Abstract
Purpose of review
The development of a myelodysplastic syndrome (MDS) or acute myeloid leukemia (AML) in patients with congenital neutropenia is now the major cause of mortality. Treatment options are limited and there are no effective prevention strategies. This review focuses on mechanisms of leukemic transformation in severe congenital neutropenia (SCN) and Shwachman Diamond syndrome (SDS), the two most common types of congenital neutropenia.
Recent findings
AML/MDS that develops in the setting of congenital neutropenia has distinct molecular features. Clonal hematopoiesis due to TP53 mutations is seen in nearly 50% of patients with SDS, but is not seen in patients with SCN. Accordingly, there is a very high frequency of TP53 mutations in AML/MDS arising in the setting of SDS but not SCN. The rate of mutation accumulation in hematopoietic stem cells (HSCs) from patients with congenital neutropenia is not increased.
Summary
Both HSC cell-intrinsic and non-cell intrinsic changes contribute to the development of clonal hematopoiesis in congenital neutropenia and likely accounts for the high rate of leukemic transformation. In SCN, the persistently high levels of G-CSF drive expansion of HSCs carrying truncation mutations of CSF3R. In SDS, impaired ribosome biogenesis induces p53-mediated growth inhibition and drives expansion of HSCs carrying TP53 mutations.
Keywords: Severe congenital neutropenia, Shwachman Diamond syndrome, TP53, CSF3R, G-CSF, AML, MDS
INTRODUCTION
Bone marrow failure syndromes are characterized by a deficiency of one or more hematopoietic lineage. A common feature of both congenital and acquired forms of bone marrow failure is an increased risk to develop acute myeloid leukemia (AML) or a myelodysplastic syndrome (MDS). Significantly increased risks of AML/MDS have been reported in Fanconi anemia, dyskeratosis congenita, Diamond Blackfan syndrome, Shwachman Diamond syndrome (SDS), and severe congenital neutropenia (SCN). Treatment options for patients with bone marrow failure syndromes who develop AML/MDS are limited, and there are no reliable biomarkers that predict progression. Thus, there is a pressing clinical need to develop strategies to prevent, diagnose early, and treat AML/MDS in patients with bone marrow failure syndromes. This review will focus on mechanisms of leukemic transformation in the two most common types of congenital neutropenia, SCN and SDS.
LEUKEMIC TRANSFORMATION IN SCN
SCN is a congenital bone marrow failure syndrome characterized by severe neutropenia present from birth, an arrest of myeloid differentiation at the promyelocyte/myelocyte stage, and frequent infections. SCN is a genetically heterogeneous disorder. Mutations of ELANE are the most common cause of SCN, accounting for approximately 50% of cases (1–3). Other genes mutated in SCN include HAX1, G6PC3, SRP54, GFI1, CSF3R, VPS45, WAS, JAGN1, and TCIRG1; the genetic cause of approximately 30% of SCN cases remains unknown (4, 5). As expected given the genetic heterogeneity, diverse mechanisms of disease pathogenesis have been proposed for SCN. Although controversial, there is evidence that mutated ELANE (which encodes for neutrophil elastase) results in the production of misfolded neutrophil elastase, which in turn, induces the unfolded protein response and apoptosis in promyelocytes (6, 7). Treatment with granulocyte colony-stimulating factor (G-CSF) is the standard of care for SCN, as it increases the level of circulating neutrophils and reduces infection-related mortality (8). Indeed, since the widespread use of G-CSF, the major cause of mortality in SCN is the development of AML/MDS. The French Neutropenia Registry reported a cumulative incidence of MDS or AML in patients with SCN of 10.8% at 20 years of age (9). A prospective study of 374 patients with SCN on long-term G-CSF enrolled in the Severe Chronic Neutropenia International Registry (SCNIR) showed that the cumulative risk of developing AML/MDS is 22% after 15 years on G-CSF (10). Moreover, no plateau in the incidence of AML or MDS was observed, suggesting that the cumulative risk of progression may be even higher. Patients requiring higher doses of G-CSF and who have a reduced neutrophil response to G-CSF have the highest rate of leukemic transformation (10, 11). Of note, AML or MDS has been reported in SCN associated with mutations in ELANE, HAX1, G6PC3, and WAS (12, 13).
AML/MDS arising in the setting of SCN is associated with distinct molecular features compared with de novo AML/MDS (Table 1). Secondary AML/MDS in patients with SCN is frequently associated with monosomy 7 and abnormalities of chromosome 21 (14–16). In the largest published series to date, candidate gene sequencing of 15 genes in 30 cases of SCN-AML/MDS showed that 27 (90%) had truncation mutations of CSF3R, encoding the G-CSF receptor (G-CSFR), and 19 (63%) had mutations in RUNX1 (17). These data are consistent with prior studies showing that truncations mutations of CSF3R are present in approximately 80% of cases of SCN-AML/MDS (18, 19). In contrast, monosomy 7, RUNX1, and CSF3R mutations are relatively uncommon in de novo AML (18). Studies reporting results of whole genome or exome sequencing of SCN-AML/MDS are limited, with only a single case reported (20). Thus, the full spectrum of somatic mutations contributing to leukemic transformation in SCN has not been fully characterized.
