Abstract
The way in which a balanced vaginal microbiome helps prevent gynecological diseases in women and maintain health remains to be fully elucidated. In the present study, the potential effect of aberrations in the vaginal flora on unexplained recurrent miscarriage (RM) was investigated. The vaginal bacterial communities of 10 patients with unexplained RM and 10 healthy volunteers were sampled and subjected to sequencing analysis of the V3-V4 regions of the bacterial 16S ribosomal RNA gene using the Illumina MiSeq platform. Beta diversity analysis/principal component analysis indicated that bacterial community structures were different between the RM and control groups. A lower microbiota diversity in samples from RM patients was revealed by alpha diversity estimation. Taxonomic analysis demonstrated that abundance of three types of phyla (Firmicutes, Actinobacteria and Bacteroidetes) was significantly different between the RM and the normal control group. Furthermore, at the genus level, Lactobacillus was the most dominant genus in the two groups. Statistically significant differences were observed in 5 genera between the two groups. In the RM group, 3 bacterial taxa (Atopobium, Prevotella and Streptococcus) were significantly more abundant, while only 2 taxa were overrepresented in the control group (Lactobacillus and Gardnerella). In conclusion, the present results provide experimental evidence supporting dysbiosis of the vaginal flora in women with RM.
Keywords: unexplained recurrent miscarriage, vaginal microbiome, bacterial diversity, 16S ribosomal RNA
Introduction
Unexplained recurrent miscarriage (RM) is defined as ≥3 consecutive idiopathic miscarriages prior to the gestational age of 12 weeks of pregnancy. RM affects 1–2% of females attempting to conceive and represents a considerable challenge for physicians (1,2). RM may be explained by genetic abnormalities, maternal immunological causes and endocrine dysfunction only in a minority of cases, while the majority of cases are diagnosed as unexplained RM (3). The vagina harbors different species of bacteria in variable quantities and relative proportions that have important effects on the health of women (4). A healthy vaginal flora, dominated by Lactobacillus species, has an important role in the protection against genital infections, which are considered as a common cause of miscarriage (5). A change in the bacterial flora of the vagina is represented by a decrease of Lactobacillus and overgrowth of several anaerobic or facultative bacteria, including Mobiluncus species, Prevotella species, Gardnerella vaginalis and genital Mycoplasm: Mycoplasma hominis and Ureaplasma urealyticum (6). At present, bacterial vaginosis is thought to be mainly caused by vaginal dysbacteriosis, which may lead to premature birth, premature rupture of membranes, low birth weight at premature birth, RM, chorioamnionitis and a series of diseases (7).
Advances in sequencing technologies and analytical methods have enabled the exploration of the human microbiota (8). At the end of 2007, the US National Institutes of Health invested hundreds of millions of dollars to start a five-year human microbial genome project, mainly using high-throughput sequencing technology analysis of different parts of the body's bacterial community structure and the female vaginal micro-ecological environment is an important part of this program (9,10). Theoretically, 16S ribosomal (r)RNA gene sequencing is designed to detect most pathologival bacteria. Via this method, bacteria that are not detectable by cultivation and empirical antibiotic treatments are identifiable. In addition, this method may be utilized to identify novel pathogens that have not been previously recognized as etiologic agents (11).
The diversity of the microflora in the female reproductive tract is affected by various drugs, as well as changes in the environment, and the host's hormone levels and immune system (12). Numerous studies have demonstrated that the microbial balance in the reproductive tract is closely associated with a poor outcome of pregnancy (13–16). However, due to the complex etiology and pathogenesis of RM, the association of aberrations in the vaginal flora and RM has remained to be sufficiently elucidated. In the present study, the diversity of the vaginal microbial community in patients with unexplained RM was analyzed in order to determine the involvement of the vaginal flora in the mechanisms of RM.
Materials and methods
Sample collection
Vaginal secretions of 10 women with RM (case group) and 10 healthy volunteers (control group) were collected at Shaoxing Women and Children's Hospital (Shaoxing, China) between September 2016 and March 2017. The samples were collected in accordance with the relevant guidelines and all subjects provided written informed consent for use of their samples in the present study. The present study was approved by the Ethics Committee of Shaoxing Women and Children's Hospital (Shaoxing, China). None of the women were pregnant, while they were of reproductive age (case group, 20–35 years, median age, 28 years; control group, 26–34 years; median age, 30 years; P<0.05), had a regular menstrual cycle and a history of sexual activity, and had not taken any antibiotics or antimycotic drugs in the past 30 days. Women were asked to refrain from sexual activity for 3 days prior to sampling. Women were excluded from the study if they had used douches, vaginal medications or suppositories, feminine sprays, genital wipes or contraceptive spermicides, or had reported vaginal discharge in the past 48 h (12). The secretions were scraped from the vaginal walls of the RM patients and healthy controls using aseptic swabs. From each subject, two vaginal swabs were collected. The sample centrifuge tubes were immediately placed in a prepared ice box or in a foam box filled with ice packs to maintain a low temperature. The samples were then transferred to the laboratory and stored at −80°C.
DNA extraction and polymerase chain reaction (PCR) amplification
Microbial DNA was extracted from vaginal discharge using the QIAamp DNA Mini Kit (Qiagen, Hilden, Germany) according to manufacturer's protocols. The V3-V4 region of the bacterial 16S rRNA gene was amplified by PCR (ABI GeneAmp® 9700). The thermocycling protocol was as follows: 95°C for 2 min, followed by 25 cycles at 95°C for 30 sec, 55°C for 30 sec and 72°C for 30 sec and a final extension at 72°C for 5 min. The primers used were 341F 5′-barcode-CCTACGGGNGGCWGCAG-3′ and 805R 5′-barcode-GACTACHVGGGTATCTAATCC-3′, where the barcode is an eight-base sequence unique to each sample. PCR was performed in triplicate in 20 µl mixtures containing 4 µl 5X Fast Pfu Buffer, 2 µl 2.5 mM deoxynucleoside triphosphates, 0.8 µl of each primer (5 µM), 0.4 µl FastPfu Polymerase and 10 ng template DNA (17). All samples were subjected to the same conditions and each sample was assessed in duplicate. The PCR products of the same sample were mixed and examined by 2% agarose gel electrophoresis. The PCR products were recovered using AxyPrepDNA Gel Recovery Kit (Axygen; Corning, Inc., Corning, NY, USA) Tris-HCl elution (18).
