Summary
KIR3DL1 is an inhibitory killer cell Ig-like receptor (KIR) that negatively regulates natural killer (NK) cell cytotoxicity. The KIR3DL1 cytoplasmic region (3DL1-cyto) is disordered and can be dissected into three segments: (I) H340-V351; (II) M352-D371; and (III) P372-P423. NMR studies indicate that segment II can dynamically adopt a loop-like conformation, and segments I and III can form dynamic helices that may mediate binding to membranes, particularly in the region around the N-terminal (N) immunoreceptor tyrosine-based inhibitory motif (ITIM), consistent with its role in signaling. Furthermore, individual SH2 domains of SHP-2 strongly engage with the unphosphorylated N-ITIM of 3DL1-cyto, while binding of the tandem SHP-2 SH2 domains to the bis-phosphorylated ITIMs results in more extensive conformational changes in segments I and III. The findings enhance our understanding of KIR function and how ITIMs in a target receptor operate in concert to engage the tandem SH2 domains of SHP-2.
Keywords: killer cell Ig-like receptor (KIR), immune receptors, SHP-2, immunoreceptor tyrosine-based inhibitory motif (ITIM), SH2 domain, inhibitory receptor, CON spectrum NMR, intrinsically disordered protein (IDP), protein-lipid interaction, tyrosine phosphorylation
In Brief
KIR3DL1 is an inhibitory receptor that regulates natural killer (NK) cell immune function. Cheng et al. describe NMR-based studies of its disordered cytoplasmic portion. The work characterized conformation dynamics as it binds with SH2 domains of the tyrosine phosphatase, SHP-2, which has broad implications in understanding inhibitory receptor function.
Introduction
Natural killer (NK) cells are innate immune effectors that can eradicate transformed, stressed, or virus-infected cells. In humans, NK cells constitute 5–15% of peripheral blood lymphocytes and are widely distributed in blood and peripheral tissues (Caligiuri, 2008; Campbell and Hasegawa, 2013). They are particularly important in cancer prevention and therapy, recognizing, adhering to and mediating direct spontaneous cytotoxicity of certain tumor cells, as well as releasing the pro-inflammatory cytokines, IFN-γ and TNF-α (Campbell and Hasegawa, 2013; Mentlik James et al., 2013).
NK cell activation is controlled by a balance of opposing signals from activating receptors, such as natural cytotoxicity receptors, NKG2D, CD16, DNAM-1, and 2B4, and inhibitory receptors, such as CD94/NKG2A and the killer cell immunoglobulin-like receptors (KIR; also known as CD158). KIR are a family of transmembrane glycoproteins expressed on NK cells and a subset of T cells encoded by 14 polymorphic genes (KIR2DL1–5, KIR3DL1–3, KIR2DS1–5, and KIR3DS1) (Campbell and Purdy, 2011). They play important roles in human NK cell development, tolerance, and activation by recognizing major histocompatibility complex class I (MHC-I) molecules expressed on normal cells as ligands. Activating receptors initiate target cell lysis and cytokine production when an NK cell encounters an abnormal target cell lacking MHC-I, whereas inhibitory KIR suppress responses toward normal cells through recognition of MHC-I expressed on their surfaces (Biron, 1997; Storkus et al., 1987).
When inhibitory KIR engage with MHC-I on normal cells, tyrosine residues in two immunoreceptor tyrosine-based inhibitory motifs (ITIMs; I/V/LxYxxL/V) are phosphorylated by Src-family protein tyrosine kinases (PTKs; such as Lck and Fyn), which creates specific docking sites for the cytosolic Src Homology Region 2 (SH2)-containing Protein Tyrosine Phosphatases (PTPs), SHP-1 and SHP-2 (PTPN6 and PTPN11, respectively). Recruitment of SHP-1/2 leads to the dominant suppression of activating receptor signals transduced via PTKs, thereby tolerizing NK cells toward normal MHC-I-expressing cells in the body (MacFarlane and Campbell, 2006; Matalon et al., 2016). Each KIR family member has specificity for a distinct subset of the MHCI molecules. KIR3DL1 (3DL1) is a common inhibitory receptor that recognizes HLA-Bw4-containing MHC-I (Litwin et al., 1994). The 3DL1 cytoplasmic region (3DL1-cyto: aa 340–423) contains a pair of ITIM motifs, N-ITIM and C-ITIM (Fig. 1A), which are similar in most inhibitory KIR (Campbell and Purdy, 2011).
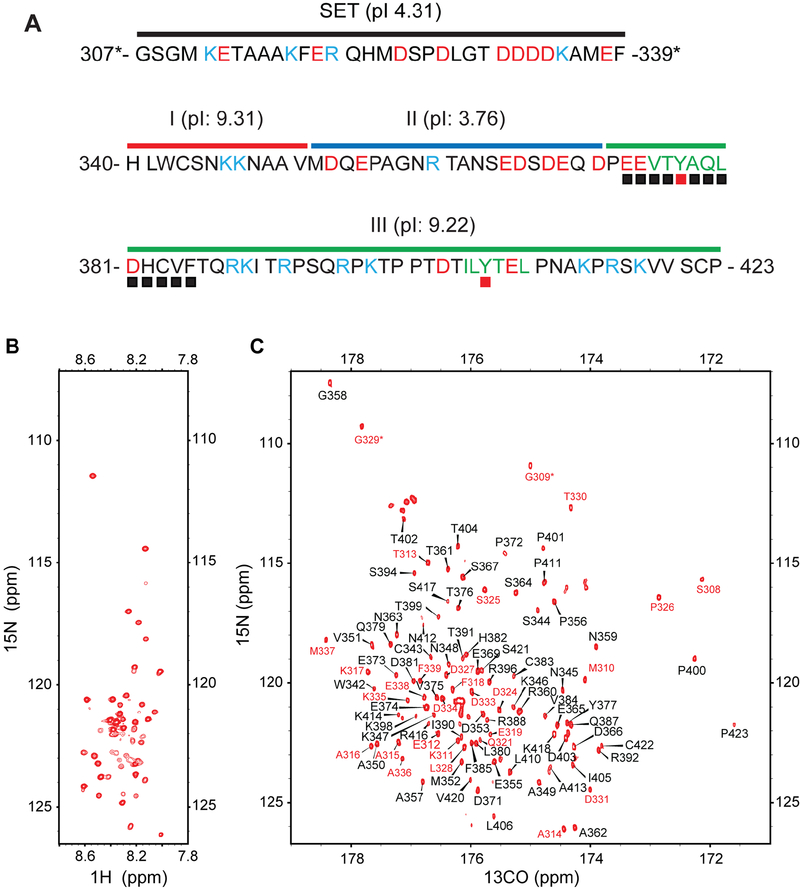
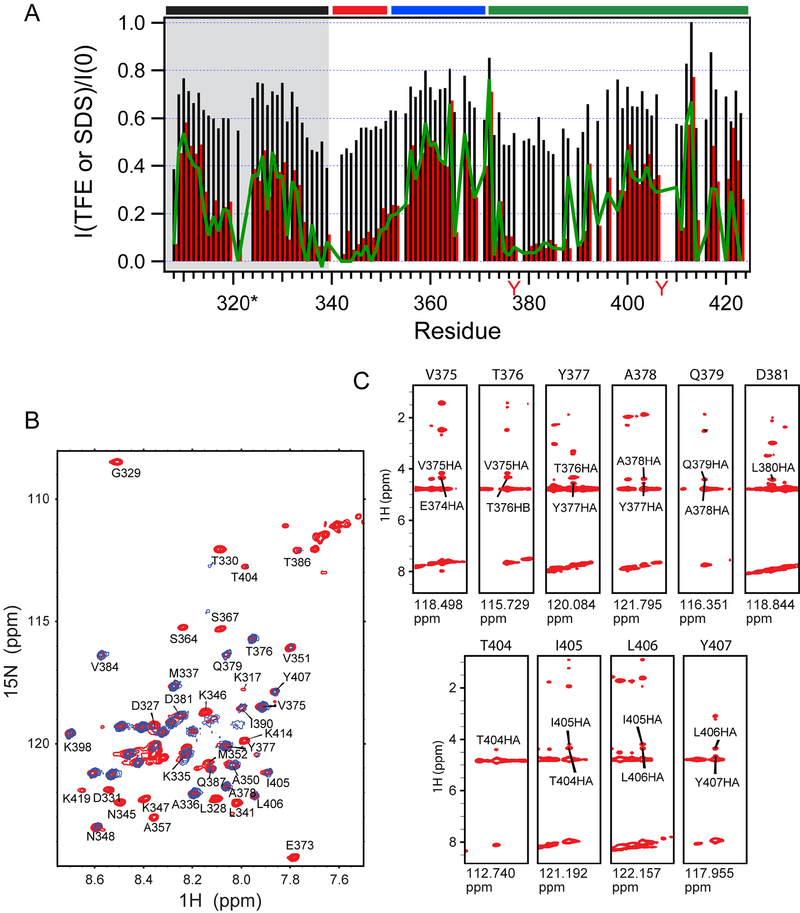
Figure 1. NMR spectra of KIR3DL1 cytoplasmic region.
