Abstract
Flax (Linum usitatissimum) is a cool season crop commercially cultivated for seed oil and stem fibre production. A comprehensive characterization of the heat shock factor (HSF) candidate genes in flax can accelerate genetic improvement and adaptive breeding for high temperature stress tolerance. We report the genome-wide identification of 34 putative HSF genes from the flax genome, which we mapped on 14 of the 15 chromosomes. Through comparative homology analysis, we classified these genes into three broad groups, and sub-groups. The arrangement of HSF-specific protein motifs, DNA-binding domain (DBD) and hydrophobic heptad repeat (HR-A/B), and exon-intron boundaries substantiated the phylogenetic separation of these genes. Orthologous relationships and evolutionary analysis revealed that the co-evolution of the LusHSF genes was due to recent genome duplication events. Digital and RT-qPCR analyses provided significant evidence of the differential expression of the LusHSF genes in various tissues, at various developmental stages, and in response to high-temperature stress. The co-localization of diverse cis-acting elements in the promoters of the LusHSF genes further emphasized their regulatory roles in the abiotic stress response. We further confirmed DNA-binding sites on the LusHSF proteins and designed guide RNA sequences for gene editing with minimal off-target effects. These results will hasten functional investigations of LusHSFs or assist in devising genome engineering strategies to develop high-temperature stress tolerant flax cultivars.
Introduction
The impact of global warming on crop productivity is alarming and presumed to decrease global crop yield by 1.5% per decade. The effects of high-temperature (HT) stress are detrimental to plant growth and development, physiological processes, and crop yield per se1. At the cellular level, basic stresses, such as temperature, drought, and salinity, result in cell injury due to osmotic and oxidative stresses. Being immobile, plants respond through a variety of adaptive, avoidance, and/or acclimation mechanisms to mitigate HT stress. These responses include the activation of various physiological and biochemical processes, antioxidant defences, and metabolite synthesis pathways. Similarly, several genetic components, such as structural and regulatory genes perform essential roles in HT stress alleviation.
Heat shock proteins (HSPs) are molecular chaperones that execute crucial functions in response to HT stress. These proteins respond by the folding, accumulation, and degradation of other protective proteins when cells are exposed to HT stress2. The expression of these HSP-coding genes is regulated by a group of DNA-binding transcription factors, known as heat shock factors (HSFs). Therefore, HSFs are the primary regulators of the HT stress-responsive gene expression pathway, which operates by modulating a cascade of signal transduction networks3. Structurally, HSF proteins comprise an N-terminal conserved DNA-binding domain (DBD) of helix-turn-helix motifs that specifically interact with the heat shock elements (HSEs) of HSP gene promoters. Adjacent to DBD exists the oligomerization domain (OD) with the characteristic heptad hydrophobic repeat (HR-A/B) motif. Variations in the amino acid residues of HR-A/B motifs and the distance between the DBD and the OD facilitate the grouping of HSF proteins. Plant HSFs are grouped together within HSFA, B, and C with further sub-groups existing within the respective groups. In addition, HSF proteins also comprise a C-terminal activation domain (CTAD) with an AHA motif, often nuclear localization (NLS) and nuclear export signals (NES)4. The intra-HSF protein domain interactions usually regulate the activation and cellular localization of HSF proteins. Under natural conditions, the HSFA monomer, containing one C-terminal and three N-terminal leucine zipper repeats, is suppressed by an association with HSPs to inactivate this protein in the cytosol. A bi-partite NLS sequence flanking the N-terminal zippers confers nuclear localization. An interaction between the N- and C-terminal zippers in the HSFA monomer masks the NLS sequence. In addition, the interaction of HR-A/B motifs maintains HSFA in a monomeric form and negatively regulates CTAD under normal conditions. Upon HT stress, several proteins in the cell misfold, to which HSPs interact and become dissociated from HSFA. This dissociation allows HSFA to form trimers, expose the NLS sequence and translocate to the nucleus to trigger transcription. The DBD of the trimeric HSF recognizes at least three copies of a typical penta-nucleotide sequence, 5′-nGAAn-3′, in the HSE to regulate HSP transcription3. With much sequenced plant genome data available, a large number of HSFs were characterized in several plant species, and their putative roles were predicted through gene expression studies3. The genome-wide analysis of HSF genes in various plants has revealed their regulatory roles not only in HT stress but also in other abiotic stress responses. This finding emphasizes their possible involvement in a complex crosstalk among the different stress response pathways4. Hence, HSFs are excellent candidate genes for genetic engineering, gene editing or breeding of climate-resilient crops. A thorough delineation of these key factors at the genome-scale is indispensable to the target species before they can be harnessed in any genetic improvement programme.
Flax (Linum usitatissimum L.) is an important global cash crop producing seed oil (linseed) and bast tissue-derived fibre (linen) as economic products. For various reasons, there is a renewed interest in the cultivation and advanced scientific study of flax. Seed oil from flax is an abundant source of alpha-linolenic acid (ALA) and omega-3 fatty acid. It serves as an exceptional food, feed and industrial feedstock for several purposes. In addition, the cellulosic stem fibre serves as a source for fine textile-grade fabric, the geotextile industry, the composite industry, and the paper and pulp industry5,6. Since flax is a rabi season crop, HT stress is one of the major limiting factors of flax cultivation, especially at the terminal stages. A cold temperature over an extended period is essential for fibre maturation. Thus, the adaptability of elite fibre flax genotypes to warmer climates is extremely poor. The HT stress of 40 °C, over five to seven days, affects pollen viability, boll formation, and seed setting7,8. However, the genetically variable superior alleles of HSPs and HSFs can be harnessed to breed flax varieties with an enhanced capacity to adapt to warm climatic conditions. A comprehensive analysis of the HSF genes from the flax genome9 is thus apt for the genetic improvement of flax with an enhanced resilience to adverse climatic conditions. In this study, we identified and characterized HSFs from the flax genome. The characterization included phylogeny, evolutionary time, and gene expression analysis in tissues and with HT stress treatment. We also identified guide RNA (gRNA) sequences from the LusHSFs to be used in functional studies and genetic improvement through gene editing with the aim of obtaining minimal off-target genomic effects.
Results
HSF gene identification in the flax genome and their sequence features
A search for HSFs Hidden Markov Model (HMM)-based Pfam ID PF00447 in the genome of L. usitatissimum (cv. CDC Bethune) hosted in the Phytozome database produced 40 sequences. The individual HSF protein sequences were further supported by scanning against the Pfam-A database at the E-value threshold of 10−3 and the Simple Modular Architecture Research Tool (SMART) web server for the presence of the characteristic HSF-DBD and coiled-coil structures. Finally, the 40 putative HSF protein sequences were analysed in the HEATSTER database, revealing six loci (Lus10005925, Lus10016634, Lus10022546, Lus10026819, Lus10029852, and Lus10038874) consisting of incomplete domains that are essential for classification as HSF proteins (Supplementary Table S1). These six loci were removed from further analysis because they lacked the essential ‘coiled coil’ oligomeric domain (HR-A/B region), which functions through trimerization upon HT response. The remaining 34 HSF sequences consisting of characteristic DBD, HR-A, and HR-B motifs were named LushsfA1a to LushsfC1b based on their classification in the HEATSTER database (Table 1). Other domains, such as NLS, NES, activator motifs (AHA), and tetrapeptide repressor domain (RD), were also located on the LusHSF proteins. As per Table 1, the length of the LusHSF genes and their CDS ranged from 912 bp (LushsfA4c) to 3585 bp (LushsfA1d) and 273 bp (LushsfB5b) to 1473 bp (LushsfA4d), respectively. The amino acid sequence length of the LusHSF proteins varied from 200 (LushsfB5b) to 822 (LushsfB1a) amino acids. The molecular weight (Mw) and isoelectric points (pI) of the LusHSF proteins ranged from 23.19 (LushsfB5b) to 55.15 (LushsfA4d) kDa and 4.78 (LushsfA8b) to 9.32 (LushsfB5a), respectively. The GRAVY score of each LusHSF protein was found to be negative, ranging from −0.995 to −0.499, indicating that these proteins are highly polar molecules. Subcellular localization predictions of the LusHSF proteins based on the k-nearest neighbour classifier of the WoLF PSORT program showed that most of these proteins are localized in the nucleus.
Table 1.
Features of LusHSF genes and proteins in the flax genome.
| HSF ID | Phytozome | HSF | Location | Strand | Length | pI | GRAVY | Mw (kD) | Subcel loc and no. NN | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| locus | group | Chr* | Start* | End* | Gene | CDS | Protein | ||||||
| LushsfA1a | Lus10011065 | A1a | Lu08 | 18101211 | 18103152 | reverse | 1941 | 1377 | 458 | 4.82 | −0.578 | 50.06 | nu: 14 |
| LushsfA1b | Lus10040911 | A1b | Lu15 | 8448889 | 8450468 | forward | 1579 | 1377 | 458 | 5.65 | −0.583 | 50.64 | nu: 14 |
| LushsfA1c | Lus10000312 | A1c | Lu08 | 16643985 | 16645926 | reverse | 1941 | 1377 | 458 | 4.95 | −0.602 | 50.01 | nu: 14 |
| LushsfA1d | Lus10030956 | A1d | Lu09 | 5719143 | 5722728 | reverse | 3585 | 1416 | 471 | 4.90 | −0.778 | 52.43 | nu: 14 |
| LushsfA1f | Lus10040091 | A1f | Lu07 | 4032070 | 4035485 | forward | 3415 | 1416 | 471 | 4.72 | −0.770 | 52.11 | nu: 14 |
| LushsfA2a | Lus10013797 | A2a | Lu11 | 2671577 | 2672762 | reverse | 1185 | 1026 | 341 | 5.01 | −0.682 | 38.43 | nu: 14 |
| LushsfA2b | Lus10039134 | A2b | Lu14 | 18116147 | 18117412 | forward | 1265 | 1110 | 369 | 5.22 | −0.649 | 41.46 | nu: 13, cp: 1 |
| LushsfA3a | Lus10023866 | A3a | Lu08 | 4723718 | 4725601 | forward | 1883 | 1326 | 441 | 4.73 | −0.570 | 49.81 | nu: 14 |
| LushsfA3b | Lus10014369 | A3b | Lu06 | 14835072 | 14836968 | forward | 1896 | 1341 | 446 | 4.71 | −0.561 | 50.31 | nu: 14 |
| LushsfA4a | Lus10007318 | A4a | Lu15 | 2083596 | 2084949 | reverse | 1353 | 1242 | 413 | 4.97 | −0.648 | 46.38 | nu: 14 |
| LushsfA4b | Lus10015237 | A4b | Lu12 | 2940951 | 2942258 | reverse | 1307 | 1212 | 403 | 5.75 | −0.880 | 45.68 | nu: 14 |
| LushsfA4c | Lus10029269 | A4c | Lu03 | 8165193 | 8166105 | reverse | 912 | 801 | 266 | 6.01 | −0.623 | 30.58 | nu: 13, cp: 1 |
| LushsfA4d | Lus10005420 | A4d | Lu07 | 17858515 | 17860349 | forward | 1834 | 1473 | 490 | 7.92 | −0.602 | 55.15 | nu: 5, cp: 1, cy: 2, mt: 2, vac: 2, er: 1, gb:1 |
| LushsfA6a | Lus10006618 | A6a | Lu08 | 22068530 | 22070272 | forward | 1742 | 1185 | 394 | 5.56 | −0.734 | 44.47 | nu: 14 |
| LushsfA6b | Lus10039376 | A6b | Lu10 | 3252307 | 3253927 | forward | 1620 | 1122 | 373 | 5.11 | −0.650 | 42.35 | nu: 13, cp: 1 |
| LushsfA7a | Lus10000492 | A7a | Lu14 | 4577433 | 4578881 | forward | 1448 | 1125 | 374 | 5.52 | −0.737 | 42.24 | nu: 13 |
| LushsfA7b | Lus10014698 | A7b | Lu01 | 26411220 | 26412688 | reverse | 1468 | 1137 | 378 | 5.51 | −0.780 | 42.96 | nu: 14 |
| LushsfA7c | Lus10022083 | A7c | unmapped | - | - | reverse | 1410 | 1131 | 376 | 5.75 | −0.744 | 43.11 | nu: 14 |
| LushsfA8a | Lus10003707 | A8a | Lu03 | 4843561 | 4845792 | reverse | 2231 | 1122 | 373 | 4.97 | −0.703 | 42.42 | nu: 13, per: 1 |
| LushsfA8b | Lus10001591 | A8b | Lu04 | 8648138 | 8650354 | reverse | 2216 | 1122 | 373 | 4.78 | −0.724 | 42.72 | nu: 13, per: 1 |
| LushsfA9a | Lus10027627 | A9a | Lu11 | 14143005 | 14144526 | forward | 1521 | 1422 | 473 | 5.03 | −0.697 | 53.45 | nu: 14 |
| LushsfA9b | Lus10011941 | A9b | Lu10 | 7901682 | 7903160 | reverse | 1478 | 1377 | 458 | 5.19 | −0.696 | 51.76 | nu: 14 |
| LushsfB1a | Lus10019348 | B1a | Lu02 | 1244140 | 1247030 | forward | 2890 | 273 | 822 | 4.81 | −0.995 | 30.15 | nu: 14 |
| LushsfB1c | Lus10009351 | B1c | Lu13 | 5055375 | 5058490 | forward | 3115 | 852 | 283 | 4.84 | −0.991 | 31.44 | nu: 11, cp: 1, cy: 1, pm: 1 |
| LushsfB2a | Lus10014994 | B2a | Lu14 | 19043189 | 19044240 | forward | 1051 | 924 | 307 | 5.58 | −0.694 | 33.51 | nu: 14 |
| LushsfB2b | Lus10008007 | B2b | Lu07 | 6384378 | 6385552 | reverse | 1174 | 1098 | 365 | 4.80 | −0.554 | 39.28 | nu: 13, per: 1 |
| LushsfB2d | Lus10024508 | B2d | Lu09 | 7452072 | 7454461 | reverse | 2389 | 1227 | 408 | 5.27 | −0.499 | 44.34 | nu: 13, per: 1 |
| LushsfB4a | Lus10042646 | B4a | Lu09 | 17698210 | 17699546 | reverse | 1336 | 1233 | 410 | 8.25 | −0.656 | 45.07 | nu: 14 |
| LushsfB4b | Lus10036062 | B4b | Lu01 | 503457 | 505257 | reverse | 1800 | 951 | 316 | 6.21 | −0.899 | 36.54 | nu: 14 |
| LushsfB4c | Lus10001133 | B4c | Lu10 | 13207020 | 13208344 | reverse | 1324 | 1215 | 404 | 7.84 | −0.704 | 44.83 | nu: 14 |
| LushsfB5a | Lus10011185 | B5a | Lu15 | 837963 | 839813 | forward | 1850 | 615 | 204 | 9.32 | −0.812 | 23.78 | nu: 13.5, cy_nu: 0.5 |
| LushsfB5b | Lus10018488 | B5b | Lu08 | 23772105 | 23774084 | reverse | 1979 | 603 | 200 | 9.27 | −0.668 | 23.19 | nu: 12.5, cy_nu: 7, cp 1 |
| LushsfC1a | Lus10023636 | C1a | Lu11 | 15833912 | 15834932 | forward | 1020 | 921 | 306 | 7.78 | −0.715 | 34.63 | nu: 14 |
| LushsfC1b | Lus10034907 | C1b | Lu12 | 8134272 | 8135285 | forward | 1013 | 924 | 307 | 6.98 | −0.695 | 34.46 | nu: 14 |
Chromosomal location estimated as per You et al.10.
