Abstract
Many bacteria can assemble functional amyloid fibers on their cell surface. Most bacterial amyloids contribute to biofilm or other community behaviors where cells interact with a surface or with other cells. Bacterial amyloids, like all functional amyloids, share structural and biochemical properties with disease-associated eukaryotic amyloids. The general ability of amyloids to bind specific dyes, like Congo red and Thioflavin T, and their resistance to denaturation have provided useful tools for scoring and quantifying bacterial amyloid formation. Here, we present basic approaches to study bacterial amyloids by focusing on the well-studied curli amyloid fibers expressed by Enterobacteriaceae. These methods exploit the specific tinctorial and biophysical properties of amyloids. The methods described here are straightforward and can be easily applied by any modern molecular biology lab for the study of other bacterial amyloids.
Keywords: Bacterial amyloids, Curli, Congo red dye, Western blot analysis, Plug Western blot analysis, Overlay assay, Interbacterial complementation
1. Introduction
Amyloids are traditionally associated with protein misfolding and neurodegenerative diseases including Alzheimer’s Disease, Parkinson’s Disease, and Creutzfeldt-Jacob Disease [1–3]. However, a rapidly growing class of “functional amyloids” have recently been described, which indicates that the amyloid fold can be utilized by all walks of cellular life for a variety of functions [4–8]. So far, most functional amyloids in bacteria perform physiological tasks on the cell surface including biofilm formation, adhesion, invasion of host cells, and host-pathogen interactions [9–21]. For example, curli produced by Escherichia coli and Salmonella spp. [22, 23], chaplins formed by Streptomyces spp. [24, 25], and TasA fibers produced by Bacillus subtilis [17] are all functional amyloids utilized by microbes to promote interbacterial interactions. Unlike disease-associated amyloids, functional bacterial amyloids are assembled by highly regulated biosynthetic pathways [4]. Bacterial amyloids share the structural and biochemical properties of disease-associated amyloids. Like all amyloids, functional amyloids bind dyes such as Congo red (CR) and Thioflavin T [17, 22, 25, 26]. Structural analysis of bacterial amyloid fibers indicates a beta-sheet-rich secondary structure [17, 22, 24, 27, 28]. Amyloid fibers are extraordinarily stable, being resistant to denaturation by SDS and digestion by proteases [23, 24, 29]. These properties afford researchers an array of powerful tools for studying bacterial amyloid assembly and function. Here we use curli, one of the best characterized bacterial amyloids, as an example to describe a few basic approaches in the study of bacterial amyloids.
Curli are extracellular amyloid fibers produced by many Enterobacteriaceae including E. coli and Salmonella spp. [30–33]. Purified curli fibers bind CR and induce a spectral red shift in absorbance [22]. Colonies of curli-producing E. coli K-12 stain red on agar plates containing CR, whereas curli-deficient mutants do not [22]. Once CR interacts with curli, it also produces a bright red fluorescence that can be quantified with an excitation wavelength of 485 nm and an emission wavelength of 612 nm.
Curli fibers are composed of two structural components: the major curli subunit CsgA (csg: curli specific gene) and the minor subunit CsgB. The secretion of CsgA and CsgB requires the outer membrane lipoprotein CsgG and the periplasmic accessory factors CsgE and CsgF [34–40]. CsgE tempers CsgA amyloid formation in the periplasm and is hypothesized to guide CsgA to CsgG to allow the unstructured CsgA protein to be secreted [35]. CsgC is another periplasmic accessory protein that inhibits inappropriate CsgA amyloid assembly within the periplasm [41]. CsgB, with the assistance of CsgF, functions as a nucleator by templating the polymerization of CsgA in vivo [36, 39, 42]. Without CsgB, CsgA proteins are secreted to the extracellular space in a SDS-soluble, unstructured form that can be detected in the agar [37, 42]. Once incorporated into curli fibers, CsgA and CsgB are no longer soluble in detergents such as SDS [22].
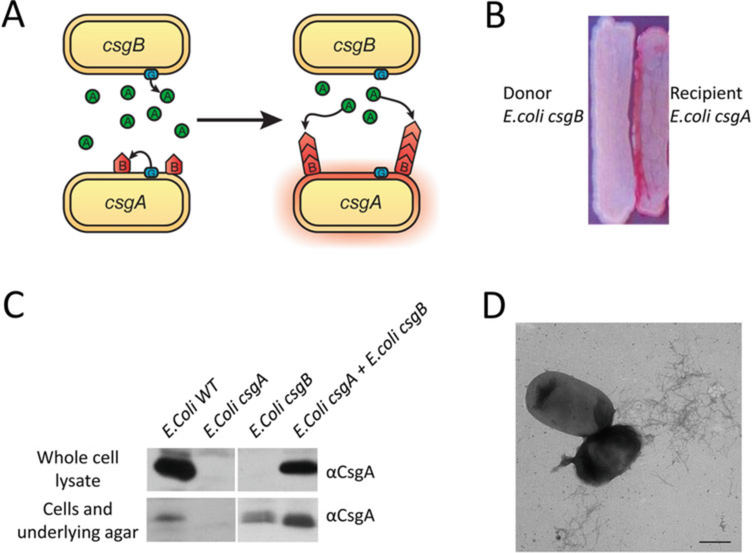
In this chapter, we describe basic approaches for analyzing the presence and/or integrity of curli fibers under physiological conditions in vivo and in vitro. The CR-based assays described here are amenable to high-throughput screens that assess curli production. CR indicator plates can be used to screen for curli deficient mutants and to identify genes important for curli regulation and assembly [43, 44]. Western blot analysis of whole cell lysates is also useful to sort factors involved in curli amyloidogenesis [45–47]. Curli produced by wild type E. coli are cell associated and remain intact even after boiling in SDS-sample buffer [22]. Treatment of whole cell lysates with formic acid (FA) or hexafluoro-2-propanol (HFIP) dissociates the curli fibers into monomers of the major subunit CsgA. After chemical denaturation, CsgA can migrate into an SDS-PAGE gel and can be visualized as a band that runs at 17.5 kDa using anti-CsgA antibodies [22]. We will also detail how a “plug” Western blot assay can be used to differentiate between curli subunits that are un-polymerized from those that are cell-associated and polymerized [22, 42, 46]. CsgA can also be purified and studied in vitro. Finally, an overlay assay and interbacterial complementation provide ways to test CsgA polymerization templated by CsgB in vivo on the bacterial surface. Freshly purified CsgA or CsgA secreted by a csgB mutant assembles on a csgA mutant that presents CsgB on the cell surface (Figs. 2a and 5). The assays described in this chapter can be carried out using common equipment and can be adapted to study other bacterial amyloids.
Fig. 2.