(A) Sequence of SET-3DL1-cyto with basic residues in light blue, acidic residues in red, ITIMs in green, and ITIM tyrosines with red filled squares. SET sequence is under black bar. We divided 3DL1-cyto into three segments: Segment I (red bar; H340-V351), Segment II (blue bar; M352-D371), and Segment III (green bar; P372-P423). Residues denoted by black filled boxes are involved in binding to N-SH2 (from Fig. 7). (B) 1H-15N HSQC spectrum of 1 mM SET-3DL1-cyto recorded at 37°C in 20 mM HEPES (pH 7.4), 20 mM NaCl, 3 mM EDTA, and 3 mM DTT. (C) 13C-detected CON spectrum of 1 mM SET-3DL1-cyto recorded at 37°C in 20 mM HEPES (pH 7.4), 20 mM NaCl, 3 mM EDTA, and 3 mM DTT. Peaks are labeled based on nitrogen chemical shifts. Positions of prolines are likely folded inside the spectrum due to spectral width used in the nitrogen dimension. The 28 peaks assigned to residues in the SET have red numbering, and 70 peaks assigned from 3DL1-cyto are in black.
SHP-1 and SHP-2 are members of a small subfamily of PTPs that play vital regulatory roles in cell biology by modulating protein-tyrosine phosphorylation patterns in concert with PTKs. They have high structural and sequence homology (~60% amino acid sequence identity), and both contain tandem SH2 domains (N-SH2 and C-SH2) and a C-terminal catalytic PTP domain. Numerous studies have established a role for SHP-1 in KIR inhibitory function (Binstadt et al., 1996; Burshtyn et al., 1996; Campbell et al., 1996; Fry et al., 1996; Olcese et al., 1996), but involvement of SHP-2 is less clear. Several reports described SHP-2 association with tyrosine phosphorylated KIR (Bruhns et al., 1999; Kikuchi-Maki et al., 2003; Yusa and Campbell, 2003; Yusa et al., 2004). Also, expression of dominant negative (DN)-SHP-1 (C459S) or DN-SHP-2 (C453S) in NK cell lines can block KIR-mediated inhibition of cytotoxicity (Yusa and Campbell, 2003), indicating that both play important functional roles. However, these results do not rule out competition with other negative effector proteins that might be physiologically recruited to mediate inhibition. SHP-2 often contributes to activation signals in other receptor systems (Matozaki et al., 2009; Poole and Jones, 2005), which further complicates data interpretation. We previously performed biochemical and structure/function analysis of 3DL1(Yusa and Campbell, 2003) to show: (1) whereas SHP-1 only binds when both KIR ITIMs are phosphorylated, SHP-2 can constitutively associate with the unphosphorylated NITIM; (2) the N-ITIM was essential and sufficient to impart strong inhibitory function to 3DL1 via SHP-2; and (3) the degree of inhibition directly correlated with SHP-2 binding to wild-type (WT) and tyrosine mutant forms of 3DL1. Furthermore, replacing the cytoplasmic domain of 3DL1 with the PTP domain of SHP-2 resulted in a receptor that inhibits target cell cytotoxicity by NK cells. Although these cell biology results provide strong evidence for involvement of SHP-2 in NK cell inhibitory function, the structural basis of SHP-2 binding to KIR ITIMs in the unphosphorylated and phosphorylated states is unknown.
Despite our observations that both sequence-based predictions and NMR spectra showed 3DL1-cyto is highly flexible and represents an intrinsically disordered protein (IDP), we have assigned backbone and side-chain NMR 13C and 15N chemical shifts for 83% of residues. The conformational properties of the disordered 3DL1-cyto were explored by NMR and structural calculations. Furthermore, we mapped conformational changes in 3DL1-cyto in the presence of TFE and SDS and upon binding to SHP-2 SH2 domains with unphosphorylated or phosphorylated ITIMs. Our findings shed new light on structural and dynamic features involved in these protein-protein and protein-membrane interactions and refine our mechanistic understanding of how the two SH2 domains of SHP-2 work in concert to mediate binding to unphosphorylated and phosphorylated tyrosine motifs in target receptors.
Results
13C-detected CON spectrum and resonance assignment
To generate an aqueous-soluble form of 3DL1-cyto, we fused a polar and intrinsically disordered solubility enhancement tag (SET) containing 33 amino acids to the N-terminus (Fig. 1A). For clarity, we use residue numbers based on 3DL1-cyto (H340-P423) and mark SET residues with an asterisk in the same numbering scheme (G307*-F339*). Results from our NMR studies (see below) indicate that this SET-3DL1-cyto fusion does not affect binding to SH2 domains of SHP-2 (Figs. 2, S3) or tyrosine phosphorylation of 3DL1-cyto by FYN kinase (Fig. 6 and S2).
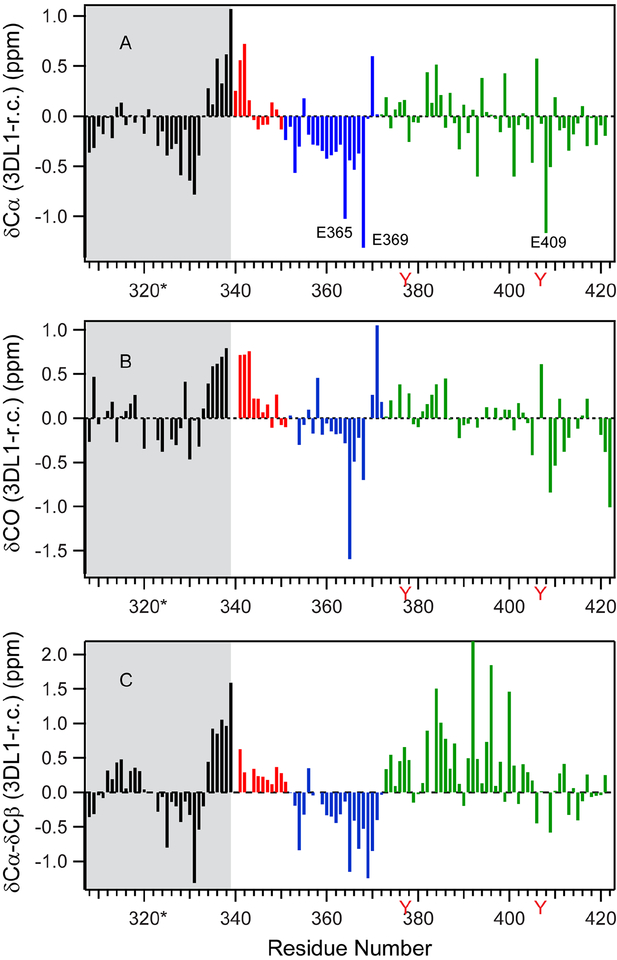
Figure 2. Secondary chemical shift plots.
Histogram plots are shown for (A) δCα, (B) δCO, and (C) δCα-δCβ values. Black denotes SET (grey shaded), red is segment I, blue designates segment II, and green is segment III. Tyrosines (Y) are denoted in red lettering to mark the positions of ITIMs.
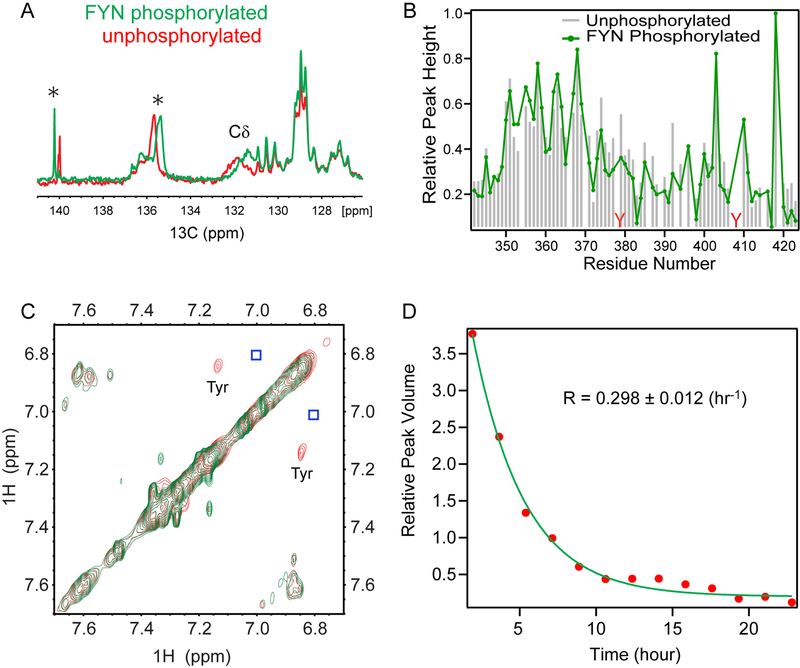
Figure 6. Impact of FYN-mediated tyrosine phosphorylation on 3DL1-cyto.
(A) Aromatic carbon region (126–141 ppm) of 1D 13C NMR spectra of [13C] SET-3DL1-cyto (500 μL and ~1 mM) in 50 mM HEPES, 10 mM MgCl2, 5.5 mM ATP, pH 7.5, recorded at 30°C either before (red) or 40 hrs after adding FYN (final concentration: ~0.16 uM, ~800 units; green). (B) Changes in relative peak height in CON spectra of individual amino acids before (grey bars) and after phosphorylation by FYN (green line) as measured using NMR conditions as (A). (C) Superimposed portions of 1H NOESY spectra in the aromatic region of 0.5 mM unphosphorylated (red) and phosphorylated 3DL1-cyto (green). Spectra were recorded using ~1 mM protein, and identical conditions as in (A). Strong crosspeaks arising from tyrosine Hε-Hδ protons are in red and the invisible weak crosspeaks are positioned in blue boxes. (D) Kinetics of tyrosine phosphorylation plotted as peak volumes of the strong tyrosine Hε-Hδ proton NOE crosspeaks in (C) vs. reaction time (hrs). Rate of phosphorylation was calculated as 0.298± 0.120 h−1.
