Abstract
A network of pattern recognition receptors (PRRs) is responsible for the detection of invading viruses and acts as the trigger for the host antiviral response. Central to this apparatus is stimulator of interferon genes (STING), which functions as a node and integrator of detection signals. Owing to its role in both intrinsic and adaptive immunity, STING has become a focus for researchers in the field of oncolytic virotherapy. In this review, we consider the function of the cGAS-STING axis and its regulation, both by cellular mechanisms and as a result of viral interference.
Keywords: oncolytic virotherapy, immunotherapy, STING, cGAS, IFI16, HSV, interferon, cancer
Main Text
In order to maintain homeostasis, organisms must monitor for potentially harmful changes in their environment. This is true not only on the macroscopic but also on the microscopic level. To this end, cells have evolved specialized sensors that rapidly identify molecules that differ from those produced by their normal physiology. This network of pattern recognition receptors (PRRs) plays an essential role in the intracellular immune response by initiating interferon and pro-inflammatory signaling upon detection of conserved non-self-molecular patterns. Host PRRs detect a variety of these pathogen-associated molecular patterns (PAMPS), including lipopolysaccharide and peptidoglycan (associated with bacteria), chitin and ergosterol (associated with fungi), and nucleic acid complexes (associated with bacterial and viral genomes as well as viral replication intermediates).1, 2 In this review, we focus upon stimulator of interferon genes (STING), an adaptor protein that plays an indispensable role in coordinating the cellular antiviral response by way of integrating signals from numerous DNA and RNA-sensing PRRs. Owing to its role in stimulating both the intrinsic antiviral response and the cellular adaptive immune response, STING has garnered the interest of researchers working to untangle the cellular mechanisms relevant to radiation therapy,3 oncolytic virotherapy,4, 5, 6 and immunotherapy.7, 8
Overview of RNA and DNA Sensors
Host PRRs are positioned to detect viral-associated nucleic acids from both the extracellular and intracellular environments. Toll-like receptors (TLRs), located along the plasma membrane and within endosomal compartments, monitor the extracellular environment for single- and double-stranded RNA (dsRNA) as well as unmethylated CpG DNA motifs that are released from pathogens.9 While normally protected by capsid proteins, viral nucleic acids can be liberated following endocytosis and lysosomal digestion10 and can ultimately escape from dying cells.11 In contrast to TLRs, intracellular receptors monitor for de novo synthesized nucleic acids produced by intracellular pathogens. The retinoic acid-inducible gene receptors (RIG-I-like receptors), including RIG-I and melanoma differentiation associated protein 5 (MDA5), monitor for cytosolic double- and single-stranded RNAs.12
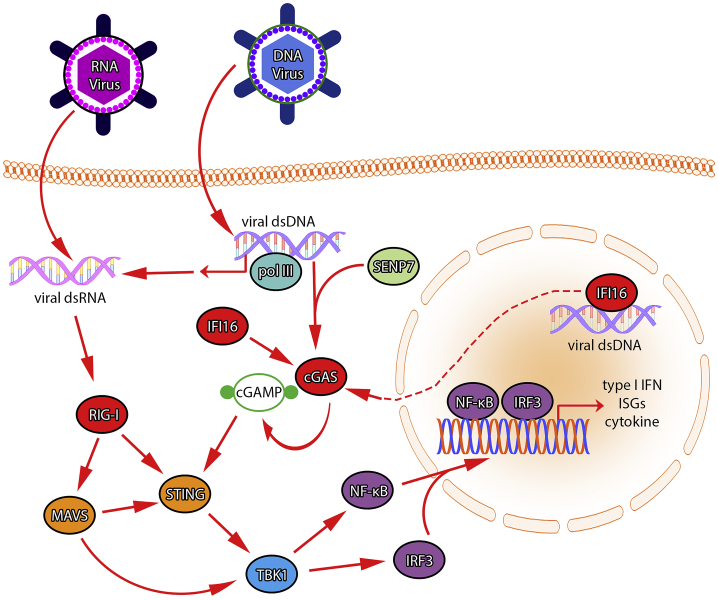
Numerous PRRs also monitor for pathogen- and tumor-derived DNA13 and include absent in melanoma 2 (AIM2),14 DEAD-box helicase 41 (DDX41),15 DNA-dependent activator of interferon regulatory factors (DAI),16 interferon-gamma inducible protein 16 (IFI16),17 and cyclic GMP-AMP synthase (cGAS).18 Upon activation, these DNA sensors stimulate a common signal relay protein encoded by the TMEM173 gene: STING, also known as mediator of IRF3 activation (MITA). STING is an adaptor protein which sits downstream of multiple RNA- and DNA-based PRRs and acts as a signal integrator, relaying tumor- and pathogen-stimulated signals to downstream transcriptional machinery. STING is an ER-resident protein which also localizes to mitochondria-associated ER membranes (MAMs) via its N-terminal transmembrane domain.19 Upon stimulation, STING translocates, via the Golgi, to sec-5-containing perinuclear vesicles, along with TANK-binding kinase 1 (TBK1).20 Upon activation, STING undergoes a conformational change that allows for homodimerization, followed by TBK1 binding and interaction with interferon regulatory factor 3 (IRF3) and nuclear factor kappa-light-chain-enhancer of activated B cells (NF-κB). This leads to phosphorylation of these transcription factors, enabling type I interferon, interferon-stimulated genes (ISGs), and inflammatory cytokines production (Figure 1).19, 21, 22, 23
Figure 1.
The Antiviral Apparatus
Double-stranded DNA from a replicating virus can be bound by cGAS, triggering production of cGAMP—a process that is stimulated by de-SUMOylation of cGAS by SENP7. IFI16 also feeds into this process and participates in cGAS activation and downstream signaling on multiple levels. Signaling through STING begins with cGAMP binding and results in downstream phosphorylation of TBK1 and transcription factors NF-kB and IRF3. NF-kB and IRF3 participate in transcription of type I interferons, interferon-stimulated genes, and inflammatory cytokines. Detection of viral DNA in the nuclear compartment is mediated by IFI16, which triggers signaling on the cGAS/STING axis. RNA polymerase III (pol III) can bind viral DNA and transcribe RNA, thus acting as a DNA pattern recognition receptor. Viral double-stranded RNA can be bound by RIG-I, which in conjunction with MAVS, can activate signaling through STING and TBK1.
