Abstract
With the advancement of microbiological discovery, it is evident that many infections, particularly bloodstream infections, are polymicrobial in nature. Consequently, new challenges have emerged in identifying the numerous etiologic organisms in an accurate and timely manner using the current diagnostic standard. Various molecular diagnostic methods have been utilized as an effort to provide a fast and reliable identification in lieu or parallel to the conventional culture-based methods. These technologies are mostly based on nucleic acid, proteins, or physical properties of the pathogens with differing advantages and limitations. This review evaluates the different molecular methods and technologies currently available to diagnose polymicrobial infections, which will help determine the most appropriate option for future diagnosis.
Keywords: polymicrobial infection, bacterial infection, bloodstream infection, sepsis, drug resistance, molecular diagnostics, fingerprinting, sequencing, broad testing, target-specific
1. Introduction & Background
In nature, microbes rarely exist in pure culture forms, thriving instead in complex communities shared with other species. Human microbiota in both healthy and disease states are no exceptions. With the advent of culture-independent analytical strategies, infections previously classified as monomicrobial have now been shown to be associated with a considerably diverse microbial population. The growing recognition of polymicrobial diseases not only reveals the intricate microbe-microbe and microbe-host dynamics, (Layeghifard et al., 2017; Peters et al., 2012; Tay et al., 2016) but underscores the shortcomings in current diagnostic practices that can profoundly impact clinical outcomes.
Polymicrobial infection is defined as the disease caused by mixed infection of two or more microorganisms. Polymicrobial bloodstream infection (BSI) has been gaining epidemiologic importance. In fact, a common sequela of BSIs—sepsis, is the tenth leading cause of death in the United States with approximately 250,000 new cases each year and has been called an emerging “hidden public health disaster” (Angus, 2010; Wisplinghoff et al., 2004). Polymicrobial BSI has been reported for the past 50 years, with rates ranging from 5–38%. (Bodey et al., 1965; Hermans and Washington, 1970; Hochstein et al., 1965; Kiani et al., 1979; Reuben et al., 1989) These infections increase the risk of mortality (21–63%) by 2–3 folds and extend the length of hospital stay as compared to their monomicrobial counterparts across all age groups in both ICU and non-ICU settings (Cooper et al., 1990; Downes et al., 2008; Fanaroff et al., 1994; Hermans and Washington, 1970; Kiani et al., 1979; Mackowiak et al., 1980; Pammi et al., 2014; Pavlaki et al., 2013; Sancho et al., 2012; Weinstein et al., 1986) The mechanisms for increased mortality in polymicrobial BSI are not clear. The increased mortality has been associated with inadequate and inappropriate antimicrobial treatments;(Cooper et al., 1990; Elting et al., 1986; Harbarth et al., 2003; Reuben et al., 1989; Roselle and Watanakunakorn, 1979) it may also arise from inherent host vulnerability or synergism of one infection by another. Predisposing factors which may compound mortality risk include the presence of intravascular catheter, gastrointestinal or genitourinary diseases, malignancies, immunocompromised, recent surgical procedures, reliance on parenteral nutrition, and chronic co-morbid conditions.(Hermans and Washington, 1970; Ing et al., 1981; Kiani et al., 1979; Rello et al., 1993; Sutter et al., 2008) These predisposing factors with varying sources of infections, patient populations, and clinical settings can all influence the heterogeneity in the microbial composition of BSI. Despite poor outcome from polymicrobial BSI, comprehensive characterization of its microbial composition and its impact on diagnosis, prognosis, and treatment are still ill-defined.
Time is of the essence in BSI-related sepsis. Every hour delay in appropriate treatment decreases survival by 7.6%.(Kumar et al., 2006) For patients with septic shock, mortality risk increases by 5-fold within the first 6 hours without appropriate antibacterials.(Kumar et al.) Prompt recognition is critical to saving lives. Since the spectrum of BSI pathogens is wide and the associated clinical presentations are variable, the high mortality risk warrants empiric broad-spectrum antimicrobial therapy to be initiated immediately in suspected cases (Beekmann et al., 2003; Rhodes et al., 2017) Unfortunately, current diagnostic standard based on the blood culture has failed to provide accurate and comprehensive microbial profiles quickly enough to guide the early management of suspected patients. Prolonged multi-step processing to detect positive microbial growth, followed by the identification of pure isolates and antimicrobial susceptibility testing (AST) makes blood culture very time-consuming (2–5 days) and labor-intensive.(Loonen et al., 2014) Administration of the antibiotic therapy prior to sampling can also compromise the culture sensitivity.(Struelens, 2009) Although BSI are predominantly caused by non-fastidious organisms, slow growing, fastidious, inert, or non-cultivatable yet disease-causing organisms may be underreported. In polymicrobial BSI, differential inherent microbial fitness and co-culture conditions may favor one species/strain over another, prohibiting a comprehensive survey of all organisms involved. Quantitative microbiology for BSI may provide clinically valuable information, such as differentiating pathogens from colonizer/contaminant, stratifying disease severity, or monitoring treatment response.(Hall and Lyman, 2006; Yagupsky and Nolte, 1990) Unfortunately, the value of this metric may be under-appreciated due to broth culture’s inability to measure microbial load routinely.
During the prolonged delay in obtaining blood culture results, patients may continue receiving ineffective or unnecessary broad-spectrum antimicrobials, leading not only to poor patient outcome, but also adverse iatrogenic effects (e.g. Clostridium difficiles colitis) and increased selection for multi-drug-resistant (MDR) pathogens.(Alm et al., 2008; Boucher et al., 2009; Cotten et al., 2009) Consequently, clinicians have fewer treatment options particularly in the direst patients. Antimicrobial therapies have been found to be insufficient for approximately 2 out of every 3 cases of polymicrobial BSIs and 1 out of every 3 cases of monomicrobial BSIs.(Retamar et al., 2012) Despite broad initial empiric antimicrobial coverage, escalation in antimicrobial spectrum − e.g. double covering for Pseudomonas spp, adding colistin for Acinetobacter spp or antifungal for candidemia − may be required if these pathogens are involved as the primary or co-infectants. The emphasis to de-escalate antimicrobial therapy early in order to reduce drug side effects and the selective pressure for resistance is often based on the poorly sensitive blood culture results. However, de-escalation is less likely when time to AST result is longer or when all pathogens involved in a polymicrobial BSI are not reliably identified.(Munson et al., 2003; Trenholme et al., 1989)
The burden of polymicrobial BSIs corroborates the need to re-examine current clinical microbiological practices and explore better identification techniques for more sensitive, rapid, and reliable diagnosis, prognosis, and treatment of the disease. Accurate identification of the pathogens and their relative abundances is imperative for effective monitoring, treatment, and understanding of the disease. Improved diagnostics provide clinicians with more information on a microbial community’s composition and antibiotic susceptibility, allowing for 1) earlier detection of infections and the identification of all pathogens involved; 2) determination of the clinical significance and the underlying source of the detected organisms;(Seifert, 2009) 3) assessment of the disease stage and progression; 4) more personalized and targeted treatment plans for patients; 5) feedback on the effectiveness of the treatment; and 6) greater insight into interspecies interactions contributing to the disease progression.
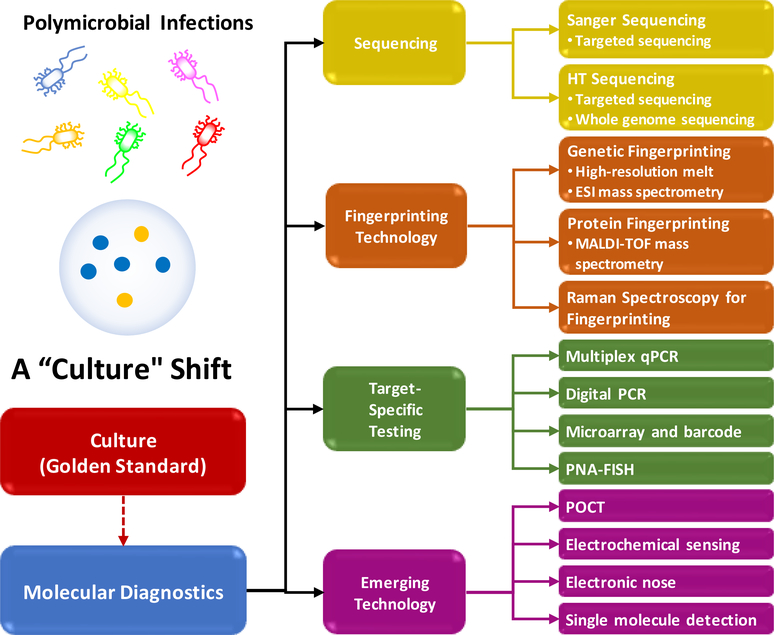
Advances in molecular technologies now provide tools to examine complex microbial communities in a high-throughput manner. The cultivation-independent molecular approaches are now being used to reveal “new” pathogens, as well as the polymicrobial nature of some infections. This review will evaluate the different molecular methods available to potentially diagnose a polymicrobial infection (Fig. 1), particularly the BSI.(Daniels, 2011; Hermans and Washington, 1970; Kumar et al., 2006; Pavlaki et al., 2013; Weinstein et al., 1986) An optimal diagnostic technique should not only be accurate, inexpensive, and easy to use, but also possesses additional features such as quantification of relative abundances, unbiased breadth of detection with species level identification, and determine antimicrobial susceptibility profile, with results available within the acute-care timeframe to impact clinical care. Based on the recommendations of Surviving Sepsis Campaign(Rhodes et al., 2017), the optimal window of opportunity for a diagnostic to optimize the antimicrobial stewardship should be within 6 hours after the initial empiric treatment, during which the clinical response is assessed prior to second scheduled drug dosing.(Daniels, 2011) Collectively assessing these techniques will provide a clearer understanding of their benefits and drawbacks. This will help determine the most appropriate option for a polymicrobial infection diagnostic technique that can significantly lower healthcare costs and save lives by providing timely and accurate diagnoses to guide clinical decisions.
Fig. 1.
A “culture” shift towards molecule diagnostics of polymicrobial infections.
The diagnostic technologies summarized in Fig. 1 is loosely categorized into detection technology and identification technology in this review. The main purpose of the detection technologies, such as the traditional blood culture and PCR, is to determine the if the pathogens are present in the blood sample. The main purpose of the identification technologies, such as the sequencing, fingerprinting and microarray, is to determine the pathogen species. The nucleic acid-based identification technologies are usually coupled with a pre-amplification process by PCR. The sensitivity of these identification technologies hence is limited by PCR. In many studies, the performance of the identification technologies is evaluated primarily based on their ability to correctly determine the pathogen species, but the detection sensitivity is not assessed. There are some overlaps between the two categories. For example, the target-specific PCR and digital PCR are able to perform both pathogen detection and identification as the same time.
In the following sections, we will evaluate well-established molecular detection techniques with an emphasis on their multiplexing capability for polymicrobial infection diagnostics. Next, we will look into several emerging technologies that shed light on possible new pathogen sensing strategies. In the end, we will discuss problems that remain to be resolved and share our view on the future development required to advance polymicrobial infection diagnostics.
2. Molecular Techniques for Pathogen Detection and Antimicrobial Susceptibility Testing
2.1. Sequencing
Existing sequencing techniques could be broadly categorized into the low-throughput sequencing (LTS, e.g. Sanger sequencing and pyrosequencing) and the high-throughput sequencing (HTS, e.g. the next generation sequencing (NGS) and the third-generation single molecule sequencing). Because the sequencing technologies analyze the DNA and not live bacteria, it is capable of identifying slow-growing bacteria or anaerobic bacteria that are incapable of growing in culture.(Liderot et al., 2010)
2.1.1. Low-Throughput Sequencing
The LTS is well-suited for the small-scale analysis. Common LTS technologies include Sanger sequencing and pyrosequencing. Compared to Sanger sequencing, pyrosequencing does not require the post-reaction analysis by electrophoresis. In pyrosequencing, the sequence information is obtained in real-time. Therefore, pyrosequencing is more commonly used for HTS via large-scale parallelization nowadays. The LTS technologies are mainly used for targeted sequencing. Instead of mapping out the entire genome, LTS only unveils the sequence of several specific loci of interest. In current clinical settings, LTS platforms such as the Sanger sequencing remains the standard sequencing method for genotyping. (Dewey et al., 2012) The throughput of Sanger sequencing is significantly lower compared to the HTS, but it could satisfy the needs of diagnostic laboratories which only analyze limited number of samples. The LTS has several unparalleled advantages over the HTS. First of all, the cost per sample of LTS is substantially lower than the HTS for the low number of targets tested in clinical laboratories. As a result, LTS such as Sanger sequencing has been widely adopted by the clinical laboratories, whereas the HTS is not an affordable option for the routine diagnostics. The main disadvantage of LTS is its low throughput. The LTS is often used to read a DNA fragment rather than the full-length sequence of the target gene or the genome. The partial sequence would limit the phylogenic resolution in pathogen identification. Another major drawback of LTS is its limited capability in analyzing polymicrobial samples. The complexity of the chromatogram for polymicrobial infection limits Sanger sequencing to analyzing only three or four bacterial species in one sample before the specificity starts to decrease. (Kommedal et al., 2009)
2.1.2. High-Throughput Sequencing
Common HTS technologies include Illumina’s reversibly terminated dye technology, 454 pyrosequencing technology, Ion Torrent’s semiconductor sequencing technology and Pacific Bioscience’s single molecule sequencing technology. The HTS for bacterial identification remains costly to run on a regular basis and not within the means of most laboratories. In addition, the entire sequencing workflow takes several days to complete, which is unpragmatic as a routine diagnostic tool. (Gyarmati et al., 2015) Nevertheless, with the advent of less expensive and more rapid HTS technologies, genomic analysis could become a viable option for diagnostic laboratories. Successful diagnostics of bacteremia in septic patients by the HTS has already been demonstrated in a clinically relevant timeframe. (Grumaz et al., 2016) The HTS could easily sequence the entire genome, providing a massive set of data that unveils any genetic and genomic abnormalities. Besides its high-throughput, the greatest advantage of HTS is its high degree of multiplexing, (Dubin et al., 2016; Huttenhower et al., 2012; Langille et al., 2013; Qin et al., 2010; Qin et al., 2012) which makes HTS an ideal tool for diagnosing polymicrobial infections. HTS has been demonstrated to differentiate tens to hundreds of bacterial species present in the microbiota.(Qin et al., 2010; Qin et al., 2012) A very limited number of studies have been reported on the diagnostics of infectious diseases using clinical samples with HTS, among which HTS shows a 85%−99% concordance with the culture-based assays in terms of species identification and antibiotic resistance profiling. (Anis et al., 2018; Stoesser et al., 2013) In terms of the disadvantage, the major drawback of the HTS is the burden of big data. The HTS generates an enormous amount of data that may be excessive for the simple task of pathogen identification. Clinical laboratories might be able to afford the sequencing equipment, but would lack the necessary computational power and specialized bioinformatician to make full sense of these data for diagnostics. This issue could be mitigated by the development of more user-friendly bioinformatic tools and cloud computing service. Compared with Sanger sequencing, the error rate of the HTS is slightly higher. (Dewey et al., 2012) To address this issue, high-fidelity DNA polymerase has been optimized specifically for HTS to lower the chance of introducing mutation and reduce the GC bias, hence increase the accuracy of the sequencing data. (Oyola et al., 2012; Quail et al., 2012) The library prepared using such as Kappa HiFi DNA polymerase is able be achieve a high performance similar to that of an amplification-free library. (Quail et al., 2012)
Targeted HTS identifies bacterial species by profiling a specific DNA sequence instead of the mapping out the entire genome. The 16S ribosome RNA (rRNA) gene is a widely used marker sequence. Several bioinformatic databases are dedicated to the rRNA. (Cole et al., 2008; Cole et al., 2013; Pruesse et al., 2007; Quast et al., 2012) These databases contain the full sequence of the 16S rRNA gene of a large number of strains. The 16S rRNA gene has several attractive features that make it a desirable biomarker for pathogen identification. First, the 16S rRNA gene is a well-characterized gene that contains the unique signature of the bacteria. The 16S sequence information alone is often sufficient to identify the bacterial species and determine the phylogenetic relationship between species. (Clarridge, 2004; Conlan et al., 2012) Second, the 16S rRNA gene contains highly conserved regions, which could serve as the flanking region for the universal amplification of polymicrobial samples. Studies of microbiome often use this approach to identify the bacterial species that are present in the microbial community.(Huttenhower et al., 2012; Langille et al., 2013; Qin et al., 2010; Qin et al., 2012)
Although the 16S rRNA gene has been used extensively in the bacterial classification, some bacteria cannot be resolved beyond the genus level using this gene, such as certain Escherichia and Shigella. (Janda and Abbott, 2007; Mignard and Flandrois, 2006) Sequencing the 16S rRNA gene alone may not provide enough precision for clinical diagnostics where the pathogenicity can depend on the species or even subspecies. Other sequencing approaches make use of the 23S rRNA gene, the rpoB (RNA polymerase subunit B) gene, the intergenic spacer region (ISR) between the 16S rRNA and 23S rRNA, or the ISR between rpoB and rpoC.(Gürtler and Stanisich, 1996) These target sequences have their own advantages. For example, rpoB has been shown to provide more details and allow for sub-species identification.