Consistent with the computer prediction that 3DL1-cyto is disordered, the 1H-15N HSQC spectrum (Fig. 1B) exhibits limited chemical shift dispersion in the NH dimension (8–8.6 ppm). We identified only ~50 out of 106 expected backbone NH crosspeaks in the HSQC spectrum, even at relatively low pH (6.3), where exchange broadening due to solvent exchange is not expected to be a major factor. The poor quality of this spectrum indicates that it would be difficult or impossible to obtain comprehensive backbone resonance assignments by only relying on conventional proton-detected multi-dimensional NMR.
To overcome these problems, we used multidimensional NMR techniques involving direct detection of 13C signals, which were recently shown to be especially well suited for studies of IDPs (Felli and Pierattelli, 2014). Fig. 1C shows a 13C-detected CON spectrum for [13C, 15N] SET-3DL1-cyto, which sequentially connects amide 15N resonances and the preceding 13CO nuclei, recorded on a sample of 3DL1-cyto using a conventional Bruker TCI cryoprobe (12-hr acquisition time). In contrast to the intraresidue HN and N correlated 1H-15N HSQC spectrum, the interresidue 13CO and 15N correlated CON spectrum is well resolved. By adopting the published sequential assignment strategies (Felli and Pierattelli, 2014) using multidimensional 13C-detected NMR experiments, such as CANCO, CANCOi, CBCACO, CBCANCO, and CCCON, we were able to assign 70 (labeled in black numbering) out of 84 expected crosspeaks in the CON spectrum to specific peptide bonds (including X-Pro) of 3DL1-cyto along with 28 additional peaks (in red numbering) from the SET (Fig. 1C). The fact that fewer crosspeaks are observable in HSQC than in CON spectra can be attributed to more severe conformational exchange broadening for NH proton resonances in the HSQC compared to CO carbon resonances in the CON experiment.
Local conformational propensities
Chemical shifts are very sensitive to peptide bond conformation and commonly used for secondary structure determination and amino acid dihedral angle prediction (Cornilescu et al., 1999; Spera and Bax, 1991; Wishart and Sykes, 1994; Wishart et al., 1992). To assess any propensities in 3DL1-cyto for transient secondary-structure formation, we calculated secondary chemical shifts from our NMR data for Cα, CO, and Cβ by taking the difference between observed chemical shifts and those predicted for a random coil (fully disordered) conformation of the polypeptide (Fig. 2). Consecutive positive values of δCα, δCO, and δCα -δCβ values indicate a conformational propensity for α-helix, and negative scores for extended β-strand structure (Wishart and Sykes, 1994). Since secondary chemical shifts are usually small for IDPs, to obtain reliable secondary structure propensities, we chose the chemical shift library of Tamiola et al. as a random-coil reference, which accounts for nearest-neighbor effects, based on a representative database of 14 IDPs (Gibbs et al., 2017; Tamiola et al., 2010; Wishart et al., 1992). As in Fig. 2, the secondary Cα, CO, and δCα-δCβ chemical shifts range between −2 to +2. Interestingly, three Glu residues, E365, E369 and E409, show unusually large negative Cα and CO secondary chemical shifts, even after accounting for neighbor effects. Both E365 and E369 are adjacent to Glu, Asn or Gln residues, suggesting local electrostatic effects on the chemical shifts may be responsible for these anomalies.
The secondary chemical shifts of SET-3DL1-cyto, especially for δCα-δCβ, suggested three segments in 3DL1-cyto with distinct conformational and dynamic properties: (I) H340-V351; (II) M352-D371; and (III) P372-P423 (Figs. 1A and2C). This may reflect the fact that, due to lack of stable long-range interactions, the simplified spin Hamiltonian of IDP secondary chemical shifts consist of only local interactions, i.e., local dipolar interactions, local electrostatic interactions, and chemical shift anisotropy-dipolar interactions (Saito et al., 2010; Smith, 1992; Wishart and Case, 2001). Several residues at the N-terminus of segment I show positive secondary chemical shifts for Cα, CO, and δCα-δCβ (Fig. 2, red) indicative of a small, but significant, α-helix propensity. Interestingly, the seven residues at the C-terminus of the SET, appended to the N-terminal segment I, also show a tendency to form α-helix with positive secondary chemical shifts (Fig. 2, black). This structural feature of the SET is consistent with a strong prediction of helical secondary structure for the transmembrane (TM) region and the first eight residues in the cytoplasmic region of 3DL1 (aa. 321–347), as assessed using NetSurfP (data not shown). Segment II has propensity to adopt an extended β-strand as most residues exhibit negative secondary chemical shifts. Segment III exhibits both positive and negative Cα and CO secondary chemical shifts, but (δCα-δCβ) values are predominantly positive, making conformation preference difficult to predict. The average backbone dihedral angles, ϕ and ψ, for assigned residues were calculated based on chemical shifts using TALOS+ (Cornilescu et al., 1999). Most residues in segment I fall in the α-helical region of the Ramachandran plot while residues in segments II and III tend to be in or near the extended (β-strand) region (Fig. 3), which is consistent with the qualitative analysis of secondary chemical shift (Fig. 2).
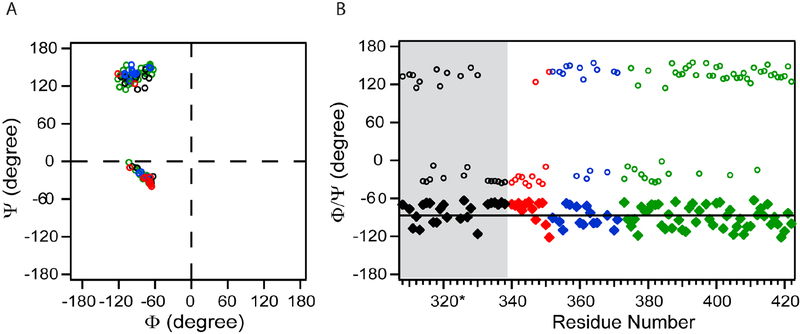
Figure 3. Average ϕ and ψ dihedral angles calculated by TALOA+.
Red points are residues in segment I, blue from segment II, green from segment III, and black from SET (grey shaded in B). (A) Ramachandran Plot of amino acids in 3DL1-cyto. (B) Dihedral angles plotted versus residue number: diamonds: ϕ; circles: ψ.
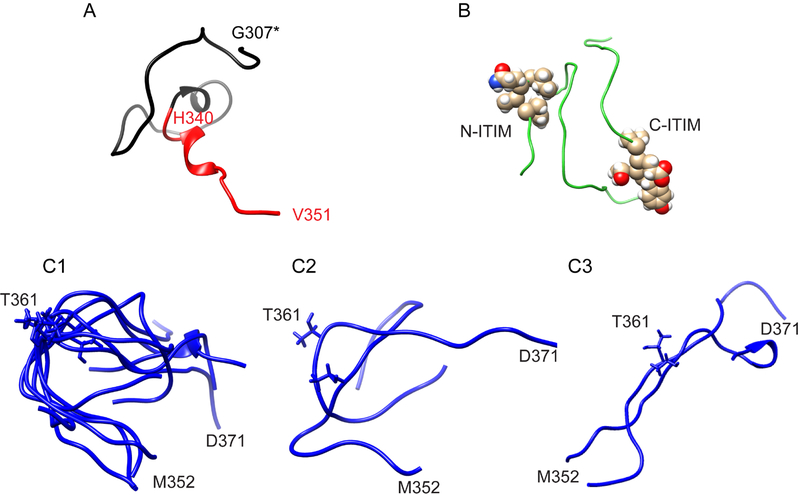
While the dihedral angles of most amino acids in both segments II and III are consistent with extended or β-structure (Fig. 3), secondary chemical shifts from these two segments exhibit different patterns (Fig. 2). To assess the local conformational preferences of these segments, we calculated protein structural ensembles by using CS-ROSETTA with constraints of Cα, Cβ, and N (except for Pro residues) chemical shifts (Shen et al., 2008), which generated 3000 structure predictions. The averaged Cα-rmsd for the ten best structures with lowest-energy is 12.0±1.3 Å. Fig. 4 shows representative structures for each segment. The C-terminal SET and segment I can assume helix-like conformations by this analysis (Fig. 4A). Interestingly, Segment II is predicted to dynamically change between loop-like and extend conformations. Six out of the ten best structures adopt a loop conformation centered at T361, including residues 357-AGNRTANSE-365, with a rmsd of 4.2 Å for both backbone and side-chain atoms (Fig. 4C1). Of the remaining top ten segment II structures, two adopt alternative dynamic loop conformations (Fig. 4C2) and two show completely extended conformations (Fig. 4C3). The conformational ensemble of segment III is much more heterogeneous without defined conformational propensities (Fig. 4B).
Figure 4. Best structures of CS-Rosetta for each segment.
(A) SET and segment I. One predicted structure is shown in ribbon diagram. SET is in black, and segment I in red. (B) Segment III shows collapsed coils. The ITIMs in one representative structure are shown. (C) Segment II (352-MDQEPAGNRTANSEDSDEQD-371). C1: six structures show a matched loop; C2: two structures with alternative loop conformations; C3: two structures with extended β-strand. Common orientations of end residues M352 and D371 in the majority of structures are marked. T361 is also marked.
Conformational Flexibility
An immunoreceptor tyrosine-based activation motif (ITAM) has also been identified in the cytoplasmic tails of many activating immune receptors, including B and T cell antigen receptors (TCR and BCR) and Fc receptors (Cambier, 1995; Campbell and Purdy, 2011; Flaswinkel et al., 1995). The ITAM consists of two conserved YxxI/L sequences separated by 6–8 amino acids (YxxI/Lx(6–8)YxxI/L), which is analogous to a similar single tyrosine containing element within the ITIM. The ITAM tyrosines become phosphorylated following receptor interaction with ligands, leading to recruitment of Syk or ZAP-70 PTKs for downstream signaling. Several biophysical studies using CD (Laczko et al., 1998) and NMR (Duchardt et al., 2007; Xu et al., 2008) have demonstrated that lipid binding induces the ITAM to form dynamic α-helices inserted in the membrane.
