collection time with regard to the deltamethrin fogging (pre-fogging, post first round of fogging or second round of fogging) was analyzed through a multiple correspondence analysis (MCA).35 A second MCA was performed to analyze the association between specific blood meal sources, Leishmania spp. infection and vector species. Multiple correspondence analysis was chosen given its ability to measure the association between categorical variables.
Briefly, information for each one of the study objects, in our study the sand fly pools, is organized in rows forming a table where columns are the different factors whose association wants to be studied. This matrix then goes through a singular value decomposition that allows to represent the data in two dimensions by projecting the original data into the vectors associated with the largest singular values from the decomposition.35 Centroids for the different levels from the categorical variables can be plotted in simple two-dimensional graphs where proximity between levels of different variables allows the evaluation of their association, which is stronger as levels from different variables lie together but farther apart from the origin, that is, when the x and y axis equal 0, the point where levels that associate randomly are expected to appear.35 All analyses were performed with the statistical software R. (University of Auckland, New Zealand).
RESULTS
A total of 5,628 individuals of Phlebotomine sand flies (23 species from the genus Lutzomyia spp., and one species from the genus Brumptomyia spp.) were collected in the 24 houses evaluated, 2,357 inside the houses and 3,271 in the peridomicile.27 Dominant anthropophilic species incriminated in ACL transmission in Panamá were further analyzed: Lu. trapidoi (1,151 = 20%), Lu. gomezi (1,146 = 20%), and Lu. panamensis (967 = 17%). The most abundant zoophilic species were also analyzed: Lu. triramula (1,150 = 20%), and Lu. dysponeta (490 = 8%).
From 455 pools tested, 166 pools from anthropophilic species resulted positive by the PCR technique of the ITS-1 gene: Lu. trapidoi (89 pools), Lu. gomezi (48 pools), and Lu. panamensis (29 pools). Meanwhile 89 pools were positive for the most abundant zoophilic species: Lu. triramula (74 pools), and Lu. dysponeta (15 pools). The overall Leishmania spp. prevalence in Lutzomyia spp. before the insecticide thermal fogging application was 0.096 (Table 1). Although the infection rate was a little higher after the intervention, 0.116, this increase was not significant (Table 1). Among the anthropophilic species, Lu. trapidoi had the highest Leishmania infection rate (0.135) prior to the insecticide thermal fogging application. Interestingly, this prevalence significantly decreased to 0.033 after the intervention (Table 1). Among the zoophilic vectors analyzed, Lu. dysponeta presented a higher infection rate prior to the intervention, while Lu. triramula stayed the same (Table 1).
Table 1.
Prevalence of Leishmania spp. infection in pools of dominant Lutzomyia spp. before and after applying two rounds with deltamethrin insecticide fogging (6 mg a.i. m−2) in Trinidad de Las Minas, Panamá
| Species | Pre-fogging | Post-fogging | Change | ||||
|---|---|---|---|---|---|---|---|
| Prevalence [95% CI] | Pools | Sand fly abundance | Prevalence [95% CI] | Pools | Sand fly abundance | ||
| Lutzomyia gomezi (Nitzulescu) | 0.022 [0.010–0.040] | 45 | 399 | 0.037 [0.020–0.060] | 59 | 405 | NS |
| Lutzomyia trapidoi (Fairchild and Hertig) | 0.135 [0.044–0.291] | 10 | 36 | 0.033 [0.022–0.046] | 106 | 891 | Decrease* |
| Lutzomyia panamensis (Shannon) | 0.037 [0.021–0.059] | 49 | 445 | 0.044 [0.025–0.069] | 53 | 405 | NS |
| Lutzomyia dysponeta (Fairchild and Hertig) | 0.091 [0.051–0.147] | 25 | 193 | 0.040 [0.007–0.117] | 18 | 51 | NS |
| Lutzomyia triramula (Fairchild and Hertig) | 0.213 [0.160–0.279] | 67 | 622 | 0.209 [0.123–0.323] | 23 | 111 | NS |
| Total | 0.096 [0.080–0.115] | 196 | 1,695 | 0.116 [0.098–0.136] | 259 | 1,863 | NS |
CI = confidence interval; NS = nonsignificant; SF = pools and sand flies. Maximum likelihood prevalence, and 95% CI, was estimated from pools of variable size. The number of SFs for each estimate are also presented. The column “Change” indicates whether there were NS changes, an increase or decrease in Leishmania spp. prevalence when comparing SFs caught during the pre- and post-fogging periods. Raw data used in this analysis is presented in Supplemental Table 1.
Statistically significant (P < 0.05).
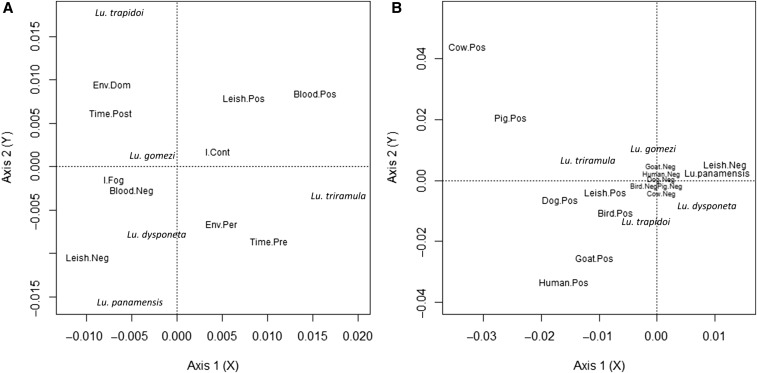
Results from the MCA considering Leishmania spp. infection, blood meal presence, and variables about the deltamethrin fogging intervention (Figure 2A) indicated that sand fly pools with blood were more likely to be Leishmania spp. positive, and tended to occur more often in the control houses than in the fogged ones. The sand fly species that were more likely to have Leishmania spp. positive pools were Lu. trapidoi sampled inside houses after the deltamethrin fogging and Lu. triramula sampled in peridomiciles before the deltamethrin fogging. By contrast, Lu. gomezi, Lu. dysponeta, and Lu. panamensis pools were more likely to be parasite free and to have no blood, especially when sampled in/around fogged houses. These inferences are based on the proximity between the label for the different categories in the studied variables as observed in Figure 2A.
Figure 2.