There is a debate on whether to use targeted HTS or the whole genome sequencing (WGS) for pathogen identification. The main advantage of targeted HTS is its cost effectiveness. It is well suited for studies that require analyzing samples from a large cohort. However, targeted HTS offers limited phylogenetic resolution. Because it only analyzes a number of short DNA segments, the degree of sequence variation in these regions may not be adequate to distinguish closely related species.(Větrovský and Baldrian, 2013) In several studies that compared targeted HTS and the WGS, the number of species identified by targeted HTS was significantly lower than the WGS.(Jovel et al., 2016; Ranjan et al., 2016) Furthermore, the selection of the target regions and the design of flanking primers may cause bias during amplification and lead to discordant results. In contrast, WGS uses random primers and degenerate oligonucleotides, thus eliminating the need for designing specific primers if a sufficiently conserved region cannot be found. Compared to targeted HTS, WGS is less cost effective, but it is able to identify more species from the metagenome because WGS covers more variable sites that could be used for the phylogenic classification. In addition, WGS is able to concurrently identify bacteria, viruses, fungi and other organisms present in the metagenome. In polymicrobial BSI, in addition to bacteria, fungi such as Candida species, viruses such as Herpes Simplex Virus or Cytomegalovirus (particularly in immunocompromised patients), and other organisms such as Plasmodium species or Trypanosoma cruzi may be present in the blood sample. Only the WGS can the identify other microorganisms in addition to bacteria. It can also predict antimicrobial susceptibility profiles based on the presence of resistance genes.(Hasman et al., 2014)
In the diagnostic context, the accuracy and the cost are the primary concerns. Low-throughput sequencing, e.g. Sanger sequencing, meets most of the current diagnostic needs, which usually only requires targeted sequencing instead of WGS and does not need the throughput level of HTS after taking the cost into consideration. However, Sanger sequencing has limited success in characterizing polymicrobial samples. The HTS certainly has an edge in diagnosing polymicrobial infection. While both targeted HTS and WGS show a high degree of multiplexing capability, only the WGS could identify coinfections by microbes of different kingdoms, such as bacteria-fungi coinfection and fungi-parasites coinfection. The technical advantages of WGS in diagnosing polymicrobial infections are unparalleled. Yet, WGS is the technique that faces the greatest challenge to enter the diagnostic laboratory due to its high cost. A compromise must be made, at least for now, between the cost and the test accuracy to benefit the patients most.
2.2. Fingerprinting Technology
Fingerprinting refers to a set of technologies that identify pathogens by capturing their unique genetic, proteinic or extrinsic optical signatures. Fingerprinting technologies do not intend to obtain the entire genomic or proteomic information. Instead, only selected information unique to the species is collected for the purpose of pathogen identification and differentiation. This information, which is often an indirect spectral profile, i.e. the spectral information that does not contain the DNA sequence or protein structure, is compiled into a database. Unknown samples are identified by matching their spectral profiles against the database.
2.2.1. Genetic Fingerprinting
Genetic fingerprinting for the identification of bacterial pathogens usually starts with a broad PCR that uses flanking primers to amplify certain hypervariable regions. Amplicons of these hypervariable regions carry the signature of the pathogens. Various techniques are implemented to transform the pathogen-specific information embedded in the amplicons to a unique fingerprint spectral profile which is then matched against a spectral database for a possible hit. In this section, we will discuss two well-established genetic fingerprinting technologies that use the high-resolution melting curve (HRM) (Andini et al., 2017; Athamanolap et al., 2014; Fraley et al., 2016; Fraley et al., 2013; Hardick et al., 2012; Jeng, Kevin et al., 2012; Masek et al., 2014; Reed et al., 2007; Velez et al., 2017; Won et al., 2010; Yang et al., 2009) and the electrospray ionization mass spectrometry (ESI-MS) (Ecker et al., 2005; Ecker et al., 2008; Jacob et al., 2012; Kaleta et al., 2011a; Pierce et al., 2012; Sampath et al., 2012) to analyze the broad PCR amplicons for the bacterial pathogen identification.
2.2.1.1. Genetic fingerprinting by HRM
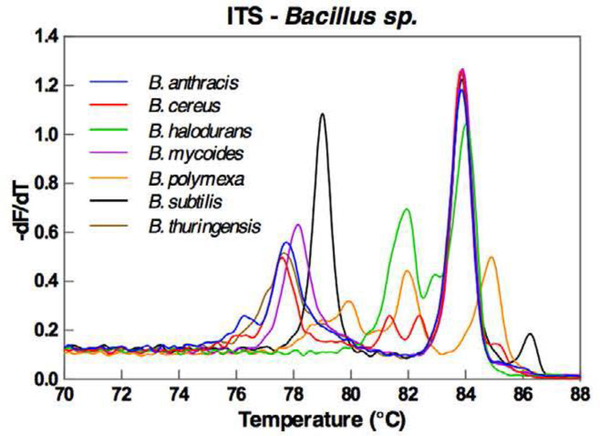
Broad PCR followed by the HRM analysis generates melt curve profiles that are unique to the pathogens. The melting curve profile is dependent on the size and the sequence of the PCR amplicon.(Reed et al., 2007) By targeting the hypervariable regions of the 16S rRNA, each bacterial species would result in a unique PCR amplicon thus a distinctive melt curve profile. This melting curve profile allows us to differentiate bacterial species based on the sequence variations without having to determine the exact sequence. Furthermore, the HRM analysis is a single-tube assay that uses a small volume of reagents and the sample. Hence, it is a low-cost and reliable assay that only takes a few hours to complete, falling within the optimal time window for the urgent care of infectious diseases. Several works have demonstrated the successful identification of bacterial species in blood samples by amplifying the hypervariable regions of the 16S rRNA gene followed by the HRM fingerprinting of the amplicons.(Andini et al., 2018; Won et al., 2010) To increase the phylogenic resolution of the HRM fingerprinting technology, three strategies have been proposed to enhance the sequence variation of the amplicons. One of the early strategies is to produce multiple amplicons, each of which targets one hypervariable region of the 16S rRNA gene.(Yang et al., 2009) As a result, multiple melting curve profiles are generated for each sample, and all the melting curve profiles must match the database to qualify for a hit. This approach is relatively tedious and resource-demanding as each sample must be analyzed multiple times. To simplify this assay, later an alternative strategy has been proposed to cover multiple hypervariable regions with a long amplicon.(Andini et al., 2017; Fraley et al., 2016; Velez et al., 2017) The long amplicon would generate highly complex melting curve profiles with more distinctive features. (Fig. 2) Machine learning programs have been developed to match these highly intricate melting curve profiles against the database. Using this approach, Fraley and colleagues has identified 37 clinically relevant bacterial in positive blood culture samples by targeting the 16S region with an accuracy of 100%. (Fraley et al., 2016) By targeting the internal transcribed spacer (ITS) region, the same group has achieved 95% accuracy in identifying 89 bacterial isolates and 90% accuracy in identifying pathogens in 59 positive blood culture samples at species level. (Andini et al., 2017) Later, the same approach has been extended to the whole blood sample. (Andini et al., 2018) The authors have achieved a detection sensitivity of 1 CFU/mL in the whole blood sample for four nosocomial organisms (Bcinetobacter baumannii, Escherichia coli, Klebsiella pneumoniae, and Staphylococcus aureus) using the broad PCR to target the ITS region. All four bacterial species have been successfully identified in the whole blood samples.(Andini et al., 2018) Yet another unconventional strategy uses molecular beacon instead of the intercalating dye as the melting probe.(El-Hajj et al., 2009) A set of “sloppy” molecular beacons labeled with different fluorescent dyes would hybridize to the amplicon. The sequence variation leads to different mismatch patterns between the molecular beacons and the amplicon, generating multiple melting curves in different fluorescent channels, each of which represents a melting curve from one molecular beacon. Although it has the potential for a higher degree of multiplexing compared to the intercalating dye-based HRM fingerprinting, the molecular beacon-based HRM is less cost-effective and more resource demanding. Unfortunately, there is no follow-up clinical study to demonstrate the capability of the “sloppy” molecular beacon in pathogen identification. In the context of molecular diagnostics, the intercalating-dye based HRM fingerprinting is more widely used for pathogen identification.
Fig. 2.
The complex melting curve profile of several bacterial species after the broad-PCR amplification that targets the ITS region. Reproduced from Andini et al., 2017 with the permission from Nature-Springer under the agreement of the creative common license.
A commercially available assay known as the SeptiFast MG (Straub et al., 2017; Warhurst et al., 2015) is capable of detecting a panel of 25 different fungal, Gram-positive, and Gram-negative pathogens within six hours directly from blood based on the HRM fingerprinting technology. It uses both universal flanking primers and specific primers to amplify the ITS region of the bacteria and fungi. In addition, sequence specific melting probes are added to enhance the distinguishability of the amplicons.(Lehmann et al., 2008) The analytical sensitivity of Septifast ranges from 3 CFU/mL to 100 CFU/mL depending on the microorganism. (Lehmann et al., 2008) In spiked whole blood samples, the authors successfully identified all 25 species at 100 CFU/mL with a 100% hit rate. At 30 CFU/mL, all 25 species were correctly identified, and 20 out of 25 had a 100% hit rate. At 3 CFU/mL, 21 out 25 species were correctly identified, and 15 out 25 species had a hit rate over 75%. The authors also tested over 1500 clinical isolates, and there was only 1.2% discrepancy from the conventional PCR and the culture-based approach. In clinical validation studies, the overall agreement between Septifast and blood culture is between 70%−80%. (Casalta et al., 2009; Westh et al., 2009)
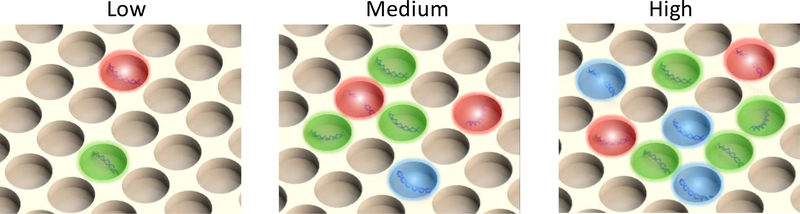
A significant drawback of the conventional HRM fingerprinting technology is its difficulty in distinguishing individual pathogens in the polymicrobial sample based on the melting curve profile alone.(Corless et al., 2000; García et al., 2004; Tong and Giffard, 2012; Weinstein et al., 1997) Although the complex melting curve profile of a polymicrobial sample can be deconvoluted to identify the individual components as demonstrated with the SeptiFast MG, it is limited to 2–3 microorganisms.(Mancini et al., 2009) The deconvolution requires that the melting curve profile of each microorganism is significantly different from each other. Even though, the deconvolution process becomes exponentially more difficult with increasing number of microorganisms in polymicrobial infections. This issue could be resolved with the digital HRM fingerprinting (dHRM) technology (Fig. 3). In dHRM, the DNA sample is partitioned by limited dilution so that each PCR compartment contains only one target DNA template.(Fraley et al., 2013; Quail et al., 2012) Essentially, each PCR compartment only generates one melting curve profile according to the type of DNA template it contains. The combined melting curve profiles from all the digitized PCR compartments would indicate all the pathogens in the polymicrobial infection. The digital HRM technology shows a great promise in both the quantification and identification of bacterial pathogens in polymicrobial samples. Nonetheless, it remains a proof-of-concept technology at current stage with demonstration performed with only bacterial isolates.
Fig. 3.
Digitization. Samples are partitioned into individual PCR reactions so that no single partition contains more than 1 copy of the DNA template. The quantification is accomplished by counting the number of positive partitions. The digitization also applies to cells by partitioning a single cell into each compartment. By digitization, polymicrobial samples are physically deconvoluted to “monomicrobial” samples. There is only one pathogen in each partition, and the combined information from all partitions reveals the composition of the polymicrobial sample.
