Abstract
Jiangquhai pigs are one of the 42 representative local breeds listed in the national livestock genetic resources conservation project of China. This breed is known for its prolificacy, desirable meat quality, and excellent adaptability to crude feed and local environments. In this study, we genotyped 105 Jiangquhai pigs from the state conservation farm using GeneSeek GGP Porcine 80K SNP chip, and explored the SNP data to unravel genetic diversity, evolutionary phylogeny, signatures of selection, and population structure of Jiangquhai pigs in a context of 33 global breeds. Five indices of observed heterozygosity, expected heterozygosity, effective population size, runs of homozygosity, and linkage disequilibrium extent indicate that the Jiangquhai breed are still rich in genetic diversity in comparison with other breeds also from East China despite the recent decline of its population size. Phylogenetic, principal component, TreeMix, and admixture analyses show that Jiangquhai pigs represent an authentic genetic resource and have close genetic relationships with East Chinese breeds, their geographical neighbors. A genome scan unravels a list of reproduction-related genes potentially under selection in Jiangquhai pigs. Using the neighbor-joining clustering approach, we reconstructed the family structure of the conservation population of Jiangquhai pigs. This finding allowed us to suggest a rotational mating scheme across the reconstructed families to reduce the risk of inbreeding depression in the population.
Keywords: conservation, inbreeding depression, Jiangquhai pigs, population genetics
INTRODUCTION
Jiangsu province, located in the East China, is rich in indigenous pig genetic resources. Eight local pig breeds are originally distributed in Jiangsu province, including Dongchuan, Erhualian, Fengjing, Huai, Jiangquhai, Meishan, Mi, and Shawutou. These pigs are reputed not only for their prolificacy but also for their excellent meat quality and adaptability to crude feed and local environments (National Commission of Animal Genetic Resources of China, 2011). During the past few decades, European modern breeds (mainly Large White, Landrace, and Duroc) of high lean meat percent, fast growth rate, and excellent food conversion efficiency have been continuously introduced to China and now dominate Chinese pig industry, leading to remarkable reduction of Chinese indigenous pigs and even extinction of some breeds such as Hengjing and Dahulian pigs in Jiangsu province (National Commission of Animal Genetic Resources of China, 2011). Given this, the Chinese central government has launched a national conservation program for 42 Chinese indigenous pig breeds (National Commission of Animal Genetic Resources of China, 2011). To better conserve these valuable germplasm, researchers have recently used genome-wide genetic markers to investigate genetic diversity and population structure of Chinese indigenous pigs (Wang et al., 2015, 2018).
Jiangquhai pigs were originated from historical cross between Huai and the extinct Dahualian pigs, which are particularly known for their fecundity and are now distributed in Jiangyan, Qutang and Haian cities, Jiangsu province. This breed and five other breeds (Erhualian, Huai, Meishan, Mi, and Shawutou) from Jiangsu province have been listed in the national conservation program (National Commission of Animal Genetic Resources of China, 2011). Recently, the population size of Jiangquhai pigs has also declined considerably, presumably resulting in an increase of inbreeding coefficient. It is hence imperative to well manage the conservation population of Jiangquhai pigs. To achieve this goal, we herein evaluated the genetic diversity of the current nucleus population of Jiangquhai pigs using genome-wide SNP markers. We then explored genome-wide SNPs to infer evolutionary history and phylogeny, genomic signature of selection and population structure of this breed in a global perspective. The results not only allow us to suggest a reasonable conservation scheme for Jiangquhai pigs, but also show that Jiangquhai pigs represent an authentic genetic resource and have close genetic relationships with East Chinese breeds, and a list of genes functionally related to reproduction could have been preferentially selected in this prolific breed.
MATERIALS AND METHODS
All procedures were approved by the ethics committee of Jiangsu Agri-animal Husbandry Vocational College.
Animals
Ear tissue samples of 105 pigs including 12 boars and 93 sows were collected from the nucleus population of Jiangquhai pigs raised in the national conservation farm in Taizhou city, Jiangsu province. The 105 individuals included all boars and nearly all sows of Jiangquhai pigs in this farm. DNA was extracted from ear samples using a routine phenol/chlorofrom way. A concentration of 50 ng/μL DNA of each sample was used for SNP genotyping. Moreover, 662 unrelated pigs from 28 Chinese, 3 European, and 1 hybrid breeds (Supplementary Table S1) were explored in this study. These pigs have no common ancestor within three generations and cover nearly all existing consanguinities of the 28 Chinese breeds as previously reported (Ai et al., 2013; Wang et al., 2018).
SNP Genotyping
The 105 Jiangquhai pigs were genotyped using the GeneSeek GGP Porcine 80K SNP chips via a professional company (Compass Biotechnology, China). The SNP chip contains 68,528 SNPs across the pig genome. The genotype input was converted into a PLINK (v1.9) (Purcell et al., 2007) input file. The porcine 60K chip SNP data of the 662 pigs (Ai et al., 2013; Wang et al., 2018) were downloaded publicly. The two data set were then merged to a common set of 42,276 SNPs. These SNPs were further filtered under the criteria of individual call rate ≥90%, SNPs call rate ≥90%, and minor allele frequency (MAF) ≥0.01. All unmapped SNPs and those on sex chromosomes were discarded. A final set of 34,744 informative SNPs and all 767 pigs were remained for subsequent analyses. Ten Jiangquhai individuals separated from the major cluster of Jiangquhai pigs in the neighbor-joining (NJ) tree (Supplementary Figure S1). The 10 most likely admixed animals (referred hereafter as to JQH2 individuals) should be removed from the conservation population and were discarded in the following statistical analyses, unless otherwise specified.
