Abstract
Prothymosin‐α (PTMA) is a small, acidic protein that is usually transported into the nucleus and involves many cellular and immunological functions. Previous studies demonstrated that aberrant location of PTMA expression exists in human bladder cancer, but the role of PTMA protein expression remains elusive. In this study, we created ectopic nuclear or cytoplasmic PTMA expression in human bladder cancer cells by infecting lentiviruses carrying wild type or deleted nuclear localization signal of the PTMA gene. The in vivo tumorigenesis assay showed PTMA protein with deleted nuclear localization signal promotes J82 xenograft tumor growth in mice and shortens their survival more so than the wild type. Chromatin immunoprecipitation showed that wild‐type PTMA protein binds to the PTEN promoter and enhances phosphatase and tensin homolog (PTEN) expression. Through immunoblot proteomics and in vivo ubiquitination studies, PTMA protein can bind with tripartite motif‐containing protein 21 (TRIM21) and block its ubiquitination. Also, TRIM21 can downregulate both forms of PTMA protein. In human bladder tumors, loss of nuclear PTMA expression was an unfavorable prognostic indicator for shorter disease‐free survival (hazard ratio, 1.54; P = 0.009). Our data support that nuclear PTMA protein serves as a tumor suppressor in bladder cancer through upregulating PTEN and orchestrating TRIM21 for the regulation of Nrf2 signaling.
Keywords: bladder cancer, nuclear factor erythroid 2‐related factor 2 protein, phosphatase and tensin homolog protein, prothymosin‐α, tripartite motif‐containing protein 21 protein
1. INTRODUCTION
Bladder cancer is the fourth most common male cancer and the ninth female cancer in Western countries and the USA, and the highest incidence rates in men are noted in countries from Southern Europe, North America, Northern Africa and Western Asia.1, 2 In Taiwan, there is an unusually high incidence of bladder cancer in the black‐foot‐disease endemic area located on the southwest coast due to chronic arsenism, 3 and upper tract urothelial carcinoma (UTUC) due to aristolochic acid nephropathy.4 At initial diagnosis, approximately 70%‐80% of bladder cancer cases are non‐muscle invasive. Most of these (~70%) easily recur, and approximately 15% may progress to muscle‐invasive diseases despite endoscopic resection and adjuvant intravesical therapy.5 In contrast, 20%‐30% of bladder cancer cases are muscle‐invasive, advanced or metastatic tumors upon diagnosis. Progression of cancer to metastatic disease is accompanied by an altered antioxidant expression profile, resulting in a net increase in the production of reactive oxygen species (ROS).6 Owing to the limited therapeutic progress in recent decades, choices of radical cystectomy or bladder‐sparing tri‐modal therapy were the mainstream for patients with muscle‐invasive disease. Despite these, half of all patients with advanced disease die of metastatic disease within 5 years. Until recently, several immune checkpoint inhibitors have gained US Food and Drug Administration approval for treating advanced or metastatic urothelial carcinoma,7, 8 there is still 70%‐80% patients presenting with primary, adaptive and acquired resistance to cancer immunotherapy.9 Therefore, it is necessary to explore more regulatory mechanisms involved in human bladder cancer.
Prothymosin‐α (PTMA) (usually isoform 1 or 2) is a small, acidic, nuclear protein that plays a pivotal role in a number of biological activities, including cellular (i.e. apoptosis, proliferation, chromatin decondensation or transportation) or immunological functions (i.e. tumor‐specific T‐lymphocyte response, interleukin‐2 receptor expression or T[H]1‐type immune responses).10, 11, 12, 13, 14, 15 Recently, two new PTMA variants (iso b and pseudogene 7) lack of nuclear localization signal (NLS) were isolated and identified from CD8+ T cells and cervicovaginal lavage with potent anti‐HIV‐1 activity, which were suggested to be innate immune mediators involved in immune surveillance.16, 17, 18, 19, 20 PTMA can induce the import of Kelch‐like ECH‐associated protein 1 (Keap1) into the nucleus to inhibit nuclear factor erythroid 2‐related factor 2 (Nrf2)‐mediated antioxidant response through Nrf2 degradation.21 From our previous studies, we found that not only PTMA could be detected in the nucleus of the tumor cell, but also in the cytoplasm. The loss of nuclear PTMA (cytoplasmic or null expression pattern) predicts shorter progression‐free survival in superficial bladder cancer and UTUC.22, 23 These findings prompted us to explore the molecular and regulatory mechanisms of PTMA expression involved in human urothelial carcinoma.
Phosphatase and tensin homolog (PTEN) tumor suppressor protein is a multifunctional phosphatase that is involved in several cellular signaling pathways.24 PTEN can negatively regulate the phosphatidylinositol 3‐kinase (PI3K) pathway and then work as a tumor suppressor.25, 26 PTEN can also regulate Nrf2 expression through glycogen synthase kinase‐3 (GSK‐3)‐mediated phosphorylation of Nrf2 and subsequent β‐transducin repeat‐containing protein (β‐TrCP)‐dependent, degradation, which is Keap1‐independent.27 PTEN not only executes pivotal functions at the cell membrane, but it also plays some important roles in the nucleus, including acting as a chromosomal stabilizer and collaborating with nuclear p53.28, 29, 30 In human bladder cancer, a somatic mutation of the PTEN gene and a homozygous deletion have been detected.31, 32, 33 Inactivation of PTEN‐related signaling allows T24 cells to be more invasive and urothelial cells to become hyperplasia and urothelial carcinoma in PTEN‐deficient mice.33, 34, 35 Tripartite motif‐containing protein 21(TRIM21) is an E3 ubiquitin‐protein ligase that involves ubiquitination of IKBκB, CDKN1B, IRF3 and IRF8 ubiquitination.36 Recently, TRIM21 was reported to be able to interact with and ubiquitinate p62 and prevent p62 dimerization and Keap1 sequestration, which downregulates the Nrf2 redox pathway.37
In the current study, we demonstrated that the loss of nuclear PTMA is an unfavorable prognostic factor for disease relapse in human bladder cancer, in which nuclear PTMA could enhance PTEN transcription and mRNA expression through binding to the PTEN promoter and interacting with TRIM21, which can downregulate p62 and then Nrf2 expression in human bladder cancer.
2. MATERIALS AND METHODS
2.1. Cells
Seven human cell lines, including immortalized urothelial cell line (SVHUC), bladder cancer cell lines (TSGH8301, T24, J82, TCCSUP, BFTC905 and HT1197), renal pelvis tumor cell line (BFTC909) and prostate cancer cell line (PC3) were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum, 2 mM l‐glutamine and 50 μg/mL gentamicin, except minimum essential medium for HT1197. Once cell growth reached 80%‐90% confluence, cells were trypsinized for subsequent experiments, including total RNA extraction, western blotting and xenograft growing in SCID mice.
2.2. Plasmid construction and lentivirus production
The full‐length fragment and deleted nuclear localization signal (∆NLS) of the human PTMA gene were cloned into the lentiviral vector (pWPXL‐enhancer‐humanWTPTMA and pWPXL‐enhancer‐humanΔNLS PTMA). The human PTMA protein has a bipartite NLS, KKQK (residues 101‐104).38 After co‐transfection of these two plasmids with the pMD2.G vector into 293 cells, the supernatants containing the lentiviruses were collected and stored for future experiments. After that, J82 cells were infected with the lentivirus carrying the indicated gene, and then three J82 transfectants were collected with cell sorting of the existence of green fluorescent protein.
