Abstract
CDKL5 disorder is a devastating neurodevelopmental disorder associated with epilepsy, developmental retardation, autism and related phenotypes. Mutations in the CDKL5 gene, encoding CDKL5, have been identified in this disorder. CDKL5 is a protein with homology to the serine-threonine kinases and incompletely characterized function. We generated and validated a murine model bearing a floxed allele of CDKL5 and polyclonal antibodies to CDKL5. CDKL5 is well expressed in the cortex, hippocampus and striatum, localized to synaptosomes and nuclei and developmentally regulated in the hippocampus. Using Cre- mediated mechanisms, we deleted CDKL5 from excitatory CaMKIIα positive neurons or inhibitory GABAergic neurons. Our data indicate that loss of CDKL5 in excitatory neurons of the cortex or inhibitory neurons of the striatum differentially alters expression of some components of the mTOR signaling pathway. Further loss of CDKL5 in excitatory neurons of the cortex or inhibitory neurons of the striatum leads to alterations in levels of synaptic markers in a neuron-type specific manner. Taken together, these data support a model in which loss of CDKL5 alters mTOR signaling and synaptic compositions in a neuron type specific manner and suggest that CDKL5 may have distinct functional roles related to cellular signaling in excitatory and inhibitory neurons. Thus, these studies provide new insights into the biology of CDKL5 and suggest that the molecular pathology in CDKL5 disorder may have distinct neuron-type specific origins and effects.
Introduction:
CDKL5 disorder is a devastating neurodevelopmental disorder associated with neurodevelopmental phenotypes, autism [1], intellectual disability and epilepsy [2,3]. CDKL5 encodes CDKL5, a protein with homology to the serine-threonine kinases. Structurally, the protein has a kinase domain and putative nuclear localization and nuclear export signals. The functional roles of CDKL5 are incompletely characterized, but include roles in regulation of synaptic density, architecture and stability [4-6], postsynaptic localization of NMDA receptors [7], surface expression of AMPA receptors [8], neuronal polarization [9], microtubule dynamics [10], RNA splicing [11], synaptic connectivity in the cortex [12], dendritic spine stability [13] and dendritic architecture [14]. Loss of CDKL5 in mouse models leads to phenotypes associated with CDKL5 disorder, including autistic phenotypes [15], memory impairment [16], increased seizure susceptibility [7] and sleep apnea [17]. Thus, CDKL5 is a critical regulator of neural circuit function and disruption of these functional roles in CDKL5 disorder likely contribute to neural circuit deficits and behavioral outcomes associated with the disorder.
Neurodevelopmental disorders with phenotypes similar to those observed in CDKL5 disorder vary in their origin and etiology and several genetic mouse models recapitulate core features of these disorders [18]. However, aberrations in mTOR signaling pathways [19-21] and synaptic density, function and architecture are commonly observed in a variety of disorders associated with similar phenotypes [22,23].
To begin to address the functional roles of CDKL5 in vivo, we generated and validated a mouse model bearing a floxed allele of CDKL5. We also generated a rabbit polyclonal antibody to CDKL5 and validated this and two commercial antibodies on tissue from Cre-mediated recombination. We examined the brain region distribution of CDKL5. Our data indicate that CDKL5 is well expressed in the cortex, hippocampus and striatum, with little expression in the olfactory bulb and cerebellum. Further, CDKL5 is localized in synaptosomes and nuclei and developmentally regulated in the hippocampus. By taking advantage of Cre-mediated recombination, we examined the effects of loss of CDKL5 in excitatory neurons (CaMKIIα-positive) or inhibitory (GAD65-positive) neurons on components of the mTOR signaling pathway and loss of CDKL5 in excitatory (CaMKIIα-positive) or inhibitory (GAD65-positive) neurons on excitatory synaptic markers. These data support a model in which loss of CDKL5 alters mTOR signaling and synaptic compositions in a neuron type-specific manner and suggest that CDKL5 may have distinct functional roles in excitatory and inhibitory neurons.
Methods:
Experimental Procedures:
CDKL5 conditional knockout mouse:
The mouse CDKL5 gene consists of 22 exons [24]of which the fourth exon was targeted for creating a conditional knockout allele. The targeting construct was commercially synthesized that contained a left and right homology arms of 7.3 and 6 kilobases respectively along with the upstream LoxP site in intron 3, and a Frt-Neo-Frt-LoxP cassette in intron 4. If a truncated protein is expressed from the upstream exons, it will produce only about 33 amino acids polypeptide, along with another 29 amino acids originating from frameshifted reading of the exon 6. Upon Cre-mediated deletion of the exon 4, the transcript will undergo nonsense mediated decay due to frameshift in the protein coding sequence of the downstream exons. The targeting construct was linearized and electroporated into C57BL6/J derived ES cells [25], the positive clones were screened by long range PCRs and confirmed by southern blotting. The ES cell clones were injected into Albino C57BL6/J (www.jax.org/strain/000058) strain derived blastocysts, to generate Chimeras, at the mouse genome engineering core facility, UNMC. A genotyping PCR assay was developed for detecting the conditional knockout allele. The primer pairs were CDKL5 Flox F TGCTCTTGGAGTATGTTGATTGAC and CDKL5 Flox R ACTTGGAATCATAATACTGTATACCTTG. The expected amplicons sizes are 204 and 267 base pairs, for wild type and conditional KO alleles respectively. The floxed mice were bred to Cre mice to generate neuron specific conditional knockout allele for CDKL5. The loss of CDKL5 protein expression, in the target tissue, was confirmed by Western blotting.
Animals
All animal experiments were approved by Institutional Animal Care and Use Committee of the University of Nebraska Medical Center. Mice were housed with 12/12 hr dark/light cycle with free access to food and water. The heterozygous CDKL5fl females were mated with homozygous CaMKIIα-Cre (the Jackson Laboratory, Stock No. 005359) or Gad2-IRES-Cre (the Jackson Laboratory, Stock No. 019022) male. Three offspring from homozygous CDKL5fl female and heterozygous Gad2-IRES-Cre male were also included and no significant difference were observed. Only male mice were included in this study.
Immunoblotting
Mice were anesthetized by isoflurane and brain tissues from cortex and striatum were dissected out in cold 1xPBS on ice. Tissue were quick frozen on dry ice and stored at −80 °C. For tissue lysate preparation, cold lysis buffer, including 50 mM Tris pH 7.4, 107 mM NaCl, 1% Triton X-100, 0.1% SDS, 1:100 protease inhibitor (Sigma, P8340), 1:100 phosphatase inhibitor (Sigma, P0044), 5 mM EDTA and 5 mM EGTA, were added to the samples before thawing, to minimize nonspecific cleavage by protease. Tissue were then homogenized by sonication (22-25% amplitude, 2 s sonication for 3-8 times according to the size of the tissue, with at least 60 s interval between each sonication) on ice and centrifuged 15,000 rpm for 5 min at 4 °C. Supernatant were loaded on 7.5% or 10% hand-casted SDS-PAGE, with 15 – 50 μg of protein per well. The dilutions of antibodies for immunoblot are as follows: 1:100 NR2B (NeuroMab, clone N59/36, 73-101), 1:100 PSD-95 (NeuroMab, clone K28/43, 73-028), 1:1000 Arc (Santa Cruz Biotechnology C-7, sc-17839), 1:8000 VGLUT1 (Millipore, AB5905), 1:5000 N-cadherin (BD Bioscience, 610920), 1:2000 CaMKIIα (Millipore, clone 6G9, 05-532), 1:1000 mTOR (Cell Signaling Technology, 2983), 1:1000 p-mTOR(S2448) (Cell Signaling Technology, 5536), 1:1000 p-mTOR(S2481) (Cell Signaling Technology, 2974), 1:1000 p-TSC2(S1387) (Cell Signaling Technology, 5584), 1:1000 p-p70S6K(T389) (Cell Signaling Technology, 9234), 1:1000 p70S6K (Cell Signaling Technology, 9202), 1:1000 TSC2 (Cell Signaling Technology, 4308), 1:1000 Rag C (Cell Signaling Technology, 5466), 1:1000 Rag B (Cell Signaling Technology, 8150), 1:1000 Rag A (Cell Signaling Technology, 4357), 1:1000 p-TSC2(S939) (Cell Signaling Technology, 3615), 1:10000 p62 (Abcam, ab109012), 1:200 β-tubulin (DHSB, E-7), 1:200 actin (DHSB, JLA20), 1:16000 HRP-conjugated anti-rabbit IgG (Jackson ImmunoResearch 711-035-152), 1:16000 HRP-conjugated anti-mouse IgG (Jackson ImmunoResearch 711-035-151), 1:6000 HRP-conjugated anti-guinea pig IgG (Invitrogen, 614620). CDKL5 antibody, Rabbit 6680 was generated using a peptide spanning aa indicated in Fig 1 using a commercial vendor.