pI: isoelectric point, GRAVY: Grand average of hydropathicity, Mw(kD): Molecular weight in kilo Dalton, Subcel loc and no. NN: Subcellular location and number of nearest neighbours, nu: nucleus, mt: mitochondria, cp: chloroplast, per: peroxisome, cy: cytosol, vac: vacuole, ER: endoplasmic reticulum, gb: golgi body, cy_nu: dual localization cytosol and nucleus, pm: plasma membrane.
Chromosomal distribution and gene duplications
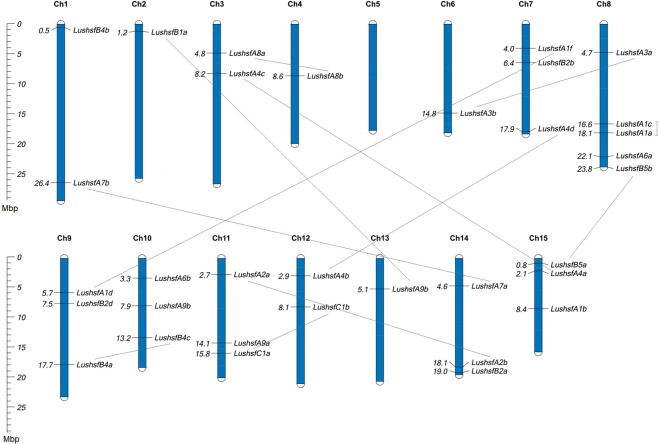
The genomic coordinates of the LusHSF genes on the scaffolds and flax chromosomes10 allowed us to estimate the physical location of these genes. Except for one gene, we found all LusHSF genes were randomly distributed on 14 out of the 15 flax chromosomes (Fig. 1). LushsfA7c, which is located on scaffold 87, remains unmapped because the entire scaffold has yet to be mapped on any chromosome. Not a single LusHSF gene was mapped on chromosome 5, while chromosome 8 consists of a maximum of five LusHSF genes. Four chromosomes, viz. 2, 4, 6 and 13, consisted of one LusHSF gene each. Gene expansion by duplication of the LusHSFs was checked through sequence homology analysis and their distribution patterns on the chromosomes. These analyses disclosed that twelve LusHSF genes have homologous gene pairs with >70% sequence identity and >90% query coverage. Eleven LusHSF genes have their duplicate counterparts (paralogues) distributed in separate chromosomes, while one pair, viz. LushsfA1c and LushsfA1a, are located on the same chromosome (Fig. 1). To further investigate whether this interspersed pattern of gene duplication resulted from segmental gene duplications, we compared LusHSF genes and their adjacent genomic regions using the GEvo tool of the CoGe database. Most of the putative LusHSF paralogues and their adjacent regions evolved because of local genomic rearrangements or microcolinearity (see Supplementary Fig. S1). This result indicates that segmental duplication played a significant part in the expansion of the LusHSF genes.
Figure 1.
Chromosomal locations and duplication of LusHSF genes. Each bar represents the flax chromosome with the chromosome number shown above the bars. Chromosomal lengths are represented in Mbp. All 34 LusHSF genes are mapped on 14 out of the 15 flax chromosomes. The numbers on the left side of the chromosomes represent their physical positions in Mbp from top to bottom. Putative paralogous LusHSF genes are depicted through connected lines.
Phylogenetic relationships of LusHSFs
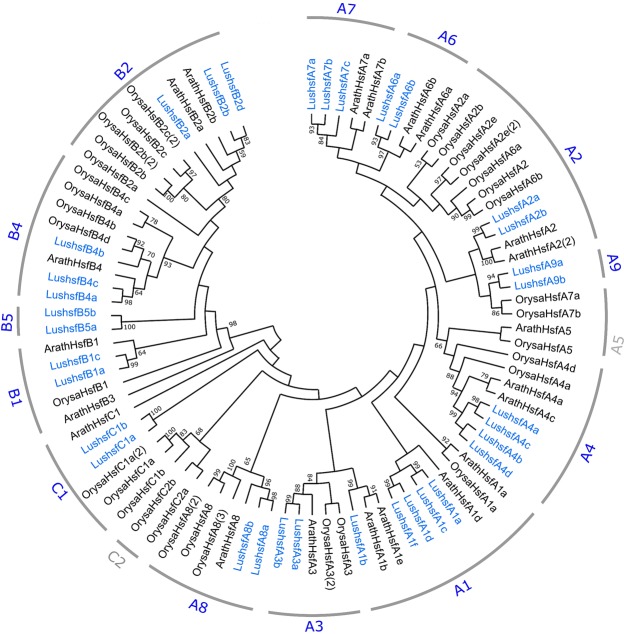
Employing multiple sequence alignment to Arabidopsis thaliana HSF (AtHSF) and Oryza sativa (OsHSF) proteins, the LusHSF proteins were classified into diverse groups, and a Maximum Likelihood (ML) tree was constructed based on highest log likelihood score (−4581.19) (Fig. 2). The best amino acid substitution model was found Jones-Taylor-Thornton (JTT) with lowest Bayesian Information Criterion (BIC) score of 14963.17. As per the phylogenetic tree, the LusHSFs were clustered into three broad groups, A, B, and C, and a total of 13 sub-groups, A1, A2, A3, A4, A6, A7, A8, A9, B1, B2, B4, B5 and C1, according to the HSF proteins grouped in clusters. These groupings were supported by high bootstrap values (>90%). All of the LusHSFs in the phylogenetic tree corroborated the classifications obtained from the HEATSTER database (Table 1). Neither of the LusHSF proteins was clustered in A5 and C2 sub-groups. Two LusHSF proteins, LushsfB5a and LushsfB5b, clustered separately in B5 sub-group, whereas AtHSFs and OsHSFs lack members from B5. Sub-group A1 comprised the most LusHSF proteins (five), followed by sub-group A4 (four). In the comparative phylogenetic analysis of HSFs with other plant species from the Malpighiales order and other fibre crops (such as cotton and jute), LusHSFs were grouped distinctly from the other proteins (see Supplementary Fig. S2).
Figure 2.
Phylogenetic clustering of LusHSFs, AtHSFs and OsHSFs. The phylogenetic relationship tree was inferred from the Maximum Likelihood (ML) method and JTT + G + I matrix-based model in MEGA-X. Domain-centric alignment of amino acid sequences from DBD and OD domains were performed using the MUSCLE algorithm with maximum 16 iterations. Thirty-four LusHSF, 21 AtHSF and 33 OsHSF proteins were clustered into 3 broad classes A, B, and C and 15 sub-classes within. Sub-groups marked in grey did not consist any LusHSFs. Bootstrap support values of >50% are shown on the nodes.
Organization of gene structure
The gene structure pattern, including the exon and introns on the LusHSF genes, was analysed by comparing the respective coding sequence and genomic sequences (Fig. 3). Introns were found in all 34 LusHSF genes, which ranged from one to three. The pattern of occurrence, position and length of the introns were found similar among the LusHSFs grouped under different sub-categories. The closely associated members of the same HSF group shared similar intron numbers and lengths, except for the LushsfA4d and LushsfB2d. A maximum of three introns was observed in the LushsfA4d sequence. The longest intron sequence (2.263 kbp) was found in LushsfB1c, followed by LushsfA1d (2.169 kbp). The smallest intron length, 76 bp, was observed in the LushsfB2d gene. In all LusHSF genes, an intron sequence was observed within the HSF-DBD, thus splitting the domain into two. The splicing phase class of all the introns within the HSF-DBD was observed as ‘0’ (i.e., between two codons resulting in unchanged frames or intact codon), except in LushsfA4d, where an intron splicing phase in one of the three introns was observed as ‘1’ (i.e., splitting codons between the first and second nucleotides). The presence of a single intron within the HSF-DBD region is one of the common features of plant HSFs that might have a possible role in mediating alternate splicing in genes that encode diverse protein products.
Figure 3.
Gene structure showing the distribution of exons and introns of LusHSF genes. A phylogenetic ML tree rectangular diagram of LusHSFs genes is shown on the left. The lengths of the boxes and lines were scaled based on gene length. Blue bars represent exons, while thin black lines indicate introns. The green bars denote the position of the HSF DBD on exons. The numbers indicate splicing phases of LusHSF genes: 0 for phase 0 and 1 for phase 1.