Multiple correspondence analyzes (MCA). A MCA represents the association between categorical variables. Distance from the origin (coordinates 0.00, 0.00) indicate the degree of association between categories from the studied factors. Categories that are close, but far from the origin, indicate a strong association, whereas categories from the origin and the other indicate a weak association. Meanwhile, categories near the origin indicate a random pattern of association with categories from other factors. (A) Consider dominant sand fly species Lutzomyia (Lu), sampled environment (Env, Per = peridomiciliary; Dom = Domicile), time in relation to the deltamethrin fogging (Pre = before fogging; Post = after fogging), focal house intervention status (Cont = Control; Fog = Fogged), PCR diagnostic Leishmania spp. (Leish, Pos = positive; Neg = negative), the blood-feeding of any vertebrate species (blood, Pos = positive; Neg = negative) of the pool of Lutzomyia analyzed. The cumulative variance explained by the two dimensions projected is 22%. (B) Consider the species Lutzomyia (Lu), PCR diagnosis of Leishmania spp. (Leish) and blood meals from the following hosts: human, cow, dog, goat, pig and poultry. Results come from 455 pools of sand flies. The cumulative variance explained by the two dimensions projected is 13%. To ease visualization the coordinates of categories near the origin were altered to ensure their reading.
Table 2 shows the blood meals found in the sand fly pools by molecular methods. Pools from all species fed on birds and dogs. None of the Lu. trapidoi pools had blood from cows, none of the Lu. gomezi pools had blood from goats, and none of the Lu. dysponeta pools had blood from cows or pigs. In accordance with their description as zoophilic species, Lu. dysponeta and Lu. triramula did not have blood from humans.
Table 2.
Number of pools with different blood sources for each of the dominant sand fly species at Trinidad de Las Minas, Panamá
| Species | Vertebrate | Dog | Cow | Pig | Human | Goat | Poultry |
|---|---|---|---|---|---|---|---|
| Lutzomyia gomezi (Nitzulescu) | 34 | 7 | 6 | 14 | 4 | 0 | 10 |
| Lutzomyia trapidoi (Fairchild and Hertig) | 22 | 3 | 0 | 1 | 11 | 4 | 7 |
| Lutzomyia panamensis (Shannon) | 14 | 4 | 1 | 3 | 3 | 7 | 2 |
| Lutzomyia dysponeta (Fairchild and Hertig) | 11 | 1 | 0 | 0 | 0 | 8 | 2 |
| Lutzomyia triramula (Fairchild and Hertig) | 29 | 4 | 3 | 20 | 0 | 5 | 2 |
| Total | 110 | 19 | 10 | 38 | 18 | 24 | 23 |
The result from the MCA between blood-feeding sources and Leishmania spp. infection detected by PCR is shown in Figure 2B. Lutzomyia trapidoi and Lu. triramula were more likely to have Leishmania spp. parasites in their pools. Lutzomyia trapidoi pools were mainly associated with blood meals from dogs, birds, and humans, whereas Lu. triramula and Lu. gomezi pools were associated with pig blood. Most pools from Lu. panamensis and Lu. dysponeta had no blood meals or fed on goats, and had no parasites detected by PCR. According to the MCA analysis, pools from all sand fly species had a random pattern with respect to not having a specific blood source, as pools negative for most blood sources were close to the origin (Figure 2B). A point worth highlighting here is that some of the Leishmania spp.–positive DNA samples were from sand fly pools without blood meals (Table 3) which indicates that parasites were associated with the sand flies and that amplified Leishmania spp. DNA did not come from blood meals.
Table 3.
Leishmania spp. ITS-1 and blood meal Cyt b PCR amplification in pools from dominant sand fly species at Trinidad de Las Minas, Panamá
| Species | ITS-1 (+) Cyt b (+) | ITS-1 (+) Cyt b (−) | ITS-1 (−) Cyt b (+) | ITS-1 (−) Cyt b (−) |
|---|---|---|---|---|
| Lutzomyia gomezi (Nitzulescu) | 24 | 24 | 0 | 56 |
| Lutzomyia trapidoi (Fairchild and Hertig) | 14 | 75 | 0 | 27 |
| Lutzomyia panamensis (Shannon) | 11 | 18 | 0 | 73 |
| Lutzomyia dysponeta (Fairchild and Hertig) | 27 | 47 | 0 | 16 |
| Lutzomyia triramula (Fairchild and Hertig) | 9 | 6 | 0 | 28 |
Cyt b = cytochrome b; ITS-1 = internal transcribed spacer region 1. (+) indicates the number of pools that have PCR amplifications and (−) indicates the number of pools without PCR amplifications
BLAST analysis of ITS-1 sequences suggested the presence of L. panamensis in two pools of Lu. trapidoi (99% identity, GenBank Accession Numbers MH195209 and MH195212, Table 4). One of the Lu. trapidoi pools and one of the Lu. tiramula pools suggested the presence of Leishmania naiffi (99% identity, Accession Numbers MH195211 and MH195205, Table 4). Lutzomyia gomezi, Lu. panamensis and Lu. dysponeta pools had DNA that suggested infections with Leishmania (Viannia) spp. In Lu. triramula, we also detected Trypanosomatidae DNA that could not be identified at the genus level.
Table 4.
ITS-1 DNA sequence BLAST analysis results
| Sandfly species | Analyzed pools | Genbank ID code | Potential parasite species | Genbank ID code of closest sequence, parasite species and identity (%) |
|---|---|---|---|---|
| Lutzomyia gomezi (Nitzulescu) | 2 | MH195204 | Leishmania (Viannia) spp. | DQ182543.1, L. naiffi, 89 |
| MH195210 | Leishmania (Viannia) spp. | DQ182543.1, L. naiffi, 88 | ||
| Lutzomyia trapidoi (Fairchild and Hertig) | 4 | MH195208 | Leishmania (Viannia) spp. | DQ182543.1, L. naiffi, 93 |
| MH195209 | Leishmania (Viannia) panamensis | CP009396.1, Leishmania panamensis, 99 | ||
| MH195211 | Leishmania (Viannia) naiffi | DQ182543, L. naiffi, 99 | ||
| MH195212 | Leishmania (Viannia) panamensis | CP009396.1, Leishmania panamensis, 99 | ||
| Lutzomyia panamensis (Shannon) | 1 | MH195206 | Leishmania (Viannia) spp. | DQ182543.1, L. naiffi, 93 |
| Lutzomyia dysponeta (Fairchild and Hertig) | 1 | MH195203 | Leishmania (Viannia) spp. | DQ182543, L. naiffi, 89 |
| Lutzomyia triramula (Fairchild and Hertig) | 2 | MH195205 | L. (Viannia) naiffi | DQ182543.1, L. naiffi, 99 |
| MH195207 | Trypanosomatidae spp. | JN673400.1, Trypanosomatidae sp., 97 |
ITS-1 = internal transcribed spacer region 1. Analyzed pools indicates the number of pools that were sequenced, Genbank ID Code indicates the accession code for the sequences generated from the analyzed pools. Potential parasite species indicates the most likely species based on a sequence identity. For this we identified to the species level when sequences were over 95% similar, otherwise to the genus level. Finally, Genbank ID code of closest sequence indicates the accession number for the most similar ITS-1 DNA sequence according to the BLAST analysis, the parasite species and identity (%) indicate the percent identity between the sequences from sand fly pools that we analyzed and the sequences in the National Center for Biotechnology Information Genbank.