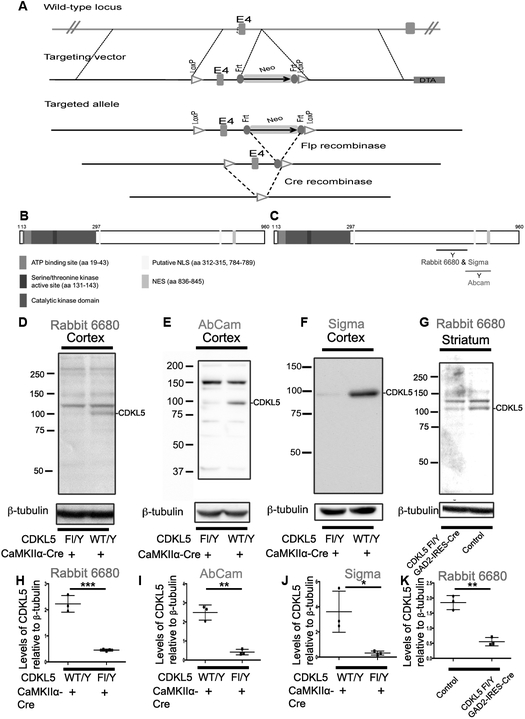
Fig 1: Generation and validation of CDKL5 mouse model and CDKL5 antibodies.
A. Schematic of targeted locus and locus after Cre-mediated recombination.
B. Schematic of CDKL5 with different known domains indicated – sequence based on human CDKL5.
C. Schematic of CDKL5 with location of three antibody epitopes. The Sigma and Abcam are commercial antibodies and Rbt 6680 is a custom polyclonal peptide antibody.
D. Western blots of cortical lysates from CDKL5 Fl/Y +/− CaMKIIα-Cre with anti-CDKL5 - Rbt6680
E. Western blots of cortical lysates from CDKL5 Fl/Y +/− CaMKIIα-Cre with anti-CDKL5 - Abcam
F. Western blots of cortical lysates from CDKL5 Fl/Y +/− CaMKIIα-Cre with anti CDKL5 – Sigma
G. Western blots of striatal lysates from CDKL5 Fl/Y +/− Gad2-IRES-Cre with anti CDKL5 – Rbt 6680
(D-G-Data N=3 for each genotype)
H. Levels of CDKL5 relative to β-tubulin in cortical lysates from CDKL5 Fl/Y +/− CaMKIIα-Cre with anti CDKL5 – Rbt 6680
I. Levels of CDKL5 relative to β-tubulin in cortical lysates from CDKL5 Fl/Y +/− CaMKIIα-Cre with anti CDKL5 - Abcam
J. Levels of CDKL5 relative to β-tubulin in cortical lysates from CDKL5 Fl/Y +/− CaMKIIα-Cre with anti CDKL5 – Sigma
K. Levels of CDKL5 relative to β-tubulin in striatal lysates from CDKL5 Fl/Y +/− Gad2-IRES-Cre with anti CDKL5 – Rbt 6680
(H-K-Data presented ± SEM, *=P<0.05, **=P<0.005, ***=P<0.0005, Student’s t-test assuming equal variances).
Quantitation of westerns:
Blots were detected by SuperSignal West Dura Chemiluminescent Substrate (Thermo, 34075), imaged by FluorChem HD2 (Cell Biosciences) and quantified on AlphaView (ProteinSimple) or ImageJ. After background subtraction, band intensity was normalized to housekeeping protein (β-tubulin or actin accordingly). Individual values were then normalized to the geometric mean of the litter so that unpaired statistical tests can be performed. For every western blot band that was quantitated, we ensured that the signal was not saturated using tools available with the AlphaView software.
Primary neuron culture from E18 rats:
As previously described [26]
Nuclear preparation:
Primary rat neurons were washed with 1X PBS. Culture dishes were incubated on a shaker with 600 μL per 100 mm dish of 1X Buffer A (0.5M HEPES, 2M KCl, 0.5M EDTA, water), 4% NP-40, and 1% protease and phosphatase inhibitors for 15 minutes at 4°C on ice. Cells were scraped using a disposable cell scraper. The homogenate was centrifuged at 15,000 × g for 3 minutes at 4°C. Tubes were placed on ice and the supernatant was collected as the cytosolic fraction. Three washes of 250 μL of 1X Buffer A (-NP-40) were performed with centrifugation at 15,000 × g for 3 minutes at 4°C between washes. The pellet was resuspended in 1X Buffer B (0.5M HEPES, 5M NaCL, 0.5M EDTA, 40% glycerol, water), and 1% protease and phosphatase inhibitors and vortexed for 10 s. The homogenate was placed on ice on a shaker in a cold room for 2 hours. Tubes were centrifuged at 15,000 × g for 5 minutes at 4°C and the supernatant was collected as the nuclear soluble fraction. Three washes of 100 μL of 1X Buffer B were performed with centrifugation at 15,000 × g for 3 minutes at 4°C between washes. The final pellet was suspended in RIPA buffer and sonicated to produce lysates for Western blot analysis.
Synaptosome preparation:
Dissected brain tissue (hippocamus ~20 mg; cortex ~40 mg form mouse P21 brain), was homogenized in 10 volumes of the Syn-PER Reagent (ThermoScientific.#87785, both protein phosphatase inhibitors and protease inhibitors included) using a 2 mL Dounce tissue grinder with 13 up-and-down even strokes (all procedures on ice). The homogenate was centrifuged at 1200 × g for 10 minutes to pellet synaptoromes. To further remove residual cell debris, the pellet was resuspended in Syn-PER Reagent and centrifuged again (1200×g ,10 minutes). For Western blot, the pellets, containing synaptosomes, were suspended in the RIPA buffer. When needed, to the supernatant (after the inital 1200×g centrifugation), 5X RIPA buffer was added and briefly sonicated to produce lysates for Western blot analysis (synptosome-minus fraction). To get cytosol fraction, the supernatant (after the inital 1200×g centrifugation) was centrifuged for 20 min at 15000×g. When needed, the remaining pellet (enriched for big organelles and membrane) was resupsended in RIPA buffer for Western Blot analysis.
Statistics
All the statistical tests were performed on Prism 7 (GraphPad). Normality of every group was first tested by D'Agostino & Pearson normality test and statistical tests were performed accordingly. Groups with too few data points for the normality test were treated as normally distributed.
Results:
Generation and validation of Floxed CDKL5 mice and CDKL5 antibodies:
We generated a conditional murine allele (Fig 1A) using standard techniques by generating a floxed allele of CDKL5 flanking exon 4. Cre mediated recombination is likely to generate a null allele (see methods). We generated a rabbit polyclonal peptide antibody to the C-terminal region of the mouse CDKL5 (Fig 1B), including amino acids 636-758 (Fig 1C). In addition to this antibody, two commercial antibodies to CDKL5 are also available (Sigma – aa 636-758 and Abcam – aa750-850). We generated mouse models in which CDKL5 is deleted from the cortical excitatory neurons by crossing the CDKL5 floxed allele into the CaMIIα-Cre line or from GABAergic neurons by crossing the CDKL5 floxed allele into the Gad2-IRES-Cre line. We examined the expression of CDKL5 in the CDKL5 Fl/Y or CDKL5 Fl/Y-CaMKIIα-Cre cortex by Western blot analysis with all the 3 antibodies. The levels of CDKL5 detected by all three antibodies, Rbt 6680 (Fig 1D, H), Abcam (Fig 1 E, I) and Sigma (Fig 1 F, J), were significantly reduced in the CDKL5 Fl/Y Cre mice in comparison to control. Rbt 6680 also reacts with an additional higher molecular weight non-specific band. We similarly examined the expression of CDKL5 in the striatum from the CDKL5 Fl/Y-Gad2-IRES-Cre mice. The striatum is predominantly composed of GABAergic neurons. Similar to the results observed with the CDKL5 Fl/Y-CaMKIIα-cre mice, the levels of CDKL5 in the CDKL5 Fl/Y-Gad2-IRES-Cre striatum were significantly reduced (Fig 1,G, K). These results indicate that (1) Cre-mediated recombination of the floxed allele of CDKL5 results in loss of CDKL5 protein and (2) the three antibodies are specific to CDKL5.
