Abstract
A direct role for cholesterol signaling in mammals is clearly established; yet, the direct role in signaling for a plant sterol or sterol precursor is unclear. Fluctuations in sitosterol and stigmasterol levels during development and stress conditions suggest their involvement in signaling activities essential for plant development and stress compensation. Stigmasterol may be involved in gravitropism and tolerance to abiotic stress. The isolation of stigmasterol biosynthesis mutants offers a promising tool to test the function of sterol end products in signaling responses to developmental and environmental cues.
Keywords: stigmasterol, sterol end products, cell signaling, physiology, abiotic, sterol pathway regulation
Introduction
Unlike mammals and fungi, plants produce mixtures of sterols, including sitosterol, stigmasterol, campesterol, and cholesterol (Figure 1). The interaction of sterols with phospholipids helps plant cells to maintain plasma membrane fluidity and permeability during stress conditions (Grunwald, 1971; Hartmann, 1998). In addition, sterols are precursors in the synthesis of steroid hormones, e.g., testosterone, estrogen, glucocorticoids, and mineral corticoids in mammals, ecdysteroids in insects and crustaceans, antheridiol and oogoniol (mating hormones of fungi), and brassinosteroids (BR) in plants (Fujioka et al., 1997; Noguchi et al., 1999; Nomura et al., 1999; Clouse, 2002).
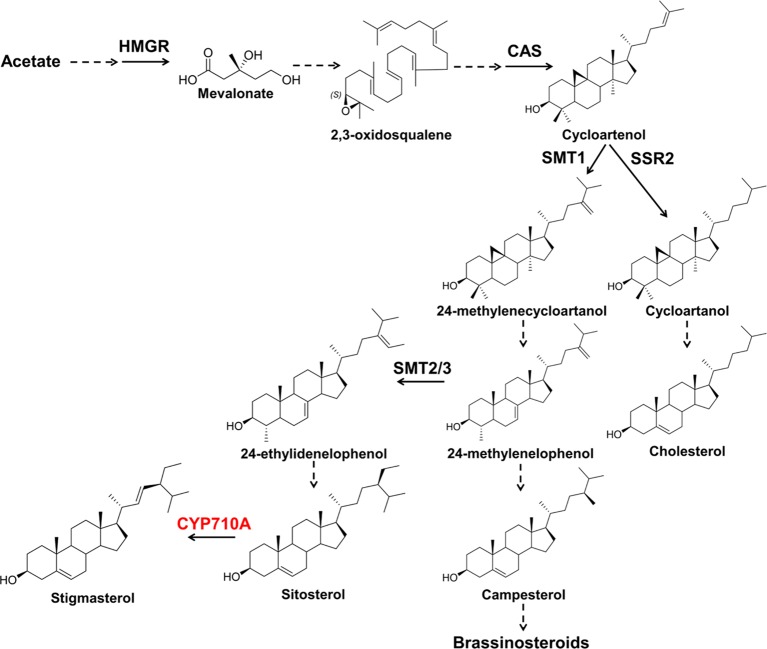
Figure 1.
The plant sterol pathway leading to stigmasterol. Plants produce a mixture of sterols, campesterol (24-methyl) and sitosterol and stigmasterol (24-ethyl sterols). Stigmasterol is derived from sitosterol by the action of sterol C-22 desaturases. Campesterol is the preferred precursor of brassinosteroids (BR). Dashed arrows indicate multiple steps and solid arrows denote single step in the pathway. HMGR, 3-hydroxy-3-methylglutaryl-CoA reductase; CAS, cycloartenol synthase; SMT1, sterol methyltransferase 1; SMT2/3, sterol methyltransferase2/3; SSR2, sterol sidechain reductase2; CYP710A, sterol C-22 desaturase.
Campesterol is the precursor of BR (Fujioka and Yokota, 2003), and the crucial role of BR in plant growth and development is well established (Fujioka et al., 1997; Choe et al., 1998, 1999; Noguchi et al., 1999; Nomura et al., 1999; Clouse, 2002), while sitosterol is implicated in cellulose synthesis (Peng et al., 2002; Schrick et al., 2012). It is, however, unclear whether fluctuations in stigmasterol concentration observed during development and conditions of stress are responsible for cellular signaling. Nonetheless, evidence pointing to stigmasterol as a potential signal for cellular defense and gravitropic responses is emerging (Griebel and Zeier, 2010; Dalal et al., 2016). Functional characterization of genes controlling stigmasterol biosynthesis might help increase our understanding of the direct role of this sterol in plant development and stress responses. Therefore, our purpose is to examine genetic, developmental, and environmental conditions affecting stigmasterol production and suggest experimental approaches to investigate the role of stigmasterol in cell signaling.
The Biosynthesis of Stigmasterol
Stigmasterol is produced in the mevalonate pathway following a series of enzyme-catalyzed reactions leading to the generation of 2,3-oxidosqualene (Schaller, 2003; Bach, 2016). Subsequently, 2,3-oxidosqualene is converted to cycloartenol by cycloartenol synthase (Schaller, 2003; Gas-Pascual et al., 2014; Sonawane et al., 2016). Cycloartenol is the target of branch-point enzymes including sterol side chain reductase 2 (SSR2) and sterol methyl transferase 1 (SMT1). SSR2 channels cycloartenol to the cholesterol branch, while SMT1 catalyzes the alkylation of cycloartenol to produce precursors for plant sterols (Benveniste, 1986; Nes and Venkatramesh, 1999; Diener et al., 2000; Schaeffer et al., 2001; Sonawane et al., 2016). Downstream of SMT1, other branching enzymes SMT2/3, directs carbon toward sitosterol and stigmasterol (Carland et al., 2010). Besides biosynthesis, free stigmasterol content can also be modulated by converting it to sterol conjugates such as steryl esters, steryl glucosides, and acyl steryl glucosides. Steryl esters are conjugated by acyl transferases (Chen et al., 2007; Bouvier-Navé et al., 2010) and the steryl glucosides by UDP-glucose: sterol glucosyltransferase (DeBolt et al., 2009).
Structurally, stigmasterol is similar to sitosterol but differs from sitosterol due to a double bond at position C-22 introduced by the sterol C-22 desaturase (Benveniste, 2002; Morikawa et al., 2006). Arabidopsis contains four genes encoding sterol C-22 desaturases belonging to the cytochrome P450, CYP710A superfamily (Benveniste, 2004; Morikawa et al., 2006) and two sterol C-22 desaturases are responsible for stigmasterol biosynthesis in Physcomitrella patens, a moss in which the major sterol is stigmasterol (Morikawa et al., 2009). The number of genes encoding/predicted to encode sterol C-22 desaturase, however, varies across species (Supplementary Table S1). It is noteworthy that SMT2/3 and CYP710A1 are the only two unique enzymes leading to the biosynthesis of stigmasterol in the stigmasterol branch, while both campesterol and stigmasterol branches (Figure 1) share intermediate enzymes. This is a case similar to cholesterol synthesis in plants, where the pathway involves both unique and shared enzymes with the campesterol branch (Sonawane et al., 2016).