DISCUSSION
In TM, we have observed that vector control insecticide interventions significantly reduced sand fly species diversity and abundance.26,27 To complement these observations, here we evaluated Leishmania spp. infection rate and blood meals in the most abundant anthropophilic and zoophilic Lutzomyia spp. captured inside and around selected houses from this community before and after deltamethrin fogging. Studies by Arias et al.,64 Miranda et al.,65 and Silva and Castellon,66 in ACL-endemic areas of Panamá, found low infection rates in sand flies, ranging between 0 and 3%, with observations coming from areas that have been subjected to forest clearance and fragmentation, like TM. In that sense, our infection rate results are within the range of what has been observed at similar sites and suggest deltamethrin fogging decreased the abundance of vectors26 but did not change the overall proportion of sand flies infected with Leishmania spp. parasites.
Lu. trapidoi and Lu. gomezi were the most abundant sand fly species inside the studied households, the presence of both species being the major risk factor for clinical cutaneous leishmaniasis cases in this community.25 Likewise, Lu. trapidoi and Lu. gomezi infection with Leishmania spp. parasites and blood meal sources, which include domestic animals, unidentified vertebrates, and humans, suggest an important role in ACL transmission to humans. We think Lu. trapidoi is the main ACL vector in TM because Lu. trapidoi sand flies captured inside households were the most frequently engorged with blood, suggesting an ability to feed and/or rest after a blood meal inside rural houses, thus, increasing potential contacts with humans. Moreover, Lu. trapidoi was the species that accounted for most of the detected human blood meals. Similar observations were made in an earlier study in wooded areas of Limbo and Aguacate, in Central Panamá, using human baits to sample adult sand flies.67 That study also recorded 70% of Lu. trapidoi feeding on rodents, primates, and edentates, all species commonly infected with Leishmania braziliensis.67 Also, for Lu. trapidoi we observed a high prevalence of infection with Leishmania spp. (0.135) before the deltamethrin fogging. The prevalence significantly (P < 0.05) decreased after the deltamethrin fogging (0.033). Nevertheless, the net effect of the intervention suggests that the reduction in infection could have been compensated by an increased abundance of Lu. trapidoi after the deltamethrin fogging,26 which has the potential to lead to similar transmission levels.68,69 However, these observations require further research because the relationship between vector abundance and transmission seems nonlinear for ACL.6 The vectorial importance of Lu. trapidoi at TM is furtherly supported by the observation of L. panamensis and L. naiffi in Lu. trapidoi pools, suggesting that Lu. trapidoi might be the main vector involved in the transmission of multiple Leishmania spp.8,25,26,66
Meanwhile, Lu. gomezi at TM had pigs as its most common blood source, followed by poultry and dogs, then humans. These observations for Lu. gomezi contrast with previous reports about humans being the most common blood meal source in human residences and deforested areas, where vegetation is sparse.70–73 This vector species has likely adapted to new environments by altering its feeding behavior, so that ACL transmission patterns might have concomitantly changed. Populations of Lu. gomezi are able to bite a wide range of vertebrates, in both domiciliary and peridomiciliary areas, making its control complicated,25,71 and suggesting an important role in Leishmania spp. parasite circulation among its community of vertebrate host species, as well as vectors.
By contrast to Lu. trapidoi and Lu. gomezi, Lu. panamensis was more prevalent and with higher abundance outside houses, in peridomiciliary areas.27 Lu. panamensis showed a greater proportion of blood meals from goats, compared with humans, even though it is considered anthropophilic.75–77 The Lu. panamensis abundance pattern, in addition to its low Leishmania spp. infection prevalence and diversity of blood meals, suggests a small role on ACL transmission to humans at our study site.