Expression pattern of CDKL5:
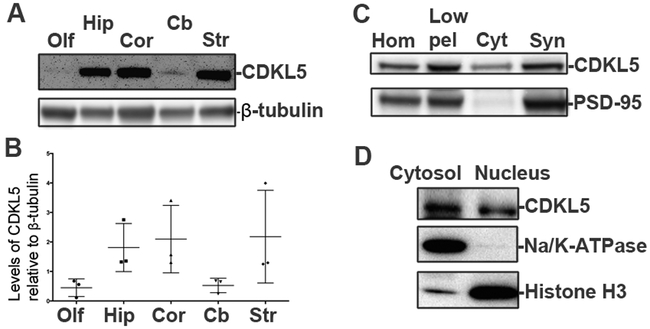
We examined the expression of CDKL5 in different regions of the mouse brain using Western blot analysis (Fig 2A, B). CDKL5 is predominantly expressed in the cortex, striatum and hippocampus with little expression in the cerebellum and olfactory bulb. To examine the subcellular distribution of CDKL5, we examined the expression of CDKL5 in synaptosomal preparations from the mouse cortex. CDKL5 is well expressed in synaptosomal fractions as indicated by coenrichment with PSD-95, an excitatory postsynaptic marker (Fig 2C). This is consistent with literature. We also examined the subcellular distribution of CDKL5 in the nucleus by Western blot analysis of cytosolic and nuclear fractions from cultured primary rat neurons. Nuclear fractions are enriched in histone H3. CDKL5 is localized in nuclear fractions (Fig 2D). Taken together, these results suggest that CDKL5 is predominantly expressed in the cortex, hippocampus and striatum and localized to synaptosomes and nuclei.
Fig 2: Regional and temporal expression patterns for CDKL5.
A. Western blots for CDKL5 (Sigma) in different regions of the mouse brain as indicated (P28, male, N=3).
B. Levels of CDKL5 relative to β-tubulin in different regions of the mouse brain (P28, male, N=3) (Data ± SEM).
C. Western blot for CDKL5 (Sigma) and PSD-95 (excitatory synaptic marker) in synaptosomal preparations from mouse cortex. (Adult, N=3)
D. Western blot for CDKL5 (Sigma), histone H3 and Na+/K+-ATPase in cytosol and nuclear preparations from primary rat neurons.
Developmental expression of CDKL5:
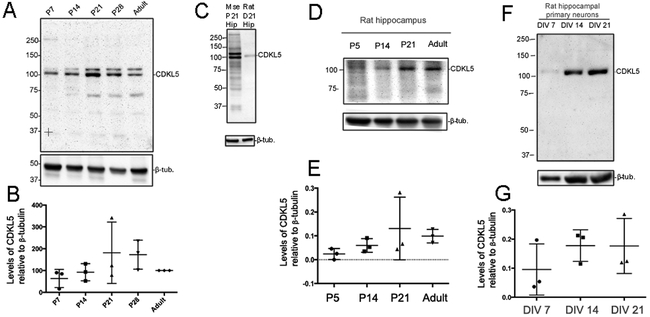
We examined the expression pattern of CDKL5 in the hippocampus across development (Fig 3A). CDKL5 was well expressed at all stages during development and in the adult. Our data suggest that there may be trends towards expression peaks around P21-P28, however, these did not reach statistical significance (Fig 3B). We compared the molecular weights of the mouse CDKL5 from hippocampal tissue (p21) and rat CDKL5 from in vitro primary cultures by Western blot analysis with Rbt 6680 (Fig 3C). Unlike the mouse lysates, Rbt 6680 only detects a single CDKL5 specific band in rat lysates. We also examined developmental expression of CDKL5 in the rat hippocampus (Fig 3D, E). Similar to the mouse hippocampus, CDKL5 is well expressed at time points coincident with neural circuit formation and refinement. We further examined the expression of CDKL5 in primary rat neurons at DIV 7, 14 and 21 (Fig 3F, G). Similar to the in vivo data, we observe expression of CDKL5 in primary neurons at time points coincident with synapse formation and maturity. Taken together, these results indicate that the expression of CDKL5 is developmentally regulated in the hippocampus, both in vivo and in vitro.
Fig 3: Developmental expression of CDKL5 in rat and mouse hippocampus.
A. Western blot for CDKL5 in hippocampal lysates at indicated time points (N=3).
B. Levels of CDKL5 relative to β-tubulin at indicated time points in the mouse hippocampus. Levels were normalized to percentage of adult levels.
C. Western blot for CDKL5 (Rbt 6680) in P21 mouse hippocampal lysate and rat primary neuron lysates.
D. Western blot for CDKL5 (Rbt 6680) in hippocampal lysates from P7 to adult rat tissue (N=3).
E. Levels of CDKL5 relative to β-tubulin at P7 to adult in the rat hippocampus.
F. Western blot for CDKL5 (Rbt 6680) in hippocampal lysates from primary rat neurons at DIV 7, 14 and 21 (N=3).
G. Levels of CDKL5 relative to β-tubulin in hippocampal lysates from primary rat neurons at DIV 7, 14 and 21.
Expression of components of the mTOR signaling pathway with loss of CDKL5 in excitatory neurons of the mouse cortex:
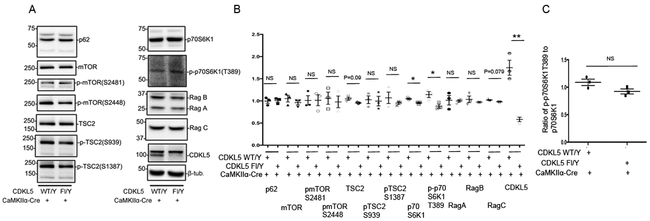
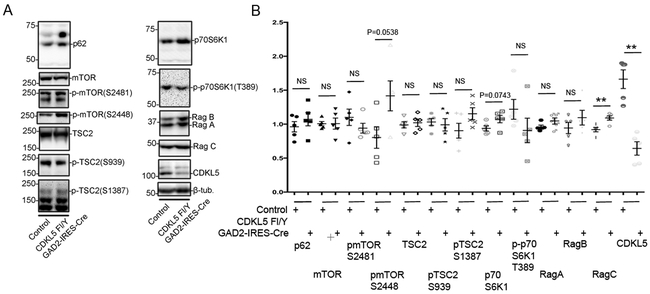
The mTOR (mechanistic target of rapamycin) pathway coordinates cellular signaling and metabolic pathways with environmental inputs [27]. Alterations in components of the mTOR signaling pathway have been observed in a variety of models of neurodevelopmental disorders associated with phenotypes common to CDKL5 disorder [28,29,19]. We examined the levels of components of the mTOR pathway by Western blot analyses in cortical lysates from CDKL5 Fl/Y CaMKIIα-cre mice. The levels of, p62, mTOR, pmTORS2448, pmTORS2481, pTSC2S1387, pTSC2S939, Rag A and Rag B in the CDKL5 Fl/Y CaMKIIα-Cre lysates were not significantly altered in comparison to control mice. The levels of TSC2 and Rag C showed trends towards a decrease, however, these did not reach significance. Interestingly, the levels of pp70S6K1T389 were significantly reduced in lysates from the CDKL5 Fl/Y CaMKIIα-Cre mice in comparison to control lysates (Fig 4A, B). These alterations were accompanied by similar change in the level of p70S6K1, suggesting that the reduction in the levels of the phosphorylated form is likely a secondary consequence of reduction of total levels of p70S6K1 (Fig 4C). These data indicate that selective loss of CDKL5 in excitatory neurons of the cortex leads to deficits in expression of components of the mTOR signaling pathway.
