Abstract
Pancreatic ductal adenocarcinoma (PDAC) is the most life‐threating disease among all digestive system malignancies. We developed a blood mRNA PDAC screening system using real‐time detection PCR to detect the expression of 56 genes, to discriminate PDAC from noncancer subjects. We undertook a clinical study to assess the performance of the developed system. We collected whole blood RNA from 53 PDAC patients, 102 noncancer subjects, 22 patients with chronic pancreatitis, and 23 patients with intraductal papillary mucinous neoplasms in a per protocol analysis. The sensitivity of the system for PDAC diagnosis was 73.6% (95% confidence interval, 59.7%‐84.7%). The specificity for noncancer volunteers, chronic pancreatitis, and patients with intraductal papillary mucinous neoplasms was 64.7% (54.6%‐73.9%), 63.6% (40.7%‐82.8%), and 47.8% (26.8%‐69.4%), respectively. Importantly, the sensitivity of this system for both stage I and stage II PDAC was 78.6% (57.1%‐100%), suggesting that detection of PDAC by the system is not dependent on the stage of PDAC. These results indicated that the screening system, relying on assessment of changes in mRNA expression in blood cells, is a viable alternative screening strategy for PDAC.
Keywords: clinical trial, in vitro diagnostics, mRNA screening system, pancreas cancer, whole blood cells
Abbreviations
- CA
cancer antigen
- CEA
carcinoembryonic antigen
- CI
confidence interval
- Cp
crossing point
- IPMN
intraductal papillary mucinous neoplasm
- NPV
negative predictive value
- PDAC
pancreatic ductal adenocarcinoma
- PPV
positive predictive value
- RTD‐PCR
real‐time detection PCR
1. INTRODUCTION
Pancreatic ductal adenocarcinoma is among the most difficult malignancies to diagnose and treat, resulting in poor prognosis.1, 2 Approximately 10% of PDAC patients are diagnosed in an early phase (ie, stage I and II), when radical treatment involving surgical removal of the PDAC lesions is available.3, 4 Chemotherapy has only limited efficacy in terms of prolonging the survival of PDAC patients.5 The overall survival after diagnosis of PDAC is less than 5 years, which is the poorest among all digestive system malignancies. Therefore, novel diagnostic tools capable of detecting PDAC in the early stages, as well as sufficiently effective treatment options to substantially improve the prognosis of PDAC, are required.
Previously, we reported that the gene expression profile of peripheral blood showed characteristic alterations in the digestive system of cancer patients.6 The local cancer tissues of PDAC were substantially infiltrated with inflammatory cells, including immune‐suppressing cells as well as lymphocytes expressing the immune checkpoint molecule, programmed cell death 1 (PD‐1). In addition, certain genes expressed in whole peripheral blood, as well as CD4+ cells and CD14+ cells, were found to be important in the inflammation seen in PDAC.7 Therefore, peripheral blood cells reflect the host inflammatory condition in association with local cancer tissues in PDAC.
We developed a blood mRNA PDAC screening system based on determining changes in the mRNA expression levels of 56 genes in whole blood by RTD‐PCR. We undertook a multi‐institutional clinical study to test the performance of this system. The system performance was assessed in 53 PDAC patients, 102 noncancer subjects, 22 chronic pancreatitis patients, and 23 IPMN patients. The system showed sensitivity of 73.6% (95% CI, 59.7%‐84.7%). The specificity for noncancer volunteers, chronic pancreatitis patients, and IPMN patients was 64.7% (54.6%‐73.9%), 63.6% (40.7%‐82.8%), and 47.8% (26.8%‐69.4%), respectively. Thus, the screening system, based on examination of changes in gene expression in blood, could be useful.
2. MATERIALS AND METHODS
2.1. Complementary DNA microarray analysis to determine gene expression in whole blood
Twenty‐eight PDAC patients and 27 noncancer subjects (Table S1) were enrolled for gene expression analysis of whole blood cells, to select candidate genes for establishing a blood PDAC screening system. For blood collection, we used PAXgene blood RNA tubes (PreAnalytix, Hombrechtikon, Switzerland) in which the RNA of whole blood cells was immediately stabilized after collection and mixed with reagents in the tubes. The study was approved by the Institutional Review Board of Kanazawa University (Kanazawa, Japan) and its related hospitals.
We used a cDNA microarray Whole Human Genome 4× 44K Array kit (G4112F; Agilent Technologies, Santa Clara, CA, USA) for analysis of gene expression of whole blood obtained from 17 PDAC patients and 13 noncancer subjects, as described previously.7 Briefly, RNA was extracted from PAXgene blood RNA tubes using a PAXgene Blood RNA System kit (PreAnalytix) in accordance with the manufacturer's protocol, followed by RNA quantification using a NanoDrop ND‐1000 Spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA). Cy‐3‐labeled cRNA was synthesized from 300 ng total RNA using a Quick Amp Labeling kit, and then hybridized to the cDNA microarray. The hybridized microarray was scanned using a cDNA Microarray Scanner (Model G2505B; Agilent Technologies), and the obtained image was processed using Feature Extraction software version 9.5.3.1 (Agilent Technologies) to obtain the value data. The data, which were normalized using GeneSpring GX software version 11.0.2 (Agilent Technologies), were used to select candidate genes for RTD‐PCR analysis. The cDNA microarray data were deposited in Gene Expression Omnibus (NBCI, Bethesda, MD, USA) with the number GSE125158.
2.2. Real‐time detection PCR
Gene expression analysis by RTD‐PCR for candidate genes selected from cDNA microarray data was undertaken as described previously.7 Briefly, cDNA was synthesized from total RNA extracted from samples in PAXgene Blood RNA tubes using an RT2 HT First Strand Kit (Qiagen, Hilden, Germany). Synthesized cDNA was measured using Custom RT2 Profiler PCR Arrays (Qiagen) according to the manufacturer's instructions. Quantitative PCR was carried out using a LightCycler 480 Instrument II version 1.5.0.39 (Roche, Basel, Switzerland).
2.3. Selection of genes for blood mRNA screening system
To establish the mRNA blood screening system, we used blood RNA from 28 PDAC patients and 27 noncancer subjects. Using RTD‐PCR, we measured the mRNA expression levels of candidate genes whose expression was assessed by cDNA microarray. For the selection of genes, we used a support vector machine with the linear kernel and sequential variable erasing methods.8 Discrimination between PDAC and noncancer subjects was undertaken using Fisher's linear discriminant with a penalized matrix for gene expression correlation analysis. We applied leave‐one‐out cross‐validation, using the RTD‐PCR data of 1 case as a test dataset for both gene selection and classifier building. Discrimination ability was assessed according to the sensitivity and specificity. The number of genes used for classification was based on cross‐validated sensitivity and specificity. Finally, we developed a final classifier by applying the whole algorithm with the determined number of genes to all 55 cases to be mounted in the blood mRNA screening system.
2.4. Clinical study design to assess performance of blood mRNA screening system
The clinical study to test the system performance was designed in consultation with the Japanese Pharmaceutical and Medical Device Agency, which is a Japanese regulatory agency, working together with the Ministry of Health, Labour, and Welfare to protect public health by assuring the safety, efficacy, and quality of pharmaceuticals and medical devices, and was approved by each participating institutional ethical committee (UMIN000020758). The primary end‐points were sensitivity and specificity of the blood mRNA PDAC screening system. The secondary end‐points were the PPV and NPV for the blood mRNA PDAC screening system, and the sensitivity, specificity, PPV, and NPV for CEA and CA19‐9. The clinical study was undertaken in 50 PDAC patients and 100 noncancer subjects. We also enrolled patients with IPMN and chronic pancreatitis, whose clinical data were not available. The inclusion criteria were as follows: PDAC patients, diagnosis by imaging studies or pathologically; chronic pancreatitis and IPMN patients, diagnosis by imaging studies; and non‐cancer subjects, considered normal based on medical records for the previous year or on current medical check‐up. The subjects were aged between 40 and 90 years. All subjects provided written informed consent prior to participating in the study. Exclusion criteria are shown in Table S2. The participating institutions were as follows (all based in Japan): Department of Gastroenterology, Kanazawa University Hospital; Department of Internal Medicine, Toyama Prefectural Central Hospital; Department of Gastroenterology, Toyama Municipal Hospital; Department of Gastroenterology, Tonami General Hospital; Department of Gastroenterology, Kanazawa Medical Center; Ishikawa Saiseikai Kanazawa Hospital; Department of Gastroenterology, Matto Ishikawa Central Hospital; Fukui Saiseikai Hospital; Department of Gastroenterology, Tsuruga Hospital; and Department of Internal Medicine, Keijyu General Hospital.
Blood was collected using PAXgene blood RNA tubes (PreAnalytiX) for mRNA expression measurement. Sera were also collected for CEA and CA19‐9 measurement by the standard chemiluminescent enzyme immunoassay. Results of CEA greater than 5.0 ng/mL and CA19‐9 greater than 37.0 U/mL were judged to be positive.
