Abstract
Loop-mediated isothermal amplification (LAMP) has gained wide popularity in the detection of Salmonella in foods owing to its simplicity, rapidity, and robustness. This multi-laboratory validation (MLV) study aimed to validate a Salmonella LAMP-based method against the United States Food and Drug Administration (FDA) Bacteriological Analytical Manual (BAM) Chapter 5 Salmonella reference method in a representative animal food matrix (dry dog food). Fourteen independent collaborators from seven laboratories in the United States and Canada participated in the study. Each collaborator received two sets of 24 blind-coded dry dog food samples (eight uninoculated; eight inoculated at a low level, 0.65 MPN/25 g; and eight inoculated at a high level, 3.01 MPN/25 g) and initiated the testing on the same day. The MLV study used an unpaired design where different test portions were analyzed by the LAMP and BAM methods using different preenrichment protocols (buffered peptone water for LAMP and lactose broth for BAM). All LAMP samples were confirmed by culture using the BAM method. BAM samples were also tested by LAMP following lactose broth preenrichment (paired samples). Statistical analysis was carried out by the probability of detection (POD) per AOAC guidelines and by a random intercept logistic regression model. Overall, no significant differences in POD between the Salmonella LAMP and BAM methods were observed with either unpaired or paired samples, indicating the methods were comparable. LAMP testing following preenrichment in buffered peptone water or lactose broth also resulted in insignificant POD differences (P > 0.05). The MLV study strongly supports the utility and applicability of this rapid and reliable LAMP method in routine regulatory screening of Salmonella in animal food.
Keywords: LAMP, Salmonella, multi-laboratory, validation, animal food
Introduction
Salmonella is a ubiquitous human and animal pathogen, with human outbreak-related illnesses broadly attributed to multiple food categories of plant and animal origins (Interagency Food Safety Analytics Collaboration [IFSAC], 2018). The presence of Salmonella in animal food (e.g., pet food, animal feed, and raw materials and ingredients) is also well documented (Li et al., 2012; Ge et al., 2013; Hsieh et al., 2014; Nemser et al., 2014; Food and Agriculture Organization [FAO]/World Health Organization [WHO], 2015; Jiang, 2016; Molina et al., 2016; Magossi et al., 2018), which impacts not only animal health but also human food safety due to consumption of animal-derived food or direct contact with pet food (Crump et al., 2002; Food and Agriculture Organization [FAO]/World Health Organization [WHO], 2015). The FDA Food Safety Modernization Act (FSMA) prioritizes preventive controls for human and animal foods, emphasizing vigilant product testing and environmental monitoring for zoonotic pathogens such as Salmonella (Food and Drug Administration [FDA], 2017a,b). Rapid and reliable methods are thus in great need to effectively support such efforts.
Current Salmonella testing in foods relies on microbiological culturing, which consists of time-consuming and labor-intensive procedures that require days or weeks for a definitive result (International Organization for Standardization [ISO], 2017; United States Department of Agriculture [USDA], 2017; Andrews et al., 2018). Rapid, sensitive, and specific nucleic acid amplification tests (NAATs), including PCR, real-time quantitative PCR (qPCR), and loop-mediated isothermal amplification (LAMP), have been developed and applied in the detection and identification of Salmonella in foods (Malorny et al., 2009; Balachandran et al., 2012; Lofstrom and Hoorfar, 2012; Cheng et al., 2015; Yang et al., 2016; Domesle et al., 2018; Hu et al., 2018). The isothermal LAMP method, in particular, has gained wide popularity as highlighted in a recent comprehensive review (Yang et al., 2018). Two distinct advantages of LAMP over PCR are running at a constant temperature (Notomi et al., 2000) and high tolerance to matrix inhibitors (Kaneko et al., 2007), which obviate the need for a sophisticated thermocycler or a complicated DNA extraction protocol. These attractive features have led to the development of many new Salmonella LAMP assays, portable microfluidic devices, and commercially available systems (Yang et al., 2018).
Validation plays a critical role in the life cycle of a method from development to implementation. Despite the growing enthusiasm in developing new Salmonella LAMP assays, limited effort has been devoted to validate the assay performance against well-established reference methods (International Organization for Standardization [ISO], 2017; United States Department of Agriculture [USDA], 2017; Andrews et al., 2018). These validation studies, performed at single laboratory, independent laboratory, and collaborative study (multi-laboratory) levels, represent rigorous evaluations of an alternative method’s performance compared with that of the reference method in a food matrix when conducted per international guidelines (Association of Official Analytical Chemists [AOAC], 2012; International Organization for Standardization [ISO], 2016). Similar FDA guidelines have been established for the validation of microbiological methods in foods (Food and Drug Administration [FDA], 2015). Methods that have successfully gone through multi-laboratory validation (MLV) are thus suitable for routine regulatory use. We previously developed a LAMP assay specifically targeting the Salmonella invasion gene invA and showed it to be rapid, reliable, and robust in multiple food matrices (Chen et al., 2011; Yang et al., 2013, 2014, 2015, 2016; Domesle et al., 2018). The method was 100% specific among 300 strains (247 Salmonella of 185 serovars and 53 non-Salmonella) tested and was capable of detecting <1 CFU/25 g in animal food (Domesle et al., 2018). Following FDA guidelines (Food and Drug Administration [FDA], 2015), we recently completed a stringent single-laboratory validation of the method in six animal food matrices including cattle feed, chicken feed, horse feed, swine feed, dry cat food, and dry dog food (Domesle et al., 2018).
This MLV collaborative study aimed to validate the invA-based Salmonella LAMP assay as performed on the Genie II or Genie III platform (OptiGene Ltd., West Sussex, United Kingdom) (alternative method) against the FDA BAM Chapter 5 Salmonella (reference method) in a representative animal food matrix (dry dog food) for future incorporation into the FDA’s compendium of analytical laboratory methods for food and feed safety. MLV participants included 14 independent collaborators from seven FDA, state, and academic laboratories in the United States and Canada. The MLV study also compared the effects of two preenrichment buffers used in LAMP and BAM on Salmonella detection in animal food.
Materials and Methods
Study Design
Figure 1 shows a diagram of the MLV study design. The main component (panels 2 and 3) used an unpaired design, where different test portions were analyzed by the reference FDA BAM method (panel 2) and the alternative LAMP method (panel 3) following preenrichment in different buffers (lactose broth [LB] for BAM and buffered peptone water [BPW] for LAMP). All LAMP samples were confirmed by BAM culturing (panel 4, i.e., BPW-BAM). BAM samples were also tested by LAMP following LB preenrichment (panel 1, i.e., LB-LAMP), essentially forming paired samples (panels 1 and 2).
FIGURE 1.
A schematic diagram of the MLV study design comparing the LAMP alternative method and the FDA BAM Chapter 5 reference method for the detection of Salmonella Infantis ATCC 51741 in 25 g dry dog food test portions. RV, Rappaport-Vassiliadis medium; TT, tetrathionate broth; BS, bismuth sulfite agar; XLD, xylose lysine desoxycholate agar; HE, Hektoen enteric agar; TSI, triple sugar iron agar; LIA, lysine iron agar.
Fourteen independent collaborators (or independent teams), two each from seven FDA, state, and academic laboratories participated in the MLV. The FDA laboratories were from the Office of Regulatory Science at FDA’s Center for Food Safety and Applied Nutrition, and Northeast Food and Feed Laboratory, Pacific Southwest Food and Feed Laboratory, and San Francisco Laboratory at FDA’s Office of Regulatory Affairs. Other participants were the Food and Drug Laboratory Branch at California Department of Public Health, Office of Indiana State Chemist, and Animal Health Laboratory at University of Guelph (ON, Canada).
Sample Inoculation, Storage, and Shipment
Inoculated samples were prepared by Q Laboratories (Cincinnati, OH, United States). Briefly, bulk dry dog food in kibble form was obtained from a local pet store and screened for the presence of Salmonella by the BAM Chapter 5 reference method (Andrews et al., 2018) and the iQ-Check Salmonella II Real-Time PCR detection kit (Bio-Rad, Hercules, CA, United States) to confirm negative results.