HSV-I Infection as a Template for STING Signaling Activation
In this review, we will consider the STING-mediated interferon (IFN) response to viral infection, cellular regulation of this response, and the viral mechanisms that directly counter it. We will also look briefly at related findings which predict the efficacy of oncolytic DNA viruses. Although HSV-I (a double-stranded DNA [dsDNA] α-herpes virus) has been utilized extensively to study the STING-mediated response to DNA virus infection, adenoviruses,24 and poxviruses25 have been shown capable of stimulating the same response. Interestingly, although STING is also required to restrict the replication of a diverse group of RNA viruses, this restriction does not always depend upon downstream IFN signaling, in some cases relying upon a form of STING-mediated translation inhibition.26
Though many sensors were previously identified that detect cytosolic DNA during viral infection, most of these were cell-type specific or redundant. A universal activator of STING had remained elusive27 until the work of a group led by Dr. Zhijian Chen,18 which identified cyclic GMP-AMP (cGAMP) as the signal molecule responsible for STING activation28 and identified the cytosolic DNA sensor responsible for catalyzing production of cGAMP: GMP-AMP synthase (cGAS). The dsDNA-activated signaling molecule, cGAMP, is similar to a dsRNA-activated antiviral signaling molecule, 2′-5′-oligoadenylate, in that both are characterized by uncommon 2′-5′ phosphodiester bonded nucleotides.29 In addition to activating STING in the infected cell, cGAMP can also translocate via gap junctions to neighboring uninfected cells, priming their STING-mediated antiviral response.30
Recent work has suggested that cGAS and STING play a critical role in HSV-I resistance. In microglia, the HSV-I-induced type-I IFN response is dependent upon cGAS-STING signaling. Mice with defective cGAS or STING functions were found to be more susceptible to herpes simplex encephalitis (HSE).31 This work echoes previous findings that show that STING knockout increases HSV disease in mice and is required for effective type-I IFN production in MEFs, dendritic cells, and macrophages when infected with HSV-I, or when transfected with cytosolic DNA.20 While these works provided evidence to support that the cGAS-STING axis is instrumental in the cellular response to HSV-I infection, they were not able to explain how a DNA virus that replicates in the nucleus is able to activate a cytosolic DNA sensor (such as cGAS).
An answer to this question began to emerge when PYHIN (pyrin and HIN200-domain containing proteins) family member IFI16 was first described as a DNA-sensing protein that can stimulate the IFN response in a STING-dependent manner.17 While previous work had identified that IFI16 can bind dsDNA via its two C-terminal HIN domains,32 Unterholzner et al.17 found that IFI16 could also recruit STING and that its activity was required for efficient IFN-β induction by dsDNA in monocytic leukemia (THP-1) cells. Later, Li et al.33 showed that acetylation of IFI16’s nuclear localization signal (NLS) regulates its cellular distribution and ability to detect HSV-1 DNA in the nucleus. Taken together, these studies suggested a model whereby IFI16 detects viral DNA in the nucleus, followed by re-localization to the cytosol, where it could then activate IFN signaling through the STING-TBK1 axis. However, this left questions regarding the role of cGAS-STING signaling.
While the DNA-sensing pathways described thus far utilize TBK1-mediated IRF3 phosphorylation for IFN-β upregulation, more recent work by Diner suggests that a TBK1-independent mechanism may also stimulate IFN-β production.34 In HFF (human foreskin fibroblast) cells, CRISPR knockout of cGAS or STING abrogated TBK1 activation, while a similar IFI16 knockout did not. However, given that IFI16 knockout was shown to strongly inhibit induction of IFN-β, this suggested a model wherein IFN-β may be induced by a canonical pathway involving cGAS-STING-TBK1 in the cytoplasm or by a non-canonical pathway involving IFI16 detection of viral DNA in the nucleus. For the canonical pathway, instead of placing IFI16 in the cytosolic role of STING activation, Diner suggested that IFI16 might operate in a purely nuclear role, as a downstream effector of IRF3.
The seemingly variable behavior of IFI16 during DNA virus infection is consistent with its role as a functionally diverse gene product whose localization is cell-type specific and thus influenced by a range of conditions, including gene polymorphisms, post-translational modifications, hormonal regulation, and protein-protein interactions.35, 36, 37, 38, 39, 40 Similarly, cGAS function and cellular localization may also vary. In HFF and normal oral keratinocyte (NOK) cells, cGAS was observed to localize both to the cytosol and nuclear compartment.41 It was also found that cGAS stabilizes IFI16 levels and that both IFI16 and cGAS are required for robust IFN-β and ISG induction during HSV-I infection of HFFs.
Recent works have therefore sought to elucidate the combined role of IFI16 and cGAS during HSV-I infection. In one such work, it was found that cGAS, STING, and IFI16 are each necessary for type-I IFN production in HSV-I infected THP-1 cells42 and that IFI16 integrates into the cGAS-STING signaling axis at multiple levels. IFI16 enhances cGAS production of cGAMP, is required for cGAMP to stimulate STING dimerization and activation, and works to recruit TBK1 to STING, enabling IRF3 activation. Concurrent work in immortalized human keratinocytes (HaCaT cells) echoes that IFI16 is required for the IFN response to HSV-I infection, as well as for the DNA-induced activation of STING.43 In agreement with other recent findings, it was shown that DNA sensing in HaCaT keratinocytes also requires cGAS, that IFI16 interacts with cGAS in a DNA-dependent manner, and that IFI16 enhances phosphorylation of STING, leading to activation of TBK1 and IRF3.