Developmental Regulation of Stigmasterol
In mammalian systems, cholesterol biosynthesis is regulated via negative feedback suppression of several key genes (Goldstein and Brown, 1990). Thus, analysis of gene transcripts may help increase our understanding of the regulation of sterol biosynthesis during plant development (Schrick et al., 2011; Sonawane et al., 2016; Suza and Chappell, 2016). For instance, in the developing seeds of tobacco (N. tabacum), pea (Pisum sativum), rape (Brassica napus), and seedlings of N. benthamiana and B. napus, increased gene expression and enzyme activities coincides with sterol accumulation (Harker et al., 2003; Schrick et al., 2011; Suza and Chappell, 2016). In addition, apical tissues of B. campestris contain high levels of cholesterol but exhibit a decline in cholesterol and a rise in sitosterol at later stages of development (Hobbs et al., 1996). Moreover, varying concentrations of stigmasterol and its precursor are noticeable at both the seed and whole plant developmental stages. For instance, during germination of tobacco seed, stigmasterol increases two-fold (Bush and Grunwald, 1972), and in mung bean (Vigna radiata) seedlings, younger sections of hypocotyls contain higher levels of stigmasterol compared to sitosterol (Stalleart and Geuns, 1994).
Stigmasterol content also increases in tomato (Solanum lycopersicon) during fruit ripening and is associated with an increase in CYP710A11 gene expression (Whitaker and Gapper, 2008). In addition, in maize (Zea mays) seedlings, the concentration of stigmasterol is higher in roots than in shoots (Kemp et al., 1967). Similar to the findings of Kemp et al. (1967), N. benthamiana seedlings display striking differences in sterol composition between organs, with higher stigmasterol content in roots than in leaves (Suza and Chappell, 2016). In contrast, stigmasterol concentration is elevated in P. sativum leaves but lower in seeds (Schrick et al., 2011). Taken together, the developmental profile of sterols and gene expression data from Arabidopsis (Supplementary Figure S1) suggests highly coordinated regulation of stigmasterol metabolism in plants.
Impact of Biotic and Abiotic Stress on Stigmasterol
In Solanaceous plants, e.g., potato (Solanum tuberosum), cholesterol production rises to match the demand for the synthesis of steroid glycoalkaloids in response to wounding or pathogen infection (Choi et al., 1992; Hartmann, 1998; Arnqvist et al., 2003). Similarly, pathogenic bacteria and reactive oxygen species stimulate the biosynthesis of stigmasterol in Arabidopsis (Griebel and Zeier, 2010; Sewelam et al., 2014). Furthermore, genes encoding sterol C-22 desaturase are responsive to phytohormones, suggesting a role for stigmasterol in various stress responses (Supplementary Figure S1). Indeed, the overexpression of one of the Arabidopsis stigmasterol biosynthesis genes resulted in enhanced resistance to bacterial pathogens (Wang et al., 2012). Recently, Gamir et al. (2017) reported that PATHOGENESIS-RELATED PROTEIN 1 (PR-1) can bind sterols including stigmasterol in vitro. The authors conclude that PR-1 inhibits pathogen growth by sequestering sterols from pathogens (Gamir et al., 2017). However, it remains to be demonstrated whether the predicted ability of PR-1 to bind stigmasterol has a real biological significance.
Stigmasterol concentration increases in roots of wheat (Triticum aestivum) exposed to salt (Magdy et al., 1994). In addition, salt-induced increase in stigmasterol is associated with salt exclusion capacity of citrus (Citrus medica) rootstocks (Douglas and Walker, 1983), possibly due to the activation of the plasma membrane H+-ATPase by stigmasterol (Grandmougin-Ferjani et al., 1997). The plasma membrane H+-ATPase is the primary transporter of protons out of the cell (Muramatsu et al., 2002), and its activity is essential for maintaining ion homeostasis (Niu et al., 1995). Indeed, stigmasterol treatment of germinating seeds improved salt tolerances of faba beans (Vicia faba L.) and flax (Linum usitatissimum) (Hashem et al., 2011; Hassanein et al., 2012).
Plants grown in saline conditions experience retardation of root growth, but Ca2+ supply ameliorates these deleterious effects of salinity stress (Shabala et al., 2003). The beneficial effect of Ca2+ in the context of salinity stress is associated with the stabilization of plasma membrane and enhanced exchange of cations such as Na+ (Hirschi, 2004). It appears that Ca2+ may stimulate stigmasterol production in roots (Pilar et al., 1993; Magdy et al., 1994), and possibly, the stigmasterol induced by Ca2+ affects the plasma membrane H+-ATPase (Grandmougin-Ferjani et al., 1997), leading to enhanced extrusion of Na+ from the cell (Qiu et al., 2003).
Stigmasterol is elevated at the expense of sitosterol in tomato (Lycopersicon esculentum) when stored at 15°C (Whitaker, 1991). Indeed, analysis of Arabidopsis over-expressing AtCYP710A1 and Atcyp710a1 mutant lines suggests a role for stigmasterol in tolerance to unfavorable temperatures (Senthil-Kumar et al., 2013). Higher levels of sitosterol are detected in etiolated barley (Hordeum vulgare) tissues compared to stigmasterol, but the two sterols are detected in equal amounts in green tissues (Bush et al., 1971). Similar to etiolated barley, soybean plants grown under filtered sunlight conditions accumulate sitosterol, while stigmasterol levels decrease (Izzo and Navari-Izzo, 1981). The fluctuations in stigmasterol content in response to various environmental cues suggest that the conversion of sitosterol to stigmasterol may modulate plant response to environmental stimuli.
Potential Role for Stigmasterol in Cell Signaling
Cholesterol modulates its own biosynthesis in mammalian cells via negative feedback (Marigo and Tabin, 1996; Edwards and Ericsson, 1999). Research in Solanum species suggested the existence of analogous cholesterol feedback mechanisms in plants (Bhatt and Bhatt, 1984); however, the idea that cholesterol modulates sterol biosynthesis in plants did not escape skepticism, since unlike mammals, plants synthesize an array of sterol end products (Hartmann, 1998). Production of several sterol end products presents a challenge in elucidating role of sterol end products in cell signaling in plants.
Analysis of Arabidopsis sterol biosynthesis mutants suggests that sterols play critical roles in plant development independent of BR (Lindsey et al., 2003) by influencing position-dependent cell fate during embryogenesis (Jang et al., 2000; Schrick et al., 2000; Clouse, 2002). For example, the fackel mutants lacking a functional sterol C-14 reductase display embryonic defects and dwarfism at the seedling stage and produce less BR, but exogenous BR fails to complement the mutant (Mayer et al., 1991; Jang et al., 2000; Schrick et al., 2000), whereas the loss of SMT1 function in smt1/cph plants results in the accumulation of cholesterol, defective embryo development, and increased sensitivity to Ca2+. Similar to fackel, the defective phenotype of smt1/cph plants cannot be rescued by exogenous BR (Diener et al., 2000).