Surprisingly, Lu. triramula and Lu. dysponeta, zoophilic species, which were frequently caught in peridomiciles, were likely infected with Leishmania spp. In Panamá, these two species are zoophilic and have been captured in peridomiciliary and domiciliary environments, as well as in primary and secondary forests.10,26,78 Until now these species have not been implicated in the transmission of Leishmania spp. in the New World. Lutzomyia triramula belongs to the subgenus Trichopygomyia. This subgenus is characterized by species that are attracted to light.28 The medical importance of the subgenus is unknown because females are not anthropophilic. Nevertheless, Hashiguchi et al.79 captured Trichopygomyia members using human baits in a hyperendemic ACL area in Paraguay. However, to date, no Lu. triramula individuals have been captured using human landing catch, raising questions about their ability to transmit ACL parasites to humans. Interestingly, in Lu. triramula we also found DNA likely belonging to L. naiffi, a common ACL etiologic agent in South America,80–82 highlighting the need to better understand the role of this sand fly as a potential ACL vector. Although L. naiffi has never been reported in human cases from Panamá, it has been previously described infecting both Lu. trapidoi and Lu. gomezi collected in Barro Colorado, an island located relatively near our mainland study area.83
The blood meal analysis of Lu. dysponeta showed goats as the main blood source whereas Lu. triramula preferred pigs. Thus, the significance of our findings about Lu. triramula and Lu. dysponeta for the eco-epidemiology of the ACL in Panamá should be further investigated. Our evidence is exclusively based on Leishmania spp. parasite DNA detection by PCR, lacking individual based information about the blood foraging and the confirmation of infection by metacyclic Leishmania spp. parasites in these two sand fly species, as well as vectorial competence experiments.8
To further contextualize our parasite infection and blood meal results, during the study period we found domestic dogs to be present in almost all houses and to have seroprevalence patterns indicative of endemic transmission15 at TM. Nevertheless, we were unable to detect or isolate parasites from dogs,15 which does not allow us to implicate this species as a Leishmania spp. reservoir in the studied area. By contrast, we found high parasitemia in two-toed sloths, Choloepus hoffmani, from which we were able to isolate L. panamensis parasites, a parasite species commonly causing ACL in Panamá8 and the parasite whose DNA was present in Lu. trapidoi pools, and for which circumstantial evidence suggest a reservoir role in TM.14 All domestic animals whose blood was found in our samples were common in the study area, as well as spiny rats Proechymis spp.,14,25 but more definitive linkages in transmission will only be possible using more specific and sensitive tools for blood meal identification, for example, developing next generation sequencing techniques84 that will also allow the quantification of multiple blood sources.85
Regarding the limitations of our study, it is important to highlight that the probability of detecting Leishmania spp. in engorged female is increased by the potential presence of parasites in undigested blood. By contrast the detection of Leishmania spp. in non-engorged females implies the presence of parasites that survived the blood meal digestion, and which became metacyclic infective forms.86,87 In this study we did not separate engorged and non-engorged females for Leishmania spp. detection. This might slightly change the infection rates we estimated, assuming infections in positive pools were due to engorged females with parasites in blood, but for all species we mainly had positive pools without blood meals (Table 3), ensuring the validity of our inferences. Our infection estimates also have the limitation of being an approximation by coming from pools.34 For Lu. triramula it is also important to consider that our PCR results suggested that infection could have been also due to a non-Leishmania spp. Trypanosomatidae, given the detection of 500–900 bp band sizes in some pools. These sizes are unusual because Leishmania spp. normally amplify a 300–350-bp product with this methodology, and it is known that ITS-1 cannot discriminate between groups of Leishmania spp. and other trypanosomes in a contaminated sample.88 One possibility is that we amplified fragments that correspond to common protozoa belonging to the intestinal microbiota of sand flies.89 This limitation about parasite identification could be overcome by evaluating other molecular markers for Leishmania spp. detection.87 Similarly, our finding of L. naiffi high identity DNA in Lu. trapidoi and Lu. triramula pools opens questions for further research, specially to elucidate whether this parasite species has become an ACL etiologic agent in Panamá, and whether L. naiffi has a complex transmission cycle where Lu. triramula might act as a bridge vector90 between wildlife, in Brazil L. naiffi reservoirs are armadillos, Dasypus novemcinctus, and Dasypus kapplieri,80 species we have not observed at the study site,14 and domestic hosts although Lu. trapidoi might be the main vector transmitting parasites to humans as suggested by the blood-feeding patterns of these two sand fly species. Unfortunately, due to the lack of resources we did not study infections and blood meals at the individual sand fly level, which would have been ideal to better understand the association between specific blood meal source species and infection. Future studies should be focused on studying infections and blood meals at the individual level, also using more refined tools for Leishmania spp. identification to better understand the circulation and transmission of ACL parasites between humans, domestic, and wildlife animals.
Finally, Leishmania spp. infections were common in the five most abundant species at our study site, which included both anthropophilic (Lu. trapidoi, Lu. gomezi, and Lu. panamensis) and zoophilic species (Lu. triramula and Lu. dysponeta), highlighting the need to consider the role that blood feeding by a community of vector species has on a community of vertebrate host species. The synergy between these two types of feeding behaviors might play an important role in the transmission dynamics of leishmaniasis, for example, with zoophilic species introducing Leishmania spp. parasites in synanthropic environments, and anthropophilic species spreading the infection to humans,90 highlighting the importance of the sand fly species community for the transmission of leishmaniasis,91 something that should be studied in further detail. In conclusion, our results furtherly support the suggestion that Lu. trapidoi is the main sand fly vector species transmitting Leishmania spp. to humans in Panamá. This species is strongly associated with humans and domestic animals, for example, dogs,15 infected by (or exposed to) ACL parasites, and its abundance was associated with transmission to humans at the household level.25 Our results also highlight the importance of implementing active surveillance looking at both anthropophilic and zoophilic species, to better understand the eco-epidemiology of ACL parasite transmission. This study and our previous results,26 call for more detailed knowledge about pesticide impacts in sand fly vector communities, to better understand the trade-offs that might emerge by applying insecticides, where some sand fly species can increase their proportional and total abundance,26 as we observed for Lu. trapidoi.
Supplementary Material
Acknowledgments:
We thank Roberto Rojas for his support in the identification of specimens and José Montenegro for his support with fieldwork; Nathan Gundacker for English edition; Vanessa Vásquez and Leyda Abrego for DNA sequencing analysis; Ana Rosa Caballero for helping with laboratory analysis; and Caitlin Mertzlufft for helping with the mapping.
Note: Supplemental figure and table appear at www.ajtmh.org.
REFERENCES
- 1.Miranda A, Carrasco R, Paz H, Pascale JM, Samudio F, Saldaña A, Santamaría G, Mendoza Y, Calzada JE, 2009. Molecular epidemiology of American tegumentary leishmaniasis in Panama. Am J Trop Med Hyg 81: 565–571. [DOI] [PubMed] [Google Scholar]
- 2.Ministerio de Salud de Panamá (Minsa) , 2014. Informe Anual de Epidemiología de la Leishmaniasis. Panamá: Depto. De Epidemiologia, Dirección de Salud Pública. [Google Scholar]
- 3.Chaves LF, Cohen JM, Pascual M, Wilson ML, 2008. Social exclusion modifies climate and deforestation impacts on a vector-borne disease. PLoS Negl Trop Dis 2: e176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alvar J, Vélez ID, Bern C, Herrero M, Desjeux P, Cano J, Jannin J, den Boer M; WHO Leishmaniasis Control Team , 2012. Leishmaniasis worldwide and global estimates of its incidence. PLoS One 7: e35671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Salomón OD, Quintana MG, Mastrángelo AV, Fernández MS, 2012. Leishmaniasis and climate change—case study: Argentina. J Trop Med 2012: 601242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chaves LF, Calzada JE, Valderama A, Saldaña A, 2014. Cutaneous leishmaniasis and sand fly fluctuations are associated with El Niño in Panamá. PLoS Negl Trop Dis 8: e3210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yamada K, Valderrama A, Gottdenker N, Cerezo L, Minakawa N, Saldaña A, Calzada JE, Chaves LF, 2016. Macroecological patterns of American cutaneous leishmaniasis transmission across the health areas of Panamá (1980–2012). Parasite Epidemiol Control 1: 42–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Christensen HA, Fairchild GB, Herrer A, Johnson CM, Young DG, Vasquez AM, 1983. The ecology of cutaneous leishmaniasis in the Republic of Panama. J Med Entomol 20: 463–484. [DOI] [PubMed] [Google Scholar]
- 9.Vásquez AM, Paz H, Méndez E, Alvar J, 1994. Leishmaniasis en Panamá. Panamá: Ministerio de Salud, 12.