2.3. Analysis of the cDNA differential array
We performed a cDNA differential array using three J82 transfectants, including J82‐wild‐type prothymosin‐α (WTPTMA) expression, J82‐ΔNLSPTMA clones and J82‐control clones following mRNA isolation, amplification and gene expression profiling. The total RNA from each clone was extracted using standard procedures (TRIzol reagent; Invitrogen, Carlsbad, CA, USA). Labeling of cDNA, hybridization and scanning of the microarrays was done according to the manufacturer's protocols (Asia BioInnovations, Taipei, Taiwan) and the previous study.39 To identify affected pathways, these groups of genes were mapped to the MetaCore pathway database (GeneGo, St Joseph, MI, USA). The MetaCore analysis identified the functional link between differentially expressed genes associated with each condition.
2.4. Quantitative reverse transcription polymerase chain reaction (qRT‐PCR), western blot analysis, enzyme‐linked immunosorbent assay (ELISA) and immunoprecipitation assay
Total RNA was extracted using the TRIzol® (Invitrogen) method according to the manufacturer's protocol and then reverse transcribed with high capacity cDNA reverse transcription kits (Applied Biosystems, Waltham, CA, USA). The resulting cDNA was used for PCR in triplicate, and data collection was performed using a Smart Cycler 2 PCR system (Cepheid, Sunnyvale, CA, USA). All samples were amplified simultaneously in duplicate in a single assay run. Primers for human PTMA and PTEN are shown in Table S1. The −ΔΔC t method40 was utilized to measure relative changes in mRNA levels examined by qRT‐PCR experiments, after normalizing transcript levels of each gene by levels of β‐actin as internal controls.
Western blotting and immunoprecipitation assays were performed as per our previous studies.41 Protein (30 μg) from each sample was subjected to sodium dodecyl sulfate‐polyacrylamide gel electrophoresis (SDS‐PAGE), transferred onto nitrocellulose membrane filter, and subsequently immunoblotted with anti‐human PTMA (4F4, ALX‐804‐487; 2F11, ALX‐804‐486; Alexis, Lausen, Switzerland), anti‐human TRIM21 (GTX113554, Genetex, Irvine, CA, USA), anti‐human PTEN (6H2.1, #04‐035; Millipore, Burlington, MA, USA), anti‐human Nrf2 (C‐20, sc‐722; Santa Cruz Biotechnology, Santa Cruz, CA, USA), anti‐human Keap1 (E‐20, sc‐15246, Santa Cruz), anti‐human p62 (D‐3, sc‐28359, Santa Cruz), anti‐Ubiquitin (FL‐76, sc‐9133; Santa Cruz Biotechnology), anti‐superoxide dismutase‐2 (GTX116093; GeneTex) and anti‐heme oxygenase‐1 (GTX101147; Genetex). β‐Actin protein served as an internal control. The content of vascular endothelial growth factor (VEGF) was measured with a VEGF ELISA kit (R&D Systems, Minneapolis, MI, USA) with total protein normalization.
The interaction between the protein and PTMA was investigated by performing proteomics for PTMA‐immunoblotted protein. In brief, total lysate from BFTC‐905 cells was immunoblotted with PTMA (clone 2F11), and with immunoglobulin (Ig)G as a control. The obtained band (~17 kDa) was analyzed with the liquid chromatography‐mass spectrometry/mass spectrometry (LC‐MS/MS) assay. The result was obtained from the Mascot search website.
2.5. In vivo ubiquitination assay
This assay was similar to that in our previous study.42 In brief, input fractions as indicated were prepared using RIPA buffer. His‐tagged protein was pulled down using Dynabeads® His‐Tag Isolation, Pulldown kit (#10103D; Life Technologies, Carlsbad, CA, USA). Bound material was eluted from the beads, collected and subjected to SDS‐PAGE and western blot analyses.
2.6. In vivo tumor growth study
Female (NOD‐SCID, 6‐8 weeks old) mice were s.c. injected with 1 × 106 J82 transfectants as indicated in 100 μL serum‐free medium. Tumor formation and growth were recorded every other day for at least 60 days according to the “Guidelines for the Welfare of Animals in Experimental Neoplasia” (1998). Tumor volumes were calculated using the formula: length × (width)2 × 0.45. Once tumor burden grows more than 2500 mm3, it is viewed as death from tumor. At the end of this experiment, tumors were harvested for PTMA immunostaining.
2.7. PTEN promoter cloning and reporter analysis
To clone PTEN promoter for the PTEN promoter activity analysis, we used PCR to amplify DNA fragments containing PTEN promoter genomic sequences from J82 cell genomic DNA. The primers used were PTEN_‐1344/Kpn I (5′‐ATGGTACCCTCGAGTCAGTGACACTGCTC‐3′) and PTEN_‐821/XhoI (5′‐AACTCGAGCGCCGCCGTCTCTCATCT‐3′) and derived from human genomic PTEN from ‐821 to ‐1344 according to GenBank accession number AF067844. Amplified DNA fragments were subcloned into the luciferase reporter vector pGL3 (Promega, Madison, WI, USA) according to the standard procedures. The in‐frame integrity of the construct was confirmed by DNA sequencing. Cells were transfected using Lipofectamine (GIBCO BRL, Gaithersburg, MD, USA) in the presence of trace amounts of pCMV‐β‐gal. Luciferase assays were performed using the Luciferase Assay System (Promega), and activities were normalized to β‐galactosidase activity.
2.8. Chromatin immunoprecipitation (ChIP) assay
Cells lysates were prepared from three J82 transfectants. In brief, the specific antibodies used for immunoprecipitations were the anti‐PTMA antibody (2F11; Alexis Biochemicals, San Diego, CA, USA) and a control antibody (rabbit IgG). After protein‐DNA cross‐links in the immunoprecipitates were reversed, the purified DNA was analyzed by PCR (35 cycles; 30 seconds at 95°C, 45 seconds at 58°C, 60 seconds at 72°C) with primers that detect sequences containing PTEN promoter (nucleotide number −523 to transcription starting site). The PCR products were visualized on an ethidium bromide gel.
2.9. Patient population and study samples
The study was undertaken with the approval and institutional oversight of the Institutional Review Board for the Protection of Human Subjects at both Chia‐Yi Christian Hospital (IRB‐101014) and National Cheng Kung University Hospital (ER‐95‐49). The tumor specimens from 151 bladder cancer patients were retrospectively retrieved, as well as two‐tissue microarray. All the patients were managed according to the bladder cancer treatment guideline, modified from NCCN Clinical Practice Guidelines in Oncology.