Dry dog food confirmed negative for Salmonella was separated into two sets and inoculated with a lyophilized culture of Salmonella enterica serovar Infantis ATCC 51741 at two target levels: a high level of ca. 2 to 5 CFU/25 g test portion and a low level of ca. 0.2 to 2 CFU/25 g test portion. An uninoculated control set (0 CFU/25 g test portion) was also included. After inoculation, the three sets of bulk samples were homogenized and held at room temperature for 2 weeks for aging to simulate storage. Replicate samples (5–10; 25 g each) from the two inoculated sets were evaluated at three time points (immediately after inoculation and homogenization, after 1 week of aging, and after 2 weeks of aging) by BAM and iQ-Check methods to verify the target levels and homogeneity.
On the day of shipment, a five-tube three-level most probable number (MPN) analysis was performed by evaluating 5 × 50 g replicates, 5 × 25 g replicates, and 5 × 10 g replicates to obtain final inoculation levels in the dry dog food sample sets. The samples were apportioned (25 g each), packaged, labeled (with randomized, blind-coded, three-digit numbers), and shipped overnight to the seven participating laboratories. For each laboratory, four sets of eight samples from each of the three inoculation levels were sent, along with two sets of samples from the uninoculated control set reserved for aerobic plate count (APC).
Overview of Sample Analysis
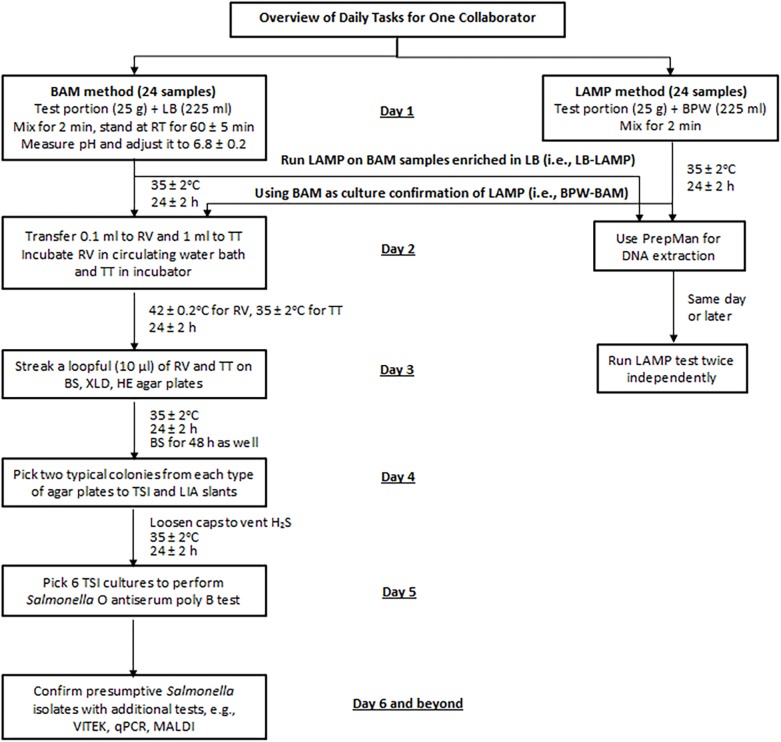
All collaborators (or teams) began testing on the same day. APC was performed by the pour plate method according to the FDA BAM Chapter 3 (Maturin and Peeler, 2018) or using the CompactDry plates (Hardy Diagnostics, Santa Maria, CA, United States). On day 1, each collaborator processed 24 samples following the BAM method and 24 samples following the LAMP method (Figure 2). Additionally, all LB preenrichment cultures from the BAM samples were tested by LAMP (i.e., LB-LAMP), and all BPW preenrichment cultures from the LAMP samples were processed with BAM for culture confirmation (i.e., BPW-BAM) from day 2. Therefore, a full data set from each collaborator consisted of 48 BAM and 48 LAMP results.
FIGURE 2.
Sample analysis flowchart for the MLV study. LB, lactose broth; BPW, buffered peptone water; RV, Rappaport-Vassiliadis medium; TT, tetrathionate broth; BS, bismuth sulfite agar; XLD, xylose lysine desoxycholate agar; HE, Hektoen enteric agar; TSI, triple sugar iron agar; LIA, lysine iron agar; qPCR, real-time quantitative PCR; MALDI, Matrix Assisted Laser Desorption Ionization.
The Loop-Mediated Isothermal Amplification (LAMP) Method
DNA extraction was performed by using the PrepMan Ultra sample preparation reagents (Thermo Fisher Scientific, Waltham, MA, United States). Briefly, aliquots (1 ml) of BPW or LB preenrichment cultures were first centrifuged at 900 × g for 1 min to remove large particles followed by another centrifugation at 16,000 × g for 2 min. The pellets were suspended in 100 μl of PrepMan Ultra reagent, heated at 100°C for 10 min, cooled to room temperature, and centrifuged again at 16,000 × g for 2 min. The supernatants (sample DNA extracts) were stored at -20°C until use.
The LAMP assay was carried out as described previously (Domesle et al., 2018). A positive control (S. enterica Typhimurium ATCC 19585 [LT2] at 1.7 × 104 CFU/reaction) and no template control (molecular grade water) were included in each LAMP run. Briefly, the reagent mixture in a total volume of 25 μl contained 1× isothermal master mix ISO-001 (consisting of a strand-displacing GspSSD DNA polymerase large fragment from Geobacillus spp., thermostable inorganic pyrophosphatase, reaction buffer, MgSO4, dNTPs, and a double-stranded DNA binding dye; OptiGene Ltd.), 1× primer mix (0.1 μM each outer primer Sal4-F3 [GAACGTGTCGCGGAAGTC] and Sal4-B3 [CGGCAATAGCGTCACCTT], 1.8 μM each inner primer Sal4-FIP [GCGCGGCATCCGCATCAATATCTGGATGGTATGCCCGG] and Sal4-BIP [GCGAACGGCGAAGCGTACTGTCGCACCGTCAAAGGAAC], and 1 μM each loop primer Sal4-LF [TCAAATCGGCATCAATACTCATCTG] and Sal4-LB [AAAGGGAAAGCCAGCTTTACG]; Integrated DNA Technologies, Coralville, IA, United States), and 2 μl of sample DNA extract. The LAMP reaction was run at 65°C for 30 min followed by an annealing step from 98 to 80°C with 0.05°C decrement per second (Figure 3C) in the Genie II or Genie III real-time fluorometer (OptiGene Ltd.). Fluorescence readings were acquired using the 6-carboxyfluorescein (FAM) channel (Figure 3A) and time-to-peak values (Tmax; min) were determined when fluorescence ratios reached the maximum value of the amplification rate curve (Figure 3B). Corresponding annealing temperatures (Tm; °C) of LAMP products were obtained in the anneal derivative curve (Figure 3D). Both Tmax and Tm values were displayed in the “Results” tab at the end of the run (Figure 3E). Testing was repeated once independently.
FIGURE 3.
Representative LAMP graphs and results generated and viewed using the Genie Explorer software (version 2.0.6.3). (A) Amplification; (B) amplification rate; (C) anneal; (D) anneal derivative; and (E) results. Samples with three-digit codes correspond to dry dog food test portions with low-level inoculation of Salmonella Infantis ATCC 51741. Samples marked as PC are positive control, i.e., S. enterica Typhimurium ATCC 19585 (LT2) at 1.7 × 104 CFU/reaction. Samples marked as NTC are no template control, i.e., molecular grade water.
The Bacteriological Analytical Manual (BAM) Method
Procedures described in the BAM Chapter 5 (Andrews et al., 2018) were followed. All media and reagents were from BD Diagnostic Systems (Sparks, MD, United States) unless specified otherwise. As outlined in Figure 2, samples were processed by preenrichment in LB (day 1), selective enrichment in Rappaport-Vassiliadis (RV) medium and tetrathionate (TT) broth (day 2), selective plating on bismuth sulfite (BS) agar, xylose lysine desoxycholate (XLD) agar, and Hektoen enteric (HE) agar (day 3), biochemical confirmation on triple sugar iron agar (TSI) slant and lysine iron agar (LIA) slant (day 4), and serological identification by Salmonella O antiserum poly B (day 5). Additional confirmation tests performed included VITEK 2 Gram-negative biochemical identification method (AOAC Official Method 2011.17), Bruker MALDI Gram-negative Biotyper method (AOAC Official Method 2017.09) (Association of Official Analytical Chemists [AOAC], 2018), or real-time qPCR as specified in the BAM Chapter 5 (Andrews et al., 2018).