- 10.Valderrama A, Herrera M, Salazar A, 2008. Relacioìn entre la composicioìn de especies del geìnero de Lutzomyia frança (Diptera: Psychodidae: Phlebotominae) y los diferentes tipos de bosques en Panamaì. Acta Zool Mex 24: 67–78. [Google Scholar]
- 11.Telford SR, Herrer A, Christensen HA, 1972. Enzootic cutaneous leishmaniasis in eastern Panama. Ecological factors relating to the mammalian hosts. Ann Trop Med Parasitol 66: 173–179. [DOI] [PubMed] [Google Scholar]
- 12.Herrer A, Christensen HA, 1980. Leishmania braziliensis in the Panamanian two-toed sloth, Choloepus hoffmanni. Am J Trop Med Hyg 29: 1196–1200. [DOI] [PubMed] [Google Scholar]
- 13.Christensen H, Johnson C, Vasquez AM, 1993. Leishmaniasis cutánea en Panamá: un breve resumen [Article in Spanish]. Rev Med Panama 9: 182–187. [PubMed] [Google Scholar]
- 14.González K, et al. 2015. Survey of wild mammal hosts of cutaneous leishmaniasis parasites in Panamá and Costa Rica. Trop Med Health 43: 75–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Calzada JE, Saldaña A, González K, Rigg C, Pineda V, Santamaría AM, Rodríguez I, Gottdenker NL, Laurenti MD, Chaves LF, 2015. Cutaneous leishmaniasis in dogs: is high seroprevalence indicative of a reservoir role? Parasitology 142: 1202–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miranda A, Saldaña A, González K, Paz H, Santamaría G, Samudio F, Calzada JE, 2012. Evaluation of PCR for cutaneous leishmaniasis diagnosis and species identification using filter paper samples in Panama, central America. Trans R Soc Trop Med Hyg 106: 544–548. [DOI] [PubMed] [Google Scholar]
- 17.Kent RJ, 2009. Molecular methods for arthropod bloodmeal identification and applications to ecological and vector-borne disease studies. Mol Ecol Resour 9: 4–18. [DOI] [PubMed] [Google Scholar]
- 18.Afonso MM, Duarte R, Miranda JC, Caranha L, Rangel EF, 2012. Studies on the feeding habits of Lutzomyia (Lutzomyia) longipalpis (Lutz & Neiva, 1912) (Diptera: Psychodidae: Phlebotominae) populations from endemic areas of American visceral leishmaniasis in northeastern Brazil. J Trop Med 2012: 858657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rêgo FD, Rugani JMN, Shimabukuro PHF, Tonelli GB, Quaresma PF, Gontijo CMF, 2015. Molecular detection of Leishmania in phlebotomine sand flies (Diptera: Psychodidae) from a cutaneous leishmaniasis focus at Xakriabá Indigenous Reserve, Brazil. PLoS One 10: e0122038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Van Eys GJ, Schoone GJ, Kroon NC, Ebeling SB, 1992. Sequence analysis of small subunit ribosomal RNA genes and its use for detection and identification of Leishmania parasites. Mol Biochem Parasitol 51: 133–142. [DOI] [PubMed] [Google Scholar]
- 21.Kirstein F, Gray JS, 1996. A molecular marker for the identification of the zoonotic reservoirs of lyme borreliosis by analysis of the blood meal in its European vector Ixodes ricinus. Appl Environ Microbiol 62: 4060–4065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Aransay AM, Scoulica E, Tselentis Y, 2000. Detection and identification of Leishmania DNA within naturally infected sand flies by seminested PCR on minicircle kinetoplastic DNA. Appl Environ Microbiol 66: 1933–1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schönian G, Nasereddin A, Dinse N, Schweynoch C, Schallig HDF, Presber W, Jaffe CL, 2003. PCR diagnosis and characterization of Leishmania in local and imported clinical samples. Diagn Microbiol Infect Dis 47: 349–358. [DOI] [PubMed] [Google Scholar]
- 24.Quaresma PF, de Lima Carvalho GM, Ramos MCNF, Andrade Filho JD, 2012. Natural Leishmania spp. reservoirs and phlebotomine sand fly food source identification in Ibitipoca State Park, Minas Gerais, Brazil. Mem Inst Oswaldo Cruz 107: 480–485. [DOI] [PubMed] [Google Scholar]
- 25.Saldaña A, Chaves LF, Rigg CA, Wald C, Calzada JE, 2013. Clinical cutaneous leishmaniasis rates are associated with household Lutzomyia gomezi, Lu. panamensis, and Lu. trapidoi abundance in Trinidad de Las Minas, western Panama. Am J Trop Med Hyg 88: 572–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chaves LF, Calzada JE, Rigg C, Valderrama A, Gottdenker NL, Saldaña A, 2013. Leishmaniasis sand fly vector density reduction is less marked in destitute housing after insecticide thermal fogging. Parasit Vectors 6: 164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Calzada JE, Saldaña A, Rigg C, Valderrama A, Romero L, Chaves LF, 2013. Changes in phlebotomine sand fly species composition following insecticide thermal fogging in a rural setting of western Panamá. PLoS One 8: e53289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Young D, Duncan M, 1994. Guide to the Identification and Geographic Distribution of Lutzomyia Sand Flies in Mexico, the West Indies, Central and South America (Diptera: Psychodidae). Memories of the American Entomological Institute. Gainesville, FL: Associated Publishers, 54.