2.2.1.2. Genetic fingerprinting by ESI-MS
The broad PCR amplicons can also be fingerprinted using ESI-MS. ESI-MS analyzes the base composition of the amplicon (i.e. the number of A, T, C and G) using the mass information of the nucleotide bases. Usually, multiple amplicons that target the variable regions are analyzed using ESI-MS. The base compositions of all the amplicons of an unknown sample are subsequently matched against the reference MS database.(Jordana-Lluch et al., 2014) An automated ESI-MS platform (PLEX-ID) with a built-in assay for BSI pathogen identification (BAC Spectrum Assay) is commercially available (Abbott Laboratories, Lake Bluff, United States).(Jacob et al., 2012) On the PLEX-ID platform, the base composition are analyzed for the identification of pathogens at species level or even subspecies level. Results are usually available within a few hours using whole blood samples, (Laffler et al., 2013) falling within the optimal time window for the urgent care of infectious diseases. ESI-MS also provides information for antibiotic susceptibility (Balážová et al., 2014; Hrabák et al., 2013) as well as fungal(Kaleta et al., 2011a; Kaleta et al., 2011b; Simner et al., 2013) and viral(Mengelle et al., 2013; Metzgar et al., 2010; Tang et al., 2013) infections. Unfortunately, Abbot discontinued the PLEX-ID platform due to possible financial, logistic and regulatory issues. (Özenci et al., 2017)
2.2.2. Protein Fingerprinting
The total protein profile could serve as a fingerprint of a species. Early forms of total protein profiling of microbes use the high-performance liquid chromatography (HPLC), and the resulting spectral profile are analyzed and matched against a known spectral database. More recent developments such as the electrospray ion (ESI) and the matrix-assisted laser desorption ionization time-of-flight (MALDI-TOF) mass spectrometry have improved the resolution and the coverage substantially.(van Baar, 2000) ESI and MALDI are two different popular ionization techniques employed by mass spectrometry. (El-Aneed et al., 2009) ESI takes soluble samples dissolved in a polar solvent and favors molecules with multiple charges. Therefore, ESI is well-suited to analyze PCR amplicons. MALDI is the most popular ionization technique used for protein analysis. It crystalizes the sample in an appropriate solid matrix. The sample-matrix crystal absorbs the laser energy, causing the analyte to ionize and desorb from the matrix. As the ionized molecules from the whole bacteria sample fly through a vacuum tube, they are separated based on their mass-to-charge (m/z) ratio, generating a m/z profile of all the proteins in the sample. MALDI-TOF has been in existence since the 1980s. Only in recent few years, it has obtained US FDA approval and Europe CE mark for the diagnostics of infectious disease in clinical laboratories.(La Scola, 2011) MALDI-TOF MS’s exceptional identification accuracy is the primary highlight of this technology. The bacteria identification rate by MS fingerprinting is similar to that of 16S gene sequencing.(Benagli et al., 2011; Saffert et al., 2011; Tan et al., 2012) MS has been primarily used with microbes growing on the solid culture media, but it has also been implemented with microbial suspension. While the analysis itself can be completed within minutes, the requirement of culture growth is a significant limitation that contributes to the amount of processing time needed.(Mancini et al., 2010) MS is unable to perform direct analysis on clinical blood samples because of the low microbial abundance. Usually blood culture is required to enrich the bacteria to a detectable level. Low reproducibility is also an issue because of the variability in preparatory methods and the matrix composition. (van Baar, 2000) Another limitation of MS-based fingerprinting is its limited capability in classifying multiple pathogens from the polymicrobial samples.(Chen et al., 2013; Lagacé-Wiens et al., 2012; Tadros and Petrich, 2013) Compared to the spectral profiles generated by the genetic fingerprinting techniques, MS’s spectral profile is more complex, which makes it problematic to deconvolute the composite spectra collected simultaneously from multiple bacterial species in the polymicrobial sample.
2.2.3. Fingerprinting by Surface Enhanced Raman Spectroscopy (SERS)
SERS measures the Raman scattering of the target molecules on the surface made of metal or other materials such as graphene.(Campion and Kambhampati, 1998; Schlücker, 2014; Stiles et al., 2008; Yang et al., 2015; Yang et al., 2016). Raman scattering is a label-free technology for the molecule identification. When excited by the laser, the vibrational motions of each molecule generate a unique and accurate light spectrum that serves as the molecule signature. The surface plasmonic effect in SERS substantially enhances the Raman scattering signals by many orders of magnitude to achieve a high detection sensitivity. In addition to the solid surface, metal colloidal particles can also function as the solid substrate for surface plasmonic enhancement. SERS can easily detect the presence of bacterial cells on the surface and yields an information-rich spectrum that could be used as a fingerprint for pathogen identification.(Boardman et al., 2016; Dina et al., 2017; Jarvis and Goodacre, 2004; Premasiri et al., 2017; Premasiri et al., 2016; Wang et al., 2016; Wu et al., 2015) The SERS spectral profile is then matched against a reference database. Bacteria have been successfully discriminated at the strain level by SERS-based fingerprinting.(Walter et al., 2011) In one study,(Boardman et al., 2016) pathogens from the positive blood culture samples were concentrated by centrifugation after selectively lysing the blood cells and measured by SERS on a silica substrate covered with gold nanoparticles. 17 bacterial species were successfully identified based on their SERS spectral profiles. Similar to MS, the major drawback of SERS is its limited capability in analyzing polymicrobial samples. Although a composite spectrum can be deconvoluted in theory, it may prove technically challenging as the SERS spectral profile is highly complex. The single-cell sensitivity of SERS gives a silver lining. (Boardman et al., 2016; Dina et al., 2017; Wang et al., 2016) With its ability to analyze a single bacterial cell, we propose a digital SERS for the diagnostics of polymicrobial BSI. Similar to digital PCR (Fig. 3), the bacterial cells are digitalized by limited dilution so that no more than one bacterial cell is contained in each micro compartment. The SERS spectrum could be measured from each compartment, and the bacterial species could be identified one at a time to diagnose polymicrobial infections.
2.2.4. Fingerprinting Technology and Polymicrobial Infection Diagnostics
In the context of molecular diagnostics, the fingerprinting technologies have compelling advantages over HTS. Although HTS provides a detailed genetic or genomic map, it generates a massive amount of information and overwhelms the user with unnecessary data burden. The simplicity, low cost and fast turnaround time suggest that the fingerprinting technology is more well-suited for molecular diagnostics in clinical settings. Unfortunately, the fingerprinting technology has limited multiplexing capability which is the key to diagnosing polymicrobial infections. Although PCR followed by ESI-MS fingerprinting has been demonstrated to resolve pathogens in polymicrobial infections, the workflow is technically complicated by clinical diagnostic due to the requirement of multiple amplicons.(Jeng, K. et al., 2012) In theory, one could deconvolute a composite HRM, an MS or a SERS spectral profile of multiple pathogens into individual spectrum to match against the reference database. In reality, this approach proves challenging. As the number of species increases in polymicrobial infections, the composite spectral profile becomes too intertwined to be deconvoluted. In addition, the interactions between different targets may further obscure the true identity of individual species in polymicrobial infections. For example, the main concern for polymicrobial HRM fingerprinting is the distorted melting curves caused by inter-strand interactions between amplicons resulted from different species. In such a case, the resulted composite melting curve profile is not a simple convolution of individual melting curves.
We propose one potential solution to accomplish a high degree of multiplexing for the diagnostics of polymicrobial infections by fingerprinting technologies is the digitization at the single-molecule or the single-cell level (Fig. 3). In a way, the digitization physically deconvolutes the fingerprinting spectral profiles and employs the large-scale parallelization for multiplexing. At the single-molecule level, each DNA template is partitioned into an individual compartment for digital PCR. The amplicon in each partition originates from a single species and can be easily identified using the HRM or the ESI-MS fingerprinting technology. The compiled spectral profiles from all the particles paint a full picture of the microbial composition in the polymicrobial sample. At the single-cell level, each cell could be partitioned for direct PCR (i.e. without DNA extraction) and identified using the HRM or the ESI-MS fingerprinting. Alternatively, each single bacterial cell could be directly analyzed using its protein fingerprint or the SERS fingerprint. The single-cell fingerprinting techniques do not have an amplification mechanism. Therefore, the detection sensitivity is the greatest limiting factor. Although both MS and SERS show a single-cell sensitivity, these techniques are still immature for clinical implementation.
2.3. Target-Specific Testing
The sequencing and fingerprinting technologies intend to classify all pathogens in polymicrobial infections, hence are categorized as the broad testing. In contrast, the target-specific testing is designed to check if certain pathogens of interest are present, but does not examine other possible pathogens. The target recognition is accomplished either by sequence-specific primers during PCR or through the sequence-specific probe that hybridize to amplicons post-PCR or directly to RNA in situ.
2.3.1. Multiplexed PCR
Real-time quantitative PCR (qPCR) is capable of multiplexed pathogen identification of polymicrobial infections. Because real time qPCR checks for the presence of DNA, it is useful in situations where antibiotics have previously been administered, or if bacteria do not grow well in the culture. Multiplex real-time qPCR uses several different sequence-specific primers and probes to target the pathogens of interest; each primer/probe set recognizes one pathogen. The pathogen identification is based on the successful amplification by each of these primer/probe sets. The diagnoses of polymicrobial infection are made possible through this method, because each target is reported by a distinct fluorescent dye. However, the competition for PCR reagents may hinder the accuracy of real-time qPCR in the detection of polymicrobial infections.(Dierkes et al., 2009; Tsalik et al., 2010) An alternative approach is to separate each target and run several PCR in parallel. This approach avoids the complication in multiplexed PCR, but it increases the reagent consumption and demands more effort in assay preparation.
A commercial system known as the Xpert from Cepheid uses a self-contained cartridge for both the sample preparation and multiplexed real-time PCR in infectious diseases diagnsotics.(Cepheid) It uses the traditional Taqman® probe for multiplexing. Due to the limited width of the fluorescent spectrum, Xpert and other real-time qPCR systems have a limited multiplexing capability. Usually only up to 5 targets can be detected in a single reaction. As a matter of fact, most Xpert panel targets only one or two pathogens. Its Carba-R panel has the highest degree of multiplexing and detects 5 target resistant genes for carbapenemase.(Tato et al., 2016)
Another commercial product is known as the FilmArray from Biomérieux. Its blood culture panel is capable of detecting 3 antibiotic resistant genes in addition to 24 pathogen targets, including Gram positive bacteria, Gram negative bacteria and yeasts.(Altun et al., 2013) It uses a two-stage nested PCR approach. After the first amplification, the amplicons are divided into a microwell array for the second stage PCR. The multiplexing is realized by spatially separating each reaction into a micro reaction chamber, and only one fluorescent dye is required. Although the FilmArray uses the melting curve to confirm the identity of the pathogen, the primary detection strategy relies on the target-specific primers. Using the positive blood culture sample, the assay turnaround time is 1 hour with only 2 minutes of hands-on time with an automated instrument. The manufacturer claims that “you can identify pathogens in 9 out of 10 positive blood cultures”. Users’ evaluation suggests this number could be true for monomicrobial infections if all pathogens are on the panel.(Altun et al., 2013) The number drops to ~80% once microorganisms outside the panel are present in the sample. In the case of polymicrobial infections, the FilmArray is only able to detect all the target pathogens in ~70% of the positive blood culture samples.(Southern et al., 2015) The FilmArray is a closed system with dedicated instrumentation. While this standardizes the results, it also increases equipment expenses for laboratories that currently use a different machine for qPCR.
The SeptiFast MG system mentioned in the previous section uses HRM fingerprinting to identify pathogens. However, it introduces the sequence-specific probes to facilitate the melting curve analysis. These melting probes enhance the distinguishability of the melting curve profiles. In addition, several sequence-specific primers are added in addition to the universal flanking primers. Strictly speaking, SeptiFast MG is a hybrid of sequence-specific testing and genetic fingerprinting.
2.3.2. Digital PCR
Digital PCR quantifies by counting the number of DNA template molecules. The DNA template is diluted and partitioned into hundreds to thousands of separate wells or droplets, each of which contains a digital copy of the DNA template, i.e. a single DNA template molecule or no molecule at all. The wells containing the molecule of interest are then amplified by the target-specific PCR, allowing for a precise quantification of the DNA template by counting the number of positive wells.(Hindson et al., 2011) Applied to pathogen detection, the use of digital PCR eliminates the possible bias caused by the preferential amplification as there is only one target template in each reaction and no PCR competition. Digital PCR is capable of quantifying extremely low concentrations of bacterial pathogens. A multiplexing digital PCR could be used to identify pathogens in polymicrobial infections.(Whale et al., 2016) However, same as the real-time qPCR, the multiplexing capability of digital PCR is also limited by the fluorescent spectrum with a maximum of 5 optical channels available for different targets.(Buchan and Ledeboer, 2014) The substantial increase in the quantification resolution by digital PCR is a significant advantage, but the numerous wells require greater volumes of reagents and longer processing time compared to the qPCR, which contributes to a higher cost.(Yang et al., 2014) Although recent development in microfluidic digital PCR technology has substantially reduced the reagent volume and the cost associated with microfluidic chip and equipment, the additional microfluidic processing time brings new challenges. By analyzing the circulating DNA in the plasma from the sputum smear-positive pulmonary tuberculosis patients, Ushio and colleagues detected Mycobacterium tuberculosis using digital PCR and achieved 69% sensitivity and 93% specificity.(Ushio et al., 2016) In another study that detects Helicobacter pylori in the stool sample, the digital PCR assay achieved 84% sensitivity and 100% specificity when benchmarked against the serological test.(Talarico et al., 2016) As a recently developed diagnostic device, digital PCR lacks the substantive clinical records that many other techniques have. So far, digital PCR is only used as a generic technology platform. Its applications in infection disease diagnostics are being explored, but no diagnostic product has been deployed. Nevertheless, its powerful quantification capability promises great diagnostic potential for polymicrobial infections.
2.3.3. Microarray
Microarray utilizes surface-immobilized oligomeric probes (DNA, RNA and their derivatives) to capture and detect DNA/RNA of the pathogens through the sequence-specific complementary hybridization. It reduces the sample and reagent consumption and related cost. Microarray is highly accurate and able to discriminate down to the species or strain level.(Ballarini et al., 2013) Complex bacterial pathogen communities can be differentiated by microarrays. For instance, the BactoChip is capable of distinguishing six different Staphylococcus species from a mixed sample.(Ballarini et al., 2013) Its fluorescent probes can detect and quantify up to 54 distinct species, and its accuracy does not decrease with more complex samples.(Ballarini et al., 2013) Several other commercially available microarray platforms read the scattering signals from the gold/silver nanoparticle-labeled probes that recognize specific bacterial pathogen targets. The Verigene platform from Luminex Corporation has two separate panels for Gram-negative (Wojewoda et al., 2013) and Gram-positive (Dodémont et al., 2014) blood culture samples. The Gram-positive panel detects 9 bacterial species and 3 antibiotic resistant genes against methicillin and vancomycin. The concordance with the blood culture is generally above 90% for monomicrobial infections.(Beal et al., 2013; Samuel et al., 2013; Wojewoda et al., 2013) The platform has very limited ability in diagnosing polymicrobial infections. In one study, it only correctly identified 3/9 polymicrobial infections, although 8/9 detected at least one target in the polymicrobial sample.(Beal et al., 2013) The Verigene’s Gram-negative panel detects 5 species and 6 antibiotic resistant genes for carbapenemase and extended-spectrum beta-lactamases. The performance of the Gram-negative panel is satisfactory for monomicrobial infections with a concordance rate generally over 90%. (Bork et al., 2015; Dodémont et al., 2014; Mancini et al., 2014) For polymicrobial infections, the panel correctly detected all targets in only ~50% of the cases, but identified at least one target in ~95% out of 40 cases. In these 40 cases, 22 contained two organisms in the panel, 5 contained three organisms in the panel, and 13 contained two organisms with one of which outside the panel.(Ledeboer et al., 2015) The company Genmark has a microarray platform known as the ePlex which measures electrochemical signals instead of optical signals of the probes. Three blood culture panels for Gram-positive, Gram-negative and fungal pathogens have been recently announced.(GenMark) These panels use samples from the blood culture bottle as the input and cover a wider range of targets than the Verigene and the FilmArray panels. So far, there is no independent evaluation of these newly launched blood culture panels. However, independent evaluation of other ePlex panels, such as the respiratory infection panel and the pharmacogenetic panel, has demonstrated satisfactory performance.(Guerendiain et al., 2016)
These microarray technologies are able to identify pathogens 24 hours earlier than the conventional culture-based approach because it bypasses the sub-culture.(Bork et al., 2015; Huang et al., 2019) While the traditional high-density microarray remains expensive, the low-density microarray that targets a relatively small number of pathogens has a reasonable price tag for clinical diagnostics. In theory, the microarray has a high degree of multiplexing capability. In reality, its ability to correctly identify all pathogens in polymicrobial infections is far from ideal.(Samuel et al., 2013) The unsatisfactory performance may be attributed to the competition of multiple targets during the labeling reaction. So far, no commercial microarray platform has been optimized to diagnose polymicrobial infections.