Conserved protein domain and motif predictions
A systematic examination of all 34 LusHSF protein sequences revealed positions and sequences of discrete conserved motifs and domains (Table 2 and Fig. 4). Six types of conserved domains, DBD, OD (HR-A/B), NLS, NES, AHA, and RD, were identified within the LusHSF protein sequences. Except in one protein (LushsfB2d), the DBD was found at the N-terminus of all the LusHSF proteins, followed by the HR-A/B motif and other conserved motifs. In LushsfB2d, RD and NLS motifs precede the DBD. This finding indicated that the DBD and OD, comprising the HR-A/B motif, are the highly conserved domains on LusHSF proteins, followed by the NLS and NES domains. The NLS and NES domains, which are responsible for translocating HSF proteins to the nucleus, were found on most of the LusHSFs either individually or together, except in three proteins, LushsfB1c, LushsfB5b, and LushsfB1a (Table 2). A thorough look at the multiple sequence alignments of the DBD revealed a highly structured domain of conserved motifs that forms three bundles of alpha helices (α1, α2 and α3) and four antiparallel beta strands (see Supplementary Fig. S3a). However, minor differences in the DBD length and amino acid sequence insertions were observed in the DBD alignment, notably in the LushsfA2b sequence. Compared to the amino acid sequence alignment of the DBD, the HR-A/B motif, which forms a coiled-coil structure, was found to be more variable (see Supplementary Fig. S3b). Typically, HR-A and HR-B are two conserved motifs with sequence inserts between them. The HR-A motif was absent or partial in six of the 34 LusHSFs, while ten LusHSFs consisted of partial or no HR-B motifs.
Figure 4.
Distribution of conserved domains of the LusHSF proteins. A phylogenetic ML tree rectangular diagram of LusHSFs is shown on the left. Proteins with DBD and OD (HR-A/B motif) were scaled according to their lengths. Domain and motif legends are provided below the protein-length scale. For the detailed positions of the domains and motifs, see Table 2.
Table 2.
Conserved domains and motifs on LusHSF proteins.
| LusHSF ID | Group | DBD | HR-A/B | NLS | NES | AHA1 | AHA2 | AHA3 | AHA4 | RD | NLS and NES domain sequences |
|---|---|---|---|---|---|---|---|---|---|---|---|
| LushsfA1a | A1a | 39–134 | 141–233 | 236–253 | 436–450 | — | — | — | — | — | NLS-SRRISESSKKRRLKQDGV; NES-PMENMDQLTQQMGLL |
| LushsfA1b | A1b | 9–104 | 110–202 | 205–222 | 437–451 | — | 392–410 | — | — | — | NLS-SRRIVGGTKKRRLPAHEG; NES-KVEGMNYLTEQMGLL |
| LushsfA1c | A1c | 39–134 | 141–233 | 236–253 | 436–450 | — | — | — | — | — | NLS-TRRISESSKKRRLKQDGV; NES-PMENMDQLTQQMGLL |
| LushsfA1d | A1d | 43–138 | 147–239 | 242–259 | 449–463 | — | — | — | — | — | NLS-NRRITETNKKRRLKQDRT; NES-KTEHVDQLTEQMGHL |
| LushsfA1f | A1f | 43–138 | 147–239 | 242–259 | 449–463 | — | — | — | — | — | NLS-NRRIAETNKKRRLKQDGT; NES-KSEHVDQLTEQMGHL |
| LushsfA2a | A2a | 36–142 | 158–237 | — | 326–334 | — | 298–325 | — | — | — | NES-QDLVDQMGFL |
| LushsfA2b | A2b | 36–179 | 189–268 | — | 359–364 | 316–328 | — | — | — | — | NES-QMGFL |
| LushsfA3a | A3a | 68–186 | 193–260 | 264–273 | — | — | — | — | — | — | NLS-TTRKKFIRHN |
| LushsfA3b | A3b | 68–187 | 195–262 | 273–282 | — | — | — | 403–412 | 420–426 | — | NLS-RTRKKFIRHN |
| LushsfA4a | A4a | 6–118 | 123–202 | 206–225 | 396–410 | 336–372 | 376–393 | — | — | — | NLS-RKRRLPRISCYDDPMLEDST; NES-NVNSLAGHIGPLTPA |
| LushsfA4b | A4b | 6–118 | 126–205 | 206–225 | 386–400 | 327–363 | 368–383 | — | — | — | NLS-RKRRLPRLGCIDDDPMKEDT; NES-SVNGLAGQVGPLTPA |
| LushsfA4c | A4c | 6–118 | 123–202 | 206–225 | — | — | — | — | — | — | NLS-RKRRLPRISCYDDPMLEDST |
| LushsfA4d | A4d | 6–119 | 126–205 | 210–229 | — | 331–367 | 372–387 | — | — | — | NLS-RKRRLPRLDCIDDDPMKEDT |
| LushsfA6a | A6a | 38–139 | 144–225 | 227–249 | 362–380 | — | — | — | — | — | NLS-HQKGIMRELENAITKKRRRRPID; NES-RNVDVLLEQLGFFLPPPY |
| LushsfA6b | A6b | 36–137 | 142–223 | 225–237 | 356–373 | 325–338 | — | — | — | — | NLS-HQKGIMRELENAITKKRRRRPID; NES-GNVDVLVEQLGFLDSDIM |
| LushsfA7a | A7a | 34–135 | 158–239 | 241–251 | 356–373 | 335–247 | — | — | — | — | NLS-QRKEKRKELEEALSKKRRRPID; NES-EDVNTLAEQLGYLSSSSP |
| LushsfA7b | A7b | 34–135 | 159–240 | 242–263 | 360–377 | 339–351 | — | — | — | — | NLS-QRKEKRKELEEALSKKRRRPIE; NES-EDVNTLAEQLGYLSSSSP |
| LushsfA7c | A7c | 40–141 | 162–243 | 255–265 | 347–364 | 323–335 | — | — | — | — | NLS-NNKRRRRRPIG; NES-EAVTALAEQLGYLPIRLK |
| LushsfA8a | A8a | 10–110 | 137–208 | 209–216 | — | — | — | — | — | — | NLS-SWRMAEPG |
| LushsfA8b | A8b | 10–109 | 137–208 | 209–216 | — | — | — | — | — | — | NLS-SWRMAEPG |
| LushsfA9a | A9a | 128–231 | 249–342 | — | — | 403–431 | 436–456 | — | — | — | — |
| LushsfA9b | A9b | 120–223 | 243–336 | — | — | — | — | — | — | — | — |
| LushsfB1a | B1a | 1–100 | 165–213 | — | — | — | — | — | — | 236–244 | — |
| LushsfB1c | B1c | 1–100 | 173–222 | 273–283 | 245–253 | — | — | — | — | 247–253 | NLS-GPRAKEIKICY |
| LushsfB2a | B2a | 19–136 | 167–216 | 255–264 | — | — | — | — | — | 237–254 | NLS-KKRGREEGGG |
| LushsfB2b | B2b | 37–154 | 186–235 | 296–305 | —— | — | — | — | — | 279–295 | NLS-KRARIEEEEE |
| LushsfB2d | B2d | 82–199 | 229–278 | 339–348 | — | — | — | — | — | 321–338 | NLS-KRVRIDEEEE |
| LushsfB4a | B4a | 27–126 | 221–267 | 357–363 | — | — | — | — | — | 341–356 | NLS-NNHNHN |
| LushsfB4b | B4b | 30–133 | 191–237 | 293–297 | — | — | — | — | — | 275–292 | NLS-KKRQL |
| LushsfB4c | B4c | 27–126 | 216–262 | 356–359 | 381–394 | — | — | — | — | 337–355 | NLS-HHHN; NES-SSKSHRLVLEKDDL |
| LushsfB5a | B5a | 25–136 | 156–196 | 199–204 | — | — | — | — | — | — | NLS-KNRRTC |
| LushsfB5b | B5b | 26–137 | 157–197 | — | — | — | — | — | — | — | — |
| LushsfC1a | C1a | 20–117 | 134–197 | 198–212 | — | — | — | — | — | — | NLS-KLDHRKKRCLMTSIS |
| LushsfC1b | C1b | 15–113 | 129–192 | 193–207 | — | — | — | — | — | — | NLS-KLDHRKKRCLMALVS |
DBD: DNA-bind domain; HR-A/B: heptad repeat A (N-terminus) or B (C-terminus) domain; NLS: nuclear localization signal; NES: nuclear export signal; AHA: aromatic and hydrophobic amino acid residues embedded in an acidic context; RD: repressor domain.
DNA interaction interface predictions on LusHSF proteins
Identification of protein-protein interaction sites and protein-DNA binding sites on the LusHSF proteins through the PredictProtein server showed a change in the number and location of the active sites (see Supplementary Fig. S4a). Except in LushsfA3a, all LusHSF amino acid sequences were predicted to have these macro-molecular interaction sites. Twenty-two out of the 34 LusHSFs consisted of polynucleotide binding sites. The diversity of the DNA contact points or active DNA binding sites on the LusHSF proteins was further analysed utilizing the protein model-based server TFmodeller, which revealed that most of the LusHSF proteins have DNA contact sites in the N-ring, i.e., purine or pyrimidine through six amino acid interface residues (Table 3). These DNA contact sites were predicted from a matrix of homologous interface contacts by comparing structurally related protein-DNA complexes. The six amino acid residues included serine (S), glutamine (Q), asparagine (N), threonine (T) and two arginine (R) residues. These residues are conserved in all the contact sites, except for LushsfA8b and LushsfA8a, where threonine (T) is replaced with isoleucine (I). The only notable diversity of these DNA contact sites is generally owing to the positional variance of these six amino acid residues in the protein sequence, typically residing between 60 and 221 amino acids from the N-terminus. With our findings, the template human heat shock factor protein model 5d5v_B chain was compared to reveal protein-DNA interface sites on most of the LusHSFs (see Supplementary Fig. S4b). Four LusHSF proteins found no homologous templates to model the protein-DNA interface. The specificity, which represents the evolutionary proportion of sequence-specific contacts for the complex, was almost comparable, 0.26–0.27 (except in four non-homologous LusHSFs, 0.04), in all the LusHSFs, but the level of entropy varied from 0.73 to 1.00.
Table 3.
Details of DNA binding site predictions on LusHSF proteins.