The SMT2/3 (COTYLEDON VASCULAR PATTERNING1—CVP1) locus converts 24-methylene lophenol to 24-ethylidene lophenol (Carland et al., 2002). Consequently, Arabidopsis plants overexpressing SMT2 accumulate sitosterol at the expense of campesterol and display reduced stature and growth (Schaller et al., 1998; Schaeffer et al., 2001). The smt2/cvp1 plants exhibit aberrant alignment of vascular strands and misshapen vascular cells, reduced levels of sitosterol, and higher concentration of campesterol (Schaeffer et al., 2001; Carland et al., 2002); however, the aberrant phenotype of AtSMT2 and smt2/cvp1 plants is not associated with defective BR signaling (Schaller et al., 1998; Schaeffer et al., 2001; Carland et al., 2002).
Another classic Arabidopsis sterol mutant is hydra, with defective embryonic morphogenesis, seedling cell patterning, and root growth (Lindsey et al., 2003). HYDRA1 and HYDRA2/FACKEL encode sterol isomerase and C-14 reductase, respectively (Souter et al., 2002). Similar to fackel, hydra mutants produce less campesterol, but BR application does not rescue their phenotypic defects. Interestingly, both hydra1 and hydra2/fackel mutants produce high levels of stigmasterol compared to the wild type (Souter et al., 2002). Whether dysregulation of stigmasterol is the cause for the pleiotropic defects in the hydra mutants is unclear.
The compactness in the packing of plasma membrane (PM) lipid bilayer acyl chains—referred as membrane order (or liquid-ordered)—is influenced by sterol composition (Roche et al., 2008). The separation of liquid-ordered and liquid-disordered phases in the PM is observed in vivo in tobacco cells (Gerbeau-Pissot et al., 2014). In “raft hypothesis,” stress induction can lead to the formation of larger structures (proposed lipid rafts) from liquid-ordered nanodomains enriched in sterols and sphingolipids (Lingwood and Simons, 2010). The interaction of sterols with phospholipids to form lipid rafts in mammalian membrane systems is crucial for correct signaling and activity of intrinsic membrane proteins. Lipid rafts are associated with many plant proteins involved in redox regulation, hormone transport and signaling, and ion homeostasis (Willemsen et al., 2003; Borner et al., 2005; Lefebvre et al., 2007; Zauber et al., 2014). Examples of proteins associated with lipid rafts and sterols include GLABRA2 (GL2), SCRAMBLED (SCM), and PIN-FORMED (PIN). PIN proteins are involved in the transport of auxin to mediate polar cell growth and root gravitropism (Moore, 2002). GL2 is a phospholipid/sterol-binding transcription factor involved in the regulation of root hair development (Masucci et al., 1996), whereas SCM is a receptor for positional cues to modulate expression of GL2 and other cell fate regulators during root hair development (Grierson et al., 2014). Indeed, proteome analysis of smt1/cph, with an altered plasma membrane composition, revealed a compromised cell signaling (Zauber et al., 2014). Sterol depletion in the plasma membrane by cyclodextrin and filipin suggests the sensing of modifications of cell environment at the PM is sterol dependent in plants, which can lead to adaptive cell responses through regulated signaling processes (Roche et al., 2008; Bonneau et al., 2010). In tobacco cells, the proportion of ordered phases transiently increased during the early steps of the signaling triggered by cryptogein and flagellin, two elicitors of plant defense reactions (Gerbeau-Pissot et al., 2014).
The composition of free sterols and sterol conjugates influences the liquid-ordered phase formation (Grosjean et al., 2015). Stigmasterol by itself lacks the ability to increase membrane order, whereas sitosterol and campesterol increase the order. However, by interacting together with glycosylinositolphosphoceramide, the major sphingolipid in plant, stigmasterol can increase the membrane order, while the interaction with glucosylceramide decreased the order. Sitosterol by itself induces the production of many small domains, which increases in size together with the addition of free sterol-sphingolipid and free sterol-sterylglycoside/acylsterylglycoside combination (Grosjean et al., 2015). These findings suggest a role for specific sterol species to fine tune the membrane sterol composition, thereby regulating signaling events.
The orc mutation is allelic to SMT1, and analysis of the smt1orc plants revealed trace amounts of stigmasterol and aberrant localization of PIN2 and PIN3 (Willemsen et al., 2003). Therefore, regulated membrane sterol composition is important for correct positioning of proteins, such as PIN, and physiological responses such as root gravitropism (Men et al., 2008). The hydra2/fackel plants show an ectopic expression of GL2 in trichoblasts, resulting in a glabrous root phenotype possibly due to a compromised function of GL2 (Souter et al., 2002). There is a possibility that GL2 activity in hydra/fackel plants is diminished by a sterol molecule which causes a conformational change blocking DNA interaction with certain trans-factors (Schrick et al., 2004). Conversely, a sterol or its derivative may bind GL2 and tether it to the membrane in a manner similar to the way cholesterol tethers Hedgehog in vertebrate systems (Jeong and McMahon, 2002). Since stigmasterol plays a role in cell proliferation (Hartmann, 1998) and hydra/fackel plants accumulate high levels of stigmasterol (Lindsey et al., 2003), a dysregulated metabolism of stigmasterol may interfere with various cellular processes during development. Indeed, GL2 expression is dysregulated in the developing siliques of the Arabidopsis acbp1 mutant with high stigmasterol (Lung et al., 2017). Perhaps, the aberrant SCM distribution in roots of the ugt80B1 is due to deficiency in a stigmasterol conjugate (Pook et al., 2017), offering additional support for a role for stigmasterol in cell signaling.
Stigmasterol Role in Modulating Cell Biology
Stigmasterol is one of the major sterols in plasma membranes of plant cells and plays a role in cell proliferation (Hartmann, 1998) and activation of plasma membrane H+-ATPase (Grandmougin-Ferjani et al., 1997). In plants, the plasma membrane H+-ATPase is the primary transporter of protons out of the cell, thus creating a pH and electrochemical gradient across the plasma membrane (Muramatsu et al., 2002). The activity of the plasma membrane H+-ATPase is essential for maintaining ion homeostasis, since carrier-mediated ion transport is coupled to a downhill pH gradient (Niu et al., 1995). In addition, the activity of the plasma membrane H+-ATPase promotes the adaptation of maize roots to low pH (Yan et al., 1998) and low phosphorous availability in soybeans (Shen et al., 2006).
