Abstract.
We attempted to identify Plasmodium falciparum histidine–rich protein 2/3 (pfhrp2/3) deletions among rapid diagnostic test (RDT)–negative but PCR- or microscopy-positive P. falciparum–infected individuals in areas of low transmission (Choma District, 2009–2011) and high transmission (Nchelenge District, 2015–2017) in Zambia. Through community-based surveys, 5,167 participants were screened at 1,147 households by P. falciparum histidine-rich protein 2 (PfHRP2)-based RDTs. Slides were made and dried blood spots were obtained for molecular analysis. Of 28 samples with detectable P. falciparum DNA, none from Nchelenge District were pfhrp2/3 negative. All eight samples from Choma District had detectable pfhrp3 genes, but pfhrp2 was undetectable in three. DNA concentrations of pfhrp2-negative samples were low (< 0.001 ng/μL). These findings suggest that PfHRP2-based RDTs remain effective tools for malaria diagnosis in Nchelenge District, but further study is warranted to understand the potential for pfhrp2/3 deletions in southern Zambia where malaria transmission declined over the past decade.
The availability of Plasmodium falciparum histidine-rich protein (PfHRP) 2–based rapid diagnostic tests (RDTs) has dramatically improved parasitological confirmation of suspected malaria cases in resource-limited settings. Recently, numerous studies reported P. falciparum parasites lacking pfhrp2 and pfhrp3 genes in Africa, including the Democratic Republic of the Congo, Eritrea, Ghana, Kenya, Mali, Mozambique, Rwanda, and Senegal.1–3 The pfhrp3 gene is highly homologous to pfhrp2,4 and parasites lacking both pfhrp2 and pfhrp3 genes, or substantial parts of these genes, do not express functional proteins and are not detected by PfHRP2-based RDTs.1
In Zambia, the National Malaria Control Center introduced PfHRP2-based RDTs in 2005 and achieved national-level scale-up in 2009.5 Zambia has made significant progress in reducing malaria transmission and aims to eliminate malaria by 2021. To achieve this goal, the use of PfHRP2-based RDTs has expanded beyond the diagnosis of suspected cases to also be used to screen individuals residing in proximity to symptomatic index cases through reactive case detection.6 Despite the expanded use of PfHRP2-based RDTs, the extent of pfhrp2/3 deletions in Zambia is unknown. We attempted to identify pfhrp2/3 deletions using blood samples collected during community-based, active case detection in Choma and Nchelenge districts, Zambia.
Choma District in the Southern Province has seasonal malaria transmission, and PfHRP2-based RDTs were introduced in 2007.7 As malaria transmission has declined, Choma District is considered a pre-elimination setting (malaria prevalence by RDT < 1%).8,9 By contrast, malaria transmission in Nchelenge District, Luapula Province, is high with little seasonal fluctuation. Malaria control interventions have been only modestly effective and malaria prevalence by RDT is approximately 50%.9,10 Luapula Province is one of the four focus provinces where the National Malaria Elimination Program supports additional malaria control efforts, including school-based bed net distribution, expansion of community case management, and training of health providers in the management of severe malaria.6
Random sampling based on satellite imagery was used to select households for participation in community-based surveys in between 2009 and 2011 in Choma District, and between 2015 and 2017 in Nchelenge District. Briefly, satellite images of the catchment area were used to create a sampling frame from which households were randomly selected for participation in community-based, serial cross-sectional surveys. Trained local field-workers used global positioning system coordinates to locate selected households for initial notification visits and data collection. All household residents older than the age of 3 months present at the time of the visit were eligible for enrollment, and written informed consent was obtained from all adults or caregivers of children who agreed to participate. Tympanic temperature was taken, and participants were tested for malaria by a PfHRP2-based RDT (Choma District: ICT Malaria P. falciparum [ICT Diagnostics, Cape Town, South Africa]; Nchelenge District: SD Bioline Malaria Ag P. falciparum [Standard Diagnostics, Inc., Gyeonggi-do, Republic of Korea]). Different RDTs were deployed at the two sites based on a change in the Zambian Ministry of Health guidelines in 2013. Both RDTs met World Health Organization procurement criteria and reliably detect parasite densities of 200 parasites/μL or higher.11,12 Using finger-prick blood, dried blood spots (DBS) were prepared for molecular analysis. All RDT-positive participants were offered treatment according to the guidelines of the Zambian Ministry of Health.