| HSF_ID | No of contact sites | No of contact sites in N-ring | N-ring contact sites (position and amino acid) | Specificity | Entropy | PDB_ID of model |
|---|---|---|---|---|---|---|
| LushsfA1a | 22 | 6 | 93-S, 96-R, 97-Q, 99-N, 100-T, 131-R | 0.27 | 0.73 | 5d5v_B |
| LushsfA1b | 22 | 6 | 63-S, 66-R, 67-Q, 69-N, 70-T, 101-R | 0.27 | 0.87 | 5d5v_B |
| LushsfA1c | 22 | 6 | 93-S, 96-R, 97-Q, 99-N, 100-T, 131-R | 0.27 | 0.81 | 5d5v_B |
| LushsfA1d | 22 | 6 | 97-S, 100-R, 101-Q, 103-N, 104-T, 135-R | 0.27 | 0.83 | 5d5v_B |
| LushsfA1f | 22 | 6 | 97-S, 100-R, 101-Q, 103-N, 104-T, 135-R | 0.27 | 0.86 | 5d5v_B |
| LushsfA2a | 22 | 6 | 97-S, 100-R, 101-Q, 103-N, 104-T, 135-R | 0.27 | 0.86 | 5d5v_B |
| LushsfA2b | 24 | 1 | 172-R | 0.04 | 0.88 | — |
| LushsfA3a | 24 | 1 | 176-R | 0.04 | 0.91 | — |
| LushsfA3b | 24 | 1 | 177-R | 0.04 | 0.82 | — |
| LushsfA4a | 22 | 6 | 64-S, 67-R, 68-Q, 70-N, 71-T, 102-R | 0.27 | 0.86 | 5d5v_B |
| LushsfA4b | 22 | 6 | 64-S, 67-R, 68-Q, 70-N, 71-T, 102-R | 0.27 | 0.94 | 5d5v_B |
| LushsfA4c | 22 | 6 | 64-S, 67-R, 68-Q, 70-N, 71-T, 102-R | 0.27 | 0.89 | 5d5v_B |
| LushsfA4d | 22 | 6 | 64-S, 67-R, 68-Q, 70-N, 71-T, 102-R | 0.27 | 0.84 | 5d5v_B |
| LushsfA6a | 23 | 6 | 97-S, 100-R, 101-Q, 103-N, 104-T, 135-R | 0.26 | 0.82 | 5d5v_B |
| LushsfA6b | 23 | 6 | 95-S, 98-R, 99-Q, 101-N, 102-T, 133-R | 0.26 | 0.89 | 5d5v_B |
| LushsfA7a | 22 | 6 | 93-S, 96-R, 97-Q, 99-N, 100-T, 131-R | 0.27 | 0.83 | 5d5v_B |
| LushsfA7b | 22 | 6 | 93-S, 96-R, 97-Q, 99-N, 100-T, 131-R | 0.27 | 0.87 | 5d5v_B |
| LushsfA7c | 22 | 6 | 99-S, 102-R, 103-Q, 105-N, 106-T, 137-R | 0.27 | 0.86 | 5d5v_B |
| LushsfA8a | 22 | 6 | 66-S, 69-R, 70-Q, 72-N, 73-I, 104-R | 0.27 | 0.87 | 5d5v_B |
| LushsfA8b | 22 | 6 | 66-S, 69-R, 70-Q, 72-N, 73-I, 104-R | 0.27 | 0.97 | 5d5v_B |
| LushsfA9a | 22 | 6 | 183-S, 186-R, 187-Q, 189-N, 190-T, 221-R | 0.27 | 0.89 | 5d5v_B |
| LushsfA9b | 22 | 6 | 175-S, 178-R, 179-Q, 181-N, 182-T, 213-R | 0.27 | 0.90 | 5d5v_B |
| LushsfB1a | 23 | 6 | 60-S, 63-R, 64-Q, 66-N, 67-T, 98-R | 0.26 | 0.79 | 5d5v_B |
| LushsfB1c | 24 | 1 | 111-R | 0.04 | 0.93 | - |
| LushsfB2a | 22 | 6 | 79-S, 82-R, 83-Q, 85-N, 86-T, 117-R | 0.27 | 0.88 | 5d5v_B |
| LushsfB2b | 22 | 6 | 97-S, 100-R, 101-Q, 103-N, 104-T, 135-R | 0.27 | 0.97 | 5d5v_B |
| LushsfB2d | 22 | 6 | 142-S, 145-R, 146-Q, 148-N, 149-T, 180-R | 0.27 | 0.97 | 5d5v_B |
| LushsfB4a | 23 | 6 | 83-S, 86-R, 87-Q, 89-N, 90-T, 121-R | 0.26 | 0.98 | 5d5v_B |
| LushsfB4b | 22 | 6 | 90-S, 93-R, 94-Q, 96-N, 97-T, 128-R | 0.27 | 0.97 | 5d5v_B |
| LushsfB4c | 23 | 6 | 83-S, 86-R, 87-Q, 89-N, 90-T, 121-R | 0.26 | 0.95 | 5d5v_B |
| LushsfB5a | 23 | 6 | 83-S, 86-R, 87-Q, 89-N, 90-T, 121-R | 0.26 | 1.00 | 5d5v_B |
| LushsfB5b | 23 | 6 | 84-S, 87-R, 88-Q, 90-N, 91-T, 122-R | 0.26 | 0.99 | 5d5v_B |
| LushsfC1a | 23 | 6 | 76-S, 79-R, 80-Q, 82-N, 83-T, 114-R | 0.26 | 0.89 | 5d5v_B |
| LushsfC1b | 23 | 6 | 71-S, 74-R, 75-Q, 77-N, 78-T, 109-R | 0.27 | 0.96 | 5d5v_B |
S-serine; R-arginine, Q-glutamine, N-asparagine, T-threonine, I-isoleusine ‘–’ no model.
Orthologues of LusHSFs, syntenic relationships and divergence time
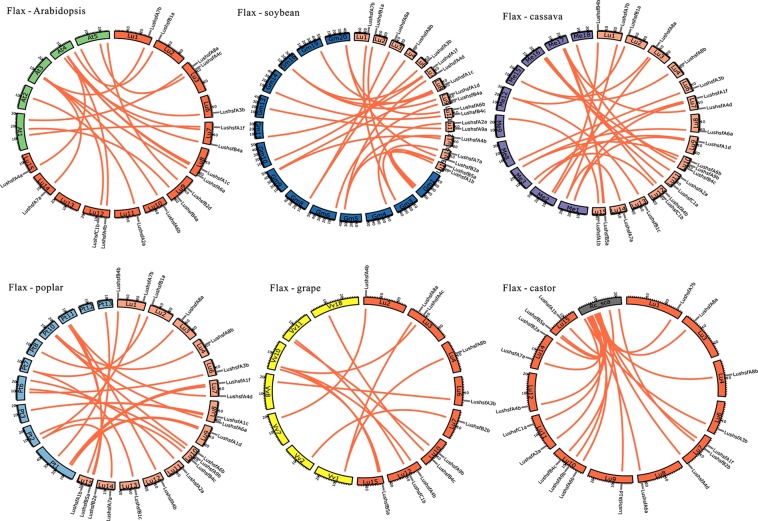
Putative orthologues of LusHSFs genes were predicted using the reciprocal protein blast approach through crb-blast and OrthoFinder software. HSF proteins from three related plant systems, such as Populus trichocarpa, Ricinus communis, Manihot esculenta, and three additional plant systems where the HSF genes are well characterized, such as A. thaliana, Vitis vinifera, and Glycine max, were compared to LusHSF proteins. The crb-blast showed that 31 LusHSF proteins matched to 87 unique HSF hits. OrthoFinder placed the 34 LusHSF proteins into eleven orthogroups and matched to 140 HSF hits. Of the 34 LusHSF proteins, thirty-one (91.2%) were consistent in both programmes and had orthologues in at least one of these six species. A maximum of 17 LusHSF orthologues was related to both P. trichocarpa HSFs (36.2%) and M. esculenta HSFs (43.6%), while a minimum of nine orthologues (47.4%) was related to the V. vinifera HSFs (Supplementary Table S2. The synteny map of the above LusHSF orthologous genes revealed that these genes are conserved and are randomly assigned in most of the chromosomes of the orthologous species (Fig. 5). To determine the evolutionary status of the putative LusHSF gene paralogues and orthologues, the ratio of substitution rates of non-synonymous (dN) versus synonymous (dS) sites was computed for each pair of duplicated genes. The dN/dS ratios computed for all the putative paralogues and orthologues varied from 0.0065 (LushsfA3a-Glyma.09G190600.1) to 0.6022 (LushsfA3b-LushsfA3a). The overall distribution of the dN/dS ratios is presented in Fig. 6a and Supplementary Table S2. The average and median dN/dS ratios were lowest, 0.096 and 0.105, for the putative LusHSFs and Arabidopsis HSF orthologues, respectively, while these values were highest, 0.268 and 0.234, for the putative LusHSF paralogues, respectively (Supplementary Table S2). In general, the dN/dS ratio was <1.0, indicating that these duplicated genes are under negative or purifying selection pressure. The dN/dS ratios were further used to predict gene duplication times in terms of million years ago (MYA) for each of these putative paralogous and orthologous gene pairs (Supplementary Table S2). The time for the gene duplication of LusHSFs (average ~12.5 MYA, median ~10.6 MYA) was observed as a more recent event than that for the divergence of the orthologues (Fig. 6b). The latest duplication time was estimated at ~6.5 MYA (LushsfA7a-LushsfA7b) and with oldest duplication time occurring ~24.5 MYA (LushsfC1a-LushsfC1b). The median values for the divergence of LusHSFs from the orthologues of P. trichocarpa HSFs were predicted as the latest (~186.2 MYA), while the earliest divergence time prediction was for orthologues from Arabidopsis HSFs (~259.7 MYA). Among the LusHSF orthologues analysed, five gene pairs, viz. LushsfB1a- AT4G36990.1, LushsfA1a- Glyma.09G206600.2, LushsfC1b- Glyma.09G190600.1, LushsfA2a-29912.m005526, and LushsfB2a-30147.m014282, showed dS values > 10 and predicted highly conserved evolutionary times, dating back >1000 MYA.
Figure 5.
Syntenic relationships among putative orthologues and LusHSF genes. The syntenic relations of LusHSF genes to Arabidopsis, soybean, cassava, poplar, grape and castor were plotted using CIRCOS v0.69-5. The chromosomal positions of the syntenic HSF gene pairs are represented with red links. LusHSF genes are labeled on flax chromosomes, and the chromosome numbers are mentioned in the karyotype chords.
Figure 6.

Box and whisker plots showing comparative distribution of (a) substitution rates of non-synonymous over synonymous site (dN/dS) and (b) estimated time of gene duplication (MYA) in putative paralogues and orthologues of LusHSFs. The top of the box or coloured region represents the 3rd quartile (Q3, maximum values) while the bottom of the box or white region represents the 1st quartile (Q1, lower values). The ends of whiskers represent maximum and minimum values 1.5 times above or lower the Q3 and Q1, respectively. The maximum and minimum outlier values are represented as open circles and star symbols, respectively. In (b), the detailed distribution of gene duplication times (in MYA) of putative LusHSF paralogous gene pairs are shown separately below the comparative figure.
Cis-acting element localization on LusHSF promoters
Since the promoter of a gene often consists of cis-acting regulatory elements that confer its functional specificity, we analysed the distribution of cis-elements in the 1000 bp upstream promoter sequence of LusHSF genes. First, our analysis with the TSSP program Softberry showed that four out of 34 LusHSF promoters comprised unverified bases, thus restricting their lengths to less than 1000 bp for the analysis. Putative promoter positions based on the transcription start site (TSS) were predicted in a total of 24 (70.6%) LusHSF upstream sequences (Table 4). Four of these sequences showed more than one putative TSS position. The location of the putative TATA box sequences in the 14–38 bp region upstream of the TSS was predicted in 23 out of 24 LusHSF promoters. Enhancer elements were predicted in 14 of the LusHSF promoters, of which two promoters consisted of more than one enhancer element. Next, our analysis of the distribution of cis-acting regulatory elements in the promoter sequences of LusHSFs demonstrated the existence of various regulatory elements related to the abiotic-stress response (Table 4). These elements include ABRE (abscisic acid responsive element), CCAAT-box, DRE/CRT/CBF (dehydration-responsive element/C-repeat/C-repeat binding factors), HSE, LTRE (low-temperature response element), MBS (MYB-binding site), and PRECONSCRHSP70 (plastid response element in the promoters of heat shock protein 70 A). Although the software programs PlantCARE and PLACE both predicted abiotic stress-related regulatory elements, in a majority of the LusHSF promoters, both programs varied in the number of predicted elements. PlantCARE predicted a smaller number of elements compared to PLACE. In agreement with the PlantCARE program, a considerable number of LusHSF promoters were found to consist of HSE and LTRE, which are linked to impart tolerance to high and low temperatures. In addition, a significant number of elements of ABRE, MBS, and TC-rich repeats were also located, which are likely to be induced under dehydration stress. Each of two LusHSF promoters consisted of DRE and CCAAT box elements; the former is responsible for dehydration stress tolerance, and the latter is involved in interactions with an HSE element to enhance heat shock promoter activity. The program PLACE predicted a considerable number of MYB/MYC transcription factor-binding sites ranging from zero to 31, followed by CRT/DRE/CBF and LTRE. A significant number of cis-elements associated with heat shock protein 70 (HSP70) were also located on the LusHSF promoters by the program PLACE. Altogether, the above results show that the LusHSF promoters are enriched with numerous potential cis-acting elements related to the abiotic-stress response.
Table 4.