Calculation of Effective Population Size and Linkage Disequilibrium (LD) Extent
Effective population size (Ne) was estimated based on the known relationship between linkage disequilibrium (r2) and the recombination rate (c) between two loci. Ne was calculated using the following equation (Sved, 1971): NeT =(1/4c) × (1/r2c − 1), where NeT is the effective population size at T = 1/r2c generation in the past, and r2c is the LD between SNPs with c being the linkage map distance (in Morgans) (Tortereau et al., 2012). LD extents were calculated using the autosomal SNP data and the ‘-r2 -ld-window-kb -ld-window-r2 0’ command in PLINK v1.9 (Purcell et al., 2007) as previously reported (Wang et al., 2018).
Identification of Heterozygosity and Homozygosity
Observed heterozygosity (HO), expected heterozygosity (HE), and runs of homozygosity (ROH) were calculated using PLINK v1.9 (Purcell et al., 2007). Considering that an occasional genotyping error or missing genotype occurring in an otherwise-unbroken homozygous segment could result in the underestimation of ROHs, we used 50 SNP sliding windows that allowed one heterozygote and five missing calls per window. To exclude the short and very common ROHs, we set 500 kb as the minimum length for an ROH. We calculated the sum of ROH values per individual using autosomal SNPs.
Estimation of Genetic Distance and Differentiation
All individuals and 34,744 qualified SNPs were used to estimate genetic distance and differentiation among individuals or breeds. PLINK v1.9 (Purcell et al., 2007) was used to calculate the average proportions of alleles shared as the Dst. The genetic distances (D) between all of the pair-wise combinations of the tested individuals were calculated as follows: D = 1 – Dst. The genetic differentiation between each pair of breeds were measured using Weir and Cockerham’s average FST (Weir and Cockerham, 1984). Neighbor joining (NJ) relationship trees were then constructed using the “neighbor” program in PHYLIP v3.695 (Felsenstein, 1989) and were displayed by FigTree v1.4.2 (http://tree.bio.ed.ac.uk/software/figtree/). GCTA (Yang et al., 2011) was used for principal component analysis (PCA). TreeMix (Pickrell and Pritchard, 2012) was further explored to investigate the genetic pattern of population splitting and admixture in Chinese domestic pigs using Western Duroc pigs as the outgroup. A subset of 10 pigs were randomly sampled from each breed to construct a maximum-likelihood tree. For Jiangquhai pigs, 10 randomly selected individuals (JQH1) and the 10 above-mentioned admixed individuals (JQH2) were analyzed.
Inferring of Population Structure
ADMIXTURE 1.23 (Alexander et al., 2009) were used to infer the most probable number of ancestral populations (K) for each breed. Results were plotted for K from 2 to 5 and 26 using in-house R scripts. As previously described (Wang et al., 2018), we randomly selected a subset of 10 pigs from each breed for the Admixture analyses. For Jiangquhai pigs, 10 randomly sampled non-admixed individuals (JQH1) and the 10 admixed individuals (JQH2) were tested in the Admixture analysis. A NJ tree was constructed for the 95 nonadmixed individuals using pairwise identical-by-state values among individuals derived from 62,331 SNPs. The NJ tree unraveled the family structure of the conservation population of Jiangquhai pigs.
Characterization of Candidate Genes Under Selection
To identify candidate genes under selection in Jiangquhai pigs, we calculated locus-specific branch length (LSBL) statistics (Shriver et al., 2004) for each polymorphic site of 34,744 qualified SNPs in the 757 individuals under a three-group contrasting model. According to the clustering pattern in the NJ tree (Figure 1), we treated the 95 Jiangquhai pigs as Group 1, 117 pigs from other four East Chinese breeds (Erhualian, Jinhua, Dongxiang, and Yushan) as Group 2, and 545 individuals from the other 27 Eurasian breeds and wild boars as Group 3. The LSBL statistics were then computed as previously reported (David et al., 2009; Ai et al., 2014). We defined outlier SNPs at which LSBL statistics surpassed 0.1% of the empirical distributions as candidate loci under selection in Jiangquhai pigs. Annotated genes within 50 kb upstream and downstream regions of the outlier SNPs were retrieved from the pig genome assembly (Sscrofa 11 available at http//www.ensembl.org). The Kyoto encyclopedia of genes and genomes (KEGG) (Minoru et al., 2012) and Gene Ontology (GO) databases (Ashburner et al., 2000) were explored for the functional enrichment analyses of the retrieved genes via Matescape (Tripathi et al., 2015).