2.5. Statistical analysis
For the clinical validation study, we used Wilcoxon's signed‐rank test or Fisher's exact test for group comparisons. In evaluating the system performance, point estimates and 95% CIs of sensitivity and specificity were calculated. Pearson's correlation analysis and t test were applied using IBM SPSS Statistics (IBM, Armonk, NY, USA).
3. RESULTS
3.1. Establishment of blood PDAC screening system
We reported previously that the gene expression profile of whole blood cells from the digestive system of cancer patients was different from that of normal noncancer subjects.6 Based on these findings, we established a blood PDAC screening system using RTD‐PCR for genes, the expression levels of which in whole blood were altered in PDAC patients. We first extracted 2 sets of 384 genes assessing the cDNA microarray data of 17 PDAC patients and 13 noncancer subjects (GSE125158). The 384 genes in 1 set were selected by their expression, which was affected at FC >1.5 and P < 0.00013. The other set of 384 genes was selected by a powerful machine learning algorithms‐random forest method,9 in which 200 bootstraps were used to find the top genes that have more power to distinguish PDAC from noncancer subjects. The 179 genes that were included in both gene sets were finally selected. From the remaining genes in this analysis, we chose 134 genes, considering their biological meanings. We also selected 51 genes, whose flags for expression were positive in 12 of 17 PDAC patients and absent in 10 of 13 noncancer subjects. Thus, we finally selected 364 genes to further construct the equation for the PDAC screening system (Figure S1). For 28 PDAC patients and 27 noncancer subjects, including the above subjects, we carried out RTD‐PCR of these 364 genes and 17 housekeeping genes including GAPDH (Table S3). We used GAPDH as a reference as its expression was substantial and showed no difference between PDAC patients and noncancer subjects. Therefore, we have used delta Cp, which was obtained by subtraction of the GAPDH Cp value from that of each gene, for mRNA expression. To determine the gene number for discrimination of PDAC and noncancer subjects, we used the statistical method applied with the support vector machine and recursive feature elimination8 during the leave‐one‐out cross‐validation among these 55 cases (Figure S2). The cross‐validated sensitivity and specificity to discriminate PDAC from noncancer subjects were sufficiently high (93% and 100%, respectively) based on 56 genes; increasing the number of genes above 56 did not increase discrimination performance. By this analysis, we found that 56 genes were sufficient for the most excellent accuracy.
Therefore, we finally selected 56 genes (Table 1) for expression analysis in whole blood cells in 28 PDAC and 27 noncancer subjects.
Table 1.
Fifty‐six genes, their annotations, and coefficient values for equation of the blood mRNA screening system
| Refseq # | Gene symbol | Gene description | Coefficient value | GOTERM_Biological process (BP) | GOTERM_Cellular component (CC) | GOTERM_Molecular function (MF) |
|---|---|---|---|---|---|---|
| NM_138340 | SLC44A1 | Solute carrier family 44 member 1 | 18.88705 |
GO:0006656~phosphatidylcholine biosynthetic process, GO:0015871~choline transport, GO:0042426~choline catabolic process, GO:0055085~transmembrane transport |
GO:0005654~nucleoplasm, GO:0005739~mitochondrion, GO:0016020~membrane, GO:0070062~extracellular exosome |
GO:0015220~choline transmembrane transporter activity |
| NM_005470 | ABHD3 | Abhydrolase domain containing 3 | 18.25234 |
GO:0006629~lipid metabolic process, GO:0046470~phosphatidylcholine metabolic process |
GO:0005886~plasma membrane, GO:0016021~integral component of membrane |
GO:0004623~phospholipase A2 activity, GO:0008970~phosphatidylcholine 1‐acylhydrolase activity, GO:0052739~phosphatidylserine 1‐acylhydrolase activity, GO:0052740~1‐acyl‐2‐lysophosphatidylserine acylhydrolase activity |
| NM_004457 | PTPRC | Protein tyrosine phosphatase, receptor type C | 15.18565 |
GO:0001915~negative regulation of T cell mediated cytotoxicity, GO:0001960~negative regulation of cytokine‐mediated signaling pathway, GO:0002244~hematopoietic progenitor cell differentiation, GO:0002378~immunoglobulin biosynthetic process, GO:0030217~T cell differentiation, GO:0030890~positive regulation of B cell proliferation, GO:0048539~bone marrow development, GO:0048864~stem cell development, GO:0050852~T cell receptor signaling pathway, GO:0050857~positive regulation of antigen receptor‐mediated signaling pathway |
GO:0005886~plasma membrane GO:0005925~focal adhesion, GO:0045121~membrane raft, GO:0070062~extracellular exosome |
GO:0005001~transmembrane receptor protein tyrosine phosphatase activity, GO:0005515~protein binding, GO:0019901~protein kinase binding |
| NM_001628 | TLE2 | Transducin like enhancer of split 2 | 14.37697 |
GO:0006351~transcription, DNA‐templated, GO:0007165~signal transduction, GO:0016055~Wnt signaling pathway, GO:0045892~negative regulation of transcription, DNA‐templated, GO:1904837~beta‐catenin‐TCF complex assembly |
GO:0005615~extracellular space, GO:0005634~nucleus, GO:0005925~focal adhesion |
GO:0003714~transcription corepressor activity, GO:0005515~protein binding |
| NM_014616 | FNDC3B | Fibronectin type III domain containing 3B | 14.19903 | — | GO:0016021~integral component of membrane | GO:0044822~poly(A) RNA binding |
| NM_032047 | LDHA | Lactate dehydrogenase A | 13.84082 |
GO:0001666~response to hypoxia, GO:0005975~carbohydrate metabolic process, GO:0006089~lactate metabolic process, GO:0006090~pyruvate metabolic process, GO:0006096~glycolytic process,GO:0007584~response to nutrient, GO:0042542~response to hydrogen peroxide, GO:0043065~positive regulation of apoptotic process, GO:0043627~response to estrogen, GO:0051591~response to cAMP, GO:0055114~oxidation‐reduction process, GO:0098609~cell‐cell adhesion |
GO:0005634~nucleus, GO:0005737~cytoplasm, GO:0005913~cell‐cell adherens junction, GO:0016020~membrane, GO:0016021~integral component of membrane, GO:0070062~extracellular exosome |
GO:0004459~L‐lactate dehydrogenase activity, GO:0016616~oxidoreductase activity, acting on the CH‐OH group of donors, NAD or NADP as acceptor, GO:0019900~kinase binding, GO:0042802~identical protein binding, GO:0051287~NAD binding, GO:0098641~cadherin binding involved in cell‐cell adhesion |
| NM_001186 | RPLP1 | Ribosomal protein lateral stalk subunit P1 | 13.75907 |
GO:0000184~nuclear‐transcribed mRNA catabolic process, nonsense‐mediated decay, GO:0002181~cytoplasmic translation, GO:0006364~rRNA processing, GO:0045860~positive regulation of protein kinase activity |
GO:0005737~cytoplasm, GO:0005840~ribosome, GO:0005925~focal adhesion, GO:0070062~extracellular exosome |
GO:0003735~structural constituent of ribosome, GO:0005515~protein binding, GO:0030295~protein kinase activator activity, GO:0070180~large ribosomal subunit rRNA binding |
| NM_001145054 | RAB10 | RAB10, member RAS oncogene family | 13.27343 |
GO:0006893~Golgi to plasma membrane transport, GO:0007264~small GTPase mediated signal transduction, GO:0007409~axonogenesis, GO:0016192~vesicle‐mediated transport, GO:0016197~endosomal transport, GO:0019882~antigen processing and presentation, GO:0030859~polarized epithelial cell differentiation, GO:0032869~cellular response to insulin stimulus, GO:0071786~endoplasmic reticulum tubular network organization, GO:0072659~protein localization to plasma membrane, GO:0097051~establishment of protein localization to endoplasmic reticulum membrane, GO:0098609~cell‐cell adhesion |
GO:0000139~Golgi membrane, GO:0000145~exocyst, GO:0005622~intracellular, GO:0005768~endosome, GO:0005789~endoplasmic reticulum membrane, GO:0005886~plasma membrane, GO:0005911~cell‐cell junction, GO:0005913~cell‐cell adherens junction, GO:0005925~focal adhesion, GO:0030670~phagocytic vesicle membrane, GO:0032593~insulin‐responsive compartment, GO:0055038~recycling endosome membrane, GO:0070062~extracellular exosome, GO:0071782~endoplasmic reticulum tubular network |
GO:0003924~GTPase activity, GO:0005515~protein binding, GO:0005525~GTP binding, GO:0019003~GDP binding, GO:0031489~myosin V binding, GO:0098641~cadherin binding involved in cell‐cell adhesion |