Details of promoter analysis in the 1000 bp upstream sequence of the LusHSF genes. Position of first nucleotide of putative TSS/TATA box/Enhancer from the 5’ end of the upstream sequence analyzed and not from the start codon. NP- No prediction; PRECON70#- PRECONSCRHSP70.
| Gene_ID | Prom_len | TSSP (SOFTBERRY) predictions* | Number of cis-elements (PlantCARE) | Number of cis-elements (PLACE | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TSS | TATA box | Enhancer | ABRE | CCAAT-box | DRE | HSE | LTR | MBS | TC-rich repeats | ABRE | MYB | LTRE | PRECON70# | CRT/DRE/CBF | ||
| LushsfA1a | 1000 | NP | — | — | 2 | 0 | 0 | 0 | 0 | 0 | 1 | 2 | 13 | 1 | 1 | 3 |
| LushsfA1b | 1000 | 749 | 725 | — | 1 | 0 | 0 | 0 | 2 | 0 | 0 | 7 | 14 | 5 | 1 | 5 |
| LushsfA1c | 1000 | 550 | 512 | — | 2 | 0 | 0 | 1 | 2 | 0 | 1 | 1 | 14 | 2 | 0 | 0 |
| LushsfA1d | 1000 | 931 | 916 | — | 0 | 0 | 0 | 1 | 0 | 2 | 0 | 2 | 15 | 2 | 1 | 2 |
| LushsfA1f | 1000 | 450 | 415 | — | 0 | 0 | 0 | 2 | 0 | 1 | 1 | 1 | 21 | 4 | 2 | 2 |
| LushsfA2a | 1000 | 892 | 855 | 761 | 1 | 0 | 0 | 0 | 0 | 3 | 0 | 0 | 18 | 0 | 0 | 2 |
| LushsfA2b | 1000 | NP | — | — | 0 | 0 | 0 | 0 | 0 | 4 | 0 | 0 | 18 | 3 | 2 | 3 |
| LushsfA3a | 1000 | 940 | 927 | 869 | 2 | 0 | 0 | 0 | 0 | 2 | 1 | 3 | 24 | 3 | 2 | 6 |
| LushsfA3b | 1000 | 941 | 927 | 869 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 3 | 24 | 2 | 1 | 2 |
| LushsfA4a | 1000 | NP | — | — | 0 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 15 | 0 | 2 | 0 |
| LushsfA4b | 1000 | NP | — | — | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 15 | 1 | 2 | 6 |
| LushsfA4c | 1000 | NP | — | — | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 16 | 0 | 1 | 1 |
| LushsfA4d | 1000 | 292 | 256 | — | 0 | 0 | 0 | 1 | 0 | 1 | 2 | 0 | 18 | 1 | 3 | 6 |
| LushsfA6a | 1000 | 895; 461 | 860; 425 | 928 | 0 | 0 | 0 | 0 | 0 | 2 | 2 | 1 | 11 | 0 | 3 | 8 |
| LushsfA6b | 1000 | 905 | 868 | 926 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 17 | 1 | 2 | 7 |
| LushsfA7a | 1000 | 828 | 795 | 155; 874 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 3 | 1 | 0 | 0 |
| LushsfA7b | 1000 | 822 | 789 | 861 | 0 | 0 | 0 | 0 | 1 | 1 | 2 | 0 | 19 | 2 | 0 | 4 |
| LushsfA7c | 748 | 438 | 412 | 696 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 2 | 11 | 0 | 1 | 1 |
| LushsfA8a | 1000 | NP | — | — | 0 | 0 | 0 | 1 | 2 | 1 | 0 | 4 | 12 | 3 | 2 | 3 |
| LushsfA8b | 1000 | NP | — | — | 0 | 0 | 0 | 2 | 0 | 6 | 0 | 2 | 27 | 2 | 2 | 11 |
| LushsfA9a | 1000 | 633 | 616 | — | 4 | 0 | 0 | 6 | 1 | 1 | 1 | 4 | 18 | 2 | 3 | 2 |
| LushsfA9b | 939 | NP | — | — | 4 | 0 | 0 | 0 | 0 | 1 | 0 | 4 | 25 | 1 | 2 | 2 |
| LushsfB1a | 1000 | 607; 192 | 575; 160 | 615 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 14 | 1 | 1 | 0 |
| LushsfB1c | 1000 | 626; 175 | 592; 156 | 632 | 0 | 0 | 0 | 2 | 0 | 1 | 1 | 0 | 9 | 1 | 4 | 0 |
| LushsfB2a | 1000 | NP | — | — | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 1 | 9 | 4 | 0 | 3 |
| LushsfB2b | 1000 | 578 | 540 | 582 | 2 | 0 | 0 | 1 | 1 | 0 | 2 | 3 | 12 | 2 | 0 | 0 |
| LushsfB2d | 1000 | 425 | 395 | 231 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 2 | 7 | 0 | 2 | 0 |
| LushsfB4a | 1000 | 223 | 203 | 922; 315 | 0 | 0 | 0 | 2 | 2 | 0 | 2 | 0 | 11 | 3 | 0 | 0 |
| LushsfB4b | 1000 | NP | — | — | 0 | 0 | 1 | 0 | 1 | 3 | 0 | 0 | 31 | 3 | 1 | 5 |
| LushsfB4c | 287 | 209 | — | — | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| LushsfB5a | 769 | 723 | 697 | — | 1 | 0 | 0 | 2 | 0 | 0 | 0 | 2 | 3 | 0 | 0 | 1 |
| LushsfB5b | 1000 | 945 | 931 | — | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 2 | 24 | 1 | 1 | 3 |
| LushsfC1a | 1000 | 948 | 913 | — | 2 | 0 | 0 | 1 | 1 | 1 | 0 | 6 | 10 | 4 | 1 | 1 |
| LushsfC1b | 1000 | 925; 108 | 892; 70 | 936 | 2 | 0 | 0 | 0 | 1 | 3 | 1 | 3 | 17 | 2 | 1 | 4 |
Gene expression dynamics of LusHSFs in different tissues
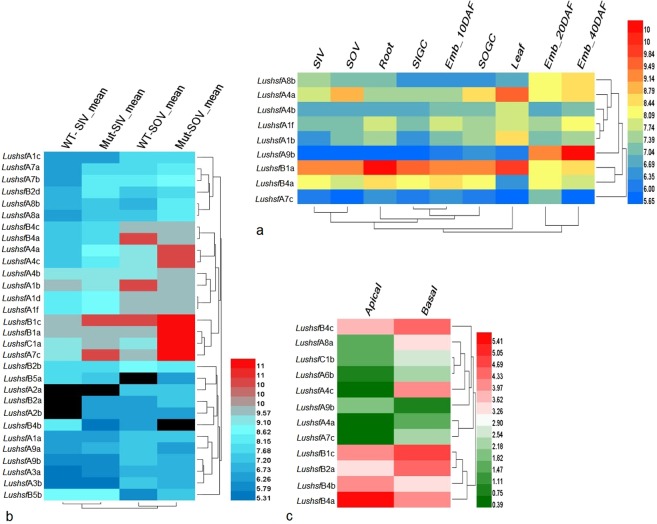
A homology search of LusHSF genes against the microarray data (Accession no. GSE21868) revealed only nine high-quality unigene hits (>95% identity). The fewer number of LusHSF hits to the microarray data could be attributed to the expressed sequence tags (ESTs) of the Hermes cultivar used to develop the array rather than the flax genome of CDC Bethune. Nonetheless, these nine LusHSF genes revealed a differential gene expression pattern in different flax tissues (Fig. 7a). On a closer look, the LushsfB1a, which belongs to the B1 group, was found to have higher gene expression in most of these tissues, while LushsfA7c was expressed at low levels. HSF genes from the B1 group are heat inducible and are known for their role in repressing other HSF genes under non-heat conditions. Interestingly, the LushsfA9b gene, which belongs to the A9 group, was less abundant in all tissues but was highly expressed in the late embryo developmental stages. HSF genes from A9 groups are known for their involvement in seed development. Similarly, in another microarray dataset (GSE61311), eight LusHSFs exhibited differential expression patterns in inner and outer stem tissues at the vegetative stage of the wild and mutant genotypes (Fig. 7b). Compared to the microarray data of LusHSFs, the differentially expressed transcriptome resources from the shoot apex of the flax variety CDC Bethune (GSE80718) showed higher hits of 27 LusHSF genes. Twelve of these LusHSF genes showed differential expression patterns in the apical and basal tissues (Fig. 7c). Five LusHSF genes were expressed in abundance and three genes showed low expression in both tissues. However, four LusHSF genes showed contrasting expression patterns in these tissues. The hierarchical clustering of the LusHSF genes in all the above digital gene expression analyses was found in accordance with their expression patterns. From the above digital gene expression analysis, we speculate that the majority of LusHSF genes differ in their expression patterns in various flax tissues and growth stages.
Figure 7.
Heat map and hierarchical clustering of digital gene expression of LusHSF genes in different flax tissues. (a) LusHSF corresponding gene IDs were derived from the microarray data under GEO accession no. GSE21868. The mean of RMA-normalized, averaged gene-level signal intensity (log2) values were plotted using the Heatmap Illustrator (HemI v.1.0). Tissue includes SIV: stem inner tissue from the vegetative stage; SOV: stem outer tissue from the vegetative stage; root; leaf; SIGC: inner stem at the green capsule stage; SOGC: outer stem at the green capsule stage; and embryo at 10, 20, and 40 days post flowering. (b) The normalized signal intensity values of the LusHSF genes derived from the transcriptome data under GEO accession no. GSE61311 is plotted as a heat map. Digital samples include WT-SIV: inner stem tissue from the vegetative stage of wild-type plants; mut-SIV: inner stem tissue from the vegetative stage of lignified bast fibre mutant plants; WT-SOV: outer stem tissue from the vegetative stage of wild type plant; and mut-SOV: outer stem tissue from the vegetative stage of lignified bast fibre mutant plant. (c) Heat map generated for the LusHSFs derived from RNA-seq data (Accession no. GSE80718) using the log2 transformed average FPKM values. In all heat map plots, the coloured bars shown on the right represent their expression levels.
Expression pattern of LusHSF genes under HT stress
We examined the expression pattern of the LusHSF genes under HT stress by comparing two different fibre flax cultivars, European Viking and Indian JRF-2, to measure the mRNA abundance in the shoot apex of 30 day-old control and HT-stressed (40 °C for 12 hrs) seedlings. From a preliminary screening, twelve LusHSF genes produced clear and consistent bands of expected size in both control and HT stressed samples. The remaining LusHSF genes either showed the presence/absence of bands or comprised non-specific amplicons (data not shown). The RT-qPCR analysis of the twelve LusHSF genes produced a differential expression pattern in the control and HT stressed plants (Fig. 8). Interestingly, in control JRF-2, the expression of a majority of the LusHSF genes was elevated when compared to that of the other samples (0.82 to 34.2-fold). In contrast, most of the LusHSF genes were down-regulated in the HT stress-treated JRF-2 (0.44 to 15.33) compared to those in the JRF-2 control plant. However, the LushsfA1c and LushsfA1a, were reasonably up-regulated in HT stress-exposed JRF-2. In HT-stressed Viking, the LushsfA7a and LushsfB2b genes were significantly up-regulated compared to those in control Viking and HT-stressed JRF-2 plants. Two genes, LushsfA1b and LushsfB4a, produced non-significant gene expression changes in all the samples compared to those in control Viking. Altogether, these differential expression patterns suggest their possible functional relevance in the HT stress response in a genotype-dependent manner.
Figure 8.
Relative quantification (RT-qPCR) of selective LusHSF genes between HT-treated and control plants of JRF-2 and Viking cultivars. (a) The upper panel shows the effect of HT stress treatments at 40 °C for 12 hrs in flax cultivars, which were used for total RNA extractions. (b) The relative gene expression fold change (2−ΔΔCt) of twelve selected LusHSF genes are represented as bar diagrams. The Ct values of each sample-HSF gene combination were normalized using the reference gene ETIF3E and calibrated with Ct values of Viking control to estimate (2−ΔΔCt) values. The statistical significance of the expression values is represented by ‘ns’ as non-significant, and ‘*‘ as significant at p > 0.05 and Bonferroni’s multiple comparisons test. One asterisk (*) represents adjusted P values between 0.01 and 0.05, and two asterisks (**) represent adjusted P values between 0.01 and 0.001, and so on. RNA samples include Vik_C: Viking under control conditions; Vik_H: Viking under HT stress conditions; JRF-2_C: JRF-2 under control conditions; JRF-2_H: JRF-2 under HT stress conditions.