- 29.Kuhls K, Mauricio IL, Pratlong F, Presber W, Schönian G, 2005. Analysis of ribosomal DNA internal transcribed spacer sequences of the Leishmania donovani complex. Microbes Infect 7: 1224–1234. [DOI] [PubMed] [Google Scholar]
- 30.Kent RJ, Norris DE, 2005. Identification of mammalian blood meals in mosquitoes by a multiplexed polymerase chain reaction targeting cytochrome B. Am J Trop Med Hyg 73: 336–342. [PMC free article] [PubMed] [Google Scholar]
- 31.Fornadel CM, Norris DE, 2008. Increased endophily by the malaria vector Anopheles arabiensis in southern Zambia and identification of digested blood meals. Am J Trop Med Hyg 79: 876–880. [PMC free article] [PubMed] [Google Scholar]
- 32.Kumar S, Stecher G, Tamura K, 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33: 1870–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Farrington CP, 1992. Estimating prevalence by group testing using generalized linear models. Stat Med 11: 1591–1597. [DOI] [PubMed] [Google Scholar]
- 34.Speybroeck N, Williams CJ, Lafia KB, Devleesschauwer B, Berkvens D, 2012. Estimating the prevalence of infections in vector populations using pools of samples. Med Vet Entomol 26: 361–371. [DOI] [PubMed] [Google Scholar]
- 35.Venables WN, Ripley BD, 2002. Modern Applied Statistics with S. Switzerland AG: Springer. [Google Scholar]
- 36.Añez N, Nieves E, Cazorla D, Oviedo M, De Yarbuh AL, Valera M, 1994. Epidemiology of cutaneous leishmaniasis in Merida, Venezuela. III. Altitudinal distribution, age structure, natural infection and feeding behavior of sandflies and their relation to the risk of transmission. Ann Trop Med Parasitol 88: 279–287. [DOI] [PubMed] [Google Scholar]
- 37.Chaves LF, Harrington LC, Keogh CL, Nguyen AM, Kitron UD, 2010. Blood-feeding patterns of mosquitoes: random or structured? Front Zool 7: 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rabinovich JE, Kitron UD, Obed Y, Yoshioka M, Gottdenker N, Chaves LF, 2011. Ecological patterns of blood-feeding by kissing-bugs (Hemiptera, Reduviidae, Triatominae). Mem Inst Oswaldo Cruz 106: 479–494. [DOI] [PubMed] [Google Scholar]
- 39.Palatnik-de-Sousa C, Day MJ, 2011. One Health: the global challenge of epidemic and endemic leishmaniasis. Parasit Vectors 4: 197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chaves LF, 2017. Climate change and the biology of insect vectors of human pathogens. Johnson S, Jones H, eds. Global Climate Change and Terrestrial Invertebrates. Hoboken, NJ: Wiley-Blackwell, 126–147. [Google Scholar]
- 41.Ruiz Márvez E, 2011. Estandarización de la Técnica de Amplificación del Gen Citocromo B, Para Identificar la Fuente de Alimento de Cx. quinquefasciatus en el Centro Agropecuario Marengo de la Universidad Nacional de Colombia Sede Bogotá. Bogotá, Colombia: Departamento de Salud Pública, Facultad de Medicina, Universidad Nacional de Colombia, 66.
- 42.Pérez JE, Ogusuku E, Inga R, Lopez M, Monje J, Paz L, Nieto E, Arevalo J, Guerra H, 1994. Natural Leishmania infection of Lutzomyia spp. in Peru. Trans R Soc Trop Med Hyg 88: 161–164. [DOI] [PubMed] [Google Scholar]
- 43.Torres M, et al. 1998. Lutzomyia nuñeztovari anglesi (Diptera: Psychodidae) as a probable vector of Leishmania braziliensis in the Yungas, Bolivia. Acta Trop 71: 311–316. [DOI] [PubMed] [Google Scholar]
- 44.Rodríguez N, De Lima H, Aguilar CM, Rodriguez A, Barker DC, Convit J, 2002. Molecular epidemiology of cutaneous leishmaniasis in Venezuela. Trans R Soc Trop Med Hyg 96 (Suppl 1): S105–S109. [DOI] [PubMed] [Google Scholar]
- 45.Jorquera A, González R, Marchán-Marcano E, Oviedo M, Matos M, 2005. Multiplex-PCR for detection of natural Leishmania infection in Lutzomyia spp. captured in an endemic region for cutaneous leishmaniasis in state of Sucre, Venezuela. Mem Inst Oswaldo Cruz 100: 45–48. [DOI] [PubMed] [Google Scholar]
- 46.De Pita-Pereira D, Alves CR, Souza MB, Brazil RP, Bertho AL, de Figueiredo Barbosa A, Britto CC, 2005. Identification of naturally infected Lutzomyia intermedia and Lutzomyia migonei with Leishmania (Viannia) braziliensis in Rio de Janeiro (Brazil) revealed by a PCR multiplex non-isotopic hybridisation assay. Trans R Soc Trop Med Hyg 99: 905–913. [DOI] [PubMed] [Google Scholar]
- 47.Córdoba-Lanús E, De Grosso ML, Piñero JE, Valladares B, Salomón OD, 2006. Natural infection of Lutzomyia neivai with Leishmania spp. in northwestern Argentina. Acta Trop 98: 1–5. [DOI] [PubMed] [Google Scholar]
- 48.Paiva BR, Secundino NF, Nascimento JC, Pimenta PF, Galati EA, Junior HF, Malafronte RS, 2006. Detection and identification of Leishmania species in field-captured phlebotomine sandflies based on mini-exon gene PCR. Acta Trop 99: 252–259. [DOI] [PubMed] [Google Scholar]
- 49.Perruolo G, Rodríguez N, Feliciangeli MD, 2006. Isolation of Leishmania (Viannia) braziliensis from Lutzomyia spinicrassa (species group Verrucarum) Morales Osorno Mesa, Osorno and Hoyos 1969, in the Venezuelan Andean region. Parasite 13: 17–22. [DOI] [PubMed] [Google Scholar]
- 50.Oliveira-Pereira YN, Rebêlo JM, Moraes JL, Pereira SR, 2006. [Molecular diagnosis of the natural infection rate due to Leishmania sp in sandflies (Psychodidae, Lutzomyia) in the Amazon region of Maranhão, Brazil] [Article in Portuguese.] Rev Soc Bras Med Trop 39: 540–543. [DOI] [PubMed] [Google Scholar]
- 51.Santamaría E, Ponce N, Zipa Y, Ferro C, 2006. Presencia en el peridomicilio de vectores infectados con Leishmania (Viannia) panamensis en dos focos endémicos en el occidente de Boyacá, piedemonte del valle del Magdalena medio, Colombia. Biomedica 26: 82–94. [PubMed] [Google Scholar]
- 52.Cochero S, Anaya Y, Díaz Y, Paternina M, Luna A, Paternina L, Eduar Elías B, 2007. [Natural infection of Lutzomyia cayennensis cayennensis with trypanosomatid parasites (Kinetoplastida: Trypanosomatidae) in Los Montes de Maria, Colombia] [article in Spanish]. Rev Cubana Med Trop 59: 35–39. [PubMed] [Google Scholar]