Statistical Analysis
MPNs were calculated for the low- and high-level inoculated dry dog food using the LCF MPN calculator version 1.6 (Least Cost Formulations, Ltd., 2008). BAM samples were considered positive when Salmonella isolates were recovered. LAMP samples with the correct Tm (approximately 90°C) and Tmax values between 5 and 30 min were considered positive. For this MLV, all LAMP testing results were reported as presumptive results (presumptive positive or presumptive negative). BAM and LAMP results for each contamination level (including uninoculated controls) were analyzed by using the probability of detection (POD) statistical model (Wehling et al., 2011) with the AOAC Binary Data Interlaboratory Study Workbook version 2.3 (Association of Official Analytical Chemists [AOAC], 2013). For each collaborator, PODs were calculated for the LAMP presumptive results (including false positive ones), LAMP confirmed by BAM results (including false negative ones), and LAMP final results (excluding false positive and false negative ones, i.e., only those LAMP samples tested positive by both LAMP and BAM confirmation), and the BAM reference results. LPOD values was determined by combining all valid collaborator-level POD data and the difference in LPOD (dLPOD) between two methods were calculated with a 95% confidence interval. The two methods were considered statistically significant when the dLPOD confidence interval did not contain zero.
Additionally, BAM and LAMP results were analyzed by using a random intercept logistic regression model for unpaired samples and Obuchowski’s modified McNemar’s test (Obuchowski, 1998) and a conditional logistic regression model for paired samples. Differences between the methods being compared were considered significant when P < 0.05.
Results
The average APC was 2.1 × 101 CFU/g (ranging from <1.0 × 101 to 1.6 × 102 CFU/g) for the uninoculated dry dog food controls. Salmonella MPNs obtained in the two inoculated sample sets, with a 95% confidence interval, were 0.65 MPN/25 g (0.30, 1.40) for the low level and 3.01 MPN/25 g (1.31, 6.89) for the high level. Two collaborators (9 and 10) mixed up sample bag sets among the four sets of samples received in their laboratory, resulting in uneven number (5–10) of samples tested per inoculation level. Nonetheless, the samples were not compromised as they were individually bagged and blindly coded, and their data were still included in the final statistical analysis for the MLV study. Data from another two collaborators (2 and 4) were excluded due to confirmed positive results among uninoculated controls. In total, there were 288 (12 collaborator × 24 samples/collaborator) data points each for LAMP and BAM in the final comparisons presented below, which include LAMP vs. BAM using unpaired samples, LAMP vs. BAM using paired samples, BPW vs. LB for use as LAMP preenrichment buffers, and BPW vs. LB for use as BAM preenrichment buffers.
Unpaired Sample Statistical Analysis: LAMP Was Comparable to BAM
Table 1 shows the collaborator-level comparative results for the detection of Salmonella Infantis ATCC 51741 in 25 g dry dog food test portions by the LAMP alternative method versus the FDA BAM Chapter 5 reference method in an unpaired study design, i.e., different portions were analyzed by LAMP and BAM using different preenrichment buffers. For the uninoculated controls, collaborators 2, 4, and 5 had LAMP presumptive positive results with the rate as high as 75% for both collaborators 2 and 4. The single sample for collaborator 5 did not confirm positive by BAM culturing, but several samples for collaborators 2 and 4 did (LAMP final). Based on these results, data from collaborators 2 and 4 were excluded from the MLV. In addition, collaborator 1 had one LAMP presumptive negative sample confirming positive by BAM and collaborator 4 had one BAM sample testing positive by BAM. Fractional recovery (i.e., 25 to 75% positive responses) was obtained for the low inoculation level by all collaborators although 2, 4, 9, and 10 achieved that by only one method (LAMP presumptive or BAM reference). All high-level inoculated samples tested positive, regardless of the method used (Table 1).
Table 1.
Comparative detection of Salmonella Infantis ATCC 51741 in 25 g dry dog food test portions by the LAMP method versus the FDA BAM Chapter 5 reference method in an unpaired study design.
| Collaboratora | Number of positive samples/number of samples tested (probability of detection, POD) among dry dog food test portionsb | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Uninoculated control (0 MPN/25 g) | Low-level inoculation (0.65 MPN/25 g) | High-level inoculation (3.01 MPN/25 g) | |||||||
| LAMP presumptive | LAMP final | BAM reference | LAMP presumptive | LAMP final | BAM reference | LAMP presumptive | LAMP final | BAM reference | |
| 1 | 0/8 (0) | 0/8 (0)c | 0/8 (0) | 4/8 (0.5) | 4/8 (0.5) | 6/8 (0.75) | 8/8 (1) | 8/8 (1) | 8/8 (1) |
| 2 | 6/8 (0.75) | 1/8 (0.13) | 0/8 (0) | 7/8 (0.88) | 5/8 (0.63) | 6/8 (0.75) | 8/8 (1) | 8/8 (1) | 8/8 (1) |
| 3 | 0/8 (0) | 0/8 (0) | 0/8 (0) | 6/8 (0.75) | 6/8 (0.75) | 2/8 (0.25) | 8/8 (1) | 8/8 (1) | 8/8 (1) |
| 4 | 6/8 (0.75) | 2/8 (0.25) | 1/8 (0.13) | 8/8 (1) | 6/8 (0.75) | 5/8 (0.63) | 8/8 (1) | 8/8 (1) | 8/8 (1) |
| 5 | 1/8 (0.13) | 0/8 (0) | 0/8 (0) | 4/8 (0.5) | 3/8 (0.38) | 6/8 (0.75) | 8/8 (1) | 8/8 (1) | 8/8 (1) |
| 6 | 0/8 (0) | 0/8 (0) | 0/8 (0) | 4/8 (0.5) | 4/8 (0.5) | 5/8 (0.63) | 8/8 (1) | 8/8 (1) | 8/8 (1) |
| 7 | 0/8 (0) | 0/8 (0) | 0/8 (0) | 2/8 (0.25) | 2/8 (0.25) | 6/8 (0.75) | 8/8 (1) | 8/8 (1) | 8/8 (1) |
| 8 | 0/8 (0) | 0/8 (0) | 0/8 (0) | 5/8 (0.63) | 5/8 (0.63) | 4/8 (0.5) | 8/8 (1) | 8/8 (1) | 8/8 (1) |
| 9 | 0/10 (0) | 0/10 (0) | 0/6 (0) | 4/5 (0.8) | 3/5 (0.6) | 5/11 (0.45) | 9/9 (1) | 9/9 (1) | 7/7 (1) |
| 10 | 0/6 (0) | 0/6 (0) | 0/10 (0) | 3/9 (0.33) | 3/9 (0.33) | 7/7 (1) | 9/9 (1) | 9/9 (1) | 7/7 (1) |
| 11 | 0/8 (0) | 0/8 (0) | 0/8 (0) | 6/8 (0.75) | 6/8 (0.75) | 5/8 (0.63) | 8/8 (1) | 8/8 (1) | 8/8 (1) |
| 12 | 0/8 (0) | 0/8 (0) | 0/8 (0) | 4/8 (0.5) | 4/8 (0.5) | 2/8 (0.25) | 8/8 (1) | 8/8 (1) | 8/8 (1) |
| 13 | 0/8 (0) | 0/8 (0) | 0/8 (0) | 5/8 (0.63) | 5/8 (0.63) | 4/8 (0.5) | 8/8 (1) | 8/8 (1) | 8/8 (1) |
| 14 | 0/8 (0) | 0/8 (0) | 0/8 (0) | 4/8 (0.5) | 3/8 (0.38) | 6/8 (0.75) | 8/8 (1) | 8/8 (1) | 8/8 (1) |
| All | 13/112 (0.12) | 3/112 (0.03) | 1/112 (0.01) | 66/110 (0.6) | 59/110 (0.54) | 69/114 (0.61) | 114/114 (100) | 114/114 (100) | 110/110 (100) |
| Finald | 1/96 (0.01) | 0/96 (0) | 0/96 (0) | 51/94 (0.54) | 48/94 (0.51) | 58/98 (0.59) | 98/98 (100) | 98/98 (100) | 94/94 (100) |
a Collaborators 1 and 2 were from the same laboratory, so were collaborators 3 and 4, 5 and 6, 7 and 8, 9 and 10, 11 and 12, 13 and 14. b The LAMP presumptive column shows the results when LAMP samples were tested by LAMP (including false positive results). The LAMP final column shows the results when combining the LAMP presumptive and LAMP confirmed by BAM results (data not shown) to exclude false positive and false negative results, i.e., only LAMP samples tested positive by both LAMP and BAM confirmation were included. The BAM reference column shows the results when BAM samples were tested by BAM. c One sample was confirmed positive by BAM. The LAMP final result excluded this sample as it was negative by LAMP. d The Final row excluded data from collaborators 2 and 4 due to culture-confirmed positives in uninoculated test portions.