In Arabidopsis, exogenous stigmasterol activates the expression of genes involved in cell expansion and division (He et al., 2003). Furthermore, exogenous stigmasterol increases flower numbers of chamomile (Chamomilla recutita L. Rausch) (Abd El-Wahed and Krima, 2004) and in vitro multiplication of shoots of Marubakaido apple rootstock (Malus prunifolia (Wild.) Borkh) (Pereira-Netto, 2012). In tobacco seeds, depletion of cycloartenol by increased activity of SMT1 was associated with elevated activity of HMGR in tobacco seeds (Holmberg et al., 2002). By contrast, studies in P. sativum showed that stigmasterol inhibits HMGR activity (Stermer et al., 1994), suggesting a role for stigmasterol in regulation of sterol biosynthesis.
In Arabidopsis, the key gene controlling stigmasterol production is CYP710A1, but Arabidopsis also produces low levels of brassicasterol from the C-22 desaturation of epi-campesterol by CYP710A2 (Benveniste, 2002; Morikawa et al., 2006). The expression of CYP710A2 mRNA responds rapidly to gravity stimulation (Kimbrough et al., 2004), suggesting a role for sterol C-22 desaturation in plants response to gravity. The recent discovery that InteractoR Of SYnaptotagmin1 (ROSY1), a regulator of cellular trafficking and gravitropic response binds stigmasterol (Dalal et al., 2016), supports the idea that CYP710A genes and stigmasterol play a role in root response to gravity. In addition, the rosy1-1 mutant is impaired in auxin transport but is more tolerant to salt stress (Dalal et al., 2016), suggesting a connection between ROSY1 and stigmasterol in regulating auxin transport and abiotic stress responses.
Stigmasterol is induced by Ca2+ (Pilar et al., 1993), and Arabidopsis mutants defective in Ca2+ uptake have a compromised cell expansion, short root hairs, and stunted roots (Foreman et al., 2003). Since gravistimulation induces Ca2+ (Monshausen et al., 2011), Ca2+ may stimulate CYP710A2 expression and stigmasterol production in roots. Therefore, CYP710A proteins might participate in a similar signaling pathway with ROSY1 to modulate plant cell response to gravity and salt stress (Dalal et al., 2016).
Concluding Remarks and Future Directions
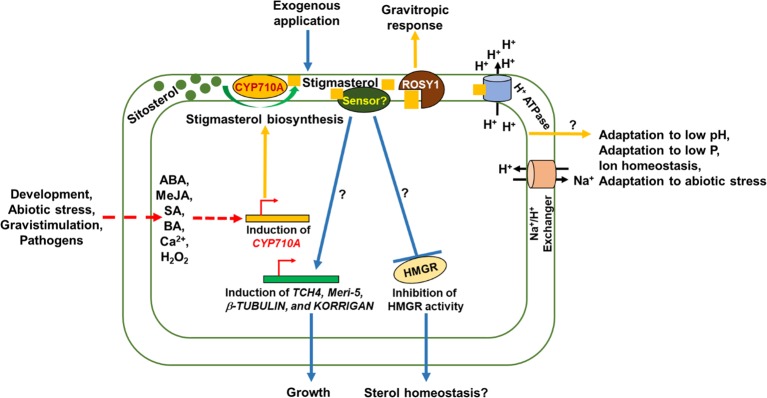
There is need to validate gene expression data from microarray experiments (Supplementary Figure S1) and correlate hormone/stress induced gene expression with stigmasterol concentration. In addition, it is intriguing that blocking BR biosynthesis affects CYP710A gene expression (Supplementary Figure S1), suggesting a role for BR in regulating stigmasterol metabolism. Based on the mechanism for cholesterol and lipid sensing in mammals and insects, the notion of plant (stigma)sterol sensor(s) is not far-fetched. Indeed, the discovery of plant proteins with sterol/lipid sensing/binding domains offers a promising avenue for testing the signaling role of stigmasterol (Figure 2).
Figure 2.
Schematic representation of functions of stigmasterol and its signaling roles in plant cells. Stigmasterol biosynthesis occurs during development, abiotic stress, gravistimulation, pathogen attack, and in response to signaling molecules such as abscisic acid (ABA), methyl jasmonate (MeJA), salicylic acid (SA), calcium (Ca2+), and hydrogen peroxide (H2O2). Stigmasterol binds to ROSY1 leading to gravitropic response. Stigmasterol activates H+-ATPase to create pH and electrochemical gradient across the plasma membrane. The pH gradient leads to activation of the Na+/H+ exchanger to exclude Na+ to adapt to salinity stress. Furthermore, the activity of H+-ATPase is necessary for ion homeostasis, adaptation to low pH, low phosphorus, and abiotic stress. Exogenous application of stigmasterol impacts growth and sterol homeostasis via an unidentified (stigma)sterol sensor.
The potential candidate for a stigmasterol sensing system would be ROSY1, which shows binding specificity for stigmasterol to regulate root response to gravity (Dalal et al., 2016). Other candidates include Arabidopsis Niemann-Pick disease type C like proteins (AtNPC1-1 and AtNPC1-2) (Feldman et al., 2015), possessing putative sterol sensing domains reminiscent of SCAP and related regulators of sterol metabolism in animals and yeast (Goldstein and Brown, 1990; Nohturfft and Losick, 2002). Evaluating the sterol binding specificity of plant NPC proteins might provide clues as to whether AtNPC1-1 and AtNPC1-2 act as sterol sensors to modulate lipid metabolism; however, testing the implication of stigmasterol interaction with plant sterol sensing proteins requires circumventing gene redundancy (Supplementary Table S1). The creation of double/triple/quadruple mutants for Arabidopsis CYP710A genes may help in overcoming the challenge. Alternatively, crop or model grass species predicted to encode single copies of CYP710A (Supplementary Table S1), and rich genetic resources, such as maize or Brachypodium, may provide an opportunity to attempt to eliminate the production of stigmasterol via insertional mutagenesis or gene editing approaches.
Sterol glucosides are synthesized at the PM (Zauber et al., 2014), while the CYP710A is predicted to localize to the apoplast (Supplementary Table S1). This begs the question of what would be the cellular site of stigmasterol synthesis, since plant sterols are believed to originate primarily within the ER (Hartmann, 1998). Experiments to test the impact of ectopic expression of CYP710A via retention to ER or vacuole may help identify the preferred site of stigmasterol synthesis. This information will be helpful in designing gene constructs to manipulate stigmasterol content in a more targeted fashion.
Data Availability
Publicly available datasets were analyzed in this study. These data can be found here: https://www.arabidopsis.org/servlets/Search?type=general&search_action=detail&method=1&show_obsolete=F&name=cyp710a&sub_type=gene&SEARCH_EXACT=4&SEARCH_CONTAINS=1.
Author Contributions
WS and SA designed the research and wrote the paper.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We apologize to those colleagues whose important contributions may have been omitted owing to space constraints.