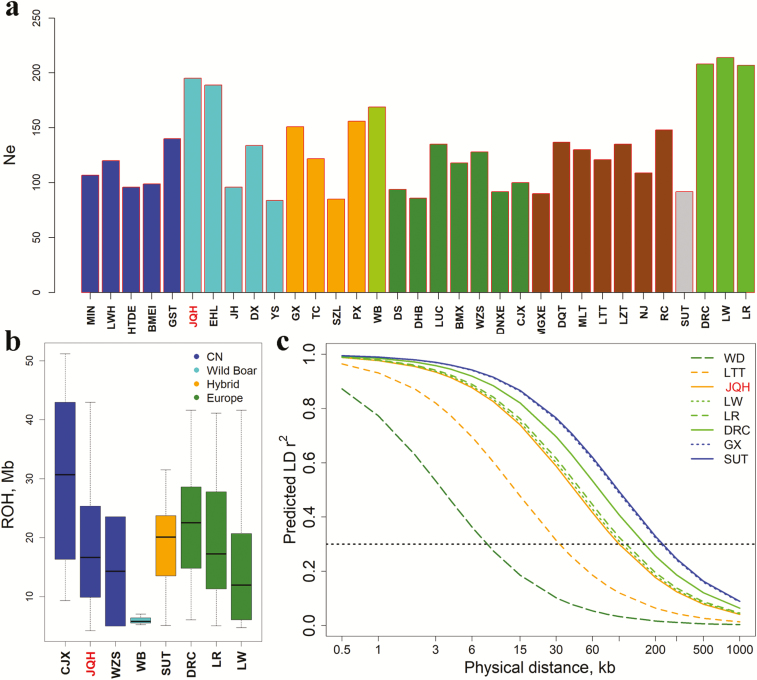
Figure 1.
Genetic diversity of Jiangquhai pigs. (a) Effective population size (Ne). Each bar plot indicates the Ne of each breed including Jiangquhai (JQH). (b) Runs of homozygosity (ROH). Each point denotes ROH value of each individual. Chinese wild boars and seven representative domestic breeds including JQH are shown. (c) LD (predicted r2) extents are plotted as a function of inter-SNP distance for Chinese wild boars and seven representative domestic breeds including JQH. The black dotted line indicates the threshold of 0.3. Abbreviations of all breeds and wild boars are given in Supplementary Table S1.
RESULTS AND DISCUSSION
Genetic Diversity of Jiangquhai Pigs
To evaluate the genetic diversity of the current conservation population of Jiangquhai pigs, we first estimated the average effective population size (Ne) in the past 20 years for each population tested in this study (Supplementary Table S1, Figure 1a). We noted that Jiangquhai pigs had the largest Ne (n =195) in Chinese local pig breeds. We also calculated HO, HE, ROH and LD extent values of Jiangquhai pigs in comparison with those of 31 Chinese and European breeds and Chinese wild boars. Jiangquhai pigs had HO of 0.23 and HE of 0.22, which were comparable with those of most Chinese indigenous breeds (Supplementary Table S1). The level of ROH reflects the recent inbreeding history of a population (Ai et al., 2013; Traspov et al., 2016). We detected a ROH value of 15.6 Mb in Jiangquhai pigs, which was lower than those of the other three East Chinese breeds (Jinhua, Dongxiang Spotted and Yushan) but higher than that of Erhualian pigs (12.7 Mb, Supplementary Table S1 and Figure 1b). We also compared ROH values among the 95 Jiangquhai pigs (Supplementary Figure S2). Three individuals had ROH values of greater than two standard deviations from the average value, indicating that the three pigs were highly inbred individuals and should be replaced by low-ROH individuals during successive generational selection.
To assess LD extent patterns, we estimated r20.3 values in all tested breeds, the physical distance at which the pair-wise genotypic association in the filtered SNP data set decays below a threshold of 0.3 (Ai et al., 2013). Jiangquhai pigs had a median extent of LD (r20.3 = 99.7 kb) in a range of 8.0 kb (Wild boar) to 227.9 kb (Sutai) in the 33 tested breeds (Supplementary Table S1). We chose Chinese wild boars, Jiangquhai pigs and six other breeds to visualize their LD patterns (Figure 1c). The six breeds included the hybrid breed (Sutai), three European modern breeds including Large White, Landrace and Duroc, and Ganxi (r20.3 = 222.3 kb) and Litang (Tibet) (r20.3 = 31.9 kb) pigs that had the highest and lowest LD extents in Chinese indigenous breeds, respectively. Jiangquhai pigs appeared to have a roughly similar LD decay pattern to Large White pigs (Figure 1c).
A microsatellite marker-based study showed that Jiangquhai pigs had the most abundant genetic diversity among five East Chinese pig breeds (Fan et al., 2002). Nowadays, Jiangquhai pigs had large Ne (n = 195), roughly comparable Ho (0.23) and HE (0.22), relatively low ROH values (15.6 Mb), and short LD extent (r20.3 = 99.7 kb) among the five East Chinese pig breeds, indicating that the Jiangquhai breed is still rich in genetic diversity in the context of its neighboring breeds despite the recent decline of its population size.