Illumina MiSeq sequencing
The PCR products were quantified using the QuantiFluor™-ST Blue Fluorescence Quantitative System (Promega Corp., Madison, WI, USA) after obtaining the initial quantitative results of the electrophoresis, followed by mixing the corresponding proportions according to the sequencing requirements of each sample. Amplicons were extracted from 2% agarose gels and purified using the AxyPrep DNA Gel Extraction kit (Axygen; Corning, Inc.) according to the manufacturer's instructions and quantified using QuantiFluor™-ST (Promega Corp.). Purified amplicons were pooled in equimolar amounts and paired-end sequenced (2×250) on an Illumina MiSeq platform according to the standard protocols.
Processing of sequencing data
Sequences produced by Miseq sequencing contain barcode sequences, and the primers and linker sequences are added at the time of sequencing. First, the primer sequences were removed using Cutadapt and the pairs were merged into a sequence according to the overlap between paired-end (PE) reads by using The PE reAd merger (19). Next the sample data were identified and distinguished according to the barcode tag sequences. Finally, quality control and filtration of each sample was performed using Prinseq (20) to obtain data.
Raw fastq files were demultiplexed and quality-filtered using FLASH (21) with the following criteria: i) The 3′ end of the sequencing primer was removed and the Read1 3′ end sequencing link was TGGAATTCTCGGGTGCCAAGGAACTC; ii) according to the overlap between PE reads, pairs of reads were merged into a sequence, and the allowed maximum mismatch ratio of the splicing sequence overlap area was 0.1; iii) data for each sample were divided from the merged data according to each sample's barcode sequence; iv) the reads were truncated at any site receiving an average quality score of <20 over a 10-bp sliding window; if the average quality value of the window was <20, the back-end base from the window was cut. v) Reads containing N sequences were removed along with the short sequence in the data, with a length threshold of 200 bp. vi) The low-complexity sequences were filtered.
Bacterial community characterization
According to the barcodes, the high-throughput pyrosequencing reads from 20 vaginal secretion samples were reassigned to samples. Operational taxonomic units (OTUs) were clustered with a 97% similarity cutoff through Usearch (version 7.0; http://drive5.com/uparse/) (22), and chimeric sequences were identified and removed using UCHIME (23). Alpha diversity analysis was performed using the OTUs that reached a similarity level of 97%, and the species diversity in a single sample was analyzed by evaluation of Chao, abundance-based coverage estimators (ACE) and Simpon parameters using Mothur (24). A Venn diagram was generated to illustrate the shared and unique OTUs among the groups, based on the occurrence of OTUs in a group of samples regardless of their relative abundance, and this was analyzed using the Venn Diagram package of R language tools (25).
Beta diversity was analyzed to investigate the similarity of the bacterial community structure among groups using Weighted Unifrac distances and visualized analysis via principal component analysis (PCoA). Circos, a sample and species association map, is a visual circle diagram that describes the correspondence between sample and species. The functional profiles of microbial communities were predicted by using PICRUSt (26). The 10,506 OTUs were normalized to the 16S rRNA copy number and their metagenomic contributions were predicted according to the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways. Based on the PICRUSt functional secondary classification results, the differences in abundance between samples were compared.
Statistical analysis
All statistical analyses were performed using R packages (V.3.2) (25). The screening criterion for analyzing differences in bacterial abundance between samples was P≤0.05. Fisher's exact test was used for comparison between samples and Welch's t-test was used for comparison between groups (27). Finally, the P-value was verified by using the false discovery rate to obtain the q-value.
Results
Alpha diversity analysis between the case and control groups
To characterize the vaginal microbiota and potential variations associated with RM, the vaginal secretions from 10 cases of RM were collected, and those from 10 healthy women were used as a control group. In total, 745,971 usable sequences from 20 samples were obtained. A total of 10,506 OTUs were delineated at a 97% similarity level. On average, >90% of reads in a sample were retained. At a 97% sequence similarity level, Good's coverage values for all sampled bacterial communities ranged from 0.989 to 0.995 when estimated from all reads (Table I).
Table I.
Summary of pyrosequencing data.