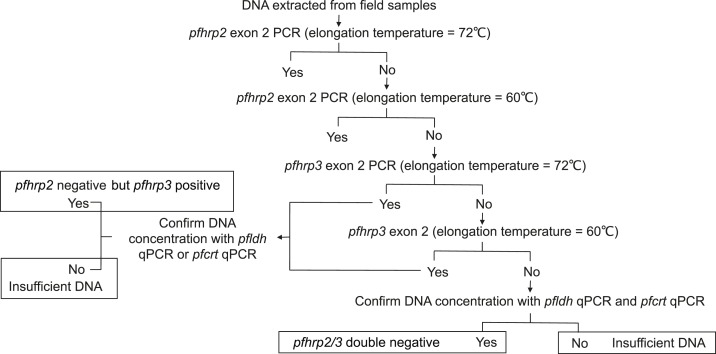
In Choma District, 2,183 residents from 414 households agreed to participate in the study. In Nchelenge District, 2,984 residents from 733 households agreed to participate. Samples were transported to laboratories at Macha Research Trust in Choma District or the Tropical Disease Research Centre in Ndola. DNA was extracted from the DBS and screened using a nested-PCR targeting the mitochondrial cytochrome b gene (cytb), which is conserved among the major human Plasmodium species.8,12 To exclude non–P. falciparum malaria infection, which results in a negative PfHRP2-based RDT, all cytb PCR-positive and RDT-negative samples from Choma District were further screened by species-specific qPCR.8,13 For samples from Nchelenge District, species were confirmed by trained microscopists. Rapid diagnostic test–negative but PCR- or microscopy-positive P. falciparum samples from both sites were analyzed by a series of PCRs targeting pfhrp2 or pfhrp3 as previously described.14 Briefly, RDT-negative but cytb PCR- or microscopy-positive samples were tested by PCR to amplify exon two of pfhrp2. Samples that failed amplification were assayed at lower elongation temperatures (Figure 1). Samples that failed pfhrp2 amplification at lower temperatures were subjected to pfhrp3 PCR. Samples that failed to amplify pfhrp3 were then assayed at lower elongation temperatures. DNA concentrations were determined by pfldh qPCR and those that failed to amplify pfhrp2 or pfhrp3 genes were further assayed using an additional single-copy gene qPCR-targeting pfcrt.15
Figure 1.
Workflow for detecting Plasmodium falciparum histidine–rich protein 2/3 (pfhrp2/3)-negative parasites.
Twenty-nine RDT-negative but P. falciparum–specific qPCR-confirmed samples from Choma District and 40 RDT-negative but PCR-positive and microcopy-confirmed P. falciparum samples from Nchelenge District were screened for pfhrp2/3 deletions (Table 1). The prevalence of fever among participants with RDT-negative but cytb PCR- or microscopy-positive results (Choma District = 3.4%; Nchelenge District = 0%) did not differ from the background population prevalence of fever on the day of the study visit (Choma District = 1.3%, P = 0.33; Nchelenge District = 3.6%, P = 0.22). All samples considered to have potential pfhrp2/3 deletions were additionally screened using a single-copy pfldh qPCR assay to ensure there was more than 0.0001 ng/μL P. falciparum DNA, the detection limit of the pfhrp2 and pfhrp3 assays.16 Samples with less than 0.0001 ng/μL parasite-specific DNA, the detection limit of the pfldh qPCR and the pfhrp2/3 PCR, were considered to have insufficient parasite DNA and were excluded from further analysis. Of those samples with sufficient DNA (Choma District = 8, Nchelenge District = 28), three samples from Choma District failed to amplify pfhrp2 (Table 1). All three samples with undetectable pfhrp2 were pfhrp3 PCR positive. Among these three samples with undetectable pfhrp2 but detectable pfhrp3 from Choma District, the presence of parasite DNA was confirmed using PCR targeting pfcrt. None of the RDT-negative but PCR-positive samples from Nchelenge District had evidence of pfhrp2 or pfhrp3 deletions.