Prediction of CRISPR/Cas9 guide sequences with minimum off-target effects in the flax genome
We screened the LusHSF genes using an online CRISPOR tool to identify unique 20 bp gRNA sequences for each LusHSF gene. These gRNA sequences, which will serve as a resource for clustered regularly interspaced short palindromic repeats/CRISPR-associated 9 (CRISPR/Cas9)-based gene editing or functional studies, were compared and aligned to the L. usitatissimum genome. The gRNA sequences with the highest specificity and those located within 12 bp adjacent to the protospacer adjacent motif (PAM) sequence (the ‘seed region’) of the gRNA were considered for assessing minimum off-target effects. The total number of gRNA predictions for each LusHSF ranged from 74 to 223 with the 3′ PAM sequence NGG (where N = A/T/G/C). At a specificity score >50 (cutoff for high specificity), the number of gRNA sequences ranged from nine to 157. The gRNA sequences with the highest specificity score and minimum off-target effects are mentioned in Supplementary Table S3. Most of these gRNA sequences had the least off-target hits, ranging from 0 to 21 at the whole genome level, which may arise from 2 to 4 nucleotide mismatches. None of the gRNA sequences was predicted to produce off-target effects (up to ≤4 nucleotide mismatches) within the seed region, i.e., within the 12 bp adjacent to the PAM sequence. The forward and reverse primers were predicted for cloning and expression of all the LusHSF gRNA sequences using the T7 RNA polymerase-based system in the popular gene editing vector DR274 (Addgene plasmid # 42250). Specific restriction enzyme sequences were also predicted within the gRNA sequence at three bp 5′ to the PAM to facilitate the screening of mutation events induced by the gRNA in CRISPR experiments. Oligonucleotides with barcodes and corresponding sequencing primers to generate lentiviral saturation mutagenesis screens with the LusHSF genes, are also shown in Supplementary Table S3.
Discussion
From past studies on HT stress in fibre flax, it is perceived that both low and HT stress, even in the absence of drought, are critical to flax growth and reproduction7,8. Seed germination, flowering, and seed setting in flax are optimum between temperatures of 16 °C to 25 °C. In a simulated experiment, HT during the initial growth phase, followed by a low temperature in the intermediate phase and HT during the late growth stages were observed as the most preferred conditions for fibre flax growth11. However, exposure to more than 40 °C for a stretch of five days during flowering in flax was detrimental, reducing seed yield and fibre quality7. Partial to complete necrosis of the ovules was the crucial limiting factor in poor seed setting due to HT stress in flax. A prolonged period of HT stress also forces the plant to undergo compensatory flowering8. This information warrants prioritized research on the genetic improvement of flax, especially fibre types, for terminal HT stress tolerance. In the long run, these findings will facilitate the acclimation of the superior fibre quality flax genotypes to a diverse climatic condition.
Among various genetic components, HSFs and HSPs play significant functions in responding to HT stress in plants. The former gene group plays the role of a regulatory partner in the functioning of the latter group, which serves as chaperones12. A fundamental knowledge of the role interplayed by these two key genetic factors is crucial beforehand to design a genetic improvement strategy for HT stress tolerance in any plant. Although the genome sequence of flax is available for the past few years9, the characterization of HSFs in flax has remained obscure until now. The present study involved the revelation of 34 true HSF sequences distributed in 14 out of the 15 flax chromosomes. A cumulative analysis of flax and other representatives from the order Malpighiales and commercial fibre crops, whose genome sequences are available, revealed a diverse HSF family size. Our report of 34 non-redundant complete LusHSFs is higher than those of Ricinus sp. (18) and Corchorus sp. (18), but lower than those of Gossypium raimondii (57), Salix purpurea (48), P. trichocarpa (47), and M. esculenta (39) (http://planttfdb.cbi.pku.edu.cn). Considering that whole genome duplication (WGD) events during ancient polyploidization and lineage-specific duplications are crucial factors for the speciation and expansion of gene family13,14, variations in the number of HSFs in flax and related plants have shed light on how this gene family has co-evolved. From WGD time estimates in Malpighiales, it is clear that flax has undergone two rounds of genome duplication: the earlier duplication occurring ~20–40 MYA and a more recent genome duplication at 5–9 MYA compared to the other plants analysed15,16. Intraspecies synteny analysis revealed that many of the LusHSF genes in flax genome constitute part of the syntenic blocks that still support their WGD origin. Genome duplication events simultaneously with gene gains or losses might have contributed to the diversity of the HSF gene family17. Through orthologue identification and dN/dS substitution-based homology analysis of the LusHSFs, we could predict that the divergence time of HSFs in other related plant species occurred much earlier than those of flax, perhaps during ancient polyploidization event. Therefore, most of the putative LusHSF paralogues that co-evolved with the recent flax genome duplication event (5–9 MYA) might also correspond to diverse gene structures and functions.
In the present study, we describe a comprehensive characterization of the LusHSF genes and amino acid sequences to identify their important domains and motifs. All 34 selected LusHSF proteins comprised conserved characteristic domains, such as DBD, HR-A/B regions, NLS, NES, and CTAD; thus qualifying these proteins as true HSF proteins. Since the promoter regions are enriched with specialized cis-acting regulatory elements that also specify their putative functions18, we queried the promoter regions of the LusHSFs. The results revealed that the LusHSF promoters are enriched with a variety of regulatory elements related to abiotic stress tolerance, including the HSE and LTRE, which confer gene expression in response to high- and low-temperature conditions, respectively. From our digital gene expression analysis, using microarray data from the national center for biotechnology information (NCBI) database, evidence of the differential expression of the LusHSF genes was detected in different tissues. Transcriptional analysis of twelve LusHSF genes was also performed in two different fibre flax cultivars, Viking and JRF-2, under control and HT stress conditions. Interestingly, the analysis reveals that the abundance of the majority of the LusHSF mRNA is significantly higher in control JRF-2 (up to 34.2-fold) compared to that in control Viking. This difference may justify the better adaptability of the Indian JRF-2 cultivar under the hot and humid conditions of India compared to that of European Viking. In a few LusHSFs, a fold change in gene expression was also observed up to 16.74 times in the HT-stressed Viking and up to 15.33 times in the HT-stressed JRF-2. Overall, we noticed that the endogenous expression of LusHSFs in control JRF-2 was higher than that in the HT-stressed JRF-2. One possibility for the down-regulation of these LusHSFs in HT-stressed JRF-2 plants could be owing to prolonged HT stress treatment for over 12 hrs. A similar down-regulation of the HSF genes under HT shock treatment was recorded in plants3. All this information suggests that the LusHSFs might produce a differential response in different flax genotypes regarding HT stress and can be selected as candidate gene resources for functional studies and genetic improvements. Genetic engineering using candidate HSF genes was reported to impart enhanced thermotolerance to crop plants, such as in wheat19.
From our analysis of LusHSFs, we assume that these proteins are involved in differential functions in various tissues and under HT-stress induced responses. Since multifunctional HSF proteins have roles in various abiotic-stress tolerance responses3,20, the involvement of LusHSFs in regulating other traits cannot be ignored. Therefore, a comprehensive functional analysis of the LusHSF genes is a prerequisite before harnessing these molecules in any genetic improvement programme. For functional studies on LusHSFs, we were interested in determining the active macro-molecular binding sites on the LusHSF proteins. Computational predictions of the active sites for protein-DNA interactions can considerably reduce the cost and time of functional assays by providing a first-hand functional annotation. These computational predictions can be addressed using information from amino acid sequences or related protein structure models21. Through both the homology model and an amino acid sequence-guided approach, our analysis of the nucleoprotein interaction sites on the interface of LusHSFs aided us in identifying the active amino acid residues that might exert an effect on their functionalities, such as the HT stress response.
Genome editing using the CRISPR/Cas9 system is rapidly emerging as a tool for targeted gene knockout in plants, thus achieving functional analysis in a precise manner and in a rapid time22,23. Nevertheless, the success of CRISPR/Cas9 technology depends primarily on the specificity of the gRNA sequences designed to perform targeted gene-knockouts. CRISPOR is a simple web tool, that permits users to design gRNA for genome-wide CRISPR and saturation screens with information on the possible off-target effects on the genome of interest24. Our screen for gRNA sequences specific to the LusHSF genes produced sequences with minimum possible off-target effects, specifically within sequences adjacent to the PAM. This tool also predicted the gRNA cloning strategy for maximum functionality of the CRISPR system. Genome-wide analysis of a few important gene families, such as the genes controlling fatty acid biosynthesis25, chalcone synthase26, β-galactosidases27, cinnamyl alcohol dehydrogenase (CAD) genes28, NBS-LRR29, aquaporin30, pectinmethylesterases (PME) and pectinmethylesterase inhibitors (PMEI)31, UDP glycosyltransferase (UGT)32 and the dirigent protein family33 were carried out in the flax genome. However, designing the gRNA sequence for functional analysis of these gene families has not been attempted in any of these studies. Our present study, in addition to identifying active protein and DNA binding sites on LusHSF proteins, predicted and designed a number of gRNA sequences with the least off-target effects for functional analysis of the candidate LusHSF genes.
In summary, we identified 34 LusHSF genes with specific DNA and amino acid sequence features, plotted these genes on flax chromosomes and phylogenetically reconfirmed them into three broad groups and 13 subgroups based on their protein domains. The putative LusHSF paralogues were estimated as a recent gene duplication event than the orthologues in terms of their evolutionary gene family expansion. Functional predictions were based on various abiotic stress-related cis-acting elements detected in the promoter regions of the LusHSFs, the dynamics of digital gene expression patterns in different tissues, and the quantitative expression patterns of these genes under control and HT stress conditions. One of the key findings of the present study embodies the design of gRNAs for individual LusHSFs to promote further functional studies of this important gene family. However, a systematic analysis of gene expression under different temperatures and at different time intervals is imperative to assign a specific role to each candidate LusHSF gene before utilizing this gene resource in the genetic improvement of fibre flax for HT stress tolerance.
Methods
Retrieval and characterization of HSF sequences
Genomic, coding, protein and promoter sequences of the HSF gene family with conserved DBDs (Pfam ID: PF00447) from the L. usitatissimum (cv. CDC Bethune) genome were retrieved from the Phytozome database v12.1 (https://phytozome.jgi.doe.gov/pz/portal.html) using the BioMart tool. The protein sequences were confirmed using the batch search tool of the Hidden Markov Models (HMMs) in the Pfam 31.0 database34 with an E-value threshold of 10−3 and the Simple Modular Architecture Research Tool (SMART) for HSF DBD35. The putative LusHSF genes and their detailed classifications were further identified against the HEATSTER platform20 (http://www.cibiv.at/services/hsf/) and MARCOIL (http://toolkit.tuebingen.mpg.de/marcoil) to determine the presence of coiled-coil structures. The isoelectric point (pI) and molecular weight of LusHSF proteins were estimated from the Compute pI/Mw tool of Expassy36 (http://www.expasy.org/). The grand average of hydropathy (GRAVY) scores, which is based on the hydropathy of all the amino acids of a protein molecule and determines whether a protein is polar or non-polar in nature37, were estimated using the GRAVY calculator (http://www.gravy-calculator.de/). Protein subcellular localization of the LusHSFs were predicted by using WoLF PSORT38 (http://www.genscript.com/wolf-psort.html).
Chromosomal mapping and analysis of gene duplication
The genomic coordinates of the 34 LusHSF genes were mapped on flax chromosome10 using the software Graphical Geno Typing v2.0 (GGT 2.0)39 and MapChart v2.340. The paralogous relationships of the LusHSF genes were identified according to their duplication patterns (tandem or block) using conditional reciprocal blast (crb-blast)41 with a stringent E-value of 1.0e−50. The paralogous partners were identified based on query coverage >90% and percentage of identical matches >70%. Patterns of genome duplication among the putative LusHSF paralogues and their adjacent genomic regions were analysed using the GEvo tool of the CoGe database (https://genomevolution.org/coge/) and the (B)LastZ: Large Regions algorithm42.
Multiple sequence alignments, phylogeny, and classification
The amino acid sequences of the conserved DBD and OD with HR-A/B motifs of LusHSFs were deduced from the HEATSTER platform20,43 for multiple sequence alignments using the online Clustal Omega tool of EMBL-EBI44 (http://www.ebi.ac.uk/Tools/msa/clustalo/) and visualized using BoxShade v3.21 (http://www.ch.embnet.org/software/BOX_form.html). For phylogenetic tree reconstruction and reconfirming the classification of the LusHSFs, the amino acid sequences from the start of DBD to the end of OD domains of flax, Arabidopsis (dicot model plant), and rice (monocot) HSFs were retrieved for multiple sequence alignments. The alignment was performed using MUSCLE algorithm and 16 maximum iterations in the MEGA-X software45. The phylogenetic tree was inferred by using the Maximum Likelihood (ML) method and the Jones-Taylor-Thornton (JTT) matrix-based amino acid substitution model46. The best model was estimated using the model selection tool in MEGA-X and from the lowest Bayesian Information Criterion (BIC) score. A discrete gamma distribution was chosen to model evolutionary rate differences among sites [16 categories (+G, parameter = 1.6109)]. The initial tree for the heuristic search were obtained automatically by applying Neighbor-Join and BioNJ algorithms to a matrix of pairwise distances estimated using a JTT model, and then selecting the topology with superior log likelihood value. All positions with less than 95% site coverage were eliminated, i.e., fewer than 5% alignment gaps, missing data, and ambiguous sequences were allowed at any position (partial deletion option). The test of phylogeny was conducted using 1000 bootstrap replications. All other parameters of phylogenetic tree reconstruction were kept default. Using similar parameters, a phylogenetic ML tree was also reconstructed using the HSFs from closely related sequenced plants of the order Malpighiales and commercial fibre crops, like cotton and Corchorus spp.