Fig 4: Alterations in levels of components of mTOR signaling pathway with loss of CDKL5 in cortical excitatory (CaMKIIα-positive) neurons.
A. Western blots for components of the mTOR signaling pathway as indicated in cortical lysates from CDKL5 Fl/Y +/− CaMKIIα-Cre tissue.
B. Levels of components of the mTOR signaling pathway relative to β-tubulin as indicated in cortical lysates from CDKL5 Fl/Y +/− CaMKIIα-Cre tissue.
C. Ratio of p70S6K1 to p70S6K1T389 from indicated genotypes.
(Data presented ± SEM, *=P<0.05, **=P<0.005, ***=P<0.0005, Student’s t-test with Welch correction. N=3 for each genotype. Age 8-11 weeks.)
Expression of components of the mTOR signaling pathway with loss of CDKL5 in inhibitory neurons of the mouse cortex:
CDKL5 is well expressed in the striatum (Fig 2A, B). It has previously been demonstrated that loss of CDKL5 in a mouse model enhances the connectivity between the parvalbumin-positive neurons and pyramidal neurons in the V1 cortex as examined by VGLUT1 immunostaining [12]. Further, Dlx5/6-Cre mediated deletion of CDKL5 in a floxed mouse model decreases home-cage activity in a sex-specific manner. These data suggest that GABAergic neurons likely contribute to the pathology of the CDKL5 disorder and loss of CDKL5 in GABAergic neurons may lead to signaling alterations relevant to the disorder. To examine levels of mTOR pathway components in absence of CDKL5 in GABAergic neurons, we generated mice bearing the CDKL5 Fl/Y and Gad2-IRES-Cre alleles[30]. Since the striatum is heavily enriched in GABAergic neurons, we examined the levels of mTOR signaling pathway markers in striatal lysates from control and mutant littermates by Western blot analysis. The levels of, p62, mTOR, pmTOR S2481, TSC2, pTSC2S1387, pTSC2S939, Rag A and Rag B in the CDKL5 Fl/Y Gad2-IRES-Cre lysates were not significantly altered in comparison to control mice. The levels of pmTORS2448 showed a trend towards an increase, however, these, did not reach significance. In contrast to the effects of loss of CDKL5 in excitatory neurons, the levels of p70S6K1 trended towards an increase in the mutants. However, these did not reach significance. In further contrast to the effects of loss of CDKL5 in excitatory neurons, the levels of Rag C were significantly increased in the mutant animals (Fig 5A, B). These data indicate that selective loss of CDKL5 in inhibitory neurons of the striatum leads to deficits in expression of components of the mTOR signaling pathway. However, these alterations do not mirror the effects observed in excitatory neurons, suggesting that loss of CDKL5 in excitatory and inhibitory neurons differentially affects signaling mediated by the mTOR pathway.
Fig 5: Alterations in levels of components of mTOR signaling pathway with loss of CDKL5 in striatal inhibitory (GAD65-positive) neurons.
A. Western blots for components of the mTOR signaling pathway as indicated in cortical lysates from CDKL5 Fl/Y +/− Gad2-IRES-Cre tissue.
B. Levels of components of the mTOR signaling pathway relative to β-tubulin as indicated in cortical lysates from CDKL5 Fl/Y +/− Gad2-IRES-Cre tissue. (Data presented ± SEM, *=P<0.05, **=P<0.005, ***=P<0.0005, Student’s t-test with Welch correction. N=5 for each genotype. Age 4-20 weeks.)
Expression of excitatory synaptic markers with loss of CDKL5 in excitatory neurons of the mouse cortex:
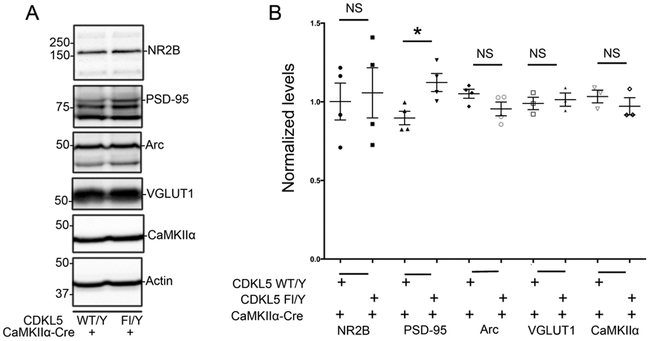
CDKL5 is localized at synapses [5] (Fig 2) and loss of CDKL5 perturbs excitatory synapse density and function. Synaptic alterations are commonly observed in several models of neurodevelopmental disorders associated with autism, intellectual disability and related deficits. Further, loss of CDKL5 in excitatory neurons via Emx-Cre-mediated deletion [31] recapitulates some behavioral features of the disorder, particularly increased hind-limb clasping, suggesting that loss of CDKL5 in excitatory neurons may perturb synaptic compositions. We examined levels of some excitatory synapse markers in cortical lysates from CDKL5 Fl/Y +/− CaMKIIα-Cre mice (Fig 6). The levels of NR2B and VGLUT1 were not significantly altered with loss of CDKL5 in cortical excitatory neurons. Interestingly, the levels of PSD-95 showed a significant increase in absence of CDKL5. We also examined the levels of Arc, a protein encoded by immediate early gene Arc, and CaMKIIα, an excitatory neuronal marker, in these lysates. The levels of these markers were not significantly altered in absence of CDKL5. Taken together, these data indicate that loss of CDKL5 in excitatory neurons in the cortex alters the levels of some excitatory synaptic markers.
Fig 6: Alterations in excitatory synaptic composition with loss of CDKL5 in cortical excitatory (CaMKIIα-positive) neurons:
A. Western blots for synaptic markers as indicated in cortical lysates from CDKL5 Fl/Y +/− CaMKIIα-Cre tissue.
B. Levels of synaptic markers as indicated relative to actin as indicated in cortical lysates from CDKL5 Fl/Y +/− CaMKIIα-Cre tissue. (Data presented ± SEM, *=P<0.05, **=P<0.005, ***=P<0.0005, Student’s t-test with Welch correction. N=4 for each genotype. Age 11-56 weeks.)
Expression of excitatory synaptic markers with loss of CDKL5 in GABAergic neurons of the mouse striatum:
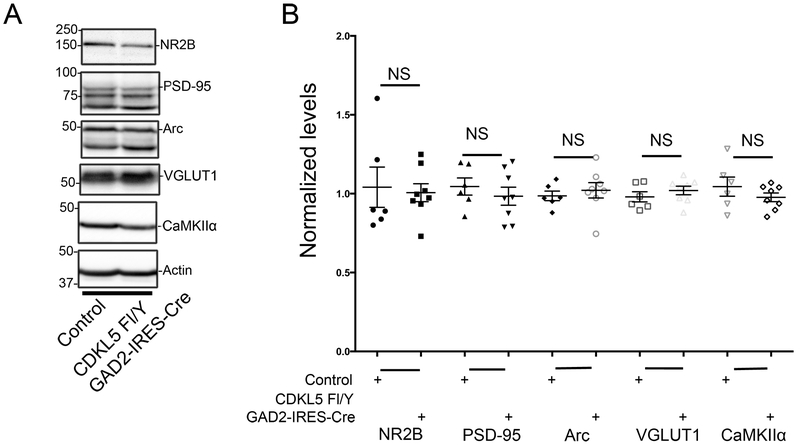
We examined the effects of loss of CDKL5 in GABAergic neurons on synaptic markers by Western blot analysis in striatal lysates from control and mutant littermates (Fig 7). The levels of NR2B, PSD-95 and VGGLUT1 were not altered with loss of CDKL5. In addition, unlike the effects observed with loss of CDKL5 in excitatory neurons, we did not observe any strong trends for alterations in levels of PSD-95 with loss of CDKL5 in GABAergic neurons. In addition, the levels of Arc, an activity regulated gene, and CaMKIIα, a marker for excitatory glutamatergic neurons, were not significantly altered. Taken together, these data suggest that loss of CDKL5 in GABAergic neurons of the striatum does not significantly alter levels of excitatory synaptic markers.
Fig 7: Synaptic composition is maintained with loss of CDKL5 in striatal inhibitory (GAD65-positive) neurons.
A. Western blots for synaptic markers as indicated in striatal lysates from CDKL5 Fl/Y +/− Gad2-IRES-Cre tissue.
B. Levels of synaptic markers as indicated relative to actin as indicated in striatal lysates from CDKL5 Fl/Y +/− Gad2-IRES-Cre tissue. (Data presented ± SEM, *=P<0.05, **=P<0.005, ***=P<0.0005, Student’s t-test with Welch correction.: N=6 for control and N=8 for CDKL5 Fl/Y - Gad2-IRES-Cre. Age 4-25 weeks.)
Discussion:
CDKL5 disorder is a disorder with neurodevelopmental phenotypes, including motor deficits, intellectual disability, autism and epilepsy. In this study, we have made several significant observations relating to the biology of CDKL5, encoded by CDKL5, that are likely relevant to dissecting the molecular pathology of the disorder.
Our data indicate that CDKL5, encoded by CDKL5, is expressed predominantly in the cortex, hippocampus and striatum with lower expression in the cerebellum and olfactory bulb. These regions are associated with higher order functions including thought, action, learning, memory and voluntary movement. The expression pattern of CDKL5 in these regions and the phenotypes associated with CDKL5 disorder suggest that CDKL5 is a critical regulator of neural circuits predominantly in these brain regions. Previous studies indicate that loss of CDKL5 in the cerebellum [32] leads to impaired motor coordination and gait, accompanied by reduction of GAD67 in the molecular layer with no alterations in the levels of VGLUT1 and reduced levels of BDNF mRNA. These results suggest that while the expression levels of CDKL5 in the cerebellum are low, they are functionally significant and that functional roles of CDKL5 in regions other than the hippocampus, cortex and striatum may also be critical to the pathology of CDKL5 disorder. Further, the developmental expression of CDKL5 coincides with the periods of neural circuit formation and refinement, consistent with a role for CDKL5 in neural circuit assembly, function and regulation.
Several mutations in CDKL5, including missense, point, frameshift and splicing mutations have been identified in CDKL5 disorder in humans. These mutations are predominantly localized in the kinase domain suggesting that the ability of the protein to function kinase is a critical. Our data demonstrating the localization of CDKL5 in synaptosomes and nuclei is consistent with data from other labs [33,34,4,35]. Thus, it is likely that the predominant kinase substrates of CDKL5 are localized to synapses and nuclei. While some of these substrates, including MeCP2, Dnmt1, and amphiphysin1 [33,36,37] have been identified, many likely remain to be identified. It is likely that the dynamic phosphorylation of these substrates in different cellular compartments contributes to the functional roles of CDKL5 in neural circuit function.
The mTOR signaling pathway [38,39] in neurons has key roles in a variety of signaling mechanisms associated with long term potentiation, long-term depression, learning and memory, neuronal survival, differentiation and morphogenesis [38]. We note that previous studies have demonstrated a reduction in the levels of pmTORS2448 in whole brain lysates of the complete null CDKL5 mouse model [15]. While our data show a slight trend towards reduction of pmTORS2448 in the CDKL5 Fl/Y CaMKIIα-Cre mouse cortex, these do not reach significance, suggesting that regional and neuronal or cell type specific mTOR signaling mechanisms may be regulated by CDKL5.
Our data indicate that loss of CDKL5 in glutamatergic neurons of the cortex leads to a significant reduction in levels of p70 ribosomal S6 protein kinase 1 (p70S6K1) and its phosphorylated form, p70S6K1T389. The ribosomal protein p70S6K1 [40] is a downstream target of the mTORC1 pathway. P70S6K1 can be phosphorylated at T389 by mTORC1. Phosphorylation of S6K1 by mTORc1 activates S6K1 and subsequently protein synthesis. One of the substrates of activated S6K1 is ribosomal protein S6, a component of the 40S ribosome. S6K1 functions have been implicated in a variety of mechanisms related to neuronal morphology, including axon regeneration [41], and dendrite morphology [42] and behaviors, including depressive behavior in the prefrontal cortex [43] and learning , memory and synaptic plasticity [44]. Interestingly, published studies indicate that loss of CDKL5 in a mouse model leads to reduced S6 Ser240-244 phosphorylation [13], suggesting that deficits in mTOR dependent translational regulation may be a key feature of CDKL5 disorder. How the loss of CDKL5 leads to an alteration in the levels of p70S6K1 remains to be identified, however, these observations are highly significant and suggest a mechanism by which CDKL5 may regulate translation via the mTOR pathway. Identifying such mechanisms and the translational products that are regulated by these mechanisms may have great significance for our understanding of the molecular pathology in CDKL5 disorder.
Our data also indicate that the loss of CDKL5 in inhibitory neurons, surprisingly, do not mirror the effects observed with loss of CDKL5 in excitatory neurons. Unlike, the alterations in glutamatergic neurons, loss of CDKL5 in GABAergic neurons of the striatum leads to a trend towards increase in the levels of p70S6K1 with a trend towards decrease of its T389 phosphorylated form, both of which were not significant. It is possible that heterogeneity of responses in subtypes of GABAergic neurons may contribute to these trends, however, these assessments require further experimental evidence. However, in stark contrast to the data from glutamatergic neurons, the levels of Rag C were significantly increased in GABAergic neurons with loss of CDKL5. The Rag family of proteins are GTPases[45]. They function as heterodimers of either Rag A or Rag B with either Rag C or Rag D and switch between engaging GTP and GDP to form part of a cellular mechanism that allows for nutrient sensing at the level of lysosomes and connect autophagy to mTORC1 signaling[46,47]. Consequently, the precise control of this signaling pathway is essential for cells to respond to changing cellular energy demands. We note that the levels of Rag A and B are unaltered with loss of CDKL5 in inhibitory neurons, but the levels of Rag C are significantly increased. At the cellular level, this is likely to lead to an imbalance in the ability of the Rag proteins to couple nutrient sensing to autophagic mechanisms, leading to deficits in neuronal metabolism that could impact multiple aspects of neuronal function relevant to the pathology of CDKL5 disorder, including synaptic physiology[48,49]. The validation of this model requires in depth assessments of nutrient sensing and neuronal responses in absence of CDKL5 in inhibitory neurons and may lead to great insights into the molecular pathology in CDKL5 disorder.
Several studies indicate that CDKL5 is localized at excitatory synapses and loss of CDKL5 perturbs excitatory synaptic structure and function. Multiple mouse models for CDKL5 have been generated and recapitulate some aspects of the CDKL5 disorder. Complete CDKL5 null mice (exon 6 deletion [50]) exhibit behavioral features similar to autism and ADHD. Complete CDKL5 null mice (exon 2 deletion [7]) have increased susceptibility to NMDA mediated seizures, enhanced anxiety, altered depressive-like and social behaviors. Another complete null mouse model for CDKL5 [15] exhibits hyperactivity, motor impairments, decreased anxiety and social behaviors reminiscent of autism. Interestingly, female heterozygous mice lacking one allele of CDKL5 also display several behavioral phenotypes including autistic-like behaviors, motor deficits and memory and breathing abnormalities[51]. These studies confirm that complete loss of CDKL5 in murine models leads to several behavioral features reminiscent of the human disorder.
CDKL5 is expressed in both glutamatergic and GABAergic neurons. Mice with selective loss of CDKL5 in glutamatergic neurons in the forebrain (using Nex-Cre [16]), exhibit a hindlimb clasping phenotype and have some deficits in hippocampal learning and memory. A similar hindlimb clasping phenotype is observed in mice in which CDKL5 is deleted from a mouse model bearing a floxed allele of CDKL5 using the Emx-Cre line. Male mice that are null for CDKL5 in GABAergic neurons generated by crossing mice bearing a CDKL5 floxed allele into the Dlx5/6 Cre line demonstrate decreased locomotion [31]. Based on these data, it is likely that CDKL5 has differential functional roles in glutamatergic and GABAergic neurons.
Our results with loss of CDKL5 in excitatory neurons of the cortex are consistent with Western blot data from CDKL5 Y/- hippocampi demonstrating no alterations in total levels of NR2B and VGLUT1 in the CDKL5 Y/- mice in comparison to control [7]. However, we do observe an increase in the levels of PSD-95 with loss of CDKL5 in glutamatergic neurons. Our results are in contrast with data from [13] demonstrating a decrease in the levels of PSD-95 in the cortex of CDKL5 Y/- mice. While our data was obtained from entire cortical lysates, the data in [13] was obtained from the somatosensory cortex. Thus, it is possible that regional differences in the levels of excitatory synaptic markers exist within the cortex in absence of CDKL5. Our data are also in contrast with data from [7] demonstrating no alterations in the levels of PSD-95 in hippocampal lysates from CDKL5 Y/- mice. Taken together, these data suggest that loss of CDKL5 may lead to regional differences in excitatory synaptic markers within the brain and cortex. Further sophisticated electrophysiological and high-resolution microscopy studies are necessary to establish such differences.
Our results indicate that loss of CDKL5 in GABAergic neurons of the striatum do not significantly alter the levels of excitatory synaptic markers. However, similar to the data from excitatory neurons, these data do not rule out the possibility that synaptic levels of some of these markers are altered while total levels are not.
Previous studies indicate that mice with complete loss of CDKL5 have reduced levels of c-fos, a protein encoded by an activity-regulated immediate early gene, in both excitatory neurons and parvalbumin-positive inhibitory neurons [12]. Our data indicate that loss of CDKL5 in either excitatory or inhibitory neurons does not alter levels of Arc, an activity regulated immediate early gene [52], suggesting that loss of CDKL5 may selectively alter expression of activity regulated genes that may be relevant to the pathology of CDKL5 disorder.
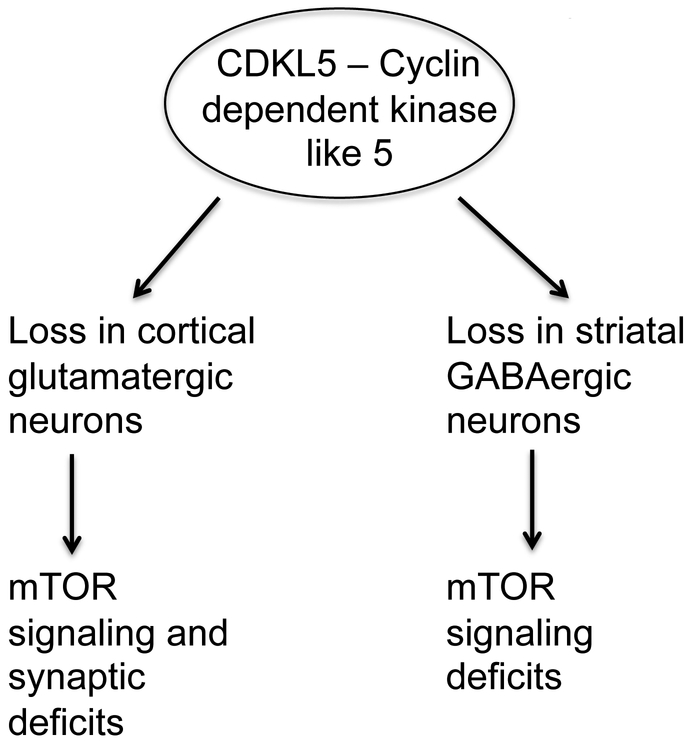
We have identified neuron-type specific alterations with loss of CDKL5 in components of the mTOR signaling pathways and synaptic proteins (Fig 8). Both of these pathways are critical for neural circuit formation and function. Taken together with published data, our results suggest that the differential functional roles of CDKL5 in excitatory and inhibitory neurons may be critical for its functional roles in neural circuit formation and function and disruptions in these functional roles with loss or mutations in CDKL5 contribute to the pathology of CDKL5 disorder.
Fig 8: Overall model.
Loss of CDKL5 in glutamatergic or GABAergic neurons perturbs expression of components of mTOR pathway and synaptic markers in a neuron-type specific manner.
Acknowledgements:
Research in the Arikkath Lab has been supported by Start up funds from the Munroe-Meyer Institute and grants from The Nebraska Research Initiative, an Institutional Development Award (IDeA) from the National Institute of General Medical Sciences of the National Institutes of Health under grant number 5P20GM103471-10, the Nebraska EPSCoR (EPS-1004094), RO3 from the National Institutes of Health (1R03MH110726-01) and the Brain and Behavior Research Foundation (#21080). Ethan Schroeder and Li Yuan were partly supported by graduate student fellowships from the University of Nebraska Medical Center and Nicholas DeKorver was supported by a grant from the National Institutes of Health (AG031158). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Conflict of interest: The authors declare that they have no conflicts of interest with the contents of this article.
References:
- 1.Posar A, Faggioli R, Visconti P (2015) Neurobehavioral phenotype in cyclin-dependent kinase-like 5 syndrome: Case report and review of literature. J Pediatr Neurosci 10 (3):258–260. doi: 10.4103/1817-1745.165685 JPN-10-258 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bodian DL, Schreiber JM, Vilboux T, Khromykh A, Hauser NS (2018) Mutation in an alternative transcript of CDKL5 in a boy with early onset seizures. Cold Spring Harb Mol Case Stud. doi:mcs.a002360 [pii] 10.1101/mcs.a002360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kilstrup-Nielsen C, Rusconi L, La Montanara P, Ciceri D, Bergo A, Bedogni F, Landsberger N (2012) What we know and would like to know about CDKL5 and its involvement in epileptic encephalopathy. Neural Plast 2012:728267. doi: 10.1155/2012/728267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.La Montanara P, Rusconi L, Locarno A, Forti L, Barbiero I, Tramarin M, Chandola C, Kilstrup-Nielsen C, Landsberger N (2015) Synaptic synthesis, dephosphorylation, and degradation: a novel paradigm for an activity-dependent neuronal control of CDKL5. J Biol Chem 290 (7):4512–4527. doi: 10.1074/jbc.M114.589762 M114.589762 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ricciardi S, Ungaro F, Hambrock M, Rademacher N, Stefanelli G, Brambilla D, Sessa A, Magagnotti C, Bachi A, Giarda E, Verpelli C, Kilstrup-Nielsen C, Sala C, Kalscheuer VM, Broccoli V (2012) CDKL5 ensures excitatory synapse stability by reinforcing NGL-1-PSD95 interaction in the postsynaptic compartment and is impaired in patient iPSC-derived neurons. Nat Cell Biol 14 (9):911–923. doi: 10.1038/ncb2566 ncb2566 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhu YC, Li D, Wang L, Lu B, Zheng J, Zhao SL, Zeng R, Xiong ZQ (2013) Palmitoylation-dependent CDKL5-PSD-95 interaction regulates synaptic targeting of CDKL5 and dendritic spine development. Proc Natl Acad Sci U S A 110 (22):9118–9123. doi: 10.1073/pnas.1300003110 1300003110 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Okuda K, Kobayashi S, Fukaya M, Watanabe A, Murakami T, Hagiwara M, Sato T, Ueno H, Ogonuki N, Komano-Inoue S, Manabe H, Yamaguchi M, Ogura A, Asahara H, Sakagami H, Mizuguchi M, Manabe T, Tanaka T (2017) CDKL5 controls postsynaptic localization of GluN2B-containing NMDA receptors in the hippocampus and regulates seizure susceptibility. Neurobiol Dis 106:158–170. doi:S0969-9961(17)301547 [pii] 10.1016/j.nbd.2017.07.002 [DOI] [PubMed] [Google Scholar]
- 8.Tramarin M, Rusconi L, Pizzamiglio L, Barbiero I, Peroni D, Scaramuzza L, Guilliams T, Cavalla D, Antonucci F, Kilstrup-Nielsen C (2018) The antidepressant tianeptine reverts synaptic AMPA receptor defects caused by deficiency of CDKL5. Hum Mol Genet 27 (12):2052–2063. doi: 10.1093/hmg/ddy1084956804 [pii] [DOI] [PubMed] [Google Scholar]
- 9.Nawaz MS, Giarda E, Bedogni F, La Montanara P, Ricciardi S, Ciceri D, Alberio T, Landsberger N, Rusconi L, Kilstrup-Nielsen C (2016) CDKL5 and Shootin1 Interact and Concur in Regulating Neuronal Polarization. PLoS One 11 (2):e0148634. doi: 10.1371/journal.pone.0148634 PONE-D-15-10186 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barbiero I, Peroni D, Tramarin M, Chandola C, Rusconi L, Landsberger N, Kilstrup-Nielsen C (2017) The neurosteroid pregnenolone reverts microtubule derangement induced by the loss of a functional CDKL5-IQGAP1 complex. Hum Mol Genet 26 (18):3520–3530. doi: 10.1093/hmg/ddx237 3876271 [pii] [DOI] [PubMed] [Google Scholar]
- 11.Ricciardi S, Kilstrup-Nielsen C, Bienvenu T, Jacquette A, Landsberger N, Broccoli V (2009) CDKL5 influences RNA splicing activity by its association to the nuclear speckle molecular machinery. Hum Mol Genet 18 (23):4590–4602. doi: 10.1093/hmg/ddp426 ddp426 [pii] [DOI] [PubMed] [Google Scholar]
- 12.Pizzo R, Gurgone A, Castroflorio E, Amendola E, Gross C, Sassoe-Pognetto M, Giustetto M (2016) Lack of Cdkl5 Disrupts the Organization of Excitatory and Inhibitory Synapses and Parvalbumin Interneurons in the Primary Visual Cortex. Front Cell Neurosci 10:261. doi: 10.3389/fncel.2016.00261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Della Sala G, Putignano E, Chelini G, Melani R, Calcagno E, Michele Ratto G, Amendola E, Gross CT, Giustetto M, Pizzorusso T (2016) Dendritic Spine Instability in a Mouse Model of CDKL5 Disorder Is Rescued by Insulin-like Growth Factor 1. Biol Psychiatry 80 (4):302–311. doi:S0006-3223(15)00727-1 [pii] 10.1016/j.biopsych.2015.08.028 [DOI] [PubMed] [Google Scholar]
- 14.Fuchs C, Trazzi S, Torricella R, Viggiano R, De Franceschi M, Amendola E, Gross C, Calza L, Bartesaghi R, Ciani E (2014) Loss of CDKL5 impairs survival and dendritic growth of newborn neurons by altering AKT/GSK-3beta signaling. Neurobiol Dis 70:53–68. doi: 10.1016/j.nbd.2014.06.006 S0969-9961(14)00170-3 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang IT, Allen M, Goffin D, Zhu X, Fairless AH, Brodkin ES, Siegel SJ, Marsh ED, Blendy JA, Zhou Z (2012) Loss of CDKL5 disrupts kinome profile and event-related potentials leading to autistic-like phenotypes in mice. Proc Natl Acad Sci U S A 109 (52):21516–21521. doi: 10.1073/pnas.1216988110 1216988110 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tang S, Wang IJ, Yue C, Takano H, Terzic B, Pance K, Lee JY, Cui Y, Coulter DA, Zhou Z (2017) Loss of CDKL5 in Glutamatergic Neurons Disrupts Hippocampal Microcircuitry and Leads to Memory Impairment in Mice. J Neurosci 37 (31):7420–7437. doi: 10.1523/JNEUROSCI.0539-17.2017 JNEUROSCI.0539-17.2017 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lo Martire V, Alvente S, Bastianini S, Berteotti C, Silvani A, Valli A, Viggiano R, Ciani E, Zoccoli G (2017) CDKL5 deficiency entails sleep apneas in mice. J Sleep Res 26 (4):495–497. doi: 10.1111/jsr.12512 [DOI] [PubMed] [Google Scholar]
- 18.Del Pino I, Rico B, Marin O (2018) Neural circuit dysfunction in mouse models of neurodevelopmental disorders. Curr Opin Neurobiol 48:174–182. doi:S0959-4388(17)30100-9 [pii] 10.1016/j.conb.2017.12.013 [DOI] [PubMed] [Google Scholar]
- 19.Yuskaitis CJ, Jones BM, Wolfson RL, Super CE, Dhamne SC, Rotenberg A, Sabatini DM, Sahin M, Poduri A (2018) A mouse model of DEPDC5-related epilepsy: Neuronal loss of Depdc5 causes dysplastic and ectopic neurons, increased mTOR signaling, and seizure susceptibility. Neurobiol Dis 111:91–101. doi:S0969-9961(17)30294-2 [pii] 10.1016/j.nbd.2017.12.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lipton JO, Sahin M (2014) The neurology of mTOR. Neuron 84 (2):275–291. doi: 10.1016/j.neuron.2014.09.034 S0896-6273(14)00892-7 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tang G, Gudsnuk K, Kuo SH, Cotrina ML, Rosoklija G, Sosunov A, Sonders MS, Kanter E, Castagna C, Yamamoto A, Yue Z, Arancio O, Peterson BS, Champagne F, Dwork AJ, Goldman J, Sulzer D (2014) Loss of mTOR-dependent macroautophagy causes autistic-like synaptic pruning deficits. Neuron 83 (5):1131–1143. doi: 10.1016/j.neuron.2014.07.040 S0896-6273(14)00651-5 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nelson SB, Valakh V (2015) Excitatory/Inhibitory Balance and Circuit Homeostasis in Autism Spectrum Disorders. Neuron 87 (4):684–698. doi: 10.1016/j.neuron.2015.07.033 S0896-6273(15)00675-3 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Contractor A, Klyachko VA, Portera-Cailliau C (2015) Altered Neuronal and Circuit Excitability in Fragile X Syndrome. Neuron 87 (4):699–715. doi: 10.1016/j.neuron.2015.06.017 S0896-6273(15)00560-7 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hector RD, Dando O, Landsberger N, Kilstrup-Nielsen C, Kind PC, Bailey ME, Cobb SR (2016) Characterisation of CDKL5 Transcript Isoforms in Human and Mouse. PLoS One 11 (6):e0157758. doi: 10.1371/journal.pone.0157758 PONE-D-16-05131 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gertsenstein M, Nutter LM, Reid T, Pereira M, Stanford WL, Rossant J, Nagy A (2010) Efficient generation of germ line transmitting chimeras from C57BL/6N ES cells by aggregation with outbred host embryos. PLoS One 5 (6):e11260. doi: 10.1371/journal.pone.0011260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Beaudoin GM 3rd, Lee SH, Singh D, Yuan Y, Ng YG, Reichardt LF, Arikkath J (2012) Culturing pyramidal neurons from the early postnatal mouse hippocampus and cortex. Nat Protoc 7 (9):1741–1754. doi: 10.1038/nprot.2012.099 nprot.2012.099 [pii] [DOI] [PubMed] [Google Scholar]
- 27.Saxton RA, Sabatini DM (2017) mTOR Signaling in Growth, Metabolism, and Disease. Cell 169 (2):361–371. doi:S0092-8674(17)30363-X [pii] 10.1016/j.cell.2017.03.035 [DOI] [PubMed] [Google Scholar]
- 28.Steinmetz AB, Stern SA, Kohtz AS, Descalzi G, Alberini CM (2018) Insulin-Like Growth Factor II Targets the mTOR Pathway to Reverse Autism-Like Phenotypes in Mice. J Neurosci 38 (4):1015–1029. doi: 10.1523/JNEUROSCI.2010-17.2017 JNEUROSCI.2010-17.2017 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Borrie SC, Brems H, Legius E, Bagni C (2017) Cognitive Dysfunctions in Intellectual Disabilities: The Contributions of the Ras-MAPK and PI3K-AKT-mTOR Pathways. Annu Rev Genomics Hum Genet 18:115–142. doi: 10.1146/annurevgenom-091416-035332 [DOI] [PubMed] [Google Scholar]
- 30.Taniguchi H, He M, Wu P, Kim S, Paik R, Sugino K, Kvitsiani D, Fu Y, Lu J, Lin Y, Miyoshi G, Shima Y, Fishell G, Nelson SB, Huang ZJ (2011) A resource of Cre driver lines for genetic targeting of GABAergic neurons in cerebral cortex. Neuron 71 (6):995–1013. doi: 10.1016/j.neuron.2011.07.026 S0896-6273(11)00679-9 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Amendola E, Zhan Y, Mattucci C, Castroflorio E, Calcagno E, Fuchs C, Lonetti G, Silingardi D, Vyssotski AL, Farley D, Ciani E, Pizzorusso T, Giustetto M, Gross CT (2014) Mapping pathological phenotypes in a mouse model of CDKL5 disorder. PLoS One 9 (5):e91613. doi: 10.1371/journal.pone.0091613 PONE-D-13-46590 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sivilia S, Mangano C, Beggiato S, Giuliani A, Torricella R, Baldassarro VA, Fernandez M, Lorenzini L, Giardino L, Borelli AC, Ferraro L, Calza L (2016) CDKL5 knockout leads to altered inhibitory transmission in the cerebellum of adult mice. Genes Brain Behav 15 (5):491–502. doi: 10.1111/gbb.12292 [DOI] [PubMed] [Google Scholar]
- 33.Kameshita I, Sekiguchi M, Hamasaki D, Sugiyama Y, Hatano N, Suetake I, Tajima S, Sueyoshi N (2008) Cyclin-dependent kinase-like 5 binds and phosphorylates DNA methyltransferase 1. Biochem Biophys Res Commun 377 (4):1162–1167. doi: 10.1016/j.bbrc.2008.10.113 S0006-291X(08)02110-4 [pii] [DOI] [PubMed] [Google Scholar]
- 34.Rusconi L, Kilstrup-Nielsen C, Landsberger N (2011) Extrasynaptic N-methyl-D-aspartate (NMDA) receptor stimulation induces cytoplasmic translocation of the CDKL5 kinase and its proteasomal degradation. J Biol Chem 286 (42):36550–36558. doi: 10.1074/jbc.M111.235630 M111.235630 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen Q, Zhu YC, Yu J, Miao S, Zheng J, Xu L, Zhou Y, Li D, Zhang C, Tao J, Xiong ZQ (2010) CDKL5, a protein associated with rett syndrome, regulates neuronal morphogenesis via Rac1 signaling. J Neurosci 30 (38):12777–12786. doi: 10.1523/JNEUROSCI.1102-10.2010 30/38/12777 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mari F, Azimonti S, Bertani I, Bolognese F, Colombo E, Caselli R, Scala E, Longo I, Grosso S, Pescucci C, Ariani F, Hayek G, Balestri P, Bergo A, Badaracco G, Zappella M, Broccoli V, Renieri A, Kilstrup-Nielsen C, Landsberger N (2005) CDKL5 belongs to the same molecular pathway of MeCP2 and it is responsible for the early-onset seizure variant of Rett syndrome. Hum Mol Genet 14 (14):1935–1946. doi:ddi198 [pii] 10.1093/hmg/ddi198 [DOI] [PubMed] [Google Scholar]
- 37.Sekiguchi M, Katayama S, Hatano N, Shigeri Y, Sueyoshi N, Kameshita I (2013) Identification of amphiphysin 1 as an endogenous substrate for CDKL5, a protein kinase associated with X-linked neurodevelopmental disorder. Arch Biochem Biophys 535 (2):257–267. doi: 10.1016/j.abb.2013.04.012 S0003-9861(13)00148-3 [pii] [DOI] [PubMed] [Google Scholar]
- 38.Switon K, Kotulska K, Janusz-Kaminska A, Zmorzynska J, Jaworski J (2017) Molecular neurobiology of mTOR. Neuroscience 341:112–153. doi:S0306-4522(16)30638-8 [pii] 10.1016/j.neuroscience.2016.11.017 [DOI] [PubMed] [Google Scholar]
- 39.Huber KM, Klann E, Costa-Mattioli M, Zukin RS (2015) Dysregulation of Mammalian Target of Rapamycin Signaling in Mouse Models of Autism. J Neurosci 35 (41):13836–13842. doi: 10.1523/JNEUROSCI.2656-15.2015 35/41/13836 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bhattacharya A, Kaphzan H, Alvarez-Dieppa AC, Murphy JP, Pierre P, Klann E (2012) Genetic removal of p70 S6 kinase 1 corrects molecular, synaptic, and behavioral phenotypes in fragile X syndrome mice. Neuron 76 (2):325–337. doi: 10.1016/j.neuron.2012.07.022 S0896-6273(12)00673-3 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang L, Miao L, Liang F, Huang H, Teng X, Li S, Nuriddinov J, Selzer ME, Hu Y (2014) The mTORC1 effectors S6K1 and 4E-BP play different roles in CNS axon regeneration. Nat Commun 5:5416. doi: 10.1038/ncomms6416 ncomms6416 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Koscielny A, Malik AR, Liszewska E, Zmorzynska J, Tempes A, Tarkowski B, Jaworski J (2018) Adaptor Complex 2 Controls Dendrite Morphology via mTOR-Dependent Expression of GluA2. Mol Neurobiol 55 (2):1590–1606. doi: 10.1007/s12035-017-0436-3 10.1007/s12035-017-0436-3 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dwyer JM, Maldonado-Aviles JG, Lepack AE, DiLeone RJ, Duman RS (2015) Ribosomal protein S6 kinase 1 signaling in prefrontal cortex controls depressive behavior. Proc Natl Acad Sci U S A 112 (19):6188–6193. doi: 10.1073/pnas.1505289112 1505289112 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Antion MD, Merhav M, Hoeffer CA, Reis G, Kozma SC, Thomas G, Schuman EM, Rosenblum K, Klann E (2008) Removal of S6K1 and S6K2 leads to divergent alterations in learning, memory, and synaptic plasticity. Learn Mem 15 (1):29–38. doi: 10.1101/lm.661908 15/1/29 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shen K, Sidik H, Talbot WS (2016) The Rag-Ragulator Complex Regulates Lysosome Function and Phagocytic Flux in Microglia. Cell Rep 14 (3):547–559. doi:S2211-1247(15)01496-5 [pii] 10.1016/j.celrep.2015.12.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Brady OA, Diab HI, Puertollano R (2016) Rags to riches: Amino acid sensing by the Rag GTPases in health and disease. Small GTPases 7 (4):197–206. doi: 10.1080/21541248.2016.1218990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pu J, Keren-Kaplan T, Bonifacino JS (2017) A Ragulator-BORC interaction controls lysosome positioning in response to amino acid availability. J Cell Biol 216 (12):4183–4197. doi: 10.1083/jcb.201703094 jcb.201703094 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nikoletopoulou V, Tavernarakis N (2018) Regulation and Roles of Autophagy at Synapses. Trends Cell Biol. doi:S0962-8924(18)30062-X [pii] 10.1016/j.tcb.2018.03.006 [DOI] [PubMed] [Google Scholar]
- 49.Cai Y, Yang L, Hu G, Chen X, Niu F, Yuan L, Liu H, Xiong H, Arikkath J, Buch S (2016) Regulation of morphine-induced synaptic alterations: Role of oxidative stress, ER stress, and autophagy. J Cell Biol 215 (2):245–258. doi:jcb.201605065 [pii] 10.1083/jcb.201605065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jhang CL, Huang TN, Hsueh YP, Liao W (2017) Mice lacking cyclin-dependent kinase-like 5 manifest autistic and ADHD-like behaviors. Hum Mol Genet 26 (20):3922–3934. doi: 10.1093/hmg/ddx279 3965512 [pii] [DOI] [PubMed] [Google Scholar]
- 51.Fuchs C, Gennaccaro L, Trazzi S, Bastianini S, Bettini S, Martire VL, Ren E, Medici G, Zoccoli G, Rimondini R, Ciani E (2018) Heterozygous CDKL5 Knockout Female Mice Are a Valuable Animal Model for CDKL5 Disorder. Neural Plast 2018:9726950. doi: 10.1155/2018/9726950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pastuzyn ED, Shepherd JD (2017) Activity-Dependent Arc Expression and Homeostatic Synaptic Plasticity Are Altered in Neurons from a Mouse Model of Angelman Syndrome. Front Mol Neurosci 10:234. doi: 10.3389/fnmol.2017.00234 [DOI] [PMC free article] [PubMed] [Google Scholar]