Completing the picture of the cGAS-STING response to viral infection, Cui et al.44 show that HSV-I infection strengthens the association of cGAS with SUMO-specific protease 7 (SENP7) and that this leads to deSUMOylation of cGAS and its subsequent activation. SENP7 inhibits cGAS SUMOylation, thus improving its ability to bind DNA. This may be a way for cells to regulate cGAS-STING signaling during HSV-I infection, as the authors show that the SENP7-cGAS association is high at the beginning of viral infection, and tapers off later. Using SENP7 knockdown mice, the authors also show that disruption of the SENP7-cGAS interaction renders mice more susceptible to HSV-I infection. Given IFI16’s multi-layered relationship with cGAS and its essential role in the response to HSV, it may be that IFI16 is also involved in this step of cGAS activation—such questions remain for future studies.
The Role of RNA Sensors in STING Signaling
While much of the literature focuses on the role of STING in response to DNA sensors, STING also has direct interactions with RIG-I (a cytosolic RNA sensor) as well as its downstream adaptor molecule, mitochondrial antiviral-signaling protein (MAVS); interactions which drive IFN-β expression.19, 21 HSV-I produces dsRNA during infection as a result of transcription from the viral genome by RNA pol II.45 These dsRNAs are recognized by protein kinase R (PKR) and trigger global translational arrest.46 Similarly, RIG-I is activated by dsRNA that is transcribed by RNA pol III47 and potentially by direct interactions with dsDNA.48 Resultant IFN-β production is mediated at least in part by MAVS,49 while the extent of STING involvement on this pathway remains to be determined. Studies that have evaluated the role of RIG-I in response to cytosolic dsDNA have found poly (dA:dT) to be a target of RNA pol III transcription and the most potent stimulator of RIG-I.47, 49 DNA containing this motif initiates signaling through both MAVS and STING; double-knockdown is necessary to abrogate poly (dA:dT)-stimulated IFN-β production in macrophages and dendritic cells (DCs).50 Finally, regardless of the downstream signaling mechanism, RIG-I may ultimately depend upon STING to restrict HSV-I. As Liu et al.51 have shown, RIG-I-mediated restriction of HSV-I depends upon its ability to upregulate the expression of STING.
Beyond merely integrating RNA sensors for the detection of DNA viruses, STING is also instrumental in the cellular response to RNA viruses. In N2A cells, Japanese encephalitis virus (JEV) genomic RNA was detected by RIG-I in concert with MAVS, which subsequently interacted with STING.52 STING knockdown decreased production of IFN-β and ISGs and increased viral loads. In contrast, a recent study by Franz et al.26 found that while STING is required to restrict the replication of vesicular stomatitis virus (VSV), Sindbis virus (SINV), Sendai virus (SeV), influenza A, and reovirus, STING is not responsible for the IFN response to these viruses in MEFs. Instead, it was found that RIG-I and MDA5 restrict RNA virus replication by triggering a MAVS-mediated IFN response that is complemented by a STING-mediated global shutdown of translation.
Cellular Regulation of STING Signaling during Viral Infection
Signaling through the STING pathway induces genes that mediate the immune response to viruses and other pathogens. While necessary to maintenance of homeostasis, prolonged activation of this pathway would lead to lethal pro-inflammatory conditions. Accordingly, host mechanisms exist to prevent this from happening.53
Early studies by Glen Barber and others identified that STING undergoes a shift in molecular weight following its activation, an observation that focused on regulatory post-translational modifications of STING. Investigators identified that STING phosphorylation (at S366) by ULK1 immediately inhibits its activation of IRF3 and ultimately contributes to the lysosomal degradation of STING.54 ULK1 phosphorylation of STING is triggered by cGAS production of cGAMP. Thus, when cGAS activates STING, it also stimulates pathways that downregulate it through a negative feedback mechanism. UKL1 knockdown inhibits this dsDNA-induced degradation of STING and leads to a sustained STING-mediated response in hTERT-BJ1 cells.
Ubiquitin-regulated degradation of STING following viral infection is also documented. One group identified that E3 ligase TRIM30α causes the K48-linked ubiquitination of STING (at Lys 275) and subsequent proteasomal degradation in response to viral infection.55 Specifically, it was shown that in mouse dendritic (D2SC) cells, knockdown of TRIM30α increased viral and cytosolic DNA-induced IFN-β and ISG expression. In these studies, the investigators identified that HSV-I infection increased TRIM30α expression and that overexpression of this protein improved viral replication. Conversely, TRIM30α knockdown decreased HSV replication in mouse fibroblast cells and TRIM30α knockout mice were less susceptible to HSV infection. Other studies have reported alternative ubiquitination sites with similar function. During SeV infection, the E3 ligase RNF5 is upregulated and causes the K48-linked ubiquitination of STING at Lys 150, also leading to proteasomal degradation.56
Separately, it has been shown that USP18 and USP20 cooperatively direct the K48-linked de-ubiquitination of STING, promoting its stability.57 This study showed that USP18 or USP20 deficiency significantly inhibited HSV-I or cytosolic DNA activation of both NF-κB and IRF3, and type-I IFN and proinflammatory cytokine expression.
Although STING’s post-translational modifications have been well-described, they are not the only cellular mechanisms which regulate it. As shown by Chen et al.,58 the expression of a STING alternative-splicing isoform known as MITA-related protein (MRP) can alter STING function. Due to a missing exon and an associated frameshift mutation, MRP has a non-functional C-terminal tail region—the same region that allows wild-type STING to function as a scaffold for the interaction of TBK1 and IRF3. However, because MRP retains a functional dimerization domain, it is still able to interact with wild-type STING. Accordingly, in 293T cells, it was found that co-expression of MRP with wild-type STING limited the STING-associated activation of an IRF3-driven reporter, while still allowing activation of an NF-κB reporter. Data regarding MRP function during viral infection is less clear-cut. When 293T cells were infected with SeV, expression of MRP restricted the activity of an IFN-β reporter. However, during HSV-I infection of colorectal cancer (HCT116) cells, MRP expression enhanced activity of IRF3 and IFN-β reporters. Future work will therefore be necessary to identify the mechanisms which control MRP modulation of STING signaling.