Interbacterial complementation between an E. coli csgA mutant and a csgB mutant. (a) A schematic representation of interbacterial complementation. A csgB mutant (the donor) secretes soluble CsgA into the media, which assembles into curli fibers on the cell surface of an adjacent csgA mutant (the recipient) expressing CsgB. (b) A csgA mutant and a csgB mutant were streaked adjacent to each other on a YESCA CR plate and incubated at 26 °C for 48 h. Colonies of the csgA mutant facing the csgB mutant stained red. (c) Western blot analysis was used to detect formation of intercellular curli fibers between an E. coli csgA mutant and a csgB mutant. Overnight cultures of csgA and csgB mutants were mixed in a 1:1 ratio and spotted onto a YESCA plate and a thin YESCA plate. Whole cell lysates and the plugs were collected for western analysis. A wild type E. coli, a csgA and a csgB mutant were used as controls. All the samples were treated with HFIP and probed with anti-CsgA antibody. (d) A mixed culture of E. coli csgA and csgB mutants was spotted onto YESCA plate and grown for 48 h. Curli were detected by EM. Scar bar is 500 nm
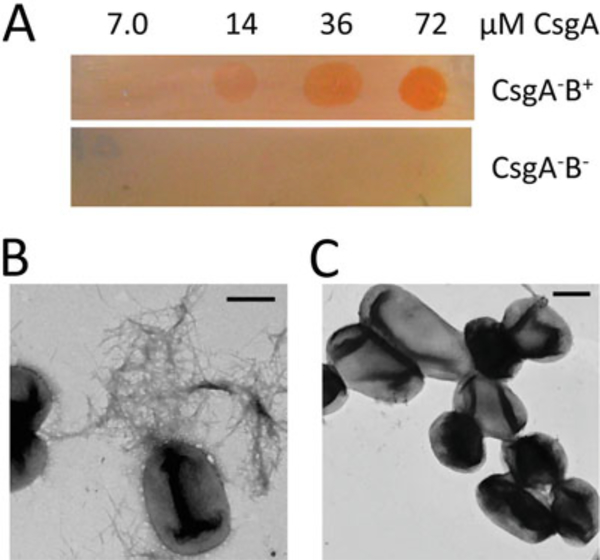
Fig. 5.
Purified CsgA assembles into curli fibers on CsgB expressing cells. (Figure adapted from [46]). (a) CR staining of CsgA–B+ mutant and CsgA–B− mutant overlaid with different concentrations of freshly purified CsgA. Only CsgA–B+ cells overlaid with CsgA protein stained red. (b) Negative-stain EM of CsgA–B+ overlaid with freshly purified CsgA. Fibers were observed on the bacterial surface. Scar bar equals to 500 nm. (c) Negative-stain EM of CsgA–B− overlaid with purified CsgA. No fibers were detected. Scar bar equals to 500 nm
2. Materials
Prepare all the solutions and media using ultrapure water. Prepare and store the reagents at room temperature (RT) unless otherwise indicated. Add antibiotics to media as needed.
2.1. Equipment
Autoclave.
Shaking and static incubators capable of incubating at 37°C and 26 °C.
Centrifuge capable of pelleting cells at 4000 × g and cell lysates at 10,000 × g.
Bench-top centrifuge.
A plate reader with temperature control, capable of intermittent lateral shaking and fluorescent reading capabilities with an excitation of 438 nm and emission of 495 nm.
Protein gel electrophoresis apparatus.
Protein transfer apparatus: wet and semi-dry.
Vacuum concentrator (such as a Savant SPD SpeedVac).
Autoradiography cassettes or fluorescent secondary antibodies.
X-Ray film developer or a scanner with fluorescent detection capabilities at 700 and 800 nm (such as an Odyssey LiCor).
Opaque flat-bottom polystyrene 96-well plates for CR fluorescence quantification.
0.22 μm polyethersulfone bottle-top filters.
Nickel-nitrilotriacetic acid agarose (Ni-NTA).
Sephadex G-25.
BCA protein assay kit.
Kontes 2.5 × 10 cm and 1 × 30 cm glass columns.
Nalgene powder funnel (150 mm with 2.5 cm spout).
Disposable polypropylene columns.
30 kDa centrifugal filter units.
Spin desalt columns.
0.02 μm Anotop filters (Whatman).
Whatman filter paper.
Polyvinylidene difluoride (PVDF) and Nitrocellulose (0.2 μM) membranes.
2.2. Media and Reagents
Luria-Bertani (LB) media: 5 g yeast extract, 10 g bacto tryptone and 10 g NaCl in 1 L water. Autoclave.
LB agar plates: LB media with 17 g agar in 1 L water. Autoclave.
YESCA agar plates: 1 g yeast extract, 10 g casamino acids and 20 g agar in 1 L water. Autoclave. Pour 25 mL per petri dish for normal YESCA plates and 15 mL per dish for thin YESCA plates (see Note 1).
Congo red stock solution: 10 mg/mL in water. Sterile filter, store at 4 °C (see Note 2).
Brilliant Blue G250 stock solution: 10 mg/mL in water. Sterile filter, store at 4 °C (see Note 3).
YESCA Congo red plates: After autoclaving, supplement YESCA agar with 5 mL CR stock (50 μg/mL final concentration) and 100 μL Brilliant Blue stock (1 μg/mL final concentration). Pour as with YESCA agar plates.
Hexafluoro-2-propanol(HFIP) or formic acid (FA) (see Note 4).
SDS sample buffer (2×): 1 mL 1.25 M Tris base pH 6.8, 1 mL β-mercaptoethanol, 200 mL 1% bromophenol blue, 2 mL glycerol, 6 mL 10% SDS and 10 mL water. Bring the final volume up to 20 mL.
SDS-polyacrylamide gel: 15% separating gel and 3% stacking gel.
SDS running buffer: 30.27 g Tris base, 144.1 g glycine and 10 g SDS in 1 L water for 10× stock solution. Dilute 1:10 with water to make 1× running buffer.
Semi-dry transfer buffer: 30.3 g Tris Base and 7.5 g glycine in 1 L water for 10× stock solution. Dilute 1:10 with water and a final concentration of 20% methanol to make 1 L of 1× semidry transfer buffer.
Wet transfer buffer: 5.53 g CAPS in 100 mL methanol and 900 mL water. Use NaOH pellets to adjust to the pH to 11.2 (see Note 5).
Tris buffered saline–Tween-20 (TBS-T): 160 g NaCl, 4 g KCl and 60 g Tris in water and adjust pH to 7.5. Add 20 mL Tween-20 and dilute to 1 L for the 20× stock solution. Dilute 1:20 with water to make a 1× working solution. Store both at 4 °C.
Blocking buffer: dissolve 1% dry fat-free milk and 1% BSA in 1× TBS-T.
Isopropyl β-D-1-thiogalactopyranoside stock (1 M): 4.76 g IPTG in 20 mL water and filter sterilize. Make 1 mL aliquots in 1.5 mL microcentrifuge tubes. Store at −20 °C.
Guanidine hydrochloride (GdnHCl) (8 M): Add 382.1 g GdnHCl (Fisher) to 125 mL 200 mM KPi pH 7.3 with mild heating on a hot plate. Add water slowly, just until the GdnHCl goes into solution. Adjust the pH to 7.2 with NaOH and bring up the final volume to 500 mL.
Potassium phosphate buffer (KPi) (50 mM, pH 7.2): dissolve 28.9 mM KH2PO4 and 21.1 mM K2HPO4 into pure water. Dilute to 1 L and filter sterilize.
100 mM imidazole in 50 mM KPi, pH 7.2: 6.8 g imidazole into 1 L 50 mM KPi and filter sterilize. Store at 4 °C.
12.5 mM imidazole in 50 mM KPi, pH 7.2: 0.85 g imidazole into 1 L 50 mM KPi and filter sterilize. Store at 4 °C.
125 mM imidazole in 50 mM KPi, pH 7.2: 8.51 g imidazole into 1 L 50 mM KPi and filter sterilize. Store at 4 °C.
2.3. Strain and Antibodies
LSR12/pMC1/pMC3: LSR12 is a csgDEFG; csgBAC deletion in E. coli C600 [28]. pMC1 was made by cloning CsgG into the NcoI and BamHI sites of pTRC99A. pMC3 was made by cloning full length C-terminal His6-tagged CsgA into the NdeI and EcoRI sites of pHL3 [22, 28].
NEB 3016/pNH11 [47]: The plasmid pNH11 was made by cloning C-terminal His6-tagged CsgA without the Sec signal sequence into the pET11d vector.