The helical conformation in peptides and proteins can be stabilized in vitro in the presence of 2,2,2-trifluoroethanol (TFE) due to a lower dielectric constant than water, or through binding to detergent micelles (Isaksson et al., 2013; Shiraki et al., 1995; Sigalov et al., 2006). To assess whether the ITIMs in 3DL1-cyto are capable of forming helices, we examined CON spectral changes of SET-3DL1-cyto in the presence of TFE. The peak intensities in CON spectra are very sensitive to TFE; a substantial intensity reduction in the CON spectrum was observed throughout the entire sequence of SET-3DL1-cyto upon addition of 1% TFE (Fig. 5A, black), which became more profound in the presence of 2% TFE (Fig. 5A, red). Crosspeaks from consecutive residues K335*-A349 (including the membrane proximal portion of segment I, Fig.1A) and V375-T391 (including the N-ITIM and downstream sequence, Fig.1A) show more than 85% loss in intensity in the presence of 2% TFE as compared to without TFE (Fig. 5A red), indicating these regions are capable of forming α-helices. A similar reduction in peak intensities for the same regions was observed in a CON spectrum (Fig. 5A, green) recorded in the presence of sodium dodecyl sulfate (SDS) at a concentration of 300 μM, which is considerably below its critical micelle concentration of 8.2 mM. SDS is an anionic detergent that interacts with positively charged residues to shield them from solvent exposure and has been used as a membrane-mimicking agent in protein lipid binding studies (Li et al., 2016). Together, these data suggest that the membrane proximal region in segment I and the N-ITIM are capable of forming α-helices. Interestingly, significantly less pronounced intensity reduction was observed for residues surrounding the C-ITIM, indicating the two ITIMs have different conformation propensities consistent with their diverse biological roles in signal transduction.
Figure 5. Relative peak intensity changes in CON spectra of 3DL1-cyto in the presence of TFE and SDS.
A. Black histogram: in 1% TFE; Red histogram: in 2% TFE; Green line: in 300 μM SDS. Residues in SET are grey shaded. Spectra were recorded for 1 mM 3DL1-cyto at 37°C in 20 mM HEPES (pH 7.4), 20 mM NaCl, 3 mM EDTA, and 3 mM DTT. (B) Superimposed contour plot for portion of 1H-15N HSQC (red) and 1H-15N NOE (only positive peaks shown, blue) spectra of SET-3DL1-cyto recorded at 37 °C in presence of 40% TFE, 20 mM HEPES (pH 7.4), 20 mM NaCl, 3 mM EDTA, and 3 mM DTT. 38 peaks are assigned. (C) Strip plots of 15NNOESY-HSQC recorded in presence of 40% TFE (red) at 37°C in 20 mM HEPES (pH 7.4), 20 mM NaCl, 3 mM EDTA, and 3 mM DTT. Sequential Hα-NH NOE are shown for peaks from residues in the N-ITIM (top) and C-ITIM (bottom).
In contrast to the CON spectra, 1H-15N HSQC NMR spectra were less sensitive to TFE. Since helical content of a peptide or protein is often enhanced with increasing concentrations of TFE (Shiraki et al., 1995), by increasing TFE to 40%, the 1H-15N HSQC spectrum became more resolved (Fig. 5B), whereas a majority of the peaks from helical residues in the CON spectra disappeared at much lower concentration of TFE (Fig. 5A). Using conventional 3D 1H-detected sequential assignment NMR methods (Ikura et al., 1990), we were able to obtain residue-specific assignments for 38 out of a total of 58 resolved backbone crosspeaks in the 1H-15N HSQC spectrum (Fig. 5B, red). Of these, 21 peaks exhibited positive 1H-15N heteronuclear Nuclear Overhauser Effects (hetNOE) (Fig. 5B, blue) with opposite sign to those from flexible side-chains (not shown in the figure). These peaks are mainly from the residues that form transient α-helices, i.e., at the C-terminal SET/N-terminal region I and within the ITIMs, suggesting that the residues become more ordered. The backbone hetNOE provides information about fast motion (ps-ns) of individual N-H bond vectors and is commonly used for identifying residues in unstructured regions of proteins, since the residues in disordered regions undergo motions faster than the overall tumbling of the molecule and yield signals of opposite sign (Renner et al., 2002). Furthermore, strong sequential Hα-NH NOE crosspeaks, an additional characteristic of α-helical conformation, were observed especially for residues around the two ITIMs (Fig. 5C), further supporting our conclusion that these regions adopt α-helical conformations at 40% TFE.
Tyrosine phosphorylation of 3DL1-cyto
Phosphorylation of the ITIMs by Src family PTKs is critical for efficient inhibitory function of KIR. We studied the phosphorylation by adding the Src family PTK, FYN, with ATP directly to the NMR tube as in Fig. 6 legend. We monitored chemical shift perturbations for aromatic ring carbon peaks in a range of 115–160 ppm by recording a series of 1D 13C NMR spectra of [13C] SET-3DL1-cyto at 30°C in real time. Three peaks of [13C] SET-3DL1-cyto experienced large changes in chemical shift by 72 hrs after introducing FYN PTK (Fig. 6A). Based on standard chemical shifts of tyrosine ring carbons (Cδ: 132 ppm, Cε: 117.7 ppm, Cζ: 155.2 ppm, and Cγ: 131.4 ppm), a broad peak centered at ~132 ppm, presumably from tyrosine Cδ carbons, shifted upfield by ~0.4 ppm (131.8 to 131.4 ppm) within 40 hours, while the origins of two other relatively narrow peaks at 135.6 ppm and 140.0 ppm (asterisks in Fig. 6A) are unknown. We also observed small chemical shift perturbations (Fig. S2) and intensity decreases for some CON peaks after FYN tyrosine phosphorylation (Fig. 6B), especially for the residues around the N-ITIM and in the linker between the two ITIMs, indicating that phosphorylation results in dynamic changes in this region. It is unlikely that these chemical shift changes surrounding the N-ITIM are due to direct binding of FYN, since the amount of kinase added was minimal (SET-3DL1-cyto to FYN molar ratio of ~6250:1). No significant spectral changes were observed for the C-ITIM tyrosine and nearby residues.
To measure kinetics of the tyrosine phosphorylation, we recorded a series of homonuclear NOESY spectra at 30°C after addition of FYN into [13C, 15N] SET-3DL1-cyto and monitored decrease in peak intensity of two crosspeaks near the expected positions of tyrosine Hε (6.86 ppm) and Hδ (7.15 ppm) ring protons. While one crosspeak with chemical shifts of 6.756 ppm and 7.088 ppm was too weak to be studied (blue boxes in Fig. 6C), we were able to measure the changes in peak volume for the stronger peaks, tentatively assigned to Y377 based on strong NOEs observed for residues around the N-ITIM, at 6.844 ppm and 7.137 ppm versus reaction time. A single-exponential fit of the observed decay yields a rate of 0.298±0.012 hr−1 (Fig. 6D). Taken together, these results indicate that the tyrosines in ITIMs of 3DL1-cyto can be efficiently phosphorylated within 12–24 hrs by FYN under the conditions of our NMR experiments.
Interactions of 3DL1 with tandem SH2 domains of SHP-2
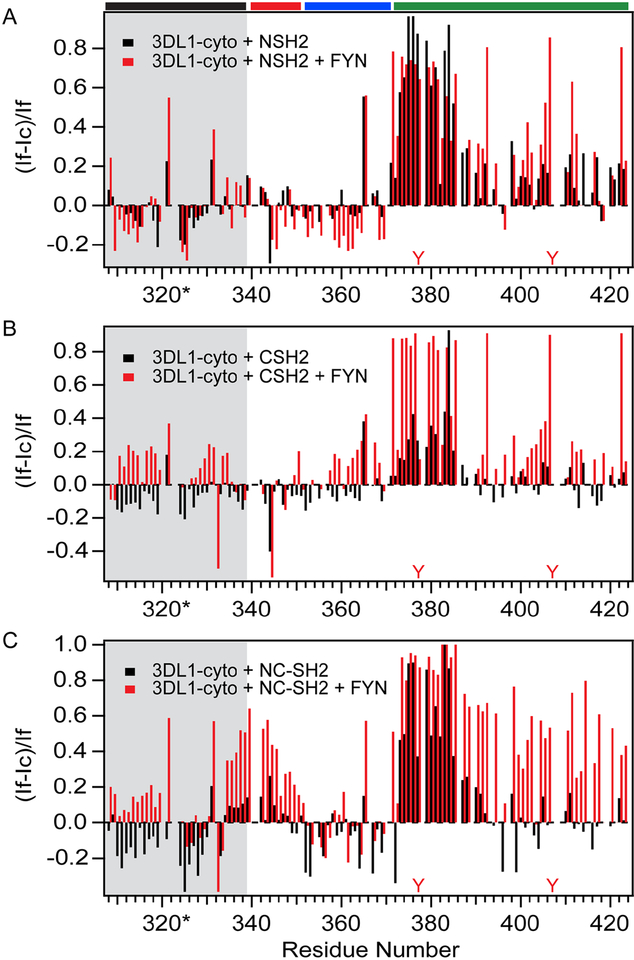
To identify residues from disordered 3DL1-cyto involved in contacts with SHP-2, we recorded CON spectra of isotopically labeled SET-3DL1-cyto in the presence of substoichiometric amounts of individual N- (M1-N103) or C-SH2 (A105-G246) or the tandem NC-SH2 (M1-G246) without or with FYN. Our previous results from 3DL1 ITIM peptide binding studies showed that the N-ITIM can bind to SHP-2 in a phosphorylation-independent manner (Yusa et al., 2002), but this work did not clarify the individual roles of the two SH2 domains of SHP-2 in mediating this interaction. The CON spectra of unphosphorylated SET-3DL1-cyto complexes with each individual SH2-domain (in molar ratios of SET-3DL1:N-SH2 or C-SH2 at 1:0.2) show binding-induced reduction of intensities for peaks mainly from residues at or surrounding the N-ITIM [371-DPEEVTYAQLPHCVF-385; Fig. S3, black (free) and red (in complex)]. These reductions in relative peak intensity (Fig. S3) give rise to an increase in relative intensity differences [(If - Ic)/If, as shown in Fig. 7], which is indicative of a binding-induced mobility decrease of these residues. Interestingly, the CON spectrum in the presence of C-SH2 shows much smaller intensity changes for peaks from the residues in the same range, as compared to N-SH2 (Figs. S3B vs. S3A, black to red; Fig. 7B vs, 7A, black). The NMR spectrum of the complex with N-SH2 also shows minor peak intensity reductions for residues 400-PPTDTILYTELPN-412 spanning the C-terminal ITIM (Fig. 7A, black; Fig. S3A, black to red) that are not evident in complex with C-SH2 (Figs. 7B and S3B).