Glossary
Abbreviations
- BR
Brassinosteroid
- CPH
Cephalopod
- CVP
Cotyledon vascular patterning
- CYP710A
Sterol C-22 desaturase
- GL2
GLABRA2
- NPC
Niemann-Pick disease type C
- PIN
PIN-FORMED
- PR-1
PATHOGENESIS-RELATED PROTEIN 1
- PM
Plasma membrane
- ROSY1
InteractoR of SYnaptotagmin
- SCM
SCRAMBLED
- SMT
Sterol methyltransferase
- SSR
Sterol sidechain reductase.
Footnotes
Funding. This work was financially supported by USAID project iAGRI (grant no. 621-A-00-11-000090-00).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2019.00354/full#supplementary-material
References
- Abd El-Wahed M. S. A., Krima M. G. E. D. (2004). Stimulation of growth, flowering, biochemical constituents and essential oil of chamomile plant (Chamomilla recutita L., Rausch) with spermidine and stigmasterol application. Bulg. J. Plant Physiol. 30, 89–102. [Google Scholar]
- Arnqvist L., Dutta P. C., Jonsson L., Sitbon F. (2003). Reduction of cholesterol and glycoalkaloid levels in transgenic potato plants by overexpression of a type 1 sterol methyltransferase cDNA. Plant Physiol. 131, 1792–1799. 10.1104/pp.102.018788, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bach T. J. (2016). Secondary metabolism: high cholesterol in tomato. Nat. Plants 3:16213. 10.1038/nplants.2016.213, PMID: [DOI] [PubMed] [Google Scholar]
- Benveniste P. (1986). Sterol biosynthesis. Annu. Rev. Plant Physiol. 37, 275–308. 10.1146/annurev.pp.37.060186.001423 [DOI] [Google Scholar]
- Benveniste P. (2002). Sterol metabolism. Arabidopsis Book 1:e0004. 10.1199/tab.0004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benveniste P. (2004). Biosynthesis and accumulation of sterols. Annu. Rev. Plant Biol. 55, 429–457. 10.1146/annurev.arplant.55.031903.141616, PMID: [DOI] [PubMed] [Google Scholar]
- Bhatt P. N., Bhatt D. P. (1984). Regulation of sterol biosynthesis in Solanum species. J. Exp. Bot. 35, 890–896. 10.1093/jxb/35.6.890 [DOI] [Google Scholar]
- Bonneau L., Gerbeau-Pissot P., Thomas D., Der C., Lherminier J., Bourque S., et al. (2010). Plasma membrane sterol complexation, generated by filipin, triggers signaling responses in tobacco cells. Biochim. Biophys. Acta 1798, 2150–2159. 10.1016/j.bbamem.2010.07.026, PMID: [DOI] [PubMed] [Google Scholar]
- Borner G. H. H., Sherrier D. J., Weimar T., Michaelson L. V., Hawkins N. D., MacAskill A., et al. (2005). Analysis of detergent-resistant membranes in Arabidopsis. Evidence for plasma membrane lipid rafts. Plant Physiol. 137, 104–116. 10.1104/pp.104.053041, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouvier-Navé P., Berna A., Noiriel A., Compagnon V., Carlsson A. S., Banas A., et al. (2010). Involvement of the phospholipid sterol acyltransferase1 in plant sterol homeostasis and leaf senescence. Plant Physiol. 152, 107–119. 10.1104/pp.109.145672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bush P. B., Grunwald C. (1972). Sterol changes during germination of Nicotiana tabacum seeds. Plant Physiol. 50, 69–72. 10.1104/pp.50.1.69, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bush P. B., Grunwald C., Davis D. L. (1971). Changes in sterol composition during greening of etiolated barley shoots. Plant Physiol. 47, 745–749. 10.1104/pp.47.6.745, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carland F., Fujioka S., Nelson T. (2010). The sterol methyltransferases SMT1, SMT2, and SMT3 influence Arabidopsis development through nonbrassinosteroid products. Plant Physiol. 153, 741–756. 10.1104/pp.109.152587, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carland F. M., Fujioka S., Takatsuto S., Yoshida S., Nelson T. (2002). The identification of CVP1 reveals a role for sterols in vascular patterning. Plant Cell 14, 2045–2058. 10.1105/tpc.003939, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q., Steinhauer L., Hamerlindl J., Keller W., Zou J. (2007). Biosynthesis of phytosterol esters: identification of a sterol O-acyltransferase in Arabidopsis. Plant Physiol. 145, 974–984. 10.1104/pp.107.106278, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choe S., Dilkes B. P., Fujioka S., Takatsuto S., Sakurai A., Feldmann K. A. (1998). The DWF4 gene of Arabidopsis encodes a cytochrome P450 that mediates multiple 22alpha-hydroxylation steps in brassinosteroid biosynthesis. Plant Cell 10, 231–243. 10.1105/tpc.10.2.231, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choe S., Noguchi T., Fujioka S., Takatsuto S., Tissier C. P., Gregory B. D., et al. (1999). The Arabidopsis dwf7/ste1 mutant is defective in the delta7 sterol C-5 desaturation step leading to brassinosteroid biosynthesis. Plant Cell 11, 207–221. 10.1105/tpc.11.2.207, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi D., Ward B. L., Bostock R. M. (1992). Differential induction and suppression of potato 3-hydroxy-3-methylglutaryl coenzyme A reductase genes in response to Phytophthora infestans and to its elicitor arachidonic acid. Plant Cell 4, 1333–1344. 10.1105/tpc.4.10.1333, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clouse S. D. (2002). Arabidopsis mutants reveal multiple roles for sterols in plant development. Plant Cell 14, 1995–2000. 10.1105/tpc.140930, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalal J., Lewis D. R., Tietz O., Brown E. M., Brown C. S., Palme K., et al. (2016). ROSY1, a novel regulator of gravitropic response is a stigmasterol binding protein. J. Plant Physiol. 196–197, 28–40. 10.1016/j.jplph.2016.03.011, PMID: [DOI] [PubMed] [Google Scholar]
- DeBolt S., Scheible W. -R., Schrick K., Auer M., Beisson F., Bischoff V., et al. (2009). Mutations in UDP-Glucose:sterol glucosyltransferase in Arabidopsis cause transparent testa phenotype and suberization defect in seeds. Plant Physiol. 151, 78–87. 10.1104/pp.109.140582, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diener A. C., Li H., Zhou W., Whoriskey W. J., Nes W. D., Fink G. R. (2000). Sterol methyltransferase 1 controls the level of cholesterol in plants. Plant Cell 12, 853–870. 10.1105/tpc.12.6.853, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas T. J., Walker R. R. (1983). 4-Desmethylsterol composition of citrus rootstocks of different salt exclusion capacity. Physiol. Plant. 58, 69–74. 10.1111/j.1399-3054.1983.tb04145.x [DOI] [Google Scholar]