2.10. Immunohistochemistry (IHC)
Serial 5‐μm sections were cut for either hematoxylin‐eosin staining or IHC as performed in our previous study.41 A total of 151 tumor specimens were collected for PTMA immunostaining and one tissue microarray of 60 patients for both PTMA and TRIM21 immunostaining. In brief, after deparaffinization and rehydration, tissue sections were autoclaved in citrate buffer (pH 6.0) at 120°C for 15 minutes and sequentially treated with 3% H2O2 in methanol for 25 minutes, then with skimmed milk in phosphate‐buffered saline for 30 minutes. Non‐specific background staining was minimized further by pre‐incubating the array with 0.3% bovine serum albumin in 0.1 M Tris‐buffered saline, pH 8.0, for 1 hour. Slides were incubated in the primary anti‐human PTMA (2F11; Alexis Biochemicals) for 45 minutes and anti‐human TRIM21 (GTX113554, Gentex; dilution, 1:200) for 1 hour. We performed parallel staining without primary antibody as a negative control and human spleen sections as a positive control. After incubation with secondary antibodies for 1 hour, immunostaining was developed with a BioGenex Super Sensitive Polymer HRP IHC System kit (Biogenex Laboratories, Fremont, CA, USA) and then counterstained with hematoxylin. Samples were analyzed blindly by one pathologist. Any tumor specimens exhibiting more than 25% immunoreactive tumor cells within one high‐power field were considered positive tumors as in our previous study.22
2.11. Statistical analysis
The disease relapse analyzed in the study was the first event of tumor recurrence in the urinary bladder or tumor progression beyond the bladder. Disease‐free survival was calculated from the time of surgery to the time of the first documented disease relapse. Statistical analysis was performed using the Statistical Package for Social Sciences, version 12.0, software. The relationships between the PTMA expression sites and clinicopathological factors were analyzed with Kaplan‐Meier plots, the log‐rank test and the multivariate Cox regression model. All P‐values reported were two‐sided and considered significant if P < 0.05.
3. RESULTS
3.1. ∆NLSPTMA protein expression promotes J82 xenograft tumor growth and shortens survival in mice
To investigate where PTMA is detected in human bladder cancer cells, PTMA expression was first examined in several bladder cancer cells, using RT‐PCR (Figure 1A), western blotting (Figure 1B) or immunohistochemical staining of xenograft sections harvested from SCID mice (Figure 1C‐E). Although PTMA is a nuclear protein, the results show that PTMA can be found not only in the nuclei but also in the cytoplasm. Immunohistochemical staining showed various expression patterns according to the subcellular localization of PTMA protein, such as low or minimal nuclear PTMA expression in J82 cells, nuclear expression in TCCSUP cells, and both nuclear and cytoplasmic PTMA expression in BFTC905 cells (Figure 1C‐E).
Figure 1.
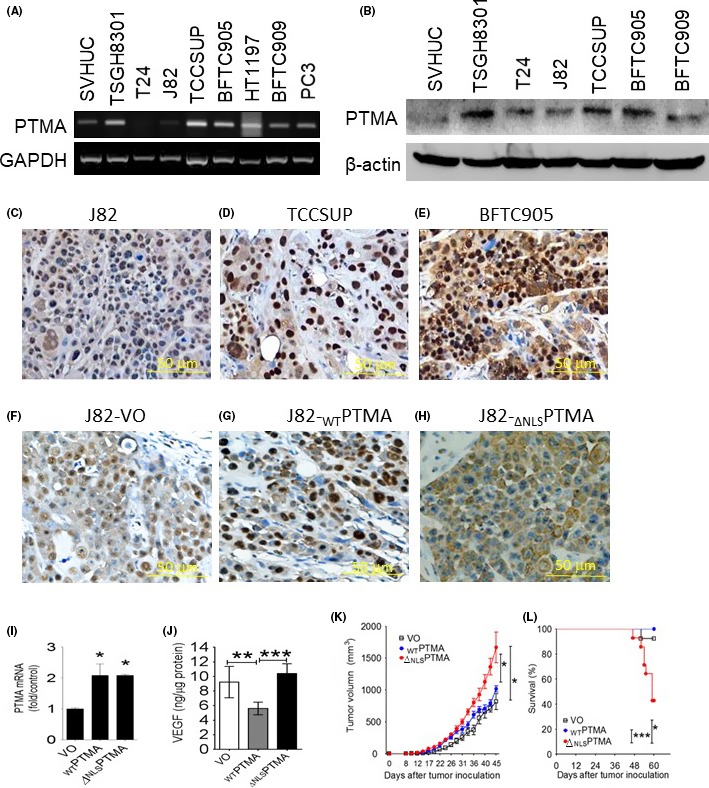
∆ NLSPTMA protein expression promotes J82 xenograft tumor growth in mice and shortens mice survival. A, RT‐PCR assay for PTMA mRNA expression in human bladder cancer cell lines and immortalized urothelial cells SVHUC. B, Western blotting analysis for PTMA protein expression. C‐E, Immunohistochemical PTMA staining showed minimal or null expression in J82 cells (C), strong nuclear expression in TCCSUP (D) and mixed nuclear and cytoplasmic expression in BFTC905 cells (E) (scale bar = 50 μm, ×320). F‐H, Immunohistochemical PTMA staining for xenograft of three J82 cell transfectant, J82‐VO (F), J82‐WTPTMA (G) and J82‐∆ NLSPTMA (H) (scale bar = 50 μm, ×320). I, Quantitative RT‐PCR assay for PTMA mRNA expression, and J, ELISA for VEGF content in three J82 cell transfectants. K, Tumor growth curves of three J82 transfectant xenografts in SCID mice, and L, mice survival. ELISA, enzyme‐linked immunosorbent assay; PTMA, prothymosin‐α; RT‐PCR, reverse transcription polymerase chain reaction; VEGF, vascular endothelial growth factor; VO, vector only; WT, wild type; ∆NLS, deletion of nuclear localization signal. *P < 0.05; **P < 0.01; ***P < 0.001
To investigate whether localization of PTMA expression can influence tumor cell growth, J82 transfectants with either ectopic WTPTMA (wild‐type, or full‐length, nuclear pattern), ∆NLSPTMA (deleted nuclear localization signal, cytoplasmic pattern) expression or control were generated by infecting J82 cells with lentiviruses carrying the full‐length PTMA gene, ∆NLS PTMA gene or control, and then cell sorting. In addition to urea‐PAGE assay for WTPTMA or ∆NLSPTMA protein expression shown in the previous study,22 all three transfectants were confirmed by immunohistochemical staining for subcellular localization of PTMA expression in the xenografts of SCID mice (Figure 1F‐H), as well as examining with qRT‐PCR for mRNA levels (Figure 1I), mimicking nuclear, cytoplasmic or null PTMA expression. Our previous study has shown that J82 cells with ectopic WTPTMA expression exhibit higher growth rate and secrete less transforming growth factor (TGF)‐β1 than do those with ∆NLSPTMA expression or control cells,22 as well as less VEGF production in the current study (Figure 1J). In addition, ∆NLSPTMA expression can promote J82 xenograft growth in SCID mice and shorten mice survival as compared with WTPTMA or null PTMA expression (Figure 1K,L).
3.2. WTPTMA protein binds to the PTEN promoter and enhances PTEN mRNA and protein expression
To further explore what kind of signaling pathway PTMA protein influences, the cDNA differential array was done with these three J82 transfectants and further analyzed by utilizing the online Metacore® software. The result showed that the top 10 signaling pathways influenced by PTMA protein include TGF‐β1/smad‐related EMT, PIP3‐Akt, GnRH, estrogen receptor and interferon (IFN)‐γ signaling (Table 1).
Table 1.