Table 2 summarizes the statistics generated using the POD model and comparisons made using this model and a random intercept logistic regression model for unpaired samples (e.g., LAMP vs. BAM) and the Obuchowski’s modified McNemar’s test and a conditional logistic regression model for paired samples (e.g., LAMP presumptive vs. LAMP confirmed). For the low inoculation level, 51 out of 94 samples were LAMP presumptive positive (LPOD of 0.54) with 48 of them confirming positive (LPOD of 0.51). No false negative results were obtained (data not shown), therefore the LAMP final LPOD was also 0.51. Among 98 samples tested by BAM, 58 produced positive results (LPOD of 0.59). A dLPOD value of -0.08 with a 95% confidence interval (-0.24, 0.08) was obtained between LAMP final and BAM, indicating they were comparable. Similarly, for the high inoculation level and uninoculated controls, no significant differences were observed between LAMP final and BAM as confidence intervals for both dLPOD values contained zero. Based on dLPOD analysis, three other comparisons (i.e., LAMP presumptive vs. BAM, LAMP presumptive vs. LAMP confirmed, and LAMP presumptive vs. LAMP final) also showed no statistical significance. The statistical insignificance for all four comparisons at all three inoculation levels were separately confirmed by using aforementioned statistical models as indicated by P-values greater than 0.05 (Table 2).
Table 2.
Summary of statistics generated using the POD model and other models for the detection of Salmonella Infantis ATCC 51741 in 25 g dry dog food test portions by the LAMP method versus the BAM reference method in an unpaired study design.
| Parameter and comparisons | Combined POD and associated statistics (lower control limit, upper control limit) among dry dog food test portions | ||
|---|---|---|---|
| Uninoculated (0 MPN/25 g) | Low-level (0.65 MPN/25 g) | High-level (3.01 MPN/25 g) | |
| Statistics generated using the POD modela | |||
| LAMP presumptive positive/total number | 1/96 | 51/94 | 98/98 |
| LPOD | 0.01 (0.00, 0.06) | 0.54 (0.44, 0.65) | 1.00 (0.96, 1.00) |
| sr | 0.10 (0.09, 0.19) | 0.51 (0.44, 0.54) | 0.00 (0.00, 0.19) |
| sL | 0.00 (0.00, 0.05) | 0.00 (0.00, 0.23) | 0.00 (0.00, 0.19) |
| sR | 0.10 (0.09, 0.12) | 0.51 (0.45, 0.54) | 0.00 (0.00, 0.27) |
| P-value | 0.4336 | 0.6048 | 1.0000 |
| LAMP confirmed positive/total number | 1/96 | 48/94 | 98/98 |
| LPOD | 0.01 (0.00, 0.06) | 0.51 (0.41, 0.62) | 1.00 (0.96, 1.00) |
| sr | 0.10 (0.09, 0.19) | 0.51 (0.44, 0.54) | 0.00 (0.00, 0.19) |
| sL | 0.00 (0.00, 0.05) | 0.00 (0.00, 0.22) | 0.00 (0.00, 0.19) |
| sR | 0.10 (0.09, 0.12) | 0.51 (0.45, 0.54) | 0.00 (0.00, 0.27) |
| P-value | 0.4336 | 0.6070 | 1.0000 |
| LAMP final positive/total number | 0/96 | 48/94 | 98/98 |
| LPOD | 0.00 (0.00, 0.04) | 0.51 (0.41, 0.62) | 1.00 (0.96, 1.00) |
| sr | 0.00 (0.00, 0.19) | 0.51 (0.44, 0.54) | 0.00 (0.00, 0.19) |
| sL | 0.00 (0.00, 0.19) | 0.00 (0.00, 0.22) | 0.00 (0.00, 0.19) |
| sR | 0.00 (0.00, 0.27) | 0.51 (0.45, 0.54) | 0.00 (0.00, 0.27) |
| P-value | 1.0000 | 0.6070 | 1.0000 |
| BAM positive/total number | 0/96 | 58/98 | 94/94 |
| LPOD | 0.00 (0.00, 0.04) | 0.59 (0.47, 0.72) | 1.00 (0.96, 1.00) |
| sr | 0.00 (0.00, 0.19) | 0.48 (0.41, 0.53) | 0.00 (0.00, 0.19) |
| sL | 0.00 (0.00, 0.19) | 0.14 (0.00, 0.33) | 0.00 (0.00, 0.19) |
| sR | 0.00 (0.00, 0.27) | 0.50 (0.44, 0.53) | 0.00 (0.00, 0.27) |
| P-value | 1.0000 | 0.0974 | 1.0000 |
| Comparisons based on the POD model and other statistical modelsb | |||
| dLPOD (LAMP final vs. BAM) | 0.00 (-0.04, 0.04) | -0.08 (-0.24, 0.08) | 0.00 (-0.04, 0.04) |
| dLPOD (LAMP presumptive vs. BAM) | 0.01 (-0.03, 0.06) | -0.05 (-0.21, 0.11) | 0.00 (-0.04, 0.04) |
| dLPOD (LAMP presumptive vs. LAMP confirmed) | 0.00 (-0.05, 0.05) | 0.03 (-0.12, 0.18) | 0.00 (-0.04, 0.04) |
| dLPOD (LAMP presumptive vs. LAMP final) | 0.01 (-0.03, 0.06) | 0.03 (-0.12, 0.18) | 0.00 (-0.04, 0.04) |
| P-value (LAMP final vs. BAM) | N/A | 0.26 | N/A |
| P-value (LAMP presumptive vs. BAM) | 0.32 | 0.49 | N/A |
| P-value (LAMP presumptive vs. LAMP confirmed) | 1 (1) | 0.99 (f) | N/A (N/A) |
| P-value (LAMP presumptive vs. LAMP final) | 0.99 (f) | 0.96 (f) | N/A (N/A) |
a LPOD is a composite POD across collaborators and includes between-collaborator variation in addition to variation inherent in the binomial nature of the binary probabilities. sr is repeatability standard deviation, sL is among-collaborator standard deviation, sR is reproducibility standard deviation. P-value is homogeneity test of collaborator PODs. b dLPOD is the difference in LPOD between two methods. The numbers in parenthesis are 95% confidence interval (lower control limit [LCL], upper control limit [UCL]) estimates on dLPOD. A confidence interval for dLPOD that does not contain 0 indicates a statistically significant difference between the two methods being compared. A random intercept logistic regression model was used for unpaired comparison and Obuchowski’s modified McNemar’s test and a conditional logistic regression model (numbers in parenthesis) were used for paired comparisons. f indicates that model fitting failed to converge. N/A, no test was done because of complete match of the results.
Paired Sample Statistical Analysis: LAMP Was Comparable to BAM
Table 3 shows the summary statistics for the LAMP and BAM methods when paired samples were used (LB-LAMP vs. BAM), i.e., the same test portions were analyzed by LAMP and BAM following preenrichment in LB. For the low inoculation level, 58 out of 98 samples were LB-LAMP positive (LPOD of 0.59) while 58 out of 98 samples were positive by BAM (LPOD of 0.59). Collaborator 13 reported one positive sample by LB-LAMP only, while collaborator 8 had one sample positive by BAM only (data not shown). A dLPOD value of 0.00 with a 95% confidence interval (-0.18, 0.18) was obtained, indicating the two methods were not significantly different. Similarly, for the high inoculation level and uninoculated controls, no significant differences were observed as confidence intervals for both dLPOD values contained zero. One uninoculated sample from collaborator 14 was positive by LB-LAMP but not BAM (data not shown). The statistical insignificance between LB-LAMP and BAM at all three inoculation levels was separately confirmed by using the Obuchowski’s modified McNemar’s test and the conditional logistic regression model (Table 3).