Table 1.
Summary of screening results to detect Plasmodium falciparum histidine–rich protein 2/3 (pfhrp2/3)-negative parasites in northern and southern Zambia
| Study area | Year | RDT positivity in the community, % (95% CI) | Prevalence of fever in the community, % (95% CI) | Samples considered for screening | Prevalence of fever among sample, % (95% CI) | Samples with P. falciparum DNA concentration > 0.0001 ng/uL | pfhrp2-negative, number (%; 95% CI) | pfhrp2/3 double negative, number (%; 95% CI) |
|---|---|---|---|---|---|---|---|---|
| Choma | 2009–2011 | 0.7 (0.4, 1.1) | 1.3 (0.8, 1.8) | 29 | 3.4 (0.1, 18) | 8 | 3 (38; 8.5, 76) | 0 (0; 0, 37)* |
| Nchelenge | 2015–2017 | 45 (43, 47) | 3.6 (2.9, 4.3) | 40 | 0 (0, 8.9)* | 28 | 0 (0; 0, 14)* | 0 (0; 0, 14)* |
RDT = rapid diagnostic test.
* One-sided, 97.5% CI.
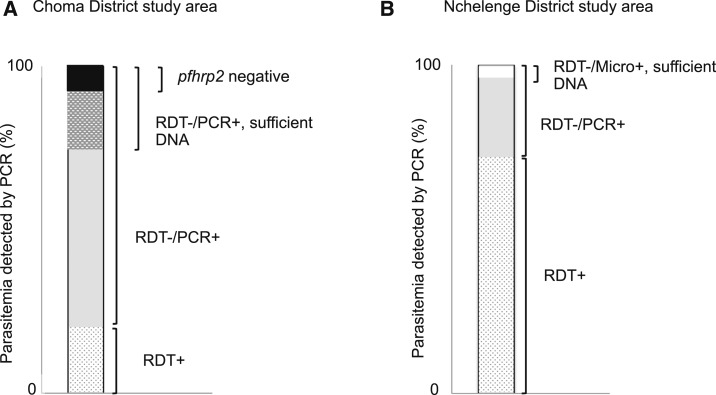
To our knowledge, this is the first report suggesting the possibility of pfhrp2 deletions in Zambia, although the level of parasite DNA was lower than those recommended by Parr et al.16 and limited our ability to make deletion calls. In our study, the range of DNA concentrations of pfhrp2-negative samples was 0.0002–0.0006 ng/μL (approximately 8–24 genomes/μL). However, all PCR protocols used to confirm the DNA concentration and subsequently amplify pfhrp2/3 had a detection limit of 0.0001 ng/μL (∼4 genomes/μL),16 and, therefore, the failure to amplify pfhrp2 in three samples was not likely because of insufficient DNA alone. It seems plausible that pfhrp2-deleted parasites are present in Choma District, Zambia, but further investigation is needed to confirm this observation. Genes encoding PfHRP2 and PfHRP3 share high homology,4 and detection of the D10 strain of P. falciparum (pfhrp2 negative but pfhrp3 positive) using PfHRP2-based RDTs at high parasitemia (≥ 1,000 parasites/μL) suggests that PfHRP3 cross-reacts with monoclonal antibodies to PfHRP2 that are used for PfHRP2-based RDTs.17 As none of three pfhrp2-negative parasites from Choma District had evidence of pfhrp3 deletions, such parasites may still be detected by PfHRP2-based RDTs if parasitemia were high. A previous study in this setting reported that most infected individuals had submicoscopic parasitemia with a low parasite load.8 These findings suggest that RDT-negative but PCR-positive infections in Choma District are mainly due to low parasitemia, with pfhrp2 deletions playing only a small role (Figure 2).