- 53.Do Nascimento JC, de Paiva BR, dos Santos Malafronte R, Fernandes WD, Galati EA, 2007. Natural infection of phlebotomines (Diptera: Psychodidae) in a visceral-leishmaniasis focus in Mato Grosso do Sul, Brazil. Rev Inst Med Trop 49: 119–122. [DOI] [PubMed] [Google Scholar]
- 54.Neitzke HC, Scodro RB, Castro KR, Sversutti AC, Silveira TG, Teodoro U, 2008. Research of natural infection of phlebotomines for Leishmania, in the state of Paraná. Rev Soc Bras Med Trop 41: 17–22. [DOI] [PubMed] [Google Scholar]
- 55.Marcondes CB, Bittencourt IA, Stoco PH, Eger I, Grisard EC, Steindel M, 2009. Natural infection of Nyssomyia neivai (Pinto, 1926) (Diptera: Psychodidae, Phlebotominae) by Leishmania (Viannia) spp. in Brazil. Trans R Soc Trop Med Hyg 103: 1093–1097. [DOI] [PubMed] [Google Scholar]
- 56.Sánchez-García L, Berzunza-Cruz M, Becker-Fauser I, Rebollar-Téllez EA, 2010. Sand flies naturally infected by Leishmania (L.) mexicana in the peri-urban area of Chetumal city, Quintana Roo, México. Trans R Soc Trop Med Hyg 104: 406–411. [DOI] [PubMed] [Google Scholar]
- 57.Kato H, Gomez EA, Cáceres AG, Vargas F, Mimori T, Yamamoto K, Iwata H, Korenaga M, Velez L, Hashiguchi Y, 2011. Natural infections of man-biting sand flies by Leishmania and Trypanosoma species in the northern Peruvian Andes. Vector Borne Zoonotic Dis 11: 515–521. [DOI] [PubMed] [Google Scholar]
- 58.Valdivia HO, et al. 2012. Natural Leishmania infection of Lutzomyia auraensis in Madre de Dios, Peru, detected by a fluorescence resonance energy transfer-based real-time polymerase chain reaction. Am J Trop Med Hyg 87: 511–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Vásquez Trujillo A, González Reina AE, Góngora Orjuela A, Prieto Suárez E, Palomares JE, Buitrago Alvarez LS, 2013. Seasonal variation and natural infection of Lutzomyia antunesi (Diptera: Psychodidae: Phlebotominae), an endemic species in the Orinoquia region of Colombia. Mem Inst Oswaldo Cruz 108: 463–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Brito VN, Almeida Ado B, Nakazato L, Duarte R, Souza CO, Sousa VR, 2014. Phlebotomine fauna, natural infection rate and feeding habits of Lutzomyia cruzi in Jaciara, state of Mato Grosso, Brazil. Mem Inst Oswaldo Cruz 109: 899–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Neitzke-Abreu HC, Reinhold-Castro KR, Venazzi MS, Scodro RB, Dias Ade C, Silveira TG, Teodoro U, Lonardoni MV, 2014. Detection of Leishmania (Viannia) in Nyssomyia neivai and Nyssomyia whitmani by multiplex polymerase chain reaction, in southern Brazil. Rev Inst Med Trop Sao Paulo 56: 391–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Moya S, Giuliani M, Manteca Acosta M, Salomón OD, Liotta DJ, 2015. First description of Migonemyia migonei (França) and Nyssomyia whitmani (Antunes & Coutinho) (Psychodidae: Phlebotominae) natural infected by Leishmania infantum in Argentina. Acta Trop 152: 181–184. [DOI] [PubMed] [Google Scholar]
- 63.Pereira Júnior AM, Garcia Teles CB, de Azevedo dos Santos AP, de Souza Rodrigues M, Marialva EF, Costa Pessoa FA, Fernandes Medeiros J, 2015. Ecological aspects and molecular detection of Leishmania DNA ross (Kinetoplastida: Trypanosomatidae) in phlebotomine sandflies (Diptera: Psychodidae) in terra Firme and Várzea environments in the middle Solimões region, Amazonas state, Brazil. Parasit Vectors 8: 180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Arias JR, Miles MA, Naiff RD, Póvoa MM, Freitas RA, Biancardi CB, Castellon EG, 1985. Flagellate infection of Brazilian sandflies (Diptera: Psychodidadae): isolation in vitro and biochemical identification of Endotrypanum and Leishmania. Am J Trop Med Hyg 34: 1098–1108. [DOI] [PubMed] [Google Scholar]
- 65.Miranda JC, Reis E, Schriefer A, Gonçalves M, Reis MG, Carvalho L, Fernandes O, Barral-Netto M, Barral A, 2002. Frequency of infection of Lutzomyia phlebotomines with Leishmania braziliensis in a Brazilian endemic area as assessed by pinpoint capture and polymerase chain reaction. Mem Inst Oswaldo Cruz 97: 185–188. [DOI] [PubMed] [Google Scholar]
- 66.Silva TM, Castellón GE, 2012. Flebotomineos (Diptera: Psychodiae) infectados naturalmente por tripanosomatídeos (Kinetoplastida: Trypanosomatidae) em fragmentos florestais urbanos em Manaus—Amazonas (Brasil). Rev Colombiana Cienc Anim 4: 121–129. [Google Scholar]
- 67.Tesh RB, Chaniotis BN, Aronson MD, Johnson KM, 1971. Natural host preferences of Panamanian phlebotomine sandflies as determined by precipitin test. Am J Trop Med Hyg 20: 150–156. [DOI] [PubMed] [Google Scholar]
- 68.Chaves LF, Hernandez MJ, 2004. Mathematical modelling of American cutaneous leishmaniasis: incidental hosts and threshold conditions for infection persistence. Acta Trop 92: 245–252. [DOI] [PubMed] [Google Scholar]
- 69.Chaves LF, Hernandez MJ, Dobson AP, Pascual M, 2007. Sources and sinks: revisiting the criteria for identifying reservoirs for American cutaneous leishmaniasis. Trends Parasitol 23: 311–316. [DOI] [PubMed] [Google Scholar]
- 70.Porter C, De Foliart G, 1981. The man-biting activity of phlebotomine sand flies (Diptera: Psychodidae) in tropical wet forest environment in Colombia. Arq Zool São Paulo 30: 81–158. [Google Scholar]
- 71.Feliciangeli MD, 1987. Ecology of sandflies (Diptera: Psychodidae) in a restricted focus of cutaneous leishmaniasis in northern Venezuela. II. Species composition in relation to habitat, catching method and hour of catching. Mem Inst Oswaldo Cruz 82: 125–131. [DOI] [PubMed] [Google Scholar]
- 72.Feliciangeli MD, 1997. Hourly activity of Lutzomyia ovallesi and L. gomezi (Diptera: Psychodidae), vectors of cutaneous leishmaniasis in northcentral Venezuela. J Med Entomol 34: 110–115. [DOI] [PubMed] [Google Scholar]
- 73.Valderrama A, Tavares M, Andrade D, 2014. Phylogeography of the Lutzomyia gomezi (Diptera: Phlebotominae) on the Panama Isthmus. Parasit Vectors 7: 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Young DG, Arias JR, 1992. Flebótomos Vectores de Leishmaniosis en las Américas. Washington, DC: OPAS, 33.