Table 3.
Summary of statistics generated using the POD model and other models for the detection of Salmonella Infantis ATCC 51741 in 25 g dry dog food test portions by the LAMP method (with LB preenrichment) versus the BAM reference method in a paired study design.
| Parameter and comparisons | Combined POD and associated statistics (lower control limit, upper control limit) among dry dog food test portions | ||
|---|---|---|---|
| Uninoculated (0 MPN/25 g) | Low-level (0.65 MPN/25 g) | High-level (3.01 MPN/25 g) | |
| Statistics generated using the POD modela | |||
| LB-LAMP positive/total number | 1/96 | 58/98b | 94/94 |
| LPOD | 0.01 (0.00, 0.06) | 0.59 (0.46, 0.72) | 1.00 (0.96, 1.00) |
| sr | 0.10 (0.09, 0.19) | 0.47 (0.41, 0.53) | 0.00 (0.00, 0.19) |
| sL | 0.00 (0.00, 0.05) | 0.15 (0.00, 0.35) | 0.00 (0.00, 0.19) |
| sR | 0.10 (0.09, 0.12) | 0.50 (0.44, 0.53) | 0.00 (0.00, 0.27) |
| P-value | 0.4336 | 0.0726 | 1.0000 |
| BAM positive/total number | 0/96 | 58/98b | 94/94 |
| LPOD | 0.00 (0.00, 0.04) | 0.59 (0.47, 0.72) | 1.00 (0.96, 1.00) |
| sr | 0.00 (0.00, 0.19) | 0.48 (0.41, 0.53) | 0.00 (0.00, 0.19) |
| sL | 0.00 (0.00, 0.19) | 0.14 (0.00, 0.33) | 0.00 (0.00, 0.19) |
| sR | 0.00 (0.00, 0.27) | 0.50 (0.44, 0.53) | 0.00 (0.00, 0.27) |
| P-value | 1.0000 | 0.0974 | 1.0000 |
| Comparisons based on the POD model and other statistical modelsc | |||
| dLPOD (LB-LAMP vs. BAM) | 0.01 (-0.03, 0.06) | 0.00 (-0.18, 0.18) | 0.00 (-0.04, 0.04) |
| P-value (LB-LAMP vs. BAM) | 0.99 (f) | 1 (1) | N/A (N/A) |
a LPOD is a composite POD across collaborators and includes between-collaborator variation in addition to variation inherent in the binomial nature of the binary probabilities. sr is repeatability standard deviation, sL is among-collaborator standard deviation, sR is reproducibility standard deviation. P-value is homogeneity test of laboratory (collaborator) PODs. b Collaborator 13 had one positive sample by LB-LAMP only, while collaborator 8 had one positive sample by BAM only. c dLPOD is the difference in LPOD between two methods. The numbers in parenthesis are 95% confidence interval (lower control limit [LCL], upper control limit [UCL]) estimates on dLPOD. A confidence interval for dLPOD that does not contain 0 indicates a statistically significant difference between the two methods being compared. Obuchowski’s modified McNemar’s test and a conditional logistic regression model (numbers in parenthesis) were used for LB-LAMP vs. BAM comparisons. f indicates that model fitting failed to converge. N/A, no test was done because of complete match of the results.
Preenrichment With BPW vs. LB Did Not Affect Salmonella Detection
Table 4 shows the statistics generated when unpaired samples were tested by either LAMP or BAM using different enrichment broths (LAMP vs. LB-LAMP and BPW-BAM vs. BAM). When tested by the LAMP method, 51 out of 94 low-level inoculated samples were positive (LPOD of 0.54) following BPW preenrichment, while 58 out of 98 samples produced positive results (LPOD of 0.59) following LB preenrichment. A dLPOD value of -0.05 with a 95% confidence interval (-0.22, 0.12) was obtained, indicating LAMP and LB-LAMP were not significantly different. Similarly, for the high inoculation level and uninoculated controls, no significant differences were observed as confidence intervals for both dLPOD values contained zero. Two different uninoculated samples were positive by either LAMP (for collaborator 5) or LB-LAMP (collaborator 14); neither was confirmed by BAM culturing (data not shown). The statistical insignificance at all three inoculation levels were separately confirmed by using the random intercept logistic regression model (Table 4). Therefore, preenrichment in BPW or LB did not significantly influence the LAMP results. The same held true for the BAM method when either BPW or LB were used as preenrichment buffers, i.e., there were no statistical significant differences for all three inoculation levels (Table 4).
Table 4.
Summary of statistics generated using the POD model and other models for the detection of Salmonella Infantis ATCC 51741 in 25 g dry dog food test portions by LAMP or BAM when different preenrichment buffers were used for each one in an unpaired study design.
| Parameter and comparisons | Combined POD and associated statistics (lower control limit, upper control limit) among dry dog food test portions | ||
|---|---|---|---|
| Uninoculated (0 MPN/25 g) | Low-level (0.65 MPN/25 g) | High-level (3.01 MPN/25 g) | |
| Statistics generated using the POD modela | |||
| LAMP positive/total number | 1/96b | 51/94 | 98/98 |
| LPOD | 0.01 (0.00, 0.06) | 0.54 (0.44, 0.65) | 1.00 (0.96, 1.00) |
| sr | 0.10 (0.09, 0.19) | 0.51 (0.44, 0.54) | 0.00 (0.00, 0.19) |
| sL | 0.00 (0.00, 0.05) | 0.00 (0.00, 0.23) | 0.00 (0.00, 0.19) |
| sR | 0.10 (0.09, 0.12) | 0.51 (0.45, 0.54) | 0.00 (0.00, 0.27) |
| P-value | 0.4336 | 0.6048 | 1.0000 |
| LB-LAMP positive/total number | 1/96b | 58/98 | 94/94 |
| LPOD | 0.01 (0.00, 0.06) | 0.59 (0.46, 0.72) | 1.00 (0.96, 1.00) |
| sr | 0.10 (0.09, 0.19) | 0.47 (0.41, 0.53) | 0.00 (0.00, 0.19) |
| sL | 0.00 (0.00, 0.05) | 0.15 (0.00, 0.35) | 0.00 (0.00, 0.19) |
| sR | 0.10 (0.09, 0.12) | 0.50 (0.44, 0.53) | 0.00 (0.00, 0.27) |
| P-value | 0.4336 | 0.0726 | 1.0000 |
| BPW-BAM positive/total number | 1/96 | 48/94 | 98/98 |
| LPOD | 0.01 (0.00, 0.06) | 0.51 (0.41, 0.62) | 1.00 (0.96, 1.00) |
| sr | 0.10 (0.09, 0.19) | 0.51 (0.44, 0.54) | 0.00 (0.00, 0.19) |
| sL | 0.00 (0.00, 0.05) | 0.00 (0.00, 0.22) | 0.00 (0.00, 0.19) |
| sR | 0.10 (0.09, 0.12) | 0.51 (0.45, 0.54) | 0.00 (0.00, 0.27) |
| P-value | 0.4336 | 0.6070 | 1.0000 |
| BAM positive/total number | 0/96 | 58/98 | 94/94 |
| LPOD | 0.00 (0.00, 0.04) | 0.59 (0.47, 0.72) | 1.00 (0.96, 1.00) |
| sr | 0.00 (0.00, 0.19) | 0.48 (0.41, 0.53) | 0.00 (0.00, 0.19) |
| sL | 0.00 (0.00, 0.19) | 0.14 (0.00, 0.33) | 0.00 (0.00, 0.19) |
| sR | 0.00 (0.00, 0.27) | 0.50 (0.44, 0.53) | 0.00 (0.00, 0.27) |
| P-value | 1.0000 | 0.0974 | 1.0000 |
| Comparisons based on the POD model and other statistical modelsc | |||
| dLPOD (LAMP vs. LB-LAMP) | 0.00 (-0.05, 0.05) | -0.05 (-0.22, 0.12) | 0.00 (-0.04, 0.04) |
| dLPOD (BPW-BAM vs. BAM) | 0.01 (-0.03, 0.06) | -0.08 (-0.24, 0.08) | 0.00 (-0.04, 0.04) |
| P-value (LAMP vs. LB-LAMP) | 1 | 0.49 | N/A |
| P-value (BPW-BAM vs. BAM) | 0.32 | 0.26 | N/A |
a LPOD is a composite POD across collaborators and includes between-collaborator variation in addition to variation inherent in the binomial nature of the binary probabilities. sr is repeatability standard deviation, sL is among-collaborator standard deviation, sR is reproducibility standard deviation. P-value is homogeneity test of collaborator PODs. b Collaborator 5 had one positive sample by LAMP and collaborator 14 had one positive sample by LB-LAMP; neither were confirmed by BAM culturing. c dLPOD is the difference in LPOD between two methods. The numbers in parenthesis are 95% confidence interval (lower control limit [LCL], upper control limit [UCL]) estimates on dLPOD. A confidence interval for dLPOD that does not contain 0 indicates a statistically significant difference between the two methods being compared. A random intercept logistic regression model was used for LAMP vs. LB-LAMP and BPW-BAM vs. BAM comparisons. N/A, no test was done because of complete match of the results.