Figure 7. Relative peak intensity changes in spectra of 3DL1-cyto upon SHP-2 SH2 domain binding alone or with FYN phosphorylation.
Black histogram: unphosphorylated; red histogram: phosphorylated conditions after 48-hour co-incubation with FYN (~800 units). (A) CON spectra for the complex of 3DL1-cyto and N-SH2 were assessed (molar ratio of 1:0.2). (B) CON spectra for 3DL1-cyto complexed with C-SH2 (molar ratio of 1:0.2). (C) (HCA)CON spectra for complex of 3DL1-cyto and NC-SH2 (molar ratio of 1:0.1). Black histogram indicates SH2 domain constructs added alone and red histogram denotes SH2 constructs added with FYN. Positions of the Y residues in ITIMs are denoted in red. Residues in SET are shaded in grey, and regions I-III are denoted by colored bars at the top of (A).
Similar to the N-SH2, the tandem NC-SH2 predominantly impacted residues in the NITIM (SET-3DL1:NC-SH2 at 1:0.1, Fig. 7C, black; Fig. S3C, black to red). These results indicate that each of the isolated and the tandem SH2 domains preferentially recognize the NITIM in the unphosphorylated state, but N-SH2 binds more tightly, and thus imparts a greater impact on residues surrounding either ITIM than C-SH2. The preferential binding of N-SH2 to unphosphorylated 3DL1-cyto observed by NMR also confirms and extends our previous biochemical and functional data (Yusa and Campbell, 2003; Yusa et al., 2002).
We next examined binding of the three SH2 domain constructs to tyrosine phosphorylated SET-3DL1-cyto by introducing FYN and analyzing changes in peak intensities in CON spectra for individual amino acids. Binding of C-SH2 to FYN-phosphorylated SET-3DL1-cyto induces substantially greater intensity reduction for residues encompassing both the N- and C-ITIMs (Fig. 7B, grey; Fig. S3B, blue), and binding of N-SH2 exhibits more pronounced intensity reduction for residues near the C-ITIM (Fig. 7A grey; Fig. S3A, blue), indicating increased SH2 domain contacts to both phosphorylated motifs. In striking contrast, the spectrum of the complex with the tandem NC-SH2 protein shows intensity reductions for peaks from nearly all residues in segment III (residues D371-P423), encompassing both the N-and C-ITIMs (Fig. 7C, grey and S2C blue). Based on the observations that the single N-SH2 domain preferentially binds to the unphosphorylated and phosphorylated N-ITIM (Figs. 7A and S3A), we propose that the orientation of the bivalent complex between NC-SH2 and 3DL1-cyto is parallel, i.e., N-SH2 pairs up with N-ITIM and C-SH2 with C-ITIM, which is consistent with previous work (Eck et al., 1996). In addition, considerable intensity decreases were observed for peaks within the entire length of segment I (Fig. 7C and S3C; residues H340-V351) upon binding to the tandem SH2 domains, suggesting that the bivalent interaction induces transient interactions between segment I and the pair of ITIM motifs in segment III and/or the bound tandem SH2 domains. Structure calculation from CS-Rosetta for the intact SET-3DL1-cyto also suggests that segments I and III dynamically approach each other in the absence of binding partners, e.g., one of the structures shows that Cα of N345 is less than 6 Å away from Cα of Y407 (Fig. S4A). We also studied intramolecular interactions by paramagnetic relaxation enhancement (PRE) measurements. Because the carbon gyromagnetic ratio is 4 times smaller than that of the proton, it is expected that much smaller PRE will be observed in 13C-detected NMR spectra based on dipole-dipole interactions between the unpaired electron and CO carbons, which is conventionally described by the Solomon-Bloembergen equation (Clore and Iwahara, 2009). Despite this, we observed small, but significant, changes in the CON spectra (Fig. S4B and C), especially for region I, due to PREs after site directed nitroxide spin labeling [3-(2-iodoacetamido)-2,2,5,5-tetramethyl-1-pyrrolidinyloxy (3-(2-iodoacetamido)-PROXYL] in two sites, C383 and C422, in SET-3DL1-cyto variants with a single cysteine (C343A/C422A and C343A/C383A).
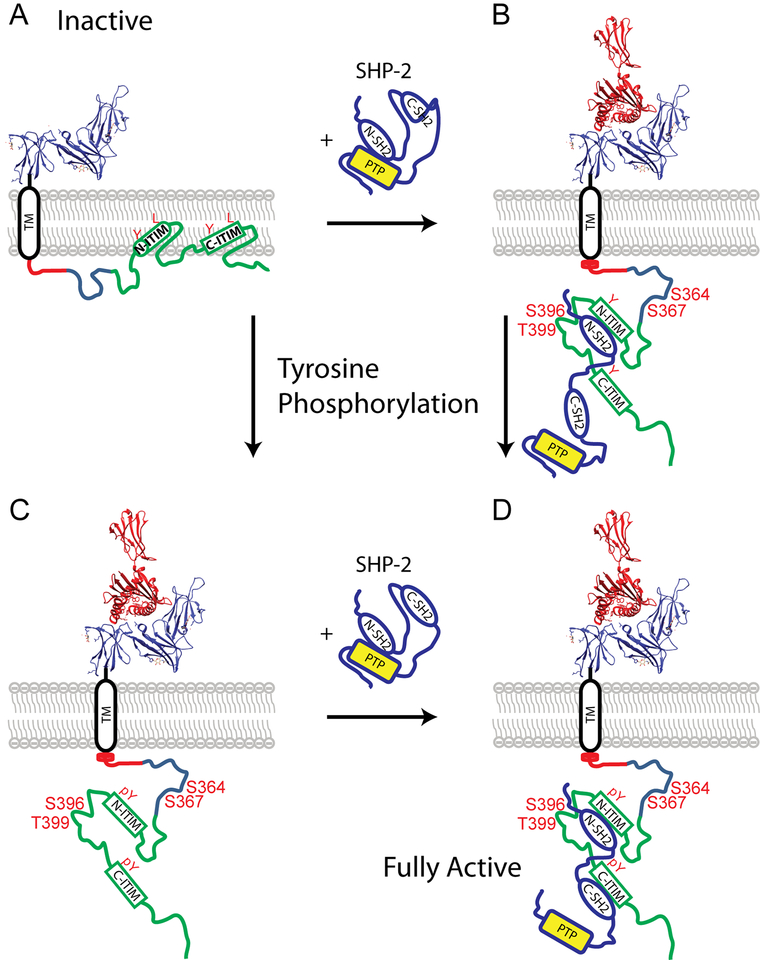
In summary, our results suggest a model (Fig. 8) in which: (1) interaction with N-SH2 of SHP-2 is capable of inducing significant changes in the unmodified N-ITIM of 3DL1-cyto and has a weaker impact on the unphosphorylated C-ITIM; (2) tyrosine phosphorylation enhances engagement of both individual N- and C-SH2 domains to 3DL1-cyto at the N- and C-ITIMs, respectively; (3) the occupancy of the N-SH2 with the N-ITIM, even in the unphosphorylated state, appears to serve as a precursor to a more intimate and specific bivalent interaction of the tandem NC-SH2 domains with the bis-phosphorylated ITIMs, which is ultimately necessary for full activation of SHP-2 and inhibitory function of the receptor.
Figure 8. Model diagram of 3DL1 association with SHP-2.
The extracellular domain of 3DL1 is shown as a ribbon diagram of the x-ray structure (3vh8) in blue, and part of MHC-I in red. (A) Inactive form, in which the basic amino acids in segments I and III associate with acidic phospholipids of the plasma membrane bilayer, thereby facilitating N-ITIM to form dynamic α-helices in association with the hydrophobic membrane interface. (B,C) MHC-I binding changes the local membrane environment and triggers release of 3DL1-cyto into the cytoplasm, so unphosphorylated N-ITIM can interact with N-SH2 of SHP-2 (B) or the ITIM tyrosines (Y) become phosphorylated by Src family PTKs (C). (D) Bisphosphorylation of both ITIM tyrosines results in association of the tandem SH2 domains of SHP-2 and full catalytic activity of the phosphatase near the plasma membrane. Known phosphorylated serine/threonine (S/T) residues that potentially influence tyrosine phosphorylation or association with membrane phospholipids are also labeled (B,C,D).