Phylogenic Relationships Between Jiangquhai Pigs and Other Chinese Breeds
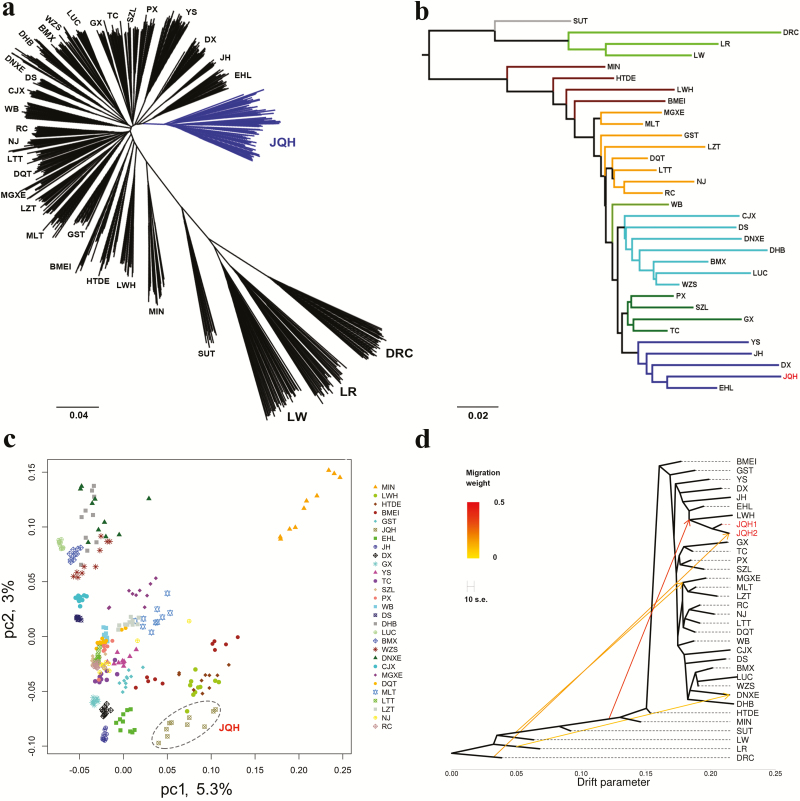
To uncover phylogenic relationships between Jiangquhai pigs and other Chinese breeds, we first constructed the NJ tree using genome-wide allele sharing among individuals. In the NJ tree, Jiangquhai, Erhualian, Jinhua, Dongxiang, and Yushan pigs, five breeds all from East China, were grouped into a major clade (Figure 2a). We then measured the genetic differentiation (FST) between each pair of breeds. Jiangquhai exhibited the least divergence with its geographical neighbor—Erhualian—in the same province (Jiangsu) as shown in the FST estimate-derived dendrogram (Figure 2b). In this dendrogram, the five East Chinese breeds defined a branch separating from the other Chinese breeds (Figure 2b). The two clustering patterns are well consistent with each other, reflecting that breeds in a neighboring region tend to have close phylogenetic relationships among each other likely due to a more recent common ancestor, such as Jiangquhai and Erhualian.
Figure 2.
Evolutionary phylogeny of Jiangquhai pigs. (a) Neighbor-joining phylogenetic tree of 757 pigs from 32 Eurasian breeds and Chinese wild boars. (b) FST estimate-derived dendrogram of 32 Eurasian breeds and Chinese wild boars. (c) Principal component (PC) plots of 28 Chinese indigenous breeds and Chinese wild boars. The first (PC1) and second component (PC2) are shown. (d) Population splits and mixture of the tested Eurasian breeds and Chinese wild boars inferred using TREEMIX. In this panel, JQH1 represents 10 non-admixted Jiangquhai pigs and JQH2 indicates 10 admixed Jiangquhai pigs revealed by the neighbor-joining phylogenetic tree (Supplementary Figure S1). Abbreviations of all breeds and wild boars are given in Supplementary Table S1.
Furthermore, we conducted PCA on the 28 Chinese indigenous breeds and Chinese wild boars tested in this study. The PCA plots also identified Jiangquhai and East Chinese breeds, especially Jiangquhai and Erhualian, as closely related breeds (Figure 2c). In the PCA plots, Jiangquhai pigs also showed a close relationship with Laiwu pigs that were distributed in a North Chinese region adjacent to (~500 km) the habitat of Jiangquhai pigs (Supplementary Figure S3).
To investigate historical split and admixture in the tested breeds, we conducted the TreeMix analysis to infer a maximum-likelihood population tree and potential migration events for these breeds. We set the migration events as 4, at which nearly all (99.98%) of the variance in the relatedness between breeds were explained. In the inferred maximum-likelihood tree (Figure 2d), the five East Chinese breeds including Jiangquhai and Laiwu (LWH) formed one major grouping, which is consistent with the NJ and PCA results. Of the four migration events, all introgression routes were from European modern breeds to Chinese indigenous breeds. The strong signal suggests a genetic contribution from the ancestry of three European breeds (Duroc, Large White, and Landrace) to Laiwu pigs (migration weight (w) = 35.1%) and from Duroc to the 10 admixed Jiangquhai (JQH2) pigs (w = 15.2%). This may explain why the 10 individuals clustered closely with European breeds in the NJ tree (Supplementary Figure S1).