CsgA antibody (Proteintech, Chicago, IL) was raised in rabbits against a synthetic peptide fragment corresponding to the C-terminal half of R4 through the N-terminal half of R5 in CsgA.
CsgB antibody (Proteintech, Chicago, IL) was raised in rabbits against a peptide fragment of the second repeating units of CsgB. Store the antibodies at −20 °C. Dilute antibodies 1:10,000 for CsgA and 1:7000 for CsgB into blocking solution for primary probing (see Note 6).
Anti-rabbit secondary antibody: peroxidase antibody produced in goat or IR-dye fluorescent secondary (we use LiCor). Dilute the HRP antibody by 1:7000 or the IR-dye 1:15,000 in blocking buffer before use.
3. Methods
3.1. Congo Red Assays
Curliated bacteria stain red when grown on plates amended with CR. Therefore, it is easy to differentiate curli-producing strains from curli-deficient strains by assessing the colony color on plates containing CR [22].
3.1.1. CR Staining of Bacterial Colonies (Fig. 1a)
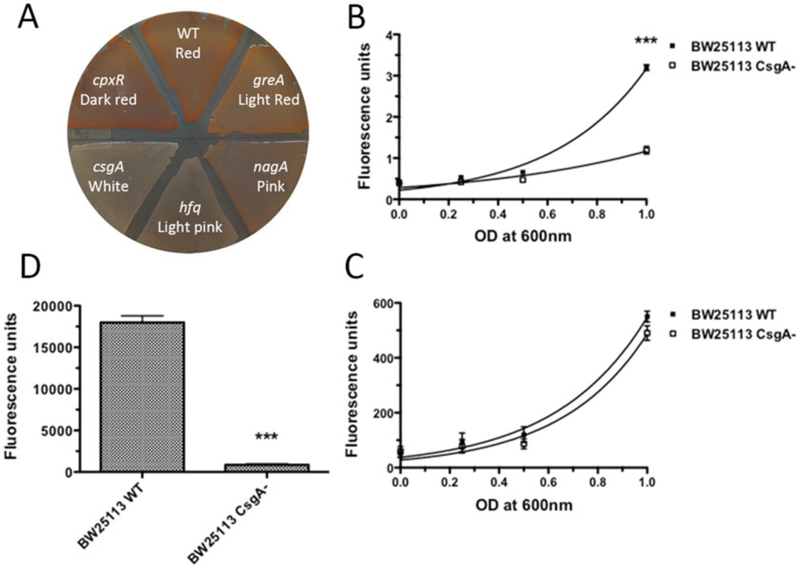
Fig. 1.
CR staining and fluorescence quantification. (a) Use of CR staining to screen for curli mutants. Colonies of the wide type E. coli stained red on a YESCA CR agar plate while the curli defective mutant, csgA, remained white. Curli mutants [56] showed dark red, pink or light pink color on the YESCA CR plate. (b–d) Measurements of the fluorescence associated with bacteria for CR (Em/Ex: 485/612 nm). BW25113 wild type and the csgA deletion mutant strains were recovered from YESCA plates after 48 h of growth at 26 °C and resuspended in 1 mL of 50 mM KPi (pH 7.2) containing 0.5 μg/mL CR or 4.5 μg/mL DAPI. After wash, serial base two dilutions are prepared and the fluorescence was measured in a 96-well plates by triplicate. As a reference, 100 μL of 50 mM KPi were used. (b) Comparison of CR fluorescence of curli producing wild type and curli deficient csgA strains. Non-linear (exponential fit): R2 = 0.970 and 0.778 for BW25113 and csgA, respectively. (c) To show that the BW25113 csgA mutant was present in the same amount as the wild type BW25113 strain, bacteria were post-labeled with the unspecific DNA 4′,6-diamidino-2-phenylindole (DAPI) fluorochrome and the DAPI fluorescence is measured (Em/Ex: 350/460 nm). Non-linear (exponential fit): R2 = 0.865 and 0.867 for BW25113 and csgA, respectively. (d) Quantification of CR fluorescence for bacteria pre-stained on YESCA CR plates. The CR fluorescence of the wild type strain was significantly higher (around three times) compared to the BW25113 csgA mutant. ***: p < 0.0001 using student’s t test. Error bars: average SEM of at least 6 wells per sample. The plate reader used in this determination was an Infinite 200 with the Tecan-I application and automatic optimization of the gain
Streak E. coli strains from −80 °C frozen stock onto an LB plate and grow at 37 °C overnight.
Pick single colonies, streak out on a YESCA CR agar plate (see Note 7).
Grow bacteria at 26 °C for 2 days to induce curli production (see Note 8).
Check the color of colonies on the YESCA CR plate. Curliated bacteria stain red on YESCA CR agar while curli-deficient mutants are usually pink or white (see Note 9).
3.1.2. Congo Red Fluorescence (See Notes 7 and 9): Quantification of CR Fluorescence by Post-CR Staining (Fig. 1b, c)
Streak E. coli strains from −80 °C frozen stock to a LB plate and grow at 37 °C overnight.
Inoculate single colonies in LB liquid broth and grow at 37 °C overnight with shaking.
Spot 4 mL drops of overnight cultures on a YESCA agar plate without CR. Incubate the plate at 26 °C for 48 h to induce curli production.
Recover bacteria cells from the YESCA plate and suspend them in 1 mL 50 mM KPi. Adjust the optical density at 600 nm to one unit in 1 mL of 50 mM KPi.
Centrifuge bacteria at 16,000 × g for 1.5 min and resuspend them in 1 mL 50 mM KPi containing 0.5 μg/mL CR. Incubate at RT with shaking for 20 min.
Wash bacterial cells by centrifugation and resuspend them in1 mL of 50 mM KPi buffer.
Prepare serial dilutions of bacterial suspension. Load 100 mLof each sample onto a 96-well opaque plate. Measure the CR fluorescence using a plate reader. Set the excitation wavelength at 485 nm and the emission at 612 nm (see Note 10). Use 100 mL of KPi as the blank.
3.1.3. Quantification of CR Fluorescence for Bacteria Prestained on YESCA CR Plates (Fig. 1d)
Spot 4 mL of overnight bacterial culture on a YESCA CR plate. Incubate the plate at 26 °C for 48 h.
Recover bacteria from the YESCA CR plate using a clean innoculating loop. Resuspend the cells in 1 mL of 50 mM KPi. Centrifuge for 1.5 min and resuspend cells in 1 mL 50 mM KPi. Adjust the cell density to OD600 = 1.0.
Load 100 mL bacterial suspensions onto a 96-well plate reader. Measure the CR fluorescence with a plate reader as described above.
3.1.4. Interbacterial Complementation (See Note 11) (Fig. 2)
Unpolymerized CsgA proteins secreted by an E. coli csgB mutant assemble into amyloid fibers on the surface of adjacent E. coli csgA mutant that expresses the nucleator protein CsgB, a process called interbacterial complementation [22]. Interbacterial complementation provides a tool for analyzing the nucleation process during curli assembly. In this section, we define the E. coli csgB mutant that secretes unpolymerized CsgA as the donor strain and the csgA mutant that expresses CsgB on the cell surface as the recipient strain.
To test the ability of E. coli mutants to nucleate CsgA secreted by the donor, streak the donor from the top of a YESCA CR plate to the bottom and then streak the putative acceptor strains perpendicularly across the donor streak. Use an E. coli csgA mutant as a positive control. Incubate the plate at 26 °C for 48 h. Bacteria that are able to accept CsgA from the donor and convert them to curli fibers stain red on a CR plate when cross-streaked.