| Sample_ID | Sequence_no. | OTUs | Filtered OTUs | Shannon_index | ACE_index | Chao1_index | Coverage | Simpon |
|---|---|---|---|---|---|---|---|---|
| RM_control1 | 34948 | 447 | 37322 | 1.2204 | 641.11437 | 580.913043 | 0.994964 | 0.56009 |
| RM_control2 | 39002 | 463 | 41410 | 0.421023 | 1672.661269 | 971.683544 | 0.992718 | 0.905217 |
| RM_control3 | 36163 | 551 | 38664 | 0.883533 | 885.603703 | 787.014925 | 0.993032 | 0.717632 |
| RM_control4 | 38540 | 336 | 39876 | 0.384966 | 448.529403 | 407.90625 | 0.996938 | 0.919171 |
| RM_control5 | 33366 | 637 | 35693 | 1.586938 | 874.401789 | 812.506329 | 0.992927 | 0.478691 |
| RM_control6 | 38787 | 377 | 39998 | 1.193842 | 691.127976 | 618.325 | 0.994921 | 0.460157 |
| RM_control7 | 33174 | 421 | 34615 | 1.90784 | 639.519629 | 559.125 | 0.994876 | 0.282237 |
| RM_control8 | 38476 | 604 | 40815 | 0.605582 | 1702.873689 | 1098.161017 | 0.991111 | 0.872647 |
| RM_control9 | 38193 | 418 | 40387 | 0.349364 | 1476.004523 | 827.296296 | 0.993245 | 0.927695 |
| RM_control10 | 39055 | 556 | 41136 | 0.430429 | 1525.391993 | 1033.097087 | 0.99196 | 0.913746 |
| RM_case1 | 38861 | 530 | 40899 | 0.47373 | 1411.786593 | 889.486486 | 0.992718 | 0.901358 |
| RM_case2 | 37973 | 495 | 39464 | 0.969683 | 688.77094 | 613.106383 | 0.995181 | 0.737364 |
| RM_case3 | 32766 | 659 | 34398 | 2.121407 | 1435.948065 | 1057.649635 | 0.989898 | 0.263954 |
| RM_case4 | 38107 | 633 | 40429 | 0.559294 | 1226.25393 | 1100.12782 | 0.990737 | 0.882253 |
| RM_case5 | 36180 | 553 | 37780 | 0.964259 | 1496.239405 | 983 | 0.99168 | 0.62994 |
| RM_case6 | 39523 | 692 | 42178 | 0.492288 | 1817.054442 | 1243.338129 | 0.990082 | 0.904096 |
| RM_case7 | 34778 | 478 | 36739 | 1.20537 | 766.093643 | 675.280992 | 0.993703 | 0.446528 |
| RM_case8 | 39885 | 510 | 41821 | 0.325184 | 1492.01625 | 904.345794 | 0.992704 | 0.94037 |
| RM_case9 | 39854 | 537 | 41768 | 0.576914 | 1407.226829 | 936.794393 | 0.992648 | 0.866492 |
| RM_case10 | 38340 | 609 | 41200 | 0.486695 | 1252.493401 | 1152.128205 | 0.990689 | 0.900224 |
The OTU, Chao I, Shannon and Simpon data were calculated at a 3% distance. RM_case, disease group; RM control, control group; OTUs, operational taxonomic units; ACE, abundance-based coverage estimators.
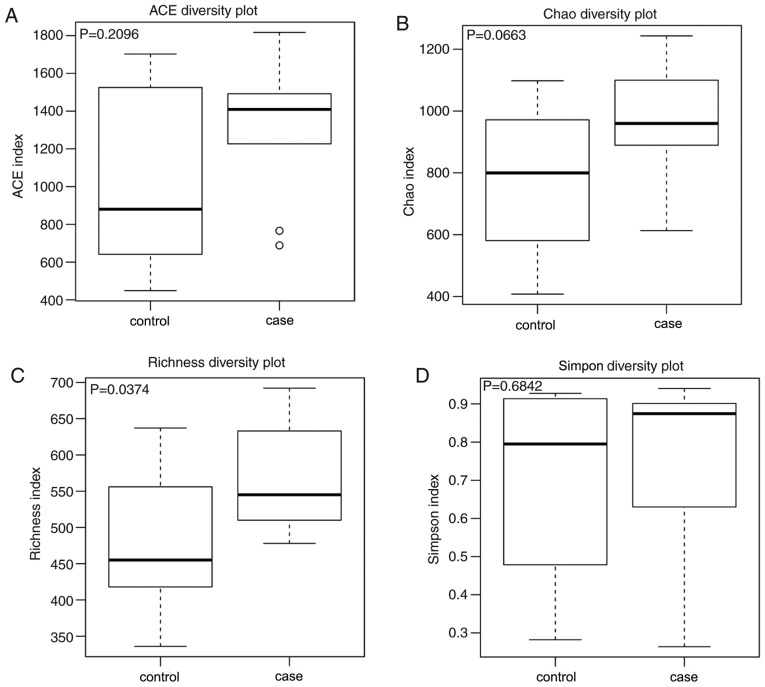
Alpha diversity was applied to analyze the complexity of the bacterial diversity of the samples. In Fig. 1, a boxplot of richness estimators was generated using the QIIME toolkit, providing a clear visualization of the diversities in the different sample groups. No significant difference in Simpon, ACE and Chao diversity was identified between the control and case groups in the box plots (P>0.05; Fig. 1). Community richness comparison indicated that the case group had a significantly increased number of observed and estimated OTUs compared with those in the control group (P=0.037). As displayed in the Venn diagram, the groups had 1,924 OTUs in common, and the proportion of unique OTUs among the total OTUs was 43.8 and 41.9% in the case and control group, respectively. However, the case group had 63 more OTUs than the control group (Fig. 2). This result demonstrated a significantly higher diversity in the case group compared with that in the control group. As presented in Fig. 3, the Simpon rarefaction curves for all samples were saturated, which indicated that the 16S rRNA gene sequence was highly abundant in the database, and that the present analysis had an adequate depth to retrieve most of the information on microbial diversity.
Figure 1.
Alpha diversity of the bacterial flora in the two group. (A) ACE diversity, (B) Chao diversity, (C) Richness diversity and (D) Simpon diversity. Box edges represent the quartiles, horizontal lines represent the medians, bars indicate edge values and circles indicate outliers. ACE, abundance-based coverage estimators.
Figure 2.