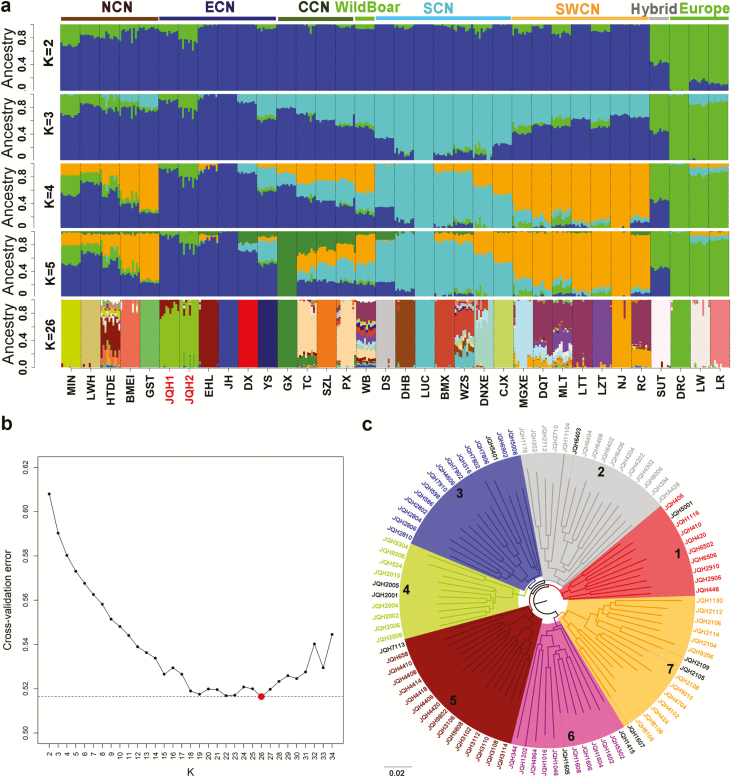
To unravel ancestral lineages and historical admixture of Jiangquhai pigs in a worldwide perspective, we conducted the Admixture analysis for the tested breeds and wild boars with K values from 2 to 5 and 26. From K = 2 to 5, ancestral lineage of Jiangquhai pigs was Jinhua-like, i.e., East Chinese lineage (Figure 3a). It should be mentioned that the 10 admixed Jiangquhai (JQH2) pigs always had a higher proportion of lineage introgressed from Duroc pigs compared with the other Jiangquhai pigs (JQH1, Figure 3a). When runs with K = 26, the cross-validation error reached the lowest level and Jiangquhai pigs formed an independent population (Figure 3b).
Figure 3.
Population structure of Jiangquhai pigs. (a) Ancestry compositions of Jiangquhai (JQH) pigs in a panel of global breeds. The ancestry compositions of 32 Eurasian breeds and Chinese wild boars were revealed by ADMIXTURE with the assumed number of ancestries from 2 to 5 and 26. Each color represents one ancestral cluster. Breeds are separated by dotted lines. JQH is highlighted by colored axis. CCN, Central China; ECN, East China; NCN, North China; SCN, South China; SWCN, Southwest China. JQH1: 10 randomly selected Jiangquhai pigs. JQH2: 10 admixed Jiangquhai pigs revealed by the neighbor-joining phylogenetic tree (Supplementary Figure S1). (b) Cross-validation errors of the admixture analysis at different K values. (c) The neighbor-joining phylogenetic tree was constructed based on pairwise identical-by-state values amongst 95 non-admixted JQH individuals. Boars are indicated in black. Abbreviations of all breeds and wild boars are given in Supplementary Table S1.
Altogether, NJ clustering trees of individuals and breeds, PCA plots, the TreeMix and Admixture results consistently illustrate that Jiangquhai pigs represent an authentic genetic resource and have close phylogenic relationships with East Chinese breeds, especially with their neighbors—Erhualian pigs. According to historical records (National Commission of Animal Genetic Resources of China, 2011), Jiangquhai pigs are hybrid descendants of Huai and the extinct Dahualian pigs, and Erhualian pigs were derived from a complex cross between Mi and Dahualian pigs. Our findings confirm that Jiangquhai has a more recent common ancestor with Erhualian compared with other Chinese breeds.
We note that Jiangquhai was clustered together with Laiwu and other East Chinese breeds, defining a major clade in the Treemix maximum-likelihood tree (Figure 2d). This contradicts to the clustering pattern in the IBS- and FST-based NJ trees (Figure 2a and b). The contradiction could be explained by the fact that European lineages (most likely Duroc), as illustrated by the Treemix and Admixture analyses, were introgressed into Laiwu pigs, resulting in a considerable fraction of genome of Duroc origin in Laiwu pigs. The introgression causes a paraphyletic clustering pattern of Laiwu in the NJ trees. Indeed, Jiangquhai, as revealed by the TreeMix analysis, has a close genetic relationship with Laiwu. The assumption is also supported by the PCA analysis, in which Jiangquhai and Laiwu grouped together in PC1 and PC2 (Figure 2c). Moreover, the Admixture analysis showed that LWH had a major genetic component (ancestral lineages) of East Chinese pigs such as Jinhua and Jiangquhai when K = 2 to 5 (Figure 3a).