Discussion
Ligand engagement of many immune receptors, including the BCR, TCR, Fc receptors, and KIRs, results in phosphorylation of tyrosines in ITAMs or ITIMs, which in turn recruit PTKs or PTPs, respectively, to phosphorylate or dephosphorylate downstream components of signaling pathways (Barrow and Trowsdale, 2006). The detailed molecular mechanisms initiating these processes are, however, not well understood. Most cytoplasmic regions of these receptors are intrinsically disordered (Sigalov, 2011), which prevents structural studies by using conventional methods, i.e., NMR and X-ray crystallography (Wright and Dyson, 2015). Here, we examined disordered 3DL1-cyto by employing a 13C-detected NMR technique. Secondary chemical shift analysis (Fig. 2) suggests the 84-residue 3DL1-cyto can be divided into three segments characterized by distinct amino acid compositions and charge distributions (Fig. 1A). Segment I (H340-V351) prefers to assume an α-helical conformation; Segment II (M352-D371) is highly acidic and appears to undergo a dynamic conformational transition between transient loop-like and extended conformations; Segment III (P372-P423) is rich in basic amino acids and contains both ITIMs. It is interesting to note that we previously identified two serine residues near the predicted dynamic loop of Segment II (S364 and 367) that can be phosphorylated by casein kinases, and alanine mutation of these serines resulted in higher KIR surface expression and enhanced inhibitory function (Alvarez-Arias and Campbell, 2007; Campbell and Purdy, 2011). Therefore, it is possible that phosphorylation of S364 and/or S367 may alter stability of the loop conformation to impact receptor expression and function (Fig. 8).
Previously, the NMR structure of the cytoplasmic domain of CD3ε from the TCR (Xu et al., 2008) showed that the two ITAM tyrosines and leucine or isoleucine in the +3 position (analogous to the ITIMS of 3DL1-cyto) inserted into the hydrophobic core of the plasma membrane bilayer. Further studies have shown that basic residues in the cytoplasmic domain of TCR-ζ promote association with the plasma membrane to influence access to ITAMs (DeFord-Watts et al., 2011; Zhang et al., 2011). Our NMR studies of 3DL1-cyto in the presence of TFE and SDS suggest that dynamic hydrophobic and charge-based interactions may similarly be occurring at the plasma membrane for KIR3DL1. As for other immune receptors, such as TCRζ and CD3ε, the membrane-proximal segment I of 3DL1-cyto contains two adjacent basic amino acids, K346 and K347 (Fig. 1A), which likely contribute to electrostatic interactions with acidic phospholipids of the plasma membrane (Sigalov, 2011; Sigalov et al., 2006; Xu et al., 2008).
Although more rigorous NMR studies in the presence of lipid mixture, such as LMPG micelles or POPG/DHPC bicelles, will be needed, our NMR experiments involving TFE and SDS suggest that both segment I and the N-ITIM may be capable of interacting with the plasma membrane, forming α-helices. The high pI value of segment III, which contains eight basic amino acids in clusters downstream from each of the ITIMs (Fig. 1A), may also promote contacts with the plasma membrane, whereas the acidic residues in segment II are expected to promote repulsion from the membrane surface. Membrane repulsion may be further enhanced by phosphorylation of S364 and S367 in Segment II and S394 and T399 between the ITIMs of Segment III (Alvarez-Arias and Campbell, 2007). In fact, we previously reported evidence that phosphorylation of S394 by protein kinase C can increase surface expression and reduce internalization/turnover of 3DL1 (Alvarez-Arias and Campbell, 2007).
Our experiments show that the NMR spectral changes caused by SHP-2 SH2-domain binding are largely limited to segment III of 3DL1-cyto (Fig. 7 and S3). We confirmed our previous biochemical finding that SHP-2 can effectively engage with the unphosphorylated NITIM (Yusa and Campbell, 2003). SH2-domains typically bind to phosphorylated tyrosine in the context of a peptide motif within a target protein, although phosphorylation-independent binding of SH2 domains to tyrosine motifs has also been reported (Ignatiuk et al., 2006; Woodside et al., 2002). Our NMR experiments show that the two ITIMs of 3DL1-cyto have distinct conformational propensities. While the N-ITIM can form α-helical structure in the presence of TFE and SDS, the C-ITIM remains more disordered. Our results convincingly demonstrate that the N-SH2 of SHP-2 is capable of strongly impacting residues surrounding the unphosphorylated N-ITIM (Fig. 7), suggesting this interaction may constitutively poise SHP-2 at the N-ITIM prior to tyrosine phosphorylation. We also previously studied the individual ITIM tyrosine residues (Y) in 3DL1 by expressing WT or phenylalanine (F) mutant forms of the receptor in human NK cells that eliminated tyrosine phosphorylation at the N-ITIM (FY), C- ITIM (YF), or both ITIMs (FF) (Yusa and Campbell, 2003). Our work demonstrated that the phosphorylated tyrosines exhibit different capacities for recruiting SHP-2 and inhibiting both target cell conjugation and cytotoxicity: WT (YY) > YF >> FY > FF (Yusa and Campbell, 2003), consistent with other studies (Bruhns et al., 1999; Fry et al., 1996; Pluskey et al., 1995; Yusa et al., 2004). Importantly, while inhibition was optimal for YY, substantial inhibitory function was also found for the N-ITIM (YF) alone, which recruited SHP-2, but not SHP-1 (Yusa and Campbell, 2003). Extending the previous studies, our NMR data directly demonstrate that each of the individual and the tandem NC-SH2 domain constructs selectively interact with the unphosphorylated and phosphorylated 3DL1-cyto primarily at N-ITIM. Notably, the tandem NC-SH2 impacts the tyrosine phosphorylated cytoplasmic region over the entire length of Segment III, encompassing both ITIMs and intervening amino acids, as well as Segment I. It is unlikely that all residues with reduced peak intensities in the CON spectrum are involved in direct contacts with the SH2 domains, but their mobility appears to be restricted as a result of the binding, resulting in line broadening and reduced peak intensity. This result suggests an extended structural reorientation or stabilization through the entire 3DL1-cyto upon binding to NC-SH2 that may include a distant interaction with Segment I.
Our study shows that 3DL1-cyto is mostly unstructured in aqueous buffer, becomes more dynamically ordered in the presence of 1–2% TFE or 300 μM SDS, and shows a greater propensity for adopting α-helical structure in 40% TFE, especially in residues in segment I and surrounding the N-ITIM. Based on these structural and previous cell biological data, we propose the following model for ligand-dependent 3DL1 inhibitory signaling: 1) In the resting state, the tyrosines in 3DL1-cyto are inaccessible to PTKs and PTPs, as they are protected via the binding to the inner leaflet of the plasma membrane (Fig. 8A). 2) Analogous to CD3ε of the TCR (Gagnon et al., 2012), 3DL1 binding to MHC-I ligand results in changes in local electrostatic interactions surrounding the TM and membrane-proximal region of 3DL1-cyto, which induces conformational changes in the N-terminal membrane-proximal region, including loss of transient α-helices, and hence liberating tyrosines from the membrane (Fig. 8B and C) to be accessible for interaction with the N-SH2 of SHP-2 (Fig. 8B) and phosphorylation by Src family PTK (Fig. 8C). Phosphorylation of both ITIM tyrosines facilitates high affinity interaction with both SH2 domains of SHP-2 (or SHP-1) to fully activate these PTPs at the plasma membrane (Fig. 8D). While our results illuminate the molecular interactions of 3DL1-cyto with SHP-2, these studies also lay the groundwork for future experiments to define the functional consequences of lipid interactions, the dynamic loop-like structure in Segment II, serine and threonine phosphorylation of 3DL1-cyto, and the additional interaction with SHP-1.
Star*Methods
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Hong Cheng (hong.cheng@fccc.edu).
METHOD DETAILS
Protein expression and purification
The cDNA encoding the 84-residue 3DL1-cyto (aa H340-P423; Fig. 1A) with an extra three amino acids (MEF) at the N-terminus into pET32b plasmid (EMD Millipore) was inserted between the NcoI and BamH I sites. This vector fuses a 109 aa TrxTag thioredoxin sequence to the N-terminus to increase expression and incorporates a His6tag for purification. The plasmid was transformed into E. coli Rosetta2 (DE3) cells (Novagen), and protein expression was induced by 1 mM IPTG when the cell density reached ~1 at A600 in LB or M9 medium. For preparing isotopically labeled protein, the cells were grown in M9 medium supplemented with 1 g/L 15N NH4 Cl and 3 g/L 13 C glucose. The protein was purified by using a HisTrap HP column (GE Healthcare), and the fused thioredoxin and His6tag was removed using thrombin (BioPharm, Lab., Utah). This yielded a highly soluble product corresponding to 3DL1-cyto with an extra 33 amino acid sequence enhancing tag (SET) peptide appended at the N-terminus (GSGMKETAAAKFERQHMDSPDLGTDDDDKAMEF, pI 4.31; Fig. 1A), which serves to improve solubility. The His-tag and extra thrombin were removed by a second Ni affinity chromatography followed by size-exclusion chromatography (Sephadex-75; GE Healthcare). Protein concentration was determined by measuring absorption at 280 nm, using an extinction coefficient of 8250 M−1cm−1.