- Edwards P. A., Ericsson J. (1999). Sterols and isoprenoids: signaling molecules derived from the cholesterol biosynthetic pathway. Annu. Rev. Biochem. 68:157–185. 10.1146/annurev.biochem.68.1.157, PMID: [DOI] [PubMed] [Google Scholar]
- Feldman M. J., Poirier B. C., Lange B. M. (2015). Misexpression of the Niemann-Pick disease type C1 (NPC1)-like protein in Arabidopsis causes sphingolipid accumulation and reproductive defects. Planta 242, 921–933. 10.1007/s00425-015-2322-4, PMID: [DOI] [PubMed] [Google Scholar]
- Foreman J., Demidchik V., Bothwell J. H. F., Mylona P., Miedema H., Torres M. A., et al. (2003). Reactive oxygen species produced by NADPH oxidase regulate plant cell growth. Nature 422, 442–446. 10.1038/nature01485, PMID: [DOI] [PubMed] [Google Scholar]
- Fujioka S., Li J., Choi Y. H., Seto H., Takatsuto S., Noguchi T., et al. (1997). The Arabidopsis deetiolated2 mutant is blocked early in brassinosteroid biosynthesis. Plant Cell 9, 1951–1962. 10.1105/tpc.9.11.1951, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujioka S., Yokota T. (2003). Biosynthesis and metabolism of brassinosteroids. Annu. Rev. Plant Biol. 54, 137–164. 10.1146/annurev.arplant.54.031902.134921, PMID: [DOI] [PubMed] [Google Scholar]
- Gamir J., Darwiche R., Van’t Hof P., Choudhary V., Stumpe M., Schneiter R., et al. (2017). The sterol-binding activity of PATHOGENESIS-RELATED PROTEIN 1 reveals the mode of action of an antimicrobial protein. Plant J. 89, 502–509. 10.1111/tpj.13398, PMID: [DOI] [PubMed] [Google Scholar]
- Gas-Pascual E., Berna A., Bach T. J., Schaller H. (2014). Plant oxidosqualene metabolism: cycloartenol synthase-dependent sterol biosynthesis in Nicotiana benthamiana. PLoS One 9:e109156. 10.1371/journal.pone.0109156, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerbeau-Pissot P., Der C., Thomas D., Anca I. -A., Grosjean K., Roche Y., et al. (2014). Modification of plasma membrane organization in tobacco cells elicited by cryptogein. Plant Physiol. 164, 273–286. 10.1104/pp.113.225755, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein J. L., Brown M. S. (1990). Regulation of the mevalonate pathway. Nature 343:425. 10.1038/343425a0, PMID: [DOI] [PubMed] [Google Scholar]
- Grandmougin-Ferjani A., Schuler-Muller I., Hartmann M. A. (1997). Sterol modulation of the plasma membrane H+-ATPase activity from corn roots reconstituted into soybean lipids. Plant Physiol. 113, 163–174. 10.1104/pp.113.1.163, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griebel T., Zeier J. (2010). A role for beta-sitosterol to stigmasterol conversion in plant-pathogen interactions. Plant J. 63, 254–268. 10.1111/j.1365-313X.2010.04235.x, PMID: [DOI] [PubMed] [Google Scholar]
- Grierson C., Nielsen E., Ketelaarc T., Schiefelbein J. (2014). Root hairs. Arabidopsis Book 12:e0172. 10.1199/tab.0172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosjean K., Mongrand S., Beney L., Simon-Plas F., Gerbeau-Pissot P. (2015). Differential effect of plant lipids on membrane organization: specificities of phytosphingolipids and phytosterols. J. Biol. Chem. 290, 5810–5825. 10.1074/jbc.M114.598805, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grunwald C. (1971). Effects of free sterols, steryl ester, and steryl glycoside on membrane permeability. Plant Physiol. 48, 653–655. 10.1104/pp.48.5.653, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harker M., Hellyer A., Clayton J., Duvoix A., Lanot A., Safford R. (2003). Co-ordinate regulation of sterol biosynthesis enzyme activity during accumulation of sterols in developing rape and tobacco seed. Planta 216, 707–715. 10.1007/s00425-002-0913-3, PMID: [DOI] [PubMed] [Google Scholar]
- Hartmann M. -A. (1998). Plant sterols and the membrane environment. Trends Plant Sci. 3, 170–175. 10.1016/S1360-1385(98)01233-3 [DOI] [Google Scholar]
- Hashem H. A., Bassuony F. M., Hassanein R. A., Baraka D. M., Khalil R. R. (2011). Stigmasterol seed treatment alleviates the drastic effect of NaCl and improves quality and yield in flax plants. Aust. J. Crop. Sci. 5, 1858–1867. [Google Scholar]
- Hassanein R. A., Hashem H. A., Khalil R. R. (2012). Stigmasterol treatment increases salt stress tolerance of faba bean plants by enhancing antioxidant systems. Plant Omics 5, 476–485. [Google Scholar]
- He J. -X., Fujioka S., Li T. -C., Kang S. G., Seto H., Takatsuto S., et al. (2003). Sterols regulate development and gene expression in Arabidopsis. Plant Physiol. 131, 1258–1269. 10.1104/pp.014605, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirschi K. D. (2004). The calcium conundrum. Both versatile nutrient and specific signal. Plant Physiol. 136, 2438–2442. 10.1104/pp.104.046490, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobbs D. H., Hume J. H., Rolph C. E., Cooke D. T. (1996). Changes in lipid composition during floral development of Brassica campestris. Phytochemistry 42, 335–339. 10.1016/0031-9422(95)00964-7 [DOI] [Google Scholar]
- Holmberg N., Harker M., Gibbard C. L., Wallace A. D., Clayton J. C., Rawlins S., et al. (2002). Sterol C-24 methyltransferase type 1 controls the flux of carbon into sterol biosynthesis in tobacco seed. Plant Physiol. 130, 303–311. 10.1104/pp.004226, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izzo R., Navari-Izzo F. (1981). Sterol composition and accumulation in Glycine max (L.) Merr leaves under different filtered sunlight conditions. Plant Physiol. 67, 1073–1077. 10.1104/pp.67.6.1073, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang J. C., Fujioka S., Tasaka M., Seto H., Takatsuto S., Ishii A., et al. (2000). A critical role of sterols in embryonic patterning and meristem programming revealed by the fackel mutants of Arabidopsis thaliana. Genes Dev. 14, 1485–1497. 10.1101/gad.14.12.1485, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong J., McMahon A. P. (2002). Cholesterol modification of Hedgehog family proteins. J. Clin. Invest. 110, 591–596. 10.1172/JCI200216506, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemp R. J., Goad L. J., Mercer E. I. (1967). Changes in the levels and composition of the esterified and unesterified sterols of maize seedlings during germination. Phytochemistry 6, 1609–1615. 10.1016/S0031-9422(00)82892-7 [DOI] [Google Scholar]