Population Structure of Jiangquhai Pigs
To investigate population structure of Jiangquhai pigs, we constructed the NJ phylogenetic trees for the nonadmixed 95 Jiangquhai individuals using pairwise identical-by-state values amongst individuals. The 95 pigs were clustered into seven branches in the NJ tree (Figure 3c). Each branch consisted of at least one boars and roughly comparable number of sows (8 to 15), thus representing seven families in the current conservation population of this breed. The reconstructed families contribute to the establishment of an effective conservation program for the endangered Jiangquhai pigs. To reduce the risk of inbreeding depression in this breed, we suggest a boar-mediated rotational crossing across the seven reconstructed families, discard highly inbred individuals and maintain the equivalent number of boars and sows in each family when close breeding in the future.
Candidate Genes Under Selection in Jiangquhai Pigs
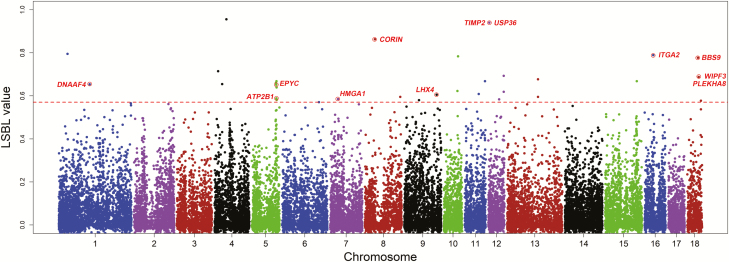
We identified 34 outlier SNPs ranking the top 0.1% of the empirical distribution. These SNPs correspond to 38 genes on 14 chromosomes (Figure 4, Supplementary Table S2). Of the 38 candidate genes under selection, 10 were overrepresented in 7 GO terms (Supplementary Table S3) related to a variety of cellular functions, such as multicellular organismal signaling, negative regulation of cellular catabolic process, mitotic cell cycle and catabolic process, and extracellular matrix organization, etc. Of note, three genes (EPYC, ITGA2, and CORIN) were enriched in the biological process of female pregnancy (P < 0.01). EPYC is highly expressed in the human placenta (Fagerberg et al., 2014). ITGA2 antigen is extensively expressed by natural killer (NK) cells and commonly used as a pan-NK-cell maker, which is reported to be important in the regulation of feto-maternal tolerance in rat pregnant uteri (Lin et al., 2009; Wang et al., 2009). CORIN is involved with preeclampsia in mice (Cui et al., 2012) and human (Stepanian et al., 2014). In addition, nine other potentially selected genes also play a role in reproduction, including ATP2B1, BBS9, DNAAF4, HMGA1, LHX4, PLEKHA8, TIMP2, USP36, and WIPF3 (Figure 4, Supplementary Table S3). ATP2B1, also known as PMCA1, is essential for embryonic development, and loss of both copies of the ATP2B1 gene causes embryolethality in mice (Okunade et al., 2004). BBS9 deleterious variants are associated with premature ovarian failure in women (Kang et al., 2008). Deleting exons 2 to 4 of DNAAF4 (previously known as DYX1C1) causes male infertility in mice (Aarti et al., 2013). HMGA1 is required for normal sperm development (Liu et al., 2003). LHX4 is involved in the formation of the pituitary and the motoneuron system and knock out of this gene results in perinatal lethality (Geng et al., 2010). PLEKHA8 knockout mice show abnormal epididymis morphology in the International Mouse Phenotyping Consortium (IMPC) data (Dickinson et al., 2016). TIMP2 is an up-regulated gene in endometrial tissues of ongoing pregnant women (Bersinger et al., 2008). Dysfunction of USP36 leads to preimplantation lethality in mice (Fraile et al., 2017). Knock out of WIPF3 (also known as CR16) causes male sterility in mice (Suetsugu et al., 2007). The findings suggest that favorable variants in these reproduction-related genes could have been preferentially selected to improve the fertility of Jiangquhai pigs, contributing to the development of this prolific breed.
Figure 4.
Genome-wide distribution of LSBL values across all 18 autosomes of Jiangquhai pigs. The 18 chromosomes are plotted along the x-axis, and locus-specific branch length (LSBL) values are plotted along the y-axis. Chromosomes are indicated by different colors, and the threshold (top 0.1%) indicating signature of selection is denoted with a dashed red line. Twelve reproduction-related candidate genes corresponding to the top SNP outliers are indicated by red circles.
Supplementary Material
Footnotes
This work was supported by the National Natural Science Foundation of China (no. 31702089), the Earmarked Fund for Jiangsu Agriculture Industry Technology System (JATS2018237), and the Phoenix Talent Project of Jiangsu Agri-animal Husbandry Vocational College (no. 201602).