- 75.Contreras MA, 2013. Lutzomyia spp. (Diptera: Psychodidae) en Zonas Cafeteras de la Región Andina Colombiana: Taxonomía e Importancia Médica. Tesis de Maestría, Facultad de Ciencias, Universidad Nacional de Colombia, Medellín, Colombia, 196. [Google Scholar]
- 76.Dutari LC, Loaiza JR, 2014. American cutaneous leishmaniasis in Panama: a historical review of entomological studies on anthropophilic Lutzomyia sand fly species. Parasit Vectors 7: 218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Feliciangeli M, 2014. Leishmaniasis en Venezuela: situación actual, acciones y perspectivas para el control vectorial en el marco de un programa de control multisectorial. Bol Mal Salud Amb 54: 1–7. [Google Scholar]
- 78.Cazorla-Perfetti D, 2015. Lista comentada de los flebotominos (Diptera: Psychodidae, Phlebotominae). Revista Multidisciplinaria Del Consejo De Investigación De La Universidad De Oriente 27: 178–231. [Google Scholar]
- 79.Hashiguchi Y, Chiller T, Inchausti A, De Arias A, Kawabata M, Alexander JB, 1992. Phlebotomine sand fly species in Paraguay and their infection with Leishmania. Ann Trop Med Parasitol 86: 175–180. [DOI] [PubMed] [Google Scholar]
- 80.Naiff R, Freitas R, Naiff M, Arias J, Barret T, Momen H, Grimaldi Júnior G, 1991. Epidemiological and nosological aspects of Leishmania naiffi Lainson & Shaw, 1989. Mem Inst Oswaldo Cruz 86: 317–321. [DOI] [PubMed] [Google Scholar]
- 81.Pratlong F, Deniau M, Darie H, Eichenlaub S, Pröll S, Garrabe E, Dedet J, 2002. Human cutaneous leishmaniasis caused by Leishmania naiffi is wide-spread in South America. Ann Trop Med Parasitol 96: 781–785. [DOI] [PubMed] [Google Scholar]
- 82.Fagundes-Silva GA, Romero GA, Cupolillo E, Yamashita EP, Gomes-Silva A, Guerra JA, Da-Cruz AM, 2015. Leishmania (Viannia) naiffi: rare enough to be neglected? Mem Inst Oswaldo Cruz 110: 797–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Azpurua J, De La Cruz D, Valderama A, Windsor D, 2010. Lutzomyia sand fly diversity and rates of infection by Wolbachia and an exotic Leishmania species on Barro Colorado Island, Panama. PLoS Negl Trop Dis 4: e627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kieran TJ, Gottdenker NL, Varian CP, Saldaña A, Means N, Owens D, Calzada JE, Glenn TC, 2017. Blood meal source characterization using illumina sequencing in the Chagas disease vector Rhodnius pallescens (Hemiptera: Reduviidae) in Panamá. J Med Entomol 54: 1786–1789. [DOI] [PubMed] [Google Scholar]
- 85.Tanure A, Peixoto JC, Afonso MM, Duarte R, Pinheiro ADC, Coelho SVB, Barata RA, 2015. Identification of sandflies (Diptera: Psychodidae: Phlebotominae) blood meals in an endemic leishmaniasis area in Brazil. Rev Inst Med Trop Sao Paulo 57: 321–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Obwaller AG, Karakus M, Poeppl W, Töz S, Özbel Y, Aspöck H, Walochnik J, 2016. Could Phlebotomus mascittii play a role as a natural vector for Leishmania infantum? New data. Parasit Vectors 9: 458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Senghor MW, et al. 2016. Transmission of Leishmania infantum in the canine leishmaniasis focus of Mont-Rolland, Senegal: ecological, parasitological and molecular evidence for a possible role of Sergentomyia sand flies. PLoS Negl Trop Dis 10: 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Mahmoudzadeh-Niknam H, Abrishami F, Doroudian M, Moradi M, Alimohammadian M, Parvizi P, Hatam G, Mohebali M, Khalaj V, 2011. The problem of mixing up of Leishmania isolates in the laboratory: suggestion of ITS1 gene sequencing for verification of species. Iran J Parasitol 6: 41–48. [PMC free article] [PubMed] [Google Scholar]
- 89.Killick-Kendrick R, Molyneux DH, Ashford RW, 1974. Leishmania in phlebotomid sandflies. I. Modifications of the flagellum associated with attachment to the mid-gut and oesophageal valve of the sand fly. Proc R Soc Lond B Biol Sci 187: 409–419. [DOI] [PubMed] [Google Scholar]
- 90.Chaves LF, Añez N, 2004. Species co-occurrence and feeding behavior in sand fly transmission of American cutaneous leishmaniasis in western Venezuela. Acta Trop 92: 219–224. [DOI] [PubMed] [Google Scholar]
- 91.Chaves LF, Añez N, 2016. Nestedness patterns of sand fly (Diptera: Psychodidae) species in a neotropical semi-arid environment. Acta Trop 153: 7–13. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.