Viral Regulation of STING Signaling
While STING can be degraded as a result of cellular safety mechanisms which prevent a persistent immune response, viruses such as HSV-I possess mechanisms which interfere with the cellular IFN response on multiple levels of the STING axis. Chief among these viral products is encoded by the γ134.5 gene, long of interest to researchers in the field of oncolytic HSVs due to its crucial role in neurovirulence.59 Among its other immunosuppressive functions, the γ134.5 gene product (ICP34.5) directly binds STING, preventing its translocation from the endoplasmic reticulum (ER) to the Golgi apparatus, thereby blocking interaction with downstream effectors, such as TBK1. Other HSV proteins also perturb STING signaling. Downstream of TBK1 activation, ICP27 interferes with the subsequent phosphorylation of IRF3.60 UL46, one of the most abundant HSV-I tegument proteins, was only recently described to have a clear function. In addition to suppression of IFI16 and STING expression in HEL cells, it was found that the UL46 protein product can directly bind to both STING and TBK1, increasing viral titers.61 Another study identified that the VHS protein (UL41) allows viral evasion of cGAS-STING signaling by selectively degrading cGAS mRNA, via its RNase activity.62 The authors of this study showed that a UL41 mutant (R2621) was replication compromised when compared to wild-type HSV-I, but that stable knockdown of cGAS was able to rescue replication. At the other end of the signaling axis, a separate study found that the HSV-I ubiquitin-specific protease (UL36) inhibits activation of NF-κB.63 Specifically, the authors found that ectopic expression of UL36 inhibited the activation of IFN-β and NF-kB promoters by overexpression of STING, TBK1, and IKKα+β (direct activators of NF-κB), but not with overexpression of NF-κB subunit p65. UL36 was found to deubiquitinate the NF-κB inhibitor, IkBα, restricting its degradation; consequently, this prevented NF-κB activation. A separate NF-kB -related study found that HSV-I UL24 blocks the translocation of NF-κB to the nucleus by interfering directly with p65 and p50,64 and this was shown to impair production of IFN-β and inflammatory cytokines.
RNA viruses also have mechanisms that target the cellular IFN response—often by disrupting RIG-I-MAVS-STING signaling. Hepatitis C virus (HCV) protein NS4B co-localizes with STING in the endoplasmic reticulum (ER) and binds STING through a structurally homologous domain. This binding prevents MAVS-STING interactions, ultimately inhibiting IFN-β production.65 The influenza A virus (IAV) hemagglutinin fusion peptide (FP) binds to STING in a highly conserved region that functions in cGAMP binding and STING dimerization. In THP-1 cells, the interaction of FP with STING inhibited a mechanism that specifically produces IFN in response to lipid membrane fusion.66 Dengue virus also produces a molecule targeting the cGAS-STING axis. Called NS2B3, this protein targets cGAS for degradation through an autophagy-dependent mechanism in addition to targeting STING for direct cleavage.67, 68 Interestingly, it was found that this virus stimulates the STING pathway through the release of mitochondrial DNA during the course of infection.
STING, Oncolytic Viruses, and Immunotherapy
The first generation of oncolytic herpes simplex viruses (oHSVs) has relied upon deletion of the γ134.5 neurovirulence gene for oncolytic selectivity. Because the gene product (ICP34.5) has direct effects upon STING signaling, researchers hypothesized that tumors with complimentary signaling defects might prove amenable to treatment with γ134.5-deleted oHSVs. Indeed, because of its role in the DNA damage response, the STING axis may often be silenced during malignant transformation, allowing cancers to escape immune surveillance.69 Thus far, carcinomas have emerged as a type of cancer in which this appears to be the case, with defective STING signaling acting as a predictor of oHSV viral productivity. Studies in ovarian cancer,4 colorectal carcinoma,6 and melanoma5 have shown decreased type-I IFN and inflammatory cytokine production in response to dsDNA stimulation and viral infection. The muted immune response in these cell types has allowed for increased oHSV titer, cell-to-cell spread, and overall oncolytic effect. In these studies, defective STING signaling was most often associated with epigenetic silencing of cGAS and/or STING, with concomitant loss of the IFN response. In vivo work has recapitulated these results, with xenografts in nude mice showing a greater reduction in volume for oHSV-treated STING-defective tumors versus tumors with normal STING function.
But while the aforementioned studies demonstrate the importance of STING signaling to oncolytic viral productivity and direct cell lysis, the larger role of this pathway in the immune surveillance machinery should also be considered. Cytosolic DNA within cancer cells can activate intrinsic STING signaling and thus, the production of inflammatory cytokines that attract phagocytic and T-cell-mediated responses.70 Meanwhile, extrinsic STING signaling within dendritic cells drives the production of type-I IFN in response to tumor-derived adjuvants, facilitating cross-priming of tumor-infiltrating CD8+ T cells.13 This paradigm places the STING axis in a pivotal role within the anti-tumor immune apparatus. In light of this, researchers have begun to target immunologically indolent tumors with STING agonists in hopes of stimulating the immune microenvironment. Using intratumoral (IT) injection of synthetic cyclic dinucleotides (CDNs), it was shown that established B16 melanoma tumors in mice were subject to profound regression, with rejection of distant metastases and lasting CD8+ T cell-mediated immunity.7 In a separate work, transfection of B16 cells with modified dsDNA species called STAVs (STING-dependent adjuvants) was used to demonstrate an effective cell-based therapy that also depended upon antigen-presenting cells (APCs) and the stimulation of a CD8+ T cell response.8 Importantly, it was found that tumor-derived DNA is likely degraded before the STING-dependent IFN response is triggered within APCs but that engulfed species such as CDNs are able to persist and act as STING adjuvants. Furthermore, it was found that within engulfing APCs, STING function (but not cGAS function) was required for activation, suggesting that CDNs (such as cGAMP) from engulfed tumor cells are chiefly responsible for activation of APCs. This finding is echoed in other work, which shows that tumor-derived cGAMP (and not tumor-derived DNA) is responsible for stimulating the natural killer (NK) cell response that mediates the rejection of certain NK-sensitive tumors.71
Work with STAVs showed that infection of cancer cells with oHSVs can also generate an APC and CD8+ T cell-mediated response.8 This leads to the question of STING’s role in the immune-mediated effects of oncolytic virotherapy. Though STING defects can increase a cancer cell’s sensitivity to viral infection, it is reasonable to ask if the same defects might also limit the ability of infected cells to stimulate an anti-tumor immune response. This question remains open to future research; however, studies involving STAVs and NK-sensitive tumors suggest that defects in tumor cGAS function—but not STING function, per se—would make engulfed tumor cells less immunostimulatory.8, 71
Conclusions
As the central adaptor protein of the cellular PRR network, STING is responsible for integrating and relaying the immune signals that are generated by a diversity of pathogens, including RNA and DNA viruses. When an invader possesses genomic DNA, cGAS and IFI16 cooperate to monitor the cytosolic and nuclear compartments for the first signs of viral replication, ultimately relying upon STING to mediate the appropriate response. In the case of RNA viruses, RIG-I performs the analogous job of nucleic acid detection, relaying signals to STING and MAVS as a cooperative pair.
This antiviral detection and response apparatus is dynamic, adapting to cellular conditions and to the specific functions accorded by cell type. In their mouse study, Reinert et al.31 found that that while STING function is redundant in the antiviral response of astrocytes and neurons in vitro, HSV-I replication was nevertheless increased in these cell types for STING-deficient mice in vivo. The study concluded that STING-mediated production of type-I IFN by microglia was required by astrocytes and neurons to stimulate an effective antiviral response. This theme of specialization is echoed in the variable localization and function of IFI16, in the potential involvement of RIG-I (an RNA sensor) in detecting DNA viruses and perhaps in the varied role of STING signaling in response to RNA viral infection.
So fundamental is this apparatus to cellular immunity that viral pathogens have evolved numerous mechanisms to rapidly silence it, while cells must work to prevent its sustained and deleterious activation. Even so, STING signaling often becomes defective during malignant transformation, allowing cancers to escape immune surveillance.4, 5, 6 Such common defects in immune signaling may prove to be the Achilles’ heel in many treatment-refractory cancers, making them amenable to oncolytic virotherapy while also shedding light upon the way to new immune-mediated therapies.
Acknowledgments
This work was supported by Alex's Lemonade Stand Foundation (ALSF) (K.A.C.), Hyundai Hope On Wheels (K.A.C.), Cancer Free Kids (M.G.G.), NIH CA222903 (K.A.C.), and UO1CA232488-01 (R.W. and K.A.C.).
References
- 1.Yu X., Feng B., He P., Shan L. From Chaos to Harmony: Responses and Signaling upon Microbial Pattern Recognition. Annu. Rev. Phytopathol. 2017;55:109–137. doi: 10.1146/annurev-phyto-080516-035649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gürtler C., Bowie A.G. Innate immune detection of microbial nucleic acids. Trends Microbiol. 2013;21:413–420. doi: 10.1016/j.tim.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baird J.R., Friedman D., Cottam B., Dubensky T.W., Jr., Kanne D.B., Bambina S., Bahjat K., Crittenden M.R., Gough M.J. Radiotherapy Combined with Novel STING-Targeting Oligonucleotides Results in Regression of Established Tumors. Cancer Res. 2016;76:50–61. doi: 10.1158/0008-5472.CAN-14-3619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.de Queiroz N.M.G.P., Xia T., Konno H., Barber G.N. Ovarian Cancer Cells Commonly Exhibit Defective STING Signaling Which Affects Sensitivity to Viral Oncolysis. Mol. Cancer Res. 2018 doi: 10.1158/1541-7786.MCR-18-0504. Published online December 26, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xia T., Konno H., Barber G.N. Recurrent Loss of STING Signaling in Melanoma Correlates with Susceptibility to Viral Oncolysis. Cancer Res. 2016;76:6747–6759. doi: 10.1158/0008-5472.CAN-16-1404. [DOI] [PubMed] [Google Scholar]
- 6.Xia T., Konno H., Ahn J., Barber G.N. Deregulation of STING Signaling in Colorectal Carcinoma Constrains DNA Damage Responses and Correlates With Tumorigenesis. Cell Rep. 2016;14:282–297. doi: 10.1016/j.celrep.2015.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Corrales L., Glickman L.H., McWhirter S.M., Kanne D.B., Sivick K.E., Katibah G.E., Woo S.R., Lemmens E., Banda T., Leong J.J. Direct Activation of STING in the Tumor Microenvironment Leads to Potent and Systemic Tumor Regression and Immunity. Cell Rep. 2015;11:1018–1030. doi: 10.1016/j.celrep.2015.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ahn J., Xia T., Rabasa Capote A., Betancourt D., Barber G.N. Extrinsic Phagocyte-Dependent STING Signaling Dictates the Immunogenicity of Dying Cells. Cancer Cell. 2018;33:862–873.e5. doi: 10.1016/j.ccell.2018.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kawasaki T., Kawai T. Toll-like receptor signaling pathways. Front. Immunol. 2014;5:461. doi: 10.3389/fimmu.2014.00461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Blaas D. Viral entry pathways: the example of common cold viruses. Wien. Med. Wochenschr. 2016;166:211–226. doi: 10.1007/s10354-016-0461-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Majde J.A., Guha-Thakurta N., Chen Z., Bredow S., Krueger J.M. Spontaneous release of stable viral double-stranded RNA into the extracellular medium by influenza virus-infected MDCK epithelial cells: implications for the viral acute phase response. Arch. Virol. 1998;143:2371–2380. doi: 10.1007/s007050050467. [DOI] [PubMed] [Google Scholar]
- 12.Loo Y.M., Gale M., Jr. Immune signaling by RIG-I-like receptors. Immunity. 2011;34:680–692. doi: 10.1016/j.immuni.2011.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Woo S.R., Fuertes M.B., Corrales L., Spranger S., Furdyna M.J., Leung M.Y., Duggan R., Wang Y., Barber G.N., Fitzgerald K.A. STING-dependent cytosolic DNA sensing mediates innate immune recognition of immunogenic tumors. Immunity. 2014;41:830–842. doi: 10.1016/j.immuni.2014.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bürckstümmer T., Baumann C., Blüml S., Dixit E., Dürnberger G., Jahn H., Planyavsky M., Bilban M., Colinge J., Bennett K.L., Superti-Furga G. An orthogonal proteomic-genomic screen identifies AIM2 as a cytoplasmic DNA sensor for the inflammasome. Nat. Immunol. 2009;10:266–272. doi: 10.1038/ni.1702. [DOI] [PubMed] [Google Scholar]
- 15.Zhang Z., Yuan B., Bao M., Lu N., Kim T., Liu Y.J. The helicase DDX41 senses intracellular DNA mediated by the adaptor STING in dendritic cells. Nat. Immunol. 2011;12:959–965. doi: 10.1038/ni.2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Takaoka A., Wang Z., Choi M.K., Yanai H., Negishi H., Ban T., Lu Y., Miyagishi M., Kodama T., Honda K. DAI (DLM-1/ZBP1) is a cytosolic DNA sensor and an activator of innate immune response. Nature. 2007;448:501–505. doi: 10.1038/nature06013. [DOI] [PubMed] [Google Scholar]
- 17.Unterholzner L., Keating S.E., Baran M., Horan K.A., Jensen S.B., Sharma S., Sirois C.M., Jin T., Latz E., Xiao T.S. IFI16 is an innate immune sensor for intracellular DNA. Nat. Immunol. 2010;11:997–1004. doi: 10.1038/ni.1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sun L., Wu J., Du F., Chen X., Chen Z.J.J. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science. 2013;339:786–791. doi: 10.1126/science.1232458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ishikawa H., Barber G.N. STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature. 2008;455:674–678. doi: 10.1038/nature07317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ishikawa H., Ma Z., Barber G.N. STING regulates intracellular DNA-mediated, type I interferon-dependent innate immunity. Nature. 2009;461:788–792. doi: 10.1038/nature08476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhong B., Yang Y., Li S., Wang Y.Y., Li Y., Diao F., Lei C., He X., Zhang L., Tien P., Shu H.B. The adaptor protein MITA links virus-sensing receptors to IRF3 transcription factor activation. Immunity. 2008;29:538–550. doi: 10.1016/j.immuni.2008.09.003. [DOI] [PubMed] [Google Scholar]
- 22.Tanaka Y., Chen Z.J. STING specifies IRF3 phosphorylation by TBK1 in the cytosolic DNA signaling pathway. Sci. Signal. 2012;5:ra20. doi: 10.1126/scisignal.2002521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Abe T., Barber G.N. Cytosolic-DNA-mediated, STING-dependent proinflammatory gene induction necessitates canonical NF-κB activation through TBK1. J. Virol. 2014;88:5328–5341. doi: 10.1128/JVI.00037-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lam E., Falck-Pedersen E. Unabated adenovirus replication following activation of the cGAS/STING-dependent antiviral response in human cells. J. Virol. 2014;88:14426–14439. doi: 10.1128/JVI.02608-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Georgana I., Sumner R.P., Towers G.J., Maluquer de Motes C. Virulent poxviruses inhibit DNA sensing by preventing STING activation. J. Virol. 2018;92 doi: 10.1128/JVI.02145-17. e02145-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Franz K.M., Neidermyer W.J., Tan Y.J., Whelan S.P.J., Kagan J.C. STING-dependent translation inhibition restricts RNA virus replication. Proc. Natl. Acad. Sci. USA. 2018;115:E2058–E2067. doi: 10.1073/pnas.1716937115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu J., Chen Z.J. Innate immune sensing and signaling of cytosolic nucleic acids. Annu. Rev. Immunol. 2014;32:461–488. doi: 10.1146/annurev-immunol-032713-120156. [DOI] [PubMed] [Google Scholar]
- 28.Wu J., Sun L., Chen X., Du F., Shi H., Chen C., Chen Z.J. Cyclic GMP-AMP is an endogenous second messenger in innate immune signaling by cytosolic DNA. Science. 2013;339:826–830. doi: 10.1126/science.1229963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ablasser A., Goldeck M., Cavlar T., Deimling T., Witte G., Röhl I., Hopfner K.P., Ludwig J., Hornung V. cGAS produces a 2′-5′-linked cyclic dinucleotide second messenger that activates STING. Nature. 2013;498:380–384. doi: 10.1038/nature12306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ablasser A., Schmid-Burgk J.L., Hemmerling I., Horvath G.L., Schmidt T., Latz E., Hornung V. Cell intrinsic immunity spreads to bystander cells via the intercellular transfer of cGAMP. Nature. 2013;503:530–534. doi: 10.1038/nature12640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Reinert L.S., Lopušná K., Winther H., Sun C., Thomsen M.K., Nandakumar R., Mogensen T.H., Meyer M., Vægter C., Nyengaard J.R. Sensing of HSV-1 by the cGAS-STING pathway in microglia orchestrates antiviral defence in the CNS. Nat. Commun. 2016;7:13348. doi: 10.1038/ncomms13348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yan H., Dalal K., Hon B.K., Youkharibache P., Lau D., Pio F. RPA nucleic acid-binding properties of IFI16-HIN200. Biochim. Biophys. Acta. 2008;1784:1087–1097. doi: 10.1016/j.bbapap.2008.04.004. [DOI] [PubMed] [Google Scholar]
- 33.Li T., Diner B.A., Chen J., Cristea I.M. Acetylation modulates cellular distribution and DNA sensing ability of interferon-inducible protein IFI16. Proc. Natl. Acad. Sci. USA. 2012;109:10558–10563. doi: 10.1073/pnas.1203447109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Diner B.A., Lum K.K., Toettcher J.E., Cristea I.M. Viral DNA Sensors IFI16 and Cyclic GMP-AMP Synthase Possess Distinct Functions in Regulating Viral Gene Expression, Immune Defenses, and Apoptotic Responses during Herpesvirus Infection. MBio. 2016;7:e01553-16. doi: 10.1128/mBio.01553-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Veeranki S., Choubey D. Interferon-inducible p200-family protein IFI16, an innate immune sensor for cytosolic and nuclear double-stranded DNA: regulation of subcellular localization. Mol. Immunol. 2012;49:567–571. doi: 10.1016/j.molimm.2011.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dawson M.J., Trapani J.A. The interferon-inducible autoantigen, IFI 16: localization to the nucleolus and identification of a DNA-binding domain. Biochem. Biophys. Res. Commun. 1995;214:152–162. doi: 10.1006/bbrc.1995.2269. [DOI] [PubMed] [Google Scholar]
- 37.Briggs L.J., Johnstone R.W., Elliot R.M., Xiao C.Y., Dawson M., Trapani J.A., Jans D.A. Novel properties of the protein kinase CK2-site-regulated nuclear- localization sequence of the interferon-induced nuclear factor IFI 16. Biochem. J. 2001;353:69–77. [PMC free article] [PubMed] [Google Scholar]
- 38.Xin H., Curry J., Johnstone R.W., Nickoloff B.J., Choubey D. Role of IFI 16, a member of the interferon-inducible p200-protein family, in prostate epithelial cellular senescence. Oncogene. 2003;22:4831–4840. doi: 10.1038/sj.onc.1206754. [DOI] [PubMed] [Google Scholar]
- 39.Hornung V., Ablasser A., Charrel-Dennis M., Bauernfeind F., Horvath G., Caffrey D.R., Latz E., Fitzgerald K.A. AIM2 recognizes cytosolic dsDNA and forms a caspase-1-activating inflammasome with ASC. Nature. 2009;458:514–518. doi: 10.1038/nature07725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Veeranki S., Duan X., Panchanathan R., Liu H., Choubey D. IFI16 protein mediates the anti-inflammatory actions of the type-I interferons through suppression of activation of caspase-1 by inflammasomes. PLoS ONE. 2011;6:e27040. doi: 10.1371/journal.pone.0027040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Orzalli M.H., Broekema N.M., Diner B.A., Hancks D.C., Elde N.C., Cristea I.M., Knipe D.M. cGAS-mediated stabilization of IFI16 promotes innate signaling during herpes simplex virus infection. Proc. Natl. Acad. Sci. USA. 2015;112:E1773–E1781. doi: 10.1073/pnas.1424637112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jønsson K.L., Laustsen A., Krapp C., Skipper K.A., Thavachelvam K., Hotter D., Egedal J.H., Kjolby M., Mohammadi P., Prabakaran T. IFI16 is required for DNA sensing in human macrophages by promoting production and function of cGAMP. Nat. Commun. 2017;8:14391. doi: 10.1038/ncomms14391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Almine J.F., O’Hare C.A., Dunphy G., Haga I.R., Naik R.J., Atrih A., Connolly D.J., Taylor J., Kelsall I.R., Bowie A.G. IFI16 and cGAS cooperate in the activation of STING during DNA sensing in human keratinocytes. Nat. Commun. 2017;8:14392. doi: 10.1038/ncomms14392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cui Y., Yu H., Zheng X., Peng R., Wang Q., Zhou Y., Wang R., Wang J., Qu B., Shen N. SENP7 Potentiates cGAS Activation by Relieving SUMO-Mediated Inhibition of Cytosolic DNA Sensing. PLoS Pathog. 2017;13:e1006156. doi: 10.1371/journal.ppat.1006156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Alwine J.C., Steinhart W.L., Hill C.W. Transcription of herpes simplex type 1 DNA in nuclei isolated from infected HEp-2 and KB cells. Virology. 1974;60:302–307. doi: 10.1016/0042-6822(74)90390-0. [DOI] [PubMed] [Google Scholar]
- 46.Lemaire P.A., Anderson E., Lary J., Cole J.L. Mechanism of PKR Activation by dsRNA. J. Mol. Biol. 2008;381:351–360. doi: 10.1016/j.jmb.2008.05.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chiu Y.H., Macmillan J.B., Chen Z.J. RNA polymerase III detects cytosolic DNA and induces type I interferons through the RIG-I pathway. Cell. 2009;138:576–591. doi: 10.1016/j.cell.2009.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Choi M.K., Wang Z., Ban T., Yanai H., Lu Y., Koshiba R., Nakaima Y., Hangai S., Savitsky D., Nakasato M. A selective contribution of the RIG-I-like receptor pathway to type I interferon responses activated by cytosolic DNA. Proc. Natl. Acad. Sci. USA. 2009;106:17870–17875. doi: 10.1073/pnas.0909545106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cheng G., Zhong J., Chung J., Chisari F.V. Double-stranded DNA and double-stranded RNA induce a common antiviral signaling pathway in human cells. Proc. Natl. Acad. Sci. USA. 2007;104:9035–9040. doi: 10.1073/pnas.0703285104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Brunette R.L., Young J.M., Whitley D.G., Brodsky I.E., Malik H.S., Stetson D.B. Extensive evolutionary and functional diversity among mammalian AIM2-like receptors. J. Exp. Med. 2012;209:1969–1983. doi: 10.1084/jem.20121960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liu Y., Goulet M.L., Sze A., Hadj S.B., Belgnaoui S.M., Lababidi R.R., Zheng C., Fritz J.H., Olagnier D., Lin R. RIG-I-Mediated STING Upregulation Restricts Herpes Simplex Virus 1 Infection. J. Virol. 2016;90:9406–9419. doi: 10.1128/JVI.00748-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nazmi A., Mukhopadhyay R., Dutta K., Basu A. STING mediates neuronal innate immune response following Japanese encephalitis virus infection. Sci. Rep. 2012;2:347. doi: 10.1038/srep00347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ahn J., Gutman D., Saijo S., Barber G.N. STING manifests self DNA-dependent inflammatory disease. Proc. Natl. Acad. Sci. USA. 2012;109:19386–19391. doi: 10.1073/pnas.1215006109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Konno H., Konno K., Barber G.N. Cyclic dinucleotides trigger ULK1 (ATG1) phosphorylation of STING to prevent sustained innate immune signaling. Cell. 2013;155:688–698. doi: 10.1016/j.cell.2013.09.