LITERATURE CITED
- Aarti T., Loges N. T., Slagle C. E., Francis R., Dougherty G. W., Tamayo J. V., Shook B., Cantino M., Schwartz D., Jahnke C., et al. 2013. DYX1C1 is required for axonemal dynein assembly and ciliary motility. Nat. Genet. 45:995 doi:10.1038/ng.2707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ai H., Huang L., and Ren J.. 2013. Genetic diversity, linkage disequilibrium and selection signatures in chinese and western pigs revealed by genome-wide SNP markers. PLoS One 8:e56001. doi:10.1371/journal.pone.0056001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ai H., Yang B., Li J., Xie X., Chen H., and Ren J.. 2014. Population history and genomic signatures for high-altitude adaptation in tibetan pigs. BMC Genomics 15:834. doi:10.1186/1471-2164-15-834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander D. H., Novembre J., and Lange K.. 2009. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 19:1655–1664. doi:10.1101/gr.094052.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner M., Ball C. A., Blake J. A., Botstein D., Butler H., Cherry J. M., Davis A. P., Dolinski K., Dwight S. S., Eppig J. T., et al. 2000. Gene ontology: tool for the unification of biology. The gene ontology consortium. Nat. Genet. 25:25–29. doi:10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bersinger N. A., Wunder D. M., Birkhäuser M. H., and Mueller M. D.. 2008. Gene expression in cultured endometrium from women with different outcomes following IVF. Mol. Hum. Reprod. 14:475–484. doi:10.1093/molehr/gan036. [DOI] [PubMed] [Google Scholar]
- Cui Y., Wang W., Dong N., Lou J., Srinivasan D. K., Cheng W., Huang X., Liu M., Fang C., Peng J., et al. 2012. Role of corin in trophoblast invasion and uterine spiral artery remodelling in pregnancy. Nature 484:246–250. doi:10.1038/nature10897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David R., Kumarasamy T., Nick P., Price A. L., and Lalji S.. 2009. Reconstructing Indian population history. Nature 461:489–494. doi:10.1038/nature08365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson M. E., Flenniken A. M., Ji X., Teboul L., Wong M. D., White J. K., Meehan T. F., Weninger W. J., Westerberg H., Adissu H., et al. ; International Mouse Phenotyping Consortium; Jackson Laboratory; Infrastructure Nationale PHENOMIN, Institut Clinique de la Souris (ICS); Charles River Laboratories; MRC Harwell; Toronto Centre for Phenogenomics; Wellcome Trust Sanger Institute; RIKEN BioResource Center. 2016. High-throughput discovery of novel developmental phenotypes. Nature 537:508–514. doi:10.1038/nature19356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagerberg L., Hallström B. M., Oksvold P., Kampf C., Djureinovic D., Odeberg J., Habuka M., Tahmasebpoor S., Danielsson A., Edlund K., et al. 2014. Analysis of the human tissue-specific expression by genome-wide integration of transcriptomics and antibody-based proteomics. Mol. Cell. Proteomics 13:397–406. doi:10.1074/mcp.M113.035600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan B., Wang Z. G., Li Y. J., Zhao X. L., Liu B., Zhao S. H., Yu M., Li M. H., Chen S. L., Xiong T. A., et al. 2002. Genetic variation analysis within and among chinese indigenous swine populations using microsatellite markers. Anim. Genet. 33:422–427. doi:10.1046/j.1365-2052.2002.00898.x. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. 1989. PHYLIP-Phylogeny interference package (version 3.2). Cladistics 5:164–166. [Google Scholar]
- Fraile J. M., Campos-Iglesias D., Rodríguez F., Astudillo A., Vilarrasa-Blasi R., Verdaguer-Dot N., Prado M. A., Paulo J. A., Gygi S. P., Martín-Subero J. I., et al. 2017. Loss of the deubiquitinase USP36 destabilizes the RNA helicase DHX33 and causes preimplantation lethality in mice. J Biol Chem 293:2183–2194. doi:jbc.M117.788430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng T., Singh U., Yu Y., Ellsworth B. S., Hemberger M., Geyer R., Stewart M. D., Behringer R. R., Fundele R.. 2010. Expression and function of the LIM-homeobox containing genes Lhx3 and Lhx4 in the mouse placenta. Dev Dyn 237:1517–1525. doi:10.1002/dvdy.21546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang H., Lee S. K., Kim M. H., Song J., Bae S. J., Kim N. K., Lee S. H., and Kwack K.. 2008. Parathyroid hormone-responsive B1 gene is associated with premature ovarian failure. Hum. Reprod. 23:1457–1465. doi:10.1093/humrep/den086. [DOI] [PubMed] [Google Scholar]
- Lin Y., Wang H., Wang W., Zeng S., Zhong Y., and Li D. J.. 2009. Prevention of embryo loss in non-obese diabetic mice using adoptive ITGA2+ISG20+ natural killer-cell transfer. Reproduction 137:943–955. doi:10.1530/REP-08-0412. [DOI] [PubMed] [Google Scholar]
- Liu J., Schiltz J. F., Ashar H. R., and Chada K. K.. 2003. Hmga1 is required for normal sperm development. Mol. Reprod. Dev. 66:81–89. doi:10.1002/mrd.10323. [DOI] [PubMed] [Google Scholar]
- Minoru K., Susumu G., Yoko S., Miho F., and Mao T.. 2012. KEGG for integration and interpretation of large-scale molecular data sets. Nucleic Acids Res. 40:D109–D114. doi:10.1093/nar/gkr988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- National Commission of Animal Genetic Resources of China 2011. Animal genetic resources in China: Pigs. China Agriculture Press, Beijing. [Google Scholar]
- Okunade G. W., Miller M. L., Pyne G. J., Sutliff R. L., O’Connor K. T., Neumann J. C., Andringa A., Miller D. A., Prasad V., Doetschman T., et al. 2004. Targeted ablation of plasma membrane Ca2+-ATPase (PMCA) 1 and 4 indicates a major housekeeping function for PMCA1 and a critical role in hyperactivated sperm motility and male fertility for PMCA4. J. Biol. Chem. 279:33742–33750. doi:10.1074/jbc.M404628200. [DOI] [PubMed] [Google Scholar]
- Pickrell J. K., and Pritchard J. K.. 2012. Inference of population splits and mixtures from genome-wide allele frequency data. PLoS Genet. 8:e1002967. doi:10.1371/journal.pgen.1002967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M. A., Bender D., Maller J., Sklar P., de Bakker P. I., Daly M. J., et al. 2007. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81:559–575. doi:10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shriver M. D., Kennedy G. C., Parra E. J., Lawson H. A., Sonpar V., Huang J., Akey J. M., and Jones K. W.. 2004. The genomic distribution of population substructure in four populations using 8,525 autosomal snps. Hum. Genomics 1:274–286. doi:10.1186/1479-7364-1-4-274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepanian A., Alcaïs A., de Prost D., Tsatsaris V., Dreyfus M., Treluyer J. M., and Mandelbrot L.; ECLAXIR Study Group 2014. Highly significant association between two common single nucleotide polymorphisms in CORIN gene and preeclampsia in caucasian women. PLoS One 9:e113176. doi:10.1371/journal.pone.0113176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suetsugu S., Banzai Y., Banzai Y., Kato M., Fukami K., Kataoka Y., Takai Y., Yoshida N., and Takenawa T.. 2007. Male-specific sterility caused by the loss of CR16. Genes Cells 12:721–733. doi:10.1111/j.1365-2443.2007.01088.x. [DOI] [PubMed] [Google Scholar]
- Sved J. A. 1971. Linkage disequilibrium and homozygosity of chromosome segments in finite populations. Theor. Popul. Biol. 2:125–141. doi:10.1016/0040-5809(71)90011-6. [DOI] [PubMed] [Google Scholar]
- Tortereau F., Servin B., Frantz L., Megens H. J., Milan D., Rohrer G., Wiedmann R., Beever J., Archibald A. L., Schook L. B., et al. 2012. A high density recombination map of the pig reveals a correlation between sex-specific recombination and GC content. BMC Genomics 13:586. doi:10.1186/1471-2164-13-586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traspov A., Deng W., Kostyunina O., Ji J., Shatokhin K., Lugovoy S., Zinovieva N., Yang B., and Huang L.. 2016. Population structure and genome characterization of local pig breeds in russia, belorussia, kazakhstan and ukraine. Genet. Sel. Evol. 48:16. doi:10.1186/s12711-016-0196-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripathi S., Pohl M. O., Zhou Y., Rodriguez-Frandsen A., Wang G., Stein D. A., Moulton H. M., DeJesus P., Che J., Mulder L. C., et al. 2015. Meta- and orthogonal integration of influenza “omics” data defines a role for UBR4 in virus budding. Cell Host Microbe 18:723–735. doi:10.1016/j.chom.2015.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Lin Y., Zeng S., and Li D. J.. 2009. Improvement of fertility with adoptive CD25+ natural killer cell transfer in subfertile non-obese diabetic mice. Reprod. Biomed. Online 18:95–103. doi:10.1016/s1472-6483(10)60430-0. [DOI] [PubMed] [Google Scholar]
- Wang X., Wang C., Huang M., Tang J., Fan Y., Li Y., Li X., Ji H., Ren J., and Ding N.. 2018. Genetic diversity, population structure and phylogenetic relationships of three indigenous pig breeds from jiangxi province, china, in a worldwide panel of pigs. Anim. Genet. 49:275–283. doi:10.1111/age.12687. [DOI] [PubMed] [Google Scholar]
- Wang Z., Chen Q., Yang Y., Liao R., Zhao J., Zhang Z., Chen Z., Zhang X., Xue M., Yang H., et al. 2015. Genetic diversity and population structure of six chinese indigenous pig breeds in the taihu lake region revealed by sequencing data. Anim. Genet. 46:697–701. doi:10.1111/age.12349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weir B. S., and Cockerham C. C.. 1984. Estimating F-statistics for the analysis of population structure. Evolution 38:1358–1370. doi:10.1111/j.1558-5646.1984.tb05657.x. [DOI] [PubMed] [Google Scholar]
- Yang J., Lee S. H., Goddard M. E., and Visscher P. M.. 2011. GCTA: a tool for genome-wide complex trait analysis. Am. J. Hum. Genet. 88:76–82. doi:10.1016/j.ajhg.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.