2.3.4. Peptide Nucleic Acid Fluorescence In Situ Hybridization (PNA-FISH)
PNA-FISH is a well-known clinical diagnostic method that detects blood pathogens through target-specific rRNA hybridization with fluorescent peptide nucleic acid probes.(Frickmann et al., 2017) It has been used in various microbiological contexts, from diagnosing polybacterial infections in the blood to polyfungal infections in the cerebrospinal fluid(Calderaro et al., 2014) and the lung tissue(Rickerts et al., 2013). Compared to MALDI-TOF mass spectrometry, PNA-FISH produces results at least one day faster.(Harris and Hata, 2013) PNA-FISH is more tolerant of impurities than PCR or microarray-based methods.(Stender et al., 2002) PNA probe hybridizes more readily than DNA probes.(Peleg et al., 2009) Studies have demonstrated that PNA-FISH can identify and quantify different species in mixed bacterial biofilm.(Almeida et al., 2011) The turnaround time for PNA-FISH following positive blood culture is rapid, completed in approximately 1.5–3 hours.(Kothari et al., 2014) Moreover, more rapid diagnostics by PNA-FISH not only helps direct more accurate antimicrobial therapies sooner, contributing to substantial pharmaceutical cost savings, but also reduces median hospital charges by nearly $20,000.(Ly et al., 2008) The time and cost savings provided by PNA-FISH make this molecular method a well-suited alternative to the conventional blood culture without a substantial loss of accuracy. Nevertheless, the drawbacks of PNA-FISH are its limited breadth of detection for pathogens, need for skilled technicians, and reliance on Gram stain results.(Afshari et al., 2012) Its lower limit of detection is 104 CFU/mL, making it unable to detect dilute microorganisms.(Peleg et al., 2009)
2.4. Molecular Antimicrobial Susceptibility Testing (AST)
The ultimate goal of a diagnosis is to guide clinical decisions and initiate clinical interventions for disease treatment and management. In the context of infectious disease, identifying the infectious pathogens alone does not guarantee the effective therapeutic regimen because the ever-increasing antibiotic resistance has greatly limited our treatment options. Conventional AST by culture-based phenotypic assays often requires an additional 1–2 days post enrichment and plating, which results in a significant delay in clinical decision making. As a consequence, empiric treatment regimens with broad-spectrum antibiotics are often administered to manage the infection during the critical clinical phase before the AST results become available. Unfortunately, the over prescription of these reserved antibiotics leads to a further spreading of multi-resistant microbes. Molecular AST predicts pathogens’ antibiotic resistance profile by analyzing the drug-resistant genes the pathogens carry using the techniques mentioned above. Compared to conventional AST, molecular AST drastically shortens the turnaround time, which enables the timely selection of antibiotics for the most effective treatment. In case of inducible antimicrobial resistance, such as inducible clindamycin resistance in Bacteriodes, in which conventional phenotypic AST may results in false susceptibility, molecular detection of resistance genes (e.g. erm genes) may prove to be more reliable.(Johnsen et al., 2017)
From the technical point of view, molecular AST shares the same characteristics as the molecular pathogen identification. In theory, any aforementioned techniques that are capable of reading gene sequences could be employed to detect antibiotic-resistant genes for AST. Although the basic research in antibiotic resistant genes has drawn substantial attention,(Allen et al., 2010; Martínez, 2008) molecular AST has yet to become a common clinical practice. Among all the aforementioned commercial molecular diagnostic platforms, only limited number of testing panels include resistant gene targets. Out of the five FilmArray molecular diagnostic panels, only the blood culture identification (BCID) panel includes 4 antibiotic resistant gene targets – mecA for the methicillin resistance, vanA and vanB for the vancomycin resistance, and KPC for the carbapenem resistance. (Fiori et al., 2016; Southern et al., 2015) ePlex’s respiratory panel does not include any resistant gene target. Although Genmark claims the ePlex blood panel would include several resistant gene targets that encode methicillin, vancomycin, carbapenem and extend spectrum β-lactam resistance (ESBL), this panel is still under development. The Gram-positive blood culture panel by Verigene includes three resistant gene targets for the methicillin and vancomycin resistance (Vareechon et al., 2018), and its Gram-negative blood culture panel includes six resistant gene targets for the ESBL and carbapenem resistance. (Ledeboer et al., 2015) Xpert system has two test kits for the detection of MRSA by detecting methicillin-resistant gene mecA and mecB in addition to the target gene for S. aureus identification.(Yarbrough et al., 2017) Xpert also has two kits for the detection of the carbapenem resistance and vancomycin resistance, but these two kits cannot identify the pathogens. Septifast MG does not test for the antibiotic resistance, but it has a separate mecA test to follow up with the positive identification of the S. aureus.(Straub et al., 2017; Warhurst et al., 2015)
Among all these systems, only FilmArray’s BCID panel, Verigene’s two blood culture panels, and Xpert MRSA kit, are capable of combined pathogen identification and AST. Other platforms separate pathogen identification and AST onto different panels and have limited multiplexing capability. The number of resistant gene targets in the FilmArray’s BCID panel is very limited. It only includes KPC, but not NDM, OXA, IMP nor VIM, which are all common carbapenem resistant genes. It does not include ESBL resistant gene either. Although the Verigene has a wider coverage of resistant genes, the blood culture identification is separated into two panels for Gram-positive and Gram-negative infections.
One key issue with combined molecular pathogen identification and AST for the polymicrobial infection is the inability to attribute the resistant gene to the pathogen. Although highly multiplexed detection platforms, such as FilmArray and Verigene, are capable of identifying all the pathogens and the antibiotic resistant genes in polymicrobial infections, they are unable to tell whether these resistant genes are carried by all the pathogens or only some of them. Likewise, these testing panels are unable to tell whether a specific pathogen carries all the antibiotic resistant genes, or only a subset of these resistant genes, or any of these resistant genes at all. The solution to this problem again is digitalization (Fig. 3). By partitioning individual pathogens into a separate chamber, the resistant genes detected by the molecular AST are associated with one pathogen, which allows us to identify what pathogens are in the polymicrobial infection and what resistant genes each pathogen carries.
While genotypic detection of a limited number of resistance genes can be performed by multiplexed PCR, it is an unreliable proxy for resistance phenotype and lacks comprehensiveness.(Evans et al., 2017; Evans et al., 2016) Antimicrobial resistance mechanisms are complex, particularly for Gram negative bacteria, and will continue to evolve.(Bard and Lee, 2018) Genotypic approaches also cannot provide quantitative susceptibility information to guide treatment with minimum inhibitory concentration (MIC). As such, phenotypic assays continue to remain as the gold standard for AST despite a lengthy assay time. To further accelerate phenotypic AST, the concept of “pheno-molecular AST” by coupling quantitative PCR with culture has been introduced recently. Quantitative PCR can be used to assess drug susceptibility with MIC reporting by enumerating changes in microbial DNA copies as a surrogate measure of microbial growth in the presence or absence of antimicrobials. Pheno-molecular AST targeting the 16S rRNA gene (rDNA), when applied to positive blood cultures, has demonstrated high overall agreement with conventional AST in a randomized controlled trial and improved turnaround time by 16 hours. (Beuving et al., 2011; Beuving et al., 2015)
More recently, our group demonstrated the use of PCR with HRM of the ITS region for broad bacterial detection, identification, and AST directly from whole blood with results available as early as 8 hours [Andini N et al, 2018]. Although promising, this approach still relies on cell division, which may be prolonged for species with longer generation time. Pheno-molecular AST has the potential to be accelerated further by rapidly detecting transient changes in levels of novel RNA biomarkers for susceptibility.(Khazaei et al., 2018)
3. Other Emerging Technology
The development of novel diagnostic tools advances mainly on two fronts. First, point-of-care diagnostic platforms with the sample-to-answer capability are still highly coveted to address the needs for rapid diagnostics in resource-limited settings. Second, novel biosensing modalities point out new ways of detecting biomarkers for infectious diseases diagnostics. These emerging technologies promise a great potential to change current clinical practice for better clinical outcomes. Nevertheless, most of these new technologies are still a proof-of-concept thus not clinically ready.
3.1. Point-of-Care Testing (POCT)
POCT for the diagnostics of infectious disease in low-resource settings or in response to an emergency outbreak is an attractive concept. The key characteristic of a true POCT is its sample-to-answer capability. A POCT must be able to process the sample (or be able to use the sample as-is) for the downstream analysis. Microfluidics has been long proposed to facilitate the sample and reagent transfer and to integrate all functional components into a lab-on-a-chip system for POCT.(Chin et al., 2007; Park et al., 2011) Several sample-to-answer diagnostic platforms have been successfully launched for infectious disease diagnostics. The Filmarray, the Xpert and the Verigene systems mentioned above all have a microfluidic component. Nevertheless, these systems are not meant for POCT because of the intricate fluidic control and the requirement for bulky instrumentation.
To avoid the complex instrumentation required by the conventional microchannel-based microfluidics, several alternatives have been proposed. One solution is to use droplets as virtual reaction chambers and to manipulate these droplets with a magnet.(Zhang and Nguyen, 2017; Zhang et al., 2013; Zhang and Wang, 2013) Such a magnetic digital microfluidic platform uses the magnet for fluidic control hence eliminates the need for the pressure source and pumps. It is able to operate in a true “power-free” manner, which is well-suited for POCT.(Zhang and Nguyen, 2017) Another promising POCT system is based on the centrifugal microfluidics. Centrifugal microfluidics uses a simple spinning motion for luidic control.(Gorkin et al., 2010) Although existing centrifugal microfluidic platform still requires relatively bulky instrument, manual centrifugal devices for clinical sample preparation, such as the egg beater, salad spinner and spin disk centrifuge,(Bhamla et al., 2017; Brown et al., 2011; Wong et al., 2008) shed some light on a new direction. Heating source is another concern for POCT. Chemical heating device that uses exothermic reactions with everyday materials or natural resource, such as the lime with water, flame, and sunlight, to control temperature for isothermal amplification completely eliminates the need for power source.(Poole et al., 2017; Snodgrass et al., 2018) A recent magnetic digital microfluidic platform developed by Shih et. al. brought the platform into the clinics.(Shin et al., 2017) This platform provided the sample-to-answer diagnostics by identifying pathogens using an isothermal amplification assay. It consisted of a cartridge pre-loaded with required reagents in the form of droplets. An automated control system used a permanent magnet to move silica magnetic particles between droplets to perform the extraction of nucleic acids. In the end, the nucleic acids were eluted in the buffer containing the reagent for the loop mediated isothermal amplification (LAMP), and the reaction was performed in droplets. The authors successfully tested samples from patients infected with Chlamydia trachomatis with a cellphone-controlled magnetic digital microfluidic platform in the emergency department and achieved 100% concordance with results obtained in centralized laboratories performed with a commercial molecular diagnostic system.
Paper is an old medium used for diagnostics. The capillary force by the paper is indeed the simplest fluid pumping mechanism. Lateral flow strips, such as the pregnancy test kit, are one of the first reliable POCT devices. However, the paper-based system is troubled with low sensitivity. Paper has experienced a resurgence in the last few years since several new methods had been proposed to pattern the paper with defined fluidic paths for making paper-based microfluidic devices.(Carrilho et al., 2009; Dungchai et al., 2011; Yetisen et al., 2013; Zhang et al., 2015) Earlier development of paper-based microfluidic devices is limited to simple one-step biochemical and enzymatic assays. Later, more complex paper platforms have been developed for AST(Deiss et al., 2014) and molecular diagnostics using PCR and isothermal amplification.(He et al., 2012; He et al., 2011) Unfortunately, none of these platforms fulfils the promise of a true POCT because they still require resource-demanding instruments for the temperature control and the optical signal readout. So far, the POCT research community has not been made aware of the pressing need for the diagnostics of polymicrobial infections. Research in related field is greatly lacking.
3.2. Novel Biosensing Strategies
3.2.1. Electrochemical Sensing
Diagnostic biosensors often take an electrochemical approach towards the target identification. They rely on species-specific probes or antibodies that emit an electrical signal after binding to their target, and the intensity of the signal corresponds to the relative amount of the target species. Not only do they identify bacteria in a matter of minutes to hours, but they also do so with a great specificity.(Lam et al., 2013) Biosensors designed for urinary tract infections are capable of detecting polymicrobial infections in just one hour. Although these devices have a high specificity, current biosensors have limits of detection of 104-106 CFU/mL. (Lam et al., 2013; Mach et al., 2009) This sensitivity may not be sufficient for detecting BSI where the concentration of bacteria in the blood can be a mere 1–10 CFU/mL (Jordana-Lluch et al., 2014). Most biosensors are limited in their scope and insufficient for the broad-range detection, but they offer a rapid method of diagnosis for specific diseases or microorganisms using only small sample volumes.(Lam et al., 2013) More work on biosensors is necessary to improve the cost of the biosensor devices, but the specificity and speed of current technologies is exceptional and may be useful in urgent situations.
3.2.2. Electronic Nose
Bacteria emit unique combinations of volatile organic compounds depending on their species, an attribute used by electronic nose (e-nose) technologies to diagnose polymicrobial infections. E-nose devices are handheld devices that sense vaporous compounds using an array of sensors coated with odor-sensitive absorbent materials. The sensory data is then matched to an existing library of microorganisms. Although the odor detection step is completed within minutes, it is performed after bacteria have been incubated on agar medium for 24 hours, thus limiting the speed of diagnosis. (Trincavelli et al., 2010) E-nose technology has been used to diagnose monomicrobial blood samples and polymicrobial samples taken from foot infections with an accuracy over 95% (Trincavelli et al., 2010; Yusuf et al., 2015), and it has also been shown that its sensors can differentiate fungi in cereal grains (Wilson and Baietto, 2011). More clinical studies are needed to establish that such artificial olfaction systems can reliably detect polymicrobial infections in blood or urine samples. E-nose technology lacks the ability to detect antibiotic resistance, but it possesses quantification abilities that would be useful in determining the relative abundances of each microbe in polymicrobial infections or in the microbiota. (Wilson and Baietto, 2011) Electronic olfactory devices have potential to become an inexpensive and portable alternative to mass spectrometry.
4. Conclusion
While the traditional culture-based approach of diagnosing infections remains the standard procedure in many clinical laboratories, molecular methods are progressively proving to be a superior option for identifying microorganisms. Blood culture misses almost one out of every two cases of bacteremia, emphasizing the urgent need for more accurate diagnostics.(Fenollar and Raoult, 2007; Laffler et al., 2013) The development of new technologies has increased the accessibility of molecular methods, especially in terms of the reproducibility and cost, but their ability in diagnosing polymicrobial infection is inadequate. Emerging research emphasizes the complexity and the severity of polymicrobial infections, calling for the use of more sensitive and multiplexed assays that can deliver diagnoses to clinicians faster. Such improvements in identification assays would lead to an earlier transition from the empirical treatment to a more informed, diagnosis-based treatment regimen for polymicrobial infections, thus improving the patient outcome. Other potential clinical uses include monitoring disease progression based on changes in microbiota composition, assessments of antibiotic resistance or pathogen virulence, and epidemiological surveillance.