Gene structures, protein domain distributions and DNA-binding site predictions
The exon/intron and splicing phase in LusHSF genes were derived by aligning the corresponding CDS and genome FASTA sequences in the Gene Structure Display Server (GSDS2.0) (gsds.cbi.pku.edu.cn/) programme47. The LusHSF DBD coordinates on the protein and the phylogenetic tree in Newick format were used as inputs to display the gene structures. The distribution of protein domains, such as DBD, OD (HR-A/B), NLS and NES, on the LusHSF amino acid sequences were determined from the online HSF prediction tool of the HEATSTER platform20,43, and Interproscan48. The conserved domains and motifs were visualized using the Illustrator for Biological Sciences (IBS) v.1.0.349 (http://ibs.biocuckoo.org/). Additionally, the LusHSF protein sequences were scanned for the prediction of protein-protein and protein-DNA binding interface identification using a FASTA sequence search approach in the online open PredictProtein server50 (https://open.predictprotein.org/) and comparative model-based TFmodeller web server51 (http://maya.ccg.unam.mx/$\sim$tfmodell/).
Orthologue identification, synteny mapping, and evolutionary analysis
Putative orthologues of LusHSF genes were identified from Arabidopsis, poplar (P. trichocarpa), castor bean (R. communis), cassava (M. esculenta), soybean (G. max), and grape (V. vinifera) HSFs, which were derived from the plant transcription factor database v.3.0 (PlantTFDB)52 (http://planttfdb.cbi.pku.edu.cn/index.php). The crb-blast program41 at an E-value of 1.0e−50 was employed for this purpose. The top query and subject BLAST hits were filtered using >70% identity and >90% query and subject coverage in the Microsoft Excel program. Orthologues were also inferred using the OrthoFinder v.1.1.853 and compared with crb-blast output. Only the consistent putative orthologues were used for synteny mapping. The corresponding genomic coordinates of the putative orthologous gene pairs were derived from the respective genomes in the Phytozome database v.12.1, and the orthologous relationships were visualized using CIRCOS v0.69-554. The dN/dS estimation of putative LusHSF homologue sequences (both orthologues and paralogues) was conducted using PAL2NAL (http://www.bork.embl.de/pal2nal/) in the codeml program in PAML55. The evolutionary time (T) or likely gene duplication event of the HSF genes was calculated in terms of million years ago (MYA) using a synonymous mutation rate of λ substitutions per synonymous site per year. A dN/dS ratio <1, >1, and = 1 indicates negative (purifying selection), positive, and neutral evolution, respectively.
Cis-acting regulatory element identifications
To predict the putative promoter region based on the transcription start site of plants (TSSP) in 1000 bp sequences upstream of LusHSF genes, the TSSP online program of SoftBerry (http://www.softberry.com/berry.phtml) was used. The unverified string of bases from the putative promoters was removed from the analysis. The cis-acting regulatory elements were searched on the putative promoter regions of LusHSF genes using plant cis-acting regulatory DNA elements (PLACE; https://www.hsls.pitt.edu/obrc/index.php?page=URL1100876009) and plant cis-acting regulatory elements (PlantCARE; http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) databases56,57. Both positive and negative promoter DNA strands were subjected to a cis-element search. Only the abiotic stress-related regulatory elements were retrieved.
Digital gene expression analysis
Gene expression data in terms of microarray and transcriptome sequences from the different flax tissues and developmental stages of the NCBI Gene Expression Omnibus (GEO) repository were downloaded to analyse the digital gene expression of LusHSF genes. Microarray data (accession number GSE21868), from inner- and outer-stems, embryo, leaves, and roots58 were subjected to a homology-based (blastn) similarity search with an E-value cutoff of 1.0e−50 to LusHSF sequences. The best LusHSF-aligned unigene sequences (≥95% identity) were considered to derive log2 values from the microarray data in robust multi-array average (RMA) values. The mean log2 values for different tissues are represented by a heatmap diagram. Similarly, other microarray data, such as GSE61311 (unpublished), with inner and outer stem tissues from a wild-type and its mutant (lignified bast fibre mutant 1) plants, and the RNA-seq data for the flax shoot apex (GSE80718)59, were searched, and corresponding fragments per kilobase of transcript per million mapped read (FPKM) values were log2 transformed before plotting in heatmaps. All heatmaps were generated using the Heatmap Illustrator (HemI v.1.0)60 and clustering was performed using the hierarchical method, average linkage, and Euclidean distance similarity metric.
Plant samples, HT stress treatment, and RT-qPCR analysis
The seeds of two different winter fibre flax cultivars, European Viking and Indian JRF-2 (Tiara), were grown under controlled glass-house conditions. The former cultivar was a French introduction, while the latter was a released variety for the conditions in India. Our initial field observation showed Viking as a heat-susceptible cultivar (deformed inflorescence, poor flowering, and seed setting) compared to JRF-2. Total RNA was extracted from the shoot apex tissues of 30-day-old control and HT stressed (40 °C for 12 hrs in a plant growth chamber) flax seedlings using TRIzol reagent (Invitrogen, Thermo Fisher Scientific, Inc., USA) according to the manufacturer’s instructions. Approximately, 5 μg/mL of DNaseI-treated total RNA was reverse transcribed using the SuperScript III First Strand cDNA synthesis system (Invitrogen Inc., USA) to generate cDNA according to the manufacturer’s protocol. Gene-specific (LusHSF) and a reference gene eukaryotic translation initiation factor (ETIF3E) primers61 were designed using the Quant Prime62 tool and synthesized at Eurofins Genomics India Private Limited, India (Supplementary Table S4). The RT-qPCR analysis was performed using PowerUpTM SYBR Green Master Mix (Applied Biosystems, Inc., USA) on a CFX Connect Real-Time PCR Detection System (Bio-Rad, Inc., USA). Each qPCR reaction (20 μL) consisted of 10 μL SYBR-Green mix, 4 μL cDNA template (120 ng), and 1.0 μL of 10 μM solution of each forward and reverse primers. The PCR cycling programme consisted of 50 °C for 2 min, 95 °C for 5 min followed by 40 cycles at 94 °C for 10 s, 55 °C for 20 s, and 68 °C for 30 s. To analyse the specificity of the amplicons, a melting curve analysis was performed at 95 °C for 30 s, 65 °C for 30 s, followed by ramping up to 95 °C with 0.5 °C increment per cycle. For each sample, three technical replicates were conducted to minimize the PCR artefacts. The relative expression of each selected gene was averaged from the differences in cycle threshold (Ct) values normalized against the reference gene and finally calibrated against the control RNA sample from the Viking accession. The relative quantification method (2−ΔΔCt)63 was plotted as fold change gene expression in all the samples utilizing the GraphPad Prism software trial version (https://www.graphpad.com/scientific-software/prism/). One-way ANOVA, followed by Bonferroni’s multiple comparisons test correction was employed to analyse the statistical hypothesis at p = 0.05.
Predictions of guide RNA sequences for gene editing and off-target effects
The web server CRISPOR v4.424 (http://crispor.tefor.net/) was employed to predict efficient gRNA sequences for CRISPR/Cas9-based gene editing experiments in LusHSFs. The genomic DNA sequences of LusHSFs were scanned as input sequences for the identification of unique 20 bp target gRNA sequences against the L. usitatissimum genome of Phytozome v.9. LusHSF genes with >2000 bp lengths were scanned up to <2000 bp from the 5′ translational start site as per the requirement of the tool. To facilitate the use of the popular CRISPR/Cas9 system and employ Streptococcus pyogenes Cas9 nuclease, the corresponding NGG trinucleotide was selected as the protospacer adjacent motif (PAM). The gRNA sequences for the respective LusHSFs were chosen based on the highest specificity score and least probable off-target cleavage sites, especially within the 12 bp region adjacent to the PAM, known as the ‘seed region’. The gRNA sequences are allowed at least four nucleotide mismatches for the probability of off-target effect predictions. Oligonucleotides for lentiviral saturation mutagenesis screening were also identified along with specific barcodes. In a saturating mutagenesis experiment, a target region of the genome is altered with many guides, to create as many DNA edits as possible followed by mutant phenotyping. The corresponding Illumina sequencing primers were also designed for each LusHSF with the Illumina adapters TCGTCGGCAGCGTCAGATGTGTATAAGAGACAG and GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAG to validate the gene sequence modifications.
Supplementary information
Acknowledgements
The authors acknowledge the ICAR-Central Research Institute for Jute and Allied Fibres for providing with the necessary funds and facilities to conduct the research under project file no. CI-JBT 10.3. The quantitative PCR studies carried out at the Faculty Centre for Integrated Rural Development and Management, Ramakrishna Mission Vivekananda Educational and Research Institute, Ramakrishna Mission Ashrama, Narendrapur, Kolkata is also acknowledged.
Author Contributions
D.S. conceptualized the study and carried out the in silico and RT-qPCR analyses. P.M. and S.D. prepared the plant samples for the RT-qPCR analysis. D.S. drafted the manuscript. K.M., S.K.S., A.B.M., T.D.G. and J.M. edited and approved the final manuscript.
Data Availability
All data generated or analysed in the study are included in this article or in its supplementary information files.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information accompanies this paper at 10.1038/s41598-019-41936-1.
References
- 1.Hasanuzzaman M, Nahar K, Alam MM, Roychowdhury R, Fujita M. Physiological, biochemical, and molecular mechanisms of heat stress tolerance in plants. Int. J. Mol. Sci. 2013;14:9643–9684. doi: 10.3390/ijms14059643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Feder ME, Hofmann GE. Heat-shock proteins, molecular chaperones, and the stress response: evolutionary and ecological physiology. Annu. Rev. Physiol. 1999;61:243–282. doi: 10.1146/annurev.physiol.61.1.243. [DOI] [PubMed] [Google Scholar]
- 3.Guo M, et al. The plant heat stress transcription factors (HSFs): structure, regulation, and function in response to abiotic stresses. Front. Plant Sci. 2016;7:114. doi: 10.3389/fpls.2016.00114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.von Koskull-Döring P, Scharf KD, Nover L. The diversity of plant heat stress transcription factors. Trends Plant Sci. 2007;12:452–457. doi: 10.1016/j.tplants.2007.08.014. [DOI] [PubMed] [Google Scholar]
- 5.Zuk M, Richter D, Matula J, Szopa J. Linseed, the multipurpose plant. Ind. Crop. Prod. 2015;75:165–177. doi: 10.1016/j.indcrop.2015.05.005. [DOI] [Google Scholar]
- 6.Hall, L. M., Booker, H., Siloto, R. M. P., Jhala, A. J. & Weselake, R. J. Flax (Linum usitatissimum L.). In McKeon, T., Hayes, D., Hildebrand, H. & Weselake, R. (eds) Industrial Oil Crops, 157–194 (AOCS Press, 2016).