| NM_152523 | HTATIP2 | HIV‐1 Tat interactive protein 2 | 12.98543 |
GO:0001525~angiogenesis, GO:0006357~regulation of transcription from RNA polymerase II promoter, GO:0006915~apoptotic process, GO:0016032~viral process, GO:0030154~cell differentiation, GO:0051170~nuclear import, GO:0055114~oxidation‐reduction process |
GO:0005634~nucleus, GO:0005635~nuclear envelope, GO:0005654~nucleoplasm, GO:0005737~cytoplasm, GO:0016020~membrane |
GO:0003713~transcription coactivator activity, GO:0005515~protein binding, GO:0016491~oxidoreductase activity |
| NM_018455 | BACH1 | BTB domain and CNC homolog 1 | 12.79754 |
GO:0000083~regulation of transcription involved in G1/S transition of mitotic cell cycle, GO:0000117~regulation of transcription involved in G2/M transition of mitotic cell cycle, GO:0006281~DNA repair, GO:0006355~regulation of transcription |
GO:0005634~nucleus, GO:0005737~cytoplasm, GO:0005829~cytosol, GO:0031463~Cul3‐RING ubiquitin ligase complex |
GO:0001078~transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence‐specific binding, GO:0001205~transcriptional activator activity, RNA polymerase II distal enhancer sequence‐specific binding, GO:0001206~transcriptional repressor activity, RNA polymerase II distal enhancer sequence‐specific binding, GO:0003700~transcription factor activity, sequence‐specific DNA binding, GO:0004842~ubiquitin‐protein transferase activity |
| NM_021023 | NFE2L2 | Nuclear factor, erythroid 2 like 2 | 11.75957 |
GO:0006351~transcription, GO:0006954~inflammatory response, GO:0010628~positive regulation of gene expression, GO:0016567~protein ubiquitination, GO:0030194~positive regulation of blood coagulation, GO:0030968~endoplasmic reticulum unfolded protein response, GO:0034599~cellular response to oxidative stress, GO:0036003~positive regulation of transcription from RNA polymerase II promoter in response to stress, GO:0036499~PERK‐mediated unfolded protein response, GO:0042149~cellular response to glucose starvation, GO:0045766~positive regulation of angiogenesis, GO:0071356~cellular response to tumor necrosis factor, GO:0071456~cellular response to hypoxia, GO:0071498~cellular response to fluid shear stress, GO:1902037~negative regulation of hematopoietic stem cell differentiation, GO:1903071~positive regulation of ER‐associated ubiquitin‐dependent protein catabolic process, GO:1903788~positive regulation of glutathione biosynthetic process |
GO:0000785~chromatin, GO:0005634~nucleus, GO:0005737~cytoplasm, GO:0005813~centrosome, GO:0005886~plasma membrane, GO:0032993~protein‐DNA complex |
GO:0000980~RNA polymerase II distal enhancer sequence‐specific DNA binding, GO:0001102~RNA polymerase II activating transcription factor binding, GO:0001205~transcriptional activator activity, RNA polymerase II distal enhancer sequence‐specific binding, GO:0003677~DNA binding, GO:0005515~protein binding |
| NM_080387 | TET2 | Tet methylcytosine dioxygenase 2 | 11.42427 |
GO:0001822~kidney development, GO:0002318~myeloid progenitor cell differentiation, GO:0006211~5‐methylcytosine catabolic process, GO:0006493~protein O‐linked glycosylation, GO:0007049~cell cycle, GO:0014070~response to organic cyclic compound, GO:0020027~hemoglobin metabolic process, GO:0030099~myeloid cell differentiation, GO:0045944~positive regulation of transcription from RNA polymerase II promoter, GO:0061484~hematopoietic stem cell homeostasis, GO:0070989~oxidative demethylation, GO:0080111~DNA demethylation, GO:0080182~histone H3‐K4 trimethylation |
GO:0005634~nucleus | GO:0000907~sulfonate dioxygenase activity,GO:0003677~DNA binding,GO:0005515~protein binding,GO:0008198~ferrous iron binding,GO:0008270~zinc ion binding,GO:0018602~2,4‐dichlorophenoxyacetate alpha‐ketoglutarate dioxygenase activity,GO:0019798~procollagen‐proline dioxygenase activity,GO:0034792~hypophosphite dioxygenase activity,GO:0043734~DNA‐N1‐methyladenine dioxygenase activity,GO:0052634~C‐19 gibberellin 2‐beta‐dioxygenase activity,GO:0052635~C‐20 gibberellin 2‐beta‐dioxygenase activity,GO:0070579~methylcytosine dioxygenase activity |
| NM_000494 | MBTPS2 | Membrane bound transcription factor peptidase, site 2 | 11.32147 |
GO:0008203~cholesterol metabolic process, GO:0031293~membrane protein intracellular domain proteolysis, GO:0034976~response to endoplasmic reticulum stress, GO:0036500~ATF6‐mediated unfolded protein response, GO:0051091~positive regulation of sequence‐specific DNA binding transcription factor activity, GO:1990440~positive regulation of transcription from RNA polymerase II promoter in response to endoplasmic reticulum stress |
GO:0000139~Golgi membrane ,GO:0005737~cytoplasm, GO:0016021~integral component of membrane |
GO:0004222~metalloendopeptidase activity, GO:0008237~metallopeptidase activity, GO:0046872~metal ion binding |
| NM_016230 | C2orf81 | Chromosome 2 open reading frame 81 | 8.747677 | — | — | — |
| NM_001195215 | PPP1R13L | Protein phosphatase 1 regulatory subunit 13 like | 8.60306 |
GO:0000122~negative regulation of transcription from RNA polymerase II promoter, GO:0006351~transcription, DNA‐templated, GO:0006915~apoptotic process, GO:0045597~positive regulation of cell differentiation, GO:0098609~cell‐cell adhesion, GO:1901796~regulation of signal transduction by p53 class mediator |
GO:0005634~nucleus, GO:0005737~cytoplasm, GO:0005913~cell‐cell adherens junction, GO:0030054~cell junction, GO:0045171~intercellular bridge |
GO:0003714~transcription corepressor activity, GO:0005515~protein binding, GO:0008134~transcription factor binding, GO:0042802~identical protein binding, GO:0098641~cadherin binding involved in cell‐cell adhesion |
| NM_024693 | AKR1B1 | Aldo‐keto reductase family 1 member B | 8.005092 |
GO:0001894~tissue homeostasis, GO:0005975~carbohydrate metabolic process, GO:0006700~C21‐steroid hormone biosynthetic process, GO:0006950~response to stress, GO:0042415~norepinephrine metabolic process, GO:0046427~positive regulation of JAK‐STAT cascade, GO:0048661~positive regulation of smooth muscle cell proliferation, GO:0055114~oxidation‐reduction process, GO:0070301~cellular response to hydrogen peroxide |
GO:0005615~extracellular space, GO:0005654~nucleoplasm, GO:0005737~cytoplasm, GO:0005829~cytosol, GO:0033010~paranodal junction, GO:0042629~mast cell granule, GO:0043220~Schmidt‐Lanterman incisure, GO:0048471~perinuclear region of cytoplasm, GO:0070062~extracellular exosome |
GO:0004032~alditol:NADP+ 1‐oxidoreductase activity, GO:0004033~aldo‐keto reductase (NADP) activity, GO:0009055~electron carrier activity, GO:0016491~oxidoreductase activity, GO:0043795~glyceraldehyde oxidoreductase activity |
| NM_001776 | COL17A1 | Collagen type XVII alpha 1 chain | 7.970873 |
GO:0007160~cell‐matrix adhesion, GO:0008544~epidermis development, GO:0031581~hemidesmosome assembly, GO:0050776~regulation of immune response |
GO:0005576~extracellular region, GO:0005578~proteinaceous extracellular matrix, GO:0005581~collagen trimer, GO:0005604~basement membrane, GO:0005788~endoplasmic reticulum lumen, GO:0005886~plasma membrane, GO:0005911~cell‐cell junction GO:0030056~hemidesmosome |
GO:0005515~protein binding |
| NM_016613 | MCMBP | Minichromosome maintenance complex binding protein | 7.570634 |
GO:0006261~DNA‐dependent DNA replication, GO:0007062~sister chromatid cohesion, GO:0007067~mitotic nuclear division, GO:0051301~cell division |
GO:0000790~nuclear chromatin, GO:0005654~nucleoplasm, GO:0005737~cytoplasm, GO:0005886~plasma membrane, GO:0042555~MCM complex |
GO:0003682~chromatin binding, GO:0005515~protein binding |
| NM_016623 | FAR1 | Fatty acyl‐CoA reductase 1 | 7.238576 |
GO:0006086~acetyl‐CoA biosynthetic process from pyruvate, GO:0008611~ether lipid biosynthetic process, GO:0010025~wax biosynthetic process, GO:0035336~long‐chain fatty‐acyl‐CoA metabolic process, GO:0046474~glycerophospholipid biosynthetic process, GO:0055114~oxidation‐reduction process |
GO:0005777~peroxisome, GO:0005778~peroxisomal membrane, GO:0005782~peroxisomal matrix, GO:0016021~integral component of membrane |
GO:0016491~oxidoreductase activity, GO:0050062~long‐chain‐fatty‐acyl‐CoA reductase activity, GO:0052814~medium‐chain‐aldehyde dehydrogenase activity, GO:0080019~fatty‐acyl‐CoA reductase (alcohol‐forming) activity |