Discussion
This collaborative study rigorously validated a LAMP-based method for the screening of Salmonella in dry dog food at the multi-laboratory level. FDA’s current method validation guidelines for microbial pathogens in foods and feeds (Food and Drug Administration [FDA], 2015) were used, which align well with those from the AOAC and ISO (Association of Official Analytical Chemists [AOAC], 2012; International Organization for Standardization [ISO], 2016). In 2016, a United Kingdom study (D’Agostino et al., 2016) reported the validation of a LAMP/ISO 6579-based method for analyzing soya meal (an animal feed ingredient) for the presence of S. enterica in ten laboratories from eight European countries. For reasons of cost and logistics, that interlaboratory study did not use centrally prepared Salmonella-contaminated soya meal samples. Instead, commercially available certified Salmonella reference materials were used for inoculation by each participating laboratory, and no aging period was incorporated. Importantly, none of the three levels tested (0, 1–5, and 14–68 CFU per test portion) produced fractional positive results (25–75%) and three uninoculated control samples were confirmed positive for Salmonella (D’Agostino et al., 2016).
Unlike the United Kingdom study which was a “paired” trial, this MLV used an unpaired study design, i.e., different test portions from the same bulk samples inoculated centrally and aged for 2 weeks were analyzed by the LAMP alternative method and the BAM reference method using different preenrichment buffers. All LAMP samples were confirmed by BAM culturing and the reported LAMP final positive results were for samples tested positive by both LAMP and BAM confirmation. For the low-level inoculation, the overall proportions of positive responses were 51% for LAMP final and 59% for BAM (Table 2), which clearly satisfies the criteria outlined in validation guidelines of the AOAC, FDA, and ISO (Association of Official Analytical Chemists [AOAC], 2012; Food and Drug Administration [FDA], 2015; International Organization for Standardization [ISO], 2016). Multiple pairwise comparisons showed insignificant differences between LAMP and BAM by using either the POD analysis or other statistical models (Table 2), which highlights the success and rigor of this MLV study. Feedback from participating laboratories showed that the LAMP method was rapid, sensitive, practical, user-friendly, and easily adoptable.
A few false positive and/or false negative results were observed across the three testing levels in this MLV. For the low-level inoculated samples, there were three LAMP false positive results (one each for collaborators 5, 9, and 14 comparing LAMP presumptive and LAMP final) and no false negative results (Table 1 excluding data from collaborators 2 and 4). In all three cases, the Tmax values were rather high (>15 min) compared to others (ca. 7 min) (data not shown), indicating the amount of target DNA in the sample DNA extracts was low. This may be attributed to low contamination levels and dead or injured cells in these samples, which failed to reach the detection limit of BAM even after enrichments. The samples were shipped without dry ice since pet food is usually stored and shipped at ambient temperature; however, this may have contributed to some of the variability observed in the study. Another possibility is there was cross-contamination introduced during DNA extraction or assay setup for LAMP. For the uninoculated controls, one false positive (collaborator 5) and one false negative (collaborators 1, noted in footnote) results were observed. The former had high Tmax values (average of 14 min), while the latter was technically true negative (false positive by BAM) as it was an uninoculated control. Cross-contamination may have occurred when the two collaborators processed the samples for LAMP or BAM. Prior to this MLV, the LAMP assay has been extensively evaluated and high specificity (100% inclusivity and exclusivity) and high sensitivity (a detection limit of <1 CFU/25 g in animal food) have been demonstrated (Chen et al., 2011; Domesle et al., 2018; Yang et al., 2013, 2014, 2015, 2016). During the MLV study, none of the positive control samples produced false negative results, and none of no-template-control samples produced false positive results. These outcomes corroborate the high specificity and sensitivity of the Salmonella LAMP assay.
Besides the main component of the MLV study (unpaired design), we also compared the performance of LAMP and BAM using paired samples, i.e., the same test portions were analyzed by LAMP and BAM following LB preenrichment (LB-LAMP vs. BAM). One false positive (collaborator 13) and one false negative (collaborator 8) results were observed in low-level inoculated samples and one false positive was observed in one uninoculated sample (Table 3). Similar reasons described above may account for these false positive or false negative results observed. Nonetheless, paired samples also confirmed the statistically insignificant differences between the two methods using either POD analysis or other models.
As LAMP is gaining popularity in clinical diagnostics and food testing, many commercially available LAMP-based systems and assays have been developed and some were validated for Salmonella detection in food (Yang et al., 2018). These include the 3M Molecular Detection Assay (MDA) Salmonella (3M Food Safety, St. Paul, MN, United States) in raw ground beef and wet dog food (Bird et al., 2013, 2014), the 3M MDA 2 – Salmonella in raw ground beef and creamy peanut butter (Bird et al., 2016), and the SAS Molecular Tests Salmonella detection kit (SA Scientific Ltd., San Antonio, TX, United States) in ground beef, beef trim, ground turkey, chicken carcass rinses, bagged mixed lettuce, and fresh spinach (Bapanpally et al., 2014). In the two studies evaluating the 3M MDA Salmonella in wet dog food against the FDA BAM method, an unpaired study design was used and fractional positive results were obtained with POD analysis showing the methods were comparable (Bird et al., 2013, 2014). It is noteworthy that platforms used for the detection of LAMP amplicons were different in these studies as compared to our study. Bioluminescence was used for the 3M MDA assays, turbidity for the SAS kit, and fluorescence was used in our MLV (Genie II or Genie III). We previously tested the Salmonella LAMP assay on all three platforms (Chen et al., 2011; Yang et al., 2013, 2014, 2015, 2016; Domesle et al., 2018) and the fluorescence-based Genie II or Genie III was chosen for its simplicity, rapidity, portability, software interface, report format, and user-friendliness, with the annealing step offering an extra checkpoint to ensure the high specificity of the assay.
Another interesting outcome of the MLV study was the comparison of preenrichment buffers used for LAMP and BAM. We chose BPW as the default preenrichment buffer for the Salmonella LAMP method since preliminary data showed that shorter Tmax values were obtained for samples pre-enriched in BPW compared to those in LB (data not shown). Comparing Tmax values generated in this MLV for low- and high-level inoculated samples showed that BPW preenrichment generated Tmax values on average 1.9 min and 1.6 min shorter than those using LB preenrichment, suggesting the amount of DNA was higher when BPW was used as the preenrichment buffer. A recent study evaluating the 3M MDA Salmonella and the ANSR (stands for amplified nucleic single temperature reaction) detection system for Salmonella (Neogen Food Safety, Lansing, MI, United States) in egg products also showed that preenrichment in BPW improved the performance of both assays compared to LB (Hu et al., 2017). Nonetheless, using POD analysis for a qualitative method, statistically significant differences were not observed between BPW and LB for either LAMP or BAM, indicating they were comparable (Table 4).
It is worth noting that the S. Infantis ATCC 51741 used for inoculation in this MLV was a non-H2S producer with uncharacteristic serological reactions to Salmonella O antiserum poly B (data not shown). As a result, multiple confirmation methods besides serotyping were performed by participating laboratories, including VITEK 2, Bruker MALDI, and real-time qPCR, extending the time of sample testing by BAM to 2 weeks in contrast to 24 h by LAMP.
In our single-laboratory validation study (Domesle et al., 2018), five other animal food types (cattle feed, chicken feed, horse feed, swine feed, and dry cat food) besides dry dog food were successfully validated following FDA’s guidelines. The LAMP method validated in this MLB study in dry dog food should be applicable to these and other animal food types, per guidelines of the AOAC, FDA, and ISO (Association of Official Analytical Chemists [AOAC], 2012; Food and Drug Administration [FDA], 2015; International Organization for Standardization [ISO], 2016). Additional matrix extension studies may be readily performed in a variety of food matrices.
Conclusion
In conclusion, the MLV study clearly demonstrated the utility and applicability of this rapid and reliable LAMP method in routine regulatory screening of Salmonella in animal food. As only LAMP-positive samples should continue with the isolation of Salmonella by the FDA BAM culture method, the LAMP method holds great potential to significantly reduce the time and labor and improve efficiency in animal food testing.
Data Availability
All datasets generated for this study are included in the manuscript.
Author Contributions
BG, KD, QRY, TH, and RP contributed to design and coordination of the study. BG, KD, and QRY performed the data analysis and interpretation. SW contributed to statistical analysis. XD, LJH, GZ, YH, XL, KC, JD, KH, SL, RR, DR, AY, PP, CW, TE, DK, JS, DW, LH, LL, CL, QY, DS, and HC contributed to sample testing and coordination of the study in seven participating laboratories. SA and SY assisted with sample analysis in the coordinating laboratory. All authors reviewed the final manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are grateful to William B. Baroletti/Lawrence E. James, Teresa Lee, and Carrie Leach/Mark Moelhman for supporting the project at FDA’s Northeast Food and Feed Laboratory, San Francisco Laboratory, and Office of Indiana State Chemist, respectively. We thank Andriy Tkachenko, Jake Guag, Sarah M. Nemser, and Renate Reimschuessel at the FDA’s Veterinary Laboratory Investigation and Response Network (Vet-LIRN) for insights and guidelines on MLV studies and recruiting participating laboratories. We also thank Claudine Kabera and Heather Tate at FDA’s National Antimicrobial Resistance Monitoring System (NARMS) for the help recruiting participating laboratories. We appreciate Maureen Davidson at the Division of Animal and Food Microbiology for supporting the project and Gordon Martin for assistance with the MLV study. Finally, we want to thank the FDA’s Microbiological Methods Validation Subcommittee (MMVS) members for their critical review of the MLV study protocols and reports.
The views expressed in this manuscript are those of the authors and do not necessarily reflect the official policy of the Department of Health and Human Services, the United States Food and Drug Administration, or the United States Government. Reference to any commercial materials, equipment, or process does not in any way constitute approval, endorsement, or recommendation by the Food and Drug Administration.
References
- Andrews W. H., Jacobson A., Hammack T. S. (2018). Bacteriological Analytical Manual (BAM) Chapter 5: Salmonella. Available at: https://www.fda.gov/Food/FoodScienceResearch/LaboratoryMethods/ucm070149.htm [Google Scholar]
- Association of Official Analytical Chemists [AOAC] (2012). Appendix J: AOAC INTERNATIONAL Methods Committee Guidelines for Validation of Microbiological Methods for Food and Environmental Surfaces. Available at: http://www.eoma.aoac.org/app_j.pdf [Google Scholar]
- Association of Official Analytical Chemists [AOAC] (2013). AOAC Interlaboratory Study Workbook - Binary Data. Available at: http://www.aoac.org/aoac_prod_imis/14AMSTAKE/Guidelines/AOAC_Member/SDCF/SDGRCF/SDGRG.aspx?hkey=961edb9e-0824-473f-b898-d2a9bc0c7ee9 [Google Scholar]
- Association of Official Analytical Chemists [AOAC] (2018). Official Methods of Analysis of AOAC INTERNATIONAL (OMA). Available at: http://www.eoma.aoac.org/ [Google Scholar]
- Balachandran P., Friberg M., Vanlandingham V., Kozak K., Manolis A., Brevnov M., et al. (2012). Rapid detection of Salmonella in pet food: design and evaluation of integrated methods based on real-time PCR detection. J. Food Prot. 75 347–352. 10.4315/0362-028X.JFP-11-210 [DOI] [PubMed] [Google Scholar]
- Bapanpally C., Montier L., Khan S., Kasra A., Brunelle S. L. (2014). SAS molecular tests Salmonella detection kit. Performance tested method 021202. J. AOAC Int. 97 808–819. 10.5740/jaoacint.13-326 [DOI] [PubMed] [Google Scholar]
- Bird P., Fisher K., Boyle M., Huffman T., Benzinger M. J., Jr., Bedinghaus P., et al. (2013). Evaluation of 3M molecular detection assay (MDA) Salmonella for the detection of Salmonella in selected foods: collaborative study. J. AOAC Int. 96 1325–1335. 10.5740/jaoacint.13-227 [DOI] [PubMed] [Google Scholar]
- Bird P., Fisher K., Boyle M., Huffman T., Benzinger M. J., Jr., Bedinghaus P., et al. (2014). Evaluation of modification of the 3M molecular detection assay (MDA) Salmonella method (2013.09) for the detection of Salmonella in selected foods: collaborative study. J. AOAC Int. 97 1329–1342. 10.5740/jaoacint.14-101 [DOI] [PubMed] [Google Scholar]
- Bird P., Flannery J., Crowley E., Agin J. R., Goins D., Monteroso L. (2016). Evaluation of the 3M molecular detection assay (MDA) 2 - Salmonella for the detection of Salmonella spp. in select foods and environmental surfaces: collaborative study, first action 2016.01. J. AOAC Int. 99 980–997. 10.5740/jaoacint.16-0085 [DOI] [PubMed] [Google Scholar]
- Chen S., Wang F., Beaulieu J. C., Stein R. E., Ge B. (2011). Rapid detection of viable salmonellae in produce by coupling propidium monoazide with loop-mediated isothermal amplification. Appl. Environ. Microbiol. 77 4008–4016. 10.1128/AEM.00354-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng C. M., Doran T., Lin W., Chen K. S., Williams-Hill D., Pamboukian R. (2015). Interlaboratory validation for a real-time PCR Salmonella detection method using the ABI 7500 FAST real-time PCR system. J. Food Prot. 78 1119–1124. 10.4315/0362-028X.JFP-14-244 [DOI] [PubMed] [Google Scholar]
- Crump J. A., Griffin P. M., Angulo F. J. (2002). Bacterial contamination of animal feed and its relationship to human foodborne illness. Clin. Infect. Dis. 35 859–865. 10.1086/342885 [DOI] [PubMed] [Google Scholar]
- D’Agostino M., Robles S., Hansen F., Ntafis V., Ikonomopoulos J., Kokkinos P., et al. (2016). Validation of a loop-mediated amplification/ISO 6579-based method for analysing soya meal for the presence of Salmonella enterica. Food Anal. Methods 9 2979–2985. 10.1007/s12161-016-0602-7 [DOI] [Google Scholar]
- Domesle K. J., Yang Q., Hammack T. S., Ge B. (2018). Validation of a Salmonella loop-mediated isothermal amplification assay in animal food. Int. J. Food Microbiol. 264 63–76. 10.1016/j.ijfoodmicro.2017.10.020 [DOI] [PubMed] [Google Scholar]
- Food and Agriculture Organization [FAO]/World Health Organization [WHO] (2015). Executive Summary Report of the Joint FAO/WHO Expert Meeting on Hazards Associated With Animal Feed. Available at: http://www.fao.org/feed-safety/resources/resources-details/en/c/451737/ [Google Scholar]
- Food and Drug Administration [FDA] (2015). Guidelines for the Validation of Analytical Methods for the Detection of Microbial Pathogens in Foods and Feeds, 2nd Edn. Available at: https://www.fda.gov/downloads/ScienceResearch/FieldScience/UCM298730.pdf [Google Scholar]
- Food and Drug Administration [FDA] (2017a). 21 CFR Part 117: Current Good Manufacturing Practice, Hazard Analysis, and Risk-Based Preventive Controls for Human Food. Silver Spring, MD: FDA. [Google Scholar]
- Food and Drug Administration [FDA] (2017b). 21 CFR Part 507: Current Good Manufacturing Practice, Hazard Analysis, and Risk-Based Preventive Controls for Food for Animals. Silver Spring, MD: FDA. [Google Scholar]
- Ge B., LaFon P. C., Carter P. J., McDermott S. D., Abbott J., Glenn A., et al. (2013). Retrospective analysis of Salmonella, Campylobacter, Escherichia coli, and Enterococcus in animal feed ingredients. Foodborne Pathog. Dis. 10 684–691. 10.1089/fpd.2012.1470 [DOI] [PubMed] [Google Scholar]
- Hsieh Y. C., Lee K. M., Poole T., Runyon M., Jones B., Herrman T. J. (2014). Detection and isolation of Salmonella spp. in animal feeds from 2007-2011. Int. J. Regul. Sci. 2 14–27. [Google Scholar]
- Hu L., Ma L. M., Zheng S., He X., Hammack T. S., Brown E. W., et al. (2018). Development of a novel loop-mediated isothermal amplification (LAMP) assay for the detection of Salmonella ser. Enteritidis from egg products. Food Control 88 190–197. 10.3168/jds.2017-12740 [DOI] [PubMed] [Google Scholar]
- Hu L., Ma L. M., Zheng S., He X., Wang H., Brown E. W., et al. (2017). Evaluation of 3M molecular detection system and ANSR pathogen detection system for rapid detection of Salmonella from egg products. Poult. Sci. 96 1410–1418. 10.3382/ps/pew399 [DOI] [PubMed] [Google Scholar]
- Interagency Food Safety Analytics Collaboration [IFSAC] (2018). Foodborne Illness Source Attribution Estimates for 2016 for Salmonella, Escherichia coli O157 Listeria monocytogenes, and Campylobacter Using Multi-Year Outbreak Surveillance Data, United States. Available at: https://www.cdc.gov/foodsafety/ifsac/pdf/P19-2016-report-TriAgency-508.pdf [Google Scholar]
- International Organization for Standardization [ISO] (2016). ISO 16140-2:2016 Microbiology of the Food Chain – Method Validation – Part 2: Protocol for the Validation of Alternative (Proprietary) Methods Against a Reference Method. Geneva: ISO. [Google Scholar]
- International Organization for Standardization [ISO] (2017). ISO 6579-1:2017 Microbiology of the Food Chain – Horizontal Method for the Detection, Enumeration and Serotyping of Salmonella – Part 1: Detection of Salmonella spp. Geneva: ISO. [Google Scholar]
- Jiang X. (2016). Prevalence and characterization of Salmonella in animal meals collected from rendering operations. J. Food Prot. 79 1026–1031. 10.4315/0362-028X.JFP-15-537 [DOI] [PubMed] [Google Scholar]
- Kaneko H., Kawana T., Fukushima E., Suzutani T. (2007). Tolerance of loop-mediated isothermal amplification to a culture medium and biological substances. J. Biochem. Biophys. Methods 70 499–501. 10.1016/j.jbbm.2006.08.008 [DOI] [PubMed] [Google Scholar]
- Least Cost Formulations, Ltd. (2008). MPN Calculator-Version 1.6. Available at: http://www.lcfltd.com/customer/LCFMPNCalculator.exe [Google Scholar]
- Li X., Bethune L. A., Jia Y., Lovell R. A., Proescholdt T. A., Benz S. A., et al. (2012). Surveillance of Salmonella prevalence in animal feeds and characterization of the Salmonella isolates by serotyping and antimicrobial susceptibility. Foodborne Pathog. Dis. 9 692–698. 10.1089/fpd.2011.1083 [DOI] [PubMed] [Google Scholar]
- Lofstrom C., Hoorfar J. (2012). Validation of an open-formula, diagnostic real-time PCR method for 20-h detection of Salmonella in animal feeds. Vet. Microbiol. 158 431–435. 10.1016/j.vetmic.2012.02.026 [DOI] [PubMed] [Google Scholar]
- Magossi G., Cernicchiaro N., Dritz S., Houser T., Woodworth J., Jones C., et al. (2018). Evaluation of Salmonella presence in selected United States feed mills. Microbiologyopen 10.1002/mbo3.711 [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malorny B., Huehn S., Dieckmann R., Kramer N., Helmuth R. (2009). Polymerase chain reaction for the rapid detection and serovar identification of Salmonella in food and feeding stuff. Food Anal. Methods 2 81–95. 10.1007/s12161-008-9057-9 [DOI] [Google Scholar]
- Maturin L., Peeler J. T. (2018). Bacteriological Analytical Manual Chapter 3: Aerobic Plate Count. Available at: https://www.fda.gov/Food/FoodScienceResearch/LaboratoryMethods/ucm063346.htm [Google Scholar]
- Molina A., Granados-Chinchilla F., Jimenez M., Acuna-Calvo M. T., Alfaro M., Chavarria G. (2016). Vigilance for Salmonella in feedstuffs available in Costa Rica: prevalence, serotyping and tetracycline resistance of isolates obtained from 2009 to 2014. Foodborne Pathog. Dis. 13 119–127. 10.1089/fpd.2015.2050 [DOI] [PubMed] [Google Scholar]
- Nemser S. M., Doran T., Grabenstein M., McConnell T., McGrath T., Pamboukian R., et al. (2014). Investigation of Listeria, Salmonella, and toxigenic Escherichia coli in various pet foods. Foodborne Pathog. Dis. 11 706–709. 10.1089/fpd.2014.1748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Notomi T., Okayama H., Masubuchi H., Yonekawa T., Watanabe K., Amino N., et al. (2000). Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 28:E63 10.1093/nar/28.12.e63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obuchowski N. A. (1998). On the comparison of correlated proportions for clustered data. Stat. Med. 17 1495–1507. [DOI] [PubMed] [Google Scholar]
- United States Department of Agriculture [USDA] (2017). Microbiology Laboratory Guidebook. 4.09: Isolation and Identification of Salmonella from Meat, Poultry, Pasteurized Egg, and Siluriformes (Fish) Products and Carcass and Environmental Sponges. Available at: https://www.fsis.usda.gov/wps/wcm/connect/700c05fe-06a2-492a-a6e1-3357f7701f52/mlg-4.pdf?MOD=AJPERES [Google Scholar]
- Wehling P., LaBudde R. A., Brunelle S. L., Nelson M. T. (2011). Probability of detection (POD) as a statistical model for the validation of qualitative methods. J. AOAC Int. 94 335–347. [PubMed] [Google Scholar]
- Yang Q., Chen S., Ge B. (2013). Detecting Salmonella serovars in shell eggs by loop-mediated isothermal amplification. J. Food Prot. 76 1790–1796. 10.4315/0362-028X.JFP-13-140 [DOI] [PubMed] [Google Scholar]
- Yang Q., Domesle K. J., Ge B. (2018). Loop-mediated isothermal amplification for Salmonella detection in food and feed: current applications and future directions. Foodborne Pathog. Dis. 15 309–331. 10.1089/fpd.2018.2445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q., Domesle K. J., Wang F., Ge B. (2016). Rapid detection of Salmonella in food and feed by coupling loop-mediated isothermal amplification with bioluminescent assay in real-time. BMC Microbiol. 16:112. 10.1186/s12866-016-0730-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q., Wang F., Jones K. L., Meng J., Prinyawiwatkul W., Ge B. (2015). Evaluation of loop-mediated isothermal amplification for the rapid, reliable, and robust detection of Salmonella in produce. Food Microbiol. 46 485–493. 10.1016/j.fm.2014.09.011 [DOI] [PubMed] [Google Scholar]
- Yang Q., Wang F., Prinyawiwatkul W., Ge B. (2014). Robustness of Salmonella loop-mediated isothermal amplification assays for food applications. J. Appl. Microbiol. 116 81–88. 10.1111/jam.12340 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All datasets generated for this study are included in the manuscript.