Table 1.
Characteristics of AML/MDS in SCN and SDS
CSF3R MUTATIONS IN SCN
Truncation mutations of the CSF3R were first identified in a patient with SCN who developed AML (21). The great majority of these mutations are nonsense mutation that truncate the cytoplasmic domain of the G-CSFR. Of note, the truncation mutations are distinct from the transmembrane proximal missense CSF3R mutations that are associated with chronic neutrophilic leukemia (22, 23) or the extracellular loss-of-function biallelic CSF3R mutations that are a rare cause of SCN (24). The truncated G-CSFR found in SCN, while remaining dependent on G-CSF, displays enhanced signaling due to impaired internalization and disturbed lysosomal targeting (25–28). Transgenic mice carrying targeted mutations of the Csf3r reproducing these truncation mutations show that it results in enhanced myeloid progenitor proliferation, due at least in part, to increased STAT5 activation (29, 30). In the largest published series, CSF3R truncation mutations were detected in 43 of 125 (34%) patients with SCN without evidence of leukemic transformation versus 18 or 23 (78%) with MDS/AML, monosomy 7, or other clonal hematopoietic malignancy (31, 32). Interestingly, CSF3R mutations were detected in some patients prior to starting G-CSF. Serial analysis of CSF3R mutations over time show that, in some cases, CSF3R mutations are present prior to the development of MDS/AML (17, 20, 33). Of note, CSF3R mutations can persist for years without the development of MDS/AML and can occasionally spontaneously disappear, limiting their usefulness as a biomarker for the development of AML/MDS (34, 35).
MECHANISMS OF LEUKEMIA TRANSFORMATION IN SCN
The accumulation of mutations in hematopoietic stem cells (HSCs) with age results in the production of a genetically heterogeneous cell population, with each HSC possessing its own unique set of private mutations (36). HSCs that acquire somatic mutations that confer a competitive fitness advantage relative to their normal counterparts may clonally expand, resulting in the presence of clonal hematopoiesis in healthy individuals (37–39). Factors that increase the rate at which mutations accumulate in HSCs may increase the frequency of clonal hematopoiesis and ultimately MDS/AML. The common mutations causing SCN are not known to be directly involved in DNA repair, suggesting the possibility that non-cell autonomous mechanisms may contribute to the high rate of leukemic transformation. For example, granulocyte colony stimulating factor (G-CSF) expression is induced by neutropenia and may increase the rate at which HSCs accumulate mutations by inducing their replication and/or by inducing reactive oxygen species (40, 41). To test this possibility, we measured the mutation burden in individual hematopoietic stem/progenitor cells (HSPCs) in patients with SCN (42). Surprisingly, the number of somatic mutations in the exome of HSPCs from patients with ELANE mutated SCN (3.6 ± 1.2) was similar to that observed in age-matched healthy controls (3.9 ± 0.4). Thus, current data suggest that the rate of mutation accumulation in HSCs with age is not increased in patients with SCN.
There is emerging evidence that hematopoietic stressors may select for HSCs carrying certain mutations, leading to their clonal expansion. For example, recent studies show that exposure to chemotherapy results in the expansion of HSC clones carrying TP53 or PPM1D mutations (43–45). Patients with SCN (or other congenital neutropenia syndromes) are often exposed to repeated bouts of infection, resulting in the production of inflammatory cytokines, including G-CSF, that might contribute to the development of clonal hematopoiesis. We recently reported the frequency of clonal hematopoiesis in 41 patients with SCN (42). As expected, clonal hematopoiesis due to truncation mutations of CSF3R were frequently detected in patients with SCN but not in healthy controls or patients with SDS. Importantly, no increase in clonal hematopoiesis due to any other mutation was detected.