Nonetheless, these molecular methods primarily serve as supplements to culture rather than replacements. Many techniques still require the pre-enrichment by culture. There is also a need to establish a consistent procedure, because the preparation method, the equipment, and the computer software analysis differ across protocols, and identification results may occasionally vary. Reproducibility still comprises a major issue in the standardization of any molecular method. Inter-laboratory variations must be resolved in order to make molecular-based methods a standardized test for polymicrobial identification and differentiation. Ongoing research continues to investigate culture-free, non-amplification methods that can produce more data, such as antibiotic susceptibility profiles, in a single run. These advancements would significantly augment our understanding of diseases and lead to better patient outcomes.
We have summarized the technologies in this review (excluding the emerging technologies) in a table (Tab. 2) and compared their performance in terms of the cost, the sensitivity, the turnaround time and their multiplexing capability of diagnosing polymicrobial infections. Each category is given a score from one to three stars. One star suggests the feature is least desired (e.g. high cost, long turnaround time, and limited multiplexing capability), and three starts suggest the feature is most desired (e.g. low cost, short turnaround time and high degree of multiplexing). Although the analysis time for certain technology platforms are short (e.g. PNA-FISH and MALDI-TOF MS), bacterial culture and plating are compulsory prior to the analysis. Therefore, the overall turnaround time is still long. Similarly, the assay cost per sample for several molecular diagnostic platforms is relatively low, but the equipment (e.g. mass spectrometry) increases the overall cost substantially.
Tab. 2.
Comparison of Molecular Diagnostic Technologies for Diagnosing Polymicrobial Infections
| Technology | Cost |
Detection Sensitivity | Identification Accuracy | Turnaround Time | Multiplexing capability To Diagnose Polymicrobial Infections | |
|---|---|---|---|---|---|---|
| Equipment Cost | Assay Cost per Sample | |||||
| Sequencing | ||||||
| Sanger Sequencing | $$ | $$$ | Rely on PCR | ★★★ | ★★ | ★ |
| Targeted HT Sequencing | $ | $$ | Rely on PCR | ★★★ | ★ | ★★★ |
| HT Whole Genome Sequencing | $ | $ | Rely on PCR | ★★★ | ★ | ★★★ |
| Fingerprinting | ||||||
| Genetic Fingerprinting by High-Resolution Melting | $$ | $$$ | Rely on PCR | ★★ | ★★★ | ★ |
| Genetic Fingerprinting by ESI Mass Spectrometry | $ | $$ | Rely on PCR | ★★ | ★★ | ★★ |
| Protein Fingerprinting by MALDI-TOF Mass Spectrometry | $ | $$$ | ★ | ★★ | ★ | ★ |
| Protein Fingerprinting by Surface Enhanced Raman Spectroscopy | $ | $$$ | ★^ | ★★ | ★ | ★ |
| Target-Specific Testing | ||||||
| Multiplexed qPCR | $$ | $$$ | ★★★ | ★★ | ★★★ | ★★ |
| Digital PCR | $$ | $$ | ★★★ | ★★ | ★★★ | ★★★ |
| Microarray | $$ | $$ | Rely on PCR | ★★ | ★★ | ★★★ |
| PNA-FISH | $$ | $$$ | ★ | ★ | ★ | ★★ |
single cell being demonstrated, but not reliable
More stars suggest higher accuracy, higher sensitivity, shorter turnaround time and higher multiplexing capability
Based on the current development of molecular techniques, there are still obvious limitations indicating that solely relying on these methods may not be sufficient for disease diagnosis, prognosis, and treatment. Using combinations of genomic and/or proteomic-based methods may be an alternative to overcome these limitations. Eventually, bringing together the power of pathogen identification and host markers assays will be the most definitive way to manage polymicrobial infections. In the meantime, molecular methods like sequencing, PCR, and MS are steadily replacing conventional methods, and research has demonstrated that they might become the mainstream infectious disease diagnostic technique in the future.
Tab. 1.
Common sample types for BSI molecule diagnostic technologies
| Common Sample Type |
|||
|---|---|---|---|
| Primary Blood Sample | Blood Culture | Bacteria Isolate | |
| Low Throughput Sequencing | √# | √ | |
| High Throughput sequencing | √# | √ | √ |
| Genetic Fingerprinting HRM | √# | √ | √ |
| Mass Spectrometry | √# | √ | √ |
| Protein Fingerprinting | √ | √ | |
| Raman Spectroscopy | √ | ||
| Multiplex qPCR | √ | √ | |
| digital PCR | √ | ||
| Microarray | √ | ||
| PNA-FISH | √ | ||
Rely on pre-enrichment by PCR
The challenges in diagnosing polymicrobial bloodstream infections is to correctly identify all microorganisms with low loading.
Various existing molecular diagnostics methods have limited capability in diagnosing polymicrobial infection diagnostics.
One potential promising solution to polymicrobial infection diagnostics is to apply digitization to existing molecule diagnostics.
Acknowledgement
The authors would like to thank the funding support from the National Institute of Health (NIH/NIAID R01AI117032), the startup grant of Nanyang Technological University, Singapore Ministry of Education Tier 1 (RG49/17), and the HealthTech NTU-LKCMedicine-NHG Point of Care Technology for Infectious Diseases Grant (ID POCT/17001).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References:
- Afshari A, Schrenzel J, Ieven M, Harbarth S, 2012. Bench-to-bedside review: Rapid molecular diagnostics for bloodstream infection-a new frontier? Crit. Care 16(3), 222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen HK, Donato J, Wang HH, Cloud-Hansen KA, Davies J, Handelsman J, 2010. Call of the wild: antibiotic resistance genes in natural environments. Nat. Rev. Microbiol 8(4), 251–259. [DOI] [PubMed] [Google Scholar]
- Alm B, Erdes L, Möllborg P, Pettersson R, Norvenius SG, Aberg N, Wennergren G, 2008. Neonatal antibiotic treatment is a risk factor for early wheezing. Pediatrics 121(4), 697–702. [DOI] [PubMed] [Google Scholar]
- Almeida C, Azevedo NF, Santos S, Keevil CW, Vieira MJ, 2011. Discriminating multispecies populations in biofilms with peptide nucleic acid fluorescence in situ hybridization (PNA FISH). PLoS One 6(3), e14786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altun O, Almuhayawi M, Ullberg M, Özenci V, 2013. Clinical evaluation of the FilmArray blood culture identification panel in identification of bacteria and yeasts from positive blood culture bottles. J. Clin. Microbiol 51(12), 4130–4136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andini N, Hu A, Zhou L, Cogill S, Wang T-H, Wittwer CT, Yang S, 2018. A “Culture” Shift: Broad Bacterial Detection, Identification, and Antimicrobial Susceptibility Testing Directly from Whole Blood. Clin. Chem., clinchem 2018.290189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andini N, Wang B, Athamanolap P, Hardick J, Masek BJ, Thair S, Hu A, Avornu G, Peterson S, Cogill S, Rothman RE, RCarroll KC, Gaydos CA, Wang JT, Batzoglous S, Yang S, 2017. Microbial typing by machine learned DNA melt signatures. Sci. Rep 7, 42097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angus DC, 2010. The lingering consequences of sepsis: a hidden public health disaster? JAMA 304(16), 1833–1834. [DOI] [PubMed] [Google Scholar]
- Anis E, Hawkins IK, Ilha MRS, Woldemeskel MW, Saliki JT, Wilkes RP, 2018. Evaluation of Targeted Next-Generation Sequencing for Detection of Bovine Pathogens in Clinical Samples. J. Clin. Microbiol 56(7), e00399–00318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Athamanolap P, Parekh V, Fraley SI, Agarwal V, Shin DJ, Jacobs MA, Wang T-H, Yang S, 2014. Trainable high resolution melt curve machine learning classifier for large-scale reliable genotyping of sequence variants. PLoS One 9(10), e109094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balážová T, Makovcová J, Šedo O, Slaný M, Faldyna M, Zdráhal Z, 2014. The influence of culture conditions on the identification of Mycobacterium species by MALDI-TOF MS profiling. FEMS Microbiol. Lett 353(1), 77–84. [DOI] [PubMed] [Google Scholar]
- Ballarini A, Segata N, Huttenhower C, Jousson O, 2013. Simultaneous quantification of multiple bacteria by the BactoChip microarray designed to target species-specific marker genes. PLoS One 8(2), e55764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bard JD, Lee F, 2018. Why Can’t We Just Use PCR? The Role of Genotypic versus Phenotypic Testing for Antimicrobial Resistance Testing. Clin. Microbiol. Newsl 40(11), 87–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beal SG, Ciurca J, Smith G, John J, Lee F, Doern CD, Gander RM, 2013. Evaluation of the nanosphere verigene gram-positive blood culture assay with the VersaTREK blood culture system and assessment of possible impact on selected patients. J. Clin. Microbiol 51(12), 3988–3992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beekmann SE, Diekema DJ, Chapin KC, Doern GV, 2003. Effects of rapid detection of bloodstream infections on length of hospitalization and hospital charges. J. Clin. Microbiol 41(7), 3119–3125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benagli C, Rossi V, Dolina M, Tonolla M, Petrini O, 2011. Matrix-assisted laser desorption ionization-time of flight mass spectrometry for the identification of clinically relevant bacteria. PLoS One 6(1), e16424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beuving J, Verbon A, Gronthoud FA, Stobberingh EE, Wolffs PFG, 2011. Antibiotic Susceptibility Testing of Grown Blood Cultures by Combining Culture and Real-Time Polymerase Chain Reaction Is Rapid and Effective. PLoS One 6(12). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beuving JJ, Wolffs PP, Hansen WWLJ, Stobberingh EE, Bruggeman CA, Kessels AGH, Verbon A, 2015. Impact of same-day antibiotic susceptibility testing on time to appropriate antibiotic treatment of patients with bacteraemia: a randomised controlled trial. Eur. J. Clin. Microbiol. Infect. Dis 34(4), 831–838. [DOI] [PubMed] [Google Scholar]
- Bhamla MS, Benson B, Chai C, Katsikis G, Johri A, Prakash M, 2017. Hand-powered ultralow-cost paper centrifuge. Nat. Biomed. Eng 1, 0009. [Google Scholar]
- Boardman AK, Wong WS, Premasiri WR, Ziegler LD, Lee JC, Miljkovic M, Klapperich CM, Sharon A, Sauer-Budge AF, 2016. Rapid detection of bacteria from blood with surface-enhanced Raman spectroscopy. Anal. Chem 88(16), 8026–8035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodey GP, Nies BA, Freireich EJ, 1965. Multiple organism septicemia in acute leukemia: analysis of 54 episodes. Arch. Intern. Med 116(2), 266–272. [DOI] [PubMed] [Google Scholar]
- Bork JT, Leekha S, Heil EL, Zhao L, Badamas R, Johnson JK, 2015. Rapid Testing Using the Verigene Gram-Negative Blood Culture Nucleic Acid Test in Combination with Antimicrobial Stewardship Intervention against Gram-Negative Bacteremia. Antimicrob. Agents Chemother 59(3), 1588–1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boucher HW, Talbot GH, Bradley JS, Edwards JE, Gilbert D, Rice LB, Scheld M, Spellberg B, Bartlett J, 2009. Bad bugs, no drugs: no ESKAPE! An update from the Infectious Diseases Society of America. Clin. Infect. Dis 48(1), 1–12. [DOI] [PubMed] [Google Scholar]
- Brown J, Theis L, Kerr L, Zakhidova N, O’Connor K, Uthman M, Oden ZM, Richards-Kortum R, 2011. A hand-powered, portable, low-cost centrifuge for diagnosing anemia in low-resource settings. Am. J. Trop. Med. Hyg 85(2), 327–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchan BW, Ledeboer NA, 2014. Emerging technologies for the clinical microbiology laboratory. Clin. Microbiol. Rev 27(4), 783–822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calderaro A, Martinelli M, Motta F, Larini S, Arcangeletti M, Medici M, Chezzi C, De Conto F, 2014. Comparison of peptide nucleic acid fluorescence in situ hybridization assays with culture-based matrix-assisted laser desorption/ionization-time of flight mass spectrometry for the identification of bacteria and yeasts from blood cultures and cerebrospinal fluid cultures. Clin. Microbiol. Infect 20(8), O468–O475. [DOI] [PubMed] [Google Scholar]
- Campion A, Kambhampati P, 1998. Surface-enhanced Raman scattering. Chem. Soc. Rev 27(4), 241–250. [Google Scholar]
- Carrilho E, Martinez AW, Whitesides GM, 2009. Understanding wax printing: a simple micropatterning process for paper-based microfluidics. Anal. Chem 81(16), 7091–7095. [DOI] [PubMed] [Google Scholar]
- Casalta J, Gouriet F, Roux V, Thuny F, Habib G, Raoult D, 2009. Evaluation of the LightCycler® SeptiFast test in the rapid etiologic diagnostic of infectious endocarditis. Eur. J. Clin. Microbiol. Infect. Dis 28(6), 569–573. [DOI] [PubMed] [Google Scholar]
- Cepheid, Xpert Clinical IVD Tests. http://www.cepheid.com/us/cepheid-solutions/clinical-ivd-tests/healthcare-associated-infections.
- Chen JH, Ho P-L, Kwan GS, She KK, Siu GK, Cheng VC, Yuen K-Y, Yam W-C, 2013. Direct bacterial identification in positive blood cultures by use of two commercial matrix-assisted laser desorption ionization–time of flight mass spectrometry systems. J. Clin. Microbiol 51(6), 1733–1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin CD, Linder V, Sia SK, 2007. Lab-on-a-chip devices for global health: Past studies and future opportunities. Lab Chip 7(1), 41–57. [DOI] [PubMed] [Google Scholar]
- Clarridge JE, 2004. Impact of 16S rRNA gene sequence analysis for identification of bacteria on clinical microbiology and infectious diseases. Clin. Microbiol. Rev 17(4), 840–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole JR, Wang Q, Cardenas E, Fish J, Chai B, Farris RJ, Kulam-Syed-Mohideen A, McGarrell DM, Marsh T, Garrity GM, 2008. The Ribosomal Database Project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res. 37(suppl_1), D141–D145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole JR, Wang Q, Fish JA, Chai B, McGarrell DM, Sun Y, Brown CT, Porras-Alfaro A, Kuske CR, Tiedje JM, 2013. Ribosomal Database Project: data and tools for high throughput rRNA analysis. Nucleic Acids Res. 42(D1), D633–D642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conlan S, Kong HH, Segre JA, 2012. Species-level analysis of DNA sequence data from the NIH Human Microbiome Project. PLoS One 7(10), e47075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper GS, Havlir DS, Shlaes DM, Salata RA, 1990. Polymicrobial bacteremia in the late 1980s: predictors of outcome and review of the literature. Medicine 69(2), 114–123. [PubMed] [Google Scholar]
- Corless CE, Guiver M, Borrow R, Edwards-Jones V, Kaczmarski EB, Fox AJ, 2000. Contamination and sensitivity issues with a real-time universal 16S rRNA PCR. J. Clin. Microbiol 38(5), 1747–1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotten CM, Taylor S, Stoll B, Goldberg RN, Hansen NI, Sánchez PJ, Ambalavanan N, Benjamin DK, Network NNR, 2009. Prolonged duration of initial empirical antibiotic treatment is associated with increased rates of necrotizing enterocolitis and death for extremely low birth weight infants. Pediatrics 123(1), 58–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels R, 2011. Surviving the first hours in sepsis: getting the basics right (an intensivist’s perspective). J. Antimicrob. Chemother 66(suppl_2), ii11–ii23. [DOI] [PubMed] [Google Scholar]
- Deiss F, Funes-Huacca ME, Bal J, Tjhung KF, Derda R, 2014. Antimicrobial susceptibility assays in paper-based portable culture devices. Lab Chip 14(1), 167–171. [DOI] [PubMed] [Google Scholar]
- Dewey FE, Pan S, Wheeler MT, Quake SR, Ashley EA, 2012. DNA sequencing: clinical applications of new DNA sequencing technologies. Circulation 125(7), 931–944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dierkes C, Ehrenstein B, Siebig S, Linde HJ, Reischl U, Salzberger B, 2009. Clinical impact of a commercially available multiplex PCR system for rapid detection of pathogens in patients with presumed sepsis. BMC Infect. Dis 9, 126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dina NE, Colniță A, Leopold N, Haisch C, 2017. Rapid single-cell detection and identification of bacteria by using surface-enhanced Raman spectroscopy. Proc. Technol 27, 203–207. [DOI] [PubMed] [Google Scholar]
- Dodémont M, De Mendonça R, Nonhoff C, Roisin S, Denis O, 2014. Performance of the Verigene Gram-negative blood culture assay for rapid detection of bacteria and resistance determinants. J. Clin. Microbiol 52(8), 3085–3087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downes KJ, Metlay JP, Bell LM, McGowan KL, Elliott MR, Shah SS, 2008. Polymicrobial bloodstream infections among children and adolescents with central venous catheters evaluated in ambulatory care. Clin. Infect. Dis 46(3), 387–394. [DOI] [PubMed] [Google Scholar]
- Dubin K, Callahan MK, Ren B, Khanin R, Viale A, Ling L, No D, Gobourne A, Littmann E, Huttenhower C, 2016. Intestinal microbiome analyses identify melanoma patients at risk for checkpoint-blockade-induced colitis. Nat. Commons 7, 10391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dungchai W, Chailapakul O, Henry CS, 2011. A low-cost, simple, and rapid fabrication method for paper-based microfluidics using wax screen-printing. Analyst 136(1), 77–82. [DOI] [PubMed] [Google Scholar]
- Ecker DJ, Sampath R, Blyn LB, Eshoo MW, Ivy C, Ecker JA, Libby B, Samant V, Sannes-Lowery KA, Melton RE, 2005. Rapid identification and strain-typing of respiratory pathogens for epidemic surveillance. Proceedings of the National Academy of Sciences 102(22), 8012–8017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ecker DJ, Sampath R, Massire C, Blyn LB, Hall TA, Eshoo MW, Hofstadler SA, 2008. Ibis T5000: a universal biosensor approach for microbiology. Nat. Rev. Microbiol 6(7), 553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Aneed A, Cohen A, Banoub J, 2009. Mass spectrometry, review of the basics: electrospray, MALDI, and commonly used mass analyzers. Applied Spectroscopy Reviews 44(3), 210–230. [Google Scholar]
- El-Hajj HH, Marras SA, Tyagi S, Shashkina E, Kamboj M, Kiehn TE, Glickman MS, Kramer FR, Alland D, 2009. Use of sloppy molecular beacon probes for identification of mycobacterial species. J. Clin. Microbiol 47(4), 1190–1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elting LS, Bodey GP, Fainstein V, 1986. Polymicrobial septicemia in the cancer patient. Medicine 65(4), 218–225. [DOI] [PubMed] [Google Scholar]
- Evans SR, Hujer AM, Jiang H, Hill C, Hujer KM, Mediavilla JR, Manca C, Tran TTT, Domitrovic TN, Higgins PG, 2017. Informing Antibiotic Treatment Decisions: Evaluating Rapid Molecular Diagnostics To Identify Susceptibility and Resistance to Carbapenems against Acinetobacter spp. in PRIMERS III. J. Clin. Microbiol 55(1), 134–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans SR, Hujer AM, Jiang H, Hujer KM, Hall TA, Marzan C, Jacobs MR, Sampath R, Ecker DJ, Manca C, 2016. Rapid Molecular Diagnostics, Antibiotic Treatment Decisions, and Developing Approaches to Inform Empiric Therapy: PRIMERS I and II. Clin. Infect. Dis 62(2), 181–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fanaroff AA, Korones SB, Wright LL, Wright EC, Poland RL, Bauer CB, Tyson JE, Philips JB, Edwards W, Lucey JF, 1994. A controlled trial of intravenous immune globulin to reduce nosocomial infections in very-low-birth-weight infants. New Engl. J. Med 330(16), 1107–1113. [DOI] [PubMed] [Google Scholar]
- Fenollar F, Raoult D, 2007. Molecular diagnosis of bloodstream infections caused by non-cultivable bacteria. Int. J. Antimicrob. Agents 30, 7–15. [DOI] [PubMed] [Google Scholar]
- Fiori B, D’Inzeo T, Giaquinto A, Menchinelli G, Liotti F, De Maio F, De Angelis G, Quaranta G, Nagel D, Tumbarello M, 2016. Optimized use of the MALDI BioTyper system and the FilmArray BCID panel for direct identification of microbial pathogens from positive blood cultures. J. Clin. Microbiol 54(3), 576–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraley SI, Athamanolap P, Masek BJ, Hardick J, Carroll KC, Hsieh Y-H, Rothman RE, Gaydos CA, Wang T-H, Yang S, 2016. Nested Machine Learning Facilitates Increased Sequence Content for Large-Scale Automated High Resolution Melt Genotyping. Sci. Rep 6, 19218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraley SI, Hardick J, Jo Masek B, Athamanolap P, Rothman RE, Gaydos CA, Carroll KC, Wakefield T, Wang T-H, Yang S, 2013. Universal digital high-resolution melt: a novel approach to broad-based profiling of heterogeneous biological samples. Nucleic Acids Res. 41(18), e175–e175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frickmann H, Zautner AE, Moter A, Kikhney J, Hagen RM, Stender H, Poppert S, 2017. Fluorescence in situ hybridization (FISH) in the microbiological diagnostic routine laboratory: a review. Crit. Rev. Microbiol 43(3), 263–293. [DOI] [PubMed] [Google Scholar]
- García P, Benítez R, Lam M, Salinas AM, Wirth H, Espinoza C, Garay T, Depix MS, Labarca J, Guzmán AM, 2004. Coagulase-negative staphylococci: clinical, microbiological and molecular features to predict true bacteraemia. J. Med. Microbiol 53(1), 67–72. [DOI] [PubMed] [Google Scholar]
- GenMark, Blood Culture Identification Panels. https://www.genmarkdx.com/solutions/panels/eplex-panels/eplex-pipeline/eplex-blood-culture-identification-panels/.
- Gorkin R, Park J, Siegrist J, Amasia M, Lee BS, Park J-M, Kim J, Kim H, Madou M, Cho Y-K, 2010. Centrifugal microfluidics for biomedical applications. Lab Chip 10(14), 1758–1773. [DOI] [PubMed] [Google Scholar]
- Grumaz S, Stevens P, Grumaz C, Decker SO, Weigand MA, Hofer S, Brenner T, von Haeseler A, Sohn K, 2016. Next-generation sequencing diagnostics of bacteremia in septic patients. Genome Med. 8(1), 73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerendiain D, MacKenzie L, Templeton KE, 2016. Evaluation of point of care testing platform (ePLEX) for respiratory viral diagnosis. J. Clin. Virol 82, S120. [Google Scholar]
- Gürtler V, Stanisich VA, 1996. New approaches to typing and identification of bacteria using the 16S-23S rDNA spacer region. Microbiol. 142(1), 3–16. [DOI] [PubMed] [Google Scholar]
- Gyarmati P, Kjellander C, Aust C, Kalin M, Öhrmalm L, Giske CG, 2015. Bacterial landscape of bloodstream infections in neutropenic patients via high throughput sequencing. PLoS One 10(8), e0135756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall KK, Lyman JA, 2006. Updated review of blood culture contamination. Clin. Microbiol. Rev 19(4), 788–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harbarth S, Garbino J, Pugin J, Romand JA, Lew D, Pittet D, 2003. Inappropriate initial antimicrobial therapy and its effect on survival in a clinical trial of immunomodulating therapy for severe sepsis. Am. J. Med 115(7), 529–535. [DOI] [PubMed] [Google Scholar]
- Hardick J, Won H, Jeng K, Hsieh Y-H, Gaydos CA, Rothman RE, Yang S, 2012. Identification of Bacterial Pathogens in Ascitic Fluids from Patients with Suspected Spontaneous Bacterial Peritonitis by Use of Broad-Based PCR (16S PCR) Coupled with High-Resolution Melt Analysis (HRMA). J. Clin. Microbiol., JCM 00345–00312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris DM, Hata DJ, 2013. Rapid identification of bacteria and candida using pna-fish from blood and peritoneal fluid cultures: a retrospective clinical study. Ann. Clin. Microbiol. Antimicrob 12(1), 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasman H, Saputra D, Sicheritz-Ponten T, Lund O, Svendsen CA, Frimodt-Moller N, Aarestrup FM, 2014. Rapid whole-genome sequencing for detection and characterization of microorganisms directly from clinical samples. J. Clin. Microbiol 52(1), 139–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y, Zeng K, Zhang S, Gurung AS, Baloda M, Zhang X, Liu G, 2012. Visual detection of gene mutations based on isothermal strand-displacement polymerase reaction and lateral flow strip. Biosens. Bioelectron 31(1), 310–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y, Zhang S, Zhang X, Baloda M, Gurung AS, Xu H, Zhang X, Liu G, 2011. Ultrasensitive nucleic acid biosensor based on enzyme–gold nanoparticle dual label and lateral flow strip biosensor. Biosens. Bioelectron 26(5), 2018–2024. [DOI] [PubMed] [Google Scholar]
- Hermans PE, Washington JA, 1970. Polymicrobial bacteremia. Ann. Intern. Med 73(3), 387–392. [DOI] [PubMed] [Google Scholar]
- Hindson BJ, Ness KD, Masquelier DA, Belgrader P, Heredia NJ, Makarewicz AJ, Bright IJ, Lucero MY, Hiddessen AL, Legler TC, Kitano TK, Hodel MR, Petersen JF, Wyatt PW, Steenblock ER, Shah PH, Bousse LJ, Troup CB, Mellen JC, Wittmann DK, Erndt NG, Cauley TH, Koehler RT, So AP, Dube S, Rose KA, Montesclaros L, Wang S, Stumbo DP, Hodges SP, Romine S, Milanovich FP, White HE, Regan JF, Karlin-Neumann GA, Hindson CM, Saxonov S, Colston BW, 2011. High-throughput droplet digital PCR system for absolute quantitation of DNA copy number. Anal. Chem 83(22), 8604–8610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochstein HD, Kirkham WR, Young VM, 1965. Recovery of more than 1 organism in septicemias. New Engl. J. Med 273(9), 468–474. [DOI] [PubMed] [Google Scholar]
- Hrabák J, Chudáčková E, Walková R, 2013. Matrix-assisted laser desorption ionization–time of flight (MALDI-TOF) mass spectrometry for detection of antibiotic resistance mechanisms: from research to routine diagnosis. Clin. Microbiol. Rev 26(1), 103–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang T-D, Melnik E, Bogaerts P, Evrard S, Glupczynski Y, 2019. Evaluation of the ePlex Blood Culture Identification Panels for Detection of Pathogens in Bloodstream Infections. J. Clin. Microbiol 57(2), e01597–01518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huttenhower C, Gevers D, Knight R, Abubucker S, Badger JH, Chinwalla AT, Creasy HH, Earl AM, FitzGerald MG, Fulton RS, 2012. Structure, function and diversity of the healthy human microbiome. Nature 486(7402), 207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ing A, McLean A, Meakins J, 1981. Multiple-organism bacteremia in the surgical intensive care unit: a sign of intraperitoneal sepsis. Surgery 90(4), 779–786. [PubMed] [Google Scholar]
- Jacob D, Sauer U, Housley R, Washington C, Sannes-Lowery K, Ecker DJ, Sampath R, Grunow R, 2012. Rapid and high-throughput detection of highly pathogenic bacteria by Ibis PLEX-ID technology. PLoS One 7(6), e39928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janda JM, Abbott SL, 2007. 16S rRNA gene sequencing for bacterial identification in the diagnostic laboratory: pluses, perils, and pitfalls. J. Clin. Microbiol 45(9), 2761–2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvis RM, Goodacre R, 2004. Discrimination of bacteria using surface-enhanced Raman spectroscopy. Anal. Chem 76(1), 40–47. [DOI] [PubMed] [Google Scholar]
- Jeng K, Gaydos CA, Blyn LB, Yang S, Won H, Matthews H, Toleno D, Hsieh YH, Carroll KC, Hardick J, Masek B, Kecojevic A, Sampath R, Peterson S, Rothman RE, 2012. Comparative analysis of two broad-range PCR assays for pathogen detection in positive-blood-culture bottles: PCR-high-resolution melting analysis versus PCR-mass spectrometry. J. Clin. Microbiol 50(10), 3287–3292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeng K, Yang S, Won H, Gaydos CA, Hsieh Y-H, Kecojevic A, Carroll KC, Hardick J, Rothman RE, 2012. Application of a 16S rRNA PCR–high-resolution melt analysis assay for rapid detection of salmonella bacteremia. J. Clin. Microbiol 50(3), 1122–1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnsen BO, Handal N, Meisal R, Bjornholt JV, Gaustad P, Leegaard TM, 2017. erm gene distribution among Norwegian Bacteroides isolates and evaluation of phenotypic tests to detect inducible clindamycin resistance in Bacteroides species. Anaerobe 47, 226–232. [DOI] [PubMed] [Google Scholar]
- Jordana-Lluch E, Gimenez M, Quesada MD, Ausina V, Martro E, 2014. Improving the diagnosis of bloodstream infections: PCR coupled with mass spectrometry. Biomed. Res. Int 2014, 501214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jovel J, Patterson J, Wang W, Hotte N, O’Keefe S, Mitchel T, Perry T, Kao D, Mason AL, Madsen KL, 2016. Characterization of the gut microbiome using 16S or shotgun metagenomics. Front. Microbiol 7, 459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaleta EJ, Clark AE, Johnson DR, Gamage DC, Wysocki VH, Cherkaoui A, Schrenzel J, Wolk DM, 2011a. Use of PCR coupled with electrospray ionization mass spectrometry for rapid identification of bacterial and yeast bloodstream pathogens from blood culture bottles. J. Clin. Microbiol 49(1), 345–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaleta EJ, Clark AE, Wysocki VH, Ingram EL, Schrenzel J, Wolk DM, 2011b. Comparative analysis of PCR–electrospray ionization/mass spectrometry (MS) and MALDI-TOF/MS for the identification of bacteria and yeast from positive blood culture bottles. Clin. Chem., clinchem 2011161968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khazaei T, Barlow JT, Schoepp NG, Ismagilov RF, 2018. RNA markers enable phenotypic test of antibiotic susceptibility in Neisseria gonorrhoeae after 10 minutes of ciprofloxacin exposure. Sci. Rep 8(1), 11606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiani D, Quinn EL, Burch KH, Madhavan T, Saravolatz LD, Neblett TR, 1979. The increasing importance of polymicrobial bacteremia. JAMA 242(10), 1044–1047. [PubMed] [Google Scholar]
- Kommedal O, Kvello K, Skjastad R, Langeland N, Wiker HG, 2009. Direct 16S rRNA gene sequencing from clinical specimens, with special focus on polybacterial samples and interpretation of mixed DNA chromatograms. J. Clin. Microbiol 47(11), 3562–3568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kothari A, Morgan M, Haake DA, 2014. Emerging technologies for rapid identification of bloodstream pathogens. Clin. Infect. Dis 59(2), 272–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A, Ellis P, Arabi Y, Roberts D, Light B, Parrillo JE, Dodek P, Wood G, Kumar A, Simon D, Peters C, Ahsan M, Chateau D, Initiation of Inappropriate Antimicrobial Therapy Results in a Fivefold Reduction of Survival in Human Septic Shock. CHEST 136(5), 1237–1248. [DOI] [PubMed] [Google Scholar]
- Kumar A, Roberts D, Wood KE, Light B, Parrillo JE, Sharma S, Suppes R, Feinstein D, Zanotti S, Taiberg L, Gurka D, Kumar A, Cheang M, 2006. Duration of hypotension before initiation of effective antimicrobial therapy is the critical determinant of survival in human septic shock. Crit. Care Med 34(6), 1589–1596. [DOI] [PubMed] [Google Scholar]
- La Scola B, 2011. Intact cell MALDI-TOF mass spectrometry-based approaches for the diagnosis of bloodstream infections. Expert Rev. Mol. Diagn 11(3), 287–298. [DOI] [PubMed] [Google Scholar]
- Laffler TG, Cummins LL, McClain CM, Quinn CD, Toro MA, Carolan HE, Toleno DM, Rounds MA, Eshoo M, Stratton CW, 2013. Enhanced diagnostic yields of bacteremia and candidemia in blood specimens by PCR/electrospray ionization mass spectrometry. J. Clin. Microbiol., JCM 00876–00813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagacé-Wiens PR, Adam HJ, Karlowsky JA, Nichol KA, Pang PF, Guenther J, Webb AA, Miller C, Alfa MJ, 2012. Identification of blood culture isolates directly from positive blood cultures by use of matrix-assisted laser desorption ionization–time of flight mass spectrometry and a commercial extraction system: analysis of performance, cost, and turnaround time. J. Clin. Microbiol 50(10), 3324–3328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam B, Das J, Holmes RD, Live L, Sage A, Sargent EH, Kelley SO, 2013. Solution-based circuits enable rapid and multiplexed pathogen detection. Nat. Commons 4, 2001. [DOI] [PubMed] [Google Scholar]
- Langille MG, Zaneveld J, Caporaso JG, McDonald D, Knights D, Reyes JA, Clemente JC, Burkepile DE, Thurber RLV, Knight R, 2013. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat. Biotechnol 31(9), 814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Layeghifard M, Hwang DM, Guttman DS, 2017. Disentangling interactions in the microbiome: a network perspective. Trends Microbiol. 25(3), 217–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ledeboer NA, Lopansri BK, Dhiman N, Cavagnolo R, Carroll KC, Granato P, Thomson R, Butler-Wu SM, Berger H, Samuel L, 2015. Identification of Gram-negative bacteria and genetic resistance determinants from positive blood culture broths by use of the Verigene Gram-negative blood culture multiplex microarray-based molecular assay. J. Clin. Microbiol 53(8), 2460–2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann LE, Hunfeld KP, Emrich T, Haberhausen G, Wissing H, Hoeft A, Stuber F, 2008. A multiplex real-time PCR assay for rapid detection and differentiation of 25 bacterial and fungal pathogens from whole blood samples. Med. Microbiol. Immunol 197(3), 313–324. [DOI] [PubMed] [Google Scholar]
- Liderot K, Larsson M, Borang S, Ozenci V, 2010. Polymicrobial bloodstream infection with Eggerthella lenta and Desulfovibrio desulfuricans. J. Clin. Microbiol. 48(10), 3810–3812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loonen AJ, Wolffs PF, Bruggeman CA, van den Brule AJ, 2014. Developments for improved diagnosis of bacterial bloodstream infections. Eur. J. Clin. Microbiol. Infect. Dis 33(10), 1687–1702. [DOI] [PubMed] [Google Scholar]
- Ly T, Gulia J, Pyrgos V, Waga M, Shoham S, 2008. Impact upon clinical outcomes of translation of PNA FISH-generated laboratory data from the clinical microbiology bench to bedside in real time. Ther. Clin. Risk Manag 4(3), 637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mach KE, Du CB, Phull H, Haake DA, Shih M-C, Baron EJ, Liao JC, 2009. Multiplex pathogen identification for polymicrobial urinary tract infections using biosensor technology: a prospective clinical study. J. Urol 182(6), 2735–2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackowiak PA, Browne RH, Southern PM Jr, Smith JW, 1980. Polymicrobial sepsis: an analysis of 184 cases using log linear models. Am. J. Med. Sci 280(2), 73–80. [DOI] [PubMed] [Google Scholar]
- Mancini N, Carletti S, Ghidoli N, Cichero P, Burioni R, Clementi M, 2010. The era of molecular and other non-culture-based methods in diagnosis of sepsis. Clin. Microbiol. Rev 23(1), 235–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancini N, Carletti S, Ghidoli N, Cichero P, Ossi CM, Ieri R, Poli E, Burioni R, Clementi M, 2009. Molecular diagnosis of polymicrobial sepsis. J. Clin. Microbiol 47(4), 1274–1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancini N, Infurnari L, Ghidoli N, Valzano G, Clementi N, Burioni R, Clementi M, 2014. Potential impact of a microarray-based nucleic acid assay for rapid detection of Gram-negative bacteria and resistance markers in positive blood cultures. J. Clin. Microbiol 52(4), 1242–1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez JL, 2008. Antibiotics and antibiotic resistance genes in natural environments. Science 321(5887), 365–367. [DOI] [PubMed] [Google Scholar]
- Masek BJ, Hardick J, Won H, Yang S, Hsieh Y-H, Rothman RE, Gaydos CA, 2014. Sensitive detection and serovar differentiation of typhoidal and nontyphoidal Salmonella enterica species using 16S rRNA Gene PCR coupled with high-resolution melt analysis. J. Mol. Diagn 16(2), 261–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mengelle C, Mansuy J-M, Da Silva I, Guerin J-L, Izopet J, 2013. Evaluation of a polymerase chain reaction–electrospray ionization time-of-flight mass spectrometry for the detection and subtyping of influenza viruses in respiratory specimens. J. Clin. Virol 57(3), 222–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzgar D, Baynes D, Myers CA, Kammerer P, Unabia M, Faix DJ, Blair PJ, 2010. Initial identification and characterization of an emerging zoonotic influenza virus prior to pandemic spread. J. Clin. Microbiol 48(11), 4228–4234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mignard S, Flandrois JP, 2006. 16S rRNA sequencing in routine bacterial identification: A 30-month experiment. Journal of Microbiological Methods 67(3), 574–581. [DOI] [PubMed] [Google Scholar]
- Munson EL, Diekema DJ, Beekmann SE, Chapin KC, Doern GV, 2003. Detection and treatment of bloodstream infection: laboratory reporting and antimicrobial management. J. Clin. Microbiol 41(1), 495–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oyola SO, Otto TD, Gu Y, Maslen G, Manske M, Campino S, Turner DJ, MacInnis B, Kwiatkowski DP, Swerdlow HP, 2012. Optimizing Illumina next-generation sequencing library preparation for extremely AT-biased genomes. BMC Genomics 13(1), 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Özenci V, Patel R, Ullberg M, Strålin K, 2017. Demise of polymerase chain reaction/electrospray ionization-mass spectrometry as an infectious diseases diagnostic tool. Clin. Infect. Dis 66(3), 452–455. [DOI] [PubMed] [Google Scholar]
- Pammi M, Zhong D, Johnson Y, Revell P, Versalovic J, 2014. Polymicrobial bloodstream infections in the neonatal intensive care unit are associated with increased mortality: a case-control study. BMC Infect. Dis 14(1), 390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S, Zhang Y, Lin S, Wang T-H, Yang S, 2011. Advances in microfluidic PCR for point-of-care infectious disease diagnostics. Biotechnol. Adv 29(6), 830–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlaki M, Poulakou G, Drimousis P, Adamis G, Apostolidou E, Gatselis NK, Kritselis I, Mega A, Mylona V, Papatsoris A, Pappas A, Prekates A, Raftogiannis M, Rigaki K, Sereti K, Sinapidis D, Tsangaris I, Tzanetakou V, Veldekis D, Mandragos K, Giamarellou H, Dimopoulos G, 2013. Polymicrobial bloodstream infections: Epidemiology and impact on mortality. J. Glob. Antimicrob. Re 1(4), 207–212. [DOI] [PubMed] [Google Scholar]
- Peleg A, Tilahun Y, Fiandaca M, D’Agata E, Venkataraman L, Moellering R, Eliopoulos G, 2009. Utility of peptide nucleic acid fluorescence in situ hybridization for rapid detection of Acinetobacter spp. and Pseudomonas aeruginosa. J. Clin. Microbiol 47(3), 830–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters BM, Jabra-Rizk MA, Graeme A, Costerton JW, Shirtliff ME, 2012. Polymicrobial interactions: impact on pathogenesis and human disease. Clin. Microbiol. Rev 25(1), 193–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierce SE, Bell RL, Hellberg RS, Cheng C-M, Chen K-S, Williams-Hill DM, Martin WB, Allard MW, 2012. Detection and Identification of Salmonella enterica, Escherichia coli, and Shigella spp. via PCR-ESI-MS: Isolate Testing and Analysis of Food Samples. Appl. Environ. Microbiol., AEM 02272–02212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poole CB, Li Z, Alhassan A, Guelig D, Diesburg S, Tanner NA, Zhang Y, Evans TC Jr, LaBarre P, Wanji S, 2017. Colorimetric tests for diagnosis of filarial infection and vector surveillance using non-instrumented nucleic acid loop-mediated isothermal amplification (NINA-LAMP). PLoS One 12(2), e0169011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Premasiri W, Chen Y, Williamson P, Bandarage D, Pyles C, Ziegler L, 2017. Rapid urinary tract infection diagnostics by surface-enhanced Raman spectroscopy (SERS): identification and antibiotic susceptibilities. Anal. Bioanal. Chem 409(11), 3043–3054. [DOI] [PubMed] [Google Scholar]
- Premasiri WR, Lee JC, Sauer-Budge A, Théberge R, Costello CE, Ziegler LD, 2016. The biochemical origins of the surface-enhanced Raman spectra of bacteria: a metabolomics profiling by SERS. Anal. Bioanal. Chem 408(17), 4631–4647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruesse E, Quast C, Knittel K, Fuchs BM, Ludwig W, Peplies J, Glöckner FO, 2007. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 35(21), 7188–7196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, Nielsen T, Pons N, Levenez F, Yamada T, 2010. A human gut microbial gene catalogue established by metagenomic sequencing. nature 464(7285), 59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin J, Li Y, Cai Z, Li S, Zhu J, Zhang F, Liang S, Zhang W, Guan Y, Shen D, 2012. A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature 490(7418), 55. [DOI] [PubMed] [Google Scholar]
- Quail MA, Otto TD, Gu Y, Harris SR, Skelly TF, McQuillan JA, Swerdlow HP, Oyola SO, 2012. Optimal enzymes for amplifying sequencing libraries. Nat. Methods 9(1), 10. [DOI] [PubMed] [Google Scholar]
- Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Peplies J, Glöckner FO, 2012. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 41(D1), D590–D596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranjan R, Rani A, Metwally A, McGee HS, Perkins DL, 2016. Analysis of the microbiome: advantages of whole genome shotgun versus 16S amplicon sequencing. Biochem. Biophys. Res. Commun 469(4), 967–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed GH, Kent JO, Wittwer CT, 2007. High-resolution DNA melting analysis for simple and efficient molecular diagnostics. Pharmacogenomics 8(6), 597–608. [DOI] [PubMed] [Google Scholar]
- Rello J, Quintana E, Mirelis B, Gurgui M, Net A, Prats G, 1993. Polymicrobial bacteremia in critically ill patients. Intensive Care Med. 19(1), 22–25. [DOI] [PubMed] [Google Scholar]
- Retamar P, Portillo MM, López-Prieto MD, Rodríguez-López F, De Cueto M, García MV, Gómez MJ, del Arco A, Muñoz A, Sánchez-Porto A, 2012. Impact of inadequate empirical therapy on the mortality of patients with bloodstream infections: a propensity score-based analysis. Antimicrob. Agents Chemother. 56(1), 472–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuben AG, Musher DM, Hamill RJ, Broucke I, 1989. Polymicrobial bacteremia: clinical and microbiologic patterns. Reviews of Infectious Diseases 11(2), 161–183. [DOI] [PubMed] [Google Scholar]
- Rhodes A, Evans LE, Alhazzani W, Levy MM, Antonelli M, Ferrer R, Kumar A, Sevransky JE, Sprung CL, Nunnally ME, Rochwerg B, Rubenfeld GD, Angus DC, Annane D, Beale RJ, Bellinghan GJ, Bernard GR, Chiche JD, Coopersmith C, De Backer DP, French CJ, Fujishima S, Gerlach H, Hidalgo JL, Hollenberg SM, Jones AE, Karnad DR, Kleinpell RM, Koh Y, Lisboa TC, Machado FR, Marini JJ, Marshall JC, Mazuski JE, McIntyre LA, McLean AS, Mehta S, Moreno RP, Myburgh J, Navalesi P, Nishida O, Osborn TM, Perner A, Plunkett CM, Ranieri M, Schorr CA, Seckel MA, Seymour CW, Shieh L, Shukri KA, Simpson SQ, Singer M, Thompson BT, Townsend SR, Van der Poll T, Vincent JL, Wiersinga WJ, Zimmerman JL, Dellinger RP, 2017. Surviving Sepsis Campaign: International Guidelines for Management of Sepsis and Septic Shock: 2016. Intensive Care Med. 43(3), 304–377. [DOI] [PubMed] [Google Scholar]
- Rickerts V, McCormick Smith I, Mousset S, Kommedal O, Fredricks DN, 2013. Deciphering the aetiology of a mixed fungal infection by broad-range PCR with sequencing and fluorescence in situ hybridisation. Mycoses 56(6), 681–686. [DOI] [PubMed] [Google Scholar]
- Roselle GA, Watanakunakorn C, 1979. Polymicrobial bacteremia. JAMA 242(22), 2411–2413. [PubMed] [Google Scholar]
- Saffert RT, Cunningham SA, Ihde SM, Jobe KEM, Mandrekar J, Patel R, 2011. Comparison of Bruker Biotyper matrix-assisted laser desorption ionization–time of flight mass spectrometer to BD Phoenix automated microbiology system for identification of gram-negative bacilli. J. Clin. Microbiol 49(3), 887–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampath R, Mulholland N, Blyn LB, Massire C, Whitehouse CA, Waybright N, Harter C, Bogan J, Miranda MS, Smith D, 2012. Comprehensive biothreat cluster identification by PCR/electrospray-ionization mass spectrometry. PLoS One 7(6), e36528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuel LP, Tibbetts RJ, Agotesku A, Fey M, Hensley R, Meier FA, 2013. Evaluation of a microarray-based assay for rapid identification of Gram-positive organisms and resistance markers in positive blood cultures. J. Clin. Microbiol 51(4), 1188–1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancho S, Artero A, Zaragoza R, Camarena J, González R, Nogueira J, 2012. Impact of nosocomial polymicrobial bloodstream infections on the outcome in critically ill patients. Eur. J. Clin. Microbiol. Infect. Dis 31(8), 1791–1796. [DOI] [PubMed] [Google Scholar]
- Schlücker S, 2014. Surface‐Enhanced raman spectroscopy: Concepts and chemical applications. Angew. Chem. Int. Ed 53(19), 4756–4795. [DOI] [PubMed] [Google Scholar]
- Seifert H, 2009. The clinical importance of microbiological findings in the diagnosis and management of bloodstream infections. Clin. Infect. Dis 48 Suppl 4, S238–245. [DOI] [PubMed] [Google Scholar]
- Shin D, Athamanolap P, Chen L, Hardick J, Lewis M, Hsieh Y, Rothman R, Gaydos C, Wang T, 2017. Mobile nucleic acid amplification testing (mobiNAAT) for Chlamydia trachomatis screening in hospital emergency department settings. Sci. Rep 7(1), 4495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simner PJ, Uhl JR, Hall L, Weber MM, Walchak RC, Buckwalter S, Wengenack NL, 2013. Broad-range direct detection and identification of fungi using the PLEX-ID PCRelectrospray ionization mass spectrometry (ESI-MS) system. J. Clin. Microbiol., JCM 03282–03212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snodgrass R, Gardner A, Semeere A, Kopparthy VL, Duru J, Maurer T, Martin J, Cesarman E, Erickson D, 2018. A portable device for nucleic acid quantification powered by sunlight, a flame or electricity. Nat. Biomed. Eng 2(9), 657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southern TR, VanSchooneveld TC, Bannister DL, Brown TL, Crismon AS, Buss SN, Iwen PC, Fey PD, 2015. Implementation and performance of the BioFire FilmArray® Blood Culture Identification panel with antimicrobial treatment recommendations for bloodstream infections at a midwestern academic tertiary hospital. Diagnostic Microbiology and Infectious Disease 81(2), 96–101. [DOI] [PubMed] [Google Scholar]
- Stender H, Fiandaca MJ, Hyldignielsen JJ, Coull JM, 2002. PNA for rapid microbiology. Journal of Microbiological Methods 48(1), 1–17. [DOI] [PubMed] [Google Scholar]
- Stiles PL, Dieringer JA, Shah NC, Van Duyne RP, 2008. Surface-enhanced Raman spectroscopy. Annu. Rev. Anal. Chem 1, 601–626. [DOI] [PubMed] [Google Scholar]
- Stoesser N, Batty E, Eyre D, Morgan M, Wyllie D, Del Ojo Elias C, Johnson J, Walker A, Peto T, Crook D, 2013. Predicting antimicrobial susceptibilities for Escherichia coli and Klebsiella pneumoniae isolates using whole genomic sequence data. J. Antimicrob. Chemother 68(10), 2234–2244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straub J, Paula H, Mayr M, Kasper D, Assadian O, Berger A, Rittenschober-Böhm J, 2017. Diagnostic accuracy of the ROCHE Septifast PCR system for the rapid detection of blood pathogens in neonatal sepsis—A prospective clinical trial. PLoS One 12(11), e0187688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struelens MJ, 2009. Guidelines and indicators for methicillin-resistant Staphylococcus aureus control in hospitals: toward international agreement? Curr. Opin. Infect. Dis 22(4), 337–338. [DOI] [PubMed] [Google Scholar]
- Sutter D, Stagliano D, Braun L, Williams F, Arnold J, Ottolini M, Epstein J, 2008. Polymicrobial bloodstream infection in pediatric patients: risk factors, microbiology, and antimicrobial management. Pediatr. Infect. Dis. J 27(5), 400–405. [DOI] [PubMed] [Google Scholar]
- Tadros M, Petrich A, 2013. Evaluation of MALDI-TOF mass spectrometry and Sepsityper Kit™ for the direct identification of organisms from sterile body fluids in a Canadian pediatric hospital. Can. J. Infect. Dis. Med. Microbiol 24(4), 191–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talarico S, Safaeian M, Gonzalez P, Hildesheim A, Herrero R, Porras C, Cortes B, Larson A, Fang FC, Salama NR, 2016. Quantitative detection and genotyping of Helicobacter pylori from stool using droplet digital PCR reveals variation in bacterial loads that correlates with cagA virulence gene carriage. Helicobacter 21(4), 325–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan K, Ellis B, Lee R, Stamper P, Zhang S, Carroll K, 2012. Prospective evaluation of a matrix-assisted laser desorption ionization–time of flight mass spectrometry system in a hospital clinical microbiology laboratory for identification of bacteria and yeasts: a bench-by-bench study for assessing the impact on time to identification and cost-effectiveness. J. Clin. Microbiol 50(10), 3301–3308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Y-W, Lowery KS, Valsamakis A, Schaefer VC, Chappell JD, White-Abell J, Quinn CD, Li H, Washington CA, Cromwell J, 2013. Clinical accuracy of a PLEX-ID flu device for simultaneous detection and identification of influenza viruses A and B. J. Clin. Microbiol 51(1), 40–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tato M, Ruiz-Garbajosa P, Traczewski M, Dodgson A, McEwan A, Humphries R, Hindler J, Veltman J, Wang H, Cantón R, 2016. Multisite evaluation of Cepheid Xpert Carba-R assay for detection of carbapenemase-producing organisms in rectal swabs. J. Clin. Microbiol 54(7), 1814–1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tay WH, Chong KKL, Kline KA, 2016. Polymicrobial–host interactions during infection. J. Mol. Biol 428(17), 3355–3371. [DOI] [PubMed] [Google Scholar]
- Tong SY, Giffard PM, 2012. Clinical microbiological applications of high-resolution melting analysis. J. Clin. Microbiol., JCM 01709–01712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trenholme GM, Kaplan RL, Karakusis PH, Stine T, Fuhrer J, Landau W, Levin S, 1989. Clinical impact of rapid identification and susceptibility testing of bacterial blood culture isolates. J. Clin. Microbiol 27(6), 1342–1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trincavelli M, Coradeschi S, Loutfi A, Soderquist B, Thunberg P, 2010. Direct identification of bacteria in blood culture samples using an electronic nose. IEEE Trans. Biomed. Eng 57(12), 2884–2890. [DOI] [PubMed] [Google Scholar]
- Tsalik EL, Jones D, Nicholson B, Waring L, Liesenfeld O, Park LP, Glickman SW, Caram LB, Langley RJ, van Velkinburgh JC, Cairns CB, Rivers EP, Otero RM, Kingsmore SF, Lalani T, Fowler VG, Woods CW, 2010. Multiplex PCR to diagnose bloodstream infections in patients admitted from the emergency department with sepsis. J. Clin. Microbiol 48(1), 26–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ushio R, Yamamoto M, Nakashima K, Watanabe H, Nagai K, Shibata Y, Tashiro K, Tsukahara T, Nagakura H, Horita N, 2016. Digital PCR assay detection of circulating Mycobacterium tuberculosis DNA in pulmonary tuberculosis patient plasma. Tuberculosis 99, 47–53. [DOI] [PubMed] [Google Scholar]
- van Baar BL, 2000. Characterisation of bacteria by matrix-assisted laser desorption/ionisation and electrospray mass spectrometry. FEMS Microbiol. Rev 24(2), 193–219. [DOI] [PubMed] [Google Scholar]
- Vareechon C, Mestas J, Polanco CM, Bard JD, 2018. A 5-year study of the performance of the Verigene Gram-positive blood culture panel in a pediatric hospital. Eur. J. Clin. Microbiol. Infect. Dis, 1–6. [DOI] [PubMed] [Google Scholar]
- Velez DO, Mack H, Jupe J, Hawker S, Kulkarni N, Hedayatnia B, Zhang Y, Lawrence S, Fraley SI, 2017. Massively parallel digital high resolution melt for rapid and absolutely quantitative sequence profiling. Sci. Rep 7, 42326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Větrovský T, Baldrian P, 2013. The variability of the 16S rRNA gene in bacterial genomes and its consequences for bacterial community analyses. PLoS One 8(2), e57923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter A, März A, Schumacher W, Rösch P, Popp J, 2011. Towards a fast, high specific and reliable discrimination of bacteria on strain level by means of SERS in a microfluidic device. Lab Chip 11(6), 1013–1021. [DOI] [PubMed] [Google Scholar]
- Wang P, Pang S, Chen J, McLandsborough L, Nugen SR, Fan M, He L, 2016. Label-free mapping of single bacterial cells using surface-enhanced Raman spectroscopy. Analyst 141(4), 1356–1362. [DOI] [PubMed] [Google Scholar]
- Warhurst G, Maddi S, Dunn G, Ghrew M, Chadwick P, Alexander P, Bentley A, Moore J, Sharman M, Carlson GL, 2015. Diagnostic accuracy of SeptiFast multi-pathogen real-time PCR in the setting of suspected healthcare-associated bloodstream infection. Intensive Care Med. 41(1), 86–93. [DOI] [PubMed] [Google Scholar]
- Weinstein MP, Reller LB, Murphy JR, 1986. Clinical importance of polymicrobial bacteremia. Diagn. Microbiol. Infect. Dis 5(3), 185–196. [DOI] [PubMed] [Google Scholar]
- Weinstein MP, Towns ML, Quartey SM, Mirrett S, Reimer LG, Parmigiani G, Barth Reller L, 1997. The clinical significance of positive blood cultures in the 1990s: a prospective comprehensive evaluation of the microbiology, epidemiology, and outcome of bacteremia and fungemia in adults. Clin. Infect. Dis 24(4), 584–602. [DOI] [PubMed] [Google Scholar]
- Westh H, Lisby G, Breysse F, Böddinghaus B, Chomarat M, Gant V, Goglio A, Raglio A, Schuster H, Stuber F, 2009. Multiplex real-time PCR and blood culture for identification of bloodstream pathogens in patients with suspected sepsis. Clin. Microbiol. Infect 15(6), 544–551. [DOI] [PubMed] [Google Scholar]
- Whale AS, Huggett JF, Tzonev S, 2016. Fundamentals of multiplexing with digital PCR. BDQ 10, 15–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson AD, Baietto M, 2011. Advances in electronic-nose technologies developed for biomedical applications. Sensors 11(1), 1105–1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisplinghoff H, Bischoff T, Tallent SM, Seifert H, Wenzel RP, Edmond MB, 2004. Nosocomial bloodstream infections in US hospitals: analysis of 24,179 cases from a prospective nationwide surveillance study. Clin. Infect. Dis 39(3), 309–317. [DOI] [PubMed] [Google Scholar]
- Wojewoda CM, Sercia L, Navas M, Tuohy M, Wilson D, Hall GS, Procop GW, Richter SS, 2013. Evaluation of the Verigene Gram-positive blood culture nucleic acid test for rapid detection of bacteria and resistance determinants. J. Clin. Microbiol 51(7), 2072–2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Won H, Rothman R, Ramachandran P, Hsieh YH, Kecojevic A, Carroll KC, Aird D, Gaydos C, Yang S, 2010. Rapid identification of bacterial pathogens in positive blood culture bottles by use of a broad-based PCR assay coupled with high-resolution melt analysis. J. Clin. Microbiol 48(9), 3410–3413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong AP, Gupta M, Shevkoplyas SS, Whitesides GM, 2008. Egg beater as centrifuge: isolating human blood plasma from whole blood in resource-poor settings. Lab Chip 8(12), 2032–2037. [DOI] [PubMed] [Google Scholar]
- Wu X, Huang Y-W, Park B, Tripp RA, Zhao Y, 2015. Differentiation and classification of bacteria using vancomycin functionalized silver nanorods array based surface-enhanced Raman spectroscopy and chemometric analysis. Talanta 139, 96–103. [DOI] [PubMed] [Google Scholar]
- Yagupsky P, Nolte FS, 1990. Quantitative aspects of septicemia. Clin. Microbiol. Rev 3(3), 269–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Li P, Liu H, Tang X, Liu J, 2015. A dynamic surface enhanced Raman spectroscopy method for ultra-sensitive detection: from the wet state to the dry state. Chem. Soc. Rev 44(10), 2837–2848. [DOI] [PubMed] [Google Scholar]
- Yang R, Paparini A, Monis P, Ryan U, 2014. Comparison of next-generation droplet digital PCR (ddPCR) with quantitative PCR (qPCR) for enumeration of Cryptosporidium oocysts in faecal samples. Int. J. Parasitol 44(14), 1105–1113. [DOI] [PubMed] [Google Scholar]
- Yang S, Dai X, Stogin BB, Wong T-S, 2016. Ultrasensitive surface-enhanced Raman scattering detection in common fluids. Proceedings of the National Academy of Sciences 113(2), 268–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S, Ramachandran P, Rothman R, Hsieh Y-H, Hardick A, Won H, Kecojevic A, Jackman J, Gaydos C, 2009. Rapid identification of biothreat and other clinically relevant bacterial species by use of universal PCR coupled with high-resolution melting analysis. J. Clin. Microbiol 47(7), 2252–2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarbrough ML, Warren DK, Allen K, Burkholder D, Daum R, Donskey C, Knaack D, LaMarca A, May L, Miller LG, 2017. Multicenter evaluation of the Xpert MRSA NxG Assay for detection of methicillin resistant Staphylococcus aureus in nasal swabs. J. Clin. Microbiol., JCM 01381–01317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yetisen AK, Akram MS, Lowe CR, 2013. based microfluidic point-of-care diagnostic devices. Lab Chip 13(12), 2210–2251. [DOI] [PubMed] [Google Scholar]
- Yusuf N, Zakaria A, Omar MI, Shakaff AYM, Masnan MJ, Kamarudin LM, Rahim NA, Zakaria NZI, Abdullah AA, Othman A, 2015. In-vitro diagnosis of single and poly microbial species targeted for diabetic foot infection using e-nose technology. BMC Bioinformatics 16(1), 158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Bai J, Ying JY, 2015. A stacking flow immunoassay for the detection of dengue-specific immunoglobulins in salivary fluid. Lab Chip 15(6), 1465–1471. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Nguyen N-T, 2017. Magnetic digital microfluidics–a review. Lab Chip 17(6), 994–1008. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Shin DJ, Wang T-H, 2013. Serial dilution via surface energy trap-assisted magnetic droplet manipulation. Lab Chip 13(24), 4827–4831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Wang TH, 2013. Full‐Range Magnetic Manipulation of Droplets via Surface Energy Traps Enables Complex Bioassays. Adv. Mater 25(21), 2903–2908. [DOI] [PMC free article] [PubMed] [Google Scholar]