| NM_032228 | OSBPL8 | Oxysterol binding protein like 8 | 6.956427 |
GO:0006869~lipid transport, GO:0010891~negative regulation of sequestering of triglyceride, GO:0030336~negative regulation of cell migration, GO:0032148~activation of protein kinase B activity, GO:0045444~fat cell differentiation, GO:0046628~positive regulation of insulin receptor signaling pathway, GO:0090204~protein localization to nuclear pore |
GO:0005783~endoplasmic reticulum, GO:0016020~membrane, GO:0031965~nuclear membrane, GO:0032541~cortical endoplasmic reticulum |
GO:0001786~phosphatidylserine binding, GO:0005548~phospholipid transporter activity, GO:0015485~cholesterol binding, GO:0070273~phosphatidylinositol‐4‐phosphate binding |
| NM_006682 | ZMPSTE24 | Zinc metallopeptidase STE24 | 6.555173 |
GO:0006508~proteolysis, GO:0006998~nuclear envelope organization, GO:0030327~prenylated protein catabolic process, GO:0071586~CAAX‐box protein processing |
GO:0005637~nuclear inner membrane, GO:0005789~endoplasmic reticulum membrane, GO:0016020~membrane, GO:0070062~extracellular exosome |
GO:0004222~metalloendopeptidase activity, GO:0008235~metalloexopeptidase activity, GO:0046872~metal ion binding |
| NM_022763 | CCNYL1 | Cyclin Y like 1 | 6.300126 | GO:0000079~regulation of cyclin‐dependent protein serine/threonine kinase activity | GO:0005886~plasma membrane |
GO:0005515~protein binding, GO:0019901~protein kinase binding |
| NM_003838 | RRAGD | Ras related GTP binding D | 6.236001 |
GO:0007050~cell cycle arrest, GO:0009267~cellular response to starvation, GO:0010506~regulation of autophagy, GO:0016236~macroautophagy, GO:0032008~positive regulation of TOR signaling, GO:0071230~cellular response to amino acid stimulus |
GO:0005634~nucleus, GO:0005737~cytoplasm, GO:0005764~lysosome, GO:0005829~cytosol, GO:0034448~EGO complex, GO:1990131~Gtr1‐Gtr2 GTPase complex |
GO:0003924~GTPase activity, GO:0005515~protein binding, GO:0005525~GTP binding, GO:0046982~protein heterodimerization activity |
| NM_003774 | GALNT4 | Polypeptide N‐acetylgalactosaminyltransferase 4 | 6.227974 |
GO:0005975~carbohydrate metabolic process, GO:0016266~O‐glycan processing |
GO:0005794~Golgi apparatus, GO:0016021~integral component of membrane, GO:0048471~perinuclear region of cytoplasm, GO:0070062~extracellular exosome |
GO:0004653~polypeptide N‐acetylgalactosaminyltransferase activity, GO:0030246~carbohydrate binding, GO:0046872~metal ion binding |
| NM_015473 | UBE2W | Ubiquitin conjugating enzyme E2 W (putative) | 5.881697 |
GO:0006281~DNA repair, GO:0006513~protein monoubiquitination, GO:0071218~cellular response to misfolded protein |
GO:0005634~nucleus, GO:0005737~cytoplasm, GO:0016021~integral component of membrane |
GO:0004842~ubiquitin‐protein transferase activity, GO:0005515~protein binding, GO:0005524~ATP binding |
| NM_006410 | HEATR5A | HEAT repeat containing 5A | 5.784991 | — | — | — |
| NM_000416 | UBE4A | Ubiquitination factor E4A | 5.612217 |
GO:0000209~protein polyubiquitination, GO:0007165~signal transduction, GO:0030433~ER‐associated ubiquitin‐dependent protein catabolic process |
GO:0000151~ubiquitin ligase complex, GO:0005634~nucleus, GO:0005737~cytoplasm |
GO:0004114~3’,5’‐cyclic‐nucleotide phosphodiesterase activity, GO:0005515~protein binding, GO:0016874~ligase activity, GO:0034450~ubiquitin‐ubiquitin ligase activity |
| NM_201612 | ABI1 | Abl interactor 1 | 5.533139 |
GO:0006928~movement of cell or subcellular component, GO:0007169~transmembrane receptor protein tyrosine kinase signaling pathway, GO:0008154~actin polymerization or depolymerization, GO:0008285~negative regulation of cell proliferation, GO:0016032~viral process, GO:0018108~peptidyl‐tyrosine phosphorylation, GO:0035855~megakaryocyte development, GO:0038096~Fc‐gamma receptor signaling pathway involved in phagocytosis, GO:0048010~vascular endothelial growth factor receptor signaling pathway, GO:0061098~positive regulation of protein tyrosine kinase activity, GO:0072673~lamellipodium morphogenesis, GO:0098609~cell‐cell adhesion |
GO:0005622~intracellular, GO:0005634~nucleus, GO:0005737~cytoplasm, GO:0005783~endoplasmic reticulum, GO:0005856~cytoskeleton, GO:0005913~cell‐cell adherens junction, GO:0030027~lamellipodium, GO:0030175~filopodium, GO:0030426~growth cone, GO:0031209~SCAR complex, GO:0045211~postsynaptic membrane, GO:0070062~extracellular exosome |
GO:0005515~protein binding, GO:0008092~cytoskeletal protein binding, GO:0017124~SH3 domain binding, GO:0030296~protein tyrosine kinase activator activity, GO:0032403~protein complex binding, GO:0098641~cadherin binding involved in cell‐cell adhesion |
| NM_005566 | PPIH | Peptidylprolyl isomerase H | 5.40687 |
GO:0000398~mRNA splicing, via spliceosome, GO:0000413~protein peptidyl‐prolyl isomerization, GO:0006457~protein folding, GO:0006461~protein complex assembly, GO:0045070~positive regulation of viral genome replication |
GO:0005654~nucleoplasm, GO:0005681~spliceosomal complex, GO:0005737~cytoplasm, GO:0016607~nuclear speck, GO:0046540~U4/U6 x U5 tri‐snRNP complex, GO:0071001~U4/U6 snRNP |
GO:0003755~peptidyl‐prolyl cis‐trans isomerase activity, GO:0005515~protein binding, GO:0016018~cyclosporin A binding, GO:0043021~ribonucleoprotein complex binding |
| NM_005767 | CENPN | Centromere protein N | 4.732161 |
GO:0007059~chromosome segregation, GO:0007062~sister chromatid cohesion, GO:0007067~mitotic nuclear division, GO:0034080~CENP‐A containing nucleosome assembly, GO:0034508~centromere complex assembly |
GO:0000775~chromosome, centromeric region, GO:0000777~condensed chromosome kinetochore, GO:0005654~nucleoplasm, GO:0005829~cytosol |
— |
| NM_015884 | MIER1 | MIER1 transcriptional regulator | 4.196767 |
GO:0006351~transcription, DNA‐templated, GO:0007165~signal transduction, GO:0031937~positive regulation of chromatin silencing, GO:0043123~positive regulation of I‐kappaB kinase/NF‐kappaB signaling |
GO:0005654~nucleoplasm, GO:0005737~cytoplasm, GO:0017053~transcriptional repressor complex |
GO:0003677~DNA binding, GO:0004871~signal transducer activity, GO:0005515~protein binding |
| NM_024834 | IKBIP | IKBKB interacting protein | −4.35974 | GO:0010165~response to X‐ray |
GO:0005783~endoplasmic reticulum, GO:0016020~membrane |
GO:0005515~protein binding |
| NM_020948 | SLC22A15 | Solute carrier family 22 member 15 | −5.63841 |
GO:0006811~ion transport, GO:0055085~transmembrane transport |
GO:0016021~integral component of membrane |
GO:0022857~transmembrane transporter activity, GO:0022891~substrate‐specific transmembrane transporter activity |
| NM_001031716 | IFNGR1 | Interferon gamma receptor 1 | −6.31219 |
GO:0007165~signal transduction, GO:0009615~response to virus, GO:0060333~interferon‐gamma‐mediated signaling pathway, GO:0060334~regulation of interferon‐gamma‐mediated signaling pathway |
GO:0005783~endoplasmic reticulum, GO:0005886~plasma membrane, GO:0030425~dendrite, GO:0031982~vesicle |
GO:0004906~interferon‐gamma receptor activity, GO:0005515~protein binding, GO:0042015~interleukin‐20 binding |
| NM_006164 | CFHR3 | Complement factor H related 3 | −6.547 | — |
GO:0005615~extracellular space, GO:0070062~extracellular exosome, GO:0072562~blood microparticle |
|
| NM_020841 | ENTPD1 | Ectonucleoside triphosphate diphosphohydrolase 1 | −7.19342 |
GO:0007155~cell adhesion, GO:0007596~blood coagulation |
GO:0005886~plasma membrane, GO:0016020~membrane, GO:0070062~extracellular exosome |
GO:0005515~protein binding, GO:0005524~ATP binding, GO:0016787~hydrolase activity |
| NM_006347 | CLEC4D | C‐type lectin domain family 4 member D | −7.20897 |
GO:0002223~stimulatory C‐type lectin receptor signaling pathway, GO:0002250~adaptive immune response, GO:0002292~T cell differentiation involved in immune response, GO:0038094~Fc‐gamma receptor signaling pathway, GO:0045087~innate immune response |
GO:0005886~plasma membrane, GO:0016021~integral component of membrane |
GO:0030246~carbohydrate binding, GO:0034987~immunoglobulin receptor binding |
| NM_006663 | USP15 | Ubiquitin specific peptidase 15 | −7.21872 |
GO:0006511~ubiquitin‐dependent protein catabolic process, GO:0007179~transforming growth factor beta receptor signaling pathway, GO:0030509~BMP signaling pathway, GO:0035616~histone H2B conserved C‐terminal lysine deubiquitination, GO:0060389~pathway‐restricted SMAD protein phosphorylation |