Figure 2.
Malaria diagnostic testing results for PCR-positive subjects in Choma and Nchelenge districts, Zambia. For study participants with Plasmodium falciparum parasitemia detected by PCR in (A) the Choma District study area, the black shaded area depicts P. falciparum histidine–rich protein 2 (pfhrp2)-negative isolates among rapid diagnostic test (RDT)-negative samples with sufficient DNA concentrations for pfhrp2 PCR testing, and (B) the Nchelenge district study area, pfhrp2 was successfully amplified in all RDT-negative samples with sufficient DNA for pfhrp2 PCR testing.
It was hypothesized and recently observed that the presence of parasites with pfhrp2 deletions may be masked in high transmission settings because of co-infection with pfhrp2 wild-type parasites.1,14 A prior study within the same study population in Nchelenge District reported that polyclonal infections are common, with a range of three to seven haplotypes present in each infected individual.18 Because of the high prevalence of polyclonal infections, we cannot exclude the possibility of parasites with pfhrp2 deletions circulating in Nchelenge District, but the presence of parasite clones with wild-type pfhrp2 should prolong the use of PfHRP2-based RDTs. Furthermore, the median parasite count of samples by microscopy was 27 parasites/μL (interquartile range = 8–39) in Nchelenge District, less than the detection threshold of commercially available RDTs.11 Based on this result, we conclude that the false-negative RDTs in Nchelenge District were most likely because of low parasitemia (Figure 2). As malaria transmission in Nchelenge District decreases, parasites with pfhrp2 deletions may become more important.
In summary, pfhrp2/3 deletions were not identified in the high transmission setting in northern Zambia, but evidence suggesting the presence of pfhrp2 deletions was found in the low transmission setting in southern Zambia. Our ability to confirm these gene deletions was limited by low DNA concentrations and small sample volumes. This is the first time parasites with possible pfhrp2 deletions have been identified in Zambia, but this study highlights the challenges of identifying pfhrp2/3 deletions using low-parasite density samples by PCR-based assays. Plasmodium falciparum histidine rich–protein 2-based RDTs remain effective tools for the diagnosis of malaria in Zambia, but further study is required to ensure their validity.
Acknowledgments:
This work was supported by the Johns Hopkins Malaria Research Institute, the Bloomberg Family Foundation, and the Division of Microbiology and Infectious Diseases, National Institutes of Allergies and Infectious Diseases, National Institutes of Health as part of the International Centers of Excellence for Malaria Research (U19 AI089680). We thank the field teams and laboratory staff at Macha Research Trust and the Tropical Disease Research Centre. Most importantly, we thank the residents of the Macha community and the Nchelenge community who participated in this study.
REFERENCES
- 1.Cheng Q, Gatton ML, Barnwell J, Chiodini P, McCarthy J, Bell D, Cunningham J, 2014. Plasmodium falciparum parasites lacking histidine-rich protein 2 and 3: a review and recommendations for accurate reporting. Malar J 13: 283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gendrot M, Fawaz R, Dormoi J, Madamet M, Pradines B, 2018. Genetic diversity and deletion of Plasmodium falciparum histidine-rich protein 2 and 3: a threat to diagnosis of P. falciparum malaria. Clin Microbiol Infect. Epub ahead of print. Available at: https://www.ncbi.nlm.nih.gov/pubmed/30267926. [DOI] [PubMed] [Google Scholar]