The individual SH2 domains of SHP-2, N-SH2 (M1-N103) and C-SH2 (A105-G246), and the tandem SH2 domains of NC-SH2 (M1-G246) were cloned into BamH I and Hind III sites of expression vector pET49b. E. coli Rosetta2 (DE3) cells (Novagen) were transformed with the plasmid constructs and grown in LB or M9 medium. Protein expression was induced with 1 mM IPTG when A600 reached ~1. The proteins were purified by using a HisTrap HP column (GE Healthcare) followed by GSTrap columns (GE Healthcare) after cleavage by HRV 3C protease (EMD Millipore) according to the manufacturer’s protocols. The purified proteins contained seven extraneous residues (GPGYQDP) at their N-termini. Protein concentrations were determined on the basis of absorbance at 280 nm, and using extinction coefficients of 14650 M−1cm−1 for the N-SH2, 9650 M−1cm−1 for C-SH2, and 22900 M−1cm−1 for the tandem NCSH2.
NMR Sample Preparation
NMR samples of uniformly 13C- and 15N-labeled 3DL1-cyto (~300 μl) were prepared by dissolving ~1 mM protein in buffer containing 20 mM HEPES, 20 mM NaC1, 3 mM EDTA, and 3 mM DTT, at pH 7.4. To form the complex of this IDR with binding partners, the samples were reconcentrated to a volume of ~300 μl after adding the desired amounts of target protein (for details see figure captions).
NMR Spectroscopy
NMR spectra were recorded at 30 or 37° C (see figure caption) on a Bruker Avance II 600 MHz NMR instrument equipped with a TCI triple-resonance cryogenic probe. NMR data were analyzed using Felix2007 (Felix NMR, Inc. San Diego, CA USA) and nmrfam-Sparky (http://www.nmrfam.wisc.edu/nmrfam-sparky-distribution.htm). To tyrosine phosphorylate ITIM residues in KIR3DL1-cyto while acquiring spectra, ~800 units of FYN PTK (10 uL of 7.8 uM) were introduced directly into the NMR tube containing ~ 500 μL of 1 mM 3DL1-cyto, and NMR spectra were recorded for up to 72 hours thereafter. The data were acquired using standard Bruker pulse sequences and processed with FELEX2007 (Felix, San Diego, CA). NMR spectra analysis and peak picking were performed using Sparky (University of California, San Francisco). Chemical shifts are reported relative to DSS (external reference). Weighted chemical shift changes upon phosphorylation and ligand binding were calculated as:
using the following weight factor: ωHN= 1, ωN= 0.154, ωCα= ωCβ=0.276 and ωCO= 0.341(Popovych et al., 2006).
Structure Analysis
Protein disorder prediction was performed by using a disorder prediction program based on neural network strategies, VL3 (http://www.dabi.temple.edu/disprot/predictor.php) (Obradovic et al., 2003). The secondary structure prediction was performed by using NetSurfP v1.1 (http://www.cbs.dtu.dk/services/NetSurfP/) (Petersen et al., 2009). The conformational ensemble of SET-3DL1-cyto was calculated by using the CS-Rosetta program based on chemical shifts (https://csrosetta.bmrb.wisc.edu/csrosetta/submit) (Lange et al., 2012). The radius of gyration of protein was calculated from pdb files performed on the webserver: http://www.scfbio-iitd.res.in/software/proteomics/rg.jsp. Chimera (https://www.cgl.ucsf.edu/chimera/) was used for the visualization and analysis of protein structures.
Data availability
The NMR chemical shifts of SET-3DL1-cyto has been deposited to BMRB (accession number 27651).
Supplementary Material
Highlights.
KIR3DL1 cytoplasmic domain is intrinsically disordered with three distinct segments
TFE and SDS enhanced alpha-helical conformation around the two tyrosines (ITIMs)
SHP-2 SH2 domains decrease NMR peaks around the unphosphorylated N-terminal ITIM
Bis-phosphorylated ITIMs are more broadly impacted by the tandem SHP-2 SH2 domains
Acknowledgements:
This work was supported by NIH grants CA083859 (K.S.C.) and GM116911 (H.R.), a Cancer Center Support Grant from the NCI (CA06927), an appropriation from the Commonwealth of Pennsylvania, and the Spectroscopy Support Facility of FCCC. We thank Irina L. Shchaveleva for preparing the expression plasmid of 3DL1 C342A/C383A. We thank Drs. Erica A. Golemis and John Karanicolas for helpful comments on the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of Interests:
Hong Cheng and Heinrich Roder (Institute for Cancer Research, Fox Chase Cancer Center, 333 Cottman Ave., Philadelphia, PA 19111) filed a patent related to this work titled as “Application of directly carbon-detected NMR techniques for mapping protein-protein interactions involving intrinsically disordered regions” in 2015.
References:
- Alvarez-Arias DA, and Campbell KS (2007). Protein kinase C regulates expression and function of inhibitory killer cell Ig-like receptors in NK cells. J Immunol 179, 5281–5290. [DOI] [PubMed] [Google Scholar]
- Barrow AD, and Trowsdale J (2006). You say ITAM and I say ITIM, let’s call the whole thing off: the ambiguity of immunoreceptor signalling. Eur J Immunol 36, 1646–1653. [DOI] [PubMed] [Google Scholar]
- Binstadt BA, Brumbaugh KM, Dick CJ, Scharenberg AM, Williams BL, Colonna M, Lanier LL, Kinet JP, Abraham RT, and Leibson PJ (1996). Sequential involvement of Lck and SHP-1 with MHC-recognizing receptors on NK cells inhibits FcR-initiated tyrosine kinase activation. Immunity 5, 629–638. [DOI] [PubMed] [Google Scholar]
- Biron CA (1997). Natural killer cell regulation during viral infection. Biochem Soc Trans 25, 687–690. [DOI] [PubMed] [Google Scholar]
- Bruhns P, Marchetti P, Fridman WH, Vivier E, and Daeron M (1999). Differential roles of N- and C-terminal immunoreceptor tyrosine-based inhibition motifs during inhibition of cell activation by killer cell inhibitory receptors. J Immunol 162, 3168–3175. [PubMed] [Google Scholar]
- Burshtyn DN, Scharenberg AM, Wagtmann N, Rajagopalan S, Berrada K, Yi T, Kinet JP, and Long EO (1996). Recruitment of tyrosine phosphatase HCP by the killer cell inhibitor receptor. Immunity 4, 77–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caligiuri MA (2008). Human natural killer cells. Blood 112, 461–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cambier JC (1995). Antigen and Fc receptor signaling. The awesome power of the immunoreceptor tyrosine-based activation motif (ITAM). J Immunol 155, 3281–3285. [PubMed] [Google Scholar]
- Campbell KS, Dessing M, Lopez-Botet M, Cella M, and Colonna M (1996). Tyrosine phosphorylation of a human killer inhibitory receptor recruits protein tyrosine phosphatase 1C. J Exp Med 184, 93–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell KS, and Hasegawa J (2013). Natural killer cell biology: an update and future directions. J Allergy Clin Immunol 132, 536–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell KS, and Purdy AK (2011). Structure/function of human killer cell immunoglobulin-like receptors: lessons from polymorphisms, evolution, crystal structures and mutations. Immunology 132, 315–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clore GM, and Iwahara J (2009). Theory, practice, and applications of paramagnetic relaxation enhancement for the characterization of transient low-population states of biological macromolecules and their complexes. Chem Rev 109, 4108–4139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornilescu G, Delaglio F, and Bax A (1999). Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J Biomol NMR 13, 289–302. [DOI] [PubMed] [Google Scholar]
- DeFord-Watts LM, Dougall DS, Belkaya S, Johnson BA, Eitson JL, Roybal KT, Barylko B, Albanesi JP, Wulfing C, and van Oers NS (2011). The CD3 zeta subunit contains a phosphoinositide-binding motif that is required for the stable accumulation of TCR-CD3 complex at the immunological synapse. J Immunol 186, 6839–6847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duchardt E, Sigalov AB, Aivazian D, Stern LJ, and Schwalbe H (2007). Structure induction of the T-cell receptor zeta-chain upon lipid binding investigated by NMR spectroscopy. Chembiochem 8, 820–827. [DOI] [PubMed] [Google Scholar]
- Eck MJ, Pluskey S, Trub T, Harrison SC, and Shoelson SE (1996). Spatial constraints on the recognition of phosphoproteins by the tandem SH2 domains of the phosphatase SHPTP2. Nature 379, 277–280. [DOI] [PubMed] [Google Scholar]
- Felli IC, and Pierattelli R (2014). Novel methods based on (13)C detection to study intrinsically disordered proteins. J Magn Reson 241, 115–125. [DOI] [PubMed] [Google Scholar]
- Flaswinkel H, Barner M, and Reth M (1995). The tyrosine activation motif as a target of protein tyrosine kinases and SH2 domains. Semin Immunol 7, 21–27. [DOI] [PubMed] [Google Scholar]
- Fry AM, Lanier LL, and Weiss A (1996). Phosphotyrosines in the killer cell inhibitory receptor motif of NKB1 are required for negative signaling and for association with protein tyrosine phosphatase 1C. J Exp Med 184, 295–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagnon E, Schubert DA, Gordo S, Chu HH, and Wucherpfennig KW (2012). Local changes in lipid environment of TCR microclusters regulate membrane binding by the CD3epsilon cytoplasmic domain. J Exp Med 209, 2423–2439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs EB, Cook EC, and Showalter SA (2017). Application of NMR to studies of intrinsically disordered proteins. Arch Biochem Biophys 628, 57–70. [DOI] [PubMed] [Google Scholar]