GO:0005634~nucleus, GO:0005737~cytoplasm |
GO:0004197~cysteine‐type endopeptidase activity, GO:0004843~thiol‐dependent ubiquitin‐specific protease activity, GO:0005160~transforming growth factor beta receptor binding, GO:0005515~protein binding, GO:0046332~SMAD binding, GO:0061649~ubiquitinated histone binding |
| NM_018699 | SLFN12 | Schlafen family member 12 | −7.8338 | — | — | GO:0005524~ATP binding |
| NM_002838 | FGL2 | Fibrinogen like 2 | −8.25242 | GO:0006508~proteolysis |
GO:0005577~fibrinogen complex, GO:0070062~extracellular exosome |
GO:0008233~peptidase activity |
| NM_016131 | ACSL3 | Acyl‐CoA synthetase long‐chain family member 3 | −8.30246 |
GO:0001676~long‐chain fatty acid metabolic process, GO:0006633~fatty acid biosynthetic process, GO:0008152~metabolic process, GO:0014070~response to organic cyclic compound, GO:0034379~very‐low‐density lipoprotein particle assembly, GO:0042998~positive regulation of Golgi to plasma membrane protein transport, GO:0044539~long‐chain fatty acid import, GO:0051047~positive regulation of secretion, GO:2001247~positive regulation of phosphatidylcholine biosynthetic process |
GO:0005741~mitochondrial outer membrane, GO:0005778~peroxisomal membrane, GO:0005783~endoplasmic reticulum, GO:0005789~endoplasmic reticulum membrane, GO:0005794~Golgi apparatus, GO:0005811~lipid particle, GO:0016020~membrane, GO:0048471~perinuclear region of cytoplasm |
GO:0003824~catalytic activity, GO:0004467~long‐chain fatty acid‐CoA ligase activity, GO:0005524~ATP binding,GO:0016874~ligase activity, GO:0019901~protein kinase binding, GO:0019904~protein domain specific binding, GO:0102391~decanoate‐CoA ligase activity |
| NM_001003 | ZEB2 | Zinc finger E‐box binding homeobox 2 | −8.42855 |
GO:0000122~negative regulation of transcription from RNA polymerase II promoter, GO:0006351~transcription, DNA‐templated, GO:0048066~developmental pigmentation, GO:0050790~regulation of catalytic activity,GO:0097324~melanocyte migration |
GO:0005634~nucleus |
GO:0001227~transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence‐specific binding, GO:0003676~nucleic acid binding, GO:0005515~protein binding, GO:0019208~phosphatase regulator activity, GO:0046872~metal ion binding |
| NM_021244 | FPGT | Fucose‐1‐phosphate guanylyltransferase | −8.48534 | GO:0006004~fucose metabolic process |
GO:0005737~cytoplasm GO:0005829~cytosol |
GO:0003824~catalytic activity, GO:0005525~GTP binding, GO:0016772~transferase activity, transferring phosphorus‐containing groups, GO:0016779~nucleotidyltransferase activity, GO:0047341~fucose‐1‐phosphate guanylyltransferase activity |
| NM_018420 | ECHDC3 | Enoyl‐CoA hydratase domain containing 3 | −8.66068 | GO:0008152~metabolic process | GO:0005739~mitochondrion | GO:0003824~catalytic activity |
| NM_080546 | PRDM5 | PR/SET domain 5 | −8.77508 |
GO:0000122~negative regulation of transcription from RNA polymerase II promoter, GO:0000278~mitotic cell cycle, GO:0006351~transcription, DNA‐templated, GO:0016575~histone deacetylation, GO:0045892~negative regulation of transcription, DNA‐templated, GO:0051567~histone H3‐K9 methylation |
GO:0005634~nucleus |
GO:0001078~transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence‐specific binding, GO:0003676~nucleic acid binding, GO:0005515~protein binding, GO:0008168~methyltransferase activity, GO:0044212~transcription regulatory region DNA binding, GO:0046872~metal ion binding, GO:0070491~repressing transcription factor binding |
| NM_018042 | FAM49B | Family with sequence similarity 49 member B | −8.78217 | — |
GO:0005929~cilium, GO:0016020~membrane, GO:0070062~extracellular exosome |
GO:0005515~protein binding |
| NM_018079 | VPS51 (C11orf2) | VPS51, GARP complex subunit | −8.85243 |
GO:0006869~lipid transport, GO:0006914~autophagy, GO:0007041~lysosomal transport, GO:0015031~protein transport, GO:0032456~endocytic recycling, GO:0042147~retrograde transport, endosome to Golgi |
GO:0000938~GARP complex, GO:0005794~Golgi apparatus, GO:0005829~cytosol, GO:0016020~membrane, GO:0016021~integral component of membrane, GO:0055037~recycling endosome, GO:1990745~EARP complex |
GO:0005515~protein binding |
| NM_017628 | DENND1B | DENN domain containing 1B | −10.4299 |
GO:0015031~protein transport, GO:0032456~endocytic recycling, GO:0035745~T‐helper 2 cell cytokine production, GO:0043547~positive regulation of GTPase activity, GO:0050776~regulation of immune response, GO:0050852~T cell receptor signaling pathway |
GO:0005829~cytosol, GO:0030136~clathrin‐coated vesicle |
GO:0017112~Rab guanyl‐nucleotide exchange factor activity, GO:0017137~Rab GTPase binding |
| NM_003260 | NABP1 (OBFC2A) | Nucleic acid binding protein 1 | −11.0293 |
GO:0006281~DNA repair, GO:0006974~cellular response to DNA damage stimulus, GO:0007093~mitotic cell cycle checkpoint, GO:0010212~response to ionizing radiation, GO:0042795~snRNA transcription from RNA polymerase II promoter |
GO:0000784~nuclear chromosome, telomeric region, GO:0005737~cytoplasm, GO:0070876~SOSS complex |
GO:0003697~single‐stranded DNA binding, GO:0003723~RNA binding, GO:0005515~protein binding |
| NM_018299 | CYB5R4 | Cytochrome b5 reductase 4 | −11.661 |
GO:0003032~detection of oxygen, GO:0006091~generation of precursor metabolites and energy, GO:0006801~superoxide metabolic process, GO:0015701~bicarbonate transport, GO:0030073~insulin secretion, GO:0042593~glucose homeostasis, GO:0048468~cell development ,GO:0055114~oxidation‐reduction process |
GO:0005783~endoplasmic reticulum, GO:0048471~perinuclear region of cytoplasm |
GO:0004128~cytochrome‐b5 reductase activity, acting on NAD(P)H, GO:0016174~NAD(P)H oxidase activity, GO:0016653~oxidoreductase activity, acting on NAD(P)H, heme protein as acceptor, GO:0020037~heme binding, GO:0046872~metal ion binding, GO:0071949~FAD binding |
| NM_004788 | LPAR6 | Lysophosphatidic acid receptor 6 | −11.9776 |
GO:0035025~positive regulation of Rho protein signal transduction, GO:0051482~positive regulation of cytosolic calcium ion concentration involved in phospholipase C‐activating G‐protein coupled signaling pathway |
GO:0005886~plasma membrane, GO:0005887~integral component of plasma membrane, GO:0016021~integral component of membrane |
GO:0004930~G‐protein coupled receptor activity, GO:0070915~lysophosphatidic acid receptor activity |
| NM_006313 | SRBD1 | S1 RNA binding domain 1 | −12.4726 | GO:0006139~nucleobase‐containing compound metabolic process | — | GO:0003723~RNA binding |
| NM_013265 | B3GNT5 | UDP‐GlcNAc:betaGal beta‐1,3‐N‐acetylglucosaminyltransferase 5 | −12.6434 |
GO:0006486~protein glycosylation, GO:0007417~central nervous system development, GO:0009247~glycolipid biosynthetic process |
GO:0000139~Golgi membrane, GO:0005622~intracellular, GO:0016021~integral component of membrane |
GO:0008378~galactosyltransferase activity,GO:0008457~beta‐galactosyl‐N‐acetylglucosaminylgalactosylglucosyl‐ceramide beta‐1,3‐acetylglucosaminyltransferase activity, GO:0008499~UDP‐galactose:beta‐N‐acetylglucosamine beta‐1,3‐galactosyltransferase activity, GO:0047256~lactosylceramide 1,3‐N‐acetyl‐beta‐D‐glucosaminyltransferase activity |
| NM_015626 | FAM198B | Family with sequence similarity 198 member B | −13.2931 | — |
GO:0000139~Golgi membrane, GO:0005794~Golgi apparatus |
— |
| NM_014795 | WSB1 | WD repeat and SOCS box containing 1 | −13.4219 |
GO:0000209~protein polyubiquitination, GO:0016567~protein ubiquitination, GO:0035556~intracellular signal transduction |
GO:0005622~intracellular, GO:0005829~cytosol |
GO:0004842~ubiquitin‐protein transferase activity, GO:0005515~protein binding |
| NM_005857 | ATP11B | ATPase phospholipid transporting 11B (putative) | −16.3131 |
GO:0006811~ion transport, GO:0015917~aminophospholipid transport, GO:0034220~ion transmembrane transport, GO:0045332~phospholipid translocation |
GO:0005637~nuclear inner membrane, GO:0005769~early endosome, GO:0005783~endoplasmic reticulum, GO:0005794~Golgi apparatus, GO:0005886~plasma membrane, GO:0016020~membrane, GO:0055037~recycling endosome |
GO:0000287~magnesium ion binding, GO:0004012~phospholipid‐translocating ATPase activity, GO:0005515~protein binding, GO:0005524~ATP binding, GO:0015075~ion transmembrane transporter activity |
—, no data available.
As a final classifier, obtained through application of the classification algorithm based on 56 genes to all 55 subjects, we obtained the following equation for PDAC diagnosis using the weighted linear sum of each gene's expression level:
In the formula above, an is a coefficient value for each gene (Table 1); “a1*gene 1 + a2*gene 2 + ……… + a56**gene 56” was determined based on Fisher's linear discriminant, and its threshold was 209.018. To simplify the threshold value as zero by the formula, intercept value, −209.018, in the equation, was mounted. Thus, sum‐values 0 or higher and less than 0 were taken as positive and negative for PDAC, respectively.
3.2. Characteristics of subjects registered in the clinical study used to assess the performance of blood mRNA screening system
We designed a clinical study to assess the performance of the blood mRNA screening system. The study cohort consisted of 54 patients with PDAC, 25 with IPMN, 22 with chronic pancreatitis, and 104 noncancer subjects (Figure 1). All subjects were assessed for suitability for the safety analysis cohort and full analysis set. After applying the exclusion criteria, we finally included 53 patients with PDAC, 23 with IPMN, 22 with chronic pancreatitis, and 102 noncancer subjects.
Figure 1.
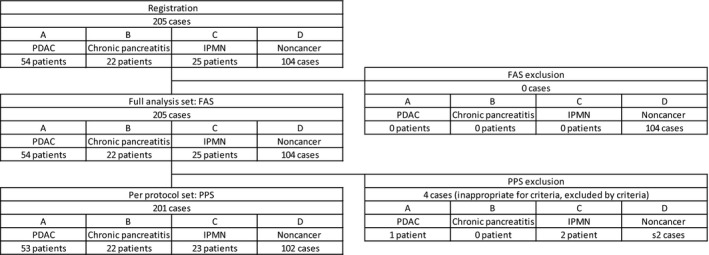
Outline of the clinical study to assess the performance of the blood mRNA screening system for pancreatic ductular adenocarcinoma (PDAC). The study cohort consisted of 54 patients with PDAC, 22 with chronic pancreatitis, 25 with intraductal papillary mucinous neoplasm (IPMN), and 104 noncancer subjects. No cases were excluded from the full analysis set. In the per protocol set, 1 PDAC patient, 2 IPMN patients, and 2 noncancer volunteers were excluded. Thus, 53 PDAC patients, 22 chronic pancreatitis patients, 23 IPMN patients, and 102 noncancer subjects were included in the per protocol analysis
Some of the clinical characteristics, such as age, gender, and body mass index, were different between PDAC patients and noncancer subjects (Table 2). The frequency of alcohol drinking was not different between PDAC patients and noncancer subjects (Table 2).
Table 2.
Clinical characteristics of study subjects to assess the blood mRNA screening system for pancreatic ductal adenocarcinoma (PDAC)
| PDAC | Chronic pancreatitis | IPMN | Noncancer | |
|---|---|---|---|---|
| Objective number | 53 | 22 | 23 | 102 |
| Age (y) | ||||
| Average | 69.6 | 61.7 | 67.7 | 63.4 |
| SD | 10.7 | 10.8 | 8.2 | 11.4 |
| Number | 53 | 22 | 23 | 102 |
| Wilcoxon signed‐rank test | Ref | P = .008 | P = .399 | P = .003 |
| — | Ref | P = .031 | P = .484 | |
| — | — | Ref | P = .122 | |
| Gender | ||||
| Male | 26 (49.1%) | 19 (86.4%) | 11 (47.8%) | 69 (67.6%) |
| Female | 27 (50.0%) | 3 (13.6%) | 12 (52.2%) | 33 (32.4%) |
| Fisher's exact test | Ref | P = .004 | P = 1.000 | P = .036 |
| — | Ref | P = .011 | P = .119 | |
| — | — | Ref | P = .093 | |
| Height (cm) | ||||
| Average | 159.63 | 167.93 | 159.82 | 162.99 |
| SD | 8.63 | 5.50 | 8.08 | 9.31 |
| N | 52 | 21 | 22 | 94 |
| Wilcoxon signed‐rank test | Ref | P < .001 | P = .717 | P = .027 |
| — | Ref | P = .002 | P = .015 | |
| — | — | Ref | P = .156 | |
| Body weight (kg) | ||||
| Average | 54.18 | 59.11 | 57.90 | 61.61 |
| SD | 12.26 | 10.18 | 9.59 | 11.49 |
| N | 51 | 21 | 22 | 95 |
| Wilcoxon signed‐rank test | Ref | 0.132 | P = .146 | P < .001 |
| — | Ref | P = .884 | P = .287 | |
| — | — | Ref | P = .203 | |
| BW loss >5 kg over 6 months | ||||
| No (N) | 29 | 16 | 12 | 75 |
| Yes (N) | 10 | 1 | 0 | 1 |
| Fisher's exact test | Ref | P = .144 | P = .092 | P < .001 |
| — | Ref | P = 1.000 | P = .334 | |
| — | — | Ref | P = 1.000 | |
| Body mass index | ||||
| Average | 21.04 | 20.94 | 22.55 | 23.09 |
| SD | 3.34 | 3.45 | 2.72 | 3.27 |
| N | 51 | 21 | 22 | 93 |
| Wilcoxon signed‐rank test | Ref | P = .536 | P = .100 | P = .002 |
| — | Ref | P = .037 | P = .004 | |
| — | — | Ref | P = 0.477 | |
| Smoking | ||||
| Nonsmoker | 31 (58.5%) | 6 (27.3%) | 9 (39.1%) | 63 (61.8%) |
| Past smoker | 13 (24.5%) | 5 (22.7%) | 8 (34.8%) | 22 (21.6%) |
| Current smoker | 6 (11.3%) | 8 (36.4%) | 2 (8.7%) | 12 (11.8%) |
| Fisher's exact test | Ref | P = .020 | P = .419 | P = .933 |
| — | Ref | P = .119 | P = .005 | |
| — | — | Ref | P = .226 | |
| Alcohol drinking | ||||
| Nondrinker | 33 (62.3%) | 6 (27.3%) | 6 (26.1%) | 42 (41.2%) |
| Past drinker | 3 (5.7%) | 4 (18.2%) | 0 (0.0%) | 4 (3.9%) |
| Current drinker | 17 (32.1%) | 9 (40.9%) | 13 (56.5%) | 49 (48.0%) |
| Fisher's exact test | Ref | P = .032 | P = .027 | P = .055 |
| — | Ref | P = .144 | P = .051 | |
| — | — | Ref | P = .398 | |
| Familial history of PDAC | ||||
| No | 24 (45.3%) | 14 (63.6%) | 14 (60.9%) | 67 (65.7%) |
| Yes | 3 (5.7%) | 0 (0.0%) | 1 (4.3%) | 3 (2.9%) |
| Unknown | 26 (49.1%) | 8 (36.4%) | 8 (34.8%) | 32 (31.4%) |
| Fisher's exact test | Ref | P = .539 | P = 1.000 | P = .344 |
| — | Ref | P = 1.000 | P = 1.000 | |
| — | — | Ref | P = .547 | |
—, not applicable; BW, body weight; IPMN, intraductal papillary mucinous neoplasm; Ref, reference.
3.3. Performance of blood mRNA screening system
The sensitivity and specificity of the blood mRNA PDAC screening system for 53 PDAC patients and 102 noncancer subjects was 73.6% (95% CI, 59.7%‐84.7%) and 64.7% (54.6%‐73.9%), respectively. The specificity for chronic pancreatitis and IPMN patients was 63.6% (95% CI, 40.7%‐82.8%) and 47.8% (95% CI, 26.8%‐69.4%), respectively (Table 3). The sensitivity for the conventional tumor markers CEA and CA19‐9 was 41.5% (95% CI, 28.1%‐55.9%) and 71.7% (95% CI, 57.7%‐83.2%), respectively (Tables 4 and 5). In noncancer subjects, the specificity for CEA and CA19‐9 was 96.1% (95% CI, 90.3%‐98.9%) and 97.1% (95% CI, 91.6%‐99.4%), respectively (Tables 4 and 5). The PPVs of the mRNA blood screening system, CEA, and CA19‐9 for chronic pancreatitis and IPMN were 76.5%‐83.0%, 71.9%, and 95.1%, respectively (Tables 3, 4, 5); the respective values for noncancer subjects were 50.6%, 82.1%, and 92.9%, respectively. The NPVs of the mRNA blood screening system, CEA, and CA19‐9 for all cases were 86.1%, 81.1%, and 90.4%, respectively (Tables 3, 4, 5). Pearson's correlation analysis was applied between these conventional tumor markers and scores of the mRNA screening system. The correlation coefficients for CEA and CA19‐9 with scores of the mRNA screening system were 0.072 and 0.14, respectively, with P‐values of .308 and .048, respectively. These results indicate no correlation between CEA and the mRNA screening system, and very weak correlation between CA19‐9 and the mRNA screening system (Figure 2).
Table 3.
Senstivity and specificity of blood mRNA screening system for pancreatic ductal adenocarcinoma (PDAC)
| PDAC | Chronic pancreatitis | IPMN | Noncancer | |
|---|---|---|---|---|
| Objective number | 53 | 22 | 23 | 102 |
| mRNA system result | ||||
| Positive | 39 (73.6%) | 8 (36.4%) | 12 (52.2%) | 36 (35.3%) |
| Negative | 14 (26.4%) | 14 (63.6%) | 11 (47.8%) | 66 (64.7%) |
| Sensitivity | ||||
| Point estimation | 73.6% | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) |
| 95% CI | 59.7%‐84.7% | — | — | — |
| Specificity | ||||
| Point estimation | — | 63.6% | 47.8% | 64.7% |
| 95% CI | — | 40.7%‐82.8% | 26.8%‐69.4% | 54.6%‐73.9% |
—, not applicable; CI, confidence interval; IPMN, intraductal papillary mucinous neoplasm.
Table 4.
Senstivity and specificity of carcinoembryonic antigen (CEA) for pancreatic ductal adenocarcinoma (PDAC)
| PDAC | Chronic pancreatitis | IPMN | Noncancer | |
|---|---|---|---|---|
| Objective number | 53 | 22 | 23 | 102 |
| Result | ||||
| Positive | 22 (41.5%) | 9 (40.9%) | 2 (8.7%) | 4 (3.9%) |
| Negative | 31 (58.5%) | 13 (59.1%) | 21 (91.3%) | 98 (96.1%) |
| Sensitivity | ||||
| Point estimation | 41.5% | — | — | — |
| 95% CI | 28.1%‐55.9% | — | — | — |
| Specificity | ||||
| Point estimation | — | 59.1% | 91.3% | 96.1% |
| 95% CI | — | 36.4%‐79.3% | 72.0%‐98.9% | 90.3%‐98.9% |
—, not applicable; CI, confidence interval; IPMN, intraductal papillary mucinous neoplasm.
Table 5.
Senstivity and specificity of cancer antigen (CA)19‐9 for pancreatic ductal adenocarcinoma (PDAC)
| PDAC | Chronic pancreatitis | IPMN | Noncancer | |
|---|---|---|---|---|
| Objective number | 53 | 22 | 23 | 102 |
| Result | ||||
| Positive | 38 (71.7%) | 2 (9.1%) | 2 (8.7%) | 3 (2.9%) |
| Negative | 15 (28.3%) | 20 (90.9%) | 21 (91.3%) | 99 (97.1%) |
| Sensitivity | ||||
| Point estimation | 71.7% | — | — | — |
| 95% CI | 57.7%‐83.2% | — | — | — |
| Specificity | ||||
| Point estimation | — | 90.9% | 91.3% | 97.1% |
| 95% CI | — | 70.8%‐98.9% | 72.0%‐98.9% | 91.6%‐99.4% |
—, not applicable; CI, confidence interval; IPMN, intraductal papillary mucinous neoplasm.
Figure 2.
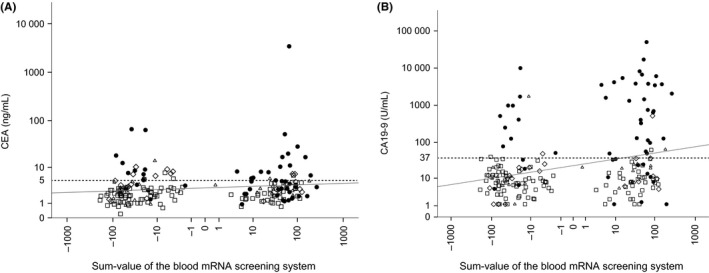
Scatter plots of carcinoembryonic antigen (CEA), cancer antigen (CA)19‐9, and sum‐values of blood mRNA screening system for pancreatic ductular adenocarcinoma. Serum concentration values of CEA (A) and serum unit values of CA19‐9 (B) with sum‐values of the blood mRNA screening system of 53 patients with pancreatic ductular adenocarcinoma (filled circle), 23 with intraductal papillary mucinous neoplasm (open triangle), 22 with chronic pancreatitis (open diamond), and 102 noncancer subjects (open square). The interval of y‐axis and x‐axis values is expressed in logarithmic scale. Solid line indicates regression lines, dashed line indicates the cut‐off value for tumor markers (A, B). R 2 values are 0.021 (A) and 0.08 (B)
Regarding disease stages, the blood mRNA screening system showed a sensitivity of 78.6% for stages I and II, and a similar value of 71.8% for stages III and IV (Table 6).
Table 6.
Sensitivity of mRNA screening system for pancreatic ductal adenocarcinoma (PDAC) staging
| Stage | ||
|---|---|---|
| I, II | III, IV | |
| Number | 14 | 39 |
| Positive by mRNA screening system | 11 | 28 |
| Sensitivity (%) | 78.6 | 71.8 |
3.4. Clinical characteristics and blood mRNA screening system
In terms of clinical parameters and sum‐values of the blood mRNA screening system, we applied correlation analyses for continuous data (age, height, and body weight) and t tests for category data (gender, body weight loss of more than 5 kg over 6 months, smoking, alcohol drinking, and familial history of PDAC) with sum‐values of the blood mRNA screening system (Table S4). Although the P value for the age correlation analysis was less than .05, correlation efficiency was weak (0.145). Average values of the sum‐values of the blood mRNA screening system for nonsmoker and current smoker were significantly different (P = .047), but both values were less than zero. Although the sum‐value of past drinker was significantly higher than those of nondrinker and current drinker (P = .045 and P = .043, respectively), the sum‐value for the current drinker was not higher than current drinker or nondrinker.
4. DISCUSSION
In this study, we developed a blood mRNA PDAC screening system measuring the expression of 56 genes in whole blood by RTD‐PCR, followed by weighted linear summation. We undertook a multicenter clinical study to validate the performance of this blood mRNA screening system for PDAC detection, by gene expression analysis using RTD‐PCR. This system showed that sensitivity was 73.6% and the specificity for healthy subjects was 64.7%.
The principle of this system is unique, being based on changes in gene expression in the peripheral blood.6, 7, 10, 11 Previously, we reported that PDAC is related to inflammatory changes in local tumor tissue, as well in peripheral blood.7, 10 Therefore, we assumed that alterations in the gene expression profile in the peripheral blood of PDAC patients would be associated with systemic inflammatory responses of the host. Based on this principle, we selected genes that showed differences in expression between the whole blood from PDAC patients and noncancer subjects.
The prognosis of PDAC is extremely poor due to in part to difficulty of diagnosis in the early stages.12 At present, there is no definitive screening method for PDAC, and various methods are used clinically, including ultrasonography and computed tomography, for imaging‐based diagnosis and determination of tumor markers in blood.13 With regard to blood test tumor markers, CA19‐9,14 Span‐1,15 CA50, CA242, DUPAN‐2, and TAG‐7216, 17, 18 are categorized as glucose‐chain antigens; CEA is an embryonic antigen19 that is typically present in high levels in PDAC, although not in all cases. In addition to these conventional tumor markers, a new approach has been developed using excreted small particles, called exosomes; these are believed to include molecules specific for cancers,20 although the analysis methods are still under development.
As diagnosis of PDAC in the early stages is extremely rare, there are insufficient data regarding the performance of the mRNA screening system for early‐stage PDAC. The PDAC patients in this study were mainly in advanced stages (ie, III or IV); only 14 patients were in stage I or II. The sensitivity of the mRNA screening system for these PDAC patients was 78.6%, illustrating the potential benefit of this system for early detection of PDAC. We previously disclosed that PDAC local tissues were associated with inflammation, even if they were still small.7 Thus, the high sensitivity of the mRNA screening system, capable of detecting PDAC in early stages, might be an important beneficial feature, one that has proven difficult using other conventional tumor marker systems. Based on this previous study, the principle of the mRNA screening system relies on gene expression alteration of peripheral blood cells affected by development of PDAC. This is the feature that distinguishes the system from conventional tumor markers (ie, CEA and CA19‐9), which are produced by cancer tissues. In this context, the novel mRNA system would be a useful screening method as the examination involves a blood test, which can be undertaken without the need for specific medical devices and is minimally invasive.