- 3.Berhane A, et al. 2018. Major threat to malaria control programs by Plasmodium falciparum lacking histidine-rich protein 2, Eritrea. Emerg Infect Dis 24: 462–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wellems TE, Howard RJ, 1986. Homologous genes encode two distinct histidine-rich proteins in a cloned isolate of Plasmodium falciparum. Proc Natl Acad Sci U S A 83: 6065–6069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yukich JO, et al. 2012. Reductions in artemisinin-based combination therapy consumption after the nationwide scale up of routine malaria rapid diagnostic testing in Zambia. Am J Trop Med Hyg 87: 437–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.PMI , 2018. FY 2018 Zambia Malaria Operational Plan—President’s Malaria Initiative. Available at: https://www.pmi.gov/docs/default-source/default-document-library/malaria-operational-plans/fy-2018/fy-2018-zambia-malaria-operational-plan.pdf?sfvrsn=7. Accessed October 23, 2018. [Google Scholar]
- 7.Stresman GH, Kamanga A, Moono P, Hamapumbu H, Mharakurwa S, Kobayashi T, Moss WJ, Shiff C, 2010. A method of active case detection to target reservoirs of asymptomatic malaria and gametocyte carriers in a rural area in Southern Province, Zambia. Malar J 9: 265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Laban NM, Kobayashi T, Hamapumbu H, Sullivan D, Mharakurwa S, Thuma PE, Shiff CJ, Moss WJ; Southern Africa International Centers of Excellence for Malaria R , 2015. Comparison of a PfHRP2-based rapid diagnostic test and PCR for malaria in a low prevalence setting in rural southern Zambia: implications for elimination. Malar J 14: 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kanyangarara M, et al. Southern Africa International Centers of Excellence for Malaria R , 2018. Malaria knowledge and bed net use in three transmission settings in southern Africa. Malar J 17: 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pinchoff J, Chaponda M, Shields TM, Sichivula J, Muleba M, Mulenga M, Kobayashi T, Curriero FC, Moss WJ; Southern Africa International Centers of Excellence for Malaria R , 2016. Individual and household level risk factors associated with malaria in Nchelenge district, a region with perennial transmission: a serial cross-sectional study from 2012 to 2015. PLoS One 11: e0156717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.WHO , 2017. Malaria Rapid Diagnostic Test Performance–Results of WHO Product Testing of Malaria RDTs: Round 7 (2015–2016). Available at: https://www.who.int/malaria/publications/atoz/978924151268/en/. Accessed October 23, 2018. [Google Scholar]
- 12.Steenkeste N, et al. 2009. Towards high-throughput molecular detection of Plasmodium: new approaches and molecular markers. Malar J 8: 86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Snounou G, Viriyakosol S, Jarra W, Thaithong S, Brown KN, 1993. Identification of the four human malaria parasite species in field samples by the polymerase chain reaction and detection of a high prevalence of mixed infections. Mol Biochem Parasitol 58: 283–292. [DOI] [PubMed] [Google Scholar]
- 14.Parr JB, et al. 2017. Pfhrp2-Deleted Plasmodium falciparum parasites in the Democratic Republic of the Congo: a national cross-sectional survey. J Infect Dis 216: 36–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Akter J, Thriemer K, Khan WA, Sullivan DJ, Jr., Noedl H, Haque R, 2012. Genotyping of Plasmodium falciparum using antigenic polymorphic markers and to study anti-malarial drug resistance markers in malaria endemic areas of Bangladesh. Malar J 11: 386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Parr JB, Anderson O, Juliano JJ, Meshnick SR, 2018. Streamlined, PCR-based testing for pfhrp2-and pfhrp3-negative Plasmodium falciparum. Malar J 17: 137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Baker J, McCarthy J, Gatton M, Kyle DE, Belizario V, Luchavez J, Bell D, Cheng Q, 2005. Genetic diversity of Plasmodium falciparum histidine-rich protein 2 (PfHRP2) and its effect on the performance of PfHRP2-based rapid diagnostic tests. J Infect Dis 192: 870–877. [DOI] [PubMed] [Google Scholar]
- 18.Pringle JC, Carpi G, Almagro-Garcia J, Zhu SJ, Kobayashi T, Mulenga M, Bobanga T, Chaponda M, Moss WJ, Norris DE, 2018. RTS,S/AS01 malaria vaccine mismatch observed among Plasmodium falciparum isolates from Southern and Central Africa and globally. Sci Rep 8: 6622. [DOI] [PMC free article] [PubMed] [Google Scholar]