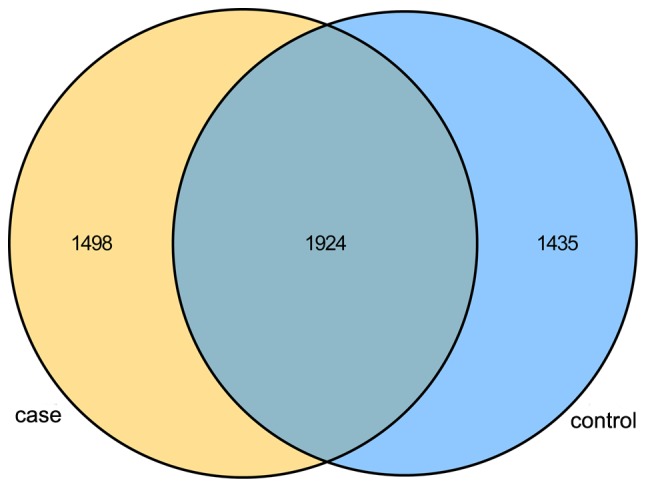
Venn diagram of OTUs for different group samples. The yellow area was used to represent the case group and light blue area was used to represent the control group, while dark blue was used to represent the common OTUs of these two different groups. OTUs, operational taxonomic units.
Figure 3.
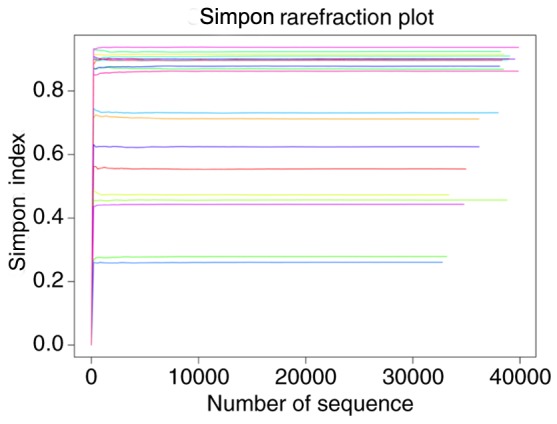
Simpon rarefraction of all samples. The horizontal axis was used to represent the number of sequences, the vertical axis was used to represent the Simpon index and the curves of different colors were used to represent different samples.
Characteristics of beta diversity analysis between the case and control groups
In the PCoA plot (Fig. 4), each dot represents one sample. PCoA based on 10,506 OTUs (grouped at a sequence identity of 97%) revealed a slight separation of the two different groups on the first two PCo scores, PCo1 and PCo2, which accounted for 36.8 and 29.9% of the total variations, respectively. This indicated that the development of RM may contribute to microbiota imbalance. Furthermore, microbiota disturbances may further promote the development of RM (Fig. 4).
Figure 4.
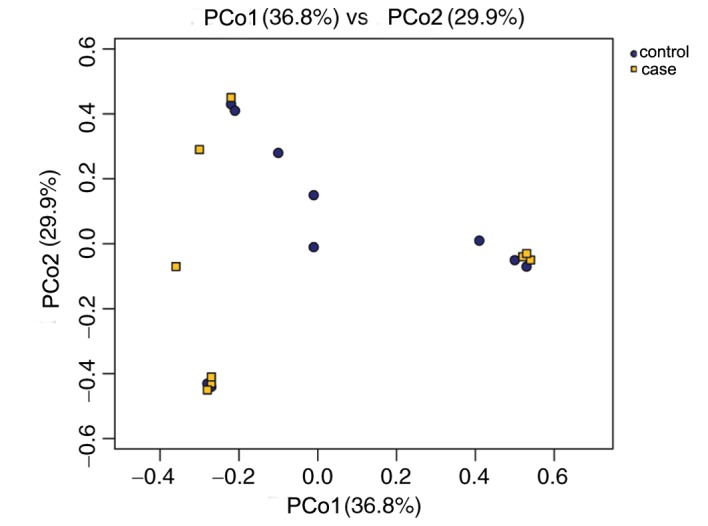
PCoA plots based on Bray-Curtis metrics for the control and case groups. The X-axis represents the PCo1 (accounting for 36.8% of the total variations) and the Y-axis represents the PCo2 (accounting for 29.9% of the total variations). Each data-point represents one sample. PCoA, principal component analysis.
Comparison of bacterial communities in the case and control groups at the phylum level
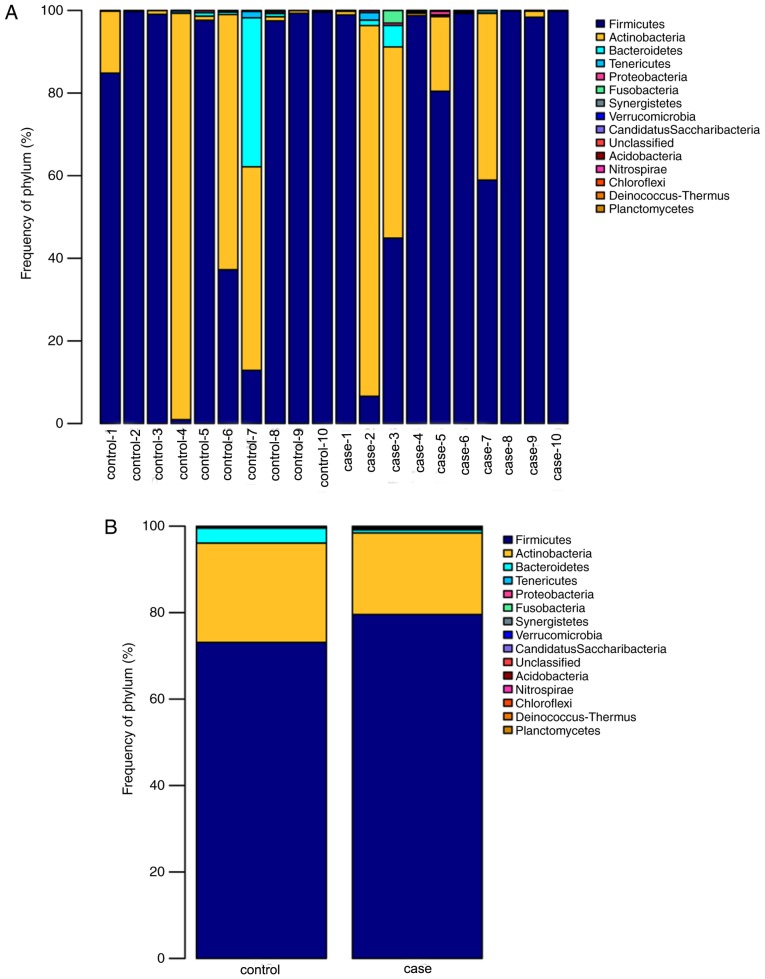
When comparing the alpha diversity between the groups, no significant differences in diversity were observed except for a higher richness index in the case group compared with that in the control group. However, the PCoA plot based on Weighted Unifrac data displayed distinct bacterial community structures in the case and the control group. To investigate the specific changes of the microbiota in the samples from the case group, the relative abundance of taxa in the different groups was assessed. Fig. 5 presents taxonomic distributions of the predominant bacteria (relative abundance, >1% of the total sequences) at the phylum level. The bacterial flora analysis indicated that Firmicutes was the most predominant phylum, accounting for 79.57 and 73.12% of the microbiota in case and control group, respectively. The second most dominant phylum was Actinobacteria (18.86% in the case group and 22.95% in the control group). Changes at the phylum level were mainly in the three most common types of bacteria (Firmicutes, Bacteroidetes and Actinobacteria). Compared with that in the control group, the relative abundance of Firmicutes in the case group was increased by 5.72%, while the Actinobacteria and the Bacteroidetes were reduced by 3.08 and 3.07%, respectively.
Figure 5.
Relative abundance of the predominant bacteria at the phylum level. The predominant taxa (>1% relative abundance) is presented. (A) Distribution of community components at the phylum level in each individual sample. (B) Group merge of community composition at the phylum level in all samples. The X-axis represents the different samples and the Y-axis represents the relative abundance ratio. Different colors represent the different phylums, and the different color block heights indicate the relative abundance ratio of different species.
Comparison of bacterial communities between case and control groups at the genus level
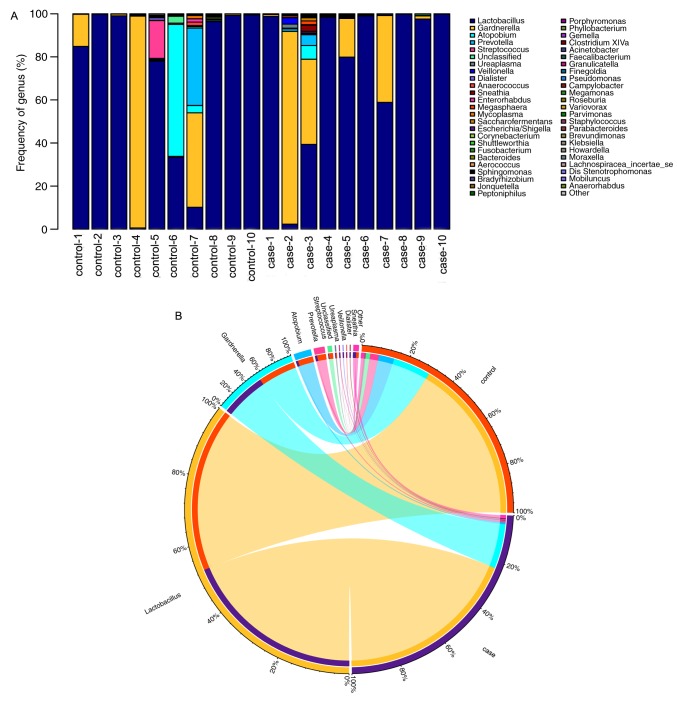
The bacterial diversity and relative abundance in all samples at the genus level are presented in Fig. 6. Fig. 6A displays the composition of the vaginal flora of the patients with RM and healthy women. In the stacked bar chart, each bar represents the average relative abundance of each bacterial taxon. The top 50 taxa with high relative abundance are illustrated. All core bacteria in the vagina, including Lactobacillus, Gardnerella, Atopobium and Prevotella, were identified in the present study. Of all the detected genera, Lactobacillus of the phylum Firmicutes were the most abundant ones. Circos was then used to identify bacterial taxa that were significantly different between the groups. A Circos presentation of bacterial taxa that are differentially represented between the different groups is provided in Fig. 6B. In the case group, 3 bacterial taxa were significantly more abundant (Atopobium, Prevotella and Streptococcus), while only 2 taxa were overrepresented in the control group (Lactobacillus and Gardnerella).
Figure 6.
Relative abundance of the predominant bacteria at the genus level. (A) Distribution of microbial community components in each sample at the genus level. The height of the bars for each genus indicates its relative abundance in the sample. (B) The data were visualized in a Circos presentation.
Functional annotation
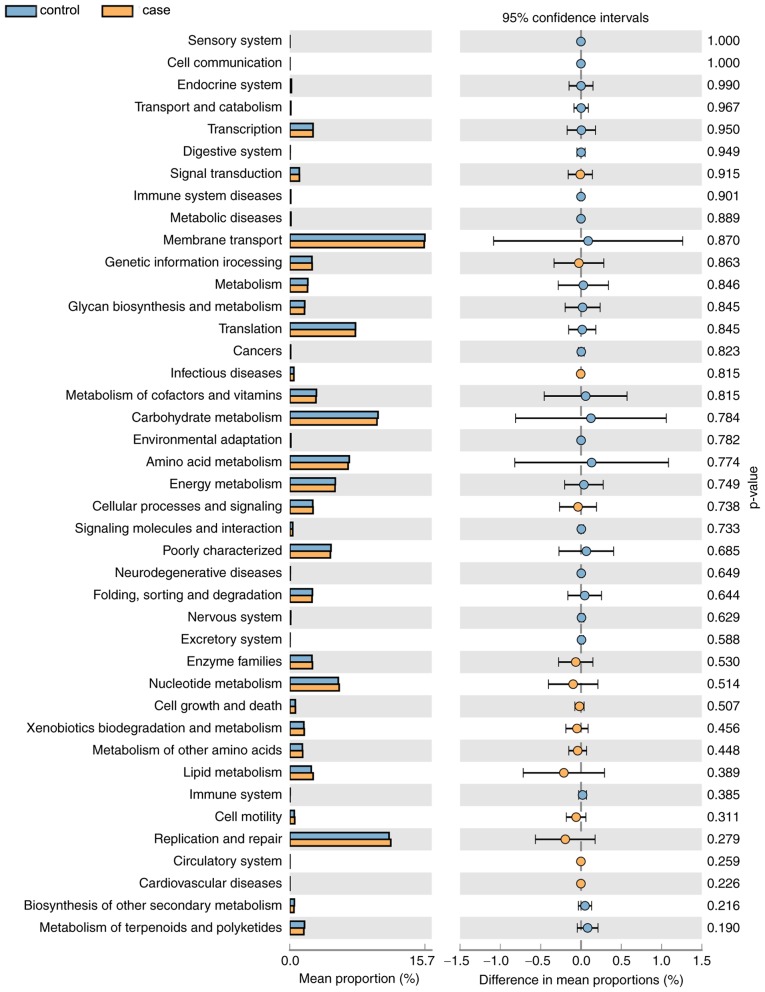
As displayed in Fig. 7, a total of 14 KEGG pathways (e.g. ‘Signal Transduction’ and ‘Cell Motility’) were more abundant in the case group and 27 pathways (e.g. ‘Metabolism of Cofactors and Vitamins’ and ‘Carbohydrate Metabolism’) were more abundant in the control group. However, the differences in the mean proportion of the abundance of the functional pathways between the groups were not significant (P>0.05). In the future, differences in the functional pathways specific for the vaginal microbial flora of the two groups will be further experimentally verified.
Figure 7.
Enriched Kyoto Encyclopedia of Genes and Genomes pathways of bacteria in the two groups. The left panel indicates the abundance ratio of the different functional pathways in the two groups, and the right panel indicates the difference in the mean proportion of the abundance of the functional pathways and the 95% confidence interval, as well as the P-values.
Discussion
Previous studies on vaginal bacterial communities suggested that the human vaginal microbiota has a key role in preventing a number of urogenital diseases, including bacterial vaginosis, yeast infections, sexually transmitted infections and urinary tract infections (12,28). However, studies indicating a direct association between RM and vaginal microbiota are currently lacking. Compared with the intestinal flora, the vaginal flora is less diverse, and women of childbearing age may have ~40 species of bacteria and facultative anaerobic bacteria, including Lactobacillus, Bacteroides, Coccidioides, Corynebacterium, Escherichia coli, Velveti and Gardneria (29). A previous study sampled and analyzed 396 women from four ethnic groups and identified that the common constituents of the vaginal flora may be roughly divided into five categories, of which four are able to produce a large quantity of lactic acid to generate the acidic environment in the vagina (30). Bacterial vaginosis is characterized by a complete loss of lactobacilli and a concomitant increase in Gram-variable and Gram-negative rods, with Gardnerella vaginalis, as well as Bacteroidetes, Prevotella and Mobiluncus species being primary among them. The presence of an abnormal vaginal microbiota in early pregnancy is a recognized risk factor for preterm delivery and low birth weight (31).
In the present study, the vaginal microbiome of the RM group exhibited an increased richness in species as well as a significant shift in the overall microbial diversity. A statistically significant increase in richness was observed in the RM group (P=0.037). These results suggested the presence of disorders in the profile of the vaginal microbiome in patients with RM and miscarriage may be either a cause or an effect of the altered composition of the vaginal microbiome. The PCo coefficients obtained in the PCoA analysis of the present study indicated that RM may be the most important factor contributing to changes in the vaginal bacterial composition. It was suggested that RM may be associated with imbalances in the microbiota. Local microbiota disturbances may further promote the development of RM (32).
When comparing the alpha diversity between samples, significant differences in diversity were observed in richness indices. Furthermore, PCoA indicated distinct bacterial community structures between the two groups. To further clarify the specific differences in the intestinal microflora between the case and control groups, taxonomic analysis at the phylum and genus level, as well as beta diversity analysis were performed. At the phylum level, Firmicutes was the most predominant phylum, followed by Actinobacteria and then Bacteroidetes. Furthermore, at the genus level, Lactobacillus was the most dominant genus. Statistically significant differences in five genera were observed. In the case group, three bacterial taxa were significantly more abundant (Atopobium, Prevotella and Streptococcus), while only two taxa were overrepresented in the control group (Lactobacillus and Gardnerella). Kuon et al (33) reported that RM patients with elevated peripheral and uterine natural killer cells suffer more frequently from colonization by Gardnerella vaginalis and gram-negative anaerobes. McDonald et al (34) demonstrated that group B streptococcus is a key pathogen in intrauterine infection and a frequent cause of spontaneous midgestation abortions. Lactobacillus is one of the diverse and phylogenetically heterogeneous orders of lactic acid-producing bacteria that includes the genus Lactobacillus, as well as the genera Facklamia, Granulicatella, Leuconostoc, Pediococcus and Streptococcus (35). The healthy human vagina is dominated by a variety of Lactobacillus species, which have an essential role in protecting women from genital infection. A deficiency in Lactobacilli may disturb the microbial balance in the vagina, frequently resulting in the syndrome of bacterial vaginosis, which is associated with a quantitative and qualitative shift from normally occurring Lactobacilli to a mixed microflora dominated by anaerobic bacteria (31). In the present study, the three anaerobic bacteria of Atopobium, Prevotella and Streptococcus were more abundant in the case group compared with those in the control group, which was consistent with the results of previous studies.
In conclusion, the present study defined a structural imbalance in the vaginal microbiota of patients with RM, represented by an increased incidence of Atopobium, Prevotella and Streptococcus and reduction of Lactobacillus, which was identified by comparing the vaginal microbiota composition of RM patients with that of healthy individuals. PCoA analysis suggested that changes in vaginal flora may be the cause of/associated with RM. The present results point towards a novel strategy aimed at preventing the development of RM through restoring the homeostasis of the vaginal microbiome, by improvement in lifestyle or early intervention with drugs or probiotics.
Acknowledgements
Not applicable.
Funding
The present study was supported by the National Natural Science Foundation of China (grant no. 81701522); the National Natural Science Foundation of Zhejiang Province, China (grant nos. LY15H040001 and LY16H040008); the Science Technology Department of Zhejiang Province, China (grant no. 2015C33280, 2016C33222, 2016C33223 and 2018C37102); the Medical and Health Project of Zhejiang Province, China (grant nos. 2014KYA275, 2014KYA276, 2017KY669, 2017KY670, 2017KY672, 2018KY844, 2018KY845, 2018KY846 and 2018KY848); the Health and Family Planning Commission of Shaoxing, China (grant nos. 2017QN008 and 2017CX011); and the Science Technology Department of Shaoxing, China (grant no. 2017B70004).
Availability of data and material
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.
Authors' contributions
FZ, TZ and HD conceived and designed the current study. YM and ZH obtained samples; YH performed the experiments; HP collected the data; MF and HD analysed and interpreted the data. All authors approved the final version of the manuscript for publication.
Ethical approval and consent to participate
All subjects provided written informed consent for use of their samples in the present study. The present study was approved by the Ethics Committee of Shaoxing Women and Children's Hospital (Shaoxing, China) and was performed in compliance with the Declaration of Helsinki.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Sugiura-Ogasawara M, Ozaki Y, Suzumori N. Management of recurrent miscarriage. J Obstet Gynaecol Res. 2014;40:1174–1179. doi: 10.1111/jog.12388. [DOI] [PubMed] [Google Scholar]
- 2.Dempsey MA, Flood K, Burke N, Murray A, Cotter B, Mullers S, Dicker P, Fletcher P, Geary M, Kenny D, Malone FD. Platelet function in patients with a history of unexplained recurrent miscarriage who subsequently miscarry again. Eur J Obstet Gynecol Reprod Biol. 2015;188:61–65. doi: 10.1016/j.ejogrb.2015.02.003. [DOI] [PubMed] [Google Scholar]
- 3.Christiansen OB, Larsen EC, Egerup P, Lunoee L, Egestad L, Nielsen HS. Intravenous immunoglobulin treatment for secondary recurrent miscarriage: A randomised, double-blind, placebo-controlled tria. BJOG. 2015;122:500–508. doi: 10.1111/1471-0528.13192. [DOI] [PubMed] [Google Scholar]
- 4.Obiero JA, Waititu KK, Mulei I, Omar FI, Jaoko W, Mwethera PG. Baboon vaginal microbial flora. J Med Primatol. 2016;45:147–155. doi: 10.1111/jmp.12217. [DOI] [PubMed] [Google Scholar]
- 5.Farr A, Kiss H, Hagmann M, Machal S, Holzer I, Kueronya V, Husslein PW, Petricevic L. Role of lactobacillus species in the intermediate vaginal flora in early pregnancy: A retrospective cohort study. PLoS One. 2015;10:e0144181. doi: 10.1371/journal.pone.0144181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Romero R, Chaiworapongsa T, Kuivaniemi H, Tromp G. Bacterial vaginosis, the inflammatory response and the risk of preterm birth: A role for genetic epidemiology in the prevention of preterm birth. Am J Obstet Gynecol. 2004;190:1509–1519. doi: 10.1016/j.ajog.2004.01.002. [DOI] [PubMed] [Google Scholar]
- 7.Romero HD, Andreu DA. Bacterial vaginosis. Enfermedades Infecciosas Y Microbiol Clin. 2016;34(Suppl 3):14–18. doi: 10.1016/S0213-005X(16)30214-2. [DOI] [PubMed] [Google Scholar]
- 8.Tseng CH, Lin JT, Ho HJ, Lai ZL, Wang CB, Tang SL, Wu CY. Gastric microbiota and predicted gene functions are altered after subtotal gastrectomy in patients with gastric cancer. Sci Rep. 2016;6:20701. doi: 10.1038/srep20701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.NIH HMP Working Group. Peterson J, Garges S, Giovanni M, McInnes P, Wang L, Schloss JA, Bonazzi V, McEwen JE, Wetterstrand KA, et al. The NIH human microbiome project. Genome Res. 2009;19:2317–2323. doi: 10.1101/gr.096651.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Human Microbiome Jumpstart Reference Strains Consortium. Nelson KE, Weinstock GM, Highlander SK, Worley KC, Creasy HH, Wortman JR, Rusch DB, Mitreva M, Sodergren E, et al. A catalog of reference genomes from the human microbiome. Science. 2010;328:994–999. doi: 10.1126/science.1183605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ling Z, Liu X, Chen X, Zhu H, Nelson KE, Xia Y, Li L, Xiang C. Diversity of cervicovaginal microbiota associated with female lower genital tract infections. Microb Ecol. 2011;61:704–714. doi: 10.1007/s00248-011-9813-z. [DOI] [PubMed] [Google Scholar]
- 12.Ravel J, Gajer P, Abdo Z, Schneider GM, Koenig SS, McCulle SL, Karlebach S, Gorle R, Russell J, Tacket CO, et al. Vaginal microbiome of reproductive-age women. Proc Natl Acad Sci USA. 2011;108(Suppl 1):4680–4687. doi: 10.1073/pnas.1002611107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Leitch H, Kiss H. Asymptomatic bacterial vaginosis and intermediate flora as risk factors for adverse pregnancy outcome. Best Pract Res Clin Obstet Gynaecol. 2007;21:375–390. doi: 10.1016/j.bpobgyn.2006.12.005. [DOI] [PubMed] [Google Scholar]