To analyze the secretion of CsgA or the properties of CsgA protein variants, streak out the recipient from the top of a YESCA CR plate to the bottom and cross-streak the putative donor strains perpendicularly over the recipient. Use an E. coli csgB mutant as the positive control. Incubate the plate at 26 °C for 48 h. Bacteria donating functional CsgA stain red after cross-streaked over the recipient (see Notes 12 and 13).
3.2. Western Blot Analysis of the Whole Cell Lysate (Fig. 3)
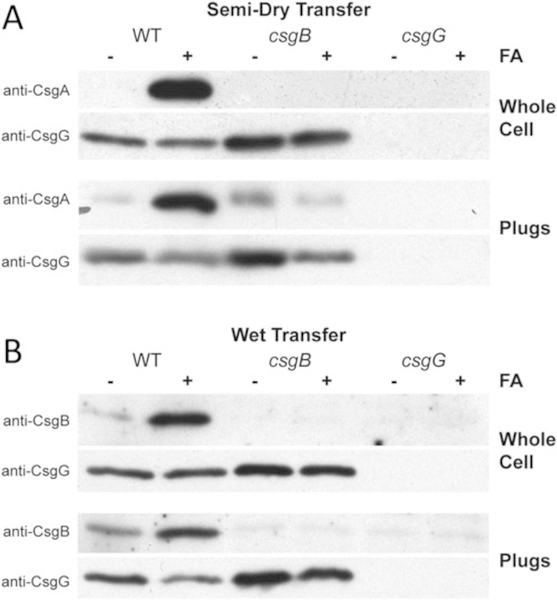
Fig. 3.
Western blot analysis of whole cell lysates and agar plugs. (a) Western blots of whole cell extracts and plugs of E. coli strains [56] that were transferred onto PVDF membranes and probed with polyclonal anti-CsgA antibodies. The csgB mutant strain did not produce curli fibers and soluble secreted CsgA was found in the agar plug. (b) Western blots of whole cell and plugs of E. coli strains [56] that were transferred onto nitrocellulose membranes and probed with polyclonal anti-CsgB antibodies. A strain lacking CsgG results in no CsgA or CsgB in whole cells or in the agar analysis
Western blotting provides another way to study curli assembly. Curli fibers are resistant to SDS. HFIP/FA treatment of curli releases monomeric CsgA and CsgB that mobilize into the SDS-PAGE gel and can be probed by anti-CsgA or anti-CsgB antibodies respectively.
Inoculate single colonies of E. coli strains in LB liquid broth and grow at 37 °C overnight with shaking. Dilute cultures to OD600= 1.0. Spot 4 mL of normalized cultures onto a thin YESCA agar plate. Grow E. coli strains on a YESCA agar plate at 26 °C for 48 h.
Scrape off bacteria cells from the YESCA agar using a sterile inoculating loop and suspend them in 1 mL 50 mM KPi. Normalize cells by optical cell density at 600 nm to OD600/mL.
Transfer 150 mL of normalized cell suspension into two separate tubes, one for HFIP/FA treatment and the other for the SDS only control.
Spin down bacteria cells at 16,000 × g for 3 min and remove the supernatant. Cell-associated curli fibers and protein aggregates should pellet.
To depolymerize curli fibers into monomers, resuspend one of the duplicate pellets in 70 μL 100% HFIP or FA briefly (see Note 14).
Immediately dry samples in a Speedvac at 45 °C for 30 min (see Note 15).
Resuspend all pellets in 150 mL 2× SDS loading buffer. FA-treated samples may contain residual acid turning the sample buffer yellow. Adjust the pH by addition of 1 μL at a time of 5 N NaOH until the loading buffer turns blue again. Boil samples at 95 °C for 10 min (see Note 15).
Run 5–7 mL of each sample on a 15% SDS-polyacrylamide gel at 25 mA per gel.
Semi-dry transfer: Use for CsgA western blotting. After electrophoresis, transfer proteins from the SDS-polyacrylamide gel to a PVDF membrane by the semi-dry transfer system. Each blotting stack should be made of three sheets of Whatman paper, the gel, a PVDF membrane followed by another three sheets of Whatman paper. The PVDF membrane needs to be pretreated with methanol, water and transfer buffer. Transfer for 20 min at 10 V (see Note 16).
Wet transfer: Use for CsgB, which does not efficiently transfer using the semi-dry transfer system, probably due to its high isoelectric point. Use a similar blotting stack as for the semi-dry transfer, except use a nitrocellulose membrane instead of PVDF and use the wet transfer buffer, pH 11.2. Transfer in a wet transfer system at 50 V for 3 h or 12 V overnight at 4 °C (see Note 16).
Block the western blot with blocking solution for at least 1 h at RT or at 4 °C overnight.
Probe the western blots with 1:10,000 diluted anti-CsgA or 1:7000 diluted anti-CsgB polyclonal antibodies in blocking buffer at RT for 1 h (see Note 6).
Wash the blot with TBS-T and then probe with anti-rabbit secondary antibody at a dilution of 1:10,000 in blocking buffer at RT for 1 h.
Develop blot with chemiluminescent substrates or scan with a fluorescence capable scanner.
3.3. Plug Western Blot Analysis (Fig. 3)
Western blotting of whole cell lysate is a powerful tool to study cell-associated fibers, however, it has its drawbacks. If no CsgA is detected by HFIP/FA treatment, it is possible that CsgA or CsgA mutants are secreted into the agar in an unpolymerized form. It is also likely that non-cell-associated fibers are formed in the agar, or CsgA is not expressed or is degraded. The plug Western blot analysis provides an approach to distinguish between those possibilities.
Inoculate single colonies of E. coli strains in LB liquid broth and grow at 37 °C overnight with shaking.
Dilute cultures to OD600= 1.0.
Spot 4 mL of normalized cultures onto a thin YESCA agar plate.
Grow bacteria on the thin YESCA plate at 26 °C for 48 h.
Use the wide end of a Pasteur pipette as a cookie cutter-like tool to remove an 8 mm circular plug including the bacteria and the underlying agar. Collect 2 plugs for each strain—one for HFIP/FA treatment and the other as an SDS only control. Put each plug into a 1.5 microcentrifuge tube.
For the HFIP/FA treatment group, suspend the plug in 100 μL HFIP or FA and briefly vortex. Dry the samples in a Speedvac at 45 °C for 40 min and then resuspend the pellets with 2× SDS loading buffer.
For the non-treated plugs, suspend plugs in 150 μL 2× SDS loading buffer.
Sonicate both HFIP/FA-treated samples and nontreated samples in a water-bath sonicator for 3 min (see Note 17).
If the SDS loading buffer turns yellow, adjust the sample pH to neutral by adding 1 or 2 μL of 5 N NaOH. Heat the samples at 95 °C for 10 min before loading onto a 15% SDS-polyacrylamide gel (see Note 15).
Electrophorese the samples, transfer proteins to a PVDF membrane for detecting CsgA or nitrocellulose for detecting CsgB, probe, and develop the blot as described in Subheading 3.2.