Top 10 postulated molecular pathways influenced by ectopic expression of wild‐type or ∆NLS prothymosin‐α protein in human bladder cancer J82 cells
| No. | Name | Min (P) | Network objects |
|---|---|---|---|
| 1 | TGF‐β‐dependent induction of EMT via MAPK | 1.65E‐10 | 24/47 |
| 2 | PIP3 signaling in cardiac myocytes | 1.05E‐09 | 24/47 |
| 3 | Regulation of EMT | 1.08E‐09 | 31/64 |
| 4 | Cell cycle‐ESR1 regulation of G1/S transition | 3.78E‐09 | 18/33 |
| 5 | HGF signaling pathway | 8.13E‐09 | 22/47 |
| 6 | TGF‐β‐dependent induction of EMT via SMAD | 9.52E‐09 | 20/35 |
| 7 | GnRH signaling | 1.07E‐08 | 27/72 |
| 8 | AKT signaling | 1.20E‐08 | 21/43 |
| 9 | TGF, WNT and cytoskeletal remodeling | 1.73E‐08 | 42/111 |
| 10 | IFN gamma signaling pathway | 1.73E‐08 | 27/54 |
AKT, Akt serine/threonine kinase; EMT, epithelial‐to‐mesenchymal transition; ESR1, estrogen receptor 1; GnRH, gonadotropin‐releasing hormone; HGF, hepatic growth factor; IFN, interferon; MAPK, mitogen‐activated protein kinase; PIP3, phosphatidylinositol 3,4,5 trisphosphate; SMAD, homologs of both proteins, mothers against decapentaplegic and SMA; TGF‐β, transforming growth factor‐β; WNT, Wnt/β‐catenin signaling pathway
To validate the findings obtained from the cDNA differential array further, we examined how PTMA protein influences PTEN expression in several cancer cell lines. We found that WTPTMA protein can increase PTEN protein expression in J82, and BFTC909 cancer cells, but ∆NLSPTMA protein expression cannot (Figure 2A,B). In contrast, PTEN expression was decreased in TCCSUP cells while knocking down PTMA expression (Figure 2C). Further, using qRT‐PCR and promoter assays, we found that J82 cells with ectopic WTPTMA expression exhibit both higher PTEN mRNA and PTEN promoter activity than those with ∆NLSPTMA protein expression or control cells (Figure 2D,E). The ChIP assay demonstrated that WTPTMA protein could bind to PTEN promoter region between ‐500 bp and transcriptional start site while ∆NLSPTMA protein cannot (Figure 2F).
Figure 2.
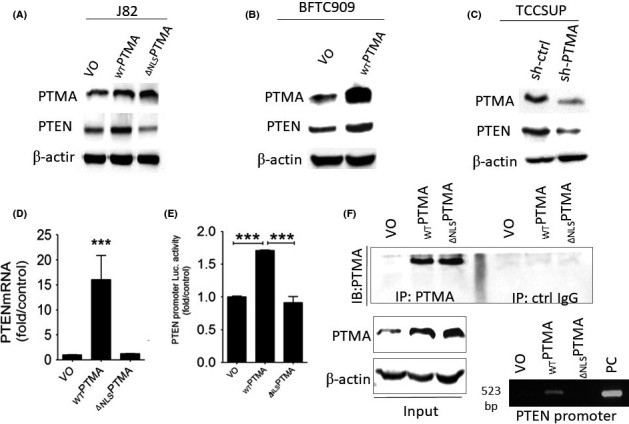
WTPTMA protein binds to PTEN promoter and enhances PTEN mRNA and protein expression. Western blotting analysis for PTMA, and PTEN in: A, J82 cells; B, BFTC909 cells; C, TCCSUP cells with the indicated transfection. D, qRT‐PCR analysis for PTEN mRNA expression and E, Luciferase assays for PTEN promoter activity in three J82 cells with indicated PTMA expression. F, Chromatin immunoprecipitation (ChIP) assay for the interaction with WTPTMA or ∆ NLSPTMA protein. PTEN, phosphatase and tensin homolog; PTMA, prothymosin‐α; qRT‐PCR, quantitative reverse transcription polymerase chain reaction; VO, vector only; WT, wild type; ∆NLS, deletion of nuclear localization signal. *P < 0.05; **P < 0.01; ***P < 0.001
3.3. PTMA binds and orchestrates with TRIM21 to regulate Nrf2 expression through p62/Keap1 signaling
To explore what kind of proteins PTMA binds to, we analyzed PTMA‐bound proteins immunoprecipitated from BFTC905 cells by utilizing the LC‐MS/MS assay. The result demonstrated that PTMA could bind to some cytoskeletal, nuclear proteins, TRIM21 (an E3‐ubiquitin ligase) and others (Figure S1). To examine the interaction between PTMA and TRIM21 protein and its associated molecular signaling, we first confirmed the physical interaction of PTMA with TRIM21 protein using an immunoprecipitation study in BFTC905 cells (Figure 3A). Also, we can detect TRIM21 protein expression in several human bladder cancer cells, as well as TRIM21‐regulated Nrf2 expression (Figure 3B). By using imaging density analysis, we found a trend of negative correlation between PTMA and Nrf2 protein (P = 0.13). Because nuclear PTMA protein can promote nuclear transportation of Keap1 and degrade Nrf2 protein, we found that WTPTMA protein expression can downregulate p62 and Nrf2 expression in a dose‐dependent manner, while either knocking down PTMA expression or increasing ∆NLSPTMA protein expression can upregulate Nrf2 expression (Figure 3C‐F). Furthermore, both WTPTMA and ∆NLSPTMA can bind with TRIM21 and regulate Keap1 and Nrf2 expression differentially (Figure S2). Furthermore, Nrf2 target protein heme oxygenase‐1 is downregulated in WTPTMA‐expressing J82 transfectant and upregulated in ∆NLSPTMA‐expressing one. In terms of another target gene superoxide dismutase‐2 (SOD2), SOD2 protein is minimally decreased in WTPTMA‐expressing J82 transfectant and significantly upregulated in ∆NLSPTMA‐expressing one (Figure 3G). Taken together, our data demonstrated PTMA can orchestrate with TRIM21 to regulate p62/Keap1/Nrf2 expression according to the presence or absence of NLS.
Figure 3.