- 14.Allsworth JE, Peipert JF. Prevalence of bacterial vaginosis: 2001–2004 National Health and Nutrition Examination Survey data. Obstet Gynecol. 2007;109:114–120. doi: 10.1097/01.AOG.0000247627.84791.91. [DOI] [PubMed] [Google Scholar]
- 15.Trabert B, Misra DP. Risk factors for bacterial vaginosis during pregnancy among African American women. Am J Obstet Gynecol. 2007;197:477.e1–e8. doi: 10.1016/j.ajog.2007.03.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Donders GG, Veerecken A, Bosmans E, Dekersmaecker A, Salembier G, Spitz B. Definition of a type of abnormal vaginal flora that is distinct from bacterial vaginosis: Aerobic vaginitis. BJOG. 2002;109:34–43. doi: 10.1111/j.1471-0528.2002.00432.x. [DOI] [PubMed] [Google Scholar]
- 17.Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Lozupone CA, Turnbaugh PJ, Fierer N, Knight R. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc Natl Acad Sci USA. 2011;108(Suppl 1):4516–4522. doi: 10.1073/pnas.1000080107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Aguirre E, Galiana A, Mira A, Guardiola R, Sánchez-Guillén L, Garcia-Pachon E, Santibañez M, Royo G, Rodríguez JC. Analysis of microbiota in stable patients with chronic obstructive pulmonary disease. APMIS. 2015;123:427–432. doi: 10.1111/apm.12363. [DOI] [PubMed] [Google Scholar]
- 19.Zhang J, Kobert K, Flouri T, Stamatakis A. PEAR: A fast and accurate Illumina Paired-End reAd mergeR. Bioinformatics. 2014;30:614–620. doi: 10.1093/bioinformatics/btt593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schmieder R, Edwards R. Quality control and preprocessing of metagenomic datasets. Bioinformatics. 2011;27:863–864. doi: 10.1093/bioinformatics/btr026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Magoč T, Salzberg SL. FLASH: Fast length adjustment of short reads to improve genome assemblies. Bioinformatics. 2011;27:2957–2963. doi: 10.1093/bioinformatics/btr507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 23.Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics. 2011;27:2194–2200. doi: 10.1093/bioinformatics/btr381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, et al. Introducing mothur: Open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol. 2009;75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Snipen L, Liland KH. micropan: An R-package for microbial pan-genomics. BMC Bioinformatics. 2015;16:79. doi: 10.1186/s12859-015-0517-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Langille MG, Zaneveld J, Caporaso JG, McDonald D, Knights D, Reyes JA, Clemente JC, Burkepile DE, Vega Thurber RL, Knight R, et al. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat Biotechnol. 2013;31:814–821. doi: 10.1038/nbt.2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Parks DH, Tyson GW, Hugenholtz P, Beiko RG. STAMP: Statistical analysis of taxonomic and functional profiles. Bioinformatics. 2014;30:3123–3124. doi: 10.1093/bioinformatics/btu494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hernándezrodríguez C, Romerogonzález R, Albanicampanario M, Figueroa-Damián R, Meraz-Cruz N, Hernández-Guerrero C. Vaginal microbiota of healthy pregnant mexican women is constituted by four lactobacillus species and, several vaginosis-associated bacteria. Infect Dis Obstet Gynecol. 2011;2011:851485. doi: 10.1155/2011/851485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wells CL, Jechorek RP, Maddaus MA. The translocation of intestinal facultative and anaerobic bacteria in defined flora mice. Microb Eco Health Dis. 1988;1:227–235. [Google Scholar]
- 30.Mendes-Soares H, Suzuki H, Hickey RJ, Forney LJ. Comparative functional genomics of Lactobacillus spp. reveals possible mechanisms for specialization of vaginal lactobacilli to their environment. J Bacteriol. 2014;196:1458–1470. doi: 10.1128/JB.01439-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Petricevic L, Domig KJ, Nierscher FJ, Sandhofer MJ, Fidesser M, Krondorfer I, Husslein P, Kneifel W, Kiss H. Characterisation of the vaginal Lactobacillus microbiota associated with preterm delivery. Sci Rep. 2014;4:5136. doi: 10.1038/srep05136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lu Y, Chen J, Zheng J, Hu G, Wang J, Huang C, Lou L, Wang X, Zeng Y. Mucosal adherent bacterial dysbiosis in patients with colorectal adenomas. Sci Rep. 2016;6:26337. doi: 10.1038/srep26337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kuon RJ, Togawa R, Vomstein K, Weber M, Goeggl T, Strowitzki T, Markert UR, Zimmermann S, Daniel V, Dalpke AH, Toth B. Higher prevalence of colonization with gardnerella vaginalis and gram-negative anaerobes in patients with recurrent miscarriage and elevated peripheral natural killer cells. J Reprod Immunol. 2017;120:15–19. doi: 10.1016/j.jri.2017.03.001. [DOI] [PubMed] [Google Scholar]
- 34.Mcdonald HM, Chambers HM. Intrauterine infection and spontaneous midgestation abortion: Is the spectrum of microorganisms similar to that in preterm labor? Infect Dis Obst Gynecol. 2000;8:220–227. doi: 10.1002/1098-0997(2000)8:5<220::AID-IDOG1022>3.0.CO;2-I. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Goldstein EJ, Tyrrell KL, Citron DM. Lactobacillus species: Taxonomic complexity and controversial susceptibilities. Clin Infect Dis. 2015;60(Suppl 2):S98–S107. doi: 10.1093/cid/civ072. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.