- Ignatiuk A, Quickfall JP, Hawrysh AD, Chamberlain MD, and Anderson DH (2006). The smaller isoforms of ankyrin 3 bind to the p85 subunit of phosphatidylinositol 3’-kinase and enhance platelet-derived growth factor receptor down-regulation. J Biol Chem 281, 5956–5964. [DOI] [PubMed] [Google Scholar]
- Ikura M, Kay LE, and Bax A (1990). A novel approach for sequential assignment of 1H, 13C, and 15N spectra of proteins: heteronuclear triple-resonance three-dimensional NMR spectroscopy. Application to calmodulin. Biochemistry 29, 4659–4667. [DOI] [PubMed] [Google Scholar]
- Isaksson L, Mayzel M, Saline M, Pedersen A, Rosenlow J, Brutscher B, Karlsson BG, and Orekhov VY (2013). Highly efficient NMR assignment of intrinsically disordered proteins: application to B- and T cell receptor domains. PLoS One 8, e62947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi-Maki A, Yusa S, Catina TL, and Campbell KS (2003). KIR2DL4 is an IL-2-regulated NK cell receptor that exhibits limited expression in humans but triggers strong IFN-gamma production. J Immunol 171, 3415–3425. [DOI] [PubMed] [Google Scholar]
- Laczko I, Hollosi M, Vass E, Hegedus Z, Monostori E, and Toth GK (1998). Conformational effect of phosphorylation on T cell receptor/CD3 zeta-chain sequences. Biochem Biophys Res Commun 242, 474–479. [DOI] [PubMed] [Google Scholar]
- Lange OF, Rossi P, Sgourakis NG, Song Y, Lee HW, Aramini JM, Ertekin A, Xiao R, Acton TB, Montelione GT, et al. (2012). Determination of solution structures of proteins up to 40 kDa using CS-Rosetta with sparse NMR data from deuterated samples. Proc Natl Acad Sci U S A 109, 10873–10878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Liew LS, Li Q, and Kang C (2016). Structure of the transmembrane domain of human nicastrin-a component of gamma-secretase. Sci Rep 6, 19522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litwin V, Gumperz J, Parham P, Phillips JH, and Lanier LL (1994). NKB1: a natural killer cell receptor involved in the recognition of polymorphic HLA-B molecules. J Exp Med 180, 537–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacFarlane AW 4th, and Campbell KS (2006). Signal transduction in natural killer cells. Current topics in microbiology and immunology 298, 23–57. [DOI] [PubMed] [Google Scholar]
- Matalon O, Fried S, Ben-Shmuel A, Pauker MH, Joseph N, Keizer D, Piterburg M, and Barda-Saad M (2016). Dephosphorylation of the adaptor LAT and phospholipase C-gamma by SHP-1 inhibits natural killer cell cytotoxicity. Sci Signal 9, ra54. [DOI] [PubMed] [Google Scholar]
- Matozaki T, Murata Y, Saito Y, Okazawa H, and Ohnishi H (2009). Protein tyrosine phosphatase SHP-2: a proto-oncogene product that promotes Ras activation. Cancer Sci 100, 1786–1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mentlik James A, Cohen AD, and Campbell KS (2013). Combination immune therapies to enhance anti-tumor responses by NK cells. Front Immunol 4, 481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obradovic Z, Peng K, Vucetic S, Radivojac P, Brown CJ, and Dunker AK (2003). Predicting intrinsic disorder from amino acid sequence. Proteins 53 Suppl 6, 566–572. [DOI] [PubMed] [Google Scholar]
- Olcese L, Lang P, Vely F, Cambiaggi A, Marguet D, Blery M, Hippen KL, Biassoni R, Moretta A, Moretta L, et al. (1996). Human and mouse killer-cell inhibitory receptors recruit PTP1C and PTP1D protein tyrosine phosphatases. J Immunol 156, 4531–4534. [PubMed] [Google Scholar]
- Petersen B, Petersen TN, Andersen P, Nielsen M, and Lundegaard C (2009). A generic method for assignment of reliability scores applied to solvent accessibility predictions. BMC Struct Biol 9, 51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pluskey S, Wandless TJ, Walsh CT, and Shoelson SE (1995). Potent stimulation of SHPTP2 phosphatase activity by simultaneous occupancy of both SH2 domains. J Biol Chem 270, 2897–2900. [DOI] [PubMed] [Google Scholar]
- Poole AW, and Jones ML (2005). A SHPing tale: perspectives on the regulation of SHP-1 and SHP-2 tyrosine phosphatases by the C-terminal tail. Cell Signal 17, 1323–1332. [DOI] [PubMed] [Google Scholar]
- Popovych N, Sun S, Ebright RH, and Kalodimos CG (2006). Dynamically driven protein allostery. Nat Struct Mol Biol 13, 831–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renner C, Schleicher M, Moroder L, and Holak TA (2002). Practical aspects of the 2D 15N-[1h]-NOE experiment. J Biomol NMR 23, 23–33. [DOI] [PubMed] [Google Scholar]
- Saito H, Ando I, and Ramamoorthy A (2010). Chemical shift tensor - the heart of NMR: Insights into biological aspects of proteins. Prog Nucl Magn Reson Spectrosc 57, 181–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y, Lange O, Delaglio F, Rossi P, Aramini JM, Liu G, Eletsky A, Wu Y, Singarapu KK, Lemak A, et al. (2008). Consistent blind protein structure generation from NMR chemical shift data. Proc Natl Acad Sci U S A 105, 4685–4690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiraki K, Nishikawa K, and Goto Y (1995). Trifluoroethanol-induced stabilization of the alpha-helical structure of beta-lactoglobulin: implication for non-hierarchical protein folding. J Mol Biol 245, 180–194. [DOI] [PubMed] [Google Scholar]
- Sigalov AB (2011). Uncoupled binding and folding of immune signaling-related intrinsically disordered proteins. Prog Biophys Mol Biol 106, 525–536. [DOI] [PubMed] [Google Scholar]
- Sigalov AB, Aivazian DA, Uversky VN, and Stern LJ (2006). Lipid-binding activity of intrinsically unstructured cytoplasmic domains of multichain immune recognition receptor signaling subunits. Biochemistry 45, 15731–15739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SAP, W. E.; Gerig JT (1992). The Hamiltonians of NMR Part I. Concepts Magn Reson 4, 107–144. [Google Scholar]
- Spera S, and Bax A (1991). Empirical correlation between protein backbone conformation and C.alpha. and C.beta. 13C nuclear magnetic resonance chemical shifts. J Am Chem Soc 113, 5490–5492. [Google Scholar]
- Storkus WJ, Howell DN, Salter RD, Dawson JR, and Cresswell P (1987). NK susceptibility varies inversely with target cell class I HLA antigen expression. J Immunol 138, 1657–1659. [PubMed] [Google Scholar]
- Tamiola K, Acar B, and Mulder FA (2010). Sequence-specific random coil chemical shifts of intrinsically disordered proteins. J Am Chem Soc 132, 18000–18003. [DOI] [PubMed] [Google Scholar]
- Wishart DS, and Case DA (2001). Use of chemical shifts in macromolecular structure determination. Methods Enzymol 338, 3–34. [DOI] [PubMed] [Google Scholar]
- Wishart DS, and Sykes BD (1994). The 13C chemical-shift index: a simple method for the identification of protein secondary structure using 13C chemical-shift data. J Biomol NMR 4, 171–180. [DOI] [PubMed] [Google Scholar]
- Wishart DS, Sykes BD, and Richards FM (1992). The chemical shift index: a fast and simple method for the assignment of protein secondary structure through NMR spectroscopy. Biochemistry 31, 1647–1651. [DOI] [PubMed] [Google Scholar]
- Woodside DG, Obergfell A, Talapatra A, Calderwood DA, Shattil SJ, and Ginsberg MH (2002). The N-terminal SH2 domains of Syk and ZAP-70 mediate phosphotyrosine-independent binding to integrin beta cytoplasmic domains. J Biol Chem 277, 39401–39408. [DOI] [PubMed] [Google Scholar]
- Wright PE, and Dyson HJ (2015). Intrinsically disordered proteins in cellular signalling and regulation. Nat Rev Mol Cell Biol 16, 18–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C, Gagnon E, Call ME, Schnell JR, Schwieters CD, Carman CV, Chou JJ, and Wucherpfennig KW (2008). Regulation of T cell receptor activation by dynamic membrane binding of the CD3epsilon cytoplasmic tyrosine-based motif. Cell 135, 702–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yusa S, and Campbell KS (2003). Src homology region 2-containing protein tyrosine phosphatase-2 (SHP-2) can play a direct role in the inhibitory function of killer cell Ig-like receptors in human NK cells. J Immunol 170, 4539–4547. [DOI] [PubMed] [Google Scholar]
- Yusa S, Catina TL, and Campbell KS (2002). SHP-1- and phosphotyrosine-independent inhibitory signaling by a killer cell Ig-like receptor cytoplasmic domain in human NK cells. J Immunol 168, 5047–5057. [DOI] [PubMed] [Google Scholar]
- Yusa S, Catina TL, and Campbell KS (2004). KIR2DL5 can inhibit human NK cell activation via recruitment of Src homology region 2-containing protein tyrosine phosphatase-2 (SHP-2). J Immunol 172, 7385–7392. [DOI] [PubMed] [Google Scholar]
- Zhang H, Cordoba SP, Dushek O, and van der Merwe PA (2011). Basic residues in the T-cell receptor zeta cytoplasmic domain mediate membrane association and modulate signaling. Proc Natl Acad Sci U S A 108, 19323–19328. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The NMR chemical shifts of SET-3DL1-cyto has been deposited to BMRB (accession number 27651).