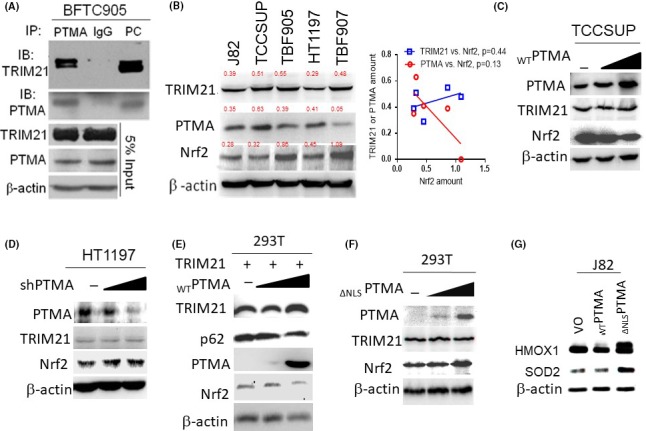
PTMA binds and orchestrates with TRIM21 to regulate Nrf2 expression through p62/Keap1 signaling. A, Immunoprecipitation study for the interaction between endogenous TRIM21 and PTMA. Total protein lysate from BFTC905 cells was immunoprecipitated either with PTMA or IgG (as a control), then immunoblotted with TRIM21. B, Western blotting for TRIM21, PTMA, and Nrf2 in several bladder cancer cells. Numeric in red indicates the ratio of protein of interest‐to‐β‐actin. C‐F, Western blotting for TRIM21 and Nrf2 expression while knocking down or overexpression of the PTMA gene in the indicated cells. G, Western blotting for heme oxygenase‐1 (HMOX1) and superoxide dismutase‐2 (SOD2) expression in J82 cells with ectopic expression of WTPTMA and ∆ NLSPTMA. IgG, immunoglobulin G; Keap1, Kelch‐like ECH‐associated protein 1; Nrf2, nuclear factor erythroid 2‐related factor 2; PTMA, prothymosin‐α; TRIM21, tripartite motif‐containing protein 21
3.4. Both TRIM21 and PTMA influence each other
Because TRIM21 is an E3 ubiquitin ligase, we further investigate whether PTMA can be regulated by TRIM21 through proteasomal ubiquitination. We found that TRIM21 can reduce both WTPTMA and ∆NLSPTMA protein expression as compared with the control Foxp3 protein, in which the ∆NLSPTMA protein is downregulated more than the WTPTMA protein (Figure 4A). The in vivo ubiquitination assay failed to show PTMA ubiquitination. In contrast, we found that both WTPTMA and ∆NLSPTMA can block TRIM21 ubiquitination (Figure 4B,C). Also, WTPTMA can promote Nrf2 ubiquitination, which can be blocked by TRIM21 overexpression (Figure 4B).
Figure 4.
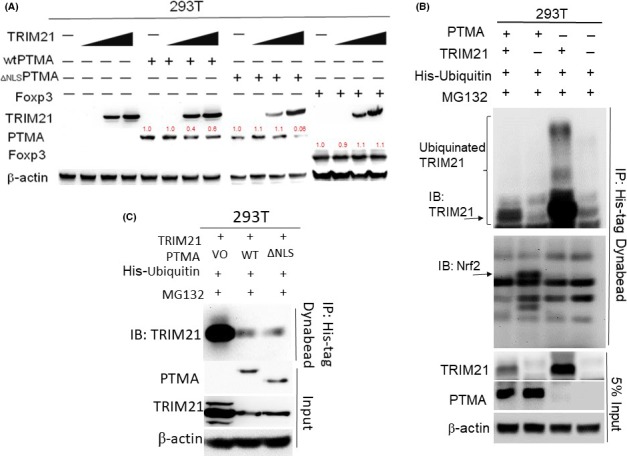
TRIM21 and PTMA influence each other. A, Western blotting studies for TRIM21, PTMA, Foxp3 expression as indicated by each condition. B, In vivo ubiquitination studies for ectopic expression of either WTPTMA, TRIM21, or both in 293 T cells. C, In vivo ubiquitination studies for the effect of ectopic expression of either WTPTMA or Δ NLSPTMA on TRIM21 ubiquitination. Immunoprecipitation with His‐tagged Nickle magnetic bead. PTMA, prothymosin‐α; TRIM21, tripartite motif‐containing protein 21
3.5. Nuclear PTMA expression predicts a favorable outcome in human bladder cancer that is associated with higher PTEN mRNA expression
A total of 151 human bladder cancer tissues were determined for PTMA immunoreactivity using immunohistochemical staining. PTMA expression can be visualized in the tumor cell nucleus, cytoplasm, both or neither (Figure 5A‐D). The clinicopathological correlates of PTMA expression in human bladder tumors is shown in Table S2 according to the subcellular localization of the PTMA protein. Among these clinicopathological factors, tumor morphology, multiplicity, staging and tumor grade were significantly associated with the PTMA expression pattern (P = 0.046, 0.039, 0.014 and 0.013, respectively; Figure 5E and Table S2). Log‐rank analysis demonstrated that nuclear PTMA is a favorable prognostic factor for disease‐free survival (Figure 5F). PTEN mRNA is significantly higher in tumors with nuclear PTMA expression than in those with cytoplasmic or null PTMA expression (Figure 5G).
Figure 5.
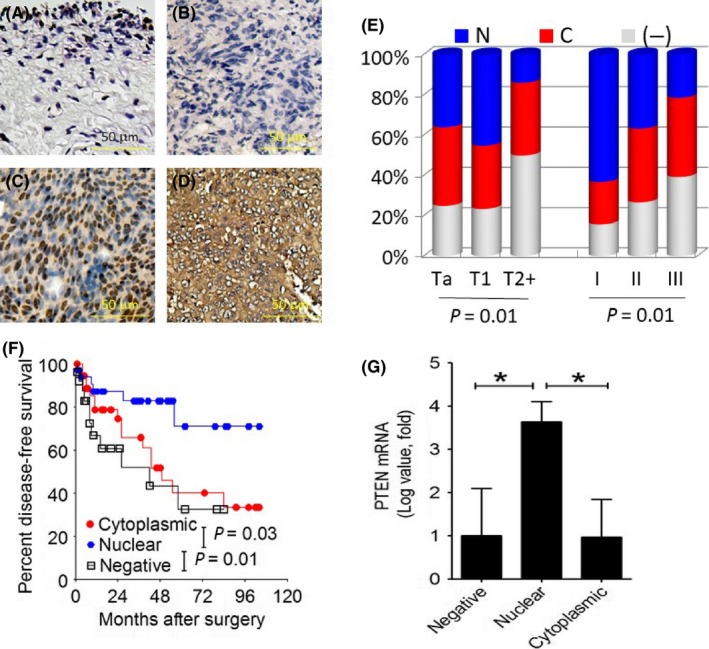
Nuclear PTMA expression predicts a favorable outcome in human bladder cancer that is associated with higher PTEN mRNA expression. A‐D, Immunohistochemical staining for PTMA expression, normal adjacent mucosa (A), negative PTMA expression (B), nuclear PTMA expression (C) and cytoplasmic PTMA expression (D) (scale bar = 50 μm, ×320). E, The association of PTMA expression pattern with tumor stage or grade. F, Kaplan‐Meier curve for disease‐free survival according to the PTMA expression pattern. G, PTEN qRT‐PCR assay using human bladder tumor specimen. N denotes nuclear; C, cytoplasmic; (−), negative PTMA expression. PTEN, phosphatase and tensin homolog; PTMA, prothymosin‐α; qRT‐PCR, quantitative reverse transcription polymerase chain reaction
3.6. Increased TRIM21 expression is associated with PTMA expression in human bladder cancer
Two serial sections of tissue microarrays containing 60 bladder tumors and 22 non‐cancerous tissues were immunostained with TRIM21 and PTMA (Figure 6A‐H). The results revealed that bladder tumors exhibit increased TRIM21 expression as compared with non‐cancerous tissues (P = 0.002) (Figure 6I). There is a weakly positive correlation between TRIM21 and PTMA expression in human bladder cancer, regardless of nuclear or cytoplasmic location (Figure 6J).
Figure 6.