- Kimbrough J. M., Salinas-Mondragon R., Boss W. F., Brown C. S., Sederoff H. W. (2004). The fast and transient transcriptional network of gravity and mechanical stimulation in the Arabidopsis root apex. Plant Physiol. 136, 2790–2805. 10.1104/pp.104.044594, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefebvre B., Furt F., Hartmann M. -A., Michaelson L. V., Carde J. -P., Sargueil-Boiron F., et al. (2007). Characterization of lipid rafts from Medicago truncatula root plasma membranes: a proteomic study reveals the presence of a raft-associated redox system. Plant Physiol. 144, 402–418. 10.1104/pp.106.094102, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsey K., Pullen M. L., Topping J. F. (2003). Importance of plant sterols in pattern formation and hormone signalling. Trends Plant Sci. 8, 521–525. 10.1016/j.tplants.2003.09.012, PMID: [DOI] [PubMed] [Google Scholar]
- Lingwood D., Simons K. (2010). Lipid rafts as a membrane-organizing principle. Science 327, 46–50. 10.1126/science.1174621 [DOI] [PubMed] [Google Scholar]
- Lung S. -C., Liao P., Yeung E. C., Hsiao A. -S., Xue Y., Chye M. -L. (2017). Acyl-CoA-binding protein ACBP1 modulates sterol synthesis during embryogenesis. Plant Physiol. 174, 1420–1435. 10.1104/pp.17.00412, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magdy M., Mansour F., Hasselt P. R., Kuiper P. J. C. (1994). Plasma membrane lipid alterations induced by NaCl in winter wheat roots. Physiol. Plant. 92, 473–478. 10.1111/j.1399-3054.1994.tb08838.x [DOI] [Google Scholar]
- Marigo V., Tabin C. J. (1996). Regulation of patched by sonic hedgehog in the developing neural tube. Proc. Natl. Acad. Sci. USA 93, 9346–9351. 10.1073/pnas.93.18.9346, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masucci J. D., Rerie W. G., Foreman D. R., Zhang M., Galway M. E., Marks M. D., et al. (1996). The homeobox gene GLABRA2 is required for position-dependent cell differentiation in the root epidermis of Arabidopsis thaliana. Development 122, 1253–1260. PMID: [DOI] [PubMed] [Google Scholar]
- Mayer U., Torres Ruiz R. A., Berleth T., Misera S., Jurgens G. (1991). Mutations affecting body organization in the Arabidopsis embryo. Nature 353, 402–407. 10.1038/353402a0 [DOI] [Google Scholar]
- Men S., Boutte Y., Ikeda Y., Li X., Palme K., Stierhof Y. -D., et al. (2008). Sterol-dependent endocytosis mediates post-cytokinetic acquisition of PIN2 auxin efflux carrier polarity. Nat. Cell Biol. 10, 237–244. 10.1038/ncb1686, PMID: [DOI] [PubMed] [Google Scholar]
- Monshausen G. B., Miller N. D., Murphy A. S., Gilroy S. (2011). Dynamics of auxin-dependent Ca2+ and pH signaling in root growth revealed by integrating high-resolution imaging with automated computer vision-based analysis. Plant J. 65, 309–318. 10.1111/j.1365-313X.2010.04423.x, PMID: [DOI] [PubMed] [Google Scholar]
- Moore I. (2002). Gravitropism: lateral thinking in auxin transport. Curr. Biol. 12, R452–R454. 10.1016/S0960-9822(02)00943-0, PMID: [DOI] [PubMed] [Google Scholar]
- Morikawa T., Mizutani M., Aoki N., Watanabe B., Saga H., Saito S., et al. (2006). Cytochrome P450 CYP710A encodes the sterol C-22 desaturase in Arabidopsis and tomato. Plant Cell 18, 1008–1022. 10.1105/tpc.105.037012, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morikawa T., Saga H., Hashizume H., Ohta D. (2009). CYP710A genes encoding sterol C22-desaturase in Physcomitrella patens as molecular evidence for the evolutionary conservation of a sterol biosynthetic pathway in plants. Planta 229, 1311–1322. 10.1007/s00425-009-0916-4, PMID: [DOI] [PubMed] [Google Scholar]
- Muramatsu Y., Harada A., Ohwaki Y., Kasahara Y., Takagi S., Fukuhara T. (2002). Salt-tolerant ATPase activity in the plasma membrane of the marine angiosperm Zostera marina L. Plant Cell Physiol. 43, 1137–1145. 10.1093/pcp/pcf139, PMID: [DOI] [PubMed] [Google Scholar]
- Nes W., Venkatramesh M. (1999). Enzymology of phytosterol transformations. Crit. Rev. Biochem. Mol. Biol. 34, 81–93. 10.1080/10409239991209219 [DOI] [PubMed] [Google Scholar]
- Niu X., Bressan R. A., Hasegawa P. M., Pardo J. M. (1995). Ion homeostasis in NaCl stress environments. Plant Physiol. 109, 735–742. 10.1104/pp.109.3.735, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noguchi T., Fujioka S., Takatsuto S., Sakurai A., Yoshida S., Li J., et al. (1999). Arabidopsis det2 is defective in the conversion of (24R)-24-methylcholest-4-En-3-one to (24R)-24-Methyl-5α-cholestan-3-one in brassinosteroid biosynthesis. Plant Physiol. 120, 833–840. 10.1104/pp.120.3.833, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nohturfft A., Losick R. (2002). Cell biology: fats, flies, and palmitate. Science 296, 857–858. 10.1126/science.1072154, PMID: [DOI] [PubMed] [Google Scholar]
- Nomura T., Takatsuto S., Reid J. B., Yokota T. (1999). Brassinosteroid/sterol synthesis and plant growth as affected by Ika and Ikb mutations of pea. Plant Physiol. 119, 1517–1526. 10.1104/pp.119.4.1517, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng L., Kawagoe Y., Hogan P., Delmer D. (2002). Sitosterol-β-glucoside as primer for cellulose synthesis in plants. Science 295, 147–150. 10.1126/science.1064281, PMID: [DOI] [PubMed] [Google Scholar]
- Pereira-Netto A. B. (2012). Stigmasterol-driven enhancement of the in vitro multiplication rate for the marubakaido apple rootstock. Trees 26, 581–586. 10.1007/s00468-011-0621-3 [DOI] [PubMed] [Google Scholar]
- Pilar C., Antonio O., Antonio C. (1993). Effects of saline stress and calcium on lipid composition in bean roots. Phytochemistry 32, 1131–1136. 10.1016/S0031-9422(00)95077-5 [DOI] [Google Scholar]
- Pook V. G., Nair M., Ryu K., Arpin J. C., Schiefelbein J., Schrick K., et al. (2017). Positioning of the SCRAMBLED receptor requires UDP-Glc:sterol glucosyltransferase 80B1 in Arabidopsis roots. Sci. Rep. 7:5714. 10.1038/s41598-017-05925-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu Q. S., Barkla B. J., Vera-Estrella R., Zhu J. K., Schumaker K. S. (2003). Na+/H+ exchange activity in the plasma membrane of Arabidopsis. Plant Physiol. 132, 1041–1052. 10.1104/pp.102.010421, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roche Y., Gerbeau-Pissot P., Buhot B., Thomas D., Bonneau L., Gresti J., et al. (2008). Depletion of phytosterols from the plant plasma membrane provides evidence for disruption of lipid rafts. FASEB J. 22, 3980–3991. 10.1096/fj.08-111070, PMID: [DOI] [PubMed] [Google Scholar]
- Schaeffer A., Bronner R., Benveniste P., Schaller H. (2001). The ratio of campesterol to sitosterol that modulates growth in Arabidopsis is controlled by STEROL METHYLTRANSFERASE 2;1. Plant J. 25, 605–615. 10.1046/j.1365-313x.2001.00994.x, PMID: [DOI] [PubMed] [Google Scholar]
- Schaller H. (2003). The role of sterols in plant growth and development. Prog. Lipid Res. 42, 163–175. 10.1016/S0163-7827(02)00047-4, PMID: [DOI] [PubMed] [Google Scholar]
- Schaller H., Bouvier-Nave P., Benveniste P. (1998). Overexpression of an Arabidopsis cDNA encoding a sterol-C24(1)-methyltransferase in tobacco modifies the ratio of 24-methyl cholesterol to sitosterol and is associated with growth reduction. Plant Physiol. 118, 461–469. 10.1104/pp.118.2.461, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrick K., Cordova C., Li G., Murray L., Fujioka S. (2011). A dynamic role for sterols in embryogenesis of Pisum sativum. Phytochemistry 72, 465–475. 10.1016/j.phytochem.2011.01.009, PMID: [DOI] [PubMed] [Google Scholar]
- Schrick K., Debolt S., Bulone V. (2012). Deciphering the molecular functions of sterols in cellulose biosynthesis. Front. Plant Sci. 3:84. 10.3389/fpls.2012.00084, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrick K., Mayer U., Horrichs A., Kuhnt C., Bellini C., Dangl J., et al. (2000). FACKEL is a sterol C-14 reductase required for organized cell division and expansion in Arabidopsis embryogenesis. Genes Dev. 14, 1471–1484. 10.1101/gad.14.12.1471, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrick K., Nguyen D., Karlowski W. M., Mayer K. F. (2004). START lipid/sterol-binding domains are amplified in plants and are predominantly associated with homeodomain transcription factors. Genome Biol. 5:R41. 10.1186/gb-2004-5-6-r41, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senthil-Kumar M., Wang K., Mysore K. S. (2013). AtCYP710A1 gene-mediated stigmasterol production plays a role in imparting temperature stress tolerance in Arabidopsis thaliana. Plant Signal. Behav. 8:e23142. 10.4161/psb.23142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sewelam N., Jaspert N., Van Der Kelen K., Tognetti V. B., Schmitz J., Frerigmann H., et al. (2014). Spatial H2O2 signaling specificity: H2O2 from chloroplasts and peroxisomes modulates the plant transcriptome differentially. Mol. Plant 7, 1191–1120. 10.1093/mp/ssu070, PMID: [DOI] [PubMed] [Google Scholar]
- Shabala S., Shabala L., Volkenburgh E. V. (2003). Effect of calcium on root development and root ion fluxes in salinised barley seedlings. Funct. Plant Biol. 30, 507–514. 10.1071/FP03016 [DOI] [PubMed] [Google Scholar]
- Shen H., Chen J., Wang Z., Yang C., Sasaki T., Yamamoto Y., et al. (2006). Root plasma membrane H+-ATPase is involved in the adaptation of soybean to phosphorus starvation. J. Exp. Bot. 57, 1353–1362. 10.1093/jxb/erj111, PMID: [DOI] [PubMed] [Google Scholar]
- Sonawane P. D., Pollier J., Panda S., Szymanski J., Massalha H., Yona M., et al. (2016). Plant cholesterol biosynthetic pathway overlaps with phytosterol metabolism. Nat. Plants 3:16205. 10.1038/nplants.2016.205, PMID: [DOI] [PubMed] [Google Scholar]
- Souter M., Topping J., Pullen M., Friml J., Palme K., Hackett R., et al. (2002). hydra Mutants of Arabidopsis are defective in sterol profiles and auxin and ethylene signaling. Plant Cell 14, 1017–1031. 10.1105/tpc.001248, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stalleart V. M., Geuns J. M. C. (1994). Phospholipid and free sterol composition of hypocotyl plasma membranes of ageing mung bean seedlings. Phytochemistry 36, 1177–1180. 10.1016/S0031-9422(00)89633-8 [DOI] [Google Scholar]
- Stermer B. A., Bianchini G. M., Korth K. L. (1994). Regulation of HMG-CoA reductase activity in plants. J. Lipid Res. 35, 1133–1140., PMID: [PubMed] [Google Scholar]
- Suza W. P., Chappell J. (2016). Spatial and temporal regulation of sterol biosynthesis in Nicotiana benthamiana. Physiol. Plant. 157, 120–134. 10.1111/ppl.12413, PMID: [DOI] [PubMed] [Google Scholar]
- Wang K., Senthil-Kumar M., Ryu C. -M., Kang L., Mysore K. S. (2012). Phytosterols play a key role in plant innate immunity against bacterial pathogens by regulating nutrient efflux into the apoplast. Plant Physiol. 158, 1789–1802. 10.1104/pp.111.189217, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitaker B. D. (1991). Changes in lipids of tomato fruit stored at chilling and non-chilling temperatures. Phytochemistry 30, 757–761. 10.1016/0031-9422(91)85247-W [DOI] [Google Scholar]
- Whitaker B. D., Gapper N. (2008). Ripening-specific stigmasterol increase in tomato fruit is associated with increased sterol C-22 desaturase (CYP710A11) gene expression. J. Agric. Food Chem. 56, 3828–3835. 10.1021/jf7037983, PMID: [DOI] [PubMed] [Google Scholar]
- Willemsen V., Friml J., Grebe M., van den Toorn A., Palme K., Scheres B. (2003). Cell polarity and PIN protein positioning in Arabidopsis require STEROL METHYLTRANSFERASE1 function. Plant Cell 15, 612–625. 10.1105/tpc.008433, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan F., Feuerle R., Schaffer S., Fortmeier H., Schubert S. (1998). Adaptation of active proton pumping and plasmalemma ATPase activity of corn roots to low root medium pH. Plant Physiol. 117, 311–319. 10.1104/pp.117.1.311, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zauber H., Burgos A., Garapati P., Schulze W. X. (2014). Plasma membrane lipid–protein interactions affect signaling processes in sterol-biosynthesis mutants in Arabidopsis thaliana. Front. Plant Sci. 5:78. 10.3389/fpls.2014.00078, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Publicly available datasets were analyzed in this study. These data can be found here: https://www.arabidopsis.org/servlets/Search?type=general&search_action=detail&method=1&show_obsolete=F&name=cyp710a&sub_type=gene&SEARCH_EXACT=4&SEARCH_CONTAINS=1.