The establishment of a PDAC screening algorithm requires further development, including the novel blood mRNA screening system described herein, along with other tumor markers and diagnostic imaging tools.21 Although ultrasonography is noninvasive, it cannot be used to determine dilatation of the pancreatic duct or tumorous lesions in the pancreas; therefore, patients also require endoscopy,22 computed tomography, and MRI examinations.23 As discussed above, conventional tumor markers are not sufficient for screening for PDAC in the early stages, which could allow the disease to be cured by surgery.
The genes included in this system were selected biostatistically, based on changes in their expression levels in the whole blood of PDAC patients forming a different cohort from that of the clinical study. The mechanisms by which these genes are affected in PDAC are intriguing; some genes are involved in immunological processes, consistent with the previous finding that PDAC is associated with both local and systemic immune reactions.7 Further investigations to determine the biological significance of these genes will provide insight into the immunopathological features of PDAC, leading to further development of diagnostic tools as well as immunological treatments for the disease.
In conclusion, we developed a novel in vitro diagnostic system based on assessment of the host immune response to PDAC. As it remains unclear how this novel system should best be used as an adjunct to conventional PDAC diagnostic or screening systems, further investigations to improve the PDAC screening algorithm, including our novel screening system, should be undertaken to improve the prognosis of PDAC patients by diagnosis in the early stages of disease progression.
DISCLOSURE STATEMENT
This work was financially supported in part by KUBIX Inc., and was assessed by our institutional review board.
Supporting information
ACKNOWLEDGMENTS
We thank all staff in the participating institutions involved in registration of the patients and noncancer subjects to the clinical study assessing the performance of the mRNA blood screening system. This study was supported in part by the Japan Ministry of Health, Labour, and Welfare, the Japan Agency for Medical Research and Development, and Kanazawa University Hospital. We also acknowledge the following members of the Hokuriku Liver Study Group: Kanazawa University Hospital; Department of Gastroenterology, Toyama Prefectural Central Hospital; Department of Internal Medicine, Toyama Municipal Hospital; Department of Gastroenterology, Tonami General Hospital; Department of Gastroenterology, National Hospital Organization Kanazawa Medical Center; Department of Gastroenterology, Ishikawa‐ken Saiseikai Kanazawa Hospital; Department of Gastroenterology, Public Central Hospital of Matto Ishikawa; Department of Gastroenterology, Fukui‐ken Saiseikai Hospital; Department of Internal Medicine, Tsuruga Municipal Hospital; Department of Gastroenterology, Keijyu General Hospital; Department of Internal Medicine, Kurobe Municipal Hospital; Department of Gastroenterology, Noto General Hospital; Department of Gastroenterology, Ushitsu General Hospital; Tsurugi Hospital, Hakui General Hospital; Department of Internal Medicine, Tsubata‐town Kahoku Central Hospital; Department of Internal Medicine, Kanazawa Municipal Hospital; Department of Gastroenterology, Kanazawa Red Cross Hospital; Department of Gastroenterology, Nomi Municipal Hospital; Department of Internal Medicine, Hokkoku Clinice, Komatsu‐Sofia Hospital; Department of Gastroenterology, Yawata Medical Center; Department of Gastroenterology, Fukui Prefectural Hospital; and Department of Gastroenterology, Kumagai Clinic. We also thank Ms. Shizuko Takahara and Ms. Ka U for data management assistance.
Sakai Y, Honda M, Matsui S, et al. Hokuriku Liver Study Group . Development of novel diagnostic system for pancreatic cancer, including early stages, measuring mRNA of whole blood cells. Cancer Sci. 2019;110:1364–1388. 10.1111/cas.13971
Clinical trial registration number: UMIN000020758
REFERENCES
- 1. Vincent A, Herman J, Schulick R, Hruban RH, Goggins M. Pancreatic cancer. Lancet. 2011;378:607‐620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Matsuda T, Ajiki W, Marugame T, et al. Population‐based survival of cancer patients diagnosed between 1993 and 1999 in Japan: a chronological and international comparative study. Jpn J Clin Oncol. 2011;41:40‐51. [DOI] [PubMed] [Google Scholar]
- 3. Buchler MW, Kleeff J, Friess H. Surgical treatment of pancreatic cancer. J Am Coll Surg. 2007;205:S81‐S86. [DOI] [PubMed] [Google Scholar]
- 4. Paulson AS, Tran CH, Tempero MA, Lowy AM. Therapeutic advances in pancreatic cancer. Gastroenterology. 2013;144:1316‐1326. [DOI] [PubMed] [Google Scholar]
- 5. Reni M, Cordio S, Milandri C, et al. Gemcitabine versus cisplatin, epirubicin, fluorouracil, and gemcitabine in advanced pancreatic cancer: a randomised controlled multicentre phase III trial. Lancet Oncol. 2005;6:369‐376. [DOI] [PubMed] [Google Scholar]
- 6. Honda M, Sakai Y, Yamashita T, et al. Differential gene expression profiling in blood from patients with digestive system cancers. Biochem Biophys Res Commun. 2010;400:7‐15. [DOI] [PubMed] [Google Scholar]
- 7. Komura T, Sakai Y, Harada K, et al. Inflammatory features of pancreatic cancer highlighted by monocytes/macrophages and CD4+ T cells with clinical impact. Cancer Sci. 2015;106:672‐686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Youn E, Koenig L, Jeong MK, Baek SH. Support vector‐based feature selection using Fisher's linear discriminant and Support Vector Machine. Expert Syst Appl. 2010;37:6148‐6156. [Google Scholar]
- 9. Breiman L. Random forests. Mach Learn. 2001;45:5‐32. [Google Scholar]
- 10. Komura T, Takabatake H, Harada K, et al. Clinical features of cystatin A expression in patients with pancreatic ductal adenocarcinoma. Cancer Sci. 2017;108:2122‐2129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sakai Y, Honda M, Fujinaga H, et al. Common transcriptional signature of tumor‐infiltrating mononuclear inflammatory cells and peripheral blood mononuclear cells in hepatocellular carcinoma patients. Cancer Res. 2008;68:10267‐10279. [DOI] [PubMed] [Google Scholar]
- 12. Kamisawa T, Wood LD, Itoi T, Takaori K. Pancreatic cancer. Lancet. 2016;388:73‐85. [DOI] [PubMed] [Google Scholar]
- 13. Kanno A, Masamune A, Hanada K, et al. Multicenter study of early pancreatic cancer in Japan. Pancreatology. 2018;18:61‐67. [DOI] [PubMed] [Google Scholar]
- 14. Koprowski H, Steplewski Z, Mitchell K, Herlyn M, Herlyn D, Fuhrer P. Colorectal carcinoma antigens detected by hybridoma antibodies. Somatic Cell Genet. 1979;5:957‐971. [DOI] [PubMed] [Google Scholar]
- 15. Kiriyama S, Hayakawa T, Kondo T, et al. Usefulness of a new tumor marker, Span‐1, for the diagnosis of pancreatic cancer. Cancer. 1990;65:1557‐1561. [DOI] [PubMed] [Google Scholar]
- 16. Kawa S, Tokoo M, Hasebe O, et al. Comparative study of CA242 and CA19‐9 for the diagnosis of pancreatic cancer. Br J Cancer. 1994;70:481‐486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Takeda S, Nakao A, Ichihara T, et al. Serum concentration and immunohistochemical localization of SPan‐1 antigen in pancreatic cancer. A comparison with CA19‐9 antigen. Hepatogastroenterology. 1991;38:143‐148. [PubMed] [Google Scholar]
- 18. Haglund C, Kuusela P, Jalanko H, Roberts PJ. Serum CA 50 as a tumor marker in pancreatic cancer: a comparison with CA 19‐9. Int J Cancer. 1987;39:477‐481. [DOI] [PubMed] [Google Scholar]
- 19. Meng Q, Shi S, Liang C, et al. Diagnostic and prognostic value of carcinoembryonic antigen in pancreatic cancer: a systematic review and meta‐analysis. Onco Targets Ther. 2017;10:4591‐4598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kadota T, Yoshioka Y, Fujita Y, Kuwano K, Ochiya T. Extracellular vesicles in lung cancer‐From bench to bedside. Semin Cell Dev Biol. 2017;67:39‐47. [DOI] [PubMed] [Google Scholar]
- 21. McAllister F, Montiel MF, Uberoi GS, Uberoi AS, Maitra A, Bhutani MS. Current status and future directions for screening patients at high risk for pancreatic cancer. Gastroenterol Hepatol (N Y). 2017;13:268‐275. [PMC free article] [PubMed] [Google Scholar]
- 22. Larghi A, Verna EC, Lecca PG, Costamagna G. Screening for pancreatic cancer in high‐risk individuals: a call for endoscopic ultrasound. Clin Cancer Res. 2009;15:1907‐1914. [DOI] [PubMed] [Google Scholar]
- 23. Tummala P, Junaidi O, Agarwal B. Imaging of pancreatic cancer: an overview. J Gastrointest Oncol. 2011;2:168‐174. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials