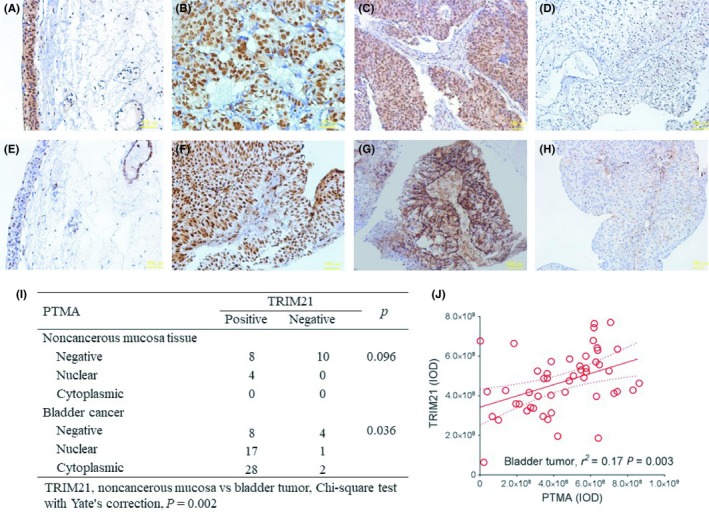
Increased TRIM21 expression is associated with PTMA expression in human bladder cancer. A‐H, Two serial sections of tissue microarrays containing 60 bladder tumors and 22 non‐cancerous tissues were immunostained with TRIM21 (A‐D) and PTMA (E‐H). Normal adjacent mucosa (A and E, scale bar = 100 μm, ×320), positive TRIM21 expression (B, scale bar = 100 μm, ×320; C, scale bar = 100 μm, ×200), nuclear PTMA expression (F, scale bar = 100 μm, ×320), cytoplasmic PTMA expression (G, scale bar = 100 μm, ×200) and negative TRIM21 or PTMA expression (D and H, scale bar = 100 μm, ×200). I, Distribution of TRIM21 expression according to the PTMA expression pattern. J, The correlation between TRIM21 and PTMA expression in terms of integrated optical density (IOD) in human bladder cancer. PTMA, prothymosin‐α; TRIM21, tripartite motif‐containing protein 21
4. DISCUSSION
In the current study, we found that cytoplasmic PTMA expression or null PTMA expression is a poor prognostic factor for disease‐free survival in bladder cancer, which is similar to the findings from the J82 cell model. Ectopic expression of WTPTMA (usually in the nucleus due to the presence of NLS) could enhance PTEN mRNA and protein expression through binding to the PTEN promoter directly, while ectopic expression of ∆NLSPTMA cannot. Nuclear PTMA‐expressing bladder tumors exhibited higher PTEN mRNA expression than did those that lacked nuclear PTMA expression (cytoplasmic PTMA or null PTMA expression). On the other hand, both WTPTMA and ∆NLSPTMA can bind to and be downregulated by an E3 ubiquitin ligase TRIM21 that can ubiquitinate p62, thereby preventing Keap1 from sequestration resulting in Nrf2 degradation.37 Also, Nrf2 expression can be both down‐ and upregulated by WTPTMA or ∆NLSPTMA protein in a TRIM21‐independent manner, respectively. Taken together, nuclear PTMA protein increases PTEN expression transcriptionally and orchestrates with TRIM21 to regulate Keap1/Nrf2 signaling in human bladder cancer. It provides direction for PTMA‐targeted therapy in human bladder cancer.
Currently, several PTMA variants have been reported, including isoforms 1 and 2, isoform B and pseudogene 7. The former two are widespread, acidic, NLS‐containing, nuclear proteins and are able to both stimulate cell proliferation and induce type I IFN immunomodulatory activity. In contrast, the latter two, detected in CD8+ T cells and cervicovaginal lavage, also exhibit anti‐HIV immunomodulatory activity despite the lack of NLS.17 Our previous study has shown WTPTMA expression can promote in vitro J82 cell proliferation than did ∆NLSPTMA or null expression,22 which is inconsistent with the in vivo J82 xenograft tumor growth shown in the current study. The probable reasons for this discrepancy can be obtained from the findings of the comparison of mRNA array using three J82 transfectants, demonstrating that the presence of NLS, in terms of PTMA protein, influences several signaling pathways, such as cell cycle regulation, PIP3‐Akt, GnRH, TGF‐β/smad, hepatic growth factor, estrogen receptor and IFN‐γ signaling. In addition, given that nuclear PTMA can inhibit TGF‐β signaling,22, 43 we further demonstrated that nuclear PTMA expression could enhance PTEN mRNA and protein expression, and decreased Nrf2 and its downstream gene expression in human bladder cancer. Taken together, our data again support that nuclear PTMA expression in human bladder cancer is a favorable prognostic factor that is associated with enhanced PTEN expression.
In human bladder cancer, alteration of the PI3K pathway is frequent, including PIK3CA, PTEN, TSC1, RHEB and LKB1 genes. As for the PTEN gene, 49% of tumors had reduced expression, including 12% loss of heterozygosity of the PTEN region and 1.4% with homozygous deletion. Reduced PTEN expression is significantly associated with tumor grade, but not with stage.44 Moreover, tumors simultaneously having altered p53 expression are associated with poor survival in human bladder cancer, in which inactivation of p53 and PTEN genes promotes invasive bladder cancer.35 There are a variety of epigenetic, transcriptional and post‐transcriptional mechanisms that regulate PTEN expression. Like several molecules (i.e. early growth response protein‐1, peroxisome proliferator‐activated receptor‐γ and p53),45 WTPTMA can directly bind to the PTEN promoter region which enhances PTEN gene expression, while ∆NLSPTMA cannot.
Tripartite motif‐containing protein 21 is a member of the tripartite motif (TRIM) family that includes three zinc‐binding domains, a RING finger domain, two B‐box zinc finger (types 1 and 2), and a coiled coil region. TRIM21 is an E3 ubiquitin ligase and plays an important role in the intracellular antibody‐mediated proteolysis pathway that forms a ubiquitin ligase complex with E2 enzyme UBE2D2 that is used not only for the ubiquitination of USP4 and IKBκB but also for its self‐ubiquitination.46 TRIM21 can monoubiquitinate IKBκB that will negatively regulate both nuclear factor‐κB signaling and IFN‐β production, and involve the ubiquitination of IRF3 and IRF8 to stimulate the transcription of cytokine genes in macrophages.36 Recently, TRIM21 was reported to be able to interact with and ubiquitylate p62 that prevent p62 dimerization and Keap1 sequestration function, resulting in Nrf2 degradation.37 In the current study, we found that ∆NLSPTMA could be more downregulated by TRIM21 than can WTPTMA, implying that TRIM21 may play an important role in the subcellular distribution of PTMA in human bladder cancer cells. Unfortunately, we failed to show PTMA ubiquitination. Nevertheless, we found that both WTPTMA and ∆NLSPTMA can block TRIM21 ubiquitination and TRIM21 can reduce PTMA‐induced ubiquitinated Nrf2. Taken together, PTMA expression may play a pivotal role in the regulation of Nrf2 signaling not only through direct binding with Keap1, but also through PTEN and TRIM21 expression.