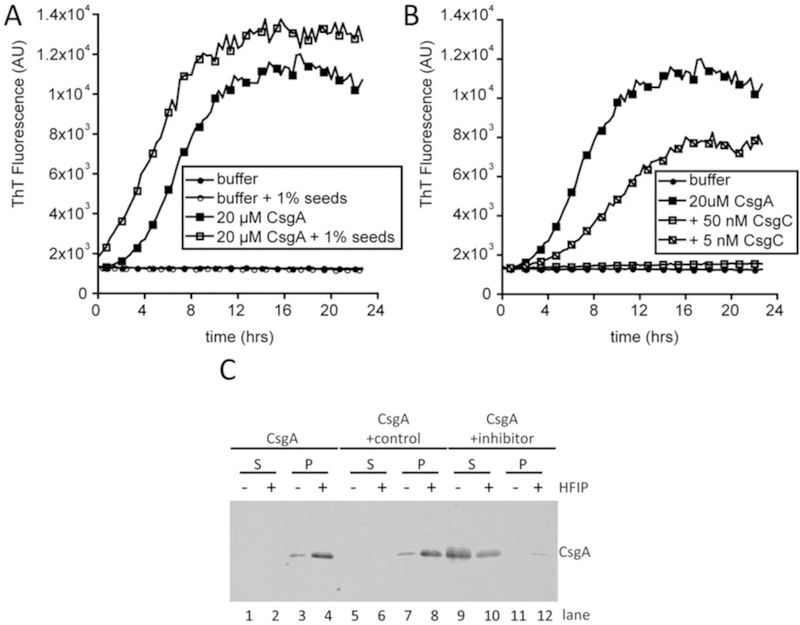
3.4. CsgA Purification, Gel Solubility, and Overlay Assay (Figs. 4 and 5)
Fig. 4.
CsgA assembles into an amyloid in vitro. (a) Polymerization of CsgA was monitored by ThT fluorescence in a plate reader over 24 h. Buffer alone (closed circles) and with sonicated CsgA fibers (seeds; open circles) do not bind ThT. CsgA alone (closed squares in a and b) rapidly polymerizes and binds ThT after approximately 2 h. The 2-h lag phase is overcome by the addition of CsgA seeds to CsgA monomers (open squares). (b) CsgA amyloid assembly can be inhibited by protein inhibitors such as CsgC. CsgC was added to purified CsgA at sub-stoichiometric molar ratios. The addition of 50 nM CsgC to 20 μM (1:400 molar ratio) completely inhibits CsgA amyloid assembly (open squares) whereas the addition of 5 μM CsgC (1:4000 molar ratio) extends the lag phase and rate of CsgA amyloid assembly (slashed squares). (c) Soluble (S) and polymerized (P) CsgA can be distinguished using a gel solubility assay. CsgA was incubated alone (lanes 1–4), with a control protein that does not affect amyloid formation (lanes 5–8) or with an inhibitory protein (lanes 9–12) prior to separating the soluble and polymerized species
There are two CsgA purification schemes in the published literature (see Note 18). In one of the approaches, both CsgA and CsgG are overexpressed in the absence of all other curli proteins. CsgA is secreted into the supernatant and is collected and purified by nickel affinity chromatography [22, 28]. In an alternative purification method, CsgA without the Sec signal sequence can be purified from the cytoplasm using a denaturing protocol [42, 48]. C-terminal His6-tagged CsgA is referred to as “CsgA” in this section.
3.4.1. CsgA Purified from Cell Supernatants (See Note 19)
Perform the purification in a 4 °C cold room or a 4 °C fridge. Store the columns and buffers at 4 °C unless otherwise stated.
Start an overnight culture of LSR12/pMC1/pMC3 in 25 mL LB-Amp-Cm per 1 L of desired prep at 37 °C.
Dilute the overnight culture into 1 L LB-Amp-Cm and incubate with shaking at 37 °C to OD600 of ~1.0.
Induce with 0.25 mM IPTG and incubate with shaking for 45 min at 37 °C.
Centrifuge the culture at 10,000 × g for 15 min at 4 °C. Collect the supernatant.
Shift purification to 4 °C.
Filter the supernatant over a 0.22 μm polyethersulfone bottle-top filter (see Note 20).
Prepare a column with 4 mL of Ni-NTA bead volume. Wash the column with 4 bed volumes (BV) of 50 mM KPi, pH 7.2.
Flow the filtrates over the column set to the maximum flow rate (see Note 21).
Wash the column with 10 BV of 50 mM KPi, pH 7.2.
Elute the protein off the column with 2 BV of 100 mM imidazole in 50 mM KPi, pH 7.2. Adjust the flow rate to 20 drops/min. Collect elutes in 1 mL fractions and store in microcentrifuge tubes.
Combine fractions containing CsgA as measured by BCA in a 15 mL conical tube.
Equilibrate a Sephadex G-25 gel filtration column with 25 mL 50 mM KPi, pH 7.2.
Load 3–4 mL of protein onto the column. Elute 1 mL fractions into micro-centrifuge tubes using 50 mM KPi, pH 7.2. Measure the protein concentration of each fraction by UV280 or BCA assay at RT. Combine the fractions that contain most of CsgA proteins, filter through a 0.02 μm Anotop filter, and measure the final concentration. Store on ice (see Notes 22 and 23).
3.4. 2. CsgA Purified from Cell Lysates (See Note 19)
Start an overnight culture of NEB C2566/pNH11 in 10 mL LB Amp-Kan-Cm per 500 mL of desired prep at 37 °C.
Dilute the overnight culture into 500 mL LB-Amp and incubate with shaking at 37 °C to an OD600 of ~1.0.
Induce with 0.5 mM IPTG and incubate for 1 h at 37 °C.
Collect bacteria by centrifugation at 5,000 × g for 20 min and discard the supernatant. The pellet can be stored at −80 °C for future use.
Lyse bacterial cells by adding 50 mL 8 M GdnHCl to the pellet and incubate at 4 °C overnight with stirring.
Centrifuge the GdnHCl bacterial suspension at 10,000 × g for 20 min to remove the insoluble portion of the lysate.
Incubate the resulting supernatant with Ni-NTA resin for 1 h at RT with rocking.
Load the supernatant with Ni-NTA onto a disposable polypropylene column.
Wash the beads with 10 BV 50 mM KPi, pH 7.2 followed by 3 BV 12.5 mM imidazole in 50 mM KPi, pH 7.2 to remove nonspecific bound proteins.
Elute the protein off the column with 6 BV 125 μM imidazole in 50 mM KPi, pH 7.2 Collect 1 mL of elutions for each fraction in 1.5 microcentrifuge tubes. Store fractions on ice (see Note 22).
Measure the protein concentration from each fraction by BCA assay. Combine the fractions with proteins and load onto a 30 kDa centrifugal filter unit. Centrifuge at 7500 × g for 10 min (see Note 24). The bottom part of the filter unit contains CsgA monomers.
Prewash a spin desalt column with 50 mM KPi by centrifugation at 1000 × g for 2 min. Collect proteins from the bottom part of the 30 kDa filter unit and load onto the desalting column. Centrifuge at 1000 × g for 2 min (see Note 25).
Store purified protein on ice (see Note 22). Measure protein concentration by BCA assay or UV280.
3.4.3. ThT Detection of CsgA Polymerization, Seeding, and Inhibition (Fig. 4a, b)
Purified, monomeric CsgA polymerizes into amyloid fibers that bind the dye ThT. ThT fluorescence can therefore be measured over time to monitor amyloid assembly of CsgA. CsgA polymerizes with kinetics characteristic of all amyloids; the early lag phase is approximately 2 h, followed by a rapid increase in ThT fluorescence and then a stationary phase. The kinetics of CsgA amyloid assembly can be altered by the addition of preformed CsgA or CsgB fibers (seeds) and protein or chemical chaperones or accelerators. Careful analysis of the kinetics of CsgA amyloid assembly in the presence of such modulators can yield insights into the mechanism of amyloid modulation.
Dilute purified CsgA to 31.25 μM in 50 mM KPi pH 7.3125 mM imidazole (or 50 mM KPi pH 7.3 without imidazole if CsgA has been desalted).
Set a plate reader to measure the fluorescence of ThT every 20 min with an excitation wavelength of 438 nm and an emission wavelength of 495 nm. Set the plate reader to shake with a lateral amplitude of 4.5 mm for 3 s prior to each reading. Set the temperature to 25 °C.
For CsgA polymerization: Add 80 μL of CsgA to each well of a black, flat-bottom, 96-well plate with 2 μL of 1 mM ThT (filtered) and 20 μL of buffer. The negative control should consist of 100 μL of buffer with 2 μL 1 mM ThT. Set up all controls and tests in triplicate. Monitor polymerization by reading ThT fluorescence as described in step 2.
For CsgA seeding: Purified and polymerized CsgA or CsgB fibers are used as the source of seeds (Fig. 4a). The purified protein should be diluted to 25 μM and stored at room temperature for 2 days in a low-retention 1.5 mL microcentrifuge tube. To generate seeds, use a probe sonicator to sonicate the CsgA or CsgB fibers on low 3 × 10 s with 30 s rest on ice in between. Add seeds to CsgA polymerization reactions (described in step 3) to a final concentration between 1 and 10% by weight. Include seeds with buffer and ThTas a negative control for polymerization. Monitor polymerization by reading ThT fluorescence as described in step 2.
For CsgA inhibition and acceleration: CsgA amyloid assembly can be modulated by both protein and chemical inhibitors and accelerators [41, 48–51]. An example of inhibition is shown in Fig. 4b. Dilute the protein or chemical stock to 5× the desired final concentration. Add 20 μL of 5× stock to 80 μL of 31.25 μM purified CsgA with 2 μL 1 mM ThT. Serially dilute the inhibitor or accelerator and add the same total volume to CsgA to test a range of molar ratios. Monitor polymerization by reading ThT fluorescence as described in step 2.
3.4. 4. Gel Solubility Assay (Fig. 4c)
Purified soluble and polymerized CsgA can be separated by centrifugation and analyzed by SDS PAGE. Amyloid aggregates can be distinguished from non-amyloid aggregates that may pellet by treating duplicate samples either with SDS or HFIP followed by SDS.
Remove samples from the 96-well plate after a ThT polymerization assay pooling triplicates together. Pipet up and down several times to wash off fibers that may adhere to the wells.
Transfer 100 μL of each sample into each of two microcentrifuge tubes. Pellet at 16,000 × g in a table top centrifuge for 10 min. Remove the supernatants to new tubes. You should now have two pellet and two supernatant fractions for each sample.
Treat one pellet and one supernatant with HFIP. Add 50 μL HFIP to pellets and 200 μL to the supernatant. Incubated at room temperature for 10 min. Dry in a vacuum evaporator or SpeedVac. Once dry, add 40 μL of 2× SDS loading buffer to each sample.
Treat the other pellet and supernatants with SDS loading buffer. Add 40 μL of 2× SDS loading buffer to the pellet and 50 μL 4× SDS loading buffer to the supernatants.
Boil samples for 10 min and load 7 μL on a 15% SDS PAGE gel. Run at 25 mA/gel.
Stain with Coomassie Blue or process as a Western blot (Fig. 4c).
3.4.5. Overlay Assay (Fig. 5)
Soluble amyloid proteins polymerize into amyloid fibers and this polymerization can be accelerated by its own fibers, a process known as seeding. The polymerization and seeding can be monitored in vitro by Thioflavin T fluorescence or transmission electron microscopy (TEM). The overlay assay provides a mechanism to test the assembly of bacterial amyloids directly on the cell surface. It can be used to determine the amyloidogenic domains of bacterial amyloid proteins [46].
Spread a lawn of 50 mL desired bacterial suspension on a YESCA plate by sterile inoculation loop and incubate for 2 days at 26 °C. Use E. coli K-12 csgA and csgBA mutant as the positive control and negative control, respectively.
Drip 10 mL of freshly purified CsgA on the bacterial lawn. Incubate at RT for 10 min.
Stain the plate with 10 mL of 0.5 mg/mL CR at RT for 5 min then wash with 50 mM KPi, pH 7.2.
A wild-type strain of curli-expressing E. coli will strain red, as will a csgA mutant that has been overlaid with purified CsgA protein, while csgBA mutant with overlaid with purified CsgA protein will remain white (Fig. 5a). Assembly of curli fibers on bacterial surfaces can also be verified by negative stained TEM (Fig. 5b, c).
Acknowledgments
We thank members of the Chapman laboratory for helpful discussions and review of this manuscript, especially DRS and LB. This work was supported by the National Institutes of Health Grant R01 GM118651 to MRC.
Abbreviations
- CR
Congo Red
- Csg
Curli specific genes
- FA
Formic acid
- HFIP
Hexafluoro-2-propanol
- RT
Room temperature
Footnotes
Other than YESCA agar, T-agar [30], colonization factor antigen (CFA) agar [23], and LB agar without salt [52] can also be used to induce curli production of Salmonella spp. and E. coli. LB agar without salt contains 5 g/L yeast extract, 10 g/L bacto-tryptone, and 1.5% agar.
Mix CR with water by stirring for at least an hour. Make sure CR is completely dissolved before filter sterilization.
Brilliant blue is used to increase the color contrast of colonies on CR agar.
HFIP is a strong solvent that disaggregates curli subunits and does not result in acid hydrolysis. FA is also used to disassociate curli subunits. FA and HFIP are corrosive and cause burns. They evaporate at room temperature. Store FA and HFIP in a cool, well-ventilated place. Always use them under a chemical fume hood and wear appropriate protective equipment.
Here, we use high pH CAPS buffer because the CsgB protein has a high pI and cannot be transferred efficiently with more neutral buffers.
1:10,000 dilution rate of anti-CsgA antibody or 1:7000–1:10,000 of anti-CsgB antibody gives good signal for western blotting. High concentrations of antibody are not recommended as this may cause nonspecific binding. The anti-CsgA antibody also binds to an unknown nonspecific protein that migrates at around 40 kDa on SDS-PAGE gels.
For CR staining and fluorescence quantification, grow a curli-producing wild type strain as a positive control and curli-defective mutant (csgA or csgB deletion strain) as a negative control.
Temperature and incubation time are important factors for CR assays. Most of the E. coli K-12 and Salmonella spp. lab strains produce curli at 26 °C, whereas some clinical isolates produce curli at 37 °C [53, 54]. Although E. coli K-12 lab strains grown at 37 °C do not produce curli, they may eventually take up CR and stain red. Incubation for longer than 2 days may also make bacteria take up or bind CR. Furthermore, defects in outer-membrane integrity may increase CR uptake.
Most E. coli K-12 isolates have CR phenotypes that are completely dependent on curli fiber production. However, many bacterial strains can produce cellulose or other extracellular polysaccharides that bind to CR. The corresponding curli-defective mutants of those strains may form pink, smooth colonies on YESCA CR plates.CR is able to interact and emit fluorescence with cellulose. Therefore, multiple approaches are recommended for the study of amyloids produced by bacteria with complex surface structures.
A wavelength scan of CR fluorescence is required to determine the ideal excitation and emission wavelength for CR quantification for the analysis of other bacterial amyloids.
Some bacteria do not participate in interbacterial complementation. Interbacterial complementation is not detected between S. enterica serovar Enteritidis 3b csgA and csgB mutants, probably due to the lipopolysaccharide O polysaccharide [55].
The curli mutants of cellulose-positive bacteria are pink on YESCA CR agar, which makes the detection of interbacterial complementation difficult by CR staining. As an alternative, a mixture of desired bacteria can be collected and the intercellular curli formation can be assessed by EM or Western blot analysis as mentioned in Subheadings 3.2 and 3.3 (Fig. 2c, d).
Another way to test interbacterial complementation is to streak the donor or recipient on a YESCA plate and then make a parallel streak 3 mm away from the first streak. Incubate the plate at 26 °C for 48 h (Fig. 2b). The recipient cells facing to donor cells will stain red if intercellular curli are formed.
FA is capable of hydrolyzing proteins including curli subunits. Long treatment with FA may result in target protein loss. Speed-dry samples immediately after the FA treatment, or use HFIP instead.
Make sure that samples are thoroughly dried after speed-vac step. If there is still liquid FA or HFIP left in the tube after 30 min in the Speedvac, dry the sample for longer. The remaining FA in bacterial pellets or plugs lowers the pH and turns the SDS-sample buffer yellow. The low pH of samples affects electrophoresis and boiling low pH sample increases the chance of acid hydrolysis. Adjust the pH of sample buffer back to around pH 7 before loading samples onto SDS-PAGE gels.
We use the semi-dry transfer apparatus from FisherBiotech and a wet transfer system from Biorad. The voltage and transfer time may vary for different transfer systems.
We found that water bath sonication makes the results for the plug assay more consistent, probably by breaking up large chunks of dried agar.
Both CsgA purification approaches have advantages and drawbacks. Purification of CsgA from bacterial supernatant does not require denaturation of the target protein. However, the whole process is time consuming and typically takes 2 days. The denaturing method takes only half a day, but the final yield is typically lower. CsgA proteins purified by both methods have the same biochemical properties and form fibers with same morphologies and with similar kinetics.
Collect ~100 mL of sample during each of the purification steps (e.g., the load, the wash, and elutes) and store on ice. Run a SDS-PAGE gel of the samples after purification to make sure that the protein is pure. In addition, an expression test is required for new expression strains to make sure that protein is expressed in the tested conditions.
To remove bacteria that remain in the supernatant after centrifugation, filter the supernatant through the 0.22 μm filter. Filtered supernatant can be stored at 4 °C overnight. However, longer storage is not recommended.
Depending on the column size and the particulates in the supernatant, it can take upward of 10 h to gravity flow 1 L of supernatant through the nickel affinity column. Use of high flow Ni-NTA and addition of a powder funnel on the top of the column both greatly accelerate the loading process.
Purified CsgA forms fibers at RT in 1–2 h, so it is important to keep the protein on ice to slow the aggregation process. Start the overlay assay immediately after the purification. CsgA cannot be stored at −20 °C or −80 °C because CsgA precipitates out of solution during the thawing process.
To purify CsgA from cell supernatants under denaturing conditions, equilibrate the Ni-NTA gel with 5BV 8 M GdnHCl in 10 mM KPi, pH 7.2 at RT after supernatant binding and washing with 5BV 10 mM KPi, pH 7.2. Elute with 3BV 8 M GdnHCl in 50 mM KPi, pH 7.3.
Use BCA assay or UV280 to detect and pool CsgA containing fractions, which can be stored at RT for several days. Desalt using Sephadex G-25 at RT using cold columns and 4 °C 50 mM KPi, pH 7.2.
Check the manuals that accompany the 30 kDa centrifuge units and desalting columns. The speed and time for centrifugation may vary from product to product.
References
- 1.Cooper GJ, Willis AC, Clark A et al. (1987) Purification and characterization of a peptide from amyloid-rich pancreases of type 2 diabetic patients. Proc Natl Acad Sci U S A 84 (23):8628–8632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Glenner GG, Wong CW (1984) Alzheimer’s disease: initial report of the purification and characterization of a novel cerebrovascular amyloid protein. Biochem Biophys Res Commun 120(3):885–890 [DOI] [PubMed] [Google Scholar]
- 3.Prusiner SB (1996) Molecular biology and pathogenesis of prion diseases. Trends Biochem Sci 21(12):482–487 [DOI] [PubMed] [Google Scholar]
- 4.Blanco LP, Evans ML, Smith DR et al. (2012) Diversity, biogenesis and function of microbial amyloids. Trends Microbiol 20(2):66–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Knowles TP, Mezzenga R (2016) Amyloid fibrils as building blocks for natural and artificial functional materials. Adv Mater 28 (31):6546–6561 [DOI] [PubMed] [Google Scholar]
- 6.Pham CL, Kwan AH, Sunde M (2014) Functional amyloid: widespread in nature, diverse in purpose. Essays Biochem 56:207–219 [DOI] [PubMed] [Google Scholar]
- 7.Schwartz K, Boles BR (2013) Microbial amyloids—functions and interactions within the host. Curr Opin Microbiol 16(1):93–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Watt B, van Niel G, Raposo G et al. (2013) PMEL: a pigment cell-specific model for functional amyloid formation. Pigment Cell Melanoma Res 26(3):300–315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Adcox HE, Vasicek EM, Dwivedi V et al. (2016) Salmonella extracellular matrix components influence biofilm formation and gallbladder colonization. Infect Immun 84 (11):3243–3251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Austin JW, Sanders G, Kay WW et al. (1998) Thin aggregative fimbriae enhance Salmonella enteritidis biofilm formation. FEMS Microbiol Lett 162(2):295–301 [DOI] [PubMed] [Google Scholar]
- 11.Gallo PM, Rapsinski GJ, Wilson RP et al. (2015) Amyloid-DNA composites of bacterial biofilms stimulate autoimmunity. Immunity 42 (6):1171–1184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gophna U, Barlev M, Seijffers R et al. (2001) Curli fibers mediate internalization of Escherichia coli by eukaryotic cells. Infect Immun 69 (4):2659–2665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gophna U, Oelschlaeger TA, Hacker J et al. (2002) Role of fibronectin in curli-mediated internalization. FEMS Microbiol Lett 212 (1):55–58 [DOI] [PubMed] [Google Scholar]
- 14.Johansson C, Nilsson T, Olsén A et al. (2001) The influence of curli, a MHC-I-binding bacterial surface structure, on macrophage-T cell interactions. FEMS Immunol Med Microbiol 30(1):21–29 [DOI] [PubMed] [Google Scholar]
- 15.Larsen P, Nielsen JL, Dueholm MS et al. (2007) Amyloid adhesins are abundant in natural biofilms. Environ Microbiol 9(12):3077–3090 [DOI] [PubMed] [Google Scholar]
- 16.Rapsinski GJ, Wynosky-Dolfi MA, Oppong GO et al. (2015) Toll-like receptor 2 and NLRP3 cooperate to recognize a functional bacterial amyloid, curli. Infect Immun 83 (2):693–701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Romero D, Aguilar C, Losick R et al. (2010) Amyloid fibers provide structural integrity to Bacillus subtilis biofilms. Proc Natl Acad Sci U S A 107(5):2230–2234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tursi SA, Lee EY, Medeiros NJ et al. (2017) Bacterial amyloid curli acts as a carrier for DNA to elicit an autoimmune response via TLR2 and TLR9. PLoS Pathog 13(4):e1006315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tükel C, Raffatellu M, Humphries AD et al. (2005) CsgA is a pathogen-associated molecular pattern of Salmonella enterica serotype Typhimurium that is recognized by Toll-like receptor 2. Mol Microbiol 58(1):289–304 [DOI] [PubMed] [Google Scholar]
- 20.Tükel C, Nishimori JH, Wilson RP et al. (2010) Toll-like receptors 1 and 2 cooperatively mediate immune responses to curli, a common amyloid from enterobacterial biofilms. Cell Microbiol 12(10):1495–1505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vidal O, Longin R, Prigent-Combaret C et al. (1998) Isolation of an Escherichia coli K-12 mutant strain able to form biofilms on inert surfaces: involvement of a new ompR allele that increases curli expression. J Bacteriol 180 (9):2442–2449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chapman MR, Robinson LS, Pinkner JS et al. (2002) Role of Escherichia coli curli operons in directing amyloid fiber formation. Science 295 (5556):851–855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Collinson SK, Doig PC, Doran JL et al. (1993) Thin, aggregative fimbriae mediate binding of Salmonella enteritidis to fibronectin. J Bacteriol 175(1):12–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Claessen D, Rink R, de Jong W et al. (2003) A novel class of secreted hydrophobic proteins is involved in aerial hyphae formation in Streptomyces coelicolor by forming amyloid-like fibrils. Genes Dev 17(14):1714–1726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Elliot MA, Karoonuthaisiri N, Huang J et al. (2003) The chaplins: a family of hydrophobic cell-surface proteins involved in aerial mycelium formation in Streptomyces coelicolor. Genes Dev 17(14):1727–1740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.de Jong W, Wösten HA, Dijkhuizen L et al. (2009) Attachment of Streptomyces coelicolor is mediated by amyloidal fimbriae that are anchored to the cell surface via cellulose. Mol Microbiol 73(6):1128–1140 [DOI] [PubMed] [Google Scholar]
- 27.Shewmaker F, McGlinchey RP, Thurber KR et al. (2009) The functional curli amyloid is not based on in-register parallel beta-sheet structure. J Biol Chem 284(37):25065–25076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang X, Smith DR, Jones JW et al. (2007) In vitro polymerization of a functional Escherichia coli amyloid protein. J Biol Chem 282 (6):3713–3719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Collinson SK, Parker JM, Hodges RS et al. (1999) Structural predictions of AgfA, the insoluble fimbrial subunit of Salmonella thin aggregative fimbriae. J Mol Biol 290 (3):741–756 [DOI] [PubMed] [Google Scholar]
- 30.Collinson SK, Emödy L, Müller KH et al. (1991) Purification and characterization of thin, aggregative fimbriae from Salmonella enteritidis. J Bacteriol 173(15):4773–4781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dueholm MS, Albertsen M, Otzen D et al. (2012) Curli functional amyloid systems are phylogenetically widespread and display large diversity in operon and protein structure. PLoS One 7(12):e51274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Olsén A, Jonsson A, Normark S (1989) Fibronectin binding mediated by a novel class of surface organelles on Escherichia coli. Nature 338(6217):652–655 [DOI] [PubMed] [Google Scholar]
- 33.Zogaj X, Bokranz W, Nimtz M et al. (2003) Production of cellulose and curli fimbriae by members of the family Enterobacteriaceae isolated from the human gastrointestinal tract. Infect Immun 71(7):4151–4158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cao B, Zhao Y, Kou Y et al. (2014) Structure of the nonameric bacterial amyloid secretion channel. Proc Natl Acad Sci U S A 111(50): E5439–E5444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Goyal P, Krasteva PV, Van Gerven N et al. (2014) Structural and mechanistic insights into the bacterial amyloid secretion channel CsgG. Nature 516(7530):250–253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hammar M, Bian Z, Normark S (1996) Nucleator-dependent intercellular assembly of adhesive curli organelles in Escherichia coli. Proc Natl Acad Sci U S A 93(13):6562–6566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Loferer H, Hammar M, Normark S (1997) Availability of the fibre subunit CsgA and the nucleator protein CsgB during assembly of fibronectin-binding curli is limited by the intracellular concentration of the novel lipoprotein CsgG. Mol Microbiol 26(1):11–23 [DOI] [PubMed] [Google Scholar]
- 38.Nenninger AA, Robinson LS, Hultgren SJ (2009) Localized and efficient curli nucleation requires the chaperone-like amyloid assembly protein CsgF. Proc Natl Acad Sci U S A 106 (3):900–905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nenninger AA, Robinson LS, Hammer ND et al. (2011) CsgE is a curli secretion specificity factor that prevents amyloid fibre aggregation. Mol Microbiol 81(2):486–499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Robinson LS, Ashman EM, Hultgren SJ et al. (2006) Secretion of curli fibre subunits is mediated by the outer membrane-localized CsgG protein. Mol Microbiol 59(3):870–881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Evans ML, Chorell E, Taylor JD et al. (2015) The bacterial curli system possesses a potent and selective inhibitor of amyloid formation. Mol Cell 57(3):445–455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hammer ND, Schmidt JC, Chapman MR (2007) The curli nucleator protein, CsgB, contains an amyloidogenic domain that directs CsgA polymerization. Proc Natl Acad Sci U S A 104(30):12494–12499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hammar M, Arnqvist A, Bian Z et al. (1995) Expression of two csg operons is required for production of fibronectin- and congo red-binding curli polymers in Escherichia coli K-12. Mol Microbiol 18(4):661–670 [DOI] [PubMed] [Google Scholar]
- 44.Weiss-Muszkat M, Shakh D, Zhou Y et al. (2010) Biofilm formation by and multicellular behavior of Escherichia coli O55:H7, an atypical enteropathogenic strain. Appl Environ Microbiol 76(5):1545–1554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang X, Chapman MR (2008) Sequence determinants of bacterial amyloid formation. J Mol Biol 380(3):570–580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang X, Hammer ND, Chapman MR (2008) The molecular basis of functional bacterial amyloid polymerization and nucleation. J Biol Chem 283(31):21530–21539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang X, Zhou Y, Ren JJ et al. (2010) Gatekeeper residues in the major curlin subunit modulate bacterial amyloid fiber biogenesis. Proc Natl Acad Sci U S A 107(1):163–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cegelski L, Pinkner JS, Hammer ND et al. (2009) Small-molecule inhibitors target Escherichia coli amyloid biogenesis and biofilm formation. Nat Chem Biol 5(12):913–919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Andersson EK, Bengtsson C, Evans ML et al. (2013) Modulation of curli assembly and pellicle biofilm formation by chemical and protein chaperones. Chem Biol 20(10):1245–1254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chorell E, Andersson E, Evans ML et al. (2015) Bacterial chaperones CsgE and CsgC differentially modulate human α-synuclein amyloid formation via transient contacts. PLoS One 10: e0140194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Evans ML, Schmidt JC, Ilbert M et al. (2011) E. coli chaperones DnaK, Hsp33 and Spy inhibit bacterial functional amyloid assembly. Prion 5(4):323–334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bokranz W, Wang X, Tschäpe H et al. (2005) Expression of cellulose and curli fimbriae by Escherichia coli isolated from the gastrointestinal tract. J Med Microbiol 54 (Pt 12):1171–1182 [DOI] [PubMed] [Google Scholar]
- 53.Bian Z, Brauner A, Li Y et al. (2000) Expression of and cytokine activation by Escherichia coli curli fibers in human sepsis. J Infect Dis 181 (2):602–612 [DOI] [PubMed] [Google Scholar]
- 54.Kikuchi T, Mizunoe Y, Takade A et al. (2005) Curli fibers are required for development of biofilm architecture in Escherichia coli K-12 and enhance bacterial adherence to human uroepithelial cells. Microbiol Immunol 49 (9):875–884 [DOI] [PubMed] [Google Scholar]
- 55.White AP, Gibson DL, Collinson SK et al. (2003) Extracellular polysaccharides associated with thin aggregative fimbriae of Salmonella enterica serovar enteritidis. J Bacteriol 185 (18):5398–5407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Baba T, Ara T, Hasegawa M et al. (2006) Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol 2:2006.0008 [DOI] [PMC free article] [PubMed] [Google Scholar]