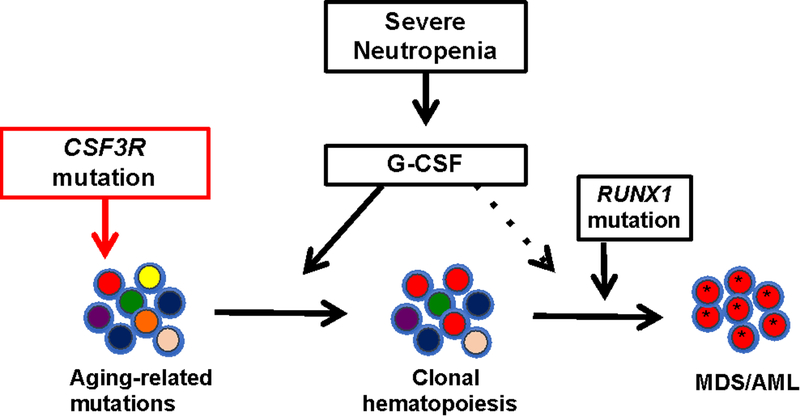
Together, these data suggest a model of leukemogenesis in which the very high level of G-CSF present in patients (either through endogenous production or pharmacologic administration) is driving the expansion of HSCs carrying CSF3R mutations (Figure 1). This is consistent with data showing that expression of a truncated G-CSFR in mice confers a clonal advantage to HSCs in a G-CSF dependent fashion (24). Although clearly not sufficient to induce AML/MDS, there is strong evidence that CSF3R truncation mutations contribute to leukemogenesis, including the following: 1) their high prevalence in SCN-AML/MDS; 2) their ability to cooperate with the PMR-RAR oncogene in mice to induced AML (46); and 3) the observation that increased G-CSFR signaling due to transmembrane proximal missense CSF3R mutations causes chronic neutrophilic leukemia. Transformation to AML/MDS requires the acquisition of additional somatic mutations, which in patients with SCN commonly includes mutations of RUNX1 and alterations of chromosome 7 (17).
Figure 1. Model of leukemic transformation in SCN.
Aging related mutations that occur during HSC replication result in the production of a genetically heterogeneous pool of HSCs. In SCN, the persistently high level of G-CSF results in the selection of HSCs that carrying truncations mutations of CSF3R (encoding the G-CSF receptor). Additional mutations, most commonly of RUNX1, are required for transformation to MDS/AML.
LEUKEMIC TRANSFORMATION IN SDS
Shwachman Diamond syndrome (SDS) is a recessive disorder characterized by hematopoietic abnormalities, exocrine pancreatic insufficiency, and skeletal abnormalities. Neutropenia is present in the majority of patients with variable severity and it may be intermittent (47). Anemia and thrombocytopenia are present in more than a third of patients. SDS is caused in the great majority of cases by bi-allelic mutations of SBDS (48). The SBDS protein facilitates the release of EIF6 from the pre-60S ribosome, allowing for the joining of 60S and 40S ribosome subunits to generate translationally active 80S ribosome (49). Consequently, the loss-of-function SBDS mutations present in patients with SDS is associated with impaired ribosome biogenesis. There also is evidence implicating SBDS in the formation and stabilization of mitotic spindles (50). Similar to SCN, patients with SDS have a marked propensity to develop MDS or AML. The French Severe Chronic Neutropenia Study Group reported a rate of transformation to AML/MDS of 18.8% at 20 years and 36.1% at 30 years based on a cohort of 55 patients with SDS (9). The Canadian Inherited Marrow Failure Registry reported that 9 of 40 patients with SDS developed AML/MDS or a clonal cytogenetic abnormality (51). Consistent with these findings, the SCNIR reported a crude rate of transformation of AML/MDS 1% per year based on a cohort of 22 patients with SDS (11). In contrast, none of the 17 patients with SDS in the NIH registry developed AML/MDS, which may reflect the relatively young (median age of 14) in this cohort (52).
AML/MDS arising in the setting of SDS also is associated with distinct molecular features. Lindsley and colleagues in their study of patients with MDS identified 7 patients carrying biallelic SBDS mutations (53). Interestingly, only two of these patients carried a diagnosis of SDS, suggesting that this syndrome may be underdiagnosed. Strikingly, mutations of TP53 were detected in all 7 cases of SDS-MDS versus an overall frequency of TP53 mutations in primary MDS of only 14%. Myers and colleagues recently reported clinical features of 37 patients with SDS who developed MDS or AML (54). Complex cytogenetic abnormalities, often involving chromosome 7, were identified in 8 of 9 cases with SDS-AML and 5 of 6 SDS patients with high-grade MDS. It is important to note that isochromosome 7q and del(20q) are common cytogenetic abnormalities associated with SDS that can persist for years and are not clearly linked to the development of AML/MDS (55, 56).
MECHANISMS OF LEUKEMIA TRANSFORMATION IN SDS
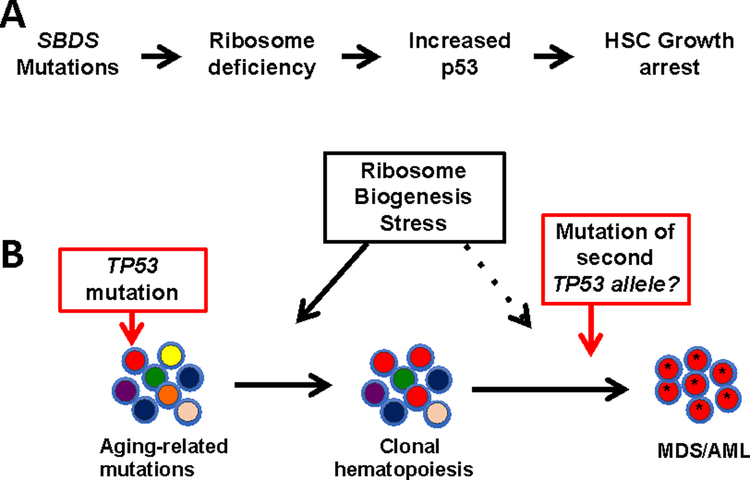
We recently reported that clonal hematopoiesis due to TP53 mutations was present 13 of 27 (48%) of patients with SDS without AML/MDS or clonal cytogenetic abnormality (42). In contrast, no TP53 mutations were detected in healthy controls or patients with SCN. As noted already, mutations of SBDS, which are present in the great majority of cases of SDS, result in impaired ribosome biogenesis (57–59). There is evidence that ribosome biogenesis stress induces p53 expression, which in turn, results in growth arrest. For example, impaired ribosome biogenesis in Diamond-Blackfan Syndrome or 5q- syndrome (which have impaired ribosome biogenesis due to mutations of RPS14 or RPS19, respectively) results in increased p53 expression that contributes to the impaired erythropoiesis in these disorders (60–64). Increased p53 expression also has been identified in hematopoietic cells from patients with SDS or in Sbds deficient murine hematopoietic cells (59, 65). Together, these observations suggest a model in which elevated p53 expression due to ribosome biogenesis stress in SDS HSCs results in impaired HSC growth and/or survival (Figure 2). Mutations of TP53 in HSCs are predicted to attenuate this growth arrest, resulting in their selective expansion in patients with SDS. Perhaps even more worrisome, this model predicts that continued ribosome biogenesis stress in TP53 mutated HSCs would provide a clonal advantage to HSCs that inactivate the second TP53 allele. We suggest that the early accumulation of TP53 mutations in HSPCs is the major reason for the increased risk of developing MDS/AML in patients with SDS. Consistent with this conclusion, a recent study showed that 7 of 7 (100%) of cases of MDS arising in the setting of SDS carried TP53 mutations (53).
Figure 2. Model of leukemic transformation in SDS.
A. Mutations of SBDS, which are found in the great majority of cases of SDS, result in impaired ribosome biogenesis. The resulting ribosome biogenesis stress induces p53 expression, which in turn, induces HSC growth arrest. B. Persistent ribosome biogenesis stress results in the selection of HSCs that carrying TP53 mutations. Continued ribosome biogenesis stress in HSCs carrying one mutated TP53 allele likely selects for clones that have mutated the second allele, leading to genomic instability and eventual MDS/AML.
CONCLUDING THOUGHTS
The development of AML/MDS is a major cause of mortality in patients with congenital neutropenia. Once AML/MDS develops, the only potentially curative therapy is allogenic hematopoietic stem cell transplantation (alloHCT). However, alloHCT is associated with significant treatment-related morbidity and mortality and risk of AML/MDS relapse. Since CSF3R truncation mutations remain dependent on G-CSF, our model of leukemogenesis predicts that therapeutic approaches in patients with SCN that eliminate or reduce the need for G-CSF would substantially lower the risk of developing AML/MDS. Moreover, since the data suggest there is no cell-intrinsic increased risk of leukemic transformation in SCN, minimal conditioning regimens resulting in mixed donor chimerism after allogeneic hematopoietic stem cell transplant are predicted to have a very low risk to develop myeloid malignancy. This is consistent with a recent report by the European which showed that no myeloid malignancies occurred after alloHCT for 136 patients with SCN (66). With respect to SDS, our model predicts that therapeutic strategies that alleviate ribosome biogenesis stress in SDS HSCs would prevent the emergence of TP53 mutated clonal hematopoiesis and reduce the risk of transformation to AML/MDS. Moreover, this model predicts that SDS HSCs would be at a competitive disadvantage compared to normal HSCs. Thus, minimal conditioning regimens for alloHCT should be sufficient to establish full donor chimerism in patients with SDS.
KEY POINTS.
AML/MDS that develops in the setting of congenital neutropenia has distinct molecular features compared to de novo AML.
The rate of mutation accumulation in HSCs from patients with SCN is similar to that observed in healthy individuals.
Persistently high levels of G-CSF drive the expansion of HSCs carrying truncation mutations of CSF3R in patients with SCN.
In SDS, impaired ribosome biogenesis induces p53-mediated growth inhibition and drives expansion of HSCs carrying TP53 mutations.
ACKNOWLEDGEMENTS
The author has no conflict of interest to report. This work was supported by NIH grants P50CA171963 and P01CA194552.
REFERENCES
- 1.Dale DC, Person RE, Bolyard AA, Aprikyan AG, Bos C, Bonilla MA, et al. Mutations in the gene encoding neutrophil elastase in congenital and cyclic neutropenia. Blood. 2000;96(7):2317–22. [PubMed] [Google Scholar]
- 2.Horwitz M, Benson KF, Person RE, Aprikyan AG, Dale DC. Mutations in ELA2, encoding neutrophil elastase, define a 21-day biological clock in cyclic haematopoiesis. Nat Genet. 1999;23(4):433–6. [DOI] [PubMed] [Google Scholar]
- 3.Xia J, Bolyard AA, Rodger E, Stein S, Aprikyan AA, Dale DC, et al. Prevalence of mutations in ELANE, GFI1, HAX1, SBDS, WAS and G6PC3 in patients with severe congenital neutropenia. Br J Haematol. 2009;147(4):535–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Donadieu J, Beaupain B, Fenneteau O, Bellanne-Chantelot C. Congenital neutropenia in the era of genomics: classification, diagnosis, and natural history. Br J Haematol. 2017;179(4):557–74. [DOI] [PubMed] [Google Scholar]
- 5.Boztug K, Klein C. Genetics and pathophysiology of severe congenital neutropenia syndromes unrelated to neutrophil elastase. Hematol Oncol Clin North Am. 2013;27(1):43–60, vii. [DOI] [PubMed] [Google Scholar]
- 6.Grenda DS, Murakami M, Ghatak J, Xia J, Boxer LA, Dale D, et al. Mutations of the ELA2 gene found in patients with severe congenital neutropenia induce the unfolded protein response and cellular apoptosis. Blood. 2007;110(13):4179–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kollner I, Sodeik B, Schreek S, Heyn H, von Neuhoff N, Germeshausen M, et al. Mutations in neutrophil elastase causing congenital neutropenia lead to cytoplasmic protein accumulation and induction of the unfolded protein response. Blood. 2006;108(2):493–500. [DOI] [PubMed] [Google Scholar]
- 8.Dale DC, Bonilla MA, Davis MW, Nakanishi AM, Hammond WP, Kurtzberg J, et al. A randomized controlled phase III trial of recombinant human granulocyte colony-stimulating factor (filgrastim) for treatment of severe chronic neutropenia. Blood. 1993;81(10):2496–502. [PMC free article] [PubMed] [Google Scholar]
- 9.Donadieu J, Leblanc T, Bader Meunier B, Barkaoui M, Fenneteau O, Bertrand Y, et al. Analysis of risk factors for myelodysplasias, leukemias and death from infection among patients with congenital neutropenia. Experience of the French Severe Chronic Neutropenia Study Group. Haematologica. 2005;90(1):45–53. [PubMed] [Google Scholar]
- 10.Rosenberg PS, Zeidler C, Bolyard AA, Alter BP, Bonilla MA, Boxer LA, et al. Stable long-term risk of leukaemia in patients with severe congenital neutropenia maintained on G-CSF therapy. Br J Haematol. 2010;150(2):196–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rosenberg PS, Alter BP, Bolyard AA, Bonilla MA, Boxer LA, Cham B, et al. The incidence of leukemia and mortality from sepsis in patients with severe congenital neutropenia receiving long-term G-CSF therapy. Blood. 2006;107(12):4628–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Desplantes C, Fremond ML, Beaupain B, Harousseau JL, Buzyn A, Pellier I, et al. Clinical spectrum and long-term follow-up of 14 cases with G6PC3 mutations from the French Severe Congenital Neutropenia Registry. Orphanet J Rare Dis. 2014;9:183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zeidler C, Vandenberghe P, Schäfer I, Hoy L, Zimmermann M, Germeshausen M, et al. Update on the risk of secondary leukemia in genetic subgroups of congenital neutropenia in europe. Blood. 2011;118(21):1106. [Google Scholar]
- 14.Freedman MH, Bonilla MA, Fier C, Bolyard AA, Scarlata D, Boxer LA, et al. Myelodysplasia syndrome and acute myeloid leukemia in patients with congenital neutropenia receiving G-CSF therapy. Blood. 2000;96(2):429–36. [PubMed] [Google Scholar]
- 15.Link DC, Kunter G, Kasai Y, Zhao Y, Miner T, McLellan MD, et al. Distinct patterns of mutations occurring in de novo AML versus AML arising in the setting of severe congenital neutropenia. Blood. 2007;110(5):1648–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kalra R, Dale D, Freedman M, Bonilla MA, Weinblatt M, Ganser A, et al. Monosomy 7 and activating RAS mutations accompany malignant transformation in patients with congenital neutropenia. Blood. 1995;86(12):4579–86. [PubMed] [Google Scholar]
- 17.Skokowa J, Steinemann D, Katsman-Kuipers JE, Zeidler C, Klimenkova O, Klimiankou M, et al. Cooperativity of RUNX1 and CSF3R mutations in severe congenital neutropenia: a unique pathway in myeloid leukemogenesis. Blood. 2014;123(14):2229–37.**This study performed targeted sequencing on a large cohort of SCN AML/MDS cases. They report a very high frequency of CSF3R, RUNX1, and chromosome 7 abnormalities.
- 18.Germeshausen M, Ballmaier M, Welte K. Incidence of CSF3R mutations in severe congenital neutropenia and relevance for leukemogenesis: Results of a long-term survey. Blood. 2007;109(1):93–9. [DOI] [PubMed] [Google Scholar]
- 19.Aref S, El-Ghonemy M, Abouzeid T, El-Sabbagh A, El-Baiomy M. Prevalence and impact of colony stimulating factor 3 receptor (CSF3R) mutations among Egyptian acute myeloid leukemia patients. Leuk Res. 2014;38(6):722–5. [DOI] [PubMed] [Google Scholar]
- 20.Beekman R, Valkhof MG, Sanders MA, van Strien PM, Haanstra JR, Broeders L, et al. Sequential gain of mutations in severe congenital neutropenia progressing to acute myeloid leukemia. Blood. 2012;119(22):5071–7. [DOI] [PubMed] [Google Scholar]
- 21.Dong F, Brynes RK, Tidow N, Welte K, Lowenberg B, Touw IP. Mutations in the gene for the granulocyte colony-stimulating-factor receptor in patients with acute myeloid leukemia preceded by severe congenital neutropenia. N Engl J Med. 1995;333(8):487–93. [DOI] [PubMed] [Google Scholar]
- 22.Pardanani A, Lasho TL, Laborde RR, Elliott M, Hanson CA, Knudson RA, et al. CSF3R T618I is a highly prevalent and specific mutation in chronic neutrophilic leukemia. Leukemia. 2013;27(9):1870–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maxson JE, Gotlib J, Pollyea DA, Fleischman AG, Agarwal A, Eide CA, et al. Oncogenic CSF3R mutations in chronic neutrophilic leukemia and atypical CML. N Engl J Med. 2013;368(19):1781–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu F, Kunter G, Krem MM, Eades WC, Cain JA, Tomasson MH, et al. Csf3r mutations in mice confer a strong clonal HSC advantage via activation of Stat5. J Clin Invest. 2008;118(3):946–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Aarts LH, Roovers O, Ward AC, Touw IP. Receptor activation and 2 distinct COOH-terminal motifs control G-CSF receptor distribution and internalization kinetics. Blood. 2004;103(2):571–9. [DOI] [PubMed] [Google Scholar]
- 26.Irandoust MI, Aarts LH, Roovers O, Gits J, Erkeland SJ, Touw IP. Suppressor of cytokine signaling 3 controls lysosomal routing of G-CSF receptor. EMBO J. 2007;26(7):1782–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hunter MG, Avalos BR. Deletion of a critical internalization domain in the G-CSFR in acute myelogenous leukemia preceded by severe congenital neutropenia. Blood. 1999;93(2):440–6. [PubMed] [Google Scholar]
- 28.Ward AC, van Aesch YM, Schelen AM, Touw IP. Defective internalization and sustained activation of truncated granulocyte colony-stimulating factor receptor found in severe congenital neutropenia/acute myeloid leukemia. Blood. 1999;93(2):447–58. [PubMed] [Google Scholar]
- 29.Hermans MH, Antonissen C, Ward AC, Mayen AE, Ploemacher RE, Touw IP. Sustained receptor activation and hyperproliferation in response to granulocyte colony-stimulating factor (G-CSF) in mice with a severe congenital neutropenia/acute myeloid leukemia-derived mutation in the G-CSF receptor gene. J Exp Med. 1999;189(4):683–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McLemore ML, Poursine-Laurent J, Link DC. Increased granulocyte colony-stimulating factor responsiveness but normal resting granulopoiesis in mice carrying a targeted granulocyte colony-stimulating factor receptor mutation derived from a patient with severe congenital neutropenia. J Clin Invest. 1998;102(3):483–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cancer Genome Atlas Research N, Ley TJ, Miller C, Ding L, Raphael BJ, Mungall AJ, et al. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med. 2013;368(22):2059–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Papaemmanuil E, Gerstung M, Bullinger L, Gaidzik VI, Paschka P, Roberts ND, et al. Genomic Classification and Prognosis in Acute Myeloid Leukemia. The New England journal of medicine. 2016;374(23):2209–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tschan CA, Pilz C, Zeidler C, Welte K, Germeshausen M. Time course of increasing numbers of mutations in the granulocyte colony-stimulating factor receptor gene in a patient with congenital neutropenia who developed leukemia. Blood. 2001;97(6):1882–4. [DOI] [PubMed] [Google Scholar]
- 34.Ancliff PJ, Gale RE, Liesner R, Hann I, Linch DC. Long-term follow-up of granulocyte colony-stimulating factor receptor mutations in patients with severe congenital neutropenia: implications for leukaemogenesis and therapy. Br J Haematol. 2003;120(4):685–90. [DOI] [PubMed] [Google Scholar]
- 35.Bernard T, Gale RE, Evans JP, Linch DC. Mutations of the granulocyte-colony stimulating factor receptor in patients with severe congenital neutropenia are not required for transformation to acute myeloid leukaemia and may be a bystander phenomenon. Br J Haematol. 1998;101(1):141–9. [DOI] [PubMed] [Google Scholar]
- 36.Welch JS, Ley TJ, Link DC, Miller CA, Larson DE, Koboldt DC, et al. The origin and evolution of mutations in acute myeloid leukemia. Cell. 2012;150(2):264–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Genovese G, Kahler AK, Handsaker RE, Lindberg J, Rose SA, Bakhoum SF, et al. Clonal Hematopoiesis and Blood-Cancer Risk Inferred from Blood DNA Sequence. New Engl J Med. 2014;371(26):2477–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jaiswal S, Fontanillas P, Flannick J, Manning A, Grauman PV, Mar BG, et al. Age-Related Clonal Hematopoiesis Associated with Adverse Outcomes. New Engl J Med. 2014;371(26):2488–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xie MC, Lu C, Wang JY, McLellan MD, Johnson KJ, Wendl MC, et al. Age-related mutations associated with clonal hematopoietic expansion and malignancies. Nat Med. 2014;20(12):1472–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wilson A, Laurenti E, Oser G, van der Wath RC, Blanco-Bose W, Jaworski M, et al. Hematopoietic stem cells reversibly switch from dormancy to self-renewal during homeostasis and repair. Cell. 2008;135(6):1118–29. [DOI] [PubMed] [Google Scholar]
- 41.Zhu Q-S, Xia L, Mills GB, Lowell CA, Touw IP, Corey SJ. G-CSF induced reactive oxygen species involves Lyn-PI3-kinase-Akt and contributes to myeloid cell growth. Blood. 2006;107(5):1847–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xia J, Miller CA, Baty J, Ramesh A, Jotte MRM, Fulton RS, et al. Somatic mutations and clonal hematopoiesis in congenital neutropenia. Blood. 2018;131(4):408–16.**This study showed clonal hematopoiesis due to TP53 mutations is present in nearly 50% of cases of SDS but not in SCN. This study also showed that mutation burden in HSPCs from patients with SCN and SDS is similar to that observed in age-matched healthy controls.
- 43.Kahn JD, Miller PG, Silver AJ, Sellar RS, Bhatt S, Gibson C, et al. PPM1D-truncating mutations confer resistance to chemotherapy and sensitivity to PPM1D inhibition in hematopoietic cells. Blood. 2018;132(11):1095–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wong TN, Miller CA, Jotte MRM, Bagegni N, Baty JD, Schmidt AP, et al. Cellular stressors contribute to the expansion of hematopoietic clones of varying leukemic potential. Nat Commun. 2018;9(1):455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gibson CJ, Lindsley RC, Tchekmedyian V, Mar BG, Shi J, Jaiswal S, et al. Clonal Hematopoiesis Associated With Adverse Outcomes After Autologous Stem-Cell Transplantation for Lymphoma. J Clin Oncol. 2017:JCO2016716712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kunter G, Woloszynek JR, Link DC. A truncation mutant of Csf3r cooperates with PML-RARalpha to induce acute myeloid leukemia in mice. Exp Hematol. 2011;39(12):1136–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ginzberg H, Shin J, Ellis L, Morrison J, Ip W, Dror Y, et al. Shwachman syndrome: phenotypic manifestations of sibling sets and isolated cases in a large patient cohort are similar. J Pediatr. 1999;135(1):81–8. [DOI] [PubMed] [Google Scholar]
- 48.Boocock GR, Morrison JA, Popovic M, Richards N, Ellis L, Durie PR, et al. Mutations in SBDS are associated with Shwachman-Diamond syndrome. Nature Genetics. 2003;33(1):97–101. [DOI] [PubMed] [Google Scholar]
- 49.Finch AJ, Hilcenko C, Basse N, Drynan LF, Goyenechea B, Menne TF, et al. Uncoupling of GTP hydrolysis from eIF6 release on the ribosome causes Shwachman-Diamond syndrome. Genes Dev. 2011;25(9):917–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Austin KM, Gupta ML Jr., Coats SA, Tulpule A, Mostoslavsky G, Balazs AB, et al. Mitotic spindle destabilization and genomic instability in Shwachman-Diamond syndrome. J Clin Invest. 2008;118(4):1511–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cada M, Segbefia CI, Klaassen R, Fernandez CV, Yanofsky RA, Wu J, et al. The impact of category, cytopathology and cytogenetics on development and progression of clonal and malignant myeloid transformation in inherited bone marrow failure syndromes. Haematologica. 2015;100(5):633–42.*This longitudinal study of 327 patients with inherited bone marrow failure sydromes provides information on the rate of transformation to MDS/AML
- 52.Alter BP, Giri N, Savage SA, Peters JA, Loud JT, Leathwood L, et al. Malignancies and survival patterns in the National Cancer Institute inherited bone marrow failure syndromes cohort study. Br J Haematol. 2010;150(2):179–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lindsley RC, Saber W, Mar BG, Redd R, Wang T, Haagenson MD, et al. Prognostic Mutations in Myelodysplastic Syndrome after Stem-Cell Transplantation. N Engl J Med. 2017;376(6):536–47.*This study sequenced a large number of MDS cases and correlated the genetic data with outcomes after HCT. They report that all 8 cases of MDS with SBDS mutations carryed mutations of TP53.
- 54.Myers KC, Furutani EM, Siegele B, Fleming MD, Elghetany MT, Arsenault V, et al. MDS and AML in Shwachman-Diamond Syndrome: Clinical Features and Outcomes. Blood. 2017;130(Suppl 1):1177.28882833 [Google Scholar]
- 55.Minelli A, Maserati E, Nicolis E, Zecca M, Sainati L, Longoni D, et al. The isochromosome i(7)(q10) carrying c.258+2t>c mutation of the SBDS gene does not promote development of myeloid malignancies in patients with Shwachman syndrome. Leukemia. 2009;23(4):708–11. [DOI] [PubMed] [Google Scholar]
- 56.Pressato B, Valli R, Marletta C, Mare L, Montalbano G, Lo Curto F, et al. Deletion of chromosome 20 in bone marrow of patients with Shwachman-Diamond syndrome, loss of the EIF6 gene and benign prognosis. Br J Haematol. 2012;157(4):503–5. [DOI] [PubMed] [Google Scholar]
- 57.Menne TF, Goyenechea B, Sanchez-Puig N, Wong CC, Tonkin LM, Ancliff PJ, et al. The Shwachman-Bodian-Diamond syndrome protein mediates translational activation of ribosomes in yeast. Nat Genet. 2007;39(4):486–95. [DOI] [PubMed] [Google Scholar]
- 58.Wong CC, Traynor D, Basse N, Kay RR, Warren AJ. Defective ribosome assembly in Shwachman-Diamond syndrome. Blood. 2011;118(16):4305–12. [DOI] [PubMed] [Google Scholar]
- 59.Zambetti NA, Bindels EM, Van Strien PM, Valkhof MG, Adisty MN, Hoogenboezem RM, et al. Deficiency of the ribosome biogenesis gene Sbds in hematopoietic stem and progenitor cells causes neutropenia in mice by attenuating lineage progression in myelocytes. Haematologica. 2015;100(10):1285–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dutt S, Narla A, Lin K, Mullally A, Abayasekara N, Megerdichian C, et al. Haploinsufficiency for ribosomal protein genes causes selective activation of p53 in human erythroid progenitor cells. Blood. 2011;117(9):2567–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ebert BL, Pretz J, Bosco J, Chang CY, Tamayo P, Galili N, et al. Identification of RPS14 as a 5q- syndrome gene by RNA interference screen. Nature. 2008;451(7176):335–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Caceres G, McGraw K, Yip BH, Pellagatti A, Johnson J, Zhang L, et al. TP53 suppression promotes erythropoiesis in del(5q) MDS, suggesting a targeted therapeutic strategy in lenalidomide-resistant patients. Proc Natl Acad Sci U S A. 2013;110(40):16127–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Danilova N, Sakamoto KM, Lin S. Ribosomal protein S19 deficiency in zebrafish leads to developmental abnormalities and defective erythropoiesis through activation of p53 protein family. Blood. 2008;112(13):5228–37. [DOI] [PubMed] [Google Scholar]
- 64.McGowan KA, Li JZ, Park CY, Beaudry V, Tabor HK, Sabnis AJ, et al. Ribosomal mutations cause p53-mediated dark skin and pleiotropic effects. Nat Genet. 2008;40(8):963–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Elghetany MT, Alter BP. p53 protein overexpression in bone marrow biopsies of patients with Shwachman-Diamond syndrome has a prevalence similar to that of patients with refractory anemia. Arch Pathol Lab Med. 2002;126(4):452–5. [DOI] [PubMed] [Google Scholar]
- 66.Fioredda F, Iacobelli S, van Biezen A, Gaspar B, Ancliff P, Donadieu J, et al. Stem cell transplantation in severe congenital neutropenia: an analysis from the European Society for Blood and Marrow Transplantation. Blood. 2015;126(16):1885–92; quiz 970. [DOI] [PubMed] [Google Scholar]