The transcription factor Nrf2 (also called NFE2L2) regulates various critical cellular processes, including cellular antioxidant response, cellular detoxification and drug uptake/efflux. In human invasive bladder cancer, 11 tumors (8%) had deleterious missense mutations in the Neh2 domain of the Nrf2 gene, which showed markedly increased expression of genes involved in genotoxic metabolism and the ROS response.47 In terms of the role of Nrf2, some inconsistencies exist in bladder cancer. Hayden et al48 reported that Nrf2 expression in cell models contributed to resistance to cisplatin in bladder cancer. In contrast, Iida et al49 reported that Nrf2 and p53 cooperatively protect against N‐butyl‐N‐(4‐hydroxybutyl)‐nitrosamine‐induced urinary bladder carcinogenesis. Nrf2 has been reported to be regulated by PTEN‐mediated phosphorylation, PTMA/Keap1‐mediated degradation. In the current study, we demonstrated that Keap1/Nrf2 signaling could be regulated inversely by PTMA according to the presence or absence of NLS.
There were some limitations in the current study. First, it is not easy to do external validation with other series because subcellular localization of the PTMA protein could not be identified based on mRNA or protein levels. Second, J82 transfected with ∆NLSPTMA expression may not exactly represent a cytoplasmic PTMA‐expressing tumor, because the C‐terminal NLS may exert some immunological function.50 Third, because TRIM21, PTEN and PTMA can regulate Nrf2 expression in the in vitro cell model in the current study, it is difficult to identify which one is the key factor that influences Nrf2 expression in human bladder cancer.
In conclusion, we demonstrated in the current study that PTMA protein could be expressed not only in the cell cytoplasm but also in the cell nuclei of human bladder cancer cells, in which the presence of nuclear PTMA expression is a favorable prognostic factor for disease‐free survival in bladder cancer. Also, WTPTMA protein but not ∆NLSPTMA can bind to the PTEN promoter and enhance PTEN expression, which is compatible with the effect of nuclear PTMA expression in human bladder cancer on PTEN expression. Moreover, an E3 ubiquitin ligase TRIM21 can downregulate the ∆NLSPTMA more than the WTPTMA protein, which can influence Nrf2 expression differentially. In human bladder cancer, TRIM21 is overexpressed together with PTMA expression. Collectively, PTMA can enhance PTEN expression through increasing PTEN promoter activity and orchestrate with TRIM21 to regulate Keap1/Nrf2 expression in human bladder cancer, which can provide a direction for Nrf2‐targeted therapy in human bladder cancer.
CONFLICTS OF INTEREST
Authors declare no conflict of interest.
Supporting information
ACKNOWLEDGMENTS
This study is supported by a Ministry of Science and Technology grant (MOST 106‐2314‐B‐006‐047‐MY3). Pathway analyses and data mining were done using the system provided by the Bioinformatics Core for Genomic Medicine and Biotechnology Development at the National Cheng‐Kung University, supported by a National Science Council grant (NSC 97‐3112‐B‐006‐011).
Tsai Y‐S, Jou Y‐C, Tsai H‐T, Shiau A‐L, Wu C‐L, Tzai T‐S. Prothymosin‐α enhances phosphatase and tensin homolog expression and binds with tripartite motif‐containing protein 21 to regulate Kelch‐like ECH‐associated protein 1/nuclear factor erythroid 2‐related factor 2 signaling in human bladder cancer. Cancer Sci. 2019;110:1208–1219. 10.1111/cas.13963
Drs Tsai and Jou contributed equally to this work.
REFERENCES
- 1. Antoni S, Ferlay J, Soerjomataram I, et al. Bladder cancer incidence and mortality: a global overview and recent trends. Eur Urol. 2017;71:96‐108. [DOI] [PubMed] [Google Scholar]
- 2. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017. CA Cancer J Clin. 2017;67:7‐30. [DOI] [PubMed] [Google Scholar]
- 3. Chen CJ, Chuang YC, Lin TM, et al. Malignant neoplasms among residents of a blackfoot disease‐endemic area in Taiwan: high‐arsenic artesian well water and cancers. Cancer Res. 1985;45:5895‐5899. [PubMed] [Google Scholar]
- 4. Chen CH, Dickman KG, Moriya M, et al. Aristolochic acid‐associated urothelial cancer in Taiwan. Proc Natl Acad Sci USA. 2012;109:8241‐8246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Gerharz EW, Mansson A, Hunt S, et al. Quality of life after cystectomy and urinary diversion: an evidence based analysis. J Urol. 2005;174:1729‐1736. [DOI] [PubMed] [Google Scholar]
- 6. Hempel N, Ye H, Abessi B, Mian B, Melendez J. Altered redox status accompanies progression to metastatic human bladder cancer. Free Radic Biol. 2009;46:42‐50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Alfred Witjes J, Lebret T, Comperat EM, et al. Updated 2016 EAU guidelines on muscle‐invasive and metastatic bladder cancer. Eur Urol. 2017;71:462‐475. [DOI] [PubMed] [Google Scholar]
- 8. Sharma P, Retz M, Siefker‐Radtke A, et al. Nivolumab in metastatic urothelial carcinoma after platinum therapy (CheckMate 275): a multicentre, single‐arm, phase 2 trial. Lancet Oncol. 2017;18:312‐322. [DOI] [PubMed] [Google Scholar]
- 9. Sharma P, Hu‐Lieskovan S, Wargo JA, et al. Primary, adaptive, and acquired resistance to cancer immunotherapy. Cell. 2017;168:707‐723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Letsas KP, Frangou‐Lazaridis M. Surfing on prothymosin alpha proliferation and anti‐apoptotic properties. Neoplasma. 2006;53:92‐96. [PubMed] [Google Scholar]
- 11. George EM, Brown DT. Prothymosin alpha is a component of a linker histone chaperone. FEBS Lett. 2010;584:2833‐2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gomez‐Marquez J. Function of prothymosin alpha in chromatin decondensation and expression of thymosin beta‐4 linked to angiogenesis and synaptic plasticity. Ann N Y Acad Sci. 2007;1112:201‐209. [DOI] [PubMed] [Google Scholar]
- 13. Ioannou K, Derhovanessian E, Tsakiri E, et al. Prothymosin alpha and a prothymosin alpha‐derived peptide enhance T(H)1‐type immune responses against defined HER‐2/neu epitopes. BMC Immunol. 2013;14:43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Cordero OJ, Sarandeses CS, Lopez JL, et al. Prothymosin alpha enhances interleukin 2 receptor expression in normal human T‐lymphocytes. Int J Immunopharmacol. 1991;13:1059‐1065. [DOI] [PubMed] [Google Scholar]