- 7.Gusta LV, O’Connor BJ, Bhatty RS. Flax (Linum usitatissimum L.) responses to chilling and heat stress on flowering and seed yield. Can. J. Plant Sci. 1997;77:97–99. doi: 10.4141/P95-205. [DOI] [Google Scholar]
- 8.Cross RH, McKay SAB, McHughen AG, Bonham-Smith PC. Heat-stress effects on reproduction and seed set in Linum usitatissimum L. (flax) Plant Cell Environ. 2003;26:1013–1020. doi: 10.1046/j.1365-3040.2003.01006.x. [DOI] [Google Scholar]
- 9.Wang Z, et al. The genome of flax (Linum usitatissimum) assembled de novo from short shotgun sequence reads. Plant J. 2012;72:461–473. doi: 10.1111/j.1365-313X.2012.05093.x.. [DOI] [PubMed] [Google Scholar]
- 10.You FM, et al. Chromosome-scale pseudomolecules refined by optical, physical and genetic maps in flax. Plant J. 2018;95:371–384. doi: 10.1111/tpj.13944. [DOI] [PubMed] [Google Scholar]
- 11.Du GH, Liu FH, Rowland G. Fiber cell development and fiber yield of flax (Linum usitatissimum L.) affected by the seasonal temperature pattern. Can. J. Plant Sci. 2015;95:1215–1220. doi: 10.4141/cjps-2014-185. [DOI] [Google Scholar]
- 12.Wang W, Vinocur B, Shoseyov O, Altman A. Role of plant heat-shock proteins and molecular chaperones in the abiotic stress response. Trends Plant Sci. 2004;9:244–252. doi: 10.1016/j.tplants.2004.03.006. [DOI] [PubMed] [Google Scholar]
- 13.MacKintosh C, Ferrier DEK. Recent advances in understanding the roles of whole genome duplications in evolution. F1000Res. 2017;6:1623. doi: 10.12688/f1000research.11792.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Panchy N, Lehti-Shiu M, Shiu SH. Evolution of gene duplication in plants. Plant Physiol. 2016;171:2294–2316. doi: 10.1104/pp.16.00523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cai, L. et al. Widespread ancient whole-genome duplications in Malpighiales coincide with Eocene global climatic upheaval. New Phytol., 10.1111/nph.15357 (2018). [DOI] [PMC free article] [PubMed]
- 16.Ren R, et al. Widespread whole genome duplications contribute to genome complexity and species diversity in angiosperms. Mol. Plant. 2018;11:414–28. doi: 10.1016/j.molp.2018.01.002. [DOI] [PubMed] [Google Scholar]
- 17.Guo YL. Gene family evolution in green plants with emphasis on the origination and evolution of Arabidopsis thaliana genes. Plant J. 2013;73:941–951. doi: 10.1111/tpj.12089. [DOI] [PubMed] [Google Scholar]
- 18.Srinivasan, R. & Saha, D. Promoter trapping in plants using T-DNA mutagenesis. In Jain, S. & Brar, D. (eds) Molecular Techniques in Crop Improvement, 545–577 (Springer, 2010).
- 19.Ni Z, et al. Genetic improvement of heat tolerance in wheat: recent progress in understanding the underlying molecular mechanisms. Crop. J. 2018;6:32–41. doi: 10.1016/j.cj.2017.09.005. [DOI] [Google Scholar]
- 20.Scharf KD, Berberich T, Ebersberger I, Nover L. The plant heat stress transcription factor (HSF) family: structure, function and evolution. Biochim. Biophys. Acta. 2012;1819:104–119. doi: 10.1016/j.bbagrm.2011.10.002. [DOI] [PubMed] [Google Scholar]
- 21.Motion GB, Howden AJ, Huitema E, Jones S. DNA-binding protein prediction using plant specific support vector machines: validation and application of a new genome annotation tool. Nucleic Acids Res. 2015;43:e158. doi: 10.1093/nar/gkv805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Feng Z, et al. Efficient genome editing in plants using a CRISPR/Cas system. Cell Res. 2013;23:1229–1232. doi: 10.1038/cr.2013.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.He F, Zhang F, Sun W, Ning Y, Wang GL. A versatile vector toolkit for functional analysis of rice genes. Rice. 2018;11:27. doi: 10.1186/s12284-018-0220-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Haeussler M, et al. Evaluation of off-target and on-target scoring algorithms and integration into the guide RNA selection tool CRISPOR. Genome Biol. 2016;17:148. doi: 10.1186/s13059-016-1012-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.You FM, Li P, Kumar S, Ragupathy R, Li ZEA. Genome-wide identification and characterization of the gene families controlling fatty acid biosynthesis in flax (Linum usitatissimum L) J. Proteomics Bioinform. 2014;7:310–326. doi: 10.4172/jpb.1000334. [DOI] [Google Scholar]
- 26.Seung HE, Tae KH. Genome-wide identification and transcriptional expression analysis of chalcone synthase in flax (Linum usitatissimum L.) Gene Reports. 2016;5:51–56. doi: 10.1016/j.genrep.2016.08.011. [DOI] [Google Scholar]
- 27.Hobson N, Deyholos MK. Genomic and expression analysis of the flax (Linum usitatissimum) family of glycosyl hydrolase 35 genes. BMC Genomics. 2013;14:344. doi: 10.1186/1471-2164-14-344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Seung EH, Heekyu K, Tae HK. The cinnamyl alcohol dehydrogenase (CAD) gene family in flax (Linum usitatissimum L.): Insight from expression profiling of cads induced by elicitors in cultured flax cells. Arch. Biol. Sci. 2016;68:603–612. doi: 10.2298/ABS150921050E. [DOI] [Google Scholar]
- 29.Kale SM, Pardeshi VC, Barvkar VT, Gupta VS, Kadoo NY. Genome-wide identification and characterization of nucleotide binding site leucine-rich repeat genes in linseed reveal distinct patterns of gene structure. Genome. 2013;56:91–99. doi: 10.1139/gen-2012-0135. [DOI] [PubMed] [Google Scholar]
- 30.Shivaraj SM, et al. Genome-wide identification, characterization, and expression profile of aquaporin gene family in flax (Linum usitatissimum) Sci. Reports. 2017;7:46137. doi: 10.1038/srep46137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pinzon-Latorre D, Deyholos MK. Pectinmethylesterases (PME) and pectinmethylesterase inhibitors (PMEI) enriched during phloem fiber development in flax (Linum usitatissimum) PLoS One. 2014;9:e105386. doi: 10.1371/journal.pone.0105386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Barvkar VT, Pardeshi VC, Kale SM, Kadoo NY, Gupta VS. Phylogenomic analysis of UDP glycosyltransferase 1 multigene family in Linum usitatissimum identified genes with varied expression patterns. BMC Genomics. 2012;13:175. doi: 10.1186/1471-2164-13-175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Corbin C, et al. Genome-wide analysis of the flax (Linum usitatissimum L.) dirigent protein family: from gene identification and evolution to differential regulation. Plant Mol. Biol. 2018;97:73–101. doi: 10.1007/s11103-018-0725-x. [DOI] [PubMed] [Google Scholar]
- 34.Finn RD, et al. The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res. 2016;44:279–285. doi: 10.1093/nar/gkv1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Letunic I, Doerks T, Bork P. SMART: recent updates, new developments and status in 2015. Nucleic Acids Res. 2014;43:257–260. doi: 10.1093/nar/gku949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Artimo P, et al. ExPASy: SIB bioinformatics resource portal. Nucleic Acids Res. 2012;40:597–603. doi: 10.1093/nar/gks400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kyte J, Doolittle RF. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 38.Horton P, et al. WoLF PSORT: protein localization predictor. Nucleic Acids Res. 2007;35:585–587. doi: 10.1093/nar/gkm259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.van Berloo R. GGT 2.0: versatile software for visualization and analysis of genetic data. J. Hered. 2008;99:232–236. doi: 10.1093/jhered/esm109. [DOI] [PubMed] [Google Scholar]
- 40.Voorrips RE. MapChart: software for the graphical presentation of linkage maps and QTLs. J. Hered. 2002;93:77–78. doi: 10.1093/jhered/93.1.77. [DOI] [PubMed] [Google Scholar]
- 41.Aubry S, Kelly S, Kümpers BMC, Smith-Unna RD, Hibberd JM. Deep evolutionary comparison of gene expression identifies parallel recruitment of trans-factors in two independent origins of C4 photosynthesis. PLoS Genet. 2014;10:e1004365. doi: 10.1371/journal.pgen.1004365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lyons E, Freeling M. How to usefully compare homologous plant genes and chromosomes as DNA sequences. Plant J. 2008;53:661–673. doi: 10.1111/j.1365-313X.2007.03326.x. [DOI] [PubMed] [Google Scholar]
- 43.Berz J, et al. HEATSTER: a database and web server for identification and classification of heat stress transcription factors in plants. Bioinforma. Biol. Insights. 2019;13:1–5. doi: 10.1177/1177932218821365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sievers F, Higgins DG. Clustal Omega, accurate alignment of very large numbers of sequences. Methods Mol. Biol. 2014;1079:105–116. doi: 10.1007/978-1-62703-646-7_6. [DOI] [PubMed] [Google Scholar]
- 45.Kumar S, et al. molecular evolutionary genetics analysis acrossc computing platforms. Mol. Biol. Evol. 2018;35:1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jones DT, Taylor WR, Thornton JM. The rapid generation of mutation data matrices from protein sequences. Comput. Appl. Biosci. 1992;8:275–282. doi: 10.1093/bioinformatics/8.3.275. [DOI] [PubMed] [Google Scholar]
- 47.Hu B, et al. GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics. 2015;31:1296–1297. doi: 10.1093/bioinformatics/btu817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Finn RD, et al. InterPro in 2017 - beyond protein family and domain annotations. Nucleic Acids Res. 2017;45:D190–D199. doi: 10.1093/nar/gkw1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu W, et al. IBS: an illustrator for the presentation and visualization of biological sequences. Bioinformatics. 2015;31:3359–3361. doi: 10.1093/bioinformatics/btv362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yachdav G, et al. PredictProtein–an open resource for online prediction of protein structural and functional features. Nucleic Acids Res. 2014;42:W337–W343. doi: 10.1093/nar/gku366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Contreras-Moreira B, Branger PA, Collado-Vides J. TFmodeller: comparative modelling of protein-DNA complexes. Bioinformatics. 2007;23:1694–1696. doi: 10.1093/bioinformatics/btm148. [DOI] [PubMed] [Google Scholar]
- 52.Jin JP, Zhang H, Kong L, Gao G, Luo JC. PlantTFDB 3.0: a portal for the functional and evolutionary study of plant transcription factors. Nucleic Acids Res. 2014;42:D1182–D1187. doi: 10.1093/nar/gkt1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Emms DM, Kelly S. OrthoFinder: solving fundamental biases in whole genome comparisons dramatically improves orthogroup inference accuracy. Genome Biol. 2015;16:157. doi: 10.1186/s13059-015-0721-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Krzywinski M, et al. Circos: an information aesthetic for comparative genomics. Genome Res. 2009;19:1639–1645. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Suyama M, Torrents D, Bork P. PAL2NAL: robust conversion of protein sequence alignments into the corresponding codon alignments. Nucleic Acids Res. 2006;34:W609–W612. doi: 10.1093/nar/gkl315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Higo K, Ugawa Y, Iwamoto M, Korenaga T. Plant cis-acting regulatory DNA elements (PLACE) database. Nucleic Acids Res. 1999;27:297–300. doi: 10.1093/nar/27.1.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lescot M, et al. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002;30:325–327. doi: 10.1093/nar/30.1.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fenart S, et al. Development and validation of a flax (Linum usitatissimum L.) gene expression oligo microarray. BMC Genomics. 2010;11:592. doi: 10.1186/1471-2164-11-592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhang N, Deyholos MK. RNA-Seq analysis of the shoot apex of flax (Linum usitatissimum) to identify phloem fiber specification genes. Front. Plant Sci. 2016;7:950. doi: 10.3389/fpls.2016.00950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Deng W, Wang Y, Liu Z, Cheng H, Xue Y. HemI: a toolkit for illustrating heatmaps. PLoS One. 2014;9:e111988. doi: 10.1371/journal.pone.0111988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Huis R, Hawkins H, Neutelings G. Selection of reference genes for quantitative gene expression normalization in flax (Linum usitatissimum L.) BMC Plant Biol. 2010;10:71. doi: 10.1186/1471-2229-10-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Arvidsson S, Kwasniewski M, Riano-Pachon DM, Mueller-Roeber B. QuantPrime - a flexible tool for reliable high-throughput primer design for quantitative PCR. BMC Bioinforma. 2008;9:465. doi: 10.1186/1471-2105-9-465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analysed in the study are included in this article or in its supplementary information files.