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang Y., Lian Q., Yang B., Yan S., Zhou H., He L., Lin G., Lian Z., Jiang Z., Sun B. TRIM30α Is a Negative-Feedback Regulator of the Intracellular DNA and DNA Virus-Triggered Response by Targeting STING. PLoS Pathog. 2015;11:e1005012. doi: 10.1371/journal.ppat.1005012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhong B., Zhang L., Lei C., Li Y., Mao A.P., Yang Y., Wang Y.Y., Zhang X.L., Shu H.B. The ubiquitin ligase RNF5 regulates antiviral responses by mediating degradation of the adaptor protein MITA. Immunity. 2009;30:397–407. doi: 10.1016/j.immuni.2009.01.008. [DOI] [PubMed] [Google Scholar]
- 57.Zhang M., Zhang M.X., Zhang Q., Zhu G.F., Yuan L., Zhang D.E., Zhu Q., Yao J., Shu H.B., Zhong B. USP18 recruits USP20 to promote innate antiviral response through deubiquitinating STING/MITA. Cell Res. 2016;26:1302–1319. doi: 10.1038/cr.2016.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chen H., Pei R., Zhu W., Zeng R., Wang Y., Wang Y., Lu M., Chen X. An alternative splicing isoform of MITA antagonizes MITA-mediated induction of type I IFNs. J. Immunol. 2014;192:1162–1170. doi: 10.4049/jimmunol.1300798. [DOI] [PubMed] [Google Scholar]
- 59.Martuza R.L., Malick A., Markert J.M., Ruffner K.L., Coen D.M. Experimental therapy of human glioma by means of a genetically engineered virus mutant. Science. 1991;252:854–856. doi: 10.1126/science.1851332. [DOI] [PubMed] [Google Scholar]
- 60.Christensen M.H., Jensen S.B., Miettinen J.J., Luecke S., Prabakaran T., Reinert L.S., Mettenleiter T., Chen Z.J., Knipe D.M., Sandri-Goldin R.M. HSV-1 ICP27 targets the TBK1-activated STING signalsome to inhibit virus-induced type I IFN expression. EMBO J. 2016;35:1385–1399. doi: 10.15252/embj.201593458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Deschamps T., Kalamvoki M. Evasion of the STING DNA-Sensing Pathway by VP11/12 of Herpes Simplex Virus 1. J. Virol. 2017;91:e00535-17. doi: 10.1128/JVI.00535-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Su C., Zheng C. Herpes Simplex Virus 1 Abrogates the cGAS/STING-Mediated Cytosolic DNA-Sensing Pathway via Its Virion Host Shutoff Protein, UL41. J. Virol. 2017;91:e02414-16. doi: 10.1128/JVI.02414-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ye R., Su C., Xu H., Zheng C. Herpes Simplex Virus 1 Ubiquitin-Specific Protease UL36 Abrogates NF-κB Activation in DNA Sensing Signal Pathway. J. Virol. 2017;91:e02417-16. doi: 10.1128/JVI.02417-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Xu H., Su C., Pearson A., Mody C.H., Zheng C. Herpes Simplex Virus 1 UL24 Abrogates the DNA Sensing Signal Pathway by Inhibiting NF-κB Activation. J. Virol. 2017;91:e00025-17. doi: 10.1128/JVI.00025-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Nitta S., Sakamoto N., Nakagawa M., Kakinuma S., Mishima K., Kusano-Kitazume A., Kiyohashi K., Murakawa M., Nishimura-Sakurai Y., Azuma S. Hepatitis C virus NS4B protein targets STING and abrogates RIG-I-mediated type I interferon-dependent innate immunity. Hepatology. 2013;57:46–58. doi: 10.1002/hep.26017. [DOI] [PubMed] [Google Scholar]
- 66.Holm C.K., Rahbek S.H., Gad H.H., Bak R.O., Jakobsen M.R., Jiang Z., Hansen A.L., Jensen S.K., Sun C., Thomsen M.K. Influenza A virus targets a cGAS-independent STING pathway that controls enveloped RNA viruses. Nat. Commun. 2016;7:10680. doi: 10.1038/ncomms10680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yu C.Y., Chang T.H., Liang J.J., Chiang R.L., Lee Y.L., Liao C.L., Lin Y.L. Dengue virus targets the adaptor protein MITA to subvert host innate immunity. PLoS Pathog. 2012;8:e1002780. doi: 10.1371/journal.ppat.1002780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Aguirre S., Luthra P., Sanchez-Aparicio M.T., Maestre A.M., Patel J., Lamothe F., Fredericks A.C., Tripathi S., Zhu T., Pintado-Silva J. Dengue virus NS2B protein targets cGAS for degradation and prevents mitochondrial DNA sensing during infection. Nat. Microbiol. 2017;2:17037. doi: 10.1038/nmicrobiol.2017.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kondo T., Kobayashi J., Saitoh T., Maruyama K., Ishii K.J., Barber G.N., Komatsu K., Akira S., Kawai T. DNA damage sensor MRE11 recognizes cytosolic double-stranded DNA and induces type I interferon by regulating STING trafficking. Proc. Natl. Acad. Sci. USA. 2013;110:2969–2974. doi: 10.1073/pnas.1222694110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ho S.S., Zhang W.Y., Tan N.Y., Khatoo M., Suter M.A., Tripathi S., Cheung F.S., Lim W.K., Tan P.H., Ngeow J., Gasser S. The DNA Structure-Specific Endonuclease MUS81 Mediates DNA Sensor STING-Dependent Host Rejection of Prostate Cancer Cells. Immunity. 2016;44:1177–1189. doi: 10.1016/j.immuni.2016.04.010. [DOI] [PubMed] [Google Scholar]
- 71.Marcus A., Mao A.J., Lensink-Vasan M., Wang L., Vance R.E., Raulet D.H. Tumor-Derived cGAMP Triggers a STING-Mediated Interferon Response in Non-tumor Cells to Activate the NK Cell Response. Immunity. 2018;49:754–763.e4. doi: 10.1016/j.immuni.2018.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]