- 15. Baxevanis CN, Gritzapis AD, Spanakos G, et al. Induction of tumor‐specific T lymphocyte responses in vivo by prothymosin alpha. Cancer Immunol Immunother. 1995;40:410‐418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Teixeira A, Yen B, Gusella GL, et al. Prothymosin alpha variants isolated from CD8+ T cells and cervicovaginal fluid suppress HIV‐1 replication through type I interferon induction. J Infect Dis. 2015;211:1467‐1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gusella GL, Teixeira A, Aberg J, et al. Prothymosin‐alpha variants elicit anti‐HIV‐1 response via TLR4 dependent and independent pathways. PLoS ONE. 2016;11:e0156486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Mosoian A, Teixeira A, Burns CS, et al. Prothymosin‐alpha inhibits HIV‐1 via Toll‐like receptor 4‐mediated type I interferon induction. Proc Natl Acad Sci USA. 2010;107:10178‐10183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Mosoian A, Teixeira A, Burns CS, et al. Influence of prothymosin‐alpha on HIV‐1 target cells. Ann N Y Acad Sci. 2007;1112:269‐285. [DOI] [PubMed] [Google Scholar]
- 20. Mosoian A, Teixeira A, High AA, et al. Novel function of prothymosin alpha as a potent inhibitor of human immunodeficiency virus type 1 gene expression in primary macrophages. J Virol. 2006;80:9200‐9206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Khan H, Cino EA, Brickenden A, et al. Fuzzy complex formation between the intrinsically disordered prothymosin alpha and the Kelch domain of Keap1 involved in the oxidative stress response. J Mol Biol. 2013;425:1011‐1027. [DOI] [PubMed] [Google Scholar]
- 22. Tsai YS, Jou YC, Tung CL, et al. Loss of nuclear prothymosin‐alpha expression is associated with disease progression in human superficial bladder cancer. Virchows Arch. 2014;464:717‐724. [DOI] [PubMed] [Google Scholar]
- 23. Jou YC, Tung CL, Tsai YS, et al. Prognostic relevance of prothymosin‐alpha expression in human upper urinary tract transitional cell carcinoma. Urology. 2009;74:951‐957. [DOI] [PubMed] [Google Scholar]
- 24. Maehama T, Dixon JE. The tumor suppressor, PTEN/MMAC1, dephosphorylates the lipid second messenger, phosphatidylinositol 3,4,5‐trisphosphate. J Biol Chem. 1998;273:13375‐13378. [DOI] [PubMed] [Google Scholar]
- 25. Salmena L, Carracedo A, Pandolfi PP. Tenets of PTEN tumor suppression. Cell. 2008;133:403‐414. [DOI] [PubMed] [Google Scholar]
- 26. Di Cristofano A, Pandolfi PP. The multiple roles of PTEN in tumor suppression. Cell. 2000;100:387‐390. [DOI] [PubMed] [Google Scholar]
- 27. Rojo AI, Rada P, Mendiola M, et al. The PTEN/NRF2 axis promotes human carcinogenesis. Antioxid Redox Signal. 2014;21:2498‐2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Freeman DJ, Li AG, Wei G, et al. PTEN tumor suppressor regulates p53 protein levels and activity through phosphatase‐dependent and ‐independent mechanisms. Cancer Cell. 2003;3:117‐130. [DOI] [PubMed] [Google Scholar]
- 29. Trotman LC, Pandolfi PP. PTEN and p53: who will get the upper hand? Cancer Cell. 2003;3:97‐99. [DOI] [PubMed] [Google Scholar]
- 30. Lei Q, Jiao J, Xin L, et al. NKX3.1 stabilizes p53, inhibits AKT activation, and blocks prostate cancer initiation caused by PTEN loss. Cancer Cell. 2006;9:367‐378. [DOI] [PubMed] [Google Scholar]
- 31. Aveyard JS, Skilleter A, Habuchi T, et al. Somatic mutation of PTEN in bladder carcinoma. Br J Cancer. 1999;80:904‐908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Cairns P, Evron E, Okami K, et al. Point mutation and homozygous deletion of PTEN/MMAC1 in primary bladder cancers. Oncogene. 1998;16:3215‐3218. [DOI] [PubMed] [Google Scholar]
- 33. Tsuruta H, Kishimoto H, Sasaki T, et al. Hyperplasia and carcinomas in Pten‐deficient mice and reduced PTEN protein in human bladder cancer patients. Cancer Res. 2006;66:8389‐8396. [DOI] [PubMed] [Google Scholar]
- 34. Gildea JJ, Herlevsen M, Harding MA, et al. PTEN can inhibit in vitro organotypic and in vivo orthotopic invasion of human bladder cancer cells even in the absence of its lipid phosphatase activity. Oncogene. 2004;23:6788‐6797. [DOI] [PubMed] [Google Scholar]
- 35. Puzio‐Kuter AM, Castillo‐Martin M, Kinkade CW, et al. Inactivation of p53 and Pten promotes invasive bladder cancer. Genes Dev. 2009;23:675‐680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. McEwan WA, Tam JC, Watkinson RE, et al. Intracellular antibody‐bound pathogens stimulate immune signaling via the Fc receptor TRIM21. Nat Immunol. 2013;14:327‐336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Pan JA, Sun Y, Jiang YP, et al. TRIM21 ubiquitylates SQSTM1/p62 and suppresses protein sequestration to regulate redox homeostasis. Mol Cell. 2016;62:149‐151. [DOI] [PubMed] [Google Scholar]
- 38. Rubtsov YP, Zolotukhin AS, Vorobjev IA, et al. Mutational analysis of human prothymosin alpha reveals a bipartite nuclear localization signal. FEBS Lett. 1997;413:135‐141. [DOI] [PubMed] [Google Scholar]
- 39. Chen YC, Chang MY, Shiau AL, et al. Mitochondrial ribosomal protein S36 delays cell cycle progression in association with p53 modification and p21(WAF1/CIP1) expression. J Cell Biochem. 2007;100:981‐990. [DOI] [PubMed] [Google Scholar]
- 40. Schmittgen TD, Livak KJ. Analyzing real‐time PCR data by the comparative C(T) method. Nat Protoc. 2008;3:1101‐1108. [DOI] [PubMed] [Google Scholar]
- 41. Tsai YS, Jou YC, Lee GF, et al. Aberrant prothymosin‐alpha expression in human bladder cancer. Urology. 2009;73:188‐192. [DOI] [PubMed] [Google Scholar]
- 42. Tsai YS, Lai CL, Lai CH, et al. The role of homeostatic regulation between tumor suppressor DAB2IP and oncogenic Skp2 in prostate cancer growth. Oncotarget. 2014;5:6425‐6436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Su BH, Tseng YL, Shieh GS, et al. Over‐expression of prothymosin‐alpha antagonizes TGFbeta signalling to promote the development of emphysema. J Pathol. 2016;238:412‐422. [DOI] [PubMed] [Google Scholar]
- 44. Platt FM, Hurst CD, Taylor CF, et al. Spectrum of phosphatidylinositol 3‐kinase pathway gene alterations in bladder cancer. Clin Cancer Res. 2009;15:6008‐6017. [DOI] [PubMed] [Google Scholar]
- 45. Bermudez Brito M, Goulielmaki E, Papakonstanti EA. Focus on PTEN regulation. Front Oncol. 2015;5:166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Mallery DL, McEwan WA, Bidgood SR, et al. Antibodies mediate intracellular immunity through tripartite motif‐containing 21 (TRIM21). Proc Natl Acad Sci USA. 2010;107:19985‐19990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Comprehensive molecular characterization of urothelial bladder carcinoma. Nature. 2014;507:315‐322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Hayden A, Douglas J, Sommerlad M, et al. The Nrf2 transcription factor contributes to resistance to cisplatin in bladder cancer. Urol Oncol. 2014;32:806‐814. [DOI] [PubMed] [Google Scholar]
- 49. Iida K, Itoh K, Maher JM, et al. Nrf2 and p53 cooperatively protect against BBN‐induced urinary bladder carcinogenesis. Carcinogenesis. 2007;28:2398‐2403. [DOI] [PubMed] [Google Scholar]
- 50. Kijogi CM, Khayeka‐Wandabwa C, Sasaki K, et al. Subcellular dissemination of prothymosin alpha at normal physiology: immunohistochemical vis‐a‐vis western blotting perspective. BMC